Abstract
MicroRNAs (miRNAs/miRs) are a class of small non-coding RNAs that maintain the precise balance of various physiological processes through regulating the function of target mRNAs. Dysregulation of miRNAs is closely associated with various types of human cancer. miR-222-3p is considered a canonical factor affecting the expression and signal transduction of multiple genes involved in tumor occurrence and progression. miR-222-3p in human biofluids, such as urine and plasma, may be a potential biomarker for the early diagnosis of tumors. In addition, miR-222-3p acts as a prognostic factor for the survival of patients with cancer. The present review first summarizes and discusses the role of miR-222-3p as a biomarker for diverse types of cancers, and then focuses on its essential roles in tumorigenesis, progression, metastasis and chemoresistance. Finally, the current understanding of the regulatory mechanisms of miR-222-3p at the molecular level are summarized. Overall, the current evidence highlights the crucial role of miR-222-3p in cancer diagnosis, prognosis and treatment.
Keywords: miR-222-3p, biomarker, cell signaling pathway, exosome, therapeutic target
1. Introduction
MicroRNAs (miRNAs/miRs) are endogenous, 19-23-nucleotide-long, non-coding single-stranded RNA molecules that act as regulators of gene expression by associating with the multiprotein RNA-induced silencing complex (RISC) (1,2). RISC silences specific mRNA species by pairing with the 3′-untranslated region (3′-UTR) of the target mRNAs to impact their expression (3-5). The biogenesis of miRNAs is composed of multiple steps. First, primary miRNAs are cleaved into stem-loop precursor structures of ~70 nucleotides, known as precursor-miRNAs (premiRNAs), by the Drosha enzyme (6,7). Ultimately, premiRNAs are digested to mature 22-nucleotide-long miRNAs by the RNase III enzyme Dicer (8).
miR-222, a member of the miR-221/222 family, is located on the X chromosome p11.3 of the human genome (9). Mature miR-222 sequences have a hairpin precursor with different arms called the 5' or 3' arm, which are also known as -5p or -3p, respectively (10,11). Dysregulated miR-222-3p expression has been reported in various human diseases, such as in cataract pathogenesis and chordomas (12-15) and appears to be a promising biomarker for cancer diagnosis and prognosis (16-18).
The development of tumors is a multistep process that includes continuous proliferation signaling, evading growth inhibitors and inducing angiogenesis, invasion and metastasis (19). miRNAs have important roles in the initiation, development and progression of various types of cancer, including breast and prostate cancer (20-22). A growing number of studies have indicated that miR-222-3p has multiple functions in tumorigenesis (23), cancer cell proliferation and apoptosis (24), cancer cell invasion and migration (16,25), therapeutic resistance (26) and the tumor microenvironment (27,28).
A number of studies have recently manifested the association of miR-222-3p dysregulation with cancer initiation and progression (29-31). Despite recent developments in cancer diagnosis and treatment, there are still numerous problems associated with the pathogenesis of cancer progression, disease recurrence and drug resistance. The present review thoroughly discusses the clinical value of miR-222-3p in cancer and whether miR-222-3p acts as an oncogene or a cancer suppressor by reviewing and summarizing studies covering cells, cancer tissues and biofluids. A comprehensive overview of these findings and the implications for molecular research are provided.
2. Diagnostic and prognostic value of miR-222-3p in cancer
Increasing studies have suggested that miR-222-3p expression may be a potential predictor of tumor type, tumor grade and lymph node metastasis in multiple types of human tumors, such as prostate cancer, uveal melanoma, papillary thyroid carcinoma and gastric cancer (32-35). Circulating miR-222-3p, alone or in combination with other miRNAs in plasma/serum, may act as a candidate biomarker for the early detection of cancer (12,22,36). Moreover, tumor cell-derived miR-222-3p serves as a prognostic factor for the survival of patients with hepatocellular carcinoma and epithelial ovarian cancer (23,37). The expression pattern of miR-222-3p has been extensively studied and compared in non-tumor and tumor tissues of different types of human cancer (Table I).
Table I.
Expression pattern of miR-222-3p in different types of human cancer.
| A, Downregulated miR-222-3p expression
| |
|---|---|
| Type of cancer | (Refs.) |
| Oral cancer | (50) |
| Ovarian carcinoma | (36) |
| Prostate cancer | (51,79-82) |
|
| |
| B, Upregulated miR-222-3p expression
| |
| Type of cancer | (Refs.) |
|
| |
| Bladder cancer | (53) |
| Breast cancer | (46,56) |
| Colorectal cancer | (18,27,57,71,88) |
| Endometrial carcinoma | (26,66) |
| Gastric carcinoma | (35,44,45,49) |
| Glioblastoma | (60) |
| Hepatocellular carcinoma | (23,43) |
| Lung carcinoma | (16,55,77) |
| Melanoma | (33) |
| Ovarian carcinoma | (97) |
| Osteosarcoma | (25) |
| Thyroid carcinoma | (38,40) |
| miR, microRNA. | |
Clinical value of miR-222-3p in cancer tissues
Aberrant miR-222-3p expression is closely associated with the clinical characteristics of patients with cancer. For instance, miR-222-3p was found to be overexpressed in papillary thyroid carcinoma (PTC) compared with in normal thyroid and benign cancer tissues (38). Additionally, Di Fazio et al (39) reported that miR-222-3p could be used to distinguish patients with typical and atypical lung carcinoid. Furthermore, miR-222-3p combined with a panel of miRNAs (miR-7-5p and miR-146b-5p) exhibited high sensitivity and specificity for identifying different subtypes of PTC (40). This panel of miRNAs could distinguish non-invasive follicular thyroid neoplasms from papillary-like nuclear features, follicular adenomas and infiltrative follicular variants of PTC (40-42). miR-222-3p was reported to be differentially expressed in some types of cancer, as shown in Table I. For example, miR-222-3p expression is decreased in prostate cancer, while it is increased in other types of cancer, including bladder and breast cancer; however, miR-222-3p expression in ovarian carcinoma remains controversial (Table I).
Recently, Wang et al (23) investigated the functions of the clustered miRNAs hsa-miR-221/222-3p in hepatocellular carcinoma (HCC) using a human miRNA tissue atlas, revealing that these two miRNAs and their target genes had potential prognostic value for HCC, especially miR-222-3p, which functioned as a tumor promotor in hepatic tumorigenesis (23). The oncogenic function of miR-222-3p has also been confirmed in non-alcoholic steatohepatitis (NASH)-associated liver carcinogenesis (43), indicating that miR-222-3p may serve as an indicator for the development of NASH-derived HCC.
In addition, dysregulated miR-222-3p expression was found to be closely associated with tumor stage, invasion and metastasis. In gastric carcinoma, high miR-222-3p expression was positively associated with advanced clinical stage and lymph node metastasis (44), and could predict the survival of patients who were unable to receive chemotherapy after surgery (45). Rinnerthaler et al (46) reported that miR-222-3p expression was associated with the progesterone receptor, and elevated miR-222-3p expression was involved in breast cancer development, tumor spread, proliferation and drug resistance. Moreover, miR-222-3p was found to be highly associated with the tumor stage and lymph node metastasis in estrogen receptor α (ERα)-negative patients with endometrial cancer (EC), and lower miR-222-3p expression was detected in ERα-negative EC with lower grades (P=0.0145) and earlier stages (I vs. II, P=0.05; II vs. III, P=0.0043; I vs. III, P=0.0002) (26). By contrast, miR-222-3p upregulation exhibited a positive association with overall survival in patients with epithelial ovarian cancer (EOC), and its expression level was negatively associated with tumor growth in an EOC mouse model (37).
miR-222-3p as a non-invasive diagnostic biomarker in cancer
miR-222-3p serves as a specific biomarker for various types of tumors in body fluids, such as serum, plasma and urine (13,47,48) indicating that miR-222-3p has the potential to be developed into a non-invasive diagnostic biomarker for tumors. High expression levels of circulating miR-222-3p were significantly associated with lymph node metastasis (P=0.009) and clinical stages (P<0.001) in gastric cancer in an analysis of 38 plasma samples (49). Chang et al (50) revealed that miR-222-3p was negatively associated with clinical staging and lymph node metastasis status in plasma samples collected from patients with oral carcinoma, and miR-222-3p may be a useful diagnostic biomarker for the differentiation of oral squamous cell carcinoma and oral leukoplakia plaque. Furthermore, Fredsoe et al (51) developed a three-microRNA ratio model (miR-222-3p*/miR-24-3p/miR-30c-5p), which provided accurate markers in the differential diagnosis of benign prostatic hyperplasia and prostate cancer (PCa). Several studies have found that serum miRNAs could be used to predict the risk of non-muscle invasive bladder cancer, and risk scores were generated according to the combination of the three miRNA ratios (miR-29a-3p/miR-222-3-p, miR-150-5-p/miR-331-3p and miR-409-3-p/miR-433-5-p) (52,53). Similar results were obtained in PTC (38,54). As aforementioned, dysregulated miR-222-3p expression has been observed in numerous types of human cancer, thus providing powerful rationales for its application as a non-invasive diagnostic biomarker.
miR-222-3p as a non-invasive prognostic biomarker in cancer
As well as having a strong potential in the diagnosis of cancer, miR-222-3p also serves an important role in predicting the prognosis of patients with cancer. An increasing number of studies has demonstrated that dysregulated miR-222-3p expression can predict the progression and poor prognosis of patients with cancer, including prostate cancer and papillary thyroid carcinoma (30,31).
