Abstract
The angiotensin-converting enzyme (ACE)-related carboxypeptidase, ACE2, is a type I integral membrane protein of 805 amino acids that contains one HEXXH + E zinc-binding consensus sequence. ACE2 has been implicated in the regulation of heart function and also as a functional receptor for the coronavirus that causes the severe acute respiratory syndrome (SARS). To gain further insights into this enzyme, the first crystal structures of the native and inhibitor-bound forms of the ACE2 extracellular domains were solved to 2.2- and 3.0-Å resolution, respectively. Comparison of these structures revealed a large inhibitor-dependent hinge-bending movement of one catalytic subdomain relative to the other (∼16°) that brings important residues into position for catalysis. The potent inhibitor MLN-4760 ((S,S)-2-{1-carboxy-2-[3-(3,5-dichlorobenzyl)-3H-imidazol4-yl]-ethylamino}-4-methylpentanoic acid) makes key binding interactions within the active site and offers insights regarding the action of residues involved in catalysis and substrate specificity. A few active site residue substitutions in ACE2 relative to ACE appear to eliminate the S2′ substrate-binding subsite and account for the observed reactivity change from the peptidyl dipeptidase activity of ACE to the carboxypeptidase activity of ACE2.
The angiotensin-converting enzyme (ACE)1 -related carboxypeptidase, ACE2, is a type I integral membrane protein of 805 amino acids that contains one HEXXH + E zinc-binding consensus sequence (1, 2). The catalytic domain of ACE2 is 42% identical to that of its closest homolog, somatic angiotensin-converting enzyme (sACE; EC 3.4.15.1), a peptidyl dipeptidase that plays an important role in the renin angiotensin system for blood pressure homeostasis. The loss of ACE2 in knockout mice has no effect on blood pressure, but reveals ACE2 as an essential regulator of heart function (3). In a recent discovery, ACE2 was identified as a functional receptor for the coronavirus that is linked to the severe acute respiratory syndrome (SARS) (4, 5).
The physiological differences observed in the phenotypes of ACE (6, 7) and/or ACE2 (3) knockout mice presumably reflect the significant differences in substrate specificity and reactivity between these enzymes. Many substrates for ACE2 were identified by screening biologically active peptides (8). In all cases, only carboxypeptidase activity was found. Of the seven best in vitro peptide substrates identified (k cat/K m > 105 m -1 s-1), proline and leucine are the preferred P1 residues, with a partiality for hydrophobic residues in the P1′ position, although basic residues at P1′ are also cleaved (peptide-binding subsites in proteins are as previously defined (9)). Some of the best in vitro peptide substrates are apelin-13, des-Arg9-bradykinin, angiotensin II, and dynorphin A-(1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13). The longest peptide substrate identified is a 36-residue peptide, apelin-36 (8). An examination of the ACE2 and ACE literature may be found in recently published reviews (10, 11, 12).
We report here the first crystal structures of the extracellular metallopeptidase domain of ACE2 in its native and inhibitor-bound states and discuss the influence of these structures in understanding the substrate specificity and catalytic mechanism of the enzyme. While preparing these ACE2 structures for publication, two reports on the crystal structures of testicular angiotensin-converting enzyme (tACE) and Drosophila ACE appeared in the literature (13, 14).
EXPERIMENTAL PROCEDURES
Protein Expression and Purification—A truncated extracellular form of human ACE2 (residues 1-740) was expressed in baculovirus and purified as described previously (8). The signal sequence (residues 1-18) is presumably removed upon secretion from Sf9 cells. The molecular mass of the purified enzyme is 89.6 kDa by matrix-assisted laser desorption ionization time-of-flight mass spectrometry, which is greater than the theoretical molecular mass of 83.5 kDa expected from the sequence (residues 19-740). The difference of ∼6 kDa is believed to be due to glycosylation at the seven predicted N-linked glycosylation sites for this protein.
Crystallization—Briefly, 2 μl of purified ACE2 (5 mg/ml) was combined with an equal volume of reservoir solution, and crystals were grown by hanging drop vapor diffusion at 16-18 °C. The best crystallization reservoir solution conditions for native ACE2 were found to be 100 mm Tris-HCl (pH 8.5), 200 mm MgCl2, and 14% polyethylene glycol 8000. Under these conditions, it took ∼2 weeks to grow single crystals suitable for x-ray diffraction. Similarly, diffraction-quality ACE2 crystals were also grown in the presence of an ACE2 inhibitor, MLN-4760 (ML00106791; (S,S)-2-{1-carboxy-2-[3-(3,5-dichlorobenzyl)-3H-imidazol-4-yl]-ethylamino}-4-methylpentanoic acid). Compound MLN-4760 corresponds to compound 16 of Dales et al. (15). Crystallization trials used 2 μl of reservoir solution plus 2 μl of ACE2 at 5.9 mg/ml containing 0.1 mm inhibitor. The best diffracting ACE2-inhibitor complex crystals were grown in the presence of 19% polyethylene glycol 3000, 100 mm Tris-HCl (pH 7.5), and 600 mm NaCl.
Data Collection and Structure Determination—The best data set for native ACE2 was at 2.2-Å resolution and was collected at the Advanced Photon Source (Argonne National Laboratory). A total of 44 x-ray data sets were collected for native ACE2, including a large number of heavy atom soaks of atoms that had good anomalous signals. The data sets for each derivative were collected at different wavelengths to maximize the anomalous signals for the bound heavy atoms. Native ACE2 data were collected to 2.2-Å resolution at λ = 1.28 Å to maximize the anomalous signal at the zinc absorption edge.
The heavy atom positions were determined and confirmed by a combination of visual inspection of Patterson maps and automatic search procedures, which included SHAKE 'N BAKE (16) and SHELXD (17). The heavy atom parameters were refined and optimized using the computer programs SHARP (18), MLPHARE (19), and XHEAVY (20). The experimental phases were improved by solvent flattening and histogram matching.
Once the native ACE2 structure was determined, it was used to solve the inhibitor-bound structure of ACE2 to 3.0-Å resolution using molecular replacement methods that employed the program AMoRe in the CCP4 software suite (21). The native structure was split into two subdomains: subdomains I and II (see Fig. 3 for definition). Subdomain II was used for molecular replacement and refined in REFMAC5, which resulted in the appearance of electron density for subdomain I. Subdomain I was then fitted into the density by hand, and the structure was refined as a whole. Final refinement was accomplished using the software suite CNX (22).
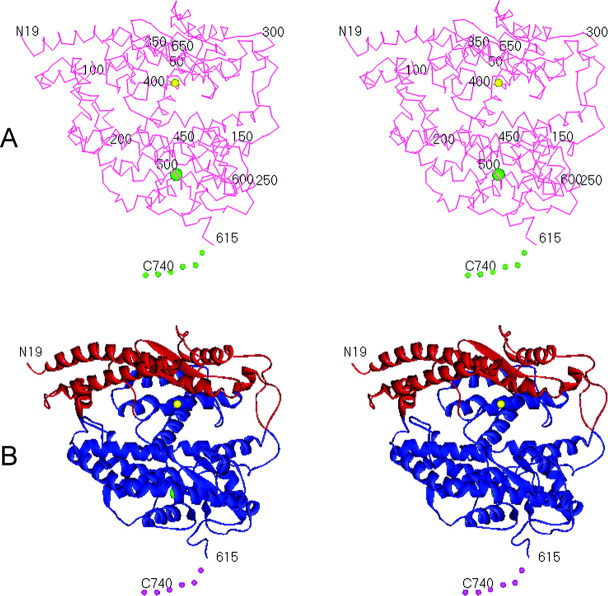
Fig. 3.
Overview of the native ACE2 crystal structure.A, α-carbon trace of the native ACE2 structure looking down into the metallopeptidase active site cleft. The metallopeptidase catalytic domain is colored red. The active site zinc ion is shown as a yellow sphere, and the single bound chloride ion is shown as a green sphere. The S1′ subsite for inhibitor and substrate binding is to the right of the zinc ion, and the S1 subsite is to the left. The collectrin homology domain at the C terminus is disordered and denoted by the green dotted line. B, ribbon diagram of native ACE2 showing the secondary structure and also the two subdomains (I and II) that form the two sides of the active site cleft. The two subdomains are defined as follows: the N terminus- and zinc-containing subdomain I (red), composed of residues 19-102, 290-397, and 417-430; and the C terminus-containing subdomain II (blue), composed of residues 103-289, 398-416, and 431-615. This definition is based on motion observed upon inhibitor binding (see Fig. 4). Zinc and chloride ions are denoted as described for A.
