Abstract
Campylobacter jejuni is recognised as the leading cause of bacterial gastroenteritis in industrialised countries. Although the majority of Campylobacter infections are self-limiting, antimicrobial treatment is necessary in severe cases. Therefore, the development of antimicrobial resistance (AMR) in Campylobacter is a growing public health challenge and surveillance of AMR is important for bacterial disease control. The aim of this study was to predict antimicrobial resistance in C. jejuni from whole-genome sequencing data. A total of 516 clinical C. jejuni isolates collected between 2014 and 2017 were subjected to WGS. Resistance phenotypes were determined by standard broth dilution, categorising isolates as either susceptible or resistant based on epidemiological cutoffs for six antimicrobials: ciprofloxacin, nalidixic acid, erythromycin, gentamicin, streptomycin, and tetracycline. Resistance genotypes were identified using an in-house database containing reference genes with known point mutations and the presence of resistance genes was determined using the ResFinder database and four bioinformatical methods (modified KMA, ABRicate, ARIBA, and ResFinder Batch Upload). We identified seven resistance genes including tet(O), tet(O/32/O), ant(6)-Ia, aph(2″)-If, blaOXA, aph(3′)-III, and cat as well as mutations in three genes: gyrA, 23S rRNA, and rpsL. There was a high correlation between phenotypic resistance and the presence of known resistance genes and/or point mutations. A correlation above 98% was seen for all antimicrobials except streptomycin with a correlation of 92%. In conclusion, we found that WGS can predict antimicrobial resistance with a high degree of accuracy and have the potential to be a powerful tool for AMR surveillance.
Electronic supplementary material
The online version of this article (10.1007/s10096-020-04043-y) contains supplementary material, which is available to authorized users.
Keywords: Campylobacter jejuni, Antimicrobial resistance, Whole-genome sequencing, Public health, AMR surveillance
Introduction
Campylobacter is a foodborne bacterial pathogen, which is able to cause gastroenteritis in humans and is found in the environment where it is widely distributed in the gastrointestinal tract of most warm-blooded animals [1]. Foodborne transmission is believed to be the main route of human infection [2, 3]. In 2018, Campylobacter continued to be the most common foodborne pathogen causing infections in Denmark with more than 4500 registered cases; most of these were caused by C. jejuni (85–95%) [4]. Clinical symptoms during infection often include diarrhoea, fever, abdominal pain, nausea, headache, and/or vomiting [5, 6] and in some cases result in Guillain-Barré syndrome (GBS) or reactive arthritis [7, 8]. Most patients recover without any antimicrobial treatment, whereas more severe and immunocompromised cases may need antibiotic treatment. Due to the extensive use of antimicrobials worldwide, within the production of food animals and human medicine, antimicrobial resistance (AMR) against clinically relevant antibiotics is emerging in Campylobacter spp. presenting a serious health concern [2, 9, 10]. In Denmark, AMR surveillance in Campylobacter jejuni has been in place since 1996 based on phenotypic susceptibility testing against ciprofloxacin (CIP), nalidixic acid (NAL), erythromycin (ERY), gentamicin (GEN), streptomycin (STR), and tetracycline (TET). The data on clinical isolates is reported as part of The Danish Integrated Antimicrobial Resistance Monitoring and Research Programme (DANMAP) (https://www.danmap.org/).
Campylobacter has developed several mechanisms for antibiotic resistance including point mutations, acquisition of resistance genes, and efflux systems [11–13]. Resistance towards fluoroquinolones (FQs) in Campylobacter is mainly caused by point mutations in the DNA gyrase A (GyrA) region at multiple positions [10, 14]. The mechanisms involved in erythromycin resistance include ribosomal target modification and efflux of the antibiotic from the cell and this resistance is often associated with point mutation in domain V of the 23S rRNA, which is the target of macrolides, or the ribosomal proteins L4 and L22 [10, 15]. Campylobacter harbour three copies of the 23S rRNA gene, but only two copies need to be affected by point mutations to result in erythromycin resistance [16, 17]. Streptomycin resistance in Campylobacter is associated with substitution within the ribosomal RpsL protein or by expression of an aminoglycoside-modifying enzyme (ANT(6)-I) [18, 19]. Resistance towards gentamicin and tetracycline is in general conferred by the acquisition of resistance-associated genes. Resistance to tetracycline in Campylobacter is mainly conferred by the ribosomal protection protein tet(O), a plasmid-encoded gene, whereas the resistance gene aph(2″)-If often is associated with gentamicin resistance [20–22].
The whole-genome sequencing (WGS) technology provides complete genomic DNA sequence of a bacterium, and WGS has proved to be an effective surveillance tool for foodborne pathogens [23, 24]. This method provides a large amount of information generated from a single assay [25] and allows multiple tests to be performed in silico simultaneously, whereas phenotypic susceptibility tests can be time-consuming, labour intensive, and are highly dependent on standardised laboratory procedures. Several studies have demonstrated that WGS can be used to identify resistant genotypes for different foodborne pathogens [23, 24, 26]. The aim of this study was to evaluate the concordance between WGS-based AMR prediction and the phenotypes identified from conventional antimicrobial susceptibility in a set of clinical C. jejuni isolates. The focus of the evaluation is to allow the replacement of phenotypic testing with a WGS-based surveillance of AMR in Campylobacter.
Materials and methods
Bacterial isolates
Clinical C. jejuni isolates representing common and less common resistance profiles were included as the study population. In total, 516 clinical C. jejuni isolates from Danish patients were analysed. The isolates were submitted to SSI from 2014 to 2017. Of the submitted isolates, 422 were from domestically acquired infections, 84 were travel related, and 10 isolates had no available travel information. At SSI, the isolates were subjected to MALDI-TOFF for species confirmation, and antimicrobial susceptibility was tested as described below.
