Abstract
Hepatocellular carcinoma (HCC) is a malignant tumor located in the liver. Secreted frizzled-related protein 4 (sFRP-4) is associated with cancer occurrence, but the relationship between sFRP-4 and HCC is not completely understood. The present study aimed to investigate the role and mechanism underlying sFRP-4 in HCC. sFRP-4 mRNA expression levels were determined via reverse transcription-quantitative PCR and immunohistochemistry. The Cell Counting Kit-8 assay was performed to evaluate HCCLM3 and Huh7 cell viability. Moreover, HCCLM3 and Huh7 cell proliferation were assessed using the BrdU ELISA assay kit, and cell apoptosis was measured via flow cytometry. Western blotting was conducted to measure β-catenin and GSK-3β protein expression levels. The results demonstrated that sFRP-4 expression was significantly downregulated in HCC tissues and cells compared with adjacent healthy tissues and MIHA cells, respectively. Moreover, the results indicated that compared with the control group, sFRP-4 overexpression inhibited HCC cell viability and proliferation, and accelerated HCC cell apoptosis. Furthermore, the results suggested that sFRP-4 inhibited the Wnt/β-catenin signaling pathway by upregulating GSK-3β expression and downregulating β-catenin expression, thus restraining the malignant behavior of HCC cells. In conclusion, the present study indicated that sFRP-4 served a tumor suppressor role in HCC cells by restraining the Wnt/β-catenin signaling pathway.
Keywords: secreted frizzled-related protein 4, hepatocellular carcinoma, Wnt/β-catenin
Introduction
Liver cancer is the third leading malignant tumor worldwide, with a mortality rate of 8.2% and ~840,000 new cases each year (1). As the most common form of liver cancer, hepatocellular carcinoma (HCC) accounts for 90% of all cases of primary liver cancer (2). HCC is considered as a severe malignant tumor in China due to 55% morbidity rate (3). Moreover, HCC morbidity has rapidly increased on a global scale in the last five decades, partly due to hepatitis B or C virus infection and cirrhosis associated with poor lifestyle (4–6). At present, the best treatment strategies available for HCC include surgical resection and liver transplantation, but the survival rate (6.9%) of patients with HCC is poor due to its high recurrence and metastasis (7–9). The molecular mechanism underlying HCC carcinogenesis is not completely understood. Therefore, improving the current understanding of the molecular mechanism underlying HCC may aid with the development of novel therapeutic strategies to improve the prognosis of the disease.
Located on chromosome 7p14.1, secreted frizzled-related protein 4 (sFRP-4) consists of a total of six exons (10). sFRP-4 belongs to the SFRP family and contains a cysteine-rich domain that is homologous to the assumed Wnt-binding site of the frizzled proteins (11). Moreover, sFRP-4 can regulate the Wnt signaling pathway by directly binding to Wnt, thus preventing Wnt from binding to its receptor (12,13). Increasing evidence has suggested that sFRP-4 could inhibit the canonical Wnt signaling pathway, form silencing complexes and suppress human cancer, including gastric (14), ovarian (15) and cervical cancer (16), as well as mesothelioma (17) and cemento ossifying fibroma (18). Several studies also reported that sFRP-4 protein was highly expressed in other types of cancer, including prostate cancer (19), pancreatic ductal adenocarcinoma (20), colon carcinoma (21) and colorectal carcinoma (22). Therefore, the aforementioned studies suggested that sFRP-4 was differentially expressed in different types of cancer. Our preliminary work demonstrated that sFRP-4 expression levels were upregulated in the serum obtained from patients with HCC. In addition, the combined use of sFRP-4 and α-fetoprotein (AFP; a standard serum marker of HCC) improved the accuracy of HCC diagnosis (23). However, the functions and mechanism underlying sFRP-4 in HCC development were not investigated in our previous study. Therefore, the present study explored the exact functions and mechanism underlying sFRP-4 in HCC using HCC clinical tissues and cell lines.
Furthermore, the Wnt signaling pathway can be classified into two major types: Classical (β-catenin-dependent) and non-classical (β-catenin-independent) (24,25). As a key component of the classical pathway, β-catenin is strictly regulated by a variety of protein complexes, including GSK-3β (26). In the absence of Wnt ligands, the Wnt/β-catenin signaling pathway is inactive and β-catenin is degraded after sustained phosphorylation to maintain low levels in the cytoplasm. Following Wnt induction, the Wnt/β-catenin signaling pathway is activated and the degradation process of β-catenin phosphorylation is inhibited, resulting in intracellular accumulation of β-catenin (27,28). As a key member of the Wnt/β-catenin signaling pathway, GSK-3β can participate in the degradation of β-catenin, but when the Wnt/β-catenin signaling pathway is activated, GSK-3β remains in an inhibitory state (29). Abnormal activation of the Wnt/β-catenin signaling pathway results in the carcinogenesis and development of various types of cancer, including gastric (30), colorectal (31), esophageal (32) and pancreatic cancer (33), as well as HCC (34). Furthermore, it has been reported that 30% of patients with HCC exhibited excessive activation of the Wnt/β-catenin signaling pathway (35). Therefore, exploring the molecular mechanism underlying the Wnt/β-catenin signaling pathway in HCC is important for the identification of novel effective therapeutic targets.
After performing preliminary research on the clinical significance and mechanism underlying sFRP-4 in HCC, as well as the regulatory mechanism underlying the Wnt/β-catenin signaling pathway in HCC, it was hypothesized that sFRP-4 could restrain HCC progression, potentially via suppression of the Wnt/β-catenin signaling pathway. To verify the hypothesis, sFRP-4 expression levels in HCC tissues and cells were assessed. Therefore, the results of the present study may aid with the identification of novel therapeutic targets for HCC.
Materials and methods
Clinical tissue samples
Tissue samples (47 paired HCC and adjacent non-cancerous tissues) were collected from patients (age range, 41–76 years) who were diagnosed with HCC at the General Hospital of the Central Theater Command of the People's Liberation Army (Wuhan, China) between June 2015 and December 2019, and 59.6% patients were >60 years old. All patients had not received treatment with anticancer drugs before surgical resection, and incomplete data were excluded. After performing surgical resection, samples of HCC tissue and samples of adjacent non-cancerous tissue (>3 cm away from the tumor site; cirrhosis tissue was excluded) were immediately labeled and frozen until further analysis. All cases were diagnosed histopathologically. The clinical characteristics of the patients are presented in Table I. The present study was approved by the Ethics Committee of the General Hospital of the Central Theater Command of the People's Liberation Army (approval no. [2020]024-2). All patients provided written informed consent.
Table I.
Association between sFRP-4 expression and the clinical characteristics of patients with hepatocellular carcinoma.
| sFRP-4 expression | ||||
|---|---|---|---|---|
| Characteristic | n (%) | High (n=20) | Low (n=27) | P-value |
| Age (years) | 0.959 | |||
| >60 | 19 (40.4%) | 8 (40.0%) | 11 (40.7%) | |
| ≤60 | 28 (59.6%) | 12 (60.0%) | 16 (59.3%) | |
| Sex | 0.824 | |||
| Male | 36 (76.6%) | 15 (75.0%) | 21 (77.8%) | |
| Female | 11 (23.4%) | 5 (25.0%) | 6 (22.2%) | |
| Tumor diameter (cm) | 0.057 | |||
| >5 | 39 (83.0%) | 14 (70.0%) | 25 (92.6%) | |
| ≤5 | 8 (17.0%) | 6 (30.0%) | 2 (7.4%) | |
| Differentiation | 0.133 | |||
| Well | 7 (14.9%) | 5 (25.0%) | 2 (7.4%) | |
| Moderate | 25 (53.2%) | 11 (55.0%) | 14 (51.9%) | |
| Poor | 15 (31.9%) | 4 (20.0%) | 11 (40.7%) | |
| TNM stage | 0.002a | |||
| I | 5 (10.6%) | 4 (20.0%) | 1 (3.7%) | |
| II | 18 (38.3%) | 12 (60.0%) | 6 (22.2%) | |
| III | 22 (46.8%) | 4 (20.0%) | 18 (66.7%) | |
| IV | 2 (4.3%) | 0 (0.0%) | 2 (7.4%) | |
| Metastasis | ||||
| Positive | 26 (55.3%) | 6 (30.0%) | 20 (74.1%) | 0.003a |
| Negative | 21 (44.7%) | 14 (70.0%) | 7 (25.9%) | |
| AFP (IU) | ||||
| ≤400 | 20 (42.6%) | 13 (65.0%) | 7 (25.9%) | 0.007a |
| >400 | 27 (57.4%) | 7 (35.0%) | 20 (74.1%) | |
For sample sizes ≥5, the data were analyzed using the χ2 test. For sample sizes <5, the data were analyzed using the Fisher's exact test.
