Graphical abstract
Keywords: SARS-CoV-2, Nucleocapsid gene, Indel mutation, Viral genome sequencing, Different geographical origins
Abstract
Commercially available reverse transcription-polymerase chain reaction (RT-PCR) kits are being used as an important tool to diagnose SARS-CoV-2 infection in clinical laboratories worldwide. However, some kits lack sufficient clinical evaluation due to the need for emergency use caused by the current COVID-19 pandemic. Here we found that a novel insertion/deletion mutation in the nucleocapsid (N) gene of SARS-CoV-2 samples is a cause of negative results for the N gene in a widely used assay that received emergency use authorization (EUA) from US FDA and Conformite Europeenne-in vitro diagnostics (CE-IVD) from EU. Although SARS-CoV-2 is diagnosed positive by other target probes in the assay, our findings provide an evidence of the genetic variability and rapid evolution of SARS-CoV-2 as well as a reference in designing commercial RT-PCR assays.
RT-PCR assays, which have received EUA from the united states (US) Food and Drug Administration (FDA) and/or CE-IVD mark from European Union (EU) and/or EUA from regulatory agencies at their country, are being used to diagnose severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) under the coronavirus disease (COVID-19) pandemic. Currently, clinical performances of existing commercially available assays are limited, and it remain unclear whether it is closely associated with genetic variability of SARS-CoV-2. Recent study has shown that numerous mutations occur on the SARS-CoV-2 diagnostic targets being widely used worldwide (Wang et al., 2020a, 2020b). For this reason, false negative or presumptive-positive results are often observed when mutations occur in the viral target genes, including the N gene (Ziegler et al., 2020; Angeletti et al., 2020; Su et al., 2020; Farfour et al., 2020). Therefore, efforts are being made to find reference sequences for accurately diagnosing the virus through variation analysis of SARS-CoV-2 (Wang et al., 2020a, 2020b). Therefore, it is needed to find reference sequences for accurately diagnosing SARS-CoV-2 through the viral variation analysis. Here, we tested clinical performance of a commercial RT-PCR assay kit targeting the E, RdRp, and N genes using the viral isolates from different geographical origins, and then performed the viral genome sequencing to identify whether mutations occurred in the samples in which only the N gene was not detected in the assay.
A total of 18 nasopharyngeal samples isolated from 3 Finns, 10 North Americans, and 5 Koreans who tested positive by RT-PCR at Seoul Clinical Laboratories (SCL) in South Korea were collected from March to July 2020. Finnish samples originate from those sent directly from Finland to SCL via Finnair flight for the SARS-CoV-2 test, and North American samples originate from a US military hospital located in Korea to SCL for the viral test. Of the 10 American samples, 3 were collected in April and the remaining 7 were collected in July. Three samples from Finn and 5 from Korean were collected in April and March, respectively. This study was approved by the Institutional Review Board of Seoul Clinical Laboratories (SCL-IRB-20−076). Written informed consent was waived by the Institutional Ethical Review Board because all personal identifiers were removed in the whole study process and only samples that were tested positive in RT-PCR diagnostic test performed at the SCL in South Korea were used.
Extraction of SARS-CoV-2 RNA from nasopharyngeal samples was performed using MagNA Pure 96 DNA and Viral NA Small Volume Kit (Roche Diagnostics, Mannheim, Germany). All samples used in this study were tested positive by Allplex 2019-nCoV (APX) assay targeting the E, RdRp, and N genes (Seegene Inc., Seoul, South Korea) that has received EUA from US FDA and Korea MFDS, and CE-IVD from EU, which was conducted at SCL in Korea. All samples tested with the APX assay were detected positive with Ct values between 9.0 and 33.0 for the E gene, and between 11.0 and 34.0 for the RdRp gene, and between 15.0 and 37.0 for the N gene, except for the 6 samples tested negative for only the N gene (Table 1 ).
Table 1.