Antimicrobial susceptibility testing
Isolates were grown on modified charcoal cefoperazone deoxycholate agar (mCCDA) plates. After overnight incubation, one colony was selected and inoculated on blood agar plates. This culture was used for AMR testing and DNA purification for WGS. Incubation was performed in 18 to 24 h under microaerophilic conditions (85% nitrogen, 10% carbon dioxide, 5% oxygen) at 41 °C.
Minimum inhibitory concentration (MIC) values were determined against six antimicrobials: CIP, NAL, ERY, GEN, STR, and TET using the Trek Sensititre Broth Microdilution setup using the EUCAMP plate design (ThermoFisher Scientific) that includes the test ranges recommended by ECDC [27]. MIC values were interpreted as susceptible (wild-type) or resistant (non-wild-type) using epidemiological cutoff (ECOFF) defined by EUCAST: CIP (> 0.5 μg/ml), NAL (> 16 μg/ml), ERY (> 4 μg/ml), GEN (> 2 μg/ml), STR (> 4 μg/ml), and TET (> 1 μg/ml) [27].
Whole-genome sequencing
Genomic DNA was purified from pure cultures using QIAGEN DNeasy Blood & Tissue Kit (https://www.qiagen.com). The DNA concentration was measured using an Invitrogen Qubit Fluorometer (ThermoFisher Scientific). After adjustment of the concentration, the isolates were sequenced using Illumina sequencing technology (https://www.illumina.com) on either Miseq or Nextseq sequencing machines using the Nextera XT Library preparation protocol for paired-end reads of 150 bp or 250 bp. The sequence data was evaluated by an in-house quality-control pipeline (https://github.com/ssi-dk/bifrost) to check for contaminations (maximum 5% of other genus allowed), correct species identification, and read depth and genome size. Sequences representing C. jejuni isolates with a genome size outside the range of 1.6–1.9 mbp were excluded as well as if the number of contigs exceeded 500. Coverage depth of the included genomes ranged from 32 to 506 (median 111). Data analysis was performed in BioNumerics version 7.6 (Applied Maths) where 7 locus MLST was assigned and phylogenetic minimum spanning trees were generated.
AMR prediction
The presence of acquired resistance genes was determined by four approaches: two based on prediction from methods using de novo assemblies (ResFinder Batch Upload (web tool) [28] and ARIBA [29]) and two methods using reference mapping (KMA [30] and ABRicate, available at https://github.com/tseemann/abricate). All methods utilised the ResFinder gene database for detection and for all four approaches, hits were only considered for gene lengths ≥ 90% and identities of ≥ 60%. When alleles were not represented in the database (i.e., novel alleles of a present resistance gene), reference mapping by the use of KMA splits the mapping between two or more references in the database. To alleviate this in our setup, an additional step was added to perform another mapping against the reference gene with the highest coverage in order to obtain a result with a full-length gene variant (modified KMA; https://github.com/ssi-dk/modified_kma).
For detection of point mutations conferring antimicrobial resistance, a database containing reference genes (gyrA, rplD, rplV, rpsL, and 23S) and associated point mutations known from the literature to be coupled with antimicrobial resistance in C. jejuni was constructed from C. jejuni strain NCTC 11168 (Table 1). Point mutations were detected using a local wrapper (https://github.com/ssi-dk/punktreskma) for KMA [30]. For 23S rRNA, where multiple gene copies (3) are present in the genome, the detection of a certain nucleotide at a certain position was done by considering the particular depth for each nucleotide, specifically by assuming a uniform distribution of reads over gene copies. Thus, the count of each nucleotide was assumed to be the fraction of reads it was observed on. Of note, this method does not consider the co-occurrence of point mutations on single reads.
Table 1.
Chromosomal point mutations resulting in AMR in C. jejuni
| Antibiotic class | Gene(s) | Point mutation | Reference(s) | ||
|---|---|---|---|---|---|
| Verifieda | Not verifiedb | Not foundc | |||
| (Fluoro)quinolone | gyrA | T86I, D90N, T86A | P104S | A70T, D85Y, T86K, T86V, D90A, D90T, D90Y | [31–35] |
| Macrolide (ERY) |
rplD rplV |
D72N, A103V |
G74D, G67V, A71D, R72I G86E, A88E, A103C A2074G, A2074C |
[36] |
|
| 23S | A2075G, A2074T | ||||
| Aminoglycoside (STR) | rpsL | K43R, K88R | K88E, K88Q | [38, 39] | |
aVerified: point mutations were found in the study population and these correlated with the relevant phenotype
bNot verified: point mutations found in study population but not possible to connect to phenotype
cNot found in data: point mutations not found within study population
In addition, we included in the database the point mutations G86A and C696T in the promotor region (PcmeABC) of the efflux system cmeABC. These point mutations have been hypothesised to increase the level of resistance towards CIP /NAL, ERY, and TET [31, 40, 41].
For each isolate, phenotypic susceptibility to a given antimicrobial agent was compared with the presence of acquired resistance genes and resistance-associated point mutations found by WGS. The correlation between resistant phenotypes and genotypes was calculated by dividing the number of isolates considered resistant based on their genotype by the number of isolates exhibiting resistant phenotypes.
Susceptibility against the six antimicrobials was retested for 62 isolates and new sequence data was obtained for 47 of these to consolidate observed discrepancies between phenotype and predicted genotype.
Results
In vitro antimicrobial susceptibility testing
Phenotypic susceptibility testing showed that 277 of the 516 clinical isolates (54%) were sensitive to all six antimicrobials (Table 2). Among isolates from domestically acquired infections, 61% (258/422) were fully sensitive compared with 18% of isolates from travel-related infections (15/84).
