Abstract
Simple Summary
Acute Myeloid Leukemia is an aggressive disease with poor outcomes. New targeted therapies that can boost the effects of currently used chemotherapy medications without added toxicity are needed. Targeting an overactive kinase, called the protein Kinase CK2 in AML, helps leukemia cells undergo cell death and helps certain chemotherapy drugs work better. Here, we present evidence that CX-4945, a CK2 inhibitor drug, effectively kills leukemia cells in mouse models and shows the mechanism of action responsible for these effects. Leukemia cells are more sensitive to a decrease in CK2 kinase levels than normal cells. Our results show that inhibiting CK2 kinase makes AML cells more susceptible to anthracycline-induced cell death. Anthracyclines like daunorubicin and doxorubicin are widely used to treat leukemia in children and adults. A rational combination of protein kinase CK2 inhibitors with the standard of care chemotherapy may help treat AML more effectively.
Abstract
Protein Kinase CK2 (Casein Kinase 2 or CK2) is a constitutively active serine-threonine kinase overactive in human malignancies. Increased expression and activity of CK2 in Acute Myeloid Leukemia (AML) is associated with a poor outcome. CK2 promotes AML cell survival by impinging on multiple oncogenic signaling pathways. The selective small-molecule CK2 inhibitor CX-4945 has shown in vitro cytotoxicity in AML. Here, we report that CX-4945 has a strong in vivo therapeutic effect in preclinical models of AML. The analysis of genome-wide DNA-binding and gene expression in CX-4945 treated AML cells shows that one mechanism, by which CK2 inhibition exerts a therapeutic effect in AML, involves the revival of IKAROS tumor suppressor function. CK2 phosphorylates IKAROS and disrupts IKAROS’ transcriptional activity by impairing DNA-binding and association with chromatin modifiers. Here, we demonstrate that CK2 inhibition decreases IKAROS phosphorylation and restores IKAROS binding to DNA. Further functional experiments show that IKAROS negatively regulates the transcription of anti-apoptotic genes, including BCL-XL (B cell Lymphoma like–2 like 1, BCL2L1). CX-4945 restitutes the IKAROS-mediated repression of BCL-XL in vivo and sensitizes AML cells to apoptosis. Using CX-4945, alongside the cytotoxic chemotherapeutic drug daunorubicin, augments BCL-XL suppression and AML cell apoptosis. Overall, these results establish the in vivo therapeutic efficacy of CX-4945 in AML preclinical models and determine the role of CK2 and IKAROS in regulating apoptosis in AML. Furthermore, our study provides functional and mechanistic bases for the addition of CK2 inhibitors to AML therapy.
Keywords: protein kinase CK2, acute myeloid leukemia, ikaros, bcl-xl, anti-apoptotic gene, CX-4945, transcriptional regulation, patient-derived xenograft, preclinical model, daunorubicin
1. Introduction
Acute Myeloid Leukemia (AML) is a heterogeneous hematological malignancy with poor prognosis, despite aggressive therapy, in children and adults alike [1]. Cytotoxic chemotherapy and hematopoietic stem cell transplant remain the mainstay of treatment for AML [2]. Anthracyclines (daunorubicin and mitoxantrone) and cytarabine (Ara-C) form the backbone of AML induction therapy. Although therapies targeting recurrent genetic mutations are being developed, strategies to target non-mutation based vulnerabilities of leukemia cells are mostly unexplored in AML [3]. The therapeutic benefits of selectively targeting constitutively overactive kinases in leukemia, even in the absence of associated genetic mutation, are well established [4].
Protein Kinase CK2, formerly known as Casein Kinase 2 (CK2), is a pleiotropic, constitutively active, serine-threonine kinase essential for cell survival and development [5]. CK2 exists as a tetramer with two catalytic subunits (α or α’) and two regulatory subunits (β). There are two isoforms, CK2α and CK2α’, which are encoded by different genes and have a similar protein structure (except for C-terminus of α subunit) and catalytic activity [6]. CK2 hyperactivity is implicated in the pathogenesis of several cancers, including hematological malignancies such as AML, Acute Lymphoblastic Leukemia (ALL), Chronic Lymphocytic Leukemia (CLL), Chronic Myelogenous Leukemia (CML), and Myeloproliferative Neoplasm (MPN) [7,8,9,10,11]. CK2 represents a key anticancer target that enables leukemia cell survival and proliferation and renders tumor cells highly dependent on its activity [12,13]. CK2 is known to regulate PI3K/PTEN-AKT[14,15], NF-kB (Nuclear Factor-Kappa B) [16], Wnt-B-catenin, and Hedgehog (Hh) [9,14] signaling pathways in AML. The CK2-driven post-translational modification of transcription factors and tumor suppressors such as PTEN (Phosphatase And Tensin Homolog), P53, IKAROS, and PML (ProMyelocytic Leukemia protein) impair their transcriptional activity and promote leukemogenesis [17,18,19,20]. Among the normal karyotype AML cases, those with high CK2 expression were associated with decreased disease-free and overall survival, suggesting that the overexpression of CK2 is a negative prognostic marker in AML [14,16]. Inhibition of CK2, either by siRNAs or the specific inhibitor CX-4945, shows a strong cytotoxic activity in AML [14,15,21]. CX-4945 (Silmitasertib), known by the chemical name 5-(3-Chlorophenyl) amino- benzo[c] naphthyridine-8-carboxylic acid, is a selective, ATP competitive small molecule and an irreversible inhibitor of CK2 with activity against all isoforms [22,23,24].
Although previous studies suggested several pathways through which CK2 inhibitors might operate, the in vivo therapeutic action mechanism for CK2 inhibition in AML is not well understood [11,14,21]. In B-cell acute lymphoblastic leukemia (B-ALL), CX-4945 exerts a therapeutic effect via restoration of the tumor suppressor activity of the IKAROS protein [25]. IKAROS is a DNA-binding protein that regulates its target genes’ transcription via chromatin remodeling [26]. Recent studies demonstrated that the IKAROS tumor suppressor function involves regulating the global epigenomic landscape and chromatin accessibility in acute lymphoblastic leukemia (ALL) [26,27]. Hyperphosphorylation of IKAROS by CK2 impairs IKAROS’ DNA-binding ability and pericentromeric localization, all of which disrupts IKAROS’ functions in transcriptional regulation, cell cycle progression, and T-cell differentiation, and even promotes IKAROS degradation [28,29]. The CK2 inhibition restores IKAROS’ DNA-binding ability and its function as a transcriptional regulator of its target genes [25,30]. IKZF1 recurrent mutations are seen in AML [31]. The impaired IKAROS function is associated with the development of AML, though its potential role in AML tumor suppression is largely unknown [32,33,34,35].
Here, we report that IKAROS is hyperphosphorylated in AML cells with CK2 overexpression. The molecular or pharmacological inhibition of CK2 decreases IKAROS phosphorylation and increases IKAROS binding to target gene promoter regions. Genome-wide DNA-binding and gene expression analyses in CX-4945-treated AML cells revealed a strong IKAROS binding to the BCL-2 family genes. We performed loss of function and gain of function experiments showing that IKAROS represses the transcription of B cell Lymphoma Like–extra-large (BCL-XL) encoded by B cell Lymphoma Like 2–like 1 (BCL2L1). Furthermore, we demonstrate that the CK2 overexpression impairs IKAROS’ ability to repress BCL-XL. CX-4945 restores the IKAROS-mediated repression of BCL-XL and sensitizes AML cells to apoptosis. Finally, CX-4945 showed anti-tumor activity and prolonged survival in an AML patient-derived xenograft model. We show that in AML, the central mechanism of BCL-XL repression following the CK2 inhibition is due to the restored IKAROS transcription factor activity. These results provide a mechanistic basis to justify the clinical development of CK2 inhibitors combined with cytotoxic therapy to treat AML.
2. Methods and Materials
2.1. Cells and Cell Culture
HEK 293T, U937, K562 and THP-1 cells were obtained from the American Type Culture Collection (ATCC) and the German Collection of Microorganisms and Cell Cultures (DSMZ). De-identified patient samples were provided by Loma Linda University (Loma Linda, CA, USA) and collaborators at the Penn State Cancer Institute in compliance with institutional review board regulations. Cells were cultured with RPMI 1640 plus (Mediatech, Manassas, VA, USA) with a 10% heat-inactivated Fetal bovine serum (FBS) (HyClone, Rockford, IL, USA) and 1% penicillin-streptomycin. HEK-293T cells were cultured in DMEM (CellGro) supplemented with 10% FBS. Human AML primary cells (AML-1), previously expanded in mice, were cultured in StemSpan SFEM (Stem Cell Technologies, Cambridge, MA, USA), supplemented with a recombinant human stem cell factor (SCF, 100 ng/mL), IL3 (20 ng/mL), FMS-like tyrosine kinase ligand (FLT3L, 100 ng/mL), G-CSF (20 ng/mL), and GM-CSF (20 ng/mL; Shenandoah Biotechnology, Warwick, PA), as well as 1% penicillin-streptomycin.
2.2. Drugs and Reagents
The CK2 inhibitor CX-4945 sodium salt was a gift from Senhwa Biosciences. CX-4945 and daunorubicin hydrochloride were purchased from MedChem Express (Monmouth Junction, NJ, USA).
2.3. Human Leukemia Mouse Xenograft Models
U937-GFP-luc cells were transplanted into NRG-S (NOD.Cg-Rag1tm1Mom Il2rgtm1Wjl Tg (CMV-IL3, CSF2, KITLG)1Eav/J) mice via tail vein injection at a dose of 25,000 cells per mouse. The treatment was initiated when a bioluminescence intensity (BLI) signal of 100,000 photons/sec was detected. Mice were randomized based on the average BLI signal and treated with CX-4945 100 mg/kg or a vehicle, given via an oral gavage twice daily for 7 days. Bioluminescence imaging (IVIS 100; analysis with the Living Image software (PerkinElmer)) was used to track the leukemia progression. AML patient samples were selected based on their increased CK2 protein expression (mRNA and protein). Irradiated NRG-S mice were injected with 2 million cells per mouse via the tail vein. Engraftment was defined as 2–5% AML cells (human CD 45+, CD13+, and CD33+, BioLegend, San Diego, CA, USA) in peripheral blood (PB). We started the CX-4945 treatment of 100 mg/kg twice daily after confirming the engraftment, and continued for a total of 21 days. After the treatment period, mice were sacrificed, and mononuclear cells were isolated from the bone marrow, spleen, and peripheral blood. The cell count and flow cytometry analysis were performed (Fortessa; BD, San Jose, CA, USA) using AML antibodies against human CD45+, CD13+, and CD33+ cells. One cohort was monitored for moribund signs, and the survival time was noted.
