Abstract
Oxidized low-density lipoprotein (ox-LDL)-induced endothelial dysfunction contributes to the progression of atherosclerosis. Interferon regulatory factor 2-binding protein 2 (IRF2BP2) attenuates macrophage-mediated inflammation and susceptibility to atherosclerosis. However, the effects of IRF2BP2 on vascular endothelial cells in atherosclerosis have not been fully elucidated. In the present study, the effects of IRF2BP2 on cell viability, inflammation and endothelial-to-mesenchymal transition (EMT) of human umbilical vein endothelial cells (HUVECs) were assessed using Cell Counting Kit-8 (CCK-8) assays, ELISA kits and western blot analysis, respectively. In addition, the expression levels of Krüppel-like factor 2 (KLF2) were determined by reverse transcription-quantitative PCR and immunofluorescence assays. A Nitrate/Nitrite assay kit was utilized to detect the production of nitric oxide (NO). The results demonstrated that ox-LDL induced inflammation and EMT of HUVECs, and decreased the NO levels. Furthermore, IRF2BP2 overexpression protected HUVECs against ox-LDL-induced inflammation, EMT and endothelial dysfunction, and resulted in upregulated expression of KLF2. Additionally, IRF2BP2 was shown to bind to KLF2, and KLF2 knockdown reversed the protective effects of IRF2BP2 on ox-LDL-exposed HUVECs. These findings indicated that IRF2BP2 may prevent ox-LDL-induced endothelial damage via upregulating KLF2 expression.
Keywords: oxidized low-density lipoprotein, human umbilical vein endothelial cells, Krüppel-like factor 2, interferon regulatory factor 2-binding protein 2
Introduction
Cardiovascular diseases are the most common cause of disease-associated mortality worldwide (1). Hypertension, hypercholesterolemia, obesity and smoking pose serious threats to the cardiovascular system, contributing to vascular inflammation and endothelial cell dysfunction (2-4). Atherosclerosis is a lipid-driven progressive inflammatory systemic disease, which primarily affects the aorta, and the carotid and coronary arteries (5,6). The accumulation of plasma lipoproteins is considered as the main factor driving formation of atherosclerotic lesions. Despite the progress that has been made in reducing the concentration of lipoproteins, there are residual risks to these cardiovascular events (7). Anti-inflammatory approaches, including reducing the levels of pro-inflammatory cytokines, are considered a promising strategy to supplement the existing lipid-lowering strategies to decrease the residual risks (8).
Interferon regulatory factor 2-binding protein 2 (IRF2BP2) is a transcriptional repressor that is extensively involved in the regulation of the expression of a range genes (9-11). A recent study reported that IRF2BP2 overexpression improves cardiomyopathy via suppressing the infiltration of inflammatory cells and the secretion of inflammatory cytokines (12). Deletion of IRF2BP2 aggravated a high fat diet-induced increase in systematic and central nervous inflammation via inhibition of the release of interleukin (IL)-1β and tumor necrosis factor (TNF)-α (13). IRF2BP2 is a negative regulator of inflammation via regulation of macrophage polarization. A study revealed that IRF2BP2-deficient macrophages exacerbated atherosclerosis in apolipoprotein E null mice by reducing the expression of the anti-inflammatory transcription factor Krüppel-like factor 2 (KLF2) in macrophages and disrupting lipid homeostasis (14). KLFs are members of the zinc finger family of DNA-binding transcription factors, which serve a range of roles in several biological processes, including quiescence, proliferation, differentiation, development, growth and inflammation (15). Amongst the members of KLFs, KLF2 is considered as a central regulator of endothelial biology through regulation of proinflammatory activation in endothelial cells. Previous studies have shown that KLF2 signaling served an important role in mediating endothelial inflammation and atherosclerosis (16,17). For example, KLF2 was reported to negatively regulate inflammation and reduce proinflammatory activity of nuclear factor-κB (NF-κB) (18). Endothelial KLF2 also exhibited an atheroprotective role as a novel inhibitor of endothelial inflammasome activation in atherogenesis in vivo (19). However, the effects of the regulation of IRF2BP2 gene expression in endothelial cells on atherosclerosis remains elusive.
In the present study, it was hypothesized that IRF2BP2 may also serve a role in endothelial cell inflammation and dysfunction via regulation of KLF2. The aim of the present study was to investigate whether IRF2BP2 overexpression in endothelial cells exhibited a beneficial effect on ox-LDL-induced endothelial cell injury, and further elucidate the underlying regulatory mechanisms.
Materials and methods
Cell culture
Human umbilical vein endothelial cells (HUVECs) were provided by The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences and maintained at 37˚C in a humidified incubator with 5% CO2. HUVECs were cultured in F12K medium (Boster Biological Technology Co., Ltd.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 µg/ml streptomycin and 100 IU/ml penicillin (Boster Biological Technology Co., Ltd.). HUVECs were treated with 100 µg/ml oxidized low-density lipoprotein (ox-LDL; UnionBiol Biotechnologies Co., Ltd.) for 24 h at 37˚C to simulate atherosclerosis, as previously described (20).
