Abstract
Background
Anti-programmed death (PD)-1 therapy has recently been used in recurrent or metastatic (R/M) nasopharyngeal carcinoma (NPC). The long-term survival and its biomarkers responding to anti-PD-1 treatment in patients with R/M NPC remain unclear.
Methods
Patients with R/M NPC were enrolled between March 2016 and January 2018 from two phase I clinical trials. The median follow-up period was 24.7 months. Eligible patients progressed on standard chemotherapy had measurable disease by Response Evaluation Criteria in Solid Tumor V.1.1. Non-obligatory contemporaneous tumor samples were collected for whole-exome sequencing. The primary outcome was objective response rate (ORR). Duration of response (DOR), progression-free survival (PFS), and overall survival (OS) were secondary outcomes assessed in all patients.
Results
Among 124 evaluable patients, anti-PD-1 therapy achieved an ORR of 29.8% and a durable clinical benefit rate of 60.5%. The median OS (mOS) was 17.1 months (95% CI 14.2 to 24.7), median PFS (mPFS) was 3.8 months (95% CI 3.4 to 6.0), and median DOR was 9.5 months. Significant OS benefit from treatment was observed in patients without liver metastasis (23.8 vs 13.3 months, p=0.006). Copy number deletion in genes encoding granzyme B or granzyme H (GZMB/H) was associated with poor treatment outcome (mPFS altered vs wildtype: 1.7 vs 3.6 months, p=0.03; mOS altered vs wildtype: 10.1 vs 18 months, p=0.012).
Conclusions
Anti-PD-1 treatment provided promising clinical benefit in pretreated patients with R/M NPC. Copy number loss in either GZMB or GZMH genes was associated with reduced survival.
Keywords: tumor biomarkers, immunotherapy, head and neck neoplasms
Background
Nasopharyngeal carcinoma (NPC) is a head and neck cancer arising from the epithelial lining of the nasopharynx,1 2 with a high prevalence in Southeast Asian populations.3 Unlike other head and neck cancer, NPC is characterized by extensive Epstein-Barr virus (EBV) infection,4 which is implicated in driving tumorigenesis via its evasion of host innate immunity.5 Currently, gemcitabine plus cisplatin is used as standard first-line treatment for recurrent or metastatic (R/M) NPC.6 7 Patients who progressed on first-line treatment have poor prognosis and few available follow-up treatment options. Active agents used in second line include paclitaxel, docetaxel, gemcitabine, capecitabine and so on, either as single agent or in combination with platinum. Nonetheless, second-line treatments often yield unsatisfactory results, with estimated median progression-free survival (mPFS) at 5.2 months and estimated median overall survival (mOS) at 12.5 months.8
The last decade has seen significant development in immune-checkpoint inhibitors (ICIs), which have shown promising antitumor activity in a range of tumors including NPC.9–11 Additionally, previous studies reported the short-term efficacy and safety of ICIs in NPC.12–14 Among these, we have published preliminary results from two phase I clinical trials composing the largest cohort of R/M NPC treated with anti-PD-1 to date12 14 that demonstrated anti-PD-1 therapy have comparable immediate benefit to chemotherapies. Tumor mutation burden (TMB) was promising biomarkers of immune-oncology (IO) therapy in other cancer types.15 16 However, others reported that the prognostic value of TMB was minimal since patients with NPC generally had lower mutation burden compared with non-small cell lung cancer (NSCLC) or melanoma. Therefore, NPC-specific molecular markers that can predict patients’ response to IO therapy are urgently needed.
Granzyme B (GZMB) is a serine protease that plays an important role in T cell and NK cell mediated tumor killing.17 It has been reported that granzyme H (GZMH) has a similar capability.18 Recently, GZMB has emerged as a predictor of IO therapy response in different cancer types. One study found that the expression of GZMB in tumor samples from patients with melanoma was significantly correlated with response, and the level of GZMB expression increased during treatment in responders.19 Another study in patients with stage IV NSCLC found that high serum GZMB level was associated with better survival and patients with germline variants in GZMB had reduced survival compared with wildtype.20 Given the significance of granzyme function in lymphocyte cytotoxicity and implication in IO therapy outcome, we hypothesize that the granzyme family is predictive of response to anti-PD-1 therapy in NPCs and loss of function in this pathway is associated with reduced survival.