Primary RT-PCR results in the APX assay, and comparison with the DPXQ assay.
| No. of samples (origins/collection date)a | Allplex 2019-nCov Assay Kit |
DiaPlexQ Novel 2019-nCoV Detection Kit | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| E gene |
RdRp gene |
N gene |
ICc |
Result | Result | |||||
| Result | Ct | Result | Ct | Result | Ct | Result | Ct | |||
| #1 (US, Apr 2020) | Positive | 32.15 | Positive | 33.89 | Positive | 36.45 | Positive | 24.47 | Positive | Positive |
| #2 (US, Apr 2020) | Positive | 17.1 | Positive | 18.18 | Positive | 20.38 | Negative | 28.96 | Positive | Positive |
| #3 (US, Apr 2020) | Positive | 24.53 | Positive | 25.9 | Positive | 27.2 | Positive | 25.24 | Positive | Positive |
| #4 (FI, Apr 2020) | Positive | 18.55 | Positive | 19.97 | Positive | 21.99 | Positive | 22.02 | Positive | Positive |
| #5 (FI, Apr 2020) | Positive | 28.94 | Positive | 31.88 | Positive | 32.78 | Positive | 23.38 | Positive | Positive |
| #6 (FI, Apr 2020) | Positive | 26.36 | Positive | 27.90 | Positive | 29.18 | Positive | 23.67 | Positive | Positive |
| #7 (KR, Mar 2020) | Positive | 25.25 | Positive | 27.35 | Positive | 27.84 | Positive | 23.30 | Positive | Positive |
| #8 (KR, Mar 2020) | Positive | 30.94 | Positive | 31.76 | Positive | 35.54 | Positive | 25.09 | Positive | Positive |
| #9 (KR, Mar 2020) | Positive | 13.11 | Positive | 13.63 | Positive | 15.99 | Negative | 32.57 | Positive | Positive |
| #10 (KR, Mar 2020) | Positive | 27.0 | Positive | 28.33 | Positive | 29.37 | Positive | 25.25 | Positive | Positive |
| #11 (KR, Mar 2020) | Positive | 15.89 | Positive | 17.49 | Positive | 19.91 | Positive | 32.83 | Positive | Positive |
| #12 (US, Apr 2020) | Positive | 29.51 | Positive | 30.46 | Positive | 30.95 | Positive | 26.3 | Positive | Positive |
| #13 (US, Apr 2020) | Positive | 11.58 | Positive | 12.94 | Negative | – | Positive | 38.85 | Positiveb | Positive |
| #14 (US, Apr 2020) | Positive | 9.55 | Positive | 11.47 | Negative | – | Negative | – | Positiveb | Positive |
| #15 (US, Apr 2020) | Positive | 13.46 | Positive | 15.55 | Negative | – | Negative | – | Positiveb | Positive |
| #16 (US, Apr 2020) | Positive | 13.31 | Positive | 15.36 | Negative | – | Positive | 39.21 | Positiveb | Positive |
| #17 (US, Apr 2020) | Positive | 20.03 | Positive | 21.31 | Negative | – | Positive | 24.00 | Positiveb | Positive |
| #18 (US, Apr 2020) | Positive | 12.23 | Positive | 14.37 | Negative | – | Negative | – | Positiveb | Positive |
US, FI, and KR indicate North America, Finland, and Korea, respectively.
According to the manufacturer’s recommendations, these profiles were considered positive.
Internal control.
The viral whole-genome sequencing (WGS) was performed using next-generation sequencing (NGS) to identify whether there are mutations in the N gene of samples that were tested negative in the APX assay. The cDNA libraries were constructed using an Illumina RNA Prep with Enrichment with Illumina Respiratory Virus Oligos Panel v2 (Illumina, USA). Preparation steps were confirmed by Agilent TapeStation 4200 system (Agilent, USA). The genome sequencing was performed on an Illumina Miseq system (Illumina Inc., San Diego, USA) according to the manufacturer’s protocols using a 300 cycle MiSeq Reagent Kit v3 (Illumina, USA). Sequence reads with an average Phred quality score of less than 30 were removed and the remaining reads were used for the viral sequence analysis.
Sequence data were assembled to each viral sequence using customized version of the DRAGEN RNA pipeline (Illumina-Edico Genome Inc., San Diego, USA). DRAGEN RNA Pathogen Detection App v3.5.7 using a combined human (gh38) with SARS-CoV-2 reference genome (NC_045512.2) was used to create consensus sequences. Assembled viral sequences were used as mutational characteristics analysis of the singularity of sequence and then were compared with the samples tested positive for 2 or 3 targets, through the MegaX program (Kumar et al., 2018).
For cross-validation and confirmatory test, all samples were reanalyzed with the DiaPlexQ Novel Coronavirus (2019-nCoV) (DPXQ) Detection Kit (Solgent Inc., Daejeon, South Korea) targeting the ORF1a and N genes of SARS-CoV-2 that has received EUA from US FDA and Korea MFDS, and CE-IVD from EU, which showed positive results for all samples including the 6 samples tested negative for only the N gene, indicating an overall agreement of 100 % between the APC and DPXQ assays (Table 1).
