Abstract
Corona Virus Disease (COVID-19) has been announced as a pandemic and is spreading rapidly throughout the world. Early detection of COVID-19 may protect many infected people. Unfortunately, COVID-19 can be mistakenly diagnosed as pneumonia or lung cancer, which with fast spread in the chest cells, can lead to patient death. The most commonly used diagnosis methods for these three diseases are chest X-ray and computed tomography (CT) images. In this paper, a multi-classification deep learning model for diagnosing COVID-19, pneumonia, and lung cancer from a combination of chest x-ray and CT images is proposed. This combination has been used because chest X-ray is less powerful in the early stages of the disease, while a CT scan of the chest is useful even before symptoms appear, and CT can precisely detect the abnormal features that are identified in images. In addition, using these two types of images will increase the dataset size, which will increase the classification accuracy. To the best of our knowledge, no other deep learning model choosing between these diseases is found in the literature. In the present work, the performance of four architectures are considered, namely: VGG19-CNN, ResNet152V2, ResNet152V2 + Gated Recurrent Unit (GRU), and ResNet152V2 + Bidirectional GRU (Bi-GRU). A comprehensive evaluation of different deep learning architectures is provided using public digital chest x-ray and CT datasets with four classes (i.e., Normal, COVID-19, Pneumonia, and Lung cancer). From the results of the experiments, it was found that the VGG19 +CNN model outperforms the three other proposed models. The VGG19+CNN model achieved 98.05% accuracy (ACC), 98.05% recall, 98.43% precision, 99.5% specificity (SPC), 99.3% negative predictive value (NPV), 98.24% F1 score, 97.7% Matthew's correlation coefficient (MCC), and 99.66% area under the curve (AUC) based on X-ray and CT images.
Keywords: COVID-19 detection, Pneumonia, Lung cancer, Chest X-ray, CT images, Deep learning
Graphical abstract
1. Introduction
As 2019 ended, coronavirus disease, known as COVID-19, started proliferating all over the world and has created an alarming situation worldwide. The virus originated in Wuhan, a town in Eastern China, in December 2019. In 2020, it was declared by the World Health Organization (WHO) as a “Public health emergency of international concerns”, and by March 2020 they classified the disease as a pandemic [1]. The disease has affected about 118.7 million people around the world, and 2.6 million deaths were confirmed by March 2021. This virus causes pneumonia with other symptoms, such as fatigue, dry cough, and fever. One of the primary methods of testing coronavirus is reverse transcription polymerase China reaction (RT-PCR), which is performed on respiratory samples, and the testing results are produced within a few hours to two days. This method of detection is expensive and time-consuming [2]. Therefore, designing other methods for virus detection is currently an important challenge for researchers. Specifically, until now, there has been no definite medical treatment for COVID-19 [3].
Automating the diagnosis of many diseases nowadays has been based on artificial intelligence, which has proven its efficiency and high performance in automatic image classification problems through different machine learning approaches. Moreover, machine learning defines models that have the ability to learn and make decisions by using large amounts of input data examples. Artificial intelligence makes calculations and predictions based on analyzing the input data, then performs tasks that require human intelligence such as speech recognition, translation, visual perception, and more. Deep learning is a combination of machine learning methods that mostly focus on the automatic feature extraction and classification of images and have shown great achievement in many applications, especially in health care [4,5].
Deep learning efficiently generates models that produce more accurate results in predicting and classifying different diseases using images as in breast cancer [6], liver diseases [7], colon cancer [8], brain tumor [9], skin cancer [10], lung cancer [11], pneumonia [12], and recently COVID-19 diagnosis, without requiring any human intervention. The main reason for using deep learning is that deep learning techniques learn by creating a more abstract representation of data as the network grows deeper (not like classical machine learning). As a result, the model automatically extracts features and yields higher accuracy results. Unlike traditional machine learning algorithms, deep learning algorithms specify features through a series of nonlinear functions that are collated in a combinatorial fashion to maximize the model accuracy.
The literature has many researches about the use of deep learning as a classification model for COVID-19 using chest X-rays, as in Refs. [[13], [14], [15]] and using computed tomography (CT) images, as in Refs. [[16], [17], [18], [19], [20], [21], [22]]. Other work was interested in detecting and diagnosing COVID-19 based on lung datasets, as in Refs. [5,23]. Moreover, some studies applied convolutional neural networks (CNN) using limited datasets for classifying and detecting COVID-19 from chest X-ray images, as in Refs. [24,25]. Additionally, several studies have focused on detecting COVID-19 and distinguishing it from other chest diseases like pneumonia, as in Refs. [[26], [27], [28], [29], [30]]. Furthermore, authors in Ref. [31] demonstrated that a chest X-ray is less impetus in the initial stages, although a CT scan of the chest is useful even before symptoms appear. One of the problems associated with chest CT or X-ray images is the possible overlap between the diagnosis of COVID-19, pneumonia, and chest cancer, especially if the person diagnosing has little experience or the patient history file is not presently at hand. This necessitates the automation of such a process in a manner that can accurately confirm the existence of one of those three diseases. Up to now, there has been no classification model for classifying these three lung diseases, which encouraged us to introduce such model a in this paper.
To this end, and to benefit from the privileges introduced by deep learning approaches, this paper introduces a multi-classification model based on deep learning techniques for detecting COVID-19 from both chest X-ray and CT images. A combination of CT and X-ray images was used for two reasons: First, to increase the dataset size. Second, since chest X-ray is less impetus in the initial stages, although a CT scan of the chest is useful even before symptoms appear, the two types, CT and X-ray images, were used, and this can precisely detect the abnormal features that are identified in the images. The study provides a detailed description for each of the architectures, and through the results, the best of them is concluded, which can achieve superior detection accuracy. Furthermore, we provide a comprehensive evaluation of different deep learning architectures using public digital chest X-ray and CT datasets with four classes: Normal, COVID-19, Pneumonia, and Lung cancer.
The rest of this paper is organized as follows: Section 2 presents the recent related work regarding COVID-19, pneumonia, and lung cancer detection methods based on deep neural networks. Materials and Methods, including the chosen datasets for the study, data pre-processing, and the proposed deep learning models are illustrated in Section 3. In Section 4, the experiments parameters and the performance metrics for our multi-classification model are explained with the experimental results comparisons. Discussions of the results are drawn in Section 5, and finally, conclusions with possible ongoing future work are drawn in Section 6.
2. Related works and background
Chest X-ray and computed tomography (CT) images are quite easy to obtain for patients and are a low-cost procedure, which makes using them in the recognition of COVID-19 applicable in most countries. In Ref. [32], three different convolutional neural network models: ResNet50, InceptionV3, and Inception-ResNetV2 were used for the detection of corona virus pneumonia in infected patients using chest X-ray radiographs. No feature extraction or selection phase were required. They achieved 98% classification accuracy using the ResNet50 model, 97% accuracy for InceptionV3, and 87% accuracy for InceptionResNetV2. To avoid overfitting, they performed around 30 epochs in their training phase for all the models. However, they used a few images in their study, which were available for them to use at that time.
Digital X-ray images were also used to automatically distinguish between COVID-19 and pneumonia patients in Ref. [33] using four different pre-trained convolutional neural networks (CNNs): ResNet18, AlexNet, SqueezeNet, and DenseNet201. Image augmentation approaches were used (namely, rotation, scaling, and translation) to generate a 20-fold training set of COVID-19 images. The results of the models were obtained with and without augmentation. To increase the detection accuracy, the images were resized to a specific size as a pre-processing step. Finally, a public database was introduced as a collection of three public databases recently published in the literature. The database contains a mixture of 1345 viral pneumonia, 190 COVID-19, and 1341 normal chest X-ray images. They obtained an accuracy of 98%.
