Summary Paragraph
All known recently emerged human coronaviruses likely originated in bats1. Here, we used a single experimental platform based on human lung-only mice (LoM) to demonstrate efficient in vivo replication of all recently emerged human coronaviruses (SARS-CoV, MERS-CoV, SARS-CoV-2) and two highly relevant endogenous pre-pandemic SARS-like bat coronaviruses. Virus replication in this model occurs in bona fide human lung tissue and does not require any type of adaptation of the virus or the host. Our results indicate that bats harbor endogenous coronaviruses capable of direct transmission into humans. Further detailed analysis of pandemic SARS-CoV-2 in vivo infection of LoM human lung tissue showed predominant infection of human lung epithelial cells, including type II pneumocytes present in alveoli and ciliated airway cells. Acute SARS-CoV-2 infection was highly cytopathic and induced a robust and sustained Type I interferon and inflammatory cytokine/chemokine response. Finally, we evaluated a therapeutic and pre-exposure prophylaxis strategy for coronavirus infection. Our results show that therapeutic and prophylactic administration of EIDD-2801, an oral broad spectrum antiviral currently in phase II-III clinical trials, dramatically inhibited SARS-CoV-2 replication in vivo and thus has significant potential for the prevention and treatment of COVID-19.
The recently emerged human pandemic coronavirus Severe Acute Respiratory Syndrome coronavirus-2 (SARS-CoV-2), the causative agent of coronavirus disease (COVID)-19, has resulted in substantial morbidity and mortality worldwide2. Bats are the presumed source of SARS-CoV-2 and the highly pathogenic human coronaviruses SARS-CoV and Middle East Respiratory Syndrome (MERS)-CoV1. Transmission of coronaviruses from bats to other species is well-documented and adaptation in an intermediary host can facilitate their transmission to humans1. While it is possible that SARS-CoV-2 was transmitted to humans via an intermediate host, phylogenic analysis indicates that the SARS-CoV-2 lineage has circulated in bats for decades and evolved in bats into a virus capable of replicating in human cells3. Given the repeated and accelerating emergence of highly pathogenic coronaviruses, it is increasingly important to monitor and characterize bat coronaviruses and to identify the viral determinants of human infection, disease, and global spread as well as to develop effective therapeutic interventions. Animal models are useful in studying highly pathogenic human coronaviruses, the emergence potential of zoonotic coronaviruses, and to evaluate the in vivo inhibitory activity of novel agents4–15. However, human coronaviruses do not replicate in mice without either extensive virus adaptation, genetic editing of the host receptor, or introduction of the appropriate human receptor genes into the host4,6–13,15. Although existing rodent models of coronavirus infection have made several important contributions, none possesses the diverse set of primary human lung cells that serve as targets for viral infection16. Here, we show that human lung-only mice (LoM), immune deficient mice implanted with authentic human lung tissue17, allow for the in vivo study of SARS-CoV, MERS-CoV, and SARS-CoV-2 in a single platform permitting direct comparison of experimental outcomes. Using LoM, we also show efficient replication of bat coronaviruses in vivo without the need for virus adaptation. In addition, we performed an in-depth in vivo analysis of acute SARS-CoV-2 infection in the human lung which revealed robust virus replication, pathogenesis, and sustained activation of the innate host immune response. Finally, we used this platform to show that EIDD-2801, an orally administered broad spectrum antiviral currently in phase II-III clinical trials for COVID-19 treatment, efficiently inhibited SARS-CoV-2 replication in human lung tissue when administered therapeutically and prevented SARS-CoV-2 infection when administered as pre-exposure prophylaxis strongly supporting its further clinical development for COVID-19.
Coronavirus replication in LoM
LoM are constructed by subcutaneous implantation of human lung tissue into the back of immune deficient mice (Fig. 1a). This tissue expands to form a palpable implant17 (Fig. 1a). Lung implants contain human fibroblast, epithelial, endothelial, and mesenchymal cells that form cartilaginous and non-cartilaginous bronchial airways lined with ciliated and non-ciliated epithelium, alveolar sac structures, and extensive vasculature17(Extended Data Fig. 1a,b). The human lung tissue in LoM has been shown to support replication of a diverse set of emerging and clinically relevant human pathogens including MERS-CoV17.
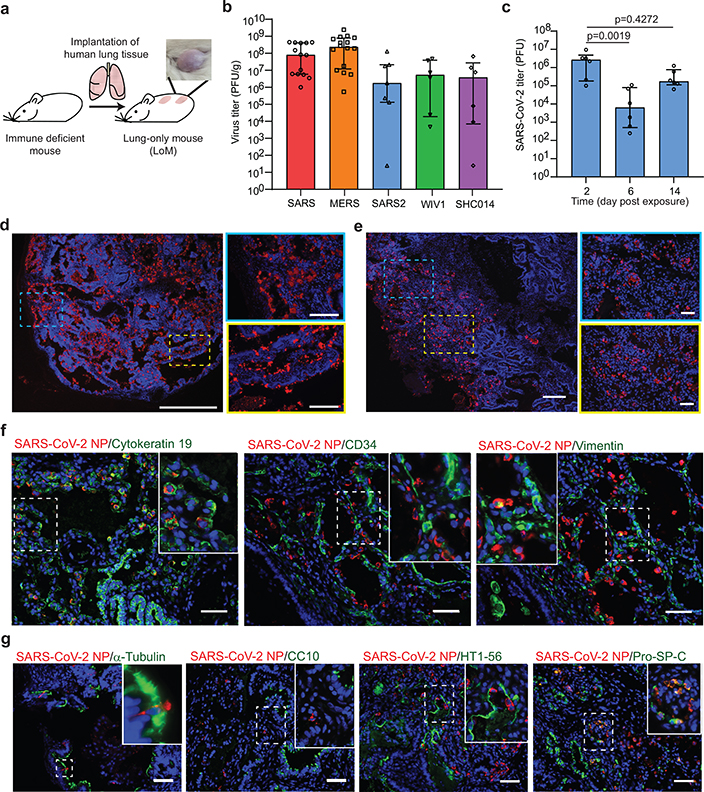
Fig. 1. Robust replication of recently emerged human and bat coronaviruses in LoM demonstrate the potential of bat coronaviruses for direct transmission to humans and the predilection of SARS-CoV-2 for infection of human epithelial cells.
a, LoM construction and image of a human lung implant. b, Viral titers in the human lung tissue of LoM injected with SARS-CoV (n=14, red), MERS-CoV (n=16, orange), SARS-CoV-2 (n=7, blue), WIV1-CoV (n=6, green), or SHC014 (n=6, purple) as determined by plaque assay (PFU, plaque forming units). c, SARS-CoV-2 titers in the human lung tissue of LoM at days 2 (n=6), 6 (n=6), and 14 (n=6) post-exposure were compared with a two-sided Kruskal-Wallis with Dunn’s multiple comparisons test. d, SARS-CoV-2 RNA in LoM human lung tissue 2 days post-exposure (SARS-CoV-2 RNA+, red; nuclei, blue; scale bars, 750 um [left image] and 250 um [right images], n=3). e, Virus nucleoprotein in LoM human lung tissue two-days post-exposure (positive cells, red; nuclei, blue; scale bars 200 um [left image] and 50 um [right images], n=6). f, Co-staining of LoM human lung tissue two days following SARS-CoV-2 exposure for virus nucleoprotein (red) and cytokeratin 19 (epithelial cells, green, n=6), CD34 (endothelial cells, green, n= 4), or vimentin (mesenchymal cells, green, n=4). Nuclei, blue; scale bars 50 um. g, Co-staining of LoM human lung tissue two days following SARS-CoV-2 exposure for virus nucleoprotein (red) and acetylated alpha-tubulin IV (ciliated cells, green, n=6), CC10 (club cells, green, n=6), HT1–56 (alveolar type 1 cells, green, n=6), or Pro-SP-C (alveolar type 2 cells, green, n=3). Nuclei, blue; scale bars 50 um. In b and c, horizontal and vertical lines represent the median and interquartile range respectively. n= number of biologically independent lung tissues analyzed.
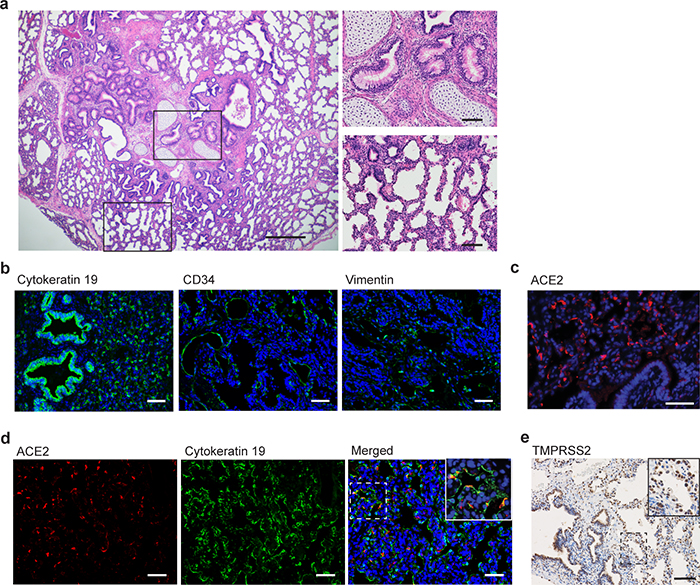
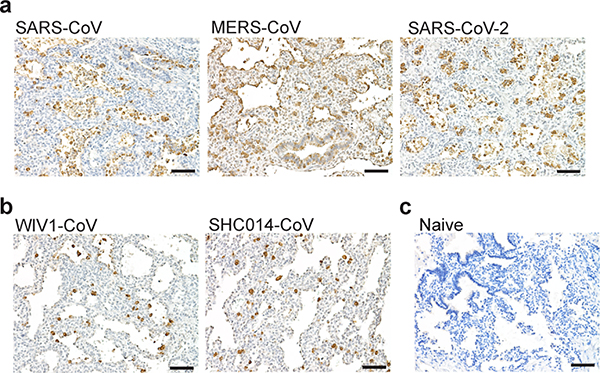
We evaluated the potential of LoM to serve as a single platform to study all known recently emerged human coronaviruses and the potential of endogenous bat coronaviruses for human emergence. Human angiotensin converting enzyme-2 (ACE2), the receptor for SARS-CoV and SARS-CoV-218–21, is expressed on human epithelial cells (cytokeratin 19+) in the human lung tissues of LoM (Extended Data Fig. 1c,d). Expression of transmembrane protease serine 2 (TMPRSS2), which primes the spike protein of SARS-CoV and SARS-CoV-218, was also confirmed (Extended Data Fig. 1e). LoM were inoculated with SARS-CoV, MERS-CoV, or SARS-CoV-2 (Extended Data Table 1). SARS-CoV and SARS-CoV-2 infection resulted in mean virus titers of 1.76×108 and 2.42×107 PFU/g respectively at 2 days post-infection (Fig. 1b). Viral nucleoprotein antigen was abundantly observed in the human lung tissues of SARS-CoV and SARS-CoV-2 infected LoM (Extended Data Fig. 2a). Consistent with our previous results17, MERS-CoV replicated to mean titers of 4.79×108 PFU/g in LoM human lung tissues at 2 days post-infection (Fig. 1b) and abundant viral antigen was observed (Extended Data Fig. 2a).
