Abstract
Long non-coding RNA LincIN has been reported to be overexpressed and to be involved in the metastasis of breast cancer. However, the expression and role of LincIN in esophageal squamous cell carcinoma (ESCC) remain unsolved. In the present study, LincIN expression was examined in ESCC by RT-qPCR, and the roles of LincIN in ESCC were determined using cell growth, migration and invasion assays. In addition, the effects of LincIN on nuclear factor 90 (NF90) and microRNA/miR (miR)-7 were examined by RNA immunoprecipitation assay, RT-qPCR, dual-luciferase reporter assay and western blot analysis. The results revealed that LincIN expression was significantly increased in ESCC tissues and cell lines. The increased expression of LincIN was positively associated with invasion depth, lymph node metastasis, TNM stage and a poor prognosis. Functional assays revealed that the overexpression of LincIN promoted ESCC cell growth, migration and invasion. Mechanistic analysis revealed that LincIN physically bound to NF90, enhanced the binding between NF90 and primary miR-7 (pri-miR-7), and further enhanced the inhibitory effects of NF90 on miR-7 biogenesis. Therefore, LincIN downregulated miR-7 expression in ESCC. The expression of miR-7 inversely correlated with that of LincIN in ESCC tissues. By downregulating miR-7, LincIN increased the expression of HOXB13, a target of miR-7. The overexpression of miR-7 or the depletion of HOXB13 both attenuated the tumor-promoting roles of LincIN in ESCC cell growth, migration and invasion. On the whole, the findings of the present study suggest that LincIN is overexpressed and plays an oncogenic role in ESCC via the regulation of the NF90/miR-7/HOXB13 axis. Thus, LincIN may prove to be a promising prognostic biomarker and therapeutic target for ESCC.
Keywords: non-coding RNA, esophageal carcinoma, proliferation, migration, invasion, nuclear factor 90, microRNA biogenesis
Introduction
Esophageal carcinoma is one of the most aggressive gastrointestinal malignancies and accounts for the sixth leading cause of cancer-related mortality worldwide; there were 572,000 estimated new cases and 509,000 estimated deaths in 2018 worldwide (1). Esophageal squamous cell carcinoma (ESCC) is the major histological type, which accounts for approximately 90% of all esophageal carcinoma cases (2). Due to the difficulty in the early diagnosis of ESCC, the majority of patients with ESCC are not suitable for radical surgical resection (3). The prognoses of patients with ESCC are extremely poor (4). Further revealing the critical molecular mechanisms underlying the initiation and development of ESCC would promote the findings of novel therapeutic targets and prognostic biomarkers for ESCC.
Genome and transcriptome sequencings have found that the majority of the human genome is transcribed; however, only approximately 2% of the human genome encodes for proteins (5). Non-coding RNAs comprise >90% of the human transcriptome (6). Long non-coding RNAs (lncRNAs) and microRNAs (miRNAs or miRs) are two classes of important regulatory non-coding RNAs, which have been revealed to play critical roles in various pathophysiological statuses (6,7). lncRNAs are a class of RNAs with >200 nucleotides in length and limited protein-coding potential (8). Accumulating evidence has indicated that a number of lncRNAs are dysregulated and play important roles in various diseases, including cancers (9-17). lncRNAs have been revealed to regulate various biological functions of cancer cells, such as growth, cycle, apoptosis, migration, invasion, senescence, drug resistance and others (18-25). The mechanisms of action of lncRNAs are diverse. lncRNAs can directly bind proteins, mRNAs, miRNAs, or DNAs, and further modulate the location, expression and function of the interacted partners (26-29).
miRNAs are another class of regulatory non-coding RNAs with 19-25 nucleotides in length (30). Consistent with lncRNAs, a number of miRNAs have been revealed to be dysregulated and to play important roles in various diseases (31-35). miRNAs negatively regulate the expression of their targets by binding to the 3′-untranslated regions (3′UTRs) of target mRNAs, and further causing translational inhibition and/or target mRNAs degradation (36-39).
lncRNA LincIN (long intergenic non-coding RNA between ITGB1 and NRP1) was recently identified to be overexpressed in human breast tumors and to be associated with a poor prognosis of patients with breast cancer (40). Furthermore, LincIN was revealed to promote breast tumor cell migration and invasion by binding nuclear factor 90 (NF90) (40). However, the expression, clinical significance, biological role and mechanisms of action of LincIN in ESCC remain unknown.
In the present study, LincIN expression was measured in ESCC by RT-qPCR, and the roles of LincIN in ESCC were detected using cell growth, migration and invasion assays. In addition, and the mechanisms of action of LincIN were investigated by RNA immunoprecipitation assay, RT-qPCR, dual-luciferase reporter assay and western blot analysis.
Materials and methods
Clinical specimens
A total of 56 pairs of ESCC tissues and adjacent noncancerous tissue specimens were acquired from patients with ESCC who received surgical resection at Nanfang Hospital, Southern Medical University (Guangzhou, China) between February, 2016 and December, 2017. The clinical specimens were immediately frozen in liquid nitrogen and stored at −80°C after surgery. All clinical specimens were diagnosed by histopathological detection. Written informed consent was acquired from all participants. The use of clinical specimens was reviewed and approved by the Nanfang Hospital Institutional Review Board (Guangzhou, China).
Cells and cell culture
The human immortalized normal esophageal epithelial cell line, Het-1A, was acquired from the American Type Culture Collection (ATCC, cat. no. CRL-2692). The ESCC cell lines, TE-1 (cat. no. TCHu 89), KYSE150 (cat. no. TCHu 236) and Eca-109 (cat. no. TCHu 69), were acquired from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Het-1A cells were maintained in BEBM (Lonza Group, Ltd.). ESCC cell lines were maintained in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.). All cells were cultured in medium supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at a humidified incubator with 5% CO2 at 37°C.
Plasmid construction
LincIN full-length sequences were PCR-amplified using the Phusion Flash High-Fidelity PCR Master Mix (Thermo Fisher Scientific, Inc.) and the primers, 5′-CCC AAG CTT ACT CTG TAG TCA CCC AGG CT-3′ (sense) and 5′-GCT CTA GAT TCT GTA AAT TAA GTT TAA TGC TG-3′ (antisense). Subsequently, the PCR products were sub-cloned into the HindIII and XbaI sites of the pcDNA™3.1(+) plasmid (Invitrogen; Thermo Fisher Scientific, Inc.). In total, 2 independent oligonucleotides inhibiting LincIN expression were synthesized and inserted into the SuperSilencing shRNA (short hairpin RNA) expression plasmid pGPU6/Neo (GenePharma, Inc.), named LincIN-shRNA1 and LincIN-shRNA2. The targeted sequences of the LincIN shRNAs were as follows: 5′-GAC ATT ATG CAA GGA GAT GGC A-3′ (LincIN-shRNA1) and 5′-CAG TTG GTC ACT CTA CTC AGT-3′ (LincIN-shRNA2) (40). The HOXB13 3′UTR containing miR-7 target sites were PCR-amplified using the Phusion Flash High-Fidelity PCR Master Mix (Thermo Fisher Scientific, Inc.) and the primers, 5′-CGA GCT CCC CTT CCA TTA CAC CTC TCA C-3′ (sense) and 5′-GCT CTA GAT CCT CCT CCT CGT CCT CTT-3′ (antisense). The PCR products were then sub-cloned into the SacI and XbaI sites of pmirGLO plasmid (Promega Corporation).
Transfection of plasmids and miRNAs
miR-7 mimics and miRNAs negative control (NC) were purchased from GenePharma, Inc. HOXB13 specific shRNA (HOXB13-shRNA) and scrambled shRNA which was used as a negative control were purchased from GeneCopoeia, Inc. The transfections of plasmids (2.5 µg) and miRNAs (10 pmol) were carried out using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the protocol. At 48 h following transfection, subsequent experimentation was performed.