RESULTS AND DISCUSSION
Native ACE2 Structure—The three-dimensional structure of the extracellular region of native ACE2 was determined by multiple isomorphous replacement with anomalous scattering and refined to a crystallographic R-factor of 23.5% (R free = 28.7%) at 2.2-Å resolution. The heavy atom data statistics are summarized in Table I , and the refinement statistics for native ACE2 are summarized in Table II .
Table I.
Heavy atom data statistics for human native ACE2
pCMB, p-chloromercuribenzoate; PIP, di-μ-iodobis(ethylenediamine) diplatinum(II) nitrate; NA, not applicable.
| Derivative | Native (Zu) | pCMB | HgCl2 | PIP | K2PtCl4 |
|---|---|---|---|---|---|
| Heavy atom | Zn | Hg | Hg | Pt | Pt |
| Molarity (mm) | NA | 1 | 1 | 1 | 1 |
| Length of soak (day(s)) | NA | 3.5 | 30 | 1 | 30 |
| No. sites/asymmetric unita | 1 | 1 | 1 | 2 | 2 |
| Wavelength (Å)b | 1.2824 | 1.009 | 1.009 | 1.072 | 1.072 |
| Unique reflections | 49,286c | 21,652c | 17,421 | 13,152 | 14,087 |
| Resolution (Å) | 40-2.2 | 30-2.9 | 30-3.0 | 30-3.4 | 30-3.3 |
| Completeness (%) | 96.3 | 96.6 | 90.6 | 95.4 | 94.2 |
| Rsym (%)d | 5.7 | 10.5 | 10.4 | 9.7 | 11.6 |
| Rmergee | NA | 21.3 | 37.6 | 20.6 | 21.8 |
| Rcullisf | 0.94 | 0.73 | 0.93 | 0.96 | 0.97 |
| Phasing powerg |
1.57 |
1.51 |
0.66 |
0.45 |
0.39 |
Each asymmetric unit contains one human ACE2 protein.
Data were collected at the Brookhaven National Laboratory (NSLS, beam line X25) or at the Argonne National Laboratory (APS, beam line sector 32, COM-CAT). The wavelength for the native (zinc) data set was 1.2824 Å to maximize the anomalous signal at the zinc absorption edge.
Values do not include Bijvoet pairs. Inclusion of Bijvoet pairs increases the number of reflections to 91,550 for native (zinc) ACE2 and 41,716 for the p-chloromercuribenzoate derivative.
, where Ii is the intensity of the measured reflection and Im is the mean intensity of all symmetry-related reflections.
.
.
Phasing power = FH/ERMS, where ERMS is the residual lack of closure.
Table II.
Refinement statistics for native and inhibitor-bound ACE2 structures
Numbers in parentheses represent final shell of data. PDB, Protein Data Bank.
| Native ACE2 (PDB code 1R42) | Inhibitor-bound ACE2 (PDB code 1R4L) | |
|---|---|---|
| Resolution (Å) | 46.7-2.20 (2.34-2.20) | 43.3-3.0 (3.19-3.00) |
| No. reflections | 47,465 (5982) | 17,228 (2250) |
| Rsym (%)a | 5.7 (40.8) | 7.0 (20.4) |
| Completeness (%) | 96.3 (81.8) | 96.8 (85.1) |
| Space group | C2 | C2 |
| a | 103.64 | 100.53 |
| b | 89.46 | 86.51 |
| c | 112.40 | 105.86 |
| β | 109.15 | 103.65 |
| Unit cell volume (Ås) | 986,854 | 894,383 |
| Solvent content (%)b | 53 | 53 |
| Molecules/asymmetric unit | 1 | 1 |
| Reflections used in Rfree | 4798 | 1723 |
| No. protein atoms | 5165 | 5147 |
| No. solvent atoms | 302 | 13 |
| No. zinc atoms | 1 | 1 |
| No. chloride atoms | 1 | 1 |
| No. sugar atoms | 42 | 28 |
| R-factor (%) | 23.5 (37.9) | 25.3 (37.9) |
| Rfree (%) | 28.7 (39.8) | 33.7 (46.0) |
| r.m.s. deviations from ideal stereochemistry | ||
| Bond lengths (Å) | 0.008 | 0.008 |
| Bond angles | 1.4° | 1.5° |
| Dihedrals | 21.7° | 22.2° |
| Impropers | 0.92° | 0.97° |
| Mean B-factor (all atoms; Å2) |
59.9 |
74.5 |
See Footnote d of Table I.
Vsolvent = 1-1.23/Vm, where Vm is the volume of protein in the unit cell/volume of unit cell, assuming one molecule/asymmetric unit and four asymmetric units in the monoclinic unit cell.
The extracellular region of the human ACE2 enzyme is composed of two domains. The first is a zinc metallopeptidase domain (residues 19-611), which is ∼42% identical to the corresponding domains of human sACE and tACE (Fig. 1 ). Electron density near the active site of native ACE2 is shown in Fig. 2a . An α-carbon trace for this metallopeptidase domain of ACE2 is shown in Fig. 3A. The second domain is located at the C terminus (residues 612-740) and is ∼48% identical to human collectrin (23). Unfortunately, the electron density map for much of the collectrin homology domain is weak. Only half of this domain is visible in the electron density map, and what can be seen is ambiguous due to topology and connectivity issues.
Fig. 1.

Sequence alignment of the metallopeptidase domains of human ACE2, sACE, and tACE. The sequences of the catalytic domain of tACE and the C-terminal catalytic domain of sACE are identical (48, 49). The human sACE and ACE2 sequences were obtained from the GenBank™/EBI Data Bank (accession numbers P12821 and AAF99721, respectively). The ClustalW Alignment Tool was used for these sequence alignments (50). The secondary structural elements for native ACE2 were assigned using STRIDE software (51) and are denoted as follows: α-helical segments,- - ▸; 310 helical elements, - - >; and β-structural segments, - - •. These secondary structural elements are color-coded red for subdomain I and blue for subdomain II (subdomains are defined in Fig. 3). Identical residues for all three enzymes are colored red. The zinc-binding residues in all three enzymes are shown in green, and the chloride ion-binding residues in all enzymes are shown in orange. Residues in ACE2 within H-bonding distance of the inhibitor MLN-4760 or in tACE within H-bonding distance of lisinopril are colored blue (13). The Cys residues conserved between ACE2 and ACE are colored magenta. The six predicted N-linked glycosylation sites for the metallopeptidase region of ACE2 are shown (gray N). The beginning of the collectrin homology domain (23) in ACE2 is indicated (▾).
Fig. 2.
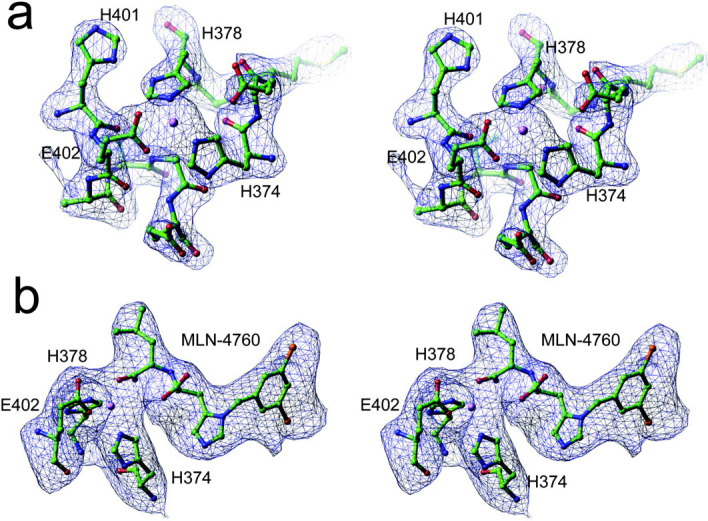
Experimental electron density maps for native and inhibitor-bound ACE2 structures.a,an omit electron density map of the zinc-binding site of the native ACE2 structure (His374-His378, His401-Glu406), calculated with phases from the refined model at 2.2-Å resolution. The map is contoured at 3σ. b, an omit electron density map of MLN-4760, zinc, and the three metal-binding ligands of the protein (His374, His378, and Glu402), calculated with phases from the refined model at 3.0-Å resolution. The map is contoured at 3σ.
The metallopeptidase domain of ACE2 can be further divided into two subdomains (I and II) (Fig. 3B), which form the two sides of a long and deep cleft with dimensions of ∼40 Å long by ∼15 Å wide by ∼25 Å deep. The two catalytic subdomains are connected only at the floor of the active site cleft. One prominent α-helix (helix 17, residues 511-531) connects the two subdomains and forms part of the floor of the canyon. The deeply recessed and shielded proteolytic active site of ACE2 is a common structural feature of proteases and exists to avoid hydrolysis of correctly folded and functional proteins (24, 25) and is also consistent with the profiles of binding of tethered inhibitors to sACE (26, 27).