Table 2.
Phenotypic resistance profiles of the 516 clinical isolates based on the test panel, including CIP, NAL, ERY, GEN, STR, and TET
| Resistance profile | Domestic | Travel | N/A | Total |
|---|---|---|---|---|
| Fully sensitive | 258 | 15 | 4 | 277 |
| CIP-NAL | 92 | 21 | 1 | 114 |
| CIP-NAL-TET | 49 | 34 | 4 | 87 |
| TET | 14 | 1 | 15 | |
| CIP-NAL-ERY-TET | 3 | 4 | 7 | |
| CIP-NAL-STR-TET | 1 | 3 | 1 | 5 |
| STR | 3 | 3 | ||
| CIP-NAL-ERY-STR | 2 | 2 | ||
| CIP-NAL-ERY-GEN-TET | 1 | 1 | ||
| CIP-NAL-ERY-STR-TET | 1 | 1 | ||
| CIP-NAL-GEN-TET | 1 | 1 | ||
| CIP-STR-TET | 1 | 1 | ||
| CIP-TET | 1 | 1 | ||
| NAL-TET | 1 | 1 | ||
| Total | 422 | 84 | 10 | 516 |
Thirteen different resistance profiles were identified and the most frequent was CIP/NAL (114/516; 22%), followed by CIP/NAL-TET (87/516; 17%) (Table 2). No isolates were resistant to all six antimicrobials.
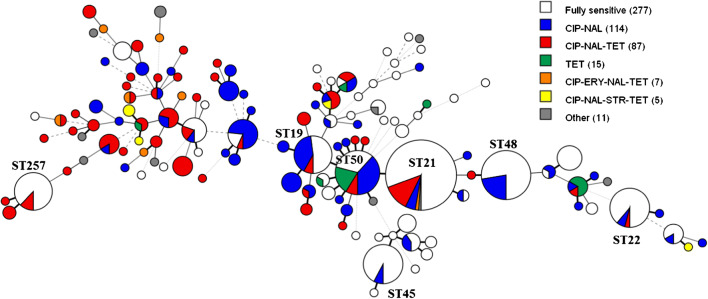
The distribution of phenotypic resistance in the study population is illustrated in Fig. 1. All phenotypic resistance profiles were widely represented in the population and the 43% quinolone-resistant isolates were found in most major sequence types (ST).
Fig. 1.
Minimum spanning three generated from 7 locus MLST showing phenotypic resistance profiles of the 516 C. jejuni isolates. Each node represents an ST and the size of the node correlates to the number of isolates. Isolates resistant to one or more of the six antimicrobials are coloured. Grey nodes (other) represent less common resistance profiles seen for three or less isolates. Isolates susceptible to all six antimicrobials are uncoloured. Numbers in the parentheses indicate number of isolates with the respective resistance profile
Concordance between phenotypic and genotypic AMR
In the study population, known resistance-associated point mutations were observed for gyrA, 23S, and rpsL mediating resistance to CIP/NAL, ERY, and STR, respectively. Some resistance-associated point mutations were not found in the study population (Table 1) and therefore, it was not possible to verify these. After retesting, it was found that most deviations were due to an error in the phenotypic resistance profile and only seven isolates ended up with a new sequence after WGS rerun (possible mix-up of isolates). An overview of the detected resistance-associated point mutations and acquired resistance genes is shown in Table 3.
Table 3.
Correlation of resistance phenotype and predicted genotype for 516 Danish clinical C. jejuni isolates. The result of identified resistance genes is found based on the modified KMA
| Drug class | Tested drugs | No. of isolates with R or S phenotype | Detected genes and mutations | Correlation between phenotype and genotype (%) |
|---|---|---|---|---|
| (Fluoro)quinolone | CIP/NAL | R (n = 218)) |
gyrA T86I (n = 199) gyrA T86I + P104S (n = 13) gyrA T86I + D90N (n = 1) gyrA D90N (n = 1) None (n = 4) |
98 (214/218) |
| NAL | R (n = 1 | |||
| gyrA T86A (n = 1) | ||||
| CIP | R (n = 2) | gyrA T86I (n = 2) | ||
| S (n = 295) | None (n = 295)a | 100 (295/295) | ||
| Macrolide | ERY | R (n = 11) |
23S rRNA A2075G (n = 8)b 23S rRNA A2074T (n = 3) |
100 (11/11) |
| S (n = 505) | None (n = 505)c | 100 (505/505) | ||
| Aminoglycoside | GEN | R (n = 2) | aph(2″)-If (n = 2) | 100 (2/2) |
| S (n = 514) | None (n = 514) | 100 (514/514) | ||
| Aminoglycoside | STR | R (n = 12) |
rpsL K43R (n = 4) rpsL K88R (n = 1) ant(6)-Ia (n = 6) None (n = 1) |
92 (11/12) |
| S (n = 504) | None (n = 504) | 100 (504/504) | ||
| Tetracycline | TET | R (n = 120) |
tet(O) (n = 90) tet(O/32/O) (n = 27) None (n = 3)d |
98 (117/120) |
| S (n = 396) |
None (n = 385) Non-functional tet(O) (n = 11) |
100 (396/396) |
Markers not verified in this study: agyrA P104S (n = 1); badditionally A103V in rplV (n = 3); cA103V in rplV (n = 65) and D72N in rplV (n = 1); dtet(O) was identified in a phenotypic TET resistant isolate with a gene length just below the cutoff (89.06%)
The most frequently observed point mutation was the gyrA T86I, which was detected in 215/221 (97%) of the quinolone-resistant isolates. In 13 of these isolates, the gyrA P104S point mutation was also present. However, the importance of P104S in relation to CIP/NAL resistance could not be verified in this study as P104S was also found in a CIP/NAL sensitive isolate (MICCIP = 0.25 μg/ml, MICNAL = 8 μg/ml). We only found one CIP/NAL resistant isolate harbouring the gyrA D90N mutation.