Methods
Study design and patients
Patients with R/M NPC were consecutively enrolled in two phase I clinical trials for advanced solid tumors (camrelizumab and nivolumab, ClinicalTrials.gov identifiers: NCT02721589 and NCT02593786) between March 2016 and January 2018.12 14 The details of studies design of dose escalation and expansion phases were already reported. Camrelizumab trial enrolled 33 patients with NPC in dose escalation phase, and 60 patients with NPC in dose expansion phase (200 mg fixed dose every 2 weeks). Nivolumab trial enrolled 33 patients with NPC in dose escalation phase. Sample sizes in dose escalation phases were based on safety (dose-limiting toxicity), and in dose expansion phase were decided for overall response rate (ORR) consideration compared with historical control. The distribution of patients’ treatment in the current study was shown in online supplemental figure S1. Baseline tumor samples and matched peripheral blood were provided non-obligatory right before the initiation of anti-PD-1 treatment. Written informed consent were provided by all patients.
jitc-2020-002014supp002.pdf (20.1KB, pdf)
Treatment and evaluation
Eligible patients received intravenous infusion of camrelizumab at dosages range from 1 mg/kg, 3 mg/kg, 200 mg and 10 mg/kg every 2 weeks; or nivolumab at dosage of 3 mg/kg, 240 mg every 2 weeks and 360 mg every 3 weeks. Radiographic tumor assessments were taken at baseline and approximately every 6 weeks. Treatment response was assessed by investigators per Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1. ORR was the sum of complete response (CR) and partial response (PR). Durable clinical benefit (DCB) was defined as the percentage of patients who achieved CR or PR or stable disease (SD) lasted >6 months; non-durable clinical benefit (NDB) was defined as PD or SD that lasted ≤6 months. Duration of response (DOR) was define as the duration from objective response to progression disease (PD) in responders. Treatment continued until confirmed PD, intolerable toxicities, death or withdrawal of consent. PFS was defined as the time from first dose to PD, or prior death. Censored data documented last radiographic assessment before cut-off, loss of follow-up or change of treatment. Treatment beyond initial RECIST disease progression was permitted as long as patients satisfied the criteria in protocols. Survival follow-up was approximately every 3 months by clinic visits or telephone calls. Overall survival (OS) was duration from first dose to death, patients who remained alive were censored at the date of their last follow-up.
WES, mutation calling and copy number analysis
Available tumor tissues from 60 patients in this cohort underwent whole-exome sequencing (WES). Genomic DNAs from Formalin-fixed, paraffin-embedded (FFPE) or biopsy tumor samples and blood samples were extracted with QIAamp DNA FFPE Tissue Kit and DNeasy Blood and tissue kit (Qiagen, USA), respectively, and quantified by Qubit V.3.0 using the dsDNA HS Assay Kit (ThermoFisher Scientific, USA). WES was performed on Illumina HiSeq4000 platform using PE150 sequencing chemistry (Illumina, USA). Trimmomatic was used for FASTQ file quality control.21 Leading/trailing low quality (quality reading below 20) or N bases were removed. Paired-end reads were then aligned to the reference human genome (build hg19), using the Burrows-Wheeler Aligner.22 PCR deduplication was performed using Picard.23 GATK3 was used for local realignment around indels and base quality score recalibration.24 Cross-sample contamination was estimated using ContEst25 (Broad Institute, contamination rate <0.02). Somatic Single Nucleotide Variant calling was generated by Mutect26 and insertion/deletions were called with Scalpel.27 Copy number alteration (CNA) analysis from sequencing data was performed using CNVKit.28 Focal level gain and loss were defined as normalized log2 depth ratio ≥1 or ≤−0.6,29 respectively.
Statistical analysis
Comparisons of proportion between groups and association between categorical variables were examined using the χ2 test. In biomarker subgroup analysis, quantitative variables such as TMB were stratified at multiple thresholds to identify association with survival. For survival analyses, Kaplan-Meier curves were compared using the log-rank test, and HRs were calculated by Cox proportional hazards model. Multivariate analysis was performed to assess potential cofounding effects among clinical variables. The focal CNA analysis was performed using a previously described pipeline.9 CNA p value was corrected for multiple hypothesis testing using the Benjamini-Hochberg method. All statistical analyses were performed in R (V.3.3.2).
Results
Survival outcomes of the entire cohort
In the current study, 124 patients with R/M NPC were retrospectively examined. OS outcome and treatment response to anti-PD-1 antibody were analyzed. Tumors from 60 patients were profiled by WES to uncover mutational profiles related to treatment outcome. The median follow-up duration was 24.7 months (95% CI 23.3 to 26.6). The majority of the patients (66.1%) failed at least two prior lines of therapy. All baseline characteristics were shown in table 1. Additionally, the baseline demographic from the 60 patients with WES data were not showing any significant differences compared with the other patients (online supplemental table S1).
Table 1.
