Abstract
BACKGROUND AND PURPOSE: Precise registration of CT and MR images is crucial in many clinical cases for proper diagnosis, decision making or navigation in surgical interventions. Various algorithms can be used to register CT and MR datasets, but prior to clinical use the result must be validated. To evaluate the registration result by visual inspection is tiring and time-consuming. We propose a new automatic registration assessment method, which provides the user a color-coded fused representation of the CT and MR images, and indicates the location and extent of poor registration accuracy.
METHODS: The method for local assessment of CT–MR registration is based on segmentation of bone structures in the CT and MR images, followed by a voxel correspondence analysis. The result is represented as a color-coded overlay. The algorithm was tested on simulated and real datasets with different levels of noise and intensity non-uniformity.
RESULTS: Based on tests on simulated MR imaging data, it was found that the algorithm was robust for noise levels up to 7% and intensity non-uniformities up to 20% of the full intensity scale. Due to the inability to distinguish clearly between bone and cerebro-spinal fluids in the MR image (T1-weighted), the algorithm was found to be optimistic in the sense that a number of voxels are classified as well-registered although they should not. However, nearly all voxels classified as misregistered are correctly classified.
CONCLUSION: The proposed algorithm offers a new way to automatically assess the CT–MR image registration accuracy locally in all the areas of the volume that contain bone and to represent the result with a user-friendly, intuitive color-coded overlay on the fused dataset.
In an increasing number of clinical cases, both X-ray CT and MR images of the head are acquired for diagnosis, surgical planning, and more recently for surgical navigation. X-ray tomography offers high resolution in the visualization of bone structures, but its soft tissue contrast is poor. Conversely, MR imaging offers high contrast for the visualization of the soft-tissue morphology, but it produces weak signal intensity in bone. Since these two imaging modalities are complementary, integration of both modalities to spatially relate the two types of structural information is desired in many clinical applications. The data integration process involves two steps: registration, the process of bringing the image data into spatial alignment, and fusion, the process of presenting the data in a common display. Fused representations of CT–MR images are valuable in clinical diagnosis, in planning surgery or radiation therapy, and in image-guided surgical interventions.
Many methods have been proposed for registering CT and MR medical images (1–3), including those using stereotactic frames, pair-point matching, surface measurements, segmented objects, and direct use the gray-value intensities such as the widely used mutual information algorithms (4, 5). Although validation experiments with controlled CT–MR datasets have shown that certain algorithms achieve high registration accuracy (3), accuracy varies from one CT–MR pair to another. Even the best algorithms can sometimes fail, leading to errors of 6 mm or more (2). Even when overall registration accuracy is good, the registration error is nonuniform and varies in different regions of the image. The location dependence of registration error has been thoroughly analyzed for pair-point based registration (6). Besides errors inherent to the registration process itself (e.g., failure of the optimization process), local registration errors may arise from geometric distortions on the source images. On MR images, geometric distortions are caused by magnetic field inhomogeneities, image wrap-arounds, and chemical shift artifacts (7), whereas on CT, artifacts due to electron-attenuated materials can be problematic. Distortions due to patient movement or anisotropic scale miscalibrations can also severely affect local registration accuracy. Lastly, despite the available registration algorithms, image alignment is often manually performed with visual inspection; this is error prone and time-consuming.
These uncertainties and errors in the registration of CT–MR image pairs may lead to uncertainty in diagnosis, surgical planning, or surgical procedure. Therefore, visual or automatic assessment of registration accuracy is necessary before the registered or fused image pair is clinically used. A previous study has shown that even experienced observers cannot reliably detect misregistration of less than 2 mm (8). Since registration error is not uniformly distributed over a whole volume, a complete 3D plot of errors would be of great use, as Woods proposed (9).
The purpose of our study was to assess an automatic local registration algorithm for CT and MR images of the head. This algorithm classifies individual voxels as well registered or badly registered on the basis of a correspondence analysis of voxels of cortical bone structures in CT and MR datasets. The results are color coded and visualized in a fused CT–MR image volume, making it easy for the human operator to identify the regions of low and high registration accuracy.
