Graphical abstract
Abstract
Members of the genus Bifidobacterium are dominant and symbiotic inhabitants of the mammalian gastrointestinal tract. Being vertically transmitted, bifidobacterial host colonization commences immediately after birth and leads to a phase of host infancy during which bifidobacteria are highly prevalent and abundant to then transit to a reduced, yet stable abundance phase during host adulthood. However, in order to reach and stably colonize their elective niche, i.e. the large intestine, bifidobacteria have to cope with a multitude of oxidative, osmotic and bile salt/acid stress challenges that occur along the gastrointestinal tract (GIT). Concurrently, bifidobacteria not only have to compete with the myriad of other gut commensals for nutrient acquisition, but they also require protection against bacterial viruses. In this context, Next-Generation Sequencing (NGS) techniques, allowing large-scale comparative and functional genome analyses have helped to identify the genetic strategies that bifidobacteria have developed in order to colonize, survive and adopt to the highly competitive mammalian gastrointestinal environment. The current review is aimed at providing a comprehensive overview concerning the molecular strategies on which bifidobacteria rely to stably and successfully colonize the mammalian gut.
1. Introduction
The gastrointestinal tract (GIT) of mammals is home to a vast and highly complex community of microorganisms which collectively form the so-called gut microbiota. Though the mammalian intestine is inhabited by hundreds of different bacterial species, disproportional scientific attention has been given to one particular symbiotic taxon, i.e. the genus Bifidobacterium, due to its widely claimed ability to exert various health-promoting effects upon its host [1], [2], [3], [4]. For example, convincing evidence has accumulated showing that the presence of bifidobacteria in the mammalian intestine supports the development of the host immune system by improving gut homeostasis and functionality, promoting the intestinal barrier integrity, limiting the onset of certain gut diseases and providing protection against pathogen proliferation [2], [3], [4], [5], [6]. At the same time, bifidobacteria are able to produce various metabolites, such as vitamins, polyphenols, conjugated linoleic acids and short-chain fatty acids (SCFAs) that are believed to elicit a beneficial impact on both epithelial host cells and gut microorganisms [1], [7], [8]. Furthermore, bifidobacteria significantly contribute to host metabolism by participating, through saccharolytic fermentation, in the breakdown of a wide variety of complex host- and diet-derived glycans. This is a feature that not only guarantees the successful colonization of the mammalian gut by bifidobacteria, but also provides accessible nutrients to both the host and other intestinal microorganisms through cross-feeding strategies [9], [10], [11], [12].
The abundance and biodiversity of bifidobacteria in the mammalian gut depends on several environmental factors. In this context, bifidobacterial prevalence and abundance is known to be influenced by mode of delivery (natural birth or C-section delivery), type of feeding (formula or breast milk) and antibiotic exposure [3], [11], [13], [14]. Indeed, since bifidobacteria may be maternally inherited through vertical transmission events and they possess, in their genetic heritage, specific genes involved in the degradation of oligosaccharides typical of breast milk, natural delivery and breast-feeding are generally associated with a higher relative abundance and prevalence of bifidobacteria in the infant gut when compared to C-section delivered and/or formula-fed infants [10], [13], [15], [16]. Bifidobacteria are regarded as keystone microorganisms in the gut microbiota associated with early life. However, a decline of their relative abundance and biodiversity changes are recorded when the host advances from infancy to adolescence and adulthood, at which point the bifidobacterial community stabilizes over time until old age, thus suggesting its relevant role during the entire lifespan [3], [17]. However, current knowledge pertaining to the precise molecular strategies on which bifidobacteria rely to ensure their effective colonization and survival in the competitive intestinal environment remains incomplete. This review will highlight genetic and metabolic features that allow bifidobacteria to successfully colonize the mammalian gut. Specifically, we will discuss how bifidobacteria are able to reach the lower parts of the intestine despite the hostile and adverse conditions of the upper GIT and how they interact with host cells to facilitate gut colonization. Furthermore, this review will outline the ability of bifidobacteria to utilize various complex glycans as an adaptation to the gut environment and their genetic strategies to counter phage attack.
General features and ecology of the genus Bifidobacterium. Taxonomically classified as members of the Bifidobacteriaceae family, bifidobacteria are Gram-positive, non-motile, non-spore forming, anaerobic, saccharolytic microorganisms with a Y-shaped or ‘bifid’ morphology, and a high G + C DNA content [18]. Starting from the description of the first bifidobacterial species, isolated by Tissier in 1899 from the fecal sample of a breast-fed infant [19], the number of species belonging to the genus Bifidobacterium has increased over time with several species being identified only recently, including Bifidobacterium erythrocebi, Bifidobacterium moraviense, Bifidobacterium oedipodis, Bifidobacterium olomucense, Bifidobacterium panos, Bifidobacterium cebidarum, Bifidobacterium leontopitheci, Bifidobacterium saimiriisciurei and Bifidobacterium platyrrhinorum, all isolated from the feces of various monkey species [20], [21], [22], and Bifidobacterium canis and Bifidobacterium choloepi, isolated from the feces of a dog and sloth, respectively [23], [24]. The genus Bifidobacterium represents the deepest branching lineage within the phylum Actinobacteria, and currently comprises 94 recognized (sub)species [22], [25].
Bifidobacteria have been isolated from various diverse ecological niches, such as human blood [26], oral cavity [27], sewage [28], fermented or raw milk [29], [30] and hindgut of birds and social insects [31], [32] (Fig. 1). However, the vast majority of currently characterized bifidobacterial species originate from the gastrointestinal tract of humans and other mammals [33], [34], [35], [36]. Members of the genus Bifidobacterium are therefore considered as abundant symbiotic microorganisms that inhabit the intestinal tract of a wide range of animals, especially those who provide parental care to their offspring, including social insects as well as warm-blooded mammals [36], [37]. Despite their broad ecological distribution, at the beginning of the metagenomic era, it was assumed that the ability of members of the genus Bifidobacterium to colonize the ecological ecosystem of a particular host was species-dependent. Indeed, while several species appeared to enjoy a cosmopolitan lifestyle, such as Bifidobacterium animalis, Bifidobacterium adolescentis, Bifidobacterium catenulatum and Bifidobacterium dentium, other bifidobacterial taxa seemed to be adapted to inhabit the gut environment of a particular animal only, for example in the case of Bifidobacterium angulatum, Bifidobacterium cuniculi and Bifidobacterium gallinarum which seemed to be exclusively associated with cows, rabbits and chickens, respectively [38], [39]. In a similar manner, based on their peculiar genetic arsenal, a group of six bifidobacterial species, i.e. Bifidobacterium asteroides, Bifidobacterium actinocoloniiforme, Bifidobacterium bohemicum, Bifidobacterium bombi, Bifidobacterium coryneforme and Bifidobacterium indicum, had up until recently been considered to be highly specialized in colonizing the intestinal tract of social insects, such as honeybees, bumblebees and wasps [40]. Nonetheless, in recent years, rapid advances of metagenomic approaches have completely revolutionized the primordial metagenomic-based ecological classification within the genus Bifidobacterium. Specifically, the application of bifidobacterial ITS (Internally Transcribed Spacer)-based microbial profiling to a large spectrum of mammalian species has led to a revision of the “cosmopolitan bifidobacterial species list”, identifying B. adolescentis, Bifidobacterium bifidum, Bifidobacterium longum and Bifidobacterium pseudolongum as the real ubiquitous and most abundant bifidobacterial taxa [36]. Furthermore, ITS microbial profiling analyses revealed that bifidobacterial species that had previously been identified as niche-specialized are instead able to colonize a wide number of mammalian hosts [36]. Specifically, those bifidobacterial species that were previously considered as highly specialized in insect gut colonization, are actually widely distributed among various mammalian hosts [36]. At the same time, next-generation sequencing techniques highlighted that several bifidobacterial species are able to colonize the intestinal tract of carnivorous mammals, although the diet of these animals contains only a limited amount of the preferred energy source of bifidobacterial species, i.e. fermentable carbohydrates [23], [36], [41], [42], [43]. This refutes previously proposed niche/host-specific specialization behaviour, and instead indicates a strain-specific adaptation of bifidobacteria to a given environment. This successful adaptation of bifidobacteria to different ecological niches has been attributed to the specific genetic features of this bacterial taxon. In this context, comparative genome analyses revealed that members of the genus Bifidobacterium are predicted to encode a very large carbohydrate-active enzyme arsenal, which is among the largest compared to other gut commensals when taken in proportion to their genome size [9], [44]. These extensive and varied metabolic abilities allow bifidobacteria to access a plethora of simple as well as complex diet- and host-derived glycans (see below), suggesting that these genetic features of bifidobacteria have evolved over time to ensure their successful ecological adaptation to, and fitness in the mammalian GIT [44].
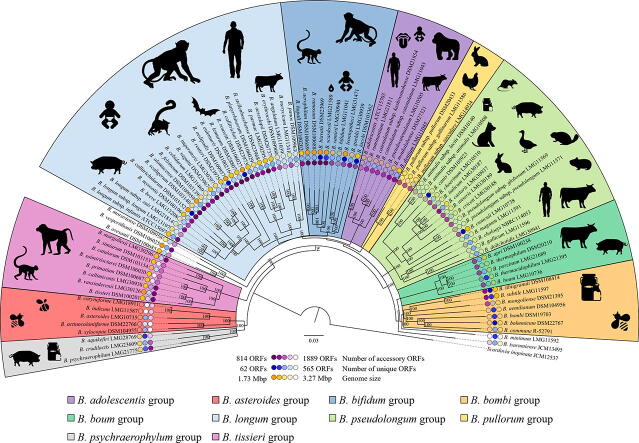
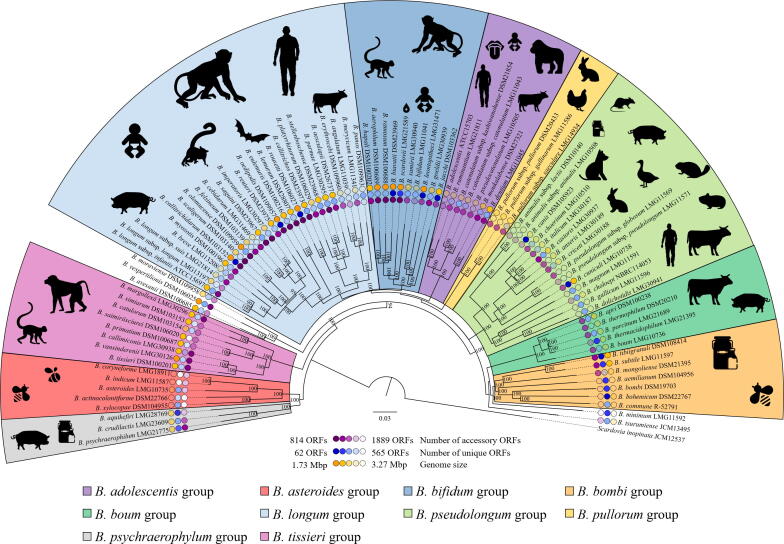
Fig. 1.
Phylogenetic tree of the genus Bifidobacterium based on the concatenation of the 169 amino acid sequences representing the Bifidobacterium core genome. The phylogenetic tree was generated by the maximum-likelihood method, and COG sequences of Scardovia inopinata JCM 12537 shared with bifidobacterial species were used as an outgroup. Bootstrap percentages above 50 are shown at node points based on 1000 replicates of the phylogenetic tree. The outer circle represents the number of accessory and unique genes as well as the genome size of each bifidobacterial type strain, while the ecological origins of bifidobacteria per each phylogenetic group are reported beside the phylogenetic tree.
Phylogenomics of the genus Bifidobacterium. Traditionally, bacterial taxonomic classification relied solely on phenotypic characteristics, such as cell morphology or metabolic features, allowing the identification of discrete phenotypic clusters [45], though completely ignoring the genetic makeup of microorganisms. Subsequently, with the advent of whole genome sequencing combined with the availability of powerful bioinformatics tools, phylogeny entered a new era, the so-called phylogenomic era, leading to a revised definition of bacterial species, i.e. “a monophyletic and genomically coherent cluster of individual organisms that show a high degree of overall similarity with respect to many independent characteristics, and is diagnosable by a discriminative phenotypic property” [46], [47]. Among the available bioinformatic tools to quantitatively measure genome relatedness, Average Nucleotide Identity (ANI) is one of the most reliable and commonly exploited method to investigate genome similarity and an ANI value of < 95% between two bacterial taxa is generally considered indicative of two separate and distinct species [48]. However, despite the NGS-based revolution in the bacterial phylogeny field, it was immediately clear that whole genome-based phylogenetic reconstruction is affected by the presence of genes acquired through horizontal gene transfer events, which prevents a reliable phylogenetic classification [49], [50]. However, assuming that the majority of genetic markers identified as distinctive of a bacterial taxon are vertically inherited, solid foundations for bacterial phylogeny reconstruction have been developed based on these particular genes [49]. In this context, although 16S rRNA gene-based comparative analysis has been and continues to be exploited as the gold standard approach for bacterial phylogenetic investigations, it suffers from several limitations that prevent assessment of the real degree of genetic (dis)similarity between two microbial genomes [51], [52], [53]. Indeed, it does not guarantee accurate discrimination between very recently diverged species or amongst two bacterial taxa belonging to distant bacterial groups if they possess highly similar 16S rRNA gene sequences, thus not capturing phylogenetic differences [54]. To overcome these limitations, a multi-gene approach relying on comparative analyses of multiple conserved molecular markers, such as the clpC, dnaB, dnaG, dnaJ1, purF, and rpoC genes, has been proposed as a more reliable approach for species discrimination [53], [55]. In this context, such a multi-gene method has offered a higher level of discriminatory resolution between closely related bifidobacterial taxa compared to 16S rRNA-sequencing and therefore represents a reliable tool to assess phylogenetic relationships among members of the genus Bifidobacterium [56], [57]. However, the advent of Next-Generation Sequencing (NGS) techniques coupled with the development of new bioinformatic tools, has allowed easy access to bacterial genome sequences, which in turn has led to a revolution in phylogenetic investigations with major improvements of associated phylogenetic tree robustness [40], [58]. In this context, in order to shed light on the phylogenetic relatedness among bifidobacterial species, a recent comparative genome investigation, involving 69 of the currently recognized bifidobacterial taxa, allowed the identification of 261 clusters of orthologous genes (COGs) shared among all analyzed genomes, representing the so-called bifidobacterial core-genome [59]. Subsequent concatenation of sequences corresponding to the proteins encoded by the core genome allowed the generation of a bifidobacterial phylogenetic supertree which highlights the division of the genus Bifidobacterium into 10 distinct clusters corresponding to the B. adolescentis, B. asteroides, B. bifidum, B. bombi, B. boum, B. longum, B. pseudolongum, Bifidobacterium pullorum, Bifidobacterium psychraerophilum, and Bifidobacterium tissieri group. Notably, regardless of the method used, the closest position to the root of the generated tree is occupied by the B. asteroides group, thus supporting the previously proposed notion that this phylogenetic cluster represents the closest genetic relation to the evolutionary ancestor of the genus Bifidobacterium [60], [61] (Fig. 1). However, recent comparative genomic and phylogenomic analyses aimed at investigating the genetic relatedness of bacterial genera included in the Bifidobacteriaceae family, of which bifidobacteria are an integral part, revealed that members of the genus Bifidobacterium clustered separately from non-bifidobacterial members of the Bifidobacteriaceae family, except for Gardnerella vaginalis strains which, in contrast, are phylogenetically positioned within the genus Bifidobacterium [62], [63]. Specifically, Bifidobacterium tsurumiense was identified as the phylogenetically most closely related bifidobacterial species to G. vaginalis genotypes, thereby creating a new cluster located alongside the B. boum group [62]. These findings therefore suggest that G. vaginalis, the only species currently described in the genus Gardnerella, should be reclassified as a member of the genus Bifidobacterium, thereby undermining the notion that all bifidobacterial species represent health-promoting microorganisms [62]. Indeed, G. vaginalis is currently described as an opportunistic pathogen whose presence is tightly associated with chronic and/or acute bacterial vaginosis [62], [63], [64], [65], [66]. Furthermore, the recent large-scale application of 16S rRNA gene-based and bifidobacterial ITS expediate microbial profiling approaches for microbiota analyses has revealed the presence of putative novel members of the genus Bifidobacterium [36], [41], [67]. All together, these observations suggest the need for a re-evaluation of the currently known taxonomy of the genus Bifidobacterium and make a further expansion and refinement of bifidobacterial phylogenetic classification predictable.
