Abstract
Nasopharyngeal carcinoma (NPC) is an epithelial carcinoma that arises from the lining of the nasopharyngeal mucosa. The efficacy of radiation therapy is limited due to radiation resistance, particularly in the advanced stages of NPC. The S100P protein is a small isoform of the S100 protein family, which is involved in the regulation of various intracellular and extracellular processes, including proliferation, differentiation and intracellular signaling. The aim of the current study was to investigate the significance of the S100P-RAGE axis in NPC progression. The expression levels of S100P and receptor for activated glycation end-products (RAGE) in NPC specimens were determined by western blotting. In addition, the effect of the S100P-RAGE axis on NPC was evaluated in vitro by proliferation and migration assays using C666-1 cells treated with S100P or the RAGE inhibitor FPS-ZM1. The underlying mechanism was also investigated by western blotting. The expression of S100P and RAGE was detected in clinical specimens from 15 patients with NPC and 15 patients with benign nasopharyngeal inflammation, and was observed to be higher in NPC tissues compared with inflamed tissues. Furthermore, the interaction of S100P with RAGE increased the proliferation and migration potential of C666-1 cells, and activated mitogen-activated protein kinase and NF-κB signaling. These results indicate that the S100P-RAGE axis exerts a promoting effect on the progression of NPC. Therefore therapeutic strategies targeting S100P-RAGE merit further exploration for the treatment of NPC.
Keywords: S100P, RAGE, proliferation, migration, C666-1 cells
Introduction
Nasopharyngeal carcinoma (NPC) is an epithelial carcinoma that arises from the lining of the nasopharyngeal mucosa (1). The tumors are usually observed at the pharyngeal recess in the nasopharynx. Despite originating from similar cell or tissue lineages, NPC is distinct from other epithelial head and neck tumors (2). NPC is characterized by poorly or undifferentiated carcinomas. It differs from other head and neck squamous cell carcinomas in several ways, including its association with the Epstein-Barr virus (EBV), increased radio- and chemosensitivity, and a greater propensity for distant metastases (3). Compared with other cancers, NPC is relatively uncommon. The incidence of NPC has a marked geographical distribution; it is most prevalent in South China, with an annual incidence of ~30 cases per 100,000 population (4). NPC is associated with multiple risk factors, including genetic susceptibility and environmental factors, such as EBV infection and dietary intake (5,6).
Proteins of the S100 family are small Ca2+-binding proteins with a helix-loop-helix (EF-hand type) binding motif, and have been demonstrated to participate in the regulation of various intracellular and extracellular processes, including cell proliferation and differentiation, and intracellular signaling (7). Receptor for activated glycation end products (RAGE) is a receptor for multiple ligands that belongs to the immunoglobulin family. The dysregulation of RAGE has been indicated to be a critical step in the development and metastasis of numerous types of tumors, including head and neck cancer (8,9). RAGE is able to interact with structurally diverse ligands, and although these interactions appear to involve oligomerization of the receptor on the cell surface (10), the exact mechanism is unknown. Members of the S100 protein family are known to be RAGE ligands (10). Several S100 proteins, including S100A12 and S100B, are released from cells and have the ability to activate RAGE, suggesting that this may be an important mechanism underlying the extracellular effects of S100 proteins (11). The S100P protein is a small isoform of the S100 protein family, the ‘P’ in its name reflecting its original isolation from the placenta by Becker et al (12) in 1992. S100P is a 95-amino-acid protein, which is encoded by a gene located on chromosome 4pl. S100P has been reported to interact with several proteins, both extracellularly and intracellularly, and its upregulation in various types of human cancer has been shown to be associated with disease progression, the acquisition of chemoresistance and poor prognosis (13,14).
In our previous study, the differential expression of S100P between NPC tissues and the tissues of patients with benign inflammation was identified using immunohistochemistry, and S100P was demonstrated to be associated with proliferation and migration in the C666-1 NPC cell line (15). However, the role of RAGE activation in S100P signaling has not yet been examined in C666-1 cells and the mechanism remains unclear.
In the present study, to determine whether the activation of RAGE by S100P is required for S100P to exert its effects on cell growth and survival, the activation of RAGE by S100P was blocked and the effects on cell function and signaling were investigated using a variety of methods. Mechanisms involving the activation of extracellular signal-related kinase 1/2 (ERK1/2), mitogen-activated protein kinase 7 (MAPK7), p38 MAPK, ERK1/2, NF-κB1 and NF-κB p65 were investigated.
Materials and methods
Patients and tumor samples
In total, 30 patients were included in the study, 15 with NPC and 15 with benign nasopharyngeal inflammation. The samples were obtained from patients who were hospitalized for nasopharyngeal biopsy between April 2018 and September 2019 at the Department of Otolaryngology, Jinshan Hospital, Fudan University (Shanghai, China). The patients were diagnosed with NPC or benign nasopharyngeal inflammation through pathological examination. None of the patients had undergone chemotherapy or radiotherapy, or had any other tumors. Tissue samples were frozen immediately in liquid nitrogen and stored at -80˚C until used. The clinical characteristics of the patients are shown in Table I. The study cohort comprised 16 men (53.3%) and 14 women (46.7%) with an age range of 25-79 years (mean age, 50.3±14.9 years; Table I). The NPC group comprised 10 men and 5 women (mean age, 55.13±15.19 years). The Benign nasopharyngeal inflammation group consisted of 6 men and 9 women (mean age, 45.4±13.88 years). When the collection of tissue samples took place, the participants signed informed consent forms to confirm that they agreed to the use of their tissues in scientific research.
