Abstract
The 22q11.2 deletion syndrome (22q11DS) is associated with a 20–25% risk of schizophrenia. In a cohort of 962 individuals with 22q11DS, we examined the shared genetic basis between schizophrenia and schizophrenia-related early trajectory phenotypes: sub-threshold symptoms of psychosis, low baseline intellectual functioning and cognitive decline. We studied the association of these phenotypes with two polygenic scores, derived for schizophrenia and intelligence, and evaluated their use for individual risk prediction in 22q11DS. Polygenic scores were not only associated with schizophrenia and baseline intelligence quotient (IQ), respectively, but schizophrenia polygenic score was also significantly associated with cognitive (verbal IQ) decline and nominally associated with sub-threshold psychosis. Furthermore, in comparing the tail-end deciles of the schizophrenia and IQ polygenic score distributions, 33% versus 9% of individuals with 22q11DS had schizophrenia, and 63% versus 24% of individuals had intellectual disability. Collectively, these data show a shared genetic basis for schizophrenia and schizophrenia-related phenotypes and also highlight the future potential of polygenic scores for risk stratification among individuals with highly, but incompletely, penetrant genetic variants.
Although schizophrenia is typically diagnosed in late adolescence or early adulthood, it is now well established that the first psychotic episode is, in fact, a manifestation of an advanced stage of this disease1. Early behavioral, cognitive and neuroanatomic changes are measurable before the first psychotic episode2–6. Both lower cognitive ability early in life (the estimated premorbid deficit is 8 IQ points)7 and cognitive decline in early adolescence (estimated IQ change equal to −1.09 s.d.)8 are associated with schizophrenia, with effect sizes in the range of 0.4–0.5 (ref.7–13). In addition, sub-threshold psychotic symptoms in youth also index increased risk for schizophrenia14–16. These observations raise an important question: Do early cognitive phenotypes and sub-threshold symptoms of psychosis share a substantial genetic basis with either schizophrenia or intellectual ability?
Early schizophrenia-related phenotypes and trajectories are difficult to study, requiring longitudinal follow-up of large cohorts to capture a sufficient number of schizophrenia cases. At-risk populations facilitate such studies, as fewer individuals need to be followed to obtain the same number of cases. The 22q11DS, which is increasingly identified around birth, provides one such at-risk population17, given the associated 20–25% risk to develop schizophrenia18,19.
Findings from 22q11DS studies reproduce observations related to schizophrenia in the general population, thereby supporting 22q11DS as a genetic model of schizophrenia, including its early trajectory20. In 22q11DS, as in the general population, sub-threshold psychotic symptoms21, low baseline intellectual ability and increasing cognitive deficits over time, particularly in verbal IQ (VIQ)22, are all associated with increased risk of subsequent psychotic illness.
A large fraction of the heritability of schizophrenia comes from a polygenic burden of multiple common variants, each of small effect23,24. Increasingly, polygenic scores derived from genome-wide association studies (GWASs) have been used to study the genetic relationship between phenotypes25. For schizophrenia, a polygenic score using a recent GWAS explains up to 7% of the variance on the liability scale. Similarly, polygenic scores for general cognitive function, or proxies thereof, explain 2.5–4.3% of its variance26,27. Polygenic scores can also be used for phenotype prediction and, hence, risk stratification28. In the general population, they are not yet particularly effective as individual risk predictors29, given the relatively low population prevalence of phenotypes, such as schizophrenia and intellectual disability (ID), and the still modest effect sizes conferred by polygenic scores30. However, in high-risk populations, such as individuals with 22q11DS, the same effect size acts upon a higher baseline prevalence (for example, 25% for schizophrenia), which might allow for more substantial differences in absolute risk31.
The International Brain Behavior Consortium (IBBC) in 22q11.2 Deletion Syndrome has assembled the largest genotype–phenotype data set of individuals with 22q11DS20. Previously, the IBBC reported on genetic associations of both common and rare variants in 520 individuals with 22q11DS, exclusively focusing on schizophrenia32. The current study presents several novel analyses, conducted in a substantially larger cohort of individuals with 22q11DS (n = 962) and including longitudinal IQ data. Our main objectives were two-fold. First, we studied the genetic relationships between schizophrenia and schizophrenia-related phenotypes of low baseline intellectual ability, cognitive decline and sub-threshold positive psychotic symptoms. Second, we examined the use of polygenic scores for schizophrenia and IQ for individual risk prediction of schizophrenia and ID (IQ <70) in individuals with 22q11DS.
Results
Description of data set.
After applying phenotypic classification and performing genotype quality control, data from 962 IBBC cohort members were available for analysis (Table 1 and Methods). Within this cohort, we distinguished individuals with schizophrenia spectrum disorder (SSD; n = 207)20, individuals with sub-threshold psychotic symptoms (n = 158) and individuals with neither phenotype, grouped into ‘putative controls’ (age <25 years (‘putative’ given the typical age at onset of schizophrenia)33, n = 382) and ‘definite controls’ (age ≥25 years, n = 215). Subsequently, we refer to all controls, regardless of age, as ‘merged controls’ (n = 597). Baseline Full Scale Intelligence Quotient (FSIQ) was transformed to z score as previously described22, with an average near 0 (0.03; Table 1). VIQ decline, operationalized as exceeding −1 s.d. (binary), occurred in 5.9% of the cohort.
Table 1 |.
Clinical characteristics of sample cohort
| SSD | Sub-threshold psychosis | Putative control | Definite control | All | Pa | |
|---|---|---|---|---|---|---|
| n max | 207 | 158 | 382 | 215 | 962 | N/A |
| Sex, % male | 49 | 49 | 54 | 39 | 49 | 0.008 |
| Age at last assessment (years)b | 31.6 (12.7) [205], {7,64} | 17.9 (5) [158] {8,36} | 15.2 (4.6) [382] {5,24} | 36.8 (9.9) [215], {25,67} | 24 (12.4) [960], {5,67} | 2.0 × 10−167 |
| Baseline FSIQb,c | −0.34 (0.87) [145], {−2,2} | 0.13 (0.96) [127] {−2,3} | 0.07 (1.03) [308] {−3,2} | 0.24 (0.85) [120], {−2,2} | 0.03 (0.97) [700], {−3,3} | 1.8 × 10−6 |
| Binary VIQ declined,e | 11.9% [59] | 5.7% [87] | 4.5% [198] | 4.7% [43] | 5.9% [387] | 0.21 |
| Comorbid mood disordersd | 41% [144] | 29.2% [154] | 16% [363] | 38.6% [153] | 27.1% [814] | 1.7 × 10−10 |
P values are from an analysis of variance of phenotype by group and are two sided and not corrected for multiple comparisons.
Quantitative phenotypes are given as mean (s.d.) [n] {range}.
Baseline FSIQ is given as a z score using a previously defined normalization procedure22.
Binary phenotypes are given as percent true [n].
Binary VIQ decline is operationalized as VIQ decline ≤1 s.d. (that is, 1 z score), as defined by the reliable change index53.
Polygenic scores and relationships among schizophrenia, IQ and associated phenotypes.
First, we examined known associations. We constructed polygenic scores for schizophrenia24 (PS_SZ) and polygenic scores for intellectual ability34 (PS_IQ) using standard methods and performed statistical analyses using either linear or logistic regression as appropriate, adjusting for age, sex and the first five principal components from the imputed genotypes32 (Methods). We observed a significant association between SSD cases versus controls and PS_SZ (n = 802, P = 4.37 × 10−8, marginal Nagelkerke pseudo r2 = 0.053; P values reported in the text are nominal) and a similar result when including definite controls only (n = 420, P = 1.89 × 10−6, r2 = 0.071) (Table 2 and Extended Data Fig. 1), corroborating previous reports from the comprehensive IBBC genetic analyses related to schizophrenia in 22q11DS32. We also observed a significant association between baseline FSIQ and PS_IQ (P = 1.08 × 10−7), and, consistent with the known genetic correlation between schizophrenia and IQ (r2g = −0.234)26, we observed a nominal association between baseline FSIQ and PS_SZ (P = 0.018) as well as a significant association between SSD and PS_IQ (P = 7.15 × 10−4).
Table 2 |.
Key regression results
| Dependent variable | IV | n | β | r2 | P |
|---|---|---|---|---|---|
| SSD | PS_SZ | 802 | 0.56 | 0.053 | 4.37 × 10−8* |
| Sub-threshold psychosis | 755 | 0.24 | 0.01 | 0.0247 | |
| Baseline FSIQ | 720 | −0.096 | 0.0077 | 0.018 | |
| VIQ decline | 396 | 0.66 | 0.051 | 0.00509* | |
| SSD | PS_IQ | 802 | −0.30 | 0.020 | 7.15 × 10−4* |
| Sub-threshold psychosis | 755 | −0.18 | 0.0072 | 0.056 | |
| Baseline FSIQ | 720 | 0.20 | 0.038 | 1.08 × 10−7* | |
| VIQ decline | 396 | −0.096 | 0.0013 | 0.658 |
Results are adjusted for standard covariates as described in Methods. β is the standard regression effect size estimate. r2 denotes the difference between model fit with or without independent variable (IV), using either standard r2 from linear regression or Nagelkerke pseudo r2. Nominal P values are reported. Asterisk (*) indicates a significant result after Bonferroni correction for eight independent main analyses in this study (two polygenic scores and four phenotypes).