Pre-pandemic bat coronaviruses WIV1-CoV and SHC014-CoV have high sequence homology to SARS-CoV, use ACE2 to infect human cells, and grow modestly in primary human airway cultures on liquid interface12,13. WIV1-CoV and SHC014-CoV efficiently replicated in the human lung tissue of LoM (Extended Data Table 1) with mean titers of 1.58×107 and 1.48×107 PFU/g, respectively at two days post-exposure (Fig. 1b) and viral antigen was readily detected in human lung tissues (Extended Data Fig. 2b). No viral antigen was detected in human lung tissues of naïve LoM (Extended Data Fig. 2c). Collectively, these results demonstrate that LoM serve as single platform where recently emerged human coronaviruses SARS-CoV, MERS-CoV, and SARS-CoV-2 replicate efficiently in human lung tissue. Importantly, the efficient replication of SARS-like bat coronaviruses WIV1-CoV and SHC014-CoV observed is in agreement with previous in vitro data12,13 suggesting that bats harbor coronaviruses capable of direct transmission to humans, bypassing the need for further adaptation in an intermediary host.
SARS-CoV-2 replication in LoM
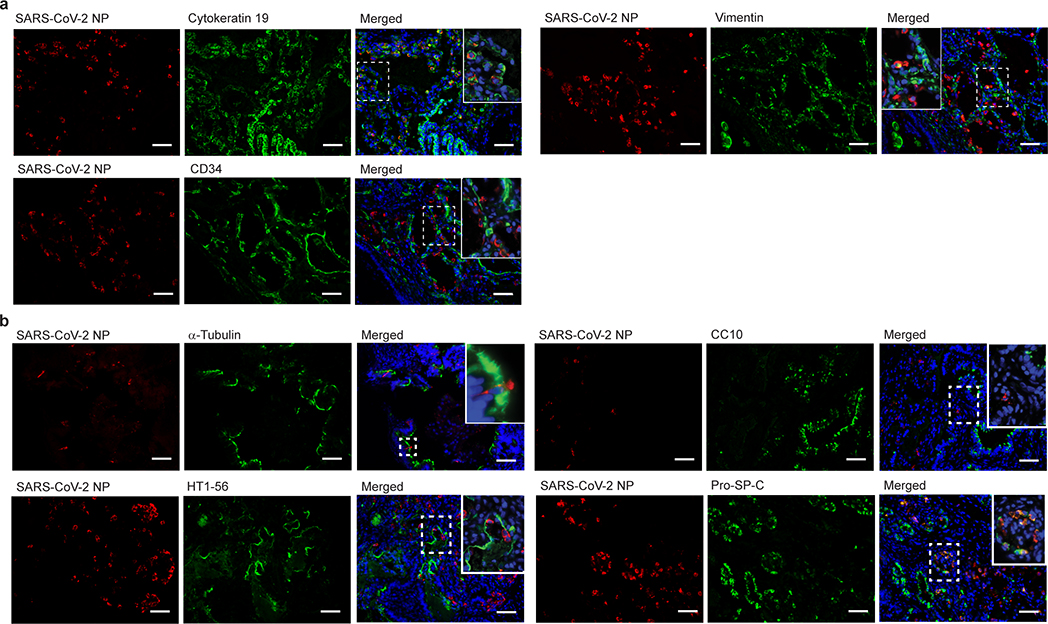
Human lung tissues of LoM were inoculated with SARS-CoV-2 and titers of replication competent virus determined 2, 6, and 14 days post-exposure (Fig. 1c, Extended Data Table 2). High titers of replication competent virus were noted at all time points although they were highest 2 days post-infection (Fig. 1c). Infection was widely distributed throughout the tissue with large numbers of cells positive for viral RNA (Fig. 1d) and nucleoprotein (Fig. 1e). Co-staining for human cytokeratin 19 demonstrated that SARS-CoV-2 predominantly infects human epithelial cells in the lung (Fig. 1f, Extended Data Fig. 3a). Viral antigen was not detected in human CD34 expressing (endothelial) cells and in only a few human vimentin expressing (mesenchymal) cells (Fig. 1f, Extended Data Fig. 3a). We further identified the types of infected epithelial cells. Virus antigen was clearly identified in cells which expressed pro-SP-C (alveolar type II [AT2] pneumocytes) or acetylated alpha-tubulin IV (ciliated cells); virus antigen was not detected in HT1–56 (alveolar type I [AT1] pneumocytes) or CC10 positive cells (club cells) (Fig. 1g, Extended Data Fig. 3b). These results demonstrate that SARS-CoV-2 has limited tropism in the lung with AT2 pneumocytes and ciliated airway epithelial cells being the predominant cells infected.
SARS-CoV-2 pathogenesis in LoM
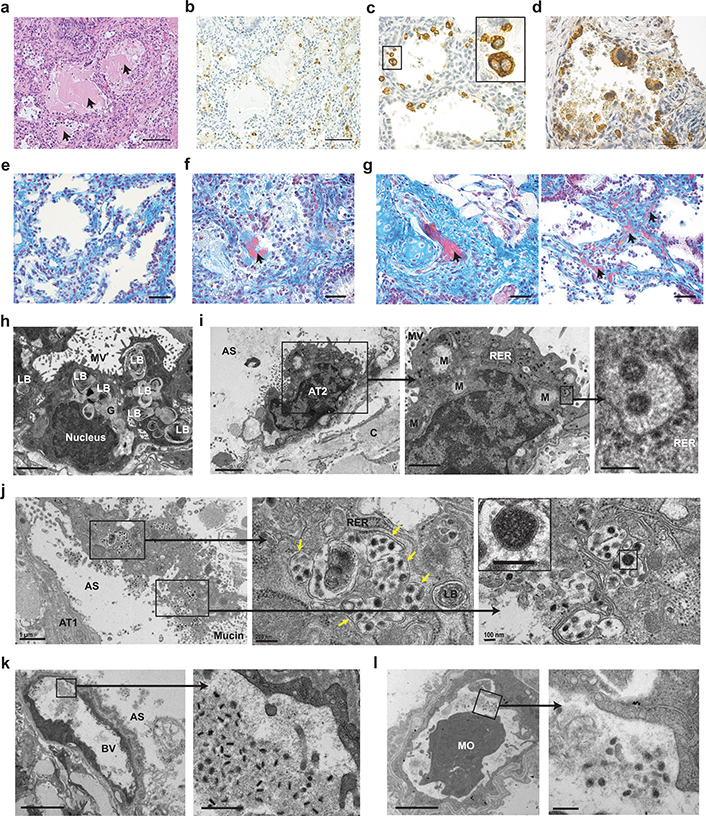
Histopathologic analysis revealed several features of early diffuse alveolar damage that have been described in lung tissues of COVID-19 patients including the accumulation of proteinaceous exudate and fibrin in alveolar spaces, desquamation of pneumocytes, multi-nucleated cell formation, and the appearance of fibrin thrombi in small vessels (Fig. 2)22–24. Proteinaceous exudate, including large protein globules, was observed in alveolar spaces, which overlapped with areas of virus accumulation (Fig. 2a,b). As early as 2 days post-infection, desquamation of pneumocytes was noted; a large number of virally infected cells were fully detached or detaching from the alveolar basement membrane (Fig. 2c,d). Infected multi-nucleated cells were also observed (Fig. 2c). While hyaline membranes were not noted, in contrast to naïve LoM (Fig. 2e), fibrin was detected in alveolar spaces (Fig. 2f). Importantly, we observed multiple occluded vessels containing fibrin thrombi as reported in COVID-19 patient lungs (Fig. 2g)22–24. Electron microscopy demonstrated the normal architecture and integrity of uninfected AT2 pneumocytes in human lung tissue obtained from LoM two days post-infection (Fig. 2h). In contrast, AT2 cells containing virus particles in the same sample had swollen mitochondria with loss of matrix and cristae as well as rough endoplasmic reticula (RER) with distended cisternae, protein accumulation, and virus particles (Fig. 2i). Degenerative SARS-CoV-2 infected AT2 cells detached from the alveolar basal membrane were observed in the alveolar luminal space (Fig. 2j). Higher magnification revealed subcellular accumulation of virus containing vesicles indicative of virus replication and egress. Virions with electron dense nucleocapsids and distinctive crown-like spikes were observed (Fig. 2i,j). Consistent with previous reports23,25, virions produced by human lung cells were pleomorphic in size (69 to 112 nm) and shape. Despite the extensive damage observed in the lung tissue, the endothelium in the majority of blood vessels was intact with tight junctions, numerous pinocytotic vesicles, and normal mitochondria and endoplasmic reticulum (Fig. 2k,l). Virions were not detected within endothelial cells in agreement with our immunofluorescence analysis (Fig. 1f and Fig. 2k,l). However, pleomorphic virions were present in capillary lumen surrounded by fibrillar protein deposits and cell debris (Fig. 2k,l). Together, these results demonstrate that acute SARS-CoV-2 infection of LoM closely resembles lung infection in humans and is highly cytopathic resulting in significant injury to the fragile alveolar lung structures.