Patients’ demographics and baseline clinical characteristics
| No. (%) | All patients (124) |
| Median age, years (range) | 46 (23–73) |
| Sex (male/female) | |
| Male | 95 (76.6) |
| Female | 29 (23.4) |
| ECOG performance status score | |
| 0 | 35 (28.2) |
| 1 | 89 (71.8) |
| Smoking | |
| Never | 85 (68.6) |
| Former/current | 35 (28.2) |
| Unknown | 4 (3.2%) |
| Stage | |
| Primary metastasis | 13 (10.5) |
| Recurrent with distant metastasis | 111 (89.5) |
| WHO histological classification | |
| Undifferentiated non-keratinised | 96 (77.4) |
| Differentiated non-keratinised | 14 (11.3) |
| Keratinised squamous carcinoma | 14 (11.3) |
| Distant metastasis sites | |
| Lung | 69 (55.6) |
| Liver | 69 (55.6) |
| Bone | 50 (40.3) |
| Distant lymph | 46 (37.1) |
| Others | 17 (13.7) |
| None | 8 (6.5) |
| Prior lines for advanced disease | |
| 1 | 42 (33.9) |
| 2 | 35 (28.2) |
| 3 | 25 (20.2) |
| 4 or more | 22 (17.7) |
| Prior chemotherapy for advanced disease | |
| Paclitaxel | 74 (59.7) |
| Gemcitabine | 69 (55.6) |
| 5-Fluoracial | 52 (41.9) |
| Docetaxel | 37 (29.8) |
| Tegafur | 25 (20.2) |
| Cetuximab | 23 (18.5) |
| Capecitabine | 19 (15.3) |
| Ipilimumab | 8 (6.5) |
| Comorbidity | |
| Yes | 83 (66.9) |
| No | 41 (33.1) |
| Prior radiation | |
| Yes | 111 (89.5) |
| No | 13 (10.5) |
| WES detection | |
| Yes | 60 (48.4) |
| No | 64 (51.6) |
ECOG, Eastern Cooperative Oncology Group; WES, whole-exome sequencing; WHO, world health organization.
jitc-2020-002014supp010.pdf (262.1KB, pdf)
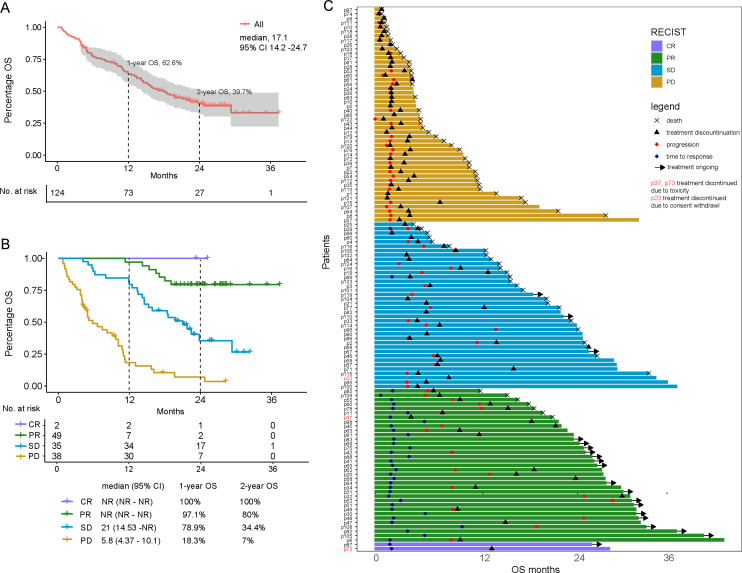
Treatment by anti-PD-1 in all enrolled patients achieved a median OS of 17.1 months (95% CI 14.2 to 24.7) and a median PFS of 3.8 months (95% CI 3.4 to 6.0). One-year OS rate was 62.6% (95% CI 54.5% to 71.9%), and the 2-year OS rate was 39.7% (95% CI 31.5% to 50%; figure 1A). The ORR for all patients was 29.8% (2 CR and 35 PR), and the DCB rate was 60.5% (online supplemental table S2). The median DOR was 9.5 months (95% CI 6.0 to 13.0).
Figure 1.
Overall survival (OS) outcome and association with clinical variables. (A) Kaplan-Meier curve of OS in all patients with 95% CI band represented by the shaded area.CI, confidence interval. (B) OS in all treated patients by best overall response. CR, complete remission; PR, partial remission; SD, stable disease; PD, progression disease; NR, not reached. (C) Swimmer plot that shows time to first response and duration of response. The length of the bars represents OS month. The first timepoint of response or progression, treatment discontinuation, and time of death were labeled for patients with events. Patients that continue treatment are labeled with an arrow.
The patient’s best response strongly predicted OS (mOS CR: not reached (NR); PR: NR; SD: 19.8 months; PD: 5.8 months; figure 1B). In univariate analysis (figure 2), prior radiation was associated with better survival, whereas the number of distant metastatic sites and liver metastasis were associated with reduced survival (online supplemental figure S2). Other baseline clinical variables were not associated with survival in univariate and multivariate models (figure 2, online supplemental table S3).
