Abstract

Non-viral delivery systems are generally of low efficiency, which limits their use in gene therapy and editing applications. We previously developed a technology termed glycosaminoglycan (GAG)-binding enhanced transduction (GET) to efficiently deliver a variety of cargos intracellularly; our system employs GAG-binding peptides, which promote cell targeting, and cell penetrating peptides (CPPs), which enhance endocytotic cell internalization. Herein, we describe a further modification by combining gene delivery and magnetic targeting with the GET technology. We associated GET peptides, plasmid (p)DNA, and iron oxide superparamagnetic nanoparticles (MNPs), allowing rapid and targeted GET-mediated uptake by application of static magnetic fields in NIH3T3 cells. This produced effective transfection levels (significantly higher than the control) with seconds to minutes of exposure and localized gene delivery two orders of magnitude higher in targeted over non-targeted cell monolayers using magnetic fields (in 15 min exposure delivering GFP reporter pDNA). More importantly, high cell membrane targeting by GET-DNA and MNP co-complexes and magnetic fields allowed further enhancement to endocytotic uptake, meaning that the nucleic acid cargo was rapidly internalized beyond that of GET complexes alone (GET-DNA). Magnetofection by MNPs combined with GET-mediated delivery allows magnetic field-guided local transfection in vitro and could facilitate focused gene delivery for future regenerative and disease-targeted therapies in vivo.
Keywords: magnetofection, magnetic nanoparticles (MNPs), GAG-binding enhanced transduction (GET), cell penetrating peptide (CPP), magnetic targeting
1. Introduction
Non-viral vectors engineered as nanoparticles or complexes are an attractive gene delivery method due to their safety, high gene carrying capacity, and scalable mass production.1,2 Unfortunately, non-viral vectors are in general much less efficient compared to viral alternatives. This has been attributed, among other reasons, to their inability to overcome extra- and intracellular barriers and to poor targeting of the cell membrane for subsequent endocytosis-mediated internalization.
In order for a delivery vector to efficiently transfect transgenes either in vitro or in vivo, nanoparticle formulations must first come into contact with and bind the cell membrane, rapidly enter the cell, in the case of endosomal entrapment avoid lysosomal and cytosolic degradation, and for plasmid (p)DNA-based therapeutics enter the cell nucleus to be transcribed.3 There are ongoing efforts to design non-viral vectors capable of efficiently overcoming these limitations.4 Slow vector accumulation and therefore low pDNA concentration on the cell membrane are a major barrier for most gene delivery methods; therefore, any approach capable of accelerating the pDNA-vector interaction with target cells could be hypothesized to result in enhanced gene delivery and transgene expression.5,6 Furthermore, a method to promote vector accumulation that could be remotely controlled and localized would be the most desirable. All these requirements have led to a relatively new technology termed magnetofection.7 This acronym (first mentioned in 20007) loosely refers to any magnetically guided or enhanced nucleic acid delivery, the most common approach involving the association of vectors (viral and non-viral) with magnetic carriers, such as magnetic nanoparticles (MNPs). These accumulate on the cells by the application of magnetic field gradients. In the past years, magnetofection has shown very promising results both in vivo and in vitro.8−13 Additionally, magnetofection provides the opportunity not only to enhance targeted nucleic acid delivery in vivo, but it can also facilitate cell targeting of nanoparticle formulations to the area of interest in the context of cell therapy through MRI focusing or static magnets.14−16
One of the most common non-viral vectors used for magnetofection is polyethylenimine (PEI).17−22 PEI is a branched polymer containing primary, secondary, and tertiary amines capable of complexing pDNA and delivering it in vitro and in vivo. However, transfection efficiency of PEI as well as other non-viral vectors remains low compared to their viral counterparts, and improvement in the field is still slow and limited. Additionally, despite being the current gold standard, PEI presents certain disadvantages such as elevated toxicity and lack of consistency and reproducibility in terms of transfection efficiency.23
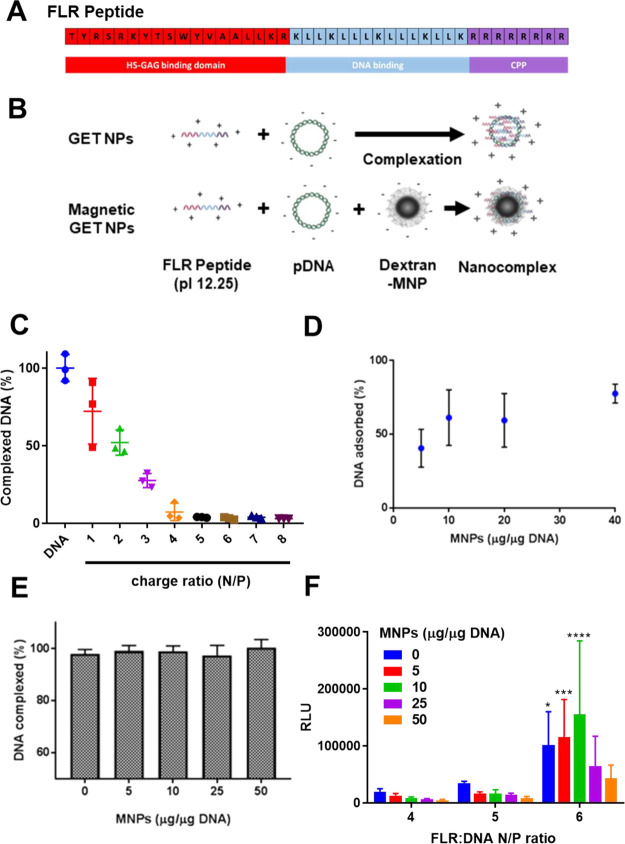
Previous work by our group has developed the glycosaminoglycan (GAG)-binding enhanced transduction (GET) system based on combining the activities of peptide-cell membrane interaction with GAGs and cell penetrating peptides (CPPs). A second-generation modified GET peptide, FGF2B-LK15-8R (FLR), has shown efficient gene delivery in vitro and in vivo with superior transfection efficiencies generating nanoparticles of GET-pDNA to current gold standard branched polymers or PEI.24 We have exploited this technology in vivo for bone repair25 and lung gene delivery,24 both based on delivery of transgenes expressed from pDNA. However, the system can also transfect mRNA and oligonucleotides, making it a generic vector for nucleic acid-nanoparticle-based delivery technologies.26 The FGF2B-LK15-8R (FLR) peptide is formed of three domains: a fibroblast growth factor 2 (FGF2B) heparin-binding domain (TYRSRKYTSWYVALKR) with high affinity for heparan sulfate proteoglycans present on the cell surface, which acts as a membrane docking domain;27 LK15 (KLLKLLLKLLLKLLK), an amphipathic sequence able to complex DNA with endosomal escape activity;28 and a cell penetrating peptide (CPP) 8R (RRRRRRRR), which further enhances endocytosis26 (Figure 1A). FLR-pDNA nanocomplexes rapidly bind to cell membranes and are internalized; however, they cannot be physically focused to transfect specific cells in vitro or tissues in vivo, with the duration of cell exposure dictating absolute and local levels of gene transfection activity.24,29 In this study, we focused on developing an FLR-DNA-MNP co-formulation complex for efficient magnetically mediated gene delivery of pDNA. Understanding the advantages and limitations of magnetofection is key for the development of effective delivery systems, and therefore, we characterized complex binding, uptake, and transfection activities dynamically under static magnetic fields. The cellular entry mechanism of FLR-DNA-MNP nanocomplexes in the presence or absence of a magnetic field was determined through the inhibition of specific uptake pathways. The predominant mechanism of uptake was dictated by the presence of the magnetic field, with caveolae-mediated endocytosis playing a more dominant role under magnetic fields. Uptake kinetics, endosomolysis, extra- and intracellular pDNA degradation, and confocal microscopy were also utilized to assess the combined effect of magnetic and GET-mediated gene delivery. We were able to demonstrate exceptional levels of faithfully localized gene expression with combination of our nanocomplex systems, which enabled more rapid (almost instantaneous, 5 s) cell membrane binding and subsequent uptake of FLR-DNA-MNPs when targeted with magnets in NIH3T3 cells. Use of such technologies will allow focused gene delivery to be translated for next-generation regenerative and disease-targeting augmentation and editing approaches. Rapid and effective gene delivery systems such as GET magnetofection clearly demonstrate the significance of magnetic field application in the future of drug delivery using nanocomplex formulations.
Figure 1.
GET magnetofection complex formulation. (A) FLR is formed of a heparan sulfate glycosaminoglycan binding domain, composed of the FGF2 heparin binding domain B (red), an amphipathic region termed LK15 (blue), and a cell penetrating peptide (CPP), 8R (purple). (B) Nanoparticle complexation. The positively charged residues (N) in the FLR peptide sequence interact electrostatically with the negatively charged (P) phosphate groups of pDNA or dextran-coated MNP forming nanocomplexes. (C) Percentage of YO-PRO-1-DNA fluorescence at increasing peptide ratios (N/P). The graph represents mean ± s.d. (n = 3 technical replicates). (D) Rhodamine (Rh)-pDNA conjugated with FLR (N/P of 6) was incubated with MNPs (5, 10, 20, and 40 μg MNPs/1 μg DNA) in water. Percentage of DNA absorbed was calculated relative to the total amount of DNA. Dots represent mean percentage of DNA adsorbed ± s.d. (n = 9 technical repeats). (E) Percentage of pDNA complexed by FLR at increasing concentrations of MNPs (5, 10, 25, and 50 μg MNPs/1 μg DNA). YO-PRO-DNA complexed with FLR (without MNPs) taken as 100% complexation. Bars represent mean complexed DNA ± s.d. (n = 3 technical repeats). (F) Gaussia luciferase expression in NIH3T3 cells after transfection with FLR-DNA-MNPs at N/P of 4, 5, and 6 at increasing concentrations of MNPs (5, 10, 25, and 50 μg of MNPs per μg of DNA). Cells treated with FLR-DNA only at N/P of 4, 5, and 6 were used as controls (0 μg MNPs/μg pDNA). Bars represent relative light units, RLU ± s.d. (*p < 0.05, ***p < 0.001, and ****p < 0.0001, significance between treatments at an N/P ratio of 6 compared with the same treatment at N/P of 4 and 5. Tukey’s multiple comparison test, n = 3 biological replicates).
2. Materials and Methods
2.1. Physicochemical Analysis
Nanomag-D MNPs (Fe3O4 core; 250 nm; 09-02-252) were purchased from MicroMod (Germany). The size and zeta potential of the bare, FLR, or FLR-DNA-functionalized MNPs were measured in water (distilled H2O) using a Malvern Nanosizer Nano ZS.
2.1.1. Dynamic Light Scattering (DLS)
Measurements consisted of 3 repeats (12–15 sub-runs per repeat) of the same sample to estimate the error in the measurements. Measurements were recorded at room temperature.
2.1.2. Zeta Potential
Measurements consisted of 3 repeats (12–15 sub-runs per repeat) of the same sample to estimate the error in the measurements. The measurements were recorded at room temperature. As zeta potential measurement was performed in an aqueous solution, the Smoluchowski approximation was used to calculate the zeta potentials from the measured electrophoretic motilities.