2.4. Apoptosis Assay
Apoptosis assays were performed following the manufacturer’s instructions using the Annexin V-7AAD apoptosis detection kit (Luminex, MCH100105) and analyzed on the Muse Cell Analyzer (Luminex, Chicago, IL, USA). Briefly, 100 μL of cells were added to 100 μL of reagent, incubated for 20 min at room temperature, and analyzed.
2.5. Proliferation and Cytotoxicity Assays
The colorimetric WST-1 cell proliferation assay (Roche Diagnostics GmbH, Manheim, Germany) was performed in 96-well white clear-bottom plates (Costar, 3610) in quadruplicate experiments, according to the manufacturer’s instructions. Absorbance at 440 nm (reflects the number of viable cells) was measured using a BioTek Synergy Mx plate reader.
2.6. Western Blot
Cells were treated with 5 and 10 µM of CX-4945 or the vehicle control for 48 h and the whole-cell lysate was collected. Briefly, samples were prepared on ice using lysis buffer containing 10 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 1% Triton X-100, 100 mM NaCl, 10 mM NaF, 1 mM Na3VO4, and a protease inhibitor cocktail. The protein was quantified using the Bradford assay and used for Western blot analysis and immunoblotting. The antibodies used for the Western blot assay are listed in Table S1.
2.7. In Vitro Phospho-IKAROS Labeling
Cells were treated with different doses (5 or 10 μM) of CX-4945 for 48 h. Cells were washed twice with phosphate-free RPMI 1640 and incubated with 0.5 mCi/ml [32P] orthophosphate (PerkinElmer, Waltham, MA, USA) in a phosphate-free RPMI 1640 medium for 6 h. Cells were collected and washed twice with cold PBS and the nuclear protein was extracted using the NE-PER Nuclear and Cytoplasmic extraction reagent kit (Thermo Fisher Scientific, Waltham, MA, USA). The lysis buffer was supplemented with a protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). Then, IKAROS was immuno-precipitated using the Dynabead Protein G Immunoprecipitation Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. Briefly, lysates were incubated with the Dynabeads Protein G and IKAROS antibody (Proteintech, Rosemont, IL, USA) complex for 2 h on a rotator-mixer. IKAROS was eluted, separated by SDS-PAGE, transferred to a PVDF membrane, and imaged by radiography.
2.8. Colony Formation Assay
Cells were pretreated with CX-4945 alone or combined with daunorubicin, or the vehicle control for 48 h. Cells were washed with PBS and plated in triplicate in a 6-well plate containing the 1.1 ml MethoCult H4100 medium (STEMCELL Technologies, Vancouver, Canada) supplemented with RPMI-1640 and 10% FBS for U937 cells, as well as a MethoCult H4034 Optimum (STEMCELL Technologies) medium for AML-1 cells. Colonies were propagated for 7–14 days in an incubator at 37 °C and 5% CO2. Colonies were counted under an inverted light microscope. Colonies that contained around 50 cells or more were counted for analysis.
2.9. Gene Expression Analysis by qRT-PCR
Total RNA (ribonucleic acid) was isolated from cells using the QIAshredder and RNeasy Mini Kit (QIAGEN, Germantown, MD, USA). Complementary DNA (cDNA) was generated from 1 μg of total RNA using the Superscript First-Strand Synthesis System (Invitrogen). The qRT-PCR was performed using a StepOne Plus real-time PCR machine (Applied Biosystems) with PerfeCTa SYBR Green FastMix (Quanta Biosciences). Values were normalized to 18 s RNA and the relative expression values were determined by the 2-ΔΔCt method.
Primers used for target gene studies include:
BCL2L1-5′-TTGGATGGCCACTTACCTGAAT-3′; BCL2L1-Rev 5′-CCGCCGTTCTCCTGGAT-3′; CK2α-5′-AGCGATGGGAACGCTTTG-3′; CK2α-Rev 5′-AAGGCCTCAGGGCTGACAA-3’; 18s-For 5’-GTAACCCGTTGAACCCCATT-3′ 18s-Rev5′-CCATCCAATCGGTAGTAGCG-3′.
2.10. Luciferase Assay
The luciferase assay was performed using the LightSwitch Luciferase Assay System (SwitchGear Genomics). Human embryonic kidney (HEK)-293T cells were seeded into 24-well plates. HEK-293T cells do not have endogenous IKAROS. After 24 h, cells were transiently transfected with 0.15 μg of indicated promoter-reporter constructs or pROM vector and 0.15 μg of pcDNA3.1-IKAROS or pcDNA3.1 vector in triplicate experiments for each group using lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s instructions. According to the manufacturer’s instructions, 24 h after transfection, cells were lysed in 100 μL of a LightSwitch Assay Solution (SwitchGear Genomics, Carlsbad, CA, USA) and rocked at RT for 30 min. Lysates were measured by a luminometer (Promega GloMax 20/20 Luminometer). Luciferase activities were calculated as a fold change relative to the vector-only cells and normalized to pcDNA3.1 vector readings. All transfection and reporter assays were performed in triplicate experiments.
2.11. Quantitative Chromatin Immunoprecipitation
IKAROS qChIP assays were performed as described previously[30]. Primers used for the qChIP experiments are listed in the Table S2.
2.12. Statistical Analysis
Graphed data are represented as mean values with bars representing the standard deviation (mean ± SD) of three technical replicates and at least two independent experiments. Statistical analysis was performed using GraphPad Prism 9.0.0. P-value summaries are as follows: p > 0.05 (ns); p ≤ 0.05 (*); p < 0.01 (**); p < 0.001 (***); p < 0.0001 (****). Statistical significance in Figure 3B,E was performed using the one-way analysis of variance (ANOVA). Statistical analysis for all other column graphs used multiple two-tailed t-tests by the Holm-Sidak method, with alpha = 0.05. Each row (representing a cell line or drug concentration) was analyzed individually, without assuming a consistent Standard Deviation (SD). The number of t-tests per analysis was dependent on the number of conditions. The qChIP values where the signal was more than 2-fold greater than the background anti-Immunoglobulin G (anti-IgG) level were analyzed. The Kaplan-Meier method and the log-rank test was used to perform the survival analysis and compare survival differences (Figures 2G and 3C).
3. Results
3.1. High Baseline Expression of CK2, Bcl-xl, and p-IKAROS in Myeloid Leukemia Cells Compared to Normal Hematopoietic Stem Cells
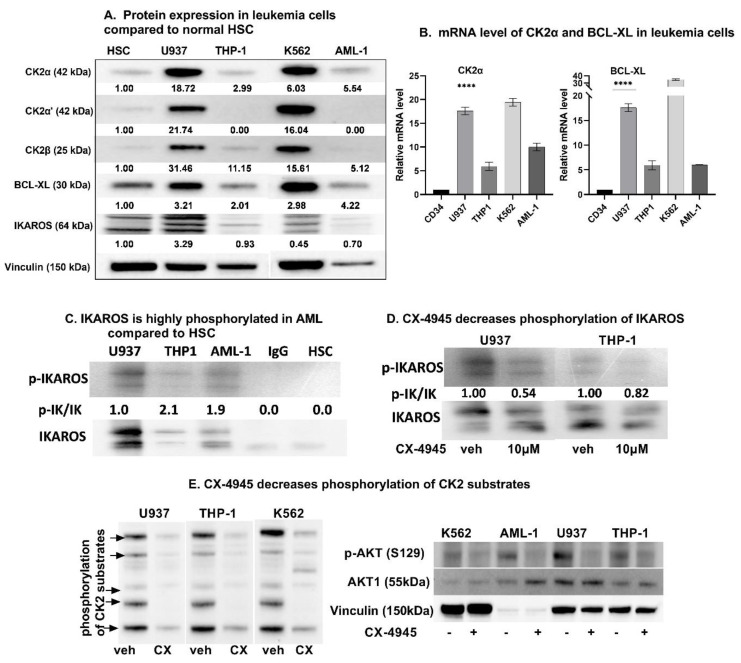
In line with the previously published data [15,16,36], we observed that CK2 is overexpressed in myeloid leukemia cells and primary cells with various cytogenetic features compared to normal hematopoietic cells (CD34+ HSC). A table with descriptions of each cell type is included in supplemental file (Table S3). We examined protein and mRNA levels of CK2α, CK2α’, CK2β, and BCL-XL in a panel of myeloid leukemia cells, using Western blot (Figure 1A) and qRT-PCR, respectively (Figure 1B). The CK2α and BCL-XL expression were higher in AML cells as compared to CD34+ HSC (Figure 1A and Figure S1). The expression of CK2, BCL-XL, and IKAROS were variable across different AML cells. AML cells showing high BCL-XL expression in the absence of high CK2 expression (as in AML-5 and AML-6 in Figure S1) suggest other possible mechanisms influencing the expression of BCL-XL in AML cells such as cytogenetic alterations, relapse disease, and chemotherapy exposure. CD34+ HSCs were obtained from different sourses such as the umbilical cord blood (UCB) and granulocyte colony stimulating factor (GCSF) treated peripheral blood collection (PB). The CK2 and BCL-XL expression were noted to be significantly and uniformly lower in CD34+ from PB and UCB compared to the leukemia cells. We chose U937, THP1, and AML-1 (primary AML cell) for our experiments due to the high expression of CK2α. Phosphorylated-IKAROS was also measured in the studied cells using a radio-immunoblot. IKAROS was observed to be highly phosphorylated in AML cells compared to HSC (Figure 1C).
Figure 1.
CK2 overexpression in AML cells causes IKAROS phosphorylation which is reversed by CK2 inhibitor, CX-4945. (A) Baseline protein levels of CK2α, CK2α’, CK2β, IKAROS and BCL-XL in the myeloid leukemia cell panel [U937, THP-1, K562, and primary AML cells (labelled AML-1) were measured by western blot and compared to CD34+ Hematopoietic Stem Cells (HSC). Multiple IKAROS bands represent Ikaros isoforms 1 and 2. (B) qRT-PCR showing mRNA level of CK2α and BCL-XL in various AML cells compared to CD34+ HSC. **** (p < 0.0001). (C) Radio-blot showing increased Phosphorylated IKAROS (p-IKAROS) in AML cells compared to HSC. (D) Radio-immunoblot showing dose-dependent decrease in Phospho-IKAROS level following CX-4945 treatment. U937 and THP-1 were treated with 10 µM and AML-1 was treated with 5μM (IC50–3.791 μM) CX-4945 for 48 h. IC50 values of CX-4945 treated cells are shown in Figure S2. (E) Western blot showing decrease in amount of phosphorylated CK2 substrates with molecular masses of approximately 175, 120, 80, 70 and 56 kDa as indicated by arrows (left panel) and western blot showing phosphorylation extent of specific CK2 substrate, AKT1 at Ser129 (right panel).
