Abstract
BACKGROUND AND PURPOSE: Alzheimer disease (AD) is a neurodegenerative disease characterized by progressive dementia. The hippocampus is particularly vulnerable to damage at the very earliest stages of AD. This article seeks to evaluate critical AD-associated regional changes in the hippocampus using machine learning methods.
MATERIALS AND METHODS: High-resolution MR images were acquired from 19 patients with AD and 20 age- and sex-matched healthy control subjects. Regional changes of bilateral hippocampi were characterized using computational anatomic mapping methods. A feature selection method for support vector machine and leave-1-out cross-validation was introduced to determine regional shape differences that minimized the error rate in the datasets.
RESULTS: Patients with AD showed significant deformations in the CA1 region of bilateral hippocampi, as well as the subiculum of the left hippocampus. There were also some changes in the CA2–4 subregions of the left hippocampus among patients with AD. Moreover, the left hippocampal surface showed greater variations than the right compared with those in healthy control subjects. The accuracies of leave-1-out cross-validation and 3-fold cross-validation experiments for assessing the reliability of these subregions were more than 80% in bilateral hippocampi.
CONCLUSION: Subtle and spatially complex deformation patterns of hippocampus between patients with AD and healthy control subjects can be detected by machine learning methods.
Alzheimer disease (AD) is a neurodegenerative disease characterized by progressive dementia. Neurofibrillary tangles and amyloid plaques in the brain of patients with AD can be identified by histologic examination.1–3 These pathologic changes are typically associated with neuronal loss and volume reductions. The hippocampus, part of the mesial temporal lobe memory system,4 is particularly vulnerable to damage at the very earliest stages of AD.3,5
MR imaging-driven volumetric studies have shown hippocampal atrophy in mild cognitive impairment (MCI) and AD.6–9 These volumetric measures proved to be more consistent than currently used mental state examinations and clinical rating scales.10 In addition, voxel-based morphometry (VBM),11 as an automated unbiased analysis of the differences in tissue concentration throughout the brain on structural MR imaging scans, has been widely used in brain tissue loss studies in AD.12-16 These VBM studies reported bilateral gray matter loss in the hippocampus. However, the above 2 methods cannot accurately detect regional abnormalities of the hippocampal atrophy.
Many recent studies17–20 have focused on characterizing regional abnormalities of hippocampal atrophy using computational anatomy mapping and statistical analysis methods. These regional surface measures of the hippocampus can provide more subtle indexes compared with the volume and tissue concentration differences in discriminating between patients with AD and healthy control subjects. They were used not only to distinguish subjects with very mild AD from nondemented subjects18 but also to track the progression of AD in drug trials.19 Usually, regional shape abnormalities of the hippocampus are statistically analyzed by univariate methods based on some previous hypotheses. However, statistical comparisons using these approaches had limitations in identifying subtle differences between 2 populations.21 Discriminative analysis based on the classifier function can potentially be used to improve understanding of detected differences between populations, as well as to identify possible dependencies in the features.22,23 Therefore, the purpose of this study was to use machine learning methods to characterize the hippocampal shape changes in AD and to construct a classifier function that could differentiate patients with AD from healthy control subjects.
Materials and Methods
Subjects
Subjects included 19 patients with AD and 20 healthy control subjects. The age, sex, and education level were matched in the 2 groups. The patients with AD were recruited after clinical diagnostic examinations, whereas the control subjects were recruited from the local community. Patients with AD were submitted to clinical, physical, and neurologic examination, as well as a battery of neuropsychiatric and laboratory tests. The examination results fulfilled all of the National Institute of Neurologic and Communicative Disorders and Stroke-Alzheimer's Disease and Related Disorders Association work group criteria.24 The cognitive status of each subject was evaluated using the Mini-Mental State Examination.25 Subjects were excluded if they presented with symptoms typically observed for other neuropsychiatric disorders. All of the subjects were right-handed. Informed consent was first acquired from each subject before the examination. Table 1 depicts the demographic details of the subjects.
Table 1:
Demographics and clinical findings
| Variable | AD (n = 19) | Controls (n = 20) | P |
|---|---|---|---|
| Sex, F/M | 9/10 | 10/10 | >.95* |
| Age, mean ± SD | 72.6 ± 6.9 | 70.7 ± 6.4 | .38† |
| Education, mean ± SD | 11.3 ± 4.0 | 10.9 ± 4.5 | .76† |
| MMSE score, mean ± SD | 18.9 ± 3.9 | 29.5 ± 0.9 | <.0001† |
Note:—MMSE indicates Mini-Mental State Examination; AD, Alzheimer disease; F, female; M, male.
The P value was obtained by Pearson χ2 2-tailed test, with continuity correction for n < 5.
