Abstract
Pandemics are caused by novel pathogens to which pre-existing antibody immunity is lacking. Under these circumstances, the body must rely on innate interferon-mediated defenses to limit pathogen replication and allow development of critical humoral protection. Here, we highlight studies on disease susceptibility during H1N1 influenza and COVID-19 (SARS-CoV-2) pandemics. An emerging concept is that genetic and non-genetic deficiencies in interferon system components lead to uncontrolled virus replication and severe illness in a subset of people. Intriguingly, new findings suggest that individuals with autoantibodies neutralizing the antiviral function of interferon are at increased risk of severe COVID-19. We discuss key questions surrounding how such autoantibodies develop and function, as well as the general implications of diagnosing interferon deficiencies for personalized therapies.
Keywords: pandemic virus, influenza, COVID-19, disease severity, interferon, host genetics, autoantibodies
The human interferon system is a critical protective barrier against pathogens
To protect against invading pathogens, the human body has evolved a multitiered set of physical, innate, and adaptive immune barriers that act synergistically to limit infection and reduce disease burden. Critical among these are the innate interferon type I (mainly α, β, or ω) and III (λ) systems (Box 1 ), which constitute a nonspecific cytokine-mediated response to infection that invokes immediate and broad-spectrum intracellular defenses to suppress pathogens such as viruses (reviewed in [1]). These systems generally protect infected individuals from uncontrolled pathogen replication, thereby limiting disease progression and ‘buying time’ for the body to develop adaptive immune responses (see Glossary), which both resolve the infection and create a potent immunological memory against future incursions with the same pathogen [1].
Box 1. The human interferon system and viral disease susceptibility.
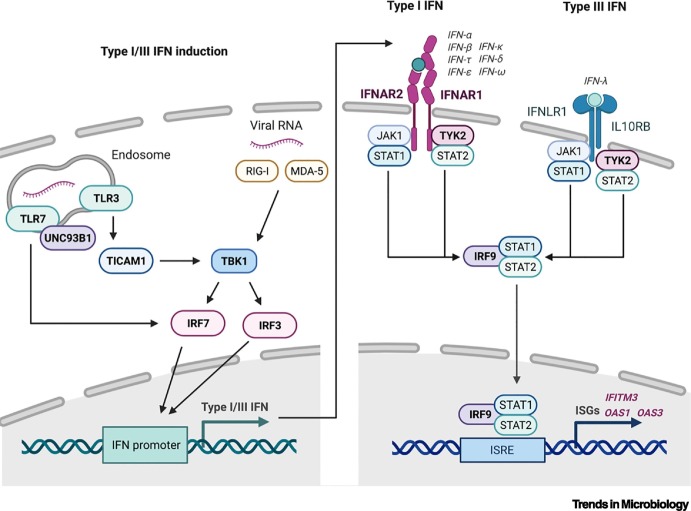
The interferon (IFN) system is set in motion when cells detect virus infections by sensing pathogen-associated molecular patterns (PAMPs), such as viral RNA, with host-encoded pattern-recognition receptors (PRRs). For RNA viruses, such as influenza virus and SARS-CoV-2, viral RNA is sensed in the cytoplasm by PRRs, including the RIG-I-like receptors (RLRs), MDA-5 and RIG-I, or in the endosome by the Toll-like receptors (TLRs) TLR3 and TLR7. Upon sensing of PAMPs, each PRR initiates a unique signaling cascade that leads to activation of the kinase, TBK1, and subsequent mobilization of transcription factors IRF3 and IRF7, which induce type I and type III interferon gene expression. Notably, TLR3 signals via an adapter protein, TICAM1 (also known as TRIF), and TLR7 signaling requires the trafficking chaperone, UNC93B1. Interferon cytokines are secreted from cells and act in a paracrine and autocrine manner to alert surrounding cells to viral infection by binding to their cognate receptors and triggering a signaling cascade leading to the transcription of several hundred antiviral genes (interferon-stimulated genes, ISGs). In the case of type I interferons (mainly α, β, or ω), the receptor consists of two subunits encoded by the IFNAR1 and IFNAR2 genes, whereas the IFNLR1 and the IL10RB genes encode the type III interferon (λ) receptor subunits. Both receptors transduce the signal via the kinases JAK1 and TYK2 to activate the transcription factor complex of STAT1, STAT2, and IRF9. This transcription factor complex binds to interferon-stimulated response elements (ISREs) in the promoters of a large set of antiviral ISGs, stimulating their expression and leading to a generalized antiviral state in cells that protects against virus infection. Prominent examples of relevant ISGs include IFITM3, OAS1, and OAS3. These pathways are summarized in Figure I, where genes with rare loss-of-function variants that have been reported to exacerbate severe influenza or COVID-19 disease are highlighted in bold.
Figure I.
Overview of the type I and type III interferon (IFN) responses.
Simplified signaling cascades leading to transcription of type I/III interferon genes following infection with an RNA virus (left panel). Simplified signaling cascades leading to transcription of interferon-stimulated genes (ISGs) following type I/III interferon stimulation (right panel). RIG-I, MDA-5, TLR3, TLR7, UNC93B1, TICAM1, TBK1, IRF3, IRF7, IFNAR1, IFNAR2, IFNLR1, IL10RB, JAK1, TYK2, STAT1, STAT2, and IRF9 are human interferon system components involved in recognizing virus infections, stimulating interferon production, or mediating antiviral signaling; ISRE, interferon-stimulated response element.