3.2. CK2 Inhibition Decreases IKAROS Phosphorylation and CK2 Cellualr Activity
In B-ALL, IKAROS is hyperphosphorylated by CK2 [37,38]. We tested the effect of CK2 inhibition with CX-4945 on IKAROS phosphorylation in AML cells. Compared to the untreated cells, the treatment with CX-4945 for 48 h resulted in a reduction in IKAROS phosphorylation (Figure 1D).
We measured the CK2 activity indirectly by using an antibody against the phospho-CK2 substrate (motif pS/pTDXE) to measure the amount of phosphorylated CK2 substrates [39,40,41,42,43]. The CK2 substrates contain multiple acidic residues (Asp and Glu) located downstream of the phosphorylated serine or threonine residues. The consensus sequence for CK2 substrates is pS/pTD/EXD/E with the most crucial residue at the +3 position followed by the residue at the +1 position. CX-4945 reduced the CK2 activity in a dose-dependent manner 48 h post-treatment as determined by immune-blotting with an antibody that recognizes phosphorylated CK2 sites in multiple proteins (Figure 1E, left panel). AML cell lines showed a reduction in multiple phospho-CK2 substrates with molecular masses of approximately 175, 120, 80, 70, and 56 kDa as indicated by arrows in Figure 1E, left panel. We also performed the Western blot analysis to evaluate the phosphorylation extent of a specific CK2 substrate, AKT1 at Ser129 [44,45]. Results showed a decrease in the p-AKT(S129) level following the CX-4945 treatment (Figure 1E, right panel).
3.3. Inhibition of CK2 Represses BCL-XL Expression
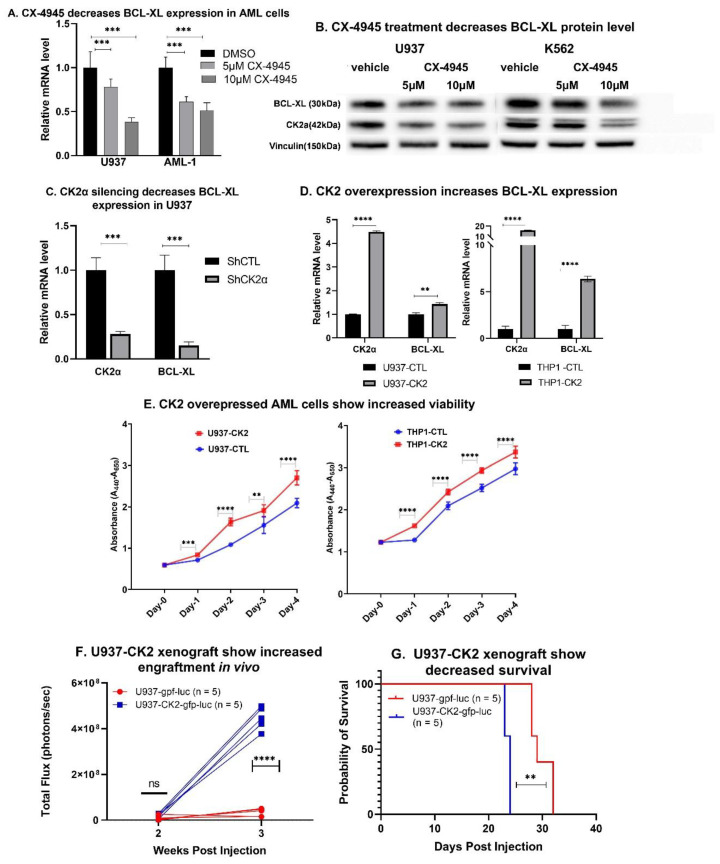
We studied the effect of CK2α downregulation and pharmacological inhibition of CK2α on BCL-XL expression. We achieved a pharmacological inhibition of CK2 using a specific inhibitor, CX-4945. The treatment of U937 and AML-1 with CX-4945 showed dose-dependent decreases in BCL-XL mRNA (Figure 2A) and protein levels (Figure 2B). Downregulation of the CK2α was achieved by treating U937 cells with a CK2α specific short hairpin (sh) RNA (Figure S3). CK2α silencing decreased the expression of BCL-XL at mRNA (Figure 2C) and protein levels (Figure S4), as well as decreased IKAROS phosphorylation (Figure S5). Next, we transduced U937 and THP-1 cells with a retrovirus expressing CK2α or the empty vector (as a negative control). Increased CK2α mRNA (Figure 2D) and protein were confirmed (Figure S6). The overexpression of CK2α showed increased BCL-XL mRNA levels in U937 and THP-1 (Figure 2D). We did not observe a significant change in IKAROS phosphorylation or the BCL-XL protein level in CK2 overexpressing U937 cells (Figures S4 and S5). This may be due to the already high level of IKAROS phosphorylation by high baseline CK2 in these cells. Phenotypically, CK2α overexpressing U937 and THP-1 cells showed increased cell viability and proliferation compared to the control cells (Figure 2E). We developed a cell line derived xenograft mouse model by transplanting luciferase labeled U937 cells transduced with CK2α-gfp-luc or the control vector (ctl-gfp-luc) into immunocompromised (NRG-S) mice via tail vein injection. AML (U937-CK2α-gfp-luc) cells overexpressing CK2α showed a robust engraftment as shown by the increased bioluminescence intensity (Figure 2F). Moreover, mice engrafted with CK2α overexpressing AML cells showed decreased survival compared to the control group (Figure 2G).
Figure 2.
CK2 inhibition suppresses BCL-XL expression in AML cells and xenograft model. (A) U937 and AML-1 were treated with 5 and 10 µM concentration of CX-4945 for 48 h before RNA and protein extraction. mRNA level of BCL-XL was measured by qRT-PCR. (B) U937 and K562 cells were treated with 5 and 10 µM of CX-4945 for 48 hours and protein was extracted. BCL-XL protein level was measured by western blot. (C) Downregulation of CK2α in U937 was achieved using shRNA. Validation of CK2α protein knockdown in U937-shCK2α cell line is shown in Figure S3. qRT-PCR shows mRNA level of CK2α and BCL-XL in CK2α shRNA treated U937 cells. (D) Overexpression of CK2α was achieved by retroviral transduction of U937 cells. Validation of CK2α protein overexpression is shown in Figure S6. qRT-PCR showed increased BCL-XL mRNA level in CK2α overexpressed U937 (left panel) and THP-1 cells (right panel). (E) WST assay showing increased cell viability in CK2α overexpressing U937 and THP-1 cells. U937 cells transduced with retroviral vector CK2α-gfp-luc were transplanted into NRG-S mice. 25,000 cells were injected via the tail vein. Bioluminescence imaging using IVIS 100 was obtained weekly following transplant. (F) Bioluminescence signal in xenograft mice engrafting U937-gfp-luc cell or control (U937-ctl-gfp-luc) shown at week 2 and 3 post transplantation. (G) Kaplan Maier plot showing survival probability in U937-CK2-gfp-luc cells vs control. P-value summaries are as follows: p > 0.05 (ns-not significant); p < 0.01 (**); p < 0.001 (***); p < 0.0001 (****).
These results demonstrate that the CK2α downregulation and pharmacological inhibition of CK2 results in decreased IKAROS phosphorylation and BCL-XL repression in AML cells. The overexpression of CK2α promotes cell viability in vitro, and accelerates leukemia progression in vivo likely by increasing the expression of anti-apoptotic BCL-XL.
3.4. CX-4945 Treatment Shows Therapeutic Efficacy in AML Patient-Derived Xenografts
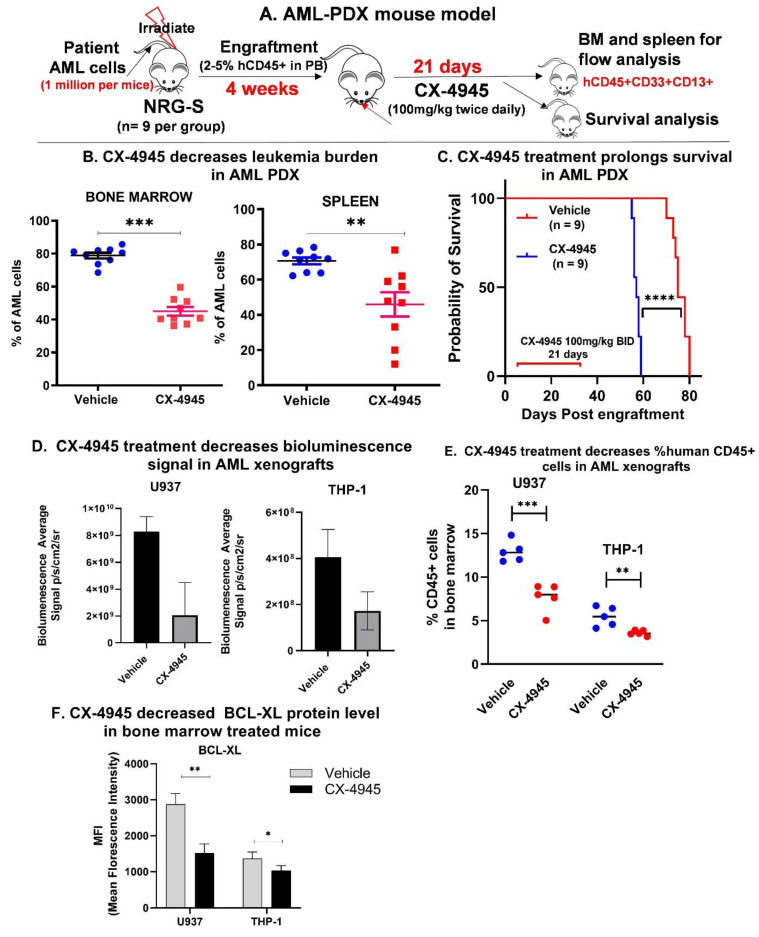
We developed a patient-derived xenograft (PDX) model by transplanting previously expanded human primary AML cells (AML-1) into irradiated NRG-S mice via tail vein injection at a dose of 2 million cells per mouse. These mice were treated with CX-4945 orally at 100 mg/kg twice daily for 21 days (Figure 3A). After the treatment period, one cohort of mice was sacrificed to measure the human leukemia burden in the bone marrow and spleen using antibodies against human AML immunophenotypic cell surface markers (hCD45+, CD13+, CD33+). The CX-4945 treated mice showed significantly less leukemia burden (Figure 3B) and prolonged survival (Figure 3C) than the untreated group.
Figure 3.