Figure 2.
Associations of baseline demographic and clinical characteristics with (A) overall survival (OS) and (B) progression-free survival (PFS) in univariate analysis. All variables are categorical. HR greater than 1 indicates association with reduced survival. P values are labeled on the right side of each individual plot. ECOG, Eastern Cooperative Oncology Group; NR, not reached.
jitc-2020-002014supp003.pdf (55.8KB, pdf)
Molecular markers associated with response
Among the 60 patients with WES data, the median TMB was 52. The distribution of TMB was not significantly different between patients with PR, SD, or PD (online supplemental figure S3A). Patients were grouped binarily as TMB high versus low based on four different cut-off values (20%, 40%, 60%, and 80% of TMB ranking), neither PFS nor OS differed between groups (online supplemental figure S3B, C). We divided patients into TMB high and low group by median and showed neither PFS nor OS were different between groups (online supplemental figure S3D, E), which was probably due to the overall low TMB levels within this cohort (data not shown). Additionally, the proportions of TMB high group among patients who received camrelizumab and nivolumab are not statistically different (28.6% vs 27.8%, Fisher’s exact test, p=1, detail data not shown). Likewise, the tumor neoantigen burden showed no impact on neither PFS nor OS (online supplemental figure S4A, B).
jitc-2020-002014supp004.pdf (84.6KB, pdf)
jitc-2020-002014supp005.pdf (34.9KB, pdf)
Next, we sought to characterize key immune-related pathways in the tumor samples, including 17 previously reported immune-related pathways from the Immunology Database and Analysis Portal (ImmPort, https://www.immport.org/), and the gasdermin, granzyme pathways. Because mutations were rare in these immune genes in this cohort, the distribution of CNA in key immune-related pathways was examined. Within these 19 immune-related pathways, only interferon (IFN), gasdermin and granzyme gene deletions occurred in more than 4 patients (online supplemental table S4), and these 3 pathways were therefore selected for downstream analysis.
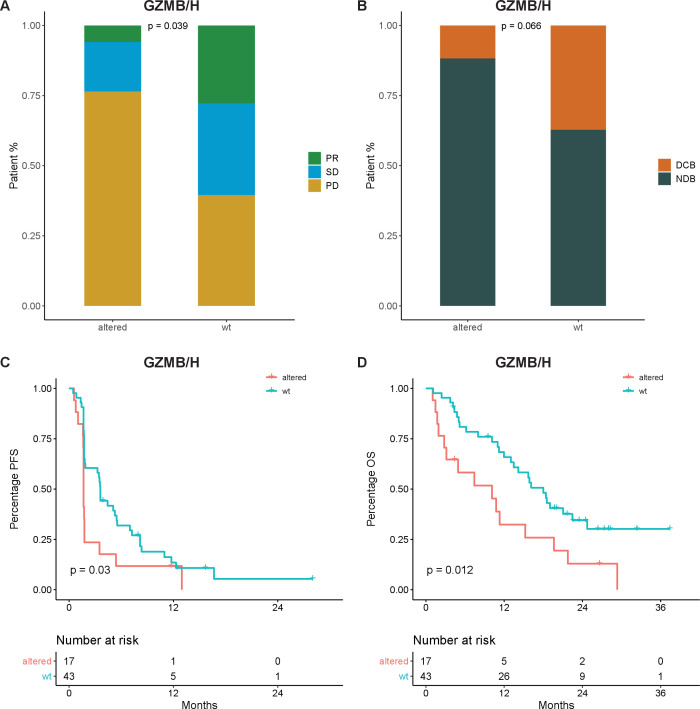
Frequent deletion in granzyme genes was observed in this cohort, among which copy number loss in GZMB and GZMH occurred at the highest frequency (figure 3, online supplemental table S5). Deletion was also observed in gasdermins and type I IFN but at lower frequencies compared with granzymes. Mutations and deletions on other genes were also analyzed as shown in online supplemental tables S6 and S7). We examined the effect of GZMB and GZMH deletion on survival and found patients with copy number loss in either gene had worse PFS (1.7 vs 3.6 months, p=0.03) and OS (10.1 vs 18 months, p=0.012) compared with the wildtype, as shown in figure 4. Fisher’s exact test confirmed that the proportions of GZMB/H loss were not statistically different among the camrelizumab and nivolumab group (47.6% vs 61.1%, p=0.405, detail data not shown). Furthermore, multivariate analysis using treatment agents and GZMB/H status confirms that GZMB/H loss have significant attribution towards patient PFS (p=0.027) and OS (p=0.013), while different treatment reagents had no significant attribution, as shown in online supplemental figure S5A, B. Similarly, these patients also had lower PR ratio (p=0.039) and DCB ratio (p=0.066) compared with the wildtype, as shown in figure 4A, B.