Fig. 2. Acute SARS-CoV-2 infection is highly cytopathic and causes extensive damage to human lung structures.
a, H&E staining of a SARS-CoV-2 infected LoM human lung tissue 2 days post-exposure (scale bar, 100 um, n=6). Arrows indicate protein globules. b-d, Immunohistochemical staining for virus nucleoprotein in LoM human lung tissue 2 days following SARS-CoV-2 exposure (positive cells, brown; b scale bars, 100 um; c and d scale bars, 50 um, n=6). e-g, Martius Scarlet Blue staining of human lung tissue from e, naïve LoM (n=6) and f and g, SARS-CoV-2 infected LoM 2 days post-exposure (scale bars, 50 um; fibrin, red; collagen, blue, n=6). Arrows indicate the presence of fibrin (red) in f, alveoli or in g, thrombi of occluded vessels. h-l, Electron microscopy analysis of SARS-CoV-2 infected human lung tissue two days post-exposure (n=3). h, Uninfected AT2 cells in an alveolus-like structure. Scale bars, 2 um. i, SARS-CoV-2 infected AT2 cell. Higher magnification images of boxed areas show virus particles with dense nucleocapsids in RER. Scale bars, 2 um (left image), 1 um (middle image), and 200 nm (right image). j, Degenerative SARS-CoV-2 infected cell in the alveolar space. Arrows indicate virus filled vesicles. Scale bars, 1 um (left image), 200 nm (middle image), and 100 nm (right image). k and l, Blood vessels containing virions, fibrillar protein and cell debris. In k, scale bars, 5 um (left image) and 500 nm (right image). In l, scale bars, 2 um (right image) and 200 nm (left image). AS, alveolar space; AT1, alveolar type 1 cells; AT2, alveolar type 2 cells; BV, blood vessel; C, collagen; G, glycogen; LB, lamellar body; M, mitochondria; MO, monocyte; MV, microvilli; RER, rough endoplasmic reticulum. In c, i-l, black boxes indicate areas of higher magnification images. n= number of biologically independent lung tissues analyzed.
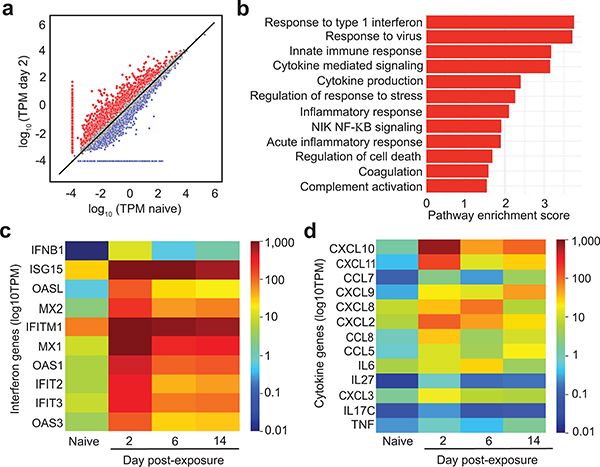
We performed RNA-sequencing analysis of human lung tissues collected from animals 2, 6 and 14 days post-infection. Abundant viral transcripts were detected, ranging from 0.55% to 3.6% of the total reads at 2 days post-infection (Extended Data Table 3). Viral transcripts were abundant but lower at 6 days and 14 days post-infection (Extended Data Table 3). Sequencing data identified canonical SARS-CoV-2 transcripts26 and confirmed maintenance of the furin cleavage site in the spike protein. Analysis of human gene transcripts revealed 1,504 differentially expressed cellular genes between naïve and infected human lung tissue at 2 days post-exposure, the peak of infection (Fig. 3a, Supplementary Tables 1 and 2). Of these genes, differential expression analysis suggested 1,043 were increased and 461 were decreased in expression in the infected human lung tissue relative to naïve controls (Supplementary Tables 1 and 2). Three patterns of note: the expression of most genes did not change with infection, many genes with increased expression in SARS-CoV-2 infected LoM were lowly or moderately expressed in naïve LoM, and a handful of genes went from either undetectable expression in naïve LoM to moderate/high expression in infected LoM or from expression in naïve LoM to undetectable expression in infected LoM. As expected, ACE2 and TMPRSS2 were expressed across the lung tissues (Extended Data Table 4). Notably, numerous interferon-stimulated genes (ISGs) and inflammatory cytokine genes, including pro-inflammatory cytokines genes IL6, CXCL8 (IL-8), CXCL10 (IP-10), TNF, and CCL5 (RANTES) were potently induced in infected lung tissue (Supplementary Tables 1 and 2). We also observed dramatic upregulation of IFNB1, IFNL1, IFNL2, and IFNL3 expression (>1,000 fold for all) at 2 days post-exposure, suggesting that these cytokines play a key role in the antiviral response to SARS-CoV-2 (Supplementary Tables 1 and 2). Gene set enrichment analysis (GSEA) showed over 840 gene pathways significantly upregulated (p<0.05) including response to type 1 interferon (p=0.0011), response to virus (p=0.0010), innate immune response (p=0.0010), cytokine mediated signaling (p=0.0010), cytokine production (p=0.0010), response to stress (p=0.0010), inflammatory response, (p=0.0010), NIK NF-KB signaling (p=0.0011), acute inflammatory response (p=0.0035), regulation of cell death (p=0.0030), and coagulation pathways (p=0.0453) (Fig. 3b). Complement activation, which contributes to SARS-CoV pathogenesis in mouse models14, was also increased (p=0.0470) (Fig. 3b). Importantly, analysis of host gene expression at later time points demonstrated a sustained upregulation of antiviral and inflammatory genes that in some instances (e.g. ISG15, IFITM1, TNF, CXCL9) persisted for up to 14 days post-infection (last time analyzed) (Fig. 3c,d, Extended Data Table 5, Supplementary Tables 1 and 2). These results demonstrate that acute SARS-CoV-2 infection causes a potent and sustained upregulation of innate immune responses in virus-infected human lung tissue.
Fig. 3. SARS-CoV-2 infection induces a strong and sustained host innate immune response in human lung tissue.
a-d, RNA-sequencing analysis of human lung tissue collected from SARS-CoV-2 infected LoM. a, Log10 gene transcripts per million (TPM) in the human lungs of naïve LoM (n=2, x-axis) and SARS-CoV-2 infected (n=2, y-axis) LoM day 2 post-exposure. Genes of interest from Supplementary Tables 1 and 2 that appear increased (red) or decreased (blue) at least two-fold in SARS-CoV-2 infected LoM are shown. Genes having zero (0) mean TPM in either naïve or infected LoM are set at a minimum of log10 (0.0001) TPM in order to visualize. b, Gene set enrichment analysis (GSEA) identified gene sets enriched in SARS-CoV-2 infected LoM human lungs. The pathway enrichment score is shown on the x-axis. The statistical significance of enrichment scores was determined using a two-sided empirical phenotype-based permutation test and adjusted for multiple testing using a False Discovery Rate (red, p<0.05). Heatmaps illustrating the expression of human c, interferon genes and d, cytokine/chemokine genes in human lung tissue collected analyzed from SARS-CoV-2 infected LoM days 2 (n=2), 6 (n=3), and 14 (n=3) post-exposure and naïve LoMs (n=4). Color scale indicates the mean log10 TPM. n= number of biologically independent lung tissues analyzed.