CX-4945 treatment shows therapeutic efficacy in AML Patient Derived Xenograft (PDX). (A) Schema showing AML PDX generation and treatment. AML-1 PDX was developed by injecting NRG-S mice with one million cells per mouse via the tail vein. Treatment was started when 2–5% human CD45+ cells were detected in peripheral blood (PB). Mice received 100 mg/kg of CX-4945 via gavage twice daily for 21 days. (B) Percent AML (human CD45+, CD13+,CD33+) in bone marrow (BM) and spleen of treated and untreated AML-1 PDX mice by flow cytometry. (C) Kaplan Maier plot showing survival probability of treated and untreated AML-1 PDX mice. Cell line derived xenograft (CDX) mouse models was developed using luciferase, and green fluro-protein (gfp) labeled U937 cells (U937-gfp-luc) or THP-1 cells (THP1-gfp-luc) transplanted into immunocompromised NRG-S mice as described in methods. Mice were treated with vehicle or CX-4945 at a dose of 100 mg/kg twice daily via gavage for up to 7 days. (D) Engraftment was monitored using bioluminescence imaging shown as the mean of the total flux in photons/second of mice in each group. (E) Following treatment, mice were sacrificed, and bone marrow mononuclear cells were collected. Human CD45+ cells in bone marrow were measured using flow cytometry. Flow cytometry using conjugated BCL-XL antibody was used to quantify intracellular BCL-XL protein level in FACS enriched human CD45+cells in the bone marrow of treated and untreated mice. Figure S6 shows histogram of BCL-XL and CK2α protein level in vehicle and CX-4945 treated U937 and THP-1 CDX. (F) Mean fluorescence intensity(MFI) is graphed, showing decreased BCL-XL protein level. P-value summaries are as follows: p > 0.05 (ns-not significant); p ≤ 0.05 (*); p < 0.01 (**); p < 0.001 (***).
3.5. CX-4945 Treatment Decreases BCL-XL Expression In Vivo
We developed a cell line derived xenograft mouse model by transplanting luciferase labeled U937 and THP-1 cells (U937-GFP-luc and THP-1-GFP-luc) into NRG-S mice via tail vein injection. Engraftment was confirmed by bioluminescence imaging. Mice were treated with CX-4945 orally at 100 mg/kg twice daily for 7 days. The CX-4945 treated mice showed decreased BLI (Figure 3D). After the treatment period, mice were sacrificed, and human CD45+ cells were collected from the bone marrow. The CX-4945 treated mice showed decreased total human CD45+ cells compared to the untreated mice (Figure 3E). Intracellular protein quantification of BCL-XL and CK2α in leukemia cells from the bone marrow of treated and untreated mice was done using flow cytometry. AML cells from the mice treated with CX-4945 showed decreased BCL-XL protein levels compared to cells from the untreated mice (Figure 3F and Figure S7) in both U937 and THP-1 xenograft models. We also observed a mild decrease in the CK2α level following the CX-4945 treatement which may suggest that BCL-XL depletion by CX-4945 may be detrmined by the CK2 protein amount (Figure S7). These findings of CK2α protein depletion were not noted in the AML-PDX treated with CX-4945, as shown later. Interestingly, the complete blood count obtained from these mice did not show thrombocytopenia (Table S4). These results confirm that CK2 inhibition by CX-4945 decreases BCL-XL expression in vivo and demonstrates the in vivo anti-leukemia effect of CX-4945 in an AML xenograft model.
3.6. CK2 Inhibitor Increases IKAROS DNA-Binding to BCL-XL
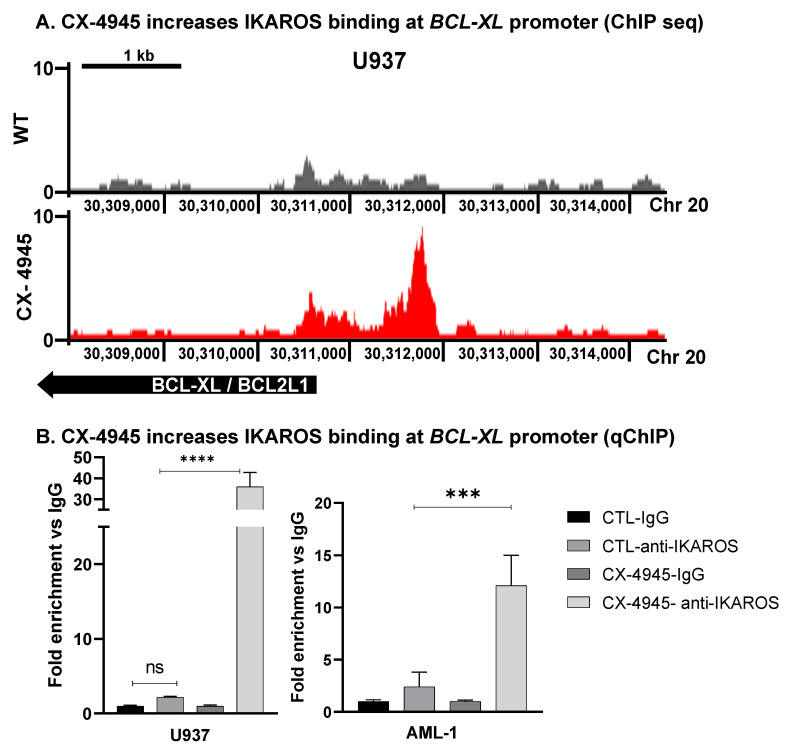
The IKAROS transcription factor binds to upstream regulatory elements of target genes and activates or represses their expression via chromatin remodeling [26,27]. Regulatory functions of the IKAROS transcription factor in AML are not known. In ALL, CK2 is the major kinase that phosphorylates IKAROS [37,38]. In AML, where the IKZF1 loss of function mutations is rare, hyperphosphorylation by CK2 can impair IKAROS’ DNA-binding ability and subsequent regulatory functions. Using chromatin immunoprecipitation followed by highly parellel DNA sequencing (ChIP-seq), we did a global analysis of changes in the genome-wide DNA-binding of IKAROS in AML cells following the treatment with CX-4945. U937 cells were treated with 10 µM of CX-4945 for 72 h to assess IKAROS mediated epigenomic changes. A link to access the ChIP-seq data in the UCSC browser is provided in the supplemental material. Here, we show that the binding of IKAROS to the promoter region of the BCL2L1 (BCL-XL) gene increases after the CX-4945 treatment (Figure 4A). We confirmed IKAROS binding at the BCL-XL promoter region using a quantitative chromatin immunoprecipitation assay (qChIP). The CX-4945 treatment showed increased binding of IKAROS to the BCL-XL promoter in U937 (Figure 4B) and primary AML cells following 72 h of treatment (Figure 4B, right panel).
Figure 4.
CK2 inhibitor increases IKAROS DNA-binding to BCL-XL. Chromatin immunoprecipitation (ChIP) followed by next-generation sequencing (ChIP-seq) and analysis of genome-wide occupancy of IKAROS was performed on U937 following the CX-4945 treatment at 10 µM concentration for 72 h. A change in IKAROS binding to promoter regions of the BCL-XL gene was analyzed following the CX-4945 treatment. (A) A chIP-seq signal map for IKAROS binding to the BCL-XL/BCL-2L1 promoter region in U937-untreated labeled as WT (wild-type) (top panel) and CX-4945 treated U937 (bottom panel). Y-axis represents the log-2-fold change enrichment of IKAROS binding (** p < 0.01). (B) U937 and primary AML cells (AML-1) cells were treated with 10 and 5 μM of CX-4945, respectively (based on the IC50 value) for 48 and 72 h. IKAROS binding to the BCL-XL promoter region was confirmed using the qChIP assay in WT and CX-4945 treated cells. Binding at 72 h was not significantly increased compared to the 48 h treatment (not shown in the graph). Results are the mean +/– SD of three independent experiments. P-value summaries are as follows: p > 0.05 (ns-non significant); p < 0.001 (***); p < 0.0001 (****).
3.7. IKAROS Represses BCL-XL Expression
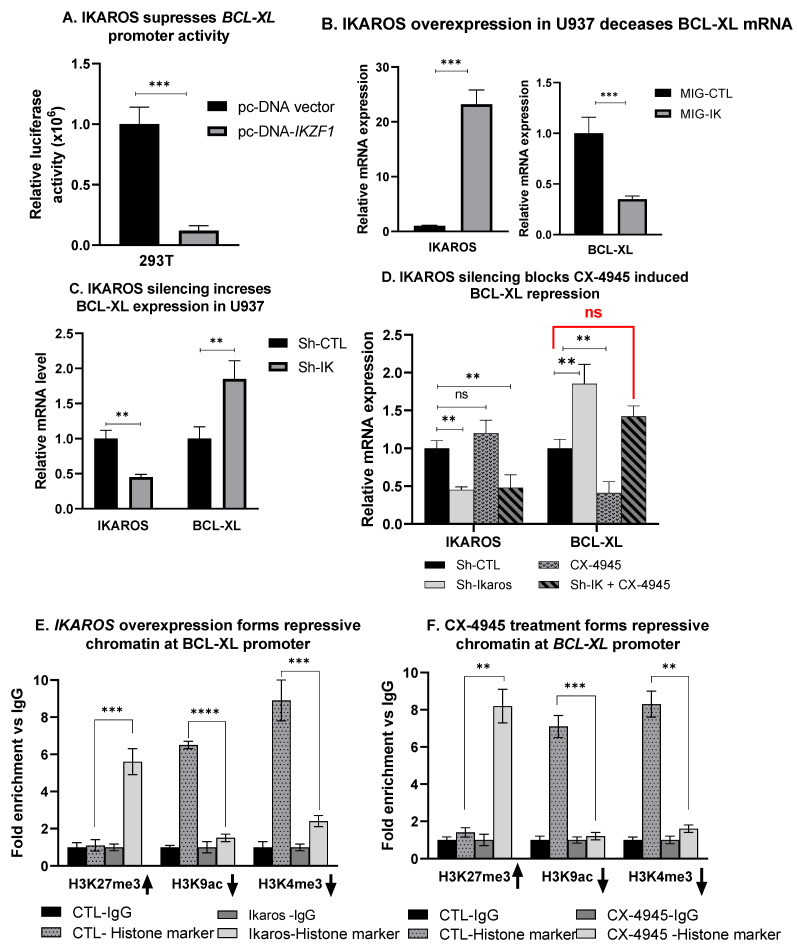
We used a luciferase reporter assay to determine if IKAROS binding to the BCL-XL promoter region alters BCL-XL gene expression. The transient co-transfection of the BCL-XL promoter region fused with the reporter gene and IKZF1 in HEK 293T cells. HEK293T cells do not have endogenous IKAROS making it an ideal system to study the effect of IKZF1 on our promoters of interest. The human embryonic kidney 293T in human cell line cells stably express the SV40 large T antigen, increasing transfection, and transduction efficiency. The results show that IKAROS represses the BCL-XL promoter activity (Figure 5A). Next, we performed the IKAROS loss of function and gain of function experiments to confirm the effect of IKAROS on BCL-XL expression in AML cells. The overexpression of IKAROS in U937 cells was achieved by transduction of the retrovirus expressing wild type IKZF1 and empty vector as a negative control (Figure 5B, left panel). Increased IKAROS mRNA (Figure 5B) and protein (Figure S8) were confirmed. The overexpression of IKAROS in U937 cells resulted in decreased BCL-XL mRNA levels (Figure 5B, right panel) but not the protein level (Figure S8). This could be due to the high level of CK2 present in U937 cells that phosphorylate the overexpressed IKAROS protein. IKAROS silencing was achieved by treating U937 cells with IKZF1 shRNA (Figure 5C, left panel). The U937 treated with scramble shRNA (sh CTL) was used as a control. IKAROS knockdown resulted in increased mRNA levels of BCL-XL (Figure 5C, right panel). These results establish the role of IKAROS as a transcriptional repressor of BCL-XL in AML.