The P value was obtained by a 2-sample 2-tailed t test.
Image Acquisition and Preprocessing
All of the MR imaging was carried out using a 1.5T MR scanner (Signa 1.5T Twinspeed; GE Healthcare, Milwaukee, Wis) equipped with shielded magnetic field gradients of up to 40 mT/m. A standard head coil was used for radio-frequency transmission and reception of the nuclear MR signal intensity. Head motion was minimized with restraining foam pads supplied by the manufacturer. High-resolution 3D T1-weighted images (TR, 11.3 ms; TE, 4.2 ms; inversion time, 400 ms; flip angle, 15°; FOV, 24 × 24 cm; matrix,256 × 224; section thickness, 1.8 mm; NEX, 2) were acquired by a spoiled gradient-recalled sequence with axial volume excitation.
All of the images were preprocessed in 2 steps. Firstly, MRIcro software (http://www.psychology.nottingham.ac.uk/staff/cr1/mricro.html) was used to manually align the scans, which made the coronal direction perpendicular to the long axis of the hippocampus (maximum anatomic delineation of the hippocampal formation). Before manual delineation, each image was corrected for inhomogeneity in the magnetic field.26
Hippocampal Delineation
The subjects were randomly divided into 2 groups. One rater blind to the diagnosis and demographics of the subject population manually traced the bilateral hippocampi in each group. Boundaries of the hippocampus were drawn on coronal MR images in a plane perpendicular to the long axis of the hippocampus according to a standard neuroanatomic atlas of the hippocampus.27 The delineation of the hippocampus included the cornu ammonis (CA), the subiculum, and the dentate gyrus. Hippocampal contours were delineated in the contiguous coronal brain sections. This process took approximately 1 hour per scan (including the left and right hippocampi). Anatomic landmarks were labeled and linked in all 3 of the orthogonal viewing planes using Iris software (http://www.cs.unc.edu/∼gerig/). Boundaries were drawn on magnified images (×4) to allow subvoxel precision and faithful tracking of small-scale features.
To estimate the reliability of measures based on manual outlining, 2 raters traced the hippocampi on 6 randomly selected brain volumes. Interrater correlation coefficient for hippocampal volume measures was 0.92.
Surface-Based Mesh Modeling
To pinpoint hippocampal regional changes in morphology, we used a simple but effective surface-based anatomic mesh modeling method28 that matched homologous hippocampal surface points between individuals. A brief description of the method used is as follows: 1) 2 landmark points of maximal geodesic distance on the surface mesh of the hippocampus were identified (referred to as the head and tail landmarks, respectively); 2) isolatitude circles (shown in cyan in Fig 1A) and an axis (shown in white in Fig 1A) were obtained using a heat conduction model; 3) the dateline (shown in pink in Fig 1A) was constructed by connecting the points of origin of the isolatitude circles; 4) each isolatitude circle was parameterized from its origin point using the normalized arc length (the blue-red hue scale indicating the changes from the head to the tail of hippocampus in Fig 1B); 5) all of the subjects were aligned in terms of the isolatitude circle area along latitude direction29 to refine their correspondences (the 2 curves on the left of Fig 1C show the area distribution of isolatitude circle from head to tail; the red is the template, and the green is one subject before transformation). The result after transformation is shown on the right of Fig 1C. The template of the hippocampus was built by averaging the hippocampal models across all of the subjects.
Fig 1.

The steps of parameterization and correspondence of hippocampus.
A, Hippocampus shape characters; H and T represent the head and tail landmarks, respectively; the cyan shows isolatitude circles; the pink shows the extracted dateline.
B, The parameterized mesh; the blue-red hue scale indicates the changes from the head to the tail of hippocampus.
C, The alignment in terms of the isolatitude circle areas along latitude direction. The 2 curves on the left show the area distribution of isolatitude circles from head to tail; the red is the template and the green is 1 subject before transformation. The results after transformation are shown at the right.
Each hippocampal surface mesh was constructed as a regular parametric grid (200 × 400 surface points) by matching 2 landmarks and a dateline so that homologous grid points from different hippocampal surfaces had correspondences. The matching procedures made it possible to statistically analyze the measurements at corresponding surface locations among different subjects.
