Abstract
Phytophthora sojae is a pathogen that causes stem and root rot in soybean (Glycine max [L.] Merr.). We previously demonstrated that GmBTB/POZ, a BTB/POZ domain-containing nuclear protein, enhances resistance to P. sojae in soybean, via a process that depends on salicylic acid (SA). Here, we demonstrate that GmBTB/POZ associates directly with soybean LIKE HETEROCHROMATIN PROTEIN1 (GmLHP1) in vitro and in vivo and promotes its ubiquitination and degradation. Both overexpression and RNA interference analysis of transgenic lines demonstrate that GmLHP1 negatively regulates the response of soybean to P. sojae by reducing SA levels and repressing GmPR1 expression. The WRKY transcription factor gene, GmWRKY40, a SA-induced gene in the SA signaling pathway, is targeted by GmLHP1, which represses its expression via at least two mechanisms (directly binding to its promoter and impairing SA accumulation). Furthermore, the nuclear localization of GmLHP1 is required for the GmLHP1-mediated negative regulation of immunity, SA levels and the suppression of GmWRKY40 expression. Finally, GmBTB/POZ releases GmLHP1-regulated GmWRKY40 suppression and increases resistance to P. sojae in GmLHP1-OE hairy roots. These findings uncover a regulatory mechanism by which GmBTB/POZ-GmLHP1 modulates resistance to P. sojae in soybean, likely by regulating the expression of downstream target gene GmWRKY40.
Subject terms: Plant breeding, Plant molecular biology
Zhang et al characterise the mechanism by which GmBTB/POZ enhances resistance to P. sojae in soybean. They show that GmBTB/POZ directly associates with the heterochromatin protein GmLHP1 and mediates its degradation to regulate the salicylic acid signaling pathway, providing insights into the defence against a common soybean disease.
Introduction
Plants have sophisticated cell-autonomous defense mechanisms that combat microbial pathogens, including a waxy cuticle, anti-microbial compounds, and plant innate immunity systems1,2. In general, the waxy cuticle and preformed anti-microbial compounds provide passive protection against pathogens rather than attacking a specific host1, whereas plants rely on innate immunity to defend themselves against widespread diseases3,4. These immunity responses arise via a regulatory network coordinating immune response proteins, transcriptional regulators, and other structural components5–7. Regulation occurs at every level, from differential transcript accumulation and processing to protein modification and turnover5,8. Thus, research on the regulatory components of plant defense responses can provide insights into the complex processes involved in plant immunity.
Ubiquitination is a common post-translational modification in which ubiquitin (Ub) is covalently bound to lysine residues in target proteins9,10. Ubiquitination is carried out by Ub-activating (E1), Ub-conjugating (E2), and Ub-ligase (E3) enzymes, and often leads to target protein degradation mediated by the 26S proteasome11. The BTB/POZ domain (Broad Complex, Tramtrack, Bric-a-brac/Pox virus and Zinc finger) is an evolutionarily conserved, NH3-terminal protein–protein interaction motif present in a variety of cytoskeletal modifiers and Ub ligase substrate recognition factors12–14. Substrate specificity factors associate with cullin 3-based E3 ligases through BTB/POZ proteins15. Therefore, BTB/POZ proteins function as a bridge between CRL3 (CUL3-RING E3 ligase) and substrate proteins and are essential for the ubiquitination process16,17.
HP1 (HETEROCHROMATIN PROTEIN1) was first described in Drosophila melanogaster as a non-histone chromosomal protein that preferentially binds to constitutive heterochromatin on polytene chromosomes18. HP1 orthologs are present in organisms ranging from yeasts to humans19,20. Plants possess a single-copy gene for HP1, LIKE HETEROCHROMATIN PROTEIN1 (LHP1)21, which was initially identified in screens for inflorescence meristem function in Arabidopsis thaliana and is also referred to as TERMINAL FLOWER2 (refs. 22,23). To date, many plant LHP1 homologs have been identified24–26. LHP1 encodes a highly evolutionarily conserved protein containing a chromo domain and a chromo shadow domain21,24.
LHP1 proteins regulate several important growth and development processes in plants27,28. Mutations in AtLHP1 cause a range of developmental defects, including reduced stability of the vernalized state, conversion of the shoot apical meristem to a terminal flower, curled leaves, and reduced root growth21,29. LHP1 is also involved in auxin biosynthesis in Arabidopsis30. In general, LHP1 proteins also function as transcriptional repressors, which play crucial roles in maintaining the transcriptionally silenced state of their targets31–33. For example, AtLHP1 directly represses the expression of the floral promoter FLOWERING LOCUS T (FT) in vascular tissue before dusk and at night34. The early-flowering phenotype of Arabidopsis lhp1 mutants results from increased expression of FT23. These findings indicate that LHP1 represses the transcription of genes that function during different stages of reproductive development. Nevertheless, most studies of LHP1 performed to date in plants other than Arabidopsis were limited to examining the differences in protein expression profiles, whereas no in-depth study of gene expression, functions, or molecular mechanisms of plant LHP1s have been performed. In particular, the role of LHP1 in soybean (Glycine max [L.] Merr.) in response to biotic stress has not yet been evaluated.
GmBTB/POZ positively regulates the response of soybean to Phytophthora sojae, a destructive pathogen that causes stem and root rot in soybean; this response primarily depends on the salicylic acid (SA) signaling pathway35. In the current study, we focused on soybean LIKE HETEROCHROMATIN PROTEIN1 (GmLHP1; NCBI protein no. XP_003548606), a GmBTB/POZ-interacting partner involved in the response to P. sojae infection. GmLHP1 was degraded in soybean inoculated with P. sojae, primarily through the 26S proteasome. Further analysis showed that GmBTB/POZ promotes the ubiquitination and degradation of GmLHP1 in vitro and in vivo. In addition, GmLHP1 inhibits the expression of GmWRKY40, a SA-inducible gene that functions downstream of SA biosynthesis. Therefore, we uncovered a potential role of the GmBTB/POZ–GmLHP1 regulatory module in plant pathogen resistance, providing insights into the mechanism underlying defense responses against P. sojae infection in soybean.
Results
GmLHP1 interacts with GmBTB/POZ
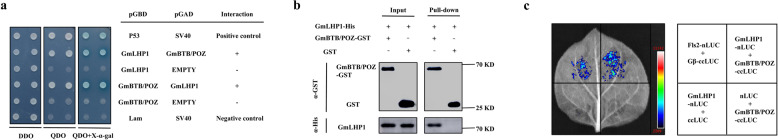
We previously demonstrated that GmBTB/POZ positively regulates the response of soybean to P. sojae infection and GmBTB/POZ interacted with GmLHP1 (LIKE HETEROCHROMATIN PROTEIN1) in a bimolecular fluorescence complementation (BiFC) assay35. In soybean, there are two genes encoding copies of LHP1 (LHP1-1 and LHP1-2)36. In the current study, we focused on LHP1-1, namely GmLHP1 (NCBI protein no. XP_003548606; Glyma.16G079900) which contains two highly conserved structural domains: a chromo domain and a chromo shadow domain (Supplementary Fig. 1). Firstly, in a Y2H assay, yeast cells co-expressing pGBD-GmLHP1 + pGAD-GmBTB/POZ or pGBD-GmBTB/POZ + pGAD-GmLHP1, but not pGBD-GmLHP1 + pGAD or pGBD-GmBTB/POZ + pGAD, grew well on SD/-Trp/-Leu/-His/-Ade (QDO) screening medium and showed α-galactosidase activity (Fig. 1a), indicating that GmLHP1 interacts with GmBTB/POZ in yeast cells.
Fig. 1. GmLHP1 interacts with GmBTB/POZ.
a GmLHP1 interacts with GmBTB/POZ in yeast cells. The yeast cells were selected on SD medium lacking Leu and Trp (DDO), and interaction was assessed based on their ability to grow on selective SD medium lacking Leu, Trp, His, and Ade (QDO) or SD medium lacking Leu, Trp, His, and Ade (QDO) but containing X-α-Gal for 3 days at 30 °C. The combination of pGBD-P53 + pGAD-SV40 was used as a positive control and pGBD-Lam + pGAD-SV40 as a negative control. X-α-Gal represents 5-bromo-4-chloro-3-indolyl-α-d-galactoside. b In vitro pull-down assays showing the interactions of GmLHP1 with GmBTB/POZ. His-tagged proteins were incubated with immobilized GST or GST-tagged proteins, and immunoprecipitated fractions were detected by anti-His antibody. c Interaction between GmLHP1 and GmBTB/POZ in LCI assays. The combination of Fls2-nLUC + Gβ-ccLUC was used as a positive control.
We performed an in vitro pull-down assay to validate the interaction between GmLHP1 and GmBTB/POZ. GmLHP1-His, GmBTB/POZ-GST, and GST alone were detected in whole-cell lysates (Input). GmLHP1 fused with a His tag was not detected in the control sample (GST protein alone), whereas GmLHP1-His was pulled down via GmBTB/POZ-GST (Fig. 1b), suggesting that GmLHP1 directly interacts with GmBTB/POZ. We further confirmed the interaction between GmLHP1 and GmBTB/POZ using firefly luciferase complementation imaging (LCI). The results confirmed that GmLHP1 interacts with GmBTB/POZ in planta (Fig. 1c). Furthermore, these assays indicated that GmLHP1 interacts with GmBTB/POZ in the nucleus (Fig. 1c). Therefore, these three different methods indicated that GmLHP1 directly interacts with GmBTB/POZ both in vitro and in vivo.
GmBTB/POZ promotes the ubiquitination and degradation of GmLHP1
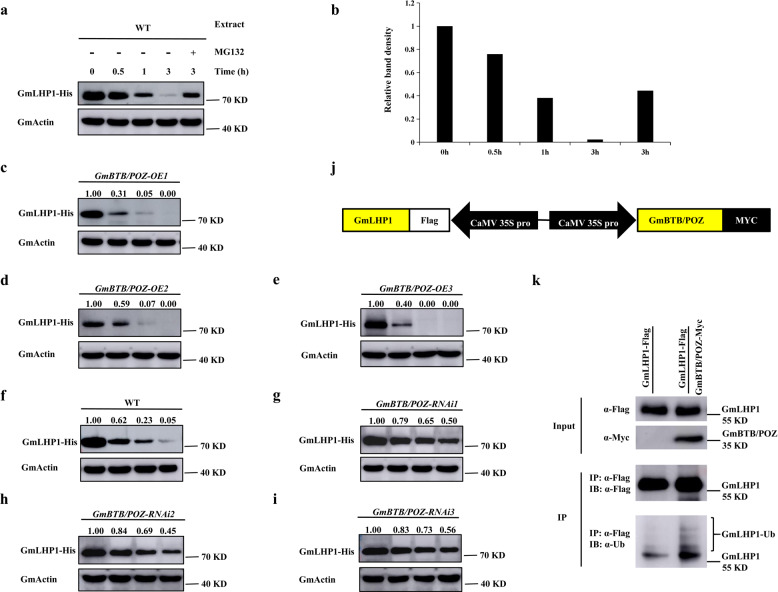
BTB/POZ proteins are a bridge between CUL3-RING E3 ligase and substrate proteins, and they are essential for the Ub process17,37. Since our protein interaction assays between GmLHP1 and GmBTB/POZ suggested that GmLHP1 is a potential substrate of GmBTB/POZ, we speculated that GmBTB/POZ plays a role in the ubiquitination and degradation of GmLHP1. To explore this possibility, we performed in vitro protein degradation assays. Specifically, protein extracts from the WT soybean were incubated with the His-tagged GmLHP1 (GmLHP1-His) proteins purified from Escherichia coli Rosetta (DE3) cells at 22 °C. Then, we performed an immunoblot assay using anti-His antibody to measure the abundance of GmLHP1-His protein. GmLHP1-His was unstable in WT soybean protein extracts; clear GmLHP1-His degradation was observed beginning at 0.5 h, and it was almost completely degraded by 3 h (Fig. 2a). However, treating the samples with 100 μM of the proteasome inhibitor MG132 significantly repressed the degradation process (Fig. 2a). This observation suggests that GmLHP1 is normally degraded, and it points to the possible involvement of the 26S proteasome pathway and ubiquitination.
