Abstract
Lipid droplets (LD) are dynamically-regulated organelles that originate from the endoplasmic reticulum (ER), and function in the storage, trafficking and metabolism of neutral lipids. In mammary epithelial cells (MEC) of lactating animals, intact LD are secreted intact into milk to form milk lipids by a novel apocrine mechanism. The secretion of intact LD and the relatively large amounts of lipid secreted by lactating MEC increase demands on the cellular processes responsible for lipid synthesis and LD formation. As yet these processes are poorly defined due to limited understanding of LD-ER interactions. To overcome these limitations, we used rapid-freezing and freeze-substitution methods in conjunction with 3D electron tomography and high resolution immunolocalization to define interactions between LD with ER in MEC of pregnant and lactating rats. Using these approaches, we identified distinct ER domains that contribute to lipid droplet formation and stabilization and which possess unique features previously unrecognized or not fully appreciated. Our results show nascent lipid droplets within the ER lumen and the association of both forming and mature droplets with structurally unique regions of ER cisternae, characterized by the presence of perilipin-2, a protein implicated in lipid droplet formation, and enzymes involved in lipid synthesis. These data demonstrate that milk lipids originate from LD-ER domains with novel structural features and suggest a mechanism for initial droplet formation in the ER lumen and subsequent maturation of the droplets in association with ER cisternae.
Keywords: Mammary gland, Lipid droplet, Endoplasmic reticulum domain, Electron microscopy, Tomography, Differentiation
Introduction
Neutral lipids, triglycerides and cholesteryl esters, are major milk constituents and singularly important sources of calories and bioactive molecules required for neonatal growth and development in most mammalian species [1]. In eukaryotic cells, neutral lipids are synthesized by enzymes located on the endoplasmic reticulum (ER) membrane and are packaged into cytoplasmic lipid droplets (LD), which are dynamically-regulated, organelles that function in neutral lipid storage, transport and metabolism [2]. Unlike conventional organelles such as the endoplasmic reticulum, Golgi, nucleus, and mitochondria, in which functional compartments are bounded by bilayer membranes, LD are uniquely composed of a neutral lipid core surrounded by a monolayer of phospholipids and abundant amounts of associated proteins, which serve as determinants of LD formation and function [3–5]. LD formation and function have become active areas of investigation due to increasing recognition of their contributions to the pathophysiology of many metabolic diseases [6].
In most cells, LD serve as temporary storage sites for neutral lipids for use in production of metabolic energy and membrane biosynthesis. However, alveolar epithelial cells of mammary, meibomian and sebaceous glands are unique in their abilities to secrete intact LD from the cytoplasm directly into the acinar lumen in conjunction with the production of milk, meibum, and sebum respectively [7–9]. The large amounts of lipid secreted into milk during lactation by many species of mammals [10] and developmentally-linked activation of lipogenesis and biogenesis of LD and other organelles in mammary epithelial cells (MEC) during their functional differentiation [11–14], make this system an important physiologically relevant model for investigating LD formation.
The concept that LD are derived from the ER originated with time course studies, which combined radiotracer and ultrastructural analyses of lactating mouse mammary epithelial cells, that showed that neutral lipids are initially synthesized on rough ER (rER) and rapidly transfer to LD, which appear to exist in continuity with ER cisternae [15]. Subsequent ultrastructural studies identified lipid filled distensions of ER cisternae and ER membrane bilayers [16] prompted the idea that LD originate from sites of neutral lipid accumulation on, or within, ER membranes [17]. Intimate associations of LD and ER were further supported by proteomic studies that identified the presence of numerous ER luminal proteins on LD isolated free of membranous material from lactating mice [4]. Studies in yeast [18], certain cell culture models [2], and in vitro biophysical experiments [19] provide evidence that LD originate from the accumulation of neutral lipids between ER membrane leaflets. However, the process is not well understood [20] and other mechanisms of LD biogenesis have been proposed [21–23]. Importantly, details about LD-ER interactions in intact cells of highly lipogenic organs, such as the mammary gland, are limited and it is unknown if such interactions are influenced by functional status. However, the significant expansion of ER and Golgi membranes and activation of lipogenic processes that occur in MEC in response to induction of secretory functions [11, 12] suggest the hypothesis that LD-ER interactions in MEC may also be influenced by functional status.
The available information about the ultrastructural properties of LD in mammary epithelial cells was obtained using conventional two-dimensional (2D) electron microscopy (EM) [17] and preparative techniques that were sub-optimal for preservation of membranes and major lipid-containing structures in their native states, which almost certainly distorted their ultrastructural properties [24, 25]. To overcome these limitations and accurately define LD ultrastructural properties in greater detail, we analyzed MEC from pregnant and lactating rats by 3D-electron tomography and immuno-EM in mammary glands prepared by cryopreservation techniques optimized for minimal extraction of lipids [26] and accurate preservation of cell structures in nearly native states [27].
Materials and Methods
Animals and Ethical Approval
Pregnant rats were purchased from Harlan, Inc. (Indianapolis, IN). Mammary acini were isolated from 2 to 4 10-day lactating or 5- and 10-day pregnant rats as described previously [28]. Animal experiments were approved by the University of Colorado School of Medicine’s Institutional Animal Care and Use Committee. Rats were housed within the University of Colorado Denver Anschutz Medical Campus, AAALAC-accredited, Center for Comparative Medicine. Experimental animals were singly housed at 22 °C in microisolator cages equipped with automated air and water on a 10:14 h dark:light cycle with ad libitum access to food.
Materials
Guinea pig antibodies against Plin2 were purchased from Research Diagnostics, Inc. (Flanders, NJ), and rabbit antibodies against PDI were purchased from Stressgen Biotechnologies Corp. (San Diego, CA). HEK293 that constitutively express Plin2 were described previously [29]. Antibodies against Dgat1 and Dgat2 antibodies were a generous gift from Dr. Robert Farese Jr., Harvard School of Public Health. Acat1 antibodies were a gift of Dr. T.Y. Chang, Dartmouth Medical School.
Electron Microscopy and Tomography
Samples of mammary tissue were prepared for EM as described previously [30]. Briefly, small blocks of tissue (<1 mm3) were placed in medium containing 10% 70 kD Ficoll as an extracellular cryoprotectant (Sigma-Aldridge) and rapidly frozen in a HPM 010 high-pressure freezer (BAL-TEC, AG, Liechtenstein). The samples were then freeze-substituted into acetone at −90 °C, first with 0.1% glutaraldehyde, 0.01% tannic acid for 3 days, then with 2% OsO4, 0.1% uranyl acetate in acetone at −20 °C for 12 h. The fixed, anhydrous tissue was then embedded in Epon-araldite resin. At least 5 tissue blocks were examined from each animal. Each block yielded ~50 projection micrographs and ~5–10 tomographic 3D reconstructions. Thin (40 nm) sections of mammary tissue were viewed in a Philips CM10 electron microscope to evaluate preservation and to select appropriate cells for high-resolution study. Evidence of LD-ER interactions was seen in nearly all samples and could be identified in a majority of micrographs where both cytoplasmic ER and LD were present. Such regions were sought and located specifically for 3D tomographic study. For tomography, ribbons of 8–10 semi-thick (~200 nm) sections were cut with an UltraCut-UCT microtome (Leica, Inc.), transferred to Formvar-coated copper-rhodium 1 mm slot grids (Electron Microscopy Sciences, Port Washington PA.) and stained with uranyl acetate and lead citrate. Colloidal gold particles (15 nm diameter) were placed on both sides of the grid to serve as fiducial markers for subsequent image alignment. Samples were imaged with a Tecnai F20 (FEI) IVEM operating at 200 kV using a pixel size corresponding to ~1 nm at the specimen plane. Tilt-series were collected over 120° at 1° intervals. The grid was then rotated 90° and a similar series was collected about the orthogonal axis. Tomographic reconstructions were calculated, analyzed and modeled using the IMOD software package [31].