The N terminus-containing subdomain I and the C terminus-containing subdomain II are defined in Fig. 3B. This subdomain definition is based on the subdomain movement that was observed upon inhibitor binding (see below). The secondary structure of the metallopeptidase domain of ACE2 is composed of 20 α-helical segments and nine 310 helical segments that together make up ∼62% of the structure (Figs. 1 and 3). This contrasts with just six short β-structural segments that make up ∼3.5% of the structure. Glycosylation is suggested by the presence of electron density at all six potential N-linked sites: Asn53, Asn90, Asn103, Asn322, Asn432, and Asn546 (2). The highest density was observed at Asn90, Asn103, and Asn546 and allowed the building of three N-acetylglucosamine groups. Three disulfide bonds of ACE2 (Cys133-Cys141, Cys344-Cys361, and Cys530-Cys542) are conserved in sACE and tACE (Fig. 1).
The zinc-binding site is located near the bottom and on one side of the large active site cleft (subdomain I side), nearly midway along its length. The zinc is coordinated by His374, His378, Glu402, and one water molecule (in the native structure). These residues at the zinc-binding site of ACE2 make up the HEXXH + E motif conserved in the zinc metallopeptidase clan MA (28).
A chloride ion (Cl-) is bound in native ACE2, coordinated by Arg169, Trp477, and Lys481 in subdomain II. The larger electron density compared with water supports the assignment of this density to a Cl- ion, as does as the larger than expected distances between the coordinating side chains and the Cl- ion (compared with water H-bonds): N-ϵ and N-η1 of Arg169 are both 3.2 Å from Cl-, indole ring nitrogen of Trp477 is 3.5 Å from Cl-, and N-ϵ of Lys481 is 5.0 Å from Cl-.
Inhibitor-bound ACE2 Structure—The structure of ACE2 with an inhibitor bound at the active site was solved by molecular replacement to a resolution of 3.0 Å using the native ACE2 structure. Refinement statistics for the inhibitor-bound ACE2 structure are shown in Table II. The bound compound MLN-4760 potently inhibits human ACE2 (IC50 = 0.44 nm), but weakly inhibits tACE (IC50 > 100 μm) and carboxypeptidase A (IC50 = 27 μm) (15). The structure of the bound inhibitor is shown in Fig. 2b along with the experimental electron density map near the active site.
Ligand-dependent Subdomain Hinge-bending Movement—There is a clear difference between the native and inhibitor-bound ACE2 structures with respect to the distance separating the two subdomains (Fig. 4A ). These two subdomains undergo a large inhibitor-dependent hinge-bending movement of one catalytic subdomain relative to the other (∼16°) that causes the deep open cleft in the native form of the enzyme to close around the inhibitor. This movement can be viewed when subdomain II from the native and inhibitor-bound ACE2 structures are superimposed (root mean square (r.m.s.) deviation of 1.41 Å for 409 residues) as shown in Fig. 4A. In this view, subdomain II remains essentially unchanged, but subdomain I moves to close the gap, essentially mimicking the action of a closing clam shell. The α-carbon atoms of some residues near the outer edge of the subdomain gap move as much as ∼13 Å, whereas residues lying near or on the hinge axis (residues 99 and 100, 284-293, 396 and 397, 409 and 410, 433 and 434, 539-548, and 564-568) are nearly stationary.
Fig. 4.
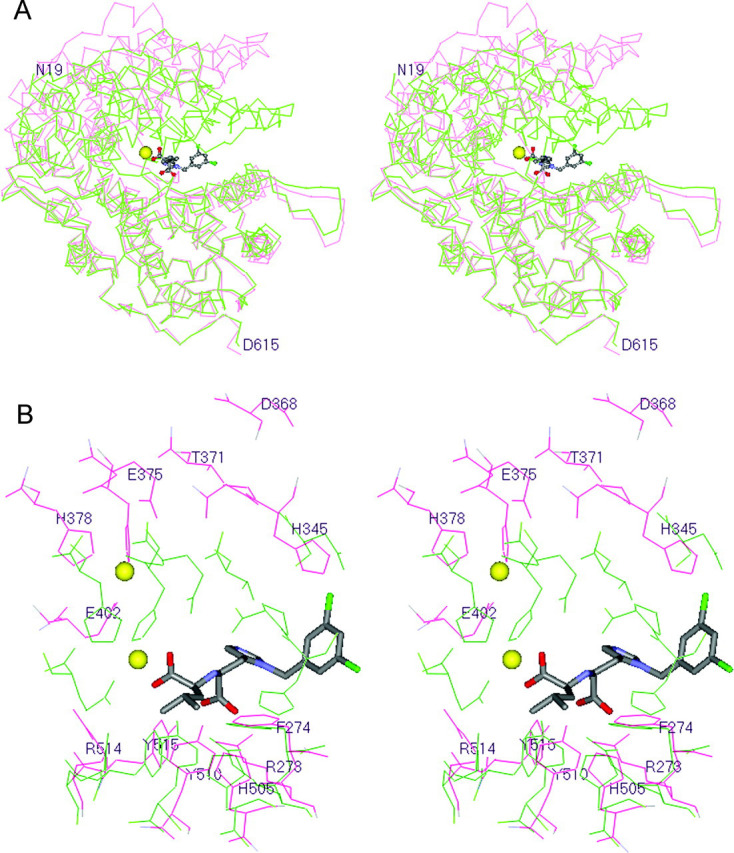
Superposition of the native and inhibitor-bound ACE2 structures.A, the 409 α-carbon atoms corresponding to subdomain II of the native and inhibitor-bound ACE2 structures were superimposed with an r.m.s. deviation of 1.41 Å. Native ACE2 is colored red, and inhibitor-bound ACE2 is colored green. The zinc ion is shown as a yellow sphere, and the inhibitor MLN-4760 is shown in a ball-and-stick rendering with default atom coloring: gray, carbon; blue, nitrogen; red, oxygen; green, chlorine. This view is looking down the length of the active site cleft and is rotated 90° from that shown in Fig. 3. This perspective illustrates the ∼16° hinge-bending movement of subdomain I relative to subdomain II that occurs upon inhibitor binding to ACE2. B, shown is a close-up view of the active sites of the superimposed native (red) and inhibitor-bound (green) ACE2 structures. This is the same superposition of subdomain II for both structures as described for A. In this perspective, the residues of subdomain I within the active site are shown to move upon inhibitor binding relative to those in subdomain II. The inhibitor MLN-4760 is shown in stick rendering with the same atom color code as described for A. The average movement for residues near the active site is 6-9 Å. The yellow spheres are the two positions of the zinc atom in the native and inhibitor-bound structures. This figure was prepared using MOE 2003.02 software (Chemical Computing Group, Inc.).
The movement of residues within the active site is shown in Fig. 4B. This view is a close-up of Fig. 4A with subdomain II of the native and inhibitor-bound ACE2 structures superimposed. Many of these residues move as much as 6-9 Å after binding of the inhibitor. Similar subdomain hinge-bending motions have also been observed for other zinc metalloproteases (29, 30). The largest previously observed ligand-dependent movement for metalloproteases is a 14° hinge-bending subdomain motion demonstrated for Pseudomonas aeruginosa elastase (open versus closed) that resulted in an ∼2-Å movement to close the N-terminal/C-terminal subdomain gap (29). Domain closure movements in proteins are a common mechanism for the positioning of critical groups around substrates and inhibitors (31, 32, 33) and also for the trapping of substrate and reaction intermediates (34).
Inhibitor Binding Interactions and Implications for Substrate Specificity and Catalysis—Both metallopeptidase subdomains of ACE2 are nearly equally involved in binding of the inhibitor MLN-4760. Inspection of the interactions between MLN-4760 and ACE2 revealed important residues responsible for inhibitor binding and presumably for substrate binding and catalysis (Fig. 5 ). The inhibitor MLN-4760 has two carboxylate groups, one of which binds to the zinc atom by displacing the bound water molecule present in the native ACE2 structure. The zinc coordination sphere (His374, His378, and Glu402) is a subset of the 21 residues of ACE2 that are located within 4.5 Å of the bound inhibitor and that make up the greater part of the active site. Six of the most important of these residues contribute specific H-bonding interactions with MLN-4760 (Fig. 5). The side chains of Arg273, His505, and His345 are H-bonded to the terminal carboxylate of the inhibitor. The carbonyl oxygen atom of Pro346 and the N-ϵ atom of His345 are within H-bonding distance of the secondary amine group of the inhibitor. Thr371 is within H-bonding distance of the imidazole ring of the 3,5-dichlorobenzylimidazole group of MLN-4760. The phenolic group of Tyr515 donates an H-bond to the zinc-bound carboxylate group of the inhibitor. The carboxyl group of Glu375 is within H-bonding distance of the other oxygen atom of the zinc-bound carboxylate of MLN-4760, but is presumably not protonated until peptide hydrolysis occurs (see below).