Fig. 2. GmBTB/POZ promotes the ubiquitination and degradation of GmLHP1.
a GmLHP1 protein was degraded, which likely occurred primarily through the 26S proteasome. For MG132 treatment, WT soybean plant extracts were treated with 100 µM MG132 for 1 h and incubated with GmLHP1-His protein for the indicated time. GmActin was used as a loading control. b Relative band density of GmLHP1-His. GmLHP1-His was quantified using ImageJ software. c–i In vitro cell-free degradation assays of GmLHP1-His in protein extracts from GmBTB/POZ transgenic soybean plants. Protein extracts from transgenic (GmBTB/POZ-OE and GmBTB/POZ-RNAi) and WT soybean plants were incubated with GmLHP1-His for the indicated time. GmLHP1-His levels were visualized by immunoblotting using anti-His antibody. GmActin was used as a loading control. The protein level of 0 h was set to 1.00. j Diagram of the plant binary expression vector system (p35S: Flag-GmLHP1 + p35S: GmBTB/POZ-Myc). k GmBTB/POZ promotes the ubiquitination of GmLHP1 in vivo. GmLHP1-Flag was immunoprecipitated using anti-Flag-Tag Mouse mAb (Agarose Conjugated) from GmLHP1-OE and GmLHP1-OE/GmBTB/POZ-OE transgenic soybean hairy roots by high-efficiency A. rhizogenes-mediated transformation. The transgenic hairy roots were treated with 100 µM MG132 for 8 h before extraction. The immunoprecipitated protein was examined using anti-Flag and anti-ubi antibodies.
To determine whether GmBTB/POZ improves the ubiquitination and degradation of GmLHP1, we identified three T4 GmBTB/POZ-OE soybean plants and three T4 GmBTB/POZ-RNAi soybean plants using immunoblot analysis, QuickStix Kit for LibertyLink (bar) strips, and quantitative reverse-transcription PCR (qRT-PCR), respectively (Supplementary Fig. 2a–d) and subjected them to degradation experiments. Compared to the WT, the degradation rate of GmLHP1-His significantly increased in GmBTB/POZ-OE plant extract (Fig. 2c–e), whereas its stability increased in GmBTB/POZ-RNAi extract (Fig. 2g–i). These results indicate that GmBTB/POZ enhances the degradation of GmLHP1 in vitro. To further explore this notion, we performed in vivo ubiquitination assays. We transformed soybean hairy roots with the plant binary expression vector system p35S: Flag-GmLHP1 + p35S: GmBTB/POZ-Myc (Fig. 2j), immunoprecipitated GmLHP1-Flag and GmBTB/POZ-Myc from proteins extracted from the plants using anti-Flag antibody, and probed the eluted proteins with anti-Flag and anti-Ubi antibodies. In hairy roots overexpressing GmBTB/POZ, much more ubiquitinated GmLHP1-Flag protein was detected compared to the WT (Fig. 2k). Taken together, these results indicate that GmBTB/POZ likely promotes the ubiquitination of GmLHP1 in vitro and in vivo.
To explore the active region of GmBTB/POZ involved in interaction of GmLHP1, different regions of the GmBTB/POZ cDNA encoding the full length (amino acids 1–258), the N-terminal part (amino acids 1–83), the BTB/POZ domain part (amino acids 84–188), or the C-terminal part (amino acids 189–258) of the protein were inserted into yeast vectors pGBD (Supplementary Fig. 3a, b). The Y2H assay showed that both the full-length, the N-terminal part (amino acids 1–83) and the C-terminal part (amino acids 189–258) of the GmBTB/POZ protein were able to interact with the GmLHP1 protein, but not the BTB/POZ domain part (amino acids 84–188) of GmBTB/POZ (Supplementary Fig. 3b). Consistent results were obtained by the BiFC assay (Supplementary Fig. 3c). Furthermore, it is reported that members of the BTB/POZ protein family use the BTB/POZ domain to bind the cullin-based E3 ligases and the other regions to recruit substrate proteins38–40. In this study, the interaction analyses suggested that the N-terminal or the C-terminal of the GmBTB/POZ may play a role in recruiting GmLHP1 in GmBTB/POZ-mediated ubiquitination. To test the hypothesis, we firstly constructed the (domain+C) and (N+domain) regions of GmBTB/POZ and verified that both the (domain+C) and (N+domain) regions still interact with GmLHP1 by the Y2H assay and BiFC (Supplementary Fig. 3e–g). Then, we further analyzed that whether the (domain+C) and (N+domain) region proteins can still function as a bridge in the ubiquitination of GmLHP1 by in vitro cell-free degradation assay and in vivo ubiquitination assay. The results showed that both the (domain+C) and (N+domain) region proteins can promote the ubiquitination and degradation of GmLHP1 in vitro and in vivo (Supplementary Fig. 3d, h). These findings suggest that the N-terminal and C-terminal of GmBTB/POZ could be the active region of GmBTB/POZ involved in interaction and recruitment of GmLHP1 in the protein ubiquitination.
GmLHP1 negatively regulates plant immunity
LHP1 plays an important role in plant responses to environmental stimuli41. In addition, LHP1 interacts with various proteins to perform distinct roles in different cell types42,43; for example, LHP1 interacts with GmPHD6 to regulate the expression of genes involved in salt tolerance36. GmBTB/POZ plays an integral role in the response of soybean to P. sojae attack, which mainly depends on the SA signaling pathway35. The interaction and ubiquitination between GmBTB/POZ and GmLHP1 raised the question of whether GmLHP1 plays a role in a GmBTB/POZ-mediated SA and immune signaling pathway. To explore the biological function of GmLHP1, we produced transgenic soybean plants expressing p35S: Flag-GmLHP1 (GmLHP1OE) or p35S: GmLHP1-RNA interference (GmLHP1RNAi). Immunoblotting analysis confirmed the expression of recombinant GmLHP1-Flag protein in three independently selected T4 GmLHP1-OE lines (GmLHP1OE5, GmLHP1OE10, and GmLHP1OE14) using anti-Flag antibody (Supplementary Fig. 2e). Southern blot and qRT-PCR analyses confirmed the reduced expression of GmLHP1 in the three independently selected T4 GmLHP1-RNAi transgenic soybean lines (GmLHP1RNAi4, GmLHP1RNAi5, and GmLHP1RNAi6), which were integrated into the genomes of the three lines in a single copy (Supplementary Fig. 2g, h).
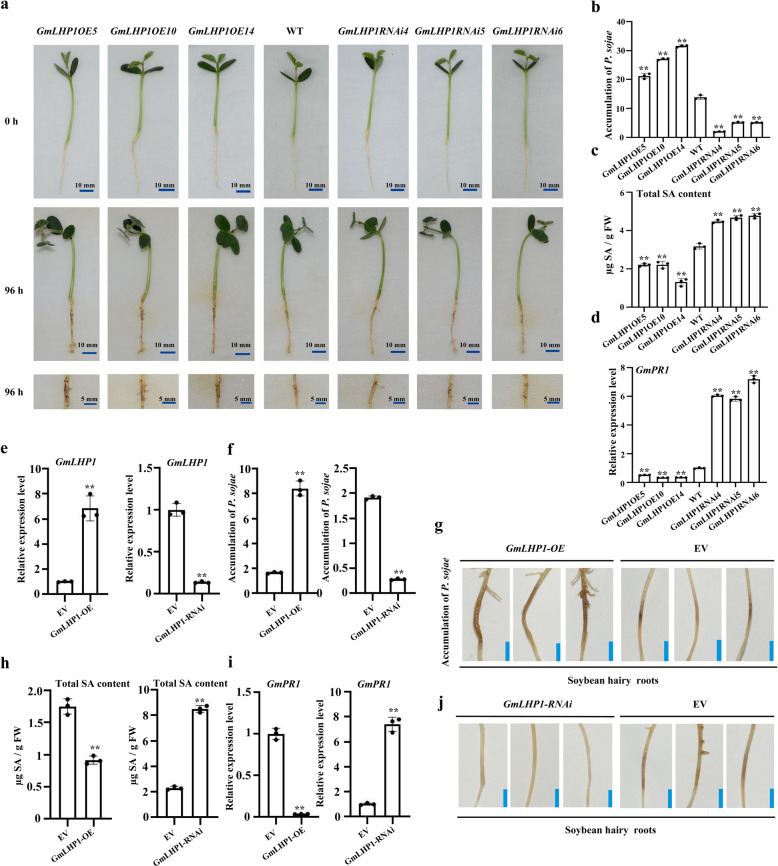
We investigated P. sojae resistance in the roots of these transgenic plants. At 96 h of post-inoculation (hpi), the roots of all three GmLHP1OE soybean lines exhibited more serious symptoms than WT roots, including watery and rotting lesions (Fig. 3a). By contrast, the three GmLHP1RNAi soybean lines displayed almost no visible lesions compared to WT roots (Fig. 3a). We analyzed the relative biomass of P. sojae in soybean roots based on the transcript level of P. sojae TEF1 (EU079791). P. sojae biomass was significantly higher (**P < 0.01) in the GmLHP1OE lines and significantly lower (**P < 0.01) in the GmLHP1RNAi lines compared to WT plants (Fig. 3b). Similar results were obtained for GmLHP1-OE and GmLHP1-RNAi transgenic soybean hairy roots, which were generated by high-efficiency Agrobacterium rhizogenes-mediated transformation (Fig. 3f, g, j)44,45. These results indicate that overexpressing GmLHP1 in soybean increases susceptibility to P. sojae and that silencing this gene improves resistance to P. sojae.
Fig. 3. GmLHP1 negatively regulates plant immunity.
a Disease symptoms in the roots of wild-type (WT), GmLHP1OE, and GmLHP1RNAi soybean plants at 96 h after inoculation with P. sojae. b Relative biomass of P. sojae in WT, GmLHP1OE, and GmLHP1RNAi soybean plants based on P. sojae TEF1 (EU079791) transcript levels. c SA contents in leaves of transgenic and WT soybean. FW, fresh weight. d Relative GmPR1 expression levels in transgenic and WT soybean plants. The expression level of the control sample (WT plants) was set to 1. e qRT-PCR analysis of relative GmLHP1 expression in transgenic soybean hairy roots. Empty vector (EV) transgenic hairy roots were used as controls, and the expression level of the control sample (EV) was set to 1. f Relative biomass of P. sojae in GmLHP1-transgenic hairy roots based on P. sojae TEF1 (EU079791) transcript levels. g Typical infection symptoms of GmLHP1-OE and EV soybean hairy roots at 48 h after P. sojae inoculation. Bars, 0.5 cm. h SA contents in GmLHP1-OE, GmLHP1-RNAi, and EV hairy roots. FW, fresh weight. i Relative GmPR1 expression levels in GmLHP1-OE, GmLHP1-RNAi, and EV hairy roots. The expression level of the control sample (EV) was set to 1. j Typical infection symptoms of GmLHP1-RNAi and EV hairy roots at 48 h after P. sojae inoculation. Bars, 0.5 cm. The housekeeping gene GmEF1 was used as an internal control to normalize the data. The experiment was performed on three biological replicates, each with three technical replicates, and the results were statistically analyzed using Student’s t-test (*P < 0.05, **P < 0.01). Bars indicate the standard deviation of the mean (n = 3).
SA plays major roles in regulating basal defense responses during plant immunity46 and acts as a crucial signaling element in systemic acquired resistance (SAR) signaling pathways47,48. SA mediates SAR, which limits the growth of biotrophic and necrotrophic virulent pathogens and favors long-term protection against a broad spectrum of microorganisms49,50. Increased endogenous SA levels trigger SAR by inducing the expression of pathogenesis-related (PR) genes, such as PR1, which is considered to be an effector gene for SAR48. To determine whether GmLHP1 also regulates the SA signaling pathway, we analyzed the SA contents and expression levels of GmPR1 (AF136636) in GmLHP1OE, WT, and GmLHP1RNAi soybean plants. Both SA levels and GmPR1 expression levels were significantly lower (**P < 0.01) in GmLHP1OE plants and higher (**P < 0.01) in GmLHP1RNAi plants compared to WT (Fig. 3c, d). In addition, both SA levels and GmPR1 expression levels were significantly reduced (**P < 0.01) in GmLHP1-OE transgenic hairy roots vs. the control. However, SA levels and GmPR1 expression levels were significantly higher (**P < 0.01) in GmLHP1-RNAi vs. control hairy roots (Fig. 3h, i). The results suggest that GmLHP1 regulates defense responses against P. sojae by affecting SA levels and GmPR1 expression.
