Abstract
Since the human brain functional mechanism has been enabled for investigation by the functional Magnetic Resonance Imaging (fMRI) technology, simultaneous modeling of both the spatial and temporal patterns of brain functional networks from 4D fMRI data has been a fundamental but still challenging research topic for neuroimaging and medical image analysis fields. Currently, general linear model (GLM), independent component analysis (ICA), sparse dictionary learning, and recently deep learning models, are major methods for fMRI data analysis in either spatial or temporal domains, but there are few joint spatial-temporal methods proposed, as far as we know. As a result, the 4D nature of fMRI data has not been effectively investigated due to this methodological gap. The recent success of deep learning applications for functional brain decoding and encoding greatly inspired us in this work to propose a novel framework called spatio-temporal convolutional neural network (ST-CNN) to extract both spatial and temporal characteristics from targeted networks jointly and automatically identify of functional networks. The identification of Default Mode Network (DMN) from fMRI data was used for evaluation of the proposed framework. Results show that only training the framework on one fMRI dataset is sufficiently generalizable to identify the DMN from different datasets of different cognitive tasks and resting state. Further investigation of the results shows that the joint-learning scheme can capture the intrinsic relationship between the spatial and temporal characteristics of DMN and thus it ensures the accurate identification of DMN from independent datasets. The ST-CNN model brings new tools and insights for fMRI analysis in cognitive and clinical neuroscience studies.
Keywords: fMRI, functional brain networks, deep learning
I. Introduction
INVESTIGATIONS of the human brain’s functional mechanism have been enabled by in-vivo Functional Magnetic Resonance Imaging (fMRI) technology. FMRI decomposition methods (e.g., independent component analysis (ICA) [1], [2], sparse representation [3]) and deep learning methods [4], [5]) have significantly facilitated the analytics of the spatial and temporal features in fMRI data [6]. As fMRI data are the acquisition of series of 3D brain volumes during imaging scan procedure to recording functional temporal dynamics of 3D spatial brain volumes, the intrinsic spatio-temporal relationships are characterized in the form of 4D data. Thus, a comprehensive characterization and description of 4D fMRI data encoding of both spatial and temporal characteristics is significant for the understandings inside the human brain’s organizational functional architecture.
In the current literature, methods of spatio-temporal analysis of fMRI data can be categorized into two groups from the perspectives of 3D spatial or 1D temporal dimensions of fMRI data. The first group performs the conjugate analysis on single domain, and then performs regression of the variation patterns in the other domain in an alternative manner. For example, temporal ICA [1], [2], [7] extracts the independent non-gaussian temporal elements in the 4D fMRI data, and then regresses out the spatial patterns of the corresponding temporal elements. In another recent research, a deep learning-based Convolutional Auto-Encoder (CAE) model [8] are explored to characterize temporal features, and corresponding spatial features are generated through regression from temporal features. On the other hand, dictionary learning and sparse representation methods extracts the sparse spatial maps of the fMRI data, while the temporal correspondences of these components are obtained through linear combinational regression. Moreover, the work proposed in [9] utilizing Restricted Boltzmann Machine (RBM) also focusing analysis on spatial features first and then the characteristics of temporal features.
Other than focusing on single dimension analysis, methods in the second group tend to perform simultaneous analysis on both spatial and temporal domains. Realizing the intrinsic spatio-temporal interactions within fMRI data, this group aims to perform analysis of the spatio-temporal features of fMRI data comprehensively. For example, the work in [10] exploited Hidden Process Model with spatio-temporal “prototypes” for modeling both domains, and it can disentangle overlapping mental processes evoked by stimuli. However, the spatio-temporal “prototypes” are limited to a small “region of influence” spatial prior for specific stimulus analysis. Another research also proposed an effective approach using Recurrent Neural Network (RNN) to incorporate temporal dynamics (and between-time-frames correlations) into the intrinsic network (IN) modeling [11]. However, the RNN used in that research [11] is still a prior-like constraint for the ICA analysis. No comprehensive spatio-temporal analysis for whole-brain analysis is available in the abovementioned research. Thereafter, inspired by better interpretability of the simultaneous intrinsic spatio-temporal modeling concept and the superior performance of deep learning frameworks, we proposed a whole brain level spatio-temporal deep convolutional neural network (ST-CNN) for 4D fMRI data modelling. We aim to pinpoint or extract the spatial and temporal features of targeted functional networks (e.g., Default Mode Network (DMN) in this work) directly from the 4D fMRI data without any template matching/searching processes involved, through the ST-CNN model. The ST-CNN model composes two simultaneous characterizations: the characterization on the spatial pattern of the targeted network from the whole brain signals using a 3D U-Net [12], and the characterization on the temporal dynamic patterns of the targeted network, using a 1D CAE [8]. Losses from the two mappings are merged and simultaneously back-propagated to the two networks in an integrated framework, fulfilling simultaneous modeling process of both spatial and temporal domains based on the level of whole brain signals. In the evaluation part, our experimental results show that, the ST-CNN, without hyper-parameter tuning, can extract dynamics of both spatial and temporal pattern of the DMN accurately, even with the presence of remarkable variabilities of cortical structures and functions from different individuals. Further evaluations demonstrated the sufficient generalizability of ST-CNN framework for the network identification task, in that only training the ST-CNN on motor task-evoked fMRI (tfMRI) dataset, reproducible results can be achieved on other datasets (other tasks-evoked or resting-state fMRI). ST-CNN can also serve for cognitive or clinical neuroscience studies as a useful tool, with the capability of identifying network in a pin-point way. Furthermore, with the ability of modelling the spatio-temporal variation patterns of the data corresponding to their intrinsic intertwined nature within one integrated framework, ST-CNN shows great potentials to offer refreshing perspectives for understanding human brain functional organization from 4D fMRI data. It is noted that this work is an significant extension from a recently accepted MICCAI paper [13]. We extensively evaluated and validated our ST-CNN model with larger datasets from HCP 900 release. Besides, the resting-state fMRI were also tested via ST-CNN and results show that both the spatial and temporal characteristics can be modeled for the targeted network (DMN).
II. Method and Materials
Our designed ST-CNN framework takes 4D fMRI data (either task-evoked or resting-state), and then generates both of spatial map and temporal time series of brain network as outputs simultaneously. Unlike popular CNN structures for natural image classifications (e.g., [14]), the proposed ST-CNN can perform convolution operations in both spatial and temporal domains simultaneously for spatial and temporal features, making our ST-CNN a spatio-temporal convolution framework for 4D fMRI data modelling. The overview of this ST-CNN framework is illustrated in Fig. 1. To train the ST-CNN, the ground-truth DMN spatial network volumes and DMN temporal dynamics are provided via a dictionary learning and sparse coding method [3], [15], which will be explained in detail in the following sections.
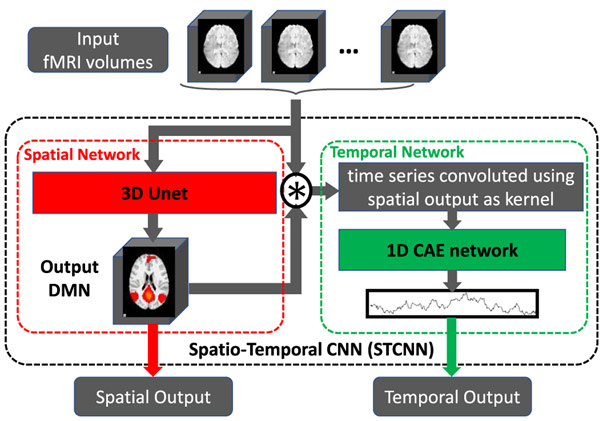
Fig. 1.
ST-CNN framework. ST-CNN consists of two subnetworks: spatial network (red part) and temporal network (green part). Combination of the two subnets is defined using “⊛”.