Cell transfection
To overexpress IRF2BP2, a pcDNA3.1-IRF2BP2 plasmid was synthesized by Shanghai GenePharma Co., Ltd.; empty pcDNA3.1 vector was used as a negative control. A small interfering RNA (siRNA) targeting KLF2 (5'-AUUUUGAAAAACAAAACUCGUGAGUUUUGUUUUUCAAAAUGG-3') was synthesized by Shanghai GenePharma Co., Ltd and transfected into HUVECs to knockdown KLF2 expression. Scrambled siRNA (5'-CCUGGCGCCUUCGGUCUUUUU-3') served as a negative control. HUVECs were grown to 60-70% confluence and 20 µg pcDNA3.1 or siRNA plasmids were transfected into cells using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) and incubated for 6 h at 37˚C. A total of 48 h post-transfection, transfected cells were selected for subsequent experiments.
Cell viability
HUVECs at a density of 5x103 cells/well were plated into 96-well plates and then incubated with 100 µg/ml ox-LDL for 24 h at 37˚C. Subsequently, 10 µl Cell Counting Kit-8 reagent (CCK-8; Beijing Solarbio Science & Technology Co., Ltd.) was added to each well for 1 h. The absorbance values were measured at 450 nm using a microplate reader (BioTek Instruments, Inc.).
ELISA
The levels of pro-inflammatory cytokines in the cell medium were determined using TNF-α (cat. no. SEKH-0047), IL-1β (cat. no. SEKH-0002) and IL-6 (cat. no. SEKH-0013) ELISA kits (Beijing Solarbio Science & Technology Co., Ltd.). Briefly, the cell culture medium was transferred to a sterile centrifuge tube and centrifuged (1,000 x g) at 4˚C for 10 min. The supernatants were then supplemented with the corresponding ELISA kit reagents, according to manufacturer's protocol. The absorbance values were measured at a wavelength of 450 nm using a microplate reader (BioTek Instruments, Inc.).
Western blot analysis
HUVECs were lysed to extract total protein and the concentration was quantified using a BCA Protein assay kit (Beijing Solarbio Science & Technology Co., Ltd.). Protein samples were loaded on a 12% SDS-gel, resolved using SDS-PAGE and transferred to a PVDF membrane (EMD Millipore). Following blocking with 5% bovine serum albumin (BSA; Thermo Fisher Scientific, Inc.) for 2 h at room temperature, the membranes were then incubated with primary antibodies against IRF2BP2 (cat. no. 18847-1-AP; 1:1,000), GAPDH (cat. no. 10494-1-AP; 1:5,000), vimentin (cat. no. 10366-1-AP; 1:1,000; all from ProteinTech Group, Inc.), KLF2 (cat. no. PA5-40591; 1:1,000), phosphorylated endothelial nitric oxide synthase (p-eNOS; cat. no. PA5-104858; 1:500), eNOS (cat. no. PA1-037; 1:1,000; all from Invitrogen; Thermo Fisher Scientific, Inc.) and VE-cadherin (cat. no. 2500; 1:1,000; Cell Signaling Technology, Inc.) at 4˚C overnight. The following day, the membranes were incubated with HRP-conjugated Affinipure goat anti-mouse IgG (cat. no. SA00001-1; 1:5,000) or HRP-conjugated Affinipure goat anti-rabbit IgG (cat. no. SA00001-2; 1:5,000; both from ProteinTech Group, Inc.) for 2 h at room temperature. Signals were visualized using an ECL Western Blotting Substrate (Thermo Fisher Scientific, Inc.), and densitometry analysis was performed using Image Lab system (Bio-Rad Laboratories, Inc.; version 1.52). GAPDH was used as the internal control.
Immunofluorescence assay
HUVECs were seeded into 24-well plates at a density of 1x105 cells/well. When cells reached 60-70% confluence, they were fixed with 4% paraformaldehyde at 4˚C for 15 min and incubated with 0.5% Triton X-100 for 20 min. Subsequently, 5% BSA was added to block non-specific antigen at room temperature for 30 min, after which cells were incubated with the primary antibody against KLF2 (cat. no. MA5-24300; Invitrogen; Thermo Fisher Scientific, Inc.) at 4˚C overnight. Following incubation with anti-KLF2, cells were incubated with fluorescein isothiocyanate-conjugated Affinipure goat anti-mouse IgG (cat. no. SA00003-1; ProteinTech Group, Inc.) for 1 h in the dark at room temperature. Cell nuclei were stained with DAPI for 5 min at room temperature, and images were captured under a fluorescence microscope (Carl Zeiss AG; magnification, x200).
