Abstract
Human brains develop across the life span and largely vary in morphology. Adolescent collision-sport athletes undergo repetitive head impacts over years of practices and competitions, and therefore may exhibit a neuroanatomical trajectory different from healthy adolescents in general. However, an unbiased brain atlas targeting these individuals does not exist. Although standardized brain atlases facilitate spatial normalization and voxel-wise analysis at the group level, when the underlying neuroanatomy does not represent the study population, greater biases and errors can be introduced during spatial normalization, confounding subsequent voxel-wise analysis and statistical findings. In this work, targeting early-to-middle adolescent (EMA, ages 13–19) collision-sport athletes, we developed population-specific brain atlases that include templates (T1-weighted and diffusion tensor magnetic resonance imaging) and semantic labels (cortical and white matter parcellations). Compared to standardized adult or age-appropriate templates, our templates better characterized the neuroanatomy of the EMA collision-sport athletes, reduced biases introduced during spatial normalization, and exhibited higher sensitivity in diffusion tensor imaging analysis. In summary, these results suggest the population-specific brain atlases are more appropriate towards reproducible and meaningful statistical results, which better clarify mechanisms of traumatic brain injury and monitor brain health for EMA collision-sport athletes.
Subject terms: Computational neuroscience, Data integration, Databases, Image processing, Statistical methods
Introduction
Adolescent collision-sport (e.g., American football and soccer) athletes bear high risk of mild traumatic brain injury (mTBI), a complex pathophysiological process that can arise from either single or repetitive head acceleration events1–4. The lack of sensitive biomarkers hinders the development of preventive strategies, allowing this vulnerable population to continue participating at greater risk. Multi-modal magnetic resonance imaging (MRI) can non-invasively characterize the structure and function of the human brain in healthy and disease states, thus showing promise for prospective screening and early detection of mTBI in adolescent athletes. Nevertheless, one of the critical steps in MRI processing is to spatially normalize brain images to a stereotaxic atlas, i.e., a coordinate reference system for neuroimaging studies. When the spatial normalization onto an atlas has poor accuracy, voxel-based analysis exhibits low sensitivity in detecting differences at the group level5, 6. Therefore, it is vital to ensure most of the anatomical identities pertinent to adolescent athletes are retained during spatial normalization.
In general, human brain atlases are either standardized or population-specific; each comes with a set of templates (representative spatial maps) and labels (parcellated regions). The two standardized brain atlases long established and well known by the neuroimaging community come from Talairach and the Montreal Neurological Institute7. The Talairach atlas is derived from the dissection of one hemisphere of the brain from a 60-year-old French woman8, whereas the ICBM152 template is derived from T1 scans of 152 subjects aged 18.5–43.5, averaged together after high-dimensional linear and nonlinear registration into the Talairach space9. In the same space as ICBM152, FMRIB58 is a standardized diffusion tensor imaging (DTI) template derived from 58 high-resolution volumes of fractional anisotropy (FA) from healthy male and female adults aged 20–50 (FMRIB, Oxford, UK). Other popular standardized human brain atlases include Brainnetome10, IIT11, SRI2412, etc. Recently, two systematic evaluations of DTI templates showed that the IIT standard template outperformed population-specific DTI templates11, 13, but the findings were based on healthy adults and may not generalize in younger populations. Many scientific publications pointed out the age-related changes in volumes of gray and white matter14–19. Although there are several age-specific atlases for adolescents20–25, the number is limited compared to adult atlases14.
The existing brain atlases are handy tools for various types of neuroimaging analyses, but considering the various pathological conditions and the developing nature of human brain, they often are not best suited for studying specific populations. In a multiple sclerosis population, Van Hecke et al.26 showed that choosing a non-specific template can negatively impact the final results of tract-based spatial statistics (TBSS)27, one of the standard approaches for DTI analysis5, 28. Using both simulated and real DTI data, Van Hecke et al. observed that a population-specific DTI template resulted in more reliable voxel-based analysis, as well as higher sensitivity and specificity of detecting DTI changes, compared to the standardized template26. However, developing a study-specific template from a large population is time consuming, computationally inefficient28, and may result in suboptimal quality11, 26, making the use of existing brain atlases a more pragmatic option for the time being.
To date, an unbiased brain atlas targeting adolescent collision-sport athletes does not exist, to the best of our knowledge. DTI literature of sports-related mTBI and repetitive head impacts in adolescents either manually defined their own regions of interest (ROIs)29–31, or more often did not employ a population-specific template to spatially normalize each individual brain image32–44. This may confound statistical analyses and contribute to varied DTI findings that make it difficult to interpret axonal pathology45. It is critical for studies of mTBI and repetitive head impacts, especially for adolescent collision-sport athletes, to minimize bias and errors in each pre-processing stage, because the magnitudes of changes are often subtle, and brains of this age bracket are rapidly growing46–49. Such studies may benefit from using an unbiased brain atlas created from their study cohort, as opposed to normalizing brains of adolescent collision-sport athletes to an atlas generated from adults or healthy adolescents.
Therefore, the purpose of this work is to develop population-specific brain atlases for early-to-middle adolescent (EMA) collision-sport athletes. Based on the Purdue Neurotrauma Group (PNG) longitudinal MRI database50, we aim to develop:
One T1 template, based on the images from 215 EMA collision-sport athletes,
T1-based semantic labels of cortical and white matter parcellations, and
One DTI template, based on 64 EMA football athletes in a single competition season51, 52.
Regarding evaluation of the templates, our hypothesis is that compared to using a non-specific template, the PNG templates can reduce potential bias when normalizing brain images of local adolescent athletes and improve the statistical power in detecting small differences in population studies using local datasets. The evaluation includes voxel-based morphometry that characterizes the extent of shape changes of the T1 images during spatial normalization, and sensitivity of detecting longitudinal DTI changes in high school football athletes over a single season, which has been reported in previous work51, 52 that utilized the standardized FMRIB58 template. The brain atlases have been made available for download53.
Results
Atlas construction
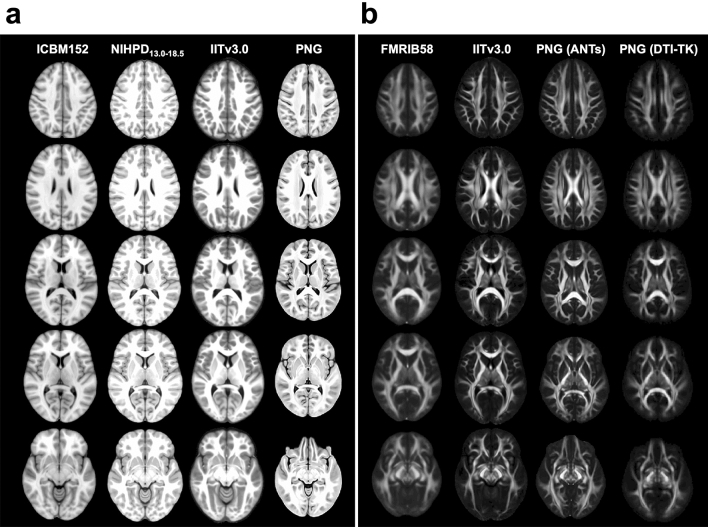
The total computation time for constructing the PNG T1 template was about 28.5 h, where ~ 6.5 h accounted for creating each individual template at the Open Science Grid, and 22 h for creating the final T1 template when fully using one node (24 cores) of the high-performance computing clusters. The comparison of shape and size between the standardized and PNG T1 templates is shown in Fig. 1a.
Figure 1.
Visual illustration of the differences in shape and size between the standardized and population-specific templates, including (a) the standardized ICBM152 (ages 18.5–43.5)9, NIHPD13.0–18.521, IITv3.011, and PNG T1 templates; (b) the standardized FMRIB58 (FMRIB, Oxford, UK), IITv3.011, and PNG DTI templates constructed by ANTs57 and DTI-TK95.
The total computation time of constructing the PNG (ANTs) DTI template was about 12.5 h; warping each of the Pre FA images (N = 33, with qualified T1) to the space of the PNG T1 template took ~ 2 min at the Open Science Grid, and the majority of time was spent on constructing the final DTI template when fully using one node (24 cores). The comparison of shape and size between the standardized and PNG DTI templates is shown in Fig. 1b.