Alt-text: Box 1
Here, we discuss selected key research findings on disease severity during the 2009 H1N1 influenza virus pandemic and the ongoing SARS-CoV-2 (COVID-19) pandemic. These studies reveal a critical contribution of genetic and non-genetic interferon system deficiencies to pandemic virus susceptibility in a subset of individuals. This concept is paving the way to a better understanding of fundamental mechanisms that protect against viral disease, and provides exciting opportunities in the field of personalized medicine to define and stratify at-risk individuals for tailored therapies.
Human genetics reveals the importance of interferon to viral disease protection
The critical nature of the human interferon system in protecting against severe infections is evidenced by groundbreaking studies on individuals with inborn errors of immunity (reviewed in [2]). Such individuals have rare genetic lesions that compromise the function of key factors in the interferon pathway and lead to an increased susceptibility to infection. Commonly, but not always, these inborn errors of immunity clinically manifest themselves in infancy, sometimes leading to death of the individual following exposure to a new, but otherwise common, pathogen [2]. For those affected who have survived into adulthood, it can be speculated that their first early exposures to these common pathogens were at relatively low doses, that exposure occurred while they still possessed some pathogen-specific passive immunity derived from maternal antibodies, or that redundant aspects of innate or intrinsic immunity temper infection. Such mechanisms could act to delay infection, thereby ‘compensating’ for the interferon deficiency and ‘buying time’ for an acquired immune response to develop. It might therefore be hypothesized that once an individual has generated a protective antibody response to a specific pathogen, the absolute importance of a fully functional interferon system as a first-line barrier to that pathogen is diminished.
Are loss-of-function variants in interferon-system genes associated with severe pandemic virus disease?
Acute pandemic viral infections are caused by antigenically novel pathogens to which widespread pre-existing antibody responses cannot protect either the individual or the population. Under these circumstances, like exposure to new common pathogens in infancy, the body’s ability to control initial virus replication using the interferon system is critical. Thus, factors that compromise functionality of interferon system components are now recognized as determinants of individual susceptibility to severe pandemic viral disease, even in adults who have not previously exhibited overt disease with common virus infections. The advent of two recent viral pandemics (the 2009 H1N1 influenza virus pandemic and the ongoing SARS-CoV-2 pandemic) in the age of advanced human genome sequencing technologies has led to a critical appreciation of both the prevalence of genetic deficiencies in contributing to severe disease as well as the mechanistic bases for such susceptibilities. Specifically, rare loss-of-function variants in human interferon system genes involved in recognizing virus infections and stimulating interferon production or antiviral signaling (TLR7, TLR3, UNC93B1, TICAM1, TBK1, IRF3, IRF7, IFNAR1, IFNAR2, and TYK2) have been associated with the development of severe COVID-19 in individual patients [3., 4., 5.] and may account for as many as 3.5% of severe life-threatening pandemic virus infections [3] (Box 1). Rare inherited deficiencies in some of these genes (TLR3, IRF7), or related ones (IRF9), have also been reported to lead to severe influenza [6., 7., 8.] (Box 1). Furthermore, increased susceptibility to pandemic viral disease has been linked to genetic variation in individual antiviral effectors (known as interferon-stimulated genes, ISGs), with IFITM3 defects contributing to life-threatening 2009 H1N1 [9., 10., 11.] (Box 2 ), and new data emerging on OAS1 and OAS3 variants in COVID-19 [5]. Until now, most reported genetic links to viral susceptibility involving the interferon system have focused on genes previously identified experimentally to play functional roles. However, unbiased genetic analyses will no doubt uncover new human factors in this system, thereby increasing knowledge on basic mechanisms of interferon-mediated protection.
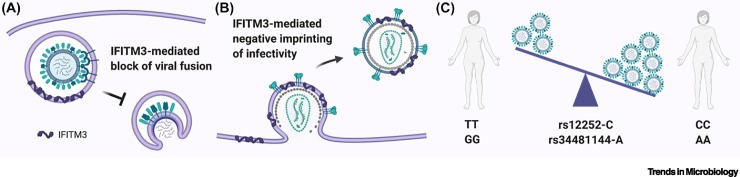
Box 2. IFITM3 single-nucleotide polymorphisms and viral disease susceptibility.
IFITM3 (interferon-induced transmembrane protein 3) is a potent antiviral protein induced by interferons that localizes to endosomal compartments [48]. Cell-based assays have revealed that IFITM3 can increase membrane rigidity, thereby impairing virus–host membrane fusion and restricting cell entry of enveloped viruses that enter via the endosome, such as influenza A, SARS-CoV-1, SARS-CoV-2, and Ebola virus [49., 50., 51., 52.] (Figure IA). Moreover, a ‘negative imprinting’ of virus infectivity function has been reported for IFITM3: HIV-1, measles, and Ebola virus particles budding from IFITM3-expressing cells appear to be less infectious [53., 54., 55.], possibly due to their increased membrane rigidity (Figure IB). Given its potent antiviral activity, and its general mode of action as an early-stage broad-spectrum inhibitor of enveloped virus infection, IFITM3 is recognized as a critical first barrier against zoonotic and pandemic viruses. This was underlined by the striking finding that some individuals carry a single-nucleotide polymorphism (SNP) in the IFITM3 locus (rs12252-C), which appears to create a novel splice acceptor site resulting in the production of a truncated, and possibly unstable/inactive, IFITM3 protein [9]. Individuals who are homozygous for this rs12252-C SNP make up only ~0.3% of European Caucasians, but a landmark study from the UK found that ~5.7% of patients hospitalized with severe pandemic H1N1 influenza in 2009 were homozygous for the deleterious IFITM3 SNP, suggesting that loss of IFITM3-mediated viral control exacerbated the disease caused by this antigenically novel pathogen [9] (Figure IC). Presumably, individuals homozygous for rs12252-C did not previously suffer from severe ‘seasonal’ influenza due to some acquired humoral immunity to these viruses. Notably, IFITM3 rs12252 allele frequencies differ markedly across populations, with homozygosity of rs12252-C being reported in around 25% of Chinese, and 44% of Japanese, individuals [11]. While there is still some debate about how precisely the IFITM3 rs12252-C SNP results in functional deficiency in IFITM3, the impact of this SNP on viral disease outcome has now been confirmed in some, but not all, studies [11,56,57]. Moreover, other IFITM3 alleles (e.g., rs34481144-A) have begun to be associated with severe influenza morbidity and mortality [10], as well as COVID-19 severity [58,59] (Figure IC). It will be interesting to assess whether differences in IFITM3 allele frequencies between human populations play a role in the distinct preliminary mortality rates observed for COVID-19 across the world [60].