Construction of stable cell lines
The LincIN overexpression plasmid and control pcDNA3.1 plasmid were transfected into TE-1 cells. TE-1 cells were established from a well-differentiated human squamous cell carcinoma of the esophagus and had a male karyotype (41). TE-1 has a mutation at codon 272 of p53, leading to p53 activity impairment (42). At 48 h following transfection, the cells were selected with neomycin for 4 weeks to obtain LincIN stably overexpressed and control TE-1 cells. LincIN-specific shRNAs (2.5 µg) and control shRNA (2.5 µg) were transfected into the Eca-109 cells. The Eca-109 cells were derived from a human oesophageal carcinoma in 1973 (43). Eca-109 harbors wild-type p53 (44). At 72 h following transfection, the cells were selected with neomycin for 4 weeks to obtain LincIN stably-depleted and control Eca-109 cells. 2×106 transducing units of miR-7 overexpression lentivirus (GeneCopoeia, Inc.) was infected into LincIN stably overexpressing TE-1 cells. At 96 h following infection, the cells were selected with neomycin and puromycin for 4 weeks to obtain LincIN and miR-7 simultaneously stably overexpressed TE-1 cells. HOXB13-specific shRNAs (2.5 µg) were transfected into LincIN stably overexpressing TE-1 cells. At 72 h following transfection, the cells were selected with neomycin and puromycin for 4 weeks to obtain HOXB13 stably depleted and simultaneously LincIN stably overexpressing TE-1 cells.
Total RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's instructions. The RNA was then used to carry out reverse transcription using the M-MLV Reverse Transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.). The first-stand cDNA generated from reverse transcription was used to carry out quantitative polymerase chain reaction (qPCR) with SYBR Premix Ex Taq™ II (Takara Biotechnology Co., Ltd.) on an ABI7500 System (Applied Biosystems; Thermo Fisher Scientific, Inc.) following the standard SYBR-Green PCR protocols. The qPCR cycling conditions were as follows: 95°C for 30 sec, then 40 cycles at the conditions of 95°C for 5 sec, 60°C for 20 sec, and 72°C for 20 sec. The RT-qPCR primers were as follows: LincIN, 5′-AAG AGG GGA GTG CGG AAC A-3′ (forward) and 5′-TCA CCA GGG ACA CGA GAT G-3′ (reverse); HOXB13, 5′-TGA CTC CCT GTT GCC TGT G-3′ (forward) and 5′-GAA CTT GTT AGC CGC ATA CTC-3′ (reverse); β-actin, 5′-GGG AAA TCG TGC GTG ACA TTA AG-3′ (forward) and 5′-TGT GTT GGC GTA CAG GTC TTT G-3′ (reverse). For the quantification of the expression of miR-7 and pri-miR-7, RT-qPCR was carried out using the TaqMan microRNA assays (Applied Biosystems; Thermo Fisher Scientific, Inc.) and TaqMan pri-miRNA assays (Applied Biosystems; Thermo Fisher Scientific, Inc.) respectively following the manufacturer's protocols. β-actin was used as a reference gene for the quantification of LincIN, HOXB13 and pri-miR-7. U6 was used as a reference gene for the quantification of miR-7. The quantification of RNA expression was calculated using the 2−ΔΔCq method (45).
Western blot analysis
Total proteins were extracted from the TE-1 and Eca-109 cells using RIPA buffer (Beyotime Institute of Biotechnology) supplemented with protease inhibitors (Beyotime Institute of Biotechnology). Protein concentrations were determined by bicinchoninic acid (BCA) assay with the BCA Protein Assay kit (Beyotime Institute of Biotechnology) following the manufacturer's protocol. Equal amounts of proteins (10 µg) were separated by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subsequently transferred onto PVDF membranes (EMD Millipore). After blocking with 5% non-fat milk in Tris-buffered saline with 0.1% Tween-20 (TBST) at room temperature for 2 h, the membranes were incubated with primary antibodies against HOXB13 (1:1,000, cat. no. 90944, Cell Signaling Technology, Inc.) or β-actin (1:10,000, cat T0022, Affinity Biosciences) overnight at 4°C. After washing with TBST buffer, the membranes were incubated with IRDye 680RD goat anti-mouse IgG secondary antibody (1:10,000, cat. no. 925-68070, LI-COR Biosciences) or IRDye 800CW goat anti-rabbit IgG secondary antibody (1:10,000, cat. no. 926-32211, LI-COR Biosciences) at room temperature for 1 h, followed by detection on an Odyssey infrared scanner (LI-COR Biosciences). β-actin was used as a loading control.
Cell growth assays
Cell growth was assessed using a Cell Counting kit-8 (CCK-8) and 5-ethynyl-2-deoxyuridine (EdU) incorporation assays (27). For CCK-8 assay, the TE-1 and Eca-109 cells were seeded at 2×103 cells/well into 96-well plates. After being incubated for 24, 48, or 72 at 37°C, 10 µl CCK-8 solution (Dojindo Laboratories, Inc.) were added to each well. The optical density values at 450 nm were measured with an enzyme-linked immune detector to indicate cell viability. EdU incorporation assay was performed using the Cell-Light™ EdU Apollo® 643 In Vitro Imaging kit (Guangzhou RiboBio Co., Ltd.) following the manufacturer's instructions. The results were detected using a Zeiss photomicroscope (Carl Zeiss AG) and counting was performed based on at least 10 random visual fields.
Transwell cell migration and invasion assays
Cell migration and invasion were assessed using Transwell assays (46). The TE-1 and Eca-109 cells suspended in serum-free medium were plated in the upper chamber of Transwell inserts (8 µm-pore size, EMD Millipore) with Matrigel (for invasion assay) or without Matrigel (for migration assay). Medium containing 20% fetal bovine serum was plated into the lower chamber. Following incubation for 48 h at 37°C, cells remaining on the upper chamber were wiped away using a cotton swab. Cells migrating into the lower surfaces of the Transwell inserts were fixed in methanol and stained with 0.5% crystal violet (Beyotime Institute of Biotechnology) at room temperature for 30 min. The results were detected using a Zeiss photomicroscope and counted based on at least 10 random visual fields.
RNA immunoprecipitation (RIP) assay
RIP assays were carried out in the TE-1 and Eca-109 cells using the EZ-Magna RIP™ RNA Binding Protein Immunoprecipitation kit (EMD Millipore) and primary antibody against NF90 (5 µg, ab131004, Abcam). The enriched RNA (LincIN and pri-miR-7) was quantified by RT-qPCR as described above.
Dual-luciferase reporter assay
Luciferase reporter plasmid pmirGLO containing HOXB13 3′UTR (50 ng) was co-transfected with LincIN overexpression plasmid (50 ng) and miR-7 mimics (1 pmol) into TE-1 cells. Luciferase reporter plasmid pmirGLO containing HOXB13 3′UTR (50 ng) was co-transfected with LincIN specific shRNAs (50 ng) into Eca-109 cells. Following incubation for 48 h at 37°C, the Firefly luciferase and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega Corporation).
Statistical analysis
GraphPad Prism software (version 5.0) was employed to carry out statistical analyses. For comparisons, the Wilcoxon signed-rank test, Pearson's Chi-squared test, Log-rank test, one-way ANOVA followed by Dunnett's multiple comparison test, the Student's t-test, and Pearson's correlation analysis were performed as indicated. A value of P<0.05 was considered to indicate a statistically significant difference.
Results
LincIN expression is increased in ESCC and is associated with a poor prognosis of patients with ESCC
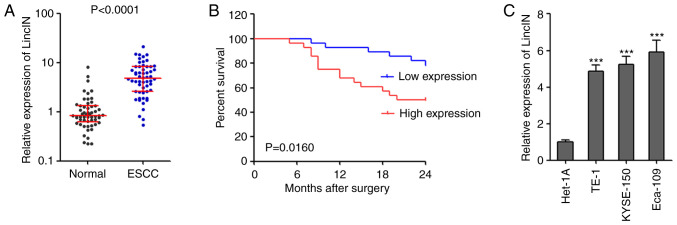
To confirm the expression pattern of LincIN in ESCC, the expression of LincIN was examined in 56 pairs of ESCC tissues and adjacent non-cancerous tissues by RT-qPCR. The results revealed that the expression of LincIN was significantly increased in ESCC tissues compared with adjacent non-cancerous tissues (Fig. 1A). The analyses of the clinicopathological characteristics of these 56 ESCC cases revealed that the increased expression of LincIN was positively associated with an advanced tumor invasion depth, lymph node metastasis and TNM stage (Table I). Patients with ESCC with a higher expression of LincIN had a shorter survival time than those with a lower expression of LincIN (Fig. 1B). In addition, the expression of LincIN was significantly increased in the ESCC cell lines, TE-1, KYSE150 and Eca-109, compared with the normal esophageal epithelial cell line, Het-1A (Fig. 1C). Collectively, these results suggested that LincIN exprewssion was increased in ESCC tissues and cell lines, and was associated with an advanced clinical stage and a poor prognosis of patients with ESCC.