EIDD-2801 treatment and prophylaxis
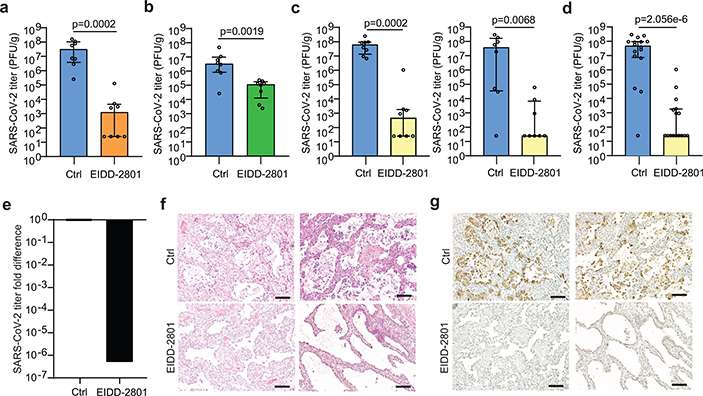
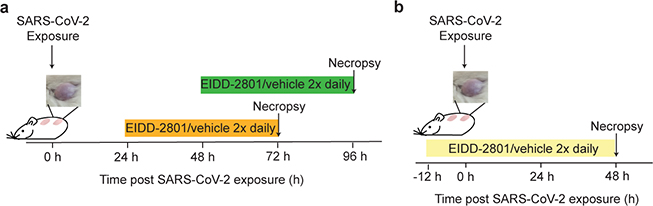
The ribonucleoside analog β-D-N4-hydroxycytidine (NHC) broadly inhibits coronavirus infection in vitro in human airway epithelial cell cultures15. Prophylactic and therapeutic administration of its oral pro-drug EIDD-2801 (also known as molnupiravir or MK-4482) reduced SARS-CoV and MERS-CoV replication and pathogenesis in mice15. 5’-triphosphate NHC acts as a competitive alternate substrate for the viral RNA-dependent RNA polymerase allowing it’s incorporation into viral RNA resulting in the accumulation of mutations within the viral RNA genome leading to error catastrophe15. As an orally bioavailable agent, EIDD-2801 could be much more readily administered to patients compared to remdesivir and other antivirals and biologics (e.g. convalescent plasma and monoclonal antibodies) which require administration by infusion in a clinical setting. We tested the ability of therapeutic EIDD-2801 to inhibit SARS-CoV-2 replication in vivo using a dose which is also similar to the human dose in clinical trials27. LoM were administered EIDD-2801 starting 24 h or 48 h post SARS-CoV-2 exposure and every 12 h thereafter (Extended Data Fig. 4a, Extended Data Table 6). Our results show that EIDD-2801 had a remarkable effect on virus replication after only two days of treatment (Fig. 4a,b). EIDD-2801 dramatically reduced the number of infectious particles in human lung tissue by 4.4 logs (>25,000 fold decrease) when treatment was initiated 24 h post-exposure (p=0.0002) (Fig. 4a). When treatment was started 48 h post-exposure, virus titers were significantly reduced by 96% (1.5 logs, p=0.0019) (Fig. 4b). Next, we tested the efficacy of EIDD-2801 pre-exposure prophylaxis. LoM were administered EIDD-2801 starting 12 h prior to SARS-CoV-2 exposure and every 12 h thereafter (Extended Data Fig. 4b, Extended Data Table 6). EIDD-2801 pre-exposure prophylaxis significantly reduced virus titers in the human lung tissues of LoM by over 100,000 fold in two independent experiments (p=0.0002 and p=0.0068, Fig. 4c–e). Furthermore, in contrast to EIDD-2801 treated mice, abundant cell debris and nucleoprotein positive cells could be readily observed in the alveolar lumen of vehicle control treated mice consistent with the extensive pathogenic effects inflicted on the lung by SARS-CoV-2 (Fig.4f,g). These results demonstrate that prophylactic administration EIDD-2801 is highly effective at preventing SARS-CoV-2 infection and pathogenesis in vivo.
Fig. 4. Treatment and pre-exposure prophylaxis with EIDD-2801, a broad-spectrum anti-coronavirus drug, potently inhibit SARS-CoV-2 infection in vivo.
a, SARS-CoV-2 titers in the human lung tissue of LoM administered EIDD-2801 (n=8) or vehicle (n=8) 24 h post virus exposure. b, SARS-CoV-2 titers in the human lung tissue of LoM administered EIDD-2801 (n=8) or vehicle (n=8) 48 h post virus exposure. c and d, SARS-CoV-2 titers in the human lung tissue of LoM administered EIDD-2801 (n=8 per experiment, yellow) or control vehicle (Ctrl, n=8 per experiment, blue) at 2 days post-exposure in two independent experiments shown c, separately and d, combined. e, Fold difference in SARS-CoV-2 titers in the human lung tissue of LoM relative to vehicle controls. f, H&E staining and g, immunohistochemical staining for virus nucleoprotein (positive cells, brown) of human lung tissue of LoM administered EIDD-2801 (n=8) or control vehicle (Ctrl, n=8) at 2 days post-exposure (scale bars, 100 um). a-d, Titers were compared with a two-tailed Mann-Whitney U test. Horizontal and vertical lines represent the median and interquartile range respectively. n= number of biologically independent lung tissues analyzed.
Discussion
Our results demonstrate replication of all known recently emerged human coronaviruses in LoM. Importantly, in agreement with in vitro studies12,13, our results demonstrate relatively efficient replication of two pre-pandemic endogenous bat viruses in human lung tissue in vivo indicating that coronaviruses circulating in bats have pandemic potential without the need for further adaptation to humans. Acute SARS-CoV-2 infection of LoM resulted in significant lung injury and exhibited key features of the extensive lung pathology observed in severe COVID-19 patients22,24,28,29. In agreement with analyses of bronchoalveolar fluid obtained from COVID-19 patients and post-mortem patient lung samples5,30, multiple ISGs were significantly increased in SARS-CoV-2 infected human lung tissue of LoM. We observed robust induction of IFNB1 expression during acute infection followed by a decline in expression. Interestingly, an analysis of post-mortem COVID-19 patient lungs5 did not reveal increased IFNB1 expression and it has been shown in vitro that its expression is blocked during SARS-CoV infection31,32. These results suggest that in human lung tissue IFNB1 expression is induced during acute SARS-CoV-2 infection. While this study was not designed to evaluate the effect of type I IFN on SARS-CoV-2 replication in vivo, in vitro studies and in vivo mouse studies showed that type I IFN treatment restricts SARS-CoV-2 replication7,33. We also observed increased expression of several human cytokine genes in SARS-CoV-2 infected LoM, many of which were also increased in analyses of COVID-19 patient serum and post-mortem lung tissue further establishing the similarities between LoM and human SARS-CoV-2 infection5,34.
Currently, the FDA approved SARS-CoV-2 vaccines are highly effective at preventing disease. However, it may take considerable time to reach target vaccination levels needed for herd immunity, especially in resource limited settings, due to manufacturing capabilities, required vaccine shipping and storage conditions, and public acceptance. Therefore, alternative treatments and preventive approaches are still urgently needed. Remdesivir, is limited to use in hospitalized patients and its effectiveness at reducing disease and mortality late in infection is not clear35,36. While a carefully controlled NIH sponsored clinical trial reported that remdesivir significantly shortened the time of recovery35, the WHO SOLIDARITY trial revealed no significant reduction in the duration of hospitalization or mortality36. Early administration of monoclonal antibody therapies bamlanivimab and REGN-CoV2 significantly reduced viral loads in COVID-19 patients with mild to moderate disease37,38. However, they must be administered by infusion in a clinical setting limiting their potential for widespread use. A highly efficacious antiviral that is administered orally has potential for more widespread use and for administration to patients with mild to moderate disease39. Here, we show that EIDD-2801 administered therapeutically for only two days significantly reduced infectious virus titers in vivo by over 4 logs. The sooner EIDD-2801 treatment was initiated after SARS-CoV-2 exposure, the greater the reduction in virus replication. We also observed that prophylactic administration of EIDD-2801 efficiently prevents SARS-CoV-2 infection in vivo highlighting its potential utility as an effective prophylactic and therapeutic agent against SARS-CoV-2 and other past and future zoonotic coronaviruses. NHC belongs to a class of ribonucleoside analogs known to often affect mitochondrial replication and function. However, NHC did not cause significant mitochondrial toxicity or impair mitochondrial function in vitro40. Furthermore, no significant increase in transition frequencies in nuclear or mitochondrial message was detected in the lung tissue of ferrets dosed with EIDD-2801 for seven days consistent with the observed good tolerability of the drug41. Phase II and Phase II-III clinical trials are ongoing to evaluate EIDD-2801 safety and its effect on viral shedding using doses up to 1,600 mg/day twice daily, a dose that is expected to provide similar intracellular 5’-triphosphate NHC levels as a 300–500 mg/kg dose in mice. As with any antiviral, its therapeutic/prophylactic use will be dictated by risk/benefit. Limitations of our study include the absence of human nasal airway structures in LoM which are thought to be early sites of SARS-CoV-2 replication in humans42. Since LoM do not have an autologous human adaptive immune system they reflect the direct effect of viruses on their targets and bystander cells as well as their innate immune response to infection. Collectively, our results demonstrate the utility of LoM as a single in vivo platform to evaluate and compare the replication and pathogenesis of past, present, and future pre-emergent, epidemic, and pandemic coronaviruses accelerating the development and testing of therapeutic and pre-exposure prophylaxis agents such as EIDD-2801.
Methods
Ethics statement
Animal studies were carried out according to protocols approved by the Institutional Use and Care Committee at UNC-Chapel Hill and in adherence to the NIH Guide for the Care and Use of Laboratory Animals. Mice were kept on a 12 h light/12 h dark cycle and housed in a temperature (20–23°C) and humidity (30–70%) controlled vivarium maintained by the Division of Comparative Medicine at the University of North Carolina-Chapel Hill.
Experimental design
Human lung-only mice (LoM) were used as an in vivo model to evaluate infection of lung tissue with recombinant coronaviruses SARS-CoV, MERS-CoV, and SARS-CoV-2 as well as full length bat coronaviruses WIV1 and SHC01412,13,43,44. Viruses were directly injected into the human lung tissue of LoMs and lung tissue collected either 2, 6, or 14 days post-exposure for virus titer determination and/or analysis by histology, electron microscopy, or RNA-seq.
Construction of humanized mice
LoMs were constructed with 1–2 human lung implants by surgically implanting human lung tissue (Advanced Bioscience Resources) subcutaneously into the upper and lower back of male and female 12–21 week old NOD.Cg-Prkdcscid ll2rgtm1Wjl/SzJ mice [NSG mice; The Jackson Laboratory] as previously described17. Engraftment of lung tissue was assessed by palpation and by 8 weeks post-surgery animals were ready for experimentation.