Next, we assessed relationships between schizophrenia-related phenotypes and the polygenic scores. We observed a decreasing trend of PS_SZ for phenotypes of SSD (mean, 0.23), sub-threshold psychosis (mean, 0.16), putative controls (mean, −0.05) and definite controls (mean, −0.27) (Fig. 1). PS_SZ was nominally significantly higher in individuals with sub-threshold psychosis compared to the merged control groups (n = 755, P = 0.0247, r2 = 0.01; Table 2 and Fig. 1). Finally, we observed a significant association between VIQ decline and PS_SZ (P = 5.09 × 10−3; Fig. 2). Neither the association between sub-threshold psychosis and PS_IQ (P = 0.056) nor between VIQ decline and PS_IQ (P = 0.658) reached statistical significance.
Fig. 1 |. PS_SZs among phenotypic subgroups.
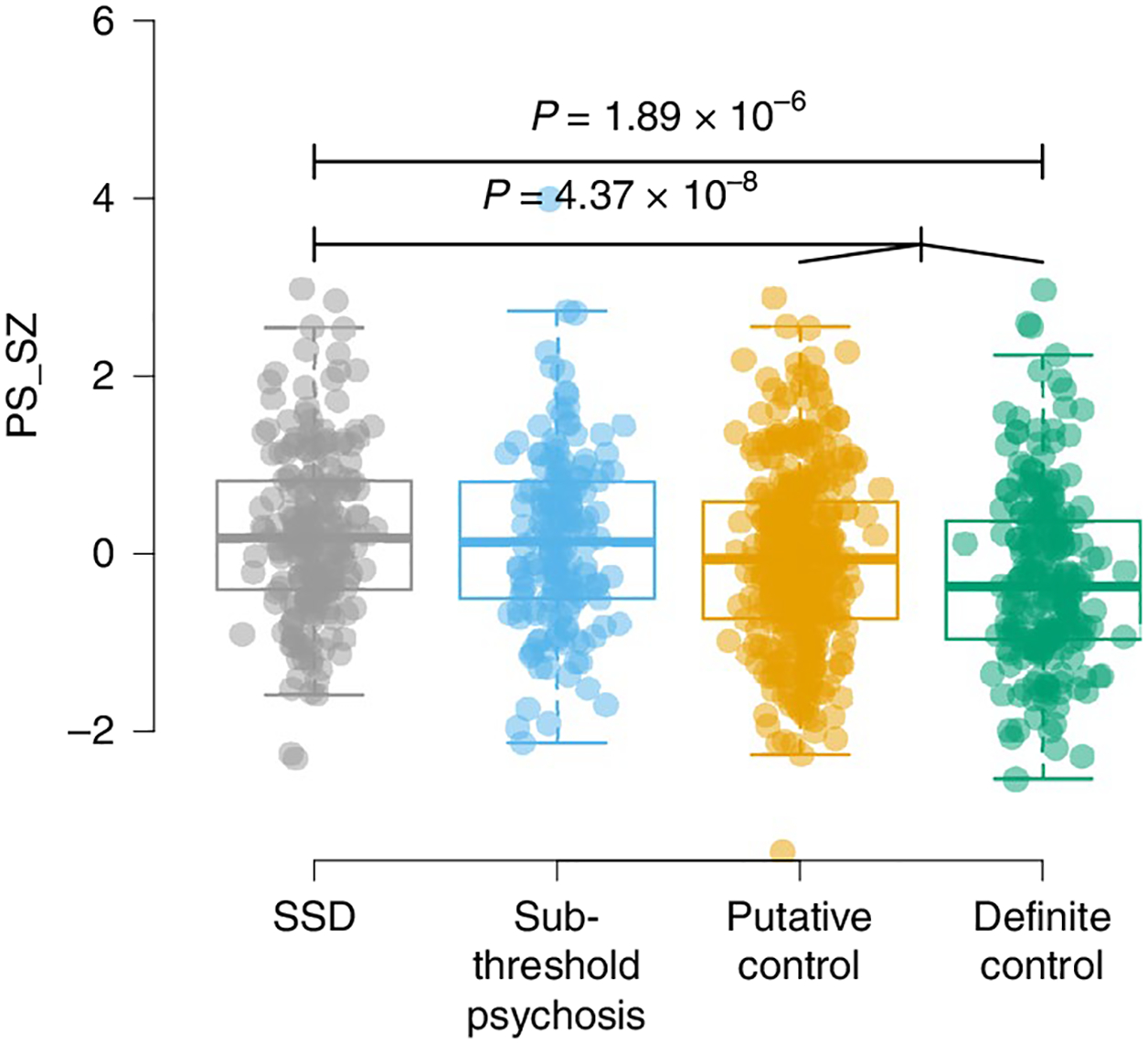
Results show per-individual values as well as summaries per group, where minimum and maximum values are directly observable from the plot; the box plot center is the median; the box plot edges represent the 25th and 75th percentiles; and the whiskers represent the lesser of the distance to the minimum or maximum value or 1.5 times the interquartile range. Associations of PS_SZ in 22q11DS with SSD, sub-threshold psychosis, putative controls and definite controls are shown. Results for PS_SZ and SSD were reported previously32 and are included in this figure for completeness. Total sample sizes for the highlighted associations are n = 423 (SSD versus putative controls) and n = 802 (SSD versus merged controls). P values are reported for select comparisons using two-sided logistic regression analyses uncorrected for multiple testing using covariates as specified in the Methods.
Fig. 2 |. Relationship between polygenic scores and novel phenotypes.
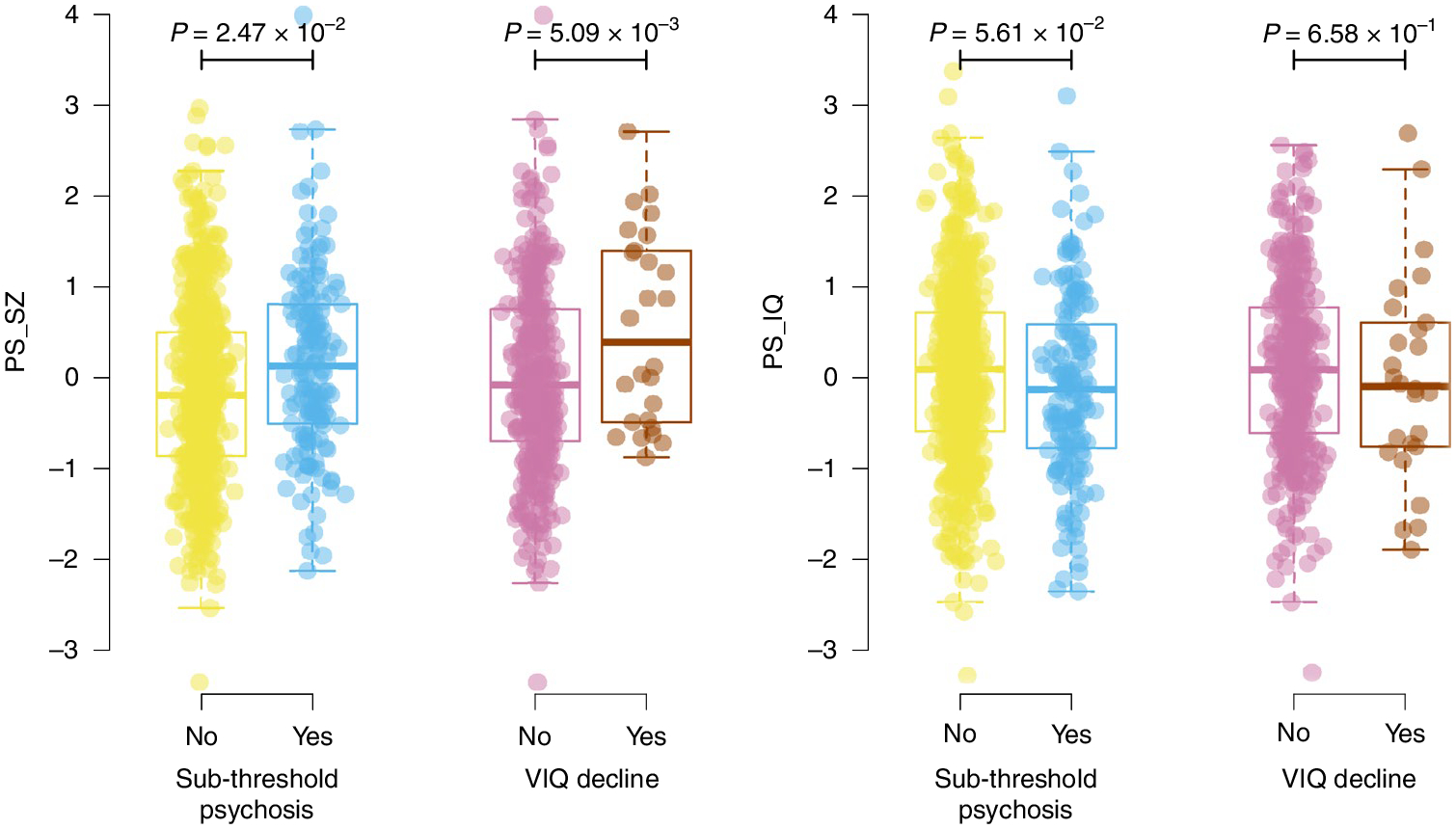
Results show per-individual values as well as summaries per group, where minimum and maximum values are directly observable from the plot; the box plot center is the median; the box plot edges represent the 25th and 75th percentiles; and the whiskers represent the lesser of the distance to the minimum or maximum value or 1.5 times the interquartile range. Results are shown for regressions of sub-threshold psychosis versus merged controls (n = 755) and VIQ decline (n = 396) for both PS_SZ (left) and PS_IQ (right). P values are reported for two-sided logistic regression analyses uncorrected for multiple testing using covariates as specified in the Methods.