P<0.05. sFRP-4, secreted frizzled-related protein 4; AFP, α-fetoprotein.
Cell lines and cell culture
The cell lines used in the present study were purchased from BeNa Culture Collection (Beijing Beina Chunglian Biotechnology Research Institute). The following HCC cell lines were used: HCCLM3 (cat. no. BNCC338460), Hep3B (cat. no. BNCC337952) and Huh7 (cat. no. BNCC337690). The MIHA immortalized normal human liver cell line (cat. no. BNCC340123) was also used. MIHA cells were cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 100 U/ml streptomycin/penicillin (Invitrogen; Thermo Fisher Scientific, Inc.). HCCLM3, Hep3B and Huh76 cells were cultured in DMEM-H (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS and 100 U/ml streptomycin/penicillin. All cells were cultured at 37°C with 5% CO2.
Cell transfection and IM-12 treatment
sFRP-4 small interfering (si)RNA vector (si-sFRP-4; forward: 5′-GGAGGAUCACAGAAAUGUAGA-3′ and reverse: 5′-UACAUUUCUGUGAUCCUCCUA-3′), sFRP-4 overexpression vector (OE; sFRP-4-OE; forward: 5′-ATGCAAGCTTTTCCTCTCCATCCTAGTGGCG-3′, reverse: 5′-TCACGGGATCCTTCTTGGGACTGGCTGGTTT-3′), empty vector (pcDNA3.1) and non-targeting siRNA (si-NC) were purchased from Guangzhou Funeng Gene Co., Ltd. Cells in the blank group were cultured under normal conditions. A total of 5×104 HCCLM3 and Huh7 cells were transfected with 100 nM si-sFRP-4, 1 µg/ml sFRP-4-OE or negative control (NC) vectors containing 1 µg/ml empty vector and 100 nM si-NC using Lipofectamine RNAiMax (Thermo Fisher Scientific, Inc.) at room temperature according to the manufacturer's protocol. After 48 h transfection, the transfection efficiency of si-sFRP-4 and sFRP-4-OE was assessed via reverse transcription-quantitative PCR (RT-qPCR). At 12 h post-transfection, cells were treated with 1 µM IM-12 (cat. no. SML0084; Sigma-Aldrich; Merck KGaA) for 36 h at 37°C, and the control group was treated with 1% DMSO.
RT-qPCR
Total RNA was isolated from frozen tissues or cultured cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). RNA concentration was measured using a spectrophotometer. Total RNA was reverse transcribed into cDNA using SuperScript IV VILO Master Mix (cat. no. 11756050; Invitrogen; Thermo Fisher Scientific, Inc.) at 50°C for 10 min and 85°C for 5 min. Subsequently, RT-qPCR was performed using PowerUp SYBR™ Green Master Mix (cat. no. A25779; Applied Biosystems; Thermo Fisher Scientific, Inc.) and an ABI 7500 Real Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: Initial denaturation at 95°C for 4 min, followed by 40 cycles of denaturation at 95°C for 15 sec and annealing at 60°C for 60 sec. The following primers were used for qPCR: GAPDH forward, 5′-CACCGTAGCCTTCCGAGTA-3′ and reverse, 5′-GCCCTTGATGAGCTGTTGA-3′; and sFRP-4 forward, 5′-CAGAGGAGTGGCTGCAATGA-3′ and reverse, 5′-CTTGATGGGGCAGGATGTGT-3′. sFRP-4 mRNA expression levels were quantified using the 2−∆∆Cq method (36) and normalized to the internal reference gene GAPDH.
Immunohistochemistry (IHC) analysis
sFRP-4 expression in 47 paired HCC and adjacent non-cancerous tissues was assessed by performing IHC staining. Tissues was imbedded in paraffin after 12 h of 10% neutral formalin fixation at room temperature. Then, 5-µm thickness tissue sections were gradually dewaxed and hydrated with xylene and descending ethanol. Sections were then treated with 10 mM citrate buffer (cat. no. AP-9003-125, Lab Vision Corp.) at 95°C for 5 min to repair antigens via heat treatment. Then, slides washed in deionized water three times for 2 min. Sections were incubated in 3% hydrogen peroxide for 10 min at room temperature to inhibit endogenous peroxidase activity. Subsequently, non-specific sites were blocked with 5% goat serum (Chemicon International Inc.) for 30 min at room temperature. Tissue sections were incubated with an anti-sFRP-4 primary antibody (1:500; cat. no. ab217180; Abcam) at 4°C overnight. Following primary incubation, tissue sections were incubated with a corresponding HRP-conjugated secondary antibody (1:1,000; cat. no. ab6721; Abcam) at 37°C for 1 h. Protein expression was visualized using the Pierce DAB Substrate kit (cat. no. 34002; Thermo Fisher Scientific, Inc.). Finally, tissue sections were counterstained with hematoxylin for 2 min at room temperature. Stained sections were observed using an objective lens of a computer-aided light microscope imager (magnification, ×200; Openlab; PerkinElmer, Inc.). Images were scanned using ImageScope Version 10 software (Aperio Technologies, Inc.).
Cell viability assay
HCCLM3 and Huh7 cell viability was assessed using the Cell Counting Kit-8 (CCK-8) assay (cat no. CK04-13; Dojindo Molecular Technologies, Inc.) according to the manufacturer's protocol. After transfection for 24 h, HCCLM3 and Huh7 cells were seeded (5×103 cells/well) into 96-well plates. Following incubation for 0, 24, 48 or 72 h at 37°C with 5% CO2, 10 µl CCK-8 solution was added to each well and incubated for 2 h. Subsequently, cell viability was assessed by measuring absorbance at a wavelength of 450 nm using a microplate reader.
Cell proliferation assay
After 48-h transfection, HCCLM3 and Huh7 cell proliferation was assessed by performing the BrdU ELISA assay using the CytoSelect™ BrdU Cell Proliferation ELISA kit (cat. no. CBA-251, Cell Biolabs, Inc.) according to the manufacturer's protocol. HCCLM3 and Huh7 cells were seeded (1×104 cells/well) into 96-well plates for 48 h. Subsequently, 10 µl BrdU solution (X10) was added to each well for 4 h at 37°C. Following washing with PBS, cells were fixed and cellular DNA was denatured by incubation with 100 µl Fix/Denature Solution for 30 min at room temperature. Cells were then incubated with 100 µl anti-BrdU antibody for 1 h at room temperature, followed by incubation with 100 µl secondary antibody for 1 h at room temperature. Cell proliferation was assessed by measuring absorbance at a wavelength of 450 nm using a microplate reader.