Adaptation of bifidobacteria to the mammalian gastrointestinal environment. In order to reach, colonize and survive in their main ecological niche, i.e. the large intestine, bifidobacteria have to counter the hostile and adverse conditions that are typical of the upper compartments of the mammalian GIT. Exposure to oxygen or oxygen-derived free radicals, organic and bile acids or osmotic stress are only some of the detrimental factors that may undermine bifidobacterial cell viability and functionality [8]. To cope with these stressful conditions, bifidobacteria have evolved adaptive responses that rely on a strictly controlled expression of several enzymes and proteins, encompassing molecular chaperons, bile efflux transporters or bile salt hydrolases coupled with ATPases and two component systems to promptly counteract the multitude of challenging gut conditions [68], [69], [70], [71]. Induction and assembly of the stress-induced response factors are controlled by a universal system whose regulation is driven by an interactive regulatory network that is highly conserved among bifidobacterial species [8], [72], [73], [74]. To limit oxygen or oxygen-derived free radical-associated insults, many anaerobic bacteria produce specific enzymes, such as NADH oxidase or peroxidase, catalase and/or superoxide dismutase, directly involved in the inactivation of reactive oxygen species (ROS) [75], [76]. Despite their anaerobic nature, bifidobacterial genomes lack genes encoding NADH peroxidase, catalase or superoxide dismutase activities, except for the oxygen-tolerant B. asteroides strains [61], [77], thus suggesting that an alternative mechanism is activated by these microorganisms to protect DNA and proteins against oxidative stress-induced damage. Actually, it has been observed that, when exposed to aerobic conditions, Bifidobacterium longum NCC2705 produces an alkyl hydroperoxide reductase, while Bifidobacterium animalis subsp. lactis strains upregulate genes encoding thioredoxin peroxidases and NADH oxidase, suggesting that these enzymes represent critical bifidobacterial actions to deal with oxidative stress [78], [79], [80], [81]. Similarly, to avoid oxidative stress-associated accumulation of unfolded and/or misfolded proteins, bifidobacteria respond by producing various molecular chaperones and proteases [72], [73].
In addition to oxidative stress, bifidobacteria have to cope with acid stress principally due to the low pH of the stomach and/or pH reduction that may occur along the intestine due to carbohydrate fermentation-associated production of organic acid by gut bacteria [82]. As a consequence, bifidobacteria may experience accumulation of protons (H+) in the intracellular environment leading to reduced cytoplasmic pH and compromised cellular transport systems and proton motive force [1]. To tolerate and react to these threatening conditions, bifidobacterial species have evolved acid resistance features by activating an F0F1-ATPase that ensures maintenance of pH homeostasis through active proton extrusion [83], [84]. At the same time, members of the genus Bifidobacterium may improve their acid resistance response by modifying cell membrane fatty acid composition or cell wall peptidoglycan biosynthesis [85], [86].
Beyond oxygen, ROS or acid exposure, the presence of primary bile salts and/or secondary bile acids in the small intestine represents another major stress-inducing factor to bacteria because of their bactericidal properties. These compounds, acting as lipid emulsifiers, target the (bifido)bacterial membrane phospholipid bilayer, thus affecting membrane integrity and provoking bile accumulation in the cytoplasm [87], [88]. In this context, to avoid cell envelope damage, bifidobacteria may react to bile exposure by modulating membrane lipid composition through changes in their fatty acid metabolism to ultimately reduce cell membrane permeability [89], [90], [91]. At the same time, they may activate membrane-associated efflux pumps able to extrude bile salts and bile acids accumulated in the cytoplasm. Specifically, several multidrug transporters belonging to the ATP-binding cassette or the major facilitator superfamily have been identified as mediators of bile tolerance in B. breve and B. longum strains [70], [92], [93]. However, other defence mechanisms may be exploited by bifidobacterial taxa to limit the harmful effects of bile, i.e. production of bile salt hydrolases and biofilm formation. Specifically, bile salt hydrolases confer protection by catalysing the de-conjugation of glycine and taurine from bile salts generating unconjugated acids that can be metabolized by other intestinal bacteria [94], [95]. In addition, biofilm formation has been proposed as a crucial adaptive strategy in response to bile stress relying on a multi-factorial process involving exopolysaccharide (EPS) production as well as protein and extracellular DNA release, as observed in the bifidobacterial prototype B. breve UCC2003 [96].
Furthermore, along the GIT, bifidobacteria can encounter host-derived proteases, such as eukaryotic-type serine proteases, especially in case of intestinal inflammation caused by bacterial infection or intestinal tissue damage related to inflammatory bowel diseases or ulcerative colitis [97], [98]. To protect themselves from host-derived proteolytic action and survive in a competitive environment, certain bifidobacterial species are supplied with a two component regulatory system which, when activated, is able to selectively induce the expression of a serine protease inhibitor (serpin), thereby inhibiting the activity of particular host proteases [3], [99], [100].
Bifidobacteria and their interaction with the host environment. Once the challenges imposed by upper part of the host GIT have been successfully overcome, bifidobacteria have to face yet other challenges. Indeed, despite having reached their preferred ecological environment, they have to employ several strategies to interact with the host and ensure their fitness and survival in the ecosystem of the caecum and large intestine. Although the molecular mechanisms through which bifidobacteria communicate with their host and adhere to the host intestinal epithelium are still under scrutiny, the combination of in vitro investigations with the availability of whole genome sequences of currently recognized bifidobacterial taxa has been instrumental in predicting and identifying various genes and associated macromolecules that drive microbe-host interactions [1], [101], [102]. In this context, certain extracellular structures, bioactive molecules and/or excreted enzymes play a crucial role in promoting the successful gut colonization by bifidobacterial species (Fig. 2) (Table 1).
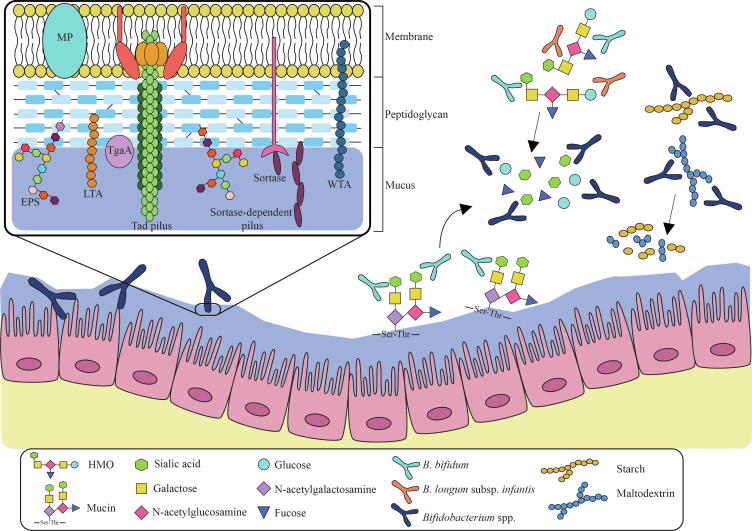
Fig. 2.
Bifidobacterial strategies to successfully colonize and survive in the human intestine. A schematic overview of the macromolecular structures exposed on the bifidobacterial surface and involved in host-microbe interactions are reported on the left: membrane protein (MP), exopolysaccharide (EPS), TgaA, wall and lipoteichoic acids (WTA and LTA) coupled with sortase-dependent and Tad pili. In addition, on the right, bifidobacterial degradative activities toward different diet- and host-derived complex carbohydrates are depicted. Hydrolysis of complex sugars by certain bifidobacterial species produces simple glycans that can be directly utilize as carbon sources by the same bifidobacterial species and/or metabolized by other members of the Bifidobacterium genus trough cross-feeding.
Table 1.
List of genes whose product is directly involved in bifidobacteria-host interaction.
| Genes | Gene product | Product function | |
|---|---|---|---|
| Sortase-dependent pili | fimA/P | Major pilin subunit | Structural element of sortase-dependent pili. |
| fimB/Q | Minor or ancillary pilin subunit | ||
| strA | Sortase (transpeptidase enzyme) | Sortase-dependent pilus assembly and anchoring to the cell surface. | |
| Tight Adherence pili | tadA | ATPase | |
| tadB | Integral membrane protein | Formation of hetero- and homo-polymers involved in the secretion apparatus of the pilus subunits. | |
| tadC | integral membrane protein | ||
| tadE | Pseudopilin | Structural pilus components synthetized as prepilins. | |
| tadF | Pseudopilin | ||
| Flp | Prepilin | ||
| tadV | Peptidase (enzyme) | Post-translational processing of prepilins to remove their hydrophobic leader peptide to obtain structural pilus components. | |
| Quorum sensing | luxS | S-ribosylhomocysteine lyase | It catalyzes the transformation of S-ribosylhomocysteine to homocysteine and 4,5-dihydroxy-2,3-pentadione, the latter known as the precursor of the autoinducer-2 (AI-2). |
| sEPS | Conserved gene set involved in sEPS production | Priming glycosyl transferase | It catalyzes the addition of the first saccharidic moiety to a lipophilic carrier molecule for initiation of sEPS repeating unit assembly. |
| Glycosyl transferases | They are involved in the assembly of sEPS repeating units. | ||
| Flippases and/or ABC transporters | Enzymes involved in the transport of sEPS subunits from the cytoplasm to the outer membrane environment. | ||
| Subunit polymerase | It catalyzes the polymerization of sEPS subunits. | ||
| Chain length determinant proteins | They determine the polymer chain length. | ||
| Carbohydrate precursor biosynthesis/modification enzymes | They catalyze the biosynthesis of sEPS precursor and the conversion of the latter into sEPS subunits. |
1.1. Pili
Described as long hair-like proteinaceous appendages protruding from the extracellular cell surface, pili and fimbriae have been identified as one of the principal extracellular structures directly involved in host-microbe interactions by promoting adhesion to the intestinal epithelium surface [103], [104]. Pilus proteins are not constitutively produced by bifidobacterial species, but rather their assembly is specifically induced under certain growth conditions, such as in vivo colonization, in vitro co-culture with human cell lines and/or exposure to the extracellular matrix [102], [105], [106], [107]. Specifically, two different pilus types have been described in bifidobacteria, one dependent on the enzymatic activity of a transpeptidase (sortase) and for this reason defined as sortase-dependent pili, while the other resembles type IVb pili, and in particular the so-called Tight Adherence pili (Tad pili) [106], [107].
Sortase-dependent pili owe their name to the peculiar mechanism by which they are assembled. Indeed, in addition to two different structural proteins represented by the so-called major pilin and ancillary pilin, sortase-dependent pilus assembly requires a pilus-specific sortase, a transpeptidase which, by cross-linking the pilus building blocks and covalently anchoring the resulting polymer to the cell surface wall, ensures correct pilus shaft formation [104], [108]. Genes encoding the sortase-dependent pilus structural and assembly proteins are organized in the same genetic locus, and include the fimA or fimP genes coding for the major pilin subunit, fimB and/or fimQ encoding for one or two minor (ancillary) pilin subunits and the sortase-encoding gene strA [106], [109]. However, comparative genome analysis revealed that sortase-dependent encoding loci are not evenly distributed among members of the genus Bifidobacterium. Indeed, while the genome of certain B. actinocoloniiforme, B. longum subsp. infantis and B. longum subsp. longum strains completely lack the genetic repertoire necessary for the production of this pilus type, the genome of B. dentium Bd1 possesses seven different pilus-encoding loci, the largest number so far recorded for a bifidobacterial species [106], [109]. Beyond this varying number of loci, also the genetic sequences of the sortase-dependent pilus-encoding genes are characterized by a high interspecies or inter-strain variability coupled with G + C content deviation and different codon usage, which suggests that these genetic elements have been acquired through horizontal gene transfer [106]. This notion has been corroborated by the observation that sortase-dependent pilus-encoding loci are frequently localized near transposon elements [106].
Despite their important role in mediating bifidobacterial adhesion to host surfaces, sortase-dependent pili have additional functions. They are involved in bifidobacteria-host immune dialogue as observed for B. bifidum PRL2010 whose sortase-dependent pili can trigger an increased level of TNF-α cytokines and a parallel reduction of the proinflammatory cytokine IL-12, thus moderating immune cells to avoid a detrimental inflammatory response [102]. At the same time, bifidobacterial sortase-dependent pili are involved in microbe-microbe interactions. Indeed, B. bifidum PRL2010 pili have been described as physical bridges able to promote aggregation between bacterial cells of a heterogeneous population, such as that of the gut microbiota [105].
The second pilus type produced by bifidobacteria, i.e. the Tad pilus, was originally described in the pathogenic Gram-negative Actinobacillus actinomycetemcomitans where it is involved in adhesion to the host cell surface, thereby promoting colonization and pathogenesis through biofilm formation [110], [111]. All genes required for the production and assembly of this pilus type are part of the same genetic locus, except for the tadV gene which is not located within the tad locus. Within the tad locus, the tadA gene encodes an ATPase which is believed to be localized at the periphery of the cytoplasmic membrane, while the TadB and TadC proteins are predicted integral membrane proteins to form homo- or hetero-oligomeric structures that together with TadA constitute the proposed secretion apparatus for the pilus subunits [111], [112]. Furthermore, the tad locus contains genes which encode structural pilus components, i.e. prepilin (Flp), and two pseudopilins (TadE and TadF), all of which are synthetized as prepilins, requiring post-translationally processing by a dedicated peptidase (TadV) to remove their hydrophobic leader peptide [107], [111], [112].