Investigations into the relationship between sub-threshold psychosis and PS_SZ.
Post hoc, we performed three analyses to additionally explore the observed association between sub-threshold psychosis and PS_SZ. First, given that some fraction of individuals with sub-threshold psychosis will eventually develop SSD, we modeled what proportion would need to develop SSD to be consistent with our findings (Methods). The findings showed that observed levels of PS_SZ are consistent with a scenario in which 86% (95% confidence interval (CI), 56–100%) of individuals with sub-threshold psychosis would, in fact, represent future patients with SSD who were not yet identified as such at the time of the assessment. This is a proportion inconsistent with known rates of SSD in 22q11DS (Extended Data Fig. 2), rendering it unlikely that our result is driven by ‘future’ SSD cases. Second, we examined whether our observation could be due to confounding through psychiatric comorbidity genetically correlated with SSD (Methods). In this sample, the rate of comorbid mood disorders in the sub-threshold psychosis group was 29.2% versus 22.7% in the merged controls (Table 1). Results from this mediation analysis indicated a lack of attenuation through the mood disorder phenotype (the effect size of PS_SZ in the model without mood disorder was 0.239, P = 0.025; the effect size of PS_SZ in the model with mood disorder was 0.250, P = 0.021) (Supplementary Table 1), indicating that the observed increased PS_SZ in sub-threshold psychosis is not readily explained by the higher rate of mood disorders in this group. Third, as a source of additional evidence, we explored the use of residual quantitative variation in the measure of sub-threshold psychosis through a transformed quantitative measure of this phenotype (Structured Interview for Prodromal Syndromes (SIPS35)) (Methods). When adjusting for the previous binary indicator of sub-threshold psychosis versus control, the association between the transformed quantitative SIPS phenotype and PS_SZ was not significant (n = 347, P = 0.77, r2 = 0.0001; Supplementary Fig. 1).
Polygenic score and individual risk prediction.
Addressing the second objective of our study, we investigated the extent to which polygenic scores could be used for individualized risk prediction among individuals with 22q11DS. We divided the cohort into quantiles based on polygenic scores, and calculated positive predictive values (PPVs) in each. For SSD, 32% of individuals with scores above the median PS_SZ had SSD (that is, a PPV of 32%) versus 20% of those with scores below the median (odds ratio (OR) = 1.91; 95% CI, [1.38, 2.64]; P = 8.4 × 10−5) (Fig. 3 and Supplementary Table 2). Values at the tails showed more extreme differences, with those exceeding the 90th percentile at substantially higher risk (33%) than those in the lowest decile (9.1%). Simulating an effect for the general population, using the observed effect sizes and assuming a general population prevalence of SSD of 1%, generated substantially smaller absolute differences (Fig. 3).
Fig. 3 |. Individual risk prediction.
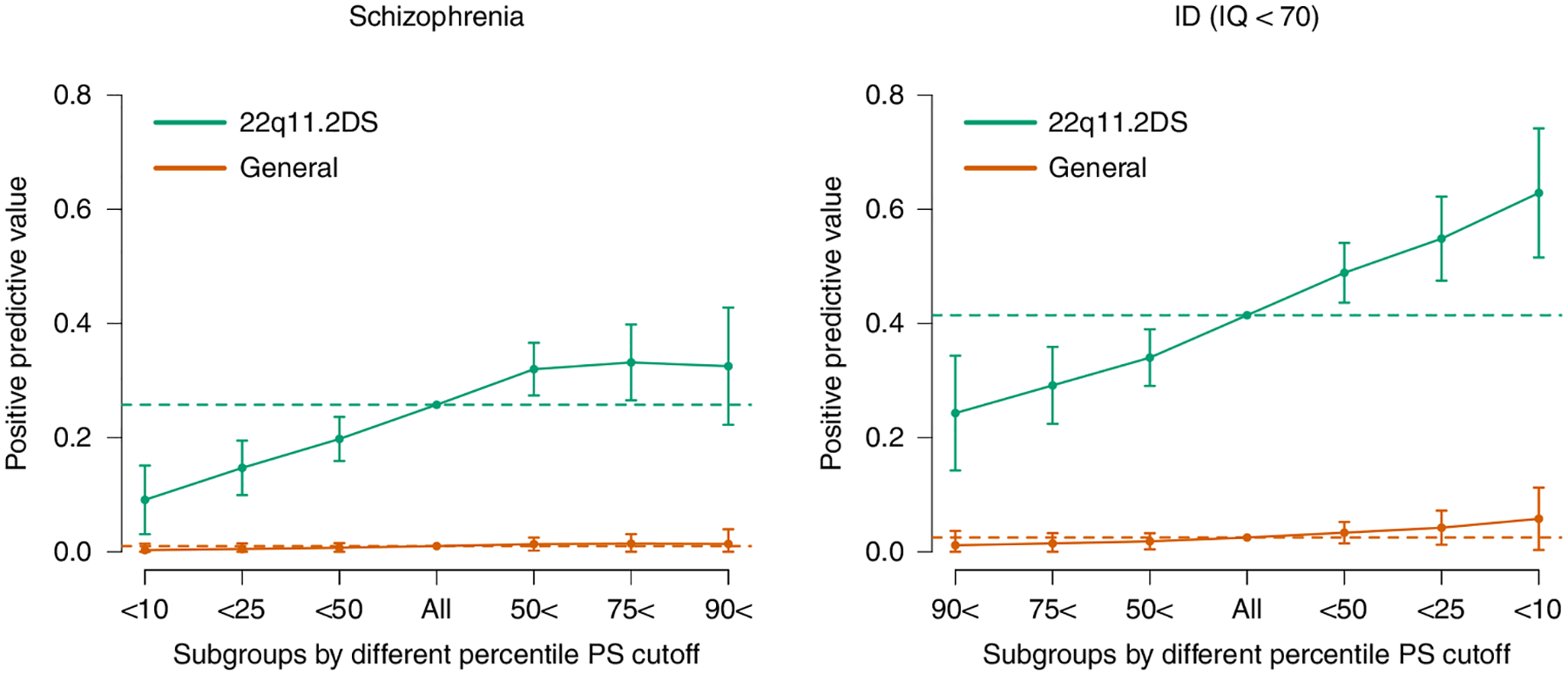
PPVs (y axis) for SSD (left) and ID (right) based on various cutoffs of PS_SZ (left) or PS_IQ (right). Colors differentiate values from the 22q11DS cohort (turquoise) versus values estimated from the general population (orange) given observed prevalences in the population (SSD = 0.01 and ID = 0.025; dotted lines) and observed ORs. Whiskers represent confidence intervals (±1.96 × standard error) around the central PPV estimate.
Similarly, using intellectual ability as a binary outcome (ID as IQ <70), we observed a higher rate of ID among individuals with a PS_IQ below the median versus above (PPV = 49% versus 34%; OR = 1.85; 95% CI,[1.37, 2.51]; P = 7.1 × 10−5). This effect is accentuated at the tails, with PPVs of 63% for individuals in the lowest decile for PS_IQ (that is, associated with lower IQ in the general population) versus 24% for individuals in the highest decile of PS_ IQ (Fig. 3 and Supplementary Table 3).