Evaluation of the population-specific T1 template
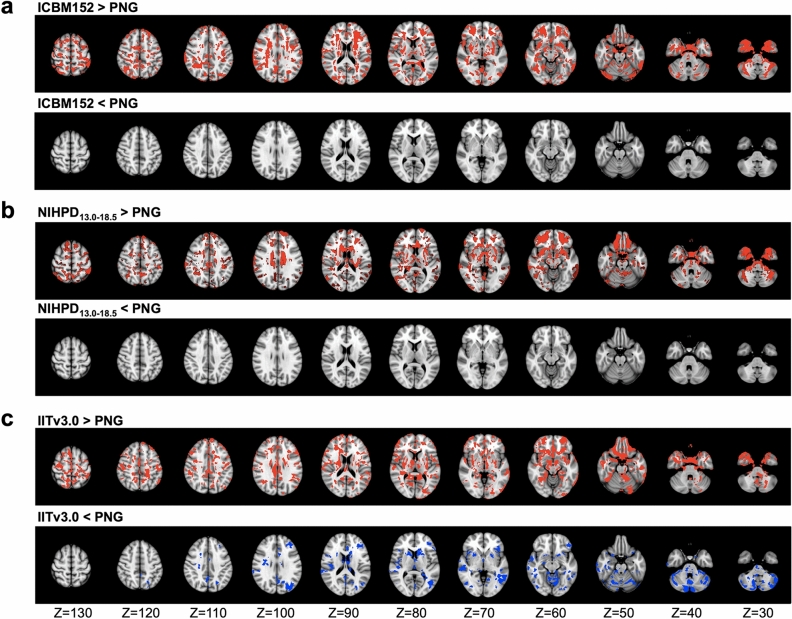
The results of deformation-based morphometry analyses are shown in Fig. 2. Compared to the ICBM152 (Fig. 2a) or NIHPD13.0–18.5 template (Fig. 2b), no significantly larger |logJ| was produced from using the PNG template for the spatial normalization. Compared to IITv3.0 (Fig. 2c), fewer voxels showed significantly larger |logJ| when using the PNG template (IITv3.0: 334,811 voxels; PNG: 109,189 voxels).
Figure 2.
Voxel-wise t-statistical maps (p < 0.05, FWE corrected) of the deformation (|logJ|) of the newly scanned subjects, compared between the PNG T1 template and (a) ICBM152 (ages 18.5–43.5)9, (b) NIHPD13.0–18.521, and (c) IITv3.011, represented in axial view in ICBM152 space. Red/blue indicates significantly larger/smaller changes of morphology during spatial normalization, compared to using the PNG template.
Evaluation of the study-specific DTI template
Similar to the deformation-based morphometry findings for T1 templates, at all the sessions (Pre, In1, In2), fewer voxels showed significantly larger |logJ| when using the PNG (ANTs) template compared to the high-quality standardized DTI templates (FMRIB58 and IITv3.0) (Table 1). Using the PNG (ANTs) template also exhibited fewer voxels of significantly larger |logJ| than the PNG (DTI-TK) template (Table 1). Illustrations of the voxel-wise t-statistical maps are provided in Supplementary Fig. S1 online.
Table 1.
Number of statistically significant (p < 0.05, FWE corrected) voxels of potential bias when normalizing the FA maps of 64 high school varsity football athletes to the PNG (ANTs) DTI template and to the other templates.
| Contrast | Pre | In1 | In2 |
|---|---|---|---|
| FMRIB58 > PNG (ANTs) | 588,188 | 594,116 | 591,845 |
| FMRIB58 < PNG (ANTs) | 126,667 | 85,782 | 153,490 |
| IITv3.0 > PNG (ANTs) | 597,688 | 598,457 | 608,754 |
| IITv3.0 < PNG (ANTs) | 69,359 | 49,774 | 81,651 |
| PNG (DTI-TK) > PNG (ANTs) | 481,365 | 464,482 | 445,434 |
| PNG (DTI-TK) < PNG (ANTs) | 172,137 | 181,668 | 189,488 |
Comparisons at multiple sessions (Pre, In1, In2) are presented. For illustrations of the voxel-wise t-statistical maps, see Supplementary Fig. S1 online.
Table 2 summarizes the number of statistically significant voxels of DTI metrics (FA, MD, AD, RD) and the total number of voxels on the TBSS skeleton. The non-parametric Friedman test did not suggest the covariate template as a significant factor (=2.370, df = 3, p = 0.499) for .
Table 2.
Summary of the ratios of the number of statistically significant voxels of DTI metrics (FA, MD, AD, RD) and the total number of voxels on TBSS skeleton.
| DTI | Contrast | Total significant voxels/total voxels | |||
|---|---|---|---|---|---|
| FMRIB58 | IITv3.0 | PNG (ANTs) | PNG (DTI-TK) | ||
| FA | Pre > In2 | 16,885/30,017 | 16,040/29,481 | 16,785/29,740 | 15,864/29,888 |
| MD | Pre < In2 | 7544/30,017 | 6791/29,481 | 6838/29,740 | 6675/29,888 |
| AD | Pre > In2 | 888/30,017 | 0/29,481 | 571/29,740 | 100/29,888 |
| RD | Pre < In2 | 12,235/30,017 | 11,598/29,481 | 12,093/29,740 | 11,889/29,888 |
The Hosmer Lemeshow Goodness-of-Fit test showed a good fit for the logistic regression models of FA, MD, and AD (all p > 0.05), except RD (p < 0.05) (Table 3). However, in all four models, the covariate template was a significant factor (p < 0.05, Wald Chi-square test) for the ratios (Table 3). Therefore indicating a strong correlation between the ratios and template selection.
Table 3.
Summary of Hosmer Lemeshow Goodness-of-Fit test and logistic analysis for the effect of the selected template on the ratio of the number of significant voxels (from the permutation-based t-statistical maps) and total number of voxels on the TBSS skeleton within the ROI (see Supplementary Tables S1-S4 online).
| DTI | Contrast | Goodness-of-fit test | Wald test | ||||
|---|---|---|---|---|---|---|---|
| df | p | Dftemplate | Ptemplate | ||||
| FA | Pre > In2 | 11.478 | 8 | 0.176 | 9.759 | 3 | 0.020 |
| MD | Pre < In2 | 7.516 | 7 | 0.377 | 12.249 | 3 | 0.007 |
| AD | Pre > In2 | 1.725 | 1 | 0.189 | 299.374 | 3 | < 0.001 |
| RD | Pre < In2 | 52.032 | 8 | < 0.001 | 20.680 | 3 | < 0.001 |
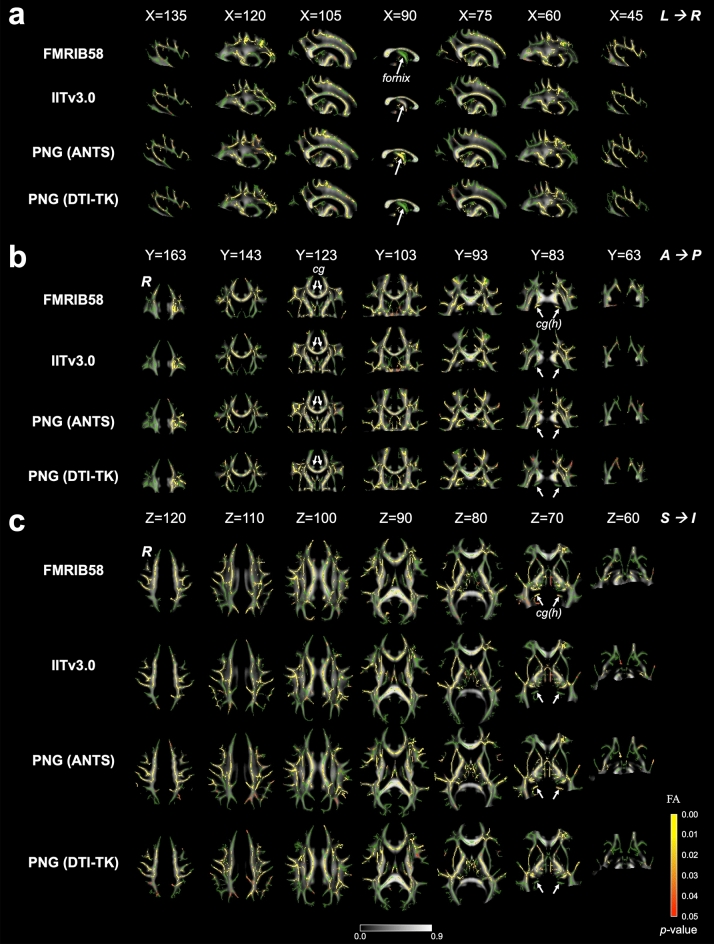
Within most of the white matter tracts, the number of significant voxels exhibiting decreased FA at In2 versus Pre was similar across the four templates (Fig. 3). For the PNG (ANTs) template, the significant voxels in the fornix were 99 mm3, much larger compared to the FMRIB58 (14 mm3), IITv3.0 (5 mm3), and the PNG (DTI-TK) templates (13 mm3) (Fig. 3a). For the PNG (DTI-TK) template, the significant voxels in the bilateral cingula were much smaller compared to the other three templates (Fig. 3b). In the bilateral cingula (hippocampi), neither the IITv3.0 nor PNG (DTI-TK) template exhibited any significant voxels of FA difference (Fig. 3b,c). Similarly, no significant voxels of FA difference were observed in the left tapetum for the FMRIB58 template.