Figure 1.
LincIN expression is increased in ESCC and is associated with a poor prognosis of patients with ESCC. (A) The expression of LincIN in 56 pairs of ESCC tissues and adjacent noncancerous tissues was detected by RT-qPCR. P<0.0001, determined by Wilcoxon signed-rank test. (B) Kaplan-Meier survival analysis of the association between LincIN expression level and the overall survival of 56 patients with ESCC. P=0.0160, determined by the log-rank test. (C) The expression of LincIN in immortalized normal esophageal epithelial cell line, Het-1A, and the ESCC cell lines, TE-1, KYSE150 and Eca-109, was detected by RT-qPCR. Results are shown as the means ± SD from 3 separate experiments. ***P<0.001, vs. control Het-1A cells, determined by one-way ANOVA followed by Dunnett's multiple comparison test vs. Het-1A group. ESCC, esophageal squamous cell carcinoma.
Table I.
Association between LincIN expression and the clinicopathological characteristics of patients with ESCC (n=56).
| Characteristics | Total | LincIN expression
|
P-value | |
|---|---|---|---|---|
| Low | High | |||
| Sex | 1.000 | |||
| Male | 30 | 15 | 15 | |
| Female | 26 | 13 | 13 | |
| Age (years) | 0.788 | |||
| ≤60 | 25 | 13 | 12 | |
| >60 | 31 | 15 | 16 | |
| Differentiation | 0.158 | |||
| Well or moderate | 37 | 21 | 16 | |
| Poor | 19 | 7 | 12 | |
| Tumor invasion depth (T) | 0.032 | |||
| T1/T2 | 30 | 19 | 11 | |
| T3/T4 | 26 | 9 | 17 | |
| Lymph node metastasis (N) | 0.029 | |||
| N0 | 34 | 21 | 13 | |
| N1-N3 | 22 | 7 | 15 | |
| TNM stage | 0.043 | |||
| I | 26 | 17 | 9 | |
| II | 17 | 8 | 9 | |
| III | 13 | 3 | 10 | |
P-values were calculated using Pearson's Chi-squared test. Values in bold font indicate statistically significant differences (P<0.05). ESCC, esophageal squamous cell carcinoma.
Overexpression of LincIN promotes ESCC cell growth, migration and invasion
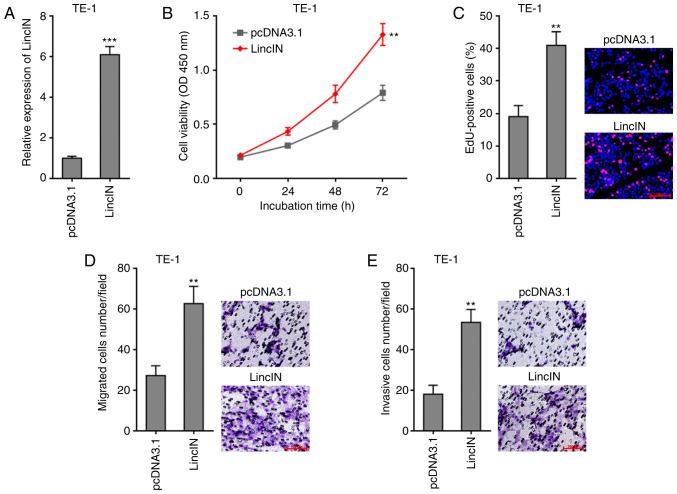
To explore the biological roles of LincIN in ESCC, LincIN was stably overexpressed in the TE-1 cells, which exhibited the lowest LincIN expression out of the 3 ESCC lines (Fig. 1C), by transfection with LincIN overexpression and control plasmids (Fig. 2A). CCK-8 assays revealed that the overexpression of LincIN promoted ESCC cell growth (Fig. 2B). EdU incorporation assays further verified the roles of LincIN overexpression in promoting ESCC cell growth (Fig. 2C). Transwell migration and invasion assays also revealed that the overexpression of LincIN promoted ESCC cell migration and invasion (Fig. 2D and E). Collectively, these results suggested that the overexpression of LincIN promoted ESCC cell growth, migration and invasion.
Figure 2.
Overexpression of LincIN promotes ESCC cell growth, migration, and invasion. (A) Expression of LincIN in LincIN stably overexpressing and control TE-1 cells detected by RT-qPCR. Empty plasmid overexpressing nothing was used as a control. (B) Cell growth of LincIN stably overexpressing and control TE-1 cells detected by CCK-8 assays. (C) Cell growth of LincIN stably overexpressing and control TE-1 cells detected by EdU incorporation assays. Red color indicates EdU-positive cells. Scale bars, 100 µm. (D) Cell migration of LincIN stably overexpressing and control TE-1 cells was detected by Transwell migration assays. Scale bars, 100 µm. (E) Cell invasion of LincIN stably overexpressed and control TE-1 cells detected by Transwell invasion assays. Scale bars, 100 µm. Results are shown as the means ± SD from 3 separate experiments. **P<0.01, ***P<0.001, vs. control group determined by the Student's t-test. ESCC, esophageal squamous cell carcinoma.
Knockdown of LincIN inhibited ESCC cell growth, migration, and invasion
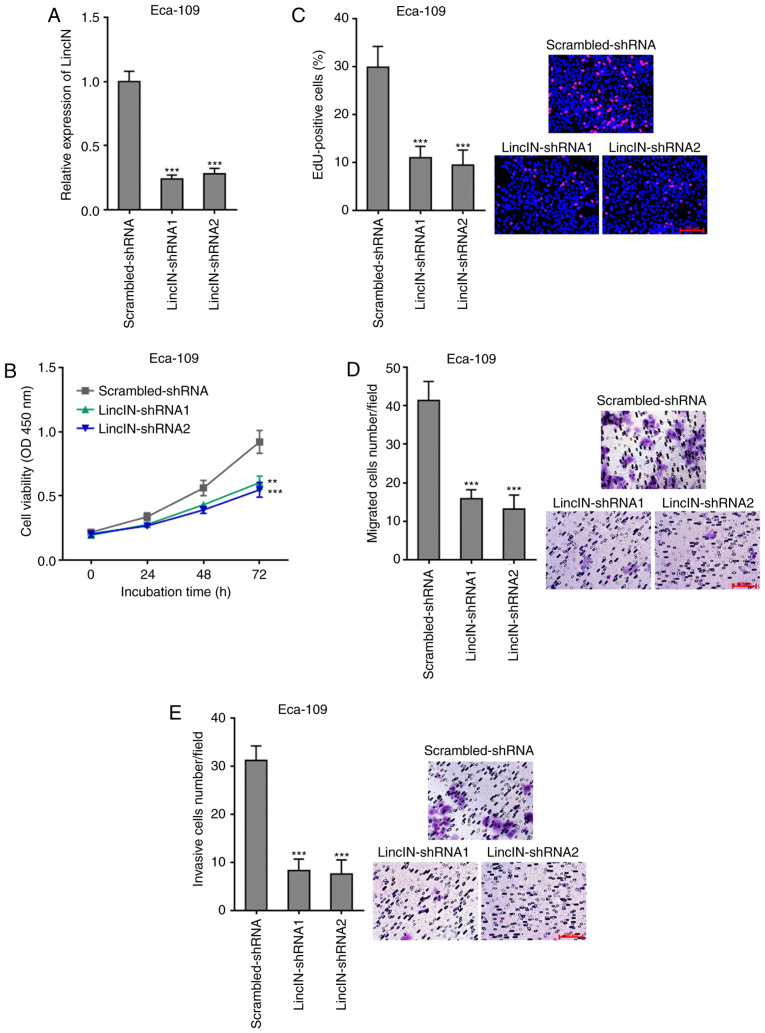
To further explore the biological roles of LincIN knockdown in ESCC, LincIN expression was stably depleted in the Eca-109 cells, which exhibited the highest expression of LincIN out of the 3 ESCC cell lines, by transfection with 2 independent LincIN-specific shRNAs (Fig. 3A). CCK-8 and EdU incorporation assays revealed that the knockdown of LincIN inhibited ESCC cell growth (Fig. 3B and C). Transwell migration and invasion assays also revealed that the knockdown of LincIN inhibited ESCC cell migration and invasion (Fig. 3D and E). Collectively, these results suggested that the knockdown of LincIN inhibited ESCC cell growth, migration and invasion.
