Graphical abstract
Keywords: Luciferase immunosorbent assay, SARS-CoV-2, Semi-quantitative, Antibody detection
Abstract
In this study, we developed and evaluated a luciferase immunosorbent assay (LISA) for quantitative detection of IgG antibody against SARS-CoV-2 nucleoprotein (NP). Anti-SARS-CoV-2 NP antibody in serum or plasma samples was captured by protein G-coated microtiter plate and detected using the crude cell lysates expressing Nanoluc luciferase (Nluc) enzyme fused with SARS-CoV-2 NP. After the addition of furimazine substrate, the levels of anti-SARS-CoV-2 NP IgG antibody were quantitatively measured as luciferase light units. As expected, SARS-CoV-2 NP showed cross-reactivity with the monoclonal antibodies against SARS-CoV NP, but not MERS-CoV NP-specific monoclonal antibodies or the monoclonal antibodies against SARS-CoV Spike protein. LISA for detecting murine monoclonal antibody against SARS-CoV NP showed a low limit of detection of 0.4 pg/μl and linear detection range from 0.4 pg/μl to 75 pg/μl. Furthermore, LISA had a sensitivity of 71 % when testing COVID-19 patients at the second week post onset and a specificity of 100 % when testing healthy blood donors.
1. Introduction
We are facing serious threats of emerging infectious diseases to public health and social security particularly when we are experiencing the pandemic of coronavirus disease 2019 (COVID-19) caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection(Chan et al., 2020a, b; Lu et al., 2020; Zhu et al., 2020). To control the epidemics of emerging infectious diseases, high-throughput serological assays are important for screening population infection and evaluating population immunity, and tracing the source and animal hosts of infection. In this study, we evaluated a luciferase immunosorbent assay (LISA), which is easily and quickly developed, semi-quantitative and is suitable for detecting the specific antibody in a wide range of species including human and animals.
2. Construction and characterization of the recombinant proteins comprised of SARS-CoV-2 nucleoprotein fused with luciferase
The gene fragment of nucleoprotein (aa 1–419) of SARS-CoV-2 (NCBI accession number MN908947) was amplified by RT-PCR using the following primers: 5′-GCGATCGCTTCCGAATTCATGTCTGATAATGGAC-3′ and 5′-CGGTTGAGCTCTGAATTCTTAGGCCTGAGTTGAG-3′. The PCR products were purified and subcloned into the pNLF1-N vector (Promega, Madison, USA) downstream of the Nluc luciferase gene with a linker of 30 base pairs. DNA sequencing was performed to confirm the integrity of the Nluc-SARS-CoV-2 NP fusion constructs. The resulting plasmid DNA was prepared by using DNA purification kit (TIANGEN Biotech, Beijing, China) and used for cell transfection. For transfection experiments, human embryonic kidney (HEK) 293 T cells were seeded in a cell culture plate (100 mm2) and cultured till the confluence of >85 %, then transfected with the recombinant plasmids using Lipofectamine™ 3000 Transfection Reagent (Invitrogen, Carlsbad, USA) according to the manufacturer's instructions. Briefly, 5 μg recombinant plasmid DNA was diluted with 240 μL Opti-MEN™ reduced serum medium (Invitrogen, Carlsbad, USA), and mixed with 10 μl P3000™ reagent (Invitrogen, Carlsbad, USA) followed by incubation at room temperature for 15 min. Then, the DNA-lipid complex was added into 293 T cells to culture for 48 h. The cells were washed twice with 0.01 M phosphate-buffered saline (PBS) and treated with 0.25 % trypsin, and then lysed on ice for 30 min in the lysis buffer developed by Burbelo et al(Burbelo et al., 2009). The lysis buffer composed of 50 mM Tris, pH7.5, 100 mM NaCl, 5 mM MgCl2, 1% Triton X-100, 50 % glycerol and protease inhibitors (Roche, Mannheim, Germany). The cell lysates containing Nluc-fusion antigen were harvested after centrifugation at 12,000 × rpm for 5 min at 4 °C and then stored at −20 °C. The expression of SARS-CoV-2 NP fragments in the fusion proteins was verified by western blotting (WB) by using anti-luciferase antibody (DingGuoChangSheng Biotech, Beijing, China) and SARS-CoV NP-specific monoclonal antibody (N-14A3, Zhujiang Hospital, Southern Medical University, Guangzhou, China), respectively (Supplementary Fig. 1). Luciferase activity of the crude cell extracts was measured by adding an equal volume of Nano-Glo Luciferase assay reagent (Promega, Madison, USA) in a Tecan infinite M200 PRO microplate luminometer (Tecan, Zurich, Switzerland). To avoid the difference in expression of the luciferase tagged antigen across lysate preparations and the potential difference in luciferase activity for each transfection, the light units (LU) of each cell lysate preparation was quantified to adjust the LU of cell lysate to 108 light units/ul.