Discussion
In this study, we used polygenic scores from large GWASs for schizophrenia and IQ both to better understand the association between schizophrenia and schizophrenia-associated phenotypes and to assess their potential for individual risk prediction. In the first part of the study, we confirmed several results known to occur in the general population and showed that known relationships between schizophrenia and IQ extend to individuals with 22q11DS. We observed that a polygenic score for IQ explained ~3.8% of the variance in IQ in 22q11DS, suggesting that the previously observed association between parental educational attainment and cognitive outcome in offspring with 22q11DS36 might be at least partly explained by common variants.
In addition, we identified two novel associations between schizophrenia-related phenotypes and schizophrenia. First, we observed a novel association between sub-threshold psychosis and PS_SZ. Given the nominal statistical significance of this observation, we performed several post hoc investigations to rule out potential confounding sources, showing that neither undiagnosed ‘future’ cases nor comorbid mood disorders in our samples can explain the observed signal. We also examined residual quantitative variation in sub-threshold psychosis and found that the association between this transformed quantitative variable and PS_SZ was not significant when adjusting for the previous binary indicator of these phenotypes. However, it is worth noting that a priori power for this analysis was limited and dependent on strong assumptions. Interestingly, studies on genetic correlations between sub-threshold psychotic symptoms and PS_SZ in the general population reported to date are conflicting37–39, impeding definite evidence in this regard. Regarding sub-threshold psychotic symptoms and schizophrenia in 22q11DS, we conclude that our findings tentatively suggest a genetic correlation, but further studies are required to provide more certainty.
In addition, we observed a novel and significant association between VIQ decline and PS_SZ, but not PS_IQ, suggesting that common risk variants for schizophrenia contribute to cognitive decline, whereas common variants associated with cognitive ability might not. A possible implication of these results is that cognitive decline before the first psychotic episode might not merely be a risk factor for schizophrenia, as reported previously for 22q11DS22 and idiopathic schizophrenia40,41 but also shares its genetic underpinnings. A previous study in a subset of this cohort showed that cognitive decline preceded the onset of the first psychotic episode by several years22, making reverse causation—that is, cognitive decline as a consequence of psychosis—a less likely explanation. The observed cognitive decline in 22q11DS could be caused by the inability of patients to keep up with peers or, alternatively, represent an absolute loss of cognitive abilities or a combination of both. The current analyses do not distinguish between these, but previous studies in 22q11DS found evidence in support of both mechanisms42,43. We cannot fully exclude the possibility that the observed cognitive decline could be affected by the negative effect of psychosis on cognitive testing. However, this is an unlikely explanation given that all study sites refrained from assessing individuals who were acutely psychotic, as is common clinical policy. Furthermore, in our data, the mean age at IQ assessment was below the age at psychosis onset for both baseline (14.8 and 20.6 years, respectively) and longitudinal IQ data (18.2 and 20.3 years, respectively). In addition, in individuals without psychosis, 55% showed an IQ decline versus 45% stable or increase (P = 0.02), indicating that, on average, a modest cognitive decline can be observed in 22q11DS regardless of the occurrence of a psychotic disorder, as previously reported22. Taken together, our findings are consistent with the notion that disruption of normal cognitive development is a core component of schizophrenia11, and investigation of high-penetrance variants for both phenotypes offers important insights into its mechanism.
In the second part of the study, we examined to what extent polygenic scores could be used for individual risk prediction of SSD and ID among individuals with 22q11DS. Whereas, in research, the existence of association between test and outcome is most relevant, in the clinic, the PPV is key, as it enables stratification of individuals into groups with different outcome probabilities that can inform clinical decision-making30,44. Previous studies showed that high-risk copy number variant carriers with schizophrenia have increased polygenic scores45,46, including, specifically, 22q11DS32,45, but these studies did not look at stratification within those groups. Importantly, PPV depends not only on the strength of association but also on the baseline prevalence. In the current study, we examined risk stratification among individuals with 22q11DS, taking advantage of the higher baseline prevalence of schizophrenia and ID compared to the general population (in our sample, 23% and 41%, respectively). Among individuals in the highest PS_SZ risk decile, 33% had schizophrenia versus 9% in the lowest decile. Applying the same effect sizes to the general population would yield estimates of 1.5% and 0.3%, respectively. Similarly, 63% of individuals in the lowest PS_IQ decile had ID versus 24% in the highest decile.
The observed differences between PPVs in our study are similar to those previously reported for BRCA1 and BRCA2 among females for breast cancer risk31 and males for prostate cancer risk47. The concept of using polygenic background to inform individual risk prediction and clinical decision-making is an area of active investigation48 and is being incorporated into clinical trials for common medical conditions (for example, ref.49). Although our findings highlight the potential clinical utility of polygenic scores in the context of a high-penetrance variant such as 22q11DS, the PPVs reported here are not yet sufficient to affect clinical decision-making. In addition, although risk prediction enables stratification within high-risk populations, it is important to note that the reduction in risk of individuals in the lowest risk strata within the 22q11DS population does not bring them to population risk levels. At present, compared to the general population, increased risk for certain outcomes remains a clinical reality for all patients with 22q11DS, regardless of polygenic score results. However, as ever-increasing GWAS size improves the strength of polygenic score associations, we suggest that polygenic score might have clinical utility in risk models in the near future29, particularly in sub-populations selected for a priori increased baseline risk, such as patients with a high-impact mutation such as 22q11DS or patients with behaviorally defined sub-threshold symptoms50. Pending more substantial polygenic score effect sizes, as well as robust replication of the findings reported here, there are several areas of potential future clinical utility. For example, in the 22q11DS population, elevated PS_SZ could be a reason to further intensify monitoring during adolescence, and PS_IQ might play a role in seeking to prevent misalignment between academic potential and demands51. Taken together, our findings highlight the potential clinical utility of polygenic scores in the context of a high-penetrance variant.
Furthermore, estimating risk raises important ethical questions that require careful consideration. For instance, in the absence of preventative interventions that can alter outcomes such as schizophrenia or ID, it will be essential to examine the balance between benefit and potential harm of exposing caregivers and patients to such information. Studies are required to examine to what extent early risk knowledge can be used to improve outcomes1. Findings like those reported here should prompt a broad societal discussion about the ethical framework in which they can be used.
Although this 22q11DS cohort is the largest ever reported, there are limitations to the work shown here. Recruitment into the IBBC cohort is not random, so there will be ascertainment biases that will affect prevalence estimates, but these are not expected to substantially affect the interpretation of the genotype–phenotype results reported here. For all analyses, given the current lack of transferability of polygenic score results across genetic backgrounds52, and that GWASs for schizophrenia and IQ are sufficiently large only within European populations to be adequately powered, our results were limited to individuals with 22q11DS who were of European descent. Future large GWASs from diverse backgrounds, as well as methodological improvements, will allow for analyses in more diverse cohorts. In addition, other uncaptured environmental variables are likely to modulate risk among 22q11DS carriers and should, therefore, be included in future studies. Finally, from a multiple testing standpoint, we intentionally restricted the main investigation of schizophrenia and associated phenotypes and polygenic scores to eight tests. Nonetheless, two of the associations, including one of the novel associations, were only nominally significant, necessitating further investigations for more definitive evidence.
In conclusion, common variants associated with schizophrenia risk and IQ variability in the general population modify expression of these phenotypes in 22q11DS. VIQ decline and sub-threshold psychosis at least partly share genetic underpinnings with schizophrenia, highlighting shared causal pathways between these phenotypes. Furthermore, in 22q11DS carriers, polygenic scores enable stratification into high- and low-risk groups substantially in excess of what would be found in a general population setting. We suggest that, in populations with high-risk rare pathogenic genetic variants such as 22q11DS, this approach is nearing a level of differentiation required for clinical utility.
Methods
Data set.
All individuals in this study were carriers of the 22q11.2 deletion, confirmed by multiplex ligation-dependent probe amplification54, as described previously32. All participants were recruited at one of 22 IBBC sites (total n = 1,789). Local research ethics boards provided appropriate study approval at all sites, and all individuals, as well as parents/guardians where appropriate, provided written informed consent regarding participation in this research.
Psychiatric assessment.