Figure I.
Schematics of IFITM3 antiviral activity and functional polymorphisms.
(A) IFITM3 at the endosomal membrane blocks entry of viruses, such as influenza A virus. (B) IFITM3 at the plasma membrane limits subsequent infectivity of budding viruses, such as HIV-1. (C) The indicated single-nucleotide polymorphisms (SNPs) in human IFITM3 impact its antiviral function and influence control of virus replication. IFITM3, interferon-induced transmembrane protein 3.
Alt-text: Box 2
Autoantibodies can neutralize the human interferon system
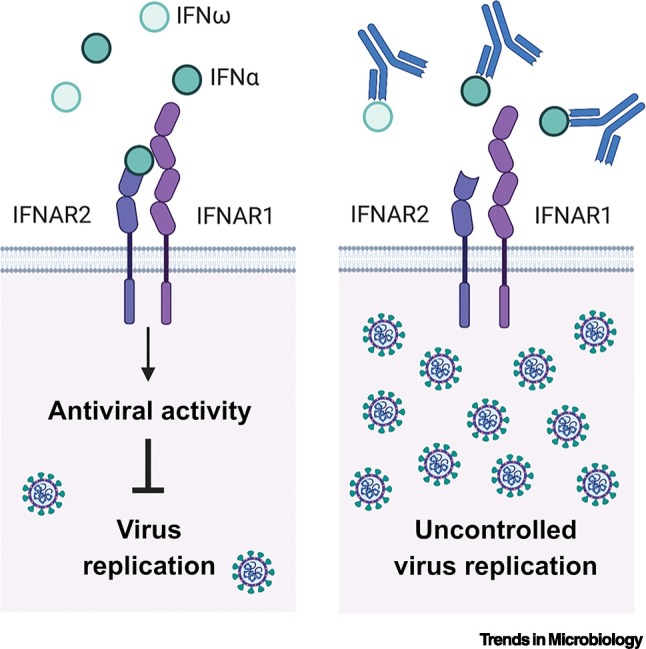
A conceptual leap was recently made with the hypothesis that non-genetic deficiencies of the human interferon system might also predispose individuals to severe viral disease, and that autoantibodies targeting and neutralizing the activity of interferons may provide such a mechanism [12] (Figure 1 ). Neutralizing anti-interferon autoantibodies have previously been described in some patients treated with type I interferons for cancers [13,14], or viral infections [15]. However, naturally occurring anti-interferon autoantibodies are not very prevalent in the general human population [16] as autoreactive induction against self-antigens should be prevented by immune-tolerance mechanisms. Thus, naturally occurring anti-interferon autoantibodies have an estimated prevalence of only 0.33% [12]. As such, they are detected sporadically [17], although their levels are high in patients with certain autoimmune diseases, such as systemic lupus erythematosus (SLE) [18] or autoimmune polyendocrinopathy syndrome type I (APS1) [19], as well as with immune disorders caused by partial RAG deficiency [20,21]. Nevertheless, it is not entirely clear whether the presence of neutralizing anti-interferon antibodies in these patients directly leads to an increased susceptibility to viral infections. For example, despite increased susceptibility to fungal infections due to neutralizing autoantibodies against several other cytokines [22], recurrent viral infections in APS1 patients are apparently rare, even though they possess relatively high serum neutralizing titers of anti-interferon-α and anti-interferon-ω antibodies [19]. This may be due to a number of reasons, including: (i) the compensatory action of other antiviral interferons that appear to be less-targeted by anti-interferon autoantibodies (e.g., interferon-β or interferon-λ); (ii) the potential low abundance of anti-interferon autoantibodies at specific sites of virus infection (e.g., mucosa); (iii) the titer of anti-interferon autoantibodies being insufficient to neutralize the substantial amounts of locally produced antiviral interferon; and (iv) the protection conferred by pre-existing humoral immunity derived from either prior infections or immunizations. Thus, in this latter regard, it might be expected that under conditions where humoral immunity is compromised (e.g., partial RAG deficiencies or in response to a new pathogen), the presence of neutralizing anti-interferon antibodies could increase susceptibility to uncontrolled infection and disease severity. Indeed, several patients with hypomorphic RAG mutations have a history of severe virus infections [20].
Figure 1.
Do neutralizing autoantibodies against type I interferons lead to uncontrolled virus replication?
Simplified representation of type I interferon (IFN)-mediated inhibition of virus replication triggered by IFNα or IFNω binding to their cognate heterodimeric receptor (IFNAR1/IFNAR2) (left panel). Autoantibodies that bind IFNα or IFNω and prevent their interaction with IFNAR1/IFNAR2 are termed ‘neutralizing’ and could limit the antiviral action of type I interferons leading to uncontrolled virus replication (right panel).