Unlike the sortase-dependent pilus gene clusters, the tad locus seems to be highly conserved among bifidobacterial species [107], [111], [113], [114]. However, this pilus type has to date only been characterized in B. breve UCC2003 where Tad pili have been shown to stimulate in vivo colonic epithelial cell proliferation thus contributing to host mucosal homeostasis, while they are also involved in host cell adhesion to ensure successful intestinal colonization and persistence [107], [115].
In addition to sortase-dependent and Tad pili, the gene cluster encoding the biosynthetic machinery for a third pilus type (type IVa pilus) was discovered in the B. adolescentis 22L genome. Transcriptomic analysis revealed that this type IVa pilus-specifying locus is upregulated when the bifidobacterial strain was cultivated on starch when compared to the control condition in glucose, suggesting that non-digestible, plant-derived glycans, in which the intestine is particularly enriched, is involved in the regulated production of these extracellular structures, in line with the notion that pili represent extracellular structures required for gut colonization and host-microbe interactions [116]. However, additional in vitro and/or in vivo studies are required to further confirm this hypothesis.
2. Extracellular polysaccharides
The cell surface of a wide spectrum of bacteria is known to be covered by one or more glycan layers, designated here as capsular polysaccharides (CPSs) to indicate that such polymers are covalently anchored to the cell surface forming a capsule, whereas they take the name of exopolysaccharides (EPSs) when they create a slimy coat that is loosely attached to the cell wall and is easily released into the growth environment [117]. However, due to the difficulties in distinguishing between EPS and CPS, since their chemical structures may be highly similar, in the present review we will use the term surface-associated exopolysaccharides or sEPSs to indicate either of these extracellular glycan layer types.
Most common monosaccharides involved in the formation of sEPSs correspond to glucose, fructose, galactose, rhamnose and fucose. However, the presence of different isomers, acetylated variants, various chemical linkage types for the monosaccharide subunit polymerization as well as multiple degrees of polymerization and branching patterns of the repeating sugar units contribute to create a large variety of possible combinations for the formation of sEPSs, thus generating a myriad of different long-chain exopolysaccharides [118]. Furthermore, according to their chemical composition and biosynthesis mechanism, sEPS may be classified either as homopolysaccharides, when composed by the repetition of a single monosaccharide unit, or heteropolysaccharides, when the sEPS structure contains two or more different monosaccharide subunits [117], [119].
From a genomic perspective, genes involved in sEPS biosynthesis are generally clustered together within a particular genetic locus (typically referred as the eps cluster) in members of the genus Bifidobacterium [116], [120], [121], [122]. Comparative genome analyses involving different bifidobacterial species revealed the presence of at least one eps locus in all analysed taxa, except for some B. bifidum strains [121], [122]. However, despite the high prevalence of this genetic locus across bifidobacterial genomes, no conserved structural organization was observed in the eps biosynthesis cluster. Rather, the latter is characterized by a consistent interspecies variability in terms of cluster length, predicted functions and number of genes, ranging from nine genes of the Bifidobacterium mongoliense type strain eps locus to the 55 genes identified in the eps region of B. dentium LMG11405, suggesting the acquisition of the eps-encoding gene cluster through horizontal gene transfer events [121], [122], [123]. This notion received support from the finding that this genetic locus is characterized by a different GC content and flanked by insertion sequence elements or transposase-encoding genes [122].
Various biosynthetic functions have been attributed to genes located in the eps locus, with certain associated genes apparently being conserved among bifidobacterial genomes, allowing in silico prediction of the presence of complete eps clusters in bifidobacterial taxa. In detail, these conserved genes encode for proteins specifically involved in the sEPS subunit assembly and attachment to the cell surface, in particular genes encoding (i) a priming glycosyl transferase together with one or more glycosyl transferases which are involved in the sEPS subunit biosynthesis, (ii) a flippase or ABC transporter for the transport of the sEPS subunits from the cytoplasm to the outer membrane environment, (iii) a polymerase able to extracellularly link transported sEPS subunits, (iv) a polysaccharide chain length determinant, and (v) monosaccharide biosynthesis or modification enzymes [121], [123]. Despite the high prevalence of eps gene clusters, there are still many strains that do not appear to produce sEPS as based on their sedimentation phenotype when they are cultivated in liquid medium, which is indicative of a EPS-negative phenotype and shows the high level of variability in sEPS production ability among individual strains that belong to the genus Bifidobacterium [122], [124], [125], [126].
Recently, the scientific community has shown increased interest in bifidobacterial sEPS producers since these complex extracellular glycans, by being involved in microbe-host interactions, have been reported to exert various beneficial host effects [101], [127], [128]. In vitro and in vivo investigations have shown that sEPS production not only favours adhesion of strains to intestinal cell lines, but also solicits a differential immune response that appears to depend on the physicochemical features of the sEPS [3], [129], [130]. Indeed, a positive correlation has been observed between the structure, composition and size of a given sEPS and the corresponding elicited immune response, as described for certain strains of B. animalis subsp. lactis, B. adolescentis and B. longum subsp. longum [126], [129], [130], [131]. Furthermore, a murine in vivo trial demonstrated that the B. breve UCC2003 sEPS acts as an ‘immunological silencer’ since this extracellular polymer protects the bifidobacterial strain from clearance by the adaptive immune system and simultaneously reduces the anti-inflammatory response [1], [101]. Similarly, the sEPS produced by B. longum subsp. longum 35624 was reported to prevent induction of an inflammatory response against this bifidobacterial strain, reinforcing the notion that sEPSs not only possess immunomodulatory activities, but that they are also involved in colonization and survival abilities of this microorganism in the intestinal environment by eluding the host immune system [132].
In addition to their ability to modulate immune responses, many other beneficial functions have been attributed to bifidobacterial sEPSs. Indeed, they are able to modulate both composition and metabolic activity of the intestinal microbial community by acting as fermentable substrates for members of the gut microbiota [133], [134], [135], [136]. At the same time, these extracellular polymers are involved in conferring protection against pathogens. In this context, a murine in vivo trail based on the administration of the EPS-producing B. breve UCC2003 (EPS+) or EPS-deficient B. breve UCC2003 (EPS-) strains with Citrobacter rodentium revealed reduced colonization levels of this murine gut pathogen in the presence of EPS+ B. breve [101]. Furthermore, in recent years, several studies have demonstrated the role of bifidobacterial sEPS in reducing apoptotic epithelial cell shedding to safeguard the integrity of the epithelial intestinal barrier or soliciting apparent antitumor activity by regulating cell cycle and cell apoptosis, thus corroborating the health-promoting effect of these glycan layers [128], [137], [138].
2.1. Other bioactive compounds
Bacteria are able to communicate with each other and coordinate appropriate response to variations in their surrounding environment by activating an intracellular communication, also known as quorum sensing, based on a system of gene expression regulation that is driven by bacterial cell density [139]. Specifically, it depends on the accumulation of signalling molecules which, when they exceed a certain concentration threshold, trigger a concerted physiological response in the microbial community regulating diverse phenomena, such as virulence, bioluminescence or biofilm formation [140]. Autoinducer-2 (AI-2) is one of the principal signal molecules regulating quorum sensing and its production relies on the LuxS enzyme involved in recycling S-adenosylhomocysteine. LuxS catalyses the cleavage of S-ribosyl-homocysteine into homocysteine and 4,5-dihydroxy-2,3-pentanedione, the precursor of AI-2 [141]. Although in silico investigations aimed at predicting the presence of the luxS gene in bifidobacteria has been limited to only a small number of species, its conservation in all analysed genomes suggests the possible presence of such a gene in all bifidobacterial species [8], [142], [143]. Despite the as yet incomplete characterization of this cell density-dependent regulatory mechanism within the genus Bifidobacterium, bifidobacterial AI-2 production seems to be involved in biofilm formation, suggesting that AI-2 plays a crucial role in influencing the colonization of these symbiotic microorganisms in the gut ecosystem [96], [142]. This hypothesis was further corroborated by the observation that a B. breve UCC2003 luxS insertion mutant strain is much more sensitive to multiple iron chelators, is not able to colonize the murine GIT and was shown to confer less protection against a pathogen infection in Caenorhabditis elegans [143]. Therefore, AI-2 appears to endow bifidobacteria with a competitive advantage to colonize and persist in the intestine.
In addition, bifidobacteria are able to synthetize teichoic acids, negatively charged polymers exposed on the cell surface. Precisely, depending on their particular attachment to the cell envelope, they can be classified as wall teichoic acid (WTA) when covalently linked to the peptidoglycan, or lipoteichoic acids (LTA) when there are linked to the diacylglycerol moiety of a membrane lipid [144], [145]. Despite the in silico predicted presence of genes encoding the biosynthetic machinery for WTA and LTA production in many bifidobacterial species, the role and function of bifidobacterial teichoic acid production remains poorly investigated [146]. Indeed, only in B. bifidum PRL2010, a study has shown that enhanced teichoic production occurs under environmental conditions simulating the human gut, suggesting a functional role for these polymers in supporting host-microbe interaction and promoting their gut colonization [146].
Another bifidobacterial surface protein, identified in B. bifidum MIMBb75 and involved in the adaptation and persistence of bifidobacteria in the mammalian gut, is represented by the TgaA protein, a peptidoglycan-lytic enzyme composed by two different conserved regions, i.e. lytic murein transglycosylase and cysteine- and histidine-dependent amidohydrolase/peptidase [147]. Specifically, in vitro experiments revealed that this TgaA protein is directly implied in cell-to-cell communication as well as in cross talk with host cells, suggesting its involvement in supporting bifidobacteria in gut colonization [147], [148]. However, as for teichoic acid, further investigations are necessary to shed light on the conservation of the genetic apparatus required for TgaA production and to understand its precise role in driving microbe- and/or host-bifidobacteria interactions.
Glycan utilization as an adaptative metabolic strategy of bifidobacteria to the gut environment. Simple sugars such as lactose or sucrose are generally metabolized by the host or by microbial community that inhabits the upper part of the host GIT, leaving complex carbohydrates to pass to more distal intestinal compartments [149]. At this site, such non-digestible glycans are represented by diet-derived carbohydrates, including cellulose, fructans, pectins, xylans or resistant starch, as well as by host-derived sugars, encompassing Human Milk Oligosaccharides (HMO) and mucins, in the latter case represented by O-linked and/or N-linked glycoproteins secreted by the intestinal goblet cells with the function of covering and protecting the gut epithelium [150] (Fig. 1). The ability to degrade a wide range of complex carbohydrates is one of the key genetic strategies that can ensure successful colonization and survival of bacteria in the intestinal environment. Since bifidobacteria are considered as symbiotic microorganisms of the mammalian GIT, it is not surprising that their genomes contain a large number of genes predicted to encode enzymes involved in carbohydrate metabolism [40], [57]. In this context, a comparative analysis involving 47 bifidobacterial type strains allowed the identification of the core of bifidobacterial genomic coding sequences (core BifCOGs) and subsequent functional annotation revealed that, apart from conserved core genes encoding housekeeping functions, quite a number of genes are associated with carbohydrate metabolism [40], [57]. In addition to assessing the bifidobacterial core-genome, the same study investigated the bifidobacterial variome (variable genome sequences) allowing the identification of so-called Truly Unique Genes (TUGs), i.e. genes present in just one of the examined bifidobacterial genomes. Also in this case, although no functional annotation was attributed to most of the predicted TUGs, some of these truly unique open reading frames were identified as genes encoding proteins involved in carbohydrate transport and metabolism, including glycosyl hydrolases, thus corroborating the central role that bifidobacterial carbohydrate degradation abilities play in driving their adaptation to the intestinal tract [40].
The bifidobacterial genetic repertoire required for the utilization of complex glycans is generally organized in glycan-specific gene clusters encompassing sequences encoding proteins and enzymes involved in sugar transport, such as permeases, proton symporters, phosphoenolpyruvate-phosphotransferase systems and carbohydrate-specific ATP-binding cassette (ABC) transporters, as well as glycosyl hydrolases (GHs) [150]. Furthermore, all bifidobacterial genomes contain the genes encoding the so-called ‘bifid shunt’, encompassing the central metabolic route that contributes to the ecological success of bifidobacteria, and generating 2.5 ATP molecules coupled with 1 Mol of lactate and 1.5 Mol of acetate per mol of glucose metabolized [151]. However, in order to recover energy from complex carbohydrates through the bifid shunt route, bifidobacteria needs the primary action of extracellular and/or intracellular GHs which, by hydrolyzing the glycosidic bond between two or more carbohydrate subunits, catalyze the breakdown of complex polysaccharide into mono- or oligo-saccharides that can then be internalized through dedicated carbohydrate uptake systems [150]. Once inside the cytoplasm, sugars may be subjected to further modification to ultimately produce phosphorylated monosaccharides which can then enter the central fermentation pathway [152].
In silico characterization of the genus Bifidobacterium pan-genome, based on the Carbohydrate-Active Enzymes (CAZy) database, identified various glycosyl hydrolases belonging to GH13 family as the most abundant and recurring GH family within the bifidobacterial glycobiome [40], [44]. Members of this enzymatic family are classified as α-glucosidases, thus able to degrade complex glycans containing α-glucopyranose units, such as starch, glycogen and related derivatives, including amylopectin, amylose, pullulan, maltodextrin and cyclomaltodextrin, representing common carbohydrates found in the adult mammalian (omnivorous and herbivorous) diet [9], [44], [153]. Together with GH13, bifidobacterial genomes possess a broad set of genes whose products belong to the GH3, GH43 and GH51 families, representing enzymes involved in the degradation of plant-derived carbohydrates, thereby suggesting the adaptation of these microorganisms to the mammalian gut environment through a generalist ecological behavior [9], [57]. Furthermore, β-galactosidase activity encoded by genes belonging to the GH2 and GH42 families is another widespread feature across the genus Bifidobacterium allowing the growth on lactose, galacto-oligosaccharides and galactan as well as the removal of galactose subunits from mucin- and/or milk-derived oligosaccharides [150], [154], [155].
However, bifidobacteria are not simply inhabitants of the gut of adult mammals, but they are overall described as pioneering and abundant colonizers of the infant intestine. From a human perspective, the prevalence and abundance levels of members of the genus Bifidobacterium in the infant gut depends on mode of delivery (natural or C-section delivery) and type of feeding (breast- or formula-feeding). Specifically, the combination of natural delivery and breast-feeding has been recognized as the condition that best promotes colonization and survival of bifidobacterial species in the infant intestinal environment. Indeed, while natural delivery promotes infant gut colonization by bifidobacteria through vertical transmission events, breast-feeding offers bifidobacteria the possibility to persist and proliferate in this ecological niche by providing complex host-derived glycans, i.e. HMOs, that represent bifidogenic compounds available at the very early stage of life in the infant intestine [10], [11], [13], [14], [15]. Produced by the mammary gland, HMOs are structurally diverse and complex unconjugated glycans, being highly abundant in and rather unique to human milk, able to reach the large intestine where they act as selective substrates for bifidobacterial growth [156]. The basic structure of these host-derived carbohydrates includes combinations of at least three of five distinct monosaccharides, including glucose, galactose, N-acetylglucosamine, fucose and sialic acid, the latter generally represented by N-acetyl-neuraminic acid [156], [157]. All HMOs contain lactose at their reducing end and are extended in various ways with these HMO building blocks through various glycolytic linkages, resulting in an immense combinatorial potential, highlighted by the fact that more than 200 structurally different human milk oligosaccharides have been described [156], [157], [158].
