Abstract
Protein metabolism plays central roles in age related decline and neurodegeneration. While a large body of research has explored age-related changes in protein degradation, alterations in the efficiency and fidelity of protein synthesis with aging are less well understood. Age-associated changes occur in both the protein synthetic machinery (ribosomal proteins and rRNA) and within regulatory factors controlling translation. At the same time, many of the interventions that prolong lifespan do so in part by pre-emptively decreasing protein synthesis rates to allow better harmonization to age-related declines in protein catabolism. Here we review the roles of translation regulation in aging, with a specific focus on factors implicated in age-related neurodegeneration. We discuss how emerging technologies such as ribosome profiling and superior mass spectrometric approaches are illuminating age-dependent mRNA-specific changes in translation rates across tissues to reveal a critical interplay between catabolic and anabolic pathways that likely contribute to functional decline. These new findings point to nodes in post-transcriptional gene regulation that both contribute to aging and offer targets for therapy.
Keywords: aging, translation, neurodegeneration, translation regulation
INTRODUCTION
Aging is characterized by a progressive decline in physiological function leading to increased disease vulnerability and decreased lifespan. Aging is also the biggest risk factor for multiple neurodegenerative disorders like Alzheimer disease (AD), Parkinson Disease (PD) and Amyotrophic Lateral Sclerosis (ALS) (Bishop, Lu, & Yankner, 2010). The aging human brain exhibits cognitive decline and structural changes observed both at the systems level using neuroimaging studies as well as at the cellular and molecular level. Functional imaging studies of the healthy aging brain report a lack of coordinated activation between brain regions, which is associated with poor cognitive performance (Andrews-Hanna et al., 2007). The hippocampus and the Prefrontal cortex (PFC) seem to be particularly vulnerable to age-associated changes (de Brabander, Kramers, & Uylings, 1998), resulting in both calcium dysregulation and changes in synaptic properties (Landfield, 1988). Thus, global declines in volume and regional connectivity coupled with region-specific changes in dendritic morphology, cellular connectivity and other factors that influence synaptic plasticity combine to impact network-level dynamics that affect cognition in normal aging (reviewed in Burke & Barnes, 2006). In addition, normal age-associated global decline in protein synthesis is a highly conserved phenomenon across multiple model organisms (Tavernarakis, 2008). Proteomic studies are just beginning to unravel these global changes in protein levels that occur in physiological brain aging (Ori et al., 2015; Somel et al., 2010; Walther et al., 2015). These and other studies show large-scale post-transcriptional regulation, mRNA-protein decoupling and changes in the stoichiometry of protein complexes with age (Georges E. Janssens et al., 2015; Kelmer Sacramento et al., 2020). Interestingly, the age-associated decoupling of the proteome and transcriptome was particularly strong for translational machinery related genes (Georges E. Janssens et al., 2015). Furthermore, recent single cell transcriptomic analyses of the aging Drosophila and mouse brains have uncovered tissue and cell type specificity in genes encoding ribosomal protein transcripts as well as other translation pathway components (Davie et al., 2018; Ximerakis et al., 2019). Together these findings point to a tissue and cell type-specific post-transcriptional change in neuronal translational machinery with aging. The most drastically affected neuronal genes in the aging human cortex are associated with synaptic function and changes at both the mRNA and protein level precede visible neuronal loss (Dillman et al., 2017). These studies suggest that age-associated alterations in the translational machinery may have downstream effects in a cell type specific manner with consequences for both normal and pathological aging.
This review highlights how alterations in protein translation and the role of the various components of the translational machinery change in the aging brain. We provide overviews on the roles of protein translational regulation within aging in general and in neurological disease where translational dysregulation has emerged as a critical driver of disease pathogenesis. We then discuss how emerging technologies might help us address shared observations between these two fields of study to reveal key drivers of brain aging. We propose a model whereby age-associated changes in translational dynamics contribute to global declines in protein homeostasis. These changes in turn result in neuronal dysfunction and an enhanced susceptibility to neurodegenerative disease. By understanding this cascade of events we hope to reveal novel targets for therapeutic development, with a goal of enhancing healthy brain aging.
1. PROTEIN SYNTHESIS
Translation is a complex multi-step process whereby the ribosome coordinates the decoding of mRNA to synthesize new proteins (Fig 1). During initiation, the mRNA to be decoded is prepared for translation by factors that assemble at its 5’ methyl −7-Guanylate (m7Gppp) cap and 3’ poly-A tail. The 5’ cap of the mature mRNA binds the eIF4F multi-subunit complex comprising the eIF4E cap binding protein, eIF4G scaffolding protein and the eIF4A helicase along with the eIF4B and 4H stimulatory subunits (see Fig 1-a) (Sachs, Sarnow, & Hentze, 1997). The 3’ poly-A tail of the mRNA binds the Poly (A)-Binding Protein I (PABP1) that also interacts with the eIF4G scaffold at the 5’ cap to circularize the mRNA presumably for efficient translation (Borman, Michel, & Kean, 2000; Hinnebusch & Lorsch, 2012; Imataka, Gradi, & Sonenberg, 1998). In parallel, the smaller 40S ribosomal subunit bound by eIF3, eIF5, eIF1 and 1A proteins assemble into a 43S preinitiation complex (PIC) by associating with the ternary complex comprised of the initiator tRNAi Met and an eIF2-GTP molecule (b) (Jackson, Hellen, & Pestova, 2010; Pestova & Kolupaeva, 2002). The PIC is then recruited by eIF4F to the mRNA (c).
Fig. 1. Schematic showing the different stages of protein synthesis.
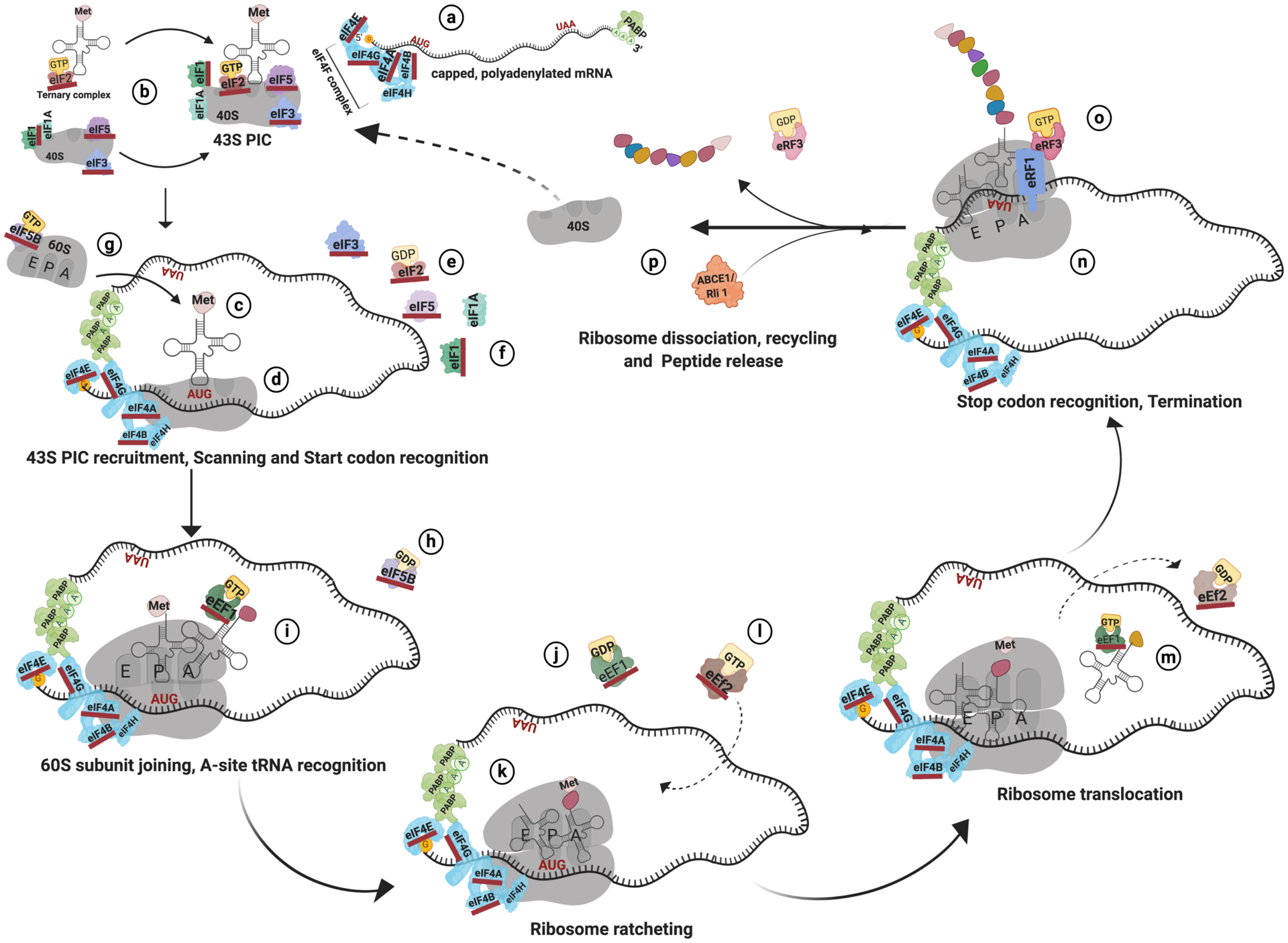
The initiation and elongation factors that are implicated in aging studies are underlined in red. The circled alphabets denote the steps described in the text.
Once assembled, the eIF4F complex and the 43S PIC scan along the mRNA in a 5’ to 3’ direction until it encounters an AUG start codon in the correct sequence, eponymously termed the “Kozak” context (Kozak, 1984). The eIF4A, 4B and 4H subunits along with additional RNA helicases help resolve mRNA secondary structures encountered during the scanning process (Hinnebusch, 2014; Pestova & Kolupaeva, 2002). The initiation complex recognizes the start codon by base pairing of the CAU anticodon of tRNAi Met to the AUG codon within the mRNA (d). This interaction triggers the irreversible hydrolysis of the eIF2 bound GTP to GDP with the assistance of the GTP-ase Activating Protein eIF5 (e), triggering a structural shift within the initiation complex. This signals the ribosome to stop scanning and causes the release of eIF1 and 1A factors that are responsible for start codon stringency (f) (Passmore et al., 2007). The Ribosome is now committed to its selected start codon and forms a stable 48S PIC complex with the mRNA. The 48S PIC recruits the larger 60S ribosomal subunit that is preassembled with eIF5B-GTP (g) (Hinnebusch, 2014). The coupling of the two ribosomal subunits is accompanied by the hydrolysis of eIF5B-GTP preventing their dissociation until the mRNA is translated (h) (Acker & Lorsch, 2008; Pestova et al., 2000; Wang et al., 2019). The released and inactive eIF2-GDP molecule is recharged by the guanine exchange factor eIF2B to allow for reassembly of the ternary complex and subsequent rounds of initiation (Webb & Proud, 1997).
The elongation-competent 80S ribosome with the tRNAi Met positioned in the Peptidyl (P) site and the next in-frame mRNA codon in the Aminoacyl/Acceptor (A) site is queried by the diffusion of ternary complexes. These complexes contain aminoacylated tRNAs bound to eEF1A-GTP (i). Elongation begins upon base pairing of the cognate tRNA with the A-site codon resulting in GTP hydrolysis and subsequent release of the eEF1A-GDP (j) (Dever & Green, 2012). The eEF1A-GDP molecules are continuously recharged by the action of the eEF1B-GTP exchange factor to bind aminoacylated tRNAs and start another cycle of elongation. Rapid peptide bond formation between the 3’ aminoacylated ends of the tRNAs in the A and the P-site is facilitated by conserved rRNA elements in the large ribosomal subunit (Beringer & Rodnina, 2007). The ribosome then performs a ratcheting movement causing the resident tRNAs to enter a P/E and A/P hybrid state (k). In this state, the anticodon loops of the P and A site tRNAs remain in place while their acceptor arms are located in the adjoining E and P sites respectively (Moazed & Noller, 1989). In order for translation elongation to proceed, the GTP-ase eEF2 hydrolyzes its bound GTP (l) to promote the irreversible translocation of the tRNAs to the E and P sites, thus freeing up the A site for subsequent diffusion of ternary complexes (m) (Ling & Ermolenko, 2016). In addition, some studies also point to a role for eIF5A in elongation possibly by association with eEF2 to stimulate reactivity of the peptidyl tRNA and facilitate ribosome transit (Schuller, Wu, Dever, Buskirk, & Green, 2017). In subsequent cycles, the conformational change of the ratcheting ribosome causes the ejection of unacylated tRNAs from the exit (E) site. As the nascent polypeptide chain elongates, it threads its way out as a linear molecule through a channel in the large ribosomal subunit and subsequently folds into its functional structure at the end of protein synthesis (Cabrita, Dobson, & Christodoulou, 2010; Javed, Christodoulou, Cabrita, & Orlova, 2017).
The termination of protein synthesis occurs when the ribosome encounters a stop codon (UAA, UAG or UGA) at its A site (n) and is facilitated by the action of eRF1 and eRF3 (Dever & Green, 2012). The eRF1 molecule resembles a tRNA molecule in shape and has an N-terminal domain that is responsible for anticodon recognition. It has been proposed to recognize and decode stop codons using the GTP-ase activity of the bound eRF3 to accelerate peptide release and increase termination efficiency (o) (Buckingham, Grentzmann, & Kisselev, 1997). The ribosome is subsequently dissociated and recycled by the ABCE1: Pelota complex for further rounds of initiation or re-initiation (p) (Skabkin, Skabkina, Hellen, & Pestova, 2013).
Most of the core components of the translation machinery are conserved among higher eukaryotes and show significant degree of homology between vertebrates and invertebrates. A number of these factors have been directly or indirectly implicated as important regulators of longevity (Gonskikh & Polacek, 2017; Hansen et al., 2007; G. E. Janssens et al., 2015; Tavernarakis, 2008) (Table 1). They also impact normal and pathological brain aging by modulating neuronal protein synthesis and synaptic activity (Schimanski & Barnes, 2010; Wyss-Coray, 2016). As such, promoting brain health both in normal and disease states could have significant impacts on cellular protein synthesis as demonstrated by intervention testing studies (Miller et al., 2011; A. C. Thompson et al., 2016).
Table 1.
Shared components of the protein synthesis machinery in aging and neurodegeneration.
| Name | Function | Aging | Neurodegeneration |
|---|---|---|---|
| eIF1 | 43S complex formation, start codon selection | 1, 2 | 17 |
| eIF2 | Ternary complex (TC) formation | 2,3,4 | Reviewed in 11 |
| eIF2B | Guanine nucleotide exchange factor for eIF2 | 4 | 10 |
| eIF3 | Binds to the 40S subunit, 43S PIC attachment to mRNA | 1,4 | Reviewed in 15 |
| eIF3a | 40S binding, eIF4B binding, TC and mRNA recruitment | 1 | - |
| eIF3b | 40S binding, TC and mRNA recruitment, scanning | 1 | - |
| eIF3c | 40S binding, TC formation and scanning | 2 | - |
| eIF3d | Binds 40S subunit | 2 | - |
| eIF3f | Possible binding for mTOR and S6 kinase | 1 | Reviewed in 15 |
| eIF3g | 40S binding, Binds eIF4B | 2 | Reviewed in 15 |
| eIF3h | Re-initiation of upstream ORFs, selective mRNA translation | 2 | - |
| eIF4A | DEAD-box RNA helicase, unwinds secondary structure | 1,2 | Reviewed in 16 |
| eIF4B | Stimulatory factor of eIF4A helicase | 2 | Reviewed in 16 |
| eIF4E | Cap- binding protein, part of the eIF4F complex | 5 | Reviewed in 16 |
| eIF4G | Scaffolding protein, part of the eIF4F complex | 1,3,6 | Reviewed in 16 |
| eIF5 | Induces hydrolysis of eIF2-GTP on start codon recognition | 2 | - |
| eIF5A | Peptide bond formation, ribosome transit during elongation | 2,7 | - |
| eIF5B | Ribosome-dependent GTP-ase, ribosome subunit joining | 2 | - |
| eEF1A1 | Isoform of eEF1A, aminoacylated (aa) tRNA delivery | 2 | - |
| eEF1A2 | Isoform of eEF1A, aa- tRNA delivery to ribosome | - | Reviewed in 8 |
| eEF1B2 | Guanine nucleotide exchange factor, delivers aa- tRNA | 2 | - |
| eEF1D | Delivery of aa- tRNAs to the ribosome for elongation | 2 | 18 |
| eEF1G | Delivery of aa- tRNAs to the ribosome for elongation | 2 | - |
| eEF2 | Catalyzes ribosome translocation after peptide bond formation | 2, 23 | 9 |
| eEF2K | Phosphorylates eEF2 to inhibit elongation | 23 | Reviewed in 24 |
| eRF1 | Terminates translation | - | 25 |
| GTBP2 | Ribosome recycling factor | - | 12, 13, 14 |
| tRNA synthetases | Catalyzes the esterification of cognate amino acid to the cognate tRNA molecule | 4, 26 | Reviewed in 8 and 19 |
| Ribosomal Proteins | Components of the ribosome | 1,2, 3,4, 20 | 21, Reviewed in 22 |
Table Legend: Numbers indicate primary literature or reviews containing primary literature.