Three isolates were phenotypically resistant to just one of the two quinolones. Two CIP-resistant isolates harboured gyrA T86I and showed high-level resistance towards CIP (MICCIP = 16 μg/ml) while being well below the breakpoint for NAL (MICNAL = 2 μg/ml and 4 μg/ml). One NAL-resistant isolate harboured the gyrA T86A mutation and showed high-level resistance towards NAL (MICNAL = 64 μg/ml) while being just below the breakpoint for CIP (MICCIP = 0.5 μg/ml). Discrepancies between the phenotype and genotypic prediction were observed for four isolates. These isolates showed a resistant phenotype against CIP/NAL but without any gyrA point mutation present.
Eleven (2%) isolates were resistant to ERY and all carried resistance-associated point mutations in the 23S genes; eight harboured the mutation A2075G, while three harboured the A2074T mutation. Most (10/11) isolates had the same point mutation in all three copies of the 23S rRNA gene. One isolate carried a mutation in only two of the three gene copies, while still displaying high-level resistance (MICERY > 128 μg/ml).
It was not possible to verify the importance of two point mutations, D72N and A103V, associated with ERY resistance (Table 1). We found that 68 isolates harboured A103V but only three isolates showed phenotypic resistance towards erythromycin (MIC > 128 μg/ml) and these isolates also harboured point mutation in 23S at position 2075. Furthermore, D72N was also found in an erythromycin sensitive isolate.
Among the 516 isolates, seven different acquired resistance genes were identified. Four of these genes are known to mediate resistance towards antimicrobials represented in the phenotypic panel: TET (tet(O) and tet(O/32/O)), STR (ant(6)-Ia), GEN (aph(2″)-If), while genes not included in the test panel also were detected: beta-lactam (blaOXA), some aminoglycosides (aph(3′)-III), and chloramphenicol (cat).
Phenotypically, 120 (23%) of the isolates were resistant to tetracycline and this was conferred by either tet(O) or tet(O/32/O). Furthermore, 11 of the 396 (3%) susceptible isolates carried tet(O) (Table 3); however, all of these were found to be a non-functional variant (frameshift mutation). For 117/120 (98%) of the phenotypically resistant isolates, a TET resistance gene was detected by modified KMA: 90 isolates harboured the tet(O) gene and 27 the mosaic gene tet(O/32/O). Figure 2 shows that TET resistance occurred widespread in the population. The less common gene, tet(O/32/O), was rare in the most common STs and the non-functional variant of tet(O) was only found in isolates of ST21 (n = 1), ST50 (n = 7), and the closely related ST8873 (n = 3).
Fig. 2.
Minimum spanning three generated from MLST showing the distribution of genotypic markers of tetracycline resistance, including the non-functional tet(O) gene found in 11 phenotypically susceptible isolates
Only two isolates in the study population were resistant to the aminoglycoside GEN, both with high-level resistance (MICGEN ≥ 16 μg/ml) and harbouring the resistance gene aph(2″)-If.
Twelve (2%) isolates were STR resistant, and 11/12 (92%) possessed either the ant(6)-Ia (n = 6) or one of the known rpsL point mutations (n = 5) (Table 3). The resistant isolate (MICSTR = 16 μg/ml) without any of these markers carried the aph(3′)-III gene, which is known to confer resistance to other aminoglycosides than STR. This gene was also found in three STR susceptible isolates.
We investigated the presence of the point mutations G86A and C696T in the promotor region of CmeABC (PcmeABC) in the study population. G86A was not found in any of the isolates, but the point mutation C696T was identified in 21/516 (4%) isolates. All 21 isolates showed resistance to CIP/NAL, 19 against TET, and two against ERY, where all isolates had a known point mutation and/or acquired resistance gene facilitating the resistant phenotype. Comparing the MIC values for these 21 isolates with the same point mutation and/or resistant gene without C696T showed significantly (P < 0.05) higher MIC values to CIP/NAL.
Evaluating modified KMA, ABRicate, ARIBA, and ResFinder Batch Upload
The detection of resistance genes was performed using four different methods (KMA, ABRicate, ARIBA, and ResFinder Batch Upload). In general, there was a high conformity between the four approaches; however, the mapping-based method modified KMA resulted in more resistance genes being identified and therefore the preferred method to use. The four methods identified four genes relevant to the phenotypic panel (Table 4).
Table 4.
Overview of identified genes related to the antimicrobial agents in the test panel, based on four different methods (modified KMA ABRicate, ARIBA, ResFinder Batch Upload)
| Gene | Mapping | Assembly | ||
|---|---|---|---|---|
| Modified KMA | ABRicate | ARIBA | ResFinder Batch Upload | |
| tet(O) | 90a | 91b | 92b | 91 |
| Non-functional tet(O) | 11 | 11 | 11 | 11 |
| tet(O/32/O) | 27 | 3 | 0 | 0 |
| ant(6)-Ia | 6 | 6 | 6 | 6 |
| aph(2″)-If | 2 | 2 | 2 | 2 |
atet(O) was identified in one isolate with a gene length just below the cutoff (89.06%) using modified KMA and therefore considered not present using this method. tet(O) was detected in this isolate by ARIBA and ResFinder Batch Upload
bOne isolate was found to carry tet(O) by ABRicate and ARIBA, but tet(O/32/O) by modified KMA
All applied methods were able to identify aph(2″)-If in the two GEN-resistant isolates and ant(6)-Ia in six streptomycin-resistant isolates with high gene coverage and identity. The tet(O) gene was detected in 90 isolates by all methods. In one additional isolate, ResFinder Batch Upload and ARIBA detected tet(O), which was not detected by ABRicate and modified KMA. However, in this isolate, modified KMA identified tet(O) with a gene length just below the cutoff (89.06%). For one isolate, the methods found different tet genes (all full-length and above 99% identity): KMA detected tet(O/32/O) whereas ABRicate and ARIBA found tet(O). It was only the mapping-based methods, KMA and ABRicate, that were able to detect tet(O/32/O) with a gene length > 90%; KMA in 27 isolates, and ABRicate in three of these isolates. Additionally, ABRicate and ARIBA identified tet(O/32/O) in 16 and 26 resistant isolates, respectively, but with insufficient gene lengths, ranging from 5 to 78%. ResFinder Batch Upload could not identify tet(O/32/O) in any isolate with the settings used.