Do anti-interferon autoantibodies lead to increased COVID-19 severity?
Building on the observation that three young APS1 patients developed severe pandemic COVID-19, Bastard et al. surveyed a cohort of nearly 1000 hospitalized COVID-19 patients without overt autoimmune disorders for the presence of anti-interferon autoantibodies in their serum [12]. Remarkably, 13.7% of these critically ill COVID-19 patients, including patients as young as 25 years, had autoantibodies against either interferon-α, interferon-ω, or both, and for 75% of these patients (10.2% of all critically ill COVID-19 patients) the autoantibodies were able to neutralize the antiviral function of interferon in vitro. Wang et al. made similar findings, identifying anti-interferon autoantibodies in 5.2% of severe COVID-19 cases [23], and other instances have recently been documented [24]. Notably, Wang et al. also found that SARS-CoV-2 viral loads in nasopharyngeal and saliva samples from patients with anti-interferon autoantibodies were generally much higher than matched patient controls without anti-interferon autoantibodies [23]. This observation, together with evidence that COVID-19 patients with anti-interferon autoantibodies had lower levels of serum interferon-α protein (and correspondingly low antiviral gene signatures) [12], is highly suggestive of a functional interferon deficiency caused by these neutralizing autoantibodies leading to uncontrolled SARS-CoV-2 replication. However, it is not yet clear whether COVID-19 severity in patients with anti-interferon autoantibodies is directly related to exacerbated virus replication per se, or is the result of disrupting other immune mechanisms [25]. Furthermore, a very recent report also suggests that interferon deficiency in some severe COVID-19 patients can be caused by the Fc portion of high-titer anti-SARS-CoV-2 antibodies triggering interferon inhibitory signaling by binding to immune cells [24], although whether this is causative of disease is unclear. It is also currently unknown whether the presence of anti-interferon autoantibodies, or other antibody-mediated mechanisms blocking interferon signaling, played any role in determining the severity of infection during the 2009 H1N1 influenza pandemic, or whether this is a COVID-19-specific phenotype.
Do anti-interferon autoantibodies pre-exist in virus-susceptible individuals?
It is apparent from multiple studies that some COVID-19 patients have autoantibodies against a diverse range of self-antigens beyond type I interferons, and that these autoantibodies can have both short- and long-term pathogenic consequences [12,23,26., 27., 28., 29.]. Thus, there is already some debate as to whether individuals with pre-existing subclinical autoimmune conditions are more likely to develop severe COVID-19, or whether severe SARS-CoV-2 infections trigger de novo autoreactivity against various antigens [12,30]. Longitudinal analyses of autoantibody reactivity in COVID-19 patient cohorts have indicated that both mechanisms may actually apply, but perhaps in an antigen-dependent manner [23,29]. Specifically, autoantibodies targeting type I interferons were found to pre-exist in several individuals who went on to develop severe COVID-19 [12,23], suggesting that the SARS-CoV-2 infection itself did not drive the primary production of anti-interferon autoantibodies. However, while the predominant isotype of anti-interferon autoantibody found in the sera of severe COVID-19 patients was IgG, low levels of IgM, IgE, and IgA anti-interferon autoantibodies have also been detected [12], with the presence of IgM anti-interferon autoantibodies a potential biomarker for an initial immune response. Notably, only IgG anti-interferon autoantibodies have so far been detected in APS1 patients [12,19], suggesting a potential immunotypic difference between acute COVID-19 and APS1 patient cohorts. Thus, it cannot be ruled out that there is heterogeneity with regard to anti-interferon autoantibody production, with SARS-CoV-2 infections possibly driving de novo breaking of interferon immune-tolerance in some individuals [30], anti-interferon autoantibodies pre-existing in other individuals [12,23], and some SARS-CoV-2 infections stimulating an increase in the titers and/or affinity of pre-existing anti-interferon autoantibodies [29].
How and why do certain individuals generate anti-interferon autoantibodies?
A striking finding by Bastard et al. was the higher-than-expected proportion of men and those over 65 years in severe COVID-19 cohorts with neutralizing antibodies against interferons [12]. This suggests that gender and age could be factors in the development of such autoantibodies, although this is not absolute as both females and individuals under 40 years were identified with anti-interferon antibodies [12], and gender and age may be confounding variables if they independently pre-dispose to severe COVID-19 [31,32]. Most individuals with anti-interferon autoantibodies did not have overt immunodeficiencies or autoimmune disorders [12], thus further studies are necessary to determine whether there is an unknown host genetic component to anti-interferon autoantibody production in these individuals, similar to the autoimmune condition APS1 that is linked to a genetic defect in AIRE [33]. It is possible that a genetic variant localizing to an immune-regulating gene on the X chromosome could account for the apparent increased likelihood of men developing autoantibodies [25]. Nevertheless, host genetics alone may not fully explain the induction of anti-interferon antibodies. Indeed, there have been reports of frequent detection of anti-interferon antibodies in acute hepatitis A or B virus-infected patients [34], or in patients with other acute viral infections [35], and enhanced levels of neutralizing anti-interferon autoantibodies have been detected in some anti-tick-borne encephalitis virus and anti-hepatitis B virus hyperimmune human IgG preparations [36]. Thus, an infectious (or at least interferon-stimulating) component to anti-interferon autoantibody production may be critical but would probably function in concert with host genetic determinants, immunosenescence, or environmental stimuli to elicit both the required antigen (interferon) as well as the breakdown of immune-tolerance necessary for autoantibody generation. In support of this ‘double-hit’ concept, a previous study concluded that a history of recurrent severe virus infections alone is not associated with generation of anti-interferon autoantibodies in humans unless immune tolerance is lost through mechanisms such as partial RAG deficiency [20]. Furthermore, studies in hypomorphic Rag mutant mice revealed enhanced autoantibody production following stimulation of the interferon-induction pathway using viral-mimetic agonists of Toll-like receptors (TLRs) and RIG-I-like receptors (RLRs) [20]. Of course, it cannot be ruled out that specific infectious agents provide both the necessary antigenic stimulus (interferon) and the ability to break immune-tolerance through currently unappreciated mechanisms: for example, (i) via unregulated activation of TLR7 [30,37]; (ii) hypothetical infection-induced changes to the structure or antigenicity of endogenous interferons to make them appear ‘non-self’; or (iii) hypothetical molecular mimicry of an agent to interferon, similar to some viruses and other host factors [38]. In addition, there are other, non-infectious, causes of chronic type I interferon upregulation that could predispose to anti-interferon autoantibody production (e.g., diabetes [39], interferonopathies [40]), and should be investigated further. However, given that such mechanisms are currently highly speculative without additional data, it may also be prudent to try and understand parallels between induction of anti-interferon autoantibodies in severe COVID-19 patients and induction in APS1 patients, who have mutations in AIRE and who likely generate anti-cytokine autoantibodies independently from exogenous infections but driven by a loss of central (T cell) immune-tolerance [33].