Cell apoptosis assay
HCCLM3 and Huh7 cell apoptosis was assessed via flow cytometry using the Annexin V-FITC Apoptosis Staining/Detection kit (cat. no. ab14085; Abcam) after transfection for 48 h. HCCLM3 and Huh7 cells were seeded (5×104 cells/well) into a 6-well plate and cultured at 37°C with 5% CO2. At 70–80% confluence, cells were incubated in 500 µl binding buffer containing 5 µl PI and 5 µl Annexin V-FITC at room temperature for 10 min in the dark. Apoptotic cells were analyzed via flow cytometry on a FACSCalibur Flow Cytometer (BD Biosciences) using the CellQuest Pro software (version 5.1; Becton, Dickinson and Company) and the rate of apoptosis was calculated as the sum of late apoptosis (Q1-UR) and early apoptosis (Q1-LR).
Western blotting
After 72-h transfection, total protein was isolated from HCCLM3 and Huh7 cells using RIPA lysis buffer (Thermo Fisher Scientific, Inc.) and the protein supernatant was collected after centrifugation at 12,000 × g for 15 min at 4°C. Total protein concentration was quantified using the BCA kit (Thermo Fisher Scientific, Inc.). Then, 30 µg proteins per lane were separated via 8% SDS-PAGE and transferred to PVDF membranes, which were blocked with 5% skimmed milk at room temperature for 1 h. Subsequently, the membranes were incubated overnight at 4°C with the following specific primary antibodies: Anti-GSK-3β (48 kDa; 1:1,000; cat. no. ab131356; Abcam), anti-β-catenin (94 kDa; 1:4,000; cat. no. ab16051; Abcam), anti-sFRP-4 (40 kDa; 1:3,000; cat. no. ab154167; Abcam) and anti-GAPDH (36 kDa; 1:1,000; cat. no. ab8245; Abcam). Following primary incubation, the membranes were incubated with the corresponding HRP-conjugated secondary antibodies (1:5,000; anti-mouse IgG, cat. no. ab197767 and anti-rabbit IgG, cat. no. ab6721; Abcam) for 1 h at room temperature. Protein bands were visualized using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific, Inc.) and the iBrightCL1000 imaging system (Thermo Fisher Scientific, Inc.). Protein expression was semi-quantified using ImageJ software (version 1.48; National Institutes of Health) with GAPDH as the loading control.
Statistical analysis
Statistical analyses were performed using GraphPad Prism software (version 8.0.1; GraphPad Software, Inc.). Data are presented as the mean ± SD from three independent experiments. The paired Student's t test was used to analyze comparisons between two groups. One way ANOVA followed by Bonferroni's post hoc test was used to analyze comparisons among multiple groups. The χ2 or Fisher's exact test was used to compare clinical characteristics between patients with HCC with lower sFRP-4 expression and patients with HCC with higher sFRP-4 expression. P<0.05 was considered to indicate a statistically significant difference.
Results
sFRP-4 expression is decreased in HCC
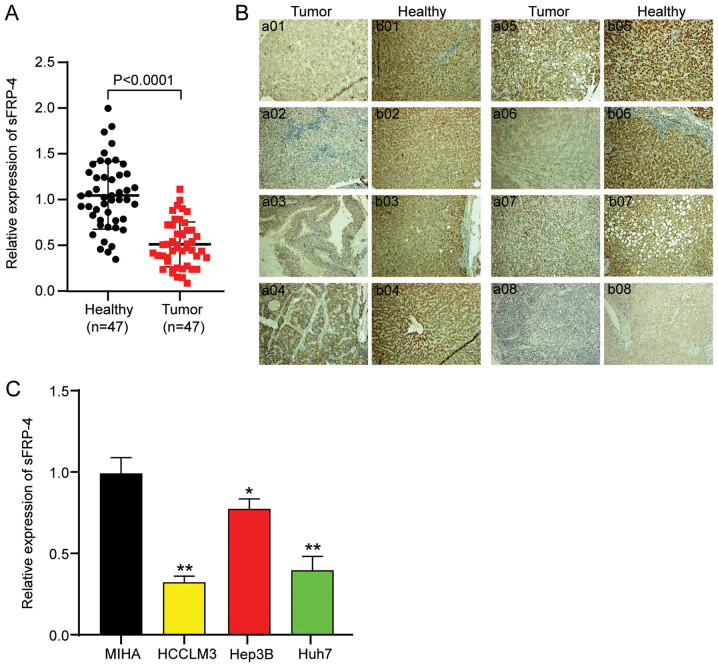
To identify the mechanism underlying sFRP-4 in HCC, the expression of sFRP-4 in 47 paired HCC and adjacent non-cancerous tissue samples was analyzed via RT-qPCR. sFRP-4 expression was significantly decreased by ~51% in HCC tissues compared with adjacent non-cancerous tissues (Fig. 1A). Subsequently, IHC analysis was performed to assess sFRP-4 expression in HCC tissues, and images from eight randomly selected paired HCC and adjacent healthy tissues are presented in Fig. 1B. sFRP-4 expression in tumor tissues was notably lower compared with adjacent healthy tissues. According to the mean value of sFRP4 (0.5) in HCC tumor tissues, as determined via RT-qPCR, patients with HCC were divided into two groups: sFRP-4 low expression (≤0.5) and sFRP-4 high expression (>0.5). The results demonstrated that the tissue expression level of sFRP-4 was significantly negatively associated with TNM stage, metastasis and AFP level in patients with HCC (Table I). Moreover, the expression levels of sFRP-4 in a normal liver cell line (MIHA) and three HCC cell lines (including HCCLM3, Hep3B and Huh7) were assessed. The results in HCC cell lines were consistent with HCC tissues. Compared with the MIHA cell line, the expression of sFRP-4 in HCC cell lines was significantly decreased. Among the HCC cell lines, sFRP-4 expression was downregulated to the lowest levels in HCCLM3 (67%) and Huh7 (60%) cells compared with MIHA cells (Fig. 1C). Therefore, HCCLM3 and Huh7 cell lines were selected for subsequent experiments. Collectively, the aforementioned results indicated that sFRP-4 expression was downregulated in HCC, which might exert tumor-suppressive functions.
Figure 1.
sFRP-4 is downregulated in HCC tissues and cells. (A) sFRP-4 mRNA expression levels in HCC tissues and adjacent healthy tissues were measured via RT-qPCR (n=47). Data were analyzed using the paired Student's t test. (B) Immunohistochemistry was performed to evaluate sFRP-4 expression in 8 paired HCC and adjacent healthy tissues. Images a01-a08 represent HCC tissues and images b01-b08 represent the corresponding adjacent healthy tissues (magnification, ×200). (C) sFRP-4 mRNA expression levels in HCC cell lines (HCCLM3, Hep3B and Huh7) and a normal human liver cell line (MIHA) were detected via RT-qPCR. Data were analyzed using one-way ANOVA followed by Bonferroni's post hoc test. Data are presented as the mean ± SD from three independent experiments. *P<0.05 and **P<0.001 vs. MIHA. sFRP-4, secreted frizzled-related protein 4; HCC, hepatocellular carcinoma; RT-qPCR, reverse transcription-quantitative PCR.