Psychiatric assessment was performed using standardized semi-structured interviews20, leading to a categorization of each participant in one of the following subgroups: SSD, sub-threshold psychosis, putative control and definite control. SSD included schizophrenia, schizo-affective disorder and related psychotic disorders, such as delusional disorder or psychotic disorder not otherwise specified, all in accordance with Diagnostic and Statistical Manual of Mental Disorders-IV criteria, based on data obtained by semi-structured in-person interviews at each site (see ref.20 for case consensus procedures). Any individual who had never met criteria for any psychotic disorder diagnosis but had endorsed clinically significant positive symptoms at any time point was included in the sub-threshold psychosis group. Supporting scores from various standardized assessment methods used across sites included symptom scores in the moderate to severe range—that is, scores of 3–5 on SIPS35, of above 2 on the Comprehensive Assessment of At-Risk Mental States (CAARMS55) or of 2 or higher (probable or definite) on any of the positive symptoms on the Schedule for Affective Disorders and Schizophrenia for School-Age Children (K-SADS56). Individuals with 22q11DS without a lifetime diagnosis of any psychotic disorder and who had never endorsed sub-threshold positive psychotic symptoms were considered controls. Given that the risk of developing schizophrenia is most elevated until age 2517,33, individuals younger than 25 years at the most recent assessment were considered ‘putative controls’, whereas individuals aged 25 years or older at the most recent assessment were classified as ‘definite controls’ (Supplementary Fig. 2 and demographics in Supplementary Table 4).
IQ values and definition of cognitive decline.
We previously found baseline FSIQ to be a significant risk factor for subsequent SSD in 22q11DS, whereas the strongest effect size for cognitive decline was observed for VIQ22. To remain consistent with our previous observations, we considered first available FSIQ as a measure of baseline intellectual ability and change in VIQ between the first and last available measurement as an index of cognitive decline. Given the moderate cognitive decline that occurs, on average, in this population42, we calculated standardized values (z scores) derived from the normative chart on which the average IQ trajectory for the 22q11DS population is mapped. Thus, a decline represents a negative deviation from the expected decline in this population.
In seeking to operationalize a cognitive decline as a binary variable, we sought a cutoff between lenient (that is, requiring less severe decline but could introduce too much noise) and conservative (more severe decline but could reduce a priori power). This task is further complicated by potential error variance inherent in the data collection across multiple sites, different versions of the Wechsler Intelligence Scale and different age groups.
We initially performed our analysis using a threshold of more than −0.5 s.d. as the cutoff for VIQ decline. Using this cutoff, the observed association with PS_SZ was not statistically significant (n = 396, P = 0.22, r2 = 0.006). Based on the literature on the reliable change index53, we subsequently revised our definition of significant change to a more stringent threshold. To minimize the chance that any observed decline was due to chance, we conservatively used the lower boundary of the reliable change index as the cutoff—that is, defining VIQ decline as a binary variable operationalized as any negative change in z scores exceeding 1 s.d. difference.
Genotyping methods and principal components analysis.
For a total of 1,789 individuals with a 22q11.2 deletion, phenotypic data were collected in a central consortium database, and available DNA samples were genotyped at Albert Einstein College of Medicine using Affymetrix Human 6.0 microarrays. We generated imputed genotypes from genotyping microarray data using standard methodological approaches as described elsewhere32. After imputation, genotype data for 992 individuals and 6,354,586 autosomal single-nucleotide polymorphisms (SNPs) were available for inclusion. We retained 4 million SNPs that had a minor allele frequency greater than 10% and were not in the major histocompatibility complex (chromosome 6, 26–34 Mbp) or in the 22q11.2 region (chromosome 22, 18,820,303–21,489,474 bp).
For principal components analysis (PCA), we then intersected this with the available GWAS SNPs described below to yield 3.2 million SNPs. We ran PCA on the 992 individuals at the 3.2 million SNPs using PLINK version 1.9, release 180612 (ref.57), which revealed between-cohort differences matching geographic ascertainment locations but no obvious outliers for quality control or non-European ancestry (Supplementary Fig. 3). Of the 992 individuals who met the criteria for subsequent analysis, 27 did not fall into one of the four pre-specified phenotype groups: 21 individuals were diagnosed with a mood disorder with psychotic features but did not meet criteria for any non-affective psychotic disorder, and six individuals had insufficient phenotypic data. We further removed three samples that overlapped with the CLOZUK cohort24, which was a component of the Psychiatric Genomics Consortium schizophrenia GWAS, yielding a total sample for analysis of n = 962 (Supplementary Fig. 4).
Polygenic score construction.
We sought out large GWASs that would enable us to generate maximally predictive polygenic scores for schizophrenia and IQ. For SSD, we used published summary statistics from a schizophrenia GWAS from the Psychiatric Genomics Consortium (max n samples = 77,096)24. For intellectual ability and IQ, we used results from Davies et al.26 from a GWAS for a general intelligence factor or ‘g-factor’58,59. However, because released GWAS statistics from this work did not contain beta coefficients, which are necessary for polygenic score construction, we used summary statistics on the largest available component—that is, based on fluid intelligence (max n samples = 108,818) from www.nealelab.is analysis extract of the UK Biobank34. In this case, fluid intelligence from the general population should capture any common genetic variants in the same fashion as FSIQ and VIQ and, thus, should serve as a suitable proxy.
We built polygenic scores using PRSice2 version 2.1.2 beta60 under default conditions—that is, using SNPs with an INFO score >0.90, r2 of 0.10 and distance of 250 kbp, where r2 was calculated on the target data (that is, this cohort). We used pre-specified P value cutoffs for SNPs for inclusion in the polygenic score based on the P value reported in the original GWAS that maximized previously reported prediction ability. For schizophrenia, we used a P value threshold of 0.05 (from Extended Data Fig. 5 in ref.24), and, for the UK Biobank Fluid Intelligence/IQ, we used 0.10 (from Supplementary Table 2 in ref.26), using 0.10 because it was the value that explained the most variance in two of three analyses). For the schizophrenia polygenic score, there were 80,496 SNPs after clumping, whereas, for the IQ polygenic score, there were 80,557 SNPs after clumping.
A priori power analyses and estimation of cohort-specific parameter values.
We conducted power analyses using simulations under a liability threshold model for our primary investigations using available sample sizes, known heritabilities, genetic correlations and assumptions regarding the nature of the relationship between schizophrenia and sub-threshold psychosis. All simulation results assume h2_g SZ = 0.46 (ref.61), h2_g SZ (PRS) = 0.08 (ref.24), h2_g IQ = 0.25 (ref.26), h2_g (PRS) IQ = 0.04 (ref.26) and r_g between SZ and IQ of −0.234 (ref.26). In addition, in the absence of pre-existing literature estimates, we assumed h2_g sub-threshold psychosis = 0.46 (based on h2_g SZ) and h2_g VIQ decline = 0.25 (based on h2_g IQ) (Supplementary Tables 5 and 6 and Supplementary Figs. 5 and 6).
To estimate cohort-specific parameters necessary for power analyses, we fit the observed data to a parametric likelihood-based model based on the liability threshold model, with parameters as follows: schizophrenia prevalence; sub-threshold psychosis prevalence; two shape parameters assuming the age distribution in the population following a beta binomial distribution; mean and s.d. for age at development of schizophrenia, assuming a normal distribution; and mean and s.d. for age at development of sub-threshold psychosis, assuming a normal distribution.
To explain the model, we considered a generative form—that is, with a population of individuals for study given the parameters above. Subsequently, we first simulated whether an individual would ever develop schizophrenia or sub-threshold psychosis, based on the prevalences of the two conditions (that is, if the prevalence was 20%, then one would simulate phenotypes under a Bernoulli distribution with probability p = 0.20). Next, independently, age was simulated, based on the shape parameters controlling the age distribution. Afterwards, age of diagnosis, conditional on ever developing the phenotype, was simulated, based on the parameters controlling the mean and s.d. age of development. From these underlying values for each simulated individual of the current age, whether they will ever develop schizophrenia or sub-threshold psychosis and the age at which they develop the phenotype, the present-day phenotype of these simulated individuals could be determined.
Using constrained optimization, we obtained the parameters that maximized the likelihood of our real data under the above-described model. To obtain a CI for each parameter, we determined the maximum values of that parameter where twice the difference in log likelihood between the maximum likelihood estimation and that point was less than the chi-squared statistic with the appropriate number of degrees of freedom. Before applying the model to real data, we first simulated under the model to verify that we could recover parameter estimates on similar-sized data sets, which confirmed the accuracy of the model (results not shown). We next generated parameter estimates on the real data (Supplementary Table 5). We used these parameter estimates and other literature-derived estimates in the power analyses that were performed.
Regression analysis.
We assessed relationships between PS_SZ and PS_IQ and binary phenotypes using logistic regression (SSD, sub-threshold psychosis and VIQ decline) and linear regression for quantitative phenotypes (baseline FSIQ), adjusting for age, sex and the first five principal components from the imputed genotypes, with the principal components calculated using PLINK. All statistical tests reported in this manuscript are two sided unless otherwise noted. r2 reported from linear regression is standard unadjusted r2, whereas, from logistic regression, it is Nagelkerke r2.
Effect of future SSD cases as a source of confounding between sub-threshold psychosis and PS_SZ.