Table I.
Clinical characteristics of the patients.
| Characteristic | Total no. of patients (n=30) | No. of NPC cases (n=15) | No. of benign inflammation cases (n=15) |
|---|---|---|---|
| Sex | |||
| Male | 16 | 10 | 6 |
| Female | 14 | 5 | 9 |
| Age (years) | |||
| <50 | 14 | 6 | 8 |
| ≥50 | 16 | 9 | 7 |
| Stage | |||
| I | 1 | ||
| II | 6 | ||
| III | 6 | ||
| IV | 2 | ||
| Histological subtype | |||
| Non-keratinized | 15 | ||
| Undifferentiated | 0 |
Cell culture and treatments
The C666-1 NPC cell line was purchased from the cell bank of the Central Laboratory of Xiangya Hospital (Central South University). The cell line was authenticated by the provider and tested for Mycoplasma contamination. The C666-1 cell line was established by Cheung et al (16) from undifferentiated NPC, and consistently carries EBV in long-term culture. The C666-1 cells were cultured in RPMI-1640 medium (Sigma-Aldrich; Merck KGaA) supplemented with 10% certified fetal bovine serum (FBS; Biological Industries) and maintained in a humidified incubator containing 5% CO2 at 37˚C. The medium was replaced with fresh culture medium every 3-4 days. The following three groups of C666-1 cells were used in subsequent assays: The S100P protein group, treated with S100P (cat. no. Ag19115; Proteintech Group, Inc.), the FPS-ZM1 group, treated with the RAGE inhibitor FPS-ZM1 (cat. no. 6237; Tocris Bioscience) (17), and the untreated control group. The S100P protein and RAGE inhibitor were separately dissolved in cell culture medium and prepared for immediate use.
Western blot analysis
Total proteins were extracted from the tissue samples and cells following lysis with sodium dodecyl sulfate (SDS) lysis buffer containing phenylmethanesulfonyl fluoride and proteinase inhibitor (100:1:1; Nanjing KeyGen Biotech Co., Ltd.) using an automatic grinding machine (JXFSTPRP-24; Jingxin Technology Co., Ltd.) at 50 Hz for 60 sec. The supernatant was collected and the protein concentration was determined using a BCA protein assay (Beyotime Institute of Biotechnology). Western blotting was performed using a standard method (18). Quantified proteins (50 µg per lane) were separated by electrophoresis using 8-12% SDS polyacrylamide gels and electroblotted onto polyvinylidene fluoride membranes. The membranes were blocked with 5% non-fat milk in tris buffer saline containing 0.05% Tween-20 for 1 h at room temperature. The membranes were then incubated sequentially at 4˚C overnight with primary antibodies. Primary antibodies targeting S100P (cat. no. 11803-1-AP; 1:1,000), RAGE (cat. no. 16346-1-AP; 1:1,000), ERK1/2 (cat. no. 66192, 1:1,000), p38 (cat. no. 66234; 1:1,000), MAPK7 (cat. no. 10036-2-AP; 1:1,000), NF-κB1 (cat. no. 14220-1-AP; 1:1,000), p65 (cat. no. 10745-1-AP; 1:1,000) and GAPDH (cat. no. 10494-1-AP; 1:5,000) were acquired from Proteintech Group, Inc. and antibodies targeting phosphorylated (p)-ERK1/2 (ab201015; 1:1,000), p-p38 (ab4822; 1:1,000) and p-MAPK7 (ab5686, 1:1,000) Abcam. Following this, the membranes were incubated with HRP-conjugated secondary antibodies (cat. no. SA00001-2; Proteintech; 1:5,000) for 1 h at room temperature. The resulting protein-antibody complexes were detected using an ECL system (Merck KGaA) and then visualized using AllDoc version 2.2.1.0 software with the Tanon 4500 imaging system (Tanon Science and Technology Co., Ltd.).
Cell Counting Kit-8 (CCK-8) assay
The C666-1 cells were seeded at a density of 3,000 cells/well in 96-well plates. After 24 h, the cell viability was determined using a CCK-8 assay (Dojindo Molecular Technologies, Inc.), by measurement of the optical density at 450 nm (OD450) according to the kit protocol. Following this, a range of concentrations (10, 100, 500, 1,000 and 5,000 ng/ml) of S100P protein or FPS-ZM1 were added to the wells, an untreated control group was established, and the cells were cultured for 6 h at 37˚C. In subsequent experiments, the S100P protein and FPS-ZM1 were used at a concentration of 1,000 ng/ml, since at this concentration, the inhibition and proliferation rates were ~50%, respectively. Cell viability was assessed using the CCK-8 assay after incubation for 6, 12 and 24 h by adding 10 µl CCK-8 solution to each well, incubating for 30 min to 4 h at 37˚C according to the manufacturer's protocol, after which the OD450 value was measured.