Additional resistance genes, not related to the phenotypic test panel, were also found and included blaOXA, aph(3′)-III and cat (results not shown). Modified KMA and ABRicate identified blaOXA, mediating resistance towards beta-lactam, within 458 and 456 isolates, respectively, whereas ARIBA and Resfinder found blaOXA in 459 and 429 isolates, respectively. For 420 of these isolates, all methods identified blaOXA genes and in the remaining isolates, not all methods identified a variant of the blaOXA gene or the gene length of blaOXA was < 90%. The aph(3′)-III gene was identified by all methods in six isolates. The cat gene, mediating chloramphenicol resistance, was found in one isolate by modified KMA, ARIBA, and ResFinder Batch Upload.
Discussion
Resistance against clinically important antibiotics in Campylobacter continues to be a public health concern worldwide. Campylobacter is exposed to antibiotics in both animal production and human medicine making development and transmission of AMR possible. To assess the ability of using WGS to identify and predict AMR in C. jejuni, a dataset of 516 clinical isolates were analysed by comparing their resistance genotypes to their respective phenotypic resistance profile. The resistance profiles were found based on susceptibility to six antimicrobials all included in the Danish antimicrobial surveillance programme, DANMAP [2]. In this study, we evaluated and optimised the analysis of WGS data to identify—with high confidence—the relevant genetic markers that enable the prediction of the phenotypic resistance against specific antimicrobials.
For the genome-based prediction of AMR, we analysed WGS data by the use of mapping- and assembly-based bioinformatics methods to detect known AMR genes listed in a curated database, ResFinder. For detection of point mutations, we constructed a database containing reference genes and associated point mutations known from the literature. For each of the six antimicrobials tested, there was a concordance between 92 and 100% between the presence of known resistance genes or mutations and MICs above the ECOFF breakpoint. To obtain this high concordance, a number of modifications were made to the raw output from the ResFinder database search as well as the detected point mutations validated before inclusion in the final output presented here.
The four approaches to identify the presence of acquired resistance genes (modified KMA, ABRicate, ARIBA, ResFinder Batch Upload) worked well and with an overall good compliance between the results obtained, however with a few important differences that made us chose a modified KMA as the preferred method. A systematic error was attained when using three of the four methods for the detection of the mosaic gene tet(O/32/O) conferring resistance to tetracycline. Although tet(O/32/O) is present in the ResFinder database, only the mapping-based KMA method was able to detect this gene in 27 isolates, whereas neither of the assembly-based methods (ARIBA and ResFinder Batch Upload) detected tet(O/32/O) with a gene length > 90% in any of the genomes. This would potentially make an under-estimation of the occurrence of TET resistance as this gene was present in 23% of the TET-resistant genomes and it was widely distributed in our C. jejuni population. For all approaches, another systematic error was found to be related to frameshift mutations resulting in non-functional genes. We discovered 11 non-functional tet(O) genes resulting in a disagreement between phenotype and genotype. For ten of the isolates, the frameshift was due to a nucleotide deletion, whereas for the last isolate, the tet gene harboured an extra nucleotide compared with the reference. As a result, 3% of the TET sensitive isolates were wrongly determined as resistant. Such frameshift-related errors can be avoided by an automated frameshift check of the matched open reading frames or by a tblastx search.
Verification of markers responsible for resistance is essential for building a reliable pipeline. In this study, it was not possible to verify the significance of some point mutations described in the literature. This was the case for the ribosomal protein L22 (rplV) and the substitution P104S in gyrA associated with resistance against ERY and CIP/NAL, respectively. One mutation in L22 (A103V) was equally common in resistant and sensitive isolates, whereas another mutation in L22 (D72N) was only detected in a sensitive isolate. Thus, our findings suggest that D72N and A103V in L22 do not contribute to erythromycin resistance although these substitutions were reported to be the responsible marker for ERY resistance in one and two isolates, respectively [37, 42]. The P104S substitution in gyrA has been found previously in clinical isolates [43, 44], but our data could not support the association to fluoroquinolone resistance, as P104S was found either together with T86I in gyrA in resistant isolates or in a phenotypically sensitive isolate. These findings, summarised in Table 1, highlight the importance of being cautious when including suggested mechanisms of resistance in a routine analysis of predicting phenotypic resistance.
The most frequent resistance profile observed among the isolates was CIP/NAL resistance. Quinolone resistance in Campylobacter is most often due to mutations in the DNA gyrase A gene, gyrA, and especially substitutions at Thr86 are associated with high-level resistance to quinolones [32]. The substitution T86I was present in 97% of the quinolone resistant isolates and none of the sensitive isolates in our study. Two isolates carrying the T86I gyrA substitution showed high-level resistance to CIP but were fully susceptible to NAL. This phenomenon is described previously [45] and shows that the exact phenotype may not be possible to predict with the current knowledge. Another mutation, the more atypical T86A gyrA substitution, was observed in one isolate that — in accordance with previous studies [45–47] — displayed high-level resistance towards NAL, but was susceptible to CIP. Double point mutations resulting in two amino acid substitutions in gyrA were also seen. In addition to the common T86I, 13 isolates had the P104S substitution and one isolate had the D90N substitution. The D90N substitution is less common and associated with moderate resistance [31–33]; however, we detected D90N as the only marker of quinolone resistance in one of the CIP/NAL-resistant isolates.