1- (Curran & Ruvkun, 2007); 2- (Ximerakis et al., 2019); 3- (Hansen et al., 2007); 4- (Chen et al., 2007); 5- (Syntichaki et al., 2007); 6- (Pan et al., 2007); 7- (Poon et al., 2006); 8- (Kapur et al., 2017); 9- (Hekman et al., 2012); 10- (Bugiani et al., 2010); 11- (Moon, Sonenberg, & Parker, 2018); 12- (Jaberi et al., 2016); 13- (Bertoli-Avella et al., 2018); 14- (Ishimura et al., 2014); 15- (Gomes-Duarte, Lacerda, Menezes, & Romao, 2018); 16- (Wolozin & Ivanov, 2019); 17- (P. Garcia-Esparcia et al., 2015); 18- (Schulte et al., 2014); 19- (Meyer-Schuman & Antonellis, 2017); 20- (Chiocchetti et al., 2007); 21- (Ding, Markesbery, Chen, Li, & Keller, 2005); 22- (Hetman & Slomnicki, 2019); 23- (Xie et al., 2019); 24- (Liu & Proud, 2016);25- (Ortega et al., 2020); 26- (Gabius et al., 1982)
2. LESSONS FROM NEURODEGENERATION
Genetic mutations in translation factors that give rise to neurological disorders have opened up new insights into their biological outcomes. These studies also extend our understanding of the possible roles of such factors in the aging nervous system. As in aging studies, many of the components of translation are implicated in neurodegenerative disorders (Table 1). Dysregulation of translation in neurological disorders have highlighted the cell-specific roles of some of these factors.
2.1 -. INITIATION FACTORS
The brain disorder Leukoencephalopathy with vanishing white matter disease (VWMD) is caused by a mutation in any one of the 5 subunits of eIF2B (Bugiani, Boor, Powers, Scheper, & van der Knaap, 2010). VWM disease is associated with the loss of astrocytes, oligodendrocytes and axons in the central nervous system. It manifests clinically as a progressive cerebellar ataxia with a tendency toward rapid neurologic deterioration when mild stressors such as a fever occurs. Interestingly, VWM mutations in eIF2B predominantly affect the brain and exclusively affect white matter with selective glial vulnerability, highlighting the specialized nature of this particular mutation. The eIF2B initiation factor is both a Guanine Exchange Factor (GEF) that recharges eIF2-GDP and a GDP dissociation-inhibitor Displacement Factor (GDF) that strips away eIF5 to restart ternary complex formation (Jennings & Pavitt, 2014). Therefore, it is a highly conserved gene with housekeeping functions in all tissues and yet shows a brain and glial specific disease pathology. Interestingly, this mutation does not affect overall translation rates in patient lymphoblast lines. However, it leads to a hyper-suppression of translation when encountering cellular stress events and prevents the recovery of the cell from the subsequent stress (Moon & Parker, 2018). This inability to reset the cellular stress response could be a plausible explanation for the episodic loss of white matter that is a classical symptom in VWMD patients.
2.2 -. ELONGATION FACTORS
Elongation factors such as eEF1A and eEF2 have also been implicated in neurodegenerative disorders such as Parkinson Disease (PD) and Alzheimer Disease (AD), though their exact role here remains unclear. The levels of both these elongation factors are lower in brains of PD and AD patients as well as in mouse models of the disease, suggesting defects in translational fidelity or efficiency (Beckelman, Zhou, Keene, & Ma, 2016; Paula Garcia-Esparcia et al., 2015; X. Li, Alafuzoff, Soininen, Winblad, & Pei, 2005). Additionally, missense mutations in the neuron and muscle specific EEF1A2 variant are associated with severe epilepsy and progressive neurodegeneration (reviewed in Kapur & Ackerman, 2018). Besides its canonical role in translation elongation, eEF1A also functions in diverse cellular activities such as nuclear export, actin cytoskeletal dynamics and apoptosis underscoring the complexity of establishing definitive mechanistic roles for such factors (Mateyak & Kinzy, 2010). Nevertheless, alterations in translational fidelity and mis-translation are widely observed in neurodegeneration.
2.3 -. tRNAs AND tRNA SYNTHETASES
Mutations in critical accessory factors such as amino acyl tRNA synthetases (ARS) have been linked to multiple neurological disorders. ARS enzymes catalyze the reaction where a cognate amino acid is covalently attached to its specific anticodon bearing tRNA molecule to give rise to a charged or aminoacylated tRNA. Thus these enzymes are critical for providing a steady supply of correctly charged tRNAs for translation. Mutations in ARS genes have been linked to peripheral neuropathies like Charcot-Marie-Tooth (CMT) disease, hereditary motor neuropathies and epileptic encephalopathies. Though some ARS mutations exhibit multi-system pathology, there is a clear neurological predisposition to most ARS disorders (reviewed in (Meyer-Schuman & Antonellis, 2017). In one study, where a spontaneous mutation in the editing domain of Alanine ARS (AARS) caused ataxia, the mice showed ubiquitinated protein aggregates and progressive Purkinje cell loss in the cerebellum (J. W. Lee et al., 2006). Further analysis showed that the mutated AARS enzyme caused mischarging of the tRNAAla with serine and increased mistranslation in neurons. Even though the increase in mischarged tRNAAla was only about 2-fold, the severe neurological outcome points to a particular vulnerability to mistranslation in neuronal cells. Interestingly, other genes that are involved in tRNA synthesis, processing and modification also cause neurodegenerative disorders by altering tRNA levels and structure to affect neuronal translation (Kapur & Ackerman, 2018; Kapur, Monaghan, & Ackerman, 2017).
Neurodegenerative disorders can also arise from epistatic mechanisms of brain-specific mutations within the translational machinery. A spontaneous mutation in the brain-specific isodecoder tRNAArg (n-Tr20) is present in the commonly used C57BL/6J (B6J) mouse strain and does not seem to have a visible phenotype. However, when combined with a second mutation in GTP-binding protein 2 (GTPBP2), it causes severe ataxia, widespread neurodegeneration and death (Ishimura et al., 2014). GTPBP2 shares domain homology with the translational GTPase family members that include eRF3 and Hbs1L. Similar to the Hbs1 gene in yeast, GTPBP2 interacts with the ribosome-recycling factor Pelota (Dom34 in yeast) to rescue and recycle ribosomes. Ribosome profiling analysis of the cerebellum indicated ribosomal pausing at arginine codons in the n-Tr20 single mutants while the double mutants showed increased ribosome stalling at these sites. This indicates that normal levels of GTPBP2 can rescue the ribosomal pauses in the n-Tr20 strain while its complete absence caused prolonged ribosome stalling and neurodegeneration in this background (Ishimura et al., 2014). Interestingly, a splice mutation in the human ortholog of GTPBP2 was described to cause neurodegeneration in one family by linkage analysis and exome sequencing (Jaberi et al., 2016). Biallelic mutation in the HBS1L gene was also recently reported in a patient with developmental delay and congenital anomalies. The absence of Hbs1L protein caused a depletion of Pelota and accumulation of 80S monosome peaks in patient cells and in a mouse model (O’Connell et al., 2019).
2.4 -. RIBOSOMAL PROTEINS AND BIOGENESIS
Defects in ribosomal proteins and rRNA processing are also linked to neurological disorders. Missense mutations in two ribosomal core proteins namely uS12 and uL16, were associated with microcephaly, intellectual disability and autism-spectrum disorder. Studies of these mutations in yeast point to reduced translational fidelity and increased mistranslation (Brooks et al., 2014; Paolini et al., 2017). In addition, transacting ribosomal factors such as proteins involved in rRNA transcription and processing have also been implicated in neurological disorders ranging from specific muscular dystrophies to progressive neurodegeneration. These factors may also cause dysregulation of ribosome biogenesis and assembly leading to either specific or more generalized defects in translation within the neuronal population (reviewed in Hetman & Slomnicki, 2019).
Interestingly, the nucleolus where ribosome biogenesis occurs is disrupted in many neurodegenerative disorders like AD, PD and in repeat expansion disorders like HD. In AD, decreased rRNA expression level is seen at an early stage in the disease similar to PD, where nucleolar integrity is disrupted in dopaminergic neurons possibly due to the association of nucleolin protein with alpha-synuclein (Pietrzak, Rempala, Nelson, Zheng, & Hetman, 2011) (reviewed in (Parlato & Kreiner, 2013). In HD, where a CAG trinucleotide repeat expansion in exon 1 of the HTT gene causes an aberrant polyglutamine tract in the protein, the transcription of rRNA genes is altered. The mutant huntingtin protein is also seen as aggregates inside the nucleolus in patient brain samples (Tsoi & Chan, 2014; Tsoi, Lau, Tsang, Lau, & Chan, 2012).
2.5 -. NON-CANONICAL TRANSLATION AND PROTEIN AGGREGATES
Aggregate prone proteins are a feature of Nucleotide Repeat Expansion Disorders (NRED). These disorders arise from unstable tandem microsatellite repeats that are ubiquitous in the genome (Rodriguez & Todd, 2019). NREDs are associated with over 50 neurological disorders (Rohilla & Gagnon, 2017), suggesting a specific neuronal vulnerability to aberrant transcription and translation from these repeat regions. These microsatellites can form stable RNA structures that confer toxicity by direct binding of proteins (Renoux & Todd, 2012; Todd & Paulson, 2010). However, they also undergo a non-canonical form of translation termed Repeat-associated Non-AUG (RAN) translation, whereby proteins are synthesized in the absence of an AUG start codon (Green, Linsalata, & Todd, 2016; Zu et al., 2011; Zu, Pattamatta, & Ranum, 2018). RAN translation occurs in many repeat expansion disorders that show an age-associated onset of motor and cognitive dysfunction, including C9ALS/FTD, the most common known inherited cause of Amyotrophic Lateral Sclerosis (ALS) and Frontotemporal Dementia (FTD) (Ash et al., 2013; Mori et al., 2013; Zu et al., 2013).
The protein products of RAN translation are generated from multiple repeat reading frames and accumulate in cells to cause toxicity (reviewed in Cleary & Ranum, 2017)(Green et al., 2016; Kearse & Todd, 2014). The mechanistic details of RAN translation were first examined for the CGG repeat expansion in Fragile X associated Tremor Ataxia Syndrome (FXTAS). In reporter-based studies, CGG RAN translation in FXTAS required the m7G cap and the eIF4E and 4A complexes similar to canonical translation even though the RAN products initiated at near-AUG or non-AUG cognate codons (Kearse et al., 2016). RAN translation at GGGGCC repeats which cause C9ALS/FTD (C9RAN) was also largely m7G cap and eIF4A scanning dependent, (Green et al., 2017; Tabet et al., 2018) from linear transcripts. However, Internal Ribosome Entry Sites (IRES)-like initiation from bicistronic constructs also occurs at lower efficiency (Cheng et al., 2018; Sonobe et al., 2018). Interestingly, activation of cellular stress pathways which trigger phosphorylation of eIF2α selectively activated RAN translation suggesting that this core stress-signaling mechanism could act as a feed forward loop to reinforce neurodegeneration in these diseases (Fig 2) (Cheng et al., 2018; Glineburg, Todd, Charlet-Berguerand, & Sellier, 2018; Green et al., 2017; Sonobe et al., 2018; Westergard et al., 2019).
Fig. 2. Repeat associated Non-AUG (RAN) translation in neurodegeneration.
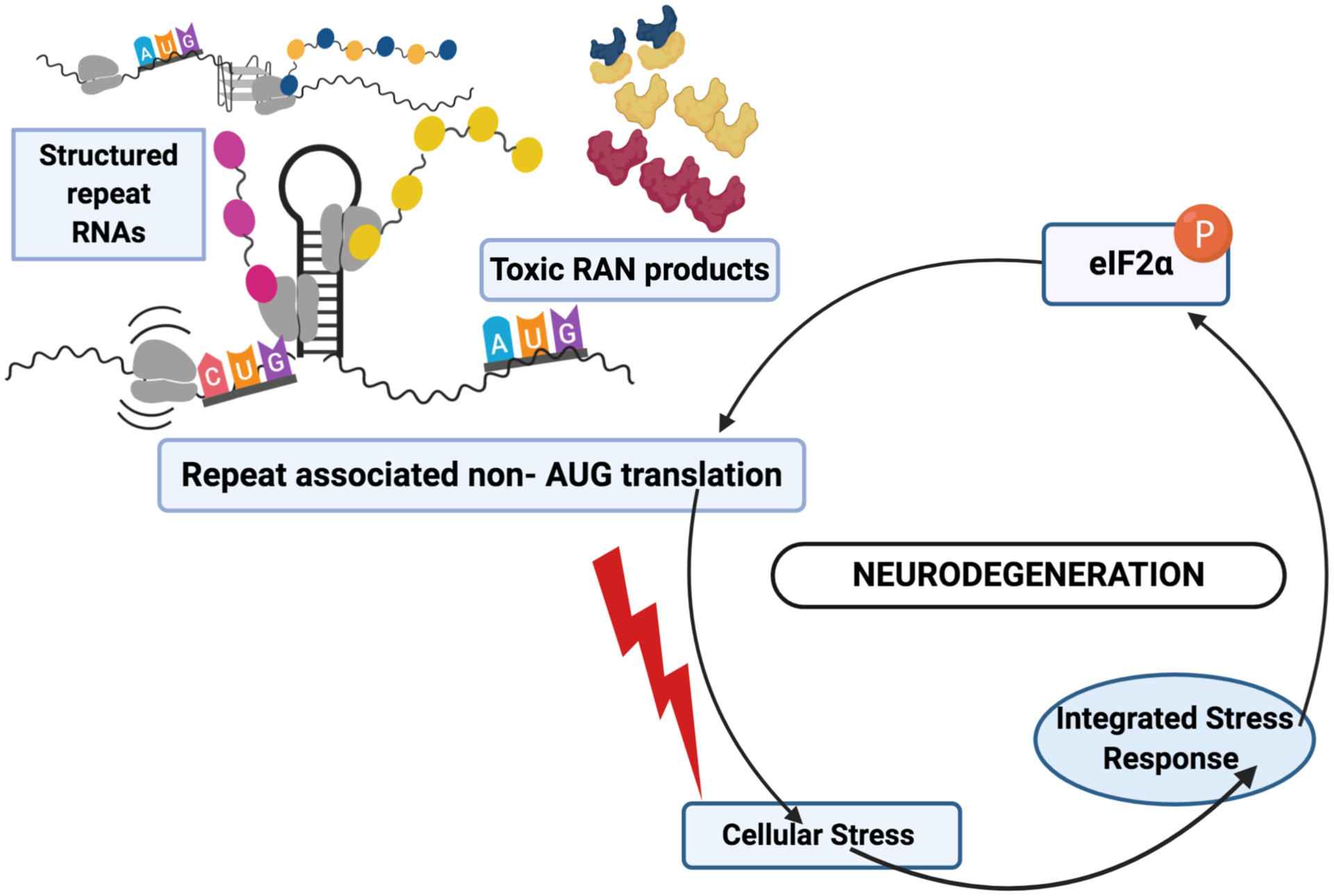
Repeat containing mRNAs form highly stable structures (hairpins or G-quadruplexes) that cause scanning pre-initiation complexes to pause at in-frame near-AUG codons or access the RNA through internal ribosomal entry. These processes lead to translation from these repeats in multiple reading frames in the absence of an AUG codon. The repeat mRNA and the resulting protein products elicit cellular stress and activate the ISR pathway, leading to phosphorylation of eIF2α. This in turn selectively activates RAN translation while suppressing global protein synthesis, creating a toxic feed-forward loop that can lead to neurodegeneration (Cheng et al., 2018; Green et al., 2017; Sonobe et al., 2018; Westergard et al., 2019).
At CGG repeats, initiation at near-AUG codons appears to be triggered by ribosome stalling at strong repeat RNA secondary structures (Kearse et al., 2016; Kochetov et al., 2007; Kozak, 1990). Helicases like DDX3, and helicase stimulatory subunits such as eIF4B and 4H that resolve secondary structures during initiation were identified as strong RAN translation modifiers in a genetic screen (Goodman et al., 2019; Linsalata et al., 2019). A second screen in C9RAN identified ribosomal protein RPS25 as an important modifier of RAN translation (Yamada et al., 2019). RPS25 is not critical for cap-dependent translation but is involved in alternative translation initiation pathways such as IRES-mediated translation and ribosomal shunting (Hertz, Landry, Willis, Luo, & Thompson, 2013). DDX3 and other factors were also implicated as modifiers of C9 RAN through a large-scale CRISPR screen (Cheng et al., 2019). Intriguingly, the impacts of DDX3 manipulation appear different across repeats-perhaps signifying mechanistic differences requiring further inquiry (Wilson, Muralidharan, & Isaacs, 2019).
These studies in NREDs indicate that non-canonical translational mechanisms could have significant implications for age-associated neurodegeneration. The rates of production of proteins from NREDs, even those initiated by AUG, strongly influence their relative toxicity and both the production and clearance of these repeat-containing proteins are influenced by age. For example, expression of a CAG repeat that generates toxic polyglutamine (polyQ) proteins in the nematode Caenorhabditis elegans (C.elegans) and mammalian cell lines shows a strong relationship between aging and polyQ aggregation. The aging-induced polyQ aggregates were distinct from those that form during osmotic stress (Moronetti Mazzeo, Dersh, Boccitto, Kalb, & Lamitina, 2012). Thus the presence of microsatellite expansions in NREDs could have a compounding effect on the aging proteome.
3. LESSONS FROM AGING
Aging shifts the balance between protein synthesis and degradation (Taylor & Dillin, 2011). Both these protein metabolism events decrease with age and contribute to age associated changes in cellular physiology. While the age associated catabolic stages are well studied, less is known about the declines in protein synthesis or anabolism with age (Anisimova, Alexandrov, Makarova, Gladyshev, & Dmitriev, 2018).