Figure 3.
Distribution of copy number loss of pyroptosis pathways in 60 patients with nasopharygeal carcinoma (NPC). Progression-free survival (PFS) and clinical characteristics of each patient were shown at the top. Loss of function (copy number loss and frame shift deletion) in interferon (IFN), gasdermins and granzyme genes were shown at the bottom. PR, partial remission; PD, progression disease; SD, stable disease.
Figure 4.
The effect of granzyme B or H (GZMB or GZMH) deletion on patients’ responses and survival. (A) Bar graph of Response Evaluation Criteria in Solid Tumors (RECIST) response in patients with alternation in either GZMB or GZMH gene versus wildtype. Fisher’s exact test. (B) Bar graph of durable clinical response in patients with alternation in either GZMB or GZMH gene versus wildtype. Fisher’s exact test. (C) Kaplan-Meier curve of progression-free survival (PFS) in patients with altered GZMB or GZMH genes versus wild type. (D) Kaplan-Meier curve of overall survival (OS) in patients with altered GZMB or GZMH genes versus wild type. DCB, durable clinical benefit; NDB, non-durable clinical benefit; PD, progression disease; PR, partial remission; SD, stable disease.
jitc-2020-002014supp006.pdf (273.2KB, pdf)
In addition, we looked at the overall effect of deletion in granzymes, gasdermins, and IFNs on survival. As shown in online supplemental figure S6A–C), the median PFS among patients with alternation in these pathways was worse compared with the wildtypes, although it was not significant for gasdermin alterations (mPFS granzyme 1.7 vs 3.6 months, p=0.012; gasdermin 1.8 vs 3.5 months, p=0.39; IFN 1.7 vs 3.5 months, p=0.023). Similarly, patients with alteration in their tumor had inferior OS compared with these without alteration (mOS granzyme 7.4 vs 18 months, p=0.0046; gasdermin 4.3 vs 15.9 months, p=0.042; IFN 8 vs 18 months, p=0.00079; online supplemental figure S7A–C). Furthermore, patients who had alteration in any of the pathways had significantly reduced PFS and OS compared with these with normal pathway functions (mPFS 1.7 vs 4.4 months, p=0.0047; mOS 7.4 vs 21 months, p<0.0001; online supplemental figure S7D).
jitc-2020-002014supp007.pdf (54.7KB, pdf)
jitc-2020-002014supp008.pdf (55.7KB, pdf)
Interestingly, in a cohort of patients with NPC treated with radiotherapy instead of checkpoint blockade and had tumor sequenced by WES, GZMB and GZMH deletion was not associated with OS nor PFS (online supplemental figure S8A, B).30 Furthermore, to test if GZMB and GZMH deletion were associated with response to checkpoint inhibitor in non-microbial-associated cancers, we examined a pan-cancer cohort including NSCLC, melanoma, bladder, and several other cancer types.10 Patients with deletion in either gene had reduced PFS and OS compared with wildtype although the difference was not significant (mPFS 2.6 vs 4.6 months, p=0.084; mOS 10.0 vs 21.7 months, p=0.11; online supplemental figure S8C, D).
jitc-2020-002014supp009.pdf (59.1KB, pdf)
Discussion
This study reported the first and largest cohort of long-term survival outcome under anti-PD-1 therapy in R/M NPC. Overall, our results demonstrated that anti-PD-l treatment achieved durable survival benefit in patients with R/M NPC, and might yielded notable improved OS compared with historical control of second-line chemotherapies (median 12.5 months).8 31 32 Furthermore, anti-PD-1 achieved durable survival benefit in responders and SD in a subset of patients. For those 2-year survivors, mDOR was as long as 19.4 months. Furthermore, several clinical biomarkers showed strong correlation with anti-PD-1 treatment outcome in R/M NPC. Patients with liver metastasis at baseline achieved reduced PFS and OS, which is also consistent with other findings in melanoma and NSCLC treated with anti-PD-1.33 34 These findings suggest the long-term survival benefit of anti-PD-1 therapy for R/M NPC and could be realized through individualized treatment selection by biomarkers.