Figure 3.
Illustrations of t-statistical maps (red–yellow, FWE corrected, p < 0.05) showing decreased FA at In2 versus Pre in (a) sagittal, (b) coronal, and (c) axial views, overlaid on TBSS skeleton (green) and mean FA image derived from FMRIB58 (FMRIB, Oxford, UK), IITv3.011, PNG (ANTs), and PNG (DTI-TK) DTI templates respectively. Major white matter tracts showing different sensitivity across the templates for detecting FA changes are highlighted in arrows. cg: cingula. cg(h): cingula (hippocampi). L/R: left/right hemisphere. A/P: anterior/posterior. S/I: superior/inferior.
Supplementary Figs. S2-S4 online provide illustrations of the t-statistical maps of significant differences of MD, AD, and RD, at In2 versus Pre. A detailed summary of the significant voxels exhibiting differences of FA, MD, AD, and RD can be found in Supplementary Tables S1-S4 online.
According to the linear mixed-effect regression analyses for the longitudinal changes of FA (Supplementary Table S5 online), model fits were similar among the regions commonly identified across the four templates. FMRIB58 exhibited the highest model fit (i.e., lowest AIC) in most of the white matter tracts, including the left cerebral peduncle, left posterior corona radiata, right superior corona radiata, right external capsule, left anterior internal capsule, right posterior internal capsule, left inferior longitudinal/fronto-occipital fasciculus, and left stria terminalis. For IITv3.0, the highest fits were observed in the right cingulum, right anterior corona radiata, and right anterior internal capsule. For the PNG (ANTs) population-specific template, higher fits were observed in the bilateral superior fronto-occipital fasciculus and the right superior longitudinal fasciculus. whereas the PNG (DTI-TK) template exhibited the highest fit in the right anterior corona radiata and the left superior corona radiata.
Supplementary Tables S6 and S7 online summarize the linear mixed-effect regression analyses for the longitudinal changes, for MD and RD respectively. Similar to FA, model fits were similar among the commonly identified regions. No table is shown for AD, since no voxels exhibited a significant difference when the IITv3.0 DTI template was used (see Table 2 and Supplementary Fig. S3 online).
Discussion
Due to repetitive head impacts experienced during practices and games, EMA collision-sport athletes may exhibit a distinct neurological trajectory that is different from those typical at the same age. Bias may be introduced when modeling the neurological consequences using the existing standardized human brain atlases based on healthy adult or adolescent populations, leading to confounding (sometimes even contradictory) findings. In this work, population-specific brain atlases were developed for EMA collision-sport athletes in the PNG longitudinal database. Compared to the standardized adult or other age-appropriate T1 templates (Fig. 1), significantly fewer biases were introduced in spatial normalization using the PNG T1 template (Fig. 2). The PNG (ANTs) DTI template resulted in minimal biases compared to the standardized or PNG (DTI-TK) DTI templates (Table 1, Supplementary Fig. S1 online), and the selection contributed to different sensitivity of detecting DTI changes in TBSS (Table 3, Fig. 3, and Supplementary Tables S1-S4 & Figs. S2-S4 online), whereas the sensitivity of detecting longitudinal change of DTI metrics from ROI-based regression analyses was relatively comparable (Supplementary Tables S5-S7 online). In summary, the main findings suggested the PNG brain atlases better characterized the neuroanatomy of EMA collision-sport athletes, reduced biases introduced during spatial normalization, and exhibited higher sensitivity in detecting regional DTI differences. As template selection is a critical strategic step towards robust and rigorous statistical findings, we expect neuroimaging and clinical researchers will benefit from the new atlases to better clarify mechanisms of mTBI and monitor brain health of EMA collision-sport athletes.
The strengths and limitations between standardized and population-specific brain atlases have been discussed5, 6. Being a pragmatic option for computational efficiency, a standardized brain atlas often comes with a comprehensive set of templates and semantic labels, facilitating the processing and analysis of brain images acquired from multiple sites or studies6. However, when the underlying neuroanatomy of the study population is different, mis-registration can lead to greater bias and errors in voxel-wise and ROI-based statistical analyses. On the other hand, the registration errors of using a population-specific template are unbiased towards the study population; however, the population-specific template usually lacks semantic labels5; therefore, subsequent transformation to a standard space (e.g., ICBM152) is required for interpreting the statistical maps54. In addition, suboptimal data quality can lead to a poorly constructed template and lowers spatial normalization accuracy6, 11, 55, so a nontrivial amount of diligence is demanded in constructing population-specific templates. The selection strategy largely depends on the specific study, including research questions to address, participants of the study, as well as the number, type, and quality of data5. Neuroimaging researchers working on clinical populations should carefully leverage these aspects to ensure rigorous and robust neuroimaging findings are reported in clinical literature and be cautious when reporting voxel-wise statistical findings based off of a non-specific brain atlas.
This work clarifies the advantages and limitations of constructing population-specific DTI templates (Fig. 1b) using scalar-based (ANTs) and tensor-based (DTI-TK) registrations. Conventionally, spatial normalization of diffusion tensor fields is achieved by aligning the b0 image to the anatomical T1 image56, and our evaluation showed that this approach introduced minimal biases in spatial normalization (Table 1, Supplementary Fig. S1 online). ANTs is a diffeomorphic registration that uses cross-correlation metrics to optimize the shape and appearance during template construction, with the underlying assumption that possibly different shapes of the same structures exist in both images57; as a result, the PNG (ANTs) template has a sharp appearance that can discern adjacent white matter tracts. Unlike ANTs, DTI-TK utilizes the six tensor components and does not include such template update procedures, and the PNG (DTI-TK) template was computed as the average of the co-registered dataset. Although the appearance was more blurred than the PNG (ANTs) template, adjacent white matter tracts can be discerned. While it is commonly believed that tensor-based registration algorithms improve the registration quality of DTI54, 58–60, the estimates of the tensor in certain biological structures (e.g., the fornix) can be inaccurate, which may adversely affect the quality of the constructed template. A combination of tensor information with a scalar-based registration method can potentially improve the quality of a population-specific template61.
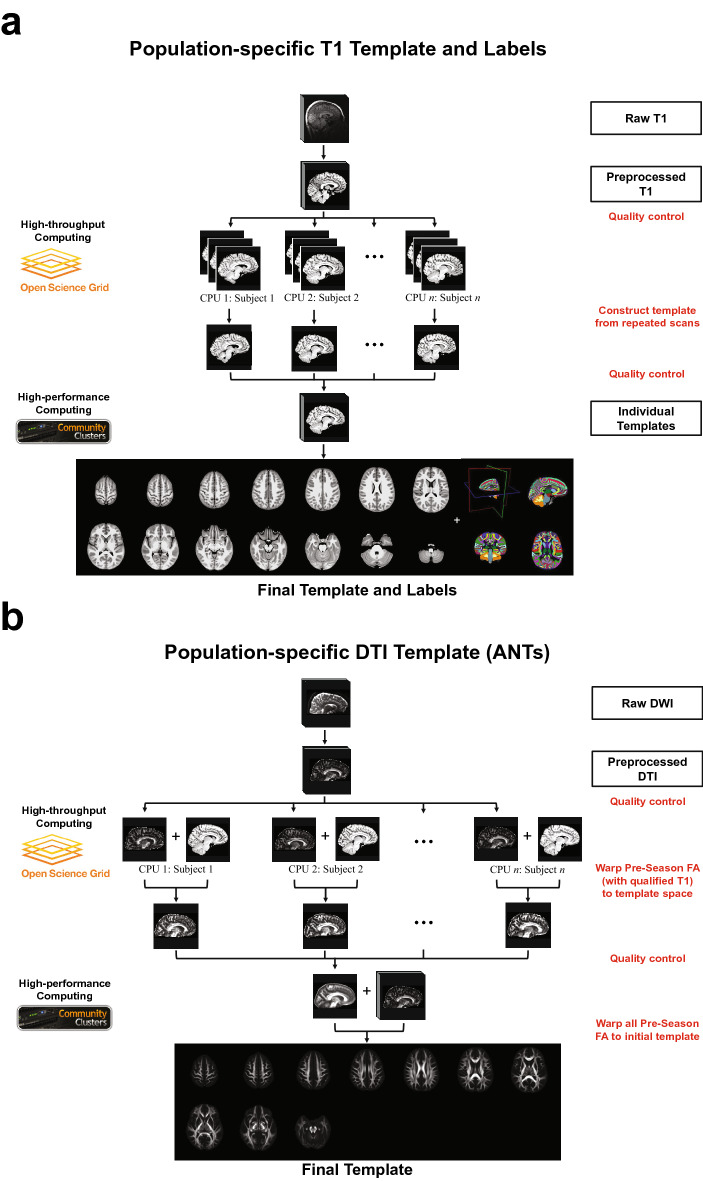
According to the voxel-based morphometric analyses (Fig. 2), using the PNG T1 template introduced minimal bias during spatial normalization of the T1 images from the EMA collision-sport athletes, even when compared to the NIHPD13.0–18.5 template (Fig. 2b). Given the NIHPD template was constructed based on healthy adolescents of a similar age range, one explanation is that the trajectory of subcortical volumes in adolescent collision-sport athletes may be different from healthy adolescents of similar ages. Previously, Narvacan et al.16 reported in a lifespan study of healthy adolescents that at age 13–17, a non-linear decrease of subcortical volumes was observed within certain regions for the male participants. Thus, it is worth exploring whether the trajectory of subcortical volumes can be driven by sports-related concussion and repetitive head impacts. Notably, future work is needed to validate the T1-based semantic labels (Fig. 4a), which may be applied to investigate the regional volumetric trajectory.