Figure 5.
IKAROS represses BCL-XL gene transcription in AML. The luciferase reporter assay was performed on HEK-293T cells transfected with IKZF1 plasmid (pcDNA-IK) or the control vector. Result in (A) shows repression of the BCL-XL luciferase promoter construct by the IKAROS-expressing vector pcDNA3.1-IK in comparison to the pcDNA3.1 empty vector control in HEK-293T cells. The luciferase activity was normalized to pcDNA3.1 and pROM empty vector controls. (B) U937 cells were transduced to express IKZF1 (MIG-IK) or with an empty vector (MIG-CTL). The relative mRNA expression of IKAROS and (left panel) BCL-XL (right panel) were assessed using qRT-PCR. (C) U937 cells were treated with IKZF1 shRNA (shIK) or scramble shRNA control (shCTL). The relative expression of IKZF1 (left panel) and BCL-XL (right panel) assessed by qRT-PCR. (D) U937 cells were treated with IKZF1 shRNA (shIK) or scramble shRNA control (shCTL). IKAROS knockdown U937 cells were then treated with 10 µM of CX-4945 for 48 h. Changes in BCL-XL gene expression were measured using qPCR. Figure 5E,F shows the qChIP assay showing an enrichment of histone markers at the BCL-XL promoter. The qChIP assay was performed using (E) IKAROS overexpressing (MIG-IKZF1) U937 cells and (F) CX-4945 treated U937 cells (10 µM for 48 h) to determine the fold enrichment of histone markers, H3K27me3, H3K9ac, and H3K4me3 at the BCL-XL promoter and compared to the control cells. P-value summaries are as follows: p > 0.05 (ns- not significant); p < 0.01 (**); p < 0.001 (***); p < 0.0001 (****). Results are the mean +/– SD of three independent experiments.
3.8. CX-4945-Induced BCL-XL Repression Is Mediated via IKAROS
The functional alteration of IKAROS due to phosphorylation by CK2 impairs its regulatory functions as a tumor suppressor in leukemia [38]. CK2 inhibition results in the redistribution of genome-wide DNA-binding of IKAROS to promoter regions of its target genes, restores IKAROS’ regulatory activity, and revives the tumor suppressor function [25]. Since CK2 has multiple substrates, IKAROS being one, we wanted to test whether silencing IKAROS would affect a CX-4945-induced decrease in the BCL-XL expression. We treated U937 cells with IKZF1 shRNA and CX-4945, and measured BCL-XL mRNA levels. IKAROS silencing blocked the CK2 inhibitor’s ability to induce BCL-XL repression (Figure 5D). This result confirms that the CK2-IKAROS axis is one of the major, if not the only, BCL-XL repression mechanisms by CX-4945.
3.9. IKAROS Represses BCL-XL via the Formation of Repressive Chromatin
Here, we investigated the mechanism by which IKAROS represses its target gene, BCL-XL, in AML. IKAROS regulates the transcription of its target genes via chromatin remodeling. Chromatin remodeling often involves chemical modifications of histone protein present in chromatin (i.e., methylation and acetylation). The modification of “histone mark” influences gene expression by changing how accessible the chromatin is to the transcription. The histone mark is a specific modification (acetylation or methylation) of a specific histone protein. Acetylation of lysine 9(K9) of the H3 histone protein is noted as H3K9Ac. Methylation of lysine 27 (K27) of H3 histone is noted as H3K27me3. Enrichment of H3K9 acetylation (H3K9Ac) is an indicator of open and active chromatin. Enrichment of H3K27 tri-methylation (H3K27me3) is a marker of closed and repressive chromatin. Repressive chromatin signature is noted as an enrichment of Histone (H) 3 lysine (K) 27 tri-methylation (H3K27me3) and loss of H3K9 acetylation (H3K9ac) and H3K4 tri-methylation (H3K4me3) [26]. We performed serial qChIP assays on CX-4945 treated U937 cells to evaluate the presence of histone markers indicating a change in the chromatin signature at the BCL-XL promoter region.
Results show that following the treatment with CX-4945, there is an enrichment of the H3K27me3, loss of H3K9ac, and H3K4me3 histone marker (Figure 5F). A similar enrichment pattern was noted at the BCL-XL promoter in U937 cells with forced IKAROS overexpression (Figure 5E). IKAROS represses the BCL-XL gene’s transcription by inducing the formation of repressive chromatin at the BCL-XL promoter.
3.10. CK2 Regulate AML Cell Sensitivity towards Daunorubicin
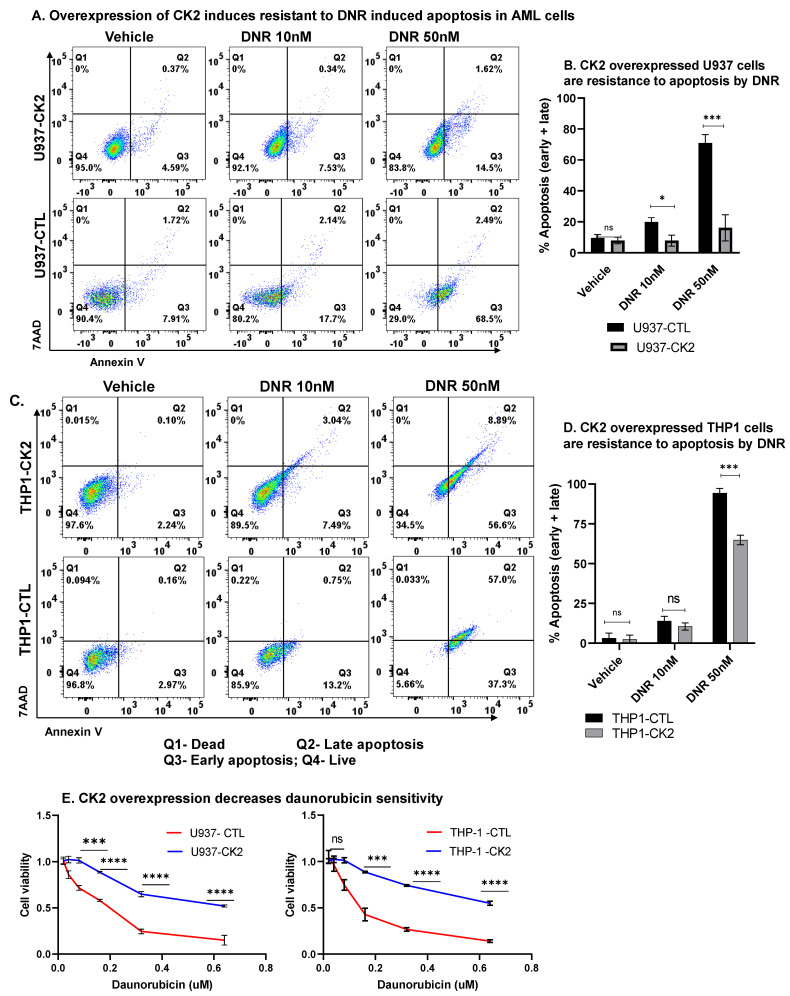
Daunorubicin (DNR) is one of the most commonly used cytotoxic therapies for AML that induces DNA damage and apoptosis. AML cells often overexpress BCL-XL and other anti-apoptotic genes as one mechanism of chemoresistance. As shown in Figure 2A,B, the CK2 inhibition decreases BCL-XL expression. Here, we tested whether the CK2 inhibition affects AML cells’ sensitivity to DNR-induced apoptosis, and, conversely, whether CK2α overexpression in AML cells confers resistance to DNR induced apoptosis. CK2α overexpressing AML cells were resistant to apoptosis (Figure 6A–D) and showed decreased drug response (Figure 6E) following the DNR treatment compared to the control cells. Overall, these results confirm the role of CK2 overexpression in inducing resistance towards cytotoxic therapy in AML.
Figure 6.
CK2 and IKAROS regulate sensitivity towards daunorubicin. CK2 overexpressing U937 (A,B) and THP-1 (C,D) were treated with 10 nM or 50 nM of daunorubicin for 48 hand stained with 7-AAD and Annexin V for flow cytometry to determine apoptosis. Flow plots (A,C) and percent apoptotic cells (early + late apoptosis) are shown in (B,D). Flow plots showing representative results from three replicates. The percentage of cells in the right upper and lower quadrant of each flow chart represents the percentage of late and early apoptotic cells, respectively. Q1-dead, Q2-late apoptosis, Q3-early apoptosis, Q4-live. (E) Cytotoxicity and drug response measured by MTT assay after treating CK2 overexpressing U937 and THP-1 cells and respective controls with various concentrations of daunorubicin for 48 h. p > 0.05 (ns); p < 0.05 (*); p < 0.001 (***); p < 0.0001 (****).