Figure 3.
Knockdown of LincIN inhibits ESCC cell growth, migration and invasion. (A) Expression of LincIN in LincIN stably depleted and control Eca-109 cells detected by RT-qPCR. Scrambled shRNA targeting nothing was used as a control. (B) Cell growth of LincIN stably depleted and control Eca-109 cells detected by CCK-8 assays. (C) Cell growth of LincIN stably depleted and control Eca-109 cells detected by EdU incorporation assays. Red color indicates EdU-positive cells. Scale bars, 100 µm. (D) Cell migration of LincIN stably depleted and control Eca-109 cells detected by Transwell migration assays. Scale bars, 100 µm. (E) Cell invasion of LincIN stably depleted and control Eca-109 cells detected by Transwell invasion assays. Scale bars, 100 µm. Results are shown as the means ± SD from 3 separate experiments. **P<0.01, ***P<0.001, vs. control-shRNA group determined by one-way ANOVA followed by Dunnett's multiple comparisons test. ESCC, esophageal squamous cell carcinoma.
LincIN enhances the suppressive effects of NF90 on miR-7 biogenesis
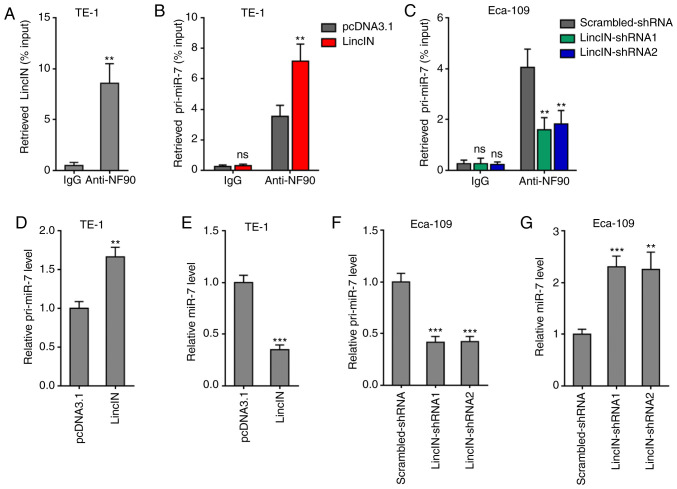
LincIN was previously identified as a NF90-binding lncRNA in breast cancer (40). Therefore, the present study further investigated whether LincIN also bound NF90 in ESCC. RIP assays revealed that LincIN was specifically enriched in the NF90 antibody group, suggesting the binding between LincIN and NF90 (Fig. 4A). NF90 has been reported to suppress miR-7 biogenesis by binding primary miR-7 (pri-miR-7) (47). miR-7 is a well-known tumor suppressor in a number of types of cancer, including ESCC (46,48,49). Therefore, the present study further explored the effects of LincIN on NF90/miR-7. RIP assays revealed that the overexpression of LincIN promoted, while the knockdown of LincIN suppressed the binding between NF90 and pri-miR-7 (Fig. 4B and C), suggesting that LincIN enhanced the binding between NF90 and pri-miR-7. Subsequently, the expression levels of pri-miR-7 and mature miR-7 in ESCC cells in which LincIN was stably overexpressed or depleted were measured by RT-qPCR. The results displayed that the overexpression of LincIN induced the accumulation of pri-miR-7 and the downregulation of mature miR-7 in ESCC cells (Fig. 4D and E). The knockdown of LincIN downregulated the accumulation of pri-miR-7 and upregulated mature miR-7 in ESCC cells (Fig. 4F and G). Collectively, these results suggested that LincIN enhanced the suppressive roles of NF90 on miR-7 biogenesis and therefore, downregulated the mature miR-7 level.
Figure 4.
LincIN enhances the suppressive effects of NF90 on miR-7 biogenesis. (A) RIP assays were performed using the TE-1 cells, followed by RT-qPCR to detect LincIN expression associated with NF90. (B) RIP assays were performed in TE-1 cells transiently overexpressing LincIN or the control, followed by RT-qPCR to detect pri-miR-7 associated with NF90. (C) RIP assays were performed in Eca-109 cells transiently depleting LincIN or control, followed by RT-qPCR to detect pri-miR-7 expression associated with NF90. (D) Expression of pri-miR-7 in LincIN stably overexpressing and control TE-1 cells detected by RT-qPCR. (E) Expression of miR-7 in LincIN stably overexpressing and control TE-1 cells detected by RT-qPCR. (F) Expression of pri-miR-7 in LincIN stably depleted and control Eca-109 cells detected by RT-qPCR. (G) Expression of miR-7 in LincIN stably depleted and control Eca-109 cells detected by RT-qPCR. Results are shown as the means ± SD from 3 separated experiments. **P<0.01, ***P<0.001, vs. vs. IgG, control, or control-shRNA group; ns, not significant, determined by the Student's t-test (A, B, D and E) or one-way ANOVA followed by Dunnett's multiple comparisons test (C, F and G). ESCC, esophageal squamous cell carcinoma.
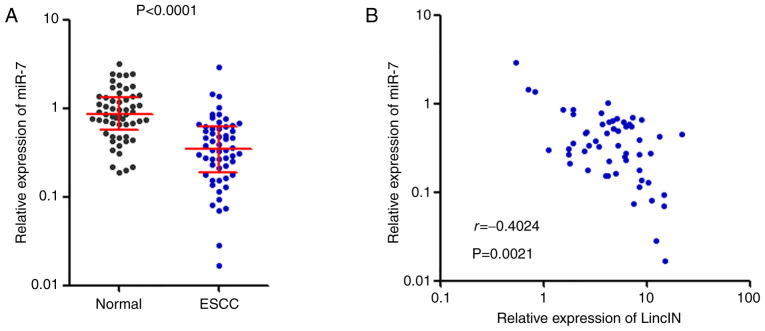
miR-7 expression is decreased and inversely correlates with LincIN expression in ESCC tissues
To determine whether the downregulation of miR-7 by LincIN exists in vivo, the expression of miR-7 was measured in the same 56 pairs of ESCC tissues and adjacent non-cancerous tissues used in Fig. 1. Conversely to the expression pattern of LincIN, miR-7 expression was significantly decreased in ESCC tissues compared with that in adjacent non-cancerous tissues (Fig. 5A). Furthermore, a statistically significant inverse correlation between miR-7 and LincIN expression levels was found in these 56 ESCC tissues (r=−0.4024, P=0.0021, Fig. 5B).
Figure 5.
miR-7 expression is decreased and inversely correlates with LincIN expression in ESCC tissues. (A) Expression of miR-7 in 56 pairs of ESCC tissues and adjacent noncancerous tissues detected by RT-qPCR. P<0.0001, determined by Wilcoxon signed-rank test. (B) Correlation between miR-7 and LincIN expression level in 56 ESCC tissues was analysed; r=-0.4024, P=0.0021, determined by Pearson's correlation analysis. ESCC, esophageal squamous cell carcinoma.