Similarly, the authors in Ref. [27] used a generative adversarial network (GAN) with fine-tuned deep transfer learning approaches, namely AlexNet, GoogLeNet, Squeeznet, and Resnet18, to detect COVID-19 using chest X-ray images for a limited dataset. Using generative adversarial network (GAN) overcome the overfitting problem and helped in generating more images from the dataset that consisted of 5863 X-ray images containing normal and infected images.
Instead of using X-ray images [34], used transverse section CT images for COVID-19 patients to detect the virus early using deep learning approaches. These images show different characteristics that can discriminate COVID-19 patients from other kinds of pneumonia, such as influenza-A. The samples were collected from three COVID-19 designated hospitals in Zhejiang Province, China and contain a total of 618 CT samples: 224 from 224 patients with influenza-A viral pneumonia, 219 samples from 110 patients identified with COVID-19, and 175 samples from healthy people. These images were segmented into multiple candidate image cubes using two three-dimensional (3D) CNN models. The first CNN type was the traditional ResNet23, while the other was a designed model based on the first network structure by concatenating the location-attention mechanism in the full-connection layer to improve the overall accuracy, which reached 86.7%.
Another important work that was based on using the CT images in detecting COVID-19 was presented in Ref. [16]. They utilized a weakly-supervised deep learning-based model to develop their own model called DeCoVNet. The lung regions, segmented using a pre-trained UNet, were fed into a 3D deep neural network to investigate the possibility of COVID-19 infectious. The masking of the lung areas helped to reduce the background information and better detect the infection. Their model consists of three stages. The first stage contained a normal 3D convolution with a kernel size of 577; a batch norm layer and a pooling layer. The second stage was composed of two 3D residual blocks (ResBlocks). In each ResBlock, a 3D fully connected (FC) layer with the SoftMax activation function was utilized. A dataset that contained 499 CT images was used for training and 131 CT image positive and COVID-negative; the accuracy of the algorithm is 90.1%, while the positive predictive value is 84%.
A CT scan COVID-19 dataset [17] has been built to assist researchers in performing the detection process of the virus. Their dataset contains 275 CT scans that are positive for COVID-19 and 195 CT scans that are negative for COVID. Moreover, they used this dataset to train a deep CNN model for detecting the presence of the COVID-19 virus. The size of their dataset is small for training the deep learning models, which results in overfitting; thus, they used a transfer learning approach that uses a large collection of chest X-ray images to pre-train a deep CNN model, then re-tune this pre-trained network on the COVID CT dataset. They also used data augmentation to increase the size of training data by creating and adding new image-label pairs to the training set. They achieved an accuracy of 84.7%. A rapid method for COVID-19 diagnosis based on artificial intelligence has been proposed in Ref. [28], where CNN applied to CT images from 108 patients with laboratory verified COVID-19 and 86 patients with pneumonia diseases. To distinguish between COVID-19 and pneumonia, the authors used several deep learning methods: VGG-19, VGG-16, SqueezeNet, AlexNet, GoogleNet, ResNet-18, MobileNet-V2, ResNet-50, ResNet-101, and Xception, where ResNet-101 achieved the highest accuracy of 99.51%.
In [35], the authors proposed a classification schema that consists of a multiclass classification and a hierarchical classification of chest X-ray images for identifying COVID-19 and pneumonia cases. The latter classification is used for pneumonia, which can be organized as a hierarchy. The RYDLS-20 dataset was used that contained chest X-ray images of pneumonia and chest X-ray images of healthy lungs. Their approach achieved 65% for F1-Score using a multiclass approach and an F1-Score of 0.89 for the COVID-19 identification in the hierarchical classification scenario. Another recent work proposed by Ref. [36] detected COVID-19 from chest X-rays using deep learning on a small dataset. They used chest X-rays of 135 patients identified with COVID-19 and about 320 chest X-rays of pneumonia patients. The experiment results showed an accuracy of 91.24% when using pre-trained ResNet50 and VGG-16 models, which, along with their CNN, were trained on a balanced set of COVID-19 and pneumonia chest X-rays.
Authors in Ref. [31], proposed a model for detecting COVID-19 using CXR and CT images based on transfer learning and Haralick features. Here, transfer learning technology can provide a fast alternative to aid in the diagnostic process and thus reduce spread. The primary purpose of this work is to provide radiologists with a less complex model that can aid in the early diagnosis of COVID-19. The proposed model produces 91% accuracy, 90% retrieval, and 93% accuracy by VGG-16 using transfet learning, which outperforms other models present in this pandemic period. A summary of these studies and more research papers using deep learning approaches, including their working methods, are illustrated in Table 1 .
Table 1.
Overview of studies using deep learning approaches with their working methods and performance metrics for COVID-19 case detection.
| Study | Method | Medical Image | Performance |
|---|---|---|---|
| [5] | ResNet + SVM | Chest x- ray | 95.38% (Acc.) |
| 97.29% (Sens.) | |||
| [29] | AlexNet | Chest x- ray | 98% (Acc.) |
| [36] | VGG-16+ CNN | Chest x- ray | 91.24% (Acc.) |
| [37] | Resnet50, VGG16 | Chest x- ray | 94.4% (Acc.) |
| [27] | GAN and CNN: AlexNet, Resnet18 | Chest x- ray | 99% (Acc.) |
| [38] | DenseNet169, ResNet50, ResNet34, Inception ResNetV2, VGG-19, RNN |
Chest x- ray | 95.72% (Acc.) |
| [39] | ResNet-50 (COVID-ResNet) | Chest x- ray | 96.23% (Acc.) |
| [40] | COVID-Net: Tailored model | Chest x- ray | 92.4% (Acc.) |
| 91% (Sens.) | |||
| 98% (Prec.) | |||
| [12] | ResNet152V2, MobileNetV2, CNN, LSTM | Chest x- ray | 99.22% (Acc.) |
| 99.43% (Prec.) | |||
| 99.44% (Sens.) | |||
| 99.77% (AUC) | |||
| [15] | COVIDx-Net | Chest x- ray | 90% (Acc.) |
| 100% (Prec.) | |||
| [40] | Tailored model | Chest x- ray | 96% (Acc.) |
| [21] | UNet++ | Chest CT | 95.2% (Acc.) |
| 100% (Sens.) | |||
| 93.6% (Spec.) | |||
| [35] | CNN | Chest CT | 89% (F1-score) |
| [22] | CNN | Chest CT | 94.1% (Sens.) |
| 95.5% (Spec.) | |||
| [28] | AlexNet, VGG-16, VGG-19, SqueezeNet, GoogleNet, MobileNet-V2, ResNet-18, ResNet-50, ResNet-101, and Xception |
Chest CT | 99.51% (Acc.) |
| [41] | ResNet- 50 | Chest CT | 86.0% (Acc.) |
| [34] | ResNet | Chest CT | 86.7% (Acc.) |
| 86.7% (Sens.) | |||
| 81.03% (Spec.) | |||
| [42] | Tailored model (COVID-19Net) | Chest CT | 87% (AUC) |
| [26] | DarkCovidNet | Chest x- ray | 90.8% (Acc.) |
| 95.13% (Sens.) | |||
| 95.3% (Spec.) | |||
| [19] | CNN | Chest CT | 82.9% (Acc.) |
| [41] | DRE-Net | Chest CT | 86% (Acc.) |
| 96% (Sens.) | |||
| 80% (Prec.) | |||
| [20] | ResNet-50 | Chest CT | 90.0% (Sens.) |
| 96.0% (Spec.) | |||
| [31] | VGG-19, ResNet-50, InceptionV3 | Chest x-ray + CT | 93.0% (Acc.) |
| 91.0% (Prec.) | |||
| 90.0% (Sens.) |
In other research papers, deep learning models only for COVID-19 as in Refs. [13,15,17,32], pneumonia only as in Ref. [12] or for both diseases as in Refs. [26,27,33,34] have been proposed. But in this paper, a multi-classification deep learning model for diagnosing COVID-19, pneumonia, and Lung Cancer from chest X-ray and CT images is developed. To the best of our knowledge, this work is the first to diagnose chest diseases. Many performance metrics will be used in our proposed framework's evaluation, such as accuracy, AUC, sensitivity or recall, specificity, precision, negative predictive value (NPV), F1 score, and Matthew's correlation coefficient range. Other researchers only used accuracy, AUC, sensitivity, or recall and specificity, as in Refs. [[43], [44], [45]].