Colony formation assay
The C666-1 cells were cultured in 6-well plates at a density of 2,000 cells/well. After 24 h, 1,000 ng/ml S100P protein or FPS-ZM1 was added to each well and the cells were cultured for a further ~14 days at 37˚C. The colonies were then stained with 0.4% crystal violet (Beyotime Institute of Biotechnology). The numbers of colonies that contained >50 cells were counted and averaged (19).
Wound healing assays
The C666-1 cells were cultured in 6-well plates until they reached confluency. The monolayers were scratched and the detached cells were washed away using PBS solution. Then, 1,000 ng/ml S100P protein or FPS-ZM1 was added to each well and the cells were cultured in serum-free RPMI-1640 for 24 h at 37˚C. The width of the wound was measured at 0 and 24 h with an inverted microscope equipped with a digital camera (Olympus Corporation).
Transwell migration assay
Transwell chambers (pore size, 8 µm; Corning Life Sciences) were used for the Transwell migration assay. A total of 2x105 cells suspended in 100 µl serum-free medium with or without S100P protein or FPS-ZM1 were added to the upper chamber, and medium containing 20% FBS was added to the lower chamber. The chambers were incubated for 24 h at 37˚C. Subsequently, the cells that had migrated through the membrane were fixed, stained with crystal violet for 20 min at room temperature and counted in 5 random fields with an inverted microscope equipped with a digital camera (Olympus Corporation).
Statistical analysis
All experimental assays were conducted as at least three independent experiments. Data are expressed as the mean ± SD. Data analysis was performed using SPSS version 26.0 (IBM Corp.). Comparison of the protein levels between the NPC and benignly inflamed tissues was performed using independent-sample t-tests. Kruskal-Wallis and Mann-Whitney U tests were used to analyze the differences in S100P expression level among or between patients according to disease stage, age and gender. Comparisons of multiple groups in other assays were performed by one-way ANOVA followed by Bonferroni post hoc tests for the multiple comparisons. P<0.05 was considered to indicate a statistically significant difference.
Results
Protein expression of S100P and RAGE in NPC
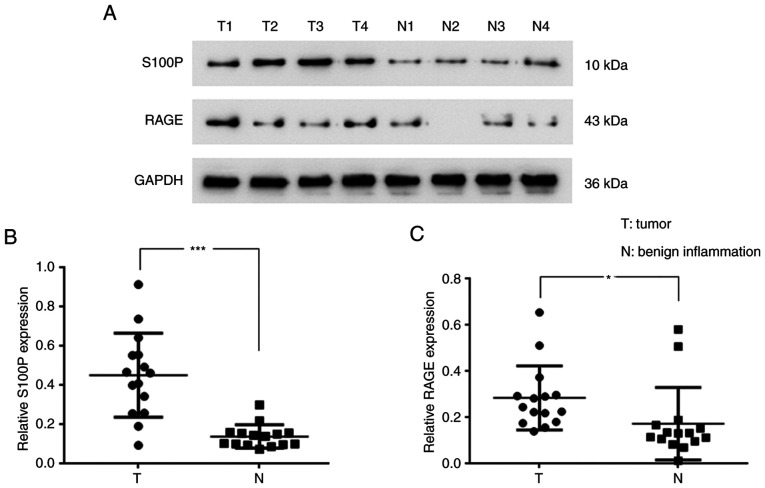
To evaluate the levels of S100P and RAGE in nasopharyngeal tumors, the tumor samples of 15 patients with NPC and tissues of 15 patients with benign nasopharygeal inflammation were analyzed. The western blotting results revealed that S100P (P<0.001; Fig. 1A and B) and RAGE (P<0.05; Fig. 1A and C) were significantly upregulated in the NPC tissues compared with the tissues with benign inflammation. The associations of the S100P expression level with disease stage and the age and gender of the patients were also analyzed. The S100P expression level was found to differ significantly according to disease stage when analyzed using a Kruskal-Wallis test (P<0.01; Table II); however, no significant difference in S100P expression level was detected according to the age and gender of the patients when analyzed using a Kruskal-Wallis and Mann-Whitney U test (Table II).
Figure 1.
Protein expression of S100P and RAGE in patients with NPC or benign nasopharyngeal inflammation. (A) The protein expression of S100P and RAGE in NPC tissues and nasopharyngeal tissues with benign inflammation by western blotting. Quantification of (B) S100P/GAPDH and (C) RAGE/GAPDH protein expression ratios. *P<0.05 and ***P<0.001. RAGE, receptor for activated glycation end products; NPC, nasopharyngeal carcinoma.
Table II.
Differences in S100P expression levels according to the disease stage, age and sex of patients.
| Characteristic | Number of cases (n=30) | Relative S100P expression | S100P expression (P-value) |
|---|---|---|---|
| Sex | 0.058 | ||
| Male | 16 | 0.327±0.179 | |
| Female | 14 | 0.254±0.264 | |
| Age (years) | 0.42 | ||
| <50 | 14 | 0.301±0.272 | |
| ≥50 | 16 | 0.308±0.176 | |
| Stage | 0.017 | ||
| Benign inflammation | 15 | 0.137±0.059 | |
| I | 1 | 0.092 | |
| II | 6 | 0.417±0.267 | |
| III | 6 | 0.493±0.138 | 0.005 |
| IV | 2 | 0.596±0.06 |
Effects of S100P-RAGE on cell proliferation
We hypothesized that the effects of S100P in NPC might be due to its binding with RAGE and the activation of autocrine signaling mechanisms. Therefore, three groups of C666-1 cells were analyzed, namely the S100P protein group, the FPS-ZM1 group and the untreated control group. These groups were analyzed in cell viability and migration assays to identify the association between S100P and RAGE.
