Abstract
We and others previously demonstrated that a type 1 diabetes genetic risk score (GRS) improves the ability to predict disease progression and onset in at-risk subjects with islet autoantibodies. Here, we hypothesized that GRS and islet autoantibodies, combined with age at onset and disease duration, could serve as markers of residual β-cell function following type 1 diabetes diagnosis. Generalized estimating equations were used to investigate whether GRS along with insulinoma-associated protein-2 autoantibody (IA–2A), zinc transporter 8 autoantibody (ZnT8A), and GAD autoantibody (GADA) titers were predictive of C-peptide detection in a largely cross-sectional cohort of 401 subjects with type 1 diabetes (median duration 4.5 years [range 0–60]). Indeed, a combined model with incorporation of disease duration, age at onset, GRS, and titers of IA–2A, ZnT8A, and GADA provided superior capacity to predict C-peptide detection (quasi-likelihood information criterion [QIC] = 334.6) compared with the capacity of disease duration, age at onset, and GRS as the sole parameters (QIC = 359.2). These findings support the need for longitudinal validation of our combinatorial model. The ability to project the rate and extent of decline in residual C-peptide production for individuals with type 1 diabetes could critically inform enrollment and benchmarking for clinical trials where investigators are seeking to preserve or restore endogenous β-cell function.
Introduction
A 2015 Consensus Statement defined the preclinical staging of type 1 diabetes based on the number of islet autoantibodies and presence of dysglycemia (1). Extensive genotyping and the development of type 1 diabetes genetic risk scores (GRS), which consolidate the complex heritable components of the disease (2–4), have further improved our ability to predict disease progression and onset among islet autoantibody–positive individuals (5). Despite this substantial progress in characterizing metabolic and immunologic events during pre–type 1 diabetes, these factors are rarely monitored beyond the recent-onset phase of the disease.
Preservation of endogenous insulin production, as measured by C-peptide in serum, is the most common benchmark for type 1 diabetes interventional trials (6), and it is well known that the production of even low levels of endogenous insulin is associated with reduced severity of long-term complications (7). Though the disease was originally thought to result in complete loss of functional β-cell mass, the development of ultrasensitive C-peptide assays has largely overturned that notion (8). Indeed, C-peptide loss after type 1 diabetes diagnosis is now known to occur in a biphasic pattern with a window of exponential fall followed by a stable period (9), and many individuals with long-standing type 1 diabetes (i.e., up to 50 years’ duration) continue to secrete small amounts of endogenous insulin (10). Accordingly, we observed small insulin-positive islets and insulin-positive single cells scattered throughout the exocrine pancreas tissue of organ donors with established disease (11,12); however, there is marked heterogeneity in the duration and degree of maintenance of β-cell function (13).
Early studies linked islet autoantibody positivity with more precipitous loss of residual β-cell function, particularly during the first year following type 1 diabetes diagnosis (14–16). After disease onset, islet autoantibody titers are known to decline with variable kinetics and often to undetectable levels, though autoantibodies against GAD (GADA) and insulinoma-associated protein-2 (IA–2A) are more persistent than zinc transporter 8 autoantibodies (ZnT8A) (17). Declining ZnT8A and IA–2A titers have been shown to generally parallel C-peptide production in the 2.5 years immediately following diagnosis (17), but to our knowledge, long-term associations have not been explored. Separately, C-peptide persistence has been associated with genetic risk at a number of individual loci (18–20) as well as in a combined GRS model (21). These studies prompted us to explore the complex relationships linking genetic composition, islet autoantibody titers, and residual β-cell function in both recent-onset and long-standing disease.
We hypothesized that islet autoantibody titers might serve as potential biomarkers for prediction and monitoring of the rate of decline in functional β-cell mass following the onset of type 1 diabetes and that this relationship may be governed by overall genetic load for the disease. Reported herein, we assessed the utility of a type 1 diabetes GRS (22), in combination with disease duration and age at onset, as well as GADA, IA–2A, and ZnT8A titers, for prediction of the probability of C-peptide detection in individuals with type 1 diabetes. We envision that our prediction model could be applied to stratify newly diagnosed subjects according to their anticipated C-peptide trajectories to inform enrollment or establish benchmarks that define therapeutic response in intervention studies.
Research Design and Methods
Sample Collection
Study subjects were enrolled from outpatient clinics at the University of Florida, Nemours Children’s Hospital (Orlando, FL), and Emory University under institutional review board approval at each institution. Peripheral blood was collected into vacutainer tubes for serum and genomic DNA isolation. Serum samples were separated by centrifugation. Genomic DNA was extracted with QIAGEN kits on a QIAcube (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. Serum and DNA were stored at −20°C in the University of Florida Diabetes Institute (UDFI) biorepository. Samples were selected from 401 individuals with type 1 diabetes of any duration at the time of blood collection (Table 1). Because the GRS model was found to be less robust for assessment of risk in non-European populations (22), only subjects with genetically imputed European ancestry who self-reported as Caucasian were considered for study inclusion.
Table 1.