3. Materials and Methods
There are many diseases that may attack the human lung system, such as pneumonia, lung cancer, and recently COVID-19. For diagnosing these diseases, chest images using CT or X-ray are needed, as they play a critical and essential role. Computer science technologies have introduced tools to help in disease diagnoses, such as deep learning-based systems. In this paper, a multi-classification deep learning model for chest disease diagnoses is developed. The objective of our study is to propose a deep learning model for chest disease classification using four different models. To the best of our knowledge, this work can be considered the first approach toward introducing a single deep learning model for detection of a collection of chest diseases. It leverages the burden of the user to use several applications for the detection of each disease separately, and hence reduce decision time.
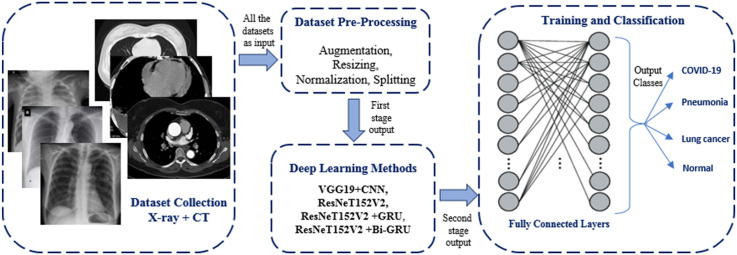
The model block diagram is shown in Fig. 1 . As clearly seen from the figure, the model consists of three main stages: data pre-processing, deep learning models for feature extraction, and classification. The proposed model uses chest images from CT and X-ray as inputs, and the final output is the classification of the input image into one of the four classes: COVID-19, Normal, Pneumonia and Lung Cancer.
Fig. 1.
Block diagram of the proposed multi-classification deep-chest model.
The first stage handles image pre-processing, such as resizing, image augmentation, and data, splitting randomly into two groups: training and validation by 70% and 30%, respectively. The dataset images are randomly split into two parts (train and validation) to insure the variety of the images. Data normalization is also used after converting the image to an array of pixels to rescale the image's pixel value to the interval [0,1]. The second and third stages are feature extraction and image classification, respectively, by using different types of deep learning approaches, as will be discussed later. The input images for our proposed model can be CT or X-ray images. These images are resized to . To increase the number of training images to produce efficient and reliable accuracy for our system, image augmentation methods are used, such as flip, rotate, and skewing, in two places: first in dataset preparing and second in data pre-processing. The use of augmentation twice increases the dataset size, which will be reflected in the system accuracy.
3.1. Datasets for the study
For the purpose of our experiments, many sources of X-rays and CT images were accessed and collated. This collection includes COVID-19, pneumonia, and lung cancer images from both types; X-ray and CT, in addition to normal images. First, for COVID-19, we selected dataset from repositories like GitHub [46,47] with about 4320 images (X-ray and CT). Secondly, some useful websites have a public and medical dataset that are considered to be the most common datasets, such as the Radiological Society of North America (RSNA), the Italian Society of Medical and Interventional Radiology (SIRM), and Radiopaedia, which are a collection of common pneumonia X-ray images with about 5856 images [[48], [49], [50]] that can be used for training our proposed deep CNN to distinguish COVID-19 from pneumonia. The third dataset source is lung cancer X-ray and CT images that are available in Ref. [51], with about 20,000 images. The last dataset that will be used in this study is normal images and contains 3500 X-ray and CT images. The total number of images from the collected datasets is 33,676 images.
Samples of chest X-ray and CT images for COVID-19, normal cases, pneumonia, and lung cancer images, with and without deep dream filter are shown in Fig. 2 . The number of patients for each disease dataset with respect to all ages, is shown in Fig. 3 . The ages were frequently between 38 and 65 for the COVID-19 dataset, 26 and 62 for the pneumonia dataset, 28 and 58 for the lung cancer dataset, and for normal patients the ages were between 33 and 58 years.
Fig. 2.
Chest X-ray and CT images: (a) COVID-19 with and without deep dream filter; (b) Normal images with and without deep dream filter; (c) Pneumonia images with and without deep dream filter; (d) Lung cancer images with and without deep dream filter.
Fig. 3.
Age-wise distribution of chest datasets related to each disease.
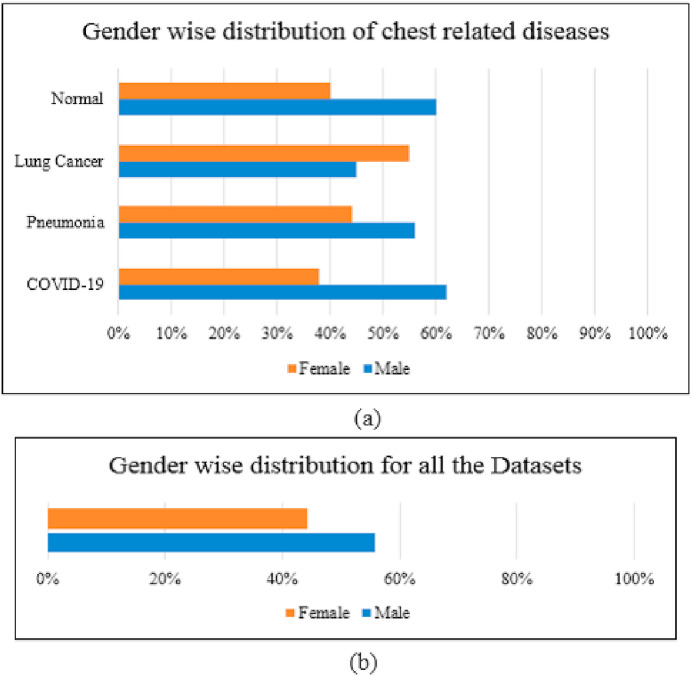
Fig. 4 illustrates the gender-wise distribution for the collected dataset. It can be realized obviously in Fig. 4 (a) which gender is affected more by each disease, i.e. COVID-19 and pneumonia affect males more than females, while lung cancer affects more females than males. Fig. 4(b) demonstrates the gender distribution for all diseases.
Fig. 4.
Gender-wise distribution: (a) for chest related to each disease; and (b) for all the datasets.
3.2. Dataset pre-processing
The used datasets are first augmented to excess the number of the used images and to balance between the four types of datasets. We have obtained around 33,676 images, as explained in the previous subsection, and after augmentation we have a total of 75,000 images. The first use of augmentation techniques will be split between the four training classes: COVID-19, Normal, Pneumonia, and Lung cancer. Then, these images are used as input for the data pre-processing stage.
The pre-processing stage is usually used for preparing the input data to satisfy the deep learning model requirements. In our model, the input images were prepared using different pre-processing steps: 1) resizing images to , 2) augmenting the resized images using augmentation methods such as rotate, flip, and skewing, 3) normalizing all images (original and augmented images), and 4) converting the images into arrays for using them as input for the next stage of the model. For training the deep learning model, the dataset must be split. The dataset images are randomly split into two parts (train and validation) 70% and 30% for training and validation, respectively, to insure the variety of the images.