Surface-Based Measures
We applied one surface-based measure23 to characterize local changes in the hippocampal surface. To reduce computational complexity, we resampled each hippocampus surface mesh (200 × 400 grids) as m × n patches by combining the neighboring (200/m) × (400/n) grids into 1 grid. The final results (as shown in the Results section) were obtained by selecting the different parameters m and n. Each surface mesh was normalized by setting the head landmark as the origin in a 3D coordinate system. We then averaged the coordinates of the corresponding vertices of the individual hippocampal mesh to construct the mean surface mesh. We represented each patch by a single summary feature as shown in the following equation23:
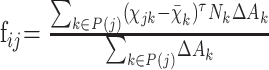 |
where fij was computed for the jth patch in the ith subject, P(j) was a set of the mesh grids belonging to the jth patch, xk and Nk were the position and approximate unit normal of the kth mesh grid in the mean shape, and ΔAk was the area element, computed as one fourth of the summarized area of all meshes that were adjacent to the kth mesh grid in the mean shape. This measure reflected the average inward or outward deformation of each patch with respect to the mean mesh.
Feature Selection for Support Vector Machine
The identification of distinguishing shape features in the hippocampal surface is important and has tremendous practical applications. In this part of the study, we selected the most critical features from the surface-based measures by the method described below.
The feature selection process included 2 steps. In the first step, feature ranking, features were ranked according to the weight magnitude of each feature in the trained linear support vector machine (SVM), which was based on the recursive feature elimination (RFE) criterion.30 In each iteration, the feature of the lowest rank (ie, associated with the smallest weight) was identified and removed. Then the SVM was trained again based on the remaining features; in this way, another least important feature was identified and removed. This process was repeated until all of the features had been used.
The second step was feature selection. The above-ranked features were selected by leave-1-out cross-validation (LOOCV).31 In each leave-1-out cycle, 1 subject was removed from the dataset and used as the test sample. The feature selection was an iterative process, and the criteria were that features were added individually from the top of the rank-ordered list to validate the test sample until the local minimum error rate on the training data was achieved. That is, the feature selection would stop when the error rate was no longer decreasing. The final feature subset was obtained by collecting all of the features selected in the LOOCV.
Validation of the Selected Features
To assess the reliability of selected features, 2 cross-validation experiments were performed on the surface-based measures of the hippocampi of patients with AD and healthy control subjects. One experiment was performed using LOOCV. In each step, 1 subject was removed from the dataset, the remaining subjects were trained to select the optimal features by SVM RFE and LOOCV, and then the selected features were used to construct an effective classifier to test the subject who had been removed. In this way, the classification accuracy was obtained. The flow chart of this experiment is shown in Fig 2, and the numbers inside of the parentheses are the sample indexes.
Fig 2.
The flow chart of LOOCV experiment; the numbers inside of the parentheses are the sample indexes.
The other experiment was to assess the reliability of selected features by using 3-fold cross-validation. The dataset was randomly divided into 3 disjointed subsets of equal size. Features were selected in 2 of these subsets by SVM RFE and LOOCV methods. Then the remaining subset (called the “validation set”) was used to estimate the predictive error of the trained classifier by using the selected features. This process was repeated 100 times. Each time, 1 subset was left out for testing, and the other 2 subsets were trained. The classification accuracy by 3-fold cross-validation experiment was obtained to average the predictive correct rates of 100 experiments.
Results
In the experiment, SVM RFE and LOOCV were used to identify the distinguishing shape features in the hippocampal surfaces of subjects with AD and healthy control subjects. These features were then used to construct an effective classifier. In the study, we adopted the linear SVM as the classifier for training and testing.
Distinguished Feature Selection Using Classification between Groups
The classification accuracies by the LOOCV experiment are shown in Table 2. The classification accuracies of the left hippocampus were more than 90%, and those of the right hippocampus were more than 80%.
Table 2:
Cross-validation accuracies in the experiment
| Patch Size | Side | Leave-1-Out Cross-Validation Accuracy, %* | 3-Fold Cross-Validation Accuracy* |
|---|---|---|---|
| 50 × 100 | Right | 84.6 | 83.4% (81.8%–84.9%)† |
| Left | 94.9 | 84.5% (81.2%–87.9%)† | |
| 25 × 50 | Right | 84.6 | 81.6% (79.8%–83.4%)† |
| Left | 94.9 | 89.6% (87.1%–92.1%)† | |
| 20 × 40 | Right | 84.6 | 82.6% (80.7%–84.6%)† |
| Left | 94.9 | 89.3% (86.8%–91.9%)† |
Note:—All of the features appearing in subsets are selected to construct the classifier.
The 95% confidence intervals of classification accuracy by 100 times cross-validation experiments.
The classification accuracies by the 3-fold cross-validation experiment were all more than 80%. At the same time, the 95% confidence intervals on the cross-validation accuracy were estimated. The detailed results are shown in Table 2.
Effects of Different Patch Sizes on the Results
To test the effects of selecting different hippocampus surface patch sizes on the classification accuracies between patients with AD and healthy control subjects, we resampled the hippocampus surface into 50 × 100, 25 × 50, and 20 × 40 patches. The results are shown in Table 2. When selecting all of the features to construct the classifier, we found that the classification results demonstrated no distinct changes by using different patch sizes.