sFRP-4 restrains HCC cell viability and proliferation, but accelerates HCC cell apoptosis
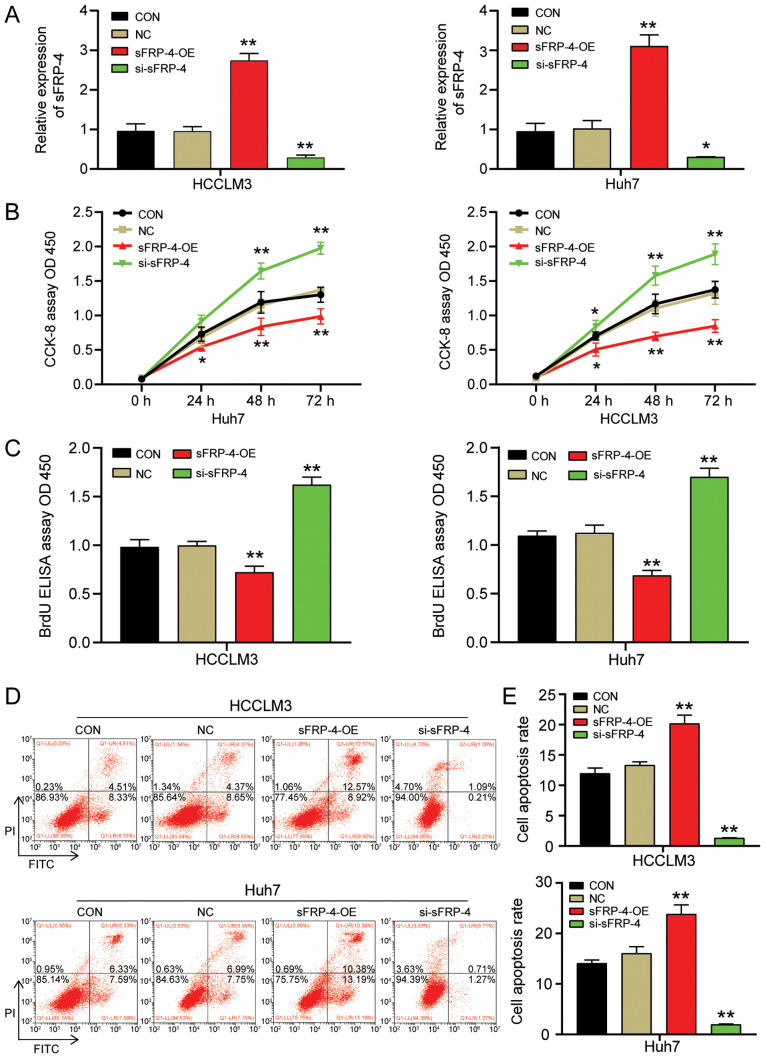
To investigate the influence of sFRP-4 on the function of HCC cells, HCCLM3 and Huh7 cells were transfected with sFRP-4-OE or si-sFRP-4. The transfection efficiencies of sFRP-4-OE and si-sFRP-4 are presented in Fig. S1. The RT-qPCR results demonstrated that sFRP-4-OE successfully increased sFRP-4 expression to a level ~3 times higher compared with the control group (Fig. 2A). By contrast, si-sFRP-4 significantly reduced the expression of sFRP-4 by ~69% compared with the control group. Subsequently, CCK-8, BrdU ELISA and flow cytometry assays were performed to evaluate the effect of sFRP-4 on HCC viability, proliferation and apoptosis, respectively. The CCK-8 assay results indicated that HCCLM3 and Huh7 cell viability was significantly decreased by sFRP-4-OE compared with the control group (Fig. 2B). However, HCCLM3 and Huh7 cell viability was significantly increased by si-sFRP-4 compared with the control group at 48 and 72 h. At 48 h post-transfection, alterations in cell viability were more pronounced compared with the control group (Fig. 2B), thus the BrdU ELISA assay was performed to assess HCCLM3 and Huh7 cell proliferation at 48 h post-transfection (Fig. 2C). Following transfection with sFRP-4-OE, the absorbance of HCCLM3 and Huh7 cells at 450 nm was reduced by ~34% compared with the control group. Moreover, sFRP-4 knockdown significantly increased the absorbance of HCCLM3 and Huh7 cells at 450 nm by ~58% compared with the control group. The results demonstrated that sFRP-4 displayed an inhibitory effect on HCC cell proliferation. Finally, flow cytometry was performed to assess cell apoptosis. Compared with the control group, HCCLM3 and Huh7 cell apoptosis was significantly increased by sFRP-4 overexpression, resulting in an apoptosis rate of 20.19%±1.39 and 23.84%±1.83, respectively (Fig. 2D and E). However, compared with the control group, HCCLM3 and Huh7 cell apoptosis was significantly decreased by sFRP-4 knockdown, resulting in an apoptosis rate of 1.32%±0.02 and 1.95%±0.12, respectively. In summary, the aforementioned results indicated that compared with the control group, sFRP-4 overexpression decreased HCC cell viability and proliferation, and accelerated HCC cell apoptosis, whereas sFRP-4 knockdown displayed the opposite effect.
Figure 2.
sFRP-4 inhibits HCC cell viability and proliferation, and promotes HCC cell apoptosis. (A) Transfection efficiency of sFRP-4-OE and si-sFRP-4 in HCCLM3 and Huh7 cells. (B) Effect of sFRP-4 on HCCLM3 and Huh7 cell viability, as evaluated by performing CCK-8 assays. (C) Effect of sFRP-4 on HCCLM3 and Huh7 cell proliferation, as evaluated by performing BrdU ELISA assays. Effect of sFRP-4 on HCCLM3 and Huh7 cell apoptosis was (D) evaluated via flow cytometry and (E) quantified. Data were analyzed using one-way ANOVA followed by Bonferroni's post hoc test. Data are presented as the mean ± SD from three independent experiments. *P<0.05 and **P<0.001 vs. CON. sFRP-4, secreted frizzled-related protein 4; HCC, hepatocellular carcinoma; OE, overexpression; si, small interfering RNA; CCK-8, Cell Counting Kit-8; CON, blank control; NC, negative control (empty vectors and si-NC); OD, optical density.
sFRP-4 inhibits the Wnt/β-catenin signaling pathway in HCC
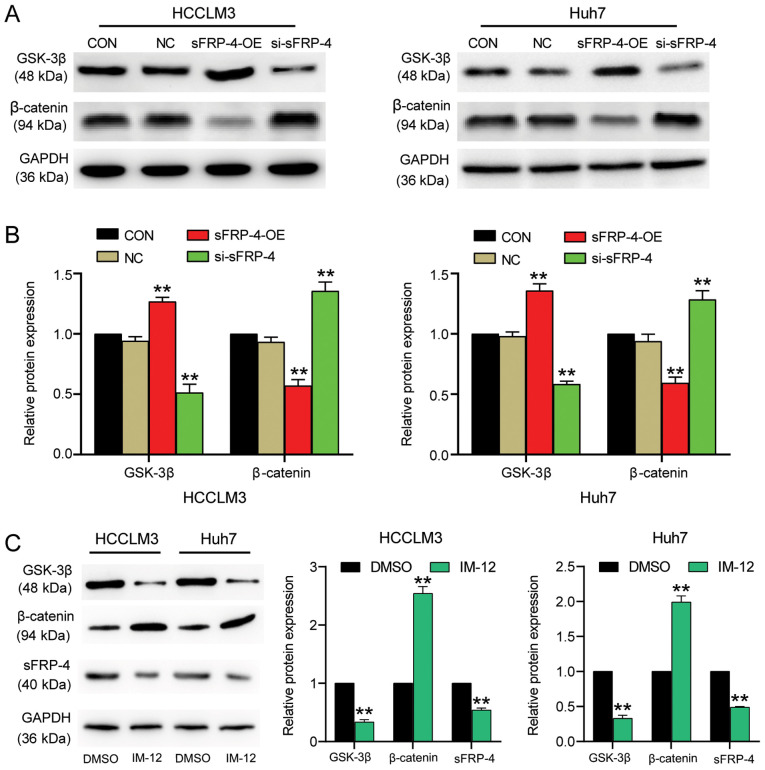
The effect of sFRP-4 on HCC carcinogenesis might alter the associated signaling pathways. The Wnt/β-catenin signaling pathway is one of the most classic carcinogenic signaling pathways (37). To explore whether sFRP-4 downregulation in HCC was mediated via regulation of the Wnt/β-catenin signaling pathway, the expression levels of key factors (GSK-3β and β-catenin) in the Wnt/β-catenin signaling pathway were measured. The western blotting results demonstrated that sFRP-4 knockdown significantly decreased GSK-3β protein expression levels by >40% compared with the control group (Fig. 3A and B). By contrast, sFRP-4 knockdown significantly increased β-catenin levels by ~30% compared with the control group. However, sFRP-4 overexpression displayed the opposite effect, significantly increasing GSK-3β expression levels by ~30% and decreasing β-catenin expression levels by ~50% compared with the control group. The results demonstrated that sFRP-4 increased GSK-3β expression and decreased β-catenin expression, suggesting that sFRP-4 might suppress the Wnt/β-catenin signaling pathway in HCC cells. To further evaluate the effect of the Wnt/β-catenin signaling pathway on sFRP-4, IM-12, a selective GSK-3β inhibitor, was used to activate the Wnt/β-catenin signaling pathway in HCCLM3 and Huh7 cells (Fig. 3C). The western blotting results demonstrated that GSK-3β protein expression levels were significantly decreased by ~70%, whereas β-catenin protein expression levels were significantly increased by >2-fold following treatment with IM-12 compared with DMSO, which indicated activation of the Wnt/β-catenin signaling pathway. Furthermore, sFRP-4 expression was significantly decreased by ~50% in cells treated with IM-12 compared with DMSO. The results suggested negative feedback regulation of sFRP-4 expression by the Wnt/β-catenin signaling pathway.