Production of coronavirus stocks and infection of humanized mice
Stocks of wild-type SARS-CoV, MERS-CoV (HCoV-EMC/2012), SHC014-CoV, and WIV1-CoV were derived from infectious virus clones and were prepared and titered on Vero E6 (SARS-CoV, SHC014-CoV, and WIV1-CoV) or Vero CCL81 cells (MERS-CoV) (American Type Culture Collection) as previously described6,12,13,17. Cell lines were authenticated by morphological identification and virus susceptibility profiles and tested for mycoplasma by the supplier. A clinical isolate of SARS-CoV-2 (2019-nCoV/USA-WA1/2020) was obtained from the U.S. Centers for Disease Control and Prevention (GenBank accession no. MN985325.1) and passaged twice in Vero E6 cells to create a passage 5 working stock42. For the infection of mice with coronavirus, the fur over the human lung tissue of anesthetized mice was shaved and virus (1–3×105 PFU in 100 ul PBS) was injected directly into the lung tissue. To evaluate the in vivo inhibitory activity of EIDD-2801 pre-exposure prophylaxis, mice were administered 500 mg/kg EIDD-2801 or vehicle control (10% PEG and 2.5% Cremophor RH40 in water) via oral gavage starting 12 h prior to SARS-CoV-2 exposure and every 12 h thereafter. The therapeutic efficacy of EIDD-2801 was assessed by administering mice 500 mg/kg EIDD-2801 or vehicle control (10% PEG and 2.5% Cremophor RH40 in water) via oral gavage starting 24 h or 48 h post exposure and every 12 h thereafter. At necropsy, human lung tissues were collected, weighed (mean weight: 0.55 grams ± 0.05 s.e.m, n=32), homogenized, and stored at −80°C until tittering on Vero E6 cells. Titers below the limit of the assay (50 PFU/mL) were assigned a value of 25 PFU/gram. No correlation was observed between SARS-CoV-2 titers (PFU/g) at 2 days post-exposure and the timing post-surgery (p=0.4474, two-tailed Spearman’s rank correlation).
H&E staining of human lung tissue
Human lung tissues collected from LoM were fixed in 10% formalin, paraffin embedded, and cut into 5 μm sections which were mounted onto Superfrost Plus slides (Fisher Scientific). Tissue sections were incubated at 60°C for 1 h, deparaffinized with xylene (2 × 3 min) and graded ethanol (100% 2 × 3 min, 95% 1 × 3 min, 80% 1× 3min, 70% 1 × 3 min), and stained with hematoxylin followed by eosin. Tissue sections were then mounted and imaged on a Nikon Eclipse Ci microscope using Nikon Elements BR software (version 4.30.01) with a Nikon Digital Sight DS-Fi2 camera. Brightness, contrast and white balance were adjusted on whole images in Adobe Photoshop (CS6).
Immunohistochemical analysis of coronavirus infection
Immunohistochemistry was performed as previously described17. Briefly, fixed (10% formalin) human lung tissues collected from coronavirus-infected LoM were paraffin embedded and sectioned (5 um). Tissue sections mounted on Superfrost Plus slides (Fisher Scientific) were deparaffinized as described above. Following antigen retrieval using Diva Decloaker (BioCare Medical), non-specific binding was blocked using Background Sniper (BioCare Medical). Tissue sections were then incubated with primary antibodies against SARS-CoV nucleocapsid (1:500 or 1:1,000), MERS-CoV nucleocapsid (1:2,000), or TMPRSS2 (1:100) overnight at 4°C. Tissue sections incubated with rabbit IgG were used as isotype controls. Tissue sections were then washed in TBST and the endogenous peroxidase activity blocked with hydrogen peroxide. Tissue sections were developed using the MACH-3 polymer system (BioCare Medical) and 3,3′-diaminobenzidine (DAB) (Vector Laboratories), counterstained with hematoxylin, and mounted. Tissue sections were imaged on a Nikon Eclipse Ci microscope using Nikon Elements BR software (version 4.30.01) with a Nikon Digital Sight DS-Fi2 camera. Adobe Photoshop (CS6) was used to adjust brightness, contrast and white balance on whole images.
Immunofluorescence analysis of SARS-CoV2 infection
Human lung tissues collected from mice were fixed in 10% formalin and paraffin embedded. Immunofluoresence staining of 5 um tissue sections was performed as previously described17. Briefly, following deparaffinization and antigen retrieval (Diva Decloaker), tissue sections were incubated with a 10% normal donkey serum solution with 0.1% Triton X-100 in 1x PBS to block non-specific binding. Tissue sections were then incubated overnight with primary antibodies at 4°C followed by incubation with fluorescent conjugated secondary antibodies. Primary antibodies were directed against SARS nucleoprotein (1:500 or 1:1,000), ACE2 (1:200), human cytokeratin 19 (1:250), CD34 (1:100), vimentin (1:100), acetylated alpha-tubulin IV, CC10 (1:500), HT1–56 (1:150), and pro-SP-C (1:500). Secondary antibodies were: Donkey anti-Mouse IgG heavy and light chains-AlexaFluor 488 (1:1,000), Donkey anti-Rabbit IgG heavy and light chains-AlexaFluor 488(1:1,000), Donkey anti-Goat IgG heavy and light chains-AlexaFluor 594 (1:1,000), Donkey anti-Mouse IgG heavy and light chains-AlexaFluor Plus 594 (1:1,000), Donkey anti-Rabbit IgG heavy and light chains-AlexaFluor Plus 594 (1:1,000), Donkey anti-Mouse IgG heavy and light chains-AlexaFluor 647 (1:1,000). Background autofluorescence was then quenched using a 0.1% Sudan Black B solution in 80% ethanol prior to staining with DAPI. Slides were mounted and then imaged using an Olympus BX61 upright wide-field microscope using Volocity software (version 6.3) with a Hamamatsu ORCA RC camera. Appropriate negative controls without primary antibodies were also imaged using the same exposure time as matching stained sections. Whole image contrast, brightness, and pseudocoloring were adjusted using ImageJ/Fiji (Version 2.0.0-rc-69/1.51w) and Adobe Photoshop (version CS6).
RNA in situ hybridization (RNA-ISH) analysis of SARS-CoV2 infection
RNA-ISH was performed on 10% formalin fixed, paraffin-embedded, 5 μm sections of human lung tissues using the RNAscope 2.5 HD Reagent Kit according to the manufacturer’s instructions (Advanced Cell Diagnostics). Briefly, tissue sections were mounted on Superfrost Plus microscope slides (Fisher Scientific), heated at 60°C for 1 h, deparaffinized in xylene (2 × 5 minutes) and 100% ethanol (2×2 minutes), and air-dried. Tissue sections were then incubated with hydrogen peroxide to block endogenous peroxidases for 10 min at RT, followed by epitope retrieval (Advanced Cell Diagnostics) for 30 min in a 95°C water bath. Subsequently, tissue sections were immediately washed in double distilled water then dehydrated in 100% ethanol for 2 min before air-drying. Tissue sections were then incubated with Protease Plus (Advanced Cell Diagnostics) for 30 min at 40°C in a HybEZ hybridization oven (Advanced Cell Diagnostics). Sections were rinsed 3 times in double distilled water and then incubated with pre-warmed target probe designed to hybridize with the spike protein of SARS-CoV-2 (Cat. Number 848561, Advanced Cell Diagnostics) for 2 h at 40°C. Tissue sections were then washed and the signal amplified according to the manufacturer’s instructions and developed using alkaline phosphatase and Fast Red substrate. Tissue sections were counterstained with DAPI, mounted with Prolong® Gold (Invitrogen), and imaged on an EVOS M5000 microscope (Invitrogen).
Electron microscopy analysis of SARS-CoV2 infection
Small pieces of human lung tissue collected from SARS-CoV-2 infected LoM at two days post-infection were fixed in 4% paraformaldehyde/2.5% glutaraldehyde in 0.15 M sodium phosphate buffer, pH 7.4, for 2 h at RT. The tissues were subsequently transferred to 10% formalin for 7 days. Specimens were washed in 0.1 M sodium cacodylate, pH 7.4, then post-fixed with 1% cacodylate-buffered osmium tetroxide for 1 h. After washing in 0.05 M sodium cacodylate buffer, pH 7.0, the samples were treated with 1% tannic acid in 0.05 M sodium cacodylate buffer for 30 min to enhance tissue contrast and preserve structure45. Tissue pieces were washed in deionized water, dehydrated in ethanol, and placed through two exchanges of propylene oxide before infiltration and embedment in PolyBed 812 epoxy resin (Polysciences). Semi-thin (1 μm) sections of tissue blocks were cut and stained with 1% toluidine blue in 1% sodium borate for examination by light microscopy. Ultra-thin (70 nm) sections were cut of selected regions of interest, mounted on 200 mesh copper grids and stained with 4% aqueous uranyl acetate and Reynolds’ lead citrate. Grids were observed on a JEOL JEM 1230 transmission electron microscope operating at 80kV (JEOL USA, Inc.) and images were acquired with a Gatan Orius SC1000 CCD Digital Camera and Gatan Microscopy Suite software (version 3.0, Gatan, Inc.). Virus particle sizes were measured in Fiji/Image J (version 2.0.0-rc-69/1.52p).
Processing of human lung tissues for RNA-sequencing analysis
Human lung tissues were collected in RNAlater and kept at 4°C for 24 h prior to storage at −80°C until further processing. To isolate RNA, samples stored in RNAlater were thawed and the tissue transferred to a new tube containing 1 mm glass beads and 1 mL Trizol. Tissues were subsequently homogenized using a MagNA Lyser (Roche) for 30 sec at 6,000 rpm. In between rounds of homogenization, tissues were incubated on ice for 1 min. Following tissue homogenization, Trizol homogenate was transferred to a new tube and stored at −80°C.