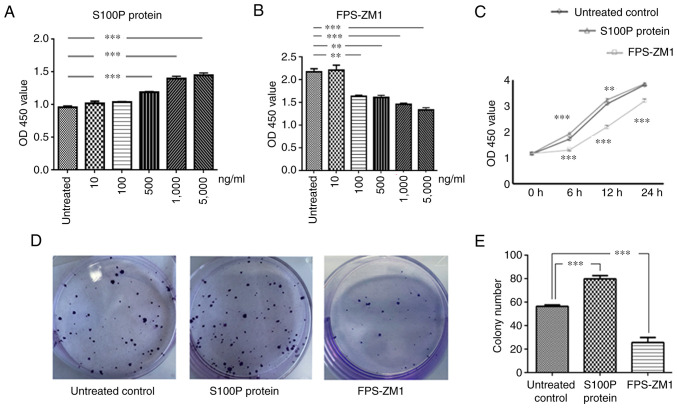
Different concentrations of S100P protein and the RAGE inhibitor FPS-ZM1 were added to C666-1 cells to test their effects on cell viability. The effects of S100P protein and FPS-ZM1 on cell viability were observed to be concentration-dependent (P<0.001 and P<0.01, respectively; Fig. 2A and B). Furthermore, the effects of S100P protein and FPS-ZM1 on cell viability were found to be time-dependent. Compared with that in the untreated control group, cell viability was significantly affected at 6 and 12 h following treatment with S100P protein or FPS-ZM1 at a concentration of 1,000 ng/ml (P<0.01 and P<0.001, respectively; Fig. 2C). The 1,000 ng/ml concentration was selected for use in subsequent experiments on the basis that it is the concentration at which the inhibition and proliferation rates for S100P and FPS-ZM1 are ~50%, respectively (data not shown). The colony forming assay revealed that the ability of C666-1 cells to form colonies was increased by treatment with S100P protein and decreased by treatment with the RAGE inhibitor FPS-ZM1 (P<0.001; Fig. 2D and E).
Figure 2.
Effects of S100P and RAGE on the proliferation and colony formation of C666-1 cells. Cell Counting Kit-8 assay results for the growth evaluation of cells treated with (A) S100P protein or (B) RAGE inhibitor FPS-ZM1 at the indicated concentrations for 6 h. The effects of S100P and FPS-ZM1 on cell viability were concentration-dependent. (C) The effects of S100P protein and FPS-ZM1 (both 1,000 ng/ml) on cell viability were also time-dependent. (D) Representative images of colony formation assay results. (E) The colony forming ability of C666-1 cells was increased in the S100P protein group and reduced in the FPS-ZM1 group. **P<0.01 and ***P<0.001 vs. untreated control. RAGE, receptor for activated glycation end products; OD450, optical density at 450 nm.
Effects of S100P-RAGE on cell migration
The effects of S100P and FPS-ZM1 on the migration of C666-1 cells were assessed by wound healing and Transwell assays. As presented in Fig. 3A and B, measurements made 24 h following wounding of the cell monolayer revealed that wound closure was induced in the S100P protein group and delayed in FPS-ZM1 group compared with the untreated control group. The migrated distances of the cells in the untreated control group, S100P protein group and FPS-ZM1 group were 746.67±55.28, 1,139.39±77.61 and 276.36±112.21 µm, respectively (P<0.001; Fig. 3A and B). The percentage of wound closure in the untreated control, S100P protein and FPS-ZM1 groups was calculated to be 41.79, 65.23 and 16.17%, respectively (P<0.001; Fig. 3C). The Transwell assay revealed that the addition of S100P protein significantly increased the migration ability of the C666-1 cells, while the addition of the RAGE inhibitor FPS-ZM1 reduced the migration ability of the C666-1 cells. The numbers of migrated cells in the untreated control, S100P protein and FPS-ZM1 groups were 91.8±6.72, 200.8±9.26 and 30.4±2.3, respectively (P<0.001; Fig. 3D and E).
Figure 3.