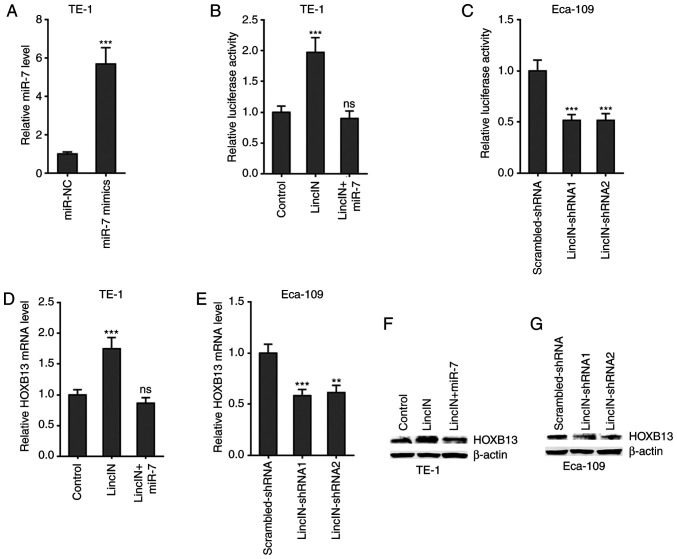
LincIN upregulates HOXB13 expression by suppressing miR-7
HOXB13 was previously identified as a critical target of miR-7 and it was found to mediate the tumor suppressive roles of miR-7 (46,50). Therefore, the present study further examined the effects of LincIN on HOXB13. Dual-luciferase reporter assays revealed that the overexpression of LincIN significantly upregulated the 3′UTR activity of HOXB13, which was reversed by the concurrent overexpression of miR-7 (Fig. 6A and B). Reciprocally, the knockdown of LincIN significantly reduced the 3′UTR activity of HOXB13 (Fig. 6C). RT-qPCR assays revealed that the overexpression of LincIN significantly upregulated the HOXB13 mRNA level, which was reversed by the concurrent overexpression of miR-7 (Fig. 6D). Reciprocally, the knockdown of LincIN significantly reduced the HOXB13 mRNA level (Fig. 6E). Western blot assays revealed that the overexpression of LincIN markedly upregulated the HOXB13 protein level, which was reversed by the concurrent overexpression of miR-7 (Fig. 6F). Reciprocally, the knockdown of LincIN markedly reduced the HOXB13 protein level (Fig. 6G). Collectively, these results suggested that LincIN upregulated HOXB13 by suppressing miR-7.
Figure 6.
LincIN upregulates HOXB13 expression by suppressing miR-7. (A) Expression of miR-7 in TE-1 cells transfected with miR-7 mimics or miRNAs negative control (NC) detected by RT-qPCR. (B) Luciferase activity in TE-1 cells co-transfected with LincIN overexpression plasmid, miR-7 mimics and luciferase reporter containing HOXB13 3′UTR. Results are shown as the relative ratio of Firefly luciferase activity to Renilla luciferase activity. (C) Luciferase activity in Eca-109 cells co-transfected with LincIN specific shRNAs and luciferase reporter containing HOXB13 3′UTR. Results are shown as the relative ratio of Firefly luciferase activity to Renilla luciferase activity. (D) Expression of HOXB13 mRNA in TE-1 cells co-transfected with LincIN overexpression plasmid and miR-7 mimics detected by RT-qPCR. (E) Expression of HOXB13 mRNA in LincIN stably depleted and control Eca-109 cells detected by RT-qPCR. (F) Expression of HOXB13 protein in TE-1 cells co-transfected with LincIN overexpression plasmid and miR-7 mimics detected by western blot analysis. (G) Expression of HOXB13 protein in LincIN stably depleted and control Eca-109 cells detected by western blot analysis. Results are shown as the means ± SD from 3 separate experiments. **P<0.01, ***P<0.001, vs. control or control-shRNA group; ns, not significant, determined by one-way ANOVA followed by Dunnett's multiple comparisons test. ESCC, esophageal squamous cell carcinoma.
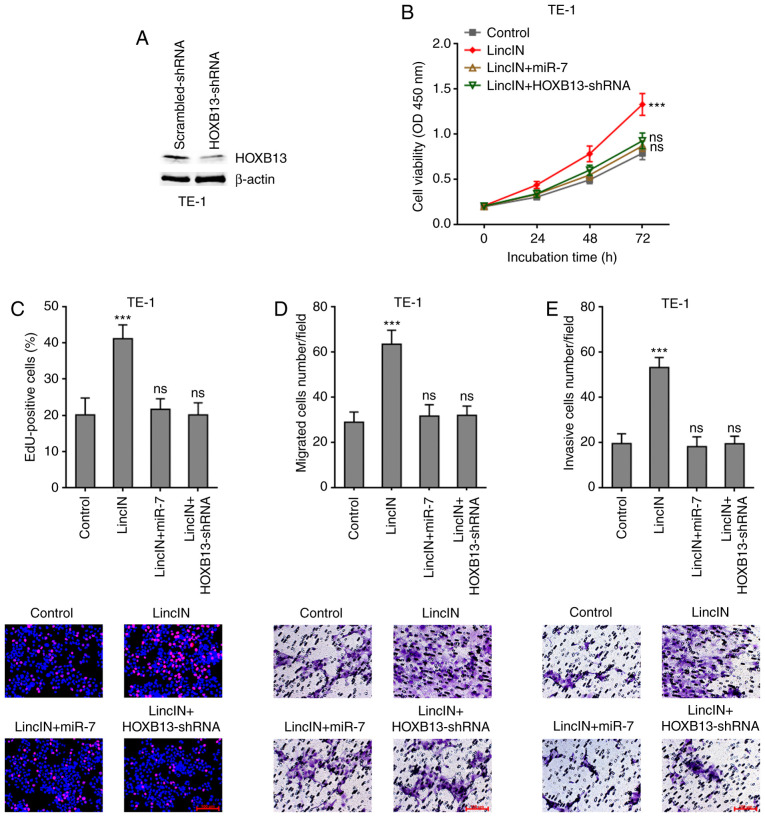
Overexpression of miR-7 or depletion of HOXB13 attenuates the promoting effects of LincIN in ESCC cell growth, migration and invasion
To examine whether miR-7/HOXB13 mediate the roles of LincIN in promoting ESCC cell growth, migration and invasion, miR-7 was stably overexpressed or HOXB13 was depleted in TE-1 cells stably overexpressing LincIN (Fig. 7A). CCK-8 and EdU incorporation assays revealed that both miR-7 overexpression and HOXB13 depletion attenuated the promoting effects of LincIN on ESCC cell growth (Fig. 7B and C). Transwell migration and invasion assays also revealed that both miR-7 overexpression and HOXB13 depletion attenuated the promoting effects of LincIN on ESCC cell migration and invasion (Fig. 7D and E). Collectively, these results suggested that the overexpression of miR-7 or depletion of HOXB13 both attenuated the promoting effects of LincIN on ESCC cell growth, migration and invasion.
Figure 7.
Overexpression of miR-7 or depletion of HOXB13 attenuates the roles of LincIN. (A) Expression of HOXB13 protein in HOXB13 stably depleted and control TE-1 cells detected by western blot analysis. (B) Cell growth of miR-7 and LincIN simultaneously stably overexpressed or HOXB13 stably depleted and simultaneously LincIN stably overexpressed TE-1 cells detected by CCK-8 assays. (C) Cell growth of miR-7 and LincIN simultaneously stably overexpressed or HOXB13 stably depleted and simultaneously LincIN stably overexpressed TE-1 cells detected by EdU incorporation assays. Red color indicates EdU-positive cells. Scale bars, 100 µm. (D) Cell migration of miR-7 and LincIN simultaneously stably overexpressed or HOXB13 stably depleted and simultaneously LincIN stably overexpressed TE-1 cells detected by Transwell migration assays. Scale bars, 100 µm. (E) Cell invasion of miR-7 and LincIN simultaneously stably overexpressed or HOXB13 stably depleted and simultaneously LincIN stably overexpressed TE-1 cells was detected by Transwell invasion assays. Scale bars, 100 µm. Results are shown as the means ± SD from 3 separate experiments. ***P<0.001, vs. control group; ns, not significant, determined by one-way ANOVA followed by Dunnett's multiple comparisons test. ESCC, esophageal squamous cell carcinoma.