Several important immune-related pathways were frequently altered in this cohort and associated with resistance to anti-PD-1 therapy. Specifically, we found that alteration in granzyme, gasdermin, and type I IFN were enriched in refractory tumors. Both GZMB and GZMH are known to induce rapid apoptosis of target cells by cytotoxic T cell or nature killer cells.18 In addition, GZMH also plays an important role in controlling viral infection. One study has shown that GZMH is important in clearing of hepatitis B virus by cytotoxic lymphocytes by direct targeting of viral proteins.35 GZMB and GZMH deletion were recurrent in this cohort and strongly correlated with poor outcome. Moreover, in patients with NPC treated with non-IO regimen, GZMB/GZMH deletion was not associated with OS. Additionally, patients with GZMB deletion also had inferior survival in IO cohort of other cancer types as reported by several studies.19 20 GZMB and GZMH are, in theory, predominantly expressed in the immune cells. Recent studies of single-cell transcriptomic analysis in patients with NPC have shown that GZMB and GZMH genes are highly expressed in the immune cells, but not expressed in the NPC cancer cells.36 37 It is likely that such signature loss in the immune cells are detected by our sequencing process, due to tumor-infiltrating immunes. Similarly, Gao et al has reported that the loss of IFN genes in the tumor cells, despite mostly expressed in immune cells, can be used as prognostic markers in patients receiving immune checkpoint therapy.38 Moreover, we noticed a difference of GZMB/H predictive power between NPC and other cancer types, which can be explained by the various levels of tumor-infiltrating lymphocytes among these tumor types. However, more experiments are needed to validate this hypothesis. Taken together, GZMB and GZMH are critical in augmenting the anti-PD-1 antibody mediated immune function and tumor killing.
Traditionally, apoptosis was considered the dominant form of programmed cell death induced by cytotoxic lymphocytes. However, recently studies have shown that non-apoptotic killing including pyroptosis is also used by these immune cells to clear infection or cancer cells.39 40 Pyroptosis, a proinflammatory form of cell death,41 is induced when gasdermin B are cleaved by granzyme A and this effect is upregulated by IFN.39 Similarly, GZMB can also activate pyroptosis in target cells by gasdermine E.42 We found alteration in granzyme, gasdermin, and IFN genes resulted in reduced IO therapy response likely due to dysfunctional pyroptosis pathway and therefore weakened cytotoxic lymphocyte function. Evidence had shown that, during EBV latency and lytic cycles, type I and II IFN Jak-STAT signaling pathways were manipulated; expression and activity of IFN regulating factors were altered and host apoptosis signaling pathways were repressed.5 Therefore, compared with other tumors, the immune system of NPC may largely depend on non-apoptotic killing such as pyroptosis to eliminate viral transformed cells. Our findings suggest that even when the immune-checkpoint brakes are released by anti-PD-1 antibodies, intact downstream effector functions of cytotoxic lymphocytes are necessary for cancer clearance. Future studies are needed to validate the prognostic value of these markers in more cancer types and to further examine their association with pyroptosis.
The present study was limited by its retrospective nature. Whether clinical and genomic variables we found were direct causes for treatment outcome is not clear and should be studied in prospective trials with larger sample size. Since only a subset of patients with both available tumor and blood samples was included in the WES profiling, results derived from these analyses should be interpreted with caution. Furthermore, the WES profiling failed to distinguish between the immune cells and cancer cells. Single-cell level transcriptomic analysis are needed to better understand the potential of GZMB/H as biomarkers. Nonetheless, our results offered new insights on potential biomarkers of anti-PD-1 therapy in patients with R/M NPC.
In summary, we reported the long-term survival of patients with R/M NPC treated with anti-PD-1 and identified baseline clinical factors that were associated with survival outcome. Our results suggested alteration in pyroptosis pathways including IFN, gasdermin, and granzyme coding genes were associated with poor survival. These findings should be validated in prospective trials.
jitc-2020-002014supp001.pdf (118.2KB, pdf)
Footnotes
YM, XC, AW and HZ contributed equally.
Contributors: YM, XC, AW and HZ contributed equally to the work.
Funding: This work was supported by Chinese National Natural Science Foundation project (No. 81572659 and 81972556 to WF) and Physician scientist training program of Sun Yat-Sen University Cancer Center (No. 14zxqk03 to WF).
Competing interests: LZ declares research grant from Lilly. AW, HB, WT, XW and YS are the employees of Nanjing Geneseeq Technology.
Patient consent for publication: Not required.