RNA-sequencing analysis
RNA was extracted from lung samples using a Trizol Plus RNA extraction kit (Thermo Fisher), quantified using a Qubit RNA assay kit and checked for quality using a Bioanalyzer RNA600 Nano kit (Agilent). RNA integrity scores were typically 7.0 and greater. 1ug of RNA was used to construct libraries for sequencing using a NEBNext Ultra II library prep kit with polyA RNA selection. Barcoded libraries were sequenced on a Novaseq 6,000 2×100 bp following manufacturer’s instructions (Illumina). Sequence quality was assessed using FASTQC (version 0.11.9). No issues were detected with the data and quality was typical for RNA extracted from fresh frozen material. A small amount of index hopping was detected (0.09%) due to the single indices used in the library preparation. Raw reads were mapped to the human, mouse, and SARS-CoV-2 reference genomes simultaneously (GRCh38.p13, GRCm38.p6 M25, NC_045512, respectively) using the BBsplit function in BBmap (version 38.86). This step minimized cross mapping of reads among genomes. There was a non-significant, negative relationship between the amount of viral and human RNA reads (R2=0.136). We then mapped and quantified on a transcript and gene model basis using STAR (version 2.7.5a) and Salmon (version 1.2.1)46,47. Reads mapping to multiple locations were dropped from analysis. On average, in the lung tissue samples from LoM mice, 80% of the reads mapped to human (standard deviation: +/− 6%), 19% mapped to mouse (standard deviation: +/− 6%) and 1% mapped to the SARS-CoV-2 genome (standard deviation: +/− 1%). The percent of virus ranged from 0.05% to 3.4% among infected mice, with day 2 mice having the most, which is consistent with the infection titers observed. Samples from naïve mice were 95% or more human data.
Statistics and reproducibility
No statistical methods were used to predetermine sample size. No randomization was used to determine allocation of samples/animals to experimental groups and downstream analysis. The investigators were not blinded to group allocation for data collection and analysis. In Fig.1b, titers for SARS-CoV and MERS-CoV represent two independent experiments and titers for SARS-CoV-2, WIV1-CoV, and SHC014-CoV represent one experiment. Virus titers between SARS-CoV-2 infected LoMs analyzed 2 post-exposure and days 6 and 14 post-exposure (Fig.1c) were compared with a two-sided Kruskal-Wallis with Dunn’s multiple comparisons test and data represents one experiment. Pieces of human lung implants obtained from LoM in Fig.1c were used for the RNAscope and immunofluorescence analysis shown in Fig. 1d–g, the histological and EM analyses in Fig.2, and the RNA-seq gene expression analyses in Fig. 3, Extended Data Fig.3, Extended Data Tables 3–5, and Supplementary Tables 1 and 2. The number of independent LoM human lung tissue samples analyzed for each parameter is indicated in the figure and table legends. RNA-sequencing data was normalized and interrogated for changes in gene expression using DESeq2 package (version 3.1.1) in R (version 3.6.3)48 and statistical tests were two-sided. We focused the analysis on the naïve controls versus LoM for days 2, 6, and 14 post-infection. Wald’s tests were performed contrasting each day versus naïve controls. Because mice were sacrificed at each time point, we treated each day independently and not as a time series. P-values were adjusted for multiple testing using a False Discovery Rate using the Benjamini & Hochberg method49. Data was analyzed both jointly and within each treatment compared to naïve controls. Differential expression of outliers was assessed and found insignificant in overall effect. Graphs and summary tables were built in R using ggplot; gene set enrichment was performed using GSEA and GO analysis (tidyverse 1.3.0; PCATools 1.2.0; Sqldf 0.4–11; na.tools 0.3.1; ggbiplot 0.55; ggplot2 3.3.1; dplyr 0.8.4). The statistical significance of GSEA calculated enrichment scores was determined using an empirical phenotype-based permutation test50 and P-values adjusted for multiple testing using a False Discovery Rate using the Benjamini & Hochberg method49. Specific gene sets of interest were then interrogated for patterns of expression across treatment and time using unsupervised clustering of normalized gene expression counts. Gene Ontology (GO) analysis and visualization were performed with Gorilla (no version)51. Data was uploaded to the NCBI GEO archive (accession: GSE155286). In Fig. 4, virus titers in vehicle control and EIDD-2801 dosed LoM were compared with a two-sided Mann-Whitney U test (GraphPad Prism, version 8.0 or R, version 3.5.3). The effect of EIDD-2801 treatment on virus titers (Fig. 4a,b) represents one experiment and the effect of EIDD-2801 prophylaxis (Fig.4c–e) represents two independent experiments. All independent attempts of repetition were completed with similar results.
Extended Data
Extended Data Fig. 1. Human epithelial cells in the human lung tissue of LoMs express ACE2.
a, H&E staining of the human lung tissue of a naïve LoM (scale bars 500 um [left image] and 100 um [right images], n=6). Boxes indicate regions shown in higher magnification images (right images) of cartilaginous airways (top) and non-cartilaginous airways and alveoli (bottom). Immunofluorescence staining for b, human cytokeratin 19 (epithelial cells, green; nuclei, blue; scale bar 50 um), CD34 (endothelial cells, green; nuclei, blue; scale bar 50 um), and vimentin (mesenchymal cells, green; nuclei, blue; scale bar 50 um). n=8 tissues analyzed. c, human ACE2 in the human lung tissue of a naïve LoM (positive cells, red; nuclei, blue; scale bar 50 um, n=9). d, Co-staining for human ACE2 (positive cells, red) and cytokeratin 19 (positive cells, green) in naïve LoM human lung tissue (nuclei, blue; scale bar 50 um, n=9). e, Immunohistochemical staining for TMPRSS2 in naïve LoM human lung tissue (positive cells, brown; scale bar, 100 um; n=6). n=number of biologically independent lung tissues analyzed.
Extended Data Fig. 2. Viral nucleoprotein in the human lung tissues of LoM infected with recently emerged human coronaviruses and bat coronaviruses.
Immunohistochemical staining for virus nucleoprotein in human lung tissue collected from a, LoM two days post exposure to recently emerged human coronaviruses SARS-CoV (n=4), MERS-CoV (n=5), or SARS-CoV-2 (n=6), b, LoM two days post exposure to bat coronaviruses WIV1-CoV or SHC014-CoV (n=10), or c, naïve LoM (n=6). Positive cells, brown. Scale bars, 100 um. n=number of biologically independent lung tissues analyzed.
Extended Data Fig. 3. SARS-CoV-2 infection of human epithelial cell types in the human lung tissue of LoM.
Single color and merged images of those shown in Fig. 1f and g depicting co-staining of LoM human lung tissue two days following SARS-CoV-2 exposure for virus nucleoprotein (red) and a, and cytokeratin 19 (epithelial cells, green, n=6), CD34 (endothelial cells, green, n= 4), or vimentin (mesenchymal cells, green, n=4) or b, acetylated alpha-tubulin IV (ciliated cells, green, n=6), CC10 (club cells, green, n=6), HT1–56 (alveolar type 1 cells, green, n=6), or Pro-SP-C (alveolar type 2 cells, green, n=3). Nuclei, blue; scale bars 50 um. n=number of biologically independent lung tissues analyzed.
Extended Data Fig. 4. Experimental design for the evaluation of EIDD-2801 as treatment or pre-exposure prophylaxis for SARS-CoV-2 infection.
a, Treatment experimental design. LoM were orally administered EIDD-2801 or vehicle at 24 h or 48 h post SARS-CoV-2 exposure and every 12 h thereafter. Virus titers were measured 2 days post-treatment initiation. b, Pre-exposure prophylaxis experimental design. LoM were orally administered EIDD-2801 or vehicle control 12 h prior to SARS-CoV-2 exposure and every 12 h thereafter. Virus titers in human lung tissues were measured 2 days post-exposure.
Extended Data Table 1.
Description of LoM used for the analysis of human and bat coronavirus replication.
| Group | Mouse | Sex | Human donor | Weeks post-surgery |
|---|---|---|---|---|
| SARS-CoV | 1 | F | 032 | 10 |
| 2 | F | 032 | 10 | |
| 3 | M | U32 | 9 | |
| 4 | M | U32 | 9 | |
| 5 | F | I33 | 14 | |
| 6 | F | L33 | 10 | |
| 7 | F | L33 | 10 | |
| 8 | F | L33 | 10 | |
| MERS-CoV | 9 | M | D33 | 9 |
| 10 | M | D33 | 9 | |
| 11 | M | F33 | 9 | |
| 12 | M | F33 | 9 | |
| 13 | F | I33 | 14 | |
| 14 | F | L33 | 10 | |
| 15 | F | L33 | 10 | |
| 16 | F | L33 | 10 | |
| SARS-CoV-2 | 17 | F | 032 | 19 |
| 18 | F | 032 | 19 | |
| 19 | M | D33 | 14 | |
| 20 | M | F33 | 14 | |
| WIV-CoV | 21 | M | F33 | 20 |
| 22 | M | F33 | 20 | |
| 23 | M | F33 | 20 | |
| SHC014-CoV | 24 | M | F33 | 20 |
| 25 | M | F33 | 20 | |
| 26 | M | F33 | 20 | |
Extended Data Table 2.
Description of LoM used for the analysis of SARS-CoV-2 replication and pathogenesis over time.
| Group | Mouse | Sex | Human donor | Weeks post-surgery |
|---|---|---|---|---|
| Day 2 | 27 | F | A31 | 59 |
| 28 | F | A31 | 59 | |
| 29 | F | F32 | 25 | |
| 30 | F | I32 | 19 | |
| Day 6 | 31 | F | R30 | 74 |
| 32 | F | A31 | 59 | |
| 33 | F | I32 | 19 | |
| 34 | F | F32 | 25 | |
| 35 | F | F32 | 25 | |
| Day 14 | 36 | F | A31 | 59 |
| 37 | F | I32 | 19 | |
| 38 | F | R30 | 74 | |
| 39 | F | F32 | 25 | |
| 40 | F | F32 | 25 | |
Extended Data Table 3.