Discussion
As one of the most lethal types of cancer, ESCC involves complex molecular mechanisms which remain unclear. Whole-exome sequencings have identified a number of mutations in ESCC, including TP53, CCND1, CDKN2A, NFE2L2, RB1, KMT2D, KMT2C, EP300, FAT1, NOTCH1 and others (51). Furthermore, transcriptome sequencing analyses have identified a number of differentially expressed mRNAs and lncRNAs in ESCC (52). The expression and roles of several lncRNAs in ESCC have also been investigated (50). lncRNA CCAT1 has been shown to be upregulated in ESCC tissues and to be associated with a poor survival (50). CCAT1 promotes ESCC cell proliferation and migration by downregulating SPRY4 and upregulating HOXB13 (50). CCAT1 binds and recruits EZH2 and SUV39H1 to epigenetically silence SPRY4 (45). Furthermore, CCAT1 upregulates HOXB13 by competitively binding miR-7 (45). lncRNA TTN-AS1 is highly expressed in ESCC, and promotes ESCC cell proliferation and metastasis via sponging miR-133b and inducing Snail1 and FSCN1 expression (53). lncRNA NMR promotes ESCC progression via NSUN2 and BPTF (54). Other lncRNAs have also been reported to have oncogenic or tumor suppressive roles in ESCC, such as PART1, CASC9, HOTTIP, LUCAT1, HNF1A-AS1, AFAP1-AS1, POU3F3 and others (55-61).
Transcriptome sequencing has identified >58,000 lncRNAs in human cells (62). Due to the large number of lncRNAs, other lncRNAs may also play biological roles in human ESCC. lncRNA LincIN, whose encoding gene is located on chromosome 10p11-12, was first identified in breast tumors (40). LincIN is overexpressed in human breast tumors, and is associated with a poor prognosis of patients with breast cancer, and is involved in the metastasis of breast cancer (40). However, the expression, clinical significance and role of LincIN in other diseases remain unknown. In the present study, it was found that LincIN expression was also significantly increased in ESCC tissues and cell lines, and was associated with an advanced clinical stage and a poor prognosis of patients with ESCC. Functional experiments revealed that the overexpression of LincIN promoted ESCC cell growth, migration and invasion. The knockdown of LincIN inhibited ESCC cell growth, migration and invasion. Thus, these data suggested LincIN as a promising prognostic biomarker and therapeutic target for ESCC.
The mechanisms of action of lncRNAs are complex. One of the main mechanisms is to bind proteins (13). In breast tumors, LincIN was revealed to bind NF90 (40). NF90, also known as ILF3, is an RNA binding protein (63). By binding double-stranded RNA, NF90 regulates gene expression and/or mRNA stabilities (64). In addition, by binding pri-miRNAs, NF90 modulates miRNAs biogenesis (47). The biogenesis of miR-7, which plays tumor suppressive roles in a number of types of cancer, has been reported to be suppressed by NF90 (46,47). Therefore, the present study further investigated the effects of LincIN on NF90/miR-7. It was verified that LincIN directly bound NF90 in ESCC cells. Moreover, LincIN promoted the binding between NF90 and pri-miR-7, thereby enhancing the suppressive roles of NF90 on miR-7 biogenesis. It was then verified that LincIN downregulated mature miR-7. Conversely to LincIN, miR-7 expression was significantly decreased in ESCC tissues. The expression of mature miR-7 was significantly and inversely associated with that of LincIN in ESCC tissues, supporting the negative regulation of miR-7 by LincIN. HOXB13 is a direct target of miR-7 and plays oncogenic roles in several types of cancer, including ESCC (46,50). By repressing miR-7, LincIN was verified to upregulate HOXB13 expression in ESCC cells. Functional rescue assays demonstrated that the overexpression of miR-7 or depletion of HOXB13 attenuated the promoting effects of LincIN on ESCC cell growth, migration and invasion, which further suggested that miR-7/HOXB13 are critical mediators of the oncogenic roles of LincIN in ESCC. Similar roles of lncRNAs in miRNAs biogenesis have also been reported in other types of cancer, such as the promotion of miR-145 biogenesis by lncRNA-ATB in bladder cancer and the disruption of miR-125b biogenesis by LINC01578 in breast cancer (63,65). The detailed mechanisms underlying the inverse effects of lncRNAs on the biogenesis of different miRNAs warrant further investigation.
Apart from miR-7, NF90 has also been reported to suppress p21 translation, upregulate hypoxia inducible factor-1α (HIF-1α) and VEGF-A, stabilize PARP1 mRNA, and facilitate DICER expression (40,64,66,67). Thus, whether LincIN modulates p21, HIF-1α, VEGF-A, PARP1 and DICER via interacting with NF90 in ESCC cells needs to be further investigated. Nevertheless, the present study identified a novel mechanism of lncRNA in ESCC and provides a novel target against ESCC.
In conclusion, the present study demonstrates that lncRNA LincIN is highly expressed in ESCC, and is associated with an advanced clinical stage and a poor prognosis of patients with ESCC. LincIN promotes ESCC cell growth, migration and invasion by binding NF90, suppressing the biogenesis of miR-7 and upregulating HOXB13. The data presented herein suggest that LincIN may be a promising prognostic biomarker and therapeutic target for ESCC.
Acknowledgments
The authors would like to thank Mr. Peng Jiang for providing assistance with language editing.
Funding Statement
The present study was supported by the National Nature Science Foundation of China (grant nos. 81672756, 81872399, 81772631 and 81572849), the Natural Science Foundation of Guangdong Province (grant no. 2018A030313530), the Chinese Scholarship Council (grant no. 201908440010), the Shenzhen Healthcare Research Project (grant no. SZFZ2017105), the Seedling Program of Shenzhen Hospital, Southern Medical University (grant no. 2016MM06) and the Sanming Project of Medicine in Shenzhen (grant no. SZSM201612041).
Availability of data and materials
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.