Ethics approval: All study protocols were approved by the ethics committee of Sun Yat-sen University Cancer Center and in accordance with international standards of good clinical practice.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: The data of this study are available upon reasonable request from the corresponding author, LZ.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1. Carioli G, Negri E, Kawakita D, et al. Global trends in nasopharyngeal cancer mortality since 1970 and predictions for 2020: focus on low-risk areas. Int J Cancer 2017;140:2256–64. 10.1002/ijc.30660 [DOI] [PubMed] [Google Scholar]
- 2. Wei WI, Sham JST. Nasopharyngeal carcinoma. Lancet 2005;365:2041–54. 10.1016/S0140-6736(05)66698-6 [DOI] [PubMed] [Google Scholar]
- 3. Yu MC, Yuan J-M. Epidemiology of nasopharyngeal carcinoma. Semin Cancer Biol 2002;12:421–9. 10.1016/s1044579x02000858 [DOI] [PubMed] [Google Scholar]
- 4. Shanmugaratnam K, Sobin LH. The world Health organization histological classification of tumours of the upper respiratory tract and ear. A commentary on the second edition. Cancer 1993;71:2689–97. [DOI] [PubMed] [Google Scholar]
- 5. Ning S. Innate immune modulation in EBV infection. Herpesviridae 2011;2:1. 10.1186/2042-4280-2-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang L, Huang Y, Hong S, et al. Gemcitabine plus cisplatin versus fluorouracil plus cisplatin in recurrent or metastatic nasopharyngeal carcinoma: a multicentre, randomised, open-label, phase 3 trial. The Lancet 2016;388:1883–92. 10.1016/S0140-6736(16)31388-5 [DOI] [PubMed] [Google Scholar]
- 7. Jin Y, Shi Y-X, Cai X-Y, et al. Comparison of five cisplatin-based regimens frequently used as the first-line protocols in metastatic nasopharyngeal carcinoma. J Cancer Res Clin Oncol 2012;138:1717–25. 10.1007/s00432-012-1219-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chua DTT, Kwong DLW, Sham JST, et al. A phase II study of ifosfamide, 5-fluorouracil and leucovorin in patients with recurrent nasopharyngeal carcinoma previously treated with platinum chemotherapy. Eur J Cancer 2000;36:736–41. 10.1016/S0959-8049(00)00008-3 [DOI] [PubMed] [Google Scholar]
- 9. Fang W, Ma Y, Yin JC, et al. Comprehensive Genomic Profiling Identifies Novel Genetic Predictors of Response to Anti–PD-(L)1 Therapies in Non–Small Cell Lung Cancer. Clin Cancer Res 2019;25:5015–26. 10.1158/1078-0432.CCR-19-0585 [DOI] [PubMed] [Google Scholar]
- 10. Miao D, Margolis CA, Vokes NI, et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet 2018;50:1271–81. 10.1038/s41588-018-0200-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ma BBY, Lim W-T, Goh B-C, et al. Antitumor activity of nivolumab in recurrent and metastatic nasopharyngeal carcinoma: an international, multicenter study of the Mayo clinic phase 2 Consortium (NCI-9742). J Clin Oncol 2018;36:1412–8. 10.1200/JCO.2017.77.0388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fang W, Yang Y, Ma Y, et al. Camrelizumab (SHR-1210) alone or in combination with gemcitabine plus cisplatin for nasopharyngeal carcinoma: results from two single-arm, phase 1 trials. Lancet Oncol 2018;19:1338–50. 10.1016/S1470-2045(18)30495-9 [DOI] [PubMed] [Google Scholar]
- 13. Hsu C, Lee S-H, Ejadi S, et al. Safety and antitumor activity of pembrolizumab in patients with programmed Death-Ligand 1–Positive nasopharyngeal carcinoma: results of the KEYNOTE-028 study. JCO 2017;35:4050–6. 10.1200/JCO.2017.73.3675 [DOI] [PubMed] [Google Scholar]
- 14. Ma Y, Fang W, Zhang Y, et al. A phase I/II open-label study of nivolumab in previously treated advanced or recurrent nasopharyngeal carcinoma and other solid tumors. Oncologist 2019;24:891–e431. 10.1634/theoncologist.2019-0284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hellmann MD, Nathanson T, Rizvi H, et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell 2018;33:843–52. 10.1016/j.ccell.2018.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD-L1–Positive Non–Small-Cell lung cancer. N Engl J Med Overseas Ed 2016;375:1823–33. 10.1056/NEJMoa1606774 [DOI] [PubMed] [Google Scholar]
- 17. Voskoboinik I, Whisstock JC, Trapani JA. Perforin and granzymes: function, dysfunction and human pathology. Nat Rev Immunol 2015;15:388–400. 10.1038/nri3839 [DOI] [PubMed] [Google Scholar]
- 18. Hou Q, Zhao T, Zhang H, et al. Granzyme H induces apoptosis of target tumor cells characterized by DNA fragmentation and Bid-dependent mitochondrial damage. Mol Immunol 2008;45:1044–55. 10.1016/j.molimm.2007.07.032 [DOI] [PubMed] [Google Scholar]