3.1-. CHANGES IN THE PROTEIN SYNTHESIS MACHINERY
Age associated changes in components of the translation machinery is widely reported. Almost all of the initiation and elongation factors involved in protein synthesis are implicated as aging modulators along with ribosomal proteins and other associated factors such as tRNA synthetases (Table 1).
3.1.1-. INITIATION FACTORS
The initiation and elongation steps of mRNA translation are important cellular checkpoints for the maintenance of protein homeostasis. Initial studies looking at the components of the translational machinery were done in C. elegans (Tavernarakis, 2008; Tissenbaum, 2015). RNA interference (RNAi) studies in adult nematode worms showed that decreasing the expression of key initiation factors such as eIF1, eIF3, eIF4A and eIF4G can extend lifespan by up to 50% (Curran & Ruvkun, 2007). Other studies looked at additional factors and found eIF2, eIF2B, eIF3 subunits and the cap-binding complex eIF4E as contributors to longevity (Chen, Pan, Palter, & Kapahi, 2007; Hansen et al., 2007; Syntichaki, Troulinaki, & Tavernarakis, 2007). All of these factors are critical to both the fidelity and efficiency of translation initiation and are subject to a highly coordinated regulation by nutrient and stress-signaling pathways as discussed below. Accordingly, multiple longevity enhancing regimens act on the core translational machinery to reduce protein synthesis.
3.1.2 -. ELONGATION FACTORS
Translational fidelity is largely regulated during elongation by ensuring cognate amino acid incorporation into the growing polypeptide chain. A comparative analysis across 17 rodent species with diverse lifespans reported a negative correlation between the frequency of amino acid mis-incorporation and longevity, suggesting that the rate of errors in translation is an important factor in determining lifespan (Ke et al., 2017). Consistent with this idea, increasing the accuracy of translation elongation by slowing down the ribosome through eEF2K activation extended lifespan in C.elegans. Activation of eEF2K decreased both codon mismatch and termination read-through errors during translation elongation (Xie et al., 2019). Surprisingly, activation of eEF2K also promoted more accurate start codon recognition in mRNAs by reducing initiation at near-AUG codons through a poorly characterized mechanism (Xie et al., 2019).
The eEF1A1 isoform of eEF1A is important during the heat shock response where it couples the transcription and translation of HSP70 mRNA. Upon stress, eEF1A1 activates both the transcription of HSP70 gene and also binds the 3’UTR of the mRNA to stabilize and transport it to the cytoplasm for translation (Vera et al., 2014). Though eEF1A1 is expressed at low levels in neurons, its expression changes in glial cells with aging (Ximerakis et al., 2019). Thus, eEF1A represents an interesting candidate factor in lifespan modulation.
3.1.3 -. tRNAs AND tRNA SYNTHETASES
The accuracy and speed of protein synthesis is determined by accessory factors such as tRNA synthetase enzymes that aminocylate cognate tRNAs. These enzymes are reported to show decreased specific activity in an organ-specific manner with age in mice (Gabius, Goldbach, Graupner, Rehm, & Cramer, 1982). Inefficient charging of tRNAs could contribute to the global decrease in protein synthesis with age. One study looked specifically at Seryl tRNA synthetase (SeRS), which has a nuclear function apart from its role in Serine tRNA aminoacylation (Xu et al., 2012). Overexpression of SeRS triggered cellular senescence by directly binding to telomeric DNA repeats and blocking the interaction with telomerase to cause progressive telomere shortening (Y. Li et al., 2019), which is a hallmark of aging (Lopez-Otin, Blasco, Partridge, Serrano, & Kroemer, 2013). Another critical determinant of protein synthesis rates is the codon composition or optimality of the mRNA transcript. Ribosome profiling studies in yeast show that ribosomes spend less time at optimal codons i.e. those that correspond to abundant tRNAs and this dynamics of the translating ribosome affects the stability of the mRNA transcript (Weinberg et al., 2016), (reviewed in Hanson & Coller, 2018). An integrative multi-omics analysis of datasets from the mouse brain indicates a strong correlation of codon sequence with mRNA abundance suggesting that codon optimality could be an important factor in brain protein homeostasis (Mandad et al., 2018). Since tRNA synthestase levels decrease during aging, it is conceivable that mRNA stability might also be affected.
3.1.4-. RIBOSOMAL PROTEINS
Ribosomal proteins (RP) and ribosomal RNA (rRNA) levels change with age (Chiocchetti et al., 2007; Curran & Ruvkun, 2007; Ximerakis et al., 2019); reviewed in (Gonskikh & Polacek, 2017; MacInnes, 2016; Tavernarakis, 2008). These changes can impact ribosomal subunit stoichiometry and protein synthesis rates both globally and selectively. A possible role for the ribosome in longevity was described in the Naked Mole Rat, which has a maximum-recorded lifespan of approximately 30 years (Buffenstein, 2005). In these animals, 28S rRNA processing generates a unique fragment that does not share homology with other regions of the genome. Using reporter-based assays, the same study showed that production of this alternative 28S rRNA is associated with increased translation fidelity in naked mole rat fibroblasts (Azpurua et al., 2013). The authors speculate that this unique cleavage event might alter the folding dynamics of the large ribosomal subunit, with detectable consequences in translation fidelity. Comparative lifespan analysis has shown that there is a strong negative correlation between amino acid misincorporation and lifespan, suggesting a coevolution of translational fidelity and longevity (Ke et al., 2017). While other factors could be involved, it is plausible that altered ribosomal dynamics that result in increased translational fidelity contribute to a more stable proteome and increased longevity. However, the relationship between this unique large ribosomal subunit and naked mole rat longevity remains associative, with a clear need for studies aimed at determining whether this alteration is necessary and sufficient for enhanced lifespan.
3.1.5 -. RIBOSOME BIOGENESIS
The importance of ribosome biogenesis and assembly in aging is highlighted by the research on signaling pathways and genetic screens that extend lifespan. Single gene deletions in yeast strains that increased replicative lifespan identified a number of RPs such as Rpl10 and Rps18, among others (Chiocchetti et al., 2007; Kaeberlein et al., 2005). Similar results were seen in RNAi-mediated knockdown of several small and large ribosome subunit proteins in C. elegans (Hansen et al., 2007). Additionally, the size of the nucleolus, which is the site for ribosomal biogenesis, increases with age and inversely correlates with longevity (Tiku et al., 2017). While the mechanistic relationship between nucleolar hypertrophy and aging is not entirely clear, there is a relationship between nucleolar size and global protein synthesis capacity (reviewed in Tiku & Antebi, 2018), (Montanaro, Trere, & Derenzini, 2008). Intriguingly, a modest downregulation of the methyltransferase fibrillarin, which functions in rRNA maturation and is a major component of the nucleolus, reduced nucleolar size and extended C.elegans lifespan (Tiku et al., 2017). Thus, evidence from a series of longevity studies and genetic screens support the view that suppressing ribosome biogenesis or altering ribosomal protein stoichiometry is sufficient to increase lifespan.
3.2 -. NUTRIENT AND STRESS SIGNALING
Some of the first reports of long-lived mutants in the nematode C. elegans identified the Insulin/IGF-1 signaling (IIS) pathway components as important genetic modulators of aging (Friedman & Johnson, 1988; Kenyon, Chang, Gensch, Rudner, & Tabtiang, 1993; Klass, 1983). The IIS pathway activates protein synthesis to control growth, development and cellular stress resistance (reviewed in Giannakou & Partridge, 2007) and mutations in this pathway increase lifespan in multiple models of aging (H. Liang et al., 2003; van Heemst, 2010). Since then a number of studies have established that protein synthesis is a major downstream target for many signaling pathways that affect aging (Hansen et al., 2007; Pan et al., 2007), (reviewed in Tavernarakis, 2008) (Fig 3). The most influential of these pathways is the mammalian Target of Rapamycin (mTOR) pathway (Kapahi et al., 2010; Nandagopal & Roux, 2015).
Fig. 3. Stress, Nutrient and Growth factor signaling pathways that affect translation.
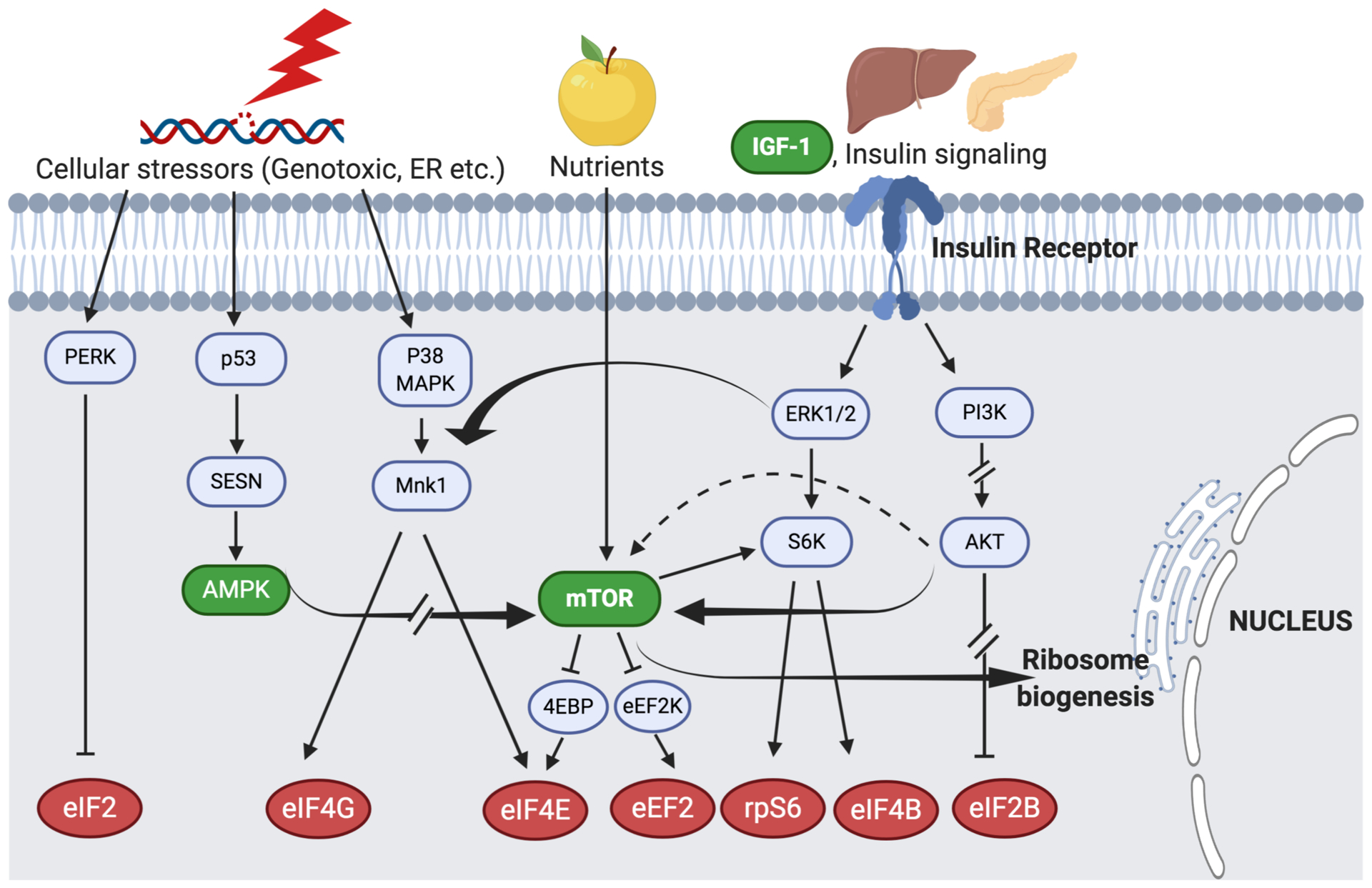
A large set of signaling cascades link translation dynamics to pathways implicated in both normal and pathological aging. Activation of mTOR by nutrient signaling causes phosphorylation and inactivation of 4EBP and eEF2K, releasing eIF4E and eEF2 respectively to activate translation. Both eIF4E and 4G are targets of Mnk1, which is controlled by p38 MAPK and ERK1/2. ERK1 and 2 respond to IGF/Insulin signaling to activate S6K, which acts on rpS6 and eIF4B to activate translation. Signaling through this pathway also activates PI3K that can directly or indirectly activate AKT. mTOR is acted on by AKT either directly (bold arrow) or indirectly (dotted arrow) via TSC2 (not shown) to inhibit eIF2B. Genotoxic stress activates sestrins (encoded by SESN) to block mTOR signaling and translation through a pathway that involves p53, AMPK and TSC2 (not shown). ER stress activates PERK to phosphorylate and inhibit eIF2. The mTOR pathway also activates ribosome biogenesis. Arrows indicate activation; bar lines indicate suppression. The molecules highlighted in green are targets and pathways for aging interventions (Longo et al., 2015). (For the purposes of clarity, other molecules such as NAD+, Sirtuins and epigenetic modulators are not shown). The downstream translation pathway targets of signaling are highlighted in red. Broken arrows signify additional activators or repressors that are upstream players in that particular cascade; Mnk1 (mitogen activated protein kinase-interacting kinase), p38 MAPK (p38 Mitogen activating kinase), ERK 1/2 (Extracellular signal-regulated kinases), S6K (S6 kinase), rpS6 (ribosomal protein S6), AKT (serine-threonine kinase AKT), GSK3 (Glycogen synthase Kinase 3), PERK (Pancreatic Endoplasmic Reticulum eIF2α Kinase), PI3K (Phosphatidylinositol 3 Kinase), 4EBP (eIF4E Binding Protein), eEF2K (eEF2 Kinase).
The mTOR pathway is an evolutionarily conserved hub that integrates signals from multiple cellular demands to ensure that growth rate and resources are adequately matched (Sossin & Costa-Mattioli, 2019). mTOR is activated by nutrients, amino acids, insulin and growth factors. This in turn activates downstream anabolic processes such as mRNA translation and ribosome biogenesis and suppression of catabolic events such as autophagy. Consequently, suppressing the mTOR pathway using the inhibitor Rapamycin helps to reverse these effects with dramatic consequences on lifespan in a host of model organisms (Garza-Lombo & Gonsebatt, 2016). Rapamycin inhibits the mTORC1 complex (mammalian Target of Rapamycin Complex 1) (Heitman, Movva, & Hall, 1991). When active, mTORC1 phosphorylates and suppresses both the eIF4E Binding Protein (4EBP) and the eEF2 Kinase, which allows their cognate targets eIF4E and eEF2 to support translation (Fig 3) (Kapahi et al., 2010). Rapamycin treatment activates both 4EBP and eEF2K with direct inhibitory effects on the initiation and elongation stages of translation. Rapamycin also inhibits the mTORC1-mediated activation of ribosome biogenesis while enhancing autophagic clearance of misfolded proteins. Surprisingly, the effects of Rapamycin on lifespan are seen even when administered at later stages in life in a mouse model of aging (Harrison et al., 2009). Additional evidence for the importance of this pathway comes from studies in long-lived mouse models of aging such as the Snell dwarf mutants that have a pituitary dysfunction and Growth hormone receptor knockout mice, both of which lack growth hormones. These mice show decreased mTORC1 activity and increased resistance to multiple forms of stress (Dominick et al., 2015).
Protein synthesis inhibition elicits improved stress resistance in multiple model systems (Giannakou & Partridge, 2007; Kourtis & Tavernarakis, 2011; H. Liang et al., 2003). For instance, knockdown of IFE-2, a somatic tissue-specific isoform of eIF4E in worms, extended their lifespan significantly by increasing their resistance to oxidative stress (Syntichaki et al., 2007). Similarly, reducing expression of the eIF4G initiation factor post-developmentally leads to an extension in organismal lifespan (Curran & Ruvkun, 2007; Smith et al., 2008). Besides reducing translation globally, this inhibition of eIF4G causes a relative increase in the expression of select mRNA transcripts including genes involved in the stress response (Rogers et al., 2011).
The net result of all these alterations is a global decrease in protein synthesis with age, suggesting that simply reducing global protein synthesis may be an important potential target for lifespan extension strategies. What remains unclear is whether such global changes are sufficient to allow these shifts in longevity in larger organisms, what costs such interventions would elicit in terms of organismal fitness, and whether some of these effects are driven by transcript-specific and tissue-specific alterations in translation dynamics.
4. PROTEIN SYNTHESIS IN THE AGING BRAIN
4.1-. mRNA-PROTEIN DECOUPLING
Global protein synthesis rates within the brain decline with age (as measured by percent of transcripts found within translating polysomes) (Fando, Salinas, & Wasterlain, 1980; Rattan, 1996). This decline occurs in the context of large-scale changes in the relative abundance of specific mRNAs. These transcripts belong to pathways that are indicative of increased inflammation, immune response and oxidative stress and reduced neurotrophic support and energy metabolism (de Magalhaes, Curado, & Church, 2009; C.-K. Lee, Weindruch, & Prolla, 2000; Zahn et al., 2007). However, the changes in mRNA levels and protein levels do not perfectly match. Instead, there is an age-associated decoupling of mRNA-protein profiles, such that steady-state protein levels increase relative to their cognate mRNA transcripts, suggesting a significant role for post-transcriptional gene regulation (G. E. Janssens et al., 2015; Wei et al., 2015). While some of these changes are likely due to altered protein degradation pathways (Taylor & Dillin, 2011), more recent single cell transcriptomic approaches suggest that some of this decoupling may result from cell-type specific alterations in the protein translational machinery that occurs with aging (Ximerakis et al., 2019). Specifically, within mature neurons of the aging mouse brain, there is a selective upregulation of many Ribosomal Proteins (RP) along with a preponderance in cellular pathways associated with ribosome assembly and cytoplasmic translation. In contrast, these genes were mostly downregulated in neuronal and oligodendrocyte precursor cells and astrocytes. This upregulation of RPs was also neuron subtype specific, occurring predominantly in Glutamatergic and GABAergic neurons but not in Dopaminergic neurons (Ximerakis et al., 2019). The cause and consequence of these cell-type specific changes in translational machinery within the aging brain are unclear.