Ulivi et al (55) analyzed the expression levels of miRNAs in early non-small cell lung cancer (NSCLC) tissues, lung squamous cell carcinoma tissues and circulating blood. Notably, miR-222-3p was significantly associated with decreased disease-free survival (DFS) and overall survival (55). Wang et al (56) revealed that downregulation of miRNAs (miR-130b-5p, miR-151a-5p, miR-206 and miR-222-3p) was closely associated with a poor prognosis after surgery and that serum miR-222-3p was an independent prognostic factor for a poor DFS time [hazard ratio (HR), 13.19; 95% CI, 1.06-163.59; P=0.045] in patients with breast cancer (56). Additionally, circulating miR-222-3p was significantly associated with the estrogen level of patients with EC, and elevated miR-222-3p expression was positively associated with tumor size, indicating that miR-222-3p may serve as a crucial indicator of prognosis in patients with EC (26).
KRAS and BRAF are poor prognostic indicators used for colorectal cancer (CRC; frequency of mutation in patients: KRAS, 38-44.9%; BRAF, 4.2-5%); KRAS and BRAF mutations were positively associated with a poor prognosis in patients with CRC (57-59). Therefore, more sensitive prognostic biomarkers are required for CRC. A panel of 16 miRNAs (including miR-222-3p) associated with improved 5-year DFS time for stage II and III CRC was identified (57). The signature was identified as an independent prognostic factor for improved 5-year DFS time by multivariate analyses (57). Similar observations were reported in gastric carcinoma (45) and thyroid cancer (54). Additionally, elevated serum miR-222-3p expression may significantly predict a poor probability of 2-year DFS time in patients with glioblastoma (60). Notably, a novel logistic regression model, comprising five urinary miRNAs (miR-151a-5p, miR-204-5p, miR-222-3p, miR-23b-3p and miR-331-3p) and serum prostate-specific antigen (PSA), was established successfully by Fredsoe et al (32) and could predict time to biochemical recurrence in 215 patients with PCa (univariate Cox regression analysis HR, 3.12; P<0.001).
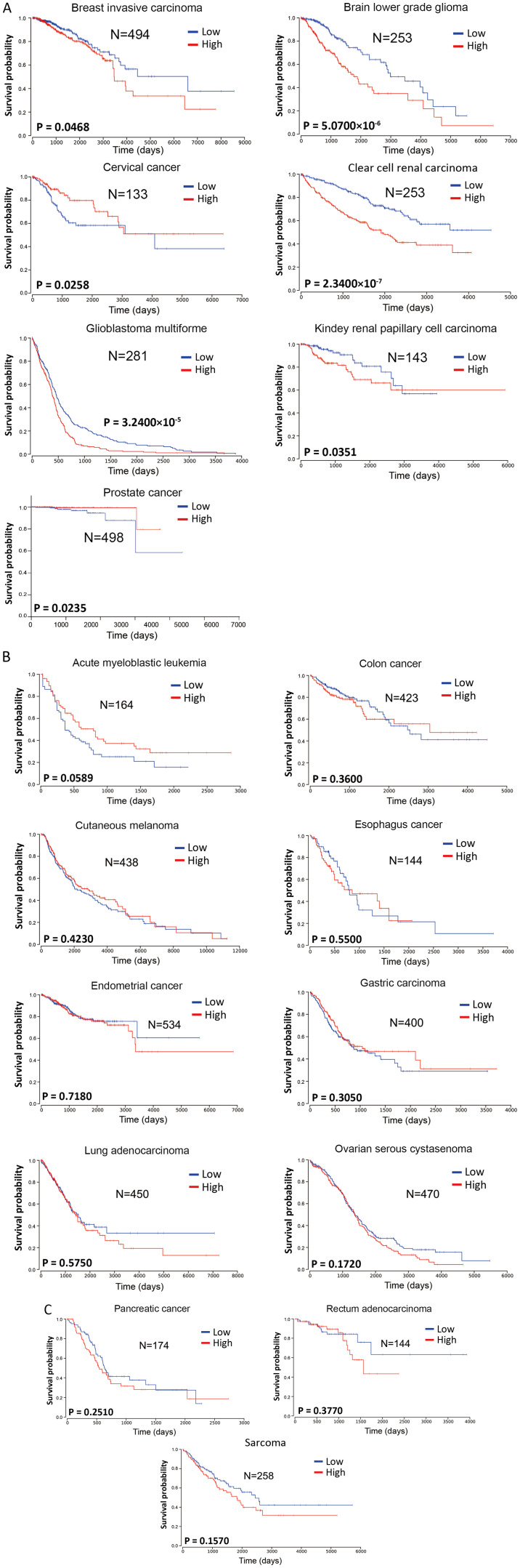
To investigate the prognostic value of miR-222-3p in the survival of patients with various types of cancer, miR-222-3p expression in human cancers was analyzed from The Cancer Genome Atlas database (https://tcga-data.nci.nih.gov/tcga/). The Kaplan-Meier analysis method was used for survival analysis using GraphPad Prism 7.00 (GraphPad Software, Inc.). Log-rank P<0.05 was considered to indicate a statistically significant difference. Considering the mid- and late-stage crossovers, the weighted method of Cramer-von Mises testing was used. The patients were divided into two groups according to the different expression levels of miR-222-3p in each tumor type, either lower or higher than the mean value.
miR-222-3p expression was significantly associated with the survival of patients with breast invasive carcinoma, brain lower grade glioma, clear cell renal carcinomas, glioblastoma multiforme or kidney renal papillary cell carcinoma (Fig. 1A), with high miR-222-3p expression predicting a poorer overall survival compared with low miR-222-3p expression. In 11 other types of cancer (acute myeloblastic leukemia, colon cancer, cutaneous melanoma, esophageal cancer, EC, gastric carcinoma, lung adenocarcinoma, ovarian serous cystadenoma, pancreatic cancer, rectum adenocarcinoma and sarcoma), the expression levels of miR-222-3p were not significantly associated with overall survival (Fig. 1B and C). Although higher miR-222-3p expression seemed to predict longer overall survival in acute myeloblastic leukemia, there was no significant difference between the two groups (P=0.0589; Fig. 1B). Additionally, higher miR-222-3p expression in cervical and prostate cancer predicted a longer overall survival (Fig. 1A). Although these data may be affected by the sample size and stability of the sequencing method in the miR-222-3p quantification, the aforementioned data suggest that miR-222-3p expression may exhibit prognostic value only in certain types of human cancer.
Figure 1.
Prognostic value of miR-222-3p in human cancer. miR-222-3p expression in different types of human cancer was analyzed from The Cancer Genome Atlas database. Patients were divided into two groups according to the expression levels of miR-222-3p, either lower or higher than the mean value. The Kaplan-Meier analysis method was used for survival analysis using the software GraphPad Prism 7. Log-rank P<0.05 was considered to indicate a statistically significant difference. Considering the mid- and late-stage crossovers, the weighted method of Cramer-von Mises testing was used. (A) Seven types of cancer in which the overall survival of patients was significantly associated with miR-222-3p expression. (B and C) Eleven types of cancer in which the overall survival of patients was not significantly associated with miR-222-3p expression. miR, microRNA.
Therapeutic resistance is a major risk factor for a poor prognosis in tumor patients who undergo chemo- and radio-therapy (61,62). A previous study indicated that miR-222-3p increased raloxifene resistance through suppressing ERα expression in EC cells (26). This mechanism is also observed in the resistance to gemcitabine, a nucleoside analogue with activity against NSCLC, with acquired gemcitabine resistance being a major obstacle in NSCLC treatment (63).
3. Functional roles of miR-222-3p in cancer
Tumorigenesis
Aberrant miR-222-3p expression serves a crucial role in numerous types of human tumors (64), and it is closely associated with certain aspects of cancer biology, including tumorigenesis (65-68). Recently, several reports have indicated that miR-222-3p exhibited an oncogenic function, including in CRC (18) and EC (69). Moreover, miR-222-3p expression drives cancer stem cell renewal in CRC, making it a potential target for therapy (70). Helicobacter pylori infection acts as a trigger in the carcinogenesis of gastric cancer (71), and increasing studies (53,72) suggest that H. pylori affects miRNA expression. Tan et al (53) revealed that miR-222-3p expression was markedly increased in the cancer group [H. pylori (+)] compared with in the normal group [H. pylori (-)]. miR-222-3p associated with H. pylori targets homeodomain-interacting protein kinases 2 (HIPK2) to promote cell proliferation and invasion, and to inhibit apoptosis in gastric cancer (72). Functional experiments demonstrated that miR-222-3p overexpression significantly enhanced the proliferative activity while inhibiting the apoptosis of SGC7901 gastric cancer cells, but miR-222-3p-knockdown exhibited the opposite effects (35). Consistent with this finding, an in vivo experiment revealed that downregulated miR-222-3p expression in AN3CA cells inhibited EC tumor growth in a mouse xenograft model (26). By contrast, Fu et al (37) demonstrated that higher miR-222-3p expression was associated with improved overall survival in patients with EOC, and its level was negatively associated with tumor growth in vivo. Thus, miR-222-3p may govern key processes during the development of various types of cancer.