- 19. Larimer BM, Wehrenberg-Klee E, Dubois F, et al. Granzyme B PET imaging as a predictive biomarker of immunotherapy response. Cancer Res 2017;77:2318–27. 10.1158/0008-5472.CAN-16-3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hurkmans DP, Basak EA, Schepers N, et al. Granzyme B is correlated with clinical outcome after PD-1 blockade in patients with stage IV non-small-cell lung cancer. J Immunother Cancer 2020;8:e000586. 10.1136/jitc-2020-000586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 2014;30:2114–20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Picard toolkit . GitHub repository. Broad Institute, 2019. Available: http://broadinstitute.github.io/picard/
- 24. Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43:11.10.1-11.10.33. 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cibulskis K, McKenna A, Fennell T, et al. Contest: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics 2011;27:2601–2. 10.1093/bioinformatics/btr446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213–9. 10.1038/nbt.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fang H, Bergmann EA, Arora K, et al. Indel variant analysis of short-read sequencing data with scalpel. Nat Protoc 2016;11:2529–48. 10.1038/nprot.2016.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Talevich E, Shain AH, Botton T, et al. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol 2016;12:e1004873. 10.1371/journal.pcbi.1004873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Poniah P, Mohd Zain S, Abdul Razack AH, et al. Genome-Wide copy number analysis reveals candidate gene loci that confer susceptibility to high-grade prostate cancer. Urol Oncol 2017;35:545.e1–545.e11. 10.1016/j.urolonc.2017.04.017 [DOI] [PubMed] [Google Scholar]
- 30. Zhang L, MacIsaac KD, Zhou T, et al. Genomic analysis of nasopharyngeal carcinoma reveals TME-Based subtypes. Mol Cancer Res 2017;15:1722–32. 10.1158/1541-7786.MCR-17-0134 [DOI] [PubMed] [Google Scholar]
- 31. Airoldi M, Pedani F, Marchionatti S, et al. Carboplatin plus taxol is an effective third-line regimen in recurrent undifferentiated nasopharyngeal carcinoma. Tumori 2002;88:273–6. 10.1177/030089160208800405 [DOI] [PubMed] [Google Scholar]
- 32. Foo K-F, Tan E-H, Leong S-S, et al. Gemcitabine in metastatic nasopharyngeal carcinoma of the undifferentiated type. Annals of Oncology 2002;13:150–6. 10.1093/annonc/mdf002 [DOI] [PubMed] [Google Scholar]
- 33. Topalian SL, Hodi FS, Brahmer JR, et al. Five-Year survival and correlates among patients with advanced melanoma, renal cell carcinoma, or non-small cell lung cancer treated with nivolumab. JAMA Oncol 2019;5:1411. 10.1001/jamaoncol.2019.2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tumeh PC, Hellmann MD, Hamid O, et al. Liver metastasis and treatment outcome with anti-PD-1 monoclonal antibody in patients with melanoma and NSCLC. Cancer Immunol Res 2017;5:417–24. 10.1158/2326-6066.CIR-16-0325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tang H, Li C, Wang L, et al. Granzyme H of cytotoxic lymphocytes is required for clearance of the hepatitis B virus through cleavage of the hepatitis B virus X protein. J.i. 2012;188:824–31. 10.4049/jimmunol.1102205 [DOI] [PubMed] [Google Scholar]
- 36. Chen Y-P, Yin J-H, Li W-F, et al. Single-Cell transcriptomics reveals regulators underlying immune cell diversity and immune subtypes associated with prognosis in nasopharyngeal carcinoma. Cell Res 2020;30:1024–42. 10.1038/s41422-020-0374-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jin S, Li R, Chen M-Y, et al. Single-Cell transcriptomic analysis defines the interplay between tumor cells, viral infection, and the microenvironment in nasopharyngeal carcinoma. Cell Res 2020;30:950–65. 10.1038/s41422-020-00402-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gao J, Shi LZ, Zhao H, et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 2016;167:397–404. 10.1016/j.cell.2016.08.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou Z, He H, Wang K, et al. Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science 2020;368:eaaz7548. 10.1126/science.aaz7548 [DOI] [PubMed] [Google Scholar]
- 40. Zychlinsky A, Zheng LM, Liu CC, et al. Cytolytic lymphocytes induce both apoptosis and necrosis in target cells. J Immunol 1991;146:393–400. [PubMed] [Google Scholar]
- 41. Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol 2009;7:99–109. 10.1038/nrmicro2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang Z, Zhang Y, Xia S, et al. Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature 2020;579:415–20. 10.1038/s41586-020-2071-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-002014supp002.pdf (20.1KB, pdf)
jitc-2020-002014supp010.pdf (262.1KB, pdf)
jitc-2020-002014supp003.pdf (55.8KB, pdf)
jitc-2020-002014supp004.pdf (84.6KB, pdf)
jitc-2020-002014supp005.pdf (34.9KB, pdf)
jitc-2020-002014supp006.pdf (273.2KB, pdf)
jitc-2020-002014supp007.pdf (54.7KB, pdf)
jitc-2020-002014supp008.pdf (55.7KB, pdf)
jitc-2020-002014supp009.pdf (59.1KB, pdf)
jitc-2020-002014supp001.pdf (118.2KB, pdf)