Reverse transcription-quantitative PCR (RT-qPCR)
HUVECs were harvested, total RNA was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and cDNA was then synthesized using a reverse transcription kit, according to the manufacturer's protocol (Takara, Bio, Inc.). Next, a SYBR Green PCR kit (Takara Bio, Inc.) was used to determine gene expression on an ABI 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The sequences of the primers used were: KLF2 forward, 5'-GCACGCACACAGGTGAGA-3' (Tm=58˚C) and reverse, 5'-CACAGATGGCACTGGAATGG-3' (Tm=62˚C), product length, 130 bp; IRF2BP2 forward, 5'-CCCTGACTGCAGTTGCAAGA-3' (Tm=62˚C) and reverse, 5'-TTGAGCCCCTCTGTGGATGT-3' (Tm=62˚C), product length, 127 bp; vascular cell adhesion molecule 1 (VCAM-1) forward, 5'-GGACCACATCTACGCTGACA-3' (Tm=62˚C) and reverse, 5'-TTGACTGTGATCGGCTTCCC-3' (Tm=62˚C), product length, 184 bp; intercellular adhesion molecule 1 (ICAM-1) forward, 5'-TCTTCCTCGGCCTTCCCATA-3' (Tm=62˚C) and reverse, 5'-AGGTACCATGGCCCCAAATG-3' (Tm=62˚C), product length, 152 bp; and GAPDH forward, 5'-ATGGGCAGCCGTTAGGAAAG-3' (Tm=62˚C) and reverse, 5'-ATCACCCGGAGGAGAAATCG-3' (Tm=62˚C), product length, 126 bp.
Co-immunoprecipitation (Co-IP)
Extraction of whole-cell lysates from HUVECs was performed using RIPA lysis buffer (Beyotime Institute of Biotechnology). For immunoprecipitation, 500 µg protein was incubated with 1-2 µg of the specific antibodies against IRF2BP2 or KLF2 (same antibodies used in western blotting; Invitrogen; Thermo Fisher Scientific, Inc.) overnight at 4˚C. Subsequently, 40 µl 50% Protein A/G PLUS-Agarose beads (Invitrogen; Thermo Fisher Scientific, Inc.) was added, and incubated for a further 2 h at room temperature. Beads were then washed three times with the lysis buffer and collected by centrifugation at 12,000 x g for 2 min at 4˚C. Following the final wash, the supernatant was aspirated and discarded, then the precipitated proteins were eluted from the beads by resuspending in 2X SDS-PAGE loading buffer and boiling for 5 min. The resulting materials from immunoprecipitation or cell lysates were analyzed using western blotting.
Detection of NO levels
The cell culture medium was collected to estimate the production of NO. Following mixing of cell culture medium with Griess Reagent I and Griess Reagent II from the Nitrate/Nitrite assay kit (Beyotime Institute of Biotechnology), the absorbance values were determined at a wavelength of 540 nm using a microplate reader (BioTek Instruments, Inc.).
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 6.0 (GraphPad Software, Inc.). All experiments were performed in triplicate. Data are presented as the mean ± standard deviation. Differences between multiple groups were analyzed using a one-way ANOVA followed by a post hoc Tukey's test. P<0.05 was considered to indicate a statistically significant difference.
Results
Protective effects of IRF2BP2 on ox-LDL-induced inflammation and EMT
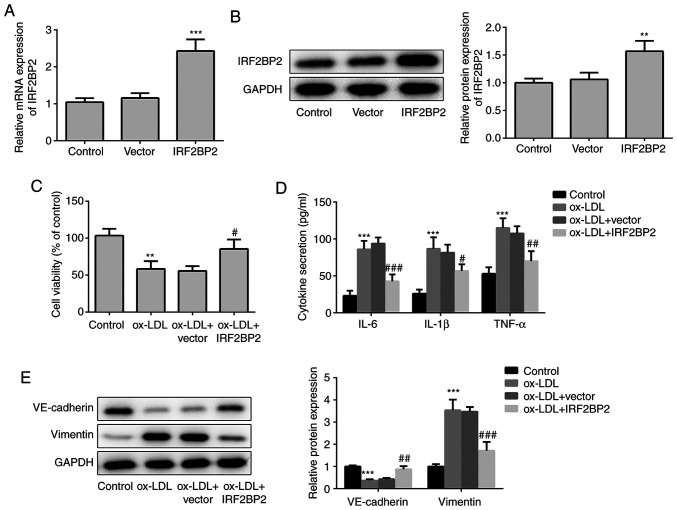
First, IRF2BP2 plasmids were transfected into HUVECs to overexpress IRF2BP2. As shown in Fig. 1A and B, transfection of HUVECs with IRF2BP2 overexpression plasmids increased the expression of IRF2BP2 at both the transcriptional (mRNA) and translational (protein) levels. Subsequently, HUVECs were treated with 100 µg/ml ox-LDL for 24 h, and the results demonstrated that ox-LDL significantly impaired cell viability, whereas IRF2BP2 overexpression effectively relieved ox-LDL-induced cell injury (Fig. 1C). In addition, treatment with ox-LDL induced inflammatory responses in HUVECs, as determined by the significant increase in pro-inflammatory cytokines, including TNF-α, IL-1β and IL-6 (Fig. 1D). HUVECs transfected with IRF2BP2 overexpression plasmid exhibited mild inflammation. As a crucial risk factor of atherosclerosis, ox-LDL promotes the secretion of inflammatory cytokines by endothelial cells, thus inducing EMT (21,22). Therefore, the decreased expression of the endothelial marker VE-cadherin, along with the upregulation of the mesenchymal marker vimentin, indicated that HUVECs underwent EMT following stimulation with ox-LDL (Fig. 1E). By contrast, IRF2BP2 overexpression inhibited EMT.