Effects of Different Strategies for Constructing the Effective Classifiers
After feature selection, we adopted different strategies to construct the effective classifiers. One strategy was to use all of the features appearing in subsets to construct the classifier. The classification results are marked with an asterisk in Table 2. The other strategy was to only select those features in subsets with repeatability greater than or equal to 2–5. The experimental results are shown in Fig 3. The horizontal axis represents the repeatability of the selected features, and the vertical axis represents the LOOCV accuracy of the corresponding repeatability. From this figure, the higher accuracy can be achieved by choosing the optimal number of features.
Fig 3.
LOOCV accuracy when selecting different repeatability of features to construct the classifier. The horizontal axis represents different repeatability of features, and the vertical axis represents the corresponding LOOCV accuracy. The different colors, respectively, represent the left and right hippocampi and different patch sizes. L represents the left hemisphere, and R represents the right hemisphere. The numbers behind L or R represent the patch sizes: 800 represents 20 × 40; 1250 represents 25 × 50; and 5000 represents 50 × 100.
Visualization of the Selected Shape Features from the Hippocampal Surface
Figure 4 provides an illustration for visualizing selected surface features by LOOCV when the patch size is 50 × 100. We divided the surface of the hippocampus into 3 zones (ie, LZ, SZ, and IMZ) according to the standard neuroanatomic atlas of the hippocampus,27 which was similar to the methods of these hippocampus-related studies.18,20 LZ represents the lateral zone of the hippocampal surface and approximates the CA1 subfield. SZ represents the superior zone approximating the combined CA2, CA3, and CA4 subfields and the gyrus dentatus, and IMZ represents the inferior-medial zone approximating the subiculum. Boundaries between the 3 zones of the hippocampal surface are drawn in black, and all 3 of the zones are labeled. The white subregions in the figure indicate that there are no differences in the hippocampal surface between patients with AD and healthy control subjects. The colors from purple to red indicate the higher repeatability of features in subsets. Here we suggest that the higher repeatability of features implicates the more significant features in differentiating 2 groups. The highest repeatability in our experiment was 39. The results showed that most of the distinct surface features were located in the CA1 and subiculum of the left hippocampus in AD. Relatively fewer surface features were found in the right hippocampus. However, those features that were observed in the right hippocampus tended to occur primarily in the CA1 subfield. There were also some changes in the CA2 to CA4 region of the left hippocampus in patients with AD compared with healthy control subjects.
Fig 4.

Visualization of selected surface features by LOOCV experiment when the patch size is 50 × 100 in patients with AD compared with healthy control subjects. The first row shows the left and right hippocampi from the top view, whereas the second row shows them from the bottom view. Boundaries between the 3 zones of the hippocampal surface (ie, LZ, SZ, and IMZ) are drawn in black. LZ represents the lateral zone of the hippocampal surface, and approximates the CA1 subfield. SZ represents the superior zone approximating the combined CA2, CA3, and CA4 subfields and the gyrus dentatus, and IMZ represents the inferior-medial zone approximating the subiculum. All 3 zones are labeled. The white subregions represent no differences in the hippocampal surfaces between patients with AD and healthy control subjects. The significance of features is measured by the repeatability in the subsets. The more significant features are shaded on the purple-red hue scale shown in the third row. The purple represents lower significance, and the red represents higher significance.
Discussion
The main purpose of this study was to present an integrated method for identifying specific subregions of the hippocampus that were prone to structural changes, significant for discriminating between patients with AD and healthy control subjects, and to build effective classifiers based on the regional changes. This was accomplished by characterizing surface deformations using surface-based measures based on the parameterization of each hippocampus. A feature selection method based on SVM RFE and LOOCV was used to identify critical changes in the 2 subject groups, that is, patients with AD versus healthy control subjects. These changes were used to construct effective classifiers to assess the reliability of the selected features. Using this method, high classification accuracies (Table 2) were obtained for the left and right hippocampi using 2 cross-validation experiments. Subregions of the hippocampus where shape changes are prominent between patients with AD and healthy control subjects are highlighted in Fig 4. Our approach was generally applicable to shape-based analysis and classification of other brain structures.
