Abstract
Toward eradicating the COVID-19 pandemic, vaccines that induce high humoral and cellular immune responses are essential. However, SARS-CoV-2 variants have begun to emerge and raise concerns, as they may potentially compromise vaccine efficiency. Here, we monitored neutralization potency of convalescent or Pfizer-BTN162b2 post-vaccination sera against pseudoviruses displaying spike proteins derived from wild-type SARS-CoV-2, or its UK-B.1.1.7 and SA-B.1.351 variants. Compared to convalescent sera, vaccination induces high titers of neutralizing antibodies, which exhibit efficient neutralization potential against pseudovirus carrying wild-type SARS-CoV-2. However, while wild-type and UK-N501Y pseudoviruses were similarly neutralized, those displaying SA-N501Y/K417N/E484K spike mutations moderately resist neutralization. Contribution of single or combined spike mutations to neutralization and infectivity were monitored, highlighting mechanisms by which viral infectivity and neutralization resistance are enhanced by N501Y or E484K/K417N mutations. Our study validates the importance of the Pfizer vaccine but raises concerns regarding its efficacy against specific SARS-CoV-2 circulating variants.
Keywords: COVID-19, SARS-CoV-2, Spike, UK and SA variants, United Kingdom and South African variants, Pfizer-BTN162b2 vaccine, neutralization antibodies
Graphical abstract
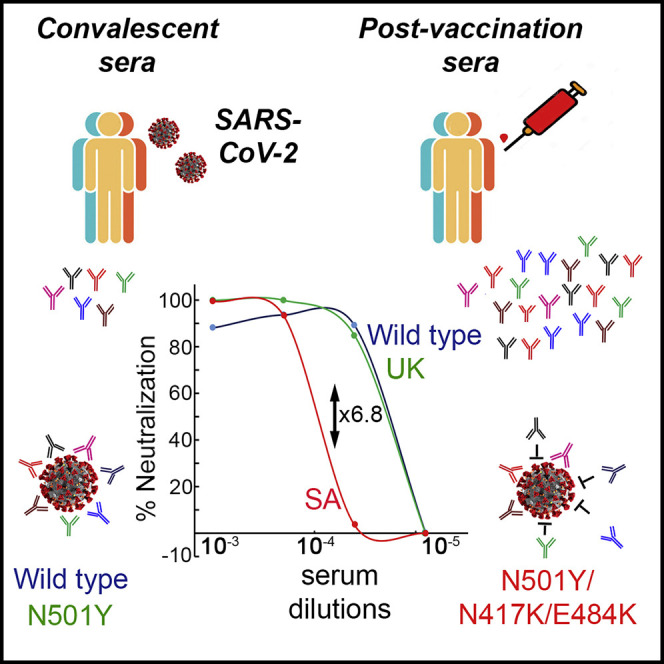
Kuzmina and colleagues monitored the neutralization potential of convalescent sera or sera from Pfizer-vaccinated individuals against pseudoviruses displaying wild-type, B.1.1.7, or B.1.351 SARS-CoV-2 spike proteins. While vaccine sera comparably neutralizes wild-type and B.1.1.7 pseudoviruses, the B.1.351 variant moderately resists vaccine-mediated neutralization, highlighting the importance of monitoring the emergence of variants.
Main text
Introduction
One year into the outbreak of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in China, it is clear that the pandemic is here to stay for the foreseeable future (Zhou et al., 2020; Zhu et al., 2020). Currently, coronavirus disease 2019 (COVID-19) has spread worldwide, infecting 117 million people and resulting in 2.6 million deaths. SARS-CoV-2 entry into cells is initiated via spike (Shang et al., 2020), a trimeric transmembrane glycoprotein with S1 and S2 subunits. While S1 subunit mediates viral attachment to the Angiotensin-Converting Enzyme 2 (ACE2) receptor expressed on target cells, S2 subunit promotes fusion. Entry is facilitated through spike priming by TMPRSS2 protease and potentially other human proteases, promoting fusion of viral and cell membranes. Cleavage of the spike at the S1/S2 boundary site also occurs via furin, facilitating cell-cell fusion during viral release and increased infectivity (Hoffmann et al., 2020a, 2020b; Letko et al., 2020; Walls et al., 2020; Wrapp et al., 2020; Zang et al., 2020).