Why are autoantibodies targeting interferon-β and interferon-λ apparently rare?
Bastard et al. found that patients with autoantibodies against interferon-α had antibodies against all 13 interferon-α subtypes, but autoantibodies against other type I interferons, including interferon-β, were much rarer and phenotypically different: for example, only 1.9% of patients with severe COVID-19 had autoantibodies against interferon-β, and only two of these sera were neutralizing [12]. Similar findings have been observed in APS1 patients, where autoantibodies against interferon-α subtypes and interferon-ω are common, but antibodies against other type I interferons are rare or at low titers [19,33]. To some extent this makes sense: interferon-α proteins are more closely related to interferon-ω than to other type I interferons [12], although it is still unknown whether the autoantibodies in COVID-19 patients are cross-reactive between these two type I interferons or whether different autoantibodies specifically recognize each interferon differently (we note that both cross-reactive and subtype-specific high-affinity anti-interferon autoantibodies have been isolated from APS1 patients [33]). Nevertheless, it does beg the question as to why loss of immune-tolerance is directed against these interferons and not others, particularly if an infectious trigger is involved which might be expected to upregulate multiple interferons. Thus, cell-type-dependent mechanisms that respond to a specific trigger and ‘favor’ the production of interferon-α proteins over interferon-β and interferon-λ may contribute to this phenomenon [41], or interferon-β and interferon-λ may simply be less ‘auto-immunogenic’, although autoantibodies against interferon-λ have been detected in APS1 patients [33], and rarely in some COVID-19 patients [23]. It is also conceivable that autoantibodies against these interferons are actually present in more individuals but that such antibodies have yet to be fully recognized because the experimental antigens used to detect them somehow fail to represent authentic interferons, or the autoantibodies are abundant at yet-unsampled sites of the body, such as mucosa. Clearly, further studies are necessary to confirm the rarity of autoantibodies targeting other interferons (such as interferon-β and interferon-λ) and to understand the mechanisms underpinning this rarity.
Why is there no protective role for interferons that are not targeted by autoantibodies?
If there were indeed a complete absence of autoantibodies against interferon-β and interferon-λ in some severe COVID-19 patients (and anti-interferon-λ autoantibodies at least seem rare according to the study by Wang et al. [23]), it is puzzling how neutralizing interferon-α and interferon-ω alone is sufficient to compromise local interferon-mediated control of SARS-CoV-2, given its sensitivity to these other interferons in human airway epithelial cells and the important protective antiviral role of interferon-λ in the respiratory tract [42,43]. One possibility is that autoantibody-mediated neutralization of the very low constitutive amounts of interferon-α proteins normally found in individuals leads to disruption of the tonic priming role that they have in maintaining expression of interferon-signaling components [44], with the consequence that signaling by other antiviral interferons (including interferon-β and interferon-λ that use the same pathways) is compromised. This will be an important mechanism to understand and dissect in the future.