2.2. Cell Culture
Unless otherwise specified, NIH3T3 cells were used in this study. NIH3T3 cells were chosen as a model to validate and characterize the FLR-DNA-MNP technology because of their consistency and robustness. All cell lines were cultured at 37 °C in 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Sigma), supplemented with 10% (v/v) fetal calf serum (FCS, Sigma), 4.5 g/L d-glucose, 2 mM l-glutamine, and 100 units/mL penicillin and 100 units/mL streptomycin (Invitrogen). All methods and reagents unless specified were detailed previously.24−26,29−31
2.3. Plasmids and Purification
The Gaussia luciferase reporter (GLuc) was expressed by delivery of the pCMV-GLuc2 (termed pGLuc) DNA (expresses secreted luciferase from the copepod Gaussia princeps under the control of a cytomegalovirus (CMV) promoter) (New England Biolabs; NEB). The enhanced green fluorescent protein (GFP) was expressed by delivery of the pEGFP-C1 pDNA (expresses enhanced GFP under the control of a CMV promoter) (Takada). pDNA was propagated in DH5α competent Escherichia coli and transformants selected for antibiotic resistance on LB agar plates. Individual colonies were picked and expanded to maxiprep volume with LB broth (with 100 μg/mL ampicillin or 50 μg/mL kanamycin). Bacterial pellets were purified using a Qiagen Plasmid Purification Maxi kit, following the manufacturer’s protocol. pDNA was diluted in nuclease free water (Sigma). Final pDNA concentration and purity were measured by a Nanodrop (NanoDrop ND-1000, Labtech International). pDNA was aliquoted and stored at −20 °C.
2.4. Plasmid Labeling
pGLuc DNA was labeled at a 1:1 ratio (v/w) of a Label IT CX Rhodamine reagent to nucleic acid according to manufacturer’s specifications (Mirus). Briefly, 5 μL of 10X Labeling Buffer A was mixed with 5 μL of 1 mg/mL pDNA and 5 μL of a Label IT CX Rhodamine Reagent in 35 μL of nuclease (DNase, RNase)-free water. The mix was incubated for 1 h at 37 °C. Labeled pDNA was purified using a G50 MicroSpin purification column. Labeled pDNA (Rh-pDNA) was stored protected from light at −20 °C. Unless otherwise specified, for in vitro studies, the following formulation was used: 1:3 of labeled pDNA diluted with unlabeled pDNA (w/w).
2.5. DNA Complexation Assays
In order to assess the interaction of FLR and pDNA, we used YO-PRO-1 iodide assays (Thermo Fisher Scientific). Briefly, for each individual repeat, 1 μg of pDNA was diluted in 6 μL of 10 mM HEPES buffer (pH 7.4). Similarly, 0.03 μL of YO-PRO-1 (1 mM) was diluted in 6 μL of the same buffer. The pDNA solution was added dropwise to the YO-PRO-1 solution and incubated for 5 h at room temperature protected from light. These quantities were scaled up proportional to the number of repeats per experiment, making up one stock solution of YO-PRO-1-DNA. The YO-PRO-1-DNA solution was diluted to a final concentration of 10 μg/mL in 10 mM HEPES buffer (pH 7.4). An increasing amount of peptides corresponding to the desired charge ratio between amine (NH2+ ) groups in the peptide and phosphate (PO3–) groups in pDNA (N/P) was added followed by mixing and further incubation for 10 min (Table 1). Fluorescence intensity was measured at ex/em 480/509 nm (Infinite 200 PRO, Tecan). Experiments were performed in triplicate, and results are expressed as percentage of fluorescence of YO-PRO-1-DNA against the charge ratio (N/P). YO-PRO-1 only was used as a blank.
Table 1. Formulation of FLR-DNA Nanoparticlesa.
| FLR/DNA ratio (N/P) | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| FLR (nmol) | 0.17 | 0.34 | 0.51 | 0.68 | 0.85 | 1.02 | 1.20 | 1.37 | 1.54 | 1.71 |
The amount of FLR peptides needed to form a peptide-DNA complex at the indicated charge ratio (N/P) for 1 μg of pDNA of approximately 5800 bp. The N/P charge ratio was calculated based on the number of positively charged amine groups of the FLR molecule (+18).
2.6. Magnetofection and Transfection
Cells (4.2 × 105 NIH3T3 cells/cm2) were seeded on a 48-well plate format (unless otherwise specified) 24 h before the treatment. Prior to transfection, the medium in the wells was replaced. FLR-DNA-MNPs were formulated as described (Table 2). For magnetofection, cells were placed on top of individual magnets arranged in an array, and the plate was fixed for the exposure duration (magnet array schematic, Figure S1). After transfection/magnetofection, cells were washed three times with PBS or heparin (first wash with 100 μg/mL Heparin in PBS then two washes with PBS). PBS was replaced with growth media followed by further 24 h incubation at 37 °C in 5% CO2. Neodymium magnets, N52 10 mm diameter and 5 mm thickness (3.2 kg pull, F645-N52-10), and N42 20 mm diameter 5 mm thickness (7.3 kg pull, F205-4) were used for magnetofection of 48 wells and targeting a specific region in 6-well plates, respectively (First for Magnets, UK).
Table 2. Formulation of FLR-DNA-MNP Nanoparticles for In Vitro Magnetofectiona.
| MNPs (μg/μg DNA) | DNA (μg) | FLR-DNAb incubation time (min) | OptiMEM (μL)c | MNPs (μL of 10 mg/mL stock)d | FLR/DNA + MNP incubation time (min) | media per well (μL) | total transfection volume (media + OptiMEM) (μL) |
|---|---|---|---|---|---|---|---|
| 5 | 0.5 | 15 | 2 × 12.5 | 0.25 | 15 | 100 | 125 |
| 10 | 0.5 | 15 | 2 × 12.5 | 0.5 | 15 | 100 | 125 |
| 25 | 0.5 | 15 | 2 × 12.5 | 1.25 | 15 | 100 | 125 |
| 50 | 0.5 | 15 | 2 × 12.5 | 2.5 | 15 | 100 | 125 |
FLR-DNA-MNP complexation for 0.5 μg of DNA per transfection on a 48-well plate format. Scalable to other well plate formats.
FLR was diluted to a total volume of 12.5 μL in OptiMEM. DNA was diluted to a total volume of 12.5 μL in OptiMEM. DNA solution was added to FLR solution and mixed thoroughly to facilitate particle formation.
An MNP volume was added straight into the FLR-DNA solution and mixed thoroughly.
2.7. Cell Viability Assays
Twenty-four hours after treatment, cells were trypsinized and diluted in trypan blue (1:1) for cell counting. Percentage of cell viability was calculated based on the total number of viable cells for each group compared to the untreated control. Half of the trypsinized cells were plated again. Proliferation was measured as the cumulative number of viable cells every 24 h for 7 days.
2.8. Reporter Gene Expression
2.8.1. Luciferase Activity
Gaussia luciferase expression was measured using a BioLux Gaussia Luciferase Assay Kit (New England Labs, UK). Briefly, 10 μL of the medium was collected from each transfection well and added onto a white 96-well plate (Corning, UK). Gaussia luciferase (GLuc) assay solution (50 μL) was added to each well (GLuc assay solution) (1:100 dilution substrate into assay buffer). Luminescence was measured using a luminometer (Infinite 200 PRO, Tecan), with integration time of 500 ms. Untransfected and no pDNA GET-MNPs were employed in each experiment as negative transfection controls, and the basic GET-pDNA system26 (FLR-DNA) was used as a positive control.
2.8.2. GFP Fluorescence
GFP-expressing cells were imaged by fluorescence (Leica DM IRB). GFP transfection efficiency (% positivity) and expression intensity were quantified by flow cytometry. Total events (50,000–100,000) were recorded per sample (Astrios EQ sorter, Beckman Coulter, US). Untreated cells were used as a control. For targeting within a single culture, cells were plated as a contiguous monolayer within wells of a 6-well plate (34.8 mm diameter, 9.5 cm2 culture area) containing a sterile coverslip (borosilicate glass, 20 mm diameter, and 3.1 cm2 culture area). Coverslips were affixed to the center of the culture surface with sterile vacuum grease, allowing them to be readily removed with forceps after seeding, exposure, and washing. On transfection, targeting to the coverslip was achieved by placing the well on the array as previously described, a 20 mm diameter magnet aligning with the coverslip. After the incubation, the array was removed, and cells were washed as described before with PBS or heparin. The coverslip was removed to a fresh well with forceps, and targeted (IN region, 3.1 cm2) and untargeted (OUT region, 6.4 cm2) cells were incubated as before analysis.
2.9. Cellular Uptake Inhibition
NIH3T3 cells were exposed to one of the following conditions for 30 min prior to transfection: (1) incubated at 4 °C (as opposed to 37 °C) or (2) addition of 0.45 M sucrose (Sigma, S9378),21,32 (3) 100 μM 5(N-ethyl-N-isopropyl) amiloride (EIPA) (Sigma, A3085),33 and (4) 5 mM methyl-B-cyclodextrin (MBCD) (Sigma, C4555)34,35 in growth medium. Cells were transfected with FLR-DNA or FLR-DNA-MNPs (formulated with Rh-pDNA) and incubated for 1 h with or without exposure to a magnetic field. Transfection was carried out at 4 °C for inhibition at low temperature. All other transfections were carried out at 37 °C. The control group was transfected at 37 °C in growth medium without inhibitors. After one hour, cells were washed with PBS or heparin (100 μg/mL). Red fluorescence in the cells was quantified by flow cytometry. Each sample was run individually through a flow cytometer; 50,000–100,000 total events were recorded per sample (Astrios EQ sorter, Beckman Coulter, US). Untreated cells were used as a control.
2.10. Cell Extraction of Transfected pDNA
After transfection/magnetofection, cells were washed with PBS. Cells were then incubated in growth medium at 37 °C, 5% CO2 until collection, immediately after transfection (0 min), 10 min, 25 min, 55 min, and 24 h (1440 min) post transfection. Briefly, cells were trypsinized for 3 min at 37 °C, 5% CO2 after which the trypsin was neutralized with a pre-warmed medium. Cells were pelleted and resuspended in 50 μL of Hirt buffer (10 mM EDTA, pH 7.5, and 0.6% SDS). This was incubated at 4 °C for 8 h after which it was stored at −20 °C until DNA extraction/purification. For pDNA purification, a QIAprep Spin Miniprep kit was used according to manufacturer’s specifications (QIAGEN, Cat: 27106) with the extract added to 500 μL of PB buffer. DH5α competent E. coli (40 μL) was added to purified DNA (2.5 μL) and incubated for 30 min on ice. Cells were then heat-shocked for 45 s in a 42 °C water bath and replaced with ice for 5 min. Cells were then incubated in a pre-warmed SOC recovery medium for 1 h at 37 °C, shaking at 225 rpm. After incubation, 50 μL of each transformation was spread on a selective LB agar plate (100 μg/mL ampicillin). Plates were then incubated overnight at 37 °C. Colonies were counted the following day and compared to an extraction control (pDNA quantity transfected, added to a cell pellet, and extracted).
2.11. Confocal Imaging
NIH3T3 cells were seeded on sterilized glass coverslips (borosilicate glass, 13 mm diameter, VWR). Cells were transfected with FLR-DNA or FLR-DNA-MNPs (formulated with Rh-pDNA) as previously described. After 30 min, 1 h, and 24 h of incubation, cells were fixed in 3.7% paraformaldehyde and permeabilized using Triton X-100 for 15 min then washed in PBS. The actin cytoskeleton was visualized by staining with Alexa Fluor 488 Phalloidin (Thermo, A12379). The coverslips were washed and sealed onto slides with a DAPI-containing Fluoroshield mounting medium (Sigma Aldrich, UK). Cells were imaged using a LSM880C confocal microscope (Zeiss, Germany). A 63x immersion objective lens was used with a 488 nm laser used for the Hoechst- and Phalloidin-stained cytoskeleton and a 561 nm diode-pumped solid-state (DPSS) laser for Rhodamine-labeled (Rh-) pDNA. Images were captured using ZEN software (Zeiss, Germany). Three-dimensional image stacks were recorded by sequential acquisition of optical sections along the z-axis with steps of 0.33–0.37 μm. The acquired digital images were merged and processed by using ImageJ version 4.