Pathologic Implications of the Selected Features
As shown in Fig 4, the most significant deformations in patients with AD were located in the CA1 region of bilateral hippocampi, as well as in the subiculum of the left hippocampus. These results were consistent with previous neuropathologic findings.32–36 Given that AD is a progressive disease, AD-mediated neuronal impairments appear in a hierarchical formation.33 Hippocampal degeneration within the CA1 subfield and subiculum appeared to be more severe compared with other components of the hippocampal formation in the early stages of AD.33–36 The main pyramidal cell layers of the hippocampus are the CA1–4 regions (primarily CA1 and CA3) and the dentate gyrus. The perforant path is the major input to the hippocampus. The axons of the perforant path arise principally in layers II and III of the entorhinal cortex, with minor contributions from the deeper layers IV and V. Axons from layers II/IV project to the granule cells of the dentate gyrus and pyramidal cells of the CA3 region, whereas those from layers III/V project to the pyramidal cells of the CA1 and the subiculum. Pathologic findings37–40 in patients with AD suggested that severe degeneration of the perforant pathway was a characteristic feature of AD. Our results showing the significant changes in the hippocampal surface were a probable consequence of this phenomenon.
Reliability of These Selected Shape Features
We assessed the reliability of these selected shape features using the permutation test. For each cross-validation experiment, we randomly selected some features from all of the features to construct a classifier to evaluate the classification accuracy between patients with AD and healthy control subjects. This process was repeated 10,000 times. We observed that in each experiment the percentage that the classification accuracies were greater than our experimental results was less than 5%. In other words, all of the P values in our cross-validation experiments were less than .05. This result indicated that our findings of the between-group differences in the shape features of the hippocampus were statistically significant.
Comparison with Related Work
Methodology.
Recently, increasing attention has focused on characterizing AD-related changes in the hippocampus using computational approaches. One popular method was to analyze the gray matter concentration of the hippocampus using VBM. Busatto et al13 found gray matter abnormalities over the entire extension of the temporal lobe in AD. However, Frisoni et al14 found that more regional changes corresponded with all parts of the left and right hippocampi. A second method6–9 was to investigate the hippocampal volume changes in AD using the region of interest method. Good et al15 compared the VBM results with region of interest measurements of temporal lobe structures and found that region of interest analyses appeared more sensitive to volume loss in the amygdalae, whereas VBM analyses appeared more sensitive to right middle temporal gyrus and regional hippocampal volume loss in patients with AD. Similarly, Testa et al41 compared the accuracy of VBM and region of interest–based hippocampal volumetry and suggested that VBM was more accurate, but the combination of both methods provided the highest accuracy for detection of hippocampal atrophy in patients with AD. A third method used the surface modeling methods to map hippocampal shape abnormality in AD. Such a surface-based method17,19,20,42,43 allows us to detect more subtle changes in the hippocampus in comparison with the VBM and region of interest–based methods. Using a 3D parametric mesh model, Thompson et al17 found that the hippocampal atrophic rates were faster in patients with AD than in the control subjects. Csernansky et al19,20 used the high-dimensional brain mapping methods to detect an AD-specific pattern in the hippocampus that was not found through the volume methods. In this study, we used surface modeling to characterize the hippocampal shape changes in AD. Compared with previous surface modeling approaches, our method provided a more specific hippocampal shape modeling, because it was particularly designed to banana-like objects, which might reduce the complexity of shape modeling. Our results also suggested that patients with AD showed significant deformations in the CA1 of bilateral hippocampi, as well as the subiculum of the left hippocampus (Fig 4), which were consistent with previous structural neuroimaging studies.18,20 In addition, in the present study, we used machine learning methods to find the abnormal subregions of the hippocampus in AD. The classification methods over the univariate statistical methods applied in previous AD-related hippocampal shape studies17,18 are that the discriminative analysis based on the classifier function can detect subtle differences between populations. The result of the analysis is a classifier function that can be used for assigning new examples and making a map over the original features indicating the extent to which each feature participates in estimating the label for any given example.22 In this study, we did not provide direct comparisons on the sensitivity of these surface-based methods applied in the present study and previous studies, because they involved different parameter selections and statistical analysis approaches.
Results.
In our study, significant deformations in AD were located in the CA1 and subiculum of the left hippocampus and the CA1 of the right hippocampus. There were also some changes in the CA2–4 subregions of the left hippocampus in AD. These results were consistent with those found in the literature.18,20 Wang et al18 found that inward deformation of the hippocampal surface in the proximity of the CA1 subfield and subiculum can be used to distinguish subjects with very mild AD from nondemented subjects. Similarly, Csernansky et al20 suggested that inward deformation of the lateral zone of the left hippocampal surface was an early predictor of the onset of AD in nondemented elderly subjects. In addition, as shown in Table 2, the selected shape features of the left hippocampus in different experiments obtain higher classification accuracies than those of the right hippocampus. Our results clearly showed that in discriminating patients with AD from healthy control subjects, an assessment of the left hippocampus proved more effective than of the right. Previous studies have alluded to this differential atrophy between the left and right hippocampi.44,45 In these studies, it was reported that bilateral hippocampal atrophy occurred to a greater extent on the left side than on the right, as evidenced in patients with AD. Moreover, several VBM studies,16,46 sulcal-warping studies,47,48 and single-photon emission CT and positron-emission tomography studies49,50 supported a laterality trend of the atrophic process and left more than right hemispheric involvement in AD.
