Abstract
In this paper, we address high performance extreme-scale molecular dynamics (MD) algorithm in the GENESIS software to perform cellular-scale molecular dynamics (MD) simulations with more than 100,000 CPU cores. It includes (1) the new algorithm of real-space non-bonded interactions maximizing the performance on ARM CPU architecture, (2) reciprocal-space non-bonded interactions minimizing communicational cost, (3) accurate temperature/pressure evaluations that allows a large time step, and (4) effective parallel file inputs/outputs (I/O) for MD simulations of extremely huge systems. The largest system that contains 1.6 billion atoms was simulated using MD with a performance of 8.30 ns/day on Fugaku supercomputer. It extends the available size and time of MD simulations to answer unresolved questions of biomacromolecules in a living cell.
Keywords: Molecular dynamics simulation, Fugaku supercomputer, ARM CPU architecture, Fast Fourier transform, parallel input/output setup
Introduction
All living matter consists of cells, in which a large number of proteins, DNAs, RNAs, and other biomacromolecules are interacting with each other. Many human diseases such as cancer and Alzheimer’s disease are caused by disruptions of such biomolecular interactions in living cells. For the treatment of such diseases, effective and safe drugs are necessary. The time and costs for developing novel drugs continue to increase, limiting progress in drug discovery. Computer-aided drug discovery is expected to greatly reduce the time and cost in drug development. Rigid-body docking of a large number of drug candidates to a target protein is carried out in the earliest stage of in-silico drug discovery. Flexible docking utilizing molecular dynamics (MD) simulations has become a popular method to predict binding modes and affinities for protein-drug complexes. Although MD simulations of proteins or other biomolecules have a history of more than 40 years,1 further developments of computational methods are still necessary to accelerate the simulations and expand the target system sizes.2–5
In most biological MD simulations, the basic theory is based on classical mechanics. Forces acting on all atoms in a given system are evaluated using an empirical energy function, the so-called “force field”.6 This energy function consists of bonded and non-bonded interactions. The bonded interactions are applied to atoms within three covalent bonds, while the non-bonded interactions are applied to all pairs separated by more than three covalent bonds or in different molecules. The computational cost required for the bonded interactions is O(N), while that of the non-bonded terms, such as van der Waals and electrostatic interactions, is, at most, O(N2). Van der Waals interaction energy and forces decay rapidly with increasing pairwise distances. The computational cost becomes O(N) when a cutoff is applied beyond which the interaction is not calculated. Electrostatic interaction energy and forces do not decay as fast with distance, so a cutoff cannot be applied in the same way as for van der Waals interactions. The smooth particle mesh Ewald (SPME) method or other Ewald-based schemes decomposes the electrostatic interactions into real-space and reciprocal-space calculations to reduce the computational cost. In this scheme, the charge points in the real space are converted to charge grid data on grid points depending on spline orders. The long-range interactions are calculated from the charge grid data in reciprocal space via fast Fourier transformation (FFT).7 These methods can reduce the computational cost to O(NlogN), but they require a greater amount of communication between CPUs.
Simulating the long-time dynamics of a single protein in solution or in a membrane has been one of the major goals of cutting-edge MD simulations to date. Specialized computer architectures for MD simulations, for instance, MDGRAPE or ANTON, provide 100~1000 times faster speed than parallel supercomputers.8,9 Efficient usage of GPUs is an alternative approach for small or medium-size systems.10,11 Other innovative MD studies have targeted large biological systems, namely, protein/DNA complexes like the ribosome, viruses, nucleosomes, chromatins, and multiple proteins in crowded cellular environments. Massively parallel supercomputers are used with optimized algorithms and reduced communication costs for a better weak-scaling performance.12–16 Simulating a large biological system including more than 100 million (M) or 1 billion (B) atoms is still very challenging. However, with better MD software and efficient parallel computing algorithms, realistic cellular effects on proteins or DNAs, which are neglected in most of the current MD simulations, can be taken into account. This might be important for in-silico drug discovery to better predict undesirable side effects and/or toxicity of drug candidates, which are now recognized only in the most costly later stages of drug discovery.
In this paper, we address a big challenge in atomistic MD simulations with explicit water molecules, namely, high-performance extreme-scale MD simulations. We already developed efficient parallelization schemes for enabling cellular-scale MD simulations on K or other supercomputers. Here we suggest a new algorithm that enhances the performance on the Fugaku supercomputer. The new algorithms here extend the available time-scale and system sizes by performing 8.30 ns/day for 1.6 B atoms system. This approach overcomes many existing problems in large-scale MD simulations and opens new possibilities in next-generation in-silico drug discovery.
Methods
1. Outline of Fugaku supercomputer
The Supercomputer Fugaku has been developed for top priority research and to continue the legacy of the K computer. The system is composed of more than 150,000 nodes. Each node is equipped with an A64FX™ CPU17 that is organized into 4 core memory groups (CMG). Each CMG has 12 cores, each of which has a 2.0 GHz clock speed. The memory of each node amounts to 32 GiB HBM2 (the second generation of high bandwidth memory). Tofu interconnect D (28 Gbps × 2 lanes × 10 ports) is used for communication between nodes.