Figure 1.
Protective effects of IRF2BP2 against ox-LDL-induced inflammation and EMT. (A) HUVECs were transfected with empty vector or pcDNA3.1-IRF2BP2. The mRNA expression levels of IRF2BP2 were measured using reverse transcription-quantitative PCR analysis. ***P<0.001 vs. the vector group. (B) The protein expression levels of IRF2BP2 were determined by western blot analysis. **P<0.01 vs. the vector group. (C) Following transfection, HUVECs were treated with ox-LDL for 24 h. The cell viability in different groups was determined using a Cell Counting Kit-8 assay. (D) The levels of TNF-α, IL-1β and IL-6 in the culture medium were assessed using ELISA kits. (E) The protein expression levels of VE-cadherin and vimentin were evaluated using western blot analysis. **P<0.01, ***P<0.001 vs. the control group; #P<0.01, ##P<0.05 and ###P<0.001 vs. the ox-LDL + vector group. IRF2BP2, interferon regulatory factor binding protein 2; ox-LDL, oxidized low-density lipoprotein; HUVECs, human umbilical vein endothelial cells; TNF-α, tumor necrosis factor α; IL-1β, interleukin 1β.
IRF2BP2 upregulates KLF2 and improves endothelial NO production
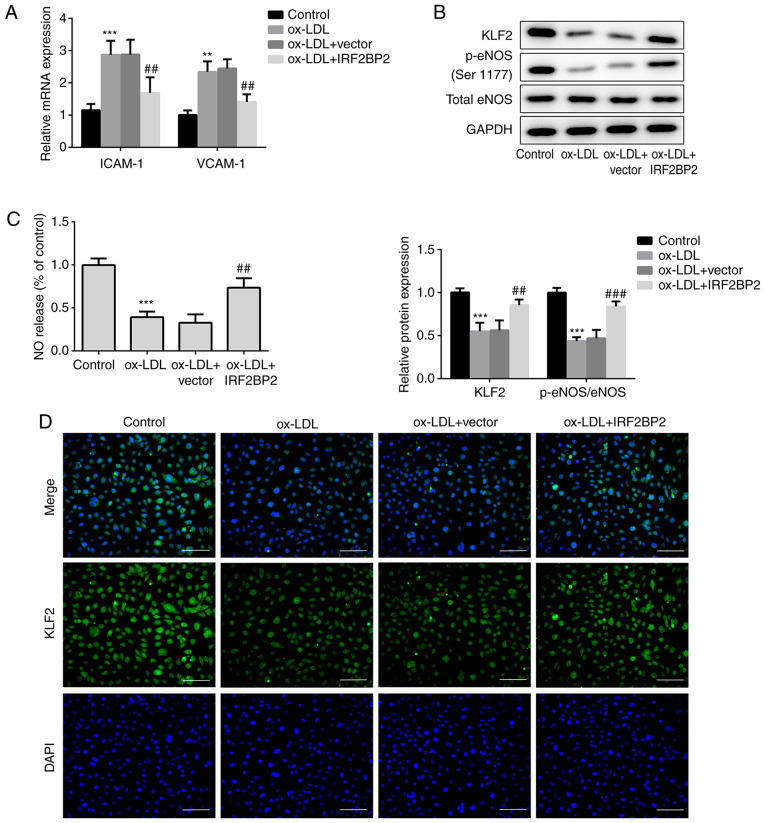
VCAM-1 and ICAM-1 are members of the endothelial adhesion molecules, which are involved in atherogenesis and may be key mediators of atherosclerosis (23,24). The cellular mRNA expression levels of VCAM-1 and ICAM-1 were upregulated by ox-LDL (Fig. 2A), whereas IRF2BP2 overexpression significantly reduced the ox-LDL-induced upregulation of both VCAM-1 and ICAM-1. Additionally, the expression of the anti-inflammatory transcription factor KLF2 was reduced in HUVECs exposed to ox-LDL. This effect was reversed following IRF2BP2 overexpression (Fig. 2B). It has been reported that the activity of eNOS maintains endothelial function (25). NO, produced by eNOS, has a profound effect on angiogenesis, vascular remodeling and endothelial permeability. Therefore, a decrease in NO release is a hallmark of endothelial cell dysfunction (24,26). In the present study, the phosphorylation of eNOS at Ser1177 was reduced after stimulation of HUVECs with ox-LDL. IRF2BP2-overexpressing cells exhibited higher eNOS phosphorylation levels compared with the empty vector group in the presence of ox-LDL (Fig. 2B). Furthermore, NO release in the ox-LDL treated group was significantly reduced, whereas IRF2BP2 overexpression partly restored its production (Fig. 2C). The expression of KLF2 was determined using immunofluorescence analysis (Fig. 2D), and the results were consistent with those of the western blot analysis.
Figure 2.