Conversely to the ubiquitous ability of bifidobacteria to degrade plant-derived polysaccharides, their access to HMOs seems to be an exclusive feature of only specific, infant-associated bifidobacterial species, i.e. B. bifidum, B. longum subsp. infantis as well as certain B. breve, B. pseudocatenulatum, Bifidobacterium catenulatum subsp. kashiwanohense, B. longum subsp. suis and B. longum subsp. longum strains [68], [159], [160], [161], [162], [163], [164], [165], [166], [167]. Not by chance, in silico investigations have revealed conserved gene clusters encoding ABC transporters with high affinity binding proteins, upstream regulatory elements and one or multiple GHs that, as a whole, represent the genetic arsenal through which the abovementioned bifidobacterial species degrade and internalize HMOs [9], [11]. Specifically, the B. longum subsp. infantis genome possesses a contiguous genomic segment encoding GHs active on the four HMO glycosidic linkages, represented by α-fucosidase, α-sialidase, β-galactosidase and β-N-hexosaminidase activities, and flanked by a cluster specifying an ABC transporter system composed of extracellular solute binding proteins and permeases, presumed to be required for HMO internalization [9], [162], [168], [169], [170]. However, due to the lack of identifiable secretion signals in the glycosidases produced by B. longum subsp. infantis, it has been inferred that HMO utilization by this bifidobacterial species relies on the initial uptake of intact HMOs which are then intracellularly metabolized by the central fructose-6-phosphate phosphoketolase pathway. Because of the apparent inability to perform extracellular HMO degradation, B. longum subsp. infantis is restricted to utilize HMOs with a relatively low degree of polymerization (DP ≤ 8), due to size limitations imposed by the ABC-type transporter [168], [171]. In contrast, the production of extracellular fucosidases and sialidases, principally encoded by genes belonging to the GH20, GH29, GH33 and GH95 families, allows B. bifidum strains to activate an initial extracellular degradation of (larger) HMOs, thereby acquiring access to highly polymerized host-derived glycans and ensuring the ecological success of this bifidobacterial species in the (breast-fed) infant gut environment [68], [158], [171], [172], [173], [174], [175]. After the initial extracellular processing, the generated mono- or di-saccharides (galactose, glucose, lactose and lacto-N-biose) are subsequently imported to be further metabolized intracellularly through the central fermentative pathway [169], [172], [173].
Apart from B. bifidum and B. longum subsp. infantis in which the ability to retrieve energy from HMO breakdown is a conserved feature, also B. longum subsp. longum and the infant gut associated B. breve can access certain HMOs [161], [176]. Indeed, all B. breve strains appear to possess metabolic abilities that allow them to grow on at least three different HMOs, i.e. lacto-N-tetraose (LNT) and lacto-N-neotetraose (LNnT) and lacto-N-biose (LNB) [160], [161], [177]. At the same time, certain B. longum subsp. longum strains encode lacto-N-biosidase activity degrading LNT to LNB and lactose [176], [178]. Furthermore, several B. breve, B. longum subsp. longum, Bifidobacterium kashiwanohense and Bifidobacterium pseudocatenulatum strains utilize some of the most abundant HMOs, such as 2′-fucosyllactose and 3′-fucosyllactose thanks to the presence, in their genomes, of a gene cluster encoding an α-fucosidase, while some B. longum subsp. suis strains are able to metabolize L-fucose [164], [167], [179], [180]. However, both B. breve and B. longum subsp. longum species are able to degrade only HMOs with a very low degree of polymerization. Therefore, to get access to these simplified host-derived glycans, they generally rely on cross-feeding events, during which other HMO-degrader bifidobacterial species deconstruct larger HMOs, to guarantee their fitness in the competitive infant gut environment [181], [182].
Another abundant class of host-derived glycans typical of the bifidobacterial elective ecological niche is represented by mucins, which are glycoproteins covering the GIT epithelium forming a thick protective layer of mucus [183]. Based on their chemical structures, mucins are described as an O-glycosylated protein component when represented by tandem repeat domains of proline, threonine and serine modified by the addition of monosaccharides such as N-acetylglucosamine, N-acetylgalactosamine, fucose and galactose that can be frequently decorated with sialic acid and sulphate, or as N-glycosylated proteins when asparagine residues are glycosylated by the addition of monosaccharide subunits to a common glycan core formed by mannose and N-acetylglucosamine [184], [185], [186].
Beyond their primary function of protecting the intestinal epithelium, mucins also represent important carbon sources for GI bacteria, especially in the distal colon where the availability and accessibility of carbohydrates is limited due to the high level of competition for nutrients among microbial gut colonizers [187]. Despite different types of glycosidic linkages that connect the mucin building blocks, the core structure of mucins is similar to that of HMOs. Not by chance, pathways activated by bifidobacteria for the degradation of these two host-derived glycans involve the same enzymes such as endo-α-N-acetylgalactosaminidases, α-L-fucosidases, sialidases, N-acetyl-β-hexosaminidases and β-galactosidases [1], [187]. Furthermore, given their structural similarity, and similar to what has been observed for HMOs, the ability to degrade mucins is restricted to a limited number of bifidobacterial species. Genome comparative analysis coupled with in vitro investigations have shown that B. bifidum have evolved the ability to efficiently metabolize mucins. Genomes of the B. bifidum species possess a conserved set of genes encoding mucin-specific glycolytic enzymes, which is indicative of how this bifidobacterial species has extensively adapted to exploit the carbon sources available in the extremely competitive gut environment, ensuring long-term colonization and survival [11], [158], [187], [188], [189]. Specifically, this conserved genetic pattern involves, as observed in the prototype B. bifidum PRL2010, both extracellular and intracellular glycosyl hydrolases, prevalently represented by fucosidases, sialidases, N-acetyl-β-hexosaminidases, β-galactosidases and an endo-α-N-acetylgalactosaminisase, coupled with carbohydrate transporters belonging to various categories, such as ATP-binding cassette (ABC) transport systems, phosphoenolpyruvate phosphotransferase system and major facilitator superfamily members, and several cell membrane-localized solute binding proteins which are presumed to bind specific oligosaccharides for their subsequent internalization via B. bifidum-associated membrane permeases [68], [187]. Furthermore, genes encoding for enzymes involved in the degradation of N-glycans were also identified in certain B. longum subsp. longum, B. longum subsp. infantis and B. breve [184]. Although all other bifidobacterial species lack the complete genetic arsenal dedicated to mucin degradation, certain bifidobacterial taxa can take advantage of the extracellular release of mucin components by the B. bifidum carbohydrate-degradative activity [181], [190]. In this context, despite lacking the complete genetic arsenal required for mucin metabolism, B. breve UCC2003 genome contains genes involved in the degradation of mucin components, such as sialic acid, sulphated amino sugars and fucose [190], [191]. In this context, co-culture experiments have shown that, through cross-feeding, the metabolically versatile B. breve UCC2003 is able to take advantage of the release of sialic acid by B. bifidum PRL2010 for its growth and proliferation, thus emphasizing the extensive adaptation of bifidobacterial species to the gut environment [192].
Genetic mechanisms adopted by bifidobacteria to counter bacteriophages. To preserve their survival in their preferred ecological niche, bifidobacteria not only have to compete for nutrients with other bacterial commensals or withstand the hostile and adverse physiological conditions typical of the mammalian GIT, but they also have to counter bacteriophage attacks [193]. Bacteriophages or phages are described as viruses infecting prokaryotic cells in order to use host transcription, replication and translation capabilities for their own proliferation, thus playing a crucial role in shaping the composition and diversity of bacterial ecosystems, favoring horizontal gene transfer and nutrient turnover, activating continuous cycles of predation and co-evolution with their host cells [194]. Based on their lifestyle, bacteriophages may be classified as virulent or temperate. The initial mode of infection is common to the two types of bacteriophages, consisting of phage genome injection into the cytoplasm of their bacterial host. Subsequently, while virulent bacteriophages take over all major bacterial cell functions to replicate their own genome, produce phage proteins and assemble phage particles that will be released upon forced lysis of the bacterial cell, temperate bacteriophages do not lyse the host bacterial cell after infection [195]. Instead, the temperate bacteriophage DNA is integrated into the chromosome of the infected bacterium creating the so-called prophage, a state in which phage genes are not expressed until prophage induction occurs with the subsequent excision of phage DNA from the bacterial chromosome followed by activation of the lytic life cycle [195].
Bacteriophages have been identified as the most abundant biological entities on the planet, so much that, in certain environments including the intestine, they far outnumber the prokaryotic cell count [196], [197]. Despite the abundance of phages in the gut ecosystem, the phageome remained poorly explored for a long time, ending up representing the “known unknown” component of the gut microbiome, due to the lack of a reliable bacteriophage isolation protocol [196]. However, the advent of NGS techniques provided powerful tools to appreciate the complexity of intestinal gut bacteriophage community, revealing that a large part of bacterial viruses inhabiting the gut is still unclassified and strictly host-specific, targeting only a single or few bacterial taxa, thereby minimizing the impact on intestinal commensal members [198]. Furthermore, molecular techniques highlighted that most identified intestinal bacteriophage sequences correspond to temperate phages, a phenomenon that may contribute to bacterial intra-species diversity through prophage acquisition by lateral transfer [199]. In addition, while phages have for a long time been regarded as microorganisms able to influence immunity only indirectly by acting on the mammalian microbiome, recent studies have demonstrated that phages actually play a direct role in interacting with the mammalian host immune system, modulating both innate and adaptive immunity [197].
Until relatively recently, bifidobacterial genomes were thought to be free from phage infections [200]. However, the recent rapid diffusion of whole bifidobacterial genome sequences have promoted large-scale comparative genome analyses which, in turn, have generated ample genetic evidence of interspersed prophage-like elements or so-called bifidoprophages in most bifidobacterial chromosomes [201], [202], [203]. In most of cases, the genetic architecture of bifidoprophages may be subdivided into functional modules resembling a typical lambdoid phage genome organization, including modules encoding DNA replication, DNA packaging, lysogeny, head and tail morphogenesis and host lysis [202]. Despite the conserved functional modules, bifidoprophages are characterized by a high sequence variability and high degree of sequence degeneration, suggesting an alteration of the bifidobacterial genomes following phage infection and genome integration [202]. In this context, prophages are a double-edged sword. On the one hand, (pro)phages may contribute to host fitness through provision of beneficial properties and activities. In this context, it has been observed that B. adolescentis prophages encode restriction/modification systems able to cleave specific recognition sequences on foreign DNA including virulent phages, thus providing protection to the bacterial host against invading genetic elements [204]. At the same time, a B. longum subsp. infantis prophage provides an advantage to the host, encoding a tRNA-encoding gene able to complement or enhance the host translational capabilities [193], [204]. On the other hand, prophages represent a real threat for bifidobacteria in case of induction from the bacterial host chromosome leading to host cell lysis [193]. In this context, to counteract bacteriophage predation and possible subsequent host lysis, bifidobacteria have evolved phage defense systems represented by restriction modification (RM) systems and so-called Cluster Regularly Interspaced Short Palindromic Repeats (CRISPR) [205], [206]. However, while the RM is a generalist defense mechanism that targets any invading foreign DNA sequence, CRISPR is considered to be a specific anti-phage system [205]. The CRISPR locus is composed of multiple repetitions of pseudo-palindromic DNA sequences creating harpin secondary structures. Furthermore, each repetition is interspersed with a DNA spacer corresponding to an acquired exogenous DNA phage sequence [207]. In addition, the CRISPR locus is flanked by cas genes encoding proteins essential for the success of the defense machinery [207]. This protective system against bacteriophages involves three different stages. In the first phase, adaptation, fragments of viral DNA are selected and integrated into the CRISPR locus as a spacer sequence to provide a memory of infection. In the second stage, i.e. expression, the CRISPR array is transcribed in order to generate the correspondent mature CRISPR RNA (crRNA). In the third or interference phase, the crRNA guides Cas nucleases to selectively recognize complementary invasive nucleic acids promoting their cleavage [193], [208], [209]. Despite CRISPR-Cas system being a powerful defense mechanism for (bifido)bacteria to limit bacteriophage predation, a very recent investigation involving 954 (representing a total of 79 species) publicly available Bifidobacterium genomes and based on the utilization of bioinformatics tools developed to specifically identify CRISPR-Cas system, revealed that just 57% of the analyzed bifidobacterial genomes possess a CRISPR-Cas machinery [208]. At the same time, the observation of distinct types of CRISPR-Cas systems among bifidobacterial strains of the same (sub)species indicates that the presence of this defense mechanism is a strain-dependent feature rather than a general characteristic of the entire (sub)species [208], [210], [211]. Furthermore, in silico analyses aimed at evaluating the nature/origin of the acquired foreign DNA segment (spacers) within bifidobacterial CRISPR arrays revealed that a large part of the spacers showed homology to prophage sequences, suggesting that they operate a powerful defense mechanism to counter phage-induced lysis and improve their fitness in the gut environment [208], [209].
3. Summary and outlook
In recent decades, the advent of NGS techniques coupled with cultivation efforts has shown that bifidobacteria are abundant symbiotic microorganisms inhabiting the GIT of mammals and exerting various health-promoting effects for their host. At the same time, the availability of hundreds of sequenced bifidobacterial genomes has allowed large-scale comparative and functional genome analyses leading to a significant expansion of our knowledge regarding the genetic features that form the basis for bifidobacteria-host interactions. Indeed, these analyses have helped us to shed light on multiple molecular strategies that bifidobacteria have evolved in order to facilitate their colonization and long-term persistence in the extremely competitive gastrointestinal environment. In this context, it has been demonstrated that bifidobacterial genomes possess a genetic arsenal encoding proteins and enzymes directly involved in coping with environmental stresses that they may encounter along the GIT, including osmotic and oxidative stress. However, once they reach the large intestine, challenges for bifidobacteria are not over. Indeed, they have to get established and ensure long-term colonization and survival in the host gut. To achieve this, bifidobacteria have the possibility to adhere to the host epithelial cells by selectively express specific genetic clusters encoding molecules able to establish direct interactions with the host cells, including pili, sEPSs, autoinducer-2, teichoic acids and serpins. At the same time, bifidobacteria have to compete for nutrients with hundreds of other commensal bacterial genera/species. In this context, comparative genome analyses revealed that many members of the genus Bifidobacterium possess, relative to their genome size, one of the largest genetic repertoire for glycosyl hydrolases and carbohydrate uptake systems involved in the degradation and internalization of plant- and host-derived glycans. This genetic arsenal endows bifidobacteria with powerful and flexible metabolic strategies to compete with other members of the gut microbiota to ensure their fitness in the intestinal environment. Moreover, bifidobacterial genomes are provided with genetic systems aimed at limiting and containing possible threats from other abundant intestinal microbiota members such as bacteriophages. Therefore, NGS techniques coupled with in vitro experiments, have very substantially expanded our knowledge on the genetic mechanisms by which bifidobacteria ensure their colonization and survival in the GIT, while it has also given us extensive insights into the molecular mechanisms by which they interact and benefit their host [212].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank GenProbio srl for financial support of the Laboratory of Probiogenomics. D.v.S. is member of APC microbiome Ireland which is funded by SFI through the Irish Government’s National Development Plan (Grant Numbers SFI/12/RC/2273-P1 and SFI/12/RC/2273-P2). The authors declare that they have no competing interests.