Cancer cell proliferation and apoptosis
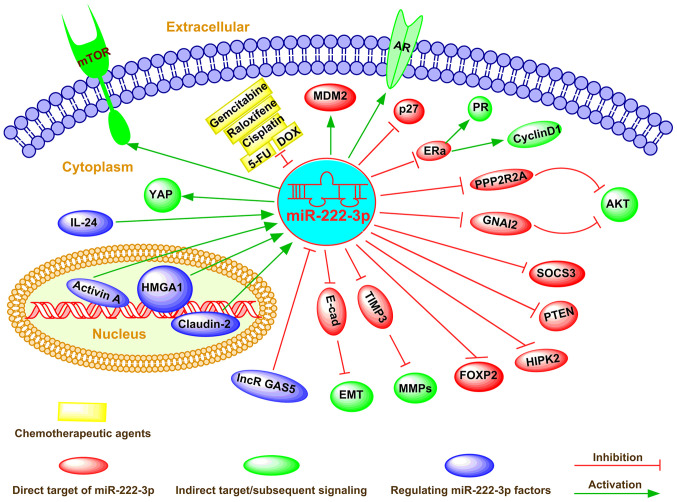
As a cellular regulator, miR-222-3p affects gene expression via direct binding to complementary sequences in the 3′-UTR of target mRNAs in numerous types of cancer cells (73). miR-222-3p determines the malignancy of cell proliferation or apoptosis in various types of cancer, including breast cancer and colorectal carcinoma (74-76). The global regulatory mechanism of miR-222-3p in determining the fate of cancer cells is shown in Fig. 2.
Figure 2.
Emerging roles and mechanisms of miR-222-3p in human cancer. miR-222-3p can directly or indirectly regulate multiple downstream pathways, such as PI3K/AKT, PTEN, JAK/STAT, TRPS1/ZEB1 and EMT, between which crosstalks usually exist, thus constituting a complex signaling network. Additionally, miR-222-3p can extensively regulate multiple cell functions, including differentiation, proliferation, apoptosis, invasion, metastasis and metabolism modulation via targeting gene expression at the post-transcriptional level. Furthermore, miR-222-3p serves an important role as either a tumor suppressor or an oncogene in different types of cancer. Dysregulated miR-222-3p expression contributes to drug resistance and has been emphasized as a new drug target. In addition, miR-222-3p expression can be regulated via both transcriptional factors and epigenetic factor-induced mechanisms in cancer cells. AR, androgen receptor; DOX, doxycycline; EMT, epithelial-mesenchymal transition; E-cad, E-cadherin; ERa, estrogen receptor α; FOXP2, forkhead box protein P2; GNAI2, G protein α inhibiting activity polypeptide 2; HIPK2, homeodomain-interacting protein kinases 2; HMGA1, high mobility group AT-hook 1; lncR GAS5, long non-coding RNA growth arrest-specific 5; MDM2, murine double minute 2; miR, microRNA; MMP, matrix metalloproteinase; PI3K, phosphoinositide 3-kinase; PPP2R2A, phosphatase 2A subunit B; PR, progesterone receptor; PTEN, phosphatase and tensin homology deleted on chromosome ten; SOCS3, suppressor cytokine signaling 3; YAP, Yes-associated protein; 5-FU, 5-fluorouracil.
In addition, it has been reported that overexpression of miR-222-3p induces alteration of the cell cycle (a high ratio of G1 to S phase) and promotes cell proliferation (26). Conversely, miR-222-3p-knockdown significantly decreases cell proliferation by upregulating cyclin D1 (26). The upregulation of miR-222-3p markedly stimulated cell proliferation and repressed apoptosis by releasing endogenous IL-24 and activating the phosphatidylinositol 3 (PI3K)/AKT signaling pathway in lung cancer (77). Further investigation indicated that activation of the PI3K/AKT signaling pathway directly suppressed high mobility group AT-hook 1 (HMGA1) expression, with the dysregulation of phosphatase and tensin homology deleted on chromosome ten (PTEN) by miR-222-3p (77,78). By contrast, Coarfa et al (17) identified a panel of 12 miRNAs (including miR-222-3p) with a proteomic footprint (using reversed-phase proteomic arrays), and the expression levels of these miRNAs were markedly decreased in metastatic PCa. This miRNA panel significantly decreased cell proliferation and targeted key tumor-associated signaling pathways involving the androgen receptor axis and the Akt/mTOR signaling pathway (17). Additionally, other studies revealed that miR-222-3p expression was decreased upon progression to high-grade PCa (Gleason score 8-10 or PSA level >20 ng/ml; clinical stage, T3a) (79-81). A similar result was obtained in a study of PCa by Tong et al (82). Ottley et al (80) reported that cyclin-dependent kinase inhibitor 1B expression was accompanied by a decrease in miR-93, miR-222-3p and miR-18a expression in activin A-treated prostate cancer LNCaP cells. Furthermore, a previous study observed higher expression levels of miR-222-3p in androgen-independent LNCaP cells than in LNCaP cells, indicating its growth-promoting role in PCa (83). A small molecule inhibitor of murine double minute 2 (MDM2), nutlin-3, selectively disrupted the interaction between MDM2 and p53 (84). Moreover, genome-wide miRNA expression analysis revealed that the expression levels of miR-34a-5p, miR-182-5p, miR-203a, miR-222-3p and miR-432-5p were upregulated following nutlin-3 treatment in a p53-dependent manner (85). Notably, miR-222-3p overexpression promoted apoptosis and suppressed proliferation in neuroblastoma cells (85).
In cancer cells, different types of mutations may mediate amplification or reduction of gene expression and lead to altered protein expression patterns (86). Although miR-222-3p is hypothesized to regulate specific genes, it may not affect some of its predicted genes (26). The function of miR-222-3p mainly affects cell fate-associated signaling pathways.
Cancer cell invasion and migration
Aberrant miRNA expression has been reported in metastatic cancers, which universally display an aggressive pathophysiology (20). Consistent with this finding, miR-222-3p has been shown to be essential for the invasion and metastasis of different types of cancer, including osteosarcoma, endometrial carcinoma and prostate cancer (25,26,87) (Fig. 2).
Guo et al (25) revealed that forced miR-222-3p expression decreased the expression levels of tissue inhibitor of metalloproteinases 3 (TIMP3) in osteosarcoma cells. TIMP3 inhibits the transfer of endogenous proteases via regulating matrix metalloproteinases 2 and 9 (MMP-2 and MMP-9), which are involved in mediating cancer progression (87). Moreover, transfection of MG-63 and U-2OS cells with miR-222-3p inhibitors significantly increased the migration and invasion of these osteosarcoma cells, suggesting that miR-222-3p may act as a cancer suppressor (25). miR-222-3p is highly expressed in ER-negative EC, and overexpression of miR-222-3p significantly promotes the migration and invasion of EC cells by targeting ERα (27). Tan et al (35) revealed that the protein HIPK2 was decreased in gastric cancer tissues [H. pylori (+) group] compared with in normal gastric tissues. Further analysis indicated that miR-222-3p could enhance the migration of gastric cancer cells by binding to the 3′-UTR of HIPK2 (35). Tricho-rhino-pharyngeal syndrome type 1 (TRPS1)/zinc finger E-box binding homeobox 1 (ZEB1) signaling is positively associated with tumor cell migration and invasion (13). TRPS1 can enhance the expression levels of E-cadherin and vimentin by upregulating miR-222-3p expression (13). Additionally, miR-222-3p targets E-cadherin-mediated ZEB1-induced epithelial-mesenchymal transition (EMT), thereby promoting chordoma progression (13).
Chemoresistance and new drug targets
The resistance of tumors to various anticancer drugs is an important factor that increases the invasiveness and metastasis of tumor cells (88,89), which depend on escaping apoptosis and increasing drug efflux (24,90). Dysregulated miR-222-3p expression may contribute to drug resistance by regulating gene expression (37), the cell cycle and apoptosis (26,37). Therefore, miR-222-3p is commonly perceived to be responsive to cancer treatment and has been emphasized as a new drug target (91) (Fig. 2).
miR-222-3p induces cisplatin resistance in EOC cells by targeting the 3′-UTR of G protein a inhibiting activity poly-peptide 2 (37). Additionally, several studies (26,34,37) revealed that the upregulation of miR-222-3p expression promoted cell survival in cisplatin-treated ovarian cancer cells. In addition, miR-222-3p increased raloxifene resistance via suppressing ERα expression in EC cells (26). Increased miR-222-3p expression directly targeted the 3'-UTR of claudin-2 mRNA to decrease apoptosis and induce CRC resistance to 5-fluo-rouracil (92). Transfection of miR-222-3p inhibitor into doxycycline (DOX)-resistant colon cancer cells (LoVo/ADR cells) decreased the expression levels of apoptotic proteins, such as poly (ADP-ribose) polymerase (PARP) and caspase 3, and significantly increased the expression levels of typical antiapoptotic proteins (BAX, cleaved PARP and cleaved caspase 3) (24). These results indicate that miR-222-3p may be a promising therapeutic target for overcoming chemotherapy resistance in human cancer.