A. Experimental Data and Preprocessing
The experimental data came from the publicly available Human Connectome Project (HCP) dataset [16] (900 release) (https://www.humanconnectome.org/study/hcp-young-adult/document/900-subjects-data-release). The 900 subjects release includes behavioral and 3T MR imaging data from over 900 healthy adult participants, which provided a systematic and comprehensive mapping of connectome-scale functional networks over a large population in the literature [17]. The detailed imaging parameters for both task-evoked and resting-state data are referred to [18]. The downloaded data were already preprocessed by a pipeline including: gradient distortion correction, motion correction, field map preprocessing, distortion correction, spline resampling to atlas space, intensity normalization and et. al. The HCP fMRI preprocessing analysis also uses FEAT in FSL for multiple regression with autocorrelation modelling and pre-whitening and spatial smoothing [19]. The preprocessing pipeline is built using FSL [20] and FreeSurfer [21].
For the experiments in this work, we used motor tfMRI, emotion tfMRI and resting-state fMRI (rsfMRI) datasets from 200 randomly selected ones of the 900 subjects. Only 160 out of 200 subjects’ motor tfMRI datasets were used for training purposes and all the rest 40 subjects’ motor tfMRI, 200 subjects’ emotion tfMRI and rsfMRI datasets were used for pure testing to validate the results using our framework. Actually, the 200 subjects for 3 tasks have intersecting subjects, as some subjects have missed scans for some tasks. A total of 282 subjects were used for motor, emotion and resting state data. We just used 200 subjects from the 282 subjects pool for each task. The ages of the 282 subjects range from 22 to 75, with a mean age range from 27.2 to 31.3. Among 282 subjects, 119 (42%) are male while 163 (58%) are female subjects. After preprocessing of the datasets following the abovementioned pipeline, all the fMRI data are normalized to 0-1 distribution (normal distribution with 0 mean and 1 standard deviation) according to (1) as inputs.
| (1) |
where vi represents voxel intensity at location i; Vin–mask represents the voxels within the brain mask; mean is the mean function, while std is the standard deviation function.
The dictionary learning and sparse representation method [3], [15] was performed to decompose the fMRI data as ground-truth for ST-CNN training for two reasons. First, using individualized DMNs to guide the ST-CNN model to learn from individualized features is the key to achieve the optimized model accommodating for individual variability. As the individual variability of each fMRI scan is huge, the extracted DMNs are very different from each other with individual variances. Thus, using universal ground truth (e.g. some DMN templates) for training is risky as the ST-CNN may just over fit the DMN templates instead of modelling the intrinsic fMRI signals. Second, we chose the work on DMNs extracted by sparse representation over ICA as the works in [22] showed that experimental results demonstrated the sparse representation could better handle network decomposition when the spatial overlap exists between functional network maps. As DMN covers substantial area of regions in the brain, this will likely have spatial overlap with other networks. Following the caveats in [22], sparse representation may have better performance (at least comparable) in constructing and interpreting DMN. The input 4D fMRI data for dictionary learning and sparse representation was flattened into a 2D matrix with t (length of 0-1 normalized time points) rows by n columns, each of which represents one brain voxel out of a total number of n flattened voxels from an individual subject. The output contains one learned dictionary and a sparse coefficient matrix , w.r.t, X = D × α + ε, as illustrated in (Fig. 2 (a)), where ε is the error term and m is the predefined dictionary size, set to 400 in this experiment. The functional networks’ temporal dynamics are obtained from the dictionary atoms: and spatial patterns are obtained from the coefficient matrix regressed using a fast implementation of the LARS algorithm [23]: are then mapped to 3D brain volume space as spatial functional network maps). To find the DMN among all the m functional networks, the network with the maximum overlap rate (2) to the well-established DMN templates [24] was taken as its correspondence, whose corresponding dictionary atoms were taken as the DMN temporal dynamics. These DMN temporal dynamics and spatial maps were then used as the training and validation/comparison sets for our framework.
| (2) |
where Vk and Wk are the activation scores of voxel k in the spatial maps V and W, respectively. ∣V∣ is the total number of the voxels in the spatial map.
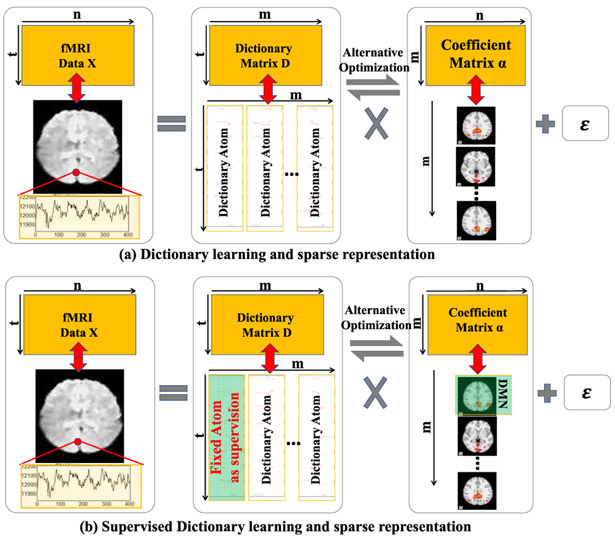
Fig. 2.
Illustration of (a) dictionary learning and sparse representation and (b) supervised version. 4D fMRI data was converted in a 2D matrix X as input. The decomposed dictionary D contains temporal dynamics in each atom (column) and the coefficient matrix α contains the corresponding spatial maps of the functional networks. For supervised version, one of the dictionary atoms is fixed as the desired temporal dynamic curves. The corresponding coefficient is the corresponding spatial map.
B. ST-CNN
As shown in Fig. 1, the ST-CNN framework consists of 2 parts: a spatial part and a temporal part. Unlike traditional ICA or dictionary learning and sparse representation methods, the inputs are 4D fMRI data without flattening the 3D volumes into a vector thus sacrificing the spatial geometric relationship between each voxel. Furthermore, rather than conjugating the updates between the spatial and temporal outputs, the ST-CNN use a spatio-temporal combination joint to process the spatio-temporal relationship inside the original input 4D fMRI data and output the spatial and temporal results simultaneously in a pin-point way for the specific function network (DMN in this case).
1). Spatial network:
The spatial network is basically inspired by the 2D Unet [12] designed for 2D biomedical image segmentation. The key innovation of the 2D Unet is the feature preserving from the contracting path to the extending path of the Unet structure. The preservation of the features generated from the contracting path will be fed back (copied) directly to the expansive path to assist the accurate segmentation by providing the original image feature information. The autoencoder-like contracting and expansive structure makes Unet an end-to-end (image-to-image) framework for image segmentation, which is suitable to be modified as an end-to-end image pixel-level regression framework while still maintaining the feature of accurate reconstruction of the original input images. Intending to preserve the 3D spatial context information of the fMRI, we finally adapted the 2D image segmentation Unet to a 3D image regression Unet as our spatial network.