IRF2BP2 increases the expression of KLF2 and improves endothelial function. (A) The expression levels of ICAM-1 and VCAM-1 were determined using reverse transcription-quantitative PCR analysis. (B) The protein expression levels of KLF2, p-eNOS, total eNOS and GAPDH were measured using western blot analysis. (C) A Nitrate/Nitrite assay kit was utilized to estimate the release of NO. (D) The expression of KLF2 was evaluated by immunofluorescence analysis (magnification, x200). Scale bar, 50 µm. **P<0.01, ***P<0.001 vs. the control group; ##P<0.05, ###P<0.001 vs. the ox-LDL + vector group. IRF2BP2, interferon regulatory factor binding protein 2; KLF2, Krüppel-like factor 2; ICAM-1, intercellular adhesion molecule 1; VCAM-1, vascular cell adhesion molecule 1; p-eNOS, phosphorylated endothelial nitric oxide synthase; NO, nitric oxide; ox-LDL, oxidized low-density lipoprotein.
IRF2BP2 attenuates inflammation and EMT via KLF2 expression
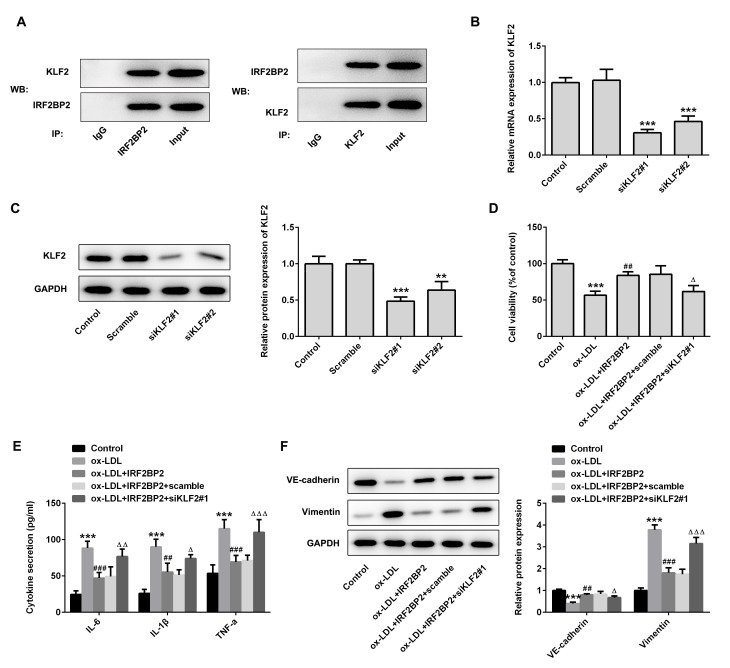
The interaction between IRF2BP2 and KLF2 was verified using Co-IP (Fig. 3A). Subsequently, two pairs of siRNAs targeting KLF2 were transfected into HUVECs, and the transfection efficiency is shown in Fig. 3B and C. The siKLF2#1 clone exhibited better knockdown of KLF2 expression compared with siKLF2#2. Cell viability assays revealed that siKLF2#1 could reduce the viability of IRF2BP2-overexpressing HUVECs following treatment with ox-LDL (Fig. 3D). Furthermore, the IRF2BP2 overexpression-mediated reduction in secretion of TNF-α, IL-1β and IL-6 were reversed following transfection with siKLF2#1 in the presence of ox-LDL (Fig. 3E). Additionally, siKLF2#1 abrogated the inhibitory effect of IRF2BP2 overexpression on EMT (Fig. 3F).
Figure 3.
IRF2BP2 attenuates inflammation and EMT via KLF2. (A) The interaction between IRF2BP2 and KLF2 was confirmed using a Co-IP assay. (B) HUVECs were transfected with scrambled siRNA, siKLF2#1 or siKLF2#2. The mRNA expression levels of KLF2 were measured using reverse transcription-quantitative PCR analysis. ***P<0.001 vs. the scramble group. (C) The protein expression levels of KLF2 were evaluated using western blot analysis. **P<0.01, ***P<0.001 vs. the scrambled group. (D) The cell viability in different groups was determined using a CCK-8 assay. (E) The levels of TNF-α, IL-1β and IL-6 in the culture medium were assessed using ELISA kits. (F) The protein expression levels of VE-cadherin and vimentin were evaluated using western blot analysis. ***P<0.001 vs. the control group; ##P<0.05, ###P<0.001 vs. the ox-LDL group; ΔP<0.05, ΔΔP<0.01, ΔΔΔP<0.001 vs. the ox-LDL + IRF2BP2 + scramble group. IRF2BP2, interferon regulatory factor binding protein 2; KLF2, Krüppel-like factor 2; EMT, endothelial-to-mesenchymal transition; HUVECs, human umbilical vein endothelial cells; siRNA, small interfering RNA; TNF-α, tumor necrosis factor α; IL-1β, interleukin 1β; ox-LDL, oxidized low-density lipoprotein; Co-IP, co-immunoprecipitation.
KLF2 is required for the effects of IRF2BP2 on endothelial cells
Subsequently, the role of KLF2 in ox-LDL-regulated endothelial dysfunction were determined. The mRNA expression levels of ICAM-1 and VCAM-1 were increased in ox-LDL-induced HUVECs. IRF2BP2 overexpression repressed the expression of both ICAM-1 and VCAM-1, which was restored following KLF2 knockdown (Fig. 4A). Additionally, the phosphorylation levels of eNOS and the production of NO were also evaluated. As shown in Fig. 4B and C, ox-LDL reduced the levels of p-eNOS and NO. IRF2BP2 overexpression significantly increased the levels of p-eNOS and NO, whereas transfection with siKLF2#1 counteracted the effects of IRF2BP2.