GmLHP1 regulates the transcription of GmWRKY40 via two mechanisms
LHP1 is a nucleus-localized protein that generally functions as a transcriptional repressor in both plants and animals21,33,34,51. To examine the subcellular localization of GmLHP1, we analyzed the expression of the GmLHP1-GFP fusion protein. GmLHP1-GFP signals were observed in the nuclei of transformed cells, like the GmBTB/POZ-GFP expression pattern reported by Zhang et al.35, indicating that GmLHP1 is a nucleus-localized protein (Supplementary Fig. 4a). In a transient expression assay in yeast cells using a GAL4-responsive reporter system, transformed yeast cells containing DBD-GmLHP1 (pGBKT7-GmLHP1) exhibited no α-gal activity, indicating that GmLHP1 did not activate the transcription of the reporter gene (Supplementary Fig. 4b).
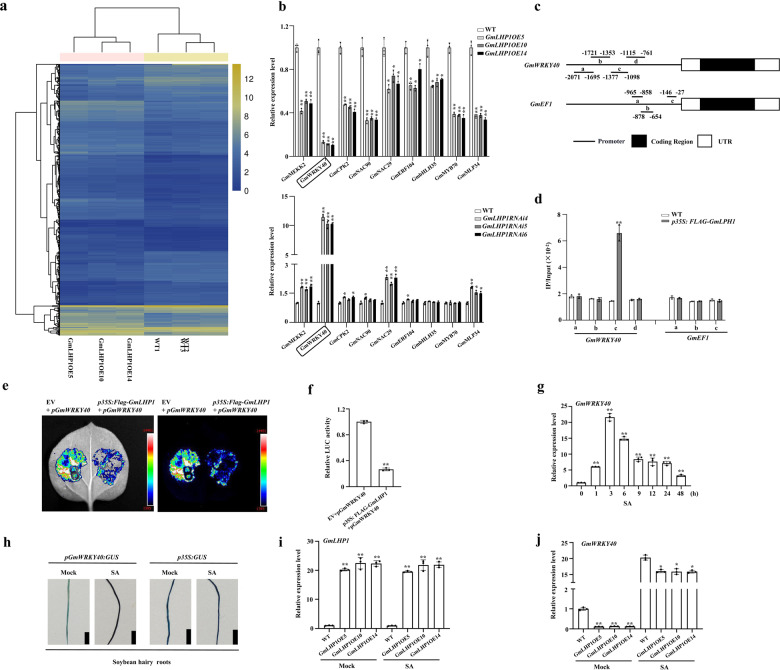
To explore the extent of the regulatory impact of GmLHP1 and to identify GmLHP1-regulated genes, we performed RNA-sequence (RNA-Seq) analysis of the transcriptomes of both WT and GmLHP1OE transgenic soybean plants after 6 weeks of growth in the field. RNA-Seq analysis identified 422 differentially expressed genes (DEGs) with >2.0-fold differences in expression in GmLHP1-OE vs. WT plants under non-stress conditions (false discovery rate (FDR) **P < 0.01). Among the 422 DEGs, 253 were significantly upregulated and 169 were significantly downregulated (Supplementary Fig. 5a and Fig. 4a). Gene ontology (GO) analysis revealed that these genes are primarily enriched in the GO terms plant response to biotic and abiotic stress, hormone stimulus, transferase activity, transport, and other metabolic processes (Supplementary Fig. 5b).
Fig. 4. GmWRKY40 is a target gene of GmLHP1.
a Heat map of the expression patterns of significantly differentially expressed genes in WT and GmLHP1OE soybean plants determined by RNA-Seq analysis. The scale bar indicates fold changes (log2 value). b Relative expression of several stress-related genes in WT, GmLHP1OE, and GmLHP1RNAi soybean plants. c, d ChIP analysis of GmLHP1 binding to the GmWRKY40 promoter region. Chromatin from LHP1-Flag transgenic soybean plants and the WT was immunoprecipitated by anti-Flag or no antibody. The precipitated chromatin fragments were analyzed by qPCR using four pairs of specific ChIP-qPCR primer sets to amplify four regions upstream of GmWRKY40 (pGmWRKY40a, pGmWRKY40b, pGmWRKY40c, and pGmWRKY40d), as indicated. One-tenth of the input (without antibody precipitation) of chromatin was analyzed and used as a control. pGmEF1 was used as a negative control. Three biological replicates, each with three technical replicates, were averaged and statistically analyzed using Student’s t-test (*P < 0.05, **P < 0.01). Bars indicate standard deviation of the mean (n = 3). e Dual-luciferase assay in N. benthamiana leaves showing that GmLHP1 represses the expression of GmWRKY40. Representative photographs are shown. f Detection of LUC/REN activity to verify that GmLHP1 represses the transcription of GmWRKY40. The combination of the reporter construct (pGmWRKY40: LUC) and the blank effector construct (empty vector) was used as a control. g Relative expression of GmWRKY40 in WT soybean plants in response to SA (0.5 mM) treatment. The relative expression levels of GmWRKY40 were compared with those of mock-treated plants. Fourteen-day-old plants were used for analysis. h GmWRKY40 promoter-driven GUS expression in transgenic soybean hairy roots under SA or mock treatment for 3 h. Bars, 0.5 cm. i Expression patterns of GmLHP1 in WT and GmLHP1OE transgenic soybean plants under SA or mock treatment for 3 h. The expression level of the control sample (mock-treated wild-type (WT) plants) was set to 1. j Expression patterns of GmWRKY40 in WT and GmLHP1OE transgenic soybean plants under SA or mock treatment for 3 h. The expression level of the control sample (mock-treated wild-type (WT) plants) was set to 1. The housekeeping gene GmEF1 was used as an internal control to normalize the data. The experiment was performed on three biological replicates, each with three technical replicates, and the results were statistically analyzed using Student’s t-test (*P < 0.05, **P < 0.01). Bars indicate the standard deviation of the mean (n = 3).
We examined the expression of several downregulated stress-related DEGs in GmLHP1OE and GmLHP1RNAi soybean plants by qRT-PCR analysis. Examples of these genes include immunity signaling genes such as GmMEKK2 (Glyma.17G173000), GmWRKY40 (Glyma.15G003300), and GmCPK2 (Glyma.11G206300) and defense-associated genes such as GmNAC90 (Glyma.11G182000), GmNAC29 (Glyma.02G109800), GmERF104 (Glyma.20G070000), GmbHLH35 (Glyma.13G101100), GmMYB70 (Glyma.17G237900), and GmMLP34 (Glyma.09G102400). GmWRKY40 expression was dramatically reduced in GmLHP1OE vs. WT plants. Notably, in GmLHP1RNAi soybean plants, GmWRKY40 expression significantly increased (**P < 0.01) compared to the WT, while none of the other genes showed markedly altered expression (Fig. 4b). These findings indicate that the regulation of GmWRKY40 expression likely plays a role in GmLHP1-mediated defense responses.
To explore how GmLHP1 regulates the expression of GmWRKY40, we performed a dual effector–reporter assay using GmLHP1 as the effector and the luciferase gene under the control of 2.0 kb of the GmWRKY40 promoter as the reporter. The effector construct harbored GmLHP1 expressed under the control of the 35S promoter (p35S: Flag-GmLHP1). We transformed the reporter construct (p35S: REN-pGmWRKY40: LUC) and the effector construct (p35S: Flag-GmLHP1) or the reporter construct (p35S: REN-pGmWRKY40: LUC) and the blank effector construct (empty vector (EV)) into healthy Nicotiana benthamiana leaves. The co-existence of GmLHP1 and the GmWRKY40 promoter significantly inhibited (**P < 0.01) luciferase expression in N. benthamiana leaves (Fig. 4e, f), suggesting that GmLHP1 significantly represses the expression of GmWRKY40.
To investigate the binding capacity of GmLHP1 to the promoter of GmWRKY40, we performed chromatin immunoprecipitation (ChIP)-qPCR assays using cell extracts from WT plants and GmLHP1OE transgenic soybean plants expressing Flag-fused GmLHP1 under the control of the constitutive 35S promoter. However, since the binding elements or target regions of LHP1 were unclear, we analyzed the enrichment of four regions in the GmWRKY40 promoter via ChIP-qPCR using four pairs of specific primers. As shown in Fig. 4c, d, the c region was significantly enriched (**P < 0.01) with GmLHP1-Flag, whereas none of the three regions in the GmEF1 promoter were enriched with GmLHP1-Flag; GmEF1 is often used as a reference gene, since it is expressed constitutively at a constant level throughout the plant and is not influenced by exogenous treatment52. These results indicate that GmLHP1 specifically associates with the regulatory regions of its target gene (Fig. 4c, d).
Some WRKY genes are SA-inducible transcription factor genes involved in disease resistance in a number of plant species53–55. Analysis of GmWRKY40 transcript levels in response to SA (0.5 mM) treatment showed that GmWRKY40 expression was significantly induced by SA in WT plants, reaching a peak at 12 h, followed by a steep decline (Fig. 4g), indicating that GmWRKY40 expression is significantly induced by SA in soybean. To further elucidate the underlying regulatory mechanism, we used the 2 kb promoter region of GmWRKY40 to drive the expression of the GUS reporter gene in the pBI121 expression vector, which we transformed into “Dongnong 50” soybean hairy roots via high-efficiency A. rhizogenes-mediated transformation. We analyzed GmWRKY40 promoter activity in hairy roots at 6 h after SA treatment. The amount of histochemical GUS staining in hairy roots was higher under SA treatment than under mock (H2O) treatment (Fig. 4h), suggesting that GmWRKY40 functions downstream of SA biosynthesis. Moreover, we demonstrated that GmLHP1 has an effect on SA accumulation (Fig. 3h).
To investigate whether GmLHP1 suppresses GmWRKY40 expression via impaired SA accumulation, we examined whether exogenous SA application would weaken the inhibition of GmWRKY40 expression in GmLHP1OE soybean plants. As shown in Fig. 4i, we analyzed the expression efficiency of GmLHP1 by qRT-PCR. GmLHP1 transcript levels were significantly higher in GmLHP1-OE plants compared to WT plants under both mock (H2O) treatment and after 6 h of SA treatment. As expected, SA-treated plants displayed clearly increased GmWRKY40 transcript abundance compared to mock-treated plants (Fig. 4j). GmWRKY40 transcript levels were markedly lower (**P < 0.01) in GmLHP1OE plants than in WT plants under mock treatment, while there was no obvious difference (*P < 0.05) in GmWRKY40 transcript level between GmLHP1OE and WT plants under SA treatment, suggesting that SA induces changes in GmWRKY40 expression in GmLHP1OE soybean plants. Taken together, these findings suggest that at least two mechanisms (direct repression of GmWRKY40 expression and impaired SA accumulation) contribute to the regulation of GmWRKY40 expression by GmLHP1.