3.3. Deep learning proposed methods
In this paper, several types of supervised deep learning techniques are utilized for developing the proposed four deep-chest classification models. We aim to explore their performance in detecting the three considered chest diseases and concluding the best of them. Each of these models consists of a combination of CNN and recurrent neural network (RNN). Two pre-trained models, VGG19 and ResNet152V2, are used with normal CNN, gated recurrent unit (GRU) and bidirectional gated recurrent unit (Bi-GRU) as types of RNN. In the following subsections, we add the details of each of the four developed models.
3.3.1. VGG19+CNN deep model
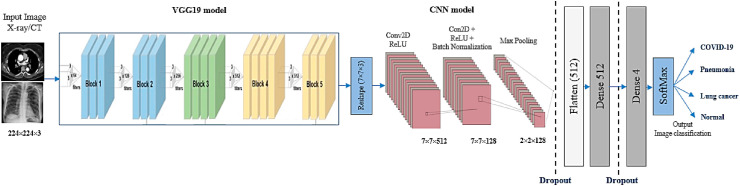
A deep model that consists of a VGG19 pre-trained model followed by CNNs is designed to diagnose chest diseases using CT and X-ray images. It employs as a feature extraction and classification. The proposed model details are illustrated in Fig. 5 . It contains input images, feature extraction, and classification layers.
Fig. 5.
VGG19+CNN proposed model architecture.
The input layer for the model receives a CT or X-ray chest image. The feature extraction consists of VGG19, followed by two CNN blocks that work as feature extraction sections. The first CNN block has a convolution layer and a ReLU layer. While the second block has two convolution layers, followed by two ReLU layers, then a batch normalization layer, followed by a maximum pooling layer, and finally a dropout layer, as shown in Fig. 5.
The feature extraction layer output is then passed to a flatten layer to convert the data shape to a one-dimensional data vector as the first task of classification. The classification part consists of a dense layer with 512 neurons followed by a dropout layer. The final output is produced from a dense layer with four neurons and SoftMax activation function, which classifies the output image into one of the chest diseases classes: COVID-19, pneumonia, lung cancer, or normal. The proposed model architecture is listed in Table 2 . The overall number of parameters is 22,337,860, which are separated into two groups; the total trainable parameters are 22,337,604, and the non-trainable parameters are 256. The difference between trainable and non-trainable parameters is that the trainable parameters are the parameters that are updated in the training processes and are needed by training to get the optimal value of these parameters, while non-trainable parameters mean the parameters that are not updated during training. In other words, non-trainable parameters of a model are those that you will not be updating and optimized during training, and that have to be defined a priori, or passed as inputs. Therefore, the non-trainable will not contribute to the classification process.
Table 2.
The proposed VGG19+CNN architecture.
| Layer (type) | Output Shape | Parameters |
|---|---|---|
| vgg19 (Functional) | (None, 7, 7, 512) | 20024384 |
| reshape (Reshape) | (None, 7, 7, 512) | 0 |
| conv2d (Conv2D) | (None, 7, 7, 128) | 1638528 |
| activation (Activation) | (None, 7, 7, 128) | 0 |
| conv2d_1(Conv2D) | (None, 7, 7, 128) | 409728 |
| activation_1 | (None, 7, 7, 128) | 0 |
| batch_normalization | ||
| (BatchNormalization) | (None, 7, 7, 128) | 512 |
| max_pooling2d | ||
| (MaxPooling2d) | (None, 2, 2, 128) | 0 |
| dropout (Dropout) | (None, 2, 2, 128) | 0 |
| flatten (Flatten) | (None, 512) | 0 |
| dense (Dense) | (None, 512) | 262656 |
| dropout_1 (Dropout) | (None, 512) | 0 |
| dense_1 (Dense) | (None, 4) | 2052 |
| Total parameters: 22,337,860 | ||
| Trainable parameters: 22,337,604 | ||
| Non-trainable parameters: 256 | ||
3.3.2. ResNet152V2 deep model
In the second model, ResNet152V2 is used as a feature extraction model, as shown in Fig. 6 , instead of the VGG19-CNN model. The model has initial weights because it is a pre-trained model, which can help to gain acceptable accuracy faster than a traditional CNN. The model architecture consists of the ResNet152V2 model followed by a reshape layer, a flatten layer, a dense layer with 128 neurons, a dropout layer, and finally a dense layer with Softmax activation function to classify the image into its corresponding class. The architecture is detailed in Table 3 . The whole parameters of the ResNet152V2 are 71,177,348, which consist of two types of parameters: the trainable parameters and the non-trainable parameters, which are 71,033,604 and 143,744, respectively.
Fig. 6.
ResNet152V2 proposed model architecture.
Table 3.
The proposed ResNet152V2 architecture.
| Layer (type) | Output Shape | Parameters |
|---|---|---|
| Resnet152v2 | (None, 7, 7, 2048) | 58331648 |
| reshape (Reshape) | (None, 7, 7, 2048) | 0 |
| flatten (Flatten) | (None, 512) | 0 |
| dense (Dense) | (None, 128) | 12845184 |
| dropout_1 (Dropout) | (None, 128) | 0 |
| dense_1 (Dense) | (None, 4) | 516 |
| Total params: 71,177,348 | ||
| Trainable params: 71,033,604 | ||
| Non-trainable params: 143,744 | ||
3.3.3. ResNet152V2 and GRU deep model
ResNet152V2 has many advantages as a pre-trained model in accelerating training and converging to high accuracy rapidly. Therefore, adding it before deep learning models can build efficient and reliable models, which results in higher accuracy. However, GRU is one of the RNN architectures that has the main advantage of its ability to maintain the information irrelevant to the prediction for a long time without removing it. It also has many other features, such as being very simple, easy to modify, and taking less training time, which makes it more efficient and suitable for many applications like this one.
The ResNet152V2, followed by GRU, was used as a feature extraction model, as shown in Fig. 7 . The model contains the ResNet152V2 followed by a reshape layer, GRU layer with 256 units, flatten layer, dense layer with 128 neurons, dropout layer, and dense layer, with Softmax activation function to classify the image into one of our four classes. The architecture is detailed in Table 4 . The total parameters of the ResNet152V2 are 69,573,252 that consist of two types of parameters: trainable parameters: 69,429,508 and non-trainable parameters: 143,744.
Fig. 7.
ResNet152V2 and GRU Model architecture.
Table 4.
The proposed ResNet152V2+GRU architecture.
| Layer (type) | Output Shape | Parameters |
|---|---|---|
| Resnet152v2 | (None, 7, 7, 2048) | 58331648 |
| reshape (Reshape) | (None, 7, 7, 2048) | 0 |
| Time_distributed_7 | (None, 7, 14336) | 0 |
| gru_6 (GRU) | (None, 256) | 11208192 |
| dense (Dense) | (None, 128) | 32896 |
| dropout_4 (Dropout) | (None, 128) | 0 |
| dense_7 (Dense) | (None, 4) | 516 |
| Total params: 71,177,348 | ||
| Trainable params: 71,033,604 | ||
| Non-trainable params: 143,744 | ||
3.3.4. ResNet152V2 and Bi-GRU deep model
The last proposed deep learning model in this paper consisted of a ResNet152V2, followed by bi-GRU, which was used as a feature extraction model, as shown in Fig. 8 . The model has a ResNet152V2, followed by a reshape layer, bi-GRU layer with 512 units, a dropout layer, and a dense layer, with a SoftMax activation function to classify the image into one of our four classes. The architecture of the model is detailed in Table 5 . The total parameters of the ResNet152V2 are 80,750,084, which consist of two types of parameters: trainable parameters: 80,606,340 and non-trainable parameters: 143,744.
Fig. 8.
ResNet152V2+Bi-GRU proposed model architecture.
Table 5.