Resistance to erythromycin is commonly mediated by mutations in 23S rRNA, especially A2075G [36, 48]. C. coli is more often resistant to macrolides than C. jejuni, and the ermB gene has been shown to mediate resistance to erythromycin in C. coli isolates of animal origin [49, 50], but recently ermB was also identified in a clinical C. jejuni isolate [51]. In this study, none of the isolates had the ermB gene. All isolates exhibiting phenotypic resistance against ERY had a mutation in the 23S rRNA genes at position 2074 or 2075, resulting in a 100% concordance between the phenotype and genotype. None of the isolates had any of the described point mutations in the ribosomal protein L4 (rplD).
When disregarding detection of the mentioned non-verified point mutations and the non-functional tet(O) genes, discrepancies between phenotype and genotypic prediction were seen for eight isolates, four for CIP/NAL, one for STR, and three for TET. In one of the TET-resistant isolates, a tet gene was clearly present as determined by the alternative methods. Otherwise, it is possible that the discrepancies were caused by unknown resistance mechanisms, sequence gaps in the genome preventing detection of known resistance genes or point mutations, or due to mixed cultures in the sample.
Based on our findings, we will establish a routine WGS-based analysis pipeline for AMR detection that we believe has a sufficiently high level of performance for replacing the phenotypic surveillance of AMR in clinical isolates of Campylobacter. Phenotypic susceptibility testing may still be optimal for clinical purposes as the WGS approach cannot deliver an MIC value. However, a sequence-based approach predicting MIC values from markers influencing the MICs may be clinically relevant and could be implemented in the database in the future. In our selected C. jejuni population, we only detected a limited number of genes and mutations that seem to be relevant for predicting the AMR profile. Thus, future routine analysis will only include the verified mutations and the acquired genes in the curated public databases with focus on avoiding the systematic errors encountered in this study, e.g. non-functional genes and detection of the mosaic tet gene. The pipeline should be maintained continuously. We were only able to detect a minority of the many reported point mutations, and therefore, it is relevant to include all potentially important mutations in an additional analysis and evaluate these by phenotypic testing of the isolates. Likewise, new genes or variants of genes may be added to the curated databases, and the performance of our detection pipeline should be evaluated for these. In addition, a subset of the isolates should be tested phenotypically to be able to detect new mechanisms, non-functional genes, and generally perform quality assurance on the analysis pipeline. This further evaluation and development can be done on small selected strain collections at regular intervals.
Electronic supplementary material
(XLSX 155 kb)
Funding
The study has received funding from the European Union’s Horizon 2020 research and innovation programme under Grant Agreement No 773830.
Data availability
Sequences are available at ENA, project numbers PRJEB40238 and PRJEB31119. The full antimicrobial resistance data and the individual accession numbers are available in the supplementary file.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Code availability
Code for modified KMA and detection of point mutations are available at https://github.com/ssi-dk/modified_kma and https://github.com/ssi-dk/punktreskma, respectively.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pal M. Campylobacter jejuni: an emerging foodborne pathogen of global significance. J Exp Food Chem. 2017;3:1–4. doi: 10.4172/2472-0542.1000130. [DOI] [Google Scholar]
- 2.DANMAP (2019) Danmap 2018 - Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. www.danmap.org
- 3.Allos BM. Campylobacter jejuni infections: update on emerging issues and trends. Clin Infect Dis. 2001;32:1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- 4.Anonymous (2019) Annual report on zoonoses in Denmark 2018. Natl. Food Institute, Tech. Univ. Denmark. 10.1128/AEM.02991-09
- 5.Blaser MJ. Epidemiologic and clinical features of Campylobacter jejuni infections. J Infect Dis. 1997;176:103–105. doi: 10.1086/513780. [DOI] [PubMed] [Google Scholar]
- 6.Skirrow MB (1977) Campylobacter enteritis: a “new” disease. Br Med J 9–11 [DOI] [PMC free article] [PubMed]
- 7.Godschalk PCR, Heikema AP, Gilbert M, Komagamine T, Wim Ang C, Glerum J, et al. The crucial role of Campylobacter jejuni genes in anti-ganglioside antibody induction in Guillain-Barré syndrome. J Clin Invest. 2004;114:1659–1665. doi: 10.1172/JCI200415707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannu T, Mattila L, Rautelin H, Pelkonen P, Lahdenne P, Siitonen A, et al. Campylobacter-triggered reactive arthritis: a population-based study. Rheumatology. 2002;41:312–318. doi: 10.1093/rheumatology/41.3.312. [DOI] [PubMed] [Google Scholar]
- 9.Giacomelli M, Salata C, Martini M, Montesissa C, Piccirillo A. Antimicrobial resistance of Campylobacter jejuni and Campylobacter coli from poultry in Italy. Microb Drug Resist. 2014;20:181–188. doi: 10.1089/mdr.2013.0110. [DOI] [PubMed] [Google Scholar]