2.12. Iron Quantification by Inductively Coupled Plasma Mass Spectrophotometry (ICP-MS)
MNPs were delivered as described above. After incubation overnight, the supernatant was removed, and cells were washed twice with PBS. Cells were trypsinized and lysed in 6 M HCl and HNO3 (65%) for 2 h at room temperature for the degradation of the particles in order to release the Fe content. Samples were then diluted in water in order to achieve a final acid concentration of less than 2% (w/v). A calibration curve was also produced at MNP concentrations up to 50 μg/mL, to account for possible matrix effects. Diluted solutions were analyzed by ICP-MS (Thermo Fisher Scientific iCAP-Q; Thermo Fisher Scientific, Bremen, Germany). Elemental analysis of diluted solutions was undertaken by ICP-MS (Thermo Fisher Scientific iCAP-Q and iCAP-TQ; Thermo Fisher Scientific, Bremen, Germany). Samples were introduced (flow rate of 1.2 mL/min) from an autosampler (Cetac ASX-520) incorporating an ASXpress rapid uptake module through a perfluoroalkoxy Microflow PFA-ST nebulizer (Thermo Fisher Scientific, Bremen, Germany). Sample processing was undertaken using Qtegra software (Thermo Fisher Scientific) utilizing external cross-calibration between pulse counting and analogue detector modes when required. Internal standards were introduced to the sample stream on a separate line via the ASXpress unit and included Ge (10 μg/L), Rh (10 μg/L), and Ir (5 μg/L) in 2% trace analysis grade (Fisher Scientific, UK) HNO3. An Fe external calibration standard (Claritas-PPT grade CLMS-2 from SPEX Certiprep, Inc., Metuchen, NJ, USA), in the range of 0–100 μg/L (0, 20, 40, and 100 μg/L), was employed, with phosphorus, boron, and sulfur calibration by in-house standard solutions (KH2PO4, K2SO4, and H3BO3). A collision cell (Q cell) using He with kinetic energy discrimination (He-cell) to remove polyatomic interferences was used to measure Fe. Sample processing was undertaken using Qtegra software (Thermo Fisher Scientific). Results were reported back in ppb (μg/L). Iron association per cell was calculated based on the doubling times of the respective cell lines to estimate the total number of cells.
2.13. Hemolysis
To assess the hemolytic activity of the FLR-DNA-MNP complex (as a proxy for endosomal membrane rupturing activity), freshly isolated human erythrocytes (6 × 106 erythrocytes/mL) were exposed to 0.5 μM FLR. FLR-DNA was formulated at an N/P ratio 6. MNPs (5 μg/μg DNA) were added to form the FLR-DNA-MNPs. Hemolysis experiments were performed in PBS for 30 min at physiological pH (pH 7.5) and late endosome pH (pH 5). After incubation, samples were centrifuged at 5000g for 5 min. The supernatant was collected, and hemoglobin content was analyzed by spectrophotometry at 544 nm (Infinite 200 PRO, TECAN). PBS was used as a control. Results are expressed as percentage lysis taking Triton X-100 as complete lysis (100%).
2.14. Statistical Analysis
For in vitro studies, n represents the number of biological replicates. Technical replicates refer to experiments carried out with different passage cells but identical experimental conditions. Data were presented as mean ± standard deviation (s.d.) and analyzed by Prism statistical analysis software (GraphPad v. 7.03).
3. Results & Discussion
3.1. Co-Complexation FLR Peptides, pDNA, and MNPs for GET Magnetofection
In order to develop a targetable GET peptide transfection system, we generated an FLR-MNP-based gene delivery nanocomplex following a step by step formulation process6 (Figure 1B). The optimal amount of FLR needed to fully complex DNA was initially confirmed using a YO-PRO-1 dye fluorescence-based assay for DNA complexation. Briefly, YO-PRO-1 (carbocyanine) becomes fluorescent through DNA binding via its positive side chain. The amount of fluorescence emitted is proportional to the amount of free non-complexed DNA. When DNA is complexed or interacts with other molecules such as cationic FLR peptides, the YO-PRO-1-DNA interaction becomes unstable, quenching fluorescence proportionally to the amount of DNA complexed. We exploited this assay to study optimal negative/positive (N/P) ratio for DNA complexation.24 Increasing concentrations of FLR were added to defined amounts of YO-PRO-1-labeled pDNA, confirming that as FLR concentration increased, YO-PRO-1-DNA fluorescence decreased, indicating direct interaction of FLR with pDNA (Figure 1C). Total pDNA complexation occurred at N/P ratios of 4 (7.4 ± 5.6% of fluorescence left) or above. We therefore focused on formulations containing a minimum FLR concentration to allow full complexation of pDNA, those having the N/P ratio of 4 or more.
Dextran-coated MNPs are approved by the Food and Drug Administration (FDA) for their use in vivo and are currently being optimized for multiple applications in biomedicine.16,36−38 We have previously shown that positive GET peptides complex to the negatively charged dextran coating of Nanomag-D MNPs (MicroMod) and enhance the cell uptake of the MNPs.26,37,39 In our previous studies,39 we focused on the characterization of the complexation of GET peptides and MNPs, establishing the minimum amount of GET peptides required in order to enhance cellular uptake of MNPs, as well as the concentration of GET at which MNPs would be saturated. We found that the optimal concentration of GET for MNP delivery (4 nmol GET per mg MNPs) was significantly inferior to the saturation maximum (40 nmol for GET per mg of MNPs). It was therefore key to assess the co-complexation of all three components and the amounts of FLR needed to both interact with MNPs and fully complex pDNA before testing the transfection ability of the nanocomplex formulations.
To confirm if pDNA can indeed be incorporated into FLR-MNP nanocomplexes, rhodamine-labeled pDNA (Rh-pDNA) was used for complexation at increasing concentrations of MNPs and at an N/P ratio of 6. After complex assembly, MNPs were separated using a static magnetic field, and unbound Rh-pDNA was measured in the supernatant using fluorimetry to define percentage absorption. As the concentration of MNPs increased, the percentage of DNA adsorbed onto the particles increased progressively toward a plateau, suggesting an adsorption mechanism of the FLR-Rh-pDNA nanoparticles onto the MNP surface (Figure 1D).
In order to assess whether the pDNA associated with FLR-MNP complexes remained directly associated with FLR after incorporation, we again exploited the YO-PRO-1 assay. YO-PRO1-labeled pDNA was incubated with FLR at an N/P ratio of 6 and then with increasing amounts of MNPs (5, 10, 25, and 50 μg MNPs/1 μg pDNA) (Figure 1E). The percentage of complexed pDNA was calculated as a function of the loss in fluorescence in solution compared to YO-PRO-1-labeled pDNA only controls. There is no significant difference in pDNA complexation in the presence of any concentration of MNPs tested. This data indicates that binding of the FLR-pDNA complexes to the MNPs does not disturb FLR-DNA interactions or at least not sufficiently to dequench YO-PRO-1 and allow its productive binding to pDNA.
3.2. Defined Monodispersed FLR-DNA-MNP Nanocomplexes
After confirmation that FLR peptides, pDNA, and MNPs can form co-complexes, we assessed their physical characteristics, using dynamic light scattering (DLS) and zeta potential analyses (Table 3). As previously confirmed, MNPs alone are negatively charged due to their dextran coating. In the presence of FLR, all MNP nanocomplexes are positively charged, indicating the disposition of FLR on the outer layer of MNPs. In contrast, when MNPs were incubated with pDNA only, particle charge became significantly more negative compared to MNPs alone (−31.6 ± 0.8 and −20.7 ± 0.5 mV, respectively), suggesting that pDNA even though negatively charged can interact with MNPs. Particle size measurement by DLS suggests that MNP complexes are mostly in the monodisperse range after functionalization with FLR and DNA-FLR (Table 3).40 FLR-DNA-MNPs (244.7 ± 8.5 nm) are larger than MNPs alone and FLR-MNPs (225.1 ± 4.4 and 228 ± 4.6 nm, respectively) and comparable to MNPs-DNA (239.1 ± 3.9 nm). This ∼20 nm increase in diameter could be attributed to pDNA absorption to the MNPs. FLR-DNA complexes are 124.6 ± 2.9 nm, suggesting that the size of FLR-DNA-MNPs did not represent the coalescing of these sized FLR-DNA complexes with MNPs. Based on these findings and the previously reported adsorption of pDNA onto the MNPs as well as the stable complexation of pDNA throughout the adsorption process, we hypothesize that upon encountering MNPs, FLR-DNA nanocomplexes interact with surface functional groups of MNPs and rearrange seeking the most stable conformation.41 The positive zeta potential of FLR-DNA-MNP complexes suggests that positively charged FLR is arranged in the outer layer of the particle shielding the negative charge provided by absorbed pDNA and the MNP dextran coating. We therefore confirmed the successful formulation of FLR-DNA-MNP nanocomplexes for further testing.
Table 3. Physical Characterization of GET-Magnetofection Complex Vectors for pDNA Deliverya.
| DH (nm)b | PDIc | zeta potential (mV) | |
|---|---|---|---|
| MNPs | 225.1 ± 4.4 | 0.18 ± 0.03 | –20.7 ± 0.5 |
| FLR-DNA | 124.6 ± 2.9 | 0.24 ± 0.002 | 49.8 ± 1.1 |
| MNPs-FLR | 228.0 ± 4.7 | 0.16 ± 0.02 | 35.3 ± 0.8 |
| MNPs-DNA | 239.1 ± 3.9 | 0.24 ± 0.01 | –31.6 ± 0.8 |
| FLR-DNA-MNPs | 244.7 ± 8.5 | 0.21 ± 0.01 | 34 ± 1 |
The size (DH) and zeta potential of bare MNPs, FLR-DNA complex, FLR-functionalized MNPs, DNA-functionalized MNPs, and MNPs functionalized with the FLR-DNA complex. Measurements were performed in distilled water (dH2O) using a Malvern Nanosizer Nano ZS. Values represent mean ± s.d.
Z-average hydrodynamic diameter extracted by cumulant analysis of the data.
The polydispersity index (PDI) from cumulant analysis.
3.3. FLR-DNA-MNP Nanocomplexes Retain High Transfection Activity
We next assessed if the inclusion of MNPs in FLR-DNA nanocomplexes was compatible with cell uptake and delivery. We undertook a series of reporter transfection experiments in NIH3T3 cells transfected with a secreted Gaussia luciferase (GLuc)-encoding plasmid (pCMV-GLuc2; termed pGLuc) comparing complexes with and without MNP inclusion. We exposed cells for 24 h using an FLR-DNA formulation at N/P of 4, 5, and 6 and increasing concentrations of co-complexation MNPs (5, 10, 25, and 50 μg MNPs/μg pDNA).