Figure 4.
High-throughput high-performance computing workflow, for constructing (a) the population-specific T1 template and labels, and (b) the population-specific DTI template using Advanced Normalization Tools (ANTs)57.
The selection of DTI templates did not lead to significantly different TBSS skeleton but was a significant covariate for the voxel-wise statistical analyses (Table 2, Table 3, Supplementary Tables S1-S4 online). Previously, utilizing the standardized FMRIB58 template in TBSS processing, abnormal DTI changes in 64 adolescent football athletes from Pre to In2 were observed, including decreased FA, decreased AD, increased MD, and increased RD51, 52. In this work, compared to FMRIB58, the PNG (ANTs) template resulted in consistent but more sensitive detection of FA decrease within the bilateral cingula adjacent to the hippocampi [cg(h)] [PNG (ANTs): 73%, FMRIB58: 65%, IITv3.0: 3%] and the fornix [PNG (ANTs): 61%, FMRIB58: 9%, IITv3.0: 62%] (Supplementary Table S1 online), whereas on the skeletons of IITv3.0 and PNG (DTI-TK), such a difference was either detected with fewer voxels or not statistically significant (Fig. 3). The fornix and cg(h) are major parts of the limbic system. Surrounded by cerebrospinal fluids, the main body of the fornix is located in the midline of the brain, with neuronal projections to the cg(h) in medial temporal lobes. The fornix is critical for normal cognitive functioning; literature reported atrophy in the fornix for neurological disorders62. The cg(h) consist of gray matter, with a thin layer of white matter on its ventricular surface63. Atrophy of the hippocampus is a common neuropathology in chronic traumatic encephalopathy64, 65. The volume of the cg(h) correlated with FA in the fornix66, and reduced fornix and hippocampal volumes have been reported in morphometric study of TBI67 (for review, see Shenton et al.68). Both structures are relatively small and prone to mis-registration in DWI, due to low spatial resolution, geometric distortions from eddy current, and partial volume effects69–72. There are multiple literature that observed changes in FA for adolescent collision-sport athletes in the fornix and cingulum (hippocampus); however, the direction of change is not consistent across literature as described in our previous work39. Myer et al.35 and Kuzminski et al.40 reported decreased FA in cingulum (hippocampus), whereas Bazarian et al.73 and Manning et al.74 reported increased FA. Similarly, both increased75 and decreased FA40 were observed in the fornix. The conflicting direction of changes in other DTI metrics (e.g. MD, AD, RD) were also observed in the fornix38 and cingulum (hippocampus)44, 74, 76. The PNG (ANTs) template potentially provided DTI insights at improved sensitivity and complemented the volumetric findings, which is likely due to repetitive head impacts experienced by this vulnerable population. Consistent with Cabeen et al.13 that the study template resulted in significantly larger number of voxels in the fornix, the higher sensitivity within these areas was perhaps attributable to the better anatomical alignment in the PNG T1 template, which is unbiased towards the study population and better captured anatomical structure than the standardized templates (Fig. 1).
In addition to fornix and cingulum (hippocampus), we observed noticeably more voxels exhibiting decreased FA for the PNG (ANTs) DTI template than the standardized FMRIB58 and IITv3.0 templates, within the right cingulum (cingulate gyrus) [PNG (ANTs): 43%, FMRIB58: 8%, IITv3.0: 42%], bilateral retrolenticular internal capsules [PNG (ANTs): 76%, FMRIB58: 70%, IITv3.0: 66%], right superior longitudinal fasciculus [PNG (ANTs): 61%, FMRIB58: 55%, IITv3.0: 51%], and bilateral tapetum [PNG (ANTs): 42%, FMRIB58: 23%, IITv3.0: 35%] (Supplementary Table S1 online). Moreover, the proportions of significant voxels with decreased FA between the bilateral cingula (cingulate gyrus) are more laterality (L: 36%, R: 43%) for the PNG (ANTs) DTI template than the FMRIB58 template (L: 45%, R: 8%) (Supplementary Table S1 online). These observations may suggest that improper selection of template in neuroimage data processing can contribute to potential bias in specific ROIs.
This work has several limitations. Considering that our recruited participants were exclusively male football or female soccer players (Table 4), we constructed brain atlases only for the combined collision-sport population and not for each sport, but sex differences may exist and warrant future investigation to determine the necessity of having sex-specific brain atlases for collision sports. The visual quality assessment used in the study can be subjective and biased; future work will involve an automated and quantitative quality assessment with more specific criteria in ground truth and landmark (e.g., in Jang et al.39). Template selection is only one out of many aspects in the image processing pipeline that contributes to the inconsistent DTI findings reported in the mTBI literature; to achieve reproducible and meaningful results, variability in study design, scanning parameters, and analytic techniques should also be considered45, 77, 78. The PNG T1 template was constructed using buildtemplateparallel.sh, which was found to have an issue of rigid-only registration and later superseded by antsMultivariateTemplateConstruction.sh. This could have an impact on the quality of individual templates based on repeated scans (Fig. 4a). There are several limitations when using ANTs for template construction5: first, due to the differences in acquisition protocols, DWI data often have artifacts such as distortions due to eddy currents, so that mis-registration can occur between b0 and T1-weighted images. Second, since white matter has rather homogeneous intensity on a T1 image, using the warping of T1 images to guide DTI alignment may lead to a mismatch of white matter alignment. This work evaluated the proposed DTI templates using real data and compared to previous findings utilizing a standardized template51, 52, but a more robust scheme of evaluation is to employ simulated data, with a priori knowledge of the pathology as a ground truth79; such a scheme is robust for modeling pathology like multiple sclerosis, where white matter degeneration is well characterized by the corresponding FA reduction80, 81, but is difficult for sports-related mTBI and repetitive head impacts in adolescents, given the conflicting DTI characterization for axonal pathology in the literature45.
Table 4.
Summary of the datasets for constructing and evaluating the population-specific T1 and DTI templates, based on the Purdue Neurotrauma Group (PNG) longitudinal MRI database50.
| Task | Season | Sessiona | Sportb (n) | Age range (mean ± std) | n Ethnicity (FB, SOC) |
|---|---|---|---|---|---|
| PNG T1 template | |||||
| Construction | 2011–2017 | Pre In Post |
FB (155) SOC (60) |
All: 13–19 (16.16 ± 1.05) FB: 13–19 (16.20 ± 1.08) SOC: 14–18 (16.04 ± 0.97) |
White: 157 (104, 53) Black or African American: 21 (21, 0) Hispanic or Latino: 11 (8, 3) Asian: 2 (2, 0) Native American: 5 (5, 0) More than one: 17 (13, 4) Unspecified: 2 (2, 0) |
| Evaluation | 2018–2019 | Pre | FB (12) |
14–19 (16.67 ± 1.37) |
White: 6 Black or African American: 2 Hispanic or Latino: 1 Native American: 1 More than one: 2 |
| PNG DTI template | |||||
| Construction | 2016–2017 | Pre | FB (64) |
14–18 (16.00 ± 1.04) |
White: 36 Black or African American: 9 Hispanic or Latino: 5 Native American: 3 More than one: 7 Unspecified: 4 |
| Evaluationc | 2016–2017 |
Pre In |
|||
aData were acquired across multiple sessions, including one approximately one month before contact practices began (Pre), one or more within competition season (In), and one or more after the season ended (Post).
bAll football athletes (FB) are male participants, and all soccer athletes (SOC) are female participants.
cSame sport, age range, and ethnicity as the dataset for constructing the DTI template.