2. Characteristics of parallelization schemes in GENESIS MD software
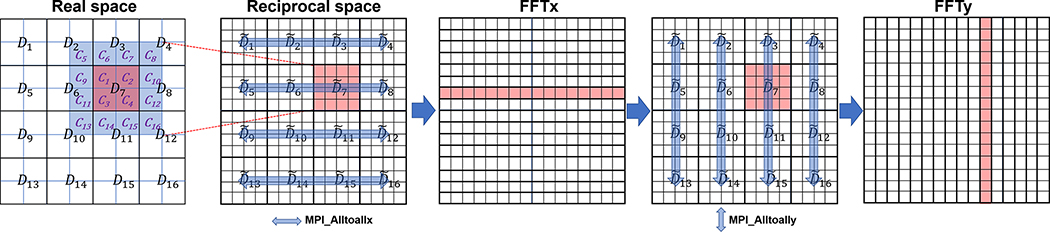
GENESIS has been developed to extend maximal system sizes in MD simulations to 1 B atoms and beyond with efficient parallelization and enhanced sampling algorithms. The source code is written in modern Fortran and the software is released to the community under the LGPLv3 license.18 Based on the parallelization scheme of GENESIS, the simulation space S is divided into subdomains, and each subdomain is again divided into cells. Interactions between particles in different subdomain are computed based on the midpoint cell scheme.19 The k-th MPI process has the data of the corresponding subdomain Dk and its margin Bk. Fig. 1 shows the two-dimensional (2D) case with 16 MPI processes. Process id 7 has the data of four cells in D7 (colored in red) and its adjacent cells: B7 (from C9 to C16 colored in blue). The charge grid data in is obtained from the charge data of atoms in D7 and B7. MPI_alltoall communications among four subdomains are done to obtain the global data in the FFT direction. For the reciprocal-space interaction, we developed two parallelization schemes of FFT; 1d_alltoall and 2d_alltoall.20 In the 1d_alltoall scheme, five one-dimensional (1D) MPI_alltoall are applied in forward and backward FFTs. In the 2d_alltoall scheme, there are two 1D MPI_alltoall and one 2D MPI_alltoall communications. Therefore, the 2d_alltoall scheme has less frequent MPI_alltoall communications, but the number of processes involved in the communications could be larger.
Fig. 1.
Domain decomposition of GENESIS in real- (first left) and reciprocal-space (second left).
3. Performance optimization of the real-space non-bonded interaction
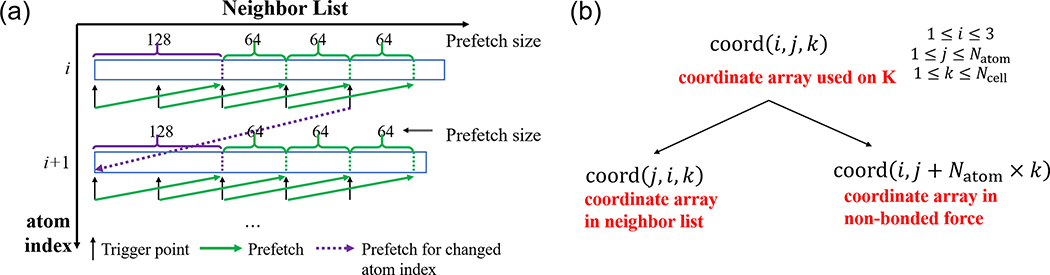
In MD, one of the main bottlenecks is the evaluation of non-bonded van der Waals and electrostatic interactions. We optimized the real-space non-bonded force calculations and the neighbor list searches, individually. First, we applied the “CONTIGUOUS” attribute to arrays in the force calculations. Second, we applied L1 cache prefetch for the neighbor list, coordinate, atom class number, charge, and force arrays (Fig. 2). The prefetch point and size are 128 and 64, respectively. If the trigger point is within 64 from the endpoint of the neighbor list in each atom index, the prefetch point is changed to the next atom index. We also changed the loop structure of the non-bonded interaction evaluation. The main reason of this change is to minimize operand/cache waiting by increasing the most inner do loop length. Third, we applied different types of array structures for coordinates in the neighbor list search and force calculations. All atom pairs are needed for the neighbor list search so that contiguous memory access is applied in the innermost loop. Thus, the Structure of Array (SoA) types are used for the coordinates. In the force evaluations, memory is not accessed in a contiguous way, and the Array of Structure (AoS) types are used for coordinates and forces. We also changed the coordinate and force array types from 3D to 2D by combining the atom and cell indexes together (Fig. 2). The force evaluation algorithm for K in GENESIS 1.0–1.4 and the new one for Fugaku are given in the Appendix, as Algorithms 2 and 3, respectively.
Fig. 2.
The prefetch scheme used in non-bonded interaction (left) and changed coordinate array on Fugaku (right).
4. Optimization of the reciprocal-space interaction
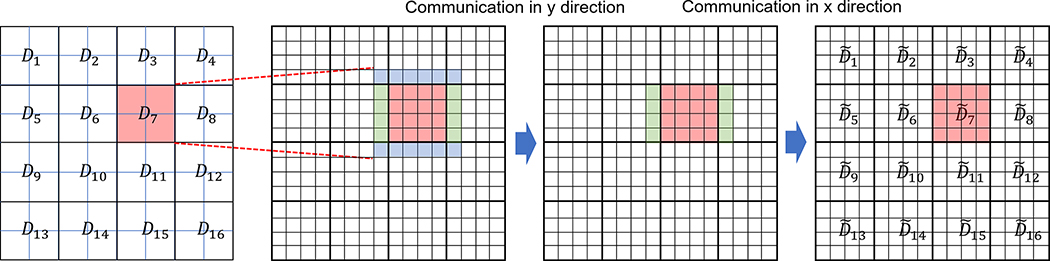
The reciprocal-space calculation consists of five steps: (i) charge grid calculation, (ii) forward FFT of charge grid data, (iii) energy calculation and convolution of charge data, (iv) backward FFT, and (v) force calculation. In GENESIS 1.0–1.4, we made use of the same domain decomposition by applying the midpoint cell method for the real space and the volumetric decomposition FFT for the reciprocal space. The k-th process first generates charge grid data from Dk and Bk. From the generated charge grid data, only the data in are saved and followed by forward FFT. This avoids communication before the FFT (Fig. 1). However, this scheme prevents the usage of a large grid spacing with a larger spline order because of the limited size of Bk. Subsequently, this scheme is called “PME_DB”, where DB means that the grid data is obtained from the union of domain Dk and boundary Bk. To improve the performance and enable a large grid spacing with a larger spline order, we devised a new scheme. First, the k-th process generates the charge grid data only from Dk. The charge grid data can be in and its margin (the amount of margin depends on the spline order in PME). The data in the margin is sent to the neighboring subdomain sequentially, and we can obtain the charge grid data in by accumulating the transferred data (Fig. 3). This is referred to as “PME_D” where D means that the grid data is only obtained from Dk. In this way, the computational cost is reduced greatly, and global communications are not required. In addition, we can assign a large PME grid spacing with a large spline order to reduce the communication cost for the FFT. Based on these developments, we further make a tool inside the program to choose the best scheme of the reciprocal-space calculation, by performing numerical tests of different algorithms for a given target system before starting production runs. Because there are two PME schemes (PME_DB and PME_D) and two parallelization schemes of the FFT itself (1d_alltoall and 2d_alltoall), there are four combinations in total:
Fig. 3.