We modeled a scenario whereby the PS_SZ signal would be driven by the presence of individuals with future, as of yet undiagnosed, SSD in the sub-threshold psychosis group. In essence, we estimated, in this scenario, what proportion of such future SSD cases would be required to explain the observed PS_SZ in the sub-threshold psychosis group.
Genetically correlated traits as a source of confounding between sub-threshold psychosis and PS_SZ.
We examined whether the observed PS_SZ results in the sub-threshold psychosis group could originate from increased rates of other psychiatric phenotypes that are genetically correlated with schizophrenia. Available IBBC data allowed us to analyze this possibility for comorbid mood disorders. Underlying assumptions for our mediation analysis were based on extrapolations of the IBBC data and include increased rates of (future) SSD (~40%) and mood disorder (49%) in the sub-threshold psychosis group, compared to ~17% rates for both phenotypes in controls.
Quantitative measure of sub-threshold psychosis as additional evidence for relationship between sub-threshold psychosis and PS_SZ.
In a subset of 347 of 962 individuals with a well-defined phenotype and imputed genotype data, we were able to obtain an integer-coded measure of sub-threshold psychosis from SIPS35. We first generated a transformation from the integer-coded, non-normally distributed quantitative SIPS score by fitting an exponential distribution using the least square estimate, yielding a transformation, in R, of ‘qnorm(pexp(q = x + 0.5, rate = 0.2238))’, where x is the original integer-coded SIPS score (Supplementary Fig. 7). This yields a more approximately normally distributed value.
We assessed power to detect an association between the quantitative SIPS-based phenotype and PS_SZ using simulations. Using the same assumptions listed before regarding heritabilities and predictive accuracies of polygenic scores, we first simulated an underlying total liability (genetic and environmental) for the quantitative sub-threshold psychosis. As before, this total liability becomes binary under a liability threshold model, giving us the binary definition of sub-threshold psychosis. In addition, using the continuous total liability, we generated an integer-coded value (representing a simulated SIPS score) as ‘round(qexp(pnorm(Y_sub), rate = 0.2238))’. We then re-transformed this to a continuous value using its inverse ‘qnorm(pexp(q = x + 0.5, rate = 0.2238))’, and from this we could calculate power for detecting an association between the quantitative sub-threshold psychosis and PS_SZ, with or without conditioning on the binary phenotype (Supplementary Fig. 8).
Calculation of PPVs.
We calculated PPV in the traditional way, given binary phenotypes schizophrenia and ID, and observed PS_SZ and PS_IQ among 22q11DS samples. We also estimated PPVs for the general population using known estimates of general population prevalence of schizophrenia and ID as well as sensitivity and specificity values derived from our analysis in this 22q11DS sample.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1 |. Relationship between polygenic scores and previously studied phenotypes.
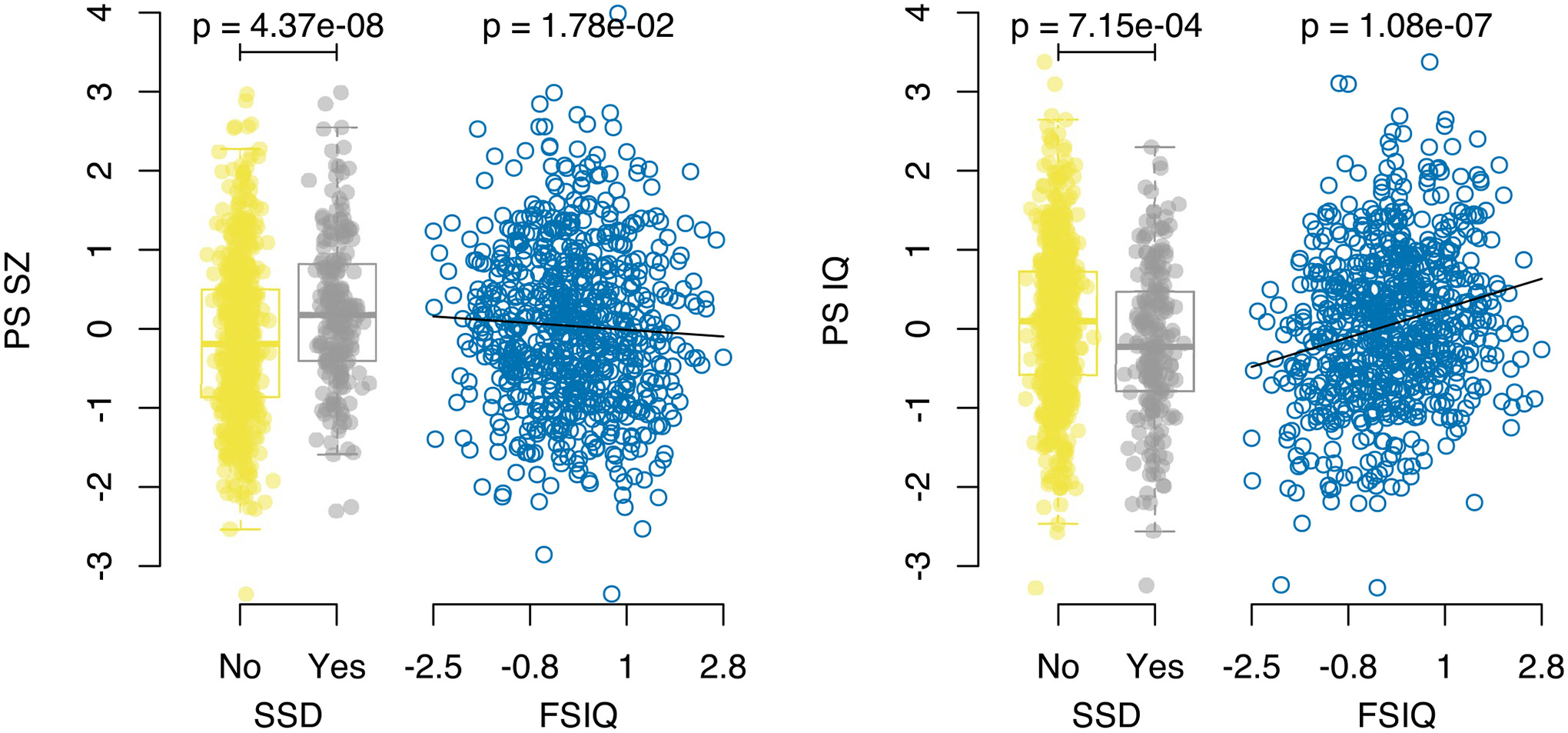
Results for the binary SSD phenotype show per-individual values as well as summaries per group, where minimum and maximum values are directly observable from the plot, the box-plot centre is the median, the boxplot edges represent the 25th and 75th percentiles, and the whiskers represent the lesser of the distance to the minimum or maximum value, or 1.5 times the inter-quartile range. Results are shown for logistic regression of SSD on controls (N = 802) and linear regression for FSIQ (N = 720), for both PS_SZ (left panel) and PS_IQ (right panel). P-values are reported from regression analyses and are two sided and are not corrected for multiple testing.
Extended Data Fig. 2 |. Inferred contribution of controls and future SSD cases given PS SZ.