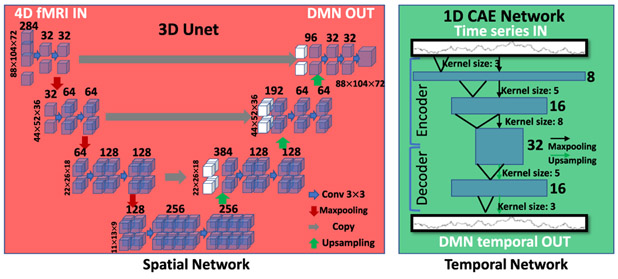
By extending and adapting the 2D classification Unet to a 3D regression network as shown in Fig. 3, we can take 4D fMRI data as input, each 3D brain volume of which along the time points is assigned with one independent channel and regress/output DMN’s spatial map. Basically, this 3D Unet is a symmetrical structure with a contracting path and an expansive path. The contracting path is a structure with successive convolutional layers, alternating with the pooling layers (red arrows in Fig. 3 in spatial network). The expansive path is arguably symmetric with the contracting path with convolutional layers alternating with up-sampling layers. This 3D u-shaped CNN structure is a fully convolutional network (FCN) without fully connected layers. For the contracting path among the structure, it follows the canonical CNN architecture, which repeated two 3*3 convolution operation, each followed by a rectified linear unit (ReLU), batch normalization layer and a down sampling max-pooling layer of pooling kernel size of 2. We will then double the size of the feature map channels following the down sampling process. The expansive path symmetrizes contracting path, except that the max-pooling layers are replaced with up-sampling layers, and that the number of the feature map channels are halved (except that the output layer has 1 channel as 1 3D map output) after each up-sampling step. There are connections between contracting path and expansive path by copying feature maps from contracting path to expansive path to preserve features and contexts from the original input images. The loss function for training the spatial network is mean squared error (MSE) to resemble the targeted training spatial maps of DMN.
Fig. 3.
Spatial network and temporal network structure inside ST-CNN.
2). Temporal network:
The temporal network (Fig. 3 temporal network) is inspired from the excellent performance of a 1D convolutional autoencoder (CAE) network to deal with the time series for fMRI modeling [8]. In this work, we adopt a 6-layers depth 1D CAE to deal with the temporal features of the input fMRI. As shown in Fig. 3 temporal network part, the 1D CAE is also a symmetric structure with a contractive and an expansive path. The contractive path, namely, the encoder starts by taking input 1D time series and convolving them using size 3 convolutional kernel, which yields 8 feature map channels, followed by a pooling layer for down-sampling. Then a size 5 convolutional layer is cascaded, which yields 16 feature map channels, also followed by a pooling layer for down-sampling. The last part of the encoder consists of a size 8 convolutional layer, which yields feature maps of 32 channels. The expansive path, namely, the decoder, takes the features output by the encoder as input and mirrors symmetrically the encoder to reconstruct the input time series. The loss function for training temporal 1D CAE network is negative Pearson correlation (3) to resemble the temporal dynamics of the training DMN. ST-CNN incorporates this 1D CAE to generate temporal dynamics of the DMN from the fMRI data.
Temporal loss
| (3) |
where x, y are output time series and ground-truth time series, and N is the length of the time series.
C. Convolutional spatio-temporal combination joint
Since we already built networks for both spatial and temporal analysis, the next question is how to connect those two parts as an entire framework. That is, the relationship between spatial and temporal features needed to be characterized. Intuitively, as the activated regions (see the red regions of the convolutional kernel in Fig. 4) shall contain concurrent signals, we designed a convolutional spatio-temporal combination joint (Fig. 4). Through this joint, the concurrent signal features will be fused corresponding to the spatial map, meanwhile preserving a fully convolutional network structure. In detail, this combination joint in Fig. 4 connects spatial network and temporal network through a convolution operator. The combination takes the 4-D fMRI data and the 3-D DMN generated from spatial network as input. The spatial network generated 3-D output is taken as a 3-D convolutional kernel to convolve with each 3-D volume along the time frames of the input 4-D fMRI data (4) in a valid way without any paddings. This valid no-padding convolutional operation will yield a single value for each time frame, resulting in a series of values as time series for the estimated DMN, namely, ts, which will be the input for temporal 1-D CAE, as abovementioned.
| (4) |
where ti represents the single convolutional value at each time frame, Vi represents each 3-D volume scanned at time frame i. DMN represents the 3-D spatial map, which is also used in the combination joint as convolution kernel. T is the total length of the time series, which is 284 in this paper.
Fig. 4.
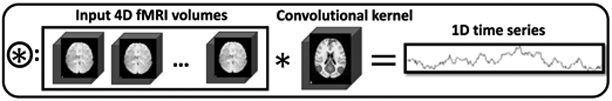
Spatio-temporal combination joint illustration. The spatial output will be used a convolutional kernel applied to the original input 4D fMRI data. The output of this combination joint is a time series reflecting the temporal dynamics from the fMRI data associated with the spatial map.
D. Model Training Scheme
As introduced in the combination joint part, the temporal network relies on the DMN output of the spatial network, and we have designed a 3-step training strategy for efficiency. At the first stage, we train spatial network only; the second stage, we freeze the weights of the spatial network and train temporal network only; as a simultaneous spatio-temporal framework, during the third stage, the entire ST-CNN is fine-tuned simultaneously. From empirical practice, we observed that the temporal network loss is around 10 times less than the spatial network loss, thus we designed a weighted total loss (10:1 for spatial:temporal) as the ST-CNN loss.
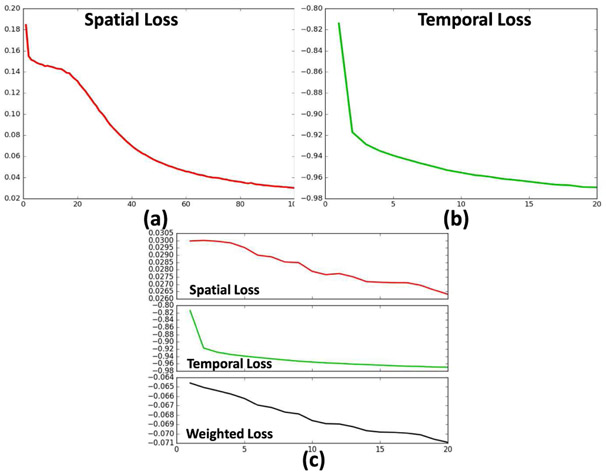
For the first stage, the spatial network was trained for 100 epochs until the spatial loss reaches 0.03 (Fig. 5. (a)), which indicated the output mean squared error with the training DMN is very small. For the second stage, the temporal network was trained for 20 epochs when the temporal loss reached −0.97 (Fig. 5. (b)), which indicated the Pearson correlation coefficient between the temporal output and the ground-truth is 0.97 (highly correlated). For the third stage (Fig. 5. (c)), we can see co-operative refinement of both spatial and temporal network. The gradient descent optimizer is Adadelta [25].
Fig. 5.
Training loss curves. (a) stage one for spatial network training. (b) stage two for temporal network training. (c) stage three for fine-tuning.
In order to demonstrate the generalizability to different task-evoked data and resting-state data of the ST-CNN, our training dataset was only based on the motor task tfMRI from 160 subjects. The rest 40 subjects’ motor task tfMRI data and all the 200 subjects’ emotion task tfMRI and rsfMRI were used for pure testing purpose.
E. Evaluation and Validation
To evaluate the performance of the framework, both the spatial and temporal similarities were quantified with the well-established DMN template spatial map and the corresponding time series of the DMNs decomposed from each individual. For the spatial similarity measurement, the overlap rate (2) between the output spatial map and the ground truth map was used, while the temporal similarity was measured by the Pearson correlation coefficient (the negative value of the temporal loss in (3)). Qualitative evaluation will also be done as the “ground-truth” DMN decomposed from dictionary learning and sparse coding and identified with spatial overlap scheme may not be perfectly reliable as “true” DMN. In the result section, we will show some qualitative cases where the dictionary learning and sparse coding failed to generate DMN while our ST-CNN can successfully pinpoint the DMN.