Concluding remarks and future perspectives
While usually considered rare, current estimates suggest that genetic and non-genetic causes of interferon system deficiencies together may contribute to almost 15% of life-threatening COVID-19 cases [3,12]. While interferon system genetics also impacted individual susceptibility during the 2009 H1N1 influenza pandemic [9], the role of anti-interferon autoantibodies in severe pandemic influenza, or indeed most other viral diseases, is unknown. However, opportunities may exist to assess this retrospectively if biobanked serum samples from appropriately documented 2009 pandemic influenza patient cohorts are available. This would determine whether functional interferon deficiencies mediated by autoantibodies are a general feature of disease severity with antigenically novel viruses. Indeed, very recent data have implicated anti-interferon autoantibodies in enhancing adverse disease reactions to an antigenically novel live-attenuated yellow fever virus vaccine [45]. The same question applies to the intriguing report of functional interferon deficiency (and possibly consequent severe disease) caused by the Fc portion of high-titer anti-SARS-CoV-2 antibodies triggering inhibitory signaling in immune cells [24], which breaks the generally perceived dogma that high titers of neutralizing anti-pathogen antibodies against a pandemic virus are a marker of protection. Furthermore, it could be worthwhile to explore the contributions of such genetic and non-genetic interferon system deficiencies to exacerbated disease in the context of future potential endemic SARS-CoV-2 or influenza virus infections, particularly if humoral immunity to these viruses wanes with time. This, and several other important research questions therefore remain to be addressed (Figure 2 , Key Figure; see Outstanding questions). Regardless, the continued identification of new interferon-system-related genetic traits, or anti-interferon antibody profiles, that predispose individuals to severe virus infections could form the basis of new personalized medicine strategies to prioritize at-risk individuals for preventative measures (such as vaccination) or appropriate treatments. For example, plasma exchange has already been trialled to remove anti-interferon autoantibodies from the blood of patients with life-threatening COVID-19 [46], and recombinant interferon-β or interferon-λ could provide a life-saving boost to innate antiviral defenses in individuals with autoantibodies that neutralize interferon-α and interferon-ω [12], mirroring a successful interferon-based strategy to treat severe COVID-19 patients with genetic deficiencies in interferon production [47]. However, the long-term consequences of such cytokine treatments will have to be considered carefully given that these individuals may be prone to raising autoantibodies that neutralize interferon-β or interferon-λ, thus leaving them more vulnerable to future infections. Therefore, as the underlying bases of rare individual, or population-specific, severe phenotypes during pandemic viral infections are identified, an emphasis should be placed on ways to use this information both to understand mechanisms of action and to develop novel, tailored therapeutic strategies that might circumvent such issues.
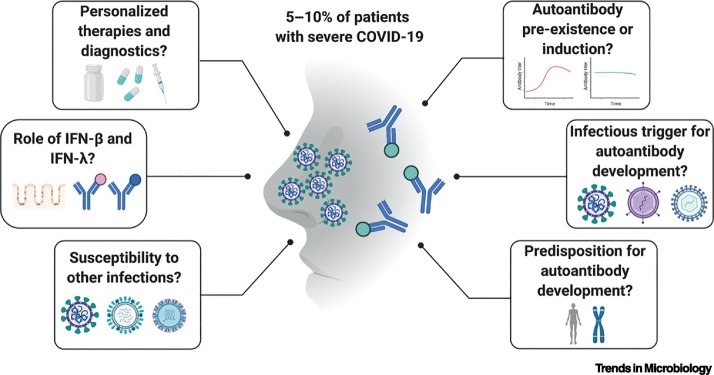
Figure 2.
Key Figure. The high proportion of severe COVID-19 patients with autoantibodies against type I interferons raises important questions.
Autoantibodies against type I interferons have been found in 5–10% of individuals who suffer from severe COVID-19. There are several important questions that still need to be addressed regarding how these autoantibodies are triggered (right side), and what these autoantibodies mean for virus susceptibility and future personalized therapies (left side).
Outstanding questions.
How can prospective targeted genetic testing be realistically deployed to identify and prioritize individuals at risk of developing severe viral disease?
How can tailored therapies be developed to treat individuals with life-threatening infections due to genetic interferon system deficiencies, particularly those with lesions in interferon-signaling genes or interferon-stimulated antiviral effectors?
Will unbiased studies on the human genetics of severe pandemic viral disease identify new factors and mechanisms involved in innate immune protection?
What stimulates the breaking of immune-tolerance and the development of anti-interferon autoantibodies? Is there a specific infectious trigger and a role for X-linked host genetics or immunosenescence?
Why does autoantibody development seem focused on interferon-α and interferon-ω, rather than on interferon-β and interferon-λ? Have prevalent autoantibodies against interferon-β and interferon-λ simply not been detected yet?
Do anti-interferon autoantibodies play a role in the pathogenesis of pandemic viral diseases other than COVID-19? If they are triggered de novo by SARS-CoV-2, how long does such a response last, and what impact will these autoantibodies have on the long-term infection susceptibility of those who have recovered from COVID-19?
Can testing for anti-interferon autoantibodies provide rapid diagnostic value for understanding the cause of a severe pandemic viral infection and help to stratify patients for treatment?
What is the molecular mechanism by which anti-interferon-α and anti-interferon-ω autoantibodies exacerbate viral disease? Is it by neutralizing virus-induced interferons or by disrupting tonic interferon signaling? Why are interferon-β and interferon-λ not able to compensate functionally for this deficiency?
What treatment options are there for individuals with anti-interferon autoantibodies other than providing exogenous interferons to which further auto-reactive antibodies could develop?
Alt-text: Outstanding questions
Acknowledgments
Acknowledgments
Figures were created with BioRender.com. The authors’ laboratories are funded by the Swiss National Science Foundation through grants 31003A_182464 to B.G.H. and 31003A_176170 to S.S.
Declaration of interests
There are no interests to declare.
Glossary
- Adaptive immune responses
refers to pathogen-specific acquired immunity, for example, antibodies.
- AIRE
autoimmune regulatory gene, the product of which acts as a transcription factor regulating T cell tolerance.
- Antigenically novel
refers to a new type of agent that is not recognized by existing antibodies.
- Autoantibodies
antibodies raised against one of the body’s own components.
- COVID-19
coronavirus disease 2019.
- Endemic
constantly maintained at a certain level in humans.
- Fc
the fragment crystallizable (‘tail’) region of an antibody that interacts with specific cell-surface receptors.
- Humoral immunity
protection conferred by substances found in bodily fluids.
- Hyperimmune human IgG
immunoglobulins from a donor with high antibody titers against a specific antigen.
- Hypomorphic mutation
a type of genetic mutation leading to reduced activity or expression of the corresponding gene product.