Based on the result of the cross-validation and confirmatory test by the DPXQ assay, we assumed that there may be potential mutation that interferes with a diagnostic probe that bind to the N gene region in the APX assay. In the APX assay, the 6 samples that were tested negative for the N gene were those originated from North Americans in July 2020 (Table 1 and Fig. 1 ).
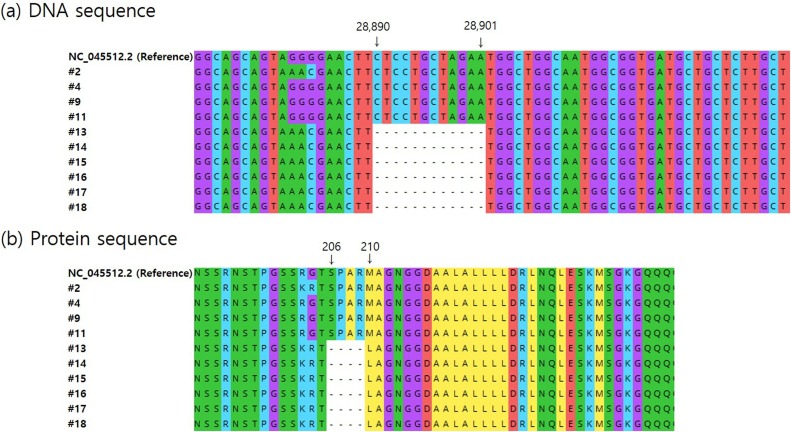
Fig. 1.
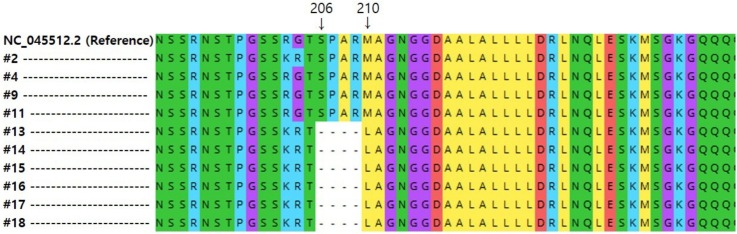
Genomic mutation analysis in the N gene region of SARS-CoV-2 strains from different geographical origins. (a) Novel 12-nt deletions in N gene region of viral strains from North Americans (#13 to #18). (b) 5 amino acid deletions from 206 to 210 positions and insertion of leucine at position 210 (p.Ser206_Met210delinsLeu). Raw sequence reads were trimmed for removal of low-quality bases and for length and assembled by mapping to the reference genome (GenBank accession number: NC_045512.2) from Wuhan, China. Among all samples shown in Table 1, only the 10 strains (7 strains from North American (#2 and #13 to #18), 1 from Finnish (#4), and 2 from Korean (#9 and #11) that were successfully analyzed by WGS.
To accurately elucidate negative results for the N gene in the assay, we performed the viral WGS with a focus on the analysis of the N gene region for all samples used in this study. Of the samples (n = 18) tested positive in the APX assay, only the 10 strains (7 out of 10 North American samples, 1 out of 3 Finnish, and 2 out of 5 Korean) extracted from nasopharyngeal samples without any viral cultivation were successfully analyzed by WGS. On the other hand, WGS data of the remaining 8 samples were not obtained due to the lack of sufficient read depth of analyzed samples. The viral sequencing data with a focus of the analysis of the N gene region revealed that 7 samples (#2 and #13 to #18) originated from North Americans had a substitution mutation at nucleotide (nt) positions 28881–28883 (GGG > AAC). This nt mutation caused 2 nonsynonymous amino acid substitutions (R203 K and G204R) (Fig. 1a). These mutations were not detected in the genomes of strains isolated from Koreans (#9, #11) collected in March 2020, and each strain isolated from Finnish (#4) and North Americans (#2) collected in April 2020. Interestingly, we also found 12 nt deletions at nt positions 28890–28901 of the viral genomes of 6 North Americans isolates (#13 to #18) collected in July 2020. The 6 samples harbored 5 amino acid deletions from 206 to 210 and a leucine insertion at amino acid position 210 (Fig. 1b).
