Abstract
Today, the whole world is facing a great medical disaster that affects the health and lives of the people: the COVID-19 disease, colloquially known as the Corona virus. Deep learning is an effective means to assist radiologists to analyze the vast amount of chest X-ray images, which can potentially have a substantial role in streamlining and accelerating the diagnosis of COVID-19. Such techniques involve large datasets for training and all such data must be centralized in order to be processed. Due to medical data privacy regulations, it is often not possible to collect and share patient data in a centralized data server. In this work, we present a collaborative federated learning framework allowing multiple medical institutions screening COVID-19 from Chest X-ray images using deep learning without sharing patient data. We investigate several key properties and specificities of federated learning setting including the not independent and identically distributed (non-IID) and unbalanced data distributions that naturally arise. We experimentally demonstrate that the proposed federated learning framework provides competitive results to that of models trained by sharing data, considering two different model architectures. These findings would encourage medical institutions to adopt collaborative process and reap benefits of the rich private data in order to rapidly build a powerful model for COVID-19 screening.
Keywords: Federated learning, Decentralized training, COVID-19 screening, X-ray images, Deep learning, CNN
1. Introduction
When we talk about machine learning and privacy, there is a sense of conflict. Indeed, machine learning generally and deep learning models specially, need to have access to very large dataset to achieve good performance. Unfortunately, this data is often stored in several organizations because of privacy concerns and liability risks. Especially in healthcare domain, most data is hard to obtain due to legal, privacy, technical, and data-ownership challenges. International regulations such as the Health Insurance Portability and Accountability Act in USA (HIPAA) and the General Data Protection Regulation in European Union (GDPR)1 completely redefine the data management policy. There is no longer any question of massively collecting client’s data without a specific service objective. The GDPR sets the legal framework for the protection of personal data within the European Union. Making companies more responsible, the GDPR gives new obligations to service operators with regard to data management, in particular making their centralization much more regulated. The respect of privacy is more than ever an important issue at the heart of data processing. These challenges create a problem for data scientists building and deploying machine-learning-based healthcare systems as a service. In short, in order to benefit from these powerful diagnostics, you have to share your data.
Federated learning (FL), introduced by Google in 2017 [1], is a distributed machine learning approach that enables multi-institutional collaboration on deep learning projects withoutsharing client data. A motivating example for FL arises when we keep the training data on local device’s users (nodes) rather than logging it to a data center. These nodes perform computations from their own data in order to update a global model.
Because each node generates its data with different patterns, the distribution of data within each node differs from node to node. For example, one client may have much more data than others. So it is impossible to define a representative sample of the overall distribution. Here, we talk about two of key properties that differentiate federated optimization from a typical distributed optimization problem: (1) Not independent and identically distributed (Non-IID) data: since each particular user has his local training data, so there is no single representation of population distribution. (2) Unbalanced data: similarly, each user has a quantity of data that differs from others.
In view of the federated learning advantages, we have exploited this technique in order to deal with a very sensitive topic in the healthcare field. Indeed, since last December 2019, a new coronavirus infection disease (named COVID-19) was first reported in Wuhan in China. Subsequently, the outbreak began to spread widely in China and most countries in the world [2]. The rapid escalation of this pandemic (with hundreds of deaths and thousands of infections) is presenting great challenges for stopping the virus.
Currently, more than one diagnostic method is possible for the detection of coronavirus but Chest X-ray images and CT scans are from most accepted standard diagnostic [3], [4], [5]. Indeed, since COVID-19 attacks the epithelial cells that line our respiratory tract, we can use Chest X-ray images to analyze the health of a patient’s lungs, and given that nearly all hospitals have X-ray imaging machines, it could be possible to use X-ray images to test for COVID-19 without the dedicated test kits. Compared to these tests, chest X-ray images analyzed with Artificial Intelligence offer a fast and cost-effective way to COVID-19 screening.
Therefore, many research works have been devoted to the COVID-19 outbreak prediction [6], [7] and diagnosis [8], [9] based on machine Learning techniques.
In the present work, we have concentrated our efforts to develop and validate a system based on federated learning for detection of COVID-19 from Chest X-ray images, which is the root of all the novelties of the article. To the best of our knowledge, this is the first study that addresses the problem of federated learning on X-ray images for COVID-19 detection. The main contributions of this paper are:
-
•
We propose a decentralized and collaborative framework that allows clinicians to reap benefits of the rich private data share while conserving privacy.
-
•
We demonstrate, that despite the decentralized data, the non-IID and unbalanced properties of the data distribution, the proposed federated learning framework remains robust and shows competitive results compared to a centralized learning process.
-
•
We conducted extensive experiments and comparisons with different variations to show the interest and significance of the proposed strategy which can be particularly useful in situations like COVID-19.
The remaining paper is organized as follows: Section 2 cites the related works. Section 3 describes an overview of our proposed framework of federated optimization procedure adapted to a detection problem of COVID-19 disease in X-ray images. Section 4 is dedicated to the experiments and results, where both the centralized and federated ways used to train our COVID-19 dataset are introduced and their results are discussed. Finally, we conclude this study in Section 5.