Cells transfected with FLR-DNA nanocomplexes alone at N/P ratios of 4, 5, and 6 were used as controls (0 μg MNPs/μgDNA) and to confirm any inhibitory effect of MNPs on transfection. Gene transfer efficiency was measured by GLuc protein expression secreted into the media by luminometry (Figure 1F). An N/P ratio of 6 showed significantly enhanced protein expression overall when compared with lower ratios. There were no significant differences in transfection efficiency between the MNP-free control (0 μg MNPs/μg of DNA) and the highest doses of MNPs (25 and 50 μg MNPs/μg of DNA) when comparing transfection at the same N/P, suggesting that transfection efficiency was not significantly affected with the inclusion of MNPs, even at the highest amounts tested. MNPs are therefore compatible with cell uptake and transfection activity. These findings further corroborate our previous hypothesis that FLR-pDNA nanocomplexes are not destabilized upon adsorption onto the MNPs (Figure 1E). The GET peptide system not only allows coupling and condensation of the pDNA cargo with MNPs but also enhances transfection efficiency. We have extensively shown that without the DNA cargo, GET-conjugated nanocomplexes have significantly improved the speed and loading of MNPs into cells,26 which also, we now show for the co-delivery of pDNA in the GET-MNP nanocomplex.
3.4. GET Magnetofection Significantly Enhances Transfection Speed
In order to determine the effect of a magnetic field on FLR-DNA-MNP co-complex gene delivery, we assessed for overall transfection efficiency and transfection speed in the presence or absence of a magnetic field.19,22,42,43
Previous data suggested that 5 and 10 μg MNPs/μg DNA were the most efficient for magnetofection at an N/P of 6. Optimal concentration of MNPs for magnetofection during short incubation periods (1 h) was established at 5 μg MNPs per μg pDNA (Figure S2). Cells were transfected with pGLuc for short or longer durations (1 or 24 h, respectively) with and without an external magnetic field (Figure 2A) using MNPs-DNA and FLR-DNA nanocomplexes as controls. FLR-DNA-MNP nanocomplex transfection was significantly enhanced (almost 2-fold) by the presence of a magnetic field during 1 h transfection (2.09 ± 0.45 × 107 RLU with a magnet compared to 1.13 ± 0.6 × 107 RLU without a magnet). Interestingly, in one hour, FLR-DNA-MNPs in the absence of an external magnetic field induced similar levels of transfection as FLR-DNA (0.80 ± 0.76 × 107 RLU), once again suggesting that MNPs did not hamper the gene transfer process. After a 24 h exposure, cells transfected with both FLR-DNA and FLR-DNA-MNP nanocomplexes showed comparable levels of protein expression independent of the magnetic field. Transgene expression after 24 h was comparable to that of FLR-DNA-MNPs in 1 h under a magnetic field.
Figure 2.
GET magnetofection allows rapid transfection without cytotoxicity. (A) Gaussia luciferase expression in NIH3T3 cells after 1 h or 24 h delivery. pDNA (pGLuc) was delivered with MNPs, FLR, and FLR-MNPs in the presence or absence of a magnet. For all formulations, 0.5 μg of DNA was delivered. MNP complexes were formulated at 5 μg MNPs/1 μg of DNA. The FLR-DNA ratio was constant at an N/P of 6 (n = 4 biological replicates, **p < 0.01, comparison between transfection at 1 h and 24 h, and Sidak’s multiple comparison test; $$p < 0.01, comparison between transfection at 1 h, and Tukey’s multiple comparison test). (B) Cumulative life cell number count and (C) cell viability after incubation with FLR-based complexes. NIH3T3 cells were treated with FLR-DNA and FLR-DNA-MNPs with and without the presence of an external magnetic field (magnet) for 1 h. Values represent mean percentage of cell viability ± s.d. (n = 3 biological replicates).
These data confirm that the FLR-DNA-MNP co-complex formulation could induce comparable protein expression to its non-magnetic counterpart FLR-DNA in the absence of a magnetic field. Importantly, under the influence of a magnetic field, FLR-DNA-MNP nanocomplexes were able to achieve maximal transfection expression after only 1 h magnetofection.
3.5. GET Magnetofection Does Not Affect Cell Viability
The effect of magnetofection on viability and growth of NIH3T3 cells was assessed using a trypan blue-based assay (Figure 2B and C, respectively) and metabolic assays (PrestoBlue, data not shown). Cells were transfected for 1 h with pGLuc incorporated within FLR-DNA and FLR-DNA-MNP nanocomplexes with or without an external magnetic field. We tested 1 h magnetofection as this generated the same transfection levels of overnight exposure with FLR-DNA complexes. Cell proliferation and viability were assessed 24 h post-transfection (day 1) and every 24 h for 7 days. There were no significant differences in cell viability or proliferation across all treatment groups compared to untreated controls as determined by the trypan blue dye exclusion assay (Figure 2C) and metabolic assessments (data not shown). Metabolic assessment post-delivery (immediately and after 24 h) showed that there was no statistically significant difference after uptake, with or without magnetic targeting. These data indicate that regardless of the rapid accumulation of nanocomplexes on the cell membranes and enhanced uptake mediated by magnetofection, the doses of pDNA, FLR peptides, and MNPs were fully cytocompatible.
We compared the magnetically targeted GET-MNP system, with or without magnets to PEI.44 PEI was slow to transfect and yielded ∼3-fold lower transfection levels in DC2.4 (dendritic cells) and ∼1.5-fold lower in HeLa cells (data not shown). GET-MNP transfection with 30 min magnetic targeting yielded higher levels of reporter expression than full transfection exposure (overnight) of the PEI-based systems (data not shown). Furthermore, there was no statistically significant effect on viability (trypan blue dye exclusion) or metabolism (PrestoBlue) with any of the GET-MNP variables, but PEI even at short exposure (>1 h) showed some effect on viability and metabolism, with full exposure showing a ∼10% increase in dead cells (trypan blue) and ∼20% reduction in cell metabolism (PrestoBlue) (data not shown).
3.6. Significant GET Magnetofection with 5 s Exposure
As we have previously shown, FLR-DNA nanocomplexes transfect cells rapidly in comparison to some other systems.24 As short 1 h exposure still generated significant transfection irrelevant of magnetic targeting, we repeated experiments with ever shorter incubation times with and without a magnetic field. Our goal was to gain further understanding of transfection kinetics mediated by FLR-DNA and FLR-DNA-MNP nanocomplexes with and without an external magnetic field. NIH3T3 cells were transfected for increasing amounts of time (from 5 to 60 min). We assessed transfection kinetics by reporter gene expression (GLuc); FLR-DNA-MNP nanocomplexes delivered under an external magnetic field were able to generate significant levels of reporter gene expression after just 5 min (1.13 ± 0.27 × 107 RLU), which was comparable to gene expression mediated by FLR-DNA or FLR-DNA-MNPs after 1 h incubation (Figure 3A). Importantly, we also conducted shortened exposure times to assess how effective partial targeting was on transgene expression. Our shortest exposure tested was 5 s, which was the minimum that was technically feasible and reproducible. In 5 s, most of the nanocomplexes were not focused onto the cell monolayer (only 8.25 ± 1.87% was cell associated by ICP-MS of iron), but this still produced significant (although variable) transfection levels (2.14 ± 1.31 × 105 RLU). This was achieved with the FLR-DNA-MNP co-complex and static magnet combination. Without magnetic targeting, we observed very low levels of MNP cell association and reporter expression in 5 s (∼0.82 ± 0.37% by ICP-MS of iron, 1.62 ± 1.31 × 103 RLU). One minute exposure was more reproducible and technically easy to standardize. This was sufficient to focus significant amounts of the FLR-DNA-MNP nanocomplexes to cell monolayers (43.47 ± 8.65% by ICP-MS) and yielded similar transfection levels to that of 5 min exposure (65.76 ± 10.03% by ICP-MS) under a magnetic field (0.41 ± 0.76 × 107 versus 0.80 ± 0.76 × 107 RLU, respectively). Therefore, short and incomplete targeting of GET magnetofection nanocomplexes can yield significant transfection levels in seconds.
Figure 3.
Rapid and efficient GET magnetofection with low exposure times. (A) Gaussia luciferase expression after 5, 15, 30, and 60 min transfection/magnetofection with FLR (FLR-DNA) and FLR and MNPs(FLR-DNA-MNPs) with/without the application of a magnetic field. MNP complexes were formulated at 5 μg MNPs/μg DNA (FLR-DNA N/P of 6). Bars represent mean values ± s.d. (1 technical replicate). (****p < 0.0001 compared to treatments in the same group, &p < 0.01 compared to FLR-DNA, Tukey’s multiple comparison test, and n = 3 biological replicates). (B) Percentage and (C) mean intensity of rhodamine (Rh)-positive NIH3T3 cells after Rh-pDNA transfection/magnetofection for 5, 15, 30, and 60 min. Rh-pDNA was delivered with FLR (FLR-DNA) and FLR and MNPs, FLR-DNA-MNPs with/without the application of a magnetic field. MNP complexes were formulated at 5 μg MNPs/μg DNA (FLR-DNA N/P of 6). Values represent mean ± s.d. n = 3 biological replicates. (D) Iron cell association in NIH3T3 cells after 5, 15, 30, and 60 min magnetofection with FLR-DNA-MNPs with/without the application of a magnetic field. MNP complexes were formulated at 5 μg MNPs/μg DNA (FLR-DNA N/P of 6). Bars represent mean values ± s.d. n = 3 biological replicates (***p < 0.001, ****p < 0.0001, comparison between magnetofection with and without magnets, and Sidak’s multiple comparison test).
3.7. pDNA Is Rapidly Cell Membrane-Associated and Uptaken with GET Magnetofection
We next assessed the location of the pDNA cargo under these conditions, defining percentage of pDNA-labeled cells (using labeled Rhodamine (Rh)-pDNA) and also the amount of pDNA delivered per cell (intensity mean). Initially, it was important to assess the effect of Rh labeling of pDNA on its ability to transfect, be a transcriptional template for the reporter, and confirm lack of toxicity (Figure S3). Delivery of Rh-pDNA pGLuc did not affect cell metabolic activity and performed similarly to unlabeled pDNA in NIH3T3 cells when transfected with FLR nanocomplexes. Next, Rh-pDNA was employed to quantify pDNA association with cells using FLR-DNA and FLR-DNA-MNPs with/without a magnet at increasing time points. After delivery, cells were washed with PBS to remove any unbound complexes and trypsinized to collect cells for flow cytometry.
Flow cytometry quantification of the percentage of Rhodamine-positive (Rh+) cells confirmed the association of Rh-pDNA to the cell as early as 5 min (70 ± 12% of positive cells with FLR-DNA-MNPs-magnet compared with 4 ± 2 and 6.4 ± 2% for FLR-DNA and FLR-DNA-MNPs, respectively) (Figure 3B). Rh-pDNA association over time followed two different trends when FLR-DNA-MNPs were delivered with or without a magnetic field. The percentage of Rh+ cells remained almost constant over 60 min (at around 80%) when Rh-pDNA was delivered in FLR-DNA-MNPs in the presence of magnetic field, suggesting a saturation of MNP cell association.45 On the other hand, in the absence of a magnetic field, Rh-pDNA association increased progressively over time (6.4 ± 2 and 39.3 ± 7.7% at 5 and 60 min, respectively). Rh-pDNA association when delivered with FLR showed a similar pattern over time to FLR-DNA-MNP nanocomplexes in the absence of a magnetic field.
Interestingly, the mean fluorescence intensity per cell remained constant or minimally increased over time with longer incubations (Figure 3C). This data suggests that a similar amount of pDNA is either membrane bound or uptaken in the same experimental conditions; however, longer exposure times increase the overall percentage of labeled cells (Figure 3B). The application of an external magnetic field on FLR-DNA-MNP nanocomplexes allows for rapid concentration of pDNA on cells; in the absence of any magnetic forces, the FLR-DNA-MNP nanocomplexes and non-magnetic FLR-DNA nanocomplexes progressively accumulate onto cell membranes over time.