Methods
Participants and data collection
This study used data collected by PNG in their ongoing longitudinal study of adolescent athletes50, which has been approved by the Biomedical IRB of Purdue’s Human Research Protection Program and was carried out in accordance with the Declaration of Helsinki. Before enrolling in the study, written informed consent was obtained from each participant, and subject assent and parental consent were obtained for participants under the age of 18. The data include athletes participating in the collision sports of American football (all males) and soccer (all females). Data were acquired across multiple sessions, including one approximately one month before contact practices began (Pre), one or more within competition season (In), and one or more after the season ended (Post). These data were grouped into different datasets to construct or evaluate the population-specific T1 or DTI templates. See Table 4 for the total number of participants and relevant details for each dataset.
Note that during the period of study, no participant was diagnosed by their athletic trainer or team physician as being concussed.
MR imaging
All data were acquired using a 3 T General Electric Signa HDx (Waukesha, WI) with a 16-channel brain array (Nova Medical; Wilmington, MA).
T1-weighted imaging data
Anatomical T1 scans were acquired using a 3D fast spoiled gradient-echo sequence (TR/TE = 5.7/2.0 ms, tip angle = 73°, 1 mm isotropic resolution). Longitudinal volumetric data from 227 athletes (167 males; 60 females) were used for construction and evaluation of the template (Table 4).
Diffusion-weighted imaging data
Diffusion-weighted imaging (DWI) data were acquired using a spin-echo echo-planar imaging sequence (TR/TE = 12,500/100 ms, 40 slices with 2.5 mm thickness), FOV of 24 × 24 cm2, a 96 × 96 acquisition matrix, in-plane resolution of 2.5 × 2.5 mm2, with 30 diffusion-encoding directions at b = 1000 s/mm2 and one at b = 0 s/mm2, and an upsampled isotropic resolution of 1 mm. Longitudinal data were from sixty-four male football athletes that participated in one competition season (Table 4). All participants completed three MRI sessions: one scan at Pre and two In-Season scans (In), with one in the first (In1) and one in the second (In2) 5-week halves of the season.
Atlas construction
To accelerate the computation time, we established a workflow integrating a high-throughput (The Open Science Grid)82, 83 and a high-performance (Purdue Community Clusters) computing platform. Specifically, the Open Science Grid integrates the computing and storage elements from over 100 individual sites spanning the United States and provides a distributed fabric of high-throughput computational services, allowing numerous individual, small, and independent tasks to run concurrently on different CPU cores. Purdue Community Clusters consists of Dell compute nodes with 16–24 cores of Intel Xeon Gold Sky Lake processors per node, at least 192 GB of RAM, and 100 Gbps InfiniBand interconnects, which processes single, large, and interdependent tasks at its fastest speed. The workflow was implemented to construct the population-specific T1 template and one of the two population-specific DTI templates, and the schematic diagrams were shown in Fig. 4.
Population-specific T1 template and labels
The population consisted of 215 EMA collision-sport athletes (ages: 13 − 19, 16.16 ± 1.05) scanned at Pre of the 2011–2017 competition seasons, including 155 high school varsity football athletes (ages: 13 − 19, 16.20 ± 1.08) and 60 soccer athletes (ages: 14 − 18, 16.04 ± 0.97) (Table 4).
The workflow of constructing the PNG (ANTs) template is summarized in Fig. 4a. T1 preprocessing included (1) denoising using an adaptive non-local (NL)-means filter to improve signal-to-noise ratio across all the spatial-frequency domains of the image84; (2) bias correction using FSL FAST to correct for spatial intensity variations and minimize the influence of intensity gradient on segmentation85, 86; (3) skull-stripping using a brain extraction program to remove non-brain part of the image (FSL BET)87, 88; and (4) intensity normalization using FSL MATH commands (fslmaths -inm) to make T1 images between subject comparable89, followed by the first visual quality assessment where preprocessed T1 images with low signals, cutoff of brain regions, motion, or observable artifacts were excluded; this resulted in 782 T1 images from 235 participants, where 547 were repeated scans from 168 participants.
Advanced Normalization Tools (ANTs)57, a top-performing registration tool, was applied to construct the T1 template. ANTs employs symmetric groupwise normalization that has been shown to retain accurate anatomical details90. Specifically, ANTs utilizes Symmetric Normalization (SyN)91 as the registration algorithm, and formulate the problem of atlas construction as “estimating a common space and set of transformations that gives the smallest parameterization of the dataset.” The size and shape of the template image is optimized to converge to the group mean via Symmetric Groupwise Normalization (SyGN)92, which is achieved mathematically by optimizing
where is the template image, is the ith individual image, is the diffeomorphism, where the initial conditions of each is denoted as . For complete explanations of the algorithm, see Avants et al.57, 92.
Using buildtemplateparallel.sh in ANTs, one individual template was created per participant on the Open Science Grid, with 30 × 50 × 20 iterations per registration, and cross correlation as the evaluation metric (buildtemplateparallel.sh -d 3 -m 30 × 50 × 20 -t GR -s CC -c 1), followed by a second visual quality assessment, which mainly focused on resemblance of neuroanatomy pertinent to each individual, including the matching of major sulci and gyri, and gray matter-CSF border; this resulted in individual templates from 215 participants with good quality. Using Purdue Community Clusters, the final population-specific template (PNG T1) was created from the individual templates, with one full node (24 cores) and one indivitual template as the target volume (buildtemplateparallel.sh -d 3 -m 30 × 50 × 20 -t GR -s CC -c 2 -j 24 -z).
Based on the final template, the semantic labels were created using the recon-all pipeline of FreeSurfer (v6.0.0)93, employing the Desikan-Killiany labeling protocol94 to assign the neuroanatomical label to each cortical region. The template and labels have been made available at the Purdue University Research Repository53.
Population-specific DTI templates
The population consisted of 64 football athletes (age: 14–18, 16.00 ± 1.04) scanned at Pre of the 2016–2017 competition season (Table 4).
Two top-performing registration tools, namely ANTs57 and DTI-TK95, was applied to construct DTI templates. DTI-TK incorporates explicit optimization of tensor orientation with piecewise affine registration95, and the problem of atlas construction is formulated as “estimating an image that requires the minimum amount of deformation to map into every image in the population.”96 Given a population of N diffusion tensor images , the template estimation is defined mathmatically as97.
where is the deformation applied to the image . The tensor metric represents the image term, which is the summation of the region-wise tensor image difference. is a metric that quantifies the amount of deformation associated with . For complete explanations of the algorithm, see Zhang et al.95, 97.
Before constructing the templates, raw DWI data were first preprocessed using FSL (FMRIB 5.0, Oxford, U.K.), including corrections for motion and eddy currents (eddy_correct), followed by the extraction of aliasing-corrected brains (BET). DTI metrics, including FA, mean diffusivity (MD), axial diffusivity (AD), and radial diffusivity (RD), were estimated for each individual (DTIFit), and all passed the first visual quality assessment for presence of motion artifact or geometrical distortion.
The workflow of constructing the PNG (ANTs) template is summarized in Fig. 4b. Based on the quality assessments when constructing the population-specific T1 template, 33 of the 64 football players had a qualified T1 image at Pre. Only the corresponding DWI images at Pre were used, considering that DTI changes were observed at In1 and In251, 52. For each subject, the b0 image served as the reference image to warp the FA image to the corresponding T1 image, and subsequently to the PNG T1 template space. All warping processes were carried out by running antsIntermodalityIntrasubject.sh. All the warped FA images passed the second visual quality assessment for inspecting whether they were normalized to the same space of the template. Then, an average map of the warped FA images was computed to serve as the initial reference image to register to, and the population-specific DTI template was constructed based on the 64 football players at Pre, with one full node (24 cores) and one indivitual template as the target volume (antsMultivariateTemplateConstruction.sh -d 3 -m 30x50x20 -t GR -s CC -c 2 -j 24 -i 4 -z), and has been made available at the Purdue University Research Repository53. The second DTI template was constructed using DTI-TK95 as a comparison to the PNG (ANTs) template, where the diffusion tensors in the native space of 64 subjects were used.
Evaluations
Population-specific T1 template
The population consisted of 12 newly scanned high school varsity football athletes (ages: 14–19, 16.67 ± 1.37) scanned at Pre of the 2018–2019 competition season (Table 4).
Deformation-based morphometry analyses were performed to compare the potential bias of using different T1 templates. The newly acquired T1 scans were normalized (via antsRegistrationSyN) to the ICBM152 template, an age-appropriate template (NIHPD13.0–18.521, IITv3.011, and the population-specific T1 template (PNG); this yielded 4 × 12 = 48 maps of deformation field. The logarithm of Jacobian determinant (logJ, representing local volume difference) was estimated (via ANTSJacobian) for each map. The maps of absolute logJ (|logJ|) were computed and transformed to the standard space of ICBM152 (1 mm spatial resolution) via antsApplyTransforms. In the standard space, voxel-wise permutation-based t-statistics were computed with 5000 permutations with a repeated ANOVA design, using the FSL Randomise program98, with threshold-free cluster enhancement99 and family wise error (FWE) of 5% used to control for type-I error.