Furthermore, to validate that the output of the ST-CNN models the correct spatio-temporal relationship from the 4D fMRI data rather than overfitting the DMN without modelling the spatio-temporal relationships, we performed a supervised dictionary learning and sparse representation method [26] (Fig. 2 (b)) to check whether our ST-CNN framework generate spatio-temporal outputs by successfully modelling the intrinsic spatio-temporal characteristics within the 4D fMRI data. The supervised dictionary learning and sparse representation method [26] takes the temporal output of the ST-CNN as the temporal supervision of the dictionary (Fig. 2 (b) green part in dictionary), and reconstruct the corresponding spatial maps (Fig. 2 (b) green part in coefficient matrix) based on the supervised dictionary to generate the corresponding spatial maps to the supervision. The spatial overlap rate was utilized to check the similarity between the spatial maps generated by ST-CNN and the supervised dictionary learning and sparse representation. By this way, we can confirm with confidence that our ST-CNN produces intrinsic spatio-temporal dynamics of the DMN.
III. Results
In this section, result analysis and performance evaluation of the spatio-temporal output of the ST-CNN from testing datasets are presented. ST-CNN was trained on the motor tfMRI data from 160 subjects, while the testing datasets includes motor tfMRI data from 40 different subjects, emotion tfMRI and resting-state fMRI from the corresponding 200 subjects in HCP Q900 release. In summary, testing results showed that ST-CNN can perform DMN identification with simultaneous intrinsic spatial and temporal characterization.
A. DMN Spatio-temporal Identification
ST-CNN is firstly tested on the same task (motor) tfMRI data from 40 different subjects. Visualizations of the identified DMN from 3 sample subjects are shown in Fig. 6. Spatial output of ST-CNN resembled ground-truth spatial maps decomposed by dictionary learning and sparse representations (SR for brevity) (overlap rate all larger than 0.2). While the spatial pattern of DMN template was never provided to ST-CNN (only subject-wise decomposition results were used for training), ST-CNN outputs are more similar or at least comparable to dictionary learning method which used DMN template as input (TABLE I). As reported in [27], networks with spatial overlap rate larger than 0.1 will be considered similar to each other. Results of motor tfMRI from 40 subjects can be found at: http://hafni.cs.uga.edu/DMN_dynamic/HCP_900/MOTOR/.
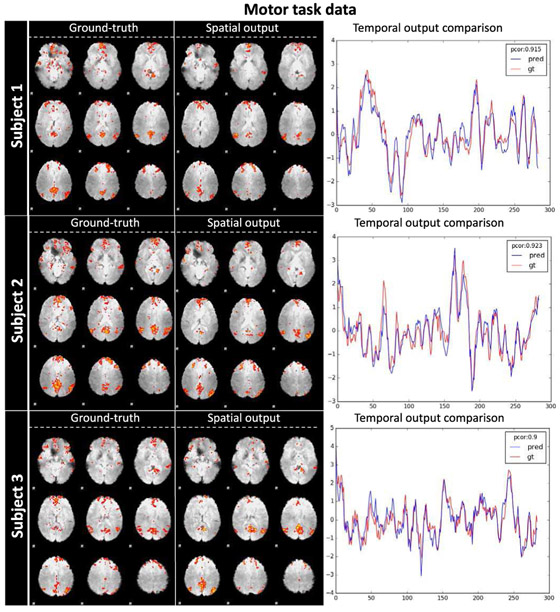
Fig. 6.
DMN identification with spatio-temporal co-learning by ST-CNN. Randomly-selected 3 subjects are visualized with their ground-truth DMN spatial map (decomposed by dictionary learning and sparse representation and identified by spatial overlap with DMN template), spatial output map from ST-CNN, and temporal dynamics of both SR and ST-CNN. For temporal dynamics, the blue curve is the output of ST-CNN and the red curve is the ground-truth dictionary atom corresponding to the ground-truth DMN spatial map. The Pearson correlation of the 2 curves are also displayed in the top right corner: 0.915, 0.923, 0.9 respectively.
TABLE I.
DMN identification quantitative analysis for 3 sample subjects. Higher values for spatial overlap rates with DMN templates are highlighted in bold texts.
| Subject | Spatial overlap between SR and ST-CNN |
Spatial overlap with DMN template |
Temporal similarity (Pearson correlation) |
|
|---|---|---|---|---|
| Sparse Representation |
ST-CNN | |||
| Subject 1 | 0.248 | 0.121 | 0.120 | 0.915 |
| Subject 2 | 0.255 | 0.120 | 0.122 | 0.923 |
| Subject 3 | 0.238 | 0.128 | 0.133 | 0.900 |
After examining all the testing results from motor tfMRI data, we found ST-CNN perform superiorly than SR in the following aspects: ST-CNN can identify DMN in a pinpoint way, rather than relying on spatial overlap measurement (such as SR/ICA). Besides, as the spatial-temporal dynamics of DMN within fMRI data are simultaneously captured by ST-CNN, and it can more accurately identify DMN comparing with unsupervised approaches such as SR (which relies on the sparsity prior of fMRI data). We measured the spatial overlap rate between the results by ST-CNN/SR and DMN template in motor fMRI data from all 40 testing subjects. The result shows that ST-CNN achieved a noticeably higher mean spatial overlap rate and lower standard deviation with DMN template (0.124±0.016) comparing with SR (0.107±0.042). In addition, temporal dynamics of the identified DMNs by ST-CNN shows high Pearson correlation (averagely 0.758 across 40 subjects) with temporal dynamics of ground-truth DMNs.
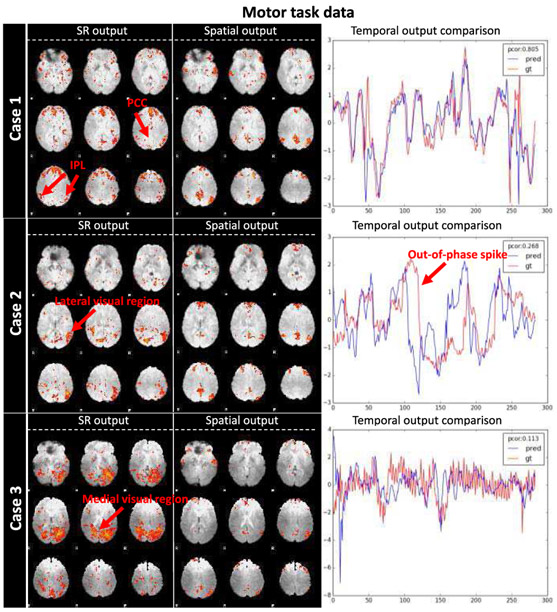
In addition to the fact that ST-CNN outperformed SR on average, we also observed cases where ST-CNN generated obviously better DMN maps than SR (Fig. 7, quantitative results are shown in TABLE III). These cases are particularly interesting, as the ST-CNN is trained based on the results of SR. Thus, if ST-CNN can obtain correct DMN identification where SR fails (which is not uncommon due to various factors, as illustrated below), that would be an indication for the superior generalizability of ST-CNN over its training data.
Fig. 7.
Superior DMN identification ability of ST-CNN than dictionary learning and sparse representation (SR). Case 1, ground-truth DMN partial spatial pattern. Case 2, ground-truth DMN mixed spatial pattern. Case 3, ground-truth DMN identification failure.
TABLE III.
ST-CNN superior performance in DMN identification
| Case | Spatial overlap between SR and ST-CNN output |
Spatial overlap with DMN template |
Temporal similarity (Pearson correlation) |
|
|---|---|---|---|---|
| Sparse Representation |
ST-CNN | |||
| Case 1 | 0.166 | 0.044 | 0.106 | 0.805 |
| Case 2 | 0.059 | 0.080 | 0.135 | 0.268 |
| Case 3 | 0.067 | 0.068 | 0.109 | 0.113 |
In case 1, only a partial DMN was identified by SR, while posterior cingulate cortex (PCC) and inferior parietal lobe (IPL) were partially inactivated. On the contrary, ST-CNN identified these two regions correctly. Temporal dynamics of the results from two models are similar (Pearson correlation 0.805), as major regions in DMN were still preserved by SR.