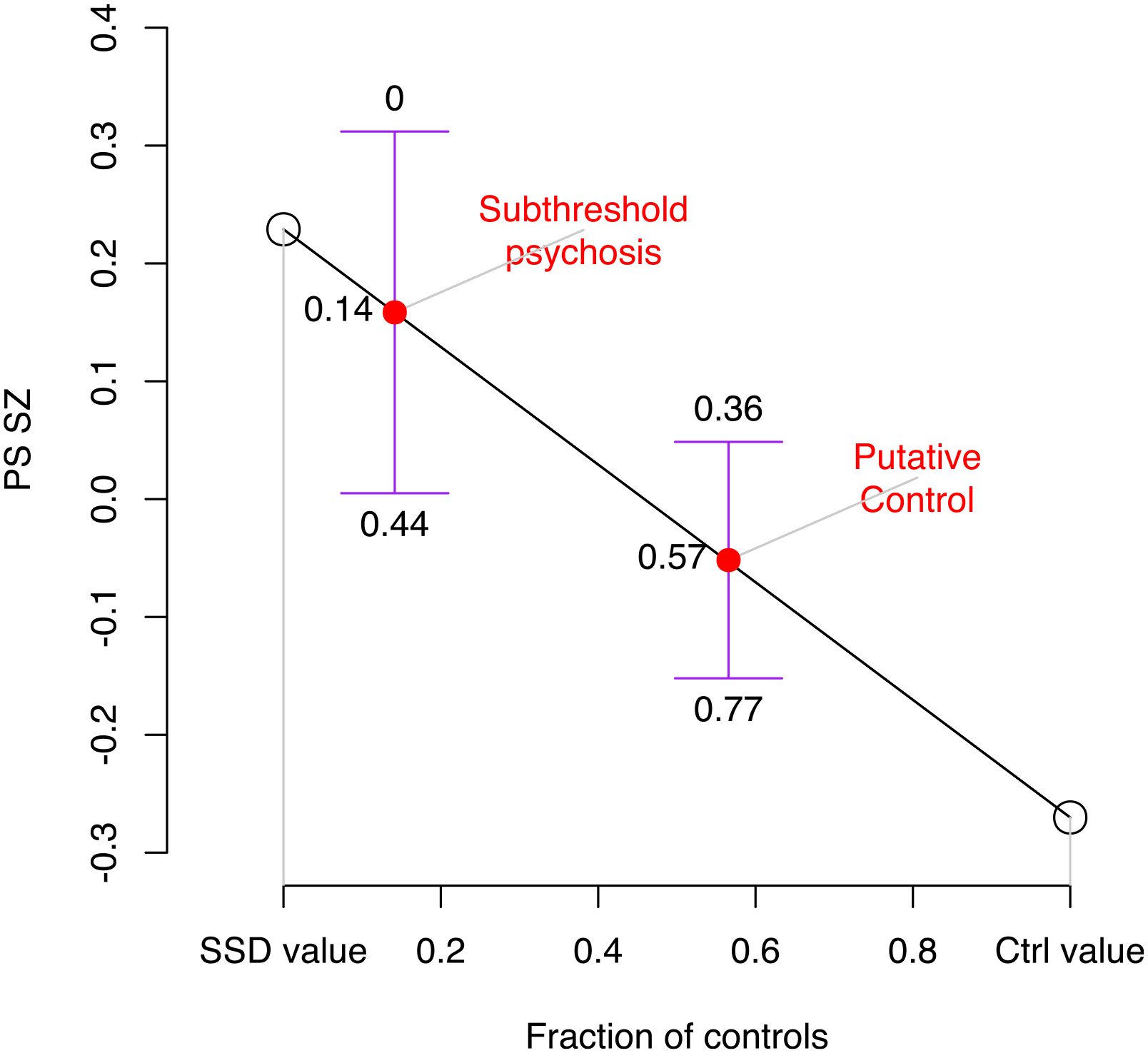
Shown on the y-axis are group means of PS_SZ, on the x-axis the fraction of controls. For SDD and controls the fractions of controls were taken as 0 and 1, respectively (open circles). For subthreshold psychosis and putative controls they were inferred through the observed PS-SZ values for each group, using linear interpolation based on fitting a straight line between SSD and control values (red circles). Confidence intervals are shown for the group mean values for subthreshold psychosis and putative controls, as the mean plus or minus 1.96 times the standard error, and above and below these confidence intervals are the inferred fraction of controls this would represent. The observed PS_SZ in the subthreshold group is consistent with a scenario in which 86% (95% CI 56 – 100%) of individuals who had subthreshold psychotic symptoms at the time of the assessment for this study would subsequently transition to SSD, a proportion inconsistent with known rates of SSD in 22q11DS.
Supplementary Material
Acknowledgements
This study was supported by the National Institute of Mental Health (NIMH) International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome (U01MH101719, U01MH0101720, U01MH0101723, U01MH101722 and EU01MH101724); the Lap-Chee Tsui Fellowship for Research Excellence and the CIHR STAGE Fellowship (to R.W.D.); the Brain and Behavior Research Foundation (to J.A.S.V.; Young Investigator Award); NIMH R01MH085953 and 1U01MH119736-01 (to C.E.B.); NIH RO1 MH064824 (to W.K.); Wellcome Trust grant 102428/Z/13/Z (to N.W. and T.M.); Canadian Institutes of Health Research grants MOP-79518, MOP-89066, MOP-97800 and MOP-111238, a McLaughlin Centre Accelerator grant, the Canada Research Chairs program and Dalglish Chair (to A.S.B); the Academic Scholars Award from the Department of Psychiatry, University of Toronto and the O’Brien Scholars Fund (to E.B.); Fondecyt 1171014 and ACT 192064 (ANID-Chile) (to G.R.); Spanish Ministry of Science and Innovation, Instituto de Salud Carlos III (SAM16PE07CP1, PI16/02012, PI19/024), CIBERSAM, Madrid Regional Government (B2017/BMD-3740 AGES-CM-2), European Union Structural Funds, European Union Seventh Framework Program (FP7-HEALTH-2013-2.2.1-2-603196 Project PSYSCAN) and European Union H2020 Program under the Innovative Medicines Initiative 2 Joint Undertaking (grant agreement 115916, Project PRISM, and grant agreement 777394, Project AIMS-2-TRIALS), Fundación Familia Alonso and Fundación Alicia Koplowitz (to C.A); Innovative Medicines Initiative 2 Joint Undertaking (#777394 for the project AIMS-2-TRIALS), the NIHR Maudsley BRC (to D.M.); and Binational Science Foundation grant 2017369 (to R.E.G. and D.G.).
Footnotes
Data availability
The data sets (raw data) generated and/or analyzed during the current study are available through the National Institute of Mental Health Data Archive repository at https://nda.nih.gov/study.html?id=938, accession number 10.15154/1519190. To construct the polygenic score for schizophrenia, we used results from the Schizophrenia Working Group of the Psychiatric Genomics Consortium24, specifically the ‘Full SNP Results’ file ckqny.scz2snpres.gz with md5 af7b9b521a196ce711d99060426fe01e, which is available after filling out an application as described at https://www.med.unc.edu/pgc/download-results/scz/. To construct the polygenic score for fluid intelligence, we used results from the Neale lab, which are available at http://www.nealelab.is/blog/2017/7/19/rapid-gwas-of-thousands-of-phenotypes-for-337000-samples-in-the-uk-biobank, specifically the file fluid_intelligence.20016. assoc.tsv.gz with md5 685d4b5e2f35c82fe29d9d9ac6e35db4, which is available through their website, http://www.nealelab.is/uk-biobank, and is additionally mirrored at https://figshare.com/articles/dataset/fluid_intelligence_20016_assoc_tsv_gz_from_2017_Neale_lab_analysis/12746570.
Code availability
Bespoke analysis code for analyses downstream of genotype generation is available. https://github.com/rwdavies/IBBC_Aim2_22Q11DS.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41591-020-1103-1.
Competing interests
M.J.O. and M.v.d.B. report grants from Takeda Pharmaceuticals outside of the submitted work. C.A. has been a consultant to or has received honoraria or grants from Acadia, Ambrosseti, Gedeon Richter, Janssen Cilag, Lundbeck, Otsuka, Roche, Sage, Servier, Shire, Schering Plough, Sumitomo Dainippon Pharma, Sunovion and Takeda. D.G.M. has provided consultation to Roche. S.R.H. has provided consultation to Novartis. O.Y.O. is a collaborator in a Biomarin Pharmaceutical study. None of the other authors has financial disclosures.
Extended data is available for this paper at https://doi.org/10.1038/s41591-020-1103-1.
Supplementary information is available for this paper at https://doi.org/10.1038/s41591-020-1103-1.