- 10.Luangtongkum T, Jeon B, Han J, Plummer P, Logue CM, Zhang Q. Antibiotic resistance in Campylobacter: emergence, transmission and persistance. Future Microbiol. 2009;4:1–18. doi: 10.2217/17460913.4.2.189.Antibiotic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wirz SE, Overesch G, Kuhnert P, Korczak BM. Genotype and antibiotic resistance analyses of Campylobacter isolates from ceca and carcasses of slaughtered broiler flocks. Appl Environ Microbiol. 2010;76:6377–6386. doi: 10.1128/AEM.00813-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Zhang M, Deng F, Shen Z, Wu C, Zhang J, et al. Emergence of multidrug-resistant Campylobacter species isolates with a horizontally acquired rRNA methylase. Antimicrob Agents Chemother. 2014;58:5405–5412. doi: 10.1128/AAC.03039-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akiba M, Lin J, Barton Y-W, Zhang Q. Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J Antimicrob Chemother. 2006;57:52–60. doi: 10.1093/jac/dki419. [DOI] [PubMed] [Google Scholar]
- 14.Sierra-Arguello YM, Furian TQ, Perdoncini G, Moraes HLS, Salle CTP, Rodrigues LB, et al. Fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli from poultry and human samples assessed by PCR-restriction fragment length polymorphism assay. PLoS One. 2018;13:1–9. doi: 10.1371/journal.pone.0199974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim S-K, Moon D-C, Chae MH, Kim HJ, Nam H-M, Kim S-R, et al. Macrolide resistance mechanisms and virulence factors in erythromycin-resistant Campylobacter species isolated from chicken and swine feces and carcasses. J Vet Med Sci. 2016;78:1791–1795. doi: 10.1292/jvms.16-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibreel A, Kos VN, Keelan M, Trieber CA, Levesque S, Michaud S, et al. Macrolide resistance in Campylobacter jejuni and Campylobacter coli: molecular mechanism and stability of the resistance phenotype. Antimicrob Agenst Chemother. 2005;49:2753–2759. doi: 10.1128/AAC.49.7.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ladely SR, Meinersmann RJ, Englen MD, Fedorka-Cray PJ, Harrison MA. 23S rRNA gene mutations contributing to macrolide resistance in Campylobacter jejuni and Campylobacter coli. Foodborne Pathog Dis. 2009;6:91–98. doi: 10.1089/fpd.2008.0098. [DOI] [PubMed] [Google Scholar]
- 18.Iovine NM. Resistance mechanisms in Campylobacter jejuni. Virulence. 2013;4:230–240. doi: 10.4161/viru.23753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hormeño L, Ugarte-Ruiz M, Palomo G, Borge C, Florez-Cuadrado D, Vadillo S, et al. ant(6)-I genes encoding aminoglycoside O-nucleotidyltransferases are widely spread among streptomycin resistant strains of Campylobacter jejuni and Campylobacter coli. Front Microbiol. 2018;9:1–8. doi: 10.3389/fmicb.2018.02515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdi-Hachesoo B, Khoshbakht R, Sharifiyazdi H, Tabatabaei M, Hosseinzadeh S, Asasi K. Tetracycline resistance genes in Campylobacter jejuni and C. coli isolated from poultry carcasses. Jundishapur J. Microbiol. 2014;7:1–5. doi: 10.5812/jjm.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao H, Liu D, Wang Y, Zhang Q, Shen Z. High prevalence and predominance of aph(2″)-If gene conferring aminoglycoside resistance in Campylobacter. Antimicrob Agents Chemother. 2017;61:1–9. doi: 10.1128/AAC.00112-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao S, Mukherjee S, Chen Y, Li C, Young S, Warren M, et al. Novel gentamicin resistance genes in Campylobacter isolated from humans and retail meats in the USA. J Antimicrob Chemother. 2014;70:1314–1321. doi: 10.1093/jac/dkv001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, et al. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother. 2016;60:5515–5520. doi: 10.1128/AAC.01030-16.Address. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Z, Tyson GH, Chen Y, Li C, Mukherjee S, Young S, et al. Whole-genome sequencing analysis accurately predicts antimicrobial resistance phenotypes in Campylobacter spp. Appl Environ Microbiol. 2016;82:459–466. doi: 10.1128/AEM.02873-15.Editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nadon C, Walle IV, Gerner-Smidt P, Campos J, Chinen I, Concepcion-Acevedo J, et al. PulseNet International: vision for the implementation of whole genome sequencing (WGS) for global food- borne disease surveillance. Euro Surveill. 2017;22:1–12. doi: 10.2807/1560-7917.ES.2017.22.23.30544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyson GH, McDermott PF, Li C, Chen Y, Tadesse DA, Mukherjee S, et al. WGS accurately predicts antimicrobial resistance in Escherichia coli. J Antimicrob Chemother. 2015;70:2763–2769. doi: 10.1093/jac/dkv186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.European Centre for Disease Prevention and Controle (2016) EU protocol for harmonised monitoring of antimicrobial resistance in human Salmonella and Campylobacter isolates - June 2016
- 28.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunt M, Mather AE, Sánchez-Busó L, Page AJ, Parkhill J, Keane JA et al (2017) ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genomics 3. 10.1099/mgen.0.000131 [DOI] [PMC free article] [PubMed]
- 30.Clausen PTLC, Aarestrup FM, Lund O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics. 2018;19:1–8. doi: 10.1186/s12859-018-2336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Payot S, Bolla JM, Corcoran D, Fanning S, Mégraud F, Zhang Q. Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect. 2006;8:1967–1971. doi: 10.1016/j.micinf.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 32.Engberg J, Aarestrup FM, Taylor DE, Gerner-Smidt P, Nachamkin I. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg Infect Dis. 2001;7:24–34. doi: 10.3201/eid0701.010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wieczorek K, Osek J (2013) Antimicrobial resistance mechanisms among Campylobacter. Biomed Res Int 1–12. 10.1155/2013/340605 [DOI] [PMC free article] [PubMed]