Methods
Outline of the Assessment Method
The proposed registration assessment method for CT–MR images of the head was based on a correspondence analysis of cortical bone structures. On CT images, dense structures such as bones appear bright and correspond to strong absorption of X-rays, whereas on MR images, the signal intensity of dense structures is low because of their low content of excitable hydrogen atoms.
The registration assessment method is summarized in five steps, as follows (Fig 1): First, the CT and MR datasets (Fig 1A and B) are registered and resampled to obtain voxel-by-voxel correspondence. Second, cortical bone structures on the CT image are segmented by using a user-adjustable threshold, initialized based on Hounsfield units (Fig 1C). The isolated volume of cortical bone structures is labeled SCT. Third, the bone volume SCT is mapped onto the MR dataset, resulting in an MR subvolume labeled SMR1. Fourth, a custom segmentation algorithm of cortical bone structures in the MR volume is applied, yielding a subvolume labeled SMR2. Voxels that belong to both SMR1 and SMR2 are classified as safe, which represents high registration accuracy, and colored green. Voxels that belong to SMR1 but not SMR2 are classified as unsafe, representing a low registration accuracy; these are colored red. Fifth, a fused CT–MR image is synthesized by overlaying the color-coded subvolume SMR1 onto the original MR image (Fig 1D).
Fig 1.
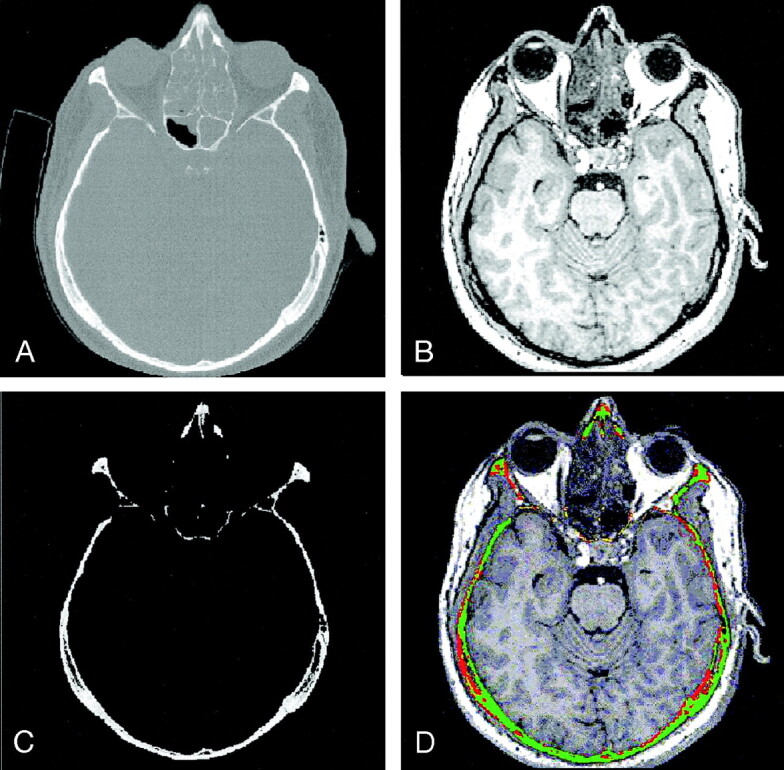
Steps in the correspondence analysis algorithm.
A and B, Pair of registered CT (A) and MR imaging (B) datasets.
C, Subvolume SCT containing the segmented cortical bone structures of the original CT image.
D, Result of registration assessment overlaid onto the original MR image.
Segmentation of Cortical Bone Structures on MR Images
The CT–MR registration assessment method requires a segmentation of bone in the MR images (step 4), which is difficult because of the low signal intensity of bone and the difficulty in distinguishing this from other low-intensity signals resulting from background, air cavities, or cerebro-spinal fluids (CSF) (10). The segmentation algorithm developed for this application consisted of a combination of thresholding and region-growing.
Thresholding.—
To find an adequate threshold to separate bone structures from soft-tissue, we used the nonparametric and unsupervised method of automatic threshold selection presented by Otsu (11). This method “selects an optimal threshold to separate objects from their background. Ideally, the histogram of the image has a deep valley between the peaks representing objects and background. However, for most real images it is difficult to detect the bottom of the valley precisely, especially when the valley is flat and broad or when the two peaks are extremely unequal in height” (11). This algorithm computes the optimal threshold k to separate object classes 0 and 1 by maximizing the between-class variance σB2(k), as follows (Eq 1).