2. Related work
Recently, a lot of work has been done to develop algorithms of deep learning for the detection of such a disease from chest X-ray images [10], [11]. Indeed, the work in [12] developed an algorithm that can detect pneumonia from chest X-ray images at a level exceeding practicing radiologists with a Dense Convolutional Network. Xu et al. [13] used an hierarchical Convolutional Neural Network (CNN) to classify X-ray images into normal and abnormal categories. A descriptive study [14] of radiology images obtained from COVID-19 cases demonstrated that these images contain useful information for diagnostics and early recognition of this disease. As consequence, many works on radiology images have been proposed for COVID-19 detection. Hemdan et al. [15] and Wang and Wong [16] used deep learning models to diagnose COVID-19 from Chest X-ray images. Nour and Cömert [17] used a CNN model to extract deep discriminative features from X-ray images and used them to feed three machine learning algorithms, which were k-nearest neighbor, support vector machine, and decision tree. Gupta et al. [18] proposed an integrated stacked deep convolutional network to detect COVID-19 and pneumonia by identifying the abnormalities in Chest X-ray images. Zhang et al. [19] developed a deep learning-based model that can detect COVID-19 based on chest X-ray images with sufficiently high sensitivity, enabling fast and reliable screening. In [20], the authors introduced a deep model for early detection of COVID-19 cases using X-ray images that can achieve good accuracy rates for binary and multi-classes. Narin et al. [21] and Chowdhury et al. [22] trained and compared multiple pre-trained CNN based models for the detection of COVID-19 infected patients using chest X-ray images. Recently, Demir [23] proposed a deep Long short-term memory (LSTM) architecture learned from scratch to automatically identify COVID-19 cases from X-ray images. Other works [24], [25], [26] focused on detecting COVID-19 positive cases from chest CT scans using CNN based models.
A drawback of these centralized models is that, in practical cases, medical organizations do not agree to devote their doctor–patient confidentiality by giving out the medical images, like X-ray images, for training purposes. In contrast, many research in healthcare [27], [28], [29], [30] demonstrated that the technique of federated learning is a good way to connect all the medical institutions and make them share their experiences with privacy guarantee. In this case, the performance of machine learning model will be significantly improved by the formed large medical dataset. As an example, Lee et al. [30] presented a privacy-preserving platform in a federated setting for patient similarity learning across institutions. Their model can find similar patients from one hospital to another without sharing patient-level information. Similarly, Huang et al. [29] sought to tackle the challenge of non-IID ICU patient data that complicated decentralized learning, by clustering patients into clinically meaningful communities and optimizing performance of predicting mortality and ICU stay time. More recently, Baheti et al. [27] used the concept of federated learning for detection of pulmonary lung nodules with CT scans.
As COVID-19 is a recent emerging infectious disease, there is no publicly available large datasets. Most of the existing data is stored privately because of concerns over privacy. So, we propose in this paper to develop a collaborative framework to avoid compromising patient privacy while promoting scientific research on large datasets to improve patient care. The goal of our work is to promote screening COVID-19 from Chest X-ray images using the federated learning. We demonstrate that a decentralized learning may address the demands for data protection without impacting the performance compared to a data-centralized learning.
3. Proposed framework
We depict in this section the details of our proposed method for Chest X-ray images classification to identify COVID-19 from non-COVID-19 cases. This section first presents the preliminaries of the federated learning context, then an overview of our proposed framework, followed by the architecture of our training model, and finally a description of the client-side model training procedure and the server-side model aggregation procedure.
3.1. Preliminaries
We consider the standard machine learning problem objective function , that is the loss of prediction on example when using a model described by a vector parameter . In a federated setting, we assume that the data points are partitioned across clients, is the set of data points on client , and designs the number of the client data points. Thus, the optimization objective is:
| (1) |
McMahan et al. [31] introduced an algorithm for federated learning: FederatedAveraging or FedAvg which aims at minimizing the objective function in Eq. (1) assuming a synchronous update scheme and a generic non-convex neural network loss function. In terms of convergence, FedAvg is practically equivalent to a central model when IID data is used. McMahan et al. [1] demonstrated that FedAvg is still robust for some examples of non-IID data. However, Zhao et al. [32] showed that the accuracy of FedAvg is significantly reduced when trained on highly skewed non-IID data even under convex optimization setting.
3.2. Our framework overview
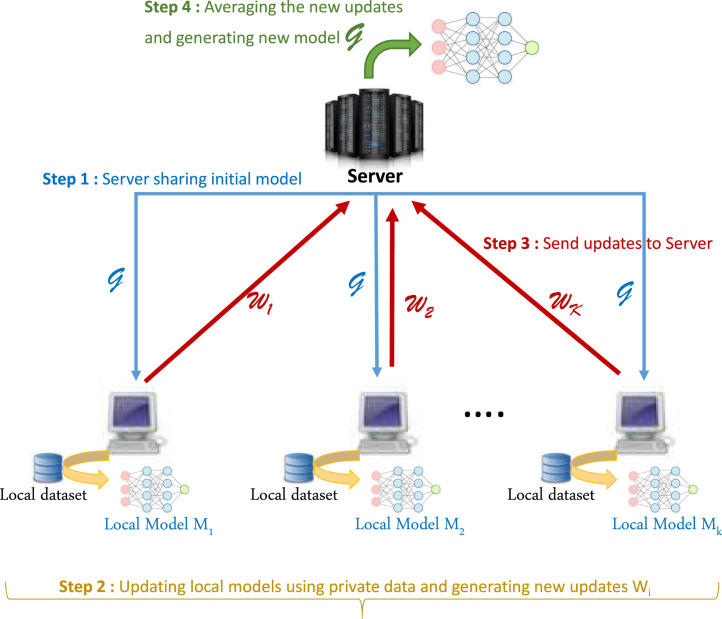
In this work, we propose to study a federated learning framework based on a client–server architecture (illustrated in Fig. 1) implementing the FedAvg algorithm in order to classify X-ray images into COVID-19 infected cases and non-COVID-19 ones. In this configuration, a centralized parameter server maintains a global model that shares with clients and then coordinates their updates. Clients coordinate to build a powerful model based on their own private datasets.