Proposed Resnet152V2+Bi-GRU architecture.
| Layer (type) | Output Shape | Parameters |
|---|---|---|
| Resnet152v2 | (None, 7, 7, 2048) | 58331648 |
| reshape (Reshape) | (None, 7, 7, 2048) | 0 |
| Time_distributed_7 | (None, 7, 14336) | 0 |
| bidirectional GRU | (None, 512) | 22416384 |
| dropout (Dropout) | (None, 512) | 0 |
| dense (Dense) | (None, 4) | 2052 |
| Total params: 80,750,084 | ||
| Trainable params: 80,606,340 | ||
| Non-trainable params: 143,744 | ||
4. Results
4.1. Experimental parameters
The models were implemented using Python 3 and the Keras framework. These were run on Google Colab pro version [52] with 2 TB storage, 25 GB RAM, and a P100 graphical processing unit (GPU) processor. To accomplish the statistical results, the images of the input classes were augmented using an API Keras Augmentor [53] in the first use of the dataset to raise the number of images in each class. The utilized augmentation methods are image rotation, skew, and shift. All images, original and augmented, were passed to the ImageDataGenerator class in Keras [53] to perform pre-processing operations, such as augmentation, resizing, and normalization. The generated images were fed into our proposed multi-classification deep learning models. For training and validating our models, an optimizer and suitable fit functions were used, where each model ran around 800 epochs with eight iterations per epoch and a batch size of 64. To train deep learning models well, we needed to increase the number of samples and epochs. When we used increased numbers of epochs, we found that the loss decreased. Therefore, we used 800 epochs.
The results were accomplished by applying the performance metric equations to the resulting validation data outputs, and the registered results represent the maximum obtained validation values. The optimizer used for our proposed models was the (Adamax) [54]. The learning rate (LR) value and optimizer for all models are listed in Table 6 . The entire code for our multi-classification deep learning models was uploaded to the GitHub website at [55].
Table 6.
Models’ training parameters.
| Model | Optimizer | Learning Rate |
|---|---|---|
| VGG19+CNN | Adamax | 0.00006 |
| ResNet152V2 | 0.00009 | |
| ResNet152V2+GRU | 0.00009 | |
| ResNet152V2+Bi-GRU | 0.00009 |
4.2. Performance metrics
The performance of the chest disease classification models was evaluated based on several metrics: accuracy (ACC), loss, recall, positive predictive value (PPV), which is commonly known as precision, specificity (SPC), negative predictive value (NPV), F1-score, Matthew's correlation coefficient (MCC), and area under curve (AUC). Correspondingly, a confusion matrix is introduced for each model. Accuracy, given in eq. (1), is the number of examples correctly predicted from the total number of examples.
| (1) |
where Tp and Tn are the true positive and negative parameters, respectively. Fp and Fn are the false positive and false negative values.
Sensitivity or recall, given in eq. (2), is the number of samples actually and predicted as positive from the total number of samples actually positive; also known as true positive rate.
| (2) |
While the true negative rate, called specificity, given in eq. (3), is the number of samples actually and predicted as negative from the total number of samples actually negative.
| (3) |
Eq. (4) shows precision, also called positive predictive value [43], which represents the number of samples actually and predicted as positive from the total number of samples predicted as positive.
| (4) |
Whereas the negative predictive value (NPV) [43], is the number of samples actually and predicted as negative from the total number of samples predicted as negative, given in eq. (5).
| (5) |
The harmonic mean of precision and recall, which is known as F1 score, is shown in eq. (6).
| (6) |
Finally, Matthew's correlation coefficient range [44], allows one to gauge how well the classification model/function is performing.
| (7) |
Moreover, recent studies support the use of confusion matrix analysis in model validation [56] since it is robust to categorize data relationships and any distribution. It provides extra information on illustrating the classification models. To analyze our models using a confusion matrix, we must understand how it is structured, and we need to define all variables and parameters that can be extracted from Table 7 .
Table 7.
Confusion matrix explanations.
| Predicted |
|||||
|---|---|---|---|---|---|
| Normal |
Pneumonia |
COVID-19 |
Lung Cancer |
||
| Actual | Normal |
Pnn |
Ppn |
Pcn |
Pln |
| Pneumonia |
Pnp |
Ppp |
Pcp |
Plp |
|
| COVID-19 |
Pnc |
Ppc |
Pcc |
Plc |
|
| Lung Cancer | Pnl | Ppl | Pcl | Pll | |
| Pcc: COVID-19 class were correctly classified as COVID-19 | |||||
| Ppc: COVID-19 class were incorrectly classified as Pneumonia | |||||
| Pnc: COVID-19 class were incorrectly classified as Normal | |||||
| Plc: COVID-19 class were incorrectly classified as Lung Cancer | |||||
| Pcp: Pneumonia class were incorrectly classified as COVID-19 | |||||
| Ppp: Pneumonia class were correctly classified as Pneumonia | |||||
| Pnp: Pneumonia class were incorrectly classified as Normal | |||||
| Plp: Pneumonia class were incorrectly classified as Lung Cancer | |||||
| Pcn: Normal class were incorrectly classified as COVID-19 | |||||
| Ppn: Normal class were incorrectly classified as Pneumonia | |||||
| Pnn: Normal class were correctly classified as Normal | |||||
| Pln: Normal class were incorrectly classified as Lung Cancer | |||||
| Pcl: Lung Cancer class were incorrectly classified as COVID-19 | |||||
| Ppl: Lung Cancer class were incorrectly classified as Pneumonia | |||||
| Pnl: Lung Cancer class were incorrectly classified as Normal | |||||
| Pll: Lung Cancer class were correctly classified as Lung Cancer | |||||
Using these parameters, we can define other variables for true positives, true negatives, false positives, and false negatives, as in Table 8.
Table 8.
True positives, true negatives, false positives, and false negatives variables definitions.
| True Positives: | True Negatives: |
|---|---|
| TP (Normal): Pnn | TN (Normal): Ppc + Ppl + Pcl + Plc + Plp + Pcp + Pcc + Pll + Ppp |
| TP (Pneumonia): Ppp | TN (Pneumonia): Pnl + Pnc + Pln + Pcn + Pnn + Pcl + Plc + Pcc + Pll |
| TP (COVID-19): Pcc | TN (COVID-19): Pll + Ppl + Pnl + Pnp + Ppp + Plp + Pln + Ppn + Pnn |
| TP (Lung cancer): Pll |
TN (Lung cancer): Pnn + Ppn + Pcn + Pnp + Ppp + Pcp + Pnc + Ppc + Pcc |
|
False Positives: |
False Negatives: |
| FP (Normal): Pnl + Pnc + Pnp | FN (Normal): Pln + Pcn + Ppn |
| FP (Pneumonia): Ppl + Ppc + Ppn | FN (Pneumonia): Plp + Pcp + Pnp |
| FP (COVID-19): Pcn + Pcp + Pcl | FN (COVID-19): Pnc + Ppc + Plc |
| FP (Lung cancer): Plc + Plp + Pln | FN (Lung cancer): Pcl + Ppl + Pnl |
4.3. Multi-classification deep learning model results
In this section, we illustrate our multi-classification model results, followed by a brief discussion and analysis for each proposed model. The confusion matrix is the major means to evaluate errors in classification problems. According to the confusion matrix explanations presented in Table 6, we built the confusion matrix for the VGG19+CNN proposed model, as shown in Fig. 9 . The figure shows that the VGG19+CNN model can successfully classify the four patient statuses (COVID-19, Pneumonia, Lung Cancer, and Normal) with the highest ratio to the normal images (0.9323), then lung cancer (0.8963), followed by pneumonia (0.88), and finally COVID-19 (0.8747). This result assures that the classification is performed correctly for the four statuses.
Fig. 9.
Confusion matrix for the proposed VGG19-CNN model.