The receptor-binding domain (RBD) in spike is a main target of neutralizing antibodies (nAbs). These are elicited by prior infection or vaccination, and inhibit SARS-CoV-2-ACE2 engagement and viral entry, and are thus likely to be key for future protection against SARS-CoV-2 (Alsoussi et al., 2020; Brouwer et al., 2020; Gaebler et al., 2021; ACTIV-3/TICO LY-CoV555 Study Group, 2021; Klasse and Sattentau, 2002; Plotkin, 2010; Rogers et al., 2020; Wu et al., 2020). As such, defining the epitopes that are recognized by nAbs is important for proper development of therapies and vaccines. To date, several vaccines against SARS-CoV-2 have been introduced. The two leading candidates are manufactured by Pfizer (BNT162b2) and Moderna (mRNA-1273) and administered worldwide. These are lipid-based nanoparticles that package mRNA coding for a stabilized form of spike (Anderson et al., 2020; Krammer, 2020; Polack et al., 2020; Walsh et al., 2020). Both vaccines elicit high nAb titers, displaying >94% efficacy at preventing disease (Baden et al., 2021). However, as SARS-CoV-2 rapidly spreads, viral variants emerge, and to some extent, resist neutralization. Numerous variants that carry multiple mutations in their spike appear worldwide and spread at a high pace. B.1.1.7 (501Y.V1) was first detected in the United Kingdom (UK) (Kemp et al., 2021; Korber et al., 2020) and displays the dominant early D614G mutation, which improves viral fitness and transmission (Hou et al., 2020; Plante et al., 2020). Additional mutations in spike, three amino acid deletions and seven missense mutations, including N501Y in RBD, make B.1.1.7 more infectious than the wild-type or the D614G strain (Kemp et al., 2021). Tighter RBD-N501Y interactions with ACE2 are considered the driving force for the increased B.1.1.7 infectivity (Chan et al., 2020; Santos and Passos, 2021; Starr et al., 2020; Xie et al., 2020). A second variant of concern, B.1.351 (501Y.V2), was first reported in South Africa (SA). Its spike harbors the N501Y mutation and includes an additional nine changes (Giandhari et al., 2021; Tegally et al., 2021). One cluster includes four substitutions and a deletion (L18F, D80A, D215G, delta 242-244, and R246I), while a second cluster contains three RBD substitutions—K417N, E484K, and N501Y. Early reports have indicated that while the N501Y spike mutation in B.1.1.7 does not compromise post-vaccine neutralization (Shi et al., 2021), E484K partly impairs neutralization resistance (Greaney et al., 2021), potentially compromising vaccines effectiveness (Baum et al., 2020; Ku et al., 2021; Liu et al., 2021; Muik et al., 2021; Wang et al., 2021b; Weisblum et al., 2020; Wibmer et al., 2021; Wu et al., 2021; Andreano et al., 2020; Xie et al., 2021). In addition, other variants have been described—B.1.1.298 in Denmark, B1.429 in California, Brazil-P1 and recently, B.1.526, in New York (Annavajhala et al., 2021; Candido et al., 2020; Sabino et al., 2021; West et al., 2021). In such a rapidly evolving pandemic, it is important to determine the nature of neutralization resistance of emerging variants to sera from both convalescent and vaccinated individuals. In this study, we compared neutralization potency of sera drawn from COVID-19 recovered patients and from individuals who received one or two doses of the Pfizer-BTN162b2 vaccine against pseudoviruses displaying spike from wild-type SARS-CoV-2 or its UK-B.1.1.7 or SA-B.1.351 variants. We also determined the contribution of each of the RBD-spike mutations that appear in these two variants, or their combinations, to viral infectivity and neutralization resistance. Our findings show that sera from the second-dose vaccinated cohort exhibited high nAb titers and a significance increase in ability to neutralize viral entry relative to convalescent sera. Both wild-type and the UK-N501Y pseudoviruses were efficiently neutralized by tested sera. However, pseudovirus carrying the SA-N501Y/K417N/E484K spike mutations moderately resisted neutralization by convalescent and post-vaccinated sera. Effects of spike RBD mutations, N501Y, K417N, and E484K (or their combinations) on viral infectivity (i.e., transduction and inhibition of viral entry) were also documented. While pseudovirus displaying the N501Y spike mutant exhibited high infectivity rates relative to wild-type SARS-CoV-2, it was efficiently neutralized by post-vaccinated sera. In contrast, pseudovirus displaying E484K spike mutation, and to a lesser extent, K417N, partly resisted neutralization by post-vaccination sera. However, such pseudoviruses displayed similar infectivity rates relative to wild-type pseudovirus. Uniquely, pseudovirus displaying the SA-N501Y/K417N/E484K spike mutations exhibited both high infectivity levels and efficient neutralization resistance, raising concerns for vaccine efficiency. Nevertheless, the substantial increase in neutralization potency upon vaccination highlighted the clinical significance of a two-dose vaccination.