Fig. 5.
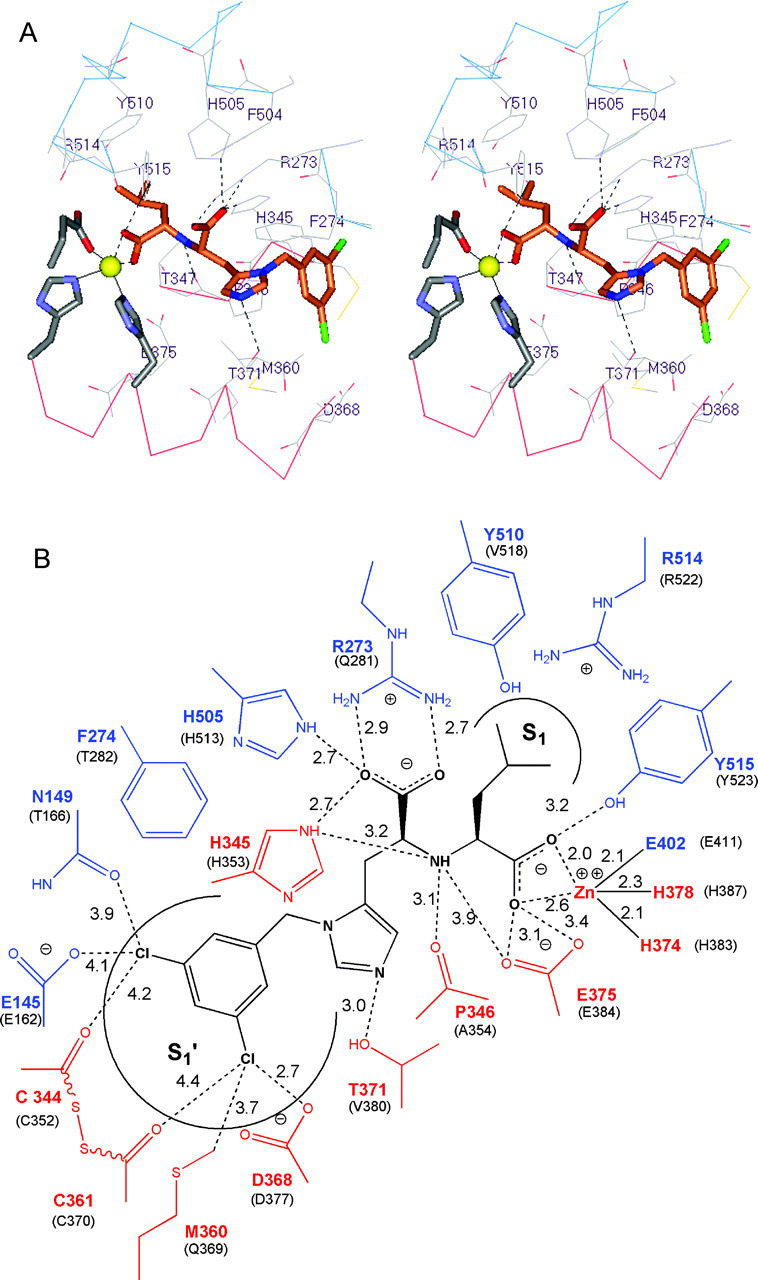
Binding interactions of the inhibitor MLN-4760 at the active site of ACE2.A, the residues of ACE2 that contribute direct binding interactions with the inhibitor MLN-4760 are shown. MLN-4760 is shown in stick rendering with the same atom color code as described in the legend to Fig. 4A, except carbon is orange. The α-helix 11 segment derived from subdomain I has the α-carbon wire colored red, and turns and β-elements derived from subdomain II have the α-carbon wire colored blue. Probable H-bonding interactions are shown as black dashed lines. The zinc ion is shown as a yellow sphere. ACE2 residues coordinating the zinc ion are shown in stick rendering. B, shown is a schematic view of MLN-4760 binding interactions. MLN-4760 is shown in black. Residues derived from subdomain I are red, and residues derived from subdomain II are blue. The equivalent residues in tACE are in given in parentheses. Distances are measured in angstroms.
The zinc-bound carboxylate of MLN-4760 appears to mimic the zinc-bound tetrahedral intermediate characteristic of nucleophilic attack of the scissile bond by the zinc-bound water during peptide hydrolysis (35). This transition state structure is usually stabilized by H-bonds donated by imidazole, phenolic, or guanidino functional groups of neighboring amino acid side chains in other zinc metalloproteases (30, 35). For ACE2, this carboxyl anion stabilization most likely occurs through the phenolic group of Tyr515 (Fig. 5).
Besides the zinc coordination sphere and potential H-bond-contributing residues, there are an additional 11 residues of ACE2 that make close contacts (<4.5 Å) with MLN-4760, many of which are shown in Fig. 5. These residues do not contribute direct H-bonding interactions with the inhibitor, but provide important electrostatic and van der Waals interactions with MLN-4760. These residues define the S1 and S1′ peptide-binding subsites, thereby providing the inhibitor, as well as substrate, binding specificity.
The isobutyl group of MLN-4760 packs nicely into the S1 subsite of ACE2 (Fig. 5A), suggesting a similar preferred fit for a leucyl side chain at the P1 position of peptide substrates. The topology and chemical environment of the S1 subsite are dictated by four residues (Tyr510, Arg514, Phe504, and Thr347) that are expected to restrict the size of substrate P1 side chains. Large substrate P1 residues such as Trp, Tyr, Phe, Arg, and Lys would require significant movement of the phenolic side chain of Tyr510, which forms a lid over the top of the S1 subsite. This observation is consistent with the reported substrate specificity data showing only medium sized residues (Pro and Leu) at the P1 position for preferred substrates with k cat/K m > 105 m -1 s-1 (8). Peptides with Phe and Tyr at the P1 position, such as angiotensin-(1, 2, 3, 4, 5, 6, 7, 8, 9) (DRVYIHPFH), bradykinin (RPPGFSPFR), Leu-enkephalin (YGGFL), Met-enkephalin (YGGFM), and angiotensin-(1, 2, 3, 4, 5) (DRVYI), are not substrates for ACE2 despite the presence of preferred hydrophobic or basic P1′ residues. Some ACE2-catalyzed hydrolysis was observed for peptides with a slightly larger residue (His) at the P1 site (i.e. angiotensin I), but the catalytic efficiency was orders of magnitude lower (k cat/K m = 4.9 × 103 m -1 s-1) despite good binding.
The limited size of the S1 subsite may also account for the observed orientation of binding of MLN-4760. The design principles leading to the identification of MLN-4760 were based on assumptions from substrate screening studies and information regarding known metallopeptidase inhibitors (36). Consequently, MLN-4760 was designed as a His-Leu mimetic, in which the isobutyl side chain was predicted to mimic the Leu side chain (P1′), and the substituted His was expected to bind in the S1-S2 subsites (15). Furthermore, the Leu carboxylate was envisioned to mimic the substrate's C-terminal carboxylate, and the His carboxylate was expected to coordinate the zinc atom. Although the inhibitor potency optimization was supported by this design hypothesis (several selective inhibitors at <10 nm), experimentally, the inhibitor-bound crystal structure reveals that MNL-4760 binds in the opposite orientation: the two carboxylates are reversed, the isobutyl side chain occupies the S1 subsite, and the 3,5-dichlorobenzylimidazole group binds in the S1′ subsite. This alternative binding mode may be the result of the small S1 subsite, which cannot accommodate the 3,5-dichlorobenzyl group. Models of MLN-4760 bound to ACE2 that were based on the tACE structure (37) did not predict the observed binding mode shown in Fig. 5. Retrospective comparison of the inhibitor structure-activity relationship with the crystal structure supports the unanticipated binding orientation. During the potency optimization studies, it appeared that the His portion of the inhibitors better tolerated changes in size and physicochemical properties compared with the Leu portion, which is consistent with the new experimental evidence (Fig. 5).