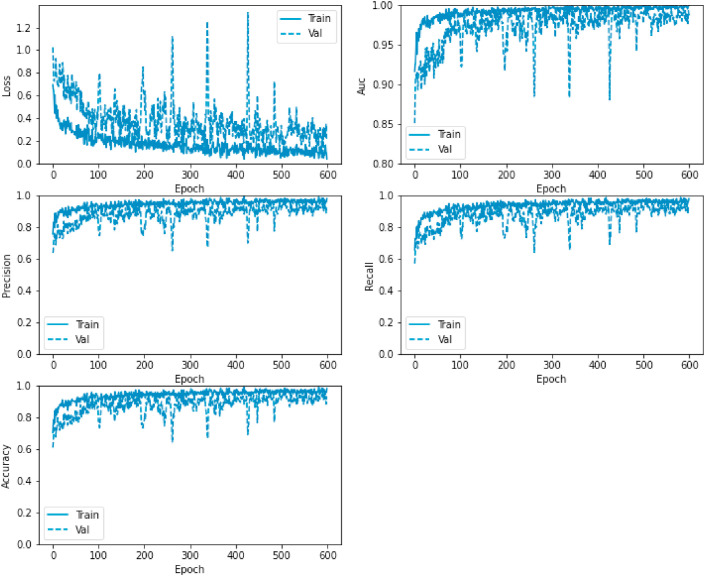
In addition, we illustrate the loss, AUC, precision, recall, and accuracy between the training and validation phases with the number of epochs in Fig. 10 .
Fig. 10.
Loss, AUC, precision, recall, and accuracy between the training and validation phases with the number of epochs for the VGG19-CNN model.
Fig. 11 displays the ResNet152V2 model confusion matrix which demonstrates that the model can classify the four patient status (COVID-19, Pneumonia, Lung Cancer, and Normal) starting with the normal images that has the highest ratio (0.9678), then COVID-19 (0.8677), followed by lung cancer (0.865), and finally pneumonia (0.818). These results are lower compared to those generated by the VGG19+CNN model, which had better values for pneumonia and lung cancer.
Fig. 11.
Confusion matrix for the proposed ResNet152V2 model.
In addition, Fig. 12 shows the loss, AUC, precision, recall, and accuracy between the training and validation phases with the number of epochs. Similarly, in Fig. 13 , the confusion matrix of the ResNet152V2+GRU model shows the classification of the four patient statuses (COVID-19, Pneumonia, Lung Cancer, and Normal) with the highest ratio to the normal images (0.9329), then lung cancer (0.8736), followed by pneumonia (0.8599), and finally COVID-19 (0.8591). These results are also lower than the VGG19+CNN model results but better than the ResNet152V2 model.
Fig. 12.
Loss, AUC, precision, recall, and accuracy between the training and validation phases, with the number of epochs for the ResNet152V2 model.
Fig. 13.
Confusion matrix for the proposed ResNet152V2+GRU model.
Also, as in Fig. 14 , loss, AUC, precision, recall, and accuracy between the training and validation phases, with the number of epochs, are illustrated. Correspondingly, the confusion matrix of the ResNet152V2+Bi-GRU model is presented in Fig. 15 . The figure demonstrates the successful classification of the four patient statuses (COVID-19, Pneumonia, Lung Cancer, and Normal) starting with the normal images, which have the highest ratio, then lung cancer, followed by COVID-19, and lastly pneumonia.
Fig. 14.
Loss, AUC, precision, recall, and accuracy between the training and validation phases, with the number of epochs for the ResNet152+GRU model.
Fig. 15.
Confusion matrix for the proposed ResNet152V2+Bi-GRU model.
Likewise, as shown in Fig. 16 , loss, AUC, precision, recall, and accuracy between the training and validation phases, with the number of epochs, are demonstrated.
Fig. 16.
Loss, AUC, precision, recall, and accuracy between the training and validation phases, with the number of epochs for the ResNet152+Bi-GRU model.
4.4. Experimental comparisons
The experimental results of the proposed models are summarized in Table 9 . The comparison between the models in terms of the different performance metrics as loss values is illustrated in Fig. 17 , and the other metrics: accuracy (ACC), recall, precision (PPV), specificity (SPC), negative predictive value (NPV), F1-score, Matthew's correlation coefficient (MCC), and area under the curve (AUC) for the proposed models are presented in Fig. 18 . As clearly seen from the figures, the VGG19+CNN model outperforms the other three proposed models in terms of accuracy, precision, F1-score, and AUC; however, it has the highest value of loss.
Table 9.
Evaluation metrics for the different models.
| Models | Loss | TP | FP | TN | FN | ACC | Recall | PPV | SPC | NPV | F1-Score | MCC | AUC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VGG19+CNN | 0.3280 | 251 | 4 | 764 | 5 | 98.05 | 98.05 | 98.43 | 99.5 | 99.3 | 98.24 | 97.7 | 99.66 |
| ResNet152V2 | 0.1693 | 244 | 12 | 756 | 12 | 95.31 | 95.31 | 95.31 | 98.4 | 98.4 | 95.31 | 93.8 | 99.17 |
| ResNet152V2+GRU | 0.1350 | 246 | 10 | 758 | 10 | 96.09 | 96.09 | 96.06 | 98.7 | 98.7 | 96.09 | 94.8 | 99.34 |
| ResNet152V2+Bi-GRU | 0.2554 | 477 | 34 | 1502 | 35 | 93.36 | 93.16 | 93.35 | 97.8 | 97.8 | 93.26 | 91.1 | 98.44 |
Fig. 17.
Loss measures for the VGG19+CNN, ResNet152V2, ResNet152V2+GRU, and ResNet152V2+Bi-GRU models.
Fig. 18.
Accuracy (ACC), recall, precision (PPV), specificity (SPC), negative predictive value (NPV), F1-score, Matthew's correlation coefficient (MCC), and area under the curve (AUC) measures for the proposed models.
5. Comparative analysis and discussion
In the present work, four deep learning architectures for lung disease detection and diagnosis in the human lung system are proposed. These architectures are used to classify between the most common chest diseases: COVID-19, pneumonia, and lung cancer. The different proposed models were compared according to accuracy, recall, precision, F1 score, and AUC. As listed in Table 8, the obtained results show that the VGG19+CNN model gave the best classification performance (98.05% accuracy) followed by ResNet152V2+GRU, with 96.09% accuracy. Contrarily, ResNet152V2 and ResNet152V2+Bi-GRU are the lowest compared to the other architectures, since these models have obtained 95.31% and 93.36% accuracy, respectively. From Table 1, it seems that our result is not better than [27,28]. The reason is the authors measured the performance of their models only by one metric, which is accuracy. In addition, the authors in Ref. [27] used only chest X-rays, and the authors in Ref. [28] used only CT images. While in our proposed model, we measured the performance based on eight metrics in addition to accuracy, as represented in Table 9.
Consequently, we recommend the VGG19+CNN model (which achieved 98.05% accuracy, 98.05% recall, 98.43% precision, 99.5% specificity, 99.3% negative predictive value, 98.24% F1 score, 97.7% MCC, and 99.66% AUC) based on X-ray and CT images be used to diagnose the health status of chest patients against COVID-19, pneumonia, and lung cancer. We hope that the introduced deep models and their results may serve as a first step toward developing a chest disease diagnosis system from CT and X-ray chest images.
6. Conclusion and future work
In this study, a multi-classification deep learning model was designed and evaluated for detecting COVID-19, pneumonia, and lung cancer from chest x-ray and CT images. This model is, to the best of our knowledge, the first attempt to classify the three chest diseases in a single model. It is important to correctly diagnose these diseases early to determine the proper treatment and apply isolation to COVID-19 patients to prevent the virus from spreading. Four architectures were presented in this study: VGG19+CNN, ResNet152V2, ResNet152V2+GRU, and ResNet152V2+Bi-GRU.