4.2 -. CHANGES IN TRANSLATIONAL COMPONENTS
In premature or accelerated aging disorders such as Hutchinson-Gilford Progeria Syndrome (HGPS), there is an increase in ribosome biogenesis (Buchwalter & Hetzer, 2017) suggesting that this could be a hallmark of aging cells. In addition, ribosomal proteins are locally translated in neuronal axons and can physically associate with the ribosomes present at that site (Shigeoka et al., 2019) through a process that occurs independent of the nucleolus and which has functional consequences for axonal branching and plasticity. The age-associated increases in ribosomal proteins (RP) could also have downstream effects on ribosome complex composition, as there is a rapid turnover of some RPs compared to other core components (Lastick & McConkey, 1976; Samir et al., 2018). A possible function for the presence of RPs in dendritic and axonal fractions could be to outfit the ribosome for immediate activity-mediated alterations in translation rate (Holt & Schuman, 2013; A. S. Lee, Burdeinick-Kerr, & Whelan, 2013). Consequently, the dysregulation of ribosomal biogenesis might lead to abnormal ribosomal assembly and stoichiometry and subsequent errors in translation within aging neuronal populations. Defects in assembly could also be an indirect contributing factor to the decrease in translating polysomes that are observed in the aging brain (Fando et al., 1980). Besides RPs, the transcripts encoding the translation initiation factors eIF1, eIF3h and eIF5b are also elevated in mature neurons in the aging brain (Ximerakis et al., 2019). eIF1 an eIF5b function in start codon recognition and ribosome subunit joining respectively (Pestova & Kolupaeva, 2002; Pestova et al., 2000; Wang et al., 2019) and alterations in their relative levels could impact these functions in aging. Additionally, a decrease in components of the translation machinery such as tRNA synthetases, could affect mRNA stability (Wu et al., 2019) and might explain the decoupling of mRNA and protein levels.
4.3 -. CROSSTALK-PROTEIN SYNTESIS AND DEGRADATION
One of the hallmarks of aging is altered proteostasis (Lopez-Otin et al., 2013; Taylor & Dillin, 2011). Global decreases in translation that occur with age are proposed to assist with maintaining a balance such that protein clearance pathways such as autophagy and the proteasome are not overwhelmed. However, both autophagy and the proteasome show age-associated declines in activity (reviewed in Martinez-Lopez, Athonvarangkul, & Singh, 2015; Saez & Vilchez, 2014) suggesting that these mechanisms could be creating a feed-forward loop. For example, declines in Proteasome activity or proteasomal inhibition are sufficient to impair ribosome function and translation (Ding, Dimayuga, Markesbery, & Keller, 2006). Recent evidence from Nothobranchius furzeri (African killifish) suggests that there is a progressive decoupling of mRNA and protein levels in the aging brain. This decoupling is accompanied by, and perhaps caused by, a widespread stoichiometric imbalance in various protein complexes including the proteasome and ribosomes (Kelmer Sacramento et al., 2020). They propose that altered proteasomal stoichiometry impedes proteasome function, which in turn triggers defects in ribosomal assembly/disassembly and a collapse in neuronal protein homeostasis later in life (Kelmer Sacramento et al., 2020; Walther et al., 2015). However, the temporal order of these events and the primacy and causality of proteosomal dysfunction is not yet clear.
4.4 -. ALTERED MITOCHONDRIA
Stoichiometric imbalances in the aging brain were also reported to affect the composition of a number of other protein complexes such as the Mitochondrial ribosome (Ori et al., 2015). The Mitochondria have their own DNA (mtDNA) and protein synthetic machinery including dedicated mito-ribosomes (Lightowlers, Rozanska, & Chrzanowska-Lightowlers, 2014). They serve as the primary generators of energy in the cell and their dysfunction contributes meaningfully to both global and neuronal aging (Wallace, 2005; Yankner, Lu, & Loerch, 2008). Age-associated decreases in mitochondrial translation occur in both mice and humans. These changes in mitochondrial translation arise independent of mtDNA mutations or copy number alterations, suggesting something intrinsic to the aging process (Takai, Inoue, Shisa, Kagawa, & Hayashi, 1995). Mitochondrial translation acts as a critical checkpoint for maintaining homeostasis within the cell, with impacts on mitochondrial fusion and fission as well as non-mitochondrial cellular functions (Battersby & Richter, 2013; Suhm et al., 2018). Interestingly, decreased mitochondrial translational fidelity impairs cytosolic translation and evokes a transcriptional stress response that leads to shortened lifespans in yeast (Suhm et al., 2018). A major contribution to aging by inefficient mitochondrial function is the generation of reactive oxygen species (ROS) that accumulate in cells. These highly reactive radicals damage proteins and lipids and are particularly detrimental to aging postmitotic cells such as neurons (Yankner et al., 2008). A recent study using induced neurons (iNs) derived from fibroblasts of individuals at different ages confirmed the mitochondrial aging seen in other systems (Kim et al., 2018). Furthermore, mitochondrial morphology based on their fission and fusion dynamics impact function and also undergo alterations with age (Sebastian, Palacin, & Zorzano, 2017; Sharma, Smith, Yao, & Mair, 2019). It remains unclear whether these effects are a consequence of age or a direct contributor to the aging process, but mitochondrial function is highly linked to neuronal function and implicated independently in myriad neurodegenerative disorders (Knott, Perkins, Schwarzenbacher, & Bossy-Wetzel, 2008).
4.5-. PROTEIN AGGREGATES IN AGING
In the backdrop of decreased protein synthesis and degradation, there is also an observed accumulation of misfolded proteins with aging leading to an increase in protein aggregates (David et al., 2010; Huang et al., 2019). Though most studies indicate that the aggregates are toxic and reduce lifespan, one report suggests that they could be protective (Walther et al., 2015). Accumulation of misfolded proteins trigger restorative proteostatic mechanisms such as molecular chaperones and stress response pathways that attempt to compensate for failures in neuronal protein quality control. Despite these compensatory measures, the aging cellular environment appears poorly adapted to correct this proteostatic imbalance (Morimoto, 1998), leading to chronic activation of stress pathways in aging neurons that contribute to dysfunction and cell death.
These studies suggest that acquired defects in proteostasis during neuronal aging might not be an isolated event triggered by impaired proteolytic mechanisms leading to the accumulation of aggregated toxic proteins, but instead may reflect failure in a more complicated feedback mechanism where protein degradation, ribosomal biogenesis and protein synthesis are coordinately affected (Fig 4).
Fig. 4. Alterations in translational dynamics with aging.
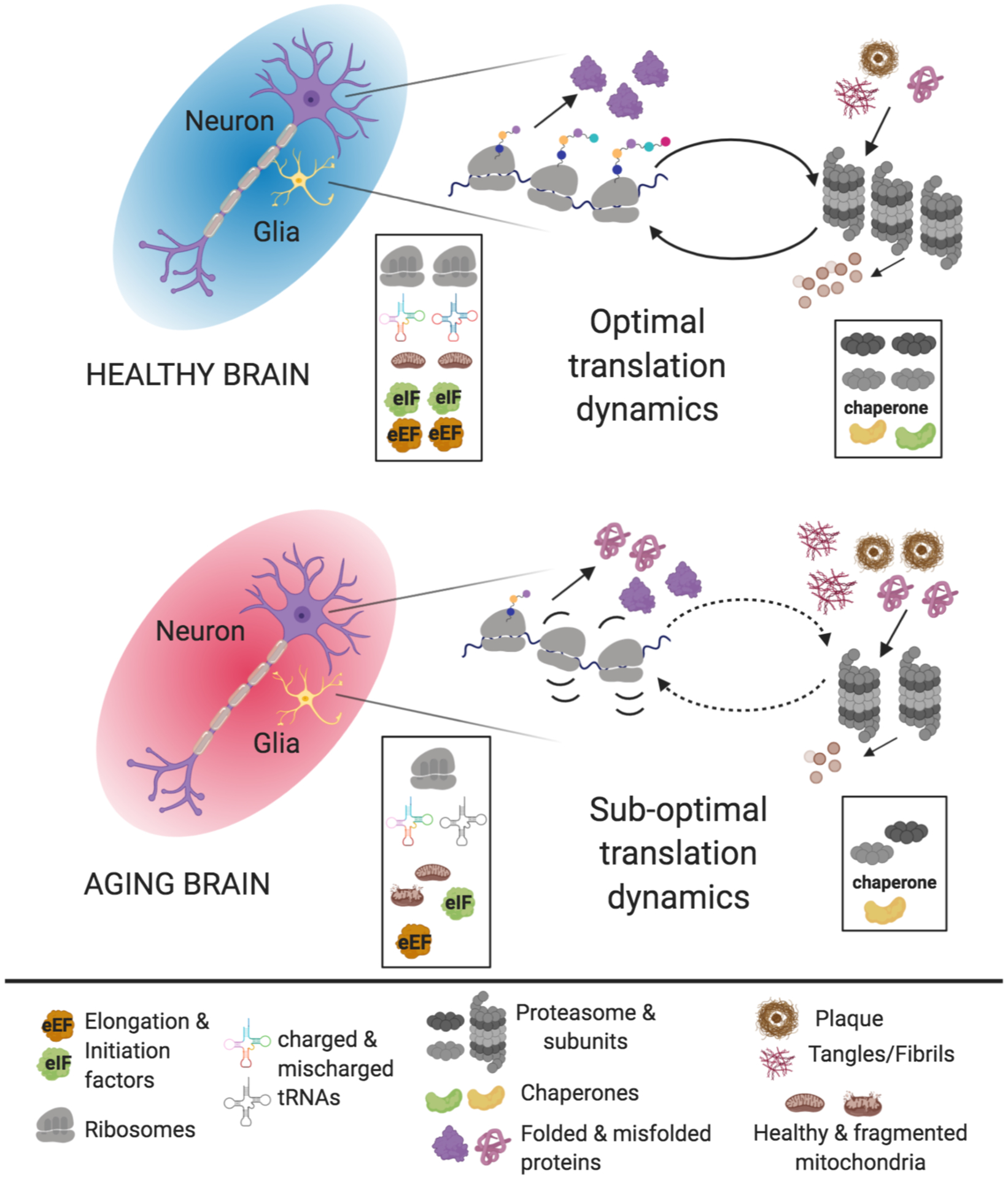
In the healthy brain (top), protein metabolism is maintained in homeostatic balance by the availability of optimal numbers of properly assembled ribosomes (healthy nucleoli), sufficient initiation factors, elongation factors, accurately charged tRNAs (properly functioning tRNA synthetases) and healthy mitochondria that provide sufficient energy for protein synthesis. At the same time, there is precise assembly of proteasomal subunits in the correct stoichiometric ratios along with adequate amounts of chaperones, which are dependent on the protein synthetic machinery for appropriate function and which directly aid in both maintaining translational fidelity and clearance of mis-translated and misfolded proteins (See legend at the bottom for illustrations). Alterations in the stoichiometry of key components of the translation machinery and decreases in the both fidelity of translation and in the corrective measures in place to address mis-translation events emerge with normal aging and are enhanced in pathological age-associated neurodegenerative conditions. These failures in translation place enhanced burdens upon proteostatic systems both directly (through generation of misfolded proteins) and indirectly (through decrements in generation of functional protein degradation factors). The combinatorial impact of these changes lead to dysregulation in neuronal proteostasis, which feed into one another and contribute to unhealthy cellular environments prone to pathogenic cascades.
4.6-. ALTERED GLIAL DYNAMICS
Astrocytes, Oligodendrocytes and other neuroglial cells are important support cells of the nervous system and are also affected in the aging brain (Palmer & Ousman, 2018; Soreq et al., 2017). The majority of these changes seem to be driven by a pro-inflammatory environment that develops in the brain with aging, perhaps through alterations in the blood-brain barrier and a loss of immune privilege with age. However, it is also likely that some of the changes in translational profiles that occur in these support cells with age are compensatory and at least partly neuroprotective. Stereological cell counts of postmortem brains show that oligodendrocyte numbers decrease with age (Pelvig, Pakkenberg, Stark, & Pakkenberg, 2008) without appreciable changes in neuronal numbers (Freeman et al., 2008). Oligodendrocytes secrete the myelin sheets that wrap axons and form the White Matter (WM) of the brain and spinal cord. Aging is associated with a significant decrease in WM and myelinated fiber thickness (Marner, Nyengaard, Tang, & Pakkenberg, 2003). Microglial cells in the aging brain show altered morphology and inefficient responses to inflammatory stimuli believed to be caused by sensitization to overproduction of proinflammatory signals (Valles et al., 2019). Interestingly, cell-type specific transcriptomic studies have identified brain specific microglial phenotypes (Olah et al., 2018) associated with pathological aging. A similar observation was made using single-nucleus RNA sequencing of astrocytes as well (Habib et al., 2020). Protein synthesis in peripheral processes of astrocytes show age-associated changes in the brain (Sakers et al., 2017; Ximerakis et al., 2019). While some of these patterns mirror what was seen in mature neurons, astrocytes exhibit a significant downregulation of most ribosomal proteins as well as pathways associated with oxidative phosphorylation and mitochondrial respiration. These changes have profound consequences in neuronal protein homeostasis. Dynamic SILAC based approaches in neuronal and glial enriched cultures show that protein turnover in neurons is highly dependent on both glial cell functions and their extracellular environment (Dorrbaum, Kochen, Langer, & Schuman, 2018). As such, age-dependent declines in glial function may drive key failures in neuronal homeostasis and contribute to neurodegeneration.
What exactly drives age-related declines in global brain translation remains enigmatic. Altered translation could be a direct consequence of altered ribosomal biogenesis or loss of protein components in key proteostatic networks, or mRNA instability elicited by mischarging of tRNAs or declines in translational fidelity. Whatever the cause; when defects in protein synthesis arise they directly impact cellular protein clearance mechanisms to derail proteostasis. These mis-coded proteins and RNA decay pathways in turn require proteosomal activation, degradation of partially synthesized proteins and re-synthesis of key factors. As the normal aging brain is already dealing with an energy crisis elicited by dysfunctional mitochondria and declines in the capabilities of supporting glial cells, these perturbations in proteostasis have synergistically deleterious impacts on neuronal function and predispose neurons to pathological feed-forward cascades which end in age-associated neurodegeneration (Fig 4).
5. LOOKING AHEAD: METHODS TO ADDRESS TRANSLATIONAL DYNAMICS IN AGING
To date, the information we have on translation dynamics in the aging brain are quite limited. Most current studies have looked at global translation levels as readout or at the select translation of a small set of transcripts. Moreover, the majority of these studies have ignored the massive complexity and cellular and subcellular heterogeneities of the brain. Understanding how translational dynamics change across brain regions and cell types and with transcript specific resolution may well reveal key elements that are not obvious from our current vantage point. Accomplishing this, however, will require a combination of both large scale and cell-specific analyses. Luckily, a number of new tools have emerged over the past decade that should accelerate these types of analyses (Table 2).
Table 2.
Overview of transcriptomic and proteomic approaches.
| Method | Purpose/ Application | References |
|---|---|---|
| RNA sequencing | A widely used high throughput sequencing technology to query all transcripts in a tissue or in cell culture. | 1 |
| Polysome profiling | Query transcript distribution in terms of number of ribosomes on a transcript to infer translation status. | 2, 3 |
| Ribosome Profiling or Ribo-seq | Query global and specific Translation efficiency of transcripts at nucleotide resolution using elongating 80S ribosome footprints. | 4, 5 |
| Ribosome Complex Profiling (RCP-seq) | Query all stages of translation initiation step using both 80S and the initiating small subunit ribosome footprints. | 6 |
| Translation Complex profiling (TCP)-seq | Query early intermediates of translation initiation using 80S ribosomes. | 7, 8 |
| Global Translation Initiation (GTI)-seq | Query translation Initiation sites using two translation inhibitors (Cycloheximide and Lactimidomycin) followed by Ribosome Profiling. | 9 |
| Quantitative Translation Initiation (QTI)-seq | Query translation Initiation dynamics using Lactimidomycin and Puromycin to enrich for translation initiation start sites followed by Ribosome Profiling. | 10 |
| RiboLace | Query translation at single nucleotide resolution by active ribosomes versus inactive/ unbound ribosomes using a Puromycin analog. | 11 |
| Ribo-Tag | Query cell type specific translation using an HA tagged Rpl22 ribosomal protein under Cre driver. | 12 |
| Translating Ribosome Affinity Purification (TRAP) | Query cell type specific translation using transgene and an L10 ribosomal protein tag. | 13, 14 |
| Surface Sensing of Translation (SUnSET) | Query global protein synthesis in single or heterogeneous cell populations. | 15 |
| Selected or Parallel Reaction Monitoring (SRM/PRM)-MS | Query absolute stoichiometries of ribosomal proteins and other protein complexes. | 16 |
| Tandem Mass Tag (TMT)-MS | Query and compare identity and relative abundances of ribosomal and proteasomal peptides. Can also be used to identify post-translational modifications of proteins. | 17 |
| BONCAT | Query nascent protein synthesis using click-chemistry based chemical tags | 18 |
| FUNCAT | Visualize nascent protein synthesis using click-chemistry based flourescent tags | 19 |
Table legend- References corresponding to the numbers listed in the last column.