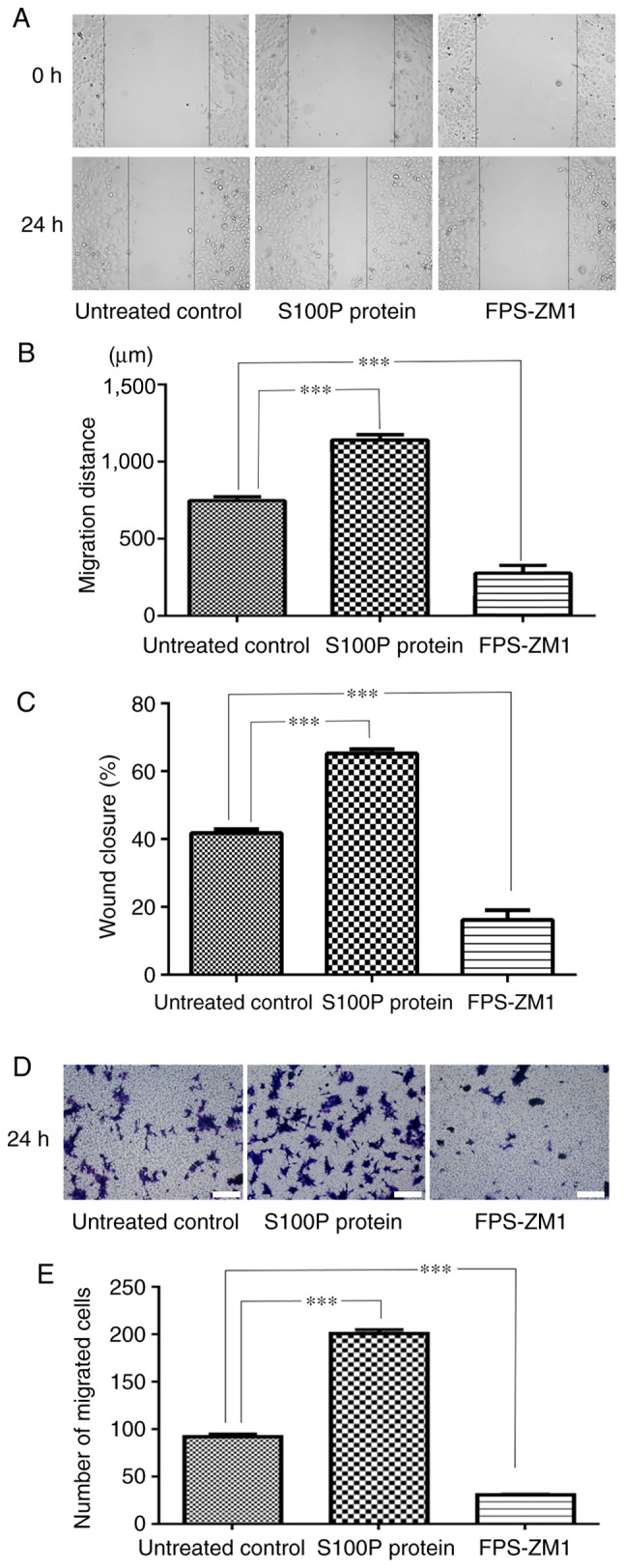
Effects of S100P and RAGE on C666-1 cell migration. (A) Representative images of the wound healing assay (magnification, x400). (B) Measurements of the wound in the cell monolayer revealed that wound closure was induced by treatment with S100P protein and delayed by treatment with the RAGE inhibitor FPS-ZM1 compared with the untreated control. (C) The percentage of wound closure was different in the S100P protein and FPS-ZM1 groups compared with the untreated control group. (D) Representative images of the Transwell assay results. Scale bar, 200 µm. (E) The Transwell assay revealed that the migration ability of C666-1 cells was significantly increased by S100P protein and significantly reduced by the RAGE inhibitor FPS-ZM1. RAGE, receptor for activated glycation end products. ***P<0.001.
S100P-RAGE activates MAPK and NF-κB signaling
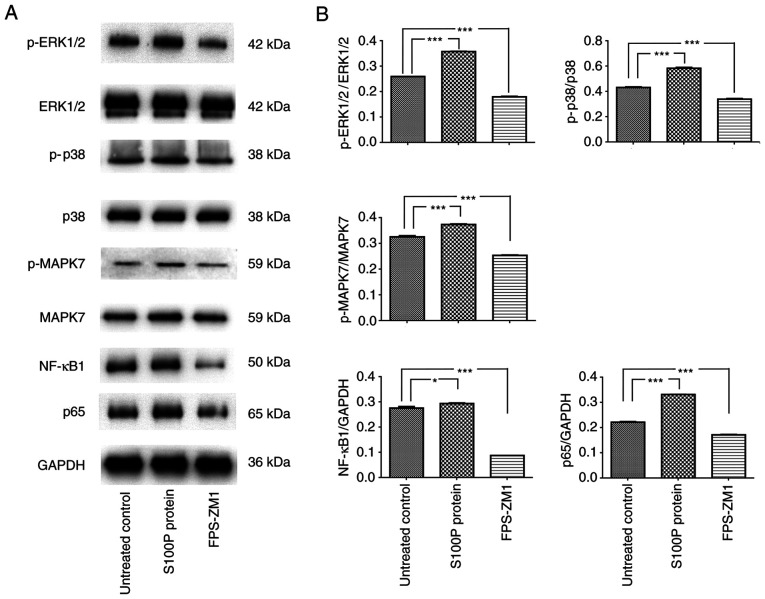
The MAPK signaling pathway is reported to be associated with cell proliferation, differentiation, migration, senescence and apoptosis (20). The activation of NF-κB is often associated with increased cell survival (21). Therefore, whether extracellular S100P activates these transcription factors was investigated. Consistent with the effects on cell proliferation and migration, blockade of the S100P-RAGE interaction inhibited the effects of S100P on MAPK and NF-κB signaling. Following treatment of the cells with S100P protein, the western blotting results revealed that the p-ERK1/2/ERK1/2, p-p38/p38 and p-MAPK7/MAPK7 ratios and NF-κB1 and p65 expression levels were significantly increased compared with those in the untreated cells (NF-κB1, P<0.05; other proteins, P<0.001). Conversely, following treatment with the RAGE inhibitor FPS-ZM1, the p-ERK1/2/ERK1/2, p-p38/p38 and p-MAPK7/MAPK7 ratios and NF-κB1 and p65 expression levels were decreased compared with those in the untreated cells (all P<0.001; Fig. 4). These results indicate that the S100P-RAGE interaction activates MAPK and NF-κB signaling in NPC cells.