3. Development of LISA-based method for detecting antibody against SARS-CoV-2 nucleoprotein
Briefly, 96-well Costar flat-bottomed luminometry plates (Corning, New York, USA) were coated with 50 μL of 5 μg/mL protein G (Genscript, Nanjing, China) in carbonate buffer (pH 9.6) overnight at 4 °C. The best concentration of protein G for coating plate was experimentally determined to be 5 ug/mL and 250 ng per well in our assay. After three washes with phosphate-buffered saline (PBS) containing 0.05 % Tween 20 (PBS-T), the plates were incubated with blocking solution (PBS containing 5% non-fat milk) for 1 h at 37 °C. The wells were then washed, and incubated with 50 μL of diluted sera (1:100 dilution in PBS containing 2% non-fat milk) for 1 h at 37 °C. After washing, the plates were incubated with 50 μL of the 1:1000 diluted crude cell extracts with Nluc-fusion proteins (5 × 106 light units) for 30 min at 37 °C followed by the addition of 50 μL luciferase substrate (Promega, Madison, USA) to each well to determine the luciferase light units according to the manufacturer’s protocol. In each assay run, a single negative serum sample and a PBS blank control were used as the negative control. The background value of the blank control was not subtracted from all the wells. The final results were expressed as the ratio of sample/cut-off value. Each sample was tested in triplicate. The cut-off value was derived from the average value plus 3 standard deviations (SD) of the negative controls that are the serum samples from healthy blood donors (median 31.0 years old, IQR 28.0–41.0).
4. The sensitivity and specificity of LISA for detecting anti-SARS-CoV-2 NP IgG antibody
A total of 492 serum samples (median 5.0 per patient, IQR, 4.0–6.0) were collected from 98 COVID-19 patients in Guangzhou Eighth People's Hospital, Guangzhou, China. Demographic and clinical outcome data were extracted from the electronic medical records. Written informed consents were obtained from the individuals enrolled in this study, which has been approved by Ethics Committees of Guangzhou Eighth People's Hospital (No. 202,002,136). The demographic and clinical characteristics of COVID-19 patients were provided in the supplementary material (Supplementary Table 1). The COVID-19 patients were all laboratory-confirmed cases by using reverse-transcription polymerase chain reaction assay targeting both nucleoprotein and ORF1a/b genes of SARS-CoV-2 according to the manufacturer’s protocols (DaAn Gene company, Guangzhou, China). 88 serum samples of healthy blood donors were used as negative controls since they were collected in 2016 when no SARS-CoV-2 infection was identified and were negative for SARS-CoV-2 RNA. Based on LISA, the percentage of anti-SARS-CoV-2 NP IgG seroconversion was 42.7 % (35/82) during 0–7 days post onset (d.p.o.), then increased to 71.2 % (104/146) 8–14 d.p.o., 96.7 % (117/121) 15–21 d.p.o., 98.4 % (61/62) 21–28 d.p.o.and 100.0 % (81/81) more than 28 d.p.o.
Furthermore, the COVID-19 patients were stratified according to the disease severity to investigate their antibody response and to evaluate the performance of LISA. The level of IgG antibodies against SARS-CoV-2 nucleoprotein was not different between severe/critical patients and moderate/mild patients during the first week after onset (p = 0.588). However, the level of antibodies against SARS-CoV-2 nucleoprotein for severe or critical patients was significantly higher than that of mild or moderate patients (p<0.001). Higher detecting sensitivity of LISA was observed among severe or critical patients (86.4 %) than mild or moderate patients (66.4 %, p = 0.0326) at the second week after onset (Fig. 1 ).
Fig. 1.