Authors' contributions
DW and ZT designed the study. ZT, WS, PZ, YW, ZZ, ZG and MS performed the experiments. DW, ZT and YX performed the statistical analysis. DW and ZT wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The use of clinical specimens was reviewed and approved by the Nanfang Hospital Institutional Review Board (Guangzhou, China). Written informed consent was acquired from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400–412. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- 3.Rustgi AK, El-Serag HB. Esophageal carcinoma. N Engl J Med. 2014;371:2499–2509. doi: 10.1056/NEJMra1314530. [DOI] [PubMed] [Google Scholar]
- 4.Sjoquist KM, Burmeister BH, Smithers BM, Zalcberg JR, Simes RJ, Barbour A, Gebski V, Australasian Gastro-Intestinal Trials Group Survival after neoadjuvant chemotherapy or chemoradiotherapy for resectable oesophageal carcinoma: An updated meta-analysis. Lancet Oncol. 2011;12:681–692. doi: 10.1016/S1470-2045(11)70142-5. [DOI] [PubMed] [Google Scholar]
- 5.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 6.Batista PJ, Chang HY. Long noncoding RNAs: Cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Berger AC, Korkut A, Kanchi RS, Hegde AM, Lenoir W, Liu W, Liu Y, Fan H, Shen H, Ravikumar V, et al. A comprehensive pan-cancer molecular study of gynecologic and breast cancers. Cancer Cell. 2018;33:690–705. doi: 10.1016/j.ccell.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan JH, Yang F, Wang F, Ma Jz, Guo Yj, Tao Qf, Liu F, Pan W, Wang Tt, Zhou Cc, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 11.Zhang C, Yuan J, Hu H, Chen W, Liu M, Zhang J, Sun S, Guo Z. Long non-coding RNA CHCHD4P4 promotes epithelial-mesenchymal transition and inhibits cell proliferation in calcium oxalate-induced kidney damage. Braz J Med Biol Res. 2017;51:e6536. doi: 10.1590/1414-431x20176536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin D, Lu X, Su J, He X, De W, Yang J, Li W, Han L, Zhang E. Long noncoding RNA AFAP1-AS1 predicts a poor prognosis and regulates non-small cell lung cancer cell proliferation by epigenetically repressing p21 expression. Mol Cancer. 2018;17:92. doi: 10.1186/s12943-018-0836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu WL, Jin L, Xu A, Wang YF, Thorne RF, Zhang XD, Wu M. GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat Cell Biol. 2018;20:492–502. doi: 10.1038/s41556-018-0066-7. [DOI] [PubMed] [Google Scholar]
- 15.Li JK, Cheng C, Liu JY, Shi JZ, Liu SP, Liu B, Wu DS, Fang ZY, Bao Y, Jiang MM, et al. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol Cancer. 2017;16:111. doi: 10.1186/s12943-017-0681-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Zheng X, Wang N, Zhao W, Zhang X, Teng S, Zhang Y, Lu Z. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17:89. doi: 10.1186/s12943-018-0837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu H, Zheng T, Yu J, Zhou L, Wang L. LncRNA XIST accelerates cervical cancer progression via upregulating fus through competitively binding with miR-200a. Biomed Pharmacother. 2018;105:789–797. doi: 10.1016/j.biopha.2018.05.053. [DOI] [PubMed] [Google Scholar]
- 18.Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang Y, Jia L, Li S, Cancer Genome Atlas Research Network. Xie W, Yang D. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. 2018;33:706–720. doi: 10.1016/j.ccell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mondal T, Juvvuna PK, Kirkeby A, Mitra S, Kosalai ST, Traxler L, Hertwig F, Wernig-Zorc S, Miranda C, Deland L, et al. Sense-Antisense lncRNA pair encoded by locus 6p223 determines neuroblastoma susceptibility via the USP36-CHD7-SOX9 regulatory axis. Cancer Cell. 2018;33:417–434. doi: 10.1016/j.ccell.2018.01.020. [DOI] [PubMed] [Google Scholar]
- 20.Lin A, Hu Q, Li C, Xing Z, Ma G, Wang C, Li J, Ye Y, Yao J, Liang K, et al. The LINK-A lncRNA interacts with PtdIns(3,4,5) P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat Cell Biol. 2017;19:238–251. doi: 10.1038/ncb3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grelet S, Link LA, Howley B, Obellianne C, Palanisamy V, Gangaraju VK, Diehl JA, Howe PH. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat Cell Biol. 2017;19:1105–1115. doi: 10.1038/ncb3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilot D, Migault M, Bachelot L, Journé F, Rogiers A, Donnou-Fournet E, Mogha A, Mouchet N, Pinel-Marie ML, Mari B, et al. A non-coding function of TYRP1 mRNA promotes melanoma growth. Nat Cell Biol. 2017;19:1348–1357. doi: 10.1038/ncb3623. [DOI] [PubMed] [Google Scholar]
- 23.Zhu XT, Yuan JH, Zhu TT, Li YY, Cheng XY. Long noncoding RNA glypican 3 (GPC3) antisense transcript 1 promotes hepatocellular carcinoma progression via epigenetically activating GPC3. FEBS J. 2016;283:3739–3754. doi: 10.1111/febs.13839. [DOI] [PubMed] [Google Scholar]
- 24.Tsai KW, Lo YH, Liu H, Yeh CY, Chen YZ, Hsu CW, Chen WS, Wang JH. Linc00659, a long noncoding RNA, acts as novel oncogene in regulating cancer cell growth in colorectal cancer. Mol Cancer. 2018;17:72. doi: 10.1186/s12943-018-0821-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Wang W, Liu G, Xie S, Li Q, Li Y, Lin Z. Long non-coding RNA HOTTIP promotes hypoxia-induced epithelial-mesenchymal transition of malignant glioma by regulating the miR-101/ZEB1 axis. Biomed Pharmacother. 2017;95:711–720. doi: 10.1016/j.biopha.2017.08.133. [DOI] [PubMed] [Google Scholar]
- 26.Michelini F, Pitchiaya S, Vitelli V, Sharma S, Gioia U, Pessina F, Cabrini M, Wang Y, Capozzo I, Iannelli F, et al. Damage-Induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat Cell Biol. 2017;19:1400–1411. doi: 10.1038/ncb3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan JH, Liu XN, Wang TT, Pan W, Tao QF, Zhou WP, Wang F, Sun SH. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 2017;19:820–832. doi: 10.1038/ncb3538. [DOI] [PubMed] [Google Scholar]
- 28.Su R, Cao S, Ma J, Liu Y, Liu X, Zheng J, Chen J, Liu L, Cai H, Li Z, et al. Knockdown of SOX2OT inhibits the malignant biological behaviors of glioblastoma stem cells via up-regulating the expression of miR-194-5p and miR-122. Mol Cancer. 2017;16:171. doi: 10.1186/s12943-017-0737-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao J, Zhang L, Zheng L, Hong Y, Zhao L. LncRNATCF7 promotes the growth and self-renewal of glioma cells via suppressing the miR-200c-EpCAM axis. Biomed Pharmacother. 2018;97:203–208. doi: 10.1016/j.biopha.2017.10.039. [DOI] [PubMed] [Google Scholar]
- 30.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 31.Mohr S, Doebele C, Comoglio F, Berg T, Beck J, Bohnenberger H, Alexe G, Corso J, Ströbel P, Wachter A, et al. Hoxa9 and meis1 cooperatively induce addiction to syk signaling by suppressing miR-146a in acute myeloid leukemia. Cancer Cell. 2017;31:549–562. doi: 10.1016/j.ccell.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bu Y, Yoshida A, Chitnis N, Altman BJ, Tameire F, Oran A, Gennaro V, Armeson KE, McMahon SB, Wertheim GB, et al. A PERK-miR-211 axis suppresses circadian regulators and protein synthesis to promote cancer cell survival. Nat Cell Biol. 2018;20:104–115. doi: 10.1038/s41556-017-0006-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan W, Wu X, Zhou W, Fong MY, Cao M, Liu J, Liu X, Chen CH, Fadare O, Pizzo DP, et al. Cancer-Cell-Secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat Cell Biol. 2018;20:597–609. doi: 10.1038/s41556-018-0083-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chernyy V, Pustylnyak V, Kozlov V, Gulyaeva L. Increased expression of miR-155 and miR-222 is associated with lymph node positive status. J Cancer. 2018;9:135–140. doi: 10.7150/jca.22181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leite KR, Reis ST, Viana N, Morais DR, Moura CM, Silva IA, Pontes J, Jr, Katz B, Srougi M. Controlling RECK miR21 promotes tumor cell invasion and is related to biochemical recurrence in prostate cancer. J Cancer. 2015;6:292–301. doi: 10.7150/jca.11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Celia-Terrassa T, Liu DD, Choudhury A, Hang X, Wei Y, Zamalloa J, Alfaro-Aco R, Chakrabarti R, Jiang YZ, Koh BI, et al. Normal and cancerous mammary stem cells evade interferon-induced constraint through the miR-199a-LCOR axis. Nat Cell Biol. 2017;19:711–723. doi: 10.1038/ncb3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu MZ, Cheng WC, Chen SF, Nieh S, O'Connor C, Liu CL, Tsai WW, Wu CJ, Martin L, Lin YS, et al. MiR-25/93 mediates hypoxia-induced immunosuppression by repressing cGAS. Nat Cell Biol. 2017;19:1286–1296. doi: 10.1038/ncb3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan JH, Yang F, Chen BF, Lu Z, Huo Xs, Zhou Wp, Wang F, Sun Sh. The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology. 2011;54:2025–2035. doi: 10.1002/hep.24606. [DOI] [PubMed] [Google Scholar]