GmWRKY40 also functions in responses to P. sojae infection and increases the expression of SA-marker gene GmPR1
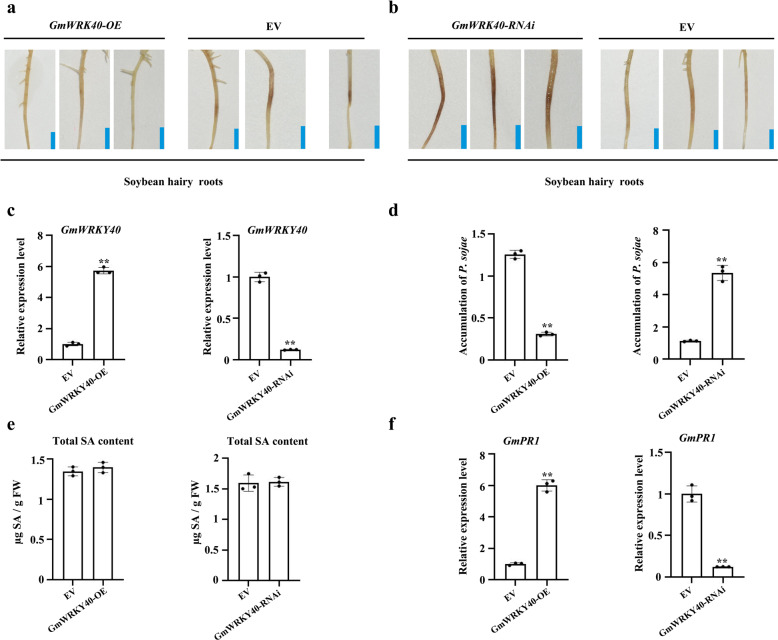
We then explored the possible role of GmWRKY40 in the response to P. sojae infection by analyzing the phenotypes of control, GmWRKY40-OE, and GmWRKY40-RNAi hairy roots after incubation with P. sojae zoospores. The GmWRKY40-OE transgenic hairy roots were examined by immunoblotting (Supplementary Fig. 2k) and qRT-PCR (Fig. 5c) and the GmWRKY40-RNAi transgenic hairy roots by analysis with QuickStix Kit for LibertyLink bar strips (Supplementary Fig. 2l) and qRT-PCR (Fig. 5c). After 2 d of incubation with P. sojae zoospores, GmWRKY40-OE hairy roots displayed almost no visible lesions (Fig. 5a), whereas the GmWRKY40-RNAi lines exhibited enhanced wilting symptoms and chlorosis compared to the control (Fig. 5b). We also analyzed the relative biomass of P. sojae in infected hairy roots after 2 days of incubation with P. sojae zoospores. The biomass of P. sojae was significantly (**P < 0.01) lower in the roots of GmWRKY40-OE lines but significantly (**P < 0.01) higher in the roots of GmWRKY40-RNAi lines compared to the control (Fig. 5d).
Fig. 5. GmWRKY40 also functions downstream of SA biosynthesis and enhances the expression of SA-marker gene GmPR1 in response to P. sojae.
a Typical phenotypes of WRKY40-OE and EV soybean hairy roots after 48 h of P. sojae inoculation. Bars, 0.5 cm. b Typical phenotypes of WRKY40-RNAi and EV soybean hairy roots after 48 h of P. sojae inoculation. Bars, 0.5 cm. c qRT-PCR analysis of relative GmLHP1 expression in transgenic soybean hairy roots. Soybean hairy roots transformed with empty vector (EV) were used as controls; the expression level of the control sample (EV) was set to 1. d Relative biomass of P. sojae in GmLHP1-transgenic soybean hairy roots based on the transcript level of P. sojae TEF1 (EU079791). e SA contents in WRKY40-OE, WRKY40-RNAi, and EV hairy roots. FW, fresh weight. f Relative expression level of GmPR1 in WRKY40-OE, WRKY40-RNAi, and EV hairy roots. The expression level of the control sample (EV) was set to 1. The housekeeping gene GmEF1 was used as an internal control to normalize the data. The experiment was performed on three biological replicates, each with three technical replicates, and the results were statistically analyzed using Student’s t-test (*P < 0.05, **P < 0.01). Bars indicate the standard deviation of the mean (n = 3).
GmWRKY40 expression was significantly induced by SA, suggesting that GmWRKY40 functions downstream of SA biosynthesis as a component of SA signaling. To determine whether GmWRKY40 also participates in the SA signaling pathway, we measured SA content and GmPR1 expression in the transgenic hairy roots. GmPR1 was expressed at significantly higher levels (**P < 0.01) in GmWRKY40-OE lines but at significantly lower levels (**P < 0.01) in GmWRKY40-RNAi lines compared to the control (Fig. 5f). However, there was no significant difference in SA level between the GmWRKY40 lines and control hairy roots (Fig. 5e). These results suggest that GmWRKY40 functions as a SA-induced gene downstream of SA biosynthesis and enhances the expression of SA-marker gene GmPR1 in response to P. sojae infection.
Nuclear localization of GmLHP1 is required for its functionality
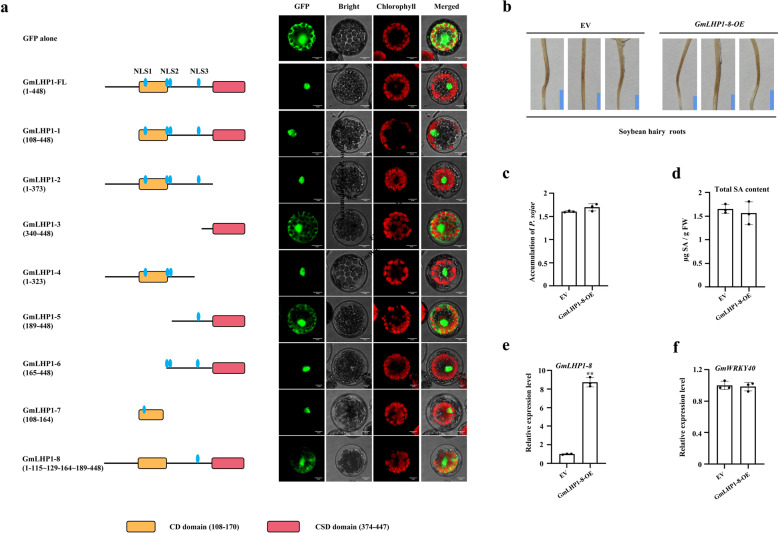
To determine the region(s) responsible for the nuclear localization of GmLHP1 and analyze whether the nuclear localization of GmLHP1 is required for its functionality, we firstly analyzed the nuclear localization signal (NLS) regions of GmLHP1 using NLS Mapper software56,57. Three putative NLS regions (NLS1 to NLS3) were identified (Fig. 6a, left column), NLS1 (IRRKR-EVQY, amino acids 116–128) is located at the conserved CD domain, and the other two are located at the hinge region: a bipartite NLS2 (GKHRK-LERS, amino acids 165–188) and NLS3 (RCRGS-VKRF, amino acids 324–339). Then, we constructed the GmLHP1 deletion mutants (GmLHP1-1 to 8), each fused with GFP at its C terminus, and analyzed its subcellular localization (Fig. 6a). Transient expression into Arabidopsis protoplasts showed that the localization of GmLHP1-1 (amino acids 108–448), containing all NLS regions but lacking the N-terminal part (amino acids 1–107), and GmLHP1-2 (amino acids 1–373), containing all NLS regions but lacking the CSD domain, both were localized in the nucleus of the transformed cell and were indistinguishable from that of the intact protein (amino acids 1–448). The results showed that the absence of N-terminal part alone or the CSD domain alone does not change the nuclear localization of GmLHP1. GmLHP1-3 (amino acids 340–448), containing the conserved CSD domain region, green fluorescent signal was dispersed in the entire cell of protoplasts similar to that displayed by GFP alone, further indicating that the conserved CSD domain has no specific nuclear targeting properties. GmLHP1-4 (amino acids 1–323), containing the conserved CD domain and NLS2 regions but lacking NLS3 and the CSD domain, was localized in the nucleus of the transformed cell, while GmLHP1-5 (amino acids 189–448), which containing NLS3 and the CSD domain, green fluorescent signal was dispersed in the entire cell of protoplasts similar to that displayed by GFP alone, suggesting that the region (amino acids 189–448) of GmLHP1 is not required for the nuclear localization of GmLHP1 and the putative NLS3 region is nonfunctional. On the basis of GmLHP1-5 deletion mutant sequence (amino acids 189–448), GmLHP1-6 (amino acids 165–448) which added NLS2 region was localized in the nucleus, suggesting NLS2 region has specific nuclear targeting properties. Furthermore, we found that the region encompassing residues 108 to 164 (GmLHP1-7), corresponding to NLS1 region, retained the nucleolus-targeting localization property, indicating the putative NLS1 may also be functional, like the NLS2 region. To verify this prediction, we finally constructed the GmLHP1-8 deletion mutants (amino acids 1–115~129–164~189–448), which deleted the NLS1 and NLS2, and after transformation, we analyzed its subcellular localization. GmLHP1-8 was detected in the entire cell of protoplasts similar to the GFP alone control. Together, the results showed that both NLS1 and NLS2 regions are required for the nuclear targeting properties of GmLHP1.
Fig. 6. Deletion analysis of GmLHP1.
a Subcellular localization of various GmLHP1 deletion mutants. Left column, scheme of GmLHP1 and its deletion mutants fused to GFP. FL, full length. NLS, nuclear localization signal. Right column, the indicated constructs were transiently expressed in Arabidopsis protoplasts and inspected with a confocal microscope. Scale bars indicate 10 µm. b Typical infection symptoms of GmLHP1-8-OE and EV soybean hairy roots at 48 h after P. sojae inoculation. Bars, 0.5 cm. c Relative biomass of P. sojae in GmLHP1-8-OE hairy roots based on P. sojae TEF1 (EU079791) transcript levels. d SA contents in GmLHP1-8-OE hairy roots. FW, fresh weight. e GmLHP1-8 transcript levels in EV and GmLHP1-8-OE hairy roots. f GmWRKY40 transcript levels in EV and GmLHP1-8-OE hairy roots. The expression level of the control sample (EV) was set to 1. The housekeeping gene GmEF1 was used as an internal control to normalize the data. The experiment was performed on three biological replicates, each with three technical replicates, and the results were statistically analyzed using Student’s t-test (*P < 0.05, **P < 0.01). Bars indicate the standard deviation of the mean (n = 3).
To analyze whether the nuclear localization of GmLHP1 is necessary for its functionality, we investigated the P. sojae resistance in GmLHP1-8-OE transgenic soybean hairy roots. After 2 days of incubation with P. sojae zoospores, there was no significant phenotype difference between EV and GmLHP1-8-OE soybean hairy roots (Fig. 6b). In accordance with this, the relative biomass of P. sojae in infected EV and GmLHP1-8-OE soybean hairy roots have no significant difference (Fig. 6c). Furthermore, the SA levels in GmLHP1-8-OE soybean hairy roots were not significantly downregulated compared to that in EV soybean hairy roots (Fig. 6d). To further determine whether the changes of GmLHP1 nuclear localization have an effect on the suppression of GmWRKY40 expression by GmLHP1, we also analyzed the expression levels of GmLHP1-8 and GmWRKY40 in GmLHP1-8-OE soybean hairy roots (Fig. 6e, f). GmWRKY40 expression was not significantly suppressed in the GmLHP1-8-OE soybean hairy roots. These results indicated that the nuclear localization of GmLHP1 is required for its functionality.
GmBTB/POZ releases GmLHP1-regulated GmWRKY40 suppression in GmLHP1-OE soybean lines
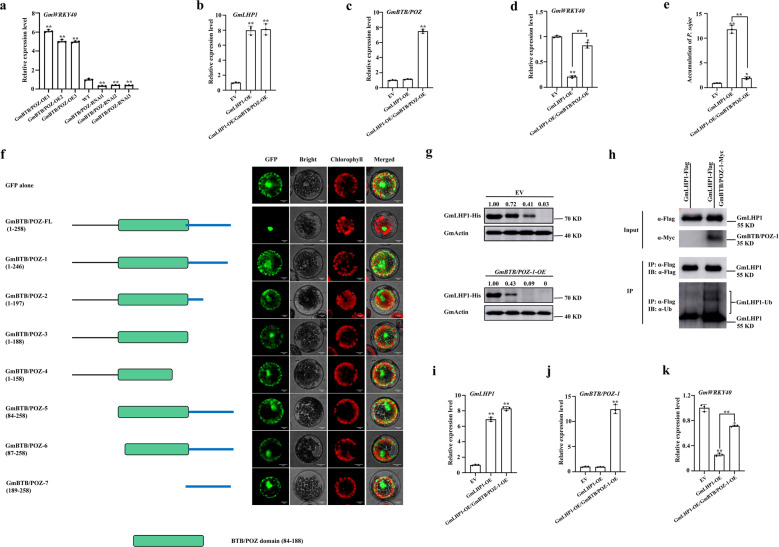
We also analyzed the expression levels of GmWRKY40 in GmBTB/POZ-OE and GmBTB/POZ-RNAi soybean plants. As shown in Fig. 7a, GmWRKY40 was upregulated in GmBTB/POZ-OE plants and downregulated in GmBTB/POZ-RNAi plants compared to the WT. These results indicate that GmBTB/POZ is also involved in regulating GmWRKY40 transcription. To further explore the role of GmBTB/POZ in GmLHP1-mediated suppression of GmWRKY40 expression, we generated GmLHP1-OE and GmBTB/POZ-OE/GmLHP1-OE transgenic soybean hairy roots and used hairy roots transformed with EV as a negative control. After measuring GmBTB/POZ and GmLHP1 transcript levels to evaluate the efficiency of GmBTB/POZ and GmLHP1 expression (Fig. 7b, c), we measured GmWRKY40 transcript levels in EV, GmLHP1-OE, and GmBTB/POZ-OE/GmLHP1-OE hairy roots by qRT-PCR. GmWRKY40 expression was significantly suppressed in the GmLHP1-OE lines, but this effect was inhibited in GmBTB/POZ-OE/GmLHP1-OE hairy roots (Fig. 7d). These results suggest that GmBTB/POZ releases GmLHP1-mediated suppression of GmWRKY40 expression, likely by inducing the degradation of GmLHP1.