Figure 4.
S100P-RAGE activates MAPK and NF-κB signaling in C666-1 cells. (A) Representative western blots of cells treated with S100P protein or RAGE inhibitor FPS-ZM1. (B) Quantified western blotting data showing that p-ERK1/2/ERK1/2, p-p38/p38 and p-MAPK7/MAPK7 ratios and NF-κB1 and p65 expression levels were significantly increased in the S100P protein group and significantly reduced in the FPS-ZM1 group compared with those in untreated cells. *P<0.05 and ***P<0.001. RAGE, receptor for activated glycation end products; MAPK, mitogen-activated protein kinase; ERK, extracellular signal-related kinase.
Discussion
The gold standard treatment for NPC is radiation therapy, which is a local treatment for the cure or palliative treatment of tumors (22). The cure rate of early-stage NPC is >90% (23). However, the efficacy of radiation therapy in advanced stages of NPC is limited owing to radiation resistance (24). Increased tumor volumes, tumor hypoxia and the dysregulation of genes can cause tumor cells to become tolerant to radiation, and thus reduce their sensitivity (25). Therefore, the early diagnosis of NPC and the development of novel treatment approaches are particularly important.
In a previous study, the effect of S100P expression on the proliferation and migration of C666-1 cells was investigated by knocking down S100P expression via infection with S100P small interfering RNA (siRNA) (15). Following S100P knockdown, the downregulation, proliferation and migration of the cells were significantly decreased (15). In addition, it was observed that RAGE expression was downregulated in the cells transfected with S100P siRNA, as compared with the untreated and negative siRNA-transfected controls (15). Notably, certain S100 proteins have been identified to serve a role in tumors by interacting with receptors, suggesting that extracellular S100 proteins have important effects (11). In addition, some S100 proteins have been demonstrated to interact with RAGE in vitro, triggering RAGE-dependent signal transduction in cell-based assays. For example, Arumugam et al (26) demonstrated that S100P triggers the activation of NF-κB in a RAGE-dependent manner through a MAPK signaling pathway in BxPC3 and SW480 adenocarcinoma cells. However, the mechanism remains incompletely explored. In the present study, to determine whether the activation of RAGE by S100P is required for the effects of S100P on the growth and migration of C666-1 cells, the activation of RAGE by S100P was blocked and its effects on cell proliferative and migratory behavior and downstream signaling were investigated. In particular, to elucidate the molecular mechanisms underlying the activity and function of S100P in NPC, the role of S100P as an activator of signaling pathways known to affect the development and progression of numerous types of cancer was assessed. The most frequently reported signaling pathways in which S100P participates include ERK1/2, NF-κB and PI3K/AKT (27). Arumugam et al (26) revealed that exogenous S100P increased the survival of NIH3T3 cells and simultaneously activated ERK in the cells. The MAPK signaling pathway comprises distinct ERK1/2, JNK1/2/3, p38 MAPK and ERK5 pathways (28). The MAPK/ERK pathway is reported to be associated with cell proliferation, differentiation, migration, senescence and apoptosis (20). The present study demonstrated that the S100P-RAGE interaction is associated with MAPK activation in C666-1 cells.
S100P has been reported to exert autocrine effects via RAGE that stimulate cell proliferation and survival via the NF-κB pathway (26). The transcription factor NF-κB is a nuclear factor that binds to the enhancer element of the immunoglobulin κ light-chain of activated B cells (29). The NF-κB transcription factor family has five members, known as p65 (RelA), RelB, c-Rel, NF-κB1 and NF-κB2. The constitutive activation of NF-κB has been demonstrated to lead to the promotion of cell proliferation, angiogenesis, invasion and metastasis (21). Consistent with this, the present study detected a significant increase in NF-κB activation in NPC.
The inhibition of S100P, using antisense mRNA retroviral transfection for example, has been shown to decrease cellular motility and metastatic potential in colon, gastric and breast cancer cell lines (30-32). The expression of S100P has been identified to be associated with the resistance of cancers to several chemotherapeutic agents, and its silencing sensitizes cancer cells to cisplatin and oxaliplatin in vitro (33,34). In the present study, the ability of extracellular S100P to activate MAPK and NF-κB signaling was investigated. Notably, blockade of the S100P-RAGE interaction inhibited the effects of S100P on MAPK and NF-κB signaling, which is similar to its effects on cell proliferation and migration.