We next assessed MNP uptake using inductively coupled plasma mass spectroscopy (ICP-MS). NIH3T3 cells were incubated with FLR-DNA-MNP nanocomplexes for increasing amounts of time (5, 15, 30, and 60 min) with and without an external magnetic field. The amount of iron per cell was quantified 24 h post-delivery by ICP-MS. Significantly more iron was associated in the cells in the presence of a magnetic field (Figure 3D). Importantly, iron content progressively increased with prolonged incubation times when FLR-DNA-MNP nanocomplexes were delivered in the presence of a magnetic field.
It is important to note that despite the accuracy of the methods used to determine kinetics of transfection and particle internalization, they potentially struggle to differentiate bound nanoparticles from internalized complexes, providing inaccurate results.46 As one of the aims of this study was to confirm the uptake mechanism, it was therefore important for us to technically distinguish between internalization and cell membrane association of nanocomplexes. In order to achieve this, cells were washed with either PBS as before—defined as cell-associated (removing unbound or loosely bound nanocomplexes) or heparin, known to destabilize the FLR-DNA interaction, preventing gene transfer if nanocomplexes were not internalized—defined as cell-internalized.26 Destabilization of the FLR-DNA complex in the presence of heparin was confirmed by YO-PRO-1 assay (Figure S4). Transfection efficiency and Rh-pDNA and MNP internalization were assessed at 5, 15, 30, and 60 min by including an additional step of heparin wash after incubation (Figures S5–S7). Overall, values of transfection efficiency, Rh-pDNA uptake, and iron internalization were lower after the cells were washed with heparin, suggesting that a significant fraction of the complexes associated with the cells (up to 60 min) are cell bound but not completely internalized with short incubation periods. However, despite the lack of internalization of both pDNA and MNPs after short incubation periods, in most of the cases, transfection with FLR-DNA-MNP nanocomplexes in the presence of a magnetic field is still significantly better than the other treatments.
Taken together, these studies suggest that even limited interaction of FLR-DNA-MNP nanocomplexes with cells is sufficient to generate significant magnetofection (and delivery of nanocomplex constituents, pDNA and MNPs). Furthermore, targeted loading of cell membranes was the most important facet for rapid gene delivery using our system.
3.8. Rapid Regional Targeting of Transgene Expression with GET Magnetofection
Since we confirmed significant enhancement of FLR-DNA-MNP nanocomplex transfection efficiency under a magnetic field, we next assessed if an external static field could target transfection to specific regions of a cell monolayer in culture. For these experiments, we transfected pDNA that expresses enhanced GFP (pEGFP-CI) allowing measurement of transfection efficiency and levels at a cell autonomous level. GFP transfection mediated by FLR-DNA-MNP nanocomplexes with and without a magnetic field aligned well with pGLuc transfection (Figure 4). Exposure of 15 min to FLR-DNA-MNP nanocomplexes yielded enhanced transfection with a magnetic field by microscopy (38.9 ± 10.4 and 4.5 ± 2.3%, with and without a magnet, respectively) (Figure 4Ai). When transfection efficiency was assessed by expression of GFP proteins, we found that 1 h transfection of FLR-DNA-MNPs in the presence of a magnet (48.0 ± 5.9%) was comparable to 24 h exposure of FLR-DNA nanocomplexes (56.9 ± 9.1%) (Figure 4Aii).
Figure 4.
Rapid and efficient transgene targeting with GET magnetofection using a static magnetic field. (A) eGFP expression after 15, 30, 60, 120, and 1440 min (24 h) transfection/magnetofection exposure. After transfection, cells were washed with PBS to remove any unbound DNA complex. DNA (0.5 μg) was delivered with FLR-DNA and FLR-DNA-MNP complexes with/without the application of a magnetic field. MNP complexes were formulated at 5 μg MNPs/1 μg of DNA. The FLR-DNA ratio was constant at an N/P of 6. n = 3 biological repeats, 3 technical replicates. Scale bar is 100 μm. (B) Quantitation of eGFP-positive percentage from (A). (Bi) Schematic of the system used to target a fixed coverslip in a larger monolayer cell culture, defining IN (under a magnetic field) and OUT (no magnetic field) regions. (Bii) Demonstration of targeting GET magnetofection in 15 min exposure with a static magnet. Scale bar is 100 μm. (Biii) Quantitation of eGFP-positive cell percentage from (Bii) by flow cytometry. n = 3 biological repeats, 3 technical replicates.
We next aimed to target specific cells in the same culture. To achieve this, we developed an assay in which an interrupted monolayer can be seeded in culture, transfected, and a specific region removed with the monolayer still intact, allowing more complete characterization to transfection efficiency such as flow cytometry. This involved temporarily affixing a coverslip (20 mm diameter) to the middle of a 6-well plate with vacuum grease, allowing it to be readily removed with forceps after exposure and washing (Figure 4Bi). Magnetic focusing of FLR-DNA-MNP nanocomplexes to cells generated significant transfection efficiency within the magnetic field area by microscopy (Figure 4Bii) and quantitatively assessed by flow cytometry (IN region, 66.8 ± 8.3%) and prevented transfection of cells outside of the targeted area (OUT region, 0.6 ± 0.3%) (two orders of magnitude increase in targeting), when compared to that without a magnetic field (4.3 ± 1.4%) (Figure 4Biii). This represented a 111-fold enrichment in transfection of the targeted area and a reduction in background transfection without targeting of 7.2-fold. Enhancing the accuracy of pDNA targeting could have implications when improving efficacy and safety of gene therapy strategies using nanocomplexes.
3.9. GET Magnetofection Occurs via Multiple Endocytotic Pathways
Most of the non-viral nanocomplex vectors are hydrophilic, which greatly inhibits their ability to passively traverse the hydrophobic cell membrane. Therefore, these systems require active, energy-dependent endocytosis processes to cross the cell membrane. There is some evidence of lipoplex-mediated pDNA delivery through fusion with the cell membrane and direct release to the cytoplasm, but there is no confirmation that this is the case for cationic peptides/polymers such as our system.47−50 The most widely researched endocytic pathways are clathrin- or caveolae-mediated endocytosis and macropinocytosis.34 GET-mediated gene delivery has been previously associated with the macropinocytotic uptake pathway as vesicular-sequestered cargos delivered with GET appear to have longer half-lives than would be expected. However, it is likely that changes in cargo size, charge, and payload could change the mode of uptake of any system.26
It was important to confirm if rapid accumulation of FLR-DNA-MNP nanocomplexes on the cell membrane mediated by a magnetic field had an effect on the uptake mechanism. We conducted a series of experiments using labeled pDNA (Rh-pDNA) to quantify pDNA uptake. These compared FLR-DNA and FLR-DNA-MNP nanocomplexes with/without a magnet in culture conditions, aiming to inhibit uptake through endocytosis: low temperature (4 °C), which rigidifies the cell membrane affecting both passive and active uptake,21 hypertonic conditions (employing sucrose) to hinder clathrin lattice formation,32 methyl-B-cyclodextrin (MBCD) to repress caveolae-mediated endocytosis through complexation of cholesterol,35 and amiloride, an inhibitor of Na+/H+ exchange required for macropinocytosis.51 The experimental conditions including effective concentrations and treatment times of low temperature (4 °C), MBCD, and amiloride had been previously validated.26 Similarly, previous literature reported that treatment of NIH3T3 cells with MBCD (0–5 mM) or amiloride (0–5 mM) does not affect cell viability.51
We dissected the effect of these inhibitory conditions on MNP cell association (by removing lightly bound complexes with PBS) and on particle uptake (by disrupting non-internalized complexes with heparin). Interestingly, only a hypertonic medium (containing high sucrose), which is known to disrupt clathrin lattices, significantly decreased overall pDNA cell association for FLR-DNA and FLR-DNA-MNP nanocomplexes with or without magnets (Figure 5A). However, all inhibitors significantly decreased pDNA internalization of the nanocomplexes (Figure 5B). FLR-DNA and FLR-DNA-MNPs without a magnet showed similar pDNA internalization patterns in response to the different inhibitors. Low temperature (4 °C) known to rigidify cell membranes and the hypertonic medium had the most significant effect on pDNA internalization, suggesting a heavy contribution of clathrin-mediated endocytosis in the uptake process. Clathrin-mediated uptake of similar size particles and magnetofection complexes was previously reported in the literature.21,34,43
Figure 5.

Effect of endocytosis inhibitors on pDNA cell association and uptake by GET magnetofection. Percentage of rhodamine (Rh)-positive NIH3T3 cells after Rh-pDNA transfection/magnetofection with FLR (FLR-DNA) and FLR and MNPs (FLR-DNA-MNPs) with/without the application of a magnetic field in the presence of endocytosis inhibitors. Final concentration of inhibitors: methyl-B-cyclodextrin (MBCD), 5 mM; 5(N-ethyl-N-isopropyl) amiloride, 100 μM; and sucrose, 0.45 M. Cells were exposed to the inhibitors/4 °C for 1 h. The effect of inhibitors was tested on DNA cell association (A) and DNA cell uptake (B) 24 h post-delivery. Bars represent average percentage of rhodamine-labeled cells ± s.d. n = 6 biological replicates (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, comparison to control, and Tukey’s multiple comparison test).
Interestingly, when FLR-DNA-MNP nanocomplexes were delivered with a magnetic field, MBCD, which inhibits caveolae-mediated endocytosis, had a greater effect on pDNA internalization than sucrose, which inhibits clathrin lattice formation (Figure 5B). One of the hypotheses that could explain this change in the uptake mechanism in the presence of a magnetic field is based on previous observations reported in the literature that suggest that high concentrations of nanocomplexes on the cell membrane could saturate binding sites specific to a particular uptake mechanism.52−54 We have previously demonstrated that in the presence of a magnetic field, FLR-DNA-MNPs are rapidly attracted toward the cell surface, increasing pDNA concentration on the cell membrane (Figure 3C). We hypothesized that the saturation of FLR-specific endocytotic pathways (previously suggested as macropinocytosis for the mechanism of GET-mediated cargo uptake) triggers rerouting toward different endocytic internalization pathways, such as caveolae-mediated endocytosis. Alternatively, aggregation of the MNPs in the presence of a magnetic field could explain the difference in the uptake mechanisms. Previous studies focusing on examining the direct effects of aggregation on magnetofection are limited; however, most seem to attribute particle aggregation to the medium composition (i.e., FBS) over the presence of a magnetic field,45,55 which would not account for the differences observed in the presence of a magnetic field.
It is important to note that FLR-DNA and FLR-DNA-MNP nanocomplexes in the absence of a magnetic field showed comparable uptake mechanisms. These results are in agreement with transfection data that shows both transgene expression and pDNA uptake profiles over time were similar for these complexes, reinforcing the hypothesis that the incorporation of FLR-DNA into MNP nanocomplexes does not prevent efficient gene delivery mediated by FLR and the mechanism of uptake is similar.45,55,56
3.10. FLR Peptide Mediates Membrane Rupturing Endosomal Escape in GET Magnetofection
We next aimed to understand how magnetofected nanocomplexes successfully navigate to the cell nucleus. After being internalized, endocytosed pDNA must be efficiently released into the cytosol and access the nucleus in order to transcribe the delivered pDNA encoded transgene. Endosomal membrane rupturing activity of FLR-DNA, FLR-MNP, and FLR-DNA-MNP nanocomplexes was assessed through hemolysis assay, in which erythrocyte membranes serve as a surrogate for the lipid bilayer membrane in endo-lysosomal vesicles.57−60
Membrane disruption activity was assessed at physiological pH (pH 7.5) and late endosome/lysosome pH (pH 5). Hemolytic activity was calculated as a percentage of total hemolysis mediated by detergent Triton X-100 (Figure S8). There were no significant differences between the hemolytic activity of FLR-DNA and FLR-DNA-MNP nanocomplexes independent of the pH (58.8 ± 14.8 and 48.7 ± 12.3%, respectively, at pH 7.5 and 53.8 ± 27.8 and 44.8 ± 20.8%, respectively, at pH 5). MNPs alone did not show any significant membrane rupturing activity. These results suggest that any endosomal escape activity, triggered by the complex, would be mostly mediated by the FLR peptide. Additionally, FLR membrane disruptive activity is pH-independent, which is consistent with the lack of carboxylic side chains on FLR molecules, which are known to mediate pH-dependent endosomal disruptive activity.57 The membrane disruptive activity of FLR could then potentially be explained by physical interaction between the peptide and the lipid bilayer, similar to that previously reported for similar peptides.61 Furthermore, in this assay, membrane disruptive activity of FLR decreases in the presence of serum and drops down to approximately ∼20% (19 ± 12.6% hemolysis) at 10% FCS (in vitro experimental conditions) (Figure S9). This would suggest that FLR should not significantly affect the cell plasma membrane integrity during transfection as observed indirectly in our cell viability and proliferation analyses.