The S1′ subsite of ACE2 is much larger than the S1 subsite and is formed by the lengthwise channel between the two subdomains. In addition to the aforementioned residues that contribute specific H-bonding interactions with MLN-4760 (Arg273, His345, His505, Thr371, and the carbonyl group of Pro346), there are a number of other residues that make close contacts with the 3,5-dichlorobenzylimidazole group of the inhibitor. The side chains of Phe274, Pro346, Thr371, Met360 and the disulfide linkage of Cys344 and Cys361 provide a hydrophobic environment for the S1′ subsite. Additional close contacts are observed for the side chains of Asp368, Glu145, Asn149, and Lys363 and the carbonyl groups of Cys344 and Cys361, which accommodate the far end of the dichlorobenzyl ring as depicted in Fig. 5B. The large size of the this S1′ subsite easily accommodates the 3,5-dichlorobenzylimidazole group of MLN-4760 and mirrors the observed preference for large hydrophobic or basic residues (Arg and Lys) at the P1′ position of peptide substrates (8).
Comparison with Structural Homologs—There are four proteins in the Protein Data Bank (38) that have overall architectural similarity to ACE2. They include the following: 1) the recently solved human tACE (code 1O86) (13), an enzyme of the M2 metallopeptidase family (EC 3.4.15.1); 2) Drosophila ACE (code 1J36), with which ACE2 shares ∼35% sequence identity (14); 3) rat neurolysin (code 1I1I) (39), an M3 metallopeptidase family member (EC 3.4.24.16) with which ACE2 shares just ∼17% sequence identity; and 4) Pyrococcus furiosus carboxypeptidase (code 1KA2) (40), a member of the M32 carboxypeptidase family.
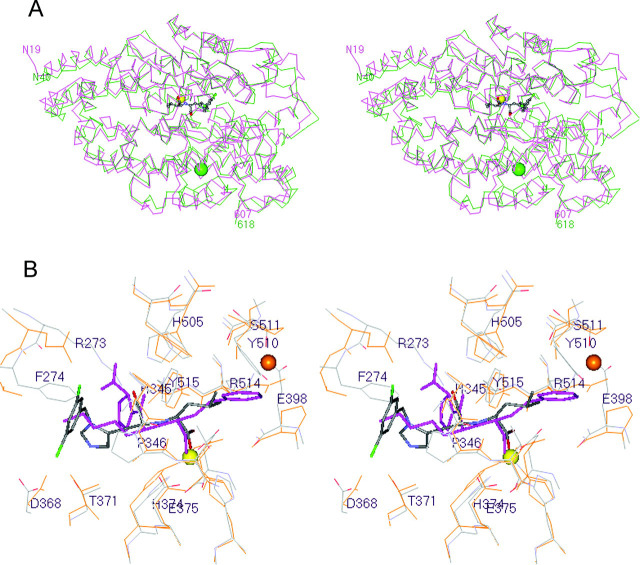
Comparison with tACE—The α-carbon atoms of lisinopril-bound tACE were superimposed onto the equivalent atoms of inhibitor-bound ACE2 (588 residues) with an r.m.s. deviation of 1.80 Å (Fig. 6A ), suggesting that these two protein structures are very similar. The largest difference between the ACE2 and tACE α-carbon traces is the insertion of a loop extension of 10 residues (Gly211-Gly220) between α-helices 7 and 8 in subdomain II of ACE2.
Fig. 6.
Superposition of the ACE2 and tACE structures.A, the α-carbon atoms in lisinopril-bound tACE (13) were superimposed onto the equivalent atoms in inhibitor-bound ACE2 (588 residues) with an r.m.s. deviation of 1.80 Å. MLN-4760-bound ACE2 is magenta, and lisinopril-bound tACE is green. MLN-4760 is shown bound to ACE2 with the same color code described in the legend to Fig. 4A. Similarly, the zinc and chloride ions are shown as described in the legend to Fig. 3. The orientation is the same as that shown for native ACE2 in Fig. 3. Structures were superimposed using MOE 2003.02 software. B, the 21 α-carbon atoms at the inhibitor-bound active site of ACE2 (residues 4.5 Å from the inhibitor) were superimposed onto the equivalent atoms of lisinopril-bound tACE (Protein Data Bank code 1O86) with an r.m.s. deviation of 0.53 Å. The active site of ACE2 and MLN-4760 are shown in default colors, with the inhibitor displayed in stick rendering. Labels are for ACE2 residues only. The active site residues of tACE are shown in orange, with the inhibitor lisinopril colored purple in stick rendering. The zinc ion is shown as a yellow sphere, and the second chloride ion of tACE (CL2) is shown as an orange sphere. This chloride ion site does not exist in ACE2 due to the Glu398 substitution for Pro407 (see “Results and Discussion”). Other important differences between ACE2 and tACE are as follows: Arg273versus Gln281, Phe274versus Thr282, and Tyr510versus Val518, respectively.
An improved superposition of the inhibitor-bound ACE2 and tACE structures (r.m.s. deviation of 0.53 Å) was obtained by considering just the equivalent α-carbon atoms of the aforementioned 21 residues of ACE2 that come within 4.5 Å of the bound MLN-4760 (Fig. 6B). Thirteen of these active site residues are conserved in the tACE and sACE enzymes: Glu145, Cys344, His345, Cys361, Asp368, His374, Glu375, His378, Glu402, Phe504, His505, Arg514, and Tyr515. These structural similarities reflect the nearly identical way that MLN-4760 and lisinopril sit in the superimposed active sites (Fig. 6B).
The remaining eight active site ACE2 residues are substituted in tACE as follows: Asn149 → Thr, Arg273 → Gln, Pro346 → Ala, Thr347 → Ser, Met360 → Gln, Lys363 → Thr, Thr371 → Val, and Tyr510 → Val. These changes at the active sites of ACE2, tACE, and sACE presumably play a significant role in the observed differences in substrate specificity and inhibitor binding profiles for these homologous enzymes. As was discussed previously, Tyr510 and Thr347 line the S1 subsite of ACE2 and confine this subsite for the accommodation of only small and medium sized side chains such as leucyl and prolyl, which is consistent with the known substrate preferences (8). The smaller side chains at the S1 subsite of tACE make this subsite somewhat larger in tACE than observed for ACE2. In a superposition of the inhibitor-bound ACE2 and lisinopril-bound tACE structures, the Tyr510 phenolic group of ACE2 occupies the space where the P1 phenylpropyl group of the lisinopril resides (Fig. 6B).
Another important difference between ACE2 and tACE is the Arg273 → Gln substitution. The guanidino group of Arg273 makes a bidentate H-bond with the terminal carboxylate of MLN-4760 (Fig. 5). Not only does the guanidino group of Arg273 stabilize the terminal carboxylate of the inhibitors and peptide substrates, but its larger size (compared with Gln in tACE) causes steric crowding at the potential S2′ binding subsite. The superposition of lisinopril-bound tACE onto MLN-4760-bound ACE2 reveals that the guanidino group of Arg273 nearly superimposes on the terminal carboxylate of the P2′ Pro residue of lisinopril, thereby severely limiting the size of the S2′ subsite in ACE2 compared with tACE (Fig. 6B). This single residue difference in ACE2 relative to tACE essentially eliminates the S2′ subsite in ACE2 and offers an explanation for the observed switch in the peptidyl dipeptidase activity of tACE to the observed carboxypeptidase activity of ACE2. This difference at the S1′ and S2′ subsites for the homologous enzymes also helps to clarify why the potent ACE inhibitors lisinopril, enalaprilat, and captopril are inactive against ACE2 (2).
The native tACE structure was reported to be nearly identical to the lisinopril-bound tACE structure (13), suggesting that no ligand-dependent conformational change occurs for tACE or at least under the conditions used to obtain these crystals (pH 4.7, 50 mm sodium acetate with an unspecified [Cl-]) (41). However, inspection of the reported native tACE structure (Protein Data Bank code 1O8A) reveals that two ligands are bound to the active site: an acetate molecule bound to the zinc atom and an unknown compound that presumably originated during protein expression and purification. This compound was modeled as an N-carboxyalanine and is isosteric with 2-methylsuccinic acid according to notes deposited with the native tACE coordinates in the Protein Data Bank file. This 2-methylsuccinate-like ligand is bound to tACE in a manner that closely resembles that observed for lisinopril. Five of the same H-bonds present in the lisinopril-bound tACE structure are also present for the 2-methylsuccinate-like ligand. Therefore, the reported native tACE structure would appear to be another ligand-bound structure, and its closed conformational state is consistent with other ligand-bound tACE and ACE2 structures.