Results
Post-vaccination sera exhibit improved efficiency in neutralizing wild-type SARS-CoV-2 pseudovirus relative to convalescent sera
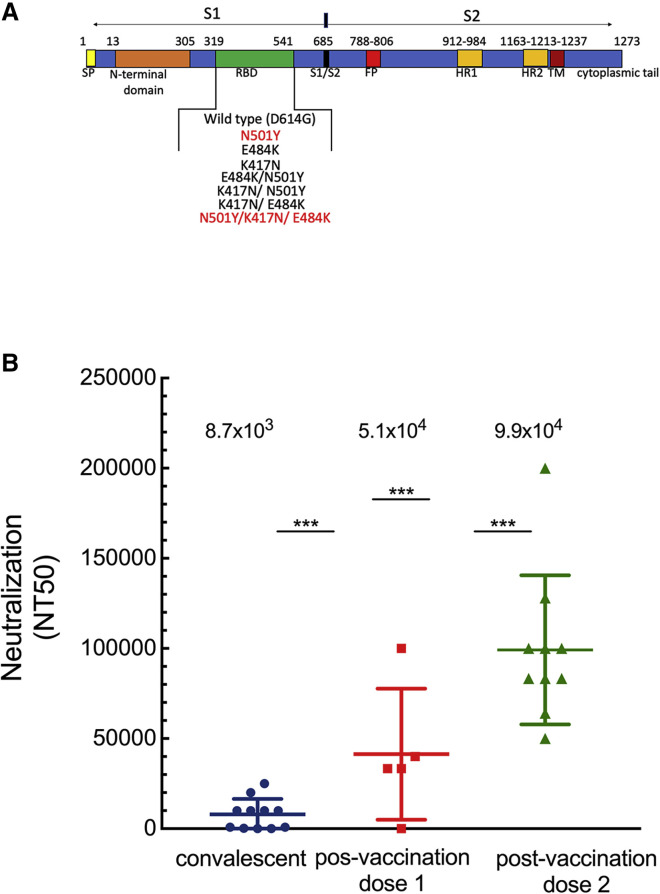
We acquired sera from a cohort of COVID-19 recovered patients who exhibited severe disease symptoms (n = 10) and assessed its ability to neutralize entry of wild-type SARS-CoV-2 pseudovirus into ACE2 target cells. Neutralization potency of convalescent sera was compared to post-vaccination sera from individuals who received one (3 weeks post-first dose; n = 5) or two doses (9–11 days post-second dose; n = 10) of the BNT162b2 vaccine. Our findings show that all sera samples exhibited detectable entry neutralization potency against the SARS-CoV-2 wild-type pseudovirus (Figure 1 B). Significantly, sera from vaccinated individuals who received the second dose exhibited a robust neutralizing potential, with a mean NT50 value of 99,000. This was an average of a 2-fold increase, relative to sera drawn from the individuals who received one dose of vaccination—mean NT50 dilution of 51,300. Moreover, an 1-fold increase was documented in mean NT50 of second dose post-vaccination sera relative to convalescent sera—mean NT50 dilution of 8,700. Importantly, a 6-fold increase in mean NT50 dilution was obtained when sera from the first vaccination dose was compared to convalescent sera (compare mean NT50 of 51,000 to 8,700). These findings imply that the second dose of vaccination is essential to achieve high neutralizing titers against wild-type SARS-CoV-2 pseudoviruses, relative to the first-dose or to convalescent sera (Figure 1B). The magnitude of neutralizing potencies of tested sera partly correlated with total anti-SARS-CoV-2 antibody levels and detected by total IgG ELISA (Tables S1 and S2).
Figure 1.
Neutralization of wild-type SARS-CoV-2 pseudovirus by convalescent or post-vaccination sera
(A) Schematic organization of SARS-CoV-2 spike indicating domains and studied RBD mutations.
(B) Convalescent or post-vaccination sera neutralize pseudoviruses carrying wild-type SARS-CoV-2 spike. Neutralization assays were performed by transducing HEK293T-ACE2 cells with pseudovirus displaying wild-type SARS-CoV-2 spike in the presence of increasing dilutions of sera drawn from convalescent or post-vaccinated individuals. 48 h post-transduction, cells were harvested and their luciferase readings were monitored. Neutralizing potency was calculated at increased serial dilutions, relative to transduced cells with no sera added. Neutralization, NT50 is defined as the inverse dilution that achieved 50% neutralization. Results are the average of two independent biological experiments. Triplicates were performed for each tested serum dilution. Black bars represent geometric mean of NT50 values, indicated at the top. Statistical significance was determined using one-tailed t test ∗∗∗p < 0.001.
Neutralizing potency of convalescent or post-vaccination sera against pseudoviruses displaying UK-N501Y and SA-N501Y/K417N/E484K spike mutants
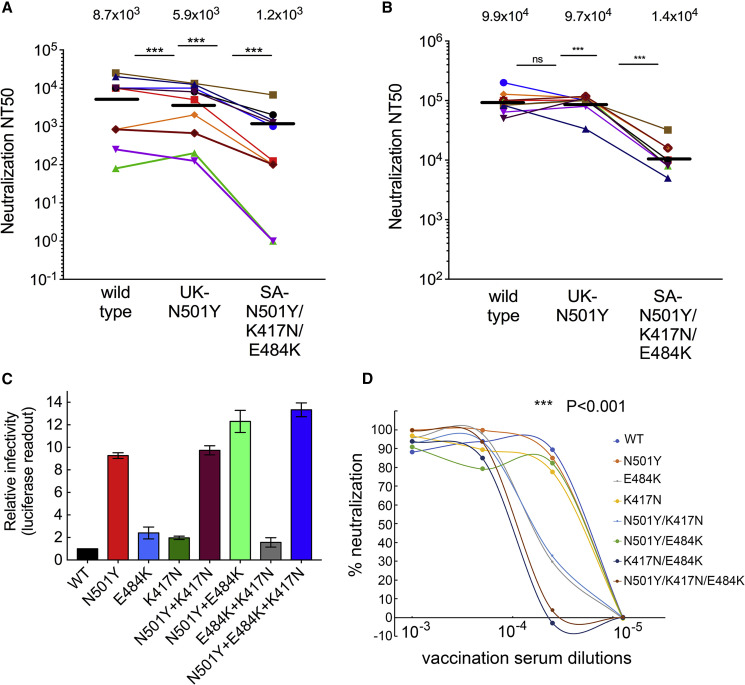
We further monitored the ability of our sera samples to neutralize entry of pseudoviruses carrying the UK-N501Y and the SA-N501Y/K417N/E484K spike mutations. NT50 values were determined by neutralization assays relative to pseudovirus displaying wild-type SARS-CoV-2 spike (Figures 2A and 2B). Our findings confirmed that convalescent sera neutralized entry of the wild-type pseudovirus with an average NT50 of 8,700. Interestingly, pseudovirus that carried the UK-N501Y spike mutation was also efficiently neutralized by convalescent sera samples, exhibiting only a mean of 1.5-fold decrease in ability to neutralize viral entry relative to wild-type SARS-CoV-2 pseudovirus. In contrast, pseudovirus displaying the SA-N501Y/K417N/E484K spike mutations exhibited a moderate resistance to neutralization by convalescent sera, with a 6.8-fold decrease in mean NT50 value relative to the wild-type SARS-CoV-2 pseudovirus (Figure 2A). Herein, there was a rather wide distribution of