In case 2, DMN identified by SR show a mixed spatial pattern of DMN and later visual network [24], possibly caused by the interdigitated functional area [22] that affected decomposition results (i.e. two networks are not decomposable based on current parameter setting of SR). Again, DMN identified by ST-CNN maintained most of the related functional networks. Correspondingly, temporal dynamics of SR show a significant out-of-phase spike comparing with network identified by ST-CNN, decreasing the Pearson correlation between these two to 0.268. This is likely caused by the extra involvement of later visual network in SR result.
Case 3, SR DMN identification failure. As cortical microcircuits overlap and interdigitate with each other [28], rather than being independent and segregated in space, the medial visual regions and DMN are spatially overlapped. Either incurred by the failure of dictionary learning and sparse representation method or the failure of spatial overlap based DMN identification process, the SR DMN turned out to be a medial visual network. As a result, the ST-CNN predicted temporal dynamics for DMN is quite different from the one corresponding to the medial visual network (Pearson correlation 0.113).
B. Generalizability for Other Task fMRI Data
We trained our ST-CNN on motor tfMRI data, since the pure testing on motor tfMRI data isn’t adequate for demonstrating the generalizability of the proposed framework as one can argue that the trained ST-CNN is overfitting to motor task data. Therefore, without any further training after purely training on motor data, we deployed a test on emotion tfMRI data to test the generalizability of ST-CNN for other tasks.
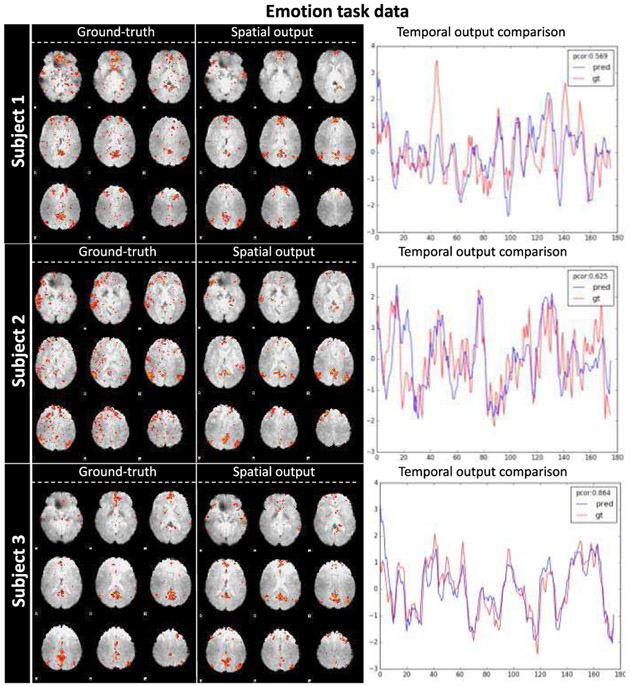
Similarly, we show the ST-CNN prediction for emotion task from 3 randomly selected subjects in Fig. 8. As we can see, the spatial output of ST-CNN successfully identified DMN spatial maps. Using the dictionary learning and sparse representation DMN output as “ground-truth”, we can also see the temporal output of ST-CNN is highly correlated with ground-truth. All 200 emotion tfMRI testing results are referred to http://hafni.cs.uga.edu/DMN_dynamic/HCP_900/EMOTION/.
Fig. 8.
DMN spatial-temporal identification generalizability to emotion task. The spatial and temporal output of three subjects are shown above. Ground-truth DMN spatial map (decomposed by dictionary learning and sparse representation and identified by spatial overlap with DMN template), spatial output map from ST-CNN, and temporal dynamics of both dictionary learning and sparse representation and ST-CNN are listed for each column respectively. For temporal dynamics, the blue curve is the output of ST-CNN and the red curve is the ground-truth dictionary atom corresponding to the ground-truth DMN spatial map. The Pearson correlation of the 2 curves are also displayed in the top right corner.
As analyzed in TABLE II, the mean spatial overlap rate of the ST-CNN outputs with well-established DMN template is still clearly larger than the sparse representation outputs, and the larger standard deviation of the sparse representation results than ST-CNN results also demonstrate that ST-CNN is much more accurate and robust in DMN identification. Still, the temporal similarity preserved the same level (mean Pearson correlation 0.751) as in motor data set. Both spatial and temporal quantitative and qualitative results support that our ST-CNN trained on one specific task has robust generalizability to other tasks.
TABLE II.
DMN identification quantitative analysis both spatially (spatial overlap rate, mean±std) and temporally (Pearson correlation) for HCP 900 release data.
| Datasets | Spatial overlap with DMN template | Temporal similarity (Pearson correlation) |
|
|---|---|---|---|
| Sparse Representation |
ST-CNN | ||
| MOTOR (40 subjects) | 0.107±0.042 | 0.124±0.016 | 0.758 |
| EMOTION (200 subjects) | 0.102±0.041 | 0.115±0.026 | 0.751 |
| RSN (200 subjects) | 0.109±0.044 | 0.118±0.030 | 0.725 |
C. Generalizability for Resting-state fMRI Data
The DMN is vastly known for its presence during resting state, namely, default mode [29], [30]. DMN will also establish or internally orient tasks, which means during task-evoked states, DMN is also present [30], [31]. Correspondences of DMN during activation and rest were also found as full repertoire of functional networks utilized by the brain in task-evoked states is continuously and dynamically active [24]. According to literature DMN related research studies [29]-[31], DMN tends to be an intrinsic network that constantly exists inside human brain no matter it is healthy brain or diseased brain [32]-[34]. Following this logic and the generalizability of the trained ST-CNN to other task data, we further tested our ST-CNN trained on task-evoked data for resting state DMN modelling, which is another important reason we designed ST-CNN to pinpoint DMN.
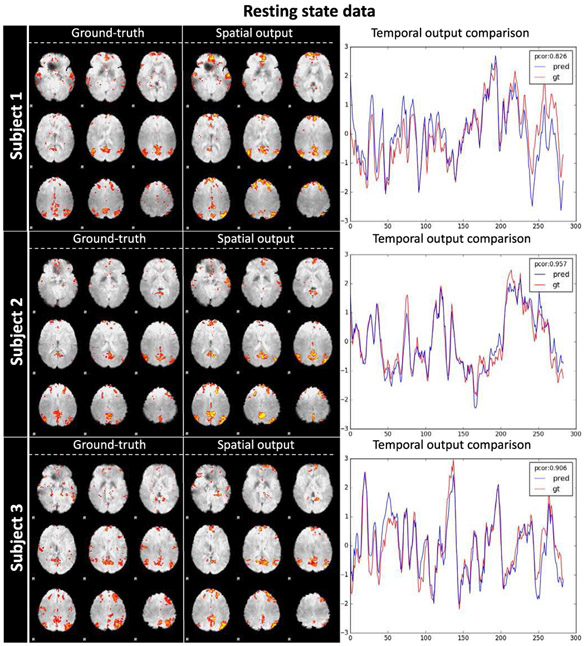
Similar to motor and emotion results sections, we randomly pick 3 subjects’ results as a qualitative illustration in Fig. 9 and put all results for DMN spatio-temporal dynamics outputs for rsfMRI at http://hafni.cs.uga.edu/DMN_dynamic/HCP_900/RSN/. As shown in Fig. 9, the DMN spatial pattern is accurately captured by our ST-CNN, with posterior cingulate cortex (PCC), medial prefrontal cortex (mPFC) and inferior parietal lobe (IPL) activated. The ground-truth DMN spatial maps decomposed by dictionary learning and sparse representation clearly have high similarity with ST-CNN outputs. It is intriguing that we still achieved high spatial overlap rate for DMN in rsfMRI. As shown in TABLE II, the average spatial overlap rate of ST-CNN output with well-established DMN templates is higher than the outputs from dictionary learning and sparse representation. The main reason is similar to the analysis for motor data, that is, dictionary learning and sparse representation method has limited power for DMN interpretation and identifying DMN using spatial overlap rate from hundreds of networks is not very robust.