Regulatory ef fect of miR-222-3p on the tumor microenvironment
The tumor microenvironment serves an important role in cancer initiation, progression and metastasis (93). The primary components of the tumor micro-environment are fibroblasts, immune cells, endothelial cells, extracellular matrix and cytokines (94,95). Among them, immune cells serve vital roles in enhancing cancer progression by secreting numerous proinflammatory factors (96). According to previous studies (97-100), miR-222-3p can modulate the function of immune cells, such as natural killer (NK) cells and cancer-associated fibroblasts, and serve an important role in the tumor microenvironment.
Ying et al (97) revealed that upregulated miR-222-3p expression activated the JAK/STAT signaling pathway by inhibiting suppressor of cytokine signaling 3 (SOCS3) expression, thereby decreasing the polarization of ovarian cancer-associated M2-like macrophages. Decreased expression levels of apoptosis-inducing ligand tumor necrosis factor and CD107a associated with liver NK cells impaired degranulation, which may contribute to the persistence of chronic hepatitis C virus (HCV) infection (101). Therefore, chronic HCV infection has different effects on the function of NK cell subsets in intrahepatic and peripheral blood chambers (101). In addition, strong inflammatory signals suppress the degranulation of NK cells, increasing the risk of disease progression in several types of patients (102,103). These results indicate that miR-222-3p may be an important factor that can regulate the NK cell activity involved in HCV-associated HCC.
4. Regulation of miR-222-3p in human cancer
miR-222-3p serves various roles in the initiation, progression, metastasis and treatment response of cancer, and its expression can be dysregulated by multiple factors in human cancer (104-106). For example, the Chinese medicinal herb andrographolide was reported to inhibit hematoma tumor growth by altering the miRNA profile (107). Mechanistically, miR-222-3p expression can be regulated via both transcriptional factors and epigenetic factor-induced mechanisms in cancer cells (70,108,109) (Fig. 2). Additionally, Ignacio et al (110) elucidated that several miRNA (including miR-222-3p) changes caused by ethanol were reversed by social activity and caused a number of novel epigenetic mechanisms; for example, prenatal alcohol exposure imposed a long-lasting effect on neuronal and, ultimately, behavioral function in adolescents.
miR-222-3p expression was significantly decreased after activin A treatment in LNCaP cells (83). Recently, miR-222-3p was found to be positively regulated by HMGA1, an architectural transcription factor that participates in the biological progression of different types of human cancer, including uveal melanoma and lung cancer (33,109). Moreover, HMGA1 overexpression exacerbated tumor progression by activating miR-222-3p via the PI3K/Akt/MMP-9 signaling pathway in uveal melanoma (33). Consistent with this finding, DOX-mediated IL-24 expression markedly decreased HMGA1 mRNA and protein expression by downregulating miR-222-3p expression, which resulted in a substantial increase in phosphatase 2A subunit B expression and a concomitant decrease in phosphorylated AKTT308/S473 expression (109). In addition, small interfering (si)RNA-mediated knockdown of HMGA1 significantly decreased AKT T308/S473 protein expression and markedly decreased cell migration and invasion by targeting miR-222-3p in lung cancer (109), suggesting that HMGA1 siRNA or miR-222-3p inhibitor may be used as effective treatments for lung cancer. Paquet-Fifield et al (70) revealed that activation of claudin-2 resulted in increased miR-222-3p expression, which activated the Yes-associated protein and promoted CRC cell self-renewal. In addition, the long non-coding RNA growth arrest-specific 5 (GAS5) activated the PTEN/AKT signaling pathway as a competing endogenous RNA of miR-222-3p in PTC (34). Coincidently, Liu et al (18) demonstrated that lncRNA GAS5 dramatically increased PTEN expression by decoying miR-222-3p, thus inhibiting CRC cell migration and invasion, and promoting cell autophagy.
5. Exosomes and miR-222-3p in human cancer
Exosomes are membranous extracellular vesicles, with a diameter of 30-100 nm, that are critical mediators of intercellular communications (111,112). miRNAs, which may act as post-transcriptional regulators of gene expression, have also been identified in exosomes (113). There is differential expression between cancer and normal exosomes with specific onco-genic and tumor suppressive miRNAs, which may provide diagnostic or prognostic potential of circulating exosomal miRNAs in cancer (114). Notably, Ostenfeld et al (27) isolated cancer-derived epithelial cell adhesion molecule-positive-exosomes from the serum and plasma of patients with CRC, and revealed that increased exosomal miR-222-3p may be an effective marker in the early stage of CRC. Higher miR-222-3p expression was also observed in serum-derived exosomes from patients with EOC compared with in healthy individuals (97). The aforementioned studies suggest that miR-222-3p, alone or in combination with a panel of other miRNAs, may serve as a non-invasive biomarker in cancer diagnosis. Ryu et al (105) reported that a panel of five candidate miRNAs (miR-320e, miR-4454, miR-222-3p, miR-21-5p and miR-25-3p) predicted poor survival outcomes in patients with extranodal NK/T-cell lymphoma (ENKTL), indicating the prognostic value of serum-derived exosomal miRNA profiles in patients with ENKTL. Circulating exosomal miR-342-5p, miR-222-3p and miR-574-5p have been reported to be the diagnostic and prognostic markers for early-stage lung adenocarcinoma (108,115). Jiang et al (31) revealed that significantly upregulated expression levels of miR-146b-5p and miR-222-3p from plasma exosomes may be potential biomarkers for lymph node metastasis in papillary thyroid carcinoma. Via high-throughput microarrays, the dysregulated miRNAs in paired serum samples from patients with breast cancer before and after surgery were screened, and miR-222-3p was identified as an independent prognostic factor for DFS (HR, 13.19; 95% CI, 1.06-163.59; P=0.045) (56). In addition, increased exosomal miR-222-3p expression tended to predict a worse prognosis in patients with NSCLC and exosomal miR-222-3p expression in serum may be used as a potential prognostic biomarker for predicting gemcitabine sensitivity in patients with NSCLC (16). Wei et al (16) revealed that gemcitabine-resistant cells contributed to the development of NSCLC tumor malignancy via exosome-mediated transfer of miR-222-3p. Moreover, exosome-derived miR-222-3p enhanced the migration and invasion of gemcitabine-resistant cells by directly targeting the promoter of SOCS3 in NSCLC (16). Therefore, gemcitabine-resistant A549 lung cancer cells could transmit their malignant phenotype to gemcitabine-sensitive A549 parental cells via exosome-derived miR-222-3p (16). In addition, HCV infection can present as an acute manifestation and can cause various complications, such as chronic hepatitis, liver fibrosis, cirrhosis and HCC (98,99). Santangelo et al (100) recently revealed that HCV-derived exosomes suppressed NK cell activity, and this is associated with high miR-222-3p expression.
6. Discussion
In studies of several types of tumors, the expression levels and function of miR-222-3p exhibited opposite results (Table I). These findings may be due to the inconsistent quality of the studies. For example, the use of miR-222-3p inhibitors and mimics should be further confirmed in functional verification studies. There are no high-quality controls for negative and positive products, which strongly affects the consistency of the data. In future miR-222-3p studies, the guidelines recommended for miRNA studies should be applied (116-118). Although the biological functions and mechanisms of miR-222-3p have been extensively studied (Fig. 2), further studies on its clinical application are required. miR-222-3p is widely involved in the regulation of numerous cellular, physiological and pathological processes. Additionally, miR-222-3p expression is aberrant during cancer progression (34,109). Thus, miR-222-3p may be used as a potential biomarker to predict cancer malignancy. Notably, it may be possible to develop an innovative strategy to treat various types of cancer by targeting miR-222-3p. miR-222-3p can directly or indirectly regulate multiple downstream molecules, which are involved in multiple tumor signaling pathways, including PI3K/AKT, PTEN, JAK/STAT, TRPS1/ZEB1 and EMT, between which crosstalks usually exist, thus constituting a complex signaling network. Additionally, miR-222-3p is extensively involved in cancer cell differentiation, proliferation, apoptosis, invasion, metastasis and metabolism modulation via targeting gene expression at the post-transcriptional level. Furthermore, miR-222-3p functions as either a tumor suppressor or an oncogene in different types of tumors, indicating its potential as a new target for cancer treatment. Despite miR-222-3p being extensively involved in cancer progression, numerous potential target mRNAs of miR-222-3p remain to be identified, and its functions and mechanisms in tumor metabolism and tumor immunity require to be further investigated. Overall, an improved understanding of miR-222-3p and its mechanism of action may provide research ideas for potentially developing a novel therapeutic intervention for cancer treatment and an increased overall survival rate.
Notably, some studies have focused on exosomal miR-222-3p (16,97,100), which has shown potential for tumor diagnosis and treatment. Although several clinical trials investigating the role of miRNA-based therapy for cancer have been initiated and may lead to novel therapeutic interventions in the future (119-121), prospects for the clinical application of miRNAs in cancer therapy are vague, indicating that more preclinical studies, especially those on toxicity and safety, should be conducted.