NT50 values between convalescent sera samples when tested against the wild-type or the SA pseudoviruses. Samples that exhibited low neutralizing titers against wild-type pseudovirus could not neutralize the SA pseudovirus. Upon vaccination, levels of nAbs substantially increased, confirming our findings. Wild-type or UK-N501Y SARS-CoV-2 spike pseudoviruses were comparably neutralized by sera from the second post-vaccination dose (Figure 2B). These findings point to a high efficacy of the vaccine against the UK-N501Y strain. However, SA-N501Y/K417N/E484K spike pseudovirus partly resisted neutralization by post-vaccinated sera, exhibiting 6.8-fold decrease in mean NT50 relative to wild-type SARS-CoV-2 spike pseudoviruses (Figure 2C). The recurrence of the same 6.8-fold decrease in neutralization potency may reflect similar epitope interactions of nAbs from convalescent or vaccination sera. In this case, the distribution of mean NT50 values in between post-vaccination sera samples was relatively small. We conclude that pseudovirus displaying SA-N501Y/K417N/E484K spike mutations exhibits a moderately increased resistance to neutralization by both convalescent and Pfizer-vaccinated sera. Nevertheless, mean NT50 of post-vaccination sera still equaled or exceeded mean NT50 of convalescent sera, emphasizing vaccine efficiency. Given that we do not have evidence of widespread reinfection with wild-type virus, it is likely that infection of immunized individuals by the SA variant would potentially be at a similarly low level.
Figure 2.
Pseudoviruses carrying SARS-CoV-2 spike variants are neutralized to a different extent by convalescent or post-vaccination sera
(A and B) Neutralization sensitivity of SARS-CoV-2 pseudoviruses displaying UK or SA spike variants by convalescent (A) or post-vaccination sera (B). Pseudoviruses displaying wild-type or mutant SARS-CoV-2 spike from UK-N50Y or SA-N501Y/K417N/E484K variants were used in neutralization assays. HEK293T-ACE2 cells were transduced with the indicated pseudoviruses in the presence of increasing dilutions of convalescent or vaccination sera. 48 h post-infection, cells were harvested and their luciferase readings were monitored. Serum neutralizing potency was calculated at serial dilutions, relative to transduced cells with no sera added. Results are the average of two independent experiments. Triplicates were performed for each tested sera and pseudovirus. Horizontal black bars represent geometric mean NT50, indicated at the top. Statistical significance was determined using one-tailed t test ∗∗∗p < 0.001.
(C) SARS-CoV-2 pseudoviruses from UK and SA variants exhibit different infectivity levels. Pseudoviruses bearing wild-type or the indicated mutant SARS-CoV-2 spike were used to transduce HEK293T-ACE2 cells. Equal viral loads were normalized based on p24 protein levels. 48 h post-transduction, cells were harvested and their luciferase readouts were monitored. Bar graphs show mean values ± SD error bars of three independent experiments. Measured statistical significance was calculated between experiments by a two-tailed Student’s t test ∗∗∗p ≤ 0.001.
(D) Neutralization sensitivity of SARS-CoV-2 pseudoviruses variants displaying combined UK and SA spike mutants. Pseudoviruses displaying wild-type or the indicated SARS-CoV-2 mutant spike were used in neutralization assays. HEK-ACE2 cells were transduced with the indicated pseudoviruses in the presence of increasing dilutions of a mixed vaccination serum drawn from vaccinated individuals that received the second dose (n = 10). 48 h post-transduction, cells were harvested and their luciferase readouts were monitored. Percentage of neutralizing potential was calculated at serial dilutions, relative to transduced cells with no sera added. Arrows indicate NT50 values obtained for each mutant. Results are the average of two independent experiments. Black arrows represent mean NT50 values. Measured statistical significance was calculated between experiments by a two-tailed Student’s t test ∗∗∗p ≤ 0.001.