- 39.Ho CS, Noor SM, Nagoor NH. MiR-378 and MiR-1827 regulate tumor invasion, migration and angiogenesis in human lung adenocarcinoma by targeting RBX1 and CRKL, respectively. J Cancer. 2018;9:331–345. doi: 10.7150/jca.18188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang Z, Slater CM, Zhou Y, Devarajan K, Ruth KJ, Li Y, Cai KQ, Daly M, Chen X. LincIN, a novel NF90-binding long non-coding RNA, is overexpressed in advanced breast tumors and involved in metastasis. Breast Cancer Res. 2017;19:62. doi: 10.1186/s13058-017-0853-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishihira T, Kasai M, Mori S, Watanabe T, Kuriya Y, Suda M, Kitamura M, Hirayama K, Akaishi T, Sasaki T. Characteristics of two cell lines (TE-1 and TE-2) derived from human squamous cell carcinoma of the esophagus. Gan. 1979;70:575–584. [PubMed] [Google Scholar]
- 42.Barnas C, Martel-Planche G, Furukawa Y, Hollstein M, Montesano R, Hainaut P. Inactivation of the p53 protein in cell lines derived from human esophageal cancers. Int J Cancer. 1997;71:79–87. doi: 10.1002/(SICI)1097-0215(19970328)71:1<79::AID-IJC14>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 43.Gao J, Xue KX, Li BG, Dong HY, Wei LS, Zhang ZH, Tian S, Xu ZG. Site-Dependence of invasiveness of ECA109 human oesophageal carcinoma cells in nude mice. Clin Exp Metastasis. 1984;2:205–212. doi: 10.1007/BF00132927. [DOI] [PubMed] [Google Scholar]
- 44.Ye Z, Fang J, Dai S, Wang Y, Fu Z, Feng W, Wei Q, Huang P. MicroRNA-34a induces a senescence-like change via the down-regulation of SIRT1 and up-regulation of p53 protein in human esophageal squamous cancer cells with a wild-type p53 gene background. Cancer Lett. 2016;370:216–221. doi: 10.1016/j.canlet.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 45.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Li RC, Ke S, Meng FK, Lu J, Zou XJ, He ZG, Wang WF, Fang MH. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis. 2018;9:838. doi: 10.1038/s41419-018-0852-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Higuchi T, Todaka H, Sugiyama Y, Ono M, Tamaki N, Hatano E, Takezaki Y, Hanazaki K, Miwa T, Lai S, et al. Suppression of microRNA-7 (miR-7) biogenesis by nuclear factor 90-nuclear factor 45 complex (NF90-NF45) controls cell proliferation in hepatocellular carcinoma. J Biol Chem. 2016;291:21074–21084. doi: 10.1074/jbc.M116.748210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su C, Han Y, Zhang H, Li Y, Yi L, Wang X, Zhou S, Yu D, Song X, Xiao N, et al. CiRS-7 targeting miR-7 modulates the progression of non-small cell lung cancer in a manner dependent on NF-κB signalling. J Cell Mol Med. 2018;22:3097–3107. doi: 10.1111/jcmm.13587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu B, Yang T, Wang Z, Zhang Y, Liu S, Shen M. CircRNA CDR1as/miR-7 signals promote tumor growth of osteosarcoma with a potential therapeutic and diagnostic value. Cancer Manag Res. 2018;10:4871–4880. doi: 10.2147/CMAR.S178213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang E, Han L, Yin D, He X, Hong L, Si X, Qiu M, Xu T, De W, Xu L, et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2017;45:3086–3101. doi: 10.1093/nar/gkw1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao YB, Chen ZL, Li JG, Hu XD, Shi XJ, Sun ZM, Zhang F, Zhao ZR, Li ZT, Liu ZY, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet. 2014;46:1097–1102. doi: 10.1038/ng.3076. [DOI] [PubMed] [Google Scholar]
- 52.Li CQ, Huang GW, Wu ZY, Xu YJ, Li XC, Xue YJ, Zhu Y, Zhao JM, Li M, Zhang J, et al. Integrative analyses of transcriptome sequencing identify novel functional lncRNAs in esophageal squamous cell carcinoma. Oncogenesis. 2017;6:e297. doi: 10.1038/oncsis.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin C, Zhang S, Wang Y, Wang Y, Nice E, Guo C, Zhang E, Yu L, Li M, Liu C, et al. Functional role of a novel long noncoding RNA TTN-AS1 in esophageal squamous cell carcinoma progression and metastasis. Clin Cancer Res. 2018;24:486–498. doi: 10.1158/1078-0432.CCR-17-1851. [DOI] [PubMed] [Google Scholar]
- 54.Li Y, Li J, Luo M, Zhou C, Shi X, Yang W, Lu Z, Chen Z, Sun N, He J. Novel long noncoding RNA NMR promotes tumor progression via NSUN2 and BPTF in esophageal squamous cell carcinoma. Cancer Lett. 2018;430:57–66. doi: 10.1016/j.canlet.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 55.Kang M, Ren M, Li Y, Fu Y, Deng M, Li C. Exosome-mediated transfer of lncRNA PART1 induces gefitinib resistance in esophageal squamous cell carcinoma via functioning as a competing endogenous RNA. J Exp Clin Cancer Res. 2018;37:171. doi: 10.1186/s13046-018-0845-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56.Wu Y, Hu L, Liang Y, Li J, Wang K, Chen X, Meng H, Guan X, Yang K, Bai Y. Up-Regulation of lncRNA CASC9 promotes esophageal squamous cell carcinoma growth by negatively regulating PDCD4 expression through EZH2. Mol Cancer. 2017;16:150. doi: 10.1186/s12943-017-0715-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin C, Wang Y, Wang Y, Zhang S, Yu L, Guo C, Xu H. Transcriptional and posttranscriptional regulation of HOXA13 by lncRNA HOTTIP facilitates tumorigenesis and metastasis in esophageal squamous carcinoma cells. Oncogene. 2017;36:5392–5406. doi: 10.1038/onc.2017.133. [DOI] [PubMed] [Google Scholar]
- 58.Yoon JH, You BH, Park CH, Kim YJ, Nam JW, Lee SK. The long noncoding RNA LUCAT1 promotes tumorigenesis by controlling ubiquitination and stability of DNA methyltransferase 1 in esophageal squamous cell carcinoma. Cancer Lett. 2018;417:47–57. doi: 10.1016/j.canlet.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 59.Yang X, Song JH, Cheng Y, Wu W, Bhagat T, Yu Y, Abraham JM, Ibrahim S, Ravich W, Roland BC, et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63:881–890. doi: 10.1136/gutjnl-2013-305266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luo HL, Huang MD, Guo JN, Fan RH, Xia XT, He JD, Chen XF. AFAP1-AS1 is upregulated and promotes esophageal squamous cell carcinoma cell proliferation and inhibits cell apoptosis. Cancer Med. 2016;5:2879–2885. doi: 10.1002/cam4.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li W, Zheng J, Deng J, You Y, Wu H, Li N, Lu J, Zhou Y. Increased levels of the long intergenic non-protein coding RNAPOU3F3 promote DNA methylation in esophageal squamous cell carcinoma cells. Gastroenterology. 2014;146:1714–1726 e1715. doi: 10.1053/j.gastro.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 62.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhuang J, Shen L, Yang L, Huang X, Lu Q, Cui Y, Zheng X, Zhao X, Zhang D, Huang R, et al. TGFβ1 promotes gemcitabine resistance through regulating the LncRNA-LET/NF90/miR-145 signaling axis in bladder cancer. Theranostics. 2017;7:3053–3067. doi: 10.7150/thno.19542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang W, Xiong Z, Wei T, Li Q, Tan Y, Ling L, Feng X. Nuclear factor 90 promotes angiogenesis by regulating HIF-1α/VEGF-A expression through the PI3K/akt signaling pathway in human cervical cancer. Cell Death Dis. 2018;9:276. doi: 10.1038/s41419-018-0334-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu J, Zhan Y, Wang J, Wang J, Guo J, Kong D. Long noncoding RNA LINC01578 drives colon cancer metastasis through a positive feedback loop with the NF-κB/YY1 axis. Mol Oncol. 2020;14:3211–3233. doi: 10.1002/1878-0261.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song D, Huang H, Wang J, Zhao Y, Hu X, He F, Yu L, Wu J. NF90 regulates PARP1 mRNA stability in hepatocellular carcinoma. Biochem Biophys Res Commun. 2017;488:211–217. doi: 10.1016/j.bbrc.2017.05.037. [DOI] [PubMed] [Google Scholar]
- 67.Barbier J, Chen X, Sanchez G, Cai M, Helsmoortel M, Higuchi T, Giraud P, Contreras X, Yuan G, Feng Z, et al. An NF90/NF110-mediated feedback amplification loop regulates dicer expression and controls ovarian carcinoma progression. Cell Res. 2018;28:556–571. doi: 10.1038/s41422-018-0016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.