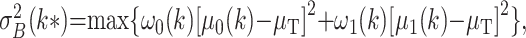 |
(1) |
where ω0(k) and ω1(k) are the respective probabilities of class 0 and 1 occurring on the image, μ0 and μ1 are the respective mean gray values of the object classes 0 and 1, and μT the mean gray level of the entire image. The between-class variance is introduced as a discriminant criterion to measure the goodness of class separation.
MR images have more than two main object classes (air, CSF, bone, gray matter, white matter, muscle and fat). When this algorithm was applied to MR images of the head, the computed threshold (t1 in Fig 2A) roughly separated the low-signal-intensity classes from the high-signal-intensity classes. The low-signal-intensity classes include air, bone, CSF, and some soft tissues, whereas the high-signal-intensity class represent soft tissues. A single application of the Otsu algorithm, as shown by the thresholded image ∀ xk ∈ MR, xk < t1 (Fig 2B), produced an overestimated threshold for the segmentation of cortical bone. It was necessary to further separate the low-intensity class into its subclasses to obtain an adequate threshold for bone segmentation.
Fig 2.
Bone Segmentation in MR Images (triple application of Otsu method).
A, Gray-value histogram of MR image in Figure 1B. Thresholds t1, t2, and t3 are obtained by triple application of the Otsu threshold selection method.
B, inverted MR image after application of the upper threshold t1, which does not clearly separate all of the soft tissues from the lower-intensity classes.
C, Zoomed view of histogram in A.
D, Inverted MR image after application of the upper threshold t3. Soft tissues are effectively removed. What remains are the low-intensity-classes, including bone structures, air, and CSF.
For this reason, Otsu’s algorithm was performed again. This time we considered only the voxels that had gray values less than the initial Otsu threshold t1. The result was a new threshold t2, which separated the lowest-intensity subclass—air background—from the rest. This threshold underestimated the intensity barrier that separated bone structures from soft-tissue (i.e., some parts of cortical bone were not segmented).
Finally, Otsu’s algorithm was performed a third time, when only voxels with intensity less than t1 and greater than t2 were considered. This yielded a third threshold value, t3. Using the new value t3 as an upper threshold on the MR images led to good separation of cortical bone and soft tissues (Fig 2D). However, some undesired areas with air and CSF were also segmented. It could be shown that no single threshold unambiguously separated bone from other structures in the head area without the use of additional segmentation steps.
Region Growing.—
To improve the quality of bone segmentation on MR images, the second Otsu threshold t2 was used for initial bone segmentation, followed by region growing up to the third Otsu threshold, t3. This procedure reduced the effect of segmenting disconnected non-bone areas (small volumes of air or CSF) that would have been segmented by direct application of a third Otsu method.
The described segmentation procedure primarily segmented bone from soft-tissues. However, it also segmented smaller regions of CSF and air cavities. Their effect on the registration assessment algorithm was small (as will be explained later), since the assessment was not performed on the whole image volume but only on the subvolume SMR1 corresponding to the segmented bone on CT.
Because voxels have finite size, partial-volume effects occurred where voxels contain a mixture of two materials, as was the case for voxels at the border between cortical bone and soft tissues. Therefore, registration uncertainty could occur for voxels at border locations between bone and soft tissue. The extent of measured misregistration (in millimeters) could be given for any point in the image for the three spatial directions by counting the number of adjacent red voxels in the x, y, and z directions.
Image Data
To evaluate the proposed registration assessment method, we first performed validation experiments with simulated and then real CT–MR image pairs.
Because no a priori knowledge of the hard and soft tissue locations was available for real patient data, we validated the algorithm by using simulated T1-weighted MR data (with 1-mm section thickness and TR/TE of 18/10) from the MR imaging simulator of McGill University, Montreal, Canada (available at www.bic.mni.mcgill.ca/brainweb/). The simulator provided complete MR imaging volumes of the head at different noise levels and intensity inhomogeneities. It also provided separate volumes of the main tissue classes that make up the anatomic model, such as bone, background, CSF, gray matter, and white matter. The bone subvolume was treated as the segmented bone from the CT data.