Immunoelectron Microscopy
Acini were prepared for immuno-EM as previously described [32]. Briefly, blocks of mammary tissue were fixed with 4% paraformaldehyde in PBS + 5% sucrose in 100 mM HEPES buffer, pH 7.2, at 4 °C for 12 h, infiltrated with 2.1 M sucrose in PBS, then frozen in liquid nitrogen. Thin (90–100 nm) cryosections were cut with a cryo-diamond knife (Diatome U.S.) at −110 °C on an UltraCut-UCT microtome equipped with a FCS cryostage (Leica Inc.). Sections were picked up with a wire loop containing a drop of 2.3 M sucrose, 1% methylcellulose in phosphate-buffered saline (PBS) and transferred to Formvar-coated, carbon-stabilized, glow-discharged 100-mesh copper-rhodium EM grids. Labeled cryosections were observed with the Tecnai F20 IVEM operating at 80 kV, and images were recorded digitally.
Immunofluorescence Microscopy
Mammary glands from rats on P5 were excised and fixed overnight in 4% paraformaldehyde, 5% sucrose, in PBS at 4 °C, followed by successive incubations at 4 °C in 30% sucrose in PBS (overnight), 30% sucrose in Tissue-Tek embedding medium (Sakura Finetek USA, Inc.) (30 min) and 100% Tissue-Tek (overnight). Cryosections (10–16 μm) were prepared and deposited on Superfrost Plus slides (Fisher Scientific, Pittsburgh, PA), post-fixed with 4% paraformaldehyde in PBS for 30 min, incubated in 0.2% Triton X-100 in PBS for 20 min, and blocked in 0.2% gelatin in PBS for 20 min. For labeling, both primary antibodies and secondary antibodies labeled with Alexa Fluor 488 (green) or Alexa Fluor 594 (red) (Molecular Probes, Eugene, OR) were incubated for 30 min at 37 °C, and coverslips were mounted onto slides with Moviol in the presence of 1 microgram/ml of 4,6-diamino-2-phenylindole (DAPI; Molecular Probes). Sections were visualized with an Axiovert 200 M deconvolution microscope (Carl Zeiss, Thornwood, NY). Images were collected and processed using SlideBook software (Intelligent Imaging Innovations, Inc., Denver CO). For double labeling, images in two channels were collected separately and then overlaid using Adobe Photoshop software (Adobe Systems Inc.). To label lipid, slides were incubated with 100 nM Nile red (Sigma) in PBS for 10 min.
Results
LD-ER Interactions in the Lactating Mammary Gland
As prior ultrastructural studies demonstrated the association of LD and ER in MEC of lactating animals, we focused initially on MEC from day 10 (L10) lactating rats in order to characterize interactions of forming droplets with other organelles under conditions of elevated neutral lipid synthesis [33]. In cryo-preserved tissue, a well-developed acinus from this stage is composed of a group of polarized epithelial cells surrounding a large lumen filled with milk fat globules (MFG) of varying sizes, apoptotic cells and other debris (Fig. 1). The epithelial cells are characterized by highly enlarged and distended Golgi membranes and the presence of numerous large lipid droplets. Some droplets are in the process of being secreted into the lumen and appear to be bound by membrane that is continuous with the apical plasma membrane. Although most cellular features are visible in this thin section, low-magnification overview, greater volume and higher resolution are required to evaluate the associations between LD and cytoplasmic membranes.
Fig. 1.

Secretory acinus at lactation day 10. Low-magnification view of a typical acinus from the mammary gland of a lactating rat. Seven epithelial cells are surrounding a lumen than contains an apoptotic cell, numerous milk fat globules (MFG) and casein micelles. The acinar cells have distinct apical micro-villi and contain lipid droplets of varying sizes, both in the cytoplasm (LD) and in the process of being secreted (LDS) at the apical plasma membrane. Voluminous Golgi structures (G) are evident in most of the epithelial cells in this thin section. The trans-Golgi cisternae are distended and contain large amounts of micellar casein (see [34]). Bar = 5 μm
Electron microscope surveys of thin sections of mammary tissue 2–4 animals identified LD-ER interactions in nearly all samples and in a majority of micrographs where both cytoplasmic ER and LD were present. A LD may be partially or fully surrounded by a single ER cisterna and/or multiple cisternae can be closely apposed to the droplet at points along its surface. Such ER cisternae form flattened, ribosome-free domains that associate with the droplet surface and with similar domains of adjacent cisternae. These ribosome-free domains are continuous with ribosome-bound cytoplasmic ER distal to the lipid droplet (Fig. 2A). Remarkably, smooth ER with the tubular morphology associated with lipid synthesis, and which is abundant in hepatocytes and in other lipogenic cells, is not observed to a significant extent in the cytosol of mammary epithelial cells [12] even though lipids are being extensively synthesized for secretion into milk. In other regions of the cell, ER with ribosomes on both surfaces appear in very close proximity to LD. This suggests that the droplet-associating ER domains are functional and not simply a result of a growing lipid droplet displacing adjacent cytoplasm (data not shown). This domaining and “layering” of multiple ER cisternae is not specific to mammary epithelial cells as a similar organization is present in hepatocytes (Fig. 2B). In this image, two ER cisternae encompass the lipid droplet with ribosomes present only on the cytoplasmic face of the outermost of the two cisternae.