Through extensive experiments and results performed on collected datasets from several sources that contained chest x-ray and CT images, the VGG19+CNN model outperformed the other three proposed models. The VGG19+CNN model achieved 98.05% accuracy, 98.05% recall, 98.43% precision, 99.5% specificity, 99.3% negative predictive value, 98.24% F1 score, 97.7% MCC, and 99.66% AUC, based on X-ray and CT images.
Ongoing work attempts to enhance the performance of the proposed model by raising the number of images in the used datasets, increasing the training epochs, and using other deep learning techniques such as GAN in both classification and augmentation.
CRediT authorship contribution statement
Dina M. Ibrahim: Conceptualization of this study, investigation, methodology, resources, validation, visualization, writing 1–original draft preparation. Nada M. Elshennawy: Conceptualization of this study, formal analysis, methodology, software, validation, writing - original draft preparation. Amany M. Sarhan: Conceptualization of this study, resources, writing1–review and editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Biographies
Dina M. Ibrahim has been a professor at Information Technology Department, College of Computer, Qassim University, KSA since 2015. In addition, Dina works as Lecturer at Computers and Control Engineering Department, Faculty of Engineering, Tanta University, Egypt. She was born in the United Arab Emirates, her B.Sc., M.Sc., and Ph.D. degrees were obtained from the Computers and Control Engineering Department, Faculty of Engineering, Tanta University in 2002, 2008, and 2014, respectively. Dina worked as a Consultant Engineer, Database Administrator, and Vice Manager on the Management Information Systems (MIS) Project, Tanta University, Egypt, from 2008 until 2014. Her research interests include networking, wireless communications, machine learning, security, and the internet of things. Dina has published more than 40 articles in various refereed international journals and conferences. She has been serving as a reviewer in Wireless Network (WINE) Journal since 2015. Dina has also acted as a co-chair of the International Technical Committee for the Middle East Region of the ICCMIT conference since 2020. (email: d.hussein@qu.edu.sa, dina.mahmoud@f-eng.tanta.edu.eg)
Nada M. Elshennawy is an assistant professor in the Computers and Control Engineering Department, Faculty of Engineering, Tanta University, Egypt. She was born in 1978 in Egypt. Her B.Sc., M.Sc., and Ph.D. degrees were obtained from Computers and Control Engineering Department, Faculty of Engineering, Tanta University in 2001, 2007, and 2014, respectively. Nada worked as a manager at Information Technology (IT) Unit, Faculty of Engineering Tanta University, Egypt from 2016 until 2018. Her research interests are in machine learning, computer vision and human behavior recognition, wireless networks, wireless sensor networks, and neural networks. (email: Nada_elshennawy@f-eng.tanta.edu.eg)
Amany M. Sarhan received her B.Sc. degree in Electronics Engineering, and M.Sc. in Computer Engineering from the Faculty of Engineering, Mansoura University, in 1990 and 1997, respectively. She was awarded her Ph.D. degree as a joint researcher between Tanta University, Egypt, and the University of Connecticut, USA. She is now working as a full professor and head of the Computers and Control Department, Tanta University, Egypt. Her interests are in the areas of: networking, distributed systems, image and video processing, GPUs, and distributed computing. (email: Amany_sarhan@f-eng.tanta.edu.eg)
References
- 1.Albahli S. Efficient gan-based chest radiographs (cxr) augmentation to diagnose coronavirus disease pneumonia. Int. J. Med. Sci. 2020;17:1439–1448. doi: 10.7150/ijms.46684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shibly K.H., Dey S.K., Islam M.T.U., Rahman M.M. medRxiv; 2020. Covid Faster R-Cnn: A Novel Framework to Diagnose Novel Coronavirus Disease (Covid-19) in X-Ray Images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basu S., Campbell R.H. medRxiv; 2020. Going by the Numbers: Learning and Modeling Covid-19 Disease Dynamics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sethy P.K., Behera S.K. Preprints; 2020. Detection of coronavirus disease (covid-19) based on deep features; p. 2020. 2020030300. [Google Scholar]
- 5.Hassantabar S., Ahmadi M., Sharifi A. Diagnosis and detection of infected tissue of covid-19 patients based on lung x-ray image using convolutional neural network approaches. Chaos, Solitons & Fractals. 2020;140:110170. doi: 10.1016/j.chaos.2020.110170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ragab D.A., Sharkas M., Marshall S., Ren J. Breast cancer detection using deep convolutional neural networks and support vector machines. PeerJ. 2019;7 doi: 10.7717/peerj.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao Z., Li J., Guan Z., Ye Y., Chen Y. Liver disease screening based on densely connected deep neural networks. Neural Network. 2020;123:299–304. doi: 10.1016/j.neunet.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Pacal I., Karaboga D., Basturk A., Akay B., Nalbantoglu U. A comprehensive review of deep learning in colon cancer. Comput. Biol. Med. 2020:104003. doi: 10.1016/j.compbiomed.2020.104003. [DOI] [PubMed] [Google Scholar]
- 9.Gao X.W., Hui R., Tian Z. Classification of ct brain images based on deep learning networks. Comput. Methods Progr. Biomed. 2017;138:49–56. doi: 10.1016/j.cmpb.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M., Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ausawalaithong W., Thirach A., Marukatat S., Wilaiprasitporn T. 2018 11th Biomedical Engineering International Conference (BMEiCON) IEEE; 2018. Automatic lung cancer prediction from chest x-ray images using the deep learning approach; pp. 1–5. [Google Scholar]
- 12.Elshennawy N.M., Ibrahim D.M. Deep-pneumonia framework using deep learning models based on chest x-ray images. Diagnostics. 2020;10:649. doi: 10.3390/diagnostics10090649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abbas A., Abdelsamea M.M., Gaber M.M. 2020. Classification of Covid-19 in Chest X-Ray Images Using Detrac Deep Convolutional Neural Network. arXiv preprint arXiv:2003.13815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asif S., Wenhui Y., Jin H., Tao Y., Jinhai S. medRxiv; 2020. Classification of Covid-19 from Chest X-Ray Images Using Deep Convolutional Neural Networks. [Google Scholar]
- 15.Hemdan E.E.-D., Shouman M.A., Karar M.E. 2020. Covidx-net: A Framework of Deep Learning Classifiers to Diagnose Covid-19 in X-Ray Images. arXiv preprint arXiv:2003.11055. [Google Scholar]
- 16.Zheng C., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Wang X. medRxiv; 2020. Deep Learning-Based Detection for Covid-19 from Chest Ct Using Weak Label. [Google Scholar]
- 17.Zhao J., Zhang Y., He X., Xie P. 2020. Covid-ct-dataset: a Ct Scan Dataset about Covid-19. arXiv preprint arXiv:2003.13865. [Google Scholar]
- 18.Gozes O., Frid-Adar M., Sagie N., Zhang H., Ji W., Greenspan H. 2020. Coronavirus Detection and Analysis on Chest Ct with Deep Learning. arXiv preprint arXiv:2004.02640. [Google Scholar]