The charge grid data before forward FFT in new development.
PME_DB_1d (PME_DB with 1d_alltoall),
PME_DB_2d (PME_DB with 2d_alltoall),
PME_D_1d (PME_D with 1d_alltoall),
PME_D_2d (PME_D with 2d_alltoall).
Timing statistics are recoded for each scheme before starting the MD, and the best is chosen for the production run.
5. Improvements of MD integration
For biological problems, we performed MD simulation under isothermal (NVT) or isothermal-isobaric (NPT) conditions. In NVT, a thermostat is applied so that the temperature of the simulation system is kept constant. In NPT, a thermostat and barostat are applied to maintain target temperature and pressure values. Therefore, accurate estimates of temperature and pressure are critical to obtain reliable results from the MD. In the conventional integration algorithm, such as the velocity Verlet or leapfrog methods, the instantaneous temperature at time t is usually evaluated in two ways:
| (1) |
| (2) |
where kB is the Boltzmann factor, f is the degree of freedom, Δt is the time step length, and K(t) is the kinetic energy at time t. These temperatures are accurate up to the first order of Δt with the following relationship:
| (3) |
| (4) |
where U are the potential energy and pi is the momentum of i-th particle.21 The Δt2 term in (3) is generally positive definite, so temperature is underestimated by Tfull and overestimated by Thalf. In the NVT condition, it changes physical properties with a large time step due to overheating or overcooling in thermostat process to assign the system in the target temperature. Using such an inaccurate estimation, we usually assign Δt = 2~2.5 fs to keep accuracy. We recently developed more accurate estimations of temperature and pressure up to O(Δt3) without any modification of the integration. Temperature is estimated as a combination of Tfull and Thalf to cancel out the Δt2 term in (3) and (4):
| (5) |
Similarly, pressure is estimated using and at time t.22 It allows the time step to be extended to 3.5 fs when the reciprocal-space interaction is performed every other step. Using such a scheme, the performance improves about 1.3 times compared to the case of Δt = 2.5 fs with the same working conditions. To avoid constraint error using Δt = 3.5 fs, hydrogen mass repartitioning scheme is used.23,24
6. Parallel file input/output (I/O) for extremely-scale MD
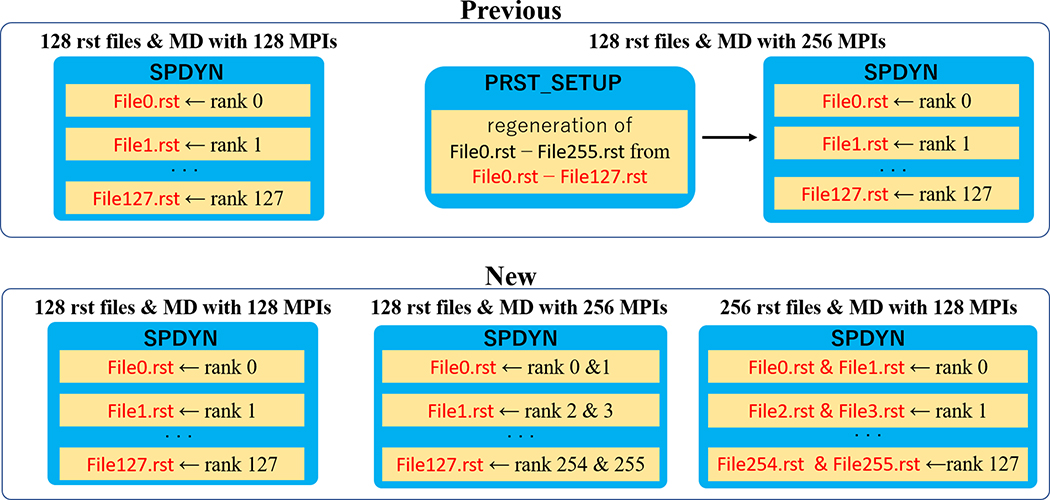
In MD simulations, information about particles between runs is exchanged via a so called ”restart file”. Conventionally, each MPI process with the domain decomposition scheme saves all the information to a restart file and selects the information only from particles in the corresponding subdomain (Algorithm 1 in Appendix). The procedure requires large memory usage and significant CPU time, especially for large-scale systems that contain more than 1 M atoms. To avoid these problems, each MPI process can read/write a set of restart and trajectory files in GENESIS 1.0–1.4. A tool named “prst_setup” generates multiple restart files for an MD simulation. The number of files is decided by the number of MPI processes in the MD simulation. Once the number of MPI processes is changed in the next run, parallel restart files need to be regenerated by prst_setup. LAMMPS supports parallel I/O restart files using the MPI-IO library.25 NAMD developed parallel I/O via dedicated input/output processes that are separate from the MPI processes and smaller in number.26
The parallel I/O scheme in GENESIS 1.0–1.4 requires regenerating multiple I/O files when the working condition is changed. However, the regeneration of the restart files is time-consuming and should be done just once for large-scale MD. Here, we changed the scheme to generate multiple restart files based on the fixed cell size condition. Let us assume that we perform an MD simulation with (Px, Py, Pz) subdomains from restart files with (Px,prev, Py,prev, Pz,prev). For each component α, one requirement is that Pα is a multiple or divisor of Pα,prev. If Pα is a multiple of Pα,prev (the case which the number of MPI tasks is less than the number of restart files), each MPI rank reads multiple restart files and obtains the parameters in the corresponding subdomain. If Pα is a divisor of Pα,prev, each MPI rank reads one restart file that contains information of the subdomain. This parallel I/O procedure does not require any additional time and is efficient. Simple examples are shown in Fig. 4.