Fig. 2.
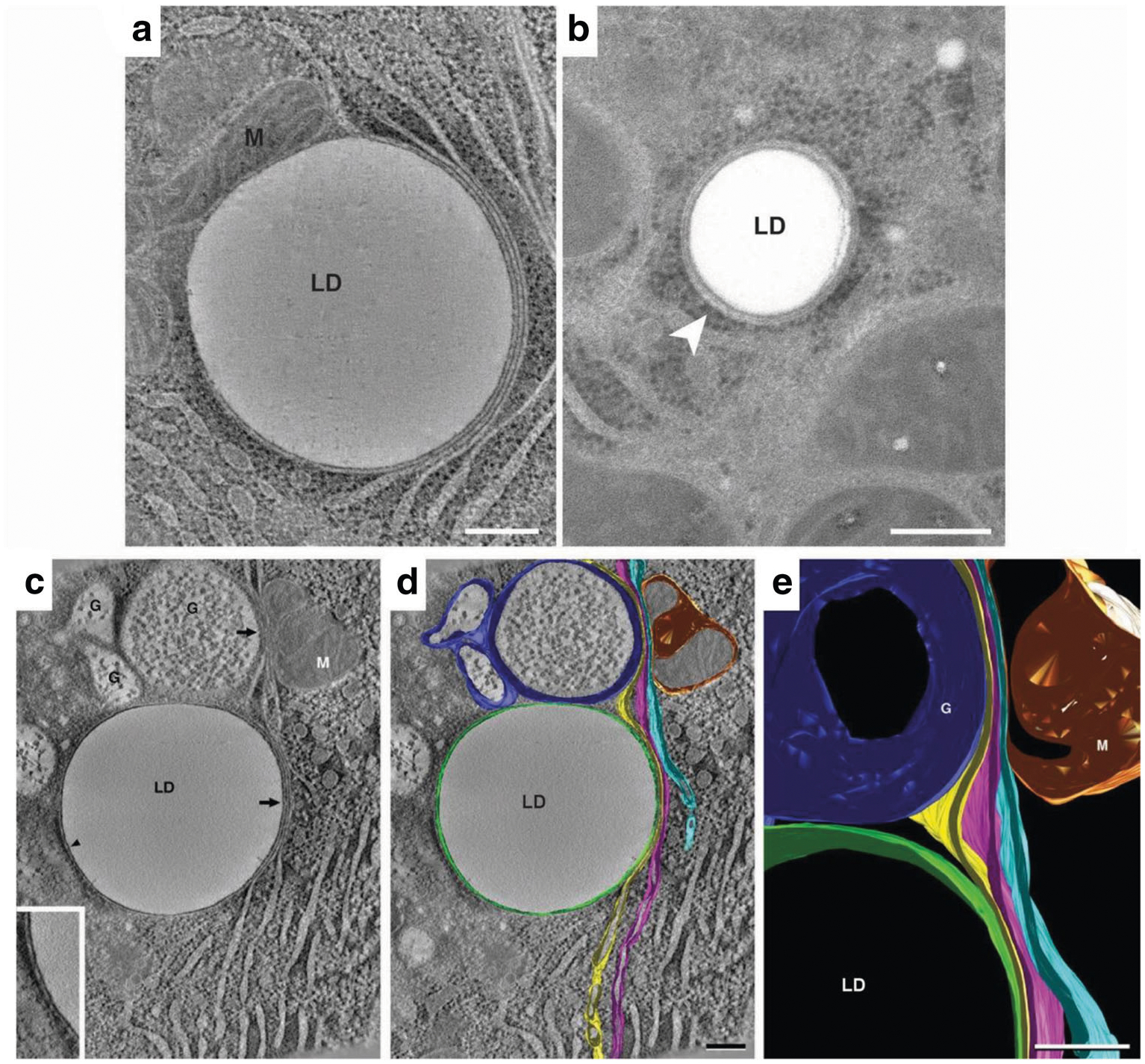
Endoplasmic reticulum association with lipid droplets in mammary epithelial cells on lactation day 10. (A) A representative composite of 15, 0.9 nm slices from a tomographic reconstruction of a MEC from an L10 mammary gland prepared by high-pressure freezing and freeze substitution. Domains of three distinct ER cisternae exist in tight association with the lipid droplet (LD). One cisterna wraps around ~60% of the droplet’s perimeter. The other two are wrapped tightly adjacent to the first cisternae on the right side of the droplet. All three cisternae extend beyond the droplet into the cytosol. Ribosomes are absent from ER domains that contact the LD surface or other cisternae but are present on domains facing the cytosol. A closely associated mitochondrion (M) is present on the left side of the LD. (B) Thin (40 nm) section of a LD in a rat hepatocyte. The droplet is smaller than those in L10 MEC, but it associates with two ER cisternae in a similar fashion and only the cytoplasmic face of the outer ER cisterna bears ribosomes. Bars = 0.2 μm. (C-E) Tomographic reconstruction of 10, 0.9 nm slices from another L10 MEC. (C) A tomographic slice shows a lipid droplet (LD) in contact (arrowhead) with single ER cisterna (inset), which is juxtaposed to three additional slender ER cisternae to form a four-layer domain that is in contour with a portion of the LD surface (arrow). The outer cisternae extend upwards from the lipid droplet to approach both the trans-region of a nearby Golgi complex (G) and a mitochondrion (M). (D) The same image data shown in panel C overlaid with filled model contours of ER cisterna, Golgi and Mitochondria. The cisterna in contact with the LD is shown in green, the three juxtaposed cisternae are shown in yellow, magenta, and light blue respectively. The yellow and light blue ER cisternae form similarly close appositions with the trans-Golgi (dark blue) and mitochondrion (gold) respectively, with the magenta cisterna between them. Domains of ER cisternae in contact with each other or with the other organelles are free of ribosomes but bear ribosomes on cisternal regions that face the cytoplasm. (C) Detail of the projected model showing specialized ER domains in close apposition to the lipid droplet, Golgi (G), and mitochondrion (M). Bars = 0.2 μm
Tomographic reconstruction and 3D analysis permit closer evaluation of ER-LD associations. A slice from a tomographic volume of an L10 MEC (Fig. 2C) shows a large lipid droplet (LD), Golgi (G; identified by the presence of casein micelles [35], a mitochondrion (M) and abundant ER. All of these structures were modeled through the tomogram volume, rendered in different colors, and superimposed over the tomographic slice to highlight the continuity of ER membranes in Fig. 2D. A single ER cisterna (green) encompasses the droplet while three other distinct ER cisternae are layered on top of one another in close apposition to the right side of the droplet. These same three cisternae are continuous with domains that appose the largest Golgi element and the mitochondrion. A detail of the projected 3D model (Fig. 2E) illustrates their close apposition to the lipid droplet and organelles. Tracing of the ER contours through the tomographic volume indicates that the region of LD association is large. This and other datasets confirm that ER cisternae associate with a large portion, but not all, of a lipid droplet’s surface. These data identify specific multidimensional interactions between LD and ER membranes in mammary epithelial cells of lactating animals that have not been described previously, and which suggest that specialized ER-LD interaction domains produce distal alterations in ER structure and functional properties.
Perilipin-2 Localizes to the ER-LD Contact Sites
LD-ER interactions are mediated by specific protein interactions [2]. Perilipin-2 (Plin2; also known as adipophilin/ADPH and adipose differentiation related protein/ADRP) is a prominent constitutively-associated LD protein in MEC that has been linked to LD accumulation during secretory differentiation [36, 37] and is implicated as mediator of LD growth [22] and LD-ER interactions [5]. In the context of this evidence, and data showing extensive association between the LD surface and the ER, we used immuno-EM localization of Plin2 in cryopreserved tissue to further delineate LD-ER interacting domains in L10 samples. Antibodies against the ER luminal enzyme, protein disulfide isomerase (PDI), were used to specifically immune-label ER in these samples. In double-labeling experiments, Plin2 (small gold particles) primarily localized to the periphery of LD while there was abundant PDI labelling (large gold particles) over the entirety of the cell’s ER network (Fig. 3A). At higher magnification, labeled lipid droplets show Plin2 localized to the surface of the droplet, often specifically to the ends of membranes that appear to terminate at (Fig. 3B, arrowhead), or within (Fig. 3B, arrow) the droplet. The presence of both Plin2 and PDI at these domains (Fig. 3C, arrows) indicate that ER membranes may penetrate the LD interior. Using the superior spatial resolution obtained with tomography, Plin2 labeling can be seen both at the limiting edge (3D) and on thin strands that traverse the middle of the droplet (3D’), which provides direct evidence that Plin2 associates with ER domains that penetrate the interior of the lipid droplet, a finding that is consistent with freeze-fracture immuno-EM data [22, 39].