Fig. 1.
Federated Learning architecture for COVID-19 detection from Chest X-ray images.
We propose to build a deep convolutional neural network (CNN) to deal with the feature extraction and the classification of X-ray images to detect the COVID-19 disease. This model takes as input an X-ray image and outputs the probability of COVID-19 infection. The details of this model architecture (CNN) are described in Section 3.2.1.
The learning phase of this CNN model consists of several communication rounds where the central server interacts synchronously with the clients. Before starting the training rounds, the CNN model is first initialized with random weights . We suppose that there are available clients having each private X-ray images stored locally. Each communication round consists of four steps:
Step 1. Initially the central server maintains a global central model , with initial weights , which is shared with a subset of clients (hospitals in our case) that are randomly selected given a fraction , with .
Step 2. Each client , receiving initial parameters , performs training steps on a mini-batch of its local own private data based on the minimization of the local objective using mini batch stochastic gradient descent (SGD) with a local learning rate and for a number of epochs . Clients optimize the model via minimizing the categorical cross entropy loss for classification.
Step 3. If local training is finished (running SGD for epochs on local data points), users from send back to the server their model updates , .
Step 4. Finally, the server receives updates from all participating clients and computes an average model according to Eq. (2) to update the global model parameters.
| (2) |
Here are parameters updated at round , are parameters sent by client at round , is number of data points stored on client , and is total number of data points participated in collaboratively training.
These four steps constitute one round of FL of our CNN model. This operation is then repeated many times (rounds). We notice that at each new round , the server re-sends the new parameters of the global model built in the previous round . We also notice that the subset of clients can be changed from one round to another if many clients are available. The client selection protocol is given in Section 3.2.2.
3.2.1. Model architecture
We propose in this study a decentralized and collaborative framework for the screening of COVID-19 from chest X-ray images. Our aim is to demonstrate that the federated learning of a deep CNN model allows to reap benefits of the rich private data sharing while conserving privacy. For this reason, the choice of the CNN architecture is not our main concern, and there are several architectural choices that can slightly increase or decrease the overall performance. For simplicity we adopt two well-known CNN architectures in image classification, namely VGG16 [33] and ResNet50 [34] as backbone network. For both architectures, we use the pre-trained CNN leaving off the fully connected layer head. Then, we add a classification head composed of global average pooling, a fully connected layer of 64 and 256 units with dropout for VGG16 and ResNet50, respectively, and a final fully connected layer composed of two units with softmax activation for classification. To optimize the classification head, we use the categorical cross-entropy loss. Our CNN takes as input an X-ray image of size 224 × 224, and outputs probability values belonging to our classes.
As shown in Fig. 1, we have two parties exchanging information: federated clients and a central server. We provide in the following sections the details of these two parties.
3.2.2. Client-side model update
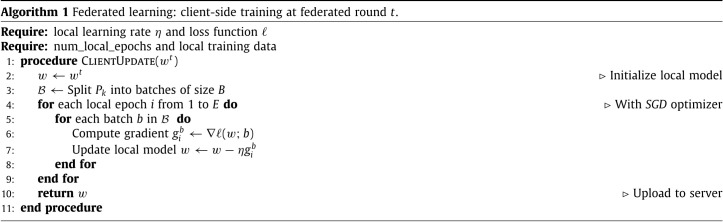
The training is performed on the client-side, indeed, each federated client has a fixed dataset and computational capabilities to run mini-batch SGD. We dispose of clients having all the same CNN architecture (described in Section 3.2.1) and loss functions. The proposed training algorithm is listed in Algorithm 1. At round , each local model is initialized by a global model coming from the server. After running a number of iterations of as many times as number of local epochs, the client computes a gradient update in order to generate the new updated model which is shared with the aggregation server. Following this training protocol, local data remains private to each client and is never shared.
3.2.3. Server-side model aggregation
The server that owns the global model, manages the overall progress of the model training and distributes the original model to all participating clients. It receives synchronized updates from all participating clients at each federated round (see Algorithm 2) and aggregates them to build a new model with updated parameters according to Eq. (2). Algorithm 2 presents the details of the server side learning process.
4. Experiments
In this work, we simulate experiments with clients (hospitals), and each client treats the full local dataset as a single mini-batch at each round.
4.1. Data preparation
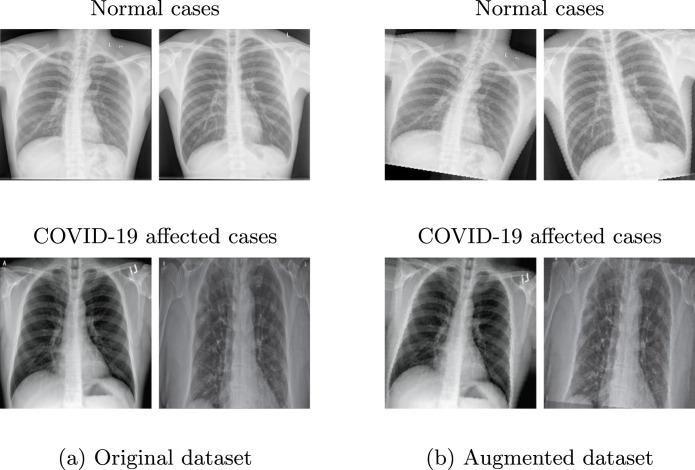
Since there are no available large public datasets belonging to COVID-19 cases, the dataset used for this work only includes chest X-ray images belonging to patients, all of which were confirmed with COVID-19, and chest X-ray images diagnosed as normal (not COVID-19) belonging to healthy patients. The COVID-19 X-ray images used for this research are available at the Github repository2 while X-ray images of normal cases are randomly selected from the public chest X-ray dataset [35], which contains normal and abnormal chest X-ray images. Fig. 2 (left) shows sample images belonging to the two classes.