1- (Lowe, Shirley, Bleackley, Dolan, & Shafee, 2017), 2- (Pan et al., 2007), 3- (Rogers et al., 2011), 4- (Ingolia et al., 2009), 5- (Sudmant et al., 2018), 6- (Giess et al., 2020), 7- (Archer, Shirokikh, Beilharz, & Preiss, 2016), 8- (Archer et al., 2016), 9- (S. Lee et al., 2012), 10- (Gao et al., 2015), 11- (Clamer et al., 2018), 12- (Sanz et al., 2019), 13- (Heiman et al., 2014), 14- (Heiman et al., 2008), 15- (Schmidt, Clavarino, Ceppi, & Pierre, 2009), 16- (Rauniyar, 2015), 17- (A. Thompson et al., 2003), 18 - (D. C. Dieterich et al., 2006), 19- (Daniela C. Dieterich et al., 2010).
5.1. TRANSCRIPTOMIC APPROACHES
5.1.1-. Polysome Profiling
Polysome profiling underlies many advanced sequencing based methods for measuring translation efficiency. It is considered the gold standard for identifying mRNAs that are being actively translated as opposed to those that are merely present in the cell at a given time. A reliable measure of active translation is the association with ribosomes. Polysome profiling is used to separate out the ribosomes along with their associated mRNAs from a whole cell lysate. This is achieved using a sucrose gradient centrifugation method to isolate the ribosomes based on their density (Masek, Valasek, & Pospisek, 2011). The mRNAs that are associated with multiple ribosomes (polysomes) will have a higher density than non-translating transcripts associated with RNA binding proteins or those associated with the 40S subunit and 80S monosomes. These distinct populations are collected separately for downstream analyses such as Northern and Western blots, qRT-PCR or RNA sequencing. A polysome profiling study in the aging rodent brain reported reduced polysomes fractions at 16 months suggesting a decline in protein synthesis as an event preceding senescence (Fando et al., 1980). Additional studies in longevity-enhancing mutants have also reported reductions in polysome fractions (Pan et al., 2007; Rogers et al., 2011). Since the separated ribosome fractions remain associated with initiation and elongation factors and other proteins, polysome profiling can also be used to evaluate differences in the absolute levels of these factors under different cellular conditions (Coudert, Adjibade, & Mazroui, 2014). However, this technique is limited by the need for purification of the diluted fractions and a large number of cells or tissue to obtain sufficient starting material for analysis (see King & Gerber, 2014). While some recent improvements in polysome profiling have addressed some of these issues (S. Liang et al., 2018), this technique lacks the type of regional, cellular and sub-cellular resolution that will be required to fully interrogate these circuits.
5.1.2 -. Translating Ribosome Affinity Purification (TRAP)
The heterogeneity of cell types in the brain along with their close proximity makes it hard to assign differences in transcript level patterns to a particular cell type. Translating Ribosome Affinity Purification (TRAP) is a way to capture cell-type specific gene expression profiles and has been used in the mammalian brain and other systems. TRAP employs an indirect mRNA tagging approach using an affinity tag to label the large L10 ribosomal subunit protein along with a transgene to drive expression in a cell type specific manner. Use of this method enabled the distinction of morphologically similar subclasses of medium spiny neurons and showed that they have different translational profiles (Heiman, Kulicke, Fenster, Greengard, & Heintz, 2014). Several Bacterial Artificial Chromosomes (BAC)-TRAP transgenic mouse lines as well as other conditional TRAP lines are now available. A study applying TRAP in Astrocytes identified local translation in the peripheral processes of these cells that could alter adjacent synapses (Sakers et al., 2017). Such effects might be altered in the aging brain and TRAP could be used to characterize the translational profiles of the astroglial classes observed in pathological aging (Olah et al., 2018). The advantage of TRAP in gene expression is that it provides a read out of the true translated mRNAs, which is closer to the protein content of the cell. These and other features of the TRAP technology are extensively reviewed in Heiman et al., 2008.
5.1.3 -. RiboTag
RiboTag is a complementary approach to TRAP that uses a mouse line with a modified allele of the 60S ribosomal protein gene Rpl22. This gene has been modified such that it will undergo Cre-lox mediated recombination to insert an HA tag in Rpl22 when crossed to a Cre-driver mouse. This generates epitope-tagged polysome populations in a cell-type specific manner enabling immunoprecipitation and downstream analyses (Sanz, Bean, Carey, Quintana, & McKnight, 2019). The advantage of the RiboTag method is the availability of a large number of Cre-driver lines and a robust endogenous expression of tagged ribosomal populations. Both TRAP and RiboTag can be used in combination with ribosome profiling (discussed below) to address cell subtype specific questions in aging. Of particular interest would be the question of neuron subtype specificity in the upregulation of ribosomal proteins that was observed in single cell transcriptomic studies of the aging mouse brain (Ximerakis et al., 2019). Neuron subtype specific Cre-driver lines such as the DAT (Dopamine Transporter)-promoter line for Dopaminergic neurons, v-glut2-Cre lines for glutamatergic neurons and a number of GABA-ergic drivers can be used here for subtype specific Ribosome tagging and profiling (Harris et al., 2014; Taniguchi et al., 2011).
5.1.4 -. Ribosome Profiling
Ribosome profiling is a powerful method to query translational efficiency transcriptome-wide at nucleotide resolution (Ingolia et al., 2014; Ingolia, Ghaemmaghami, Newman, & Weissman, 2009). Unlike Microarrays and other RNA sequencing techniques, ribosome profiling provides position specific information of ribosome occupancy. It can be used to estimate the number of ribosomes occupying different regions of the mRNA such as the 5’UTR, coding sequence, start sites or stop sites and 3’UTR regions. The principle behind ribosome profiling is similar to earlier footprinting assays used to measure initiation sites (Steitz, 1969). Ribosome profiling relies on the deep sequencing of ribosome footprints i.e. ~30nt regions of the mRNA that is enclosed within the 80S ribosome and protected from RNAse digestion. These ribosome-protected mRNA fragments (RPF) are converted to DNA and mapped back to genomic regions to find out where along a transcript a single ribosome was translating (Ingolia, Hussmann, & Weissman, 2019). Total RNA extraction and sequencing is carried out in parallel to normalize the RPFs to mRNA abundance. Thus, translation efficiency of individual transcripts can be calculated using the mRNA counts to establish steady state transcript levels and the RPF counts as readout for active translation (Brar & Weissman, 2015). Ribosome profiling has been adapted to multiple systems since the first study in budding yeast reported widespread initiation at upstream ORFs and novel non-AUG start codons (Ingolia et al., 2009). It has also been a great tool to study the mechanistic details of protein translation such as ribosomal pausing and defects in ribosome recycling (Brar & Weissman, 2015; Guydosh & Green, 2014). However, artifacts arising from cycloheximide treatments, endonuclease digestions etc. are known to occur and need to be optimized and controlled for (reviewed in Andreev et al., 2016).
In the field of aging, Ribosome profiling has already yielded some interesting observations. A study in the aging mouse and human brain found a region-specific accumulation of 3’UTR mRNA and encoded peptides (Sudmant, Lee, Dominguez, Heiman, & Burge, 2018). The generation of these 3’UTR products was associated with oxidative stress and accumulated when the ribosome-recycling factor ABCE1 was depleted. The authors propose a possible model for the biogenesis of these 3’UTR fragments where oxidative stress impairs translation termination causing ribosomal entry into 3’UTR regions and triggering endonucleolytic cleavage near the stop codon. The presence of isolated 3’UTRs has been reported in neurons in another study (Kocabas, Duarte, Kumar, & Hynes, 2015) and hints at a biological function for this phenomenon in regulating the coding sequence of some proteins. Ribosome profiling can be used to shed light on the translation dynamics at play in the aging brain. Specifically, it can be used to query the translational efficiency of particular transcripts such as chaperones in the aging brain to understand the failures in protein clearance. This could also help delineate the cross talk between the protein synthesis and degradation mechanisms that have been implied in aging and neurodegeneration (Ding, Cecarini, & Keller, 2007; Keller, Gee, & Ding, 2002).
5.1.5 -. Ribosome Complex Profiling (RCP)
Unlike Ribosome profiling, Ribosome Complex Profiling (RCP) or RCP-seq captures all the footprints of the ribosome - i.e. both the 80S and the initiating small subunit (SSU) ribosomes (Giess et al., 2020). This technique allows for the querying of each stage of translation initiation from recruitment of the SSU to scanning along the 5’UTR and conversion to an elongation-competent ribosome and could help identify regulators of this crucial rate-limiting step in the aging brain. Since RCP-seq also uses a paraformaldehyde crosslinking step, the tRNA molecules that are associated with the ribosome can also be identified. This is particularly useful to determine the prevalence of alternate translation initiation site usage with aging.
5.2. PROTEOMIC APPROACHES
Both polysome profiling and ribosome profiling provide transcript specific information on genome wide changes in translation dynamics. These techniques can be combined with emerging Mass spectrometric analyses to look at ribosome composition (Shi et al., 2017) as well as posttranslational modifications of ribosomes (Imami et al., 2018). Recent single cell transcriptomic studies show distinct transcriptional profiles driving aging with ribosomal protein dynamics as an important player (Ximerakis et al., 2019). In agreement with these observations, ribosomal stoichiometry was altered in at least one model of aging (Kelmer Sacramento et al., 2020). Changes in the composition or stoichiometry of ribosomes with age can be studied using proteomics based approaches such as Tandem Mass Tags (TMT) and Selected or Parallel Reaction Monitoring (SRM/PRM) for individual proteins. TMT based Mass Spectrometry (MS) is widely used for the relative quantitation of peptides and has been used to study ribosome heterogeneity and function (Emmott, Jovanovic, & Slavov, 2019; Slavov, Semrau, Airoldi, Budnik, & van Oudenaarden, 2015). This can be employed to analyze the non-translating monosome fractions that have been reported in aging brains (Fando et al., 1980) for instance. Additional methods to calculate absolute stoichiometries of ribosomal complexes include Selected or Parallel Reaction Monitoring (SRM/PRM) based mass spectrometry (Rauniyar, 2015), where isolated complexes are digested and mixed with a known amount of heavy-labeled isotope of the ribosomal protein of interest. The sample is then subjected to MS to determine the difference in peak intensity from the known amount to measure absolute levels. In theory, Mass spectrometry can also be used to detect mistranslation frequency that occurs above tolerable levels or as a favorable response to cellular stress in some instances (reviewed in Moghal, Mohler, & Ibba, 2014), (Netzer et al., 2009; Wiltrout, Goodenbour, Frechin, & Pan, 2012). Such analyses could prove useful in comparative lifespan studies to understand the contributions of protein synthesis fidelity to longevity differences in various long-lived species, such as the naked mole rat and the bowhead whale (Buffenstein, 2005; Seim et al., 2014).
Alternatively, identification of nascent protein translation using Bio-Orthogonal Non-Canonical Aminoacid Tagging (BONCAT) (D. C. Dieterich, Link, Graumann, Tirrell, & Schuman, 2006) and the related visualization approach Fluorescent Non-Canonical Aminoacid Tagging (FUNCAT) (Daniela C. Dieterich et al., 2010) can provide alternative means to monitor age-associated decoupling of mRNA and protein levels (Kelmer Sacramento et al., 2020; Wei et al., 2015) with potentially subcellular resolution. Both these techniques use click-chemistry for chemoselective tagging of Azidohomoalanine (AHA) that gets incorporated at methionine codons in newly synthesized proteins. These labeled proteins are analyzed using MS based approaches to identify nascent translation. The combination of ribosome profiling with peptidomic approaches can be invaluable in assessing global and transcript level changes that occur in specific regions of the aging brain.
Despite their widespread use in the protein translation field, many of the techniques have not yet been extensively applied in the study of normal and pathological brain aging. Evaluations of translational dynamics in normal and pathological aging as well as the effects of longevity enhancing drugs and genetic perturbations may reveal new insights into neuronal health. Combined with powerful genetic tools and genome-wide datasets, the findings from such studies of translation dynamics could allow for a transcript and cell-type specific dissection of the global trends in neuronal protein synthesis and its effect on protein degradation in the aging brain. This could reveal new targets for future analyses and perhaps stage-specific molecular interventions that might slow age-related functional declines.
CONCLUSIONS
Given that life expectancy is projected to increase over the next decade along with the associated costs of caring for an aging society, it is encouraging that promoting healthy aging to delay or prevent disease development is a foreseeable goal (Kennedy et al., 2014; Olshansky, Goldman, Zheng, & Rowe, 2009). The achievement of such a goal would have huge impacts on societal and economic indices worldwide and particularly in countries with aging populations (Kontis et al., 2017). Whether aging is a disease (a pathology) or a strictly normal process is still debated by the research and medical communities (Bulterijs, Hull, Björk, & Roy, 2015; Kirkwood & Austad, 2000). Importantly, even though there are physiological changes in the aging brain in the absence of disease, these changes are amenable to interventions that promote health-span.
Studies from the aging field and neurodegeneration underscore the importance of both the translation machinery and translational fidelity in maintaining cellular proteostasis. It is telling that the most impactful regimens for promoting prolonged health and lifespan such as caloric restriction have direct effects on protein synthesis and degradation (Fontana, Partridge, & Longo, 2010; Longo et al., 2015). Though loss of proteostasis is a cellular hallmark of aging in multiple organisms, the mechanisms behind this complex process and in particular the contributions from anabolic changes remain unclear. It is also unclear how or if translational dynamics change in normal or healthy aging-particularly with regards to the behavior of the ribosome but also in components of the translation machinery. Could there be changes in the stability of certain transcripts or the increased use of alternate translation factors that favor certain transcripts over others? Such transcript-and proteome-specific changes could also occur directly within the proteasomal and protein degradation machinery pathways to alter cellular translation and homeostasis.
It is counterintuitive that translation decreases with normal aging but interventions that reduce translation rates slow aging and increase lifespan. This could suggest a transcript-specific selectivity of the interventions or a balancing of the synthesis and degradation dynamics in the aging environment to sustain homeostasis. Emerging transcriptomic and proteomic approaches may help decipher how aging interventions prioritize which transcripts to regulate and broadly how the aging brain maintains this decreased translation rates without compromising cellular health and integrity.
Finally, early-stage analysis may be needed to reveal the starting points for these pathological cascades in the brain. For instance, the aging signature of reduced protein synthesis and changes in synaptic plasticity are observed in early middle age (12–18 mo) in the rat brain (Fando et al., 1980; Lynch, Rex, & Gall, 2006). The aging brain may also reveal region and cell-type specific alterations in components of the translation machinery, which in turn may explain the selective neuronal vulnerability seen in many neurological disorders (Mattson & Magnus, 2006; Ximerakis et al., 2019). By applying the vast array of recently developed tools to assess translation dynamics in the aging brain, it is hoped that we will identify critical elements, which tie together protein synthesis, degradation and aging and reveal nodes that can be targeted therapeutically.
ACKNOWLEDGEMENTS
The authors would like to thank Rich Miller, Scott Leiser and members of the Todd lab for helpful discussions. Figures were created with BioRender.com.
FUNDING
GS was supported by a post-doctoral fellowship grants from the National Ataxia Foundation and a combined grant from Pepper Center for aging (NIH P30AG024824) and Michigan Alzheimer's Disease Research Center (P30 AG053760). PKT was supported by NIH R01NS086810, R01NS099280 and the Department of Veterans Affairs BLRD I01BX004842. Funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
CONFLICT OF INTEREST DISCLOSURE
Peter Todd holds a joint patent with Ionis Pharmaceuticals. He served as a consultant at Denali Therapeutics from 2016–2019 for which he received compensation. None of these potential conflicts of interest directly pertain to this manuscript and none of these parties had any role in writing or editing of the manuscript.