Fig. 3.
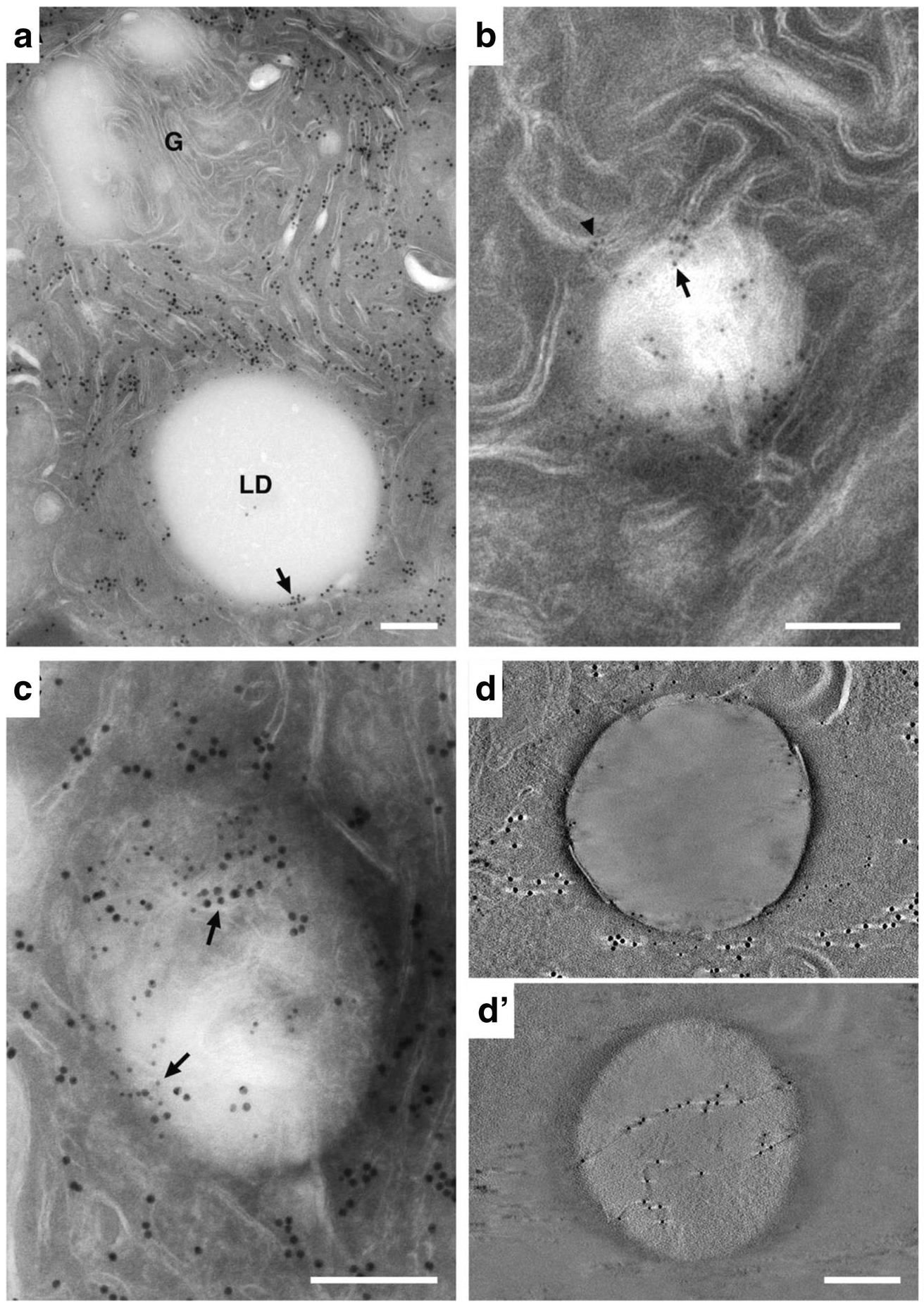
Immuno-EM localization of ER and lipid droplet markers in lactating mammary epithelial cells. (A) Double-labeled section showing the localization of PDI (15 nm gold) and Plin2 (10 nm gold). Abundant PDI is detected on ER throughout the section but is not present on the Golgi complex (G). Plin2 selectively localizes to the LD surface. PDI and Plin2 co-localize at distinct places on the surface of the LD (arrow). The inset is an enlargement of the boxed area showing 10 nm gold labelled Plin2 localizing to the LD surface (arrow) and 15 nm gold labelled PDI localizing to the ER (arrowhead). (B) Detail of a lipid droplet in a thin (100 nm) section labeled with antibodies against Plin2. Gold is localized to membranes at the periphery of the droplet (arrowhead), some of which appear to penetrate into the body of the droplet (arrow). (C) Double-labeled 100 nm section showing PDI (15 nm gold, small arrows) and Plin2 (10 nm gold, large arrows) colocalizing at the periphery and within the body of the LD (arrows). (D & D’) Tomographic reconstructions of thin sections labeled with antibodies to PDI and Plin2. (D) A 0.91 nm tomographic slice from the surface of the reconstruction showing Plin2 on the LD surface and PDI near the droplet periphery (the light streaks extending from either side of the large gold particles are artifacts of single axis tomographic reconstruction [38]). (D’) A 0.91 nm tomographic slice from within the LD interior demonstrating that Plin2 labels strands that cross through the interior of the droplet. Bars = 0.2 μm
As a test of whether the presence of ER domains within the interior of LD is a specific feature of ER-LD interactions in MEC, we defined Plin2 association with ER and LD in HEK293 cells that were engineered to constitutively express Plin2, and which have been used previously as a cell culture model to study Plin2 and LD functions [29]. LD formation was induced by culturing Plin2 expressing HEK293 cells in media containing oleic acid [29]. In this model, LD are visible within 2 h of feeding oleic acid-containing media (unpublished observation). A 100 nm thin section from cryo-fixed cells double-labeled with Plin2 and PDI show that Plin2 and PDI are present on the LD periphery and co-localize at a site within the LD interior (Fig. 4A). In agreement with results obtained from lactating MEC, a tomographic section from the LD interior demonstrates discrete co-localization of Plin2 and PDI on a structure that invaginates into the interior of the lipid droplet (Fig. 4B), which is consistent with the presence of Plin2 on ER domains that penetrate within the LD.
Fig. 4.
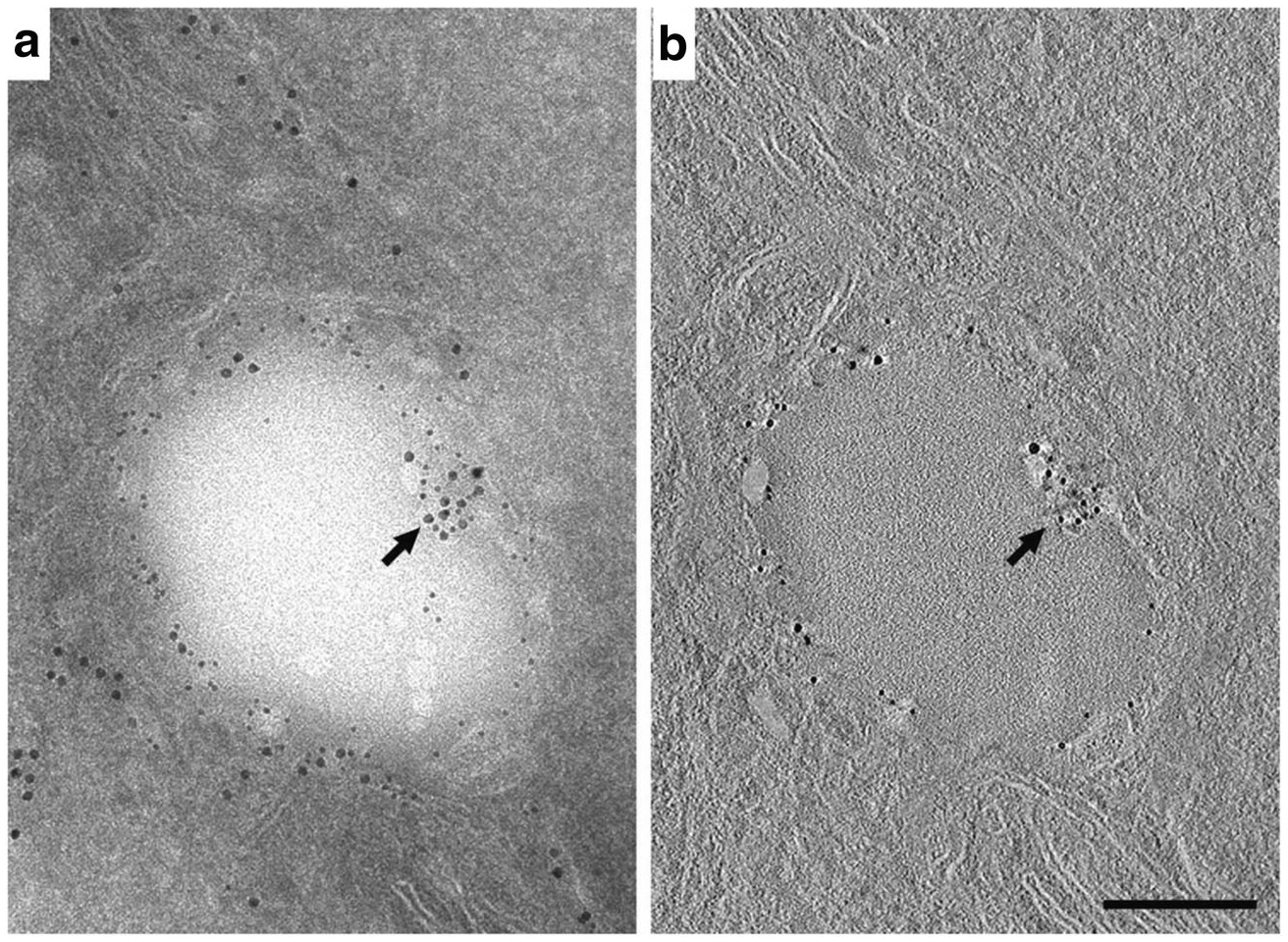
Immuno-EM localization and tomographic reconstruction of Plin2 on lipid droplets in HEK293 cells. (A) A 100 nm section of an HEK293 cell double-labeled with antibodies to PDI (15 nm gold) and Plin2 (10 nm gold). PDI localizes to regions of ER that are proximal and distal to the LD surface. Plin2 localizes to the LD surface. A portion of the ER that intercalates into the right side of the LD is labeled with antibodies against both PDI and Plin2 (arrow). (The inset shows an enlargement of this area). (B) A single 0.77 nm slice taken from the middle of the interior of the tomographic reconstruction shown in (A). Plin2 labeling is present through the interior of the section and the co-labeled region (arrow) is easily distinguished, indicating that it is indeed an intercalation and not a superimposition of a labeled structure above or below the lipid droplet (The inset shows an enlargement of this area documenting the presence of both immunogold labeled Plin2 and PDI). Bar = 0.2 μm
LD-ER Interactions in the Pregnant Mammary Gland
As LD formation in MEC is stimulated in response to secretory differentiation of the mammary gland during pregnancy [12, 36], we investigated LD-ER interactions in mammary glands of rats on pregnancy days 5 (P5) and 10 (P10) to identify organelle associations involved in the initial periods of their formation. A survey of thin section images of cryopreserved MEC at P10 identified ER cisternae with distended by 50–100 nm spherical inclusions that are similar in shape and electron density to LD, and which have a size consistent with that reported for nascent LD in other systems [40]. Nascent LD localize to cisternal domains that are continuous with, and have the same density as, the ER lumen (Fig. 5A) and are surrounded by an outer ER membrane bearing ribosomes on its surface (Fig. 5A, inset). Additionally, nascent LD with diameters in the 100–200 nm range are found in MEC from P10 animals. A composite tomographic reconstruction of a P10 cell (Fig. 5B) shows 2 nascent LD within a distinctive ribosome-free region that is penetrated by multiple ER cisternae, all of which possess abundant numbers of ribosomes on both membranes prior to the point of entering the matrix. The nascent LD within the matrix lie in close proximity (7–10 nm) to an ER cisterna and may correspond to an initial stage of LD egression from the ER [2].