Pseudoviruses carrying combination of spike UK and SA mutants exhibit enhanced infectivity levels
We next monitored the ability of psedoviruses carrying SARS-CoV-2 spike variants to infect HEK-ACE2 cells. Pseudoviruses with UK-N501Y or SA-N501Y/K417N/E484K spike mutants were tested, as were pseudoviruses carrying single or combined RBD mutations—K417N, E484K, N501Y/E484K, N501Y/K417N, and K417N/E484K (Figure 2C). As our system uses single-round pseudoviruses, the term “transduction” is more suitable than “infectivity,” which would imply the use of infections of SARS-CoV-2. Our findings show that, relative to pseudovirus bearing wild-type SARS-CoV-2 spike, the ability of pseudovirus carrying the UK-N501Y mutation to transduce its target cells significantly increased, up to 9-fold. Moreover, the SA-N501Y/K417N/E484K spike mutations further boosted transduction levels, up to 13-fold, relative to SARS-CoV-2 wild-type pseudovirus. We also evaluated the contribution of additional combinations of RBD mutations. Pseudovirus carrying the E484K single mutation exhibited only a 2-fold increase in its ability to transduce cells, relative to SARS-CoV-2 wild-type pseudovirus. Similarly, pseudovirus displaying the K417N single spike mutation, or K417N/E484K double mutations, also showed a 2-fold increase in transduction rates relative to the wild-type pseudovirus. Interestingly, pseudoviruses where the N501Y spike mutation was attached to either K417N or E484K mutations (i.e., N501Y/K417N or N501Y/E484K), exhibited high transduction rates, slightly higher than those of the UK-N501Y pseudovirus alone. Indeed, a combined N501Y/E484K mutation led to a 13-fold increase in transduction levels relative to wild-type pseudovirus, similarly to that of the SA pseudovirus. Thus, in the context of N501Y spike mutation, the contribution of the E484K mutation to viral transduction was boosted. These results suggest that the emergence and spread of viral variants may be driven by infectivity rather than resistance to neutralization, and highlight the contribution of the N501Y spike mutation for enhancement of viral infectivity into target cells (Figure 2C).
Pseudoviruses carrying combination of UK and spike SA mutants exhibit different neutralization sensitivity against post-vaccination sera
We also tested the ability of post-vaccination sera (a mixture of samples from the second dose) to neutralize entry into hACE2 target cells of pseudoviruses displaying different UK and SA combined spike mutations (Figure 2D). We confirmed our results showing that pseudovirus with the UK-N501Y spike mutation exhibited similar neutralization potency between vaccinated sera and wild-type SARS-CoV-2 pseudovirus. Pseudoviruses with K417N or N501Y/K417N mutations were also similarly neutralized by post-vaccination sera, similarly to that of wild-type pseudovirus. However, pseudoviruses displaying the E484K and N501Y/E484K spike mutations partly resisted neutralization by vaccination sera. Finally, pseudoviruses with the K417N/E484K and the SA-N501Y/K417N/E484K spike mutations exhibited the highest neutralization resistance to post-vaccination sera, emphasizing the role of E484K mutation in neutralization resistance (Figure 2D).
Discussion
The induction of nAbs that target the RBD of the SARS-CoV-2 spike through vaccination is a main goal toward complete eradication of COVID-19. Our results indicate that the overwhelming spread of SARS-CoV-2 is a combination of increased infectivity and ability to resist neutralization, thus potentially posing a risk for reduced vaccine efficacy. Indeed, SARS-CoV-2 variants UK-B.1.1.7 and SA-B.1.351 are spreading worldwide and are defined as a concern by the WHO. In this study, we employed neutralization assays to examine whether the currently administered Pfizer vaccine efficiently neutralizes entry of wild-type or mutated spike SARS-CoV-2 pseudoviruses into HEK-ACE2 cells. We also determined which of the UK or SA mutations within the RBD, or their combinations, contribute to a potential neutralization resistance and enhanced infectivity. Our results show that pseudovirus carrying the UK-N501Y mutation is highly infectious, relative to pseudovirus bearing wild-type SARS-CoV-2 (Figure 2D). Moreover, when N501Y mutation within spike was combined with other mutations—K417N (N501Y/K417N), E484K (N501Y/E484K), or N501Y/K417N/E484K—enhancement of infectivity was documented relative to the wild-type SARS-CoV-2 pseudovirus. Pseudoviruses carrying K417N or E484K single mutations, or the combined K417N/E484K mutations, exhibited infectivity rates that were similar to the wild-type pseudovirus. Oppositely, resistance to neutralization seemed to be driven by the E484K, and to a lesser extent, on the K417N mutations. Pseudoviruses carrying the E484K mutation alone, or the combined N501Y/E484K spike mutations, resisted neutralization to some extent. Similarly, pseudovirus bearing the E484K/K417N mutations also resisted neutralization by post-vaccination sera relative to wild-type SARS-CoV-2 pseudovirus (Figure 2D). However, the SA-N501Y/K417N/E484K pseudovirus was unique, as it exhibited high infectivity levels and exhibited moderate neutralization resistance from vaccination sera. Based on these results, we suggest that elevated spread of SARS-CoV-2 variants is a combination of neutralization resistance and increased infectivity. N501Y potentially enhances association of the virus with hACE2, leading to enhanced infectivity and spread (Santos and Passos, 2021; Starr et al., 2020). When combined with E484K or K417N mutations, an increase in neutralization resistance is also observed. Combination of all three mutations as seen in the SA variant results in high infectivity and neutralization resistance. However, while reduced neutralization may be a major factor, there is still insufficient data to support its potential, and additional experiments with infectious virus will be required.