Fig. 7. GmBTB/POZ weakens GmLHP1-mediated suppression of GmWRKY40 and increases GmLHP1-regulated responses to P. sojae.
a GmWRKY40 transcript levels in GmBTB/POZ-OE and GmBTB/POZ-RNAi soybean plants. b GmLHP1 transcript levels in EV, GmLHP1-OE, and GmBTB/POZ-OE/GmLHP1-OE transgenic soybean hairy roots. c GmBTB/POZ transcript levels in EV, GmLHP1-OE, and GmBTB/POZ-OE/GmLHP1-OE transgenic hairy roots. d GmWRKY40 transcript levels in EV, GmLHP1-OE, and GmBTB/POZ-OE/GmLHP1-OE transgenic hairy roots. e Relative biomass of P. sojae in EV, GmLHP1-OE, and GmBTB/POZ-OE/GmLHP1-OE transgenic hairy roots based on the transcript level of P. sojae TEF1 (EU079791) after 48 h of P. sojae inoculation. f Subcellular localization of various GmBTB/POZ deletion mutants. Left column, scheme of GmBTB/POZ and its deletion mutants fused to GFP. FL, full length. Right column, the indicated constructs were transiently expressed in Arabidopsis protoplasts and inspected with a confocal microscope. Scale bars indicate 10 µm. g In vitro cell-free degradation assays of GmLHP1-His in protein extracts from GmBTB/POZ-1-OE soybean hairy roots. h GmBTB/POZ-1 promotes the ubiquitination of GmLHP1 in vivo. GmLHP1-Flag was immunoprecipitated using anti-Flag-Tag Mouse mAb (Agarose Conjugated) from GmLHP1-OE and GmLHP1-OE/GmBTB/POZ-1-OE soybean hairy roots by high-efficiency A. rhizogenes-mediated transformation. The transgenic hairy roots were treated with 100 µM MG132 for 8 h before extraction. The immunoprecipitated protein was examined using anti-Flag and anti-ubi antibodies. i GmLHP1 transcript levels in EV, GmLHP1-OE, and GmLHP1-OE/GmBTB/POZ-1-OE transgenic soybean hairy roots. j GmBTB/POZ transcript levels in EV, GmLHP1-OE, and GmLHP1-OE/GmBTB/POZ-1-OE transgenic hairy roots. k GmWRKY40 transcript levels in EV, GmLHP1-OE, and GmLHP1-OE/GmBTB/POZ-1-OE transgenic hairy roots. The housekeeping gene GmEF1 was used as an internal control to normalize the data. The experiment was performed on three biological replicates, each with three technical replicates, and the results were statistically analyzed using Student’s t-test (*P < 0.05, **P < 0.01). Bars indicate the standard deviation of the mean (n = 3).
GmBTB/POZ increases resistance to P. sojae in GmLHP1-OE soybean lines
Since GmBTB/POZ directly interacts with GmLHP1 to induce its degradation and weakens GmLHP1-mediated GmWRKY40 suppression, we investigated whether GmBTB/POZ modifies GmLHP1-regulated P. sojae defense responses by quantifying P. sojae biomass in EV control, GmLHP1-OE, and GmBTB/POZ-OE/GmLHP1-OE transgenic soybean hairy roots at 48 hpi. As expected, P. sojae biomass was significantly (**P < 0.01) higher in GmLHP1-OE hairy roots than in EV hairy roots (Fig. 7e). However, the overexpression of GmBTB/POZ resulted in a significant reduction in P. sojae biomass (Fig. 7e). These results indicate that GmBTB/POZ modulates GmLHP1-mediated P. sojae defense responses in soybean, possibly by regulating the expression of the downstream target gene GmWRKY40.
Regulatory mechanism of GmBTB/POZ to GmLHP1 is independent of exclusive or predominant nuclear localization of GmBTB/POZ
To test whether the nuclear localization of GmBTB/POZ is required for the regulatory mechanism of GmBTB/POZ to GmLHP1, we firstly analyzed the NLS regions of GmBTB/POZ using NLS Mapper software56,57. However, no putative NLS region was identified. We further constructed the GmBTB/POZ deletion mutants, each fused with GFP at its C terminus, and analyzed its subcellular localization (Fig. 7f). Transient expression into Arabidopsis protoplasts showed that the full-length GmBTB/POZ protein (amino acids 1–258) was localized to the nucleus, which has also been demonstrated by Zhang et al.35, while all the GmBTB/POZ deletion mutants (GmBTB/POZ-1 to 7) green fluorescent signal was dispersed in the entire cell of protoplasts similar to that displayed by GFP alone. These results suggested that the integrity of GmBTB/POZ may be required for the nuclear-targeting localization of GmBTB/POZ, the nuclear localization of GmBTB/POZ may not be controlled by a specific region.
Then, we take the deletion mutant GmBTB/POZ-1, in which the nuclear localization has been changed and the protein sequence is the nearest to the full-length GmBTB/POZ protein, to analyze whether the nuclear localization of GmBTB/POZ is required for the ubiquitination-regulatory of GmBTB/POZ to GmLHP1 by in vitro cell-free degradation assay and in vivo ubiquitination assay. The results suggested that GmBTB/POZ-1 could promote the ubiquitination of GmLHP1 in vitro and in vivo (Fig. 7g, h). To further explore whether the change of GmBTB/POZ nuclear localization has an effect on the GmLHP1-mediated suppression of GmWRKY40 expression, we also measured GmWRKY40 transcript levels in EV, GmLHP1-OE, and GmLHP1-OE/GmBTB/POZ-1-OE soybean hairy roots (Fig. 7k), while GmBTB/POZ-1 and GmLHP1 transcript levels were tested to evaluate the efficiency of GmBTB/POZ-1 and GmLHP1 expression (Fig. 7i, j). GmWRKY40 expression was significantly suppressed in the GmLHP1-OE lines, but the effect was inhibited in GmLHP1-OE/GmBTB/POZ-1-OE soybean hairy roots (Fig. 7k), suggesting GmBTB/POZ-1 still can release GmLHP1-mediated suppression of GmWRKY40 expression. Taken together, these results indicated that the ubiquitination-regulatory of GmBTB/POZ to GmLHP1 may be independent of exclusive or predominant nuclear localization of GmBTB/POZ.
We further investigated the expression kinetics of GmBTB/POZ, GmLHP1, GmWRKY40, and GmPR1 in response to P. sojae. As shown in Supplementary Fig. 6, GmBTB/POZ was rapidly induced by P. sojae infection, with transcript levels peaking at 24 h. By contrast, GmLHP1 was downregulated after P. sojae infection and reached a peak within 24 h. GmBTB/POZ and GmLHP1 exhibited the opposite expression patterns in response to P. sojae. GmWRKY40 transcription was not significantly altered during the first 9 h of infection but reached a peak at 48 h. GmPR1 showed the slowest response to P. sojae infection, reaching a peak at 72 h. These findings support the notion that GmBTB/POZ and GmLHP1 play key roles in the response of soybean to P. sojae at both the transcriptional and post-translational levels.
Discussion
Many soybean genes respond to P. sojae infection35,58–62. The characterization of such genes has helped elucidate the genetic mechanisms underlying defense against P. sojae infection61–63. However, knowledge about the regulator components in plant–pathogen interaction model and plant immunity has remained fragmented. In the present study, we demonstrated that GmLHP1 is an important component of the GmBTB/POZ-mediated SA and immune signaling pathway, providing evidence that the linkage between GmBTB/POZ and GmLHP1 is involved in the response of soybean to P. sojae attack.
Protein ubiquitination is a key mechanism that regulates immune responses64. BTB/POZ functions as a Ub ligase by forming a complex with CRL3 (ref. 17). We previously demonstrated that GmBTB/POZ positively regulates disease resistance in plants, which primarily depends on SA signaling35. However, the components involved in GmBTB/POZ-mediated SA and defense response signaling had been unknown. In the current study, we demonstrated that GmBTB/POZ interacts with GmLHP1 in vitro and in vivo. In vitro protein degradation and in vivo ubiquitination assays suggested that GmBTB/POZ contributes to the Ub-mediated degradation of GmLHP1 through the 26S proteasome system (Fig. 2).
In addition to the roles of LHP1 in regulating flowering time and root development33,34,42,65,66, its potential roles in plant responses to abiotic and biotic stress have been receiving increasing attention. LHP1 interacts with different proteins in different cell types to perform distinct functions42,43. In soybean, LHP1 interacts with GmPHD6 to regulate the expression of genes involved in salt tolerance36. Along with the observation that GmBTB/POZ interacts with and ubiquitinates GmLHP1, these findings prompted us to investigate whether GmLHP1 is also involved in the response of soybean to P. sojae infection. In agreement with our speculation, overexpression and RNA interference analysis of transgenic soybean plants and hairy roots revealed that GmLHP1 negatively regulates the defense responses of soybean to P. sojae infection (Fig. 3). We also analyzed the SA content and expression levels of SA-marker gene GmPR1 in GmLHP1OE, WT, and GmLHP1RNAi soybean plants. Compared to WT plants, SA content and GmPR1 transcript levels were significantly lower in GmLHP1-OE plants but higher in GmLHP1-RNAi plants (Fig. 3c, d). Similar results were obtained for GmLHP1-OE and GmLHP1-RNAi transgenic soybean hairy roots (Fig. 3h, i).
SA mediates the plant immune response SAR, a long-lasting, broad-spectrum resistance response to a variety of pathogenic fungi, bacteria, and viruses48,50,67. SAR is characterized by increased endogenous SA levels and the increased expression of PR genes, such as PR1, which are considered to be effector genes for SAR48. Germinating soybean in red light improves resistance to Pseudomonas putida 229 by regulating SA levels and upregulating PR1 (ref. 68). Consistent with this, our findings suggest that GmLHP1 negatively regulates the response of soybean to P. sojae, possibly by suppressing SA levels and GmPR1 gene expression. Our study provides clear evidence for the linkage between a BTB/POZ-mediated ubiquitination pathway and a plant LHP1-associated defense system. Such a linkage has not been previously reported for any plant species.
LHP1 represses the transcription of numerous genes, including FLOWERING LOCUS C (FLC) and the floral organ identity genes AGAMOUS (AG) and APETALA3 (AP3)29,69,70. It has also been reported that GmPHD6 could form a complex with LHP1 to bind to the GAL4 element through BD-GmPHD6 and to activate gene expression in soybean, indicating that LHP1 could also function as the coactivator in transcriptional complex36. However, in the current study, a series of physiological and biochemical assays showed that GmLHP1 could directly target and suppress the expression of GmWRKY40. In a transient expression assay in yeast cells using a GAL4-responsive reporter system, GmLHP1 alone did not activate the transcription of the reporter gene (Supplementary Fig. 4b). RNA-Seq showed that various stress-related genes, including GmWRKY40, were significantly downregulated in GmLHP1OE transgenic soybean plants (Fig. 4a). Furthermore, qRT-PCR analysis indicated that the changes in GmWRKY40 expression were much more pronounced in GmLHP1OE and GmLHP1RNAi vs. WT plants: GmWRKY40 expression was dramatically reduced in GmLHP1OE vs. WT plants, and in GmLHP1RNAi soybean plants, GmWRKY40 expression significantly increased (**P < 0.01) compared to the WT, while none of the other genes showed markedly altered expression (Fig. 4b). A dual effector–reporter system using GmLHP1 as the effector and the luciferase gene under the control of the GmWRKY40 promoter as the reporter, as well as ChIP-qPCR assays, demonstrated that GmLHP1 directly binds to the GmWRKY40 promoter and suppress its expression (Fig. 4c–f).