Fig. 9.
DMN’s spatial-temporal identification generalizability to resting state fMRI data. The spatial and temporal output of three subjects are shown above. Ground-truth DMN spatial map (decomposed by dictionary learning and sparse representation and identified by spatial overlap with DMN template), spatial output map from ST-CNN, and temporal dynamics of both dictionary learning and sparse representation and ST-CNN are listed for each column respectively. For temporal dynamics, the blue curve is the output of ST-CNN and the red curve is the ground-truth dictionary atom corresponding to the ground-truth DMN spatial map. The Pearson correlation of the 2 curves are also displayed in the top right corner.
From the testing results from resting state data, we can conclude that our ST-CNN can successfully model the intrinsic dynamics of the DMN from fMRI data and identify DMN in a pinpoint way for different tasks as well as resting state data. Only being trained using one task dataset (motor task), the generalizability of the ST-CNN can be demonstrated for other task data and resting-state data. This result cross-validated the correspondence of DMN in both task-evoked state and resting state and suggested the ST-CNN can capture the spatial and temporal dynamics intrinsically from any given fMRI data.
D. Robustness to Noise
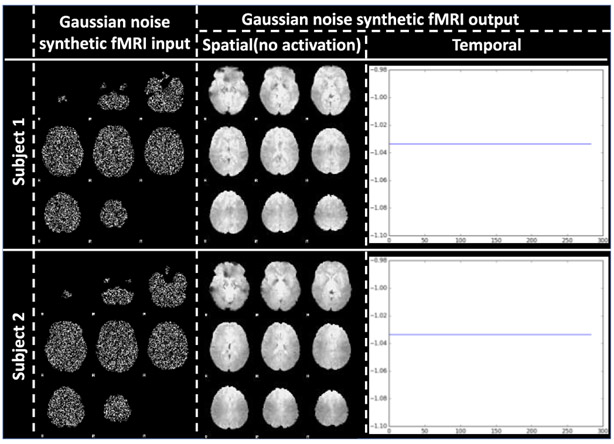
To further test the robustness to noise of the ST-CNN and to further demonstrate that the ST-CNN is not just overfitting DMN from “brain shaped” signals, we synthesized fMRI data using Gaussian noises (first column in Fig. 10) within the brain mask to test our ST-CNN framework.
Fig. 10.
ST-CNN output for synthetic fMRI data input with Gaussian noise. Visualization of synthetic fMRI data with Gaussian noise on 1st column. The synthetic data and real fMRI data have the same brain shape as boundary mask. Spatial output: only brain background is shown, not activation for the output. Temporal output: no active curve corresponding to spatial maps with no activation.
The testing results showed that the trained ST-CNN is no sensitive to noise and there is no output temporal signals and no activation for the spatial maps, as shown in Fig. 10. The results confirmed the robustness to noise of our trained ST-CNN. Further, it also demonstrated that our trained ST-CNN is not overfitting DMN according to the brain shapes, rather modelling the intrinsic signals from the fMRI data. With the confidence in the intrinsic 4D modelling from the fMRI data of ST-CNN, we further checked whether ST-CNN can model the intrinsic spatial and temporal relationships from the fMRI data in the next section.
E. Spatial and Temporal Relationship
As network spatial pattern and temporal dynamics are intertwined with each other, it is interesting to examine the relationship between spatial and temporal domains of functional networks. In order to validate that the ST-CNN can well capture the intrinsic spatial and temporal relationship from fMRI data, we performed a supervised dictionary learning method [26] (introduced in the evaluation and validation section) onto the fMRI data by taking ST-CNN temporal output as input supervision to reconstruct the corresponding spatial response of that temporal supervision to check whether the spatial response is DMN or not.
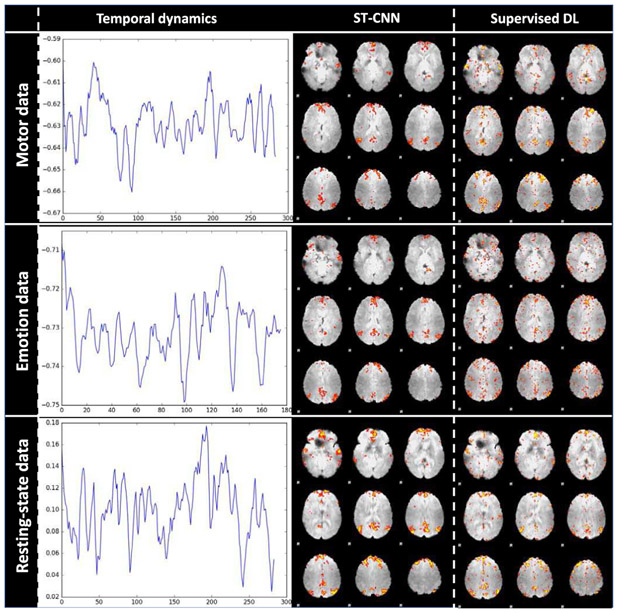
We performed supervised dictionary learning on all 3 test data sets: 40 motor task data, 200 emotion task data and 200 resting state data. We randomly picked one exemplar result per dataset to briefly illustrate the validation result for ST-CNN in Fig. 11. The first column of Fig. 11 shows temporal dynamics of ST-CNN, which was used to be the fixed dictionary atom as the supervision, and the corresponding spatial outputs of the supervised dictionary learning are shown in the third column in Fig. 11. The ST-CNN spatial outputs are in the second column of Fig. 11. We can clearly see that the ST-CNN spatial maps resemble the supervised dictionary learning spatial maps matching DMN spatial pattern, which means that ST-CNN captured intrinsic relationships between temporal and spatial dynamics of DMN. We also put all validation results for further reference: motor data validation: http://hafni.cs.uga.edu/DMN_dynamic/HCP_900/validation/MOTOR/; emotion data validation: http://hafni.cs.uga.edu/DMN_dynamic/HCP_900/validation/EMOTION/; resting-state data validation: http://hafni.cs.uga.edu/DMN_dynamic/HCP_900/validation/RSN/. Quantitatively, we calculated the spatial overlap rate as the similarity metric to measure how similar the ST-CNN outputs resemble the supervised dictionary learning outputs. As shown in TABLE IV, the mean spatial overlap rates are all larger than 0.2, which can be considered strongly similar according to [27]. The results statistically demonstrated the high similarity between the ST-CNN and the supervised dictionary learning results, which indicates that our proposed ST-CNN is effectively modelling the intrinsic spatio-temporal relationship of DMN from fMRI data. However, to achieve the same results, the supervised dictionary learning methods or other equivalent methods need to take prior knowledge such as the temporal input as supervision to obtain the spatial output, which might hamper the application of such methods given that we don’t have any prior knowledge of a specific data. On the contrary, once ST-CNN is properly trained, it can produce both temporal and spatial dynamics of DMN without any form of prior information, which paves a much broader way for applications.
Fig. 11.
Validation of ST-CNN performing supervised dictionary learning (Supervised DL) method. The first column shows temporal dynamics of ST-CNN. Using that as input, we performed supervised DL to generate the corresponding spatial response (Third column). The second column shows ST-CNN spatial maps, which resemble the supervised DL results. Both spatial map results display the DMN spatial pattern.
TABLE IV.