Our data also indicate that the Pfizer vaccine is moderately compromised against SA-N501Y/K417N/E484K pseudo-variants. Average decrease in mean neutralization potential of the vaccinated sera against this pseudovirus was 6.8-fold, relative to wild-type SARS-CoV-2 pseudovirus. This result is only partly aligned with recent conclusions from Pfizer, reporting that its vaccine is almost similarly efficient against the SA variant as wild-type SARS-CoV-2 (Xie et al., 2021). A Moderna report also documented that its vaccine is 6.4-fold less efficient in neutralizing SA-B.1.351 variant, relative to neutralization of the wild-type SARS-CoV-2. However, their conclusion indicated that such a reduction is not clinically significant (Wu et al., 2021). In our mind, the clinical significance of a 6.8-fold-reduced neutralization potency of convalescent or post-vaccination sera against the SA strain remains to be determined and raises concerns about vaccine efficiency against current or future SARS-CoV-2 variants (Wang et al., 2021a, 2021b; Wibmer et al., 2021). Overall, these results call for close attention to variant spread. Moreover, development of new vaccines with improved neutralizing potency against specific SARS-CoV-2 variants may be required. Nonetheless, our findings urge the need for a two-dose vaccination protocol, as it boosts the levels of nAbs against the virus even after recovering from COVID-19.
Limitations of the study
Importantly, our study relies on pseudoviruses, which can only model ACE2-dependent viral entry. Nonetheless, several studies demonstrated a close correlation between pseudotyping/binding assays and true neutralization assays against infectious SARS-CoV-2 (Crawford et al., 2020; Ju et al., 2020; Wang et al., 2020). At this point, in order for such a correlation to be validated, further investigation needs to be performed with the infectious clone. Moreover, the contribution of additional mutations outside of the spike may also affect resistance to neutralization, infectivity levels, or pathogenesis of SARS-CoV-2. Finally, it is worth stating that our findings are relevant only to the tested sera. However, the mid-sized cohort that was analyzed in our work, combined with other reports with similar conclusions, validates our findings.
STAR★Methods
Key resources table
| REAGENTS or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Cells Bacterial and virus strains | ||
| Competent E.Coli DH5α | NEB | Cat#18265017 |
| HEK293T cells | ATCC | Cat#3216 |
| Molecular cloning enzymes | ||
| T4 ligase | NEB | M0202S |
| XbaI | NEB | R0145S |
| SalI | NEB | R0138S |
| Recombinant DNA | ||
| pCG1-SARS-CoV-2 spike | Hoffmann et.al., 2020b | NA |
| pLenti-PGK_Luc | Addgene | Cat#19360 |
| pCMV delta 8.2 | Addgene | Cat#12263 |
| pCG1_ACE2 | Hoffmann et.al., 2020b | NA |
| pLenti_CMV_PURO | Addgene | Cat#17448 |
| Recombinant Spike protein | ACROBioSystems | SPD-C82E9 |
| Software | ||
| Prism 9.0 | GraphPad | NA |
| Photoshop | Acrobat | NA |
| Luciferase assay | Promega | E1500 |
| QuikChange Lightning Site-Directed Mutagenesis kit | Agilent Technologies | Cat#200522 |
| Oligonucleotides for quick mutagenesis | ||
| TTTCAGCCCACATATGGCGTGGGCTAT | Hylabs | FWD-N501Y |
| ATAGCCCACGCCATATGTGGGCTGAAA | Hylabs | Rev-N501Y |
| GGACAGACAGGCAACATCGCCGACTAC | Hylabs | FWD-K417N |
| GTAGTCGGCGATGTTGCCTGTCTGTCC | Hylabs | Rev-K417N |
| TGTAACGGCGTGAAAGGCTTCAACTGC | Hylabs | FWD-E484K |
| GCAGTTGAAGCCTTTCACGCCGTTACA | Hylabs | Rev-E484K |
Resource availability
Lead contact
Resources and reagents generated in this study are available from the Lead Contact with completed Materials Transfer Agreement. Further information and requests for resources and reagents should be directed to the Lead Contact, and will be fulfilled by the lead contact, Ran Taube (rantaube@bgu.ac.il).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate/analyze any dadset/code.
Experimental model and subject details
Human subject collection
The study was conducted in compliance with ethical principles of the Declaration of Helsinki and approved by the Soroka Medical Center Institutional Review Board (protocol 0281-20-SOR). Sera was collected from a 10 cohort of COVID19 recovered patients, or from 10 individuals that were vaccinated in the first or second Pfizer vaccine. Samples were from either a 21-day post first dose, or 9-11 days post second dose. Collected sera are summarized in Tables S1 and S2. To confirm total SARS-CoV-2 IgG in the collected sera, specific IgG levels were determined by Liasonx ELISA.