Limitations of the Present Method
Some limitations of the study need to be emphasized. First, the subjects with AD were not categorized clinically as early stage or advanced stage for this experiment. At this point, it should be emphasized that there is a need to detect AD as early as possible. In the near future, the methods presented in this article could potentially be applied to discriminate patients with MCI or mild AD from healthy control subjects. Second, because of the limited spatial resolution of MR imaging in our study, the segmented gray matter in the images may have contained some subcortical white matter, leading to potential contamination with respect to measurements of the hippocampus. A higher magnetic field scanner should be adopted to acquire higher spatial resolution images. Finally, a shape analysis approach for evaluating the hippocampus may not be regarded as the only tool necessary for discriminating between patients with AD and healthy control subjects. A combination of various quantitative MR techniques that measure the anatomic, biochemical, microstructural, functional, and blood flow changes may provide useful markers for early diagnosis of AD. In addition, Kantarci and Jack51 suggested that these approaches of directly imaging the pathologic substrate would need to undergo a validation process with longitudinal studies to prove their usefulness as surrogate markers in AD. In the future, our method will be validated by tracking the progression of patients with MCI. The combination of the valuable biomarkers and the results of our hippocampal shape analysis are expected to form a more effective computer-aided diagnostic tool for AD.
Conclusion
In this article we presented an integrated method for finding subregions of the hippocampus that were significant for discriminating between patients with AD and healthy control subjects and building effective classifiers based on these regional changes. The major advantage of the machine learning methods compared with the univariate method was that it could detect subtle and spatially complex deformation patterns of the hippocampus in patients with AD compared with healthy control subjects. The results were objective and reliable, because the methods were validated by permutation test, and the findings were consistent with previous studies.
In summary, the shape analysis methods presented in the article provided a useful tool for detecting regional differences of the subcortical structures. These methods can also be applied to other neuropsychiatric diseases.
Acknowledgments
We gratefully acknowledge Gusphyl Justin and Alex MacGregor for editing and proofreading assistance.
Footnotes
This work was partially supported by the Natural Science Foundation of China grants 30425004 and 60121302 and the National Key Basic Research and Development Program (973) grant 2004CB318107.
References
- 1.Arriagada PV, Growdon JH, Hedley-Whyte ET, et al. Neurofibrillary tangles but not senile plaques parallel duration and severity of Alzheimer's disease. Neurology 1992;42:631–39 [DOI] [PubMed] [Google Scholar]
- 2.Berg L, McKeel DWJ, Miller JP, et al. Clinico-pathologic studies in cognitively healthy aging and Alzheimer's disease: relation of histologic markers to dementia severity, age, sex and apolipoprotein e genotype. Arch Neurology 1998;55:326–35 [DOI] [PubMed] [Google Scholar]
- 3.Braak H, Braak E. Neuropathological staging of Alzheimer-related changes. Acta Neuropathol (Berl) 1991;82:239–59 [DOI] [PubMed] [Google Scholar]
- 4.Squire LR, Zola-Morgan S. The medial temporal lobe memory system. Science 1991;253:1380–86 [DOI] [PubMed] [Google Scholar]
- 5.West MJ, Coleman PD, Flood DG, et al. Differences in the pattern of hippocampal neuronal loss in normal ageing and Alzheimer's disease. Lancet 1994;344:769–72 [DOI] [PubMed] [Google Scholar]
- 6.Convit A, DeLeon MJ, Tarshish C, et al. Specific hippocampal volume reductions in individuals at risk for Alzheimer's disease. Neurobiol Aging 1997;18:131–38 [DOI] [PubMed] [Google Scholar]
- 7.DeLeon M, Bobinski M, Convit A, et al. Usefulness of MRI measures of entorhinal cortex versus hippocampus in AD. Neurology 2001;56:820–21 [DOI] [PubMed] [Google Scholar]