Fig. 2.
Sample Chest X-ray images from the used dataset. Left : sample images selected from the original dataset. Right : corresponding augmented images generated with random zoom and rotation augmentations.
We randomly split the dataset into a training set containing 80% of the images (76 COVID-19 images belonging to 55 patients and 76 healthy patient images) and a test set containing 20% of the images (32 COVID-19 images belonging to 21 patients and 32 healthy patient images). The training dataset is then split into sub-sets according to the appropriate testing data distribution. All our simulations are done using clients.
When we deal with IID data, we assign 38 images (19 COVID-19 cases and 19 Normal cases) for each client. All clients have the same amount of data (25%) according to the same distribution. In order to simulate non-IID training on our dataset, we use a skewed class distribution and we divide the learning data so that each client gets a different number of images from each class (44% of images of one class and 6% of images of the second one). Finally, we generate a third version of our training dataset in order to test unbalanced data distribution over clients. To do this, we spread the entire training samples over the 4 clients, so that each has more observations than others. The four clients have respectively 44%, 37%, 13%, and 6% of training dataset.
All images from the same patient only appear in either training or testing set. In addition, there is no patient overlap between the client sub-sets in order to make our federated setup realistic.
Since the dataset is small, we applied data augmentation operations in order to artificially expand the size of the training and test sub-sets by creating modified versions of the images. We used two geometric transformations, namely rotation and zoom. Rotation augmentations consist of rotating the image right or left on an axis by a random and small degree (). Zoom augmentations are done by zooming in or out the image according to a small range (). Fig. 2 (right) presents some samples of augmented images. By these operations, the number of training samples is augmented from 38 to 152 (76 COVID-19 cases and 76 normal cases) for each client. This augmented dataset is used only for one experiment, in order to demonstrate the dataset size impact on the model quality and test accuracy. Results are explained in Section 4.3.1.
4.2. Model settings and evaluation metric
On both the federated learning and the centralized learning end, we adopt the same CNN networks, with pre-trained weights on ImageNet [36], leaving off the fully connected layer head and replaced by a new classification head, for training and prediction. Two CNN architectures are tested, the details of which are provided in Section 3.2.1.
The model weights of the CNN backbone (VGG16 andResNet50) are frozen such that only the new fully connected layers head will be trained. The standard SGD optimizer is used for minimizing the loss function and then updating the network parameters. We refer to this method as FL-VGG16 and FL-ResNet50 respectively when using VGG16 and ResNet50 architectures as the model backbone. We set the local learning rate 0.001, the batch size 2, and the training epochs 10. In addition, we resize each image to a fixed size of 224 × 224 pixels.
Significant accuracy rate is required in COVID-19 diagnosis and detection system to limit the spread of the infection and to guide the patient treatment. Therefore, to evaluate the performance of our proposed method, we report accuracy rates on testing data after each round of federated learning. For each method, we repeat experiments 3 times and all the curves represent average results obtained over these 3 simulations. Since we have binary classification tests, we provide also statistical measures of performance that are widely used in medical and epidemiological research [25], namely sensitivity and specificity. Indeed, the sensitivity reflects the probability that the screening test will be positive among those who are already diseased (True Positives) and the specificity reflects the probability that the screening test will be negative among those who do not have the disease (True Negatives) [37].
To show the effectiveness of our federated learning based method, we first compare its performance with traditional learning method, where we train the same architecture network on shared and centralized data (We refer to this method asCentralized-VGG16 and Centralized-ResNet50 respectively when using VGG16 and ResNet50 architectures as the model backbone).
4.3. Results
We conduct this study of federated learning for COVID-19 detection to highlight the effectiveness of this type of decentralized and collaborative learning in such context where data is private.
First, we compare our decentralized method with the centralized one. Then, we study the effect of the parameter on the model performance after each round when we deal with IID data distribution. Finally, we compare the two distribution settings IID and non-IID, balanced and unbalanced.
4.3.1. Federated vs. data-centralized training:
We have 152 training samples and 64 testing samples in our dataset. Since there is not a natural user partitioning of this data, we considered the balanced and IID setting. So, we partition the training dataset into 4 clients each containing 38 training (25%).
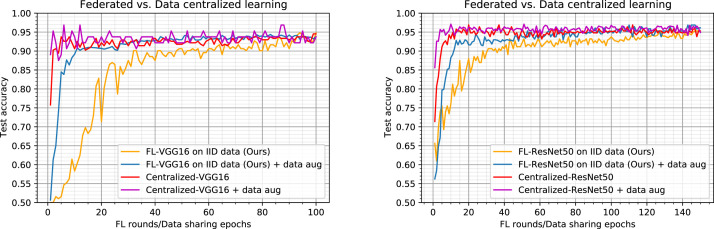
In this section, federated results are compared with a centralized learning method. Our aim is to evaluate accuracy performance of our proposed FL based method. Fig. 3 shows comparative results across data-sharing and FL for our two implementations (VGG16 and ResNet50) over our original training dataset and the augmented one. The models quality is measured by accuracy scores on a held-out test dataset, plotted against the number of communication rounds for FL based methods, and against data-sharing epochs for centralized methods.