Contributor Information
Douwe van Sinderen, Email: d.vansinderen@ucc.ie.
Marco Ventura, Email: marco.ventura@unipr.it.
References
- 1.Bottacini F., van Sinderen D., Ventura M. Omics of bifidobacteria: research and insights into their health-promoting activities. Biochem J. 2017;474(24):4137–4152. doi: 10.1042/BCJ20160756. [DOI] [PubMed] [Google Scholar]
- 2.Hidalgo-Cantabrana C., Delgado S., Ruiz L., Ruas-Madiedo P., Sanchez B., Margolles A. Bifidobacteria and their health-promoting effects. Microbiol Spectr. 2017;5(3) doi: 10.1128/microbiolspec.BAD-0010-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alessandri G., Ossiprandi M.C., MacSharry J., van Sinderen D., Ventura M. Bifidobacterial dialogue with its human host and consequent modulation of the immune system. Front Immunol. 2019;10:2348. doi: 10.3389/fimmu.2019.02348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tojo R., Suarez A., Clemente M.G., de los Reyes-Gavilan C.G., Margolles A., Gueimonde M., Ruas-Madiedo P. Intestinal microbiota in health and disease: role of bifidobacteria in gut homeostasis. World J Gastroenterol. 2014;20(41):15163–15176. doi: 10.3748/wjg.v20.i41.15163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruiz L., Delgado S., Ruas-Madiedo P., Sanchez B., Margolles A. Bifidobacteria and Their Molecular Communication with the Immune System. Front Microbiol. 2017;8:2345. doi: 10.3389/fmicb.2017.02345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aw W., Fukuda S. Protective effects of bifidobacteria against enteropathogens. Microb Biotechnol. 2019;12(6):1097–1100. doi: 10.1111/mbt2.v12.610.1111/1751-7915.13460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riviere A., Selak M., Lantin D., Leroy F., De Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol. 2016;7:979. doi: 10.3389/fmicb.2016.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bottacini F., Ventura M., Sinderen D., Motherway M. Diversity, ecology and intestinal function of bifidobacteria. Microb Cell Fact. 2014;13(Suppl 1):S4. doi: 10.1186/1475-2859-13-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turroni F., Milani C., Duranti S., Mahony J., van Sinderen D., Ventura M. Glycan utilization and cross-feeding activities by bifidobacteria. Trends Microbiol. 2018;26(4):339–350. doi: 10.1016/j.tim.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Thomson P., Medina D.A., Garrido D. Human milk oligosaccharides and infant gut bifidobacteria: Molecular strategies for their utilization. Food Microbiol. 2018;75:37–46. doi: 10.1016/j.fm.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Turroni F., Milani C., Duranti S., Ferrario C., Lugli G.A., Mancabelli L. Bifidobacteria and the infant gut: an example of co-evolution and natural selection. Cell Mol Life Sci. 2018;75(1):103–118. doi: 10.1007/s00018-017-2672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly S.M., Munoz-Munoz J., van Sinderen D. Plant Glycan Metabolism by Bifidobacteria. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.609418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milani C., Mancabelli L., Lugli G.A., Duranti S., Turroni F., Ferrario C. Exploring vertical transmission of bifidobacteria from mother to child. Appl Environ Microbiol. 2015;81(20):7078–7087. doi: 10.1128/AEM.02037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milani C., Duranti S., Bottacini F., Casey E., Turroni F., Mahony J. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. 2017;81(4) doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duranti S., Lugli G.A., Mancabelli L., Armanini F., Turroni F., James K. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome. 2017;5(1) doi: 10.1186/s40168-017-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar H., Collado M.C., Wopereis H., Salminen S., Knol J., Roeselers G. The bifidogenic effect revisited-ecology and health perspectives of bifidobacterial colonization in early life. Microorganisms. 2020;8(12):1855. doi: 10.3390/microorganisms8121855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turroni F., Foroni E., Pizzetti P., Giubellini V., Ribbera A., Merusi P. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl Environ Microbiol. 2009;75(6):1534–1545. doi: 10.1128/AEM.02216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turroni F., van Sinderen D., Ventura M. Bifidobacteria: from ecology to genomics. Front Biosci (Landmark Ed) 2009;14:4673–4684. doi: 10.2741/3559. [DOI] [PubMed] [Google Scholar]
- 19.H. Tissier: Recherches sur la flore intestinale des nourrissons: état normal et pathologique. (1900)
- 20.Duranti S., Lugli G.A., Viappiani A., Mancabelli L., Alessandri G., Anzalone R. Characterization of the phylogenetic diversity of two novel species belonging to the genus Bifidobacterium: Bifidobacterium cebidarum sp. nov. and Bifidobacterium leontopitheci sp. nov. Int J Syst Evol Microbiol. 2020;70(4):2288–2297. doi: 10.1099/ijsem.0.004032. [DOI] [PubMed] [Google Scholar]
- 21.Modesto M., Satti M., Watanabe K., Scarafile D., Huang C.-H., Liou J.-S. Phylogenetic characterization of two novel species of the genus Bifidobacterium: Bifidobacterium saimiriisciurei sp. nov. and Bifidobacterium platyrrhinorum sp. nov. Syst Appl Microbiol. 2020;43(5):126111. doi: 10.1016/j.syapm.2020.126111. [DOI] [PubMed] [Google Scholar]
- 22.Neuzil-Bunesova V., Lugli G.A., Modrackova N., Vlkova E., Bolechova P., Burtscher J. Five novel bifidobacterial species isolated from faeces of primates in two Czech zoos: Bifidobacterium erythrocebi sp. nov., Bifidobacterium moraviense sp. nov., Bifidobacterium oedipodis sp. nov., Bifidobacterium olomucense sp. nov. and Bifidobacterium panos sp. nov. Int J Syst Evol Microbiol. 2020 doi: 10.1099/ijsem.0.004573. [DOI] [PubMed] [Google Scholar]
- 23.Neuzil-Bunesova V., Lugli G.A., Modrackova N., Makovska M., Mrazek J., Mekadim C. Bifidobacterium canis sp. nov., a novel member of the Bifidobacterium pseudolongum phylogenetic group isolated from faeces of a dog (Canis lupus f. familiaris) Int J Syst Evol Microbiol. 2020;70(9):5040–5047. doi: 10.1099/ijsem.0.004378. [DOI] [PubMed] [Google Scholar]
- 24.Modesto M., Satti M., Watanabe K., Huang C.H., Liou J.S., Tamura T. Bifidobacteria in two-toed sloths (Choloepus didactylus): phylogenetic characterization of the novel taxon Bifidobacterium choloepi sp. nov. Int J Syst Evol Microbiol. 2020 doi: 10.1099/ijsem.0.004506. [DOI] [PubMed] [Google Scholar]
- 25.Ventura M., Canchaya C., Tauch A., Chandra G., Fitzgerald G.F., Chater K.F. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007;71(3):495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoyles L., Inganas E., Falsen E., Drancourt M., Weiss N., McCartney A.L. Bifidobacterium scardovii sp. nov., from human sources. Int J Syst Evol Microbiol. 2002;52(Pt 3):995–999. doi: 10.1099/00207713-52-3-995. [DOI] [PubMed] [Google Scholar]
- 27.Okamoto M., Benno Y., Leung K.P., Maeda N. Bifidobacterium tsurumiense sp. nov., from hamster dental plaque. Int J Syst Evol Microbiol. 2008;58(Pt 1):144–148. doi: 10.1099/ijs.0.65296-0. [DOI] [PubMed] [Google Scholar]
- 28.B. Biavati, V. Scardovi and M. w. E. C.: Electrophoretic Patterns of Proteins in the Genus Bifidobacterium and Proposal of Four New Species (1982) doi:10.1099/00207713-32-3-358
- 29.Watanabe K., Makino H., Sasamoto M., Kudo Y., Fujimoto J., Demberel S. Bifidobacterium mongoliense sp. nov., from airag, a traditional fermented mare's milk product from Mongolia. Int J Syst Evol Microbiol. 2009;59(Pt 6):1535–1540. doi: 10.1099/ijs.0.006247-0. [DOI] [PubMed] [Google Scholar]
- 30.Delcenserie V., Gavini F., Beerens H., Tresse O., Franssen C., Daube G. Description of a new species, Bifidobacterium crudilactis sp. nov., isolated from raw milk and raw milk cheeses. Syst Appl Microbiol. 2007;30(5):381–389. doi: 10.1016/j.syapm.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Praet J., Meeus I., Cnockaert M., Aerts M., Smagghe G., Vandamme P. Bifidobacterium commune sp. nov. isolated from the bumble bee gut. Antonie Van Leeuwenhoek. 2015;107(5):1307–1313. doi: 10.1007/s10482-015-0425-3. [DOI] [PubMed] [Google Scholar]
- 32.Trovatelli L.D., Crociani F., Pedinotti M., Scardovi V. Bifidobacterium pullorum sp. nov.: a new species isolated from chicken feces and a related group of bifidobacteria isolated from rabbit feces. Arch Microbiol. 1974;98(1):187–198. doi: 10.1007/BF00425281. [DOI] [PubMed] [Google Scholar]
- 33.Duranti S., Lugli G.A., Napoli S., Anzalone R., Milani C., Mancabelli L. Characterization of the phylogenetic diversity of five novel species belonging to the genus Bifidobacterium: Bifidobacterium castoris sp. nov., Bifidobacterium callimiconis sp. nov., Bifidobacterium goeldii sp. nov., Bifidobacterium samirii sp. nov. and Bifidobacterium dolichotidis sp. nov. Int J Syst Evol Microbiol. 2019;69(5):1288–1298. doi: 10.1099/ijsem.0.003306. [DOI] [PubMed] [Google Scholar]
- 34.Mattarelli P., Bonaparte C., Pot B., Biavati B. Proposal to reclassify the three biotypes of Bifidobacterium longum as three subspecies: Bifidobacterium longum subsp. longum subsp. nov., Bifidobacterium longum subsp. infantis comb. nov. and Bifidobacterium longum subsp. suis comb. nov. Int J Syst Evol Microbiol. 2008;58(Pt 4):767–772. doi: 10.1099/ijs.0.65319-0. [DOI] [PubMed] [Google Scholar]
- 35.Lugli G.A., Mangifesta M., Duranti S., Anzalone R., Milani C., Mancabelli L. D. van Sinderen and M. Ventura: Phylogenetic classification of six novel species belonging to the genus Bifidobacterium comprising Bifidobacterium anseris sp. nov., Bifidobacterium criceti sp. nov., Bifidobacterium imperatoris sp. nov., Bifidobacterium italicum sp. nov., Bifidobacterium margollesii sp. nov. and Bifidobacterium parmae sp. nov. Syst Appl Microbiol. 2018;41(3):173–183. doi: 10.1016/j.syapm.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Milani C., Mangifesta M., Mancabelli L., Lugli G.A., James K., Duranti S. Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J. 2017;11(12):2834–2847. doi: 10.1038/ismej.2017.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bunesova V., Vlkova E., Rada V., Killer J., Musilova S. Bifidobacteria from the gastrointestinal tract of animals: differences and similarities. Benef Microbes. 2014;5(4):377–388. doi: 10.3920/BM2013.0081. [DOI] [PubMed] [Google Scholar]
- 38.Turroni F., van Sinderen D., Ventura M. Genomics and ecological overview of the genus Bifidobacterium. Int J Food Microbiol. 2011;149(1):37–44. doi: 10.1016/j.ijfoodmicro.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 39.Lamendella R., Santo Domingo J.W., Kelty C., Oerther D.B. Bifidobacteria in feces and environmental waters. Appl Environ Microbiol. 2008;74(3):575–584. doi: 10.1128/AEM.01221-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milani C., Lugli G.A., Duranti S., Turroni F., Bottacini F., Mangifesta M. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl Environ Microbiol. 2014;80(20):6290–6302. doi: 10.1128/AEM.02308-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alessandri G., Milani C., Mancabelli L., Longhi G., Anzalone R., Lugli G.A. D. van Sinderen and M. Ventura: Deciphering the Bifidobacterial Populations within the Canine and Feline Gut Microbiota. Appl Environ Microbiol. 2020;86(7) doi: 10.1128/AEM.02875-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alessandri G., Milani C., Mancabelli L., Mangifesta M., Lugli G.A., Viappiani A. Metagenomic dissection of the canine gut microbiota: insights into taxonomic, metabolic and nutritional features. Environ Microbiol. 2019;21(4):1331–1343. doi: 10.1111/emi.2019.21.issue-410.1111/1462-2920.14540. [DOI] [PubMed] [Google Scholar]
- 43.Alessandri G., Milani C., Mancabelli L., Mangifesta M., Lugli G.A., Viappiani A. The impact of human-facilitated selection on the gut microbiota of domesticated mammals. FEMS Microbiol Ecol. 2019;95(9) doi: 10.1093/femsec/fiz121. [DOI] [PubMed] [Google Scholar]
- 44.Milani C., Lugli G.A., Duranti S., Turroni F., Mancabelli L., Ferrario C. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep. 2015;5(1) doi: 10.1038/srep15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan J.-M., Halachev M.R., Loman N.J., Constantinidou C., Pallen M.J. Defining bacterial species in the genomic era: insights from the genus Acinetobacter. BMC Microbiol. 2012;12(1):302. doi: 10.1186/1471-2180-12-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossello-Mora R., Amann R. The species concept for prokaryotes. FEMS Microbiol Rev. 2001;25(1):39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x. [DOI] [PubMed] [Google Scholar]
- 47.Caputo A., Fournier P.E., Raoult D. Genome and pan-genome analysis to classify emerging bacteria. Biol Direct. 2019;14(1):5. doi: 10.1186/s13062-019-0234-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paul B., Dixit G., Murali T.S., Satyamoorthy K., Hao W. Genome-based taxonomic classification. Genome. 2019;62(2):45–52. doi: 10.1139/gen-2018-0072. [DOI] [PubMed] [Google Scholar]
- 49.Sentausa E., Fournier P.E. Advantages and limitations of genomics in prokaryotic taxonomy. Clin Microbiol Infect. 2013;19(9):790–795. doi: 10.1111/1469-0691.12181. [DOI] [PubMed] [Google Scholar]
- 50.Methods G. Bergery's Manu System Bacteriol. 1989;4 [Google Scholar]
- 51.Wayne L.G. International Committee on Systematic Bacteriology: announcement of the report of the ad hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Zentralbl Bakteriol Mikrobiol Hyg A. 1988;268(4):433–434. doi: 10.1016/s0176-6724(88)80120-2. [DOI] [PubMed] [Google Scholar]
- 52.Fox G.E., Wisotzkey J.D., Jurtshuk P., Jr. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42(1):166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 53.Stackebrandt E., Frederiksen W., Garrity G.M., Grimont P.A.D., Kampfer P., Maiden M.C.J. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol. 2002;52(Pt 3):1043–1047. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