The algorithm was also validated with eight patient CT and MR datasets. Three samples of which are shown in Figure 3, where green voxels represent safe regions with high registration accuracy, and red voxels represent unsafe regions with low registration accuracy.
Fig 3.
Three fused CT–MR patient datasets assessed with our algorithm. Green voxels represent safe regions with high registration accuracy; red voxels represent unsafe regions with low registration accuracy.
A, Patient A.
B, Patient B.
C, Patient C.
CT data were obtained from a helical scanner with 1.25-mm section thickness, 140 kV, and 120 mA (LightSpeed Ultra, GE Medical Systems, Milwaukee, WI). MR imaging data were of the T1-weighted type obtained from a 1.5-T unit (Sonata; Siemens AG, Erlangen, Germany) with TR/TE of 2000/3.9. Since the data were acquired independent of our study, approval of the local ethic committee was not needed. The corresponding CT–MR datasets in our trials were registered by using normalized mutual information (12, 13), although other methods could be used as well.
Software Implementation
The registration assessment algorithm was implemented in C++ by using classes and routines made available by the open-source software system ITK (Insight Segmentation and Registration Toolkit, U.S. National Library of Medicine, Bethesda, MD, available at www.itk.org). The processing time was typically 3 minutes for a CT volume of 512 × 512 × 100 voxels on a standard desktop computer (Linux kernel version 2.4.20); this included loading the images to viewing the final result. Since the registration was done offline, computation time was not an issue. After the registration assessment was completed, the user could section the volume in real time in arbitrary directions and inspect the quality of the registration in the region of interest in real time.
Results
Validation with Simulated CT–MR Imaging Data
As a first step in the validation process with simulated data, we evaluated the segmentation method for bone structures on MR images. Subsequently, we validated the complete registration assessment algorithm.
Validation of the Segmentation Method for Bone Structures on MR Images.—
To evaluate the segmentation algorithm for bone structures with MR imaging data, we computed the percentage of the bone volume segmented by the algorithm (Eq 2):
 |
(2) |
where SMR is the segmented bone volume of the MR image and Sb is the a priori known bone volume, as provided by the simulator. The value of rbone is 1 if Sb is fully contained in SMR, and 0 if the two volumes do not overlap.
The bone segmentation algorithm was tested on simulated MR datasets with different levels of noise and nonuniformity of intensity, as shown in the Table. For gray-value noise levels from 0% to 7% and intensity nonuniformities from 0% to 20% of the full intensity scale, the rbone values were close to 99%, indicating good segmentation of bone structures. For higher noise and intensity nonuniformity levels the Otsu-based segmentation algorithm failed. Figure 4 illustrates the segmentation results for a simulated MR imaging volume with 3% noise and 20% intensity nonuniformity and displays the corresponding pure bone volume, as provided by the simulator. As explained in Methods, the algorithm segmented more than only pure bone. It also segmented small areas of CSF and air cavities, which represented false-positive errors. To evaluate the effect and magnitude of this error for our application, we validated the registration assessment algorithm, as described in the next section.
Assessment of the bone segmentation algorithm on simulated MR images at different noise and intensity nonuniformity levels
| Noise (%)* | Intensity Nonuniformity (%)* | rbone (%)† |
|---|---|---|
| 0 | 0 | 99.98 |
| 0 | 20 | 99.93 |
| 3 | 0 | 99.74 |
| 3 | 20 | 99.70 |
| 5 | 0 | 99.55 |
| 5 | 20 | 99.45 |
| 7 | 0 | 99.00 |
| 7 | 20 | 97.28 |
| 9 | 0 | 14.96 |
| 9 | 20 | 13.65 |
Note.—Segmentation fails above 7% noise.
Percentages refer to the full intensity scale.
Percentage of bone voxels that were segmented.