Fig. 3.
Comparison of Federated Learning to data-sharing learning using original and augmented dataset for learning. Left: results using the VGG16 as the model backbone. Right: results using the ResNet50 as the model backbone. An epoch for centralized methods is defined as a single training pass over all of the centralized data. A round for FL methods is defined as a parallel training pass of every client over their local training data.
For FL settings, we fix (all clients are considered at each round). Fig. 3 shows that the proposed FL procedure can achieve a comparable classification performance without sharing clients’ data. Fig. 3-left shows that from the round , our FL-VGG16 method trained on the original dataset (FL-VGG16: orange curve) approaches the Centralized-VGG16 method trained on the same dataset (Centralized-VGG16: red curve) but after collecting and sharing all data from the 4 clients. Fig. 3-right shows that our FL-ResNet50 (orange curve) method provides similar behavior but requires more rounds ( rounds) to approach the Centralized-ResNet50 method (red curve).
We notice that the blue and the magenta curves correspond to our FL based methods (FL-VGG16 and FL-ResNet50) and the Centralized-VGG16/ResNet50 methods respectively learned on an augmented dataset with data augmentation techniques described above. Our FL results show a remarkable consistency on the simulated distributions. Our method (FL-VGG16+data aug : blue curve) has comparable results with the two centralized methods (Centralized-VGG16 and Centralized-VGG16+data aug) after only rounds. The same result is observed with the FL-ResNet50 method which provides comparable results with the two centralized methods using the same CNN architecture after rounds. The amount of data at each client side has then a significant impact on the final result.
Another important result that can be underlined from Fig. 3-left is that after about rounds, all methods (with the VGG16 CNN) are equivalent and provide similar results. Fig. 3-right shows the same behavior but from round . This result highlights the effectiveness of our proposed FL based framework, since it allows to achieve similar results to centralized methods by iterating several rounds without never sharing data that preserve their privacy.
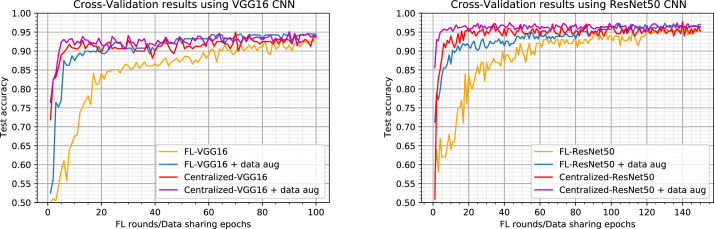
Results with 5-fold cross validation.
To further evaluate our proposed FL based method, we used 5-fold cross-validationmethod, which consists of dividing all the available data into a predefined number of folds (5 in our case), and using one fold for testing and the others for training. The training process is repeated 5 times until all folds are used as a test set. Using the cross-validation method is motivated by the fact that we have little data and our model will be tested on only few data samples. So by doing cross-validation, we use all of our data both for training and testing while evaluating our model on examples it has never seen before. At each iteration, the training and test sets are randomly divided into sub-sets each for one client while respecting the protocol described above where all images from the same patient only appear in either training or test set and there is no patient overlap between the client sub-sets.
We provide in Fig. 4 comparative results across data-sharing and FL for our VGG16 and ResNet50 implementations using a 5-fold cross-validation method. All the curves represent average results obtained over the 5 simulations for each method. Fig. 4 confirms all the results presented in Fig. 3 for both VGG16 and ResNet50 implementations. This finding confirms the generalization ability of the proposed model and the independence of our reported results of the train/test dataset splits.
Fig. 4.
Comparison of Federated Learning to data-sharing learning using original and augmented dataset for learning. Curves represent average results obtained over the 5 simulations for each method. Left: results using the VGG16 as the model backbone. Right: results using the ResNet50 as the model backbone.
We report in Table 1 the performance of the tested methods based on accuracy, sensitivity, and specificity measures after the last round for FL based methods and the last epoch for the centralized ones. The first result that we can see in Table 1 is that after 150 rounds, our FL-ResNet50 model provides the higher accuracy performance when it is trained with data augmentation achieving an accuracy of 97%. This method also provides a high sensitivity rate of 98.11% and a specificity rate of 95.89%. Another result that can be underlined from Table 1 is that all tested methods provide comparable sensitivity and specificity rates when using the same model backbone. Indeed, all VGG16 based models provide sensitivity and specificity rates ranging in and in , respectively. On the other hand, the ResNet50 based models provide higher sensitivity and specificity rates ranging in and in , respectively. This result highlights the effectiveness of our proposed FL based methods, since they provide comparable performances to centralized methods while preserving data privacy, showing their suitability for privacy-restricted applications.
Table 1.