WRKY family genes are involved in SA signaling pathways. Several WRKY genes are associated with SA biosynthesis; for example, the Arabidopsis wrky54 wrky70 double mutant has strongly increased SA levels71. In addition, several WRKYs are induced by SA and function downstream of SA the biosynthesis pathway. SA induces the rapid expression of WRKY genes in a number of plants72–74. In Arabidopsis, 49 of the 72 WRKY genes examined were differentially regulated in plants after treatment with SA72. In the current study, we determined that GmWRKY40 contains the WRKY domain, a highly conserved structural domain (Supplementary Fig. 7). GmWRKY40 expression was significantly induced by SA, and the amount of histochemical GUS staining in soybean hairy roots under SA treatment was clearly higher relative to mock (H2O) conditions (Fig. 4g, h). Moreover, whereas SA levels in GmWRKY40 transgenic hairy roots were not significantly different from those of the control (Fig. 5e), GmWRKY40 expression enhanced the expression of SA-marker gene GmPR1 (Fig. 5f). These results suggest that GmWRKY40 functions as a SA-induced gene in the SA signaling pathway downstream of SA biosynthesis. We also demonstrated that GmLHP1 participates in the SA signaling pathway and inhibits SA accumulation. Meanwhile, exogenous SA application weakened the inhibition of GmWRKY40 expression in GmLHP1OE soybean plants, suggesting that GmLHP1-mediated suppression of GmWRKY40 expression might also occur via impaired SA accumulation (Fig. 4i, g). These findings indicate that GmWRKY40 is a GmLHP1 target and that at least two types of mechanisms (directly repressed GmWRKY40 expression and impaired SA accumulation) contribute to the regulation of GmWRKY40 expression by GmLHP1.
Specific WRKY transcription factors function in plant defense responses by affecting the expression of PR1 (ref. 75). For instance, Arabidopsis WRKY18 and WRKY70 activate the expression of genes including PR1 and increase resistance to pathogens76,77. Consistent with this finding, in the current study, GmWRKY40 expression enhanced resistance to P. sojae (Fig. 5a–d) and increased the transcript level of GmPR1 (Fig. 5f). Thus, perhaps GmLHP1 represses the expression of GmWRKY40, thereby negatively regulating resistance to P. sojae. Thereinto, the nuclear localization of GmLHP1 is required for the GmLHP1-mediated negative regulation of immunity, SA levels, and the suppression of GmWRKY40 expression (Fig. 6a–f).
More importantly, GmWRKY40 was upregulated in GmBTB/POZ-OE soybean lines and downregulated in GmBTB/POZ-RNAi lines compared to WT plants, indicating that GmBTB/POZ also affects the transcription of GmWRKY40. Analysis of soybean hairy roots co-transformed with GmBTB/POZ and GmLHP1 indicated that GmBTB/POZ released GmLHP1-regulated GmWRKY40 suppression and increased resistance to P. sojae in GmLHP1-OE hairy roots.
Finally, we demonstrated that GmBTB/POZ and GmLHP1 are both involved in regulating P. sojae resistance and GmWRKY40 expression but play opposite roles in this process. Specifically, we propose that GmBTB/POZ and GmLHP1 function together in SA and immune signaling pathways and that GmBTB/POZ recruits and degrades GmLHP1, thereby regulating the expression of downstream target gene GmWRKY40 in soybean. The expressions of GmLHP1 and GmBTB/POZ are inversely regulated during P. sojae infection (Supplementary Fig. 6). These findings strongly suggest that GmBTB/POZ and GmLHP1 play key roles in the response of soybean to P. sojae at both the transcriptional and post-translational levels.
Taken together, based on previous and current findings, we propose a model explaining how the GmBTB/POZ–GmLHP1 complex regulates the response of soybean to P. sojae infection (Fig. 8). According to our model, GmBTB/POZ and GmWRKY40 act as positive regulators, but GmLHP1 acts as a negative regulator, of the response of soybean to P. sojae infection. GmLHP1 functions as an upstream regulator to repress GmWRKY40 expression by directly suppressing its promoter activity and impairing SA accumulation, thus inhibiting plant defense responses. Moreover, P. sojae induces the transcription of GmBTB/POZ, whereas GmLHP1 is downregulated during P. sojae infection. The high levels of GmBTB/POZ recruit and degrade GmLHP1, thereby releasing its suppressive effect on GmWRKY40 expression, thus increasing the defense response to P. sojae. This study provides compelling evidence for the role of the GmBTB/POZ–GmLHP1 complex in modulating the response of soybean to P. sojae infection. Furthermore, it has been previously proved that LHP1 plays a central role in regulating flowering time, and Arabidopsis loss-of-function lhp1 mutants exhibit photoperiod-independent early flowering compared to WT plants23,78. In our study, we also observed that GmLHP1RNAi soybean plants showed early flowering compared with WT plants under artificial long-day condititions (Supplementary Fig. 8). However, whether GmBTB/POZ–GmLHP1 complex is also involved in flowering-regulatory, as well as the underlying genetic and molecular mechanisms still require further exploration.
Fig. 8. Model of the role of GmBTB/POZ, GmLHP1, and GmWRKY40 expression in the response of soybean to Phytophthora sojae infection.
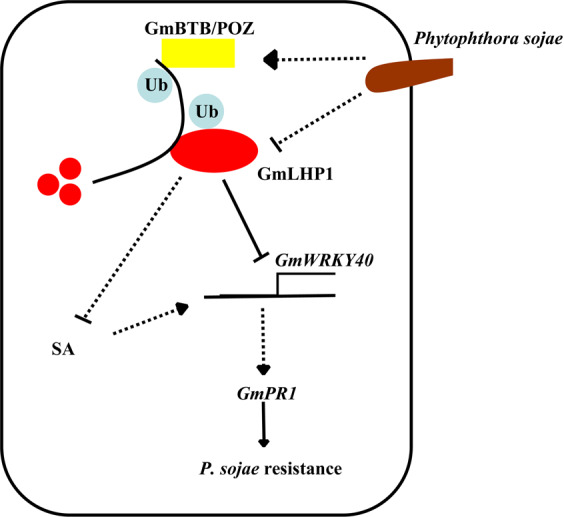
P. sojae induces the transcription of GmBTB/POZ, whereas GmLHP1 is downregulated during P. sojae infection. GmLHP1 functions as an upstream regulator to repress GmWRKY40 expression by directly suppressing its promoter activity and impairing SA accumulation, thus inhibiting plant defense responses. The increased levels of GmBTB/POZ recruit and degrade GmLHP1, thereby releasing the GmLHP1-suppressed expression of GmWRKY40 and thus increasing the defense response to P. sojae.
Methods
Plant materials and pathogen inoculation
“Dongnong 50”, a soybean (Glycine max) cultivar susceptible to P. sojae race 1, was obtained from the Key Laboratory of Soybean Biology in the Chinese Ministry of Education, Harbin, and used for gene transformation experiments and expression analysis. “Suinong 10”, a soybean cultivar with gene-for-gene resistance against P. sojae race 1, the predominant race in Heilongjiang, China79, was used for the gene isolation and gene expression kinetics experiments. The coding sequences (CDS) of GmLHP1 (Glyma.16G079900) and GmWRKY40 (Glyma.15G003300) were amplified by PCR using cDNA derived from leaves of “Suinong 10” soybean as the template. The seeds were grown in a growth chamber at 25 °C and 70% relative humidity under a 16 h light/8 h dark cycle. N. benthamiana plants for the LCI assays and dual-luciferase assays were grown at 22 °C under a 16 h light/8 h dark photoperiod with a light intensity of 120 µE m−2 s−1.
Phytophthora sojae race 1 (PSR01) was isolated from infected soybean plants in Heilongjiang, China79, and cultivated at 25 °C for 7 days on V8 juice agar in a polystyrene dish.
In vitro pull-down assay
To produce the GmLHP1-His fusion protein, the CDS of GmLHP1 was cloned into the pET29b (+) expression vector. The recombinant fusion plasmids were transformed into E. coli Rosetta (DE3) cells. The fusion proteins was purified at 4 °C and quantified according to the pET System Manual. To produce the GmBTB/POZ-GST protein, the CDS of GmBTB/POZ was inserted into the pGEX-4T-1 expression vector and expressed in Rosetta (DE3) cells. The target protein was purified with GST resin (GE Healthcare; 17-0756-01). Pull-down was performed as described by Yang et al.80. The pulled-down proteins were eluted by boiling, separated by 12% SDS-PAGE, and detected by immunoblotting using anti-GST (Abmart, code number M20007S) and anti-His antibodies (Abmart, code number M20001S), respectively.
Firefly LCI assay
The CDS of GmLHP1 and GmBTB/POZ were fused with the N-terminal and C-terminal parts of the luciferase reporter gene, respectively. Agrobacteria harboring the pCAMBIA1300-GmLHP1nLUC and pCAMBIA1300-GmBTB/POZcLUC constructs were co-infiltrated into N. benthamiana leaves, which were subsequently sprayed with luciferin (1 mM luciferin and 0.01% Triton X-100) and photographed using Chemiluminescence imaging (Tanon 5200) at 72 h after infiltration.
BiFC assays and subcellular localization analysis
For interaction studies, the gene sequences were cloned into serial pSAT6 vectors encoding N- or C-terminal-enhanced yellow fluorescent protein fragments. To determine the subcellular localization of target proteins, the target gene sequences were ligated into the pCAMBIA1302 vector under the control of the 35S promoter, generating the recombinant plasmid. The resulting constructs were used for transient assays via PEG transfection of Arabidopsis protoplasts as described by Yoo et al.81. Transfected cells were imaged using a TCS SP2 confocal spectral microscope imaging system (Leica, Solms, Germany).
In vitro cell-free degradation assays
Total proteins were extracted from WT and transgenic soybean lines with degradation buffer82. Each reaction contained 500 µg of soybean total proteins and 100 ng of GmLHP1-His proteins purified from E. coli Rosetta (DE3) cells. For the proteasome inhibitor experiments, 100 µM MG132 was added to the total proteins 60 min prior to the cell-free degradation experiment. The reactions were incubated at 22 °C. The mixed solutions were collected at the designated time point (0, 0.5, 1, and 3 h) and examined using an anti-His antibody (Abmart, code number M20001S). The quantified results were analyzed using ImageJ software (https://imagej.nih.gov/ij/index.html).
In vivo ubiquitination assay
To detect ubiquitination of GmLHP1 in vivo, a plant binary expression vector system was constructed and used to generate GmBTB/POZ-OE, GmLHP1-OE/GmBTB/POZ-OE, GmLHP1-OE/(domain+C)-OE, GmLHP1-OE/(N+domain)-OE, or GmLHP1-OE/GmBTB/POZ-1-OE transgenic soybean hairy roots by high-efficiency A. rhizogenes-mediated transformation. The transgenic hairy roots were treated with 100 µM MG132 for 8 h prior to protein extraction. GmLHP1-Flag protein was immunoprecipitated using anti-Flag-Tag Mouse mAb (Agarose Conjugated) (Abmart, code number M20018S). The eluted proteins were detected using anti-Flag antibody (Abmart, code number M20008M) and anti-Ubi antibody (Abcam, code number ab19169).
Plasmid construction and genetic transformation of soybean
To produce the GmLHP1 overexpression and GmLHP1-Flag fusion constructs, the CDS of GmLHP1 and Flag sequence (ATGGACTACAAGGATGACGATGACAAG) were cloned into the pCAMBIA3301 vector with the bar gene (as the selectable marker) and Flag tag under the control of the cauliflower mosaic virus 35S (CaMV35S) promoter to overexpress GmLHP1. To suppress GmLHP1 expression, the cDNA fragment of GmLHP1 was amplified and inserted into vector pFGC5941 (ref. 83). The p35S: Flag-GmLHP1 and p35S: GmLHP1-RNAi recombinant plasmids were transferred into Agrobacterium tumefaciens strain LBA4404 via the freeze–thaw method as described by Holsters et al.84. “Dongnong 50” soybean was used for the gene transformation experiments, and transgenic soybean plants expressing p35S: Flag-GmLHP1 and p35S: GmLHP1-RNAi were generated by Agrobacterium-mediated transformation using the cotyledonary node method85. All primers used for genotyping and vector construction are listed in Supplementary Table 1.