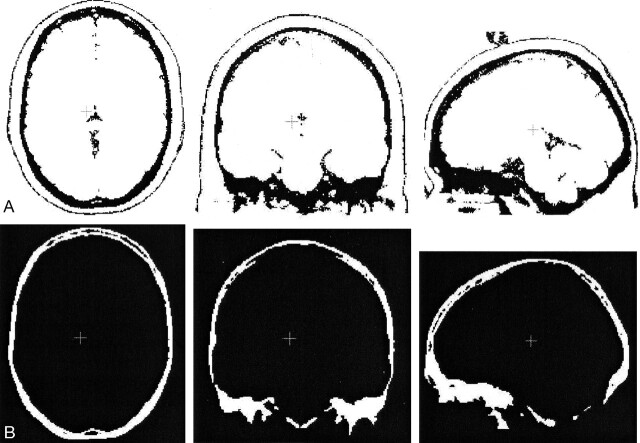
Fig 4.
Segmentation results for a simulated MR volume.
A, Segmentation result for bone regions on the MR image (axial, coronal, and sagittal).
B, True bone regions as provided by the MR simulator (axial, coronal, and sagittal).
Validation of the Registration Assessment Result.—
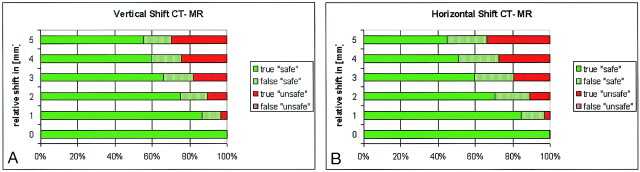
To validate the registration assessment algorithm, we evaluated a set of simulated CT–MR data pairs, which were intentionally misregistered by inducing a relative translation in horizontal and vertical directions. Given that a priori knowledge of the bone location was available for this setup, the assessment result could be verified for each voxel. Figure 5A shows for the horizontal misregistration the percentage of voxels that were classified correctly or falsely as safe (high registration accuracy = green), respectively unsafe (low registration accuracy = red). Similar results were obtained for the vertical misregistration, shown in Figure 5B. The results show that the algorithm was optimistic in the sense that a number of voxels were classified as safe, although they should not have been. However, all of the unsafe voxels, with rare exception (less than 0.07%), were correctly and consistently classified as unsafe. In other terms, the algorithm had good predictive value for unsafe voxels αunsafe ≈ 1 for all examined levels of misregistrations (Eq 3):
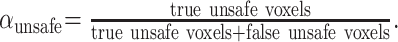 |
(3) |
Fig 5.
Validation of the registration assessment results based on simulated CT–MR data shifted purposely. Colored bars represent the percentage of correctly or falsely classified voxels as: safe (green) or unsafe (red). Results show that the algorithm is optimistic (some false-safe voxels exist), but there is practically no false detection (< 0.3%) of unsafe voxels.
A, Horizontal shift by 0–5 mm.
B, Vertical shift by 0–5 mm.
On the other hand, the predictive value of correctly registered voxels αsafe, (Eq 4) varied depending on the level of misregistration, from 1.0 to 0.65.
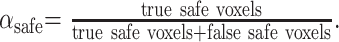 |
(4) |
This effect is well explained by the fact that the segmentation method for bone in MR segmented bone and also partially segmented some air cavities and CSF. As a result, a voxel representing bone in the CT that matches a voxel representing CSF in MR imaging was not seen as being misregistered.
Evaluation with Real CT–MR Imaging Data
Validating the registration assessment algorithm on real clinical data was difficult because of the lack of a prior knowledge of the location of the various tissues on the image volume. One possibility was to evaluate the results on a set of CT–MR pairs with controlled misregistrations. Starting from a well-registered image pair, we translated the CT dataset laterally by 5 mm and then 10 mm. With the color-coded results (Fig 6), we confirmed that the apparent area of misregistration (red) increased in the lateral direction by approximately 5 and 10 millimeters, consistent with the first and second translations. To provide a metric estimate of the registration error at a given location, the system counted the number of unsafe voxels in the x, y, and z directions and converted that number in metric units as an estimate of the registration error.
Fig 6.
Evaluation of the results of the registration assessment algorithm on real data.
A, Color-coded result of the assessment of a well-registered CT–MR image pair.
B, Same CT–MR pair, with a purposely introduced relative horizontal shift of five voxels.
C, Relative horizontal shift of 10 voxels.