Accuracy, Sensitivity, and Specificity rates after the last FL round/Data sharing epoch. Reported results are given with respect to our experiments made with the 5-fold cross-validation method. Accuracy, Sensitivity, and Specificity rates provided in this table are average results over the 5 simulations.
| Method | Accuracy | Sensitivity | Specificity |
|---|---|---|---|
| FL-VGG16 | 93.57 | 95.03 | 92.12 |
| FL-VGG16 data aug | 94.40 | 96.15 | 92.66 |
| Centralized-VGG16 | 93.75 | 95.20 | 92.3 |
| Centralized-VGG16 data aug | 94.0 | 95.01 | 93.0 |
| FL-ResNet50 | 95.4 | 96.03 | 94.78 |
| FL-ResNet50 data aug | 97.0 | 98.11 | 95.89 |
| Centralized-ResNet50 | 95.3 | 96.0 | 94.6 |
| Centralized-ResNet50 data aug | 96.5 | 96.8 | 96.2 |
4.3.2. Results on IID data
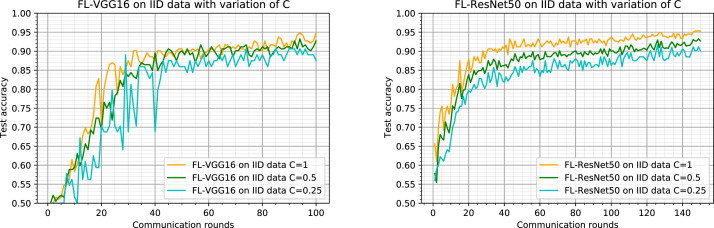
We consider here the IID and balanced data partition (same as Section 4.3.1) and we provide experiments with the client fraction , which controls the amount of multi-client parallelism. We notice that means that all available clients are selected for collaborative training at each round, and means that only one client is selected at each round. When , there is no parallelism between clients, and the learning process is considered to be sequential. In our case, is equivalent to since we have only clients. We report in this section results obtained with three values of the parameter : , 0.5, and 0.25.
Fig. 5 shows the test accuracy curves plotted against the communication rounds up to 100 and 150 for FL-VGG16 and FL-ResNet50, respectively. Fig. 5-left shows that the FL-VGG16 converges to close values faster when all clients are considered ( : orange curve) and collaborate at each round. When only half of clients are selected ( : green curve), the results are slightly worse but they approach the case when after about rounds. When only one client is selected at each round ( : cyan curve), the results are fluctuating and the convergence to close values begin only after several rounds. This behavior is justified by the fact that only one client is considered at each round, so the update model process on the server consists of replacing the old model by the one sent by the considered client and there is no collaborative learning. The quality of the new model at each round depends then on the selected client data.
Fig. 5.
Effect of the client fraction on the test accuracy of our proposed method FL-VGG16. Note corresponds to all clients are selected at each round (4 clients in our case), corresponds to half clients (2 clients in our case) and corresponds to only one client per round. Left: results using the VGG16 as the model backbone. Right: results using the ResNet50 as the model backbone.
For FL-ResNet50, Fig. 5-right illustrates the same convergence behavior for all curves but we get slightly worse accuracy rates when and . When using half of clients, the accuracy decreases in regard to use all clients and it decreases even more when using 25% of clients (only one client) per round. In the federated learning context, generally the ratio is set to 10% because it is more realistic in a practical setup where there are several available clients [38].
We can conclude that for the IID data partition, using more clients in each round increases the accuracy at convergence and the learning process requires less rounds to converge. This result is observed in our context where the number of clients is limited and the available data size is small. Such results can be specific for this context.
4.3.3. Results on non-IID data
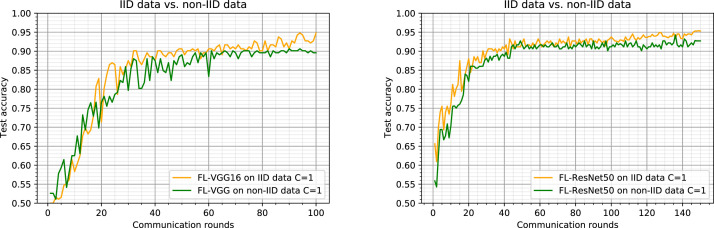
In this section, we fix and compare the two FL methods on IID data and non-IID data and provide results in Fig. 6.
Fig. 6.
Comparison of Federated Learning results on IID data and non-IID data partitions with (all clients are considered at each round). Left: results using the VGG16 as the model backbone. Right: results using the ResNet50 as the model backbone.
We show that the speedups with partitioned non-IID data (green curve) are smaller but still substantial, this implies that the performance of the model is random. We notice that despite the non-IID aspect of the data distribution, our implementation of FL based methods on non-IID data has shown their robustness by trying to achieve test-set accuracy of FL methods on IID data (94% for FL-VGG16 and 95.3% for FL-ResNet50) which in turn surpassed that of centralized learning method. This small degradation of the quality of model training is due to the fact that each client has a lot of data from one class and little data from the other. We also notice that by increasing the number of rounds, for the non-IID partition the test accuracy is almost stabilized (0.9 for FL-VGG16 and around 0.92 for FL-ReNet50), in contrast for the case of IID data partition, it continues to converge.
4.3.4. Results on unbalanced data
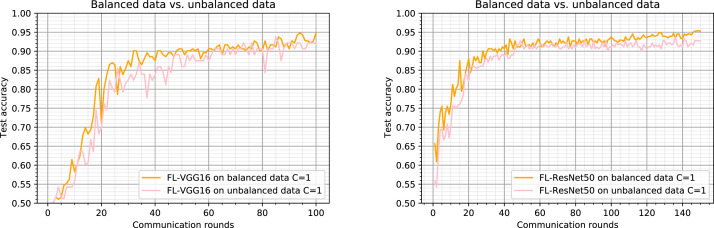
Generally, the unbalanced and non-IID distribution of such a dataset is much more representative of the type of data distribution for medical applications. And since we are manipulating a method intended for medical applications, we have adopted our implementation to converge in the case of a distribution of unbalanced data. As shown in Fig. 7-left, despite the significant imbalance in numbers of subjects per client (which are partitioned as described in Section 4.1), FL-VGG16 on unbalanced data (pink curve) achieves test-set accuracy 92% (approaching even those of the centralized learning model). The same behavior is observed with FL-ResNet50 trained on unbalanced data (pink curve in Fig. 7-right) achieving a test-set accuracy 92.7%.