Bacterial strains and cell culture
HEK-ACE2 stable cells were cultured at 37°C in a 5% CO2 incubator. Cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) high glucose (GIBCO), supplemented with 10% fetal bovine serum (FBS), 2mM GlutaMAX (GIBCO) and 100U/mL penicillin-streptomycin. HEK-ACE2 expressing cells were generated by stable transduction with lentivirus expressing human ACE2. Our pseudoviruses were standardized for equal loads by monitoring p24 levels by ELISA. E.coli DH5α bacteria were used for transformation of plasmids coding for lentivirus packaging DNA and SARS CoV-2 spike. A single colony was picked and cultured in LB broth with 50 μg penicillin at 37°C at 200 rpm in a shaker for overnight.
Method details
Generation of HEK-hACE2 stable cell line
hACE2 (received from S. Pohlmann lab, University Göttingen, Germany) was re-cloned into lentiviral expression vector. Lentiviral particles were produced as described previously (Krasnopolsky et al., 2020) Briefly, HEK293T cells were stably transduced with lentivirus expressing ACE2. Cells were analyzed for hACE2 expression by FACS, using biotinylated-labeled spike (ACROBiosystems). High ACE2 expressing cells were sorted using FACS Aria. ACE2 expression was periodically monitored by FACS.
Construction of spike mutants
QuikChange Lightening Site-Directed Mutagenesis kit was used to generate amino acid substitutions in the pCDNA spike plasmid (received from S. Pohlmann lab, University Göttingen, Germany), following the manufacturer’s instructions (Agilent Technologies, Inc., Santa Clara, CA). For each mutant the relative oligos that harbored the required mutation were employed.
Generation of pseudotyped lentivirus and neutralization assays
Pseudotyped viruses were generated in HEK293T cells. Briefly, LTR-PGK luciferase lentivector was transfected into cells together with other lentiviral packaging plasmids coding for Gag, Pol Tat Rev, and the corresponding wild type or mutate spike envelopes. Transfections were done in a 10cm format, as previously described and supernatant containing virus were harvested 72 h post transfection, filtered and stored at −80°C (Krasnopolsky et al., 2020). Neutralization assays were performed in a 96 well format, in the presence of pseudotyped viruses that were incubated with increasing dilutions of the tested sera (1:2000; 1:8000: 1;32000: 1:128000) or without sera as a control. Cell-sera were for 1 h. at 37°C, followed by transduction of HEK-ACE2 cells for additional 12 h. 72 h post transduction, cells were harvested and analyzed for luciferase readouts according to the manufacturer protocol (Promega). Neutralization measurements were performed in triplicates using an automated Tecan liquid handler and readout were used to calculate NT50 – 50% inhibitory titers concentration.
Pseudoviruses quality control and tittering
To determine the titers of pseudoviruses, 1e5 ACE2 stable HEK cells were plated in a 12-well plate. 24 h later, decreased serial dilutions of pseudovirus were used to transduce cells. 48 h post transduction, cells were harvested and analyzed for their luciferase readouts. p24 ELISA measurements were conducted to ensure equal loads.
Quantification and statistical analysis
Statistical analyses were performed using GraphPad Prism. Measured statistical significance was calculated between experiments by a two-tailed Student’s t test - P£0.001. Error bars throughout all figures represent one standard deviation. Specific details on statistical tests and experimental replicates can be found in the figure legends.
Acknowledgments
This work was supported by the Israeli Ministry of Science and Technology (MOST; grant no. 3-16897 to R.T.), the Israel Science Foundation (ISF; research grant application no. 755/17 to R.T.), and the Ben-Gurion University of the Negev COVID-19 Research Task Force.
Author contributions
A.K. and R.T. conceived the study and analyzed the data. A.K., Y.K., and O.V. performed experiments and analyzed the data. A.K., A.K.-N., Y.S.-A., L.B.-C., Y.R., and E.R. helped with obtaining blood samples. E.R. and R.T. submitted the human subject request to the Institutional Helsinki Review Board. R.T. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: March 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2021.03.008.