Structural Homology for the Catalytic Motif—ACE2 and related ACE enzymes are related to other HEXXH clan members through divergent evolution with an overall primary sequence identity that is <15% (Fig. 7A ). The two largest metallopeptidase clans are clan MA (gluzincin family) and clan MB, which include the many families of zinc metallopeptidases that have the HEXXH + E and HEXXH + H zinc-binding motifs, respectively (28). Among these clans, there is structural homology for the catalytic motif and the core of the inhibitor/substrate-binding site. Representative examples of the catalytic motifs of some members of these clans are shown in Fig. 7B. These active sites include thermolysin (clan MA) and astacin (clan MB) in complex with transition state analogs (30, 42). The ACE2 complex with MLN-4760 is shown for comparison. These transition state analogs bind to thermolysin and astacin in a manner that is similar to the way MLN-4760 binds to ACE2 and reveal a high level of structural homology between the catalytic motifs of these enzymes. Indeed, eight α-carbon atoms corresponding to residues 345, 346, 374-378, and 402 of MLN-4760-bound ACE2 were superimposed on equivalent α-carbon atoms of Z-Pro-Lys-Pheψ(PO2CH2)-dl-Ala-Pro-OMe-bound thermolysin with an r.m.s. deviation of 0.49 Å (Fig. 7C). This structural conservation of the key residues and scaffold is believed to translate into a preservation of the catalytic role of many elements of this motif.
Fig. 7.
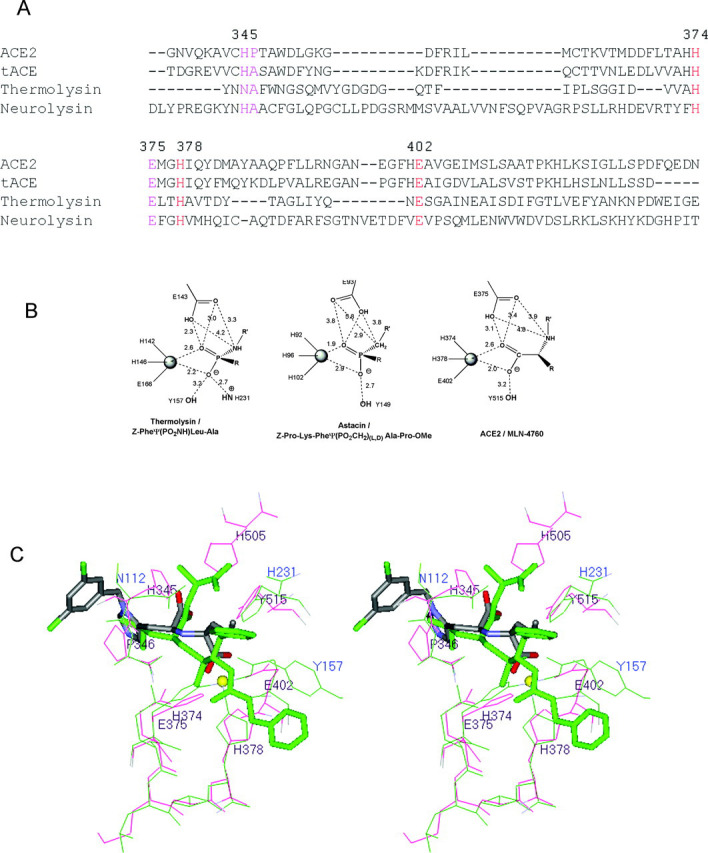
Structural homology for the ACE2 catalytic motif to other members of the HEXXH metallopeptidase clans.A, shown is the structure-based sequence alignment of ACE2, tACE, thermolysin, and neurolysin. The conserved residues correspond to the catalytic motif for these enzymes (colored red (zinc binding) and magenta). Sequence numbering is for ACE2. B, the catalytic motifs for thermolysin and astacin bound to transition state analogs Z-Pheψ(PO2NH)-Leu-Ala and Z-Pro-Lys-Pheψ(PO2CH2)-dl-Ala-Pro-OMe, respectively (30, 42), are compared with the ACE2 complex with MLN-4760. Distances are measured in angstroms. C, shown is the superposition of the catalytic motifs of ACE2 (red) and thermolysin (green). Eight α-carbon atoms corresponding to residues 345, 346, 374-378, and 402 of MLN-4760 bound ACE2 were superimposed onto the equivalent α-carbon atoms of Z-Pro-Lys-Pheψ(PO2CH2)-dl-Ala-Pro-OMe-bound thermolysin (see sequence alignment in A) with an r.m.s. deviation of 0.49 Å. Bound inhibitors are shown in stick rendering with default atom coloring for MLN-4760 and green coloring for Z-Pro-Lys-Pheψ(PO2CH2)-dl-Ala-Pro-OMe. The zinc ion is shown as a yellow sphere. ACE2 labels are black, and thermolysin labels are blue. ψ indicates replacement of the peptide bond by the group in parentheses.
Proposed Catalytic Mechanism for ACE2-mediated Peptide Hydrolysis—The strong structural homology of the active site of inhibitor-bound ACE2 to catalytic motifs of evolutionarily related enzymes in metallopeptidase clans MA and MB (Fig. 7) suggests that many of these structural elements play similar catalytic roles in ACE2. These similarities have led to a proposed catalytic mechanism for ACE2 (Fig. 8 ) that has many features in common with mechanisms proposed for other HEXXH zinc metallopeptidase clan members (30, 35, 40). The mechanism reveals a probable initial ES complex formation and generation of the tetrahedral intermediate. The formation of this ES complex causes an ∼16° subdomain hinge-bending movement of subdomain I toward subdomain II, bringing all the catalytic components into a functional orientation. These substrate and inhibitor-dependent subdomain movements are consistent with induced fit and transition state theories of catalysis (43).
Fig. 8.
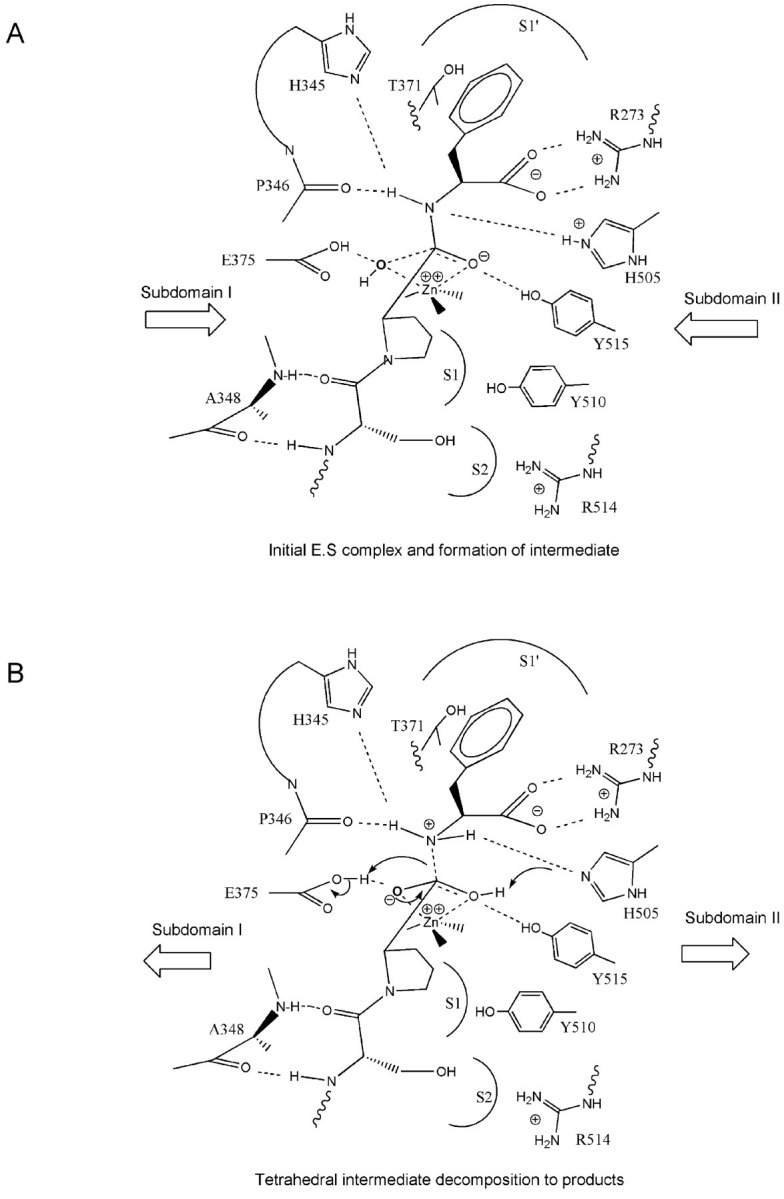
Proposed mechanism for ACE2-catalyzed hydrolysis of peptide substrates.A, shown is the ES complex and progression to the tetrahedral intermediate. Substrate binding to one subdomain induces a subdomain hinge-bending movement (indicated by large arrows) to close the active site cleft and to bring important residues into position for catalysis. This movement is followed by attack of a zinc-bound water at the carbonyl group of the scissile amide bond to form a tetrahedral intermediate, resulting in transfer of a proton from the attacking water to Glu375 (35). Simultaneously, a proton is transferred from His505 to the leaving nitrogen atom of the P1′ residue. This sp3 hybridized nitrogen is stabilized by H-bonds from Pro346, His505, and/or His345. B, collapse of the tetrahedral intermediate to the products occurs by breaking of the amide C-N bond together with abstraction of a proton from Glu375 by the emerging free nitrogen of the product amino acid. The new emerging product carboxyl group can then transfer a proton back to His505 either directly by exchange between carboxyl oxygen atoms or by exchange with solvent.