Fig. 7.
Comparison of Federated Learning results on balanced data and unbalanced data partitions with (all clients are considered at each round). Left: results using the VGG16 as the model backbone. Right: results using the ResNet50 as the model backbone.
By comparing the two curves of FL (VGG16 and ResNet50) on balanced data and FL (VGG16 and ResNet50) on unbalanced data, test accuracy of the first method is higher than test accuracy of the second one (this is justified by the fact that clients hold very different amounts of data) which tends to approach it after several rounds. The method implemented for unbalanced data shows its performance in achieving 92% and 92.7% test accuracy for the VGG16 and the ResNet50, respectively. We can then conclude that the heterogeneity of the quantity of data held by each client does not affect the model’s performance.
5. Conclusion and future work
In this paper, we presented a Federated Learning framework for COVID-19 detection from Chest X-ray images using deep convolutional neural networks (VGG16 and ResNet50). This framework operates in a decentralized and collaborative manner and allows clinicians everywhere in the world to reap benefits of the rich private medical data sharing while conserving privacy. We first presented a comparative study between two medical image machine learning scenarios: the classical centralized learning and the federated learning, using two CNN architectures as model backbone: VGG16 and ResNet50. We then demonstrated that federated learning can achieve the same performance as centralized learning, but without the obligation to share or centralize private and sensitive data. We also demonstrated that despite the decentralized data, the non-IID and unbalanced properties of the data distribution, the proposed Federated Learning framework remains robust and shows comparable performance with a centralized learning process. We note that the federated learning framework is validated on COVID-19 screening from Chest X-ray images, but could be generalized to other medical imaging applications with large, distributed, and privacy-sensitive data.
Federated learning has the potential to connect all the isolated medical institutions, hospitals or devices to make them share their experiences and collaborate with privacy guarantee. Such collaboration will improve the speed and accuracy in the COVID-19 positive cases detection. We aim in the future to provide such a federated platform, where all the hospitals can safely share data and train models by exploring the differential privacy technique. Another interesting direction for future work is to consider a more sophisticated CNN using very large-scale datasets.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
References
- 1.McMahan H.B., Moore E., Ramage D., Hampson S., . Arcas B.A. AISTATS. 2017. Communication-efficient learning of deep networks from decentralized data; p. 54. [Google Scholar]
- 2.Hui D.S., Azhar E.I., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., Mchugh T.D., Memish Z.A., Drosten C., Zumla A., Petersen E. The continuing covid-19 epidemic threat of novel coronaviruses to global health-the latest 2019 novel coronavirus outbreak in wuhan, china. Int. J. Infec. Dis.: IJID: Off. Publ. Int. Soc. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanne J.P., Little B.P., Chung J.H., Elicker B.M., Ketai L.H. 2020. Essentials for radiologists on covid-19: an update—radiology scientific expert panel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest ct for typical 2019-ncov pneumonia: relationship to negative rt-pcr testing. Radiology. 2020 doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zu Z.Y., Jiang M.D., Xu P.P., Chen W., Ni Q.Q., Lu G.M., Zhang L.J. Coronavirus disease 2019 (covid-19): a perspective from china. Radiology. 2020 doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kairon P., Bhattacharyya S. COVID-19 outbreak prediction using quantum neural networks. Intel. Enabled Res. 2021;11:3–123. [Google Scholar]
- 7.Ardabili S.F., Mosavi A., Ghamisi P., Ferdinand F., Varkonyi-Koczy A.R., Reuter U., Rabczuk T., Atkinson P.M. Covid-19 outbreak prediction with machine learning. Algorithms. 2020;13 [Google Scholar]
- 8.Schwab P., DuMon Schütte A., Dietz B., Bauer S. Clinical predictive models for covid-19: Systematic study. J. Med. Internet. Res. 2020;22 doi: 10.2196/21439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W.T., Ma J., Shende N., Castaneda G., Chakladar J., Tsai J.C., Apostol L., Honda C.O., Xu J., Wong L.M., Zhang T., Lee A., Gnanasekar A., Honda T.K., Kuo S.Z., Yu M.A., Chang E.Y., Rajasekaran M., Ongkeko W.M. Using machine learning of clinical data to diagnose covid-19: a systematic review and meta-analysis. BMC Med. Inform. Decis. Mak. 2020;1 doi: 10.1186/s12911-020-01266-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma H., Smal I., Daemen J., van Walsum T. Dynamic coronary roadmapping via catheter tip tracking in x-ray fluoroscopy with deep learning based bayesian filtering. Med. Image Anal. 2020;61 doi: 10.1016/j.media.2020.101634. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y., Miao S., Mansi T., Liao R. Unsupervised x-ray image segmentation with task driven generative adversarial networks. Med. Image Anal. 2020;62 doi: 10.1016/j.media.2020.101664. [DOI] [PubMed] [Google Scholar]
- 12.Rajpurkar P., Irvin J., Zhu K., Yang B., Mehta H., Duan T., Ding D., Bagul A., Ball R., Langlotz C., Shpanskaya K., Lungren M., Ng A. 2017. Chexnet : Radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv preprint arXiv:1711.05225v3. [Google Scholar]