Fig. 4.
The parallel I/O in GENESIS 1.0–1.4 (upper) and in the currently developed version (lower). In the new development, we can easily make use of parallel I/O with changed working conditions.
7. Performance measurement
We performed benchmark tests of MD simulations based on the elapsed time of the main loop. The elapsed time (ELAPSE) is measured using a Time Stamp Counter (TSC) of the CPU. We also measured floating-point operations per one integration step (FLOP/step), the SIMD instruction rate (SIMD IR), memory throughput (MEMORY TP), floating-point operation wait per step (FLOP_WT/step), and floating point load L1D cache access wait per step (L1D_WT/step) for the real-space non-bonded interaction using an event counter provided by Fujitsu. The wait time is measured by the number of cycles (cycle is the unit time interval) in which no instructions are completed. In the cycle, the oldest instruction type decides the wait type: for example, if it is floating-point operation, the cycle is considered as FLOP_WT. Everything was measured based on 1,200 integration steps except FLOP/step. To obtain FLOP/step in an exact way, MD simulations with 120 integration steps were carried out without any optimization options in the compiler. The performance of the neighbor list generation was measured based on one cycle of six integration steps because the neighbor list is generated at every sixth integration step. Therefore, we used FLOP/cycle, FLOP_WT/cycle, and L1D_WT/cycle. Because the neighbor list is written as an integer array, the integer operation wait time per cycle (INT_WT/cycle) is also recorded. The benchmark tests were carried out by simulating 1,200 MD integrations steps. The parallel efficiency (PE(Nk)) of weak scaling is defined as
| (6) |
where Tk and T16 are the elapsed time using Nk and 16 nodes.
8. Performance condition
The performance of MD using GENESIS was evaluated for cellular crowded systems with different sizes. We prepared a crowding system consisting of 1,578,958 atoms (CR1.58M). We also created larger systems by multiplying CR1.58M (CR1.58M×n, n = 1, 2, … 1024) for measuring weak scaling in MD. The time step in the MD was set to 3.5 fs. Long-range interactions were evaluated every other step. In all benchmark tests, the cutoff and the neighbor list cutoff distances were 12 and 13.5 Å, respectively. The neighbor list and atoms in each domain were updated every six steps. The PME grids was assigned as 256×256×256. In the case of spline order 8 with a grid size of 2 Å, the grid numbers was reduced to 128×128×128. In CR1.58M×n, we multiplied the grid numbers using the same ratio of the system size. For all cases, the NVT condition with the thermostat scheme to control temperature as suggested by Bussi et al. was used.27 The thermostat was applied every six steps.
Results and Discussion
1. Performance improvements of the real-space non-bonded interactions on Fugaku
We first compared the performance of the new non-bonded interaction scheme (Algorithm 3) with the old one for K (Algorithm 2). The target system here is CR1.58M, and 16 nodes of Fugaku were used. On each node, we assigned four MPI processes with twelve OpenMP threads. We measured the performance of the CMG corresponding to MPI rank 0. The MD integration steps are 240 and the total number neighbor list generation is 41 (one cycle of the neighbor list generation is identical to six steps of MD integration). TABLE I compares the performance of the real-space non-bonded force evaluation between the algorithm used on K (Algorithm 2) and the new one (Algorithm 3). Algorithm 3 improved the performance more than twice over Algorithm 2 while the number of floating-point operations was similar. Algorithm 2 was slower mainly due to the longer waiting times before starting floating-point operations and access to the L1D cache. Since the length of the inner loop with the cell pairs was shorter in Algorithm 2, it appears that software pipelining did not work well. In Algorithm 3, the waiting time before the floating-point operations was reduced significantly. The SIMD instruction rate and memory throughput were also increased significantly, meaning that the new algorithm used the Fugaku hardware much more efficiently.
TABLE I.