To identify if this insertion/deletion (indel) mutation has been previously reported, we further investigated publicly available genomes from the GISAID (http://gisaid.org), Nextstrain (www.nextstrain.org), and NCBI virus repository (http://www.ncbi.nlm.nih.gov/labs/virus) as of January 15, 2021. Analysis of N gene regions of the publicly available viral genomes revealed that the indel mutation identified in this study is a novel type of variation that has not been previously reported.
The APX RT-PCR assay is being widely used to diagnose SARS-CoV-2 infection in European countries including France (Farfour et al., 2020) and Germany (Ziegler et al., 2020), and in North America (Carter et al., 2020) and Korea (Sung et al., 2020). In this study, we identified the indel mutation as well as the nonsynonymous mutations in the N gene of SARS-CoV-2 isolates that were tested negative for only the viral N gene in the assay. The indel and nonsynonymous mutations were detected in only 6 strains isolated from North Americans samples that were collected from April 2020 to July 2020. In contrast, these mutations were not detected in the genomes of strains isolated from 2 Koreans and 1 Finnish, suggesting that the genotype and phenotype of SARS-CoV-2 may vary depending on a specific population. Although the 2 nonsynonymous amino acid substitutions (R203 K and G204R) have been frequently detected in other populations and other geographic origins (Badua et al., 2020; Mercatelli and Giorgi, 2020; Peng et al., 2020; Rahimi et al., 2020), the indel mutation identified in this study is a novel type of mutation that has not been previously reported. Our results suggest that the novel indel mutation in the viral N gene and its corresponding protein region may result from adapting to a newer geographical region and a specific population. Furthermore, our viral WGS analysis strongly suggests that the novel indel mutation in the viral N gene may be a cause of negative results for the viral N gene in the APX assay. SARS-CoV-2 may mutate in one or more of the target regions of the APX assay. If mutation occurs, the virus may not be detected. For this reason, performance of the APX assay has been often affected by genetic mutations that interfere with the binding of the probe in the target N gene region (Farfour et al., 2020).
The N protein involved in the viral assembly, replication, and regulation of host immune response is a major structural component of SARS-CoV-2 and plays important roles in the viral life cycle. These characteristics make the N protein an important target for the viral diagnosis and vaccine development (Peng et al., 2020; Dutta et al., 2020). A recent study revealed that the N protein has the highest mutation density among the viral structural proteins (Badua et al., 2020). It has been also found that SARS-CoV-2 has the most mutations on the targets of diverse N gene primers and probes being extensively used to diagnose the viral infection, and the N gene in the SARS-CoV-2 genome is one of the most non-conservative genes, as evidenced by the calculated mutation rate and mutation h-index value of the SARS-CoV-2 genomes (Wang et al., 2020a, 2020b).
Insertions are known to be a very rare type of mutation, which account for less than 0.1 % of detected SARS-CoV-2 mutation cases, whereas in-frame deletions that reduce the length of the viral N protein without using stop codons account for about 0.6 % of detected viral mutation cases (Mercatelli and Giorgi, 2020). Through the analysis of the publicly available viral genomes from the Nextstrain SARS-CoV-2 resources, we also identified that the novel 12 nt deletions at positions 28890–28901 are located at variable region of the viral N gene (Nextstrain SARS-CoV-2 resources, 2021).
Our study has some limitations. First, functional study of the novel indel mutation was not performed. Second, epidemiological information of all SARS-CoV-2 samples used in this study was lacking. Third, the number of the viral samples collected from different geographic origins was small, and WGS data for some of the samples collected for this study were not obtained because the viral nucleic acids extracted from nasopharyngeal samples without any cell culture were directly analyzed by the viral WGS. For these reasons, studies that further investigate frequency of the novel indel mutation worldwide will be of significance to fully understand its effect on function of the mutation.
Given that the APX assay is being used worldwide, it will be necessary to elucidate negative results resulting from mutations in the target genes of the assay, particularly in the N gene, through the viral genome analysis. Our study underlines that primers and probes used in commercial assays including the APX assay should be designed to target the most conservative regions of SARS-CoV-2 genome for a reliable diagnosis. Our findings can also help to better understand the viral pathogenesis and evolution during the current COVID-19 pandemic.
Authors’ contributions
SL & KRL conceived and coordinated the study. SL, DJW, CKK, and KRL shaped up the study design. SL, DJW, CKK, HN, YTK, and HSL performed the experiments from viral RNA extraction to WGS. SL, YL, DJW, JA, JRC, and MKL analyzed the sequences of the publicly available SARS-CoV-2 genomes. SL wrote the manuscript together with JA and KRL. SL and KRL edited the final manuscript. All authors approved the final version of the manuscript.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgment
We thank molecular diagnostics department at SCL for fully supporting the SARS-CoV-2 diagnostic test.