The present study has potential limitations, including the relatively small number of clinical samples and the use of only one cell line. However, NPC is associated with multiple risk factors, including EBV infection (5,6), and the C666-1 cell line consistently carries EBV in long-term cultures (16). Therefore, only the C666-1 cell line was used. Further studies using primary cultures of NPC cells and in vivo experiments are required in the future. Also, although significant data were obtained in the experiments in the present study, the potential of S100P-RAGE blockade as a therapeutic treatment for NPC requires further examination in a larger number of patients.
In conclusion, the present results confirm and extend those of our previous study, and suggest that extracellular S100P may participate in the proliferation and migration of C666-1 cells by binding to RAGE and activating the MAPK and NF-κB signaling pathways. Therefore, blocking S100P-RAGE function might also be expected to improve the response of NPC to therapeutic treatments.
Acknowledgements
Not applicable.
Funding Statement
Funding: This study was supported by funding from the Jinshan District Science and Technology Innovation project (grant no. 2017-3-07) and the Qihang Plan of Jinshan Hospital of Fudan University (grant no. 2018-JSYYQH-02).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HJ performed the diagnostic investigations and biopsy treatments. CW performed the experiments and wrote the manuscript. XW and CW confirm the authenticity of all the raw data. XW performed the statistical analysis and analyzed the data. AH performed and guided experiment operations. YW collected and analyzed pathological data. All the authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
All experimental procedures and protocols were approved by the Ethics Committee of Jinshan Hospital Affiliated to Fudan University (approval no. 2018-05-01). The participants signed informed consent forms for use of their tissues in scientific research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Tian Y, Tang L, Yi P, Pan Q, Han Y, Shi Y, Rao S, Tan S, Xia L, Lin J, et al. miRNAs in radiotherapy resistance of nasopharyngeal carcinoma. J Cancer. 2020;11:3976–3985. doi: 10.7150/jca.42734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394:64–80. doi: 10.1016/S0140-6736(19)30956-0. [DOI] [PubMed] [Google Scholar]
- 3.Lee AW, Ng WT, Chan YH, Sze H, Chan C, Lam TH. The battle against nasopharyngeal cancer. Radiother Oncol. 2012;104:272–278. doi: 10.1016/j.radonc.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Cao SM, Simons MJ, Qian CN. The prevalence and prevention of nasopharyngeal carcinoma in China. Chin J Cancer. 2011;30:114–119. doi: 10.5732/cjc.010.10377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tu C, Zeng Z, Qi P, Li X, Yu Z, Guo C, Xiong F, Xiang B, Zhou M, Gong Z, et al. Genome-wide analysis of 18 epstein-barr viruses isolated from primary nasopharyngeal carcinoma biopsy specimens. J Virol. 2017;91:e00301–e00317. doi: 10.1128/JVI.00301-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam WKJ, Chan JYK. Recent advances in the management of nasopharyngeal carcinoma. F1000Res. 2018;7(F1000) doi: 10.12688/f1000research.15066.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakayama H, Ohuchida K, Yonenaga A, Sagara A, Ando Y, Kibe S, Takesue S, Abe T, Endo S, Koikawa K, et al. S100P regulates the collective invasion of pancreatic cancer cells into the lymphatic endothelial monolayer. Int J Oncol. 2019;55:211–222. doi: 10.3892/ijo.2019.4812. [DOI] [PubMed] [Google Scholar]
- 8.Sasahira T, Kirita T, Bhawal UK, Yamamoto K, Ohmori H, Fujii K, Kuniyasu H. Receptor for advanced glycation end products (RAGE) is important in the prediction of recurrence in human oral squamous cell carcinoma. Histopathology. 2007;51:166–172. doi: 10.1111/j.1365-2559.2007.02739.x. [DOI] [PubMed] [Google Scholar]
- 9.Choi J, Lee MK, Oh KH, Kim YS, Choi HY, Baek SK, Jung KY, Woo JS, Lee SH, Kwon SY. Interaction effect between the receptor for advanced glycation end products (RAGE) and high-mobility group box-1 (HMGB-1) for the migration of a squamous cell carcinoma cell line. Tumori. 2011;97:196–202. doi: 10.1700/667.7783. [DOI] [PubMed] [Google Scholar]
- 10.Leclerc E, Fritz G, Vetter SW, Heizmann CW. Binding of S100 proteins to RAGE: An update. Biochim Biophys Acta. 2009;1793:993–1007. doi: 10.1016/j.bbamcr.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann MA, Drury S, Fu C, Qu W, Taguchi A, Lu Y, Avila C, Kambham N, Bierhaus A, Nawroth P, et al. RAGE mediates a novel proinflammatory axis: A central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97:889–901. doi: 10.1016/s0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- 12.Becker T, Gerke V, Kube E, Weber K. S100P, a novel Ca(2+)-binding protein from human placenta. cDNA cloning, recombinant protein expression and Ca2+ binding properties. Eur J Biochem. 1992;207:541–547. doi: 10.1111/j.1432-1033.1992.tb17080.x. [DOI] [PubMed] [Google Scholar]