Distribution of SARS-CoV-2 NP-specific antibody levels as determined by LISA among healthy blood donors and COVID-19 patients. The antibody level of COVID-19 mild or moderated patient (green) and severe or critical patient (red) was highlighted in different colors. Dash lines indicate the cut-off value for LISA. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Our preliminary data also indicated a better performance of our LISA than other reported serological assays. Van Elslande el at. evaluated the diagnostic performance of seven lateral flow assay (LFA) and the Euroimmun ELISA in COVID-19 patients and showed the sensitivity of 21.6 %∼40.5 % and 55.1 %∼71.8 % during the first and the second week post onset, respectively (Van Elslande et al., 2020). Liu et al. reported the positive rate of NP-based IgG ELISA less than 40 % during <10 d.p.o. (Liu et al., 2020). Furthermore, at more than 14 d.p.o., the performance of LISA (96.7–100 % sensitivity and 100 % specificity) is comparable with that of the luciferase immunoprecipitation system (LIPS) assays (100 % sensitivity and 100 % specificity) developed by Burbelo et al. (Burbelo et al., 2020). However, LIPS needs pre-incubation of Ruc-antigen and samples and a plate washer with vacuum to capture the antigen-antibody-beads complex(Burbelo et al., 2009). Our LISA is simplified by using protein G coated 96-well microtiter plate to capture the total IgG antibody from a wide range of species and followed by the addition of Nluc-fusion antigen and luciferase specific reagent. Therefore, it is a simple and universal testing system. More importantly, LISA is more suitable for rapid development of serological assays for detecting and diagnosing emerging infectious diseases in the light of no need for purification process when preparing target antigens.
5. Cross-reactivity of the monoclonal antibodies against MERS-CoV nucleoprotein or SARS-CoV with SARS-CoV-2 nucleoprotein
96-well white microtiter plates coated with 5 μg/mL protein G were incubated with 50 μL of serially diluted monoclonal antibodies (mAb) against MERS-CoV nucleoprotein or SARS-CoV for 1 h at 37 °C. The plates were then incubated with the diluted Nluc-SARS-CoV-2 NP for 30 min at 37 °C followed by the addition and incubation of luciferase substrate. The cross reactivity of the monoclonal antibodies against MERS-CoV NP or SARS-CoV with SARS-CoV-2 NP was determined by the luciferase light units. Each sample was tested in triplicate. The cut-off value was derived from the average value plus 3 standard deviations (SD) of the negative controls.
In this study, a total of 4 monoclonal antibodies (mAb) against SARS-CoV used, in which the two mAb (N-14A3 and N-1E8) were produced by immunizing BALB/c mice with the recombinant NP protein of SARS-CoV so that they are specifically target the SARS-CoV nucleoprotein(Che et al., 2003). In addition, the other two mAb (E26A15 and E14A9) were obtained by immunizing Balb/c mice with inactivated SARS-CoV isolates. Aforementioned four monoclonal antibodies were generously provided by the Central Laboratory, Zhujiang Hospital, Southern Medical University, Guangzhou, China, but the epitope mapping and biological function analysis of mAb E26A15 and E14A9 remain to be done. A dose-dependent reactivity was observed among the 2 mAb against SARS-CoV NP, i.e. N-14A3 (Fig. 2 A), N-1E8 (Fig. 2B), and the 2 mAb against SARS-CoV, i.e. E26A15 (Fig. 2C) and E14A9 (Fig. 2D), but not the mAb against MERS-CoV NP (Fig. 2E). LISA showed a linear range of detection from 0.4 pg/μl to 75 pg/μl (Fig. 2F). We would like to emphasize that the quantitation range of LISA was determined based on murine monoclonal antibody against SARS-CoV NP, not the human antibody against SARS-CoV-2 due to the lack of anti-SARS-CoV-2 NP antibody. However, it has been reported that the affinity of protein G for human IgG is higher than that of murine IgG, therefore, in theory, a wider quantitation range of LISA would be achieved for detecting human IgG antibodies.
Fig. 2.
Detection of cross-reactivity of SARS-CoV specific monoclonal antibodies (mAbs) with SARS-CoV-2 nucleoprotein. The binding of SARS-CoV NP-specific mouse mAbs (A, B) or SARS-CoV specific mouse mAbs (C, D) or MERS-CoV NP-specific mouse mAbs (E) to SARS-CoV-2 NP was detected by LISA, respectively. F, LISA showed a linear range of detection from 0.4 pg/μl to 75 pg/μl when detecting SARS-CoV NP-specific mouse mAbs (N-14A3, N-1E8) or SARS-CoV specific mouse mAbs (E26A15, E14A9). For all panels, the data are presented as mean LU/cutoff ± standard deviation of the mean LU/cutoff (s.e.m.) and Y-axes are labelled as "LU fold increase/cutoff" for all panels.