Figure 3.
sFRP-4 inhibits the Wnt/β-catenin signaling pathway in HCC. GSK-3β and β-catenin protein expression levels in HCCLM3 and Huh7 cells transfected with sFRP-4-OE, si-sFRP-4 or NC were (A) determined via western blotting and (B) semi-quantified. (C) GSK-3β, β-catenin and sFRP-4 protein expression levels in HCCLM3 and Huh7 cells following treatment with IM-12 or DMSO for 36 h were determined via western blotting and semi-quantified. Data were analyzed using one-way ANOVA followed by Bonferroni's post hoc test. Data are presented as the mean ± SD from three independent experiments. **P<0.001 vs. CON or DMSO. sFRP-4, secreted frizzled-related protein 4; HCC, hepatocellular carcinoma; OE, overexpression; si, small interfering RNA; NC, negative control (empty vectors and si-NC); CON, blank control.
sFRP-4 inhibits HCC cell viability and proliferation, and accelerates HCC cell apoptosis via the Wnt/β-catenin signaling pathway
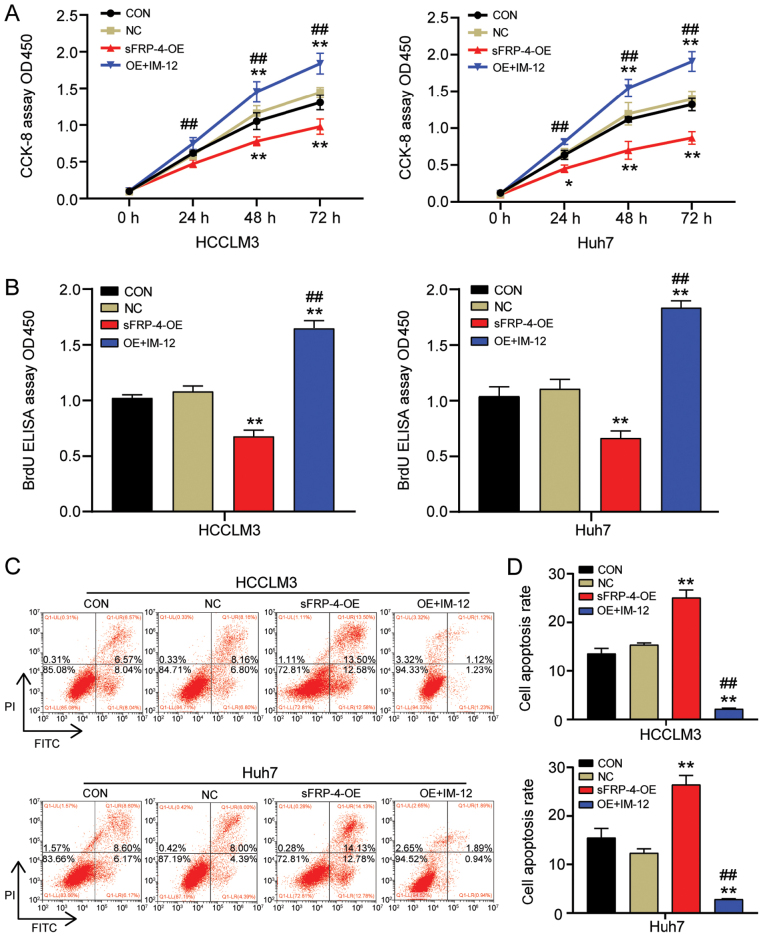
To further investigate whether sFRP-4 altered the tumor phenotype of HCC cells via the Wnt/β-catenin signaling pathway, the effect of sFRP-4 overexpression combined with IM-12 treatment on HCC cell viability, proliferation and apoptosis was assessed by performing CCK-8, BrdU ELISA and flow cytometry assays, respectively. The CCK-8 assay results demonstrated that IM-12 significantly reversed the suppressive effect of sFRP-4 overexpression on HCCLM3 and Huh7 cell viability (Fig. 4A). Similarly, the BrdU ELISA assay results demonstrated that IM-12 significantly reversed the inhibitory effects of sFRP-4 overexpression on HCCLM3 and Huh7 cell proliferation (Fig. 4B). HCCLM3 and Huh7 cell proliferation was significantly increased by 2.5-fold in the sFRP-4-OE + IM-12 group compared with the sFRP-4-OE group. Additionally, cell apoptosis was analyzed via flow cytometry. The results demonstrated that IM-12 significantly reversed sFRP-4 overexpression-mediated induction of HCCLM3 and Huh7 cell apoptosis (Fig. 4C and D). Compared with the sFRP-4-OE group, the rate of apoptosis in HCCLM3 and Huh7 cells was decreased by ~90% in the sFRP-4-OE + IM-12 group. The aforementioned results further suggested that sFRP-4 inhibited the malignant behavior of HCC cells by regulating the Wnt/β-catenin signaling pathway.
Figure 4.
sFRP-4 inhibits HCC cell viability and proliferation, and promotes HCC cell apoptosis via the Wnt/β-catenin signaling pathway. (A) HCCLM3 and Huh7 cell viability following transfection with sFRP-4-OE and treatment with IM-12 for 36 h was determined by performing CCK-8 assays. (B) HCCLM3 and Huh7 cell proliferation following transfection with sFRP-4-OE and treatment with IM-12 for 36 h was determined by performing BrdU ELISA assays. HCCLM3 and Huh7 cell apoptosis following transfection with sFRP-4-OE and treatment with IM-12 for 36 h was (C) determined via flow cytometry and (D) quantified. Data were analyzed using one-way ANOVA followed by Bonferroni's post hoc test. Data are presented as the mean ± SD from three independent experiments. *P<0.05 and **P<0.001 vs. CON; ##P<0.001 vs. sFRP-4-OE. sFRP-4, secreted frizzled-related protein 4; HCC, hepatocellular carcinoma; OE, overexpression; CCK-8, Cell Counting Kit-8; CON, blank control; NC, negative control; OD, optical density; IM-12, a selective GSK-3β inhibitor.