Fig. 5.
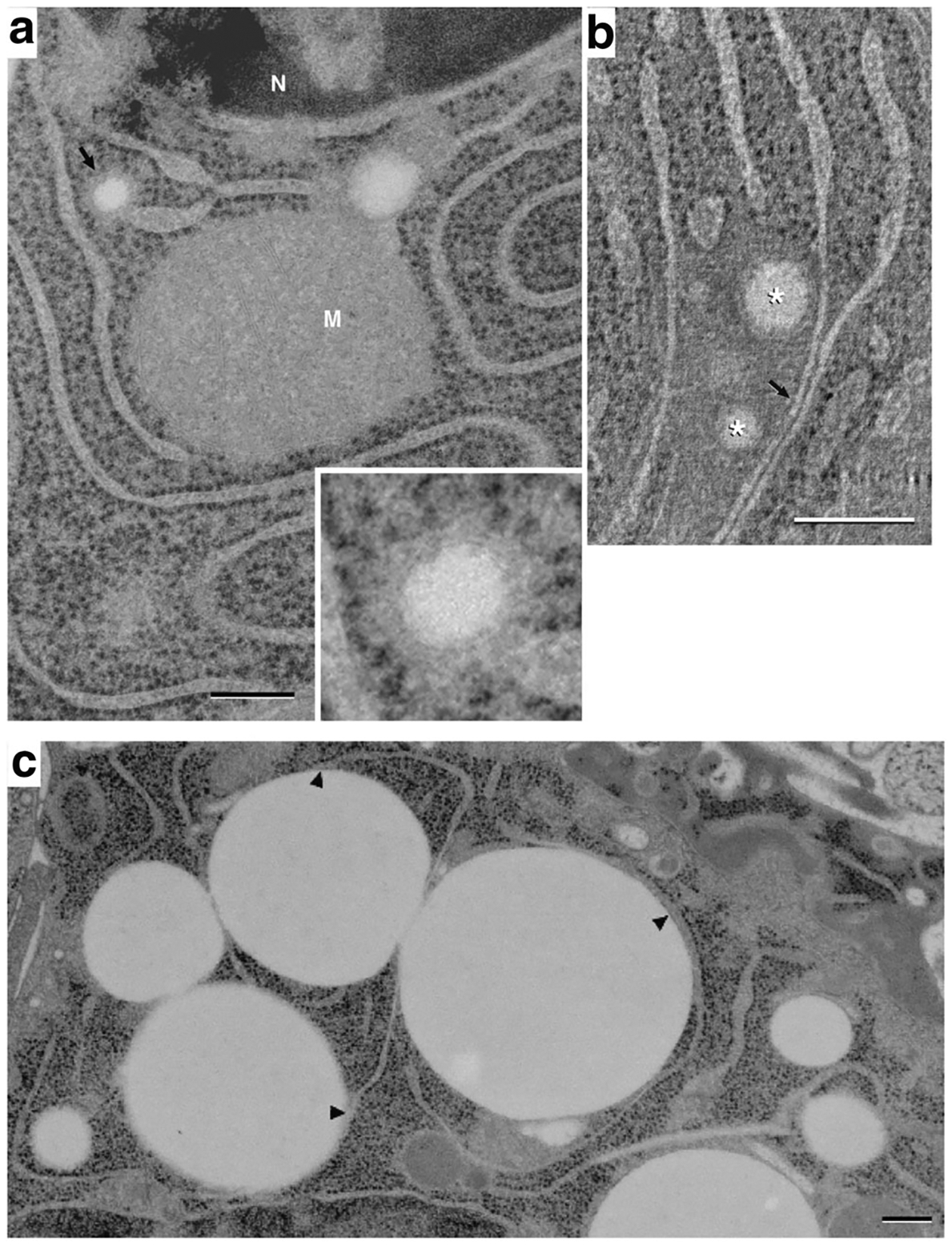
Endoplasmic reticulum association with lipid droplets in differentiating mammary epithelial cells. (A) Thin (40 nm) section image of a mammary epithelial cell on pregnancy day 10 (P10), showing a small (~100 nm) lipid droplet within the lumen of an ER cisterna (arrow and inset). The small droplet is surrounded by a dense material that is similar in appearance to, and is continuous with, the ER lumen. This density is itself surrounded by a ribosome-bearing membrane that is continuous with the ER membrane. A second, slightly larger lipid droplet is present on the upper right side of the mitochondrion (M). Although it is surrounded by dense material similar to that of the smaller droplet, its association with nearby ER cannot be determined in the plane of this thin section (N, nucleus). (B) A composite of 7, 1.1 nm slices from a tomographic reconstruction of a P10 cell, showing 2 small lipid droplets (*) amongst ER cisternae. The droplets lie within a matrix that excludes ribosomes; regions of ER cisternae bordering the matrix are smooth. Two ER cisternae near the droplets are closely apposed to one another analogously to the ER domains associating with larger droplets (arrow). Bars = 0.2 μm. (C) EM image showing a field of LD in contact with ER cisternae in a mammary epithelial cell at P5. ER cisternae are shown to contact multiple mature (1 μm) droplets at numerous points (arrowheads) while other parts of the reticulum remain distant from the droplets. Small (~100 nm) LD localize to termini of ER cisternae (asterisks)
Both nascent and mature LD were detected in MEC from rats by P5. An image from a thin section of a P5 MEC shows ER-lipid droplet domains form at numerous places along a single ER cisterna and demonstrate that a large part of the cisterna is involved in this process. Mature LD with diameters of ~ 1 μ were found to contact individual ER cisternae at multiple points (Fig. 5C). As with mature CLD from lactating animals, the ER membrane facing LD-contact sites is free of ribosomes, which are abundant on the apposing cisternal face (arrowheads, Fig. 5C). In agreement with results from P10 MEC, nascent LD with diameters in the range of 100–200 nm are found in regions of cisternae having continuity with the ER lumen.
Lipid Synthesizing Enzymes
Maturation and growth of LD has been shown to involve relocalization of neutral lipid synthesis enzymes, which are integral ER membrane proteins, from the ER to the surface of LD in some systems [2]. To determine if an analogous process occurs during the initial phase of LD accumulation in differentiating MEC, we localized enzymes that catalyze synthesis of triglycerides (acyl-CoA diacylglycerol acyltransferase (Dgat) −1 and − 2) [41] and cholesteryl esters (acyl-CoA cholesterol acyltransferase; Acat1) [42] in mammary glands of P5 rats. For this study, we used confocal immunofluorescence microscopy because the available antibodies to these enzymes unfortunately did not work on cryosections for EM. LD were identified by staining the neutral lipid core with Nile red (NR) [43], or by immunostaining with antibodies to Plin2, which has been shown previously to specifically localize to the surface of LD in MEC [36]. Double labeling of mammary gland sections from P5 rats with NR and antibodies to Plin2 verify that Plin2 specifically localizes to the periphery of NR-stained LD at this early stage of differentiation (Fig. 6A–C). Sections double labeled with Plin2 and Dgat-1 (D-F), Dgat-2 (G-I), or Acat (H-K) further demonstrate that each of these enzymes are located near LD, and in some instances are closely associated with Plin2. These results are consistent with the presence of LD-ER membrane contacts in differentiating MEC that allow neutral lipid synthesizing enzymes in ER membranes to localize near the LD surface to facilitate their maturation.