Other metallopeptidases exhibit hinge-bending motions upon ligand binding (see above), but the average ligand-dependent displacements of 6-9 Å for residues at the active site of ACE2 (Fig. 4B) are much larger than previously reported displacements (∼2 Å) (29). These relative movements at the active site of metallopeptidases are a function of the hinge-bending angle and also the size of the opposing subdomains. Another distinguishing feature of this mechanism is the proton transfer from His505 to the leaving nitrogen atom of the P1′ residue. A protonated histidine is supported by the pH-rate profile that has a maximum at 6.5 (8). Additional evidence for the role for His505 comes from site-directed mutagenesis in sACE, where mutation of the analogous His1089 to Ala results in a 100-fold drop in k cat and 4.5-fold drop in K m (44). Similarly, the His1089 → Leu mutation results in a 671-fold drop in k cat and a 6.2-fold drop in K m. The much larger impact on k cat compared with K m supports a catalytic role for this histidine rather than an important binding function (44). His505 appears to be too far from the zinc-bound carboxylate (6.3 Å) to be directly involved in H-bonding stabilization of the carbonyl tetrahedral intermediate, as was expected from sequence homology (44). Protonation and stabilization of the sp 3 hybridized nitrogen of the peptide intermediate as depicted in Fig. 8 may better explain the site-directed mutagenesis data. Many elements of this mechanism are believed to be relevant to tACE and sACE as well, except with release of dipeptide products due to the larger, more accommodating S2′ subsite in ACE.
Anion-binding Sites—An interesting aspect of the ACE2-mediated substrate hydrolysis is the activation by chloride ions and other anions (8) in much the same way that anions activate substrate hydrolysis and inhibitor binding for sACE and tACE (45, 46). A single bound chloride ion was identified in the high resolution native ACE2 structure (Fig. 3). The location of this anion-binding site in subdomain II is nearly identical to the position of a chloride ion in the tACE structure, which was designated CL1 (13). The chloride-coordinating ligands Arg169, Trp477, and Lys481 correspond to Arg186, Trp485, and Arg489, respectively, in tACE. In ACE2, this anion-binding site is ∼21 Å away from the active site zinc ion and ∼16 Å away from the dichlorobenzyl group of bound MLN-4760. A second chloride-binding site was identified for tACE (designated CL2) and found to be composed of coordinating side chains from Arg522 and Tyr224 and bracketed by Pro407 and Pro519 on two sides. Arg522 and Tyr227 are conserved in ACE2 as Arg514 and Tyr207, but the two bracketing proline residues are replaced in ACE2 with Glu398 and Ser511, respectively. This double substitution has the effect of eliminating the CL2 anion-binding site in ACE2 because it causes the projection of Glu and Ser side chains into the location of where CL2 binds to tACE (Fig. 6B).
With only a single common anion-binding site observed for ACE2 and tACE (CL1), it is possible to imagine that this site is responsible for the anion activation effect in both homologous enzymes. On the other hand, site-directed mutagenesis studies suggest that the Cl--coordinating ligand Arg1098 of sACE (Arg522 of tACE) at the second chloride ion site in tACE (CL2) plays a role in anion activation in sACE (47). The equivalent residue in ACE2 (Arg514) cannot play the same role because this chloride-binding site is replaced in ACE2 with the Glu398/Ser511 double substitution.
A simple explanation for the experimentally observed anion activation in ACE2 is that a second chloride ion site exists in substrate- or inhibitor-bound ACE2, but at a different location than observed for tACE. This second anion-binding site, specific to the inhibitor-bound form of the enzyme, may help to shift the open/closed conformational equilibrium in favor of the closed or catalytic competent closed conformer, thus ensuring a tighter grasp on the transition state for peptide hydrolysis. Kinetic models for the effect of anions on the activity and inhibition of sACE (45, 46) are consistent with chloride ions influencing catalysis through shifts in conformer equilibria. Unfortunately, the lower resolution of the MLN-4760-bound ACE2 structure (3.0 Å) limits the distinction between chloride ions and water molecules in this model. One or more of the 13 water molecules modeled into the inhibitor-bound ACE2 structure could be additional bound chloride ions, but higher resolution data are needed to confirm. Similarly, it is not known whether the second chloride ion of tACE (CL2) is unique to inhibitor-bound tACE and not bound in native tACE because no true native tACE structure is currently available.
ACE2 as the SARS Coronavirus Receptor—The recent identification of ACE2 as a functional receptor for the SARS coronavirus (4, 5) has raised questions about what binding determinants on ACE2 interact with the virus. Mutations of the ACE2 zinc-coordinating residues His374 and His378 to asparagines were found to have no effect on syncytial formation (4), suggesting that interfering with the active site has no effect on viral spike protein binding to ACE2. However, the large conformational change observed upon MLN-4760 binding to ACE2 could prove to be unfavorable for viral binding to its receptor and/or syncytial formation. Thus, metallopeptidase inhibitors such as MLN-4760 may still prove useful for prevention of viral binding to ACE2 and blockage of infection. The ACE2 structures reported here are expected to be useful in aiding efforts to identify the binding site for the SARS coronavirus spike protein. Moreover, the x-ray structures should enable the future design of more potent and specific inhibitors of ACE2, at either the metallopeptidase site or any separate viral spike protein-binding exosite that may be discovered.
Acknowledgments
We thank Rebecca Matthew and Geoff Stamper for help with the characterization of early ACE2 crystals and Larry Dick, Paul Hales, Chad Vickers, Peter Tummino, Mary Donoghue, and Susan Acton for many discussions regarding the characterization of ACE2. We are grateful to Kazumi Shiosaki, Julian Adams, Andrew Nichols, and Michael Kuranda for support.
Footnotes
The atomic coordinates and structure factors (code 1R42 and 1R4L) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The abbreviations used are: ACE, angiotensin-converting enzyme; sACE, somatic angiotensin-converting enzyme; tACE, testicular or germinal angiotensin-converting enzyme; SARS, severe acute respiratory syndrome; r.m.s., root mean square; Z, benzyloxycarbonyl.