Population-specific DTI templates
The population consisted of 64 high school varsity football athletes (ages: 14–18, 16.00 ± 1.04) scanned at Pre, In1, and In2 of the 2016–2017 competition season (Table 4).
All individual FA images were first aligned through a nonlinear transformation algorithm (FNIRT) to four DTI templates, including two standardized templates: FMRIB58 (FMRIB, Oxford, UK) and IITv3.011, and the two PNG population-specific templates constructed by ANTs and DTI-TK.
Similar to evaluating the T1 templates, deformation-based morphometry analyses were performed to compare the potential bias of using different DTI templates. At each session (Pre, In1, In2), 4 × 64 = 256 maps of |logJ| were yielded, and all were transformed to the standard space of FMRIB58 (same as ICBM152). Using the same design as evaluating the T1 templates, voxel-wise permutation-based t-statistics were computed for each session.
In the standard space of ICBM152, a skeleton representing the common white matter tracts across all the subjects was created from thinning the mean FA map that was averaged from all the aligned FA images. The skeleton was thresholded at FA > 0.2 to reduce partial volume effects between borders of different tissues. Regional maximal FA values were projected onto the skeleton according to a distance map27. Based on the mean FA skeleton, skeletons of MD, AD, and RD were obtained by projecting the corresponding DTI values onto the FA skeleton (tbss_non_FA). The processing procedure guaranteed that the variations of the TBSS results were only related to the template selection.
The resulting DTI skeletons of each subject were fed into voxel-wise permutation-based statistics with a repeated ANOVA design and with 5000 permutations among Pre, In1 and In2, using the FSL Randomise program98. The type-I error was controlled by threshold-free cluster enhancement99 and FWE of 5%. For the purpose of demonstrating the effect of template selection on subsequent statistical findings, only the contrasts comparing Pre and In2 were presented. Among all the selected contrasts of each template, the ones showing significant voxels at p < 0.05 (FA and AD: Pre > In2; MD and RD: Pre < In2) were further segmented into ROIs defined by the JHU-ICBM-DTI-81 WM label atlas100, and the corresponding voxel counts and DTI values were extracted via the FSL Cluster program.
Within each ROI overlaid on the TBSS skeletons, we counted
and
First, the non-parametric Friedman test was performed to test whether correlated with template selection, with as the response variable, template as the predictor, and ROI as the blocking variable.
Then, logistic regression was performed to test whether correlated with template selection:
where referred to the ratio from the ith template, with regard to the TBSS skeleton within the jth ROI. 4 models were established with respect to the ratio of each DTI metric (FA, MD, AD and RD). ROIs with no voxel on the TBSS skeletons were excluded from the analyses. The analyses were performed using SAS 9.4 (SAS Institute, Cary NC).
To investigate the sensitivity of different templates to the short-term changes of white matter microstructure in high-school football players51, 52, linear mixed regression analyses were performed, where timepoint and age were the fixed variables, and subject was the random variable. Models were fitted within each ROI and for each DTI metric. The Akaike information criterion (AIC) was used to evaluate model fit, and t and p values for timepoint were compared across the four templates. FDR was applied to correct for comparisons in multiple ROIs. ROIs with no voxel on the skeleton, and ROIs rejected by the Shapiro-Wilks normality test were excluded from the analyses. The analyses were performed using R version 3.5.2101.
Supplementary Information
Acknowledgements
The authors thank Dr. Frederick Damen for his intellectual contributions in early discussion. The authors extend our gratitude to all the study participants who made this research possible. The authors also thank Purdue Institute of Inflammation, Immunology and Infectious Disease for granting access to the cluster computing resources provided by Information Technology at Purdue, West Lafayette, Indiana.
Author contributions
Y.Z. conceptualized and designed the study, N.L.V., D.O.S., T.E.S., V.N.P., E.L.B., G.G.T., L.J.L., U.D., E.A.N., Y.T. and T.M.T. collected the data, Y.Z., W.Z. and H.Y. performed the data processing, Y.Z. and W.Z. analyzed the results, Y.Z., W.Z., H.Y., I.J., Y.T., T.M.T. and J.V.R. drafted the manuscript work or revised critically for important intellectual content. All authors approved the final version to be published and agreed to be accountable for the integrity and accuracy of all aspects of the work.
Funding
Publication of this article was funded in part by Purdue University Libraries Open Access Publishing Fund.
Accession codes
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-85518-6.
References
- 1.Bailes JE, Petraglia AL, Omalu BI, Nauman E, Talavage T. Role of subconcussion in repetitive mild traumatic brain injury. J. Neurosurg. 2013;119:1235–1245. doi: 10.3171/2013.7.JNS121822. [DOI] [PubMed] [Google Scholar]
- 2.Ling H, Hardy J. Neurological consequences of traumatic brain injuries in sports. Mol. Cell. Neurosci. 2015;66:114–122. doi: 10.1016/j.mcn.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Nauman EA, Talavage TM. Subconcussive trauma. Handb. Clin. Neurol. 2018;158:245–255. doi: 10.1016/B978-0-444-63954-7.00024-0. [DOI] [PubMed] [Google Scholar]
- 4.Nauman EA, Talavage TM, Auerbach PS. Mitigating the consequences of subconcussive head injuries. Annu. Rev. Biomed. Eng. 2020;22:387–407. doi: 10.1146/annurev-bioeng-091219-053447. [DOI] [PubMed] [Google Scholar]
- 5.Van Hecke W, Leemans A, Emsell L, et al. DTI analysis methods: voxel-based analysis. In: Van Hecke W, et al., editors. Diffusion Tensor Imaging: A Practical Handbook. Springer; 2016. pp. 183–203. [Google Scholar]
- 6.Zhang S, Arfanakis K. Role of standardized and study-specific human brain diffusion tensor templates in inter-subject spatial normalization. J. Magn. Reson. Imaging. 2013;37:372–381. doi: 10.1002/jmri.23842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashby FG. Statistical analysis of fMRI data. 2. Cambridge, MA: The MIT Press; 2011. [Google Scholar]
- 8.Talairach, J. & Tournoux, P. Co-Planar Stereotaxic Atlas of the Human Brain: 3-Dimensional Proportional System: An Approach to Cerebral Imaging (Thieme, 1988).