Figure 4.
KLF2 is required for IRF2BP2-regulated endothelial function. (A) The mRNA expression levels of ICAM-1 and VCAM-1 were measured by reverse transcription-quantitative PCR analysis. (B) The protein expression levels of total eNOS, p-eNOS and GAPDH were evaluated using western blot analysis. (C) A Nitrate/Nitrite assay kit was used to estimate the release of NO. ***P<0.001 vs. the control group; ##P<0.05, ###P<0.001 vs. the ox-LDL group; ΔP<0.05 vs. the ox-LDL + IRF2BP2 + scrambled group. KLF2, Krüppel-like factor 2; IRF2BP2, interferon regulatory factor binding protein 2; ICAM-1, intercellular adhesion molecule 1; VCAM-1, vascular cell adhesion molecule 1; p-eNOS, phosphorylated endothelial nitric oxide synthase; NO, nitric oxide; ox-LDL, oxidized low-density lipoprotein.
Discussion
Atherosclerosis is the most common disease of the cardiovascular system (27). Coronary heart disease, stroke and other cardiovascular diseases caused by atherosclerosis are significant causes of death worldwide (28). During the pathogenesis of atherosclerosis, endothelial cells undergo a series of pathological changes, from early dysfunction to advanced plaque transition (29). Ox-LDL triggers inflammation and EMT of endothelial cells, and facilitates the formation of atherosclerotic plaques (30,31). In the present study, treatment with ox-LDL reduced the viability of HUVECs, and IRF2BP2 overexpression ameliorated ox-LDL-induced cell injury. Furthermore, ox-LDL-induced inflammation and EMT were also impeded following IRF2BP2 overexpression.
EMT of endothelial cells promotes the expression of VCAM-1 and ICAM-1, which are key mediators of atherosclerosis, thereby recruiting monocytes to the arterial intima (32). Endothelial dysfunction usually refers to endothelium-dependent vasodilation and constriction impairment. Emerging evidence has suggested that NO, produced by eNOS, regulates vasomotor tone (29). In the present study, treatment of HUVECs with ox-LDL inhibited the activity of eNOS and reduced the synthesis of NO, and these effects were reversed following IRF2BP2 overexpression. Of note, IRF2BP2 overexpression also increased the protein expression levels of KLF2. The direct interaction between IRF2BP2 and KLF2 was also verified by Co-IP. Chen et al (14) demonstrated that IRF2BP2 could bind to the promoter of KLF2, resulting in the upregulation of KLF2 expression. Kim et al (33) also reported that IRF2BP2 overexpression increased KLF2 expression, suggesting that IRF2BP2 acts upstream of KLF2 in bone cells. As an anti-atherogenic factor in the vascular endothelium, KLF2 improves the endothelial functions and suppresses the development of atherosclerosis via regulating multiple pathways, including the NF-κB, MAPK/ERK, JNK and p38 signaling pathways (34-36). Therefore, it is hypothesized that IRF2BP2 may exert its effect through binding to KLF2 and positively regulating KLF2 expression, thereby affecting the downstream signaling transduction of KLF2; however, this requires further study. In accordance with this hypothesis, KLF2 knockdown exacerbated cell injury, inflammation and EMT. A recent report demonstrated that KLF2 orchestrates its effects on endothelial function via the eNOS/NO signaling pathway (17). In the present study, KLF2 knockdown attenuated the phosphorylation of eNOS and the generation of NO.