- 34.Zankari E, Allesøe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother. 2017;72:2764–2768. doi: 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alfredson DA, Korolik V. Antibiotic resistance and resistance mechanisms in Campylobacter jejuni and Campylobacter coli. FEMS Microbiol Lett. 2007;277:123–132. doi: 10.1111/j.1574-6968.2007.00935.x. [DOI] [PubMed] [Google Scholar]
- 36.Bolinger H, Kathariou S. The current state of macrolide resistance in Campylobacter spp.: trends and impacts of resistance mechanisms. Appl Environ Microbiol. 2017;83:1–9. doi: 10.1128/AEM.00416-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehtopolku M, Kotilainen P, Haanperä-Heikkinen M, Nakari U-M, Hänninen M-L, Huovinen P, et al. Ribosomal mutations as the main cause of macrolide resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother. 2011;55:5939–5941. doi: 10.1128/AAC.00314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller WG, Huynh S, Parker CT, Niedermeyer JA, Kathariou S. Complete genome sequences of multidrug-resistant Campylobacter jejuni strain 14980A (Turkey feces) and Campylobacter coli strain 14983A (housefly from a Turkey farm), harboring a novel gentamicin resistance mobile element. Genome Announc. 2016;4:1–2. doi: 10.1128/genomeA.01175-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olkkola S, Juntunen P, Heiska H, Hyytiäinen H, Hanninen ML. Mutations in the rpsL gene are involved in streptomycin resistance in Campylobacter coli. Microb Drug Resist. 2010;16:105–110. doi: 10.1089/mdr.2009.0128. [DOI] [PubMed] [Google Scholar]
- 40.Lin J, Akiba M, Sahin O, Zhang Q. CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob Agents Chemother. 2005;49:1067–1075. doi: 10.1128/AAC.49.3.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pumbwe L, Randall LP, Woodward MJ, Piddock LJV. Expression of the efflux pump genes cmeB, cmeF and the porin gene porA in multiple-antibiotic-resistant Campylobacter jejuni. J Antimicrob Chemother. 2004;54:341–347. doi: 10.1093/jac/dkh331. [DOI] [PubMed] [Google Scholar]
- 42.Corcoran D, Quinn T, Cotter L, Fanning S. An investigation of the molecular mechanisms contributing to high-level erythromycin resistance in Campylobacter. Int J Antimicrob Agents. 2006;27:40–45. doi: 10.1016/j.ijantimicag.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 43.Ge B, McDermott PF, White DG, Meng J. Role of efflux pumps and topoisomerase mutations in fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob Agents Chemother. 2005;49:3347–3354. doi: 10.1128/AAC.49.8.3347-3354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hakanen A, Jalava J, Kotilainen P, Jousimies-Somer H, Siitonen A, Huovinen P. gyrA polymorphism in Campylobacter jejuni: detection of gyrA mutations in 162 C. jejuni isolates by single-strand conformation polymorphism and DNA sequencing. Antimicrob. Agents Chemother. 2002;46:2644–2647. doi: 10.1128/AAC.46.8.2644-2647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bachoual R, Ouabdesselam S, Mory F, Lascols C, Soussy C-J, Tankovic J. Single or double mutational alterations of GyrA associated with fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Microb Drug Resist. 2001;7:257–261. doi: 10.1089/10766290152652800. [DOI] [PubMed] [Google Scholar]
- 46.Jesse TW, Englen MD, Pittenger-Alley LG, Fedorka-Cray PJ. Two distinct mutations in gyrA lead to ciprofloxacin and nalidixic acid resistance in Campylobacter coli and Campylobacter jejuni isolated from chickens and beef cattle *. J Appl Microbiol. 2005;100:682–688. doi: 10.1111/j.1365-2672.2005.02796.x. [DOI] [PubMed] [Google Scholar]
- 47.Beckmann L, Müller M, Luber P, Sehrader C, Bartelt E, Klein G. Analysis of gyrA mutations in quinolone-resistant and -susceptible Campylobacter jejuni isolates from retail poultry and human clinical isolates by non-radioactive single-strand conformation polymorphism analysis and DNA sequencing. J Appl Microbiol. 2004;96:1040–1047. doi: 10.1111/j.1365-2672.2004.02242.x. [DOI] [PubMed] [Google Scholar]
- 48.Bolinger HK, Zhang Q, Miller WG, Kathariou S. Lack of evidence for erm(B) infiltration into erythromycin-resistant Campylobacter coli and Campylobacter jejuni from commercial Turkey production in Eastern North Carolina: a major Turkey-growing region in the United States. Foodborne Pathog Dis. 2018;15:698–700. doi: 10.1089/fpd.2018.2477. [DOI] [PubMed] [Google Scholar]
- 49.Qin S, Wang Y, Zhang Q, Zhang M, Deng F, Shen Z, et al. Report of ribosomal RNA methylase gene erm(B) in multidrug-resistant Campylobacter coli. J Antimicrob Chemother. 2014;69:964–968. doi: 10.1093/jac/dkt492. [DOI] [PubMed] [Google Scholar]
- 50.Florez-Cuadrado D, Ugarte-Ruiz M, Quesada A, Palomo G, Domínguez L, Porrero MC. Description of an erm(B)-carrying Campylobacter coli isolate in Europe. J Antimicrob Chemother. 2016;71:841–843. doi: 10.1093/jac/dkv383. [DOI] [PubMed] [Google Scholar]
- 51.Chen JC, Tagg KA, Joung YJ, Bennett C, Watkins LF, Eikmeier D, et al. Report of erm(B) Campylobacter jejuni in the United States. Antimicrob Agents Chemother. 2018;62:1–3. doi: 10.1128/AAC.02615-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 155 kb)
Data Availability Statement
Sequences are available at ENA, project numbers PRJEB40238 and PRJEB31119. The full antimicrobial resistance data and the individual accession numbers are available in the supplementary file.