Abundance of viral transcripts detected in SARS-CoV-2 infected LoM.
| Group | Mouse | Sex | Human donor | Weeks post-surgery | %Viral transcripts |
|---|---|---|---|---|---|
| Day 2 | 1 | F | A31 | 59 | 0.55301% |
| 41 | F | I32 | 19 | 3.55979% | |
| Day 6 | 31 | F | R30 | 74 | 1.01818% |
| 32 | F | A31 | 59 | 0.46368% | |
| 33 | F | I32 | 19 | 1.38159% | |
| Day 14 | 37 | F | 132 | 19 | 0.69389% |
| 39 | F | F32 | 25 | 0.05856% | |
| 42 | F | A31 | 59 | 0.18557% | |
| Naive | 43 | F | A31 | 59 | N/A |
| 44 | F | I32 | 19 | N/A | |
| 45 | F | I32 | 19 | N/A | |
| 46 | F | I32 | 19 | N/A | |
Shown is the %viral transcripts of total transcripts sequenced from SARS-CoV-2 infected (n=8) and naïve (n=4) LoM utilized for RNA-sequencing analysis. Not applicable: N/A. Using a linear model, there was no effect of human donor on viral transcript abundance (lm() in R: estimate −0.012, p=0.103). n=number of biologically independent lung tissues analyzed.
Extended Data Table 4.
Abundance of human ACE2 and TMPRSS2 transcripts detected in SARS-CoV-2 infected LoM.
| Group | Mouse | Human donor | GAPDH (TPM) | ACE2 (TPM) | TMPRSS2 (TPM) |
|---|---|---|---|---|---|
| Day 2 | 1 | A31 | 517.41 | 1.79 | 33.54 |
| 41 | I32 | 432.19 | 4.63 | 41.96 | |
| Day 6 | 31 | R30 | 654.63 | 2.67 | 78.64 |
| 32 | A31 | 784.39 | 0.73 | 21.62 | |
| 33 | I32 | 481.94 | 1.56 | 41.07 | |
| Day 14 | 37 | I32 | 419.55 | 2.02 | 38.94 |
| 39 | F32 | 492.57 | 1.90 | 61.95 | |
| 42 | A31 | 351.54 | 1.54 | 43.50 | |
| Naive | 43 | A31 | 2384.62 | 0.19 | 15.35 |
| 44 | I32 | 363.42 | 0.82 | 38.95 | |
| 45 | I32 | 359.21 | 0.67 | 38.19 | |
| 46 | I32 | 339.77 | 0.89 | 32.93 | |
Shown are the abundance of human GAPDH, ACE2, and TMPRSS2 transcripts sequenced in the human lung tissue collected from naïve (n=4) and SARS-CoV-2 infected (n=8) LoM. Median ACE2 and TMPRSS2 expression in human lung tissue of naïve mice was 0.75 TPM and 35.6 TPM respectively, which is comparable to the median expression of ACE2 (1.01 TPM) and TMPRSS2 (43.2 TPM) observed in human lung tissue profiled by the GTEx project (https://gtexportal.org/home/; data retrieved 10/20/2020 and 11/3/2020). No effect of human donor on ACE2 expression (F = 2.06259641; p=0.1756 via two-sided ANOVA) was observed. Approximately the same amount of variation was observed within a human donor (mean variance within donors = 4.82) as across all donors (variance = 4.47). n=number of biologically independent lung tissues analyzed.
Extended Data Table 5.
Human interferon and cytokine genes upregulated during SARS-CoV-2 infection.
| Naive | Day 2 | Day 6 | Day 14 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | TPM | TPM | Log2FC | p-value | TPM | Log2FC | p-value | TPM | Log2FC | p-value | |
| Interferon genes | IFNB1 | 0.009721 | 12.89298 | 10.8264 | N/A | 0.56133 | 5.919101 | 0.000104 | 1.261203 | 6.816893 | 8.95E-07 |
| ISG15 | 29.11072 | 3529.547 | 7.5504869 | N/A | 1057.532 | 5.559945 | N/A | 680.674 | 4.441103 | 4.38E-18 | |
| OASL | 0.575651 | 117.7447 | 7.460388 | N/A | 24.14577 | 5.349643 | 4.82E-21 | 19.32386 | 4.837053 | 3.70E-40 | |
| MX2 | 2.446529 | 207.3412 | 7.1723067 | N/A | 54.58907 | 4.403233 | 4.35E-16 | 73.59163 | 4.793127 | 3.89E-23 | |
| IFITM1 | 75.8737 | 1380.718 | 6.8014838 | N/A | 841.8657 | 3.67271 | 0.027036 | 752.1089 | 3.183608 | 0.056391 | |
| MX1 | 10.01414 | 1098.343 | 6.6205208 | N/A | 282.2782 | 4.583841 | 1.03E-35 | 320.7999 | 4.690323 | 9.58E-119 | |
| OAS1 | 5.455972 | 427.8919 | 6.3944922 | N/A | 109.1991 | 4.097581 | 4.61E-13 | 128.1803 | 4.407948 | 4.03E-33 | |
| IFIT2 | 5.663018 | 327.3042 | 5.8248817 | N/A | 40.06805 | 2.678602 | 1.04E-12 | 51.76333 | 3.021741 | 3.65E-11 | |
| IFIT3 | 9.293879 | 323.3581 | 5.5590143 | N/A | 77.21678 | 2.973115 | 3.24E-09 | 90.51719 | 3.168749 | 9.62E-11 | |
| OAS3 | 4.160792 | 118.0739 | 5.1575147 | N/A | 27.98228 | 2.619502 | 5.04E-13 | 32.44652 | 3.006743 | 1.07E-25 | |
| Cytokine genes | CXCL10 | 1.334587 | 784.985 | 9.4483016 | N/A | 51.89486 | 5.060381 | 3.36E-08 | 104.6776 | 6.211797 | 2.26E-28 |
| CXCL11 | 0.368657 | 170.7106 | 9.0255049 | N/A | 13.74948 | 5.100041 | 1.73E-13 | 28.97534 | 6.20993 | 4.12E-32 | |
| CCL7 | 0.110614 | 2.114279 | 8.1032148 | N/A | 0.256785 | 0.939467 | 0.826964 | 4.136358 | 5.155454 | 0.0337375 | |
| CXCL9 | 0.31541 | 20.86055 | 6.5863786 | N/A | 13.58781 | 5.142114 | N/A | 51.63909 | 7.324334 | N/A | |
| CXCL8 | 8.329686 | 33.28712 | 6.3670255 | N/A | 88.08344 | 2.850492 | N/A | 6.213093 | −0.60592 | N/A | |
| CXCL2 | 6.093913 | 114.8106 | 5.9559545 | N/A | 51.26291 | 2.979872 | 0.018944 | 24.8723 | 2.026886 | 0.1230292 | |
| CCL8 | 0.872448 | 31.60003 | 5.7008898 | N/A | 3.819344 | 2.129537 | 0.000334 | 11.39742 | 3.657159 | 2.66E-05 | |
| CCL5 | 0.708453 | 12.48674 | 5.5750578 | N/A | 4.494502 | 2.620838 | 0.029157 | 19.43851 | 4.710446 | 7.17E-05 | |
| IL6 | 5.42304 | 12.23683 | 5.0378314 | N/A | 27.2112 | 1.542807 | N/A | 3.889728 | −0.96496 | N/A | |
| IL27 | 0.02044 | 0.802539 | 4.5555662 | N/A | 0.144501 | 2.957134 | 0.168998 | 0.166259 | 2.840334 | 0.0473019 | |
| CXCL3 | 1.665821 | 17.51558 | 4.0723405 | N/A | 7.100009 | 2.13156 | 0.03234 | 6.128888 | 1.776734 | 0.0077687 | |
| IL17C | 0.040803 | 0.246641 | 3.4322656 | N/A | 0.069498 | 1.136046 | 0.651685 | 0.079223 | 0.819111 | 0.815461 | |
| TNF | 0.252698 | 0.776601 | 2.6658632 | N/A | 0.370601 | 0.713121 | 0.693592 | 1.728727 | 2.737404 | N/A | |
Transcripts per million (TPM). Log2 fold change (Log2FC) in TPM detected in LoM human lung tissues at 2 (n=2), 6 (n=3), and 14 (n=3) days post-SARS-CoV-2 exposure compared to naïve controls (n=4). P-values were calculated in DESeq2 using a two-sided Wald’s test and adjusted for multiple testing using a False Discovery Rate. Not applicable (N/A): DESeq2 could not calculate a p-value due to sample size or the variance between samples. n=number of biologically independent lung tissues analyzed.
Extended Data Table 6.