In summary, the present study showed that IRF2BP2 overexpression could protect HUVECs from ox-LDL-induced inflammation and EMT, whilst also increasing NO production in endothelial cells by enhancing the expression of KLF2. These findings reflect the role of IRF2BP2/KLF2 in endothelial cells and may improve our understanding of the pathogenesis of atherosclerosis, and may also highlight potentially novel targets for management of this disease. However, further in-depth studies are required to elucidate the mechanisms underlying endothelium-dependent vasodilation and vasoconstriction, and the vulnerability of arterial plaques.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
QS conceived and designed the current study. QS and YJ contributed to the acquisition of data. YJ analyzed and interpreted the data. QS drafted the initial manuscript and revised it critically for important intellectual content. QS and YJ confirm the authenticity of all the raw data. Both authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Piepoli MF, Hoes AW, Agewall S, Albus C, Brotons C, Catapano AL, Cooney MT, Corrà U, Cosyns B, Deaton C, et al. 2016 European Guidelines on cardiovascular disease prevention in clinical practice: The Sixth Joint Task Force of the European Society of Cardiology and Other Societies on Cardiovascular Disease Prevention in Clinical Practice (constituted by representatives of 10 societies and by invited experts)Developed with the special contribution of the European Association for Cardiovascular Prevention & Rehabilitation (EACPR) Eur Heart J. 2016;37:2315–2381. doi: 10.1093/eurheartj/ehw106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mauricio MD, Aldasoro M, Ortega J, Vila JM. Endothelial dysfunction in morbid obesity. Curr Pharm Des. 2013;19:5718–5729. doi: 10.2174/1381612811319320007. [DOI] [PubMed] [Google Scholar]
- 3.Vanhoutte PM, Shimokawa H, Tang EH, Feletou M. Endothelial dysfunction and vascular disease. Acta Physiol (Oxf) 2009;196:193–222. doi: 10.1111/j.1748-1716.2009.01964.x. [DOI] [PubMed] [Google Scholar]
- 4.Molinaro R, Boada C, Del Rosal GM, Hartman KA, Corbo C, Andrews ED, Toledano-Furman NE, Cooke JP, Tasciotti E. Vascular inflammation: A novel access route for nanomedicine. Methodist Debakey Cardiovasc J. 2016;12:169–174. doi: 10.14797/mdcj-12-3-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seneviratne AN, Monaco C. Role of inflammatory cells and toll-like receptors in atherosclerosis. Curr Vasc Pharmacol. 2015;13:146–160. doi: 10.2174/15701611113116660160. [DOI] [PubMed] [Google Scholar]
- 6.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 7.Packard CJ. LDL cholesterol: How low to go? Trends Cardiovasc Med. 2018;28:348–354. doi: 10.1016/j.tcm.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 8.Bäck M, Hansson GK. Anti-inflammatory therapies for atherosclerosis. Nat Rev Cardiol. 2015;12:199–211. doi: 10.1038/nrcardio.2015.5. [DOI] [PubMed] [Google Scholar]
- 9.Childs KS, Goodbourn S. Identification of novel co-repressor molecules for interferon regulatory factor-2. Nucleic Acids Res. 2003;31:3016–3026. doi: 10.1093/nar/gkg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carneiro FR, Ramalho-Oliveira R, Mognol GP, Viola JP. Interferon regulatory factor 2 binding protein 2 is a new NFAT1 partner and represses its transcriptional activity. Mol Cell Biol. 2011;31:2889–2901. doi: 10.1128/MCB.00974-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stadhouders R, Cico A, Stephen T, Thongjuea S, Kolovos P, Baymaz HI, Yu X, Demmers J, Bezstarosti K, Maas A, et al. Control of developmentally primed erythroid genes by combinatorial co-repressor actions. Nat Commun. 2015;6(8893) doi: 10.1038/ncomms9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li T, Luo Q, He L, Li D, Li Q, Wang C, Xie J, Yi C. Interferon regulatory factor-2 binding protein 2 ameliorates sepsis-induced cardiomyopathy via AMPK-mediated anti-inflammation and anti-apoptosis. Inflammation. 2020;43:1464–1475. doi: 10.1007/s10753-020-01224-x. [DOI] [PubMed] [Google Scholar]
- 13.Ma YL, Xia JL, Gao X. Suppressing Irf2bp2 expressions accelerates metabolic syndrome-associated brain injury and hepatic dyslipidemia. Biochem Biophys Res Commun. 2018;503:1651–1658. doi: 10.1016/j.bbrc.2018.07.095. [DOI] [PubMed] [Google Scholar]
- 14.Chen HH, Keyhanian K, Zhou X, Vilmundarson RO, Almontashiri NA, Cruz SA, Pandey NR, Lerma Yap N, Ho T, Stewart CA, et al. IRF2BP2 reduces macrophage inflammation and susceptibility to atherosclerosis. Circ Res. 2015;117:671–683. doi: 10.1161/CIRCRESAHA.114.305777. [DOI] [PubMed] [Google Scholar]