- Immune tolerance
prevention of an immune response against a specific antigen.
- Immunological memory
the ability to recognize a previously encountered antigen and respond more quickly with an immune response.
- Immunosenescence
natural deterioration of immune system function with age.
- Inborn errors of immunity
germline mutations in a single immune-related gene resulting in a loss or gain of function.
- Interferonopathies
genetic disorders of the interferon system characterized by excessive production of interferon.
- Molecular mimicry
the ability of some foreign (non-self) antigens to mimic self-antigens, the immune response to the foreign antigen giving rise to autoimmune damage.
- Neutralizing antibody
an antibody that inhibits the function of the target it binds to.
- Pandemic
a world-wide epidemic of an infection affecting a large number of people.
- Passive immunity
short-lived immune protection usually consisting of antibodies from another source or individual.
- RAG deficiency
a severe immunodeficiency caused by loss-of-function mutations in human RAG1/2 (recombination activating genes).
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2.
- Self-antigens
antibody-inducing components originating in one’s own body.
References
- 1.Lazear H.M., et al. Shared and distinct functions of type I and type III interferons. Immunity. 2019;50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duncan C.J.A., et al. Genetic lesions of type I interferon signalling in human antiviral immunity. Trends Genet. 2021;37:46–58. doi: 10.1016/j.tig.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Q., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Made C.I., et al. Presence of genetic variants among young men with severe COVID-19. JAMA. 2020;324:1–11. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pairo-Castineira E., et al. Genetic mechanisms of critical illness in Covid-19. Nature. 2020;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 6.Lim H.K., et al. Severe influenza pneumonitis in children with inherited TLR3 deficiency. J. Exp. Med. 2019;216:2038–2056. doi: 10.1084/jem.20181621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernandez N., et al. Life-threatening influenza pneumonitis in a child with inherited IRF9 deficiency. J. Exp. Med. 2018;215:2567–2585. doi: 10.1084/jem.20180628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciancanelli M.J., et al. Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 2015;348:448–453. doi: 10.1126/science.aaa1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everitt A.R., et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen E.K., et al. SNP-mediated disruption of CTCF binding at the IFITM3 promoter is associated with risk of severe influenza in humans. Nat. Med. 2017;23:975–983. doi: 10.1038/nm.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y.H., et al. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat. Commun. 2013;4:1418. doi: 10.1038/ncomms2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bastard P., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vallbracht A., et al. Interferon-neutralizing antibodies in a patient treated with human fibroblast interferon. Nature. 1981;289:496–497. doi: 10.1038/289496a0. [DOI] [PubMed] [Google Scholar]
- 14.Steis R.G., et al. Resistance to recombinant interferon alfa-2a in hairy-cell leukemia associated with neutralizing anti-interferon antibodies. N. Engl. J. Med. 1988;318:1409–1413. doi: 10.1056/NEJM198806023182201. [DOI] [PubMed] [Google Scholar]
- 15.Antonelli G., et al. Neutralizing antibodies to interferon-alpha: relative frequency in patients treated with different interferon preparations. J. Infect. Dis. 1991;163:882–885. doi: 10.1093/infdis/163.4.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen M.B., et al. Serum-induced suppression of interferon (IFN) activity. Lack of evidence for the presence of specific autoantibodies to IFN-alpha in normal human sera. Clin. Exp. Immunol. 1992;88:559–562. doi: 10.1111/j.1365-2249.1992.tb06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pozzetto B., et al. Characteristics of autoantibodies to human interferon in a patient with varicella-zoster disease. J. Infect. Dis. 1984;150:707–713. doi: 10.1093/infdis/150.5.707. [DOI] [PubMed] [Google Scholar]
- 18.Panem S., et al. Antibodies to alpha-interferon in a patient with systemic lupus erythematosus. J. Immunol. 1982;129:1–3. [PubMed] [Google Scholar]
- 19.Meager A., et al. Anti-interferon autoantibodies in autoimmune polyendocrinopathy syndrome type 1. PLoS Med. 2006;3:e289. doi: 10.1371/journal.pmed.0030289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walter J.E., et al. Broad-spectrum antibodies against self-antigens and cytokines in RAG deficiency. J. Clin. Invest. 2015;125:4135–4148. doi: 10.1172/JCI80477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen K., et al. Autoimmunity due to RAG deficiency and estimated disease incidence in RAG1/2 mutations. J. Allergy Clin. Immunol. 2014;133:880–882. doi: 10.1016/j.jaci.2013.11.038. e810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kisand K., et al. Chronic mucocutaneous candidiasis in APECED or thymoma patients correlates with autoimmunity to Th17-associated cytokines. J. Exp. Med. 2010;207:299–308. doi: 10.1084/jem.20091669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang E.Y., et al. Diverse functional autoantibodies in patients with COVID-19. medRxiv. 2020 doi: 10.1101/2020.12.10.20247205. Published online December 12, 2020. [DOI] [PubMed] [Google Scholar]
- 24.Combes A.J., et al. Global absence and targeting of protective immune states in severe COVID-19. Nature. 2021;591:124–130. doi: 10.1038/s41586-021-03234-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meffre E., Iwasaki A. Interferon deficiency can lead to severe COVID. Nature. 2020;587:374–376. doi: 10.1038/d41586-020-03070-1. [DOI] [PubMed] [Google Scholar]
- 26.Zuo Y., et al. Prothrombotic autoantibodies in serum from patients hospitalized with COVID-19. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abd3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Consiglio C.R., et al. The immunology of multisystem inflammatory syndrome in children with COVID-19. Cell. 2020;183:968–981. doi: 10.1016/j.cell.2020.09.016. e967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gruber C.N., et al. Mapping systemic inflammation and antibody responses in multisystem inflammatory syndrome in children (MIS-C) Cell. 2020;183:982–995. doi: 10.1016/j.cell.2020.09.034. e914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang S.E., et al. New-onset IgG autoantibodies in hospitalized patients with COVID-19. medRxiv. 2021 doi: 10.1101/2021.01.27.21250559. Published online January 29, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woodruff M.C., et al. Broadly-targeted autoreactivity is common in severe SARS-CoV-2 infection. medRxiv. 2020 doi: 10.1101/2020.10.21.20216192. Published online October 23, 2020. [DOI] [Google Scholar]