Supplemental information
References
- Alsoussi W.B., Turner J.S., Case J.B., Zhao H., Schmitz A.J., Zhou J.Q., Chen R.E., Lei T., Rizk A.A., McIntire K.M., et al. A Potently Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. J. Immunol. 2020;205:915–922. doi: 10.4049/jimmunol.2000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. mRNA-1273 Study Group Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ACTIV-3/TICO LY-CoV555 Study Group A Neutralizing Monoclonal Antibody for Hospitalized Patients with Covid-19. N Engl J Med. 2021;384:905–914. doi: 10.1056/NEJMoa2033130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Piccini G., Licastro D., Casalino L., Johnson N.V., Paciello I., Monego S.D., Pantano E., Manganaro N., Manenti A., et al. SARS-CoV-2 escape <em>in vitro</em> from a highly neutralizing COVID-19 convalescent plasma. bioRxiv. 2020 doi: 10.1101/2020.12.28.424451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annavajhala M.K., Mohri H., Zucker J.E., Sheng Z., Wang P., Gomez-Simmonds A., Ho D.D., Uhlemann A.-C. A Novel SARS-CoV-2 Variant of Concern, B.1.526, Identified in New York. medRxiv. 2021 doi: 10.1101/2021.02.23.21252259. [DOI] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. COVE Study Group Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candido D.S., Claro I.M., de Jesus J.G., Souza W.M., Moreira F.R.R., Dellicour S., Mellan T.A., du Plessis L., Pereira R.H.M., Sales F.C.S., et al. Brazil-UK Centre for Arbovirus Discovery, Diagnosis, Genomics and Epidemiology (CADDE) Genomic Network Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;369:1255–1260. doi: 10.1126/science.abd2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.E.Z., Seah S.G.K., Chye D.H., Massey S., Torres M., Lim A.P.C., Wong S.K.K., Neo J.J.Y., Wong P.S., Lim J.H., et al. The Fc-mediated effector functions of a potent SARS-CoV-2 neutralizing antibody, SC31, isolated from an early convalescent COVID-19 patient, are essential for the optimal therapeutic efficacy of the antibody. bioRxiv. 2020 doi: 10.1101/2020.10.26.355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses. 2020;12 doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021 doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giandhari J., Pillay S., Wilkinson E., Tegally H., Sinayskiy I., Schuld M., Lourenço J., Chimukangara B., Lessells R., Moosa Y., et al. Early transmission of SARS-CoV-2 in South Africa: An epidemiological and phylogenetic report. Int. J. Infect. Dis. 2021;103:234–241. doi: 10.1016/j.ijid.2020.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. bioRxiv. 2021 doi: 10.1101/2020.12.31.425021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pöhlmann S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schäfer A., Nakajima N., Takahashi K., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Kemp S., Harvey W., Lytras S., Carabelli A., Robertson D., Gupta R. Recurrent emergence and transmission of a SARS-CoV-2 Spike deletion H69/V70. bioRxiv. 2021 doi: 10.1101/2020.12.14.422555. [DOI] [Google Scholar]
- Klasse P.J., Sattentau Q.J. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J. Gen. Virol. 2002;83:2091–2108. doi: 10.1099/0022-1317-83-9-2091. [DOI] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Krasnopolsky S., Kuzmina A., Taube R. Genome-wide CRISPR knockout screen identifies ZNF304 as a silencer of HIV transcription that promotes viral latency. PLoS Pathog. 2020;16:e1008834. doi: 10.1371/journal.ppat.1008834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku Z., Xie X., Davidson E., Ye X., Su H., Menachery V.D., Li Y., Yuan Z., Zhang X., Muruato A.E., et al. Molecular determinants and mechanism for antibody cocktail preventing SARS-CoV-2 escape. Nat. Commun. 2021;12:469. doi: 10.1038/s41467-020-20789-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., VanBlargan L.A., Bloyet L.-M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., et al. Landscape analysis of escape variants identifies SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. bioRxiv. 2021 doi: 10.1101/2020.11.06.372037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik A., Wallisch A.-K., Sänger B., Swanson K.A., Mühl J., Chen W., Cai H., Sarkar R., Türeci Ö., Dormitzer P.R., et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. bioRxiv. 2021 doi: 10.1101/2021.01.18.426984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020 doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin S.A. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. C4591001 Clinical Trial Group Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino E.C., Buss L.F., Carvalho M.P.S., Prete C.A., Jr., Crispim M.A.E., Fraiji N.A., Pereira R.H.M., Parag K.V., da Silva Peixoto P., Kraemer M.U.G., et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397:452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos J.C., Passos G.A. The high infectivity of SARS-CoV-2 B.1.1.7 is associated with increased interaction force between Spike-ACE2 caused by the viral N501Y mutation. bioRxiv. 2021 doi: 10.1101/2020.12.29.424708. [DOI] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P.Y., Xie X., Zou J., Fontes-Garfias C., Xia H., Swanson K., Cutler M., Cooper D., Menachery V., Weaver S., et al. Res Sq; 2021. Neutralization of N501Y mutant SARS-CoV-2 by BNT162b2 vaccine-elicited sera. [DOI] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Lessells R.J., Giandhari J., Pillay S., Msomi N., Mlisana K., Bhiman J.N., von Gottberg A., Walaza S., et al. Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat. Med. 2021;27:440–446. doi: 10.1038/s41591-021-01255-3. [DOI] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;183:1735. doi: 10.1016/j.cell.2020.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Increased Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7 to Antibody Neutralization. bioRxiv. 2021 doi: 10.1101/2021.01.25.428137. [DOI] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. bioRxiv. 2021 doi: 10.1101/2021.01.15.426911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife. 2020;9 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.P., Barnes C.O., Yang Z., Bjorkman P.J. SARS-CoV-2 lineage B.1.526 emerging in the New York region detected by software utility created to query the spike mutational landscape. bioRxiv. 2021 doi: 10.1101/2021.02.14.431043. [DOI] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv. 2021 doi: 10.1101/2021.01.18.427166. [DOI] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Moliva J.I., Koch M., Choi A., Stewart-Jones G.B.E., Bennett H., Boyoglu-Barnum S., Shi W., Graham B.S., et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv. 2021 doi: 10.1101/2021.01.25.427948. [DOI] [Google Scholar]
- Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., Liu J., Schindewolf C., Bopp N.E., Aguilar P.V., et al. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848.e3. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Liu Y., Liu J., Zhang X., Zou J., Fontes-Garfias C.R., Xia H., Swanson K.A., Cutler M., Cooper D., et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021 doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze any dadset/code.