Discussion
The present study demonstrated that sFRP-4 expression was significantly downregulated in HCC tissues and cells compared with adjacent healthy tissues and MIHA cells, respectively. Moreover, compared with the control group, sFRP-4 overexpression suppressed HCC cell viability and proliferation, but facilitated HCC cell apoptosis. In addition, sFRP-4 overexpression increased GSK-3β expression and decreased β-catenin expression compared with the control group, which suggested that sFRP-4 inhibited the Wnt/β-catenin signaling pathway. IM-12 (a selective of GSK-3β inhibitor) was used to investigate the inhibitory function of sFRP-4. The results indicated that sFRP-4 inhibited the malignant behavior of HCC cells by suppressing the Wnt/β-catenin signaling pathway. Therefore, sFRP-4 may serve a negative regulatory role during HCC carcinogenesis and development, which indicated that sFRP-4 might serve as a promising therapeutic target for HCC.
sFRPs are a class of Wnt regulatory proteins that are often downregulated in a variety of different types of cancer, such as gastric (38), oral (39) and breast cancer (40), where they are associated with tumor development and poor prognosis (41,40). sFRP-4, a member of the sFRP family, is an underlying antagonist of the Wnt signaling pathway (13). Several studies have reported that sFRP-4 functions as a tumor suppressor in various forms of cancer (42,43). For instance, in mesothelioma and glioblastoma, the tumor suppressor role of sFRP-4 has been demonstrated. Cell functional experiments demonstrated that sFRP-4 overexpression displayed an inhibitory effect on the malignant behavior of tumor cells (41,44). Furthermore, in a study investigating HCC, the sFRP-4 methylation level was evaluated in 12 HCC cell lines and 19 HCC tissue samples. The results demonstrated that sFRP-4 methylation was present in three HCC cell lines (HLF, CHC4 and CHC32) (45). The methylation of sFRP family gene promoters led to transcriptional silencing of the gene, thereby reducing its expression (46). Therefore, it was hypothesized that sFRP-4 might be abnormally expressed in HCC. In our previous work, it was demonstrated that compared with healthy patients, the expression of sFRP-4 in the serum samples of patients with HCC was obviously increased. After the diagnostic value of sFRP-4 in the serum was evaluated, the study demonstrated that the combined use of sFRP-4 and AFP (a standard serum marker of HCC) could enhance the diagnostic accuracy of HCC (23). However, whether sFRP-4 is a tumor suppressor or oncogene in HCC is not completely understood, thus the mechanism underlying sFRP-4 in HCC is unclear. In the present study, sFRP-4 expression was significantly downregulated in HCC tissues and cells compared with adjacent healthy tissues and MIHA cells, respectively. The IHC assay results further demonstrated that the expression of sFRP-4 was notably reduced in HCC tissues compared with healthy adjacent tissues. Therefore, the results of the present study were contradictory to the results of our aforementioned previous study that investigated serum expression levels of sFRP-4. Although distinct methods were used to examine sFRP4 expression in the two studies, including human antibody arrays for serum detection, and RT-qPCR and IHC for tissue detection, both results were convincing. Therefore, it was hypothesized that the distinct sFRP4 expression in HCC tissues and serum might derive from the multiple sources of serum sFRP4 according to the GeneCards description, including liver, lung, spleen, bladder and reproductive organ, which suggested that the high expression of sFRP4 in serum is the result of multiple body tissues. However, it is possible that compensatory increases in sFRP4 may be activated in other body tissues and eventually result in the upregulation of sFRP4 in the serum of patients with HCC, thus the aforementioned hypothesis requires further investigation.
The present study also demonstrated that sFRP-4 knockdown not only enhanced HCC cell viability and proliferation, but also suppressed HCC cell apoptosis compared with the control group. However, sFRP-4 overexpression displayed the opposite effect, which suggested that sFRP-4 might serve a tumor-suppressor role during HCC carcinogenesis and development. Overall, the results of the present study were consistent with the results of previous studies investigating the role of sFRP-4 in glioblastoma and mesothelioma (41,44).
In addition, numerous studies have reported that excessive activation of the Wnt/β-catenin signaling pathway is associated with different types of human cancer, including HCC (47,48). The Wnt/β-catenin signaling pathway is an intricate and precise regulation process, which is also known as the canonical Wnt signaling pathway (37). The activation of the Wnt/β-catenin signaling pathway can be identified as downregulation of GSK-3β and stabilization of β-catenin, which is negatively regulated by GSK-3β (49,50). Therefore, when the signaling pathway is inactive, GSK-3β and other proteins constitute a degradation complex that contributes to the phosphorylation of β-catenin, resulting in the degradation of β-catenin. However, when the signaling pathway is activated, this degradation complex is dissociated and β-catenin degradation is suppressed, resulting in accumulation of β-catenin in the cytoplasm (51–53). Notably, as a mediator of the canonical Wnt signaling pathway, β-catenin serves to activate a series of functional genes via recruiting other transcriptional coactivators, such as Bcl-9 and Pygopus (54,55). Therefore, the expression level of β-catenin is closely related to the active state of the Wnt/β-catenin signaling pathway (37).
Previous studies have also demonstrated that sFRP-4 overexpression in cancer could decrease the expression level of β-catenin (15,41,56). This finding was exemplified by a study on ovarian cancer. Following treatment with human recombinant sFRP-4 treatment for 72 h in vitro, sFRP-4 protein expression levels were increased, whereas β-catenin protein expression levels were decreased in the nuclei of OVCAR3 cells, thus inhibiting the Wnt signaling pathway. Furthermore, sFRP-4 inhibited OVCAR3 cell migration and EMT, and promoted cell adhesion (15). In addition, another study analyzed the protein expression levels of downstream components (GSK-3β, phosphorylated β-catenin and β-catenin) of the Wnt signaling pathway in prostate, breast and ovarian cancer stem cells. The results indicated that following demethylation of sFRP-4, the expression levels of sFRP-4, GSK-3β and phosphorylated β-catenin were enhanced, whereas the expression levels of the non-phosphorylated β-catenin were decreased, resulting in inhibition of the Wnt signaling pathway (38).
In the present study, compared with the control group, sFRP-4 overexpression significantly increased GSK-3β expression and decreased β-catenin expression, whereas sFRP-4 knockdown significantly decreased GSK-3β expression and increased β-catenin expression. In sFRP-4-overexpression HCC cells, IM-12 (an inhibitor of GSK-3β, which is the key protein of the Wnt signaling pathway) was used, and the results suggested that sFRP-4 inhibited HCC cell viability and proliferation, and induced HCC cell apoptosis by inhibiting the Wnt/β-catenin signaling pathway. In addition, the results suggested that activation of the Wnt/β-catenin signaling pathway displayed a negative feedback effect on sFRP-4 expression, thereby counteracting the suppression of sFRP-4 on the tumorous phenotype of HCC cells.
To conclude, the present study elucidated the tumor suppressor role of sFRP-4 in HCC development. The results suggested that sFRP-4 inhibited HCC cell viability and proliferation, and promoted HCC cell apoptosis by inhibiting activation of the Wnt/β-catenin signaling pathway. The results of the present study might provide novel targets and theoretical foundations for the development of HCC therapeutic strategies. However, the results of the present study were limited as only isolated clinical HCC tissues and cell lines were used; therefore, the results of the present study should be verified using in vivo experiments, and the sFRP-4 methylation level should be analyzed to further investigate the mechanism underlying sFRP-4 in HCC.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by Hubei Provincial Natural Science Foundation of China (grant no. 2019CFB101).