6. Competitive inhibition assay to validate the cross-reactivity between the monoclonal antibodies against SARS-CoV or MERS-CoV nucleoprotein and SARS-CoV-2 nucleoprotein
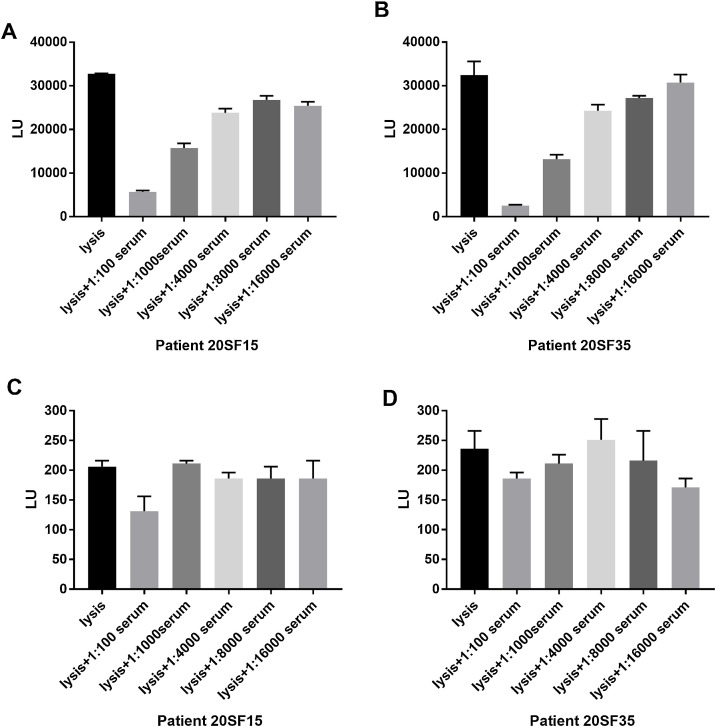
96-well white microtiter plates coated with 5 μg/mL of SARS-CoV NP-specific mAb (N-14A3, Zhujiang Hospital, Southern Medical University, Guangzhou, China) or MERS-CoV NP-specific mAb (Zhujiang Hospital, Southern Medical University, Guangzhou, China) were incubated with 25 μL of the 1:10000 diluted Nluc-SARS-CoV-2 NP and 25 μL of serially diluted sera from COVID-19 patients for 30 min at 37 °C. Then the wells were washed, and 50 μL of luciferase substrate were added to each well to determine the luciferase light units. Each sample was tested in triplicate. The cross reactivity between the mAb of anti-SARS-CoV NP and SARS-CoV-2 NP was inhibited by about 75 % using 1:100 diluted serum from CVOID-19 patient 20SF15 (Fig. 3 A) and 94 % using 1:100 diluted serum from CVOID-19 patient 20SF35 (Fig. 3B) with a dose-dependent response. However, the sera from the two COVID-19 patients could not inhibit the cross reactivity between the mAb of anti-MERS-CoV NP and SARS-CoV-2 NP (Fig. 3C and D). These results indicate the specificity of SARS-CoV-2 NP-based LISA for detecting antibodies against SARS-CoV NP and SARS-CoV-2 NP due to the 94 % homogeneity of these two NP (Chan et al., 2020a).
Fig. 3.
Dose-dependent inhibitory effect of SARS-CoV-2 NP specific antibody from serum of COVID-19 patients on the binding of Nanoluc-antigens crude cell extract to SARS-CoV NP-specific monoclonal antibody (A, B) or MERS-CoV NP-specific monoclonal antibody (C, D) immobilized on microtiter plate wells. The data are presented as mean LU ± standard deviation of the mean LU (s.e.m.).