- 15.Jha P, Das H. KLF2 in regulation of NF-κB-mediated immune cell function and inflammation. Int J Mol Sci. 2017;18(2383) doi: 10.3390/ijms18112383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Z, Tang AT, Wong WY, Bamezai S, Goddard LM, Shenkar R, Zhou S, Yang J, Wright AC, Foley M, et al. Cerebral cavernous malformations arise from endothelial gain of MEKK3-KLF2/4 signalling. Nature. 2016;532:122–126. doi: 10.1038/nature17178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu W, Geng P, Zhu J, Li JW, Zhang L, Chen WL, Zhang DF, Lu Y, Xu XH. KLF2 regulates eNOS uncoupling via Nrf2/HO-1 in endothelial cells under hypoxia and reoxygenation. Chem Biol Interact. 2019;305:105–111. doi: 10.1016/j.cbi.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 18.Das H, Kumar A, Lin Z, Patino WD, Hwang PM, Feinberg MW, Majumder PK, Jain MK. Kruppel-like factor 2 (KLF2) regulates proinflammatory activation of monocytes. Proc Natl Acad Sci USA. 2006;103:6653–6658. doi: 10.1073/pnas.0508235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhuang T, Liu J, Chen X, Zhang L, Pi J, Sun H, Li L, Bauer R, Wang H, Yu Z, et al. Endothelial Foxp1 suppresses atherosclerosis via modulation of Nlrp3 inflammasome activation. Circ Res. 2019;125:590–605. doi: 10.1161/CIRCRESAHA.118.314402. [DOI] [PubMed] [Google Scholar]
- 20.Zhao D, Sun X, Lv S, Sun M, Guo H, Zhai Y, Wang Z, Dai P, Zheng L, Ye M, Wang X. Salidroside attenuates oxidized low-density lipoprotein-induced endothelial cell injury via promotion of the AMPK/SIRT1 pathway. Int J Mol Med. 2019;43:2279–2290. doi: 10.3892/ijmm.2019.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee WJ, Ou HC, Hsu WC, Chou MM, Tseng JJ, Hsu SL, Tsai KL, Sheu WHH. Ellagic acid inhibits oxidized LDL-mediated LOX-1 expression, ROS generation, and inflammation in human endothelial cells. J Vasc Surg. 2010;52:1290–1300. doi: 10.1016/j.jvs.2010.04.085. [DOI] [PubMed] [Google Scholar]
- 22.Li H, Zhao Q, Chang L, Wei C, Bei H, Yin Y, Chen M, Wang H, Liang J, Wu Y. LncRNA MALAT1 modulates ox-LDL induced EndMT through the Wnt/β-catenin signaling pathway. Lipids Health Dis. 2019;18(62) doi: 10.1186/s12944-019-1006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cybulsky MI, Iiyama K, Li H, Zhu S, Chen M, Iiyama M, Davis V, Gutierrez-Ramos JC, Connelly PW, Milstone DS. A major role for VCAM-1, but not ICAM-1, in early atherosclerosis. J Clin Invest. 2001;107:1255–1262. doi: 10.1172/JCI11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galkina E, Ley K. Vascular adhesion molecules in atherosclerosis. Arterioscler Thromb Vasc Biol. 2007;27:2292–2301. doi: 10.1161/ATVBAHA.107.149179. [DOI] [PubMed] [Google Scholar]
- 25.Huang PL. Endothelial nitric oxide synthase and endothelial dysfunction. Curr Hypertens Rep. 2003;5:473–480. doi: 10.1007/s11906-003-0055-4. [DOI] [PubMed] [Google Scholar]
- 26.Lyons D. Impairment and restoration of nitric oxide-dependent vasodilation in cardiovascular disease. Int J Cardiol. 1997;62 (Suppl 2):S101–S109. doi: 10.1016/s0167-5273(97)00247-7. [DOI] [PubMed] [Google Scholar]
- 27.Skuratovskaia D, Vulf M, Komar A, Kirienkova E, Litvinova L. Promising directions in atherosclerosis treatment based on epigenetic regulation using MicroRNAs and long noncoding RNAs. Biomolecules. 2019;9(226) doi: 10.3390/biom9060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y, Evans MA, Allison MA, Bertoni AG, Budoff MJ, Criqui MH, Malik S, Ouyang P, Polak JF, Wong ND. Multisite atherosclerosis in subjects with metabolic syndrome and diabetes and relation to cardiovascular events: The multi-ethnic study of atherosclerosis. Atherosclerosis. 2019;282:202–209. doi: 10.1016/j.atherosclerosis.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gimbrone MA Jr, García-Cardeña G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ Res. 2016;118:620–636. doi: 10.1161/CIRCRESAHA.115.306301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Mu Q, Zhou Z, Song H, Zhang Y, Wu F, Jiang M, Wang F, Zhang W, Li L, et al. Protective effect of irisin on atherosclerosis via suppressing oxidized low density lipoprotein induced vascular inflammation and endothelial dysfunction. PLoS One. 2016;11(e0158038) doi: 10.1371/journal.pone.0158038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H, Mohamed AS, Zhou SH. Oxidized low density lipoprotein, stem cells, and atherosclerosis. Lipids Health Dis. 2012;11(85) doi: 10.1186/1476-511X-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen PY, Qin L, Baeyens N, Li G, Afolabi T, Budatha M, Tellides G, Schwartz MA, Simons M. Endothelial-to-mesenchymal transition drives atherosclerosis progression. J Clin Invest. 2015;125:4514–4528. doi: 10.1172/JCI82719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim I, Kim JH, Kim K, Seong S, Kim N. The IRF2BP2-KLF2 axis regulates osteoclast and osteoblast differentiation. BMB Rep. 2019;52:469–474. doi: 10.5483/BMBRep.2019.52.7.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atkins GB, Wang Y, Mahabeleshwar GH, Shi H, Gao H, Kawanami D, Natesan V, Lin Z, Simon DI, Jain MK. Hemizygous deficiency of Krüppel-like factor 2 augments experimental atherosclerosis. Circ Res. 2008;103:690–693. doi: 10.1161/CIRCRESAHA.108.184663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Y, Liu P, Xu S, Koroleva M, Zhang S, Si S, Jin ZG. Tannic acid as a plant-derived polyphenol exerts vasoprotection via enhancing KLF2 expression in endothelial cells. Sci Rep. 2017;7(6686) doi: 10.1038/s41598-017-06803-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novodvorsky P, Chico TJ. The role of the transcription factor KLF2 in vascular development and disease. Prog Mol Biol Transl Sci. 2014;124:155–188. doi: 10.1016/B978-0-12-386930-2.00007-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.