Peer review information Kate Gao and Joao Monteiro were the primary editors on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Contributor Information
International 22q11.2 Brain and Behavior Consortium:
Michael P. Epstein, Stephen T. Warren, Sinead Morrison, Samuel Chawner, Claudia Vingerhoets, Jeroen Breckpot, Elfi Vergaelen, Annick Vogels, Stephen Monks, Sarah E. Prasad, Corrado Sandini, Maude Schneider, Johanna Maeder, David Fraguas, Rens Evers, Flora Tassone, Jaume Morey-Canyelles, Opal Y. Ousley, Kevin M. Antshel, Wanda Fremont, Rosemarie Fritsch, Claudia Ornstein, Eileen M. Daly, Gregory A. Costain, Erik Boot, Tracy Heung, T. Blaine Crowley, Elaine H. Zackai, Monica E. Calkins, Ruben C. Gur, Kathryn L. McCabe, Tiffany Busa, Kelly Schoch, Maria Pontillo, Sasja N. Duijff, René S. Kahn, Michiel Houben, Leila Kushan, Maria Jalbrzikowski, Miri Carmel, Ehud Mekori-Domachevsky, Elena Michaelovsky, and Ronnie Weinberger
References
- 1.Sommer IE et al. Early interventions in risk groups for schizophrenia: what are we waiting for? NPJ Schizophr. 2, 16003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reichenberg A et al. Static and dynamic cognitive deficits in childhood preceding adult schizophrenia: a 30-year study. Am. J. Psychiatry 167, 160–169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bearden CE et al. A prospective cohort study of childhood behavioral deviance and language abnormalities as predictors of adult schizophrenia. Schizophr/Bull. 26, 395–410 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Rosso IM et al. Childhood neuromotor dysfunction in schizophrenia patients and their unaffected siblings: a prospective cohort study. Schizophr. Bull 26, 367–378 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Walker EF, Grimes KE, Davis DM & Smith AJ Childhood precursors of schizophrenia: facial expressions of emotion. Am. J. Psychiatry 150, 1654–1660 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Dickson H, Laurens KR, Cullen AE & Hodgins S Meta-analyses of cognitive and motor function in youth aged 16 years and younger who subsequently develop schizophrenia. Psychol. Med 42, 743–755 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Woodberry KA, Giuliano AJ & Seidman LJ Premorbid IQ in schizophrenia: a meta-analytic review. Am. J. Psychiatry 165, 579–587 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Mollon J, David AS, Zammit S, Lewis G & Reichenberg A Course of cognitive development from infancy to early adulthood in the psychosis spectrum. JAMA Psychiatry 75, 270–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zammit S et al. A longitudinal study of premorbid IQ score and risk of developing schizophrenia, bipolar disorder, severe depression, and other nonaffective psychoses. Arch. Gen. Psychiatry 61, 354–360 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Khandaker GM, Barnett JH, White IR & Jones PB A quantitative meta-analysis of population-based studies of premorbid intelligence and schizophrenia. Schizophr. Res 132, 220–227 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kahn RS & Keefe RS Schizophrenia is a cognitive illness: time for a change in focus. JAMA Psychiatry 70, 1107–1112 (2013). [DOI] [PubMed] [Google Scholar]
- 12.MacCabe JH et al. Decline in cognitive performance between ages 13 and 18 years and the risk for psychosis in adulthood: a Swedish longitudinal cohort study in males. JAMA Psychiatry 70, 261–270 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Meier MH et al. Neuropsychological decline in schizophrenia from the premorbid to the postonset period: evidence from a population-representative longitudinal study. Am. J. Psychiatry 171, 91–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin A et al. Neurocognitive predictors of functional outcome two to 13 years after identification as ultra-high risk for psychosis. Schizophr. Res 132, 1–7 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Kaymaz N et al. Do subthreshold psychotic experiences predict clinical outcomes in unselected non-help-seeking population-based samples? A systematic review and meta-analysis, enriched with new results. Psychol. Med 42, 2239–2253 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Poulton R et al. Children’s self-reported psychotic symptoms and adult schizophreniform disorder: a 15-year longitudinal study. Arch. Gen. Psychiatry 57, 1053–1058 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Insel TR Rethinking schizophrenia. Nature 468, 187–193 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Murphy KC, Jones LA & Owen MJ High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch. Gen. Psychiatry 56, 940–945 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Bassett AS & Chow EW 22q11 deletion syndrome: a genetic subtype of schizophrenia. Biol. Psychiatry 46, 882–891 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gur RE et al. A neurogenetic model for the study of schizophrenia spectrum disorders: the International 22q11.2 Deletion Syndrome Brain Behavior Consortium. Mol. Psychiatry 22, 1664–1672 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kates WR et al. Trajectories of psychiatric diagnoses and medication usage in youth with 22q11.2 deletion syndrome: a 9-year longitudinal study. Psychol. Med 49, 1914–1922 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Vorstman JA et al. Cognitive decline preceding the onset of psychosis in patients with 22q11.2 deletion syndrome. JAMA Psychiatry 72, 377–385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pardinas AF et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet 50, 381–389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wray NR et al. Research review: polygenic methods and their application to psychiatric traits. J. Child Psychol. Psychiatry 55, 1068–1087 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Davies G et al. Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat. Commun 9, 2098 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rietveld CA et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 340, 1467–1471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugrue LP & Desikan RS What are polygenic scores and why are they important? JAMA 321, 1820–1821 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Fullerton JM & Nurnberger JI Polygenic risk scores in psychiatry: will they be useful for clinicians? F1000Res 8, F1000 Faculty Rev-1293 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torkamani A, Wineinger NE & Topol EJ The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet 19, 581–590 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Kuchenbaecker KB et al. Evaluation of polygenic risk scores for breast and ovarian cancer risk prediction in BRCA1 and BRCA2 mutation carriers. J. Natl Cancer Inst 109, djw302 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cleynen I et al. Genetic contributors to risk of schizophrenia in the presence of a 22q11.2 deletion. Mol. Psychiatry 10.1038/s41380-020-0654-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marder SR & Cannon TD Schizophrenia. N. Engl. J. Med 381, 1753–1761 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Bycroft C et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller TJ et al. Prodromal assessment with the structured interview for prodromal syndromes and the scale of prodromal symptoms: predictive validity, interrater reliability, and training to reliability. Schizophr. Bull 29, 703–715 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Klaassen P et al. Explaining the variable penetrance of CNVs: parental intelligence modulates expression of intellectual impairment caused by the 22q11.2 deletion. Am. J. Med. Genet. B Neuropsychiatr. Genet 171, 790–796 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Sieradzka D et al. Are genetic risk factors for psychosis also associated with dimension-specific psychotic experiences in adolescence? PLoS ONE 9, e94398 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones HJ et al. Phenotypic manifestation of genetic risk for schizophrenia during adolescence in the general population. JAMA Psychiatry 73, 221–228 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones HJ et al. Investigating the genetic architecture of general and specific psychopathology in adolescence. Transl. Psychiatry 8, 145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuller R et al. Longitudinal assessment of premorbid cognitive functioning in patients with schizophrenia through examination of standardized scholastic test performance. Am. J. Psychiatry 159, 1183–1189 (2002). [DOI] [PubMed] [Google Scholar]
- 41.van Oel CJ, Sitskoorn MM, Cremer MP & Kahn RS School performance as a premorbid marker for schizophrenia: a twin study. Schizophr. Bull 28, 401–414 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Duijff SN et al. Cognitive development in children with 22q11.2 deletion syndrome. Br. J. Psychiatry 200, 462–468 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Chawner S et al. Childhood cognitive development in 22q11.2 deletion syndrome: case–control study. Br. J. Psychiatry 211, 223–230 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trevethan R Sensitivity, specificity, and predictive values: foundations, pliabilities, and pitfalls in research and practice. Front. Public Health 5, 307 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bergen SE et al. Joint contributions of rare copy number variants and common SNPs to risk for schizophrenia. Am. J. Psychiatry 176, 29–35 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tansey KE et al. Common alleles contribute to schizophrenia in CNV carriers. Mol. Psychiatry 21, 1085–1089 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lecarpentier J et al. Prediction of breast and prostate cancer risks in male BRCA1 and BRCA2 mutation carriers using polygenic risk scores. J. Clin. Oncol 35, 2240–2250 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gibson G On the utilization of polygenic risk scores for therapeutic targeting. PLoS Genet. 15, e1008060 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Damask A et al. Patients with high genome-wide polygenic risk scores for coronary artery disease may receive greater clinical benefit from alirocumab treatment in the ODYSSEY OUTCOMES trial. Circulation 141, 624–636 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Perkins DO et al. Polygenic risk score contribution to psychosis prediction in a target population of persons at clinical high risk. Am. J. Psychiatry 177, 155–163 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fiksinski AM et al. Understanding the pediatric psychiatric phenotype of 22q11.2 deletion syndrome. Am. J. Med. Genet. A 176, 2182–2191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin AR et al. Human demographic history impacts genetic risk prediction across diverse populations. Am. J. Hum. Genet 100, 635–649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chelune GJ, Naugle RI, Lüders H, Sedlak J & Awad IA Individual change after epilepsy surgery: practice effects and base-rate information. Neuropsychology 7, 41–52 (1993). [Google Scholar]
- 54.Vorstman JA et al. MLPA: a rapid, reliable, and sensitive method for detection and analysis of abnormalities of 22q. Hum. Mutat 27, 814–821 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yung AR et al. Mapping the onset of psychosis: the Comprehensive Assessment of At-Risk Mental States. Aust. N. Z. J. Psychiatry 39, 964–971 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Kaufman J et al. Schedule for affective disorders and schizophrenia for school-age children-present and lifetime version (K-SADS-PL): initial reliability and validity data. J. Am. Acad. Child Adolesc. Psychiatry 36, 980–988 (1997). [DOI] [PubMed] [Google Scholar]
- 57.Purcell S et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deary IJ, Johnson W & Houlihan LM Genetic foundations of human intelligence. Hum. Genet 126, 215–232 (2009). [DOI] [PubMed] [Google Scholar]
- 59.Spearman C ‘General Intelligence,’ objectively determined and measured. Am. J. Psychol 15, 201–293 (1904). [Google Scholar]
- 60.Euesden J, Lewis CM & O’Reilly PF PRSice: polygenic risk score software. Bioinformatics 31, 1466–1468 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gazal S et al. Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection. Nat. Genet 49, 1421–1427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