Agrobacterium rhizogenes-mediated transformation of soybean hairy roots
To construct the p35S: Flag-GmWRKY40 overexpression vector, the CDS of GmWRKY40 was cloned into plant expression vector pCAMBIA3301 with Flag tag as the selectable marker. To construct the GmWRKY40 RNAi vector, the cDNA fragment of GmWRKY40 was amplified and inserted into vector pFGC5941 (ref. 83). Transgenic soybean hairy roots were generated by A. rhizogenes-mediated transformation as described by Graham et al.44 and Kereszt et al.45, with some modifications.
Assessment of soybean disease responses and SA levels
For phenotypic analysis of the response of soybean to P. sojae infection, artificial inoculation was performed as described by Dou et al.86 and Ward et al.87 with minor modifications. Soybean roots and hairy roots were inoculated with P. sojae zoospores (approximately 1 × 105 spores mL−1). Disease symptoms on each root were observed after inoculation and photographed with a Nikon B7000 camera. SA levels were determined as described by Aboul et al.88 and Pan et al.89.
RNA-Seq analysis
Three independent GmLHP1-OE transgenic soybean plants and three WT “Dongnong 50” plants grown for 6 weeks in the field under non-stress conditions were used for RNA-Seq analysis. Sequencing libraries were generated using a NEB Next Ultra RNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s recommendations, and index codes were added to each sample. After cluster analysis, the RNA samples were sequenced on the Illumina HiSeq 2500 platform to generate paired-end reads. Total reads were mapped to the soybean genome using TopHat software. Read counts for each gene were generated using HTSeq in union mode. DEGs between samples were defined by DESeq using two separate models90, based on fold change >2 and FDR-adjusted P value < 0.05. GO enrichment analysis of the DEGs was performed using the GOseq R packages based on Wallenius noncentral hypergeometric distribution91, which adjusts for gene length bias in DEGs.
ChIP assay
For the ChIP assays, WT and p35S: Flag-GmLHP1 transgenic plants were subjected to chromatin extraction and immunoprecipitation as described by Saleh et al.92. Briefly, the leaves from 30-day-old plants were harvested for fixation. Nuclei were isolated and sonicated to generate DNA fragments with an average size of 500 bp. The soluble chromatin fragments were isolated and pre-absorbed with 30 μL of anti-Flag-Tag Mouse mAb (Agarose Conjugated) (Abmart, code number M20018S) to eliminate nonspecific binding and immunoprecipitated by 30 μL of anti-Flag-Tag Mouse mAb (Agarose Conjugated) (Abmart, code number M20018S). The precipitated DNA was recovered and analyzed by quantitative PCR with SYBR Premix ExTaq Mix (Takara, Japan). The ChIP-qPCR results are reported as relative binding units (IP/Input). The primers used are listed in Supplementary Table 1.
Transient transcription dual-luciferase assay
The promoter region of GmWRKY40 was amplified, cloned into the pGreenII 0800-LUC vector, and used as a reporter93. p35S: Flag-GmLHP1 was used as an effector construct. The effector and reporter constructs were cotransfected into healthy leaves of 21-day-old N. benthamiana plants by agroinfiltration94. The plants were incubated under continuous white light for 3 days after infiltration, sprayed with luciferin (1 mM luciferin and 0.01% Triton X-100), and photographed using Chemiluminescence imaging (Tanon 5200) at 72 h after infiltration. Leaf samples were collected for the dual-luciferase assay using a commercial kit (Promega; PR-E1910). Firefly luciferase (LUC) and Renilla luciferase (REN) activities were measured in the samples. The REN gene driven by the 35S promoter in the pGreenII 0800-LUC vector was used as an internal control. LUC activity was normalized to REN activity, and LUC/REN ratios were calculated. The data presented are the averages of at least three independent replicates.
Statistics and reproducibility
All statistical methods are annotated in the figure captions. The numbers of biological replicates in each assays are also indicated in the figure captions. The experiment was performed on three biological replicates, each with three technical replicates, and the results were statistically analyzed using Student’s t-test. A difference was considered to be statistically significant when *P < 0.05 or **P < 0.01. Bars indicate the standard deviation of the mean.
Reporting summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this paper.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by NSFC Projects (31671719, 31971972), Natural Science Foundation of Heilongjiang Province (ZD2019C001) and Outstanding Talents and Innovative Team of Agricultural Scientific Research.
Author contributions
S.Z., P.X., and C.Z. designed the experiments. C.Z., Q.C., H.W., and H.G. performed the experiments. C.Z., X.F., X.C., M.Z., W.W., B.S., S.L., and J.W. analyzed the data. S.Z., P.X., and C.Z. wrote the manuscript. All of the authors read and approved the final manuscript.
Data availability
All data supporting the findings of this study are available in the main text and its Supplementary Information. All the source data for graphs in Figures and Supplementary Information are presented in Supplementary Data 1 and 2. Raw images of the western blots are provided in Supplementary Fig. 9. Raw RNA sequencing data are available at the NCBI Sequence ReadArchive (SRA) under accession PRJNA702619. Gene sequences, involved in this study, were obtained from Phytozome (https://phytozome.jgi.doe.gov/). The accession numbers of genes are as follows: GmLHP1 (Glyma.16G079900), GmBTB/POZ (Glyma.04G244900), GmMEKK2 (Glyma.17G173000), GmWRKY40 (Glyma.15G003300), GmCPK2 (Glyma.11G206300), GmNAC90 (Glyma.11G182000), GmNAC29 (Glyma.02G109800), GmERF104 (Glyma.20G070000), GmbHLH35 (Glyma.13G101100), GmMYB70 (Glyma.17G237900), and GmMLP34 (Glyma.09G102400).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chuanzhong Zhang, Qun Cheng, Huiyu Wang.
Contributor Information
Shuzhen Zhang, Email: zhangshuzhen@neau.edu.cn.
Pengfei Xu, Email: xupengfei@neau.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-01907-7.
References
- 1.Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 2.Schneider DS. Plant immunity and film Noir: what gumshoe detectives can teach us about plant-pathogen interactions. Cell. 2002;109:537–540. doi: 10.1016/S0092-8674(02)00764-X. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 4.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Moore JW, Loake GJ, Spoel SH. Transcription dynamics in plant immunity. Plant Cell. 2011;23:2809–2820. doi: 10.1105/tpc.111.087346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandran D, et al. Atypical E2F transcriptional repressor DEL1 acts at the intersection of plant growth and immunity by controlling the hormone salicylic acid. Cell Host Microbe. 2014;15:506–513. doi: 10.1016/j.chom.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Cui H, Tsuda K, Parker JE. Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015;66:487–511. doi: 10.1146/annurev-arplant-050213-040012. [DOI] [PubMed] [Google Scholar]
- 8.Duplan V, Rivas S. E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front. Plant Sci. 2014;5:6. doi: 10.3389/fpls.2014.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 10.Moon J, Parry G, Estelle M. The ubiquitin-proteasome pathway and plant development. Plant Cell. 2004;16:3181–3195. doi: 10.1105/tpc.104.161220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009;10:385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 12.Oyake T, et al. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell. Biol. 1996;16:6083–6095. doi: 10.1128/MCB.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins T, Stone JR, Williams AJ. All in the family: the BTB/POZ, KRAB, and SCAN domains. Mol. Cell. Biol. 2001;21:3609–3615. doi: 10.1128/MCB.21.11.3609-3615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geyer R, Wee S, Anderson S, Yates J, Wolf DA. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell. 2003;12:783–790. doi: 10.1016/S1097-2765(03)00341-1. [DOI] [PubMed] [Google Scholar]
- 15.Pintard L, Willems A, Peter M. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 2004;23:1681–1687. doi: 10.1038/sj.emboj.7600186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figueroa P, et al. Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell. 2005;17:1180–1195. doi: 10.1105/tpc.105.031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua, Z. H. & Vierstra, R. D. in Annual Review Of Plant Biology Vol. 62 (eds Merchant, S. S. et al.) 299–334 (Annual Reviews, 2011). [DOI] [PubMed]
- 18.James TC, Elgin SC. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol. 1986;6:3862–3872. doi: 10.1128/MCB.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eissenberg JC, Elgin SC. The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 2000;10:204–210. doi: 10.1016/S0959-437X(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 20.Minc E, Allory Y, Worman HJ, Courvalin JC, Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 1999;108:220–234. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- 21.Gaudin V, et al. Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development. 2001;128:4847–4858. doi: 10.1242/dev.128.23.4847. [DOI] [PubMed] [Google Scholar]
- 22.Larsson AS, Landberg K, Meeks-Wagner DR. The TERMINAL FLOWER2 (TFL2) gene controls the reproductive transition and meristem identity in Arabidopsis thaliana. Genetics. 1998;149:597–605. doi: 10.1093/genetics/149.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotake T, Takada S, Nakahigashi K, Ohto M, Goto K. Arabidopsis TERMINAL FLOWER 2 gene encodes a heterochromatin protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol. 2003;44:555–564. doi: 10.1093/pcp/pcg091. [DOI] [PubMed] [Google Scholar]
- 24.Zemach A, et al. Different domains control the localization and mobility of LIKE HETEROCHROMATIN PROTEIN1 in Arabidopsis nuclei. Plant Cell. 2006;18:133–145. doi: 10.1105/tpc.105.036855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mimida N, Kidou SI, Kotoda N. Constitutive expression of two apple (Malus x domestica Borkh.) homolog genes of LIKE HETEROCHROMATIN PROTEIN1 affects flowering time and whole-plant growth in transgenic Arabidopsis. Mol. Genet. Genom. 2007;278:295–305. doi: 10.1007/s00438-007-0250-0. [DOI] [PubMed] [Google Scholar]
- 26.Hennig L, Derkacheva M. Diversity of polycomb group complexes in plants: same rules, different players? Trends Genet. 2009;25:414–423. doi: 10.1016/j.tig.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Valdes AE, et al. Arabidopsis thaliana TERMINAL FLOWER2 is involved in light-controlled signalling during seedling photomorphogenesis. Plant Cell Environ. 2012;35:1013–1025. doi: 10.1111/j.1365-3040.2011.02468.x. [DOI] [PubMed] [Google Scholar]
- 28.Guo L, et al. A chromatin loop represses WUSCHEL expression in Arabidopsis. Plant J. 2018;94:1083–1097. doi: 10.1111/tpj.13921. [DOI] [PubMed] [Google Scholar]
- 29.Mylne JS, et al. LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc. Natl Acad. Sci. USA. 2006;103:5012–5017. doi: 10.1073/pnas.0507427103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rizzardi K, Landberg K, Nilsson L, Ljung K, Sundas-Larsson A. TFL2/LHP1 is involved in auxin biosynthesis through positive regulation of YUCCA genes. Plant J. 2011;65:897–906. doi: 10.1111/j.1365-313X.2010.04470.x. [DOI] [PubMed] [Google Scholar]
- 31.Sung SB, et al. Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat. Genet. 2006;38:706–710. doi: 10.1038/ng1795. [DOI] [PubMed] [Google Scholar]
- 32.Derkacheva M, et al. Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J. 2013;32:2073–2085. doi: 10.1038/emboj.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turck F, et al. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007;3:855–866. doi: 10.1371/journal.pgen.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang YZ, Gu XF, Yuan WY, Schmitz RJ, He YH. Photoperiodic control of the floral transition through a distinct polycomb repressive complex. Dev. Cell. 2014;28:727–736. doi: 10.1016/j.devcel.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 35.Zhang CZ, et al. GmBTB/POZ, a novel BTB/POZ domain-containing nuclear protein, positively regulates the response of soybean to Phytophthora sojae infection. Mol. Plant Pathol. 2019;20:78–91. doi: 10.1111/mpp.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei W, et al. A histone code reader and a transcriptional activator interact to regulate genes for salt tolerance. Plant Physiol. 2017;175:1304–1320. doi: 10.1104/pp.16.01764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.An JP, et al. BTB protein MdBT2 inhibits anthocyanin and proanthocyanidin biosynthesis by triggering MdMYB9 degradation in apple. Tree Physiol. 2018;38:1578–1587. doi: 10.1093/treephys/tpy063. [DOI] [PubMed] [Google Scholar]