Funding
The present study was supported by Hubei Provincial Natural Science Foundation of China (grant no. 2019CFB101).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CX and ZR designed the study. QW and XZ performed the majority of experiments. ZZ and BY performed the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of General Hospital of the Central Theater Command of the People's Liberation Army (approval no. [2020]024-2; Wuhan, China). All patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, Gores G. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 3.Xiang X, Zhong JH, Wang YY, You XM, Ma L, Xiang BD, Li LQ. Distribution of tumor stage and initial treatment modality in patients with primary hepatocellular carcinoma. Clin Transl Oncol. 2017;19:891–897. doi: 10.1007/s12094-017-1621-6. [DOI] [PubMed] [Google Scholar]
- 4.Han J, Wang F, Lan Y, Wang J, Nie C, Liang Y, Song R, Zheng T, Pan S, Pei T, et al. KIFC1 regulated by miR-532-3p promotes epithelial-to-mesenchymal transition and metastasis of hepatocellular carcinoma via gankyrin/AKT signaling. Oncogene. 2019;38:406–420. doi: 10.1038/s41388-018-0440-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Vilchez V, Turcios L, Marti F, Gedaly R. Targeting Wnt/beta-catenin pathway in hepatocellular carcinoma treatment. World J Gastroenterol. 2016;22:823–832. doi: 10.3748/wjg.v22.i2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saran U, Humar B, Kolly P, Dufour JF. Hepatocellular carcinoma and lifestyles. J Hepatol. 2016;64:203–214. doi: 10.1016/j.jhep.2015.08.028. [DOI] [PubMed] [Google Scholar]
- 7.Ozakyol A. Global epidemiology of hepatocellular carcinoma (HCC epidemiology) J Gastrointest Cancer. 2017;48:238–240. doi: 10.1007/s12029-017-9959-0. [DOI] [PubMed] [Google Scholar]
- 8.Serper M, Taddei TH, Mehta R, D'Addeo K, Dai F, Aytaman A, Baytarian M, Fox R, Hunt K, Goldberg DS, et al. Association of provider specialty and multidisciplinary care with hepatocellular carcinoma treatment and mortality. Gastroenterology. 2017;152:1954–1964. doi: 10.1053/j.gastro.2017.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng D, Deng J, Zhang B, He X, Meng Z, Li G, Ye H, Zheng S, Wei L, Deng X, et al. lncRNA HOTAIR epigenetically suppresses miR-122 expression in hepatocellular carcinoma via DNA methylation. EBioMedicine. 2018;36:159–170. doi: 10.1016/j.ebiom.2018.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bukhari SA, Yasmin A, Zahoor MA, Mustafa G, Sarfraz I, Rasul A. Secreted frizzled-related protein 4 and its implication in obesity and type-2 diabetes. IUBMB Life. 2019;71:1701–1710. doi: 10.1002/iub.2123. [DOI] [PubMed] [Google Scholar]
- 11.Agostino M, Pohl S, Dharmarajan A. Structure-based prediction of Wnt binding affinities for Frizzled-type cysteine-rich domains. J Biol Chem. 2017;292:11218–11229. doi: 10.1074/jbc.M117.786269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Han Q, Zhao W, Bentel J, Shearwood AM, Zeps N, Joseph D, Iacopetta B, Dharmarajan A. Expression of sFRP-4 and beta-catenin in human colorectal carcinoma. Cancer Lett. 2006;231:129–137. doi: 10.1016/j.canlet.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 13.Deshmukh A, Arfuso F, Newsholme P, Dharmarajan A. Regulation of cancer stem cell metabolism by secreted frizzled-related protein 4 (sFRP4) Cancers (Basel) 2018;10:40. doi: 10.3390/cancers10020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Duan XL, Qi XL, Meng L, Xu YS, Wu T, Dai PG. Concurrent hypermethylation of SFRP2 and DKK2 activates the Wnt/β-catenin pathway and is associated with poor prognosis in patients with gastric cancer. Mol Cells. 2017;40:45–53. doi: 10.14348/molcells.2017.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ford CE, Jary E, Ma SS, Nixdorf S, Heinzelmann-Schwarz VA, Ward RL. The Wnt gatekeeper SFRP4 modulates EMT, cell migration and downstream Wnt signalling in serous ovarian cancer cells. PLoS One. 2013;8:e54362. doi: 10.1371/journal.pone.0054362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brebi P, Hoffstetter R, Andana A, Ili CG, Saavedra K, Viscarra T, Retamal J, Sanchez R, Roa JC. Evaluation of ZAR1 and SFRP4 methylation status as potentials biomarkers for diagnosis in cervical cancer: Exploratory study phase I. Biomarkers. 2014;19:181–188. doi: 10.3109/1354750X.2013.867535. [DOI] [PubMed] [Google Scholar]
- 17.He B, Lee AY, Dadfarmay S, You L, Xu Z, Reguart N, Mazieres J, Mikami I, McCormick F, Jablons DM. Secreted frizzled-related protein 4 is silenced by hypermethylation and induces apoptosis in beta-catenin-deficient human mesothelioma cells. Cancer Res. 2005;65:743–748. [PubMed] [Google Scholar]
- 18.Pereira TDSF, Diniz MG, França JA, Moreira RG, Menezes GHF, Sousa SF, Castro WH, Gomes CC, Gomez RS. The Wnt/β-catenin pathway is deregulated in cemento-ossifying fibromas. Oral Surg Oral Med Oral Pathol Oral Radiol. 2018;125:172–178. doi: 10.1016/j.oooo.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Sandsmark E, Andersen MK, Bofin AM, Bertilsson H, Drabløs F, Bathen TF, Rye MB, Tessem MB. SFRP4 gene expression is increased in aggressive prostate cancer. Sci Rep. 2017;7:14276. doi: 10.1038/s41598-017-14622-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang MW, Tao LY, Yang JY, Jiang YS, Fu XL, Liu W, Huo YM, Li J, Zhang JF, Hua R, et al. SFRP4 is a prognostic marker and correlated with Treg cell infiltration in pancreatic ductal adenocarcinoma. Am J Cancer Res. 2019;9:363–377. [PMC free article] [PubMed] [Google Scholar]
- 21.Nfonsam LE, Jandova J, Jecius HC, Omesiete PN, Nfonsam VN. SFRP4 expression correlates with epithelial mesenchymal transition-linked genes and poor overall survival in colon cancer patients. World J Gastrointest Oncol. 2019;11:589–598. doi: 10.4251/wjgo.v11.i8.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang D, Yu B, Deng Y, Sheng W, Peng Z, Qin W, Du X. SFRP4 was overexpressed in colorectal carcinoma. J Cancer Res Clin Oncol. 2010;136:395–401. doi: 10.1007/s00432-009-0669-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu C, Zeng XH, Wang L, Tao SQ, Wu QX, Zhu P, Deng GH, Wang YM. sFRP-4, a potential novel serum marker for chronic hepatitis B-related hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2015;14:164–170. doi: 10.1016/S1499-3872(15)60352-6. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Rajasekaran M, Hui KM. Atypical regulators of Wnt/β-catenin signaling as potential therapeutic targets in Hepatocellular Carcinoma. Exp Biol Med (Maywood) 2017;242:1142–1149. doi: 10.1177/1535370217705865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao TP, Kuhl M. An updated overview on Wnt signaling pathways: A prelude for more. Circ Res. 2010;106:1798–1806. doi: 10.1161/CIRCRESAHA.110.219840. [DOI] [PubMed] [Google Scholar]
- 26.Wang W, Li S, Liu P, Sideras K, van de Werken HJG, van der Heide M, Cao W, Lavrijsen M, Peppelenbosch MP, Bruno M, et al. Oncogenic STRAP supports hepatocellular carcinoma growth by enhancing Wnt/β-catenin signaling. Mol Cancer Res. 2019;17:521–531. doi: 10.1158/1541-7786.MCR-18-0054. [DOI] [PubMed] [Google Scholar]