Performance Measurements (Force Evaluation)
| Algorithm 2 | Algorithm 3 | |
|---|---|---|
| ELAPSE/step | 26.99 ms | 10.08 ms |
| FLOPS | 18.91×109 | 46.16×109 |
| FLOP/step | 0.51×109 | 0.46×109 |
| SIMD IR | 59.43 % | 86.75 % |
| MEMORY TP | 7.70 GB/s | 12.48 GB/s |
| FLOP_WT/step | 12.37 ms | 1.44 ms |
| L1D_WT/step | 6.06 ms | 4.79 ms |
For the neighbor list generation, Algorithm 3 performed twice better than Algorithm 2 (TABLE II). In Algorithm 2, no SIMD instruction is applied and the waiting time for the floating-point was the main bottleneck that prevented high performance. By changing the data layout and algorithm, the waiting time for one neighbor list generation cycle was reduced from 16.46 ms to 3.98 ms and the SIMD instruction rate was increased up to 37 %. There are several different ways of lookup tables,28,29 and in this test, we used an inverse lookup table29 for van der Waals interactions with the CHARMM force switching function30 and the real-space electrostatic interactions. The use of the lookup table increased the speed of MD based on the elapsed time, but it decreased FLOPS. When we performed the same simulation without using the lookup table, we found that the elapsed time per step is nine times longer. This increased elapsed time is mainly due to the evaluation of the complementary error function in the calculation of electrostatic interactions. If the PME scheme is not used in the electrostatic interactions, the total FLOPS without using the lookup table is increased by a factor of 1.5. However, there is not so much difference in the total elapsed time (TABLE III). In this sense, the use of a lookup table plays an important role in accelerating MD speed despite a decrease in FLOPS.
TABLE II.
Performance Measurements (Neighbor List)
| Algorithm 2 | Algorithm 3 | |
|---|---|---|
| ELAPSE/cycle | 41.46 ms | 17.56 ms |
| FLOPS | 14.32×109 | 33.58×109 |
| FLOP/cycle | 0.60×109 | 0.59×109 |
| SIMD IR | 0.00 % | 31.74 % |
| MEMORY TP | 8.23 GB/s | 17.98 GB/s |
| FLOP_WT/cycle | 16.46 ms | 3.98 ms |
| INT_WT/cycle | 0.58 ms | 0.35 ms |
| L1D_WT/cycle | 1.56 ms | 4.32 ms |
TABLE III.
Effect OF Lookup Table in PME and No PME Calculations (Force Evaluation)
| Lookup (PME) | No lookup (PME) | No lookup (NO PME) | |
|---|---|---|---|
| ELAPSE/step | 10.08 ms | 96.82 ms | 9.36 ms |
| FLOPS | 46.16×109 | 13.21×109 | 73.20×109 |
| FLOP/step | 0.46×109 | 1.28×109 | 0.68×109 |
2. Performance improvements of the reciprocal-space non-bonded interactions on Fugaku
We compared the elapsed times with four different schemes for the reciprocal-space non-bonded interactions. The performance results strongly depend on the system size. We selected CR1.58M×64 as a benchmark system. Two PME grids and spline order conditions were applied: 1024×1024×1024 grids with spline order 4 and 512×512×512 grids with spline order 8. We compared the performance just based on the CPU times for the reciprocal-space calculations, which include the computation of grid charges, forward/backward FFTs, energy and force calculations (TABLE IV). Our new scheme, named PME_D, reduces the reciprocal-space calculation time as long as the number of nodes is not too large. In addition, by reducing the PME grids with PME_D, we could further increase the computation speed of the reciprocal-space interaction. The 2d_alltoall scheme could provide good performance if the number of nodes is less than 512. We also benchmarked the FFT performance with GENESIS on Fugaku using the same grids. The torus network in Fugaku and our FFT parallelization scheme enabled us to perform the sum of forward and backward FFTs within 5 ms.
TABLE IV.
CPU Time for One Reciprocal Space Interaction (SYSTEM: CR1.5m×64, Unit: ms)
| Number of nodes | PME_D_1d | PME_DB_1d | PME_D_2d | PME_DB_2d | ||
|---|---|---|---|---|---|---|
| 1Åa | 2Åb | 1Åa | 1Åa | 2Åb | 1Åa | |
| 256 | 57.3 | 38.4 | 61.7 | 46.6 | 38.1 | 52.6 |
| 512 | 27.6 | 20.2 | 29.5 | 29.4 | 20.8 | 32.3 |
| 1024 | 18.7 | 11.7 | 18.8 | 19.0 | 12.2 | 19.5 |
| 2048 | 13.9 | 6.9 | 13.5 | 17.5 | 7.4 | 17.1 |
| 4096 | 9.5 | 4.8 | 8.7 | 14.5 | N/A | 13.6 |
10243 PME grids with PME spline order 4
5123 PME grids with PME spline order 8
3. Effect of new parallel I/O in MD simulation
To understand the effect of the new parallel I/O, we first measured the wall time for the setup procedure that involves reading files and assigning information to subdomains. Because of the large memory requirement for generating parallel restart files, we used a computer with Intel Xeon Gold 5215 CPUs (2.5 GHz clock speed) and 1.5 TB memory. Although the same task can be done without such a large memory, the execution using a hard disk would be at least tenfold slower. For CR1.58M×64 (101.05 M atoms) and CR1.58M×512 (0.8 B atoms), it takes 1,208 and 6,042 seconds for preparing the coordinates and topology information, respectively. We used a Protein Data Bank (PDB) file for coordinates and a CHARMM Protein Structure File (PSF) file to provide topology information. If we use the previous parallel I/O procedure, the setup should be done at every time when MD runs restart in different computational conditions.