REFERENCES:
- Acker MG, & Lorsch JR (2008). Mechanism of ribosomal subunit joining during eukaryotic translation initiation. Biochem Soc Trans, 36(Pt 4), 653–657. doi: 10.1042/bst0360653 [DOI] [PubMed] [Google Scholar]
- Andreev DE, O’Connor, Patrick BF, Loughran G, Dmitriev SE, Baranov PV, & Shatsky IN (2016). Insights into the mechanisms of eukaryotic translation gained with ribosome profiling. Nucleic Acids Research, 45(2), 513–526. doi: 10.1093/nar/gkw1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews-Hanna JR, Snyder AZ, Vincent JL, Lustig C, Head D, Raichle, Marcus E, & Buckner RL (2007). Disruption of Large-Scale Brain Systems in Advanced Aging. Neuron, 56(5), 924–935. doi: 10.1016/j.neuron.2007.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova AS, Alexandrov AI, Makarova NE, Gladyshev VN, & Dmitriev SE (2018). Protein synthesis and quality control in aging. Aging (Albany NY), 10(12), 4269–4288. doi: 10.18632/aging.101721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SK, Shirokikh NE, Beilharz TH, & Preiss T (2016). Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature, 535(7613), 570–574. doi: 10.1038/nature18647 [DOI] [PubMed] [Google Scholar]
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, … Petrucelli L (2013). Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron, 77(4), 639–646. doi: 10.1016/j.neuron.2013.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpurua J, Ke Z, Chen IX, Zhang Q, Ermolenko DN, Zhang ZD, … Seluanov A (2013). Naked molerat has increased translational fidelity compared with the mouse, as well as a unique 28S ribosomal RNA cleavage. Proc Natl Acad Sci U S A, 110(43), 17350–17355. doi: 10.1073/pnas.1313473110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battersby BJ, & Richter U (2013). Why translation counts for mitochondria - retrograde signalling links mitochondrial protein synthesis to mitochondrial biogenesis and cell proliferation. Journal of Cell Science, 126(19), 4331–4338. doi: 10.1242/jcs.131888 [DOI] [PubMed] [Google Scholar]
- Beckelman BC, Zhou X, Keene CD, & Ma T (2016). Impaired Eukaryotic Elongation Factor 1A Expression in Alzheimer’s Disease. Neurodegenerative Diseases, 16(1–2), 39–43. doi: 10.1159/000438925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer M, & Rodnina MV (2007). The ribosomal peptidyl transferase. Mol Cell, 26(3), 311–321. doi: 10.1016/j.molcel.2007.03.015 [DOI] [PubMed] [Google Scholar]
- Bertoli-Avella AM, Garcia-Aznar JM, Brandau O, Al-Hakami F, Yuksel Z, Marais A, … Bauer P (2018). Biallelic inactivating variants in the GTPBP2 gene cause a neurodevelopmental disorder with severe intellectual disability. Eur J Hum Genet, 26(4), 592–598. doi: 10.1038/s41431-0180097-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Lu T, & Yankner BA (2010). Neural mechanisms of ageing and cognitive decline. Nature, 464(7288), 529–535. doi: 10.1038/nature08983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman AM, Michel YM, & Kean KM (2000). Biochemical characterisation of cap-poly(A) synergy in rabbit reticulocyte lysates: the eIF4G-PABP interaction increases the functional affinity of eIF4E for the capped mRNA 5’-end. Nucleic Acids Res, 28(21), 4068–4075. doi: 10.1093/nar/28.21.4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, & Weissman JS (2015). Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol, 16(11), 651–664. doi: 10.1038/nrm4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks SS, Wall AL, Golzio C, Reid DW, Kondyles A, Willer JR, … Davis EE (2014). A Novel Ribosomopathy Caused by Dysfunction of RPL10 Disrupts Neurodevelopment and Causes X-Linked Microcephaly in Humans. Genetics, 198(2), 723–733. doi: 10.1534/genetics.114.168211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwalter A, & Hetzer MW (2017). Nucleolar expansion and elevated protein translation in premature aging. Nat Commun, 8(1), 328. doi: 10.1038/s41467-017-00322-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham RH, Grentzmann G, & Kisselev L (1997). Polypeptide chain release factors. Mol Microbiol, 24(3), 449–456. doi: 10.1046/j.1365-2958.1997.3711734.x [DOI] [PubMed] [Google Scholar]
- Buffenstein R (2005). The Naked Mole-Rat: A New Long-Living Model for Human Aging Research. The Journals of Gerontology: Series A, 60(11), 1369–1377. doi: 10.1093/gerona/60.11.1369 [DOI] [PubMed] [Google Scholar]
- Bugiani M, Boor I, Powers JM, Scheper GC, & van der Knaap MS (2010). Leukoencephalopathy With Vanishing White Matter: A Review. Journal of Neuropathology & Experimental Neurology, 69(10), 987–996. doi: 10.1097/NEN.0b013e3181f2eafa [DOI] [PubMed] [Google Scholar]
- Bulterijs S, Hull RS, Björk VC, & Roy AG (2015). It is time to classify biological aging as a disease. Front Genet, 6, 205. doi: 10.3389/fgene.2015.00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke SN, & Barnes CA (2006). Neural plasticity in the ageing brain. Nat Rev Neurosci, 7(1), 30–40. doi: 10.1038/nrn1809 [DOI] [PubMed] [Google Scholar]
- Cabrita LD, Dobson CM, & Christodoulou J (2010). Protein folding on the ribosome. Curr Opin Struct Biol, 20(1), 33–45. doi: 10.1016/j.sbi.2010.01.005 [DOI] [PubMed] [Google Scholar]
- Chen D, Pan KZ, Palter JE, & Kapahi P (2007). Longevity determined by developmental arrest genes in Caenorhabditis elegans. Aging Cell, 6(4), 525–533. doi: 10.1111/j.1474-9726.2007.00305.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Wang S, Mestre AA, Fu C, Makarem A, Xian F, … Sun S (2018). C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2α phosphorylation. Nat Commun, 9(1), 51. doi: 10.1038/s41467-017-02495-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Wang S, Zhang Z, Morgens DW, Hayes LR, Lee S, … Sun S (2019). CRISPR-Cas9 Screens Identify the RNA Helicase DDX3X as a Repressor of C9ORF72 (GGGGCC)n Repeat-Associated Non-AUG Translation. Neuron, 104(5), 885–898.e888. doi: 10.1016/j.neuron.2019.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiocchetti A, Zhou J, Zhu H, Karl T, Haubenreisser O, Rinnerthaler M, … Breitenbach-Koller L (2007). Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Exp Gerontol, 42(4), 275–286. doi: 10.1016/j.exger.2006.11.002 [DOI] [PubMed] [Google Scholar]
- Clamer M, Tebaldi T, Lauria F, Bernabò P, Gómez-Biagi RF, Marchioretto M, … Viero G (2018). Active Ribosome Profiling with RiboLace. Cell Rep, 25(4), 1097–1108.e1095. doi: 10.1016/j.celrep.2018.09.084 [DOI] [PubMed] [Google Scholar]
- Cleary JD, & Ranum LP (2017). New developments in RAN translation: insights from multiple diseases. Curr Opin Genet Dev, 44, 125–134. doi: 10.1016/j.gde.2017.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudert L, Adjibade P, & Mazroui R (2014). Analysis of translation initiation during stress conditions by polysome profiling. J Vis Exp(87). doi: 10.3791/51164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, & Ruvkun G (2007). Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet, 3(4), e56. doi: 10.1371/journal.pgen.0030056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, & Kenyon C (2010). Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol, 8(8), e1000450. doi: 10.1371/journal.pbio.1000450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie K, Janssens J, Koldere D, De Waegeneer M, Pech U, Kreft L, … Aerts S (2018). A Single-Cell Transcriptome Atlas of the Aging Drosophila Brain. Cell, 174(4), 982–998.e920. doi: 10.1016/j.cell.2018.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brabander JM, Kramers RJ, & Uylings HB (1998). Layer-specific dendritic regression of pyramidal cells with ageing in the human prefrontal cortex. Eur J Neurosci, 10(4), 1261–1269. doi: 10.1046/j.1460-9568.1998.00137.x [DOI] [PubMed] [Google Scholar]
- de Magalhaes JP, Curado J, & Church GM (2009). Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics, 25(7), 875–881. doi: 10.1093/bioinformatics/btp073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, & Green R (2012). The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol, 4(7), a013706. doi: 10.1101/cshperspect.a013706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Hodas JJL, Gouzer G, Shadrin IY, Ngo JT, Triller A, … Schuman EM (2010). In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci, 13(7), 897–905. doi: 10.1038/nn.2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Link AJ, Graumann J, Tirrell DA, & Schuman EM (2006). Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc Natl Acad Sci U S A, 103(25), 9482–9487. doi: 10.1073/pnas.0601637103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillman AA, Majounie E, Ding J, Gibbs JR, Hernandez D, Arepalli S, … Cookson MR (2017). Transcriptomic profiling of the human brain reveals that altered synaptic gene expression is associated with chronological aging. Sci Rep, 7(1), 16890. doi: 10.1038/s41598-017-17322-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Cecarini V, & Keller JN (2007). Interplay between protein synthesis and degradation in the CNS: physiological and pathological implications. Trends Neurosci, 30(1), 31–36. doi: 10.1016/j.tins.2006.11.003 [DOI] [PubMed] [Google Scholar]
- Ding Q, Dimayuga E, Markesbery WR, & Keller JN (2006). Proteasome inhibition induces reversible impairments in protein synthesis. Faseb j, 20(8), 1055–1063. doi: 10.1096/fj.05-5495com [DOI] [PubMed] [Google Scholar]
- Ding Q, Markesbery WR, Chen Q, Li F, & Keller JN (2005). Ribosome dysfunction is an early event in Alzheimer’s disease. J Neurosci, 25(40), 9171–9175. doi: 10.1523/jneurosci.3040-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominick G, Berryman DE, List EO, Kopchick JJ, Li X, Miller RA, & Garcia GG (2015). Regulation of mTOR activity in Snell dwarf and GH receptor gene-disrupted mice. Endocrinology, 156(2), 565575. doi: 10.1210/en.2014-1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrbaum AR, Kochen L, Langer JD, & Schuman EM (2018). Local and global influences on protein turnover in neurons and glia. Elife, 7. doi: 10.7554/eLife.34202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmott E, Jovanovic M, & Slavov N (2019). Ribosome Stoichiometry: From Form to Function. Trends Biochem Sci, 44(2), 95–109. doi: 10.1016/j.tibs.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fando JL, Salinas M, & Wasterlain CG (1980). Age-dependent changes in brain protein synthesis in the rat. Neurochem Res, 5(4), 373–383. doi: 10.1007/bf00964226 [DOI] [PubMed] [Google Scholar]
- Fontana L, Partridge L, & Longo VD (2010). Extending healthy life span--from yeast to humans. Science, 328(5976), 321–326. doi: 10.1126/science.1172539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman SH, Kandel R, Cruz L, Rozkalne A, Newell K, Frosch MP, … Hyman BT (2008). Preservation of neuronal number despite age-related cortical brain atrophy in elderly subjects without Alzheimer disease. J Neuropathol Exp Neurol, 67(12), 1205–1212. doi: 10.1097/NEN.0b013e31818fc72f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DB, & Johnson TE (1988). A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics, 118(1), 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabius HJ, Goldbach S, Graupner G, Rehm S, & Cramer F (1982). Organ pattern of age-related changes in the aminoacyl-tRNA synthetase activities of the mouse. Mech Ageing Dev, 20(4), 305313. doi: 10.1016/0047-6374(82)90098-7 [DOI] [PubMed] [Google Scholar]
- Gao X, Wan J, Liu B, Ma M, Shen B, & Qian S-B (2015). Quantitative profiling of initiating ribosomes in vivo. Nature methods, 12(2), 147–153. doi: 10.1038/nmeth.3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Esparcia P, Hernandez-Ortega K, Koneti A, Gil L, Delgado-Morales R, Castano E, … Ferrer I (2015). Altered machinery of protein synthesis is region- and stage-dependent and is associated with alpha-synuclein oligomers in Parkinson’s disease. Acta Neuropathol Commun, 3, 76. doi: 10.1186/s40478-015-0257-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Esparcia P, Hernández-Ortega K, Koneti A, Gil L, Delgado-Morales R, Castaño E, … Ferrer I (2015). Altered machinery of protein synthesis is region- and stage-dependent and is associated with α-synuclein oligomers in Parkinson’s disease. Acta Neuropathol Commun, 3(1), 76. doi: 10.1186/s40478-015-0257-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza-Lombo C, & Gonsebatt ME (2016). Mammalian Target of Rapamycin: Its Role in Early Neural Development and in Adult and Aged Brain Function. Front Cell Neurosci, 10, 157. doi: 10.3389/fncel.2016.00157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakou ME, & Partridge L (2007). Role of insulin-like signalling in Drosophila lifespan. Trends Biochem Sci, 32(4), 180–188. doi: 10.1016/j.tibs.2007.02.007 [DOI] [PubMed] [Google Scholar]
- Giess A, Torres Cleuren YN, Tjeldnes H, Krause M, Bizuayehu TT, Hiensch S, … Valen E (2020). Profiling of Small Ribosomal Subunits Reveals Modes and Regulation of Translation Initiation. Cell Rep, 31(3), 107534. doi: 10.1016/j.celrep.2020.107534 [DOI] [PubMed] [Google Scholar]
- Glineburg MR, Todd PK, Charlet-Berguerand N, & Sellier C (2018). Repeat-associated non-AUG (RAN) translation and other molecular mechanisms in Fragile X Tremor Ataxia Syndrome. Brain Res, 1693(Pt A), 43–54. doi: 10.1016/j.brainres.2018.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes-Duarte A, Lacerda R, Menezes J, & Romao L (2018). eIF3: a factor for human health and disease. RNA Biol, 15(1), 26–34. doi: 10.1080/15476286.2017.1391437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonskikh Y, & Polacek N (2017). Alterations of the translation apparatus during aging and stress response. Mech Ageing Dev, 168, 30–36. doi: 10.1016/j.mad.2017.04.003 [DOI] [PubMed] [Google Scholar]
- Goodman LD, Prudencio M, Srinivasan AR, Rifai OM, Lee VM, Petrucelli L, & Bonini NM (2019). eIF4B and eIF4H mediate GR production from expanded G4C2 in a Drosophila model for C9orf72-associated ALS. Acta Neuropathol Commun, 7(1), 62. doi: 10.1186/s40478-019-0711-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KM, Glineburg MR, Kearse MG, Flores BN, Linsalata AE, Fedak SJ, … Todd PK (2017). RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nat Commun, 8(1), 2005. doi: 10.1038/s41467-017-02200-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KM, Linsalata AE, & Todd PK (2016). RAN translation-What makes it run? Brain Res, 1647, 30–42. doi: 10.1016/j.brainres.2016.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh NR, & Green R (2014). Dom34 rescues ribosomes in 3’ untranslated regions. Cell, 156(5), 950–962. doi: 10.1016/j.cell.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N, McCabe C, Medina S, Varshavsky M, Kitsberg D, Dvir-Szternfeld R, … Schwartz M (2020). Disease-associated astrocytes in Alzheimer’s disease and aging. Nat Neurosci. doi: 10.1038/s41593-020-0624-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, & Kenyon C (2007). Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell, 6(1), 95–110. doi: 10.1111/j.1474-9726.2006.00267.x [DOI] [PubMed] [Google Scholar]
- Hanson G, & Coller J (2018). Codon optimality, bias and usage in translation and mRNA decay. Nature Reviews Molecular Cell Biology, 19(1), 20–30. doi: 10.1038/nrm.2017.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JA, Hirokawa KE, Sorensen SA, Gu H, Mills M, Ng LL, … Zeng H (2014). Anatomical characterization of Cre driver mice for neural circuit mapping and manipulation. Front Neural Circuits, 8, 76. doi: 10.3389/fncir.2014.00076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, … Miller RA (2009). Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature, 460(7253), 392–395. doi: 10.1038/nature08221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Kulicke R, Fenster RJ, Greengard P, & Heintz N (2014). Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP). Nat Protoc, 9(6), 1282–1291. doi: 10.1038/nprot.2014.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, … Heintz N (2008). A translational profiling approach for the molecular characterization of CNS cell types. Cell, 135(4), 738–748. doi: 10.1016/j.cell.2008.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Movva N, & Hall M (1991). Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science, 253(5022), 905–909. doi: 10.1126/science.1715094 [DOI] [PubMed] [Google Scholar]
- Hekman KE, Yu G-Y, Brown CD, Zhu H, Du X, Gervin K, … Gomez CM (2012). A conserved eEF2 coding variant in SCA26 leads to loss of translational fidelity and increased susceptibility to proteostatic insult. Hum Mol Genet, 21(26), 5472–5483. doi: 10.1093/hmg/dds392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz MI, Landry DM, Willis AE, Luo G, & Thompson SR (2013). Ribosomal protein S25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Mol Cell Biol, 33(5), 1016–1026. doi: 10.1128/mcb.00879-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetman M, & Slomnicki LP (2019). Ribosomal biogenesis as an emerging target of neurodevelopmental pathologies. J Neurochem, 148(3), 325–347. doi: 10.1111/jnc.14576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG (2014). The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem, 83, 779–812. doi: 10.1146/annurev-biochem-060713-035802 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, & Lorsch JR (2012). The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol, 4(10). doi: 10.1101/cshperspect.a011544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, & Schuman EM (2013). The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron, 80(3), 648–657. doi: 10.1016/j.neuron.2013.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wagner-Valladolid S, Stephens AD, Jung R, Poudel C, Sinnige T, … David DC (2019). Intrinsically aggregation-prone proteins form amyloid-like aggregates and contribute to tissue aging in Caenorhabditis elegans. Elife, 8. doi: 10.7554/eLife.43059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imami K, Milek M, Bogdanow B, Yasuda T, Kastelic N, Zauber H, … Selbach M (2018). Phosphorylation of the Ribosomal Protein RPL12/uL11 Affects Translation during Mitosis. Mol Cell, 72(1), 84–98.e89. doi: 10.1016/j.molcel.2018.08.019 [DOI] [PubMed] [Google Scholar]