References
- 1.Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., Breitbart R.E., Acton S. Circ. Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 2.Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 3.Crackower M.A., Sarao R., Oudit G.Y., Yagil C., Kozieradzki I., Scanga S.E., Oliveira-dos-Santos A.J., da Costa J., Zhang L., Pei Y., Scholey J., Ferrario C.M., Manoukian A.S., Chappell M.C., Backx P.H., Yagil Y., Penninger J.M. Nature. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 4.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao X., Chakraborti S., Dimitrov A.S., Gramatikoff K., Dimitrov D.S. Biochem. Biophys. Res. Commun. 2003;312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krege J.H., John S.W., Langenbach L.L., Hodgin J.B., Hagaman J.R., Bachman E.S., Jennette J.C., O'Brien D.A., Smithies O. Nature. 1995;375:146–148. doi: 10.1038/375146a0. [DOI] [PubMed] [Google Scholar]
- 7.Esther C.R., Jr., Howard T.E., Marino E.M., Goddard J.M., Capecchi M.R., Bernstein K.E. Lab. Investig. 1996;74:953–965. [PubMed] [Google Scholar]
- 8.Vickers C., Hales P., Kaushik V., Dick L., Gavin J., Tang J., Godbout K., Parsons T., Baronas E., Hsieh F., Acton S., Patane M., Nichols A., Tummino P. J. Biol. Chem. 2002;277:14838–14843. doi: 10.1074/jbc.M200581200. [DOI] [PubMed] [Google Scholar]
- 9.Schechter I., Berger A. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 10.Turner A.J., Hooper N.M. Trends Pharmacol. Sci. 2002;23:177–183. doi: 10.1016/s0165-6147(00)01994-5. [DOI] [PubMed] [Google Scholar]
- 11.Danilczyk U., Eriksson U., Crackower M.A., Penninger J.M. J. Mol. Med. 2003;81:227–234. doi: 10.1007/s00109-003-0419-x. [DOI] [PubMed] [Google Scholar]
- 12.Oudit G.Y., Crackower M.A., Backx P.H., Penninger J.M. Trends Cardiovasc. Med. 2003;13:93–101. doi: 10.1016/s1050-1738(02)00233-5. [DOI] [PubMed] [Google Scholar]
- 13.Natesh R., Schwager S.L., Sturrock E.D., Acharya K.R. Nature. 2003;421:551–554. doi: 10.1038/nature01370. [DOI] [PubMed] [Google Scholar]
- 14.Kim H.M., Shin D.R., Yoo O.J., Lee H., Lee J.O. FEBS Lett. 2003;538:65–70. doi: 10.1016/s0014-5793(03)00128-5. [DOI] [PubMed] [Google Scholar]
- 15.Dales N.A., Gould A.E., Brown J.A., Calderwood E.F., Guan B., Minor C.A., Gavin J.M., Hales P., Kaushik V.K., Stewart M., Tummino P.J., Vickers C.S., Ocain T.D., Patane M.A. J. Am. Chem. Soc. 2002;124:11852–11853. doi: 10.1021/ja0277226. [DOI] [PubMed] [Google Scholar]
- 16.Hauptman H.A. Methods Enzymol. 1997;277:3–13. doi: 10.1016/s0076-6879(97)77003-4. [DOI] [PubMed] [Google Scholar]
- 17.Abrahams J.P., de Graaff R.A. Curr. Opin. Struct. Biol. 1998;8:601–605. doi: 10.1016/s0959-440x(98)80151-6. [DOI] [PubMed] [Google Scholar]
- 18.Bricogne G., Vonrhein C., Flensburg C., Schiltz M., Paciorek W. Acta Crystallogr. Sect. D Biol. Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski Z. In: Isomorphous Replacement and Anomalous Scattering: Proceedings of the CCP4 Study Weekend. Wolf W., Evans P.R., Leslie A.G.W., editors. Daresbury Laboratory; Warrington, United Kingdom: 1991. pp. 80–86. [Google Scholar]
- 20.McRee D.E. 2nd Ed. Academic Press, Inc.; San Diego, CA: 1999. Practical Protein Crystallography. [Google Scholar]
- 21.Navaza J., Saludjian P. Methods Enzymol. 1997;276:581–594. doi: 10.1016/S0076-6879(97)76079-8. [DOI] [PubMed] [Google Scholar]
- 22.Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., Read R.J., Rice L.M., Simonson T., Warren G.L. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H., Wada J., Hida K., Tsuchiyama Y., Hiragushi K., Shikata K., Wang H., Lin S., Kanwar Y.S., Makino H. J. Biol. Chem. 2001;276:17132–17139. doi: 10.1074/jbc.M006723200. [DOI] [PubMed] [Google Scholar]
- 24.Fulop V., Bocskei Z., Polgar L. Cell. 1998;94:161–170. doi: 10.1016/s0092-8674(00)81416-6. [DOI] [PubMed] [Google Scholar]
- 25.Rockel B., Peters J., Kuhlmorgen B., Glaeser R.M., Baumeister W. EMBO J. 2002;21:5979–5984. doi: 10.1093/emboj/cdf601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pantoliano M.W., Holmquist B., Riordan J.F. Biochemistry. 1984;23:1037–1042. doi: 10.1021/bi00300a036. [DOI] [PubMed] [Google Scholar]
- 27.Bernstein K.E., Welsh S.L., Inman J.K. Biochem. Biophys. Res. Commun. 1990;167:310–316. doi: 10.1016/0006-291x(90)91766-l. [DOI] [PubMed] [Google Scholar]
- 28.Rawlings N.D., Barrett A.J. Methods Enzymol. 1995;248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- 29.Holland D.R., Tronrud D.E., Pley H.W., Flaherty K.M., Stark W., Jansonius J.N., McKay D.B., Matthews B.W. Biochemistry. 1992;31:11310–11316. doi: 10.1021/bi00161a008. [DOI] [PubMed] [Google Scholar]
- 30.Grams F., Dive V., Yiotakis A., Yiallouros I., Vassiliou S., Zwilling R., Bode W., Stocker W. Nat. Struct. Biol. 1996;3:671–675. doi: 10.1038/nsb0896-671. [DOI] [PubMed] [Google Scholar]
- 31.Gerstein M., Lesk A.M., Chothia C. Biochemistry. 1994;33:6739–6749. doi: 10.1021/bi00188a001. [DOI] [PubMed] [Google Scholar]
- 32.Gerstein M., Krebs W. Nucleic Acids Res. 1998;26:4280–4290. doi: 10.1093/nar/26.18.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teague S.J. Nat. Rev. Drug Discov. 2003;2:527–541. doi: 10.1038/nrd1129. [DOI] [PubMed] [Google Scholar]
- 34.Knowles J.R. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1991;332:115–121. doi: 10.1098/rstb.1991.0039. [DOI] [PubMed] [Google Scholar]
- 35.Matthews B.W. Acc. Chem. Res. 1988;21:333–340. [Google Scholar]
- 36.Patchett A.A., Harris E., Tristram E.W., Wyvratt M.J., Wu M.T., Taub D., Peterson E.R., Ikeler T.J., ten Broeke J., Payne L.G., Ondeyka D.L., Thorsett E.D., Greenlee W.J., Lohr N.S., Hoffsommer R.D., Joshua H., Ruyle W.V., Rothrock J.W., Aster S.D., Maycock A.L., Robinson F.M., Hirschmann R., Sweet C.S., Ulm E.H., Gross D.M., Vassil T.C., Stone C.A. Nature. 1980;288:280–283. doi: 10.1038/288280a0. [DOI] [PubMed] [Google Scholar]
- 37.Guy J.L., Jackson R.M., Acharya K.R., Sturrock E.D., Hooper N.M., Turner A.J. Biochemistry. 2003;42:13185–13192. doi: 10.1021/bi035268s. [DOI] [PubMed] [Google Scholar]
- 38.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown C.K., Madauss K., Lian W., Beck M.R., Tolbert W.D., Rodgers D.W. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3127–3132. doi: 10.1073/pnas.051633198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arndt J.W., Hao B., Ramakrishnan V., Cheng T., Chan S.I., Chan M.K. Structure. 2002;10:215–224. doi: 10.1016/s0969-2126(02)00698-6. [DOI] [PubMed] [Google Scholar]
- 41.Gordon K., Redelinghuys P., Schwager S.L., Ehlers M.R., Papageorgiou A.C., Natesh R., Acharya K.R., Sturrock E.D. Biochem. J. 2003;371:437–442. doi: 10.1042/BJ20021842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holden H.M., Tronrud D.E., Monzingo A.F., Weaver L.H., Matthews B.W. Biochemistry. 1987;26:8542–8553. doi: 10.1021/bi00400a008. [DOI] [PubMed] [Google Scholar]
- 43.Kraut J. Science. 1988;242:533–540. doi: 10.1126/science.3051385. [DOI] [PubMed] [Google Scholar]
- 44.Fernandez M., Liu X., Wouters M.A., Heyberger S., Husain A. J. Biol. Chem. 2001;276:4998–5004. doi: 10.1074/jbc.M009009200. [DOI] [PubMed] [Google Scholar]
- 45.Shapiro R., Holmquist B., Riordan J.F. Biochemistry. 1983;22:3850–3857. doi: 10.1021/bi00285a021. [DOI] [PubMed] [Google Scholar]
- 46.Shapiro R., Riordan J.F. Biochemistry. 1984;23:5234–5240. doi: 10.1021/bi00317a022. [DOI] [PubMed] [Google Scholar]
- 47.Liu X., Fernandez M., Wouters M.A., Heyberger S., Husain A. J. Biol. Chem. 2001;276:33518–33525. doi: 10.1074/jbc.M101495200. [DOI] [PubMed] [Google Scholar]
- 48.Soubrier F., Alhenc-Gelas F., Hubert C., Allegrini J., John M., Tregear G., Corvol P. Proc. Natl. Acad. Sci. U. S. A. 1988;85:9386–9390. doi: 10.1073/pnas.85.24.9386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ehlers M.R., Fox E.A., Strydom D.J., Riordan J.F. Proc. Natl. Acad. Sci. U. S. A. 1989;86:7741–7745. doi: 10.1073/pnas.86.20.7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Higgins D.G., Thompson J.D., Gibson T.J. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 51.Frishman D., Argos P. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]