7. Conclusions
It is well known that miRNAs are involved in the development of cancer and may function as promising biomarkers for early detection, diagnosis and prognosis. The present review highlighted the scientific discoveries of miR-222-3p in human cancer research and outlined the advances and challenges of miR-222-3p as a diagnostic tool for cancer, as well as providing biological and clinical insights on this topic. miR-222-3p functions as an oncogene in some tumors and as a tumor suppressor in others, suggesting that the function of miR-222-3p is tumor- and cellular context-dependent. Its biological functions are involved in the occurrence, progression, metastasis and drug resistance of cancer, indicating its potential as a new target for cancer treatment.
Acknowledgments
Not applicable.
Abbreviations
- AR
androgen receptor
- CRC
colorectal cancer
- DFS
disease-free survival
- DOX
doxycycline
- EOC
epithelial ovarian cancer
- ERα
estrogen receptor α
- EC
endometrial cancer
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HIPK2
homeodomain-interacting protein kinases 2
- HMGA1
high mobility group AT-hook 1
- H. pylori
Helicobacter pylori
- MDM2
murine double minute 2
- miRNA/miR
microRNA
- MMP
matrix metalloproteinase
- NK
natural killer
- NSCLC
non-small cell lung cancer
- PARP
poly (ADP-ribose) polymerase
- PCa
prostate cancer
- PI3K
phosphoinositide 3-kinase
- PTC
papillary thyroid carcinoma
- PTEN
phosphatase and tensin homology deleted on chromosome ten
- RISC
RNA-induced silencing complex
- SOCS3
suppressor cytokine signaling 3
- TIMP3
tissue inhibitor of metalloproteinases 3
- TRPS1
tricho-rhino-pharyngeal syndrome type 1
- ZEB1
zinc finger E-box binding homeobox 1
- 3′-UTR
3′-untranslated region
Funding Statement
The present study was supported by a grant from the National Natural Science Foundation of China Youth Science Foundation Project (grant no. 81802571) and the Zhejiang Medical and Health Science and Technology Project (grant no. 2019RC039).
Availability of data and materials
Not applicable.
Authors' contributions
DW, YS and TS wrote the manuscript. PK and YC consulted relevant literature and completed English revision. TS, LZ and YD completed the figures and tables. WL and ZT contributed to conception and design of the framework. WL completed critical revisions and proofread the manuscript. All authors have checked all the raw data to ensure its legitimacy and have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Orso F, Quirico L, Dettori D, Coppo R, Virga F, Ferreira LC, Paoletti C, Baruffaldi D, Penna E, Taverna D. Role of miRNAs in tumor and endothelial cell interactions during tumor progression. Semin Cancer Biol. 2020;60:214–224. doi: 10.1016/j.semcancer.2019.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Iwakawa HO, Tomari Y. The functions of MicroRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015;25:651–665. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 3.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 4.Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6:235–246. doi: 10.1158/2159-8290.CD-15-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20:21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20:5–20. doi: 10.1038/s41580-018-0059-1. [DOI] [PubMed] [Google Scholar]
- 7.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 8.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao JJ, Chu ZB, Hu Y, Lin J, Wang Z, Jiang M, Chen M, Wang X, Kang Y, Zhou Y, et al. Targeting the miR-221-222/PUMA/BAK/BAX pathway abrogates dexamethasone resistance in multiple myeloma. Cancer Res. 2015;75:4384–4397. doi: 10.1158/0008-5472.CAN-15-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang KW, Kao SY, Wu YH, Tsai MM, Tu HF, Liu CJ, Lui MT, Lin SC. Passenger strand miRNA miR-31* regulates the phenotypes of oral cancer cells by targeting RhoA. Oral Oncol. 2013;49:27–33. doi: 10.1016/j.oraloncology.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Ogawa T, Enomoto M, Fujii H, Sekiya Y, Yoshizato K, Ikeda K, Kawada N. MicroRNA-221/222 upregulation indicates the activation of stellate cells and the progression of liver fibrosis. Gut. 2012;61:1600–1609. doi: 10.1136/gutjnl-2011-300717. [DOI] [PubMed] [Google Scholar]
- 12.Yasmeen S, Kaur S, Mirza AH, Brodin B, Pociot F, Kruuse C. miRNA-27a-3p and miRNA-222-3p as novel modulators of phosphodiesterase 3a (PDE3A) in cerebral microvascular endothelial cells. Mol Neurobiol. 2019;56:5304–5314. doi: 10.1007/s12035-018-1446-5. [DOI] [PubMed] [Google Scholar]
- 13.Gulluoglu S, Tuysuz EC, Kuskucu A, Ture U, Atalay B, Sahin F, Bayrak OF. The potential function of microRNA in chordomas. Gene. 2016;585:76–83. doi: 10.1016/j.gene.2016.03.032. [DOI] [PubMed] [Google Scholar]
- 14.Wu C, Liu Z, Ma L, Pei C, Qin L, Gao N, Li J, Yin Y. MiRNAs regulate oxidative stress related genes via binding to the 3'UTR and TATA-box regions: A new hypothesis for cataract pathogenesis. BMC Ophthalmol. 2017;17:2–8. doi: 10.1186/s12886-017-0537-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verjans R, Peters T, Beaumont FJ, van Leeuwen R, van Herwaarden T, Verhesen W, Munts C, Bijnen M, Henkens M, Diez J, et al. MicroRNA-221/222 family counteracts myocardial fibrosis in pressure overload-induced heart failure. Hypertension. 2018;71:280–288. doi: 10.1161/HYPERTENSIONAHA.117.10094. [DOI] [PubMed] [Google Scholar]
- 16.Wei F, Ma C, Zhou T, Dong X, Luo Q, Geng L, Ding L, Zhang Y, Zhang L, Li N, et al. Exosomes derived from gemcitabine-resistant cells transfer malignant phenotypic traits via delivery of miRNA-222-3p. Mol Cancer. 2017;16:132–147. doi: 10.1186/s12943-017-0694-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coarfa C, Fiskus W, Eedunuri VK, Rajapakshe K, Foley C, Chew SA, Shah SS, Geng C, Shou J, Mohamed JS, et al. Comprehensive proteomic profiling identifies the androgen receptor axis and other signaling pathways as targets of microRNAs suppressed in metastatic prostate cancer. Oncogene. 2016;35:2345–2356. doi: 10.1038/onc.2015.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L, Wang HJ, Meng T, Lei C, Yang XH, Wang QS, Jin B, Zhu JF. lncRNA GAS5 inhibits cell migration and invasion and promotes autophagy by targeting miR-222-3p via the GAS5/PTEN-signaling pathway in CRC. Mol Ther Nucleic Acids. 2019;17:644–656. doi: 10.1016/j.omtn.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Jafri MA, Al-Qahtani MH, Shay JW. Role of miRNAs in human cancer metastasis: Implications for therapeutic intervention. Semin Cancer Biol. 2017;44:117–131. doi: 10.1016/j.semcancer.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Alves Dos Santos K, Clemente Dos Santos IC, Santos Silva C, Gomes Ribeiro H, de Farias Domingos I, Nogueira Silbiger V. Circulating exosomal miRNAs as biomarkers for the diagnosis and prognosis of Colorectal Cancer. 2020;22:346. doi: 10.3390/ijms22010346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fong M, Yan W, Ghassemian M, Wu X, Zhou X, Cao M, Jiang L, Wang J, Liu X, Zhang J, Wang SJ. Cancer-secreted miRNAs regulate amino-acid-induced mTORC1 signaling and fibroblast protein synthesis. EMBO Rep. 2020;22:e51239. doi: 10.15252/embr.202051239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Liao X, Huang K, Zeng X, Liu Z, Zhou X, Yu T, Yang C, Yu L, Wang Q, et al. Clustered microRNAs hsa-miR-221-3p/hsa-miR-222-3p and their targeted genes might be prognostic predictors for hepatocellular carcinoma. J Cancer. 2019;10:2520–2533. doi: 10.7150/jca.29207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Deng Z, Chen X, Cai J, Ma T, Zhong Q, Li R, Li L, Li T. Downregulation of miR-222-3p reverses doxorubicin-resistance in LoVo cells through upregulating forkhead box protein P2 (FOXP2) protein. Med Sci Monit. 2019;25:2169–2178. doi: 10.12659/MSM.913325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo J, Liu Q, Li Z, Guo H, Bai C, Wang F. miR-222-3p promotes osteosarcoma cell migration and invasion through targeting TIMP3. Onco Targets Ther. 2018;11:8643–8653. doi: 10.2147/OTT.S175745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu B, Che Q, Qiu H, Bao W, Chen X, Lu W, Li B, Wan X. Elevated MiR-222-3p promotes proliferation and invasion of endometrial carcinoma via targeting ERalpha. PLoS One. 2014;9:e87563. doi: 10.1371/journal.pone.0087563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ostenfeld MS, Jensen SG, Jeppesen DK, Christensen LL, Thorsen SB, Stenvang J, Hvam ML, Thomsen A, Mouritzen P, Rasmussen MH, et al. miRNA profiling of circulating EpCAM(+) extracellular vesicles: Promising biomarkers of colorectal cancer. J Extracell Vesicles. 2016;5:3402–3417. doi: 10.3402/jev.v5.31488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korabecna M, Koutova L, Tesarova P. The potential roles of vesicle-enclosed miRNAs in communication between macrophages and cancer cells in tumor microenvironment. Neoplasma. 2017;64:406–411. doi: 10.4149/neo_2017_311. [DOI] [PubMed] [Google Scholar]