Validation of ST-CNN performing supervised dictionary learning (SDL). The spatial overlap rate between ST-CNN spatial outputs and SDL spatial outputs using ST-CNN temporal outputs as input supervision. The statistics shown below are mean value ± standard deviation.
| Dataset | MOTOR | EMOTION | RS |
|---|---|---|---|
| Spatial overlap rate between ST-CNN and SDL ouputs (Mean±std) |
0.294±0.072 | 0.304±0.056 | 0.336±0.075 |
All the above qualitative and quantitative results demonstrated that our proposed ST-CNN can model the intertwined intrinsic spatio-temporal dynamics from 4D fMRI data, no matter task-evoked or resting-state data.
IV. Discussion
In this work, we proposed a novel ST-CNN to model and analyze 4D fMRI data and simultaneously generate DMN spatial and temporal dynamics in a pinpoint way. This spatio-temporal deep learning framework provided a new tool and insight for 4D fMRI analysis in future cognitive and clinical research studies. By utilizing the proposed ST-CNN, we aimed to solve the 2 challenging problems in fMRI analysis research: spatio-temporal intrinsic 4D analysis for specific functional network (DMN in this paper) and functional network identification directly from fMRI data after only basic preprocessing (e.g., gradient distortion correction, motion correction, field map preprocessing, distortion correction, spline resampling to atlas space, intensity normalization and etc). As we already discussed, the spatio-temporal and 4D simultaneous analysis for fMRI data is still an open question and many current functional network identification methods are still based on data decomposition technique (e.g., ICA, dictionary learning and sparse coding. [35]-[37]. Those techniques have randomized index for the extracted DMN among all the extracted functional networks, which will impose a burden for the DMN identification process, while ST-CNN is trained specifically for DMN, which will yield the targeted DMN directly as output without any ambiguities.) Now in the proposed ST-CNN, these two challenging open questions can be effectively handled at the same time and the DMN identification process is much more robust and reliable than traditional fMRI data decomposition and network identification techniques. More importantly, the reproducibility of the ST-CNN is clearly demonstrated by training ST-CNN on one task-evoked dataset and applied to other task-evoked datasets as well as resting-state dataset. As for DMN regression, it is logically more reasonable to use rsfMRI data for both training and testing. However, this is a relatively simpler task as training and testing are both performed on the same type of the dataset. Considering the ST-CNN framework, which is proposed to regress the DMN spatial map and temporal response within that region, while concurrent temporal response can also pose a penalty for falsely regressed spatial regions, it really doesn’t quite matter whether the DMN temporal response is positively or negatively correlated with the task design since the task design is not even utilized in training ST-CNN, as long as the temporal response is concurrent regarding to the DMN spatial response. Besides, correspondence between task-evoked state and resting-state has been found to be established in [24]. Therefore, the generalizability of the ST-CNN for different tasks/resting states fMRI data is also demonstrated. The robustness to noise and non-overfitting analysis further exhibited the robustness of the ST-CNN framework. With further validation on the relationship between spatial and temporal outputs, we further confirmed the effectiveness of the proposed ST-CNN.
In the future work, we will focus on extending the current framework on pinpointing more functional networks from raw 4D fMRI data, which can be further applied on brain disease datasets for better understanding of abnormal brain activity. As indicated by the current research results [27], [38], comprehensive resting-state networks including high-order and low-order networks [39] are necessary for brain disease analysis. Since the ST-CNN model is quite robust and reproducible for various types of data only across a small range of hyperparameter settings, we plan to use a neural architecture search scheme to investigate the optimal architecture of the ST-CNN for different types of data and applications. Other simultaneous spatio-temporal fMRI analysis models can also be inspired from ST-CNN to accelerate the investigation of the brain’s functional architecture.
Acknowledgment
We thank the Human Connectome Project (HCP) for sharing their invaluable fMRI datasets.
This work was supported by National Institute of Health (R01 DA-033393, R01 AG-042599), and National Science Foundation career award (IIS-1149260, CBET-1302089, BCS-1439051 and DBI-1564736).
Contributor Information
Yu Zhao, Cortical Architecture Imaging and Discovery (CAID) Lab, Department of Computer Science and Bioimaging Research Center, The University of Georgia, Athens, GA, USA..
Xiang Li, Massachusetts General Hospital and Harvard Medical School, Boston MA 02115, USA..
Heng Huang, School of Automation, Northwestern Polytechnical University, Xi’an, Sha’anxi 710072, China.
Wei Zhang, Cortical Architecture Imaging and Discovery (CAID) Lab, Department of Computer Science and Bioimaging Research Center, The University of Georgia, Athens, GA, USA..
Shijie Zhao, School of Automation, Northwestern Polytechnical University, Xi’an, Sha’anxi 710072, China.
Milad Makkie, Cortical Architecture Imaging and Discovery (CAID) Lab, Department of Computer Science and Bioimaging Research Center, The University of Georgia, Athens, GA, USA..
Mo Zhang, Center for Data Science, Peking University, Beijing 100080, China..
Quanzheng Li, Massachusetts General Hospital and Harvard Medical School, Boston MA 02115, USA.; Peking University, Laboratory for Biomedical Image Analysis, Beijing Institute of Big Data Research, Beijing 100871, China..
Tianming Liu, Cortical Architecture Imaging and Discovery Lab, Department of Computer Science and Bioimaging Research Center, The University of Georgia, Athens, GA, USA.
References
- [1].Cole DM, Smith SM, and Beckmann CF, “Advances and pitfalls in the analysis and interpretation of resting-state FMRI data.,” Front. Syst. Neurosci, vol. 4, p. 8, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].McKeown MJ, Hansen LK, and Sejnowsk TJ, “Independent component analysis of functional MRI: what is signal and what is noise?,” Curr. Opin. Neurobiol, vol. 13, no. 5, pp. 620–629, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lv J, Jiang X, Li X, Zhu D, Chen H, Zhang T, Zhang S, Hu X, Han J, Huang H, Zhang J, Guo L, and Liu T, “Sparse representation of whole-brain fMRI signals for identification of functional networks.,” Med. Image Anal, vol. 20, no. 1, pp. 112–34, February. 2015. [DOI] [PubMed] [Google Scholar]
- [4].Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, and Sánchez CI, “A survey on deep learning in medical image analysis.,” Med. Image Anal, vol. 42, pp. 60–88, December. 2017. [DOI] [PubMed] [Google Scholar]
- [5].Zhao Y, Dong Q, Chen H, Iraji A, Li Y, Makkie M, Kou Z, and Liu T, “Constructing fine-granularity functional brain network atlases via deep convolutional autoencoder,” Med. Image Anal, vol. 42, pp. 200–211, December. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Heeger DJ and Ress D, “WHAT DOES fMRI TELL US ABOUT NEURONAL ACTIVITY?,” Nat. Rev. Neurosci, vol. 3, no. 2, pp. 142–151, February. 2002. [DOI] [PubMed] [Google Scholar]