Description of LoM used to evaluate the efficacy of EIDD-2801 for SARS-CoV-2 pre-exposure prophylaxis and treatment.
| Group | Mouse | Sex | Human donor | Weeks post-surgery |
|---|---|---|---|---|
| Vehicle pre-exposure prophylaxis | 47 | M | F33 | 15 |
| 48 | M | F33 | 15 | |
| 49 | F | L33 | 11 | |
| 50 | F | L33 | 11 | |
| 51 | M | T33 | 13 | |
| 52 | M | T33 | 13 | |
| 53 | M | T33 | 13 | |
| 54 | M | T33 | 13 | |
| EIDD-2801 pre-exposure prophylaxis | 55 | M | F33 | 15 |
| 56 | M | F33 | 15 | |
| 57 | F | L33 | 11 | |
| 58 | F | L33 | 11 | |
| 59 | M | T33 | 13 | |
| 60 | M | T33 | 13 | |
| 61 | M | T33 | 13 | |
| 62 | M | T33 | 13 | |
| Vehicle 24h treatment | 63 | F | C34 | 16 |
| 64 | F | C34 | 16 | |
| 65 | F | C34 | 16 | |
| 66 | F | C34 | 16 | |
| EIDD-2801 24h treatment | 67 | F | C34 | 16 |
| 68 | F | C34 | 16 | |
| 69 | F | C34 | 16 | |
| 70 | F | C34 | 16 | |
| Vehicle 48h treatment | 71 | F | C34 | 16 |
| 72 | F | C34 | 16 | |
| 73 | F | C34 | 16 | |
| 74 | F | C34 | 16 | |
| EIDD-2801 48h treatment | 75 | F | C34 | 16 |
| 76 | F | C34 | 16 | |
| 77 | F | C34 | 16 | |
| 78 | F | C34 | 16 | |
Supplementary Material
Acknowledgements:
We thank current and past members of the Garcia laboratory for technical assistance. The authors also thank technicians at the UNC Animal Histopathology and Laboratory Animal Medicine Core and Division of Comparative Medicine. We also thank the UNC School of Medicine Bioinformatics and Analytics Research Collaborative (BARC) for providing technical support and K. Mollan and B. Shook-Sa of the UNC CFAR Biostatistics Core for providing statistical support.
Funding: This work was supported by funding from National Institutes of Health grants R21AI113736 (RJP), U19AI100625 and R01 AI108197 (RSB), Fast Grants (LEG), R01AI123010 (AW), R01AI111899 (JVG), R01AI140799 (JVG), R01MH108179 (JVG). This project was also supported by the North Carolina Policy Collaboratory at the University of North Carolina at Chapel Hill with funding from the North Carolina Coronavirus Relief Fund established and appropriated by the North Carolina General Assembly. The Microscopy Services Laboratory, Department of Pathology and Laboratory Medicine, is supported in part by P30 CA016086 Cancer Center Support Grant to the UNC Lineberger Comprehensive Cancer Center.
Footnotes
Competing interests: GP, MGN, GRB and AAK are employees of Emory and have a financial interest in molnupiravir (EIDD-2801).
Additional Information
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability:
Gene-expression data are available at the Gene Expression Omnibus (GEO) repository (accession: GSE155286). Source data is provided for Fig. 1b,c and Fig. 4a–d. All other data is available from corresponding authors on reasonable request. Publicly available data sets used in this study: 1) sequence of the SARS-CoV-2 strain 2019-nCoV/USA-WA1/2020 (GenBank accession: MN985325.1) and 2) human lung tissue gene expression data profiled by the GTEx project (https://gtexportal.org/home/; data retrieved 10/20/2020 and 11/3/2020).
Main References
- 1.Cui J, Li F & Shi ZL Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17, 181–192, doi: 10.1038/s41579-018-0118-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong E, Du H & Gardner L An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 20, 533–534, doi: 10.1016/S1473-3099(20)30120-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boni MF et al. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol, doi: 10.1038/s41564-020-0771-4 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Bao L et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature, doi: 10.1038/s41586-020-2312-y (2020). [DOI] [PubMed] [Google Scholar]
- 5.Blanco-Melo D et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 181, 1036–1045 e1039, doi: 10.1016/j.cell.2020.04.026 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cockrell AS et al. A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat Microbiol 2, 16226, doi: 10.1038/nmicrobiol.2016.226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinnon KH 3rd et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586, 560–566, doi: 10.1038/s41586-020-2708-8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gralinski LE et al. Complement Activation Contributes to Severe Acute Respiratory Syndrome Coronavirus Pathogenesis. mBio 9, doi: 10.1128/mBio.01753-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang RD et al. Pathogenesis of SARS-CoV-2 in Transgenic Mice Expressing Human Angiotensin-Converting Enzyme 2. Cell, doi: 10.1016/j.cell.2020.05.027 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCray PB Jr. et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol 81, 813–821, doi: 10.1128/JVI.02012-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menachery VD et al. Middle East Respiratory Syndrome Coronavirus Nonstructural Protein 16 Is Necessary for Interferon Resistance and Viral Pathogenesis. mSphere 2, doi: 10.1128/mSphere.00346-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menachery VD et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med 21, 1508–1513, doi: 10.1038/nm.3985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menachery VD et al. SARS-like WIV1-CoV poised for human emergence. Proc Natl Acad Sci U S A 113, 3048–3053, doi: 10.1073/pnas.1517719113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rockx B et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 368, 1012–1015, doi: 10.1126/science.abb7314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheahan TP et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med 12, doi: 10.1126/scitranslmed.abb5883 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franks TJ et al. Resident cellular components of the human lung: current knowledge and goals for research on cell phenotyping and function. Proc Am Thorac Soc 5, 763–766, doi: 10.1513/pats.200803-025HR (2008). [DOI] [PubMed] [Google Scholar]
- 17.Wahl A et al. Precision mouse models with expanded tropism for human pathogens. Nat Biotechnol 37, 1163–1173, doi: 10.1038/s41587-019-0225-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann M et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271–280 e278, doi: 10.1016/j.cell.2020.02.052 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454, doi: 10.1038/nature02145 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walls AC et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 181, 281–292 e286, doi: 10.1016/j.cell.2020.02.058 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan R et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448, doi: 10.1126/science.abb2762 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carsana L et al. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: a two-centre descriptive study. Lancet Infect Dis, doi: 10.1016/S1473-3099(20)30434-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menter T et al. Post-mortem examination of COVID19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings of lungs and other organs suggesting vascular dysfunction. Histopathology, doi: 10.1111/his.14134 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian S et al. Pulmonary Pathology of Early-Phase 2019 Novel Coronavirus (COVID-19) Pneumonia in Two Patients With Lung Cancer. J Thorac Oncol 15, 700–704, doi: 10.1016/j.jtho.2020.02.010 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu N et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 382, 727–733, doi: 10.1056/NEJMoa2001017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D et al. The Architecture of SARS-CoV-2 Transcriptome. Cell 181, 914–921 e910, doi: 10.1016/j.cell.2020.04.011 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Efficacy and Safety of Molnupiravir (MK-4482) in Hospitalized Adult Participants With COVID-19 (MK-4482–001), https://ClinicalTrials.gov/show/NCT04575584
- 28.Xu Z et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 8, 420–422, doi: 10.1016/S2213-2600(20)30076-X (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H et al. Histopathologic Changes and SARS-CoV-2 Immunostaining in the Lung of a Patient With COVID-19. Ann Intern Med 172, 629–632, doi: 10.7326/M20-0533 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Z et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe 27, 883–890 e882, doi: 10.1016/j.chom.2020.04.017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siu KL et al. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J Biol Chem 284, 16202–16209, doi: 10.1074/jbc.M109.008227 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spiegel M et al. Inhibition of Beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J Virol 79, 2079–2086, doi: 10.1128/JVI.79.4.2079-2086.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanderheiden A et al. Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures. J Virol 94, doi: 10.1128/JVI.00985-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin C et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis, doi: 10.1093/cid/ciaa248 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beigel JH et al. Remdesivir for the Treatment of Covid-19 - Final Report. N Engl J Med, doi: 10.1056/NEJMoa2007764 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan H et al. Repurposed antiviral drugs for COVID-19 –interim WHO SOLIDARITY trial results. 2020.2010.2015.20209817, doi: 10.1101/2020.10.15.20209817 %J medRxiv (2020). [DOI] [PMC free article] [PubMed]
- 37.Chen P et al. SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. doi: 10.1056/NEJMoa2029849 (2020). [DOI] [PMC free article] [PubMed]
- 38.Regeneron Pharmaceuticals I (TARRYTOWN, N.Y., 2020). [Google Scholar]
- 39.Kim PS, Read SW & Fauci AS Therapy for Early COVID-19: A Critical Need. JAMA, doi: 10.1001/jama.2020.22813 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Sticher ZM et al. Analysis of the Potential for N (4)-Hydroxycytidine To Inhibit Mitochondrial Replication and Function. Antimicrob Agents Chemother 64, doi: 10.1128/AAC.01719-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toots M et al. Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci Transl Med 11, doi: 10.1126/scitranslmed.aax5866 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou YJ et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell, doi: 10.1016/j.cell.2020.05.042 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 43.Scobey T et al. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci U S A 110, 16157–16162, doi: 10.1073/pnas.1311542110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yount B et al. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci U S A 100, 12995–13000, doi: 10.1073/pnas.1735582100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simionescu N & Simionescu M Galloylglucoses of low molecular weight as mordant in electron microscopy. I. Procedure, and evidence for mordanting effect. J Cell Biol 70, 608–621, doi: 10.1083/jcb.70.3.608 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patro R, Duggal G, Love MI, Irizarry RA & Kingsford C Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14, 417–419, doi: 10.1038/nmeth.4197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benjamini Y & Hochberg Y Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. 57, 289–300, doi: 10.1111/j.2517-6161.1995.tb02031.x (1995). [DOI] [Google Scholar]
- 50.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eden E, Navon R, Steinfeld I, Lipson D & Yakhini Z GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48, doi: 10.1186/1471-2105-10-48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene-expression data are available at the Gene Expression Omnibus (GEO) repository (accession: GSE155286). Source data is provided for Fig. 1b,c and Fig. 4a–d. All other data is available from corresponding authors on reasonable request. Publicly available data sets used in this study: 1) sequence of the SARS-CoV-2 strain 2019-nCoV/USA-WA1/2020 (GenBank accession: MN985325.1) and 2) human lung tissue gene expression data profiled by the GTEx project (https://gtexportal.org/home/; data retrieved 10/20/2020 and 11/3/2020).