- 13.Prica F, Radon T, Cheng Y, Crnogorac-Jurcevic T. The life and works of S100P-from conception to cancer. Am J Cancer Res. 2016;6:562–576. [PMC free article] [PubMed] [Google Scholar]
- 14.Arumugam T, Simeone DM, Van Golen K, Logsdon CD. S100P promotes pancreatic cancer growth, survival, and invasion. Clin Cancer Res. 2005;11:5356–5364. doi: 10.1158/1078-0432.CCR-05-0092. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Wang C, Shan X, Wu J, Liu H, Liu H, Zhang J, Xu W, Sha Z, He J, Fan J. S100P is associated with proliferation migration in nasopharyngeal carcinoma. Oncol Lett. 2017;14:525–532. doi: 10.3892/ol.2017.6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung ST, Huang DP, Hui AB, Lo KW, Ko CW, Tsang YS, Wong N, Whitney BM, Lee JC. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein-Barr virus. Int J Cancer. 1999;83:121–126. doi: 10.1002/(sici)1097-0215(19990924)83:1<121::aid-ijc21>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 17.Hudson BI, Lippman ME. Targeting RAGE signaling in inflammatory disease. Annu Rev Med. 2018;69:349–364. doi: 10.1146/annurev-med-041316-085215. [DOI] [PubMed] [Google Scholar]
- 18.Mahmood T, Yang PC. Western blot: Technique, theory, and trouble shooting. N Am J Med Sci. 2012;4:429–434. doi: 10.4103/1947-2714.100998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 20.Sun Y, Liu WZ, Liu T, Feng X, Yang N, Zhou HF. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res. 2015;35:600–604. doi: 10.3109/10799893.2015.1030412. [DOI] [PubMed] [Google Scholar]
- 21.Pramanik KC, Makena MR, Bhowmick K, Pandey MK. Advancement of NF-κB signaling pathway: A novel target in pancreatic cancer. Int J Mol Sci. 2018;19(3890) doi: 10.3390/ijms19123890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang T, Yin L, Wu J, Gu JJ, Wu JZ, Chen D, Yu HL, Ding K, Zhang N, Du MY, et al. MicroRNA-19b-3p regulates nasopharyngeal carcinoma radiosensitivity by targeting TNFAIP3/NF-κB axis. J Exp Clin Cancer Res. 2016;35(188) doi: 10.1186/s13046-016-0465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan AT. Current treatment of nasopharyngeal carcinoma. Eur J Cancer. 2011;47 (Suppl 3):S302–S303. doi: 10.1016/S0959-8049(11)70179-4. [DOI] [PubMed] [Google Scholar]
- 24.Yang H, Zhang G, Che X, Yu S. Slug inhibition increases radiosensitivity of nasopharyngeal carcinoma cell line C666-1. Exp Ther Med. 2018;15:3477–3482. doi: 10.3892/etm.2018.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barker HE, Paget JT, Khan AA, Harrington KJ. The tumour microenvironment after radiotherapy: Mechanisms of resistance and recurrence. Nat Rev Cancer. 2015;15:409–425. doi: 10.1038/nrc3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arumugam T, Simeone DM, Schmidt AM, Logsdon CD. S100P stimulates cell proliferation and survival via receptor for activated glycation end products (RAGE) J Biol Chem. 2004;279:5059–5065. doi: 10.1074/jbc.M310124200. [DOI] [PubMed] [Google Scholar]
- 27.Parkkila S, Pan PW, Ward A, Gibadulinova A, Oveckova I, Pastorekova S, Pastorek J, Martinez AR, Helin HO, Isola J. The calcium-binding protein S100P in normal and malignant human tissues. BMC Clin Pathol. 2008;8(2) doi: 10.1186/1472-6890-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stecca B, Rovida E. Impact of ERK5 on the hallmarks of cancer. Int J Mol Sci. 2019;20(1426) doi: 10.3390/ijms20061426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- 30.Jiang L, Lai YK, Zhang J, Wang H, Lin MC, He ML, Kung HF. Targeting S100P inhibits colon cancer growth and metastasis by Lentivirus-mediated RNA interference and proteomic analysis. Mol Med. 2011;17:709–716. doi: 10.2119/molmed.2011.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ning X, Sun S, Hong L, Liang J, Liu L, Han S, Liu Z, Shi Y, Li Y, Gong W, et al. Calcyclin-binding protein inhibits proliferation, tumorigenicity, and invasion of gastric cancer. Mol Cancer Res. 2007;5:1254–1262. doi: 10.1158/1541-7786.MCR-06-0426. [DOI] [PubMed] [Google Scholar]
- 32.Beissel B, Silva ID, Pesquero JB, Russo J, Schor N, Bellini MH. S-phase reduction in T47D human breast cancer epithelial cells induced by an S100P antisense-retroviral construct. Oncol Rep. 2007;17:611–615. [PubMed] [Google Scholar]
- 33.Zhang YW, Zheng Y, Wang JZ, Lu XX, Wang Z, Chen LB, Guan XX, Tong JD. Integrated analysis of DNA methylation and mRNA expression profiling reveals candidate genes associated with cisplatin resistance in non-small cell lung cancer. Epigenetics. 2014;9:896–909. doi: 10.4161/epi.28601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang H, Liu YJ, Liu M, Li X. Establishment and gene analysis of an oxaliplatin-resistant colon cancer cell line THC8307/L-OHP. Anticancer Drugs. 2007;18:633–639. doi: 10.1097/CAD.0b013e3280200428. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.