- 27.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36:1461–1473. doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duchartre Y, Kim YM, Kahn M. The Wnt signaling pathway in cancer. Crit Rev Oncol Hematol. 2016;99:141–149. doi: 10.1016/j.critrevonc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang R, Wu Y, Huang W, Chen W. MicroRNA-940 Targets INPP4A or GSK3beta and Activates the Wnt/beta-Catenin pathway to regulate the malignant behavior of bladder cancer Cells. Oncol Res. 2018;26:145–155. doi: 10.3727/096504017X14902261600566. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Peng Y, Zhang X, Ma Q, Yan R, Qin Y, Zhao Y, Cheng Y, Yang M, Wang Q, Feng X, et al. miRNA-194 activates the Wnt/β-catenin signaling pathway in gastric cancer by targeting the negative Wnt regulator, SUFU. Cancer Lett. 2017;385:117–127. doi: 10.1016/j.canlet.2016.10.035. [DOI] [PubMed] [Google Scholar]
- 31.Ren Y, Tao J, Jiang Z, Guo D, Tang J. Pimozide suppresses colorectal cancer via inhibition of Wnt/beta-catenin signaling pathway. Life Sci. 2018;209:267–273. doi: 10.1016/j.lfs.2018.08.027. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Hu SL, Qiao CH, Ye JF, Li M, Ma HM, Wang JH, Xin SY, Yuan ZL. lncRNA-NEF inhibits proliferation, migration and invasion of esophageal squamous-cell carcinoma cells by inactivating wnt/β-catenin pathway. Eur Rev Med Pharmacol Sci. 2018;22:6824–6831. doi: 10.26355/eurrev_201810_16150. [DOI] [PubMed] [Google Scholar]
- 33.Zhou P, Li Y, Li B, Zhang M, Liu Y, Yao Y, Li D. NMIIA promotes tumor growth and metastasis by activating the Wnt/β-catenin signaling pathway and EMT in pancreatic cancer. Oncogene. 2019;38:5500–5515. doi: 10.1038/s41388-019-0806-6. [DOI] [PubMed] [Google Scholar]
- 34.Hu Z, Wang P, Lin J, Zheng X, Yang F, Zhang G, Chen D, Xie J, Gao Z, Peng L, Xie C. MicroRNA-197 promotes metastasis of hepatocellular carcinoma by activating Wnt/β-catenin signaling. Cell Physiol Biochem. 2018;51:470–486. doi: 10.1159/000495242. [DOI] [PubMed] [Google Scholar]
- 35.Song J, Xie C, Jiang L, Wu G, Zhu J, Zhang S, Tang M, Song L, Li J. Transcription factor AP-4 promotes tumorigenic capability and activates the Wnt/β-catenin pathway in hepatocellular carcinoma. Theranostics. 2018;8:3571–3583. doi: 10.7150/thno.25194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Lv Q, Bian H, Yang L, Guo KL, Ye SS, Dong XF, Tao LL. A novel tumor suppressor SPINK5 targets Wnt/β-catenin signaling pathway in esophageal cancer. Cancer Med. 2019;8:2360–2371. doi: 10.1002/cam4.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin H, Kim JH, Lee YS, Lee YC. Change in gene expression profiles of secreted frizzled-related proteins (SFRPs) by sodium butyrate in gastric cancers: Induction of promoter demethylation and histone modification causing inhibition of Wnt signaling. Int J Oncol. 2012;40:1533–1542. doi: 10.3892/ijo.2012.1327. [DOI] [PubMed] [Google Scholar]
- 39.Pannone G, Bufo P, Santoro A, Franco R, Aquino G, Longo F, Botti G, Serpico R, Cafarelli B, Abbruzzese A, et al. WNT pathway in oral cancer: Epigenetic inactivation of WNT-inhibitors. Oncol Rep. 2010;24:1035–1041. doi: 10.3892/or.2010.1035. [DOI] [PubMed] [Google Scholar]
- 40.Deshmukh A, Arfuso F, Newsholme P, Dharmarajan A. Epigenetic demethylation of sFRPs, with emphasis on sFRP4 activation, leading to Wnt signalling suppression and histone modifications in breast, prostate, and ovary cancer stem cells. Int J Biochem Cell Biol. 2019;109:23–32. doi: 10.1016/j.biocel.2019.01.016. [DOI] [PubMed] [Google Scholar]
- 41.Perumal V, Dharmarajan AM, Fox SA. The Wnt regulator SFRP4 inhibits mesothelioma cell proliferation, migration, and antagonizes Wnt3a via its netrin-like domain. Int J Oncol. 2017;51:362–368. doi: 10.3892/ijo.2017.4011. [DOI] [PubMed] [Google Scholar]
- 42.Pawar NM, Rao P. Secreted frizzled related protein 4 (sFRP4) update: A brief review. Cell Signal. 2018;45:63–70. doi: 10.1016/j.cellsig.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 43.Bhuvanalakshmi G, Arfuso F, Millward M, Dharmarajan A, Warrier S. Secreted frizzled-related protein 4 inhibits glioma stem-like cells by reversing epithelial to mesenchymal transition, inducing apoptosis and decreasing cancer stem cell properties. PLoS One. 2015;10:e0127517. doi: 10.1371/journal.pone.0127517. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Bhuvanalakshmi G, Gamit N, Patil M, Arfuso F, Sethi G, Dharmarajan A, Kumar AP, Warrier S. Stemness, pluripotentiality, and Wnt antagonism: sFRP4, a wnt antagonist mediates pluripotency and stemness in glioblastoma. Cancers (Basel) 2018;11:25. doi: 10.3390/cancers11010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takagi H, Sasaki S, Suzuki H, Toyota M, Maruyama R, Nojima M, Yamamoto H, Omata M, Tokino T, Imai K, Shinomura Y. Frequent epigenetic inactivation of SFRP genes in hepatocellular carcinoma. J Gastroenterol. 2008;43:378–389. doi: 10.1007/s00535-008-2170-0. [DOI] [PubMed] [Google Scholar]
- 46.Yu J, Xie Y, Li M, Zhou F, Zhong Z, Liu Y, Wang F, Qi J. Association between SFRP promoter hypermethylation and different types of cancer: A systematic review and meta-analysis. Oncol Lett. 2019;18:3481–3492. doi: 10.3892/ol.2019.10709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang T, Ma Z, Liu L, Sun J, Tang H, Zhang B, Zou Y, Li H. DDX39 promotes hepatocellular carcinoma growth and metastasis through activating Wnt/β-catenin pathway. Cell Death Dis. 2018;9:675. doi: 10.1038/s41419-018-0591-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin J, Lin W, Ye Y, Wang L, Chen X, Zang S, Huang A. Kindlin-2 promotes hepatocellular carcinoma invasion and metastasis by increasing Wnt/β-catenin signaling. J Exp Clin Cancer Res. 2017;36:134. doi: 10.1186/s13046-017-0603-4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katoh M, Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res. 2007;13:4042–4045. doi: 10.1158/1078-0432.CCR-06-2316. [DOI] [PubMed] [Google Scholar]
- 51.Cai J, Guan H, Fang L, Yang Y, Zhu X, Yuan J, Wu J, Li M. MicroRNA-374a activates Wnt/β-catenin signaling to promote breast cancer metastasis. J Clin Invest. 2013;123:566–579. doi: 10.1172/JCI65871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fang L, Cai J, Chen B, Wu S, Li R, Xu X, Yang Y, Guan H, Zhu X, Zhang L, et al. Aberrantly expressed miR-582-3p maintains lung cancer stem cell-like traits by activating Wnt/β-catenin signalling. Nat Commun. 2015;6:8640. doi: 10.1038/ncomms9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng Y, Zhang X, Feng X, Fan X, Jin Z. The crosstalk between microRNAs and the Wnt/beta-catenin signaling pathway in cancer. Oncotarget. 2017;8:14089–14106. doi: 10.18632/oncotarget.12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Staal FJ, Clevers H. Tcf/Lef transcription factors during T-cell development: Unique and overlapping functions. Hematol J. 2000;1:3–6. doi: 10.1038/sj.thj.6200001. [DOI] [PubMed] [Google Scholar]
- 55.Kramps T, Peter O, Brunner E, Nellen D, Froesch B, Chatterjee S, Murone M, Züllig S, Basler K. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell. 2002;109:47–60. doi: 10.1016/S0092-8674(02)00679-7. [DOI] [PubMed] [Google Scholar]
- 56.Warrier S, Bhuvanalakshmi G, Arfuso F, Rajan G, Millward M, Dharmarajan A. Cancer stem-like cells from head and neck cancers are chemosensitized by the Wnt antagonist, sFRP4, by inducing apoptosis, decreasing stemness, drug resistance and epithelial to mesenchymal transition. Cancer Gene Ther. 2014;21:381–388. doi: 10.1038/cgt.2014.42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.