- 8.Dickerson BC, Goncharova I, Sullivan MP, et al. MRI-derived entorhinal and hippocampal atrophy in incipient and very mild Alzheimer's disease. Neurobiol Aging 2001;22:747–54 [DOI] [PubMed] [Google Scholar]
- 9.Du AT, Schuff N, Amend D, et al. Magnetic resonance imaging of the entorhinal cortex and hippocampus in mild cognitive impairment and Alzheimer's disease. J Neurol Neurosurg Psychiatry 2001;71:441–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jack CR Jr, Slomkowski M, Gracon S, et al. MRI as a biomarker of disease progression in a therapeutic trial of milameline for AD. Neurology 2003;60:253–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashburner J, Friston KJ. Voxel-based morphometry-the methods. Neuroimage 2000;11:805–21 [DOI] [PubMed] [Google Scholar]
- 12.Boxer AL, Rankin KP, Miller BL, et al. Cinguloparietal atrophy distinguishes Alzheimer disease from semantic dementia. Arch Neurol 2003;60:949–56 [DOI] [PubMed] [Google Scholar]
- 13.Busatto GF, Garrido GE, Almeida OP, et al. A voxel-based morphometry study of temporal lobe gray matter reductions in Alzheimer's disease. Neurobiol Aging 2003;24:221–31 [DOI] [PubMed] [Google Scholar]
- 14.Frisoni GB, Testa C, Zorzan A, et al. Detection of grey matter loss in mild Alzheimer's disease with voxel based morphometry. J Neurol Neurosurg Psychiatry 2002;73:657–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Good CD, Scahill RI, Fox NC, et al. Automatic differentiation of anatomical patterns in the human brain: validation with studies of degenerative dementias. Neuroimage 2002;17:29–46 [DOI] [PubMed] [Google Scholar]
- 16.Karas GB, Burton EJ, Rombouts SA, et al. A comprehensive study of gray matter loss in patients with Alzheimer's disease using optimized voxel-based morphometry. Neuroimage 2003;18:895–907 [DOI] [PubMed] [Google Scholar]
- 17.Thompson PM, Hayashi KM, De Zubicaray GI, et al. Mapping hippocampal and ventricular change in Alzheimer disease. NeuroImage 2004;22:1754–66 [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Miller JP, Gado MH, et al. Abnormalities of hippocampal surface structure in very mild dementia of the Alzheimer type. Neuroimage 2006;30:52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Csernansky JG, Wang L, Miller JP, et al. Neuroanatomical predictors of response to donepezil therapy in patients with dementia. Arch Neurol 2005;62:1718–22 [DOI] [PubMed] [Google Scholar]
- 20.Csernansky JG, Wang L, Joshi S, et al. Preclinical detection of Alzheimer's disease: hippocampal shape and volume predict dementia onset in the elderly. NeuroImage 2005;25:783–92 [DOI] [PubMed] [Google Scholar]
- 21.Lao Z, Shen D, Xue Z, et al. Morphological classification of brains via high-dimensional shape transformations and machine learning methods. NeuroImage 2004;21:46–57 [DOI] [PubMed] [Google Scholar]
- 22.Golland P, Grimson WEL, Shenton ME, et al. Deformation analysis for shape based classification. Proc IPMI 2001;2082:517–30 [Google Scholar]
- 23.Yushkevich P, Joshi S, Pizer SM, et al. Feature selection for shape-based classification of biological objects. Proc IPMI 2003;2732:114–25 [DOI] [PubMed] [Google Scholar]
- 24.McKhann G, Drachman D, Folstern M, et al. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA work group under the auspices of the Department of Health and Human Services task force on Alzheimer's disease. Neurology 1984;34:939–44 [DOI] [PubMed] [Google Scholar]
- 25.Folstein MF, Folstein SE, McHugh PR. ‘Mini mental state': a practical method of grading the cognitive state of patients for the clinician. J Psychiatr Res 1975;12:189–98 [DOI] [PubMed] [Google Scholar]
- 26.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 1998;17:87–97 [DOI] [PubMed] [Google Scholar]
- 27.Duvernoy H. The Human Hippocampus. An Atlas of Applied Anatomy. Munich: JF Bergmann Verlag;1988
- 28.Zhu LT, Jiang TZ. Parameterization of 3D brain structures for statistical shape analysis. Proc SPIE Med Imaging, February 14–19,2004; San Diego, Calif
- 29.Chui HL, Rangarajan A. A new point matching algorithm for non-rigid registration. Comput Vis Image Underst 2003;89:114–41 [Google Scholar]
- 30.Guyon I, Weston J, Barnhill S, et al. Gene selection for cancer classification using support vector machines. Mach Learn 2002;46:389–422 [Google Scholar]