Fig. 6.
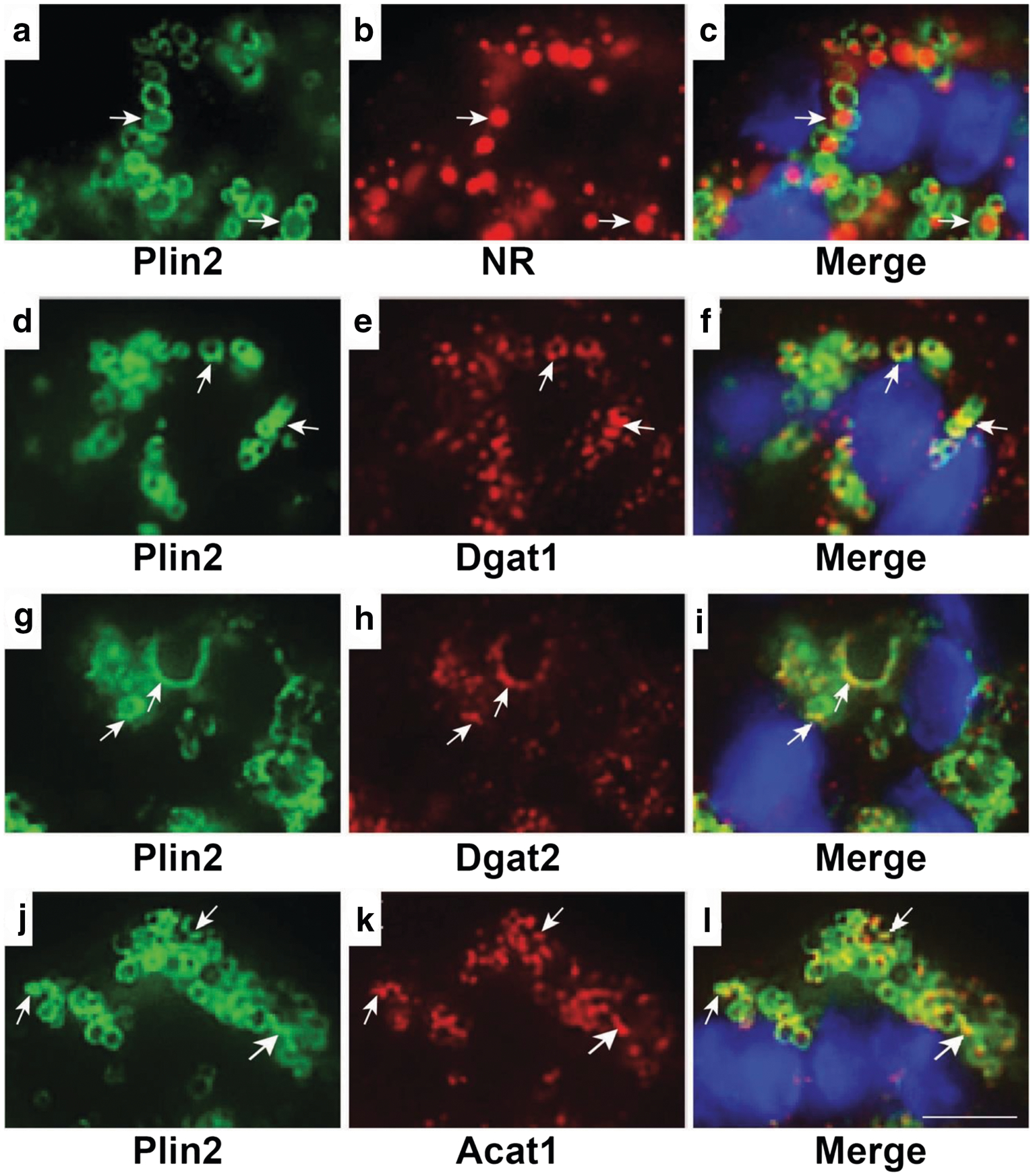
Localization of ER proteins and lipid droplets in mammary acini of pregnant rats. Representative confocal microscopy images of mammary acini from day 5 pregnant rats showing specific localization of Plin2 to the surface of Nile red (NR) stained LD (arrows, A-C). Areas of overlap of Dgat1 (arrows, D-F); Dgat2(arrows, G-I); and Acat1 (arrows, J-L) with Plin2 indicate their localization on the LD surface. DAPI-stained nuclei are shown in blue. Bar = 7 μm
Discussion
Cytoplasmic lipid droplets are organellar structures that function in the storage and trafficking of neutral lipids for use as substrates for metabolism and membrane biosynthesis in eukaryotic cells. In mammary epithelial cells of lactating animals, LD are secreted as intact structures by an apocrine mechanism to form milk lipids [28], which increases demands on the cellular processes that mediate neutral lipid synthesis and LD formation. Beginning with studies of mammary glands of lactating mice [15], there is increasing evidence from multiple organisms that LD originate from the ER, and that LD contacts with the ER and other organelles play critical roles in cellular neutral lipid storage and trafficking functions of LD [2]. Prior ultrastructural studies of MEC from lactating animals using conventional 2D-EM approaches identified contacts between LD and the ER [16]. However, subsequent technological advancements in tissue fixation that preserve cellular structures in near native states, and the introduction of 3D electron tomography [44], permit understanding of LD-ER interactions in greater detail and with greater accuracy than that achieved by conventional 2D-EM [25]. Using combinations of 3D electron tomography and immuno-EM and confocal fluorescence microscopy of mammary glands from pregnant and lactating rats prepared by high pressure freezing and freeze substitution, we identified novel features of LD-ER domains in MEC that provide new insight into their structural organization and LD biogenesis.
Distinct LD-ER Domains in Lipogenic Organs
LD-ER contacts are observed at the ultrastructural level in multiple organisms and cell types including MEC [2, 17]. However, there are considerable differences in the type and extent of contact, which can range from single or multiple contacts between discrete regions of the ER membrane and the LD surface to contacts that follow a portion of the contour of the LD surface [45]. The extent of contact between LD and ER cisternae is seldom seen to be extensive in cultured mammalian cells [46, 47], yeast [48] or in insect cells [49] under normal conditions. However, the extent of LD-ER contact can be influenced by genetic alterations that affect the composition of LD proteins [46], and by regulators of LD-ER contact and LD growth, such as seipin and the Arf1/COP1 complex [50–52]. In contrast, electron tomography of MEC and hepatocytes from lactating rats reveal continuous contact between ER cisternae and the LD surface that follow the droplet’s contour over a large portion of its circumference, and which exhibit structural features not reported for other cell types. Portions of the cisternae that contact LD in MEC and hepatocytes also contact concentrically layered stacks of flattened cisternae, which are arrayed in contour with the droplets surface. In both tissues, cisternae that wrap LD or that comprise the concentric stacks have a distinct distribution of ribosomes, which localize selectively to membrane surfaces facing the cytosol but are not found on surfaces adjoining the LD or adjacent cisternal layers, as typically found in rough ER stacks. Notably, cisternae within these stacks can also form contacts with elements of the trans-Golgi and/or mitochondria membranes in nearby regions of the cytoplasm, a property that facilitates cellular trafficking of lipids and proteins by permitting their direct inter-organelle transfer [53]. Collectively, these features suggest that LD in cells of highly lipogenic organs, such as the mammary gland and liver, form highly specialized domains with ER cisternae, which may function to accommodate lipid and protein transfer requirements associated with elevated neutral lipid synthesis and secretion during lactation. Intriguingly, the expression of seipin, which has been shown to regulate LD-ER contact and conversion of nascent to mature LD in cultured cells [20, 50], is markedly elevated in MEC of lactating mice compared to pregnant mice [54]. The extent to which seipin, or other LD or ER proteins, mediate LD-ER interactions observed in MEC remains to be determined. Moreover, LD-ER interactions are dynamic [45] and LD are able to exist independently of ER contact and to form multiple types of contact with ER membranes [2]. Consequently, it is likely that the extent of contact between LD and ER cisternae in MEC may be variable, a concept supported by previous conventional 2D EM [16] and by electron tomography of MEC from pregnant rats (Fig. 5), and that other factors, such as lipogenic activity, will influence LD-ER contacts. We did not quantify the extent of LD-ER contact observed by electron tomography in MEC due to constraints on sample number and on access to microscope time for generating statistically significant numbers of datasets. However, extensive areas of contact were observed in the majority of micrographs in which both cytoplasmic LD and ER were present, in both pregnant and lactating animals, as well as in hepatocytes, which suggest that such interactions may be a structural feature of these cells and possibly others that possess elevated lipogenic activity.