4. Performance of conventional MD using GENESIS on Fugaku
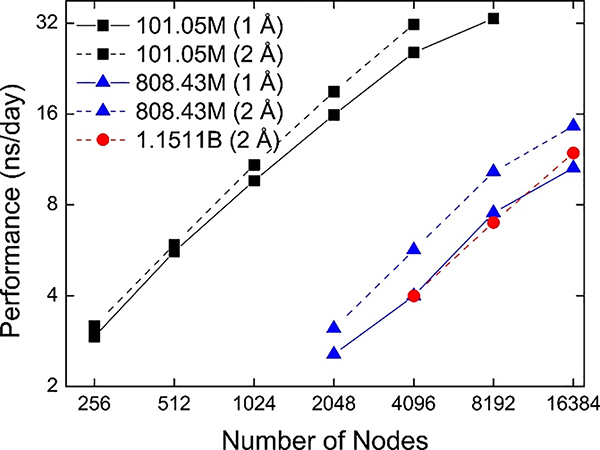
To examine the strong scaling of GENESIS on Fugaku, we performed MD simulations of the three systems: CR1.58M×64 (101.05M atoms), CR1.58M×512 (808.43 M atoms), and CR1.58M×729 (1.1511 B atoms). For all systems, two grid spacing conditions (1 Å or 2 Å) were tested. As shown in Fig. 5, we found good scalability for all systems despite communication-intensive FFTs for reciprocal-space non-bonded interactions. The best performance of GENESIS on Fugaku is significantly improved compared to that on K. For example, we obtained 33 ns/day using 8,192 nodes on Fugaku for CR1.58M×64 (101.5 M atoms) while the performance for a similar size system on 32,768 nodes of K was 8 ns/day. GENESIS on Fugaku also outperforms Trinity at Los Alamos national Laboratory and Oakforest-PACS that consist of Intel Xeon Phi (KNL) processors: We recorded 11.9 ns/day for 1.15 B atoms on Fugaku compared to slightly less than 1 ns/day for the system containing about 1 B atoms on Oakforest-PACS.3 The performance of GENESIS on Fugaku is also better than efforts using other MD software for simulating a 1B atoms system. In our understanding, the previous best performance for 1B atoms system so far is 5 ns/day using NAMD on 16,384 nodes of Oak Ridge Titan GPUs.31 Our performance results extend the time scale by about a factor of two.
Fig. 5.
Performance of MD simulations using GENESIS on Fugaku (strong scaling). The number in parentheses reflect the grid spacing.
To examine the weak scaling of GENESIS on Fugaku, we carried out MD simulations where we increased the number of processes and system sizes at the same ratio using CR1.58M on 16 nodes as a reference (TABLE V). Conventionally, due to the extensive communication in FFTs for reciprocal-space calculations, MD simulations using PME was not expected to maintain good weak scaling for a large number of nodes. However, TABLE V shows that GENESIS can achieve 80 % weak scaling up to 1,024 nodes of Fugaku with 1 Å grid spacing and up to 8,192 nodes with a 2 Å grid. Even using 16,384 nodes, the weak scaling values were kept at 53 % and 74 % for 1 Å or 2 Å grid spacings. We could simulate up to 1.62 B atoms with 8.30 ns/day, which increases both the target system size and the available time scales by a factor of 1.5 compared to the best performance of the largest system described until now (5 ns/day for a 1 B atom system31). For a system with 1.62 B atoms, we obtained 1.20 PFLOPS despite the communication-intensive FFT implications. The required MD time scale for such a large system would be greater than microseconds, and we expect our MD performance enables the MD simulations from the help of enhanced sampling schemes.
TABLE V.
Weak-Scaling Performance (Numbers in Parentheses are Parallel Efficiencies)
| Number of nodes | System size | grid spacing = 1 Å | grid spacing = 2 Å |
|---|---|---|---|
| 16 | 1.58 M | 11.62 (1.00) | 11.29 (1.00) |
| 32 | 3.16 M | 11.12 (0.96) | 11.11 (0.98) |
| 64 | 6.32 M | 11.07 (0.95) | 11.10 (0.98) |
| 128 | 12.63 M | 10.82 (0.93) | 10.99 (0.97) |
| 256 | 25.26 M | 10.01 (0.86) | 10.72 (0.95) |
| 512 | 50.53 M | 10.27 (0.88) | 10.82 (0.96) |
| 1,024 | 101.05 M | 9.26 (0.80) | 10.43 (0.92) |
| 2,048 | 202.11 M | 8.53 (0.73) | 10.26 (0.91) |
| 4,096 | 404.21 M | 7.54 (0.65) | 9.92 (0.88) |
| 8,192 | 808.43 M | 7.23 (0.62) | 9.13 (0.81) |
| 16,384 | 1.62 B | 6.19 (0.53) | 8.30 (0.74) |
Conclusions
In this study, we developed a new version of GENESIS that integrates a number of innovations and performed benchmark calculations of MD on Fugaku. The new features of GENESIS include improved real-space and reciprocal-space non-bonded interactions that maximize the performance on Fugaku and a new parallel I/O scheme that is capable of handling huge numbers of atoms in MD for large cellular-scale systems. In addition to the computational approach, we have also developed a novel MD integration scheme that allows a large time step and an efficient sampling method to examine slow biological processes. Conventional MD simulations of a 1.6 B atoms system achieved a performance of 8.30 ns/day on 16,384 nodes of Fugaku. Based on the innovations realized in the work, we are able to study structure-dynamics-function relationships of proteins, DNAs, and other biomacromolecules in a living cell. In addition to macromolecular crowding effects, there are a number of unresolved cellular effects on these biomolecules, which are difficult to explore via conventional MD or experiments. High-performance computing of large-scale MD simulations promises to contribute greatly to a better understanding of molecular and cellular biology. The developments in this study are also expected to significantly advance cellular-scale computer-aided drug discovery.
Acknowledgements
This research used computer resources of Fugaku in RIKEN Center for Computational Science. The computer resources of Oakforest-PACS were also provided through HPCI System Research project. (Project ID: hp190097, hp190181, hp200129, hp200135) The research was supported in part by MEXT as “FLAGSHIP 2020 project”, “Priority Issue on Post-K computer” (Building Innovative Drug Discovery Infrastructure Through Functional Control of Biomolecular Systems), “Program for Promoting Researches on the Supercomputer Fugaku” (Biomolecular dynamics in a living cell/MD-driven Precision Medicine), and MEXT/KAKENHI Grant Number 19H05645) (to YS). MF acknowledges support from the United States National Institutes of Health (R35 GM126948). The results obtained on the evaluation environment in the trial phase do not guarantee the performance, power and other attributes of the supercomputer Fugaku at the start of its operation.