It is important to note that this hemolysis assay only assesses membrane rupturing activity due to chemical interactions with the cell membrane, but it does not account for endosome swelling or physical alterations of the loaded endosome. Additionally, this assay has been performed in PBS or FCS, which do not represent the intracellular environment (i.e., cytosol or endosome composition). Finally, in this assay, erythrocytes were used as a surrogate for endosomal membranes; however, the lipid content and exact composition of the endosomal membranes vary between cells. Isolation and analysis of the internal structure of FLR-DNA-MNP nanocomplex-loaded endosomes may provide a better understanding of endosomal membrane composition and how they escape efficiently into the cytosol.59,62 Additional endosomal escape assays involving dye leakage or fluorescent fusion proteins would provide more information on the exact mechanism underlying pDNA translocation into the cytoplasm.63,64
3.11. GET-Magnetofected pDNA Retains Integrity when Internalized
To evaluate the ability of FLR-MNP nanocomplex vectors and magnetofection conditions to deliver fully intact pDNA inside the cells as well as its stability post-transfection, pGLuc pDNA was delivered with FLR-DNA and FLR-DNA-MNP nanocomplexes with and without a magnet for 5 min. Extrachromosomal DNA, and therefore pDNA, was isolated and quantified by bacterial transformation efficiency (a marker for un-nicked, intact pDNA) at different time points post-delivery.65 Percentage of cell bound pDNA was calculated compared to the total amount of pDNA delivered (Figure S10).
A significantly higher percentage of intact pDNA was associated with NIH3T3 cells when delivered with FLR-DNA-MNPs in a magnetic field (36.1 ± 6.3%) compared to FLR-DNA and FLR-DNA-MNPs in the absence of a magnetic field (1.3 ± 0.3 and 1.9 ± 0.3%, respectively) after 10 min incubation. The percentage of cell bound pDNA remained relatively constant during the first 60 min and decreased significantly up to 24 h post-delivery. Interestingly, the percentage of pDNA degraded over 24 h was comparable in all transfection groups (the final percentage of intact pDNA after 24 h was approximately 10% of the DNA present immediately post-delivery). Since all nanocomplex formulations were taken up through endocytotic pathways, degradation is most likely to be driven by enzymes present in lysosomal compartments or by cytosolic nucleases.66 In this context, pDNA degradation will largely depend on its presentation, where naked pDNA would be more susceptible to degradation compared to complexed FLR-associated pDNA.67 It is likely that any decomplexation of pDNA, which will be needed for its transcription, would be proportional to the amount of pDNA delivered independent of the specific uptake route.68
3.12. GET-Magnetofected Complexes Internalize Rapidly into Vesicles
We further investigated the trafficking of nanocomplexes by tracking the uptake of labeled pDNA. Intracellular localization of Rh-pDNA (pGLuc) was imaged by confocal microscopy after delivery with FLR (FLR-DNA) or FLR-MNPs under a magnetic field (FLR-DNA-MNPs) after 30 min (Figure 6A), 60 min (Figure 6B), and 24 h (Figure 6C) transfection. At the end of each incubation time, cells were washed with PBS, fixed, and stained with Alexa Fluor 488 Phalloidin and DAPI to identify the actin cytoskeleton and nucleus, respectively.69
Figure 6.

Rapid uptake of pDNA by GET magnetofection. Confocal laser scanning microscopy (CLSM) images of Rh-labeled DNA in NIH3T3 cells after (A) 30 min, (B) 60 min, or (C) 24 h delivery. Cells were transfected with FLR-DNA and FLR-DNA-MNPs in the presence of a magnetic field. Cells were treated with 1 μg of Rh-pDNA (pCMV-GLuc2) at an N/P ratio of 6 and an optimal MNPs mass ratio of 5 μg MNPs/1 μg DNA for 30 min. After incubation, the unbound complex was removed with PBS, and cells were fixed with 3.7% PFA. Images show Rh-pDNA (red), cell nuclei stained with DAPI (blue), and the actin cytoskeleton stained with Alexa Fluor 488 Phalloidin (green). Scale bar: 2 μm.
Merged fluorescence images showed minimal fluorescence within or on cell membranes after 30 min with FLR-DNA nanocomplexes; however, when delivered as FLR-DNA-MNPs under a magnetic field, large numbers of fluorescent particles could be observed localized around cell boundaries and attached to membrane surfaces (Figure 6A). After 1 h delivery, discrete fluorescent particles could be detected with FLR-DNA nanocomplexes; however, these were less abundant when compared to FLR-DNA-MNP samples (Figure 6B). After 24 h, most of the fluorescence was detected within cells concentrated around the nucleus (Figure 6C). Fluorescence was localized as discrete foci, as opposed to homogeneously distributed throughout the cytosol, suggesting the entrapment of the pDNA to intracellular vesicles.70 Even though the vast majority of pDNA delivered was visible in perinuclear endosomes, significant reporter activity from pDNA demonstrates that some must be correctly localized for nuclear expression.
We therefore demonstrated progressive interaction with and transfer through cell membranes over time, irrelevant of the complex or targeting to cells.
4. Conclusions
In this work, the use of the GET peptide, FLR, to efficiently deliver pDNA on an MNP-based vector under the influence of a magnetic field has been optimized and characterized. FLR-DNA-MNP nanocomplexes were able to significantly improve reporter gene expression after short incubations (>5 s) in the presence of a magnetic field compared with no magnetic field or FLR-DNA nanocomplexes alone. Effects of GET magnetofection on the cellular entry mechanism, pDNA stability inside the cell, and cellular viability were also assessed. The system appears to be cytocompatible, and pDNA is stable when uptaken, potentially through a variety of endocytotic pathways.
Importantly, nearly all cells could be loaded with detectable amounts of pDNA within 5 min, and some level of transgene expression was detectable even with 5 s exposure with FLR-DNA-MNP nanocomplexes in the presence of a magnetic field. The most impactful observation from our study is the rapidity and zonality of transfection using the magnetic system. We foresee translation of our system in vitro for applications where regional delivery or speed is technically important and in vivo using external static magnets or internal focusing of magnetic resonance to target tumors or specific organs or tissues.
When treated with endocytosis inhibitors, FLR-DNA and FLR-DNA-MNP nanocomplexes showed significantly lower pDNA uptake in the hypertonic medium compared to the other inhibitors, suggesting a sizable contribution of clathrin-dependent endocytosis on pDNA uptake. Interestingly, MBCD, which is involved in cholesterol depletion from the cell membrane, significantly affected pDNA uptake during GET magnetofection in the presence of a magnetic field, more so than the other inhibitors, suggesting an important role of caveolae-mediated endocytosis when complexes are targeted. Degradation of delivered pDNA seems to be consistent across the complexes over a 24 h period suggesting that the complex and uptake mechanism do not play significant roles in pDNA intracellular trafficking; more likely, the pDNA degradation rate is proportional to intracellular concentration.
Finally, confocal imaging confirmed the presence of pDNA localized around the boundaries of the cell as well as some degree of internalization at early time points (30 and 60 min) using magnetofection but very little uptake for FLR-DNA nanocomplexes. After 24 h, pDNA could be seen internalized around the cell nucleus or confined to vesicles in the cytoplasm for all complexes tested. The principle behind magnetofection is the concentration of the cargo towards a cell population either in vitro or in vivo. Our findings indeed suggest that MNP-containing nanocomplexes under a magnetic field quickly concentrate pDNA onto the cell surface and by doing so alter the uptake kinetics and mechanism; however, we found no evidence that MNPs except for targeting play any further role in gene transfer.
In summary, these results show that the GET system can efficiently be used for magnetofection. Insights into mechanisms of uptake during GET magnetofection may aid the design of future magnetic gene vectors and to develop novel approaches to target genetic therapeutics with magnetic fields. Magnetic field-guided local transfection and focused in vivo gene delivery may now be possible by combining nanocomplex magnetofection with GET-mediated non-viral gene delivery.
Acknowledgments
Research conducted in this study was funded by the European Research Council under the European Community’s Seventh Framework Programme (FP7/2007–2013)/ERC grant agreement 227845. This work was supported by the Medical Research Council (grant number MR/K026682/1), the Engineering and Physical Sciences Research Council, and the Biotechnology and Biological Sciences Research Council, acknowledged by J.E.D. for the UK Regenerative Medicine Platform Hub “Acellular Approaches for Therapeutic Delivery”. L.A.B.F. acknowledges the studentship supported by the Engineering and Physical Sciences Research Council (EP/F500491/1) in the Centre of Doctoral Training in Regenerative Medicine. The authors are grateful to Dr. Robert Markus for assistance with confocal microscopy.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsanm.0c02465.