Specialization of LD-ER Domains
The morphological features of LD-ER interacting domains identified in mammary and liver tissues are consistent with the egg cup model of LD growth proposed by the Robenek laboratory [22], which hypothesizes that extensive contact between LD and the outer ER membrane bilayer is ideally configured for transferring neutral lipids from their sites of synthesis on ER membranes to the LD core. In this model, contact between LD and the ER membrane occurs at sites enriched in Plin2, which is proposed to contribute to the mechanism of neutral lipid transfer from the ER to the LD core [22]. In agreement with this proposal, our immune-EM data demonstrate Plin2 on the surface of LD, and on ER at sites of their contact with LD. In addition, immunofluorescence microscopy demonstrated localization of Plin2 near Dgat-1 and −2 and Acat1, which are ER membrane enzymes that respectively catalyze triglyceride and cholesteryl ester synthesis [41]. Significantly, data demonstrating that Plin2 loss reduces total neutral lipid accumulation and/or decreases the size of LD in MEC and hepatocytes of mice [37, 55] support the proposed role of Plin2 in lipid transfer to LD. Although the mechanisms underlying the effects of Plin2 on LD growth are not well-understood, structural studies demonstrating that Plin2 possesses distinct LD- and membrane phospholipid-binding domains [56] suggest the possibility that it contributes to the formation of structural links between LD and ER membranes. Such interactions potentially account for the detection of Plin2, and structurally-related members of the perilipin family, within the LD interior (Figs. 3 and 4; [39]) by providing a mechanism for ER cisternae to invaginate into the LD surface. Alternatively, evidence that Plin2 modulates de novo lipid synthesis and the phospholipid composition of ER membranes in hepatocytes under conditions of elevated lipogenesis [57], suggest that it may contribute to functional specializations of LD-ER interacting domains that mediate LD growth during lactation. Additional ultrastructural and molecular studies will be required to rigorously establish the importance of Plin2, and possibly other proteins, in mediating the distinct morphological features and the functional importance of LD-ER interactions in MEC.
Sites of LD Formation
The mechanisms responsible for LD formation from the ER remain poorly understood and may differ among eukaryotic cells. The prevailing mechanism is that neutral lipid accumulation between ER membrane bilayer leaflets induces membrane distension and separation creating a LD surrounded by the outer membrane leaflet that buds into the cytoplasm [2]. Although convincing evidence for this model exists in yeast and insect cells, it has not been observed unequivocally, and other mechanisms of LD biogenesis have been proposed. The demonstration that nascent LD in differentiating MEC are found within specialized lumen domains of terminal regions of ER cisternae is consistent with results from conventional 2D-EM of MEC [16], and with biochemical and ultrastructural data from yeast [18, 21, 58], which show that neutral lipids accumulate within the lumen prior to being incorporated into LD. A luminal ER origin of LD in MEC is also consistent with proteomic evidence showing selective enrichment of ER luminal proteins on LD isolated from MEC compared to those obtained from hepatocytes [4]. Significantly, the presence of small nascent LD in lumens of terminal ER cisternae and larger mature LD that contact external membranes of ER cisternae in P5 MEC demonstrate that LD and ER form morphologically distinct domains within the same cell. How this transition occurs is not yet known and may differ between cell types and organisms. However it is consistent with concept that the ER-LD domains undergo dynamic remodeling, which include ER fission and fusion, adapted to the different requirements associated with formation and subsequent growth of LD [2]. Although we did not detect lipid accumulation as lenses between ER membrane bilayers in MEC, as demonstrated in other organisms [18], we can rule out the possibility that such structures may exist only transiently and were not effectively captured by high pressure freezing of mammary tissue. Nevertheless, lens formation occurs primarily, if not exclusively, in tubular (smooth) ER [2, 59], whereas MEC like other cells with high secretory activity are enriched in rough ER [12], which appears to be the initial location of neutral lipid formation in mammary tissue [15]. Consequently, the mechanisms of LD formation in MEC may be specifically adapted to their elevated secretory activity.
Acknowledgements
We thank Dr. Robert Farese Jr., Harvard School of Public Health for the antibodies against Dgat1 and Dgat2 and Dr. T.Y. Chang of Dartmouth Medical School for the antibodies Acat1. This work was supported by NIH grants PO1GM61306 (KEH); 5PO1HD038129 (KEH); 2PO1HD038129 (JLM); and 2R01HD45965 and R01HD093729 (JLM).
Abbreviations
- Acat1
Acyl-CoA cholesterol acyltransferase-1
- Dgat
Acyl-CoA diacylglycerol acyltransferase
- EM
Electron microscopy
- ER
Endoplasmic reticulum
- LD
Lipid droplet
- MEC
Mammary epithelial cell
- MFG
Milk fat globules
- NR
Nile red
- Plin2
Perilipin-2
- P5
Pregnancy day 5
- P10
Pregnancy day 10
- PDI
Protein disulfide isomerase
- 3D
Three-dimensional
- 2D
Two-dimensional
Footnotes
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Oftedal OT. Milk composition, milk yield and energy output at peak lactation: a comparative review. 1984;51:33–85. [Google Scholar]
- 2.Walther TC, Chung J, Farese RV Jr. Lipid Droplet Biogenesis. Annu Rev Cell Dev Biol. 2017;33:491–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J Biol Chem. 2002;277(46):44507–12. [DOI] [PubMed] [Google Scholar]
- 4.Wu CC, Howell KE, Neville MC, Yates JR 3rd, McManaman J. Proteomics reveal a link between the endoplasmic reticulum and lipid secretory mechanisms in mammary epithelial cells. Electrophoresis. 2000;21(16):3470–82. [DOI] [PubMed] [Google Scholar]
- 5.Fujimoto T, Parton RG. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb Perspect Biol 2011;3(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenberg AS, Coleman RA, Kraemer FB, McManaman J, Obin MS, Puri V, et al. The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest. 2011;121(6):2102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McManaman JL. Milk lipid secretion: recent biomolecular aspects. Biomol Concepts. 2012;3:581–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butovich IA. Meibomian glands, meibum, and meibogenesis. Exp Eye Res. 2017;163:2–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider MR. Lipid droplets and associated proteins in sebocytes. Exp Cell Res. 2016;340(2):205–8. [DOI] [PubMed] [Google Scholar]
- 10.Oftedal OT, Iverson SJ. Comparative analysis of nonhuman milks. In handbook of Milk composition (Jensen RG ed), pp 749–789, academic press; 1995. [Google Scholar]
- 11.Hollmann KH. Cytology and fine structure of the mammary gland. In lactation (Larson BL, Smith VR eds), pp 3–95, Academic Press; 1974. [Google Scholar]
- 12.Wooding FBP. Comparative mammary fine structure. In comparative aspects of lactation (Peaker M ed), pp. 1–41, Academic Press. 1977. [Google Scholar]
- 13.Mellenberger RW, Bauman DE. Metabolic adaptations during lactogenesis. Fatty acid synthesis in rabbit mammary tissue during pregnancy and lactation. Biochem J. 1974;138(3):373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson SM, et al. Key stages in mammary gland development. Secretory activation in the mammary gland: it’s not just about milk protein synthesis! Breast Cancer Res. 2007;9(1):204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stein O, Stein Y. Lipid synthesis, intracellular transport, and secretion. II Electron microscopic radioautographic study of the mouse lactating mammary gland. J Cell Biol. 1967;34(1):251–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keenan TW, Dylewski DP. Aspects of intracellular transit of serum and lipid phases of milk. J Dairy Sci. 1985;68(4):1025–40. [DOI] [PubMed] [Google Scholar]
- 17.Heid HW, Keenan TW. Intracellular origin and secretion of milk fat globules. Eur J Cell Biol. 2005;84(2–3):245–58. [DOI] [PubMed] [Google Scholar]