- 13.Xu S., Wu H., Bie R. Cxnet-m1: Anomaly detection on chest x-rays with image-based deep learning. IEEE Access. 2019;7:4466–4477. [Google Scholar]
- 14.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Radiological findings from 81 patients with covid-19 pneumonia in wuhan, china: a descriptive study. Lancet. Infect. Dis. 2020. 2020;20:425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemdan E.E.D., Shouman M.A., Karar M.E. 2020. Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv preprint arXiv:2003.11055. [Google Scholar]
- 16.Wang L., Wong A. 2020. Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. arXiv preprint arXiv:2003.09871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nour M., Cömert K. A novel medical diagnosis model for covid-19 infection detection based on deep features and bayesian optimization. Appl. Soft Comput. 2020;97 doi: 10.1016/j.asoc.2020.106580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta A., Anjum K., Gupta S., Kataryat R. Instacovnet-19: A deep learning classification model for the detection of covid-19 patients using chest x-ray. Appl. Soft Comput. 2020;97 doi: 10.1016/j.asoc.2020.106859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J., Xie Y., Li3 Y., Shen C., Xi Y. 2020. Covid-19 screening on chest x-ray images using deep learning based anomaly detection. arXiv preprint arXiv:2003.12338v1. [Google Scholar]
- 20.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of covid-19 cases using deep neural networks with x-ray images. Comput. Biol. Med. 2020 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narin A., Kaya C., Pamuk Z. 2020. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. arXiv preprint arXiv:2003.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chowdhury M.E.H., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam K.R., Khan M.S., Iqbal A., Emadi N.A., et al. Can ai help in screening viral and covid-19 pneumonia? IEEE Access. 2020;8:132665–132676. [Google Scholar]
- 23.Demir F. Deepcoronet: A deep lstm approach for automated detection of covid-19 cases from chest x-ray images. Appl. Soft Comput. 2021;103 doi: 10.1016/j.asoc.2021.107160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shan F., Gao Y., Wang J., Shi W., Shi N., Han M., Xue Z., Shen D., Shi Y. Abnormal lung quantification in chest ct images of covid-19 patients with deep learning and its application to severity prediction. Med. Phys. 2020 doi: 10.1002/mp.14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gozes O., Frid-Adar M., Greenspan H., Browning P.D., Zhang H., Ji W., Bernheim A., Siegel E. 2020. Rapid ai development cycle for the coronavirus (covid-19) pandemic: Initial results for automated detection & patient monitoring using deep learning ct image analysis. 2003.05037. [Google Scholar]
- 26.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., Cao K., Liu D., Wang G., Xu Q., Fang X., Zhang S., Xia J., Xia J. Artificial intelligence distinguishes covid-19 from community acquired pneumonia on chest ct. Radiology. 2020 doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.P. Baheti, M. Sikka, K.V. Arya, R. Rajesh, Federated learning on distributed medical records for detection of lung nodules, in: Proceedings of the 15th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications, 2020, pp. 445–451.
- 28.Brisimi T.S., Chen R., Mela T., Olshevsky A., Paschalidis I.C., Shi W. Federated learning of predictive models from federated electronic health records. Int. J. Med. Inform. 2018;112:59–67. doi: 10.1016/j.ijmedinf.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang L., Shea A.L., Qian H., Masurkar A., Deng H., Liu D. Patient clustering improves efficiency of federated machine learning to predict mortality and hospital stay time using distributed electronic medical records. J. Biomed. Inform. 2019;99 doi: 10.1016/j.jbi.2019.103291. [DOI] [PubMed] [Google Scholar]
- 30.Lee J., Sun J., Wang F., Wang S., Jun C.H., Jiang X. Privacy-preserving patient similarity learning in a federated environment: Development and analysis. JMIR Med. Inform. 2018;6 doi: 10.2196/medinform.7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMahan H.B., Moore E., Ramage D., Arcas B.A. 2016. Federated learning of deep networks using model averaging. arXiv preprint arXiv:1602.05629. [Google Scholar]
- 32.Zhao Y., Lai L., Civin N.S.D., Li M., Chandra V. 2018. Federated learning with non-iid data. arXiv preprint arXiv:1806.00582v1. [Google Scholar]
- 33.Simonyan K., Zisserman A. 2014. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556. [Google Scholar]
- 34.He K., Zhang X., Ren S., Sun J. IEEE Conference on Computer Vision and Pattern Recognition (CVPR)Deep residual learning for image recognition. 2016:770–778. [Google Scholar]
- 35.Jaeger S., Candemir S., Antani S., Wáng Y.X.J., Lu P.X., Thoma G. Two public chest x-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg. 2014;47:5–477. doi: 10.3978/j.issn.2223-4292.2014.11.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M., et al. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015;115:211–252. [Google Scholar]
- 37.Devashish O., Sharma U., Yadav P., Sharma J. The concept of sensitivity and specificity in relation to two types of errors and its application in medical reasearch. J. Reliab. Stat. Stud. 2009;2:53–58. [Google Scholar]
- 38.Leroy D., Coucke A., Lavril T., Gisselbrecht T., Dureau J. ICASSP 2019-2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) IEEE.; 2019. Federated learning for keyword spotting; pp. 6341–6345. [Google Scholar]