(Figure S1) Magnet array configuration for magnetofection, (Figure S2) additional data on optimization of GET magnetofection, (Figure S3) rhodamine labeling of pGLuc DNA not affecting its ability for protein expression or cell viability in NIH3T3 cells, (Figure S4) high concentrations of heparin destabilizing the FLR-DNA complexes, (Figure S5) efficient and fast (less than 5 min) pDNA delivery using GET magnetofection and a static magnet, (Figure S6) efficient and fast (less than 5 min) pDNA cell membrane association using GET magnetofection and a static magnet, (Figure S7) rapid cell uptake within 30 min with GET magnetofection, (Figure S8) membrane rupturing activity of GET magnetofection mediated by FLR and independent of pH, (Figure S9) FLR minimally affecting membrane integrity in cell culture conditions, and (Figure S10) stability of GET-magnetofected pDNA (PDF)
Author Contributions
L.A.B.F. and J.E.D. conceived and initiated the project; L.A.B.F. and J.E.D. designed the experiments; L.A.B.F., S.Y.C., S.V.R., and J.E.D. conducted the experiments; J.E.D. supervised the study; L.A.B.F. and J.E.D. wrote the manuscript; all authors approved the final manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Varga C. M.; Tedford N. C.; Thomas M.; Klibanov A. M.; Griffith L. G.; Lauffenburger D. A. Quantitative Comparison of Polyethylenimine Formulations and Adenoviral Vectors in Terms of Intracellular Gene Delivery Processes. Gene Ther. 2005, 12, 1023–1032. 10.1038/sj.gt.3302495. [DOI] [PubMed] [Google Scholar]
- Tros de Ilarduya C.; Sun Y.; Düzgüneş N. Gene Delivery by Lipoplexes and Polyplexes. Eur. J. Pharm. Sci. 2010, 40, 159–170. 10.1016/j.ejps.2010.03.019. [DOI] [PubMed] [Google Scholar]
- Varga C. M.; Wickham T. J.; Lauffenburger D. A. Receptor-Mediated Targeting of Gene Delivery Vectors: Insights from Molecular Mechanisms for Improved Vehicle Design. Biotechnol. Bioeng. 2000, 70, 593–605. . [DOI] [PubMed] [Google Scholar]
- Hill A. B.; Chen M.; Chen C.; Pfeifer B. A.; Jones C. H. Overcoming Gene-Delivery Hurdles : Physiological Considerations for Nonviral Vectors. Trends Biotechnol. 2016, 34, 91–105. 10.1016/j.tibtech.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank C.; Schillinger U.; Scherer F.; Bergemann C.; Rémy J. S.; Krötz F.; Anton M.; Lausier J.; Rosenecker J. The Magnetofection Method: Using Magnetic Force to Enhance Gene Delivery. Biol. Chem. 2003, 384, 737–747. 10.1515/BC.2003.082. [DOI] [PubMed] [Google Scholar]
- Mykhaylyk O.; Antequera Y. S.; Vlaskou D.; Plank C. Generation of Magnetic Nonviral Gene Transfer Agents and Magnetofection in Vitro. Nat. Protoc. 2007, 2, 2391–2411. 10.1038/nprot.2007.352. [DOI] [PubMed] [Google Scholar]
- Plank C.; Scherer F.; Schillinger U.; Anton M. Magnetofection: Enhancement and Localization of Gene Delivery with Magnetic Particles under the Influence of a Magnetic Field. J Gene Med. 2000, 2, S24. [Google Scholar]
- Plank C.; Zelphati O.; Mykhaylyk O. Magnetically Enhanced Nucleic Acid Delivery. Ten Years of Magnetofection-Progress and Prospects. Adv. Drug Deliv. Rev. 2011, 63, 1300–1331. 10.1016/j.addr.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titze de Almeida S.; Horst C.; Soto-Sánchez C.; Fernandez E.; Titze de Almeida R. Delivery of MiRNA-Targeted Oligonucleotides in the Rat Striatum by Magnetofection with Neuromag®. Molecules 2018, 23, 1825. 10.3390/molecules23071825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolders S.; Kessels S.; Smolders S. M. T.; Poulhes F.; Zelphati O.; Sapet C.; Brône B. Magnetofection Is Superior to Other Chemical Transfection Methods in a Microglial Cell Line. J. Neurosci. Methods 2018, 293, 169–173. 10.1016/j.jneumeth.2017.09.017. [DOI] [PubMed] [Google Scholar]
- Pan Z.; Shi Z.; Wei H.; Sun F.; Song J.; Huang Y.; Liu T.; Mao Y. Magnetofection Based on Superparamagnetic Iron Oxide Nanoparticles Weakens Glioma Stem Cell Proliferation and Invasion by Mediating High Expression of MicroRNA-374a. J. Cancer 2016, 7, 1487–1496. 10.7150/jca.15515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J.; Mohanty I.; Rattan S. In Vivo Magnetofection: A Novel Approach for Targeted Topical Delivery of Nucleic Acids for Rectoanal Motility Disorders. Am. J. Physiol. - Gastrointest. Liver Physiol. 2018, 314, G109–G118. 10.1152/ajpgi.00233.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y.; Li X.; Zeljic K.; Shan S.; Qiu Z.; Wang Z. Effect of PEGylated Magnetic PLGA-PEI Nanoparticles on Primary Hippocampal Neurons: Reduced Nanoneurotoxicity and Enhanced Transfection Efficiency with Magnetofection. ACS Appl. Mater. Interfaces 2019, 11, 38190–38204. 10.1021/acsami.9b15014. [DOI] [PubMed] [Google Scholar]
- Qiu Y.; Tong S.; Zhang L.; Sakurai Y.; Myers D. R.; Hong L.; Lam W. A.; Bao G. Magnetic Forces Enable Controlled Drug Delivery by Disrupting Endothelial Cell-Cell Junctions. Nat. Commun. 2017, 8, 15594–15510. 10.1038/ncomms15594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Ortega L.; Rojas J. M.; Marcos A.; Portilla Y.; Stein J. V.; Barber D. F. T Cells Loaded with Magnetic Nanoparticles Are Retained in Peripheral Lymph Nodes by the Application of a Magnetic Field. J. Nanobiotechnol. 2019, 17, 14–20. 10.1186/s12951-019-0440-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthana M.; Kennerley A. J.; Hughes R.; Fagnano E.; Richardson J.; Paul M.; Murdoch C.; Wright F.; Payne C.; Lythgoe M. F.; Farrow N.; Dobson J.; Conner J.; Wild J. M.; Lewis C. Directing Cell Therapy to Anatomic Target Sites in Vivo with Magnetic Resonance Targeting. Nat. Commun. 2015, 6, 8009–8011. 10.1038/ncomms9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer F.; Anton M.; Schillinger U.; Henke J.; Bergemann C.; Krüger A.; Gänsbacher B.; Plank C. Magnetofection: Enhancing and Targeting Gene Delivery by Magnetic Force in Vitro and in Vivo. Gene Ther. 2002, 9, 102–109. 10.1038/sj.gt.3301624. [DOI] [PubMed] [Google Scholar]
- Liu W.-M.; Xue Y.-N.; Peng N.; He W.-T.; Zhuo R.-X.; Huang S.-W. Dendrimer Modified Magnetic Iron Oxide Nanoparticle / DNA / PEI Ternary Magnetoplexes : A Novel Strategy for Magnetofection. J. Mater. Chem. 2011, 21, 13306–13315. 10.1039/c1jm11460c. [DOI] [Google Scholar]
- Ma Y.; Zhang Z.; Wang X.; Xia W.; Gu H. Insights into the Mechanism of Magnetofection Using MNPs-PEI/PDNA/Free PEI Magnetofectins. Int. J. Pharm. 2011, 419, 247–254. 10.1016/j.ijpharm.2011.07.017. [DOI] [PubMed] [Google Scholar]
- McBain S. C.; Yiu H. H. P.; El Haj A.; Dobson J. Polyethyleneimine Functionalized Iron Oxide Nanoparticles as Agents for DNA Delivery and Transfection. J. Mater. Chem. 2007, 17, 2561–2565. 10.1039/b617402g. [DOI] [Google Scholar]
- Arsianti M.; Lim M.; Marquis C. P.; Amal R. Polyethylenimine Based Magnetic Iron-Oxide Vector: The Effect of Vector Component Assembly on Cellular Entry Mechanism, Intracellular Localization, and Cellular Viability. Biomacromolecules 2010, 11, 2521–2531. 10.1021/bm100748p. [DOI] [PubMed] [Google Scholar]
- Sun S.-L.; Lo Y.-L.; Chen H.-Y.; Wang L.-F. Hybrid Polyethylenimine and Polyacrylic Acid-Bound Iron Oxide as a Magnetoplex for Gene Delivery. Langmuir 2012, 28, 3542–3552. 10.1021/la204529u. [DOI] [PubMed] [Google Scholar]
- Bono N.; Ponti F.; Mantovani D.; Candiani G. Non-Viral in Vitro Gene Delivery: It Is Now Time to Set the Bar!. Pharmaceutics 2020, 12, 183. 10.3390/pharmaceutics12020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman G.; Rodriguez J.; Chan S. Y.; Chisholm J.; Duncan G.; Kim N.; Tatler A. L.; Shakesheff K. M.; Hanes J.; Suk J. S.; Dixon J. E. PEGylated Enhanced Cell Penetrating Peptide Nanoparticles for Lung Gene Therapy. J. Controlled Release 2018, 285, 35–45. 10.1016/j.jconrel.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Awwad H. A. D. M.; Thiagarajan L.; Dixon J. E. Controlled Release of GAG-Binding Enhanced Transduction (GET) Peptides for Sustained and Highly Efficient Intracellular Delivery. Acta Biomater. 2017, 57, 225–237. 10.1016/j.actbio.2017.04.028. [DOI] [PubMed] [Google Scholar]
- Dixon J. E.; Osman G.; Morris G. E.; Markides H.; Rotherham M.; Bayoussef Z.; El Haj A. J.; Denning C.; Shakesheff K. M. Highly Efficient Delivery of Functional Cargoes by the Synergistic Effect of GAG Binding Motifs and Cell-Penetrating Peptides. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, E291–E299. 10.1073/pnas.1518634113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.; Choo J.; Choi Y.; Lee K.; Min D.; Pi S.; Seol Y.; Lee S.; Jo I.; Chung C.; Park Y. Characterization of the Surface Immobilized Synthetic Heparin Binding Domain Derived from Human Fibroblast Growth Factor-2 and Its Effect on Osteoblast Differentiation. J. Biomed. Mater. Res., Part A: Off. J. Soc. Biomater., Jpn. Soc. Biomater. Aust. Soc. Biomater. Korean Soc. Biomater. 2007, 83, 970–979. 10.1002/jbm.a. [DOI] [PubMed] [Google Scholar]
- Saleh A. F.; Aojula H.; Arthanari Y.; Offerman S.; Alkotaji M.; Pluen A. Improved Tat-Mediated Plasmid DNA Transfer by Fusion to LK15 Peptide. J. Controlled Release 2010, 143, 233–242. 10.1016/j.jconrel.2009.12.025. [DOI] [PubMed] [Google Scholar]
- Eltaher H. M.; Yang J.; Shakesheff K. M.; Dixon J. E. Highly Efficient Intracellular Transduction in Three-Dimensional Gradients for Programming Cell Fate. Acta Biomater. 2016, 41, 181–192. 10.1016/j.actbio.2016.06.004. [DOI] [PubMed] [Google Scholar]