There are several limitations in our study. First, in this study, no serum samples from patients infected with other acute respiratory infections were tested. Therefore, the potential cross-reactivity of the LISA assay with other respiratory pathogens could not be evaluated although no false positive results were observed in the healthy blood donors collected before the COVID-19 pandemic. Second, the high degree of similarity between SARS-CoV and SARS-CoV-2 NP sequences and the cross-reactivity between the specific monoclonal antibodies against SARC-CoV NP and serum samples from SARS-CoV-2 infected patients could overestimate the seroprevalence of SARS-CoV-2 infection among the population ever experienced SARS-CoV infection; however, it has been reported that SARS-CoV specific antibodies could not be detected in about 90 % (21/23) serum samples of SARS patients 6 years after infection(Tang et al., 2011). Therefore, false-positives results caused by the cross-reactivity of SARS-CoV specific antibodies presented in the population would be rare because SARS-CoV infection has not been documented since 2003. Third, in the current study, we have only detected human serum samples, and did not evaluate the performance of this SARS-CoV-2 specific serological assay in testing samples of various animal species due to the lack of these samples. However, in our previous study, we evaluated and found that the LISA assay achieved very good performance in detecting MERS-CoV specific IgG antibody in various animal samples including camel, marmoset, rhesus and mice(Wang et al., 2019).
In conclusion, a LISA assay for detection of antibody against SARS-CoV-2 nucleoprotein was developed and evaluated, and demonstrated superior performance in discriminating COVID-19 patients and uninfected subjects. Additionally, attributed to the high homogeneity between SARS-CoV NP and SARS-CoV-2 NP, SARS-CoV-2 NP showed cross-reactivity with SARS-CoV NP-specific monoclonal antibodies.
Credit authorship contribution statement
Yuanhao Liang: analyzed and interpretated data, made the tables and figures, drafted the article. Huanchang Yan: collected and analyzed data. Liping Huang: collected and analyzed data. Jianhui Zhao: collected and analyzed data. Haiying Wang: collected and analyzed data. Min Kang: collected and analyzed data. Zhengwei Wan: collected and analyzed data. Jingwei Shui: collected and analyzed data. Shixing Tang: designed the study, revised the article critically for important intellectual content.
Funding
This work was supported by the National Major Science and Technology Project (grant number 2017ZX10202101-003, 2018ZX10302103-002 and 2017ZX10202102-003-004).
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2021.114141.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Burbelo P.D., Ching K.H., Klimavicz C.M., Iadarola M.J. Antibody profiling by luciferase immunoprecipitation systems (LIPS) J. Visualized Exp.: JoVE. 2009 doi: 10.3791/1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbelo P.D., Riedo F.X., Morishima C., Rawlings S., Smith D., Das S., Strich J.R., Chertow D.S., Davey R.T., Cohen J.I. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J. Infect. Dis. 2020;222:206–213. doi: 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To K.K.-W., Yuan S., Yuen K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., Tsoi H.W., Lo S.K., Chan K.H., Poon V.K., Chan W.M., Ip J.D., Cai J.P., Cheng V.C., Chen H., Hui C.K., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet (London, England) 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che X.Y., Qiu L.W., Pan Y.X., Xu H., Hao W., Liao Z.Y., Mei Y.B., Zhang L.Y., Wan Z.Y., Yuan G.Y., Huang Z. Rapid and efficient preparation of monoclonal antibodies against SARS-associated coronavirus nucleocapsid protein by immunizing mice. Di 1 jun yi da xue xue bao = Acad. J. First Med. College of PLA. 2003;23:640–642. [PubMed] [Google Scholar]
- Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S., Xiong Z., Zheng S. Evaluation of nucleocapsid and Spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet (London, England) 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Quan Y., Xin Z.T., Wrammert J., Ma M.J., Lv H., Wang T.B., Yang H., Richardus J.H., Liu W., Cao W.C. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. (Baltimore, Md.: 1950) 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- Van Elslande J., Houben E., Depypere M., Brackenier A., Desmet S., André E., Van Ranst M., Lagrou K., Vermeersch P. Diagnostic performance of seven rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin. Microbial. Infect.: Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2020;26:1082–1087. doi: 10.1016/j.cmi.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Wang T., Deng Y., Niu P., A. R, Zhao J., Peiris M., Tang S., Tan W. A novel luciferase immunosorbent assay performs better than a commercial enzyme-linked immunosorbent assay to detect MERS-CoV specific IgG in humans and animals. J. Biosaf. Health Educ. 2019;1:134–143. doi: 10.1016/j.bsheal.2019.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.