- 54.Lugli G.A., Milani C., Duranti S., Mancabelli L., Mangifesta M., Turroni F. Tracking the taxonomy of the genus bifidobacterium based on a phylogenomic approach. Appl Environ Microbiol. 2018;84(4) doi: 10.1128/AEM.02249-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cooper J.E., Feil E.J. Multilocus sequence typing–what is resolved? Trends Microbiol. 2004;12(8):373–377. doi: 10.1016/j.tim.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 56.Ventura M., Canchaya C., Casale A.D., Dellaglio F., Neviani E., Fitzgerald G.F. Analysis of bifidobacterial evolution using a multilocus approach. Int J Syst Evol Microbiol. 2006;56(Pt 12):2783–2792. doi: 10.1099/ijs.0.64233-0. [DOI] [PubMed] [Google Scholar]
- 57.Milani C., Turroni F., Duranti S., Lugli G.A., Mancabelli L., Ferrario C. Genomics of the genus bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl Environ Microbiol. 2016;82(4):980–991. doi: 10.1128/AEM.03500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vernikos G., Medini D., Riley D.R., Tettelin H. Ten years of pan-genome analyses. Curr Opin Microbiol. 2015;23:148–154. doi: 10.1016/j.mib.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 59.Lugli G.A., Milani C., Duranti S., Alessandri G., Turroni F., Mancabelli L. Isolation of novel gut bifidobacteria using a combination of metagenomic and cultivation approaches. Genome Biol. 2019;20(1) doi: 10.1186/s13059-019-1711-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mai H., Zhou B., Liu L., Yang F., Conran C., Ji Y. Molecular pattern of lncRNAs in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38(1) doi: 10.1186/s13046-019-1213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bottacini F., Milani C., Turroni F., Sánchez B., Foroni E., Duranti S. Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS One. 2012;7(9):e44229. doi: 10.1371/journal.pone.0044229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tarracchini C., Lugli G.A., Mancabelli L., Milani C., Turroni F., Ventura M. Assessing the genomic variability of gardnerella vaginalis through comparative genomic analyses: evolutionary and ecological implications. Appl Environ Microbiol. 2020;87(1) doi: 10.1128/AEM.02188-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lugli G.A., Milani C., Turroni F., Duranti S., Mancabelli L., Mangifesta M. Comparative genomic and phylogenomic analyses of the Bifidobacteriaceae family. BMC Genomics. 2017;18(1) doi: 10.1186/s12864-017-3955-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yeoman C.J., Yildirim S., Thomas S.M., Durkin A.S., Torralba M., Sutton G. Comparative genomics of Gardnerella vaginalis strains reveals substantial differences in metabolic and virulence potential. PLoS ONE. 2010;5(8):e12411. doi: 10.1371/journal.pone.0012411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mancabelli L., Mancino W., Lugli G.A., Milani C., Viappiani A., Anzalone R. Comparative genome analyses of Lactobacillus crispatus isolated from different ecological niches reveal an environmental adaptation of this species to the human vaginal environment. Appl Environ Microbiol. 2021 doi: 10.1128/AEM.02899-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mancabelli L., Tarracchini C., Milani C., Lugli G.A., Fontana F., Turroni F. Vaginotypes of the human vaginal microbiome. Environ Microbiol. 2021 doi: 10.1111/1462-2920.15441. [DOI] [PubMed] [Google Scholar]
- 67.Turroni F., Peano C., Pass D.A., Foroni E., Severgnini M., Claesson M.J. Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE. 2012;7(5):e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Turroni F., Bottacini F., Foroni E., Mulder I., Kim J.-H., Zomer A. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci U S A. 2010;107(45):19514–19519. doi: 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alvarez-Martin P., Fernández M., O'Connell-Motherway M., O'Connell K.J., Sauvageot N., Fitzgerald G.F. A conserved two-component signal transduction system controls the response to phosphate starvation in Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2012;78(15):5258–5269. doi: 10.1128/AEM.00804-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ruiz L., Zomer A., O'Connell-Motherway M., van Sinderen D., Margolles A. Discovering novel bile protection systems in Bifidobacterium breve UCC2003 through functional genomics. Appl Environ Microbiol. 2012;78(4):1123–1131. doi: 10.1128/AEM.06060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ruiz L., O'Connell-Motherway M., Zomer A., de los Reyes-Gavilán C.G., Margolles A., van Sinderen D. Margolles and D. van Sinderen: A bile-inducible membrane protein mediates bifidobacterial bile resistance. Microb. Biotechnol. 2012;5(4):523–535. doi: 10.1111/j.1751-7915.2011.00329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zomer A., Fernandez M., Kearney B., Fitzgerald G.F., Ventura M., van Sinderen D. An interactive regulatory network controls stress response in Bifidobacterium breve UCC2003. J Bacteriol. 2009;191(22):7039–7049. doi: 10.1128/JB.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zomer A., van Sinderen D. Intertwinement of stress response regulons in Bifidobacterium breve UCC2003. Gut Microbes. 2010;1(2):100–102. doi: 10.4161/gmic.1.2.11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sanchez B., Ruiz L., Gueimonde M., Ruas-Madiedo P., Margolles A. Adaptation of bifidobacteria to the gastrointestinal tract and functional consequences. Pharmacol Res. 2013;69(1):127–136. doi: 10.1016/j.phrs.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 75.Brioukhanov A.L., Netrusov A.I. Catalase and superoxide dismutase: distribution, properties, and physiological role in cells of strict anaerobes. Biochemistry (Mosc) 2004;69(9):949–962. doi: 10.1023/b:biry.0000043537.04115.d9. [DOI] [PubMed] [Google Scholar]
- 76.Brioukhanov A.L., Netrusov A.I. Aerotolerance of strictly anaerobic microorganisms and factors of defense against oxidative stress: a review. Prikl Biokhim Mikrobiol. 2007;43(6):567–582. [PubMed] [Google Scholar]
- 77.Hayashi K., Maekawa I., Tanaka K., Ijyuin S., Shiwa Y., Suzuki I. Purification and characterization of oxygen-inducible haem catalase from oxygen-tolerant Bifidobacterium asteroides. Microbiology (Reading) 2013;159(Pt 1):89–95. doi: 10.1099/mic.0.059741-0. [DOI] [PubMed] [Google Scholar]
- 78.Ruiz L., Gueimonde M., Ruas-Madiedo P., Ribbera A., de los Reyes-Gavilán C.G., Ventura M. Molecular clues to understand the aerotolerance phenotype of Bifidobacterium animalis subsp. lactis. Appl Environ Microbiol. 2012;78(3):644–650. doi: 10.1128/AEM.05455-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zuo F., Yu R., Khaskheli G.B., Ma H., Chen L., Zeng Z. Homologous overexpression of alkyl hydroperoxide reductase subunit C (ahpC) protects Bifidobacterium longum strain NCC2705 from oxidative stress. Res Microbiol. 2014;165(7):581–589. doi: 10.1016/j.resmic.2014.05.040. [DOI] [PubMed] [Google Scholar]
- 80.Zuo F., Yu R., Xiao M., Khaskheli G.B., Sun X., Ma H. Transcriptomic analysis of Bifidobacterium longum subsp. longum BBMN68 in response to oxidative shock. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-35286-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oberg T.S., Ward R.E., Steele J.L., Broadbent J.R. Transcriptome analysis of Bifidobacterium longum strains that show a differential response to hydrogen peroxide stress. J Biotechnol. 2015;212:58–64. doi: 10.1016/j.jbiotec.2015.06.405. [DOI] [PubMed] [Google Scholar]
- 82.den Besten G., van Eunen K., Groen A.K., Venema K., Reijngoud D.J., Bakker B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54(9):2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matsumoto M., Ohishi H., Benno Y. H+-ATPase activity in Bifidobacterium with special reference to acid tolerance. Int J Food Microbiol. 2004;93(1):109–113. doi: 10.1016/j.ijfoodmicro.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 84.Ventura M., Canchaya C., van Sinderen D., Fitzgerald G.F., Zink R. Bifidobacterium lactis DSM 10140: identification of the atp (atpBEFHAGDC) operon and analysis of its genetic structure, characteristics, and phylogeny. Appl Environ Microbiol. 2004;70(5):3110–3121. doi: 10.1128/aem.70.5.3110-3121.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wei Y., Gao J., Liu D., Li Y., Liu W. Adaptational changes in physiological and transcriptional responses of Bifidobacterium longum involved in acid stress resistance after successive batch cultures. Microb Cell Fact. 2019;18(1):156. doi: 10.1186/s12934-019-1206-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu S., Ren F., Jiang J., Zhao L. Acid response of bifidobacterium longum subsp. longum BBMN68 is accompanied by modification of the cell membrane fatty acid composition. J Microbiol Biotechnol. 2016;26(7):1190–1197. doi: 10.4014/jmb.1511.11013. [DOI] [PubMed] [Google Scholar]
- 87.Merritt M.E., Donaldson J.R. Effect of bile salts on the DNA and membrane integrity of enteric bacteria. J Med Microbiol. 2009;58(Pt 12):1533–1541. doi: 10.1099/jmm.0.014092-0. [DOI] [PubMed] [Google Scholar]
- 88.Begley M., Gahan C.G., Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29(4):625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 89.Ruiz L., Sanchez B., Ruas-Madiedo P., de Los Reyes-Gavilan C.G., Margolles A. Cell envelope changes in Bifidobacterium animalis ssp. lactis as a response to bile. FEMS Microbiol Lett. 2007;274(2):316–322. doi: 10.1111/j.1574-6968.2007.00854.x. [DOI] [PubMed] [Google Scholar]
- 90.Sanchez B., Champomier-Verges M.C., Stuer-Lauridsen B., Ruas-Madiedo P., Anglade P., Baraige F. Adaptation and response of Bifidobacterium animalis subsp. lactis to bile: a proteomic and physiological approach. Appl Environ Microbiol. 2007;73(21):6757–6767. doi: 10.1128/AEM.00637-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.An H., Douillard François.P., Wang G., Zhai Z., Yang J., Song S. Integrated transcriptomic and proteomic analysis of the bile stress response in a centenarian-originated probiotic Bifidobacterium longum BBMN68. Mol Cell Proteomics. 2014;13(10):2558–2572. doi: 10.1074/mcp.M114.039156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gueimonde M., Garrigues C., van Sinderen D., de los Reyes-Gavil�n C.G., Margolles A. de los Reyes-Gavilan and A. Margolles: Bile-inducible efflux transporter from Bifidobacterium longum NCC2705, conferring bile resistance. Appl Environ Microbiol. 2009;75(10):3153–3160. doi: 10.1128/AEM.00172-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu Q., Zhai Z., An H., Yang Y., Yin J., Wang G. The MarR Family Regulator BmrR Is Involved in Bile Tolerance of Bifidobacterium longum BBMN68 via Controlling the Expression of an ABC Transporter. Appl Environ Microbiol. 2019;85(3) doi: 10.1128/AEM.02453-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ruiz L., Margolles A., Sanchez B. Bile resistance mechanisms in Lactobacillus and Bifidobacterium. Front Microbiol. 2013;4:396. doi: 10.3389/fmicb.2013.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jarocki P., Podleśny M., Glibowski P., Targoński Z., Heimesaat M.M. A new insight into the physiological role of bile salt hydrolase among intestinal bacteria from the genus Bifidobacterium. PLoS ONE. 2014;9(12):e114379. doi: 10.1371/journal.pone.0114379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kelly S.M., Lanigan N., O’Neill I.J., Bottacini F., Lugli G.A., Viappiani A. Bifidobacterial biofilm formation is a multifactorial adaptive phenomenon in response to bile exposure. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-68179-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Turroni F., Foroni E., O'Connell Motherway M., Bottacini F., Giubellini V., Zomer A. D. van Sinderen and M. Ventura: Characterization of the serpin-encoding gene of Bifidobacterium breve 210B. Appl Environ Microbiol. 2010;76(10):3206–3219. doi: 10.1128/AEM.02938-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gettins P.G.W. Serpin structure, mechanism, and function. Chem Rev. 2002;102(12):4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 99.Alvarez-Martin P., O'Connell Motherway M., Turroni F., Foroni E., Ventura M., van Sinderen D. Ventura and D. van Sinderen: A two-component regulatory system controls autoregulated serpin expression in Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2012;78(19):7032–7041. doi: 10.1128/AEM.01776-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hu Y.J., Lin D.Y., Zeng D. A general framework for studying genetic effects and gene-environment interactions with missing data. Biostatistics. 2010;11(4):583–598. doi: 10.1093/biostatistics/kxq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fanning S., Hall L.J., Cronin M., Zomer A., MacSharry J., Goulding D. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc Natl Acad Sci U S A. 2012;109(6):2108–2113. doi: 10.1073/pnas.1115621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Turroni F., Serafini F., Foroni E., Duranti S., O'Connell Motherway M., Taverniti V. D. van Sinderen and M. Ventura: Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc Natl Acad Sci U S A. 2013;110(27):11151–11156. doi: 10.1073/pnas.1303897110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Scott J.R., Zahner D. Pili with strong attachments: Gram-positive bacteria do it differently. Mol Microbiol. 2006;62(2):320–330. doi: 10.1111/j.1365-2958.2006.05279.x. [DOI] [PubMed] [Google Scholar]
- 104.Kline K.A., Dodson K.W., Caparon M.G., Hultgren S.J. A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol. 2010;18(5):224–232. doi: 10.1016/j.tim.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Turroni F., Serafini F., Mangifesta M., Arioli S., Mora D., van Sinderen D. Expression of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in response to environmental gut conditions. FEMS Microbiol Lett. 2014;357(1):23–33. doi: 10.1111/fml.2014.357.issue-110.1111/1574-6968.12509. [DOI] [PubMed] [Google Scholar]