- 29.Gasparello J, Papi C, Allegretti M, Giordani E, Carboni F, Zazza S, Pescarmona E, Romania P, Giacomini P, Scapoli C, et al. A distinctive microRNA (miRNA) signature in the blood of colorectal cancer (CRC) patients at surgery. Cancers (Basel) 2020;12:2410. doi: 10.3390/cancers12092410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pudova E, Krasnov G, Nyushko K, Kobelyatskaya A, Savvateeva M, Poloznikov A, Dolotkazin D, Klimina K, Guvatova Z, Simanovsky S, et al. miRNAs expression signature potentially associated with lymphatic dissemination in locally advanced prostate cancer. BMC Med Genomics. 2020;13(Suppl 8):S129. doi: 10.1186/s12920-020-00788-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang K, Li G, Chen W, Song L, Wei T, Li Z, Gong R, Lei J, Shi H, Zhu J. Plasma exosomal miR-146b-5p and miR-222-3p are potential biomarkers for lymph node metastasis in papillary thyroid carcinomas. Onco Targets Ther. 2020;13:1311–1319. doi: 10.2147/OTT.S231361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fredsoe J, Rasmussen AKI, Mouritzen P, Borre M, Orntoft T, Sorensen KD. A five-microRNA model (pCaP) for predicting prostate cancer aggressiveness using cell-free urine. Int J Cancer. 2019;145:2558–2567. doi: 10.1002/ijc.32296. [DOI] [PubMed] [Google Scholar]
- 33.Cheng Y, Cheng T, Zhao Y, Qu Y. HMGA1 exacerbates tumor progression by activating miR-222 through PI3K/Akt/MMP-9 signaling pathway in uveal melanoma. Cell Signal. 2019;63:5. doi: 10.1016/j.cellsig.2019.109386. [DOI] [PubMed] [Google Scholar]
- 34.Zhang XF, Ye Y, Zhao SJ. LncRNA Gas5 acts as a ceRNA to regulate PTEN expression by sponging miR-222-3p in papillary thyroid carcinoma. Oncotarget. 2018;9:3519–3530. doi: 10.18632/oncotarget.23336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan X, Tang H, Bi J, Li N, Jia Y. MicroRNA-222-3p associated with Helicobacter pylori targets HIPK2 to promote cell proliferation, invasion, and inhibits apoptosis in gastric cancer. J Cell Biochem. 2018;119:5153–5162. doi: 10.1002/jcb.26542. [DOI] [PubMed] [Google Scholar]
- 36.Ma S, Kong S, Gu X, Xu Y, Tao M, Shen L, Shen X, Ju S. As a biomarker for gastric cancer, circPTPN22 regulates the progression of gastric cancer through the EMT pathway. Cancer Cell Int. 2021;21:44. doi: 10.1186/s12935-020-01701-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu X, Li Y, Alvero A, Li J, Wu Q, Xiao Q, Peng Y, Hu Y, Li X, Yan W, et al. MicroRNA-222-3p/GNAI2/AKT axis inhibits epithelial ovarian cancer cell growth and associates with good overall survival. Oncotarget. 2016;7:80633–80654. doi: 10.18632/oncotarget.13017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosignolo F, Memeo L, Monzani F, Colarossi C, Pecce V, Verrienti A, Durante C, Grani G, Lamartina L, Forte S, et al. MicroRNA-based molecular classification of papillary thyroid carcinoma. Int J Oncol. 2017;50:1767–1777. doi: 10.3892/ijo.2017.3960. [DOI] [PubMed] [Google Scholar]
- 39.Di Fazio P, Montalbano R, Neureiter D, Alinger B, Schmidt A, Merkel AL, Quint K, Ocker M. Downregulation of HMGA2 by the pan-deacetylase inhibitor panobinostat is dependent on hsa-let-7b expression in liver cancer cell lines. Exp Cell Res. 2012;318:1832–1843. doi: 10.1016/j.yexcr.2012.04.018. [DOI] [PubMed] [Google Scholar]
- 40.Jahanbani I, Al-Abdallah A, Ali RH, Al-Brahim N, Mojiminiyi O. Discriminatory miRNAs for the management of papillary thyroid carcinoma and noninvasive follicular thyroid neoplasms with papillary-like nuclear features. Thyroid. 2018;28:319–327. doi: 10.1089/thy.2017.0127. [DOI] [PubMed] [Google Scholar]
- 41.Denaro M, Ugolini C, Poma AM, Borrelli N, Materazzi G, Piaggi P, Chiarugi M, Miccoli P, Vitti P, Basolo F. Differences in miRNA expression profiles between wild-type and mutated NIFTPs. Endocr Relat Cancer. 2017;24:543–553. doi: 10.1530/ERC-17-0167. [DOI] [PubMed] [Google Scholar]
- 42.Borrelli N, Denaro M, Ugolini C, Poma AM, Miccoli M, Vitti P, Miccoli P, Basolo F. miRNA expression profiling of 'noninvasive follicular thyroid neoplasms with papillary-like nuclear features' compared with adenomas and infiltrative follicular variants of papillary thyroid carcinomas. Mod Pathol. 2017;30:39–51. doi: 10.1038/modpathol.2016.157. [DOI] [PubMed] [Google Scholar]
- 43.de Conti A, Ortega JF, Tryndyak V, Dreval K, Moreno FS, Rusyn I, Beland FA, Pogribny IP. MicroRNA deregulation in nonalcoholic steatohepatitis-associated liver carcinogenesis. Oncotarget. 2017;8:88517–88528. doi: 10.18632/oncotarget.19774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim BH, Hong SW, Kim A, Choi SH, Yoon SO. Prognostic implications for high expression of oncogenic microRNAs in advanced gastric carcinoma. J Surg Oncol. 2013;107:505–510. doi: 10.1002/jso.23271. [DOI] [PubMed] [Google Scholar]
- 45.Zhang L, Huang Z, Zhang H, Zhu M, Zhu W, Zhou X, Liu P. Prognostic value of candidate microRNAs in gastric cancer: A validation study. Cancer Biomark. 2017;18:221–230. doi: 10.3233/CBM-160091. [DOI] [PubMed] [Google Scholar]
- 46.Rinnerthaler G, Hackl H, Gampenrieder SP, Hamacher F, Hufnagl C, Hauser-Kronberger C, Zehentmayr F, Fastner G, Sedlmayer F, Mlineritsch B, Greil R. miR-16-5p is a stably-expressed house-keeping MicroRNA in breast cancer tissues from primary tumors and from metastatic sites. Int J Mol Sci. 2016;17:156–167. doi: 10.3390/ijms17020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fredsoe J, Rasmussen AKI, Thomsen AR, Mouritzen P, Hoyer S, Borre M, Orntoft TF, Sorensen KD. Diagnostic and prognostic MicroRNA biomarkers for prostate cancer in cell-free urine. Eur Urol Focus. 2018;4:825–833. doi: 10.1016/j.euf.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 48.Fang R, Zhu Y, Hu L, Khadka VS, Ai J, Zou H, Ju D, Jiang B, Deng Y, Hu X. Plasma MicroRNA pair panels as novel biomarkers for detection of early stage breast cancer. Front Physiol. 2018;9:1879–1880. doi: 10.3389/fphys.2018.01879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu Z, Qian F, Yang X, Jiang H, Chen Y, Liu S. Circulating miR-222 in plasma and its potential diagnostic and prognostic value in gastric cancer. Med Oncol. 2014;31:164–175. doi: 10.1007/s12032-014-0164-8. [DOI] [PubMed] [Google Scholar]
- 50.Chang YA, Weng SL, Yang SF, Chou CH, Huang WC, Tu SJ, Chang TH, Huang CN, Jong YJ, Huang HD. A Three-MicroRNA signature as a potential biomarker for the early detection of oral cancer. Int J Mol Sci. 2018;19:758. doi: 10.3390/ijms19030758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fredsoe J, Rasmussen AKI, Laursen EB, Cai Y, Howard KA, Pedersen BG, Borre M, Mouritzen P, Orntoft T, Sorensen KD. Independent validation of a diagnostic noninvasive 3-MicroRNA ratio model (uCaP) for prostate cancer in cell-free urine. Clin Chem. 2019;65:540–548. doi: 10.1373/clinchem.2018.296681. [DOI] [PubMed] [Google Scholar]
- 52.Uchino K, Takeshita F, Takahashi RU, Kosaka N, Fujiwara K, Naruoka H, Sonoke S, Yano J, Sasaki H, Nozawa S, et al. Therapeutic effects of microRNA-582-5p and -3p on the inhibition of bladder cancer progression. Mol Ther. 2013;21:610–619. doi: 10.1038/mt.2012.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan X, Tang H, Bi J, Li N, Jia Y. MicroRNA-222-3p associated with Helicobacter pylori targets HIPK2 to promote cell proliferation, invasion, and inhibits apoptosis in gastric cancer. J Cell Biochem. 2018;119:5153–5162. doi: 10.1002/jcb.26542. [DOI] [PubMed] [Google Scholar]