Algorithm 1.
Setup procedure with domain decomposition (k-th process id)
| 1: | Read parameter and topology files | |
| 2: | Read atom information and save | |
| 3: | Read bond information and save | |
| 4: | Read angle information and save | |
| 5: | Read dihedral angle information and save | |
| 6: | Separate water molecules from other molecules | |
| 7: | Search for hydrogen atoms and make hydrogen groups | |
| 8: | Define {Dk} and {Bk} | ▷ subdomain and boundary |
| 9: | Define {Cα} in each Dk and {Bk} | |
| 10: | for (Cα,Cβ) ∈ cell pairs do | |
| 11: | Define M(Cα,Cβ) | ▷ midpoint cell of the cell pair |
| 12: | iα←0 for all cell index α | ▷ local atom index in each cell |
| 13: | for i ∈ atoms do | |
| 14: | if i ∈ Cα ∈ Dk then | |
| 15: | iα ← iα + 1 | |
| 16: | ▷ subdomain information | |
| 17: | else if i ∈ Cα ∈ Bk then | |
| 18: | iα ← iα + 1 | |
| 19: | ▷ boundary information | |
| 20: | else | |
| 21: | Skip | |
| 22: | iγ ← 0 | ▷ local bond index in each cell |
| 23: | for (i1,i2) ∈ bonds do | |
| 24: | Find i1 ∈ Cα ∈ Dk ∪ Bk and i 2 ∈ Cβ ∈ Dk ∪ Bk | |
| 25: | if M (Cα,Cβ) = Cγ ∈ Dk then | |
| 26: | iγ ← iγ + 1 | |
| 27: | ▷ local bond information | |
| 28: | iγ ← 0 | ▷ local angle index in each cell |
| 29: | for (i1,i2,i3) ∈ angles do | |
| 30: | Find i1 ∈ Cα ∈ Dk ∪ Bk and i3∈ Cβ ∈ Dk ∪ Bk | |
| 31: | if M (Cα,Cβ) = Cγ ∈ Dk then | |
| 32: | iγ ← iγ + 1 | |
| 33: | ▷ local angle information | |
| 34: | iγ ← 0 | ▷ local dihedral angle index in each cell |
| 35: | for (i1,i2,i3,i4) ∈ dihedral angles do | |
| 36: | Find i1 ∈ Cα ∈ Dk ∪ Bk and i4∈ Cβ ∈ Dk ∪ Bk | |
| 37: | if M (Cα,Cβ) = Cγ ∈ Dk then | |
| 38: | iγ ← iγ + 1 | |
| 39: | ▷ local dihedral angle information |
Algorithm 2.
Non-bonded interaction for K computer
| 1: | for cell pairs (Cα, Cβ) with M(Cα, Cβ) ∈ Dk do | |
| 2: | for iα ∈ Cα do | |
| 3: | ||
| 4: | ▷ charge and atom type number | |
| 5: | for jβ ∈ neighbor (iα) and jβ ∈ Cβ do | |
| 6: | ||
| 7: | ▷ LJ parameters | |
| 8: | ▷ pairwise distance | |
| 9: | ||
| 10: | ▷ electrostatic force | |
| 11: | ▷ force from LJ repulsion | |
| 12: | ▷ force from LJ dispersion | |
| 13: | ▷ sum of force values | |
| 14: | ||
| 15: | ▷ update j-th force | |
| 16: | ▷ update i-th force |
Algorithm 3.
Non-bonded interaction for Fugaku
| 1: | for Cα ∈ Dk ∪ Bk do | |
| 2: | for iα ∈ Cα do | |
| 3: | ||
| 4: | ▷ charge and atom type number | |
| 5: | for jβ ∈ neighbor (iα) do | |
| 6: | prefetch read of neighbor list | |
| 7: | prefetch read of neighbor list | |
| 8: | prefetch write of | |
| 9: | ||
| 10: | and | ▷ LJ parameters |
| 11: | ▷ pairwise distance | |
| 12: | ||
| 13: | ▷ electrostatic force | |
| 14: | ▷ force from LJ repulsion | |
| 15: | ▷ force from LJ dispersion | |
| 16: | ▷ sum of force values | |
| 17: | ||
| 18: | ▷ update j-th force | |
| 19: | ▷ update i-th force |
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Mccammon JA; Gelin BR; Karplus M Nature 1977, 267(5612), 585–590. [DOI] [PubMed] [Google Scholar]
- 2.Yu I; Mori T; Ando T; Harada R; Jung J; Sugita Y; Feig M Elife 2016, 5, e19274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jung J; Nishima W; Daniels M; Bascom G; Kobayashi C; Adedoyin A; Wall M; Lappala A; Phillips D; Fischer W; Tung CS; Schlick T; Sugita Y; Sanbonmatsu KY J Comput Chem 2019, 40(21), 1919–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao GP; Perilla JR; Yufenyuy EL; Meng X; Chen B; Ning JY; Ahn J; Gronenborn AM; Schulten K; Aiken C; Zhang PJ Nature 2013, 497(7451), 643–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singharoy A; Maffeo C; Delgado-Magnero KH; Swainsbury DJK; Sener M; Kleinekathofer U; Vant JW; Nguyen J; Hitchcock A; Isralewitz B; Teo I; Chandler DE; Stone JE; Phillips JC; Pogorelov TV; Mallus MI; Chipot C; Luthey-Schulten Z; Tieleman DP; Hunter CN; Tajkhorshid E; Aksimentiev A; Schulten K Cell 2019, 179(5), 1098–1111.e1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks BR; Brooks CL 3rd; Mackerell AD Jr.; Nilsson L; Petrella RJ; Roux B; Won Y; Archontis G; Bartels C; Boresch S; Caflisch A; Caves L; Cui Q; Dinner AR; Feig M; Fischer S; Gao J; Hodoscek M; Im W; Kuczera K; Lazaridis T; Ma J; Ovchinnikov V; Paci E; Pastor RW; Post CB; Pu JZ; Schaefer M; Tidor B; Venable RM; Woodcock HL; Wu X; Yang W; York DM; Karplus M J Comput Chem 2009, 30(10), 1545–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Essmann U; Perera L; Berkowitz ML; Darden T; Lee H; Pedersen LG J Chem Phys 1995, 103(19), 8577–8593. [Google Scholar]