- 19.Wang S., Kang B., Ma J., Zeng X., Xiao M., Guo J., Cai M., Yang J., Li Y., Meng X., et al. MedRxiv; 2020. A Deep Learning Algorithm Using Ct Images to Screen for Corona Virus Disease (Covid-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., et al. Artificial intelligence distinguishes covid-19 from community acquired pneumonia on chest ct. Radiology. 2020;296(2):65–71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J., Wu L., Zhang J., Zhang L., Gong D., Zhao Y., Chen Q., Huang S., Yang M., Yang X., et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Sci. Rep. 2020;10:1–11. doi: 10.1038/s41598-020-76282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang B., Jin S., Yan Q., Xu H., Luo C., Wei L., Zhao W., Hou X., Ma W., Xu Z., et al. Ai-assisted ct imaging analysis for covid-19 screening: building and deploying a medical ai system. Appl. Soft Comput. 2020:106897. doi: 10.1016/j.asoc.2020.106897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamac M., Ahishali M., Degerli A., Kiranyaz S., Chowdhury M.E., Gabbouj M. 2020. Convolutional Sparse Support Estimator Based Covid-19 Recognition from X-Ray Images. arXiv preprint arXiv:2005.04014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh Y., Park S., Ye J.C. IEEE Transactions on Medical Imaging; 2020. Deep Learning Covid-19 Features on Cxr Using Limited Training Data Sets. [DOI] [PubMed] [Google Scholar]
- 25.Loey M., Smarandache F., Khalifa N.E.M. Within the lack of chest covid-19 x-ray dataset: a novel detection model based on gan and deep transfer learning. Symmetry. 2020;12:651. [Google Scholar]
- 26.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of covid-19 cases using deep neural networks with x-ray images. Comput. Biol. Med. 2020:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalifa N.E.M., Taha M.H.N., Hassanien A.E., Elghamrawy S. 2020. Detection of Coronavirus (Covid-19) Associated Pneumonia Based on Generative Adversarial Networks and a Fine-Tuned Deep Transfer Learning Model Using Chest X-Ray Dataset. arXiv preprint arXiv:2004.01184. [Google Scholar]
- 28.Ardakani A.A., Kanafi A.R., Acharya U.R., Khadem N., Mohammadi A. Application of deep learning technique to manage covid-19 in routine clinical practice using ct images: results of 10 convolutional neural networks. Comput. Biol. Med. 2020:103795. doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maghdid H.S., Asaad A.T., Ghafoor K.Z., Sadiq A.S., Khan M.K. 2020. Diagnosing Covid-19 Pneumonia from X-Ray and Ct Images Using Deep Learning and Transfer Learning Algorithms. arXiv preprint arXiv:2004.00038. [Google Scholar]
- 30.Yan T., Wong P.K., Ren H., Wang H., Wang J., Li Y. Automatic distinction between covid-19 and common pneumonia using multi-scale convolutional neural network on chest ct scans. Chaos, Solitons & Fractals. 2020;140:110153. doi: 10.1016/j.chaos.2020.110153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perumal V., Narayanan V., Rajasekar S.J.S. Detection of Covid-19 using Cxr and Ct images using transfer learning and haralick features. Appli. Intell. 2020:1–18. doi: 10.1007/s10489-020-01831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narin A., Kaya C., Pamuk Z. 2020. Automatic Detection of Coronavirus Disease (Covid-19) Using X-Ray Images and Deep Convolutional Neural Networks. arXiv preprint arXiv:2003.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chowdhury M.E., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam K.R., Khan M.S., Iqbal A., Al-Emadi N., et al. 2020. Can Ai Help in Screening Viral and Covid-19 Pneumonia? arXiv preprint arXiv:2003.13145. [Google Scholar]
- 34.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., Yu L., Ni Q., Chen Y., Su J., et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6(10):1122–1129. doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pereira R.M., Bertolini D., Teixeira L.O., Silla C.N., Jr., Costa Y.M. Covid-19 identification in chest x-ray images on flat and hierarchical classification scenarios. Comput. Methods Progr. Biomed. 2020:105532. doi: 10.1016/j.cmpb.2020.105532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J., Xie Y., Li Y., Shen C., Xia Y. 2020. Covid-19 Screening on Chest X-Ray Images Using Deep Learning Based Anomaly Detection. arXiv preprint arXiv:2003.12338. [Google Scholar]
- 37.Hall L.O., Paul R., Goldgof D.B., Goldgof G.M. 2020. Finding Covid-19 from Chest X-Rays Using Deep Learning on a Small Dataset. arXiv preprint arXiv:2004.02060. [Google Scholar]
- 38.Hammoudi K., Benhabiles H., Melkemi M., Dornaika F., Arganda-Carreras I., Collard D., Scherpereel A. 2020. Deep Learning on Chest X-Ray Images to Detect and Evaluate Pneumonia Cases at the Era of Covid-19. arXiv preprint arXiv:2004.03399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farooq M., Hafeez A. 2020. Covid-resnet: A Deep Learning Framework for Screening of Covid19 from Radiographs. arXiv preprint arXiv:2003.14395. [Google Scholar]
- 40.Wang L., Lin Z.Q., Wong A. Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song Y., Zheng S., Li L., Zhang X., Zhang X., Huang Z., Chen J., Zhao H., Jie Y., Wang R., et al. medRxiv; 2020. Deep Learning Enables Accurate Diagnosis of Novel Coronavirus (Covid-19) with Ct Images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S., Zha Y., Li W., Wu Q., Li X., Niu M., Wang M., Qiu X., Li H., Yu H., et al. A fully automatic deep learning system for covid-19 diagnostic and prognostic analysis. Eur. Respir. J. 2020;56(2):1–11. doi: 10.1183/13993003.00775-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manski C.F. National Bureau of Economic Research; 2020. Bounding the Predictive Values of COVID-19 Antibody Tests, Technical Report. [Google Scholar]
- 44.Ozkaya U., Ozturk S., Barstugan M. 2020. Coronavirus (Covid-19) Classification Using Deep Features Fusion and Ranking Technique. arXiv preprint arXiv:2004.03698. [Google Scholar]
- 45.Bhandary A., Prabhu G.A., Rajinikanth V., Thanaraj K.P., Satapathy S.C., Robbins D.E., Shasky C., Zhang Y.-D., Tavares J.M.R., Raja N.S.M. Deep-learning framework to detect lung abnormality–a study with chest x-ray and lung ct scan images. Pattern Recogn. Lett. 2020;129:271–278. [Google Scholar]
- 46.KW C.D., Cheng Y.P.S.M., Hui K.P., Krishnan P., Liu Y., Ng D.Y. Deep-learning framework to detect lung abnormality–a study with chest x-ray and lung ct scan images. Clin. Chem. 2020;66:549–555. [Google Scholar]
- 47.Paul C.J., Morrison P., Dao L., Roth K., Duong T.Q., Ghassemi M. 2020. Covid-19 Image Data Collection: Prospective Predictions Are the Future. arXiv preprint arXiv:2006.11988. [Google Scholar]
- 48.Kermany D.S., Goldbaum M., Cai W., Valentim C.C., Liang H., Baxter S.L., McKeown A., Yang G., Wu X., Yan F., et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172:1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 49.Challenge R.P.D. 2018. Radiological Society of North america. [Google Scholar]
- 50.Mooney P. Chest x-ray images (pneumonia) 2020. https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia
- 51.Shiraishi J., Katsuragawa S., Ikezoe J., Matsumoto T., Kobayashi T., Komatsu K.-i., Matsui M., Fujita H., Kodera Y., Doi K. Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists' detection of pulmonary nodules. Am. J. Roentgenol. 2000;174:71–74. doi: 10.2214/ajr.174.1.1740071. [DOI] [PubMed] [Google Scholar]
- 52.Bisong E. Springer; 2019. Building Machine Learning and Deep Learning Models on Google Cloud Platform. [Google Scholar]
- 53.Gulli A., Pal S. Packt Publishing Ltd; 2017. Deep Learning with Keras. [Google Scholar]
- 54.Kingma D.P., Ba J. 2014. Adam: A Method for Stochastic Optimization. arXiv preprint arXiv:1412.6980. [Google Scholar]
- 55.GitHub Deep-chest: multi-classification deep learning model for diagnosing covid-19, pneumonia and lung cancer chest diseases. 2020. https://shorturl.at/lzAF6 [DOI] [PMC free article] [PubMed]
- 56.Ruuska S., Hämäläinen W., Kajava S., Mughal M., Matilainen P., Mononen J. Evaluation of the confusion matrix method in the validation of an automated system for measuring feeding behaviour of cattle. Behav. Process. 2018;148:56–62. doi: 10.1016/j.beproc.2018.01.004. [DOI] [PubMed] [Google Scholar]