3.11. CK2 Inhibition Augments Daunorubicin Drug Response
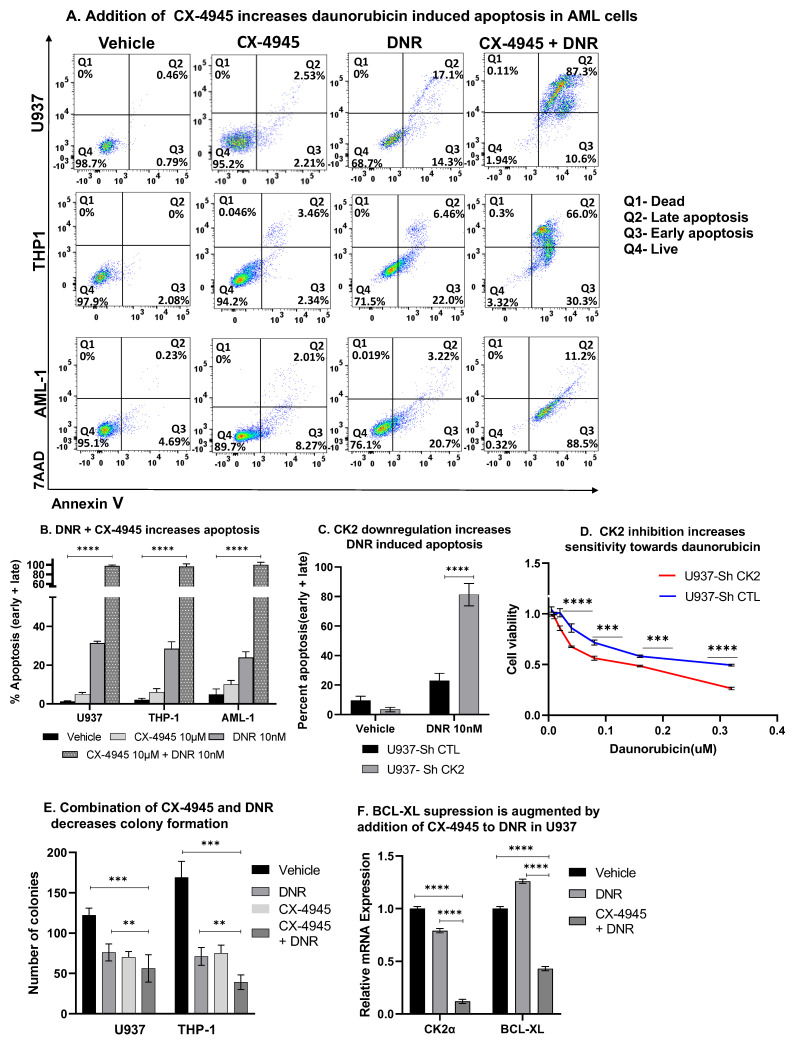
The addition of CX-4945 potentiates DNR induced apoptosis (Figure 7A,B). Downregulation of CK2α resulted in increased apoptosis (Figure 7C) and decreased cell viability (Figure 7D) when exposed to DNR compared to the control cells. A combination treatment with CX-4945 and DNR showed decreased colony formation compared to the cells treated with DNR alone (Figure 7E). The repression of BCL-XL was significantly augmented following the treatment with CX-4945 and DNR compared to the DNR single-agent treatment (Figure 7F). A combination of CX-4945 and daunorubicin showed the synergetic cytotoxic activity in primary AML cells (Figure S9). Overall, these results provide evidence of the functional synergy between CX-4945 and daunorubicin in suppressing BCL-XL expression in AML.
Figure 7.
CK2 inhibition potentiates daunorubicin-induced apoptosis in AML cells. Cells were treated with 10 µM of CX-4945 or a combination of 10 µM CX-4945 with 10 nM of daunorubicin for up to 48 h. Cells were stained with 7-AAD and Annexin V for flow cytometry to assess apoptosis. (A) Flow plots showing representative results from three replicate experiments. The percentage of cells in the right upper and lower quadrant of each flow chart represents the percentage of late and early apoptotic cells, respectively, in U937 (top row), THP-1 (middle row), and AML-1 cells (bottom row) Q1: Dead, Q2: Late apoptosis, Q3: Early apoptosis, Q4: Live. Graphed in (B) are the mean +/−SD of triplicates from two independent experiments showing the percent of apoptosis cells following the drug treatment, as indicated above. (C) CK2α silencing in U937 cells achieved using ShCK2α and sorted for GFP after 24 h. Sorted cells were treated with 10 nM of DNR for 24 h before staining for Annexin V and 7AAD to assess apoptosis. The graph shows the combined percent apoptotic cells (early + late) in each group with and without the DNR treatment. (D) Cytotoxic drug response measured by the MTT assay after treating CK2α ShRNA treated U937 cells and the respective controls with various daunorubicin concentrations for 24 h. (E) Cells were pretreated as above for 48 h and were plated in a Methocult medium. Colonies were counted under an inverted light microscope. Colonies that contained around 50 cells or more were counted for analysis. Graphed in Figure 7E is the number of colonies after 14 days as the mean of three replicates +/− SD of two independent experiments. (F) The qRT-PCR showing decreases in the mRNA level in U937 cells treated with daunorubicin alone (10 nM) or a combination of CX-4945 and daunorubicin. p-value summaries are as follows: p > 0.05 (ns); p < 0.01 (**); p < 0.001 (***); p < 0.0001 (****).
4. Discussion
Protein kinase CK2 is essential for embryonic development, cell survival, growth, and maintenance [46,47]. The overexpression of CK2 enables cancer cells to proliferate and resist apoptosis [48,49,50]. Leukemia cells are more susceptible to CK2 inhibition-triggered cytotoxicity than their normal counterparts. CK2 has emerged as a potential anti-leukemia target [10,11,51]. CX-4945 is an orally bioavailable, ATP competitive, small molecule inhibitor that has shown a favorable toxicity profile and efficacy in patients with relapsed refractory cancers [22,24,52]. CX-4945 has shown strong in vivo efficacy in lymphoid leukemia when used as a single agent and combined with cytotoxic therapy [25,53,54]. In AML, several recent studies have shown cytotoxic activity of CX-4945 as a single agent and in combination with cytotoxic therapy using the in vitro cell system and murine leukemia models and describes another CX-4945 mechanism of action to induce cell death in AML [14,15,21,36,55]. Strategically targeting a promiscuous kinase CK2, which is indispensable for several physiological cellular pathways, is challenging [56]. This nature of CK2 underscores the need for a thorough investigation of all signaling transduction pathways affected by CK2 in different leukemias. A broad understanding of the mechanisms of action of CK2 inhibitors in AML helps design rational drug combinations that would be most effective to overcome chemoresistance, sensitize cells to cytotoxic therapy, and minimize off-target effects.
In lymphoblastic leukemia, the tumor suppressor IKAROS is one of the important substrates of CK2 [37]. IKAROS (encoded by the IKZF1 gene) is a master regulator of lymphoid hematopoiesis and a tumor suppressor in leukemia [57]. Unlike the B cell acute lymphoblastic leukemia (ALL), where more than 30% of cases have IKZF1 genetic alterations, less than 10% of patients in AML have IKZF1 genetic mutations [33]. The functional inactivation of IKAROS by CK2 hyperphosphorylation impairs IKAROS’s transcriptional regulatory functions in B cell ALL, independent of the genetic alteration [37].
Chromatin immunoprecipitation coupled with next-generation sequencing (ChIP-seq) was performed to assess IKAROS binding to DNA in U937 AML cells following the treatment with CX-4945. Results showed enhanced IKAROS binding peaks at the promoter region of several genes. IKAROS binding peaks were significantly increased at the promoter region of the BCL-XL (BCL2L1) gene. Further validation by the quantitative ChIP experiment showed that treating U937 and AML primary patient cells (AML-1) with CX-4945 enhances IKAROS binding to the BCL-XL gene. Functional experiments using the loss of function or gain of the IKAROS function have shown that IKAROS negatively regulates BCL-XL transcription via chromatin remodeling (formation of repressive chromatin). IKAROS is hyperphosphorylated in AML. The CK2 inhibition can reverse this process and restore the IKAROS regulation of BCL-XL.
Here, we show that the forced overexpression of CK2 in AML cells led to increased cell proliferation, decreased apoptosis, and resistance to DNR. More importantly, CK2 overexpressing AML cells engrafted rapidly and decreased the survival time of mice. Downregulation and pharmacological inhibition of CK2 decreased the BCL-XL expression and sensitized the AML cells to DNR-induced apoptosis. Anthracyclines (DNR and mitoxantrone) are used extensively in the treatment of AML. These results provide a mechanistic basis for the use of CX-4945 in combination with DNR.
This study shows that, in AML, IKAROS and CK2 play an essential role in the transcriptional regulation of the anti-apoptotic gene BCL-XL. Other anti-apoptotic genes regulated by CK2 and IKAROS include BCL2A1 and MCL-1. Here, we focused on the transcriptional regulation of the anti-apoptotic gene BCL-XL, as this is one of the important genes regulating leukemia cell resistance to apoptosis [58]. The clinical use of BCL-XL inhibitors is limited due to the platelet destruction [59]. Here, we show that by targeting CK2, we can indirectly suppress BCL-XL expression by restoring the IKAROS transcription factor-mediated negative regulatory mechanism. Using two AML cell line (U937 and THP-1) derived xenograft models, we show that the treatment with oral CX-4945 causes decreased BCL-XL expression in vivo. Decreases in the BCL-XL expression correlated with decreases in the bone marrow engraftment suggesting an in vivo anti-leukemia effect. Interestingly, no decrease in the platelet count was noted in the CX-4945 treated AML xenograft mice (Table S4). Finally, the patient-derived AML xenograft mice treated with the CX-4945 single-agent oral therapy for 3 weeks showed significantly decreased leukemia burden and prolonged survival time compared to the vehicle-treated mice. Further preclinical studies are required to establish the in vivo efficacy of CX-4945 combined with chemotherapy such as daunorubicin in AML.
5. Conclusions
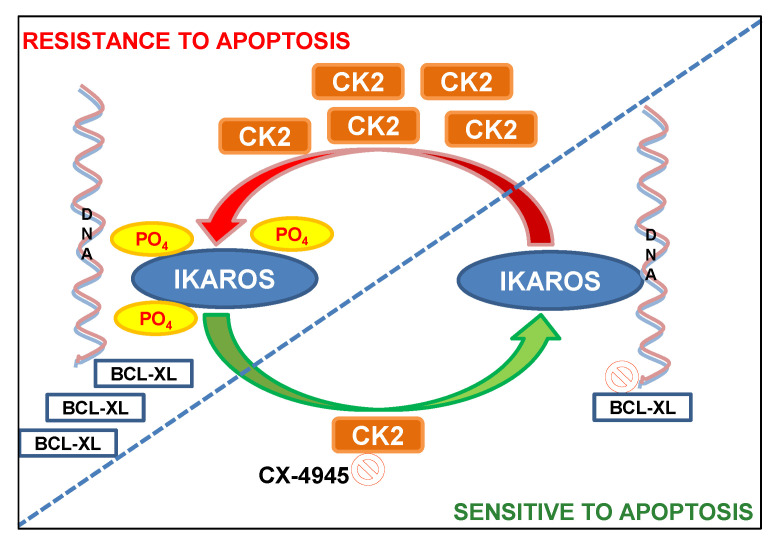
In summary, we report for the first time on the in vivo therapeutic efficacy of CX-4945 in AML patient-derived xenograft models. These results establish that one of the central mechanisms by which CX-4945 exerts an anti-leukemia effect in vivo is via the revival of IKAROS-mediated transcriptional repression of the BCL-XL gene (Figure 8). The functional synergy shown here, between the CK2 inhibitor and daunorubicin in suppressing BCL-XL provides a mechanistic basis for testing CK2 inhibitors combined with anthracycline therapy for the treatment of AML.
Figure 8.
Model illustration of the regulation of apoptosis in AML by CK2 and IKAROS via repression of BCL-XL.