References
- Angeletti S., Benvenuto D., Bianchi M., Giovanetti M., Pascarella S., Ciccozzi M. COVID-19: the role of the nsp2 and nsp3 in its pathogenesis. J. Med. Virol. 2020;92:584–588. doi: 10.1002/jmv.25719. PMID: 32083328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badua C.L.D.C., Baldo K.A.T., Medina P.M.B. Genomic and proteomic mutation landscapes of SARS-CoV-2. J. Med. Virol. 2020:1–20. doi: 10.1002/jmv.26548. PMID: 32970329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta N.K., Mazumdar K., Gordy J.T. The nucleocapsid protein of SARS-CoV-2: a target for vaccine development. J. Virol. 2020;94(13):e00647–20. doi: 10.1128/JVI.00647-20. PMID: 32546606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farfour E., Lesprit P., Visseaux B., Pascreau T., Jolly E., Houhou N., Mazaux L., Asso-Bonnet M., Vasse M. SARS-CoV-2 Foch Hospital study group, The Allplex 2019-nCoV (Seegene) assay: which performances are for SARS-CoV-2 infection diagnosis? Eur. J. Clin. Microbiol. Infect. Dis. 2020;39(10):1997–2000. doi: 10.1007/s10096-020-03930-8. PMID: 32462501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K., Mega X. Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. PMID: 29722887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 2020;11(1800) doi: 10.3389/fmicb.2020.01800. PMID: 32793182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nextstrain SARS-CoV-2 resources Genomic Epidemiology of Novel Coronavirus. https://nextstrain.org/ncov/global?gmin=28577 [Accessed: Jan 15, 2021]. Available from:
- Peng Y., Du N., Lei Y., Dorje S., Qi J., Luo T., Gao G.F., Song H. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. EMBO J. 2020;39(20) doi: 10.15252/embj.2020105938. PMID: 32914439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi A., Mirzazadeh A., Tavakolpour S. Genetics and genomics of SARS-CoV-2: a review of the literature with the special focus on genetic diversity and SARS-CoV-2 genome detection. Genomics. 2020;113(1):1221–1232. doi: 10.1016/j.ygeno.2020.09.059. PMID: 33007398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y.C.F., Anderson D.E., Young B.E., Linster M., Zhu F., Jayakumar J., Zhuang Y., Kalimuddin S., Low J.G.H., Tan C.W., Chia W.N., Mak T.M., Octavia S., Chavatte J.M., Lee R.T.C., Pada S., Tan S.Y., Sun L., Yan G.Z., Maurer-Stroh S., Mendenhall I.H., Leo Y.S., Lye D.C., Wang L.F., Smith G.J.D. Discovery and genomic characterization of a 382-nucleotide deletion in ORF7b and ORF8 during the early evolution of SARS-CoV-2. mBio. 2020;11:e01610–20. doi: 10.1128/mBio.01610-20. PMID: 32694143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H., Roh K.H., Hong K.H., Seong M.W., Ryoo N., Kim H.S., Lee J., Kim S.Y., Ryu S.W., Kim M.N., Han M.G., Lee S.W., Lee H., Yoo C.K. COVID-19 molecular testing in Korea: practical essentials and answers from experts based on experiences of emergency use authorization assays. Ann. Lab. Med. 2020;40(6):439–447. doi: 10.3343/alm.2020.40.6.439. PMID: 32539299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu Z., Chen Z., Huang X., Xu M., He T., Zhang Z. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J. Med. Virol. 2020;92(6):667–674. doi: 10.1002/jmv.25762. PMID: 32167180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Hozumi Y., Wei G.W. Mutations on COVID-19 diagnostic targets. Genomics. 2020;112(6):5204–5213. doi: 10.1016/j.ygeno.2020.09.028. PMID: 32966857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler K., Steininger P., Ziegler R., Steinmann J., Korn K., Ensser A. SARS-CoV-2 samples may escape detection because of a single point mutation in the N gene. Euro Surveill. 2020;25(39) doi: 10.2807/1560-7917.ES.2020.25.39.2001650. pii=2001650 PMID: 33006300. [DOI] [PMC free article] [PubMed] [Google Scholar]