- 31.Rydyznski Moderbacher C., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012. doi: 10.1016/j.cell.2020.09.038. e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi T., et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020;588:315–320. doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer S., et al. AIRE-deficient patients harbor unique high-affinity disease-ameliorating autoantibodies. Cell. 2016;166:582–595. doi: 10.1016/j.cell.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikeda Y., et al. Naturally occurring anti-interferon-alpha 2a antibodies in patients with acute viral hepatitis. Clin. Exp. Immunol. 1991;85:80–84. doi: 10.1111/j.1365-2249.1991.tb05686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caruso A., et al. Natural antibodies to IFN-gamma in man and their increase during viral infection. J. Immunol. 1990;144:685–690. [PubMed] [Google Scholar]
- 36.Ross C., et al. High avidity IFN-neutralizing antibodies in pharmaceutically prepared human IgG. J. Clin. Invest. 1995;95:1974–1978. doi: 10.1172/JCI117881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenks S.A., et al. Distinct effector B cells induced by unregulated Toll-like receptor 7 contribute to pathogenic responses in systemic lupus erythematosus. Immunity. 2018;49:725–739. doi: 10.1016/j.immuni.2018.08.015. e726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chuang Y.C., et al. Molecular mimicry between dengue virus and coagulation factors induces antibodies to inhibit thrombin activity and enhance fibrinolysis. J. Virol. 2014;88:13759–13768. doi: 10.1128/JVI.02166-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Op de Beeck A., Eizirik D.L. Viral infections in type 1 diabetes mellitus –why the beta cells? Nat. Rev. Endocrinol. 2016;12:263–273. doi: 10.1038/nrendo.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodero M.P., Crow Y.J. Type I interferon-mediated monogenic autoinflammation: The type I interferonopathies, a conceptual overview. J. Exp. Med. 2016;213:2527–2538. doi: 10.1084/jem.20161596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fox L.E., et al. Context is key: delineating the unique functions of IFNalpha and IFNbeta in disease. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.606874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Busnadiego I., et al. Antiviral activity of type I, II, and III interferons counterbalances ACE2 inducibility and restricts SARS-CoV-2. mBio. 2020;11 doi: 10.1128/mBio.01928-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stanifer M.L., et al. Importance of type I and III Interferons at respiratory and intestinal barrier surfaces. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.608645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gough D.J., et al. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 2012;36:166–174. doi: 10.1016/j.immuni.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Carvalho H., et al. Electrolyte imbalance in COVID-19 patients admitted to the Emergency Department: a case–control study. Intern. Emerg. Med. 2021 doi: 10.1007/s11739-021-02632-z. Published online January 23, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Prost N., et al. Plasma exchange to rescue patients with autoantibodies against type I interferons and life-threatening COVID-19 pneumonia. J. Clin. Immunol. 2021;2021:1–9. doi: 10.1007/s10875-021-00994-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levy R., et al. IFN-alpha2a therapy in two patients with inborn errors of TLR3 and IRF3 infected with SARS-CoV-2. J. Clin. Immunol. 2021;41:26–27. doi: 10.1007/s10875-020-00933-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brass A.L., et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang I.C., et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feeley E.M., et al. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Desai T.M., et al. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus–endosome hemifusion. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi G., et al. Opposing activities of IFITM proteins in SARS-CoV-2 infection. EMBO J. 2021;40 doi: 10.15252/embj.2020106501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tartour K., et al. Interference with the production of infectious viral particles and bimodal inhibition of replication are broadly conserved antiviral properties of IFITMs. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tartour K., et al. IFITM proteins are incorporated onto HIV-1 virion particles and negatively imprint their infectivity. Retrovirology. 2014;11:103. doi: 10.1186/s12977-014-0103-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Compton A.A., et al. IFITM proteins incorporated into HIV-1 virions impair viral fusion and spread. Cell Host Microbe. 2014;16:736–747. doi: 10.1016/j.chom.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Z., et al. Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proc. Natl. Acad. Sci. U. S. A. 2014;111:769–774. doi: 10.1073/pnas.1321748111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Randolph A.G., et al. Evaluation of IFITM3 rs12252 association with severe pediatric influenza infection. J. Infect. Dis. 2017;216:14–21. doi: 10.1093/infdis/jix242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y., et al. Interferon-induced transmembrane protein 3 genetic variant rs12252-C associated with disease severity in coronavirus disease 2019. J. Infect. Dis. 2020;222:34–37. doi: 10.1093/infdis/jiaa224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim Y.C., Jeong B.H. Strong correlation between the case fatality rate of COVID-19 and the rs6598045 single nucleotide polymorphism (SNP) of the interferon-induced transmembrane protein 3 (IFITM3) gene at the population-level. Genes (Basel) 2020;12:42. doi: 10.3390/genes12010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sorci G., et al. Explaining among-country variation in COVID-19 case fatality rate. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-75848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]