- 54.Rosignolo F, Sponziello M, Giacomelli L, Russo D, Pecce V, Biffoni M, Bellantone R, Lombardi CP, Lamartina L, Grani G, et al. Identification of thyroid-associated serum microRNA profiles and their potential use in thyroid cancer follow-up. J Endocr Soc. 2017;1:3–13. doi: 10.1210/js.2016-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ulivi P, Petracci E, Marisi G, Baglivo S, Chiari R, Billi M, Canale M, Pasini L, Racanicchi S, Vagheggini A, et al. Prognostic role of circulating miRNAs in Early-stage non-small cell lung cancer. J Clin Med. 2019;8:131–142. doi: 10.3390/jcm8020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y, Yin W, Lin Y, Yin K, Zhou L, Du Y, Yan T, Lu J. Downregulated circulating microRNAs after surgery: Potential noninvasive biomarkers for diagnosis and prognosis of early breast cancer. Cell Death Discov. 2018;4:2–8. doi: 10.1038/s41421-017-0003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kara M, Yumrutas O, Ozcan O, Celik OI, Bozgeyik E, Bozgeyik I, Tasdemir S. Differential expressions of cancer-associated genes and their regulatory miRNAs in colorectal carcinoma. Gene. 2015;567:81–86. doi: 10.1016/j.gene.2015.04.065. [DOI] [PubMed] [Google Scholar]
- 58.Spindler KL, Pallisgaard N, Vogelius I, Jakobsen A. Quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res. 2012;18:1177–1185. doi: 10.1158/1078-0432.CCR-11-0564. [DOI] [PubMed] [Google Scholar]
- 59.Domingo E, Camps C, Kaisaki PJ, Parsons MJ, Mouradov D, Pentony MM, Makino S, Palmieri M, Ward RL, Hawkins NJ, et al. Mutation burden and other molecular markers of prognosis in colorectal cancer treated with curative intent: Results from the QUASAR 2 clinical trial and an Australian community-based series. Lancet Gastroenterol Hepatol. 2018;3:635–643. doi: 10.1016/S2468-1253(18)30117-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao H, Shen J, Hodges TR, Song R, Fuller GN, Heimberger AB. Serum microRNA profiling in patients with glioblastoma: A survival analysis. Mol Cancer. 2017;16:59–70. doi: 10.1186/s12943-017-0628-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cooper J, Giancotti FG. Integrin signaling in cancer: Mechanotransduction, stemness, epithelial plasticity, and therapeutic resistance. Cancer Cell. 2019;35:347–367. doi: 10.1016/j.ccell.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Friedmann Angeli JP, Krysko DV, Conrad M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat Rev Cancer. 2019;19:405–414. doi: 10.1038/s41568-019-0149-1. [DOI] [PubMed] [Google Scholar]
- 63.Tooker P, Yen WC, Ng SC, Negro-Vilar A, Hermann TW. Bexarotene (LGD1069, Targretin), a selective retinoid X receptor agonist, prevents and reverses gemcitabine resistance in NSCLC cells by modulating gene amplification. Cancer Res. 2007;67:4425–4433. doi: 10.1158/0008-5472.CAN-06-4495. [DOI] [PubMed] [Google Scholar]
- 64.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 65.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 66.Garofalo M, Romano G, Di Leva G, Nuovo G, Jeon YJ, Ngankeu A, Sun J, Lovat F, Alder H, Condorelli G, et al. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med. 2011;18:74–82. doi: 10.1038/nm.2577. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Liu S, Sun X, Wang M, Hou Y, Zhan Y, Jiang Y, Liu Z, Cao X, Chen P, Chen X, et al. A microRNA 221- and 222-mediated feedback loop maintains constitutive activation of NFκB and STAT3 in colorectal cancer cells. Gastroenterology. 2014;147:847–859.e11. doi: 10.1053/j.gastro.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, Zucman-Rossi J. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 69.Li Z, Yu Z, Meng X, Zhou S, Xiao S, Li X, Liu S, Yu P. Long noncoding RNA GAS5 impairs the proliferation and invasion of endometrial carcinoma induced by high glucose via targeting miR-222-3p/p27. Am J Transl Res. 2019;11:2413–2421. [PMC free article] [PubMed] [Google Scholar]
- 70.Paquet-Fifield S, Koh SL, Cheng L, Beyit LM, Shembrey C, Molck C, Behrenbruch C, Papin M, Gironella M, Guelfi S, et al. Tight junction protein Claudin-2 promotes Self-renewal of human colorectal cancer Stem-like cells. Cancer Res. 2018;78:2925–2938. doi: 10.1158/0008-5472.CAN-17-1869. [DOI] [PubMed] [Google Scholar]
- 71.Polk DB, Peek RM., Jr Helicobacter pylori: Gastric cancer and beyond. Nat Rev Cancer. 2010;10:403–414. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ishiguro H, Kimura M, Takeyama H. Role of microRNAs in gastric cancer. World J Gastroenterol. 2014;20:5694–5699. doi: 10.3748/wjg.v20.i19.5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2016;291:31079–31086. doi: 10.1074/jbc.A116.806041. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Sun K, Wang W, Zeng JJ, Wu CT, Lei ST, Li GX. MicroRNA-221 inhibits CDKN1C/p57 expression in human colorectal carcinoma. Acta Pharmacol Sin. 2011;32:375–384. doi: 10.1038/aps.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, Ma T, Yang S, Xia M, Xu J, An H, Yang Y, Li S. High-mobility group A1 proteins enhance the expression of the oncogenic miR-222 in lung cancer cells. Mol Cell Biochem. 2011;357:363–371. doi: 10.1007/s11010-011-0907-1. [DOI] [PubMed] [Google Scholar]
- 78.Ying SY, Chang DC, Miller JD, Lin SL. The microRNA: Overview of the RNA gene that modulates gene functions. Methods Mol Biol. 2006;342:1–18. doi: 10.1385/1-59745-123-1:1. [DOI] [PubMed] [Google Scholar]
- 79.Fuse M, Kojima S, Enokida H, Chiyomaru T, Yoshino H, Nohata N, Kinoshita T, Sakamoto S, Naya Y, Nakagawa M, et al. Tumor suppressive microRNAs (miR-222 and miR-31) regulate molecular pathways based on microRNA expression signature in prostate cancer. J Hum Genet. 2012;57:691–699. doi: 10.1038/jhg.2012.95. [DOI] [PubMed] [Google Scholar]
- 80.Ottley EC, Nicholson HD, Gold EJ. Activin A regulates microRNAs and gene expression in LNCaP cells. Prostate. 2016;76:951–963. doi: 10.1002/pros.23184. [DOI] [PubMed] [Google Scholar]
- 81.Tong AW, Fulgham P, Jay C, Chen P, Khalil I, Liu S, Senzer N, Eklund AC, Han J, Nemunaitis J. MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther. 2009;16:206–216. doi: 10.1038/cgt.2008.77. [DOI] [PubMed] [Google Scholar]
- 82.Ottley EC, Nicholson HD, Gold EJ. Activin A regulates microRNAs and gene expression in LNCaP cells. Prostate. 2016;76:951–963. doi: 10.1002/pros.23184. [DOI] [PubMed] [Google Scholar]
- 83.Xu G, Wu J, Zhou L, Chen B, Sun Z, Zhao F, Tao Z. Characterization of the small RNA transcriptomes of androgen dependent and independent prostate cancer cell line by deep sequencing. PLoS One. 2010;5:e15519. doi: 10.1371/journal.pone.0015519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rihani A, Van Goethem A, Ongenaert M, De Brouwer S, Volders PJ, Agarwal S, De Preter K, Mestdagh P, Shohet J, Speleman F, et al. Genome wide expression profiling of p53 regulated miRNAs in neuroblastoma. Sci Rep. 2015;5:9027–9044. doi: 10.1038/srep09027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Narrandes S, Xu W. Gene expression detection assay for cancer clinical use. J Cancer. 2018;9:2249–2265. doi: 10.7150/jca.24744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bourboulia D, Stetler-Stevenson WG. Matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases (TIMPs): Positive and negative regulators in tumor cell adhesion. Semin Cancer Biol. 2010;20:161–168. doi: 10.1016/j.semcancer.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu W, Wang X, Wang Y, Dai Y, Xie Y, Ping Y, Yin B, Yu P, Liu Z, Duan X, et al. SGK1 inhibition-induced autophagy impairs prostate cancer metastasis by reversing. EMT J Exp Clin Cancer Res. 2018;37:73. doi: 10.1186/s13046-018-0743-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu W, Wang X, Liu Z, Wang Y, Yin B, Yu P, Duan X, Liao Z, Chen Y, Liu C, et al. SGK1 inhibition induces autophagy-dependent apoptosis via the mTOR-Foxo3a pathway. Br J Cancer. 2017;117:1139–1153. doi: 10.1038/bjc.2017.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Indran IR, Tufo G, Pervaiz S, Brenner C. Recent advances in apoptosis, mitochondria and drug resistance in cancer cells. Biochim Biophys Acta. 2011;1807:735–745. doi: 10.1016/j.bbabio.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 90.Mavrogiannis A, Kokkinopoulou I, Kontos C, Sideris DJ. Effect of vinca alkaloids on the expression levels of microRNAs targeting apoptosis-related genes in breast cancer cell lines. Curr Pharm Biotechnol. 2018;19:1076–1086. doi: 10.2174/1389201019666181112103204. [DOI] [PubMed] [Google Scholar]