- [7].Smith SM, Miller KL, Moeller S, Xu J, Auerbach EJ, Woolrich MW, Beckmann CF, Jenkinson M, Andersson J, Glasser MF, Van Essen DC, Feinberg DA, Yacoub ES, and Ugurbil K, “Temporally-independent functional modes of spontaneous brain activity,” Proc. Natl. Acad. Sci, vol. 109, no. 8, pp. 3131–3136, February. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Huang H, Hu X, Zhao Y, Makkie M, Dong Q, Zhao S, Guo L, and Liu T, “Modeling Task fMRI Data via Deep Convolutional Autoencoder,” IEEE Trans. Med. Imaging, pp. 1–1, 2017. [DOI] [PubMed] [Google Scholar]
- [9].Hjelm RD, Calhoun VD, Salakhutdinov R, Allen EA, Adali T, and Plis SM, “Restricted Boltzmann machines for neuroimaging: An application in identifying intrinsic networks,” Neuroimage, vol. 96, pp. 245–260, August. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Shen Y Mayhew SD, Kourtzi Z, and Tiňo P, “Spatial–temporal modelling of fMRI data through spatially regularized mixture of hidden process models,” Neuroimage, vol. 84, pp. 657–671, January. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hjelm RD, Plis SM, and Calhoun V, “Recurrent Neural Networks for Spatiotemporal Dynamics of Intrinsic Networks from fMRI Data,” in NIPS: Brains and Bits, 2016. [Google Scholar]
- [12].Ronneberger O, Fischer P, and Brox T, “U-Net: Convolutional Networks for Biomedical Image Segmentation,” in International Conference on Medical Image Computing and Computer Assisted Intervention, 2015. [Google Scholar]
- [13].Zhao Y, Li X, Zhang W, Zhao S, Makkie M, Zhang M, Li Q, and Liu T, “Modeling 4D fMRI Data via Spatial-temporal Convolutional Neural Networks (ST-CNN),” in Medical Image Computing and Computer Assisted Intervention Society, 2018, pp. 181–189. [Google Scholar]
- [14].Krizhevsky A, Sutskever I, and Hinton GE, ImageNet Classification with Deep Convolutional Neural Networks. 2012, pp. 1097–1105. [Google Scholar]
- [15].Zhao S, Han J, Jiang X, Huang H, Liu H, Lv J, Guo L, and Liu T, “Decoding Auditory Saliency from Brain Activity Patterns during Free Listening to Naturalistic Audio Excerpts.,” Neuroinformatics, February. 2018. [DOI] [PubMed] [Google Scholar]
- [16].Van Essen DC, Smith SM, Barch DM, Behrens TEJ, Yacoub E, Ugurbil K, and for the W.-M. H. WU-Minn HCP Consortium, “The WU-Minn Human Connectome Project: an overview.,” Neuroimage, vol. 80, pp. 62–79, October. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Barch DM, Burgess GC, Harms MP, Petersen SE, Schlaggar BL, Corbetta M, Glasser MF, Curtiss S, Dixit S, Feldt C, Nolan D, Bryant E, Hartley T, Footer O, Bjork JM, Poldrack R, Smith S, Johansen-Berg H, Snyder AZ, Van Essen DC, and WU-Minn HCP Consortium, “Function in the human connectome: task-fMRI and individual differences in behavior.,” Neuroimage, vol. 80, pp. 169–89, October. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].“WU-Minn HCP 900 Subjects Data Release: Reference Manual,” 2015. [Google Scholar]
- [19].Woolrich MW, Ripley BD, Brady M, and Smith SM, “Temporal Autocorrelation in Univariate Linear Modeling of FMRI Data,” Neuroimage, vol. 14, no. 6, pp. 1370–1386, December. 2001. [DOI] [PubMed] [Google Scholar]
- [20].Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, and Smith SM, “FSL,” Neuroimage, vol. 62, pp. 782–90, 2012. [DOI] [PubMed] [Google Scholar]
- [21].Dale AM, Fischl B, and Sereno MI, “Cortical Surface-Based Analysis,” Neuroimage, vol. 9, no. 2, pp. 179–194, February. 1999. [DOI] [PubMed] [Google Scholar]
- [22].Zhang W, Lv J, Li X, Zhu D, Jiang X, Zhang S, Zhao Y, Guo L, Ye J, Hu D, and Liu T, “Experimental Comparisons of Sparse Dictionary Learning and Independent Component Analysis for Brain Network Inference from fMRI Data,” IEEE Trans. Biomed. Eng, pp. 1–1, 2018. [DOI] [PubMed] [Google Scholar]
- [23].Tibshirani R, Johnstone I, Hastie T, and Efron B, “Least angle regression,” Ann. Stat, vol. 32, no. 2, pp. 407–499, April. 2004. [Google Scholar]
- [24].Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, and Beckmann CF, “Correspondence of the brain’s functional architecture during activation and rest.,” Proc. Natl. Acad. Sci. U. S. A, vol. 106, no. 31, pp. 13040–5, August. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zeiler MD, “ADADELTA: An Adaptive Learning Rate Method,” December. 2012. [Google Scholar]
- [26].Zhao S, Han J, Lv J, Jiang X, Hu X, Zhao Y, Ge B, Guo L, and Liu T, “Supervised Dictionary Learning for Inferring Concurrent Brain Networks,” IEEE Trans. Med. Imaging, vol. 34, no. 10, pp. 2036–2045, October. 2015. [DOI] [PubMed] [Google Scholar]
- [27].Zhao Y, Chen H, Li Y, Lv J, Jiang X, Ge F, Zhang T, Zhang S, Ge B, Lyu C, Zhao S, Han J, Guo L, and Liu T, “Connectome-scale group-wise consistent resting-state network analysis in autism spectrum disorder.,” NeuroImage. Clin, vol. 12, pp. 23–33, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Harris KD and Mrsic-Flogel TD, “Cortical connectivity and sensory coding,” Nature, vol. 503, no. 7474, pp. 51–58, November. 2013. [DOI] [PubMed] [Google Scholar]
- [29].Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, and Shulman GL, “A default mode of brain function.,” Proc. Natl. Acad. Sci. U. S. A, vol. 98, no. 2, pp. 676–82, January. 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Greicius MD, Krasnow B, Reiss AL, and Menon V, “Functional connectivity in the resting brain: a network analysis of the default mode hypothesis.,” Proc. Natl. Acad. Sci. U. S. A, vol. 100, no. 1, pp. 253–8, January. 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Vatansever D, Menon DK, and Stamatakis EA, “Default mode contributions to automated information processing,” Proc. Natl. Acad. Sci, vol. 114, no. 48, pp. 12821–12826, November. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jung M, et al. , “Default mode network in young male adults with autism spectrum disorder: relationship with autism spectrum traits,” Mol. Autism, vol. 5, no. 1, p. 35, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Iraji A, Benson RR, Welch RD, O’Neil BJ, Woodard JL, Ayaz SI, Kulek A, Mika V, Medado P, Soltanian-Zadeh H, Liu T, Haacke EM, and Kou Z, “Resting State Functional Connectivity in Mild Traumatic Brain Injury at the Acute Stage: Independent Component and Seed-Based Analyses,” J. Neurotrauma, vol. 32, no. 14, pp. 1031–1045, July. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Greicius MD, Srivastava G, Reiss AL, and Menon V, “Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI.,” Proc. Natl. Acad. Sci. U. S. A, vol. 101, no. 13, pp. 4637–42, March. 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Griffanti L, Douaud G, Bijsterbosch J, Evangelisti S, Alfaro-Almagro F, Glasser MF, Duff EP, Fitzgibbon S, Westphal R, Carone D, Beckmann CF, and Smith SM, “Hand classification of fMRI ICA noise components,” Neuroimage, vol. 154, pp. 188–205, July. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tohka J, Foerde K, Aron AR, Tom SM, Toga AW, and Poldrack RA, “Automatic independent component labeling for artifact removal in fMRI.,” Neuroimage, vol. 39, no. 3, pp. 1227–45, February. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhao Y, Dong Q, Zhang S, Zhang W, Chen H, Jiang X, Guo L, Hu X, Han J, and Liu T, “Automatic Recognition of fMRI-derived Functional Networks using 3D Convolutional Neural Networks,” IEEE Trans. Biomed. Eng, pp. 1–1, June. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhao Y, Ge F, Zhang S, and Liu T, “3D Deep Convolutional Neural Network Revealed the Value of Brain Network Overlap in Differentiating Autism Spectrum Disorder from Healthy Controls,” in MICCAI, 2018, pp. 172–180. [Google Scholar]
- [39].Liao W, Fan Y-S, Yang S, Li J, Duan X, Cui Q, and Chen H, “Preservation Effect: Cigarette Smoking Acts on the Dynamic of Influences Among Unifying Neuropsychiatric Triple Networks in Schizophrenia,” Schizophr. Bull, December. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]