- 106.Milani C., Mangifesta M., Mancabelli L., Lugli G.A., Mancino W., Viappiani A. The sortase-dependent fimbriome of the genus bifidobacterium: extracellular structures with potential to modulate microbe-host dialogue. Appl Environ Microbiol. 2017;83(19) doi: 10.1128/AEM.01295-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.O'Connell Motherway M., Zomer A., Leahy S.C., Reunanen J., Bottacini F., Claesson M.J. O'Toole and D. van Sinderen: Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci U S A. 2011;108(27):11217–11222. doi: 10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Spirig T., Weiner E.M., Clubb R.T. Sortase enzymes in Gram-positive bacteria. Mol Microbiol. 2011;82(5):1044–1059. doi: 10.1111/j.1365-2958.2011.07887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Foroni E., Serafini F., Amidani D., Turroni F., He F., Bottacini F. D. van Sinderen and M. Ventura: Genetic analysis and morphological identification of pilus-like structures in members of the genus Bifidobacterium. Microb Cell Fact. 2011;10(Suppl 1):S16. doi: 10.1186/1475-2859-10-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kachlany S.C., Planet P.J., DeSalle R., Fine D.H., Figurski D.H. Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol. 2001;9(9):429–437. doi: 10.1016/s0966-842x(01)02161-8. [DOI] [PubMed] [Google Scholar]
- 111.Tomich M., Planet P.J., Figurski D.H. The tad locus: postcards from the widespread colonization island. Nat Rev Microbiol. 2007;5(5):363–375. doi: 10.1038/nrmicro1636. [DOI] [PubMed] [Google Scholar]
- 112.Pelicic V. Type IV pili: e pluribus unum? Mol Microbiol. 2008;68(4):827–837. doi: 10.1111/j.1365-2958.2008.06197.x. [DOI] [PubMed] [Google Scholar]
- 113.Ventura M., Turroni F., Motherway M.O., MacSharry J., van Sinderen D. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol. 2012;20(10):467–476. doi: 10.1016/j.tim.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 114.Bottacini F., O’Connell Motherway M., Kuczynski J., O’Connell K., Serafini F., Duranti S. Ventura and D. van Sinderen: Comparative genomics of the Bifidobacterium breve taxon. BMC Genomics. 2014;15(1):170. doi: 10.1186/1471-2164-15-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.O'Connell Motherway M., Houston A., O’Callaghan G., Reunanen J., O’Brien F., O’Driscoll T. A Bifidobacterial pilus-associated protein promotes colonic epithelial proliferation. Mol Microbiol. 2019;111(1):287–301. doi: 10.1111/mmi.2019.111.issue-110.1111/mmi.14155. [DOI] [PubMed] [Google Scholar]
- 116.Duranti S., Turroni F., Lugli G.A., Milani C., Viappiani A., Mangifesta M. Genomic characterization and transcriptional studies of the starch-utilizing strain Bifidobacterium adolescentis 22L. Appl Environ Microbiol. 2014;80(19):6080–6090. doi: 10.1128/AEM.01993-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Welman A.D., Maddox I.S. Exopolysaccharides from lactic acid bacteria: perspectives and challenges. Trends Biotechnol. 2003;21(6):269–274. doi: 10.1016/S0167-7799(03)00107-0. [DOI] [PubMed] [Google Scholar]
- 118.Hidalgo-Cantabrana C., López P., Gueimonde M., de los Reyes-Gavilán C.G., Suárez A., Margolles A. Immune Modulation Capability of Exopolysaccharides Synthesised by Lactic Acid Bacteria and Bifidobacteria. Probiotics Antimicrob Proteins. 2012;4(4):227–237. doi: 10.1007/s12602-012-9110-2. [DOI] [PubMed] [Google Scholar]
- 119.Castro-Bravo N., Wells J.M., Margolles A., Ruas-Madiedo P. Interactions of surface exopolysaccharides from bifidobacterium and lactobacillus within the intestinal environment. Front Microbiol. 2018;9:2426. doi: 10.3389/fmicb.2018.02426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Leivers S., Hidalgo-Cantabrana C., Robinson G., Margolles A., Ruas-Madiedo P., Laws A.P. Structure of the high molecular weight exopolysaccharide produced by Bifidobacterium animalis subsp. lactis IPLA-R1 and sequence analysis of its putative eps cluster. Carbohydr Res. 2011;346(17):2710–2717. doi: 10.1016/j.carres.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 121.Hidalgo-Cantabrana C., Sanchez B., Milani C., Ventura M., Margolles A., Ruas-Madiedo P. Genomic overview and biological functions of exopolysaccharide biosynthesis in Bifidobacterium spp. Appl Environ Microbiol. 2014;80(1):9–18. doi: 10.1128/AEM.02977-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ferrario C., Milani C., Mancabelli L., Lugli G.A., Duranti S., Mangifesta M. D. van Sinderen and M. Ventura: Modulation of the eps-ome transcription of bifidobacteria through simulation of human intestinal environment. FEMS Microbiol Ecol. 2016;92(4):fiw056. doi: 10.1093/femsec/fiw056. [DOI] [PubMed] [Google Scholar]
- 123.Bottacini F., Morrissey R., Esteban-Torres M., James K., van Breen J., Dikareva E. O'Connell Motherway and D. van Sinderen: Comparative genomics and genotype-phenotype associations in Bifidobacterium breve. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-28919-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Salazar N., Ruas-Madiedo P., Prieto A., Calle L.P., de Los Reyes-Gavilan C.G. Characterization of exopolysaccharides produced by Bifidobacterium longum NB667 and its cholate-resistant derivative strain IPLA B667dCo. J Agric Food Chem. 2012;60(4):1028–1035. doi: 10.1021/jf204034n. [DOI] [PubMed] [Google Scholar]
- 125.Yan S., Yang B., Zhao J., Zhao J., Stanton C., Ross R.P. A ropy exopolysaccharide producing strain Bifidobacterium longum subsp. longum YS108R alleviates DSS-induced colitis by maintenance of the mucosal barrier and gut microbiota modulation. Food Funct. 2019;10(3):1595–1608. doi: 10.1039/C9FO00014C. [DOI] [PubMed] [Google Scholar]
- 126.Hidalgo-Cantabrana C., Nikolic M., López P., Suárez A., Miljkovic M., Kojic M. Exopolysaccharide-producing Bifidobacterium animalis subsp. lactis strains and their polymers elicit different responses on immune cells from blood and gut associated lymphoid tissue. Anaerobe. 2014;26:24–30. doi: 10.1016/j.anaerobe.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 127.Castro-Bravo N., Margolles A., Wells J.M., Ruas-Madiedo P. Exopolysaccharides synthesized by Bifidobacterium animalis subsp. lactis interact with TLR4 in intestinal epithelial cells. Anaerobe. 2019;56:98–101. doi: 10.1016/j.anaerobe.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 128.Longhi G., van Sinderen D., Ventura M., Turroni F. Microbiota and Cancer: The Emerging Beneficial Role of Bifidobacteria in Cancer Immunotherapy. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.575072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yu R., Zuo F., Ma H., Chen S. Exopolysaccharide-Producing Bifidobacterium adolescentis Strains with Similar Adhesion Property Induce Differential Regulation of Inflammatory Immune Response in Treg/Th17 Axis of DSS-Colitis Mice. Nutrients. 2019;11(4):782. doi: 10.3390/nu11040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hidalgo-Cantabrana C., Sánchez B., Moine D., Berger B., de los Reyes-Gavilán C.G., Gueimonde M. Insights into the ropy phenotype of the exopolysaccharide-producing strain Bifidobacterium animalis subsp. lactis A1dOxR. Appl Environ Microbiol. 2013;79(12):3870–3874. doi: 10.1128/AEM.00633-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.López P., Monteserín D.C., Gueimonde M., de los Reyes-Gavilán C.G., Margolles A., Suárez A. Exopolysaccharide-producing Bifidobacterium strains elicit different in vitro responses upon interaction with human cells. Food Res Int. 2012;46(1):99–107. doi: 10.1016/j.foodres.2011.11.020. [DOI] [Google Scholar]
- 132.Schiavi E., Gleinser M., Molloy E., Groeger D., Frei R., Ferstl R. D. van Sinderen and L. O'Mahony: The Surface-Associated Exopolysaccharide of Bifidobacterium longum 35624 Plays an Essential Role in Dampening Host Proinflammatory Responses and Repressing Local TH17 Responses. Appl Environ Microbiol. 2016;82(24):7185–7196. doi: 10.1128/AEM.02238-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rios-Covian D., Cuesta I., Alvarez-Buylla J.R., Ruas-Madiedo P., Gueimonde M., de Los Reyes-Gavilan C.G. Bacteroides fragilis metabolises exopolysaccharides produced by bifidobacteria. BMC Microbiol. 2016;16(1):150. doi: 10.1186/s12866-016-0773-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rios-Covian D., Sanchez B., Salazar N., Martinez N., Redruello B., Gueimonde M. Different metabolic features of Bacteroides fragilis growing in the presence of glucose and exopolysaccharides of bifidobacteria. Front Microbiol. 2015;6:825. doi: 10.3389/fmicb.2015.00825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rios-Covian D., Arboleya S., Hernandez-Barranco A.M., Alvarez-Buylla J.R., Ruas-Madiedo P., Gueimonde M. Interactions between Bifidobacterium and Bacteroides species in cofermentations are affected by carbon sources, including exopolysaccharides produced by bifidobacteria. Appl Environ Microbiol. 2013;79(23):7518–7524. doi: 10.1128/AEM.02545-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Salazar N., Gueimonde M., Hernandez-Barranco A.M., Ruas-Madiedo P., de Los Reyes-Gavilan C.G. Exopolysaccharides produced by intestinal Bifidobacterium strains act as fermentable substrates for human intestinal bacteria. Appl Environ Microbiol. 2008;74(15):4737–4745. doi: 10.1128/AEM.00325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hughes K.R., Harnisch L.C., Alcon-Giner C., Mitra S., Wright C.J., Ketskemety J. Bifidobacterium breve reduces apoptotic epithelial cell shedding in an exopolysaccharide and MyD88-dependent manner. Open Biol. 2017;7(1):160155. doi: 10.1098/rsob.160155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang L., Wang Y., Li Q., Tian K., Xu L., Liu G. Exopolysaccharide, isolated from a novel strain bifidobacterium breve lw01 possess an anticancer effect on head and neck cancer - genetic and biochemical evidences. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.01044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Atkinson S., Williams P. Quorum sensing and social networking in the microbial world. J R Soc Interface. 2009;6(40):959–978. doi: 10.1098/rsif.2009.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bassler B.L., Losick R. Bacterially speaking. Cell. 2006;125(2):237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 141.Xavier K.B., Bassler B.L. LuxS quorum sensing: more than just a numbers game. Curr Opin Microbiol. 2003;6(2):191–197. doi: 10.1016/s1369-5274(03)00028-6. [DOI] [PubMed] [Google Scholar]
- 142.Sun Z., He X., Brancaccio V.F., Yuan J., Riedel C.U., Kaufmann G.F. Bifidobacteria exhibit LuxS-dependent autoinducer 2 activity and biofilm formation. PLoS ONE. 2014;9(2):e88260. doi: 10.1371/journal.pone.0088260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Christiaen S.E.A., O'Connell Motherway M., Bottacini F., Lanigan N., Casey P.G., Huys G. Autoinducer-2 plays a crucial role in gut colonization and probiotic functionality of Bifidobacterium breve UCC2003. PLoS ONE. 2014;9(5):e98111. doi: 10.1371/journal.pone.0098111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Turroni F., Ventura M., Buttó L.F., Duranti S., O’Toole P.W., Motherway M.O. Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective. Cell Mol Life Sci. 2014;71(2):183–203. doi: 10.1007/s00018-013-1318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pyclik M., Srutkova D., Schwarzer M., Gorska S. Bifidobacteria cell wall-derived exo-polysaccharides, lipoteichoic acids, peptidoglycans, polar lipids and proteins - their chemical structure and biological attributes. Int J Biol Macromol. 2020;147:333–349. doi: 10.1016/j.ijbiomac.2019.12.227. [DOI] [PubMed] [Google Scholar]
- 146.Colagiorgi A., Turroni F., Mancabelli L., Serafini F., Secchi A., van Sinderen D. D. van Sinderen and M. Ventura: Insights into teichoic acid biosynthesis by Bifidobacterium bifidum PRL2010. FEMS Microbiol Lett. 2015;362(17):fnv141. doi: 10.1093/femsle/fnv141. [DOI] [PubMed] [Google Scholar]
- 147.Guglielmetti S., Balzaretti S., Taverniti V., Miriani M., Milani C., Scarafoni A. TgaA, a VirB1-like component belonging to a putative type IV secretion system of Bifidobacterium bifidum MIMBb75. Appl Environ Microbiol. 2014;80(17):5161–5169. doi: 10.1128/AEM.01413-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Guglielmetti S., Zanoni I., Balzaretti S., Miriani M., Taverniti V., De Noni I. Murein lytic enzyme TgaA of Bifidobacterium bifidum MIMBb75 modulates dendritic cell maturation through its cysteine- and histidine-dependent amidohydrolase/peptidase (CHAP) amidase domain. Appl Environ Microbiol. 2014;80(17):5170–5177. doi: 10.1128/AEM.00761-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.O'Callaghan A., van Sinderen D. Bifidobacteria and Their Role as Members of the Human Gut Microbiota. Front Microbiol. 2016;7:925. doi: 10.3389/fmicb.2016.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pokusaeva K., Fitzgerald G.F., van Sinderen D. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 2011;6(3):285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.V. Scardovi: The fructose-6-phosphate shunt as peculiar pattern of hexose degradation in the genus Bifidobacterium (1965)
- 152.De Vuyst L., Moens F., Selak M., Riviere A., Leroy F. Summer Meeting 2013: growth and physiology of bifidobacteria. J Appl Microbiol. 2014;116(3):477–491. doi: 10.1111/jam.12415. [DOI] [PubMed] [Google Scholar]
- 153.El Kaoutari A., Armougom F., Gordon J.I., Raoult D., Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11(7):497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 154.O'Connell Motherway M., Fitzgerald G.F., van Sinderen D. Metabolism of a plant derived galactose-containing polysaccharide by Bifidobacterium breve UCC2003. Microb Biotechnol. 2011;4(3):403–416. doi: 10.1111/j.1751-7915.2010.00218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ambrogi V., Bottacini F., O'Sullivan J., O'Connell Motherway M., Linqiu C., Schoemaker B. Schoterman and D. van Sinderen: Characterization of GH2 and GH42 beta-galactosidases derived from bifidobacterial infant isolates. AMB Express. 2019;9(1):9. doi: 10.1186/s13568-019-0735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology. 2012;22(9):1147–1162. doi: 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bode L. Human milk oligosaccharides: structure and functions. Nestle Nutr Inst Workshop Ser. 2020;94:115–123. doi: 10.1159/000505339. [DOI] [PubMed] [Google Scholar]
- 158.Katoh T., Ojima M.N., Sakanaka M., Ashida H., Gotoh A., Katayama T. Enzymatic adaptation of bifidobacterium bifidum to host glycans, viewed from glycoside hydrolyases and carbohydrate-binding modules. Microorganisms. 2020;8(4):481. doi: 10.3390/microorganisms8040481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lugli G.A., Duranti S., Milani C., Mancabelli L., Turroni F., Alessandri G. Investigating bifidobacteria and human milk oligosaccharide composition of lactating mothers. FEMS Microbiol Ecol. 2020;96(5) doi: 10.1093/femsec/fiaa049. [DOI] [PubMed] [Google Scholar]