- 91.Jacob H, Stanisavljevic L, Storli KE, Hestetun KE, Dahl O, Myklebust MP. Identification of a sixteen-microRNA signature as prognostic biomarker for stage II and III colon cancer. Oncotarget. 2017;8:87837–87847. doi: 10.18632/oncotarget.21237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tan HY, Wang N, Lam W, Guo W, Feng Y, Cheng YC. Targeting tumour microenvironment by tyrosine kinase inhibitor. Mol Cancer. 2018;17:43–52. doi: 10.1186/s12943-018-0800-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vuong L, Kotecha RR, Voss MH, Hakimi AA. Tumor microenvironment dynamics in clear-cell renal cell carcinoma. Cancer Discov. 2019;9:1349–1357. doi: 10.1158/2159-8290.CD-19-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vitale I, Manic G, Coussens LM, Kroemer G, Galluzzi L. Macrophages and metabolism in the tumor microenvironment. Cell Metab. 2019;30:36–50. doi: 10.1016/j.cmet.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 95.Zhou S, Liu R, Yuan K, Yi T, Zhao X, Huang C, Wei Y. Proteomics analysis of tumor microenvironment: Implications of metabolic and oxidative stresses in tumorigenesis. Mass Spectrom Rev. 2013;32:267–311. doi: 10.1002/mas.21362. [DOI] [PubMed] [Google Scholar]
- 96.Ying X, Wu Q, Wu X, Zhu Q, Wang X, Jiang L, Chen X, Wang X. Epithelial ovarian cancer-secreted exosomal miR-222-3p induces polarization of tumor-associated macro-phages. Oncotarget. 2016;7:43076–43087. doi: 10.18632/oncotarget.9246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cabibbo G, Celsa C, Calvaruso V, Petta S, Cacciola I, Cannavo MR, Madonia S, Rossi M, Magro B, Rini F, et al. Direct-acting antivirals after successful treatment of early hepatocellular carcinoma improve survival in HCV-cirrhotic patients. J Hepatol. 2019;71:265–273. doi: 10.1016/j.jhep.2019.03.027. [DOI] [PubMed] [Google Scholar]
- 98.Degasperi E, D'Ambrosio R, Iavarone M, Sangiovanni A, Aghemo A, Soffredini R, Borghi M, Lunghi G, Colombo M, Lampertico P. Factors associated with increased risk of de novo or recurrent hepatocellular carcinoma in patients with cirrhosis treated with direct-acting antivirals for HCV infection. Clin Gastroenterol Hepatol. 2019;17:1183–1191.e7. doi: 10.1016/j.cgh.2018.10.038. [DOI] [PubMed] [Google Scholar]
- 99.Santangelo L, Bordoni V, Montaldo C, Cimini E, Zingoni A, Battistelli C, D'Offizi G, Capobianchi MR, Santoni A, Tripodi M, Agrati C. Hepatitis C virus direct-acting antivirals therapy impacts on extracellular vesicles microRNAs content and on their immunomodulating properties. Liver Int. 2018;38:1741–1750. doi: 10.1111/liv.13700. [DOI] [PubMed] [Google Scholar]
- 100.Varchetta S, Mele D, Mantovani S, Oliviero B, Cremonesi E, Ludovisi S, Michelone G, Alessiani M, Rosati R, Montorsi M, Mondelli MU. Impaired intrahepatic natural killer cell cytotoxic function in chronic hepatitis C virus infection. Hepatology. 2012;56:841–849. doi: 10.1002/hep.25723. [DOI] [PubMed] [Google Scholar]
- 101.van der Meer AJ, Feld JJ, Hofer H, Almasio PL, Calvaruso V, Fernandez-Rodriguez CM, Aleman S, Ganne-Carrie N, D'Ambrosio R, Pol S, et al. Risk of cirrhosis-related complications in patients with advanced fibrosis following hepatitis C virus eradication. J Hepatol. 2017;66:485–493. doi: 10.1016/j.jhep.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 102.Fugier E, Marche H, Thélu MA, Macek Jilková Z, Van Campenhout N, Dufeu-Duchesne T, Leroy V, Zarski JP, Sturm N, Marche PN, Jouvin-Marche E. Functions of liver natural killer cells are dependent on the severity of liver inflammation and fibrosis in chronic hepatitis C. PLoS One. 2014;9:e95614. doi: 10.1371/journal.pone.0095614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tölle A, Jung K, Friedersdorff F, Maxeiner A, Lein M, Fendler A, Stephan C. The discriminative ability of Prostate Health Index to detect prostate cancer is enhanced in combination with miR-222-3p. Cancer Biomark. 2020 Dec 15; doi: 10.3233/CBM-201600. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ryu K, Lee J, Choi M, Yoon S, Cho J, Ko Y, Shim J, Kim W, Park C, Kim SJ. Serum-derived exosomal MicroRNA profiles can predict poor survival outcomes in patients with extranodal natural Killer/T-cell lymphoma. Cancers (Basel) 2020;12:3548. doi: 10.3390/cancers12123548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhai S, Xu Z, Xie J, Zhang J, Wang X, Peng C, Li H, Chen H, Shen B, Deng X. Epigenetic silencing of LncRNA LINC00261 promotes c-myc-mediated aerobic glycolysis by regulating miR-222-3p/HIPK2/ERK axis and sequestering IGF2BP1. Oncogene. 2021;40:277–291. doi: 10.1038/s41388-020-01525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lu B, Sheng Y, Zhang J, Zheng Z, Ji L. The altered microRNA profile in andrographolide-induced inhibition of hepatoma tumor growth. Gene. 2016;588:124–133. doi: 10.1016/j.gene.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 107.Gumbiner BM, Kim NG. The Hippo-YAP signaling pathway and contact inhibition of growth. J Cell Sci. 2014;127:709–717. doi: 10.1242/jcs.140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Panneerselvam J, Srivastava A, Muralidharan R, Wang Q, Zheng W, Zhao L, Chen A, Zhao YD, Munshi A, Ramesh R. IL-24 modulates the high mobility group (HMG) A1/miR222/AKT signaling in lung cancer cells. Oncotarget. 2016;7:70247–70263. doi: 10.18632/oncotarget.11838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ignacio C, Mooney SM, Middleton FA. Effects of acute prenatal exposure to ethanol on microRNA expression are ameliorated by social enrichment. Front Pediatr. 2014;2:103. doi: 10.3389/fped.2014.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Théry C, Zitvogel L, Amigorena S. Exosomes: Composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 111.Pant S, Hilton H, Burczynski ME. The multifaceted exosome: Biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 2012;83:1484–1494. doi: 10.1016/j.bcp.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang G, Song J, Li Z, Zhang Z, Yuan W. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17:147. doi: 10.1186/s12943-018-0897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Han Z, Li Y, Zhang J, Guo C, Li Q, Zhang X, Lan Y, Gu W, Xing Z, Liang L, et al. Tumor-derived circulating exosomal miR-342-5p and miR-574-5p as promising diagnostic biomarkers for early-stage Lung Adenocarcino. Int J Med Sci. 2020;17:1428–1438. doi: 10.7150/ijms.43500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ortega MM, Bouamar H. Guidelines on designing MicroRNA sponges: From construction to stable cell line. Methods Mol Biol. 2017;1509:221–233. doi: 10.1007/978-1-4939-6524-3_20. [DOI] [PubMed] [Google Scholar]
- 116.Wang Z. The guideline of the design and validation of MiRNA mimics. Methods Mol Biol. 2011;676:211–223. doi: 10.1007/978-1-60761-863-8_15. [DOI] [PubMed] [Google Scholar]
- 117.Arroyo J, Gallichotte E, Tewari M. Systematic design and functional analysis of artificial microRNAs. Nucleic Acids Res. 2014;42:6064–6077. doi: 10.1093/nar/gku171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ganju A, Khan S, Hafeez BB, Behrman SW, Yallapu MM, Chauhan SC, Jaggi M. miRNA nanotherapeutics for cancer. Drug Discov Today. 2017;22:424–432. doi: 10.1016/j.drudis.2016.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen Y, Gao DY, Huang L. In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv Drug Deliv Rev. 2015;81:128–141. doi: 10.1016/j.addr.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bofill-De Ros X, Gu S. Guidelines for the optimal design of miRNA-based shRNAs. Methods. 2016;103:157–166. doi: 10.1016/j.ymeth.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.