- Imataka H, Gradi A, & Sonenberg N (1998). A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. Embo j, 17(24), 7480–7489. doi: 10.1093/emboj/17.24.7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, … Weissman JS (2014). Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep, 8(5), 1365–1379. doi: 10.1016/j.celrep.2014.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, & Weissman JS (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science, 324(5924), 218–223. doi: 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Hussmann JA, & Weissman JS (2019). Ribosome Profiling: Global Views of Translation. Cold Spring Harb Perspect Biol, 11(5). doi: 10.1101/cshperspect.a032698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimura R, Nagy G, Dotu I, Zhou H, Yang XL, Schimmel P, … Ackerman SL (2014). RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science, 345(6195), 455–459. doi: 10.1126/science.1249749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaberi E, Rohani M, Shahidi GA, Nafissi S, Arefian E, Soleimani M, … Elahi E (2016). Identification of mutation in GTPBP2 in patients of a family with neurodegeneration accompanied by iron deposition in the brain. Neurobiol Aging, 38, 216.e211–216.e218. doi: 10.1016/j.neurobiolaging.2015.10.034 [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CUT, & Pestova TV (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nature Reviews Molecular Cell Biology, 11(2), 113–127. doi: 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens GE, Meinema AC, Gonzalez J, Wolters JC, Schmidt A, Guryev V, … Heinemann M (2015). Protein biogenesis machinery is a driver of replicative aging in yeast. Elife, 4, e08527. doi: 10.7554/eLife.08527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens GE, Meinema AC, González J, Wolters JC, Schmidt A, Guryev V, … Heinemann M (2015). Protein biogenesis machinery is a driver of replicative aging in yeast. Elife, 4, e08527–e08527. doi: 10.7554/eLife.08527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed A, Christodoulou J, Cabrita LD, & Orlova EV (2017). The ribosome and its role in protein folding: looking through a magnifying glass. Acta Crystallogr D Struct Biol, 73(Pt 6), 509–521. doi: 10.1107/s2059798317007446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings MD, & Pavitt GD (2014). A new function and complexity for protein translation initiation factor eIF2B. Cell Cycle, 13(17), 2660–2665. doi: 10.4161/15384101.2014.948797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, Steffen KK, Westman EA, Hu D, Dang N, … Kennedy BK (2005). Regulation of Yeast Replicative Life Span by TOR and Sch9 in Response to Nutrients. Science, 310(5751), 1193–1196. doi: 10.1126/science.1115535 [DOI] [PubMed] [Google Scholar]
- Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW, Thomas EL, & Kockel L (2010). With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab, 11(6), 453–465. doi: 10.1016/j.cmet.2010.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur M, & Ackerman SL (2018). mRNA Translation Gone Awry: Translation Fidelity and Neurological Disease. Trends Genet, 34(3), 218–231. doi: 10.1016/j.tig.2017.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur M, Monaghan CE, & Ackerman SL (2017). Regulation of mRNA Translation in Neurons-A Matter of Life and Death. Neuron, 96(3), 616–637. doi: 10.1016/j.neuron.2017.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z, Mallik P, Johnson AB, Luna F, Nevo E, Zhang ZD, … Gorbunova V (2017). Translation fidelity coevolves with longevity. Aging Cell, 16(5), 988–993. doi: 10.1111/acel.12628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, Green KM, Krans A, Rodriguez CM, Linsalata AE, Goldstrohm AC, & Todd PK (2016). CGG Repeat-Associated Non-AUG Translation Utilizes a Cap-Dependent Scanning Mechanism of Initiation to Produce Toxic Proteins. Mol Cell, 62(2), 314–322. doi: 10.1016/j.molcel.2016.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, & Todd PK (2014). Repeat-associated non-AUG translation and its impact in neurodegenerative disease. Neurotherapeutics, 11(4), 721–731. doi: 10.1007/s13311-014-0292-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller JN, Gee J, & Ding Q (2002). The proteasome in brain aging. Ageing Res Rev, 1(2), 279–293. doi: 10.1016/s1568-1637(01)00006-x [DOI] [PubMed] [Google Scholar]
- Kelmer Sacramento E, Kirkpatrick JM, Mazzetto M, Baumgart M, Bartolome A, Di Sanzo S, … Ori A (2020). Reduced proteasome activity in the aging brain results in ribosome stoichiometry loss and aggregation. Mol Syst Biol, 16(6), e9596. doi: 10.15252/msb.20209596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Berger SL, Brunet A, Campisi J, Cuervo AM, Epel ES, … Sierra F (2014). Geroscience: linking aging to chronic disease. Cell, 159(4), 709–713. doi: 10.1016/j.cell.2014.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, & Tabtiang R (1993). A C. elegans mutant that lives twice as long as wild type. Nature, 366(6454), 461–464. doi: 10.1038/366461a0 [DOI] [PubMed] [Google Scholar]
- Kim Y, Zheng X, Ansari Z, Bunnell MC, Herdy JR, Traxler L, … Gage FH (2018). Mitochondrial Aging Defects Emerge in Directly Reprogrammed Human Neurons due to Their Metabolic Profile. Cell Rep, 23(9), 2550–2558. doi: 10.1016/j.celrep.2018.04.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King HA, & Gerber AP (2014). Translatome profiling: methods for genome-scale analysis of mRNA translation. Briefings in Functional Genomics, 15(1), 22–31. doi: 10.1093/bfgp/elu045 [DOI] [PubMed] [Google Scholar]
- Kirkwood TB, & Austad SN (2000). Why do we age? Nature, 408(6809), 233–238. doi: 10.1038/35041682 [DOI] [PubMed] [Google Scholar]
- Klass MR (1983). A method for the isolation of longevity mutants in the nematode Caenorhabditis elegans and initial results. Mech Ageing Dev, 22(3–4), 279–286. doi: 10.1016/0047-6374(83)900829 [DOI] [PubMed] [Google Scholar]
- Knott AB, Perkins G, Schwarzenbacher R, & Bossy-Wetzel E (2008). Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci, 9(7), 505–518. doi: 10.1038/nrn2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocabas A, Duarte T, Kumar S, & Hynes MA (2015). Widespread Differential Expression of Coding Region and 3’ UTR Sequences in Neurons and Other Tissues. Neuron, 88(6), 1149–1156. doi: 10.1016/j.neuron.2015.10.048 [DOI] [PubMed] [Google Scholar]
- Kochetov AV, Palyanov A, Titov II, Grigorovich D, Sarai A, & Kolchanov NA (2007). AUG_hairpin: prediction of a downstream secondary structure influencing the recognition of a translation start site. BMC Bioinformatics, 8(1), 318. doi: 10.1186/1471-2105-8-318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontis V, Bennett JE, Mathers CD, Li G, Foreman K, & Ezzati M (2017). Future life expectancy in 35 industrialised countries: projections with a Bayesian model ensemble. Lancet, 389(10076), 13231335. doi: 10.1016/s0140-6736(16)32381-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis N, & Tavernarakis N (2011). Cellular stress response pathways and ageing: intricate molecular relationships. Embo j, 30(13), 2520–2531. doi: 10.1038/emboj.2011.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (1984). Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature, 308(5956), 241–246. doi: 10.1038/308241a0 [DOI] [PubMed] [Google Scholar]
- Kozak M (1990). Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci U S A, 87(21), 8301–8305. doi: 10.1073/pnas.87.21.8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landfield PW (1988). Hippocampal neurobiological mechanisms of age-related memory dysfunction. Neurobiol Aging, 9(5–6), 571–579. doi: 10.1016/s0197-4580(88)80116-7 [DOI] [PubMed] [Google Scholar]
- Lastick SM, & McConkey EH (1976). Exchange and stability of HeLa ribosomal proteins in vivo. J Biol Chem, 251(10), 2867–2875. [PubMed] [Google Scholar]
- Lee AS, Burdeinick-Kerr R, & Whelan SP (2013). A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc Natl Acad Sci U S A, 110(1), 324–329. doi: 10.1073/pnas.1216454109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-K, Weindruch R, & Prolla TA (2000). Gene-expression profile of the ageing brain in mice. Nature Genetics, 25(3), 294–297. doi: 10.1038/77046 [DOI] [PubMed] [Google Scholar]
- Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, … Ackerman SL (2006). Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature, 443(7107), 50–55. doi: 10.1038/nature05096 [DOI] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang S-X, Shen B, & Qian S-B (2012). Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci U S A, 109(37), E2424E2432. doi: 10.1073/pnas.1207846109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Alafuzoff I, Soininen H, Winblad B, & Pei JJ (2005). Levels of mTOR and its downstream targets 4E-BP1, eEF2, and eEF2 kinase in relationships with tau in Alzheimer’s disease brain. Febs j, 272(16), 4211–4220. doi: 10.1111/j.1742-4658.2005.04833.x [DOI] [PubMed] [Google Scholar]
- Li Y, Li X, Cao M, Jiang Y, Yan J, Liu Z, … Shi Y (2019). Seryl tRNA synthetase cooperates with POT1 to regulate telomere length and cellular senescence. Signal Transduct Target Ther, 4, 50. doi: 10.1038/s41392-019-0078-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Masoro EJ, Nelson JF, Strong R, McMahan CA, & Richardson A (2003). Genetic mouse models of extended lifespan. Exp Gerontol, 38(11–12), 1353–1364. doi: 10.1016/j.exger.2003.10.019 [DOI] [PubMed] [Google Scholar]
- Liang S, Bellato HM, Lorent J, Lupinacci FCS, Oertlin C, van Hoef V, … Larsson O (2018). Polysome-profiling in small tissue samples. Nucleic Acids Res, 46(1), e3. doi: 10.1093/nar/gkx940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightowlers RN, Rozanska A, & Chrzanowska-Lightowlers ZM (2014). Mitochondrial protein synthesis: figuring the fundamentals, complexities and complications, of mammalian mitochondrial translation. FEBS Lett, 588(15), 2496–2503. doi: 10.1016/j.febslet.2014.05.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C, & Ermolenko DN (2016). Structural insights into ribosome translocation. Wiley Interdiscip Rev RNA, 7(5), 620–636. doi: 10.1002/wrna.1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsalata AE, He F, Malik AM, Glineburg MR, Green KM, Natla S, … Todd PK (2019). DDX3X and specific initiation factors modulate FMR1 repeat-associated non-AUG-initiated translation. EMBO Rep, 20(9), e47498. doi: 10.15252/embr.201847498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, & Proud CG (2016). Eukaryotic elongation factor 2 kinase as a drug target in cancer, and in cardiovascular and neurodegenerative diseases. Acta Pharmacol Sin, 37(3), 285–294. doi: 10.1038/aps.2015.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD, Antebi A, Bartke A, Barzilai N, Brown-Borg HM, Caruso C, … Fontana L (2015). Interventions to Slow Aging in Humans: Are We Ready? Aging Cell, 14(4), 497–510. doi: 10.1111/acel.12338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, & Kroemer G (2013). The hallmarks of aging. Cell, 153(6), 1194–1217. doi: 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe R, Shirley N, Bleackley M, Dolan S, & Shafee T (2017). Transcriptomics technologies. PLoS computational biology, 13(5), e1005457–e1005457. doi: 10.1371/journal.pcbi.1005457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch G, Rex CS, & Gall CM (2006). Synaptic plasticity in early aging. Ageing Res Rev, 5(3), 255–280. doi: 10.1016/j.arr.2006.03.008 [DOI] [PubMed] [Google Scholar]
- MacInnes AW (2016). The role of the ribosome in the regulation of longevity and lifespan extension. Wiley Interdiscip Rev RNA, 7(2), 198–212. doi: 10.1002/wrna.1325 [DOI] [PubMed] [Google Scholar]
- Mandad S, Rahman R-U, Centeno TP, Vidal RO, Wildhagen H, Rammner B, … Fornasiero EF (2018). The codon sequences predict protein lifetimes and other parameters of the protein life cycle in the mouse brain. Sci Rep, 8(1), 16913. doi: 10.1038/s41598-018-35277-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marner L, Nyengaard JR, Tang Y, & Pakkenberg B (2003). Marked loss of myelinated nerve fibers in the human brain with age. J Comp Neurol, 462(2), 144–152. doi: 10.1002/cne.10714 [DOI] [PubMed] [Google Scholar]
- Martinez-Lopez N, Athonvarangkul D, & Singh R (2015). Autophagy and aging. Adv Exp Med Biol, 847, 73–87. doi: 10.1007/978-1-4939-2404-2_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masek T, Valasek L, & Pospisek M (2011). Polysome analysis and RNA purification from sucrose gradients. Methods Mol Biol, 703, 293–309. doi: 10.1007/978-1-59745-248-9_20 [DOI] [PubMed] [Google Scholar]
- Mateyak MK, & Kinzy TG (2010). eEF1A: thinking outside the ribosome. J Biol Chem, 285(28), 2120921213. doi: 10.1074/jbc.R110.113795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, & Magnus T (2006). Ageing and neuronal vulnerability. Nat Rev Neurosci, 7(4), 278–294. doi: 10.1038/nrn1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Schuman R, & Antonellis A (2017). Emerging mechanisms of aminoacyl-tRNA synthetase mutations in recessive and dominant human disease. Hum Mol Genet, 26(R2), R114–r127. doi: 10.1093/hmg/ddx231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RA, Harrison DE, Astle CM, Baur JA, Boyd AR, de Cabo R, … Strong R (2011). Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci, 66(2), 191–201. doi: 10.1093/gerona/glq178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, & Noller HF (1989). Intermediate states in the movement of transfer RNA in the ribosome. Nature, 342(6246), 142–148. doi: 10.1038/342142a0 [DOI] [PubMed] [Google Scholar]
- Moghal A, Mohler K, & Ibba M (2014). Mistranslation of the genetic code. FEBS Lett, 588(23), 43054310. doi: 10.1016/j.febslet.2014.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanaro L, Trere D, & Derenzini M (2008). Nucleolus, ribosomes, and cancer. Am J Pathol, 173(2), 301–310. doi: 10.2353/ajpath.2008.070752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SL, & Parker R (2018). EIF2B2 mutations in vanishing white matter disease hypersuppress translation and delay recovery during the integrated stress response. Rna, 24(6), 841–852. doi: 10.1261/rna.066563.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SL, Sonenberg N, & Parker R (2018). Neuronal Regulation of eIF2alpha Function in Health and Neurological Disorders. Trends Mol Med, 24(6), 575–589. doi: 10.1016/j.molmed.2018.04.001 [DOI] [PubMed] [Google Scholar]
- Mori K, Weng S-M, Arzberger T, May S, Rentzsch K, Kremmer E, … Edbauer D (2013). The C9orf72 GGGGCC Repeat Is Translated into Aggregating Dipeptide-Repeat Proteins in FTLD/ALS. Science, 339(6125), 1335–1338. doi: 10.1126/science.1232927 [DOI] [PubMed] [Google Scholar]
- Morimoto RI (1998). Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev, 12(24), 37883796. doi: 10.1101/gad.12.24.3788 [DOI] [PubMed] [Google Scholar]
- Moronetti Mazzeo LE, Dersh D, Boccitto M, Kalb RG, & Lamitina T (2012). Stress and aging induce distinct polyQ protein aggregation states. Proc Natl Acad Sci U S A, 109(26), 10587–10592. doi: 10.1073/pnas.1108766109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandagopal N, & Roux PP (2015). Regulation of global and specific mRNA translation by the mTOR signaling pathway. Translation (Austin), 3(1), e983402. doi: 10.4161/21690731.2014.983402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzer N, Goodenbour JM, David A, Dittmar KA, Jones RB, Schneider JR, … Pan T (2009). Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature, 462(7272), 522–526. doi: 10.1038/nature08576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell AE, Gerashchenko MV, O’Donohue MF, Rosen SM, Huntzinger E, Gleeson D, … Agrawal PB (2019). Mammalian Hbs1L deficiency causes congenital anomalies and developmental delay associated with Pelota depletion and 80S monosome accumulation. PLoS Genet, 15(2), e1007917. doi: 10.1371/journal.pgen.1007917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olah M, Patrick E, Villani A-C, Xu J, White CC, Ryan KJ, … Bradshaw EM (2018). A transcriptomic atlas of aged human microglia. Nat Commun, 9(1), 539. doi: 10.1038/s41467-01802926-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshansky SJ, Goldman DP, Zheng Y, & Rowe JW (2009). Aging in America in the twenty-first century: demographic forecasts from the MacArthur Foundation Research Network on an Aging Society. Milbank Q, 87(4), 842–862. doi: 10.1111/j.1468-0009.2009.00581.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori A, Toyama BH, Harris MS, Bock T, Iskar M, Bork P, … Beck M (2015). Integrated Transcriptome and Proteome Analyses Reveal Organ-Specific Proteome Deterioration in Old Rats. Cell Syst, 1(3), 224–237. doi: 10.1016/j.cels.2015.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega JA, Daley EL, Kour S, Samani M, Tellez L, Smith HS, … Kiskinis E (2020). Nucleocytoplasmic Proteomic Analysis Uncovers eRF1 and Nonsense-Mediated Decay as Modifiers of ALS/FTD C9orf72 Toxicity. Neuron, 106(1), 90–107.e113. doi: 10.1016/j.neuron.2020.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AL, & Ousman SS (2018). Astrocytes and Aging. Front Aging Neurosci, 10, 337. doi: 10.3389/fnagi.2018.00337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, & Kapahi P (2007). Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell, 6(1), 111–119. doi: 10.1111/j.1474-9726.2006.00266.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolini NA, Attwood M, Sondalle SB, Vieira C, van Adrichem AM, di Summa FM, … MacInnes AW (2017). A Ribosomopathy Reveals Decoding Defective Ribosomes Driving Human Dysmorphism. Am J Hum Genet, 100(3), 506–522. doi: 10.1016/j.ajhg.2017.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlato R, & Kreiner G (2013). Nucleolar activity in neurodegenerative diseases: a missing piece of the puzzle? J Mol Med (Berl), 91(5), 541–547. doi: 10.1007/s00109-012-0981-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, … Ramakrishnan V (2007). The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell, 26(1), 41–50. doi: 10.1016/j.molcel.2007.03.018 [DOI] [PubMed] [Google Scholar]