- 31.Li FM, Eun SY. Improving reliability of gene selection from microarray functional genomics data. IEEE Trans Inf Technol Biomed 2003;7:191–96 [DOI] [PubMed] [Google Scholar]
- 32.Bobinski M, de Leon MJ, Tarnawski M, et al. Neuronal and volume loss in CA1 of the hippocampal formation uniquely predicts duration and severity of Alzheimer disease. Brain Res 1998;805:267–69 [DOI] [PubMed] [Google Scholar]
- 33.Mizukami K, Ikonomovic MD, Grayson DR, et al. Immunohistochemical study of GABAA receptor alpha1 subunit in the hippocampal formation of aged brains with Alzheimer-related neuropathologic changes. Brain Res 1998;799:148–55 [DOI] [PubMed] [Google Scholar]
- 34.Muramori F, Kobayashi K, Nakamura I. A quantitative study of neurofibrillary tangles, senile plaques and astrocytes in the hippocampal subdivisions and entorhinal cortex in Alzheimer's disease, normal controls and non-Alzheimer neuropsychiatric diseases. Psychiatry Clin Neurosci 1998;52:593–99 [DOI] [PubMed] [Google Scholar]
- 35.Price JL, Morris JC. Tangles and plaques in nondemented aging and “preclinical” Alzheimer's disease. Ann Neurol 1999;45:358–68 [DOI] [PubMed] [Google Scholar]
- 36.Sheng JG, Mrak RE, Griffin WS. Progressive neuronal DNA damage associated with neurofibrillary tangle formation in Alzheimer disease. J Neuropathol Exp Neurol 1998;57:323–28 [DOI] [PubMed] [Google Scholar]
- 37.Mizutani T, Kasahara M. Degeneration of the intrahippocampal routes of the perforant and alvear pathways in senile dementia of Alzheimer type. Neurosci Lett 1995;184:141–44 [DOI] [PubMed] [Google Scholar]
- 38.Mizutani T, Kasahara M. Hippocampal atrophy secondary to entorhinal cortical degeneration in Alzheimer-type dementia. Neurosci Lett 1997;222:119–22 [DOI] [PubMed] [Google Scholar]
- 39.Mizutani T, Shimada H. Neuropathological background of twenty seven centenarian brains. J Neurol Sci 1992;108:168–77 [DOI] [PubMed] [Google Scholar]
- 40.Hyman BT, Van Hoesen GW, Kromer LJ, et al. Perforant pathway changes and the memory impairment of Alzheimer's disease. Ann Neurol 1986;20:472–81 [DOI] [PubMed] [Google Scholar]
- 41.Testa C, Laakso MP, Sabattoli F, et al. A comparison between the accuracy of voxel-based morphometry and hippocampal volumetry in Alzheimer's disease. J Magn Reson Imaging 2004;19:274–82 [DOI] [PubMed] [Google Scholar]
- 42.Bouix S, Pruessner JC, Louis Collins D, et al. Hippocampal shape analysis using medial surfaces. Neuroimage 2005;25:1077–89 [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Chiang MC, Thompson PM. Automated surface matching using mutual information applied to Riemann surface structures. Int Conf Med Image Comput Assist Interv 2005;8:666–74 [DOI] [PubMed] [Google Scholar]
- 44.Laakso MP, Soininen H, Partanen K, et al. Volumes of hippocampus, amygdala and frontal lobes in the MRI based diagnosis of early Alzheimer's disease: correlation with memory functions. J Neural Transm 1995;9:73–86 [DOI] [PubMed] [Google Scholar]
- 45.Remy F, Mirrashed F, Campbell B, et al. Verbal episodic memory impairment in Alzheimer's disease: a combined structural and functional MRI study. NeuroImage 2005;25:253–66 [DOI] [PubMed] [Google Scholar]
- 46.Baron JC, Chetelat G, Desgranges B, et al. In vivo mapping of gray matter loss with voxel-based morphometry in mild Alzheimer's disease. NeuroImage 2001;14:298–309 [DOI] [PubMed] [Google Scholar]
- 47.Thompson PM, Mega MS, Woods RP, et al. Cortical change in Alzheimer's disease detected with a disease-specific population-based brain atlas. Cereb Cortex 2001;11:1–16 [DOI] [PubMed] [Google Scholar]
- 48.Thompson PM, Hayashi KM, de Zubicaray, et al. Dynamics of gray matter loss in normal aging and Alzheimer's disease. J Neurosci 2003;23:994–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson KA, Jones K, Holman BL, et al. Preclinical prediction of Alzheimer's disease using SPECT. Neurology 1998;50:1563–71 [DOI] [PubMed] [Google Scholar]
- 50.Duara R, Grady C, Haxby J, et al. Positron emission tomography in Alzheimer's disease. Neurology 1986;36:879–87 [DOI] [PubMed] [Google Scholar]
- 51.Kantarci K, Jack CR Jr. Quantitative magnetic resonance techniques as surrogate markers of Alzheimer's disease. Neurorx 2004;1:196–205 [DOI] [PMC free article] [PubMed] [Google Scholar]