Acknowledgments
The authors would like to thank Sinisa Dovat for providing key reagents, sharing lab equipment, critical review of the manuscript, and providing feedback.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/13/5/1127/s1, Figure S1: Western blot showing baseline protein overexpression in primary AML cells, Figure S2: Cell viability of U937, THP1 and AML#1 following CX-4945 treatment for 48 h, Figure S3: Western blot showing protein level of CK2α in U937 cells treated with CK2α sh-RNA(left panel), Figure S4: Immunoblot showing BCL-XL expression in CK2α overexpressing (OE) and silenced (CK2shRNA) U937 cells compared to control, Figure S5: Radioblot (top) and immunoblot (bottom panel) showing phosphorylated Ikaros in CK2α modulated U937 cells, Figure S6: Western blot showing CK2α and p-CK2 expression in U937 and THP-1 cells transduced with retroviral vec-tor containing CK2α, Figure S7: Histogram showing CK2α and BCL-XL protein quantification in U937 (left) and THP-1 (right) xenograft mice treated with CX-4945, Figure S8: Immunoblot showing Ikaros and BCL-XL protein expression in IKAROS overexpressed U937 cells, Figure S9: Synergetic cytotoxic effect of combination of Daunorubicin and CX-4945 in primary AML #1 cells, Table S1: List of Western blot antibodies, Table S2: the qChIP primers, Table S3: Characteristics of primary AML cells used in the study, Table S4: Complete blood count showing blood parameters in vehicle and CX-4945 treated U937 xenograft mice, Additional details of methods.
Author Contributions
C.G. conceptualized the project, acquired funding, supervised studies, wrote, reviewed, and edited the manuscript; M.K. and M.A.R. performed experiments, analyzed results, wrote the first draft, and edited the manuscript; M.K. and M.A.R. contributed equally. C.S., performed experiments, edited manuscript; P.K.D., C.A., S.S., S.I., M.E., and P.B., performed experiments; Y.D., data visualisation; A.S. and D.C. provided the primary AML samples and assisted with PDX models; D.D., S.H., and D.C., manuscript review, and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by the National Center for Advancing Translational Sciences (KL2 TR002015) (C.G.); Hyundai Hope on Wheels Scholar Grant (C.G.); Four Diamonds Fund of the Pennsylvania State University College of Medicine; John Wawrynovic Leukemia Research Scholar Endowment (C.G.); St. Baldrick’s Foundation; and Rally Foundation.
Institutional Review Board Statement
All the animal experiments were conducted in the Developmental Therapeutics Preclinical Core facility at Penn State University College of Medicine under protocols approved by the Institutional Animal Care and Use Committee at Penn State Hershey, Hershey, PA (IACUC # PROTO201901034). De-identified patient samples were provided by collaborators at PennState Cancer Institute, at Penn State University College of Medicine (Hershey, PA, USA), and used in compliance with Institutional Review Board regulations.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets analyzed and novel reagents used during the current study are available from the corresponding author upon request and after material transfer agreement. The ChIP-sequencing data shown in Figure 4 is available using the link provided in the Supplemental File.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Tallman M.S., Wang E.S., Altman J.K., Appelbaum F.R., Bhatt V.R., Bixby D., Coutre S.E., De Lima M., Fathi A.T., Fiorella M., et al. Acute Myeloid Leukemia, Version 3.2019, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2019;17:721–749. doi: 10.6004/jnccn.2019.0028. [DOI] [PubMed] [Google Scholar]
- 3.Perl A.E. The role of targeted therapy in the management of patients with AML. Blood Adv. 2017;1:2281–2294. doi: 10.1182/bloodadvances.2017009829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davids M.S. Boldly Targeting Kinases without mutations. Blood. 2014;123:1119–1121. doi: 10.1182/blood-2013-12-543322. [DOI] [PubMed] [Google Scholar]
- 5.Litchfield D.W. Protein kinase CK2: Structure, regulation and role in cellular decisions of life and death. Biochem. J. 2003;369:1–15. doi: 10.1042/bj20021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Litchfield D.W., Bosc D.G., Canton D.A., Saulnier R.B., Vilk G., Zhang C. Functional specialization of CK2 isoforms and characterization of isoform-specific binding partners. Mol. Cell. Biochem. 2001;227:21–29. doi: 10.1023/A:1013188101465. [DOI] [PubMed] [Google Scholar]
- 7.Chua M.M.J., Lee M., Dominguez I. Cancer-type dependent expression of CK2 transcripts. PLoS ONE. 2017;12:e0188854. doi: 10.1371/journal.pone.0188854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ortega C.E., Seidner Y., Dominguez I. Mining CK2 in cancer. PLoS ONE. 2014;9:e115609. doi: 10.1371/journal.pone.0115609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piazza F., Manni S., Ruzzene M., Pinna L.A., Gurrieri C., Semenzato G. Protein kinase CK2 in hematologic malignancies: Reliance on a pivotal cell survival regulator by oncogenic signaling pathways. Leukemia. 2012;26:1174–1179. doi: 10.1038/leu.2011.385. [DOI] [PubMed] [Google Scholar]
- 10.Chandrika G., Mansi S., Sunil M., Malika K., Lidija P.-D., Melanie H., Yali D., Chunhua S., Jonathon L.P., Tan BH S.D. Casein kinase II (CK2) as a therapeutic target for hematological malignancies. Curr. Pharm. Des. 2016;22:1–13. doi: 10.2174/1381612822666161006154311. [DOI] [PubMed] [Google Scholar]
- 11.Buontempo F., McCubrey J.A., Orsini E., Ruzzene M., Cappellini A., Lonetti A., Evangelisti C., Chiarini F., Barata J.T., Martelli A.M. Therapeutic targeting of CK2 in acute and chronic leukemias. Leukemia. 2018;32:1–10. doi: 10.1038/leu.2017.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandato E., Manni S., Zaffino F., Semenzato G., Piazza F. Targeting CK2-driven non-oncogene addiction in B-cell tumors. Oncogene. 2016;35:6045–6052. doi: 10.1038/onc.2016.86. [DOI] [PubMed] [Google Scholar]
- 13.Ruzzene M., Pinna L.A. Addiction to protein kinase CK2: A common denominator of diverse cancer cells? Biochim. Biophys. Acta. 2010;1804:499–504. doi: 10.1016/j.bbapap.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 14.Quotti Tubi L., Canovas Nunes S., Brancalion A., Doriguzzi Breatta E., Manni S., Mandato E., Zaffino F., Macaccaro P., Carrino M., Gianesin K., et al. Protein kinase CK2 regulates AKT, NF-κB and STAT3 activation, stem cell viability and proliferation in acute myeloid leukemia. Leukemia. 2017;31:292–300. doi: 10.1038/leu.2016.209. [DOI] [PubMed] [Google Scholar]
- 15.Quotti Tubi L., Gurrieri C., Brancalion A., Bonaldi L., Bertorelle R., Manni S., Pavan L., Lessi F., Zambello R., Trentin L., et al. Inhibition of protein kinase CK2 with the clinical-grade small ATP-competitive compound CX-4945 or by RNA interference unveils its role in acute myeloid leukemia cell survival, p53-dependent apoptosis and daunorubicin-induced cytotoxicity. J. Hematol. Oncol. 2013;6:78. doi: 10.1186/1756-8722-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J.S., Eom J.I., Cheong J.W., Choi A.J., Lee J.K., Yang W.I., Min Y.H. Protein kinase CK2alpha as an unfavorable prognostic marker and novel therapeutic target in acute myeloid leukemia. Clin. Cancer Res. 2007;13:1019–1028. doi: 10.1158/1078-0432.CCR-06-1602. [DOI] [PubMed] [Google Scholar]
- 17.McKendrick L., Milne D., Meek D. Protein kinase CK2-dependent regulation of p53 function: Evidence that the phosphorylation status of the serine 386 (CK2) site of p53 is constitutive and stable. Mol. Cell. Biochem. 1999;191:187–199. doi: 10.1023/A:1006854109926. [DOI] [PubMed] [Google Scholar]
- 18.Dovat S., Song C., Payne K.J., Li Z. Ikaros, CK2 kinase, and the road to leukemia. Mol. Cell. Biochem. 2011;356:201–207. doi: 10.1007/s11010-011-0964-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres J., Pulido R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J. Biol. Chem. 2001;276:993–998. doi: 10.1074/jbc.M009134200. [DOI] [PubMed] [Google Scholar]
- 20.Ning Y., Wang C., Liu X., Du Y., Liu S., Liu K., Zhou J., Zhou C. CK2-mediated CCDC106 phosphorylation is required for p53 degradation in cancer progression. J. Exp. Clin. Cancer Res. 2019;38:131. doi: 10.1186/s13046-019-1137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arriazu E., Vicente C., Pippa R., Peris I., Martínez-Balsalobre E., García-Ramírez P., Marcotegui N., Igea A., Alignani D., Rifón J., et al. A new regulatory mechanism of protein phosphatase 2A activity via SET in acute myeloid leukemia. Blood Cancer J. 2020;10:3. doi: 10.1038/s41408-019-0270-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siddiqui-Jain A., Drygin D., Streiner N., Chua P., Pierre F., O’Brien S.E., Bliesath J., Omori M., Huser N., Ho C., et al. CX-4945, an orally bioavailable selective inhibitor of protein kinase CK2, inhibits prosurvival and angiogenic signaling and exhibits antitumor efficacy. Cancer Res. 2010;70:10288–10298. doi: 10.1158/0008-5472.CAN-10-1893. [DOI] [PubMed] [Google Scholar]
- 23.Pierre F., Chua P.C., O’Brien S.E., Siddiqui-Jain A., Bourbon P., Haddach M., Michaux J., Nagasawa J., Schwaebe M.K., Stefan E., et al. Pre-clinical characterization of CX-4945, a potent and selective small molecule inhibitor of CK2 for the treatment of cancer. Mol. Cell. Biochem. 2011;356:37–43. doi: 10.1007/s11010-011-0956-5. [DOI] [PubMed] [Google Scholar]
- 24.Battistutta R., Cozza G., Pierre F., Papinutto E., Lolli G., Sarno S., O’Brien S.E., Siddiqui-Jain A., Haddach M., Anderes K., et al. Unprecedented selectivity and structural determinants of a new class of protein kinase CK2 inhibitors in clinical trials for the treatment of cancer. Biochemistry. 2011;50:8478–8488. doi: 10.1021/bi2008382. [DOI] [PubMed] [Google Scholar]