- 8.Narumi T; Ohno Y; Okimoto N; Koishi T; Suenaga A; Futatsugi N; Yanai R; Himeno R; Fujikawa S; Taiji M Proceedings of the 2006 ACM/IEEE conference on Supercomputing, 2006, pp 49–es. [Google Scholar]
- 9.Shaw DE; Grossman JP; Bank JA; Batson B; Butts JA; Chao JC; Deneroff MM; Dror RO; Even A; Fenton CH; Forte A; Gagliardo J; Gill G; Greskamp B; Ho CR; Ierardi DJ; Iserovich L; Kuskin JS; Larson RH; Layman T; Lee L; Lerer AK; Li C; Killebrew D; Mackenzie KM; Mok SYM; Moraes MA; Mueller R; Nociolo LJ; Peticolas JL; Quan T; Ramot D; Salmon JK; Scarpazza DP; Schafer UB; Siddique N; Snyder CW; Spengler J; Tang PTP; Theobald M; Toma H; Towles B; Vitale B; Wang SC; Young C Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis (SC14), 2014, pp 41–53. [Google Scholar]
- 10.Salomon-Ferrer R; Gotz AW; Poole D; Le Grand S; Walker RC J Chem Theory Comput 2013, 9(9), 3878–3888. [DOI] [PubMed] [Google Scholar]
- 11.Eastman P; Swails J; Chodera JD; McGibbon RT; Zhao YT; Beauchamp KA; Wang LP; Simmonett AC; Harrigan MP; Stern CD; Wiewiora RP; Brooks BR; Pande VS Plos Comput Biol 2017, 13(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung J; Mori T; Kobayashi C; Matsunaga Y; Yoda T; Feig M; Sugita Y Wiley Interdisciplinary Reviews: Computational Molecular Science 2015, 5(4), 310–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi C; Jung J; Matsunaga Y; Mori T; Ando T; Tamura K; Kamiya M; Sugita Y J Comput Chem 2017, 38(25), 2193–2206. [DOI] [PubMed] [Google Scholar]
- 14.Jung J; Naurse A; Kobayashi C; Sugita Y J Chem Theory Comput 2016, 12(10), 4947–4958. [DOI] [PubMed] [Google Scholar]
- 15.Jung J; Suguita Y Journal of Computational Chemsitry 2016, 38, 1410–1418. [DOI] [PubMed] [Google Scholar]
- 16.Mei C; Sun Y; Zheng G; Bohm EJ; Kale LV; Phillips JC; Harrison C In Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis (SC11) 2011, p 1–11. [Google Scholar]
- 17. https://github.com/fujitsu/A64FX.git.
- 18. https://www.r-ccs.riken.jp/labs/cbrt.
- 19.Jung J; Mori T; Sugita Y J Comput Chem 2014, 35(14), 1064–1072. [DOI] [PubMed] [Google Scholar]
- 20.Jung J; Kobayashi C; Imamura T; Sugita Y Computer Physics Communications 2016, 200, 57–65. [Google Scholar]
- 21.Jung J; Kobayashi C; Sugita Y J Chem Theory Comput 2019, 15(1), 84–94. [DOI] [PubMed] [Google Scholar]
- 22.Jung J; Kobayashi C; Sugita Y J Chem Phys 2018, 148(16), 164109. [DOI] [PubMed] [Google Scholar]
- 23.Feenstra KA; Hess B; Berendsen HJC J Comput Chem 1999, 20(8), 786–798. [DOI] [PubMed] [Google Scholar]
- 24.Hopkins CW; Le Grand S; Walker RC; Roitberg AE J Chem Theory Comput 2015, 11(4), 1864–1874. [DOI] [PubMed] [Google Scholar]
- 25. https://lammps.sandia.gov.
- 26.Phillips JC; Schulten K; Bhatele A; Mei C; Sun Y; Bohm EJ; Kale LV In Parallel Science and Engineering Applications: The CHARMM++ Approach; Kale LV; Bhatele A, Eds.; CRC Press, 2019. [Google Scholar]
- 27.Bussi G; Donadio D; Parrinello M J Chem Phys 2007, 126(1), 014101. [DOI] [PubMed] [Google Scholar]
- 28.Nilsson L J Comput Chem 2009, 30(9), 1490–1498. [DOI] [PubMed] [Google Scholar]
- 29.Jung J; Mori T; Sugita Y J Comput Chem 2013, 34(28), 2412–2420. [DOI] [PubMed] [Google Scholar]
- 30.Steinbach PJ; Brooks BR J Comput Chem 1994, 15(7), 667–683. [Google Scholar]
- 31. http://www.ks.uiuc.edu/Research/namd/benchmarks/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.