- 9.Grabner G, et al. Symmetric atlasing and model based segmentation: an application to the hippocampus in older adults. Med. Image Comput. Comput. Assist. Interv. 2006;9:58–66. doi: 10.1007/11866763_8. [DOI] [PubMed] [Google Scholar]
- 10.Fan L, et al. The human brainnetome atlas: a new brain atlas based on connectional architecture. Cereb. Cortex. 2016;26:3508–3526. doi: 10.1093/cercor/bhw157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S, Arfanakis K. Evaluation of standardized and study-specific diffusion tensor imaging templates of the adult human brain: Template characteristics, spatial normalization accuracy, and detection of small inter-group FA differences. Neuroimage. 2018;172:40–50. doi: 10.1016/j.neuroimage.2018.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rohlfing T, Zahr NM, Sullivan EV, Pfefferbaum A. The SRI24 multichannel atlas of normal adult human brain structure. Hum. Brain Map. 2010;31:798–819. doi: 10.1002/hbm.20906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cabeen RP, Bastin ME, Laidlaw DH. A Comparative evaluation of voxel-based spatial mapping in diffusion tensor imaging. Neuroimage. 2017;146:100–112. doi: 10.1016/j.neuroimage.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickie DA, et al. Whole brain magnetic resonance image atlases: a systematic review of existing atlases and caveats for use in population imaging. Front. Neuroinform. 2017;11:1. doi: 10.3389/fninf.2017.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jernigan TL, Tallal P. Late childhood changes in brain morphology observable with MRI. Dev. Med. Child Neurol. 1990;32:379–385. doi: 10.1111/j.1469-8749.1990.tb16956.x. [DOI] [PubMed] [Google Scholar]
- 16.Narvacan K, Treit S, Camicioli R, Martin W, Beaulieu C. Evolution of deep gray matter volume across the human lifespan. Hum. Brain Map. 2017;38:3771–3790. doi: 10.1002/hbm.23604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paus T, et al. Structural maturation of neural pathways in children and adolescents: in vivo study. Science. 1999;283:1908–1911. doi: 10.1126/science.283.5409.1908. [DOI] [PubMed] [Google Scholar]
- 18.Pfefferbaum A, et al. A quantitative magnetic resonance imaging study of changes in brain morphology from infancy to late adulthood. Arch. Neurol. 1994;51:874–887. doi: 10.1001/archneur.1994.00540210046012. [DOI] [PubMed] [Google Scholar]
- 19.Richards J, Xie W. Brains for all the ages: structural neurodevelopment in infants and children from a life-span perspective. Adv. Child Dev. Behav. 2015;48:1–52. doi: 10.1016/bs.acdb.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Avants BB, et al. The pediatric template of brain perfusion. Sci. Data. 2015;2:150003. doi: 10.1038/sdata.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fonov V, et al. Unbiased average age-appropriate atlases for pediatric studies. Neuroimage. 2011;54:313–327. doi: 10.1016/j.neuroimage.2010.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gogtay N, et al. Mapping adolescent brain change reveals dynamic wave of accelerated gray matter loss in very early-onset schizophrenia. PNAS. 2004;98:11650–11655. doi: 10.1073/pnas.201243998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez CE, Richards JE, Almli CR. Age-specific MRI templates for pediatric neuroimaging. Dev. Neuropsychol. 2012;37:379–399. doi: 10.1080/87565641.2012.688900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu D, et al. Resource atlases for multi-atlas brain segmentations with multiple ontology levels based on T1-weighted MRI. Neuroimage. 2016;125:120–130. doi: 10.1016/j.neuroimage.2015.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao T, et al. Unbiased age-specific structural brain atlases for Chinese pediatric population. Neuroimage. 2019;189:55–70. doi: 10.1016/j.neuroimage.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Van Hecke W, et al. The effect of template selection on diffusion tensor voxel-based analysis results. Neuroimage. 2011;55:566–573. doi: 10.1016/j.neuroimage.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Smith SM, et al. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage. 2006;31:1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 28.Bach M, et al. Methodological considerations on tract-based spatial statistics (TBSS) Neuroimage. 2014;100:358–369. doi: 10.1016/j.neuroimage.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 29.Bartnik-Olson BL, et al. Impaired neurovascular unit function contributes to persistent symptoms after concussion: a pilot study. J. Neurotrauma. 2014;31:1497–1506. doi: 10.1089/neu.2013.3213. [DOI] [PubMed] [Google Scholar]
- 30.Maugans TA, Farley C, Altaye M, Leach J, Cecil KM. Pediatric sports-related concussion produces cerebral blood flow alterations. Pediatrics. 2012;129:28–37. doi: 10.1542/peds.2011-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nilsson KJ, et al. Repetitive head impacts in youth football: description and relationship to white matter structure. Sports Health. 2019;11:507–513. doi: 10.1177/1941738119865264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Babcock L, Yuan W, Leach J, Nash T, Wade S. White matter alterations in youth with acute mild traumatic brain injury. J. Pediatr. Rehabil. Med. 2015;8:285–296. doi: 10.3233/PRM-150347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chun IY, et al. DTI detection of longitudinal WM abnormalities due to accumulated head impacts. Dev. Neuropsychol. 2015;40:92–97. doi: 10.1080/87565641.2015.1020945. [DOI] [PubMed] [Google Scholar]
- 34.Satchell EK, et al. Use of diffusion tension imaging in the evaluation of pediatric concussions. Musculoskelet. Sci. Pract. 2019;42:162–165. doi: 10.1016/j.msksp.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Yuan W, et al. White matter alterations over the course of two consecutive high-school football seasons and the effect of a jugular compression collar: a preliminary longitudinal diffusion tensor imaging study. Hum. Brain Map. 2018;39:491–508. doi: 10.1002/hbm.23859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Virji-Babul N, et al. Diffusion tensor imaging of sports-related concussion in adolescents. Pediatr. Neurol. 2013;48:24–29. doi: 10.1016/j.pediatrneurol.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 37.Davenport EM, et al. Abnormal white matter integrity related to head impact exposure in a season of high school varsity football. J. Neurotrauma. 2014;31:1617–1624. doi: 10.1089/neu.2013.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foss KB, et al. Relative head impact exposure and brain white matter alterations after a single season of competitive football: a pilot comparison of youth versus high school football. Clin. J. Sport Med. 2019;29:442–450. doi: 10.1097/JSM.0000000000000753. [DOI] [PubMed] [Google Scholar]
- 39.Jang I, et al. Every hit matters: white matter diffusivity changes in high school football athletes are correlated with repetitive head acceleration event exposure. NeuroImage Clin. 2019;24:101930. doi: 10.1016/j.nicl.2019.101930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuzminski SJ, et al. White matter changes related to subconcussive impact frequency during a single season of high school football. Am. J. Neuroradiol. 2018;39:245–251. doi: 10.3174/ajnr.A5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayer AR, et al. Diffusion abnormalities in pediatric mild traumatic brain injury. J. Neurosci. 2012;32:17961–17969. doi: 10.1523/JNEUROSCI.3379-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Myer GD, et al. Analysis of head impact exposure and brain microstructure response in a season-long application of a jugular vein compression collar: a prospective, neuroimaging investigation in American football. Br. J. Sports Med. 2016;50:1276–1285. doi: 10.1136/bjsports-2016-096134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myer GD, et al. The effects of external jugular compression applied during head impact exposure on longitudinal changes in brain neuroanatomical and neurophysiological biomarkers: a preliminary investigation. Front. Neurol. 2016;7:74. doi: 10.3389/fneur.2016.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Myer GD, et al. Altered brain microstructure in association with repetitive subconcussive head impacts and the potential protective effect of jugular vein compression: a longitudinal study of female soccer athletes. Br. J. Sports Med. 2019;53:1539–1551. doi: 10.1136/bjsports-2018-099571. [DOI] [PubMed] [Google Scholar]
- 45.Asken BM, DeKosky ST, Clugston JR, Jaffee MS, Bauer RM. Diffusion tensor imaging (DTI) findings in adult civilian, military, and sport-related mild traumatic brain injury (mTBI): a systematic critical review. Brain Imaging Behav. 2018;12:585–612. doi: 10.1007/s11682-017-9708-9. [DOI] [PubMed] [Google Scholar]
- 46.Giorgio A, et al. Longitudinal changes in grey and white matter during adolescence. Neuroimage. 2010;49:94–103. doi: 10.1016/j.neuroimage.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 47.Lebel C, Beaulieu C. Longitudinal development of human brain wiring continues from childhood into adulthood. J. Neurosci. 2011;31:10937–10947. doi: 10.1523/JNEUROSCI.5302-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lebel C, et al. A longitudinal study of the long-term consequences of drinking during pregnancy: heavy in utero alcohol exposure disrupts the normal processes of brain development. J. Neurosci. 2012;32:15243–15251. doi: 10.1523/JNEUROSCI.1161-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simmonds DJ, Hallquist MN, Asato M, Luna B. Developmental stages and sex differences of white matter and behavioral development through adolescence: a longitudinal diffusion tensor imaging (DTI) study. Neuroimage. 2014;92:356–368. doi: 10.1016/j.neuroimage.2013.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Talavage TM, Nauman EA, Leverenz LJ. The role of medical imaging in the recharacterization of mild traumatic brain injury using youth sports as a laboratory. Front. Neurol. 2016;6:273. doi: 10.3389/fneur.2015.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zou, Y., Jang, I., Vike, N. L., Talavage, T. M. & Rispoli, J. V. Acute impacts of football competition on brain white matter microstructure in high school athletes. In Biomedical Engineering Society 2017 Annual Meeting (2017).