Another approach to evaluate the results of the registration assessment algorithm on real data was as follows: A pair of CT and MR images were registered by using mutual information and then processed by means of the registration assessment algorithm. A small 3D subvolume (50 × 50 × 50 voxels) was then extracted from homologous locations of the CT and MR volumes. This subvolume was locally reregistered by using mutual information and reprocessed with the registration assessment algorithm. The local re-registration yielded better registration for that volume of interest, as compared with a global registration performed on the whole volume. An example of the expected improvement in registration accuracy is depicted in Figure 7.
Fig 7.
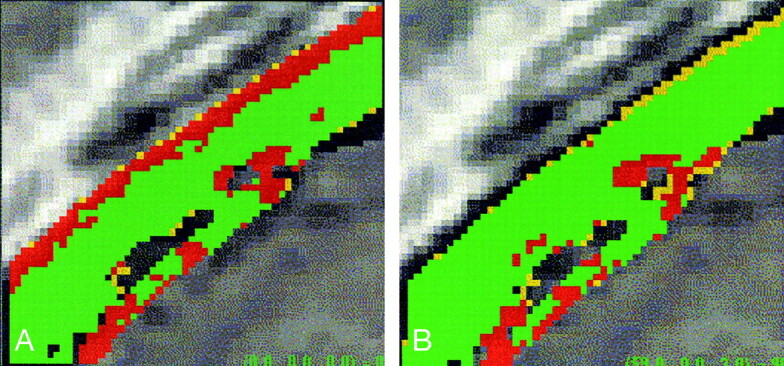
Algorithm evaluation on a CT–MR image subvolume.
A, Small 3D subvolume extracted from a larger registered CT–MR image pair. Color-coded overlay shows the registration assessment result for bone structures.
B, CT and MR subvolumes are reregistered to each other by using a mutual information algorithm. The reregistered subvolumes show an improved registration assessment result, as the low-accuracy (red) region is greatly diminished.
Discussion
In fused CT–MR image representations that result from the combination of gray values of two original images, it is difficult to assess the registration accuracy, even when semitransparency effects or different colors for each source are used. The CT–MR registration assessment method presented herein and the resulting color-code allows the user to instantly assess the quality of the registration. This information is crucial because it permits the viewer to identify areas where the correspondence between the CT and MR images cannot be trusted. For color-blind users, different colors or patterns can be used for visualization. The presented registration assessment method does not in any way account for patient-to-image registration errors, which may occur during image-guided surgery.
Although other authors have compared and evaluated the performance of different CT–MR registration techniques (2, 3, 12), our aim was to provide an automatic method to locally assess the registration errors (which are not homogeneously distributed) in every region of the image without the use of fiducials or landmarks. Our assessment of the registration accuracy is based on cortical bone tissue only. Soft tissues and bone marrow are unsuitable for correspondence analysis because they produce unspecific gray values on CT imaging. However, a well-registered hard tissue structure strengthens the reliability of the registration of other parts of the image. For the segmentation of bone in the MR images, a custom method based on Otsu’s automatic threshold selection method was used. Using simulated MR imaging data, we have shown that this method segments 99% of the bone for normal noise levels. However, it also segments some CSF and air cavities, causing some correspondence errors between the CT and MR imaging not to be picked up, leading to an optimistic registration assessment.
The use of additional T2-weighted sequences could solve the inability to distinguish between bone and CSF and might lead to even better assessment results. In this study, we focused on T1-weighted images because this is the standard sequence used in ENT imaging, as it enhances the mucosa and as it (unlike T2-weighted images) can be used with contrast medium (gadolinium based) to enhance tumors or infections. Use of fat saturation, which suppresses the intensity of fat should not affect our segmentation algorithm, but this has not been tested explicitly.
The bone segmentation algorithm in MR imaging data is robust to variations in TR and TE settings. The tests performed well on simulated as well as on clinical MR imaging data with different TR and TE indicating that the segmentation is robust to TR and TE variations of that magnitude. The algorithm, however, is not directly applicable to T2-weighted images. In the future, better morphology-oriented and model- and/or knowledge-based algorithms (10) for bone segmentation on MR images could further improve the results.