- Thiagarajan L.; Abu-Awwad H. A. D. M.; Dixon J. E. Osteogenic Programming of Human Mesenchymal Stem Cells with Highly Efficient Intracellular Delivery of RUNX2. Stem Cells Transl. Med. 2017, 6, 2146–2159. 10.1002/sctm.17-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliotopoulos A.; Ferreras L. B.; Densham R. M.; Caulton S. G.; Maddison B. C.; Morris J. R.; Dixon J. E.; Gough K. C.; Dreveny I. Discovery of Peptide Ligands Targeting a Specific Ubiquitin-like Domain–Binding Site in the Deubiquitinase USP11. J. Biol. Chem. 2019, 294, 424–436. 10.1074/jbc.RA118.004469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser J. E.; Anderson R. G. Hypertonic Media Inhibit Receptor-Mediated Endocytosis by Blocking Clathrin-Coated Pit Formation. J. Cell Biol. 1989, 108, 389–400. 10.1083/jcb.108.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B. R.; Huang Y. W.; Lee H. J. Mechanistic Studies of Intracellular Delivery of Proteins by Cell-Penetrating Peptides in Cyanobacteria. BMC Microbiol. 2013, 13, 57. 10.1186/1471-2180-13-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejman J.; Oberle V.; Zuhorn I. S.; Hoekstra D. Size-Dependent Internalization of Particles via the Pathways of Clathrin- and Caveolae-Mediated Endocytosis. Biochem. J. 2004, 377, 159–169. 10.1042/bj20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuhorn I. S.; Kalicharan R.; Hoekstra D. Lipoplex-Mediated Transfection of Mammalian Cells Occurs through the Cholesterol-Dependent Clathrin-Mediated Pathway of Endocytosis. J. Biol. Chem. 2002, 277, 18021–18028. 10.1074/jbc.M111257200. [DOI] [PubMed] [Google Scholar]
- Vasić K.; Knez Ž.; Konstantinova E. A.; Kokorin A. I.; Gyergyek S.; Leitgeb M. Structural and Magnetic Characteristics of Carboxymethyl Dextran Coated Magnetic Nanoparticles : From Characterization to Immobilization Application. React. Funct. Polym. 2020, 148, 104481. 10.1016/j.reactfunctpolym.2020.104481. [DOI] [Google Scholar]
- Markides H.; Newell K. J.; Rudorf H.; Ferreras L. B.; Dixon J. E.; Morris R. H.; Graves M.; Kaggie J.; Henson F.; El Haj A. J. Ex Vivo MRI Cell Tracking of Autologous Mesenchymal Stromal Cells in an Ovine Osteochondral Defect Model. Stem Cell Res. Ther. 2019, 10, 25–15. 10.1186/s13287-018-1123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos-Campos I.; Asín L.; Torres T. E.; Marquina C.; Tres A.; Ibarra M. R.; Goya G. F. Cell Death Induced by the Application of Alternating Magnetic Fields to Nanoparticle-Loaded Dendritic Cells. Nanotechnology 2011, 22, 205101. 10.1088/0957-4484/22/20/205101. [DOI] [PubMed] [Google Scholar]
- Blokpoel Ferreras L. A.; Scott D.; Vazquez Reina S.; Roach P.; Torres T. E.; Goya G. F.; Shakesheff K. M.; Dixon J. E. Enhanced Cellular Transduction of Nanoparticles Resistant to Rapidly Forming Plasma Protein Coronas. Adv. Biosyst. 2020, 4, 2000162–2000116. 10.1002/adbi.202000162. [DOI] [PubMed] [Google Scholar]
- Stetefeld J.; McKenna S. A.; Patel T. R. Dynamic Light Scattering: A Practical Guide and Applications in Biomedical Sciences. Biophys. Rev. 2016, 8, 409–427. 10.1007/s12551-016-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latour R. A. The Langmuir Isotherm: A Commonly Applied but Misleading Approach for the Analysis of Protein Adsorption Behavior. J. Biomed. Mater. Res. - Part A 2015, 103, 949–958. 10.1002/jbm.a.35235. [DOI] [PubMed] [Google Scholar]
- Song H. P.; Yang J. Y.; Lo S. L.; Wang Y.; Fan W. M.; Tang X. S.; Xue J. M.; Wang S. Gene Transfer Using Self-Assembled Ternary Complexes of Cationic Magnetic Nanoparticles, Plasmid DNA and Cell-Penetrating Tat Peptide. Biomaterials 2010, 31, 769–778. 10.1016/j.biomaterials.2009.09.085. [DOI] [PubMed] [Google Scholar]
- Huth S.; Lausier J.; Gersting S. W.; Rudolph C.; Plank C.; Welsch U.; Rosenecker J. Insights into the Mechanism of Magnetofection Using PEI-Based Magnetofectins for Gene Transfer. J. Gene Med. 2004, 6, 923–936. 10.1002/jgm.577. [DOI] [PubMed] [Google Scholar]
- Cruz-Acuña M.; Maldonado-Camargo L.; Dobson J.; Rinaldi C. From Oleic Acid-Capped Iron Oxide Nanoparticles to Polyethyleneimine-Coated Single-Particle Magnetofectins. J. Nanopart. Res. 2016, 18, 268. 10.1007/s11051-016-3577-9. [DOI] [Google Scholar]
- Prijic S.; Scancar J.; Romih R.; Cemazar M.; Bregar V. B.; Znidarsic A.; Sersa G. Increased Cellular Uptake of Biocompatible Superparamagnetic Iron Oxide Nanoparticles into Malignant Cells by an External Magnetic Field. J. Membr. Biol. 2010, 236, 167–179. 10.1007/s00232-010-9271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayol D.; Luciani N.; Lartigue L.; Gazeau F.; Wilhelm C. Managing Magnetic Nanoparticle Aggregation and Cellular Uptake: A Precondition for Efficient Stem-Cell Differentiation and MRI Tracking. Adv. Healthcare Mater. 2013, 2, 313–325. 10.1002/adhm.201200294. [DOI] [PubMed] [Google Scholar]
- Khalil I. A.; Kogure K.; Akita H.; Harashima H. Uptake Pathways and Subsequent Intracellular Trafficking in Nonviral Gene Delivery. Pharmacol. Rev. 2006, 58, 32–45. 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- Wattiaux R.; Laurent N.; Wattiaux-De Coninck S.; Jadot M. Endosomes, Lysosomes: Their Implication in Gene Transfer. Adv. Drug Delivery Rev. 2000, 41, 201–208. 10.1016/S0169-409X(99)00066-6. [DOI] [PubMed] [Google Scholar]
- Medina-kauwe L. K.; Xie J.; Hamm-Alvarez S. Intracellular Trafficking of Nonviral Vectors. Gene Ther. 2005, 12, 1734–1751. 10.1038/sj.gt.3302592. [DOI] [PubMed] [Google Scholar]
- Lechardeur D.; Verkman A. S.; Lukacs G. L. Intracellular Routing of Plasmid DNA during Non-Viral Gene Transfer. Adv. Drug Delivery Rev. 2005, 57, 755–767. 10.1016/j.addr.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Wadia J. S.; Stan R. V.; Dowdy S. F. Transducible TAT-HA Fusogenic Peptide Enhances Escape of TAT-Fusion Proteins after Lipid Raft Macropinocytosis. Nat. Med. 2004, 10, 310–315. 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- Doherty G. J.; McMahon H. T. Mechanisms of Endocytosis. Annu. Rev. Biochem. 2009, 78, 857–902. 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- Wilhelm C.; Gazeau F.; Roger J.; Pons J. N.; Bacri J. C. Interaction of Anionic Superparamagnetic Nanoparticles with Cells: Kinetic Analyses of Membrane Adsorption and Subsequent Internalization. Langmuir 2002, 18, 8148–8155. 10.1021/la0257337. [DOI] [Google Scholar]
- Weigel A. V.; Tamkun M. M.; Krapf D. Quantifying the Dynamic Interactions between a Clathrin-Coated Pit and Cargo Molecules. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, E4591. 10.1073/pnas.1315202110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T.-Y.; Wu J.-H.; Xu Q.-H.; Wang X.-R.; Lu J.; Hu Y.; Jo J.-i.; Yamamoto M.; Ling D.; Tabata Y.; Gao J.-Q. Design of Magnetic Gene Complexes as Effective and Serum Resistant Gene Delivery Systems for Mesenchymal Stem Cells. Int. J. Pharm. 2017, 520, 1–13. 10.1016/j.ijpharm.2017.01.041. [DOI] [PubMed] [Google Scholar]
- Dowaidar M.; Abdelhamid H. N.; Hällbrink M.; Freimann K.; Kurrikoff K.; Zou X.; Langel Ü. Magnetic Nanoparticle Assisted Self-Assembly of Cell Penetrating Peptides-Oligonucleotides Complexes for Gene Delivery. Sci. Rep. 2017, 7, 9159–9111. 10.1038/s41598-017-09803-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy N.; Robichaud J. R.; Tirrell D. A.; Stayton P. S.; Hoffman A. S. The Design and Synthesis of Polymers for Eukaryotic Membrane Disruption. J. Controlled Release 1999, 61, 137–143. 10.1016/S0168-3659(99)00114-5. [DOI] [PubMed] [Google Scholar]
- Plank C.; Oberhauser B.; Mechtler K.; Koch C.; Wagner E. The Influence of Endosome-Disruptive Peptides on Gene Transfer Using Synthetic Virus-like Gene Transfer Systems. J. Biol. Chem. 1994, 269, 12918–12924. [PubMed] [Google Scholar]
- Evans B. C.; Nelson C. E.; Yu S. S.; Beavers K. R.; Kim A. J.; Li H.; Nelson H. M.; Giorgio T. D.; Duvall C. L. Ex Vivo Red Blood Cell Hemolysis Assay for the Evaluation of PH-Responsive Endosomolytic Agents for Cytosolic Delivery of Biomacromolecular Drugs. J. Visualized Exp. 2013, 73, e50166 10.3791/50166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.-J.; Johnson G.; Pellois J.-P. Modeling of the Endosomolytic Activity of HA2-TAT Peptides With Red Blood Cells and Ghosts. Biochemistry 2010, 49, 7854–7866. 10.1021/bi1008408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won A.; Ruscito A.; Ianoul A. Imaging the Membrane Lytic Activity of Bioactive Peptide Latarcin 2a. Biochim. Biophys. Acta, Biomembr. 2012, 1818, 3072–3080. 10.1016/j.bbamem.2012.07.030. [DOI] [PubMed] [Google Scholar]
- Rivière C.; Wilhelm C.; Cousin F.; Dupuis V.; Gazeau F.; Perzynski R. Internal Structure of Magnetic Endosomes. Eur. Phys. J. E 2007, 22, 1–10. 10.1140/epje/e2007-00014-1. [DOI] [PubMed] [Google Scholar]
- Smith S. A.; Selby L. I.; Johnston A. P. R.; Such G. K. The Endosomal Escape of Nanoparticles: Toward More Efficient Cellular Delivery. Bioconjugate Chem. 2019, 30, 263–272. 10.1021/acs.bioconjchem.8b00732. [DOI] [PubMed] [Google Scholar]
- Lönn P.; Kacsinta A. D.; Cui X. S.; Hamil A. S.; Kaulich M.; Gogoi K.; Dowdy S. F. Enhancing Endosomal Escape for Intracellular Delivery of Macromolecular Biologic Therapeutics. Sci. Rep. 2016, 6, 32301–32309. 10.1038/srep32301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective Extraction of Polyoma DNA from Infected Mouse Cell Cultures. J. Mol. Biol. 1967, 26, 365–369. 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Lechardeur D.; Sohn K. J.; Haardt M.; Joshi P. B.; Monck M.; Graham R. W.; Beatty B.; Squire J.; O’Brodovich H.; Lukacs G. L. Metabolic Instability of Plasmid DNA in the Cytosol: A Potential Barrier to Gene Transfer. Gene Ther. 1999, 6, 482–497. 10.1038/sj.gt.3300867. [DOI] [PubMed] [Google Scholar]
- Raftery R. M.; Walsh D. P.; Ferreras L. B.; Castaño I. M.; Chen G.; LeMoine M.; Osman G.; Shakesheff K. M.; Dixon J. E.; O’Brien F. J. Highly Versatile Cell-Penetrating Peptide Loaded Scaffold for Efficient and Localised Gene Delivery to Multiple Cell Types: From Development to Application in Tissue Engineering. Biomaterials 2019, 216, 119277. 10.1016/j.biomaterials.2019.119277. [DOI] [PubMed] [Google Scholar]
- Kichler A.; Leborgne C.; Coeytaux E.; Danos O. Polyethylenimine-Mediated Gene Delivery: A Mechanistic Study. J. Gene Med. 2001, 3, 135–144. 10.1002/jgm.173. [DOI] [PubMed] [Google Scholar]
- Srinivasan C.; Siddiqui S.; Silbart L. K.; Papadimitrakopoulos F.; Burgess D. J. Dual Fluorescent Labeling Method to Visualize Plasmid DNA Degradation. Bioconjugate Chem. 2009, 20, 163–169. 10.1021/bc800184j. [DOI] [PubMed] [Google Scholar]
- Yameen B.; Choi W. I.; Vilos C.; Swami A.; Shi J.; Farokhzad O. C. Insight into Nanoparticle Cellular Uptake and Intracellular Targeting. J. Controlled Release 2014, 190, 485–499. 10.1016/j.jconrel.2014.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.