- Pelvig DP, Pakkenberg H, Stark AK, & Pakkenberg B (2008). Neocortical glial cell numbers in human brains. Neurobiol Aging, 29(11), 1754–1762. doi: 10.1016/j.neurobiolaging.2007.04.013 [DOI] [PubMed] [Google Scholar]
- Pestova TV, & Kolupaeva VG (2002). The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev, 16(22), 2906–2922. doi: 10.1101/gad.1020902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, & Hellen CU (2000). The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature, 403(6767), 332–335. doi: 10.1038/35002118 [DOI] [PubMed] [Google Scholar]
- Pietrzak M, Rempala G, Nelson PT, Zheng JJ, & Hetman M (2011). Epigenetic silencing of nucleolar rRNA genes in Alzheimer’s disease. PLoS One, 6(7), e22585. doi: 10.1371/journal.pone.0022585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon HF, Shepherd HM, Reed TT, Calabrese V, Stella AM, Pennisi G, … Butterfield DA (2006). Proteomics analysis provides insight into caloric restriction mediated oxidation and expression of brain proteins associated with age-related impaired cellular processes: Mitochondrial dysfunction, glutamate dysregulation and impaired protein synthesis. Neurobiol Aging, 27(7), 1020–1034. doi: 10.1016/j.neurobiolaging.2005.05.014 [DOI] [PubMed] [Google Scholar]
- Rattan SI (1996). Synthesis, modifications, and turnover of proteins during aging. Exp Gerontol, 31(1–2), 33–47. doi: 10.1016/0531-5565(95)02022-5 [DOI] [PubMed] [Google Scholar]
- Rauniyar N (2015). Parallel Reaction Monitoring: A Targeted Experiment Performed Using High Resolution and High Mass Accuracy Mass Spectrometry. Int J Mol Sci, 16(12), 28566–28581. doi: 10.3390/ijms161226120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renoux AJ, & Todd PK (2012). Neurodegeneration the RNA way. Prog Neurobiol, 97(2), 173–189. doi: 10.1016/j.pneurobio.2011.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez CM, & Todd PK (2019). New pathologic mechanisms in nucleotide repeat expansion disorders. Neurobiol Dis, 130, 104515. doi: 10.1016/j.nbd.2019.104515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers AN, Chen D, McColl G, Czerwieniec G, Felkey K, Gibson BW, … Kapahi P (2011). Life span extension via eIF4G inhibition is mediated by posttranscriptional remodeling of stress response gene expression in C. elegans. Cell Metab, 14(1), 55–66. doi: 10.1016/j.cmet.2011.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohilla KJ, & Gagnon KT (2017). RNA biology of disease-associated microsatellite repeat expansions. Acta Neuropathol Commun, 5(1), 63. doi: 10.1186/s40478-017-0468-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB, Sarnow P, & Hentze MW (1997). Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell, 89(6), 831–838. doi: 10.1016/s0092-8674(00)80268-8 [DOI] [PubMed] [Google Scholar]
- Saez I, & Vilchez D (2014). The Mechanistic Links Between Proteasome Activity, Aging and Age-related Diseases. Curr Genomics, 15(1), 38–51. doi: 10.2174/138920291501140306113344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakers K, Lake AM, Khazanchi R, Ouwenga R, Vasek MJ, Dani A, & Dougherty JD (2017). Astrocytes locally translate transcripts in their peripheral processes. Proc Natl Acad Sci U S A, 114(19), E3830–e3838. doi: 10.1073/pnas.1617782114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samir P, Browne CM, Rahul, Sun M, Shen B, Li W, … Link AJ (2018). Identification of Changing Ribosome Protein Compositions using Mass Spectrometry. Proteomics, 18(20), e1800217. doi: 10.1002/pmic.201800217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Bean JC, Carey DP, Quintana A, & McKnight GS (2019). RiboTag: Ribosomal Tagging Strategy to Analyze Cell-Type-Specific mRNA Expression In Vivo. Curr Protoc Neurosci, 88(1), e77. doi: 10.1002/cpns.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimanski LA, & Barnes CA (2010). Neural Protein Synthesis during Aging: Effects on Plasticity and Memory. Front Aging Neurosci, 2. doi: 10.3389/fnagi.2010.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, & Pierre P (2009). SUnSET, a nonradioactive method to monitor protein synthesis. Nature methods, 6(4), 275–277. doi: 10.1038/nmeth.1314 [DOI] [PubMed] [Google Scholar]
- Schuller AP, Wu CC, Dever TE, Buskirk AR, & Green R (2017). eIF5A Functions Globally in Translation Elongation and Termination. Mol Cell, 66(2), 194–205.e195. doi: 10.1016/j.molcel.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte EC, Ellwanger DC, Dihanich S, Manzoni C, Stangl K, Schormair B, … Winkelmann J (2014). Rare variants in LRRK1 and Parkinson’s disease. Neurogenetics, 15(1), 49–57. doi: 10.1007/s10048-013-0383-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian D, Palacin M, & Zorzano A (2017). Mitochondrial Dynamics: Coupling Mitochondrial Fitness with Healthy Aging. Trends Mol Med, 23(3), 201–215. doi: 10.1016/j.molmed.2017.01.003 [DOI] [PubMed] [Google Scholar]
- Seim I, Ma S, Zhou X, Gerashchenko MV, Lee SG, Suydam R, … Gladyshev VN (2014). The transcriptome of the bowhead whale Balaena mysticetus reveals adaptations of the longest-lived mammal. Aging (Albany NY), 6(10), 879–899. doi: 10.18632/aging.100699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Smith HJ, Yao P, & Mair WB (2019). Causal roles of mitochondrial dynamics in longevity and healthy aging. EMBO Rep, 20(12), e48395. doi: 10.15252/embr.201948395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Fujii K, Kovary KM, Genuth NR, Röst HL, Teruel MN, & Barna M (2017). Heterogeneous Ribosomes Preferentially Translate Distinct Subpools of mRNAs Genome-wide. Mol Cell, 67(1), 71–83.e77. doi: 10.1016/j.molcel.2017.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka T, Koppers M, Wong HH, Lin JQ, Cagnetta R, Dwivedy A, … Holt CE (2019). On-Site Ribosome Remodeling by Locally Synthesized Ribosomal Proteins in Axons. Cell Rep, 29(11), 3605–3619.e3610. doi: 10.1016/j.celrep.2019.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Hellen CU, & Pestova TV (2013). Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol Cell, 51(2), 249–264. doi: 10.1016/j.molcel.2013.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov N, Semrau S, Airoldi E, Budnik B, & van Oudenaarden A (2015). Differential Stoichiometry among Core Ribosomal Proteins. Cell Rep, 13(5), 865–873. doi: 10.1016/j.celrep.2015.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ED, Tsuchiya M, Fox LA, Dang N, Hu D, Kerr EO, … Kennedy BK (2008). Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res, 18(4), 564–570. doi: 10.1101/gr.074724.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Guo S, Fu N, Yan Z, Hu HY, Xu Y, … Khaitovich P (2010). MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res, 20(9), 12071218. doi: 10.1101/gr.106849.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonobe Y, Ghadge G, Masaki K, Sendoel A, Fuchs E, & Roos RP (2018). Translation of dipeptide repeat proteins from the C9ORF72 expanded repeat is associated with cellular stress. Neurobiol Dis, 116, 155–165. doi: 10.1016/j.nbd.2018.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soreq L, Rose J, Soreq E, Hardy J, Trabzuni D, Cookson MR, … Ule J (2017). Major Shifts in Glial Regional Identity Are a Transcriptional Hallmark of Human Brain Aging. Cell Rep, 18(2), 557–570. doi: 10.1016/j.celrep.2016.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sossin WS, & Costa-Mattioli M (2019). Translational Control in the Brain in Health and Disease. Cold Spring Harb Perspect Biol, 11(8). doi: 10.1101/cshperspect.a032912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz JA (1969). Nucleotide sequences of the ribosomal binding sites of bacteriophage R17 RNA. Cold Spring Harb Symp Quant Biol, 34, 621–630. doi: 10.1101/sqb.1969.034.01.072 [DOI] [PubMed] [Google Scholar]
- Sudmant PH, Lee H, Dominguez D, Heiman M, & Burge CB (2018). Widespread Accumulation of Ribosome-Associated Isolated 3’ UTRs in Neuronal Cell Populations of the Aging Brain. Cell Rep, 25(9), 2447–2456.e2444. doi: 10.1016/j.celrep.2018.10.094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhm T, Kaimal JM, Dawitz H, Peselj C, Masser AE, Hanzen S, … Ott M (2018). Mitochondrial Translation Efficiency Controls Cytoplasmic Protein Homeostasis. Cell Metab, 27(6), 13091322.e1306. doi: 10.1016/j.cmet.2018.04.011 [DOI] [PubMed] [Google Scholar]
- Syntichaki P, Troulinaki K, & Tavernarakis N (2007). eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature, 445(7130), 922–926. doi: 10.1038/nature05603 [DOI] [PubMed] [Google Scholar]
- Tabet R, Schaeffer L, Freyermuth F, Jambeau M, Workman M, Lee CZ, … Lagier-Tourenne C (2018). CUG initiation and frameshifting enable production of dipeptide repeat proteins from ALS/FTD C9ORF72 transcripts. Nat Commun, 9(1), 152. doi: 10.1038/s41467-017-02643-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai D, Inoue K, Shisa H, Kagawa Y, & Hayashi J (1995). Age-associated changes of mitochondrial translation and respiratory function in mouse brain. Biochem Biophys Res Commun, 217(2), 668674. doi: 10.1006/bbrc.1995.2826 [DOI] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, … Huang ZJ (2011). A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron, 71(6), 995–1013. doi: 10.1016/j.neuron.2011.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavernarakis N (2008). Ageing and the regulation of protein synthesis: a balancing act? Trends Cell Biol, 18(5), 228–235. doi: 10.1016/j.tcb.2008.02.004 [DOI] [PubMed] [Google Scholar]
- Taylor RC, & Dillin A (2011). Aging as an event of proteostasis collapse. Cold Spring Harb Perspect Biol, 3(5). doi: 10.1101/cshperspect.a004440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, … Hamon C (2003). Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem, 75(8), 1895–1904. doi: 10.1021/ac0262560 [DOI] [PubMed] [Google Scholar]
- Thompson AC, Bruss MD, Price JC, Khambatta CF, Holmes WE, Colangelo M, … Hellerstein MK (2016). Reduced in vivo hepatic proteome replacement rates but not cell proliferation rates predict maximum lifespan extension in mice. Aging Cell, 15(1), 118–127. doi: 10.1111/acel.12414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiku V, & Antebi A (2018). Nucleolar Function in Lifespan Regulation. Trends Cell Biol, 28(8), 662–672. doi: 10.1016/j.tcb.2018.03.007 [DOI] [PubMed] [Google Scholar]
- Tiku V, Jain C, Raz Y, Nakamura S, Heestand B, Liu W, … Antebi A (2017). Small nucleoli are a cellular hallmark of longevity. Nat Commun, 8, 16083. doi: 10.1038/ncomms16083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA (2015). Using C. elegans for aging research. Invertebr Reprod Dev, 59(sup1), 59–63. doi: 10.1080/07924259.2014.940470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, & Paulson HL (2010). RNA-mediated neurodegeneration in repeat expansion disorders. Ann Neurol, 67(3), 291–300. doi: 10.1002/ana.21948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi H, & Chan HY (2014). Roles of the nucleolus in the CAG RNA-mediated toxicity. Biochim Biophys Acta, 1842(6), 779–784. doi: 10.1016/j.bbadis.2013.11.015 [DOI] [PubMed] [Google Scholar]
- Tsoi H, Lau TC, Tsang SY, Lau KF, & Chan HY (2012). CAG expansion induces nucleolar stress in polyglutamine diseases. Proc Natl Acad Sci U S A, 109(33), 13428–13433. doi: 10.1073/pnas.1204089109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valles SL, Iradi A, Aldasoro M, Vila JM, Aldasoro C, de la Torre J, … Jorda A (2019). Function of Glia in Aging and the Brain Diseases. Int J Med Sci, 16(11), 1473–1479. doi: 10.7150/ijms.37769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heemst D (2010). Insulin, IGF-1 and longevity. Aging Dis, 1(2), 147–157. [PMC free article] [PubMed] [Google Scholar]
- Vera M, Pani B, Griffiths LA, Muchardt C, Abbott CM, Singer RH, & Nudler E (2014). The translation elongation factor eEF1A1 couples transcription to translation during heat shock response. Elife, 3, e03164. doi: 10.7554/eLife.03164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC (2005). A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet, 39, 359–407. doi: 10.1146/annurev.genet.39.110304.095751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther DM, Kasturi P, Zheng M, Pinkert S, Vecchi G, Ciryam P, … Hartl FU (2015). Widespread Proteome Remodeling and Aggregation in Aging C. elegans. Cell, 161(4), 919–932. doi: 10.1016/j.cell.2015.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Johnson AG, Lapointe CP, Choi J, Prabhakar A, Chen DH, … Puglisi JD (2019). eIF5B gates the transition from translation initiation to elongation. Nature, 573(7775), 605–608. doi: 10.1038/s41586-019-1561-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb BL, & Proud CG (1997). Eukaryotic initiation factor 2B (eIF2B). Int J Biochem Cell Biol, 29(10), 1127–1131. doi: 10.1016/s1357-2725(97)00039-3 [DOI] [PubMed] [Google Scholar]
- Wei YN, Hu HY, Xie GC, Fu N, Ning ZB, Zeng R, & Khaitovich P (2015). Transcript and protein expression decoupling reveals RNA binding proteins and miRNAs as potential modulators of human aging. Genome Biol, 16, 41. doi: 10.1186/s13059-015-0608-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg DE, Shah P, Eichhorn SW, Hussmann JA, Plotkin JB, & Bartel DP (2016). Improved Ribosome-Footprint and mRNA Measurements Provide Insights into Dynamics and Regulation of Yeast Translation. Cell Rep, 14(7), 1787–1799. doi: 10.1016/j.celrep.2016.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergard T, McAvoy K, Russell K, Wen X, Pang Y, Morris B, … Haeusler A (2019). Repeat-associated non-AUG translation in C9orf72-ALS/FTD is driven by neuronal excitation and stress. EMBO Mol Med, 11(2). doi: 10.15252/emmm.201809423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KM, Muralidharan B, & Isaacs AM (2019). Relax, Don’t RAN Translate It. Neuron, 104(5), 827829. doi: 10.1016/j.neuron.2019.11.014 [DOI] [PubMed] [Google Scholar]
- Wiltrout E, Goodenbour JM, Frechin M, & Pan T (2012). Misacylation of tRNA with methionine in Saccharomyces cerevisiae. Nucleic Acids Res, 40(20), 10494–10506. doi: 10.1093/nar/gks805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin B, & Ivanov P (2019). Stress granules and neurodegeneration. Nat Rev Neurosci, 20(11), 649666. doi: 10.1038/s41583-019-0222-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Medina SG, Kushawah G, DeVore ML, Castellano LA, Hand JM, … Bazzini AA (2019). Translation affects mRNA stability in a codon-dependent manner in human cells. Elife, 8. doi: 10.7554/eLife.45396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Coray T (2016). Ageing, neurodegeneration and brain rejuvenation. Nature, 539(7628), 180–186. doi: 10.1038/nature20411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, de Souza Alves V, von der Haar T, O’Keefe L, Lenchine RV, Jensen KB, … Proud CG (2019). Regulation of the Elongation Phase of Protein Synthesis Enhances Translation Accuracy and Modulates Lifespan. Curr Biol, 29(5), 737–749.e735. doi: 10.1016/j.cub.2019.01.029 [DOI] [PubMed] [Google Scholar]
- Ximerakis M, Lipnick SL, Innes BT, Simmons SK, Adiconis X, Dionne D, … Rubin LL (2019). Single-cell transcriptomic profiling of the aging mouse brain. Nat Neurosci, 22(10), 1696–1708. doi: 10.1038/s41593-019-0491-3 [DOI] [PubMed] [Google Scholar]
- Xu X, Shi Y, Zhang HM, Swindell EC, Marshall AG, Guo M, … Yang XL (2012). Unique domain appended to vertebrate tRNA synthetase is essential for vascular development. Nat Commun, 3, 681. doi: 10.1038/ncomms1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada SB, Gendron TF, Niccoli T, Genuth NR, Grosely R, Shi Y, … Gitler AD (2019). RPS25 is required for efficient RAN translation of C9orf72 and other neurodegenerative disease-associated nucleotide repeats. Nat Neurosci, 22(9), 1383–1388. doi: 10.1038/s41593-019-0455-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankner BA, Lu T, & Loerch P (2008). The aging brain. Annu Rev Pathol, 3, 41–66. doi: 10.1146/annurev.pathmechdis.2.010506.092044 [DOI] [PubMed] [Google Scholar]
- Zahn JM, Poosala S, Owen AB, Ingram DK, Lustig A, Carter A, … Becker KG (2007). AGEMAP: a gene expression database for aging in mice. PLoS Genet, 3(11), e201. doi: 10.1371/journal.pgen.0030201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, … Ranum LP (2011). NonATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A, 108(1), 260–265. doi: 10.1073/pnas.1013343108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, … Ranum LP (2013). RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A, 110(51), E4968–4977. doi: 10.1073/pnas.1315438110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Pattamatta A, & Ranum LPW (2018). Repeat-Associated Non-ATG Translation in Neurological Diseases. Cold Spring Harb Perspect Biol, 10(12). doi: 10.1101/cshperspect.a033019 [DOI] [PMC free article] [PubMed] [Google Scholar]