- 160.James K., O'Connell Motherway M., Penno C., O'Brien R.L., van Sinderen D., Cann I. Bifidobacterium breve UCC2003 employs multiple transcriptional regulators to control metabolism of particular human milk oligosaccharides. Appl Environ Microbiol. 2018;84(9):e02774-17. doi: 10.1128/AEM.02774-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.James K., Motherway M.O., Bottacini F., van Sinderen D. Bifidobacterium breve UCC2003 metabolises the human milk oligosaccharides lacto-N-tetraose and lacto-N-neo-tetraose through overlapping, yet distinct pathways. Sci Rep. 2016;6:38560. doi: 10.1038/srep38560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zabel B.E., Gerdes S., Evans K.C., Nedveck D., Singles S.K., Volk B. Strain-specific strategies of 2'-fucosyllactose, 3-fucosyllactose, and difucosyllactose assimilation by Bifidobacterium longum subsp. infantis Bi-26 and ATCC 15697. Sci Rep. 2020;10(1):15919. doi: 10.1038/s41598-020-72792-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sela D.A., Garrido D., Lerno L., Wu S., Tan K., Eom H.J. Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides. Appl Environ Microbiol. 2012;78(3):795–803. doi: 10.1128/AEM.06762-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bunesova V., Lacroix C., Schwab C. Fucosyllactose and L-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense. BMC Microbiol. 2016;16(1):248. doi: 10.1186/s12866-016-0867-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sakanaka M., Gotoh A., Yoshida K., Odamaki T., Koguchi H., Xiao J.-zhong. Varied pathways of infant gut-associated bifidobacterium to assimilate human milk oligosaccharides: prevalence of the gene set and its correlation with bifidobacteria-rich microbiota formation. Nutrients. 2019;12(1):71. doi: 10.3390/nu12010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Matsuki T., Yahagi K., Mori H., Matsumoto H., Hara T., Tajima S. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat Commun. 2016;7(1) doi: 10.1038/ncomms11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Schwab C., Ruscheweyh H.J., Bunesova V., Pham V.T., Beerenwinkel N., Lacroix C. Trophic Interactions of Infant Bifidobacteria and Eubacterium hallii during L-Fucose and Fucosyllactose Degradation. Front Microbiol. 2017;8:95. doi: 10.3389/fmicb.2017.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.LoCascio R.G., Desai P., Sela D.A., Weimer B., Mills D.A. Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl Environ Microbiol. 2010;76(22):7373–7381. doi: 10.1128/AEM.00675-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sela D.A. Bifidobacterial utilization of human milk oligosaccharides. Int J Food Microbiol. 2011;149(1):58–64. doi: 10.1016/j.ijfoodmicro.2011.01.025. [DOI] [PubMed] [Google Scholar]
- 170.Zabel B., Yde C.C., Roos P., Marcussen Jørn, Jensen H.M., Salli K. Novel Genes and Metabolite Trends in Bifidobacterium longum subsp. infantis Bi-26 Metabolism of Human Milk Oligosaccharide 2'-fucosyllactose. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-43780-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sela D.A., Mills D.A. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010;18(7):298–307. doi: 10.1016/j.tim.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kitaoka M. Bifidobacterial enzymes involved in the metabolism of human milk oligosaccharides. Adv Nutr. 2012;3(3):422S–429S. doi: 10.3945/an.111.001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Garrido D., Ruiz-Moyano S., Lemay D.G., Sela D.A., German J.B., Mills D.A. Comparative transcriptomics reveals key differences in the response to milk oligosaccharides of infant gut-associated bifidobacteria. Sci Rep. 2015;5:13517. doi: 10.1038/srep13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kiyohara M., Tanigawa K., Chaiwangsri T., Katayama T., Ashida H., Yamamoto K. An exo-alpha-sialidase from bifidobacteria involved in the degradation of sialyloligosaccharides in human milk and intestinal glycoconjugates. Glycobiology. 2011;21(4):437–447. doi: 10.1093/glycob/cwq175. [DOI] [PubMed] [Google Scholar]
- 175.Lugli G.A., Mancino W., Milani C., Duranti S., Turroni F., van Sinderen D. D. van Sinderen and M. Ventura: Reconstruction of the Bifidobacterial Pan-Secretome Reveals the Network of Extracellular Interactions between Bifidobacteria and the Infant Gut. Appl Environ Microbiol. 2018;84(16) doi: 10.1128/AEM.00796-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Garrido D., Ruiz-Moyano S., Kirmiz N., Davis J.C., Totten S.M., Lemay D.G. A novel gene cluster allows preferential utilization of fucosylated milk oligosaccharides in Bifidobacterium longum subsp. longum SC596. Sci Rep. 2016;6(1) doi: 10.1038/srep35045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ruiz-Moyano S., Totten S.M., Garrido D.A., Smilowitz J.T., German J.B., Lebrilla C.B. Variation in consumption of human milk oligosaccharides by infant gut-associated strains of Bifidobacterium breve. Appl Environ Microbiol. 2013;79(19):6040–6049. doi: 10.1128/AEM.01843-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sakurama H., Kiyohara M., Wada J., Honda Y., Yamaguchi M., Fukiya S. Lacto-N-biosidase encoded by a novel gene of Bifidobacterium longum subspecies longum shows unique substrate specificity and requires a designated chaperone for its active expression. J Biol Chem. 2013;288(35):25194–25206. doi: 10.1074/jbc.M113.484733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Arboleya S., Bottacini F., O'Connell-Motherway M., Ryan C.A., Ross R.P., van Sinderen D. Gene-trait matching across the Bifidobacterium longum pan-genome reveals considerable diversity in carbohydrate catabolism among human infant strains. BMC Genomics. 2018;19(1):33. doi: 10.1186/s12864-017-4388-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.James K., Bottacini F., Contreras J.I.S., Vigoureux M., Egan M., Motherway M.O. Metabolism of the predominant human milk oligosaccharide fucosyllactose by an infant gut commensal. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-51901-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nishiyama K., Nagai A., Uribayashi K., Yamamoto Y., Mukai T., Okada N. Two extracellular sialidases from Bifidobacterium bifidum promote the degradation of sialyl-oligosaccharides and support the growth of Bifidobacterium breve. Anaerobe. 2018;52:22–28. doi: 10.1016/j.anaerobe.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 182.Gotoh A., Katoh T., Sakanaka M., Ling Y., Yamada C., Asakuma S. Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-32080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bansil R., Turner B.S. The biology of mucus: Composition, synthesis and organization. Adv Drug Deliv Rev. 2018;124:3–15. doi: 10.1016/j.addr.2017.09.023. [DOI] [PubMed] [Google Scholar]
- 184.Garrido D., Nwosu C., Ruiz-Moyano S., Aldredge D., German J.B., Lebrilla C.B. Endo-beta-N-acetylglucosaminidases from infant gut-associated bifidobacteria release complex N-glycans from human milk glycoproteins. Mol Cell Proteomics. 2012;11(9):775–785. doi: 10.1074/mcp.M112.018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zuniga M., Monedero V., Yebra M.J. Utilization of host-derived glycans by intestinal lactobacillus and bifidobacterium species. Front Microbiol. 2018;9:1917. doi: 10.3389/fmicb.2018.01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Stanley P., Taniguchi N., Aebi M. N-Glycans. Essent Glycobiol. 2017;Chapter 9 [Google Scholar]
- 187.Turroni F., Milani C., van Sinderen D., Ventura M. Genetic strategies for mucin metabolism in Bifidobacterium bifidum PRL2010: an example of possible human-microbe co-evolution. Gut Microbes. 2011;2(3):183–189. doi: 10.4161/gmic.2.3.16105. [DOI] [PubMed] [Google Scholar]
- 188.Katoh T., Maeshibu T., Kikkawa K.-ichi., Gotoh A., Tomabechi Y., Nakamura M. Identification and characterization of a sulfoglycosidase from Bifidobacterium bifidum implicated in mucin glycan utilization. Biosci Biotechnol Biochem. 2017;81(10):2018–2027. doi: 10.1080/09168451.2017.1361810. [DOI] [PubMed] [Google Scholar]
- 189.Martín R., Bottacini F., Egan M., Chamignon C., Tondereau V., Moriez R. Smokvina and D. van Sinderen: The Infant-Derived Bifidobacterium bifidum Strain CNCM I-4319 Strengthens Gut Functionality. Microorganisms. 2020;8(9):1313. doi: 10.3390/microorganisms8091313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Egan M., O'Connell Motherway M., Ventura M., van Sinderen D., Macfarlane G.T. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2014;80(14):4414–4426. doi: 10.1128/AEM.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Egan M., Jiang H., O'Connell Motherway M., Oscarson S., van Sinderen D., Björkroth J. Glycosulfatase-Encoding Gene Cluster in Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2016;82(22):6611–6623. doi: 10.1128/AEM.02022-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Egan M., O’Connell Motherway M., Kilcoyne M., Kane M., Joshi L., Ventura M. Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiol. 2014;14(1) doi: 10.1186/s12866-014-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mahony J., Lugli G.A., van Sinderen D., Ventura M. Impact of gut-associated bifidobacteria and their phages on health: two sides of the same coin? Appl Microbiol Biotechnol. 2018;102(5):2091–2099. doi: 10.1007/s00253-018-8795-x. [DOI] [PubMed] [Google Scholar]
- 194.Sutton T.D.S., Hill C. Gut Bacteriophage: Current Understanding and Challenges. Front Endocrinol (Lausanne) 2019;10:784. doi: 10.3389/fendo.2019.00784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hobbs Z., Abedon S.T., Millard A. Diversity of phage infection types and associated terminology: the problem with 'Lytic or lysogenic'. FEMS Microbiol Lett. 2016;363(7):fnw047. doi: 10.1093/femsle/fnw047. [DOI] [PubMed] [Google Scholar]
- 196.Shkoporov A.N., Hill C. Bacteriophages of the human gut: The “Known Unknown” of the microbiome. Cell Host Microbe. 2019;25(2):195–209. doi: 10.1016/j.chom.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 197.Van Belleghem J., Dąbrowska K., Vaneechoutte M., Barr J., Bollyky P. Interactions between bacteriophage, bacteria, and the mammalian immune system. Viruses. 2018;11(1):10. doi: 10.3390/v11010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ducarmon Q.R., Zwittink R.D., Hornung B.V.H., van Schaik W., Young V.B., Kuijper E.J. Gut microbiota and colonization resistance against bacterial enteric infection. Microbiol Mol Biol Rev. 2019;83(3) doi: 10.1128/MMBR.00007-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Canchaya C., Fournous G., Chibani-Chennoufi S., Dillmann M.L., Brussow H. Phage as agents of lateral gene transfer. Curr Opin Microbiol. 2003;6(4):417–424. doi: 10.1016/s1369-5274(03)00086-9. [DOI] [PubMed] [Google Scholar]
- 200.Schell M.A., Karmirantzou M., Snel B., Vilanova D., Berger B., Pessi G. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci U S A. 2002;99(22):14422–14427. doi: 10.1073/pnas.212527599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Mancino W., Lugli G.A., van Sinderen D., Ventura M., Turroni F. Mobilome and resistome reconstruction from genomes belonging to members of the bifidobacterium genus. Microorganisms. 2019;7(12):638. doi: 10.3390/microorganisms7120638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lugli G.A., Milani C., Turroni F., Tremblay D., Ferrario C., Mancabelli L. Prophages of the genus Bifidobacterium as modulating agents of the infant gut microbiota. Environ Microbiol. 2016;18(7):2196–2213. doi: 10.1111/1462-2920.13154. [DOI] [PubMed] [Google Scholar]
- 203.Ventura M., Lee J.-H., Canchaya C., Zink R., Leahy S., Moreno-Munoz J.A. Prophage-like elements in bifidobacteria: insights from genomics, transcription, integration, distribution, and phylogenetic analysis. Appl Environ Microbiol. 2005;71(12):8692–8705. doi: 10.1128/AEM.71.12.8692-8705.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ventura M., Turroni F., Lima-Mendez G., Foroni E., Zomer A., Duranti S. Comparative analyses of prophage-like elements present in bifidobacterial genomes. Appl Environ Microbiol. 2009;75(21):6929–6936. doi: 10.1128/AEM.01112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science:1138140. [DOI] [PubMed] [Google Scholar]
- 206.Ershova A.S., Rusinov I.S., Spirin S.A., Karyagina A.S., Alexeevski A.V. Role of Restriction-Modification Systems in Prokaryotic Evolution and Ecology. Biochemistry (Mosc) 2015;80(10):1373–1386. doi: 10.1134/S0006297915100193. [DOI] [PubMed] [Google Scholar]
- 207.Hille F., Richter H., Wong S.P., Bratovic M., Ressel S., Charpentier E. The Biology of CRISPR-Cas: Backward and Forward. Cell. 2018;172(6):1239–1259. doi: 10.1016/j.cell.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 208.Pan M., Nethery M.A., Hidalgo-Cantabrana C., Barrangou R. Comprehensive Mining and Characterization of CRISPR-Cas Systems in Bifidobacterium. Microorganisms. 2020;8(5):720. doi: 10.3390/microorganisms8050720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Briner A.E., Lugli G.A., Milani C., Duranti S., Turroni F., Gueimonde M. Occurrence and Diversity of CRISPR-Cas Systems in the Genus Bifidobacterium. PLoS ONE. 2015;10(7):e0133661. doi: 10.1371/journal.pone.0133661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hidalgo-Cantabrana C., Crawley A.B., Sanchez B., Barrangou R. Characterization and Exploitation of CRISPR Loci in Bifidobacterium longum. Front Microbiol. 2017;8:1851. doi: 10.3389/fmicb.2017.01851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lugli G.A., Tarracchini C., Alessandri G., Milani C., Mancabelli L., Turroni F. Decoding the Genomic Variability among Members of the Bifidobacterium dentium Species. Microorganisms. 2020;8(11):1720. doi: 10.3390/microorganisms8111720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Audy J., Labrie S., Roy D., LaPointe G. Sugar source modulates exopolysaccharide biosynthesis in Bifidobacterium longum subsp. longum CRC 002. Microbiology (Reading) 2010;156(Pt 3):653–664. doi: 10.1099/mic.0.033720-0. [DOI] [PubMed] [Google Scholar]