Voxels representing implanted metallic objects (e.g., aneurysm clips) would be classified as bone with both modalities, and in principle, these would not appear misregistered. However, metallic objects cause distortions on MR imaging and beam-hardening artifacts, streaks, and flares on CT. These artifacts cause the algorithm to detect several misregistered voxels as a result of wrong segmentation on CT and distortions on MR imaging.
In the patient datasets used for this study, the registration appeared to be best toward the middle of the head volume, whereas sections further away from the center contained more misregistered (red) voxels. This occurrence is explained by the registration method, which uses a global optimization algorithm (12) in which the resulting errors are best averaged in the center of the volume.
Conclusion
We present a new, automatic, unsupervised, and patient-specific method for assessing CT–MR registration based on a correspondence analysis of cortical bone structures on the original images. The algorithm was successfully evaluated on simulated and real radiologic images. The color-coded result allows for immediate assessment of the overall co-registration quality as one browses through the fused volume stack and makes it simple to identify regions of the fused image, which are not reliable. Poor correspondence between CT and MR imaging means that the user should not trust the registration in this area. The presented method can potentially reduce the risk of unknowingly using misregistered CT–MR datasets in diagnostic or interventional applications.
Acknowledgments
We would like to acknowledge Mrs. Spielvogel from the Neuroradiology Department, Inselspital Bern, for providing the patient data and her valuable advice; Daniel Rueckert, Department of Computing, Imperial College, UK, for providing the tool for rigid normalized mutual information registration; and the Swiss National Science Foundation and the CO-ME (www.co-me.ch) consortium for supporting the project.
Footnotes
Funded by the Swiss National Science Foundation and the CO-ME (www.co-me.ch) consortium.
References
- 1.Maintz JB, Viergever MA. A survey of medical image registration. Med Image Anal 1998;2:1–36 [DOI] [PubMed] [Google Scholar]
- 2.West J, Fitzpatrick JM, Wang MY, et al. Comparison and evaluation of retrospective intermodality brain image registration techniques. J Comput Assist Tomogr 1997;21:554–556 [DOI] [PubMed] [Google Scholar]
- 3.Hawkes DJ. Algorithms for radiological image registration and their clinical application. J Anat 1998;193:347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viola P, Wells WM. Alignment by maximization of mutual information. Int J Compu Vision 1997;24:137–154 [Google Scholar]
- 5.Pluim JP, Maintz JB, Viergever MA. Image registration by maximization of combined mutual information and gradient information. IEEE Trans Med Imaging 2000;19:809–814 [DOI] [PubMed] [Google Scholar]
- 6.West JB, Fitzpatrick JM, Toms SA, et al. Fiducial point placement and the accuracy of point-based, rigid body registration. Neurosurgery 2001;48:810–816 [DOI] [PubMed] [Google Scholar]
- 7.Maurer Jr. CR, Rohlfing T, Dean D, et al. Sources of Error in Image Registration for Cranial Image-Guided Neurosurgery. In: Germano M ed., Advanced Techniques in Image-Guided Brain and Spine Surgery. New York: Thieme;2002. :10–36
- 8.Fitzpatrick JM, Hill DL, Shyr Y. Visual assessment of the accuracy of retrospective registration of MR and CT images of the brain. IEEE Trans Med Imaging 1998;17:571–585 [DOI] [PubMed] [Google Scholar]
- 9.Woods RP. Validation of Registration Accuracy. In: Bankman In ed., Handbook of Medical Imaging, Processing and Analysis. San Diego: Academic Press,2000. :491–498
- 10.Dogdas B, Shattuck DW, Leahy, Segmentation of the skull in 3D human MR images using mathematical morphology. Proc SPIE Medical Imaging Conf 2002;4684:1553–1562 [Google Scholar]
- 11.Otsu N. A threshold selection method from Gray-Level Histograms. IEEE Trans Syst Man Cybernet 1979; SMC-9(1):62–66
- 12.Studholme C, Hill DL, Hawkes DJ. Automated 3-D registration of MR and CT images of the head. Med Image Anal 1996;1:163–175 [DOI] [PubMed] [Google Scholar]
- 13.Rueckert D and Hawkes DJ. 3D analysis: registration of 3D biomedical images. In: Baldock R, Graham J, eds. Image Processing and Analysis—A Practical Approach. Oxford: Oxford University Press;1999