- 38.Stogios, P. J., Downs, G. S., Jauhal, J. J. S., Nandra, S. K. & Prive, G. G. Sequence and structural analysis of BTB domain proteins. Genome Biol. 6, R82 (2005). [DOI] [PMC free article] [PubMed]
- 39.Zimmerman ES, Schulman BA, Zheng N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 2010;20:714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber H, et al. Arabidopsis AtCUL3a and AtCUL3b form complexes with members of the BTB/POZ-MATH protein family. Plant Physiol. 2005;137:83–93. doi: 10.1104/pp.104.052654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Veluchamy A, et al. LHP1 regulates H3K27me3 spreading and shapes the three-dimensional conformation of the Arabidopsis genome. PLoS ONE. 2016;11:25. doi: 10.1371/journal.pone.0158936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li ZF, et al. Transcription factors AS1 and AS2 interact with LHP1 to repress KNOX genes in Arabidopsis. J. Integr. Plant Biol. 2016;58:959–970. doi: 10.1111/jipb.12485. [DOI] [PubMed] [Google Scholar]
- 43.Feng J, Lu J. LHP1 could act as an activator and a repressor of transcription in plants. Front. Plant Sci. 2017;8:7. doi: 10.3389/fpls.2017.02041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graham TL, Graham MY, Subramanian S, Yu O. RNAi silencing of genes for elicitation or biosynthesis of 5-deoxyisoflavonoids suppresses race-specific resistance and hypersensitive cell death in Pytophthora sojae infected tissues. Plant Physiol. 2007;144:728–740. doi: 10.1104/pp.107.097865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kereszt A, et al. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2007;2:948–952. doi: 10.1038/nprot.2007.141. [DOI] [PubMed] [Google Scholar]
- 46.Moon JY, Park JM. Cross-talk in viral defense signaling in plants. Front. Microbiol. 2016;7:6. doi: 10.3389/fmicb.2016.02068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong X. SA, JA, ethylene, and disease resistance in plants. Curr. Opin. Plant Biol. 1998;1:316–323. doi: 10.1016/1369-5266(88)80053-0. [DOI] [PubMed] [Google Scholar]
- 48.Durrant WE, Dong X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 49.Malamy J, Carr JP, Klessig DF, Raskin I. Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science. 1990;250:1002–1004. doi: 10.1126/science.250.4983.1002. [DOI] [PubMed] [Google Scholar]
- 50.Sanchez L, et al. Rhamnolipids elicit defense responses and induce disease resistance against biotrophic, hemibiotrophic, and necrotrophic pathogens that require different signaling pathways in Arabidopsis and highlight a central role for salicylic acid. Plant Physiol. 2012;160:1630–1641. doi: 10.1104/pp.112.201913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Libault M, et al. The Arabidopsis LHP1 protein is a component of euchromatin. Planta. 2005;222:910–925. doi: 10.1007/s00425-005-0129-4. [DOI] [PubMed] [Google Scholar]
- 52.Hu RB, Fan CM, Li HY, Zhang QZ, Fu YF. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol. Biol. 2009;10:12. doi: 10.1186/1471-2199-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hwang SH, Yie SW, Hwang DJ. Heterologous expression of OsWRKY6 gene in Arabidopsis activates the expression of defense related genes and enhances resistance to pathogens. Plant Sci. 2011;181:316–323. doi: 10.1016/j.plantsci.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 54.Chen C, Chen Z. Isolation and characterization of two pathogen- and salicylic acid-induced genes encoding WRKY DNA-binding proteins from tobacco. Plant Mol. Biol. 2000;42:387–396. doi: 10.1023/A:1006399311615. [DOI] [PubMed] [Google Scholar]
- 55.Duan Y, et al. PtrWRKY73, a salicylic acid-inducible poplar WRKY transcription factor, is involved in disease resistance in Arabidopsis thaliana. Plant Cell Rep. 2015;34:831–841. doi: 10.1007/s00299-015-1745-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kosugi S, et al. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J. Biol. Chem. 2009;284:478–485. doi: 10.1074/jbc.M807017200. [DOI] [PubMed] [Google Scholar]
- 57.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl Acad. Sci. USA. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu PF, et al. Isolation and characterization of a pathogenesis-related protein 10 gene (GmPR10) with induced expression in soybean (Glycine max) during infection with Phytophthora sojae. Mol. Biol. Rep. 2014;41:4899–4909. doi: 10.1007/s11033-014-3356-6. [DOI] [PubMed] [Google Scholar]
- 59.Dong LD, et al. Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J. Exp. Bot. 2015;66:2635–2647. doi: 10.1093/jxb/erv078. [DOI] [PubMed] [Google Scholar]
- 60.Kong GH, et al. The activation of Phytophthora effector Avr3b by plant cyclophilin is required for the nudix hydrolase activity of Avr3b. PLoS Pathog. 2015;11:22. doi: 10.1371/journal.ppat.1005139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jing MF, et al. A Phytophthora sojae effector suppresses endoplasmic reticulum stress-mediated immunity by stabilizing plant binding immunoglobulin proteins. Nat. Commun. 2016;7:17. doi: 10.1038/ncomms11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng Q, et al. The bHLH transcription factor GmPIB1 facilitates resistance to Phytophthora sojae in Glycine max. J. Exp. Bot. 2018;69:2527–2541. doi: 10.1093/jxb/ery103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yan Q, et al. GmCYP82A3, a soybean cytochrome P450 family gene involved in the jasmonic acid and ethylene signaling pathway, enhances plant resistance to biotic and abiotic stresses. PLoS ONE. 2016;11:18. doi: 10.1371/journal.pone.0162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu YF, et al. E3 ubiquitin ligase gene CMPG1-V from Haynaldia villosa L. contributes to powdery mildew resistance in common wheat (Triticum aestivum L.) Plant J. 2015;84:154–168. doi: 10.1111/tpj.12966. [DOI] [PubMed] [Google Scholar]
- 65.Cui HC, Benfey PN. Interplay between SCARECROW, GA and LIKE HETEROCHROMATIN PROTEIN 1 in ground tissue patterning in the Arabidopsis root. Plant J. 2009;58:1016–1027. doi: 10.1111/j.1365-313X.2009.03839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paquette AJ, Benfey PN. Maturation of the ground tissue of the root is regulated by gibberellin and SCARECROW and requires SHORT-ROOT. Plant Physiol. 2005;138:636–640. doi: 10.1104/pp.104.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomma BP, et al. The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 2001;13:63–68. doi: 10.1016/S0952-7915(00)00183-7. [DOI] [PubMed] [Google Scholar]
- 68.Dhakal R, et al. Soybean (Glycine max L. Merr.) sprouts germinated under red light irradiation induce disease resistance against bacterial rotting disease. PLoS ONE. 2015;10:14. doi: 10.1371/journal.pone.0117712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nakahigashi K, et al. The Arabidopsis HETEROCHROMATIN PROTEIN1 homolog (TERMINAL FLOWER2) silences genes within the euchromatic region but not genes positioned in heterochromatin. Plant Cell Physiol. 2005;46:1747–1756. doi: 10.1093/pcp/pci195. [DOI] [PubMed] [Google Scholar]
- 70.Gomez-Mena C, et al. Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development. 2005;132:429–438. doi: 10.1242/dev.01600. [DOI] [PubMed] [Google Scholar]
- 71.Li J, et al. Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 2013;200:455–472. doi: 10.1111/nph.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dong JX, et al. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 2003;51:21–37. doi: 10.1023/A:1020780022549. [DOI] [PubMed] [Google Scholar]
- 73.Kalde M, et al. Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol. Plant Microbe Interact. 2003;16:295–305. doi: 10.1094/MPMI.2003.16.4.295. [DOI] [PubMed] [Google Scholar]
- 74.Turck F, Zhou A, Somssich IE. Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to its native promoter and the defense-related gene PcPR1–1 in parsley. Plant Cell. 2004;16:2573–2585. doi: 10.1105/tpc.104.024810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ulker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 2004;7:491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 76.Chen C, Chen Z. Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 2002;129:706–716. doi: 10.1104/pp.001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li J, Brader G, Palva ET. The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 2004;16:319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Adrian J, et al. cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell. 2010;22:1425–1440. doi: 10.1105/tpc.110.074682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang SZ, et al. Races of Phytophthora sojae and their virulences on soybean cultivars in Heilongjiang, China. Plant Dis. 2010;94:87–91. doi: 10.1094/PDIS-94-1-0087. [DOI] [PubMed] [Google Scholar]
- 80.Yang L, et al. CCAAT/enhancer-binding protein alpha antagonizes transcriptional activity of hypoxia-inducible factor 1 alpha with direct protein-protein interaction. Carcinogenesis. 2008;29:291–298. doi: 10.1093/carcin/bgm262. [DOI] [PubMed] [Google Scholar]
- 81.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 82.Wang F, et al. Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell. 2009;21:2378–2390. doi: 10.1105/tpc.108.065433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kerschen A, Napoli CA, Jorgensen RA, Muller AE. Effectiveness of RNA interference in transgenic plants. FEBS Lett. 2004;566:223–228. doi: 10.1016/j.febslet.2004.04.043. [DOI] [PubMed] [Google Scholar]
- 84.Holsters M, et al. Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 1978;163:181–187. doi: 10.1007/BF00267408. [DOI] [PubMed] [Google Scholar]
- 85.Paz MM, et al. Assessment of conditions affecting Agrobacterium-mediated soybean transformation using the cotyledonary node explant. Euphytica. 2004;136:167–179. doi: 10.1023/B:EUPH.0000030670.36730.a4. [DOI] [Google Scholar]
- 86.Dou D, et al. Transgenic tobacco with NDR1 gene improved its resistance to two fungal diseases. Sci. Agric. Sin. 2003;36:1120–1124. [Google Scholar]
- 87.Ward EWB. Hypocotyl reactions and glyceollin in soybeans inoculated with zoospores of Phytophthora megasperma var. sojae. Phytopathology. 1979;69:951–955. doi: 10.1094/Phyto-69-951. [DOI] [Google Scholar]
- 88.Aboul-Soud MAM, Cook K, Loake GJ. Measurement of salicylic acid by a high-performance liquid chromatography procedure based on ion-exchange. Chromatographia. 2004;59:129–133. [Google Scholar]
- 89.Pan XQ, Welti R, Wang XM. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protoc. 2010;5:986–992. doi: 10.1038/nprot.2010.37. [DOI] [PubMed] [Google Scholar]
- 90.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:12. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:12. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Saleh A, Alvarez-Venegas R, Avramova Z. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 2008;3:1018–1025. doi: 10.1038/nprot.2008.66. [DOI] [PubMed] [Google Scholar]
- 93.Hellens RP, et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1:14. doi: 10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meng XZ, et al. Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell. 2013;25:1126–1142. doi: 10.1105/tpc.112.109074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All data supporting the findings of this study are available in the main text and its Supplementary Information. All the source data for graphs in Figures and Supplementary Information are presented in Supplementary Data 1 and 2. Raw images of the western blots are provided in Supplementary Fig. 9. Raw RNA sequencing data are available at the NCBI Sequence ReadArchive (SRA) under accession PRJNA702619. Gene sequences, involved in this study, were obtained from Phytozome (https://phytozome.jgi.doe.gov/). The accession numbers of genes are as follows: GmLHP1 (Glyma.16G079900), GmBTB/POZ (Glyma.04G244900), GmMEKK2 (Glyma.17G173000), GmWRKY40 (Glyma.15G003300), GmCPK2 (Glyma.11G206300), GmNAC90 (Glyma.11G182000), GmNAC29 (Glyma.02G109800), GmERF104 (Glyma.20G070000), GmbHLH35 (Glyma.13G101100), GmMYB70 (Glyma.17G237900), and GmMLP34 (Glyma.09G102400).