- 18.Choudhary V, Ojha N, Golden A, Prinz WA. A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J Cell Biol. 2015;211(2):261–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thiam AR, Farese RV Jr, Walther TC. The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol. 2013;14(12): 775–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H et al. Seipin is required for converting nascent to mature lipid droplets Elife 5. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choudhary V, Jacquier N, Schneiter R. The topology of the triacylglycerol synthesizing enzyme Lro1 indicates that neutral lipids can be produced within the luminal compartment of the endoplasmatic reticulum: implications for the biogenesis of lipid droplets. Commun Integr Biol. 2011;4(6):781–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robenek H, Hofnagel O, Buers I, Robenek MJ, Troyer D, Severs NJ. Adipophilin-enriched domains in the ER membrane are sites of lipid droplet biogenesis. J Cell Sci. 2006;119(Pt 20):4215–24. [DOI] [PubMed] [Google Scholar]
- 23.Ploegh HL. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature. 2007;448(7152):435–8. [DOI] [PubMed] [Google Scholar]
- 24.Al-Amoudi A, et al. Cryo-electron microscopy of vitreous sections of native biological cells and tissues. J Struct Biol. 2004;148(1): 131–5. [DOI] [PubMed] [Google Scholar]
- 25.Fujimoto T, Ohsaki Y, Suzuki M, Cheng J. Imaging lipid droplets by electron microscopy. Methods Cell Biol. 2013;116:227–51. [DOI] [PubMed] [Google Scholar]
- 26.Pfeiffer S, Vielhaber G, Vietzke JP, Wittern KP, Hintze U, Wepf R. High-pressure freezing provides new information on human epidermis: simultaneous protein antigen and lamellar lipid structure preservation. Study on human epidermis by cryoimmobilization. J Invest Dermatol. 2000;114(5):1030–8. [DOI] [PubMed] [Google Scholar]
- 27.McIntosh JR. Electron microscopy of cells: a new beginning for a new century. J Cell Biol. 2001;153(6):F25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu CC, Yates JR 3rd, Neville MC, Howell KE. Proteomic analysis of two functional states of the Golgi complex in mammary epithelial cells. Traffic. 2000;1(10):769–82. [DOI] [PubMed] [Google Scholar]
- 29.Orlicky DJ, Degala G, Greenwood C, Bales ES, Russell TD, McManaman J. Multiple functions encoded by the N-terminal PAT domain of adipophilin. J Cell Sci. 2008;121(Pt 17):2921–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morphew MK, McIntosh JR. The use of filter membranes for high-pressure freezing of cell monolayers. J Microsc. 2003;212(Pt 1):21–5. [DOI] [PubMed] [Google Scholar]
- 31.Kremer JR, Mastronarde DN, McIntosh J. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–6. [DOI] [PubMed] [Google Scholar]
- 32.Ladinsky MS, Wu CC, McIntosh S, McIntosh J, Howell KE. Structure of the Golgi and distribution of reporter molecules at 20 degrees C reveals the complexity of the exit compartments. Mol Biol Cell. 2002;13(8):2810–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mather IH, Keenan TW. Origin and secretion of milk lipids. J Mammary Gland Biol Neoplasia. 1998;3(3):259–73. [DOI] [PubMed] [Google Scholar]
- 34.Clermont Y, Xia L, Rambourg A, Turner JD, Hermo L. Structure of the Golgi apparatus in stimulated and nonstimulated acinar cells of mammary glands of the rat. Anat Rec. 1993;237:308–17. [DOI] [PubMed] [Google Scholar]
- 35.Clermont Y, Xia L, Rambourg A, Turner JD, Hermo L. Transport of casein submicelles and formation of secretion granules in the Golgi apparatus of epithelial cells of the lactating mammary gland of the rat. Anat Rec. 1993;235(3):363–73. [DOI] [PubMed] [Google Scholar]
- 36.Russell TD, Palmer CA, Orlicky DJ, Fischer A, Rudolph MC, Neville MC, et al. Cytoplasmic lipid droplet accumulation in developing mammary epithelial cells: roles of adipophilin and lipid metabolism. J Lipid Res. 2007;48(7):1463–75. [DOI] [PubMed] [Google Scholar]
- 37.Russell TD, Schaack J, Orlicky DJ, Palmer C, Chang BH, Chan L, et al. Adipophilin regulates maturation of cytoplasmic lipid droplets and alveolae in differentiating mammary glands. J Cell Sci. 2011;124(Pt 19):3247–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120(3): 343–52. [DOI] [PubMed] [Google Scholar]
- 39.Robenek H, Robenek MJ, Troyer D. PAT family proteins pervade lipid droplet cores. J Lipid Res. 2005;46(6):1331–8. [DOI] [PubMed] [Google Scholar]
- 40.Pol A, Gross SP, Parton RG. Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J Cell Biol. 2014;204(5):635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV Jr. Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res. 2008;49(11):2283–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang TY, Chang CC, Ohgami N, Yamauchi Y. Cholesterol sensing, trafficking, and esterification. Annu Rev Cell Dev Biol. 2006;22:129–57. [DOI] [PubMed] [Google Scholar]
- 43.Greenspan P, Mayer EP, Fowler SD. Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol. 1985;100(3): 965–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McIntosh R, Nicastro D, Mastronarde D. New views of cells in 3D: an introduction to electron tomography. Trends Cell Biol. 2005;15(1):43–51. [DOI] [PubMed] [Google Scholar]
- 45.Salo VT, Ikonen E. Moving out but keeping in touch: contacts between endoplasmic reticulum and lipid droplets. Curr Opin Cell Biol. 2019;57:64–70. [DOI] [PubMed] [Google Scholar]
- 46.Ozeki S, Cheng J, Tauchi-Sato K, Hatano N, Taniguchi H, Fujimoto T. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J Cell Sci. 2005;118(Pt 12):2601–11. [DOI] [PubMed] [Google Scholar]
- 47.Ohsaki Y, Cheng J, Suzuki M, Fujita A, Fujimoto T. Lipid droplets are arrested in the ER membrane by tight binding of lipidated apolipoprotein B-100. J Cell Sci. 2008;121(Pt 14):2415–22. [DOI] [PubMed] [Google Scholar]
- 48.Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Sci. 2011;124(Pt 14):2424–37. [DOI] [PubMed] [Google Scholar]
- 49.Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, et al. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell. 2013;24(4):384–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salo VT, Belevich I, Li S, Karhinen L, Vihinen H, Vigouroux C, et al. Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J. 2016;35(24):2699–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilfling F, et al. Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. Elife. 2014;3:e01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salo VT et al. Seipin facilitates triglyceride flow to lipid droplet and counteracts droplet ripening via endoplasmic reticulum contact. Dev Cell. 2019. [DOI] [PubMed] [Google Scholar]
- 53.English AR, Voeltz GK. Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb Perspect Biol. 2013;5(4):a013227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.El Zowalaty AE, et al. Seipin deficiency leads to increased endoplasmic reticulum stress and apoptosis in mammary gland alveolar epithelial cells during lactation. Biol Reprod. 2018;98(4):570–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Orlicky DJ, Libby AE, Bales ES, McMahan R, Monks J, la Rosa FG, et al. Perilipin-2 promotes obesity and progressive fatty liver disease in mice through mechanistically distinct hepatocyte and extra-hepatocyte actions. J Physiol. 2019;597(6):1565–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chong BM et al. The adipophilin C-terminus is a self-folding membrane binding domain that is important for milk lipid secretion. J Biol Chem. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Libby AE, Bales E, Orlicky DJ, McManaman J. Perilipin-2 deletion impairs hepatic lipid accumulation by interfering with sterol regulatory element-binding protein (SREBP) activation and altering the hepatic Lipidome. J Biol Chem. 2016;291(46):24231–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mishra S, Khaddaj R, Cottier S, Stradalova V, Jacob C, Schneiter R. Mature lipid droplets are accessible to ER luminal proteins. J Cell Sci. 2016;129(20):3803–15. [DOI] [PubMed] [Google Scholar]
- 59.Kassan A, Herms A, Fernández-Vidal A, Bosch M, Schieber NL, Reddy BJ, et al. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J Cell Biol. 2013;203(6):985–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]