- 25.Song C., Gowda C., Pan X., Ding Y., Tong Y., Tan B.H., Wang H., Muthusami S., Ge Z., Sachdev M., et al. Targeting casein kinase II restores Ikaros tumor suppressor activity and demonstrates therapeutic efficacy in high-risk leukemia. Blood. 2015;126:1813–1822. doi: 10.1182/blood-2015-06-651505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song C., Pan X., Ge Z., Gowda C., Ding Y., Li H., Li Z., Yochum G., Muschen M., Li Q., et al. Epigenetic regulation of gene expression by Ikaros, HDAC1 and Casein Kinase II in leukemia. Leukemia. 2016;30:1436–1440. doi: 10.1038/leu.2015.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding Y., Zhang B., Payne J.L., Song C., Ge Z., Gowda C., Iyer S., Dhanyamraju P.K., Dorsam G., Reeves M.E., et al. Ikaros tumor suppressor function includes induction of active enhancers and super-enhancers along with pioneering activity. Leukemia. 2019;33:2720–2731. doi: 10.1038/s41375-019-0474-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song C., Li Z., Erbe A.K., Savic A., Dovat S. Regulation of Ikaros function by casein kinase 2 and protein phosphatase 1. World J. Biol. Chem. 2011;2:126–131. doi: 10.4331/wjbc.v2.i6.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gowda C., Song C., Ding Y., Iyer S., Dhanyamraju P.K., McGrath M., Bamme Y., Soliman M., Kane S., Payne J.L., et al. Cellular signaling and epigenetic regulation of gene expression in leukemia. Adv. Biol. Regul. 2020;75:100665. doi: 10.1016/j.jbior.2019.100665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song C., Ge Z., Ding Y., Tan B.H., Desai D., Gowda K., Amin S.G., Gowda R., Robertson G., Yue F., et al. IKAROS and CK2 regulate expression of BCL-XL and chemosensitivity inhigh-risk B-cell acute lymphoblastic leukemia. Blood. 2020 doi: 10.1182/blood.2019002655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X., Zhang X., Li X., Lv Y., Zhu Y., Wang J., Jin J., Yu W. The specific distribution pattern of IKZF1 mutation in acute myeloid leukemia. J. Hematol. Oncol. 2020;13:140. doi: 10.1186/s13045-020-00972-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francis O.L., Payne J.L., Su R.J., Payne K.J. Regulator of myeloid differentiation and function: The secret life of Ikaros. World J. Biol. Chem. 2011;2:119–125. doi: 10.4331/wjbc.v2.i6.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Rooij J.D., Beuling E., van den Heuvel-Eibrink M.M., Obulkasim A., Baruchel A., Trka J., Reinhardt D., Sonneveld E., Gibson B.E., Pieters R., et al. Recurrent deletions of IKZF1 in pediatric acute myeloid leukemia. Haematologica. 2015;100:1151–1159. doi: 10.3324/haematol.2015.124321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Payne K.J., Huang G., Sahakian E., Zhu J.Y., Barteneva N.S., Barsky L.W., Payne M.A., Crooks G.M. Ikaros isoform x is selectively expressed in myeloid differentiation. J. Immunol. 2003;170:3091–3098. doi: 10.4049/jimmunol.170.6.3091. [DOI] [PubMed] [Google Scholar]
- 35.Jäger R., Gisslinger H., Passamonti F., Rumi E., Berg T., Gisslinger B., Pietra D., Harutyunyan A., Klampfl T., Olcaydu D., et al. Deletions of the transcription factor Ikaros in myeloproliferative neoplasms. Leukemia. 2010;24:1290–1298. doi: 10.1038/leu.2010.99. [DOI] [PubMed] [Google Scholar]
- 36.Cheong J.W., Min Y.H., Eom J.I., Kim S.J., Jeung H.K., Kim J.S. Inhibition of CK2{alpha} and PI3K/Akt synergistically induces apoptosis of CD34+CD38- leukaemia cells while sparing haematopoietic stem cells. Anticancer Res. 2010;30:4625–4634. [PubMed] [Google Scholar]
- 37.Gurel Z., Ronni T., Ho S., Kuchar J., Payne K.J., Turk C.W., Dovat S. Recruitment of ikaros to pericentromeric heterochromatin is regulated by phosphorylation. J. Biol. Chem. 2008;283:8291–8300. doi: 10.1074/jbc.M707906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Popescu M., Gurel Z., Ronni T., Song C., Hung K.Y., Payne K.J., Dovat S. Ikaros stability and pericentromeric localization are regulated by protein phosphatase 1. J. Biol. Chem. 2009;284:13869–13880. doi: 10.1074/jbc.M900209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lertsuwan J., Lertsuwan K., Sawasdichai A., Tasnawijitwong N., Lee K.Y., Kitchen P., Afford S., Gaston K., Jayaraman P.S., Satayavivad J. CX-4945 Induces Methuosis in Cholangiocarcinoma Cell Lines by a CK2-Independent Mechanism. Cancers. 2018;10 doi: 10.3390/cancers10090283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oshima T., Niwa Y., Kuwata K., Srivastava A., Hyoda T., Tsuchiya Y., Kumagai M., Tsuyuguchi M., Tamaru T., Sugiyama A., et al. Cell-based screen identifies a new potent and highly selective CK2 inhibitor for modulation of circadian rhythms and cancer cell growth. Sci. Adv. 2019;5:eaau9060. doi: 10.1126/sciadv.aau9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jung M., Park K.H., Kim H.M., Kim T.S., Zhang X., Park S.M., Beom S.H., Kim H.S., Cheong J.H., Chung H.C., et al. Inhibiting casein kinase 2 overcomes paclitaxel resistance in gastric cancer. Gastric. Cancer. 2019;22:1153–1163. doi: 10.1007/s10120-019-00971-7. [DOI] [PubMed] [Google Scholar]
- 42.Wang Z., Ma J., Miyoshi C., Li Y., Sato M., Ogawa Y., Lou T., Ma C., Gao X., Lee C., et al. Quantitative phosphoproteomic analysis of the molecular substrates of sleep need. Nature. 2018;558:435–439. doi: 10.1038/s41586-018-0218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Morgen P., Burdova K., Flower T.G., O’Reilly N.J., Boulton S.J., Smerdon S.J., Macurek L., Hořejší Z. MRE11 stability is regulated by CK2-dependent interaction with R2TP complex. Oncogene. 2017;36:4943–4950. doi: 10.1038/onc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Maira G., Salvi M., Arrigoni G., Marin O., Sarno S., Brustolon F., Pinna L.A., Ruzzene M. Protein kinase CK2 phosphorylates and upregulates Akt/PKB. Cell Death Differ. 2005;12:668–677. doi: 10.1038/sj.cdd.4401604. [DOI] [PubMed] [Google Scholar]
- 45.Ruzzene M., Di Maira G., Tosoni K., Pinna L.A. Assessment of CK2 constitutive activity in cancer cells. Methods Enzymol. 2010;484:495–514. doi: 10.1016/B978-0-12-381298-8.00024-1. [DOI] [PubMed] [Google Scholar]
- 46.Lou D.Y., Dominguez I., Toselli P., Landesman-Bollag E., O’Brien C., Seldin D.C. The alpha catalytic subunit of protein kinase CK2 is required for mouse embryonic development. Mol. Cell. Biol. 2008;28:131–139. doi: 10.1128/MCB.01119-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trembley J.H., Wang G., Unger G., Slaton J., Ahmed K. Protein kinase CK2 in health and disease: CK2: A key player in cancer biology. Cell. Mol. Life Sci. 2009;66:1858–1867. doi: 10.1007/s00018-009-9154-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martelli A.M., Paganelli F., Fazio A., Bazzichetto C., Conciatori F., McCubrey J.A. The Key Roles of PTEN in T-Cell Acute Lymphoblastic Leukemia Development, Progression, and Therapeutic Response. Cancers. 2019;11:629. doi: 10.3390/cancers11050629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J., Feng C., He Y., Ding W., Sheng J., Arshad M., Zhang X., Li P. Phosphorylation of apoptosis repressor with caspase recruitment domain by protein kinase CK2 contributes to chemotherapy resistance by inhibiting doxorubicin induced apoptosis. Oncotarget. 2015;6:27700–27713. doi: 10.18632/oncotarget.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borgo C., Ruzzene M. Role of protein kinase CK2 in antitumor drug resistance. J. Exp. Clin. Cancer Res. 2019;38:287. doi: 10.1186/s13046-019-1292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trembley J.H., Chen Z., Unger G., Slaton J., Kren B.T., Van Waes C., Ahmed K. Emergence of protein kinase CK2 as a key target in cancer therapy. Biofactors. 2010;36:187–195. doi: 10.1002/biof.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siddiqui-Jain A., Bliesath J., Macalino D., Omori M., Huser N., Streiner N., Ho C.B., Anderes K., Proffitt C., O’Brien S.E., et al. CK2 inhibitor CX-4945 suppresses DNA repair response triggered by DNA-targeted anticancer drugs and augments efficacy: Mechanistic rationale for drug combination therapy. Mol. Cancer Ther. 2012;11:994–1005. doi: 10.1158/1535-7163.MCT-11-0613. [DOI] [PubMed] [Google Scholar]
- 53.Martins L.R., Lúcio P., Silva M.C., Anderes K.L., Gameiro P., Silva M.G., Barata J.T. Targeting CK2 overexpression and hyperactivation as a novel therapeutic tool in chronic lymphocytic leukemia. Blood. 2010;116:2724–2731. doi: 10.1182/blood-2010-04-277947. [DOI] [PubMed] [Google Scholar]
- 54.Buontempo F., Orsini E., Martins L.R., Antunes I., Lonetti A., Chiarini F., Tabellini G., Evangelisti C., Melchionda F., Pession A., et al. Cytotoxic activity of the casein kinase 2 inhibitor CX-4945 against T-cell acute lymphoblastic leukemia: Targeting the unfolded protein response signaling. Leukemia. 2014;28:543–553. doi: 10.1038/leu.2013.349. [DOI] [PubMed] [Google Scholar]
- 55.Zhao Z., Wang L., Volk A.G., Birch N.W., Stoltz K.L., Bartom E.T., Marshall S.A., Rendleman E.J., Nestler C.M., Shilati J., et al. Regulation of MLL/COMPASS stability through its proteolytic cleavage by taspase1 as a possible approach for clinical therapy of leukemia. Genes Dev. 2019;33:61–74. doi: 10.1101/gad.319830.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meggio F., Pinna L.A. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 57.Mullighan C., Downing J. Ikaros and acute leukemia. Leuk. Lymphoma. 2008;49:847–849. doi: 10.1080/10428190801947500. [DOI] [PubMed] [Google Scholar]
- 58.Minn A.J., Rudin C.M., Boise L.H., Thompson C.B. Expression of bcl-xL can confer a multidrug resistance phenotype. Blood. 1995;86:1903–1910. doi: 10.1182/blood.V86.5.1903.bloodjournal8651903. [DOI] [PubMed] [Google Scholar]
- 59.Vogler M., Hamali H.A., Sun X.M., Bampton E.T., Dinsdale D., Snowden R.T., Dyer M.J., Goodall A.H., Cohen G.M. BCL2/BCL-X(L) inhibition induces apoptosis, disrupts cellular calcium homeostasis, and prevents platelet activation. Blood. 2011;117:7145–7154. doi: 10.1182/blood-2011-03-344812. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed and novel reagents used during the current study are available from the corresponding author upon request and after material transfer agreement. The ChIP-sequencing data shown in Figure 4 is available using the link provided in the Supplemental File.