- 52.Zou Y, et al. High-G head collisions are associated with short-term white matter microstructural deficits in high school football athletes. J. Neurotrauma. 2018;35:A23. [Google Scholar]
- 53.Zou Y, Zhu W, Yang H-C, Talavage TM, Rispoli JV. Population-specific brain atlas for adolescent collision-sport athletes in Purdue Neurotrauma Group longitudinal database. Purdue Univ. Res. Repos. 2020 doi: 10.4231/RTXE-0Q70. [DOI] [Google Scholar]
- 54.Van Hecke W, et al. Nonrigid coregistration of diffusion tensor images using a viscous fluid model and mutual information. IEEE Trans. Med. Imaging. 2007;26:1598–1612. doi: 10.1109/TMI.2007.906786. [DOI] [PubMed] [Google Scholar]
- 55.Wu G, Jia H, Wang Q, Shen D. SharpMean: groupwise registration guided by sharp mean image and tree-based registration. Neuroimage. 2011;56:1968–1981. doi: 10.1016/j.neuroimage.2011.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu D, Mori S, Shen D, Van Zijl PCM, Davatzikos C. Spatial normalization of diffusion tensor fields. Magn. Reson. Med. 2003;50:175–182. doi: 10.1002/mrm.10489. [DOI] [PubMed] [Google Scholar]
- 57.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Akakpo L, et al. User-independent diffusion tensor imaging analysis pipelines in a rat model presenting ventriculomegalia: a comparison study. NMR Biomed. 2017;30:e793. doi: 10.1002/nbm.3793. [DOI] [PubMed] [Google Scholar]
- 59.Park HJ, et al. Spatial normalization of diffusion tensor MRI using multiple channels. Neuroimage. 2003;20:1995–2009. doi: 10.1016/j.neuroimage.2003.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang H, et al. High-dimensional spatial normalization of diffusion tensor images improves the detection of white matter differences: an example study using amyotrophic lateral sclerosis. IEEE Trans. Med. Imaging. 2007;26:1585–1597. doi: 10.1109/TMI.2007.906784. [DOI] [PubMed] [Google Scholar]
- 61.Irfanoglu MO, et al. DR-TAMAS: diffeomorphic registration for tensor accurate alignment of anatomical structures. Neuroimage. 2016;132:439–454. doi: 10.1016/j.neuroimage.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Douet V, Chang L. Fornix as an imaging marker for episodic memory deficits in healthy aging and in various neurological disorders. Front. Aging Neurosci. 2014;6:343. doi: 10.3389/fnagi.2014.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carpenter MB, Sutin J. Human Neuroanatomy. Williams & Wilkins; 1983. [Google Scholar]
- 64.McKee AC, Abdolmohammadi B, Stein TD. The neuropathology of chronic traumatic encephalopathy. Handb. Clin. Neurol. 2018;158:297–307. doi: 10.1016/B978-0-444-63954-7.00028-8. [DOI] [PubMed] [Google Scholar]
- 65.Stern RA, et al. Long-term consequences of repetitive brain trauma: chronic traumatic encephalopathy. PM&R. 2011;3:S460–S467. doi: 10.1016/j.pmrj.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 66.Warner MA, et al. Assessing spatial relationships between axonal integrity, regional brain volumes, and neuropsychological outcomes after traumatic axonal injury. J. Neurotrauma. 2010;27:2121–2130. doi: 10.1089/neu.2010.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tate DF, Bigler ED. Fornix and hippocampal atrophy in traumatic brain injury. Learn. Mem. 2000;7:442–446. doi: 10.1101/lm.33000. [DOI] [PubMed] [Google Scholar]
- 68.Shenton ME, et al. A review of magnetic resonance imaging and diffusion tensor imaging findings in mild traumatic brain injury. Brain Imaging Behav. 2012;6:137–192. doi: 10.1007/s11682-012-9156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jin, Y., Shi, Y., Zhan, L. & Thompson, P. M. Automated Multi-Atlas Labeling of the Fornix and Its Integrity In Alzheimer’s Disease. In Proceedings. IEEE International Symposium on Biomedical Imaging2015, 140–143 (2015). [DOI] [PMC free article] [PubMed]
- 70.Jung KJ. Removal of partial volume averaging with free water in MR diffusion tensor imaging using inversion recovery for b0 image. Magn. Reson. Imaging. 2014;32:619–624. doi: 10.1016/j.mri.2014.02.020. [DOI] [PubMed] [Google Scholar]
- 71.Treit S, Steve T, Gross DW, Beaulieu C. High resolution in-vivo diffusion imaging of the human hippocampus. Neuroimage. 2018;182:479–487. doi: 10.1016/j.neuroimage.2018.01.034. [DOI] [PubMed] [Google Scholar]
- 72.Vos SB, Jones DK, Viergever MA, Leemans A. Partial volume effect as a hidden covariate in DTI analyses. Neuroimage. 2011;55:1566–1576. doi: 10.1016/j.neuroimage.2011.01.048. [DOI] [PubMed] [Google Scholar]
- 73.Bazarian JJ, Zhu T, Blyth B, Borrino A, Zhong JH. Subject-specific changes in brain white matter on diffusion tensor imaging after sports-related concussion. Magn. Reson. Imaging. 2012;30:171–180. doi: 10.1016/j.mri.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Manning KY, et al. Multiparametric MRI changes persist beyond recovery in concussed adolescent hockey players. Neurology. 2017;89:2157–2166. doi: 10.1212/WNL.0000000000004669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Borich M, Makan N, Boyd L, Virji-Babul N. Combining whole-brain voxel-wise analysis with in vivo tractography of diffusion behavior after sports-related concussion in adolescents: a preliminary report. J. Neurotrauma. 2013;30:1243–1249. doi: 10.1089/neu.2012.2818. [DOI] [PubMed] [Google Scholar]
- 76.Merchant-Borna K, et al. Novel method of weighting cumulative helmet impacts improves correlation with brain white matter changes after one football season of sub-concussive head blows. Ann. Biomed. Eng. 2016;44:3679–3692. doi: 10.1007/s10439-016-1680-9. [DOI] [PubMed] [Google Scholar]
- 77.Picciotto M. Analytical transparency and reproducibility in human neuroimaging studies. J. Neurosci. 2018;38:3375–3376. doi: 10.1523/JNEUROSCI.0424-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Poldrack RA, et al. Scanning the horizon: towards transparent and reproducible neuroimaging research. Nat. Rev. Neurosci. 2017;18:115–126. doi: 10.1038/nrn.2016.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Van Hecke W, et al. Comparing isotropic and anisotropic smoothing for voxel-based DTI analyses: a simulation study. Hum. Brain Map. 2010;31:98–114. doi: 10.1002/hbm.20848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cercignani M, Bozzali M, Iannucci G, Comi G, Filippi M. Intra-voxel and inter-voxel coherence in patients with multiple sclerosis assessed using diffusion tensor MRI. J. Neurol. 2002;249:875–883. doi: 10.1007/s00415-002-0752-y. [DOI] [PubMed] [Google Scholar]
- 81.Sbardella E, Tona F, Petsas N, Pantano P. DTI measurements in multiple sclerosis: evaluation of brain damage and clinical implications. Mult. Scler. Int. 2013;2013:1–11. doi: 10.1155/2013/671730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pordes R, et al. The open science grid. J. Phys. Conf. Ser. 2007;78:012057. doi: 10.1088/1742-6596/78/1/012057. [DOI] [Google Scholar]
- 83.Sfiligoi, I. et al. The pilot way to Grid resources using glideinWMS. In 2009 WRI World Congress Computer Science and Information Engineering CSIE 20092, 428–432 (2009).
- 84.Coupé P, Manjón JV, Robles M, Collins DL. Adaptive multiresolution non-local means filter for three-dimensional magnetic resonance image denoising. IET Image Process. 2012;6:558. doi: 10.1049/iet-ipr.2011.0161. [DOI] [Google Scholar]
- 85.Guillemaud R, Brady M. Estimating the bias field of MR images. IEEE Trans. Med. Imaging. 1997;16:238–251. doi: 10.1109/42.585758. [DOI] [PubMed] [Google Scholar]
- 86.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 87.Jenkinson, M., Pechaud, M. & Smith, S. BET2-MR-based estimation of brain, skull and scalp surfaces. In Eleventh Annual Meeting of the Organization for Human Brain Mapping 167 (2005).
- 88.Smith SM. Fast robust automated brain extraction. Hum. Brain Map. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nyúl LG, Udupa JK, Zhang X. New variants of a method of MRI scale standardization. IEEE Trans. Med. Imaging. 2000;19:143–150. doi: 10.1109/42.836373. [DOI] [PubMed] [Google Scholar]
- 90.Klein A, et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim J, et al. Structural consequences of diffuse traumatic brain injury: a large deformation tensor-based morphometry study. Neuroimage. 2008;39:1014–1026. doi: 10.1016/j.neuroimage.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Avants BB, et al. The optimal template effect in hippocampus studies of diseased populations. Neuroimage. 2010;49:2457. doi: 10.1016/j.neuroimage.2009.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fischl B. FreeSurfer. Neuroimage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Desikan RS, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 95.Zhang H, Yushkevich PA, Alexander DC, Gee JC. Deformable registration of diffusion tensor MR images with explicit orientation optimization. Med. Image Anal. 2006;10:764–785. doi: 10.1016/j.media.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 96.Joshi S, Davis B, Jomier M, Gerig G. Unbiased diffeomorphic atlas construction for computational anatomy. Neuroimage. 2004;23(Suppl 1):S151–S160. doi: 10.1016/j.neuroimage.2004.07.068. [DOI] [PubMed] [Google Scholar]
- 97.Zhang, H., Yushkevich, P. A., Rueckert, D. & Gee, J. C. Unbiased white matter atlas construction using diffusion tensor images. In Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) 211–218 (2007). [DOI] [PubMed]
- 98.Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum. Brain Mapp. 2002;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Smith SM, Nichols TE. Threshold-free cluster enhancement: addressing problems of smoothing, threshold dependence and localisation in cluster inference. Neuroimage. 2009;44:83–98. doi: 10.1016/j.neuroimage.2008.03.061. [DOI] [PubMed] [Google Scholar]
- 100.Mori, S., Wakana, S., Nagae-Poetscher, L. M. & van Zijl, P. C. M. MRI Atlas of Human White Matter. American Journal of Neuroradiology (Elsevier, 2005).
- 101.R Core Team. R: A Language and Environment for Statistical Computing. (2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.