Summary of demographic and genetic data for cross-sectional and longitudinal study subjects
| All | Cross-sectional | Longitudinal | |
|---|---|---|---|
| Total N | 401 | 382 | 19 |
| Age at sample collection, years | 19.5 ± 11.9 | 19.5 ± 11.9 | 20.4 ± 12.0 |
| Age at onset, years | 12.0 ± 9.1 | 11.9 ± 9.2 | 13.1 ± 5.4 |
| T1D duration, years | 7.5 ± 9.1 | 7.5 ± 9.0 | 7.2 ± 11.1 |
| Ethnicity, n (%) | |||
| Hispanic | 41 (10.2) | 41 (10.7) | 0 (0) |
| Non-Hispanic | 329 (82) | 311 (81.4) | 18 (94.7) |
| Not reported | 31 (7.7) | 30 (7.9) | 1 (5.3) |
| Sex, n (%) | |||
| Female | 208 (51.9) | 198 (51.8) | 10 (52.6) |
| Male | 193 (48.1) | 184 (48.2) | 9 (47.4) |
| HLA status, n (%) | |||
| DR3/3 | 38 (9.5) | 35 (9.2) | 3 (15.8) |
| DR3/4 | 102 (25.4) | 97 (25.4) | 5 (26.3) |
| DR3/X | 65 (16.2) | 63 (16.5) | 2 (10.5) |
| DR4/4 | 27 (6.7) | 26 (6.8) | 1 (5.3) |
| DR4/X | 112 (27.9) | 106 (27.7) | 6 (31.6) |
| DRX/X | 57 (14.2) | 55 (14.4) | 2 (10.5) |
| GRS | 0.278 ± 0.027 | 0.278 ± 0.027 | 0.280 ± 0.023 |
Data are means ± SD unless otherwise indicated. All subjects were Caucasian (self-reported race) and of European descent (genetically imputed race). Of 401 subjects, 382 had one blood draw, 18 had two blood draws, and 1 had four blood draws. Notably, because all samples in this study were collected from subjects diagnosed with type 1 diabetes, GRS values are high compared with those in the general population without diabetes (3,4,22); hence, both the 80th and 20th GRS percentiles examined herein reflect “high genetic risk” for type 1 diabetes. T1D, type 1 diabetes.
Autoantibody and C-Peptide Measurement
IA–2A, ZnT8A, and GADA were measured from serum with use of Islet Autoantibody Standardization Program (IASP)-validated ELISA kits (KRONUS Inc., Star, ID) according to modified protocols as previously described (23). When necessary, sera were titrated to fall within the assay’s dynamic range. C-peptide was measured from serum by ultrasensitive ELISA (lower detection limit 2.5 pmol/L; Mercodia, Uppsala, Sweden), according to the manufacturer’s instructions.
GRS Calculation
Genotyping of 975,000 genetic markers was performed on an Affymetrix GeneTitan with a custom Axiom genotyping array based on the Precision Medicine Research Array (∼790,000 single nucleotide polymorphisms [SNPs]) modified to contain the ImmunoChip (24) complement SNP set (version 2.0), as well as additional known type 1 diabetes–associated loci (25), termed the UFDIchip (Thermo Fisher Scientific, Waltham, MA). The UFDIchip included 26 of the 30 SNPs required to compute GRS (22). The remaining four were obtained from imputation (rs2069762 [R2 = 0.9962], rs1264813 [R2 = 0.9962], rs689 [R2 = 0.9486], rs3788013 [R2 = 0.9967]) to the Human Reference Consortium (version r1.1) with use of the Michigan Imputation Server (26). GRS was calculated as previously described (3,22).
Statistical Analysis
Generalized estimating equations (GEE) assuming binomial variance and logit link were used to model the effect of type 1 diabetes duration, age at onset, and GRS on the probability of C-peptide detection. GEE models were used to accommodate our mixed cohort of both cross-sectional (N = 382 subjects) and longitudinal (N = 19 subjects) samples. For significant predictors, the odds of C-peptide detection were compared for the 20th (GRS = 0.260) versus the 80th (GRS = 0.301) GRS quantiles within our cohort to compare lower versus higher genetic load. For comparison of younger versus older disease onset age, odds of C-peptide detection were compared for the 20th (6.0 years) versus the 80th (15.7 years) onset age quantiles. For these analyses, subjects were not binned into separate groups according to variable percentiles. Instead, the models were fit with use of GEE, and the values of different percentiles for age at onset, GRS, and autoantibody titers were then substituted into the models to calculate predicted probabilities of C-peptide detection. Therefore, percentiles represent not an interval but a single value. Table 2 reports unadjusted odds ratios (ORs) and 95% CI of population-average parameter estimates from the GEE model.
Table 2.
Summary of GEE logistic regression results for prediction of C-peptide detection
| Coefficient | OR | 95% CI | P |
|---|---|---|---|
| Duration | 0.697 | (0.59, 0.82) | <0.0001 |
| Onset age | 1.070 | (1.01, 1.14) | 0.033 |
| GRS | 1.180 | (0.87, 1.62) | 0.294 |
| Duration * GRS | 0.895 | (0.84, 0.96) | <0.001 |
The effects of type 1 diabetes duration, age at onset, GRS, and their interactions on detectable C-peptide were modeled using GEE with assumption of binomial variance and logit link. GEE models were used to account for the mixed cohort of both cross-sectional and longitudinal samples. Type 1 diabetes duration and onset age were found to be significantly predictive of C-peptide detection at the mean GRS (OR 0.697, P < 0.0001, and OR 1.070, P < 0.033, respectively). GRS was not found to be a direct significant predictor of C-peptide detection with duration = 0 (P = 0.294). An interaction effect between disease duration and GRS (Duration * GRS) was found to be significantly predictive of C-peptide detection (OR 0.895, P < 0.001). OR with 95% CI and corresponding P values are reported for disease duration, GRS, and the interaction effect between duration and GRS. Sex was not a significant predictor of C-peptide detection (P = 0.443) and was not included in the model.
Associations of C-peptide detection with disease duration, age at onset, GRS, titers of each autoantibody, and all two-way interactions were assessed simultaneously by logistic regression with lasso penalty. Briefly, the data set was repeatedly partitioned in half for training and testing, and the probability of model inclusion for each predictor (including all two-way interaction effects) was calculated over 1,000 iterations. No variables were weighted in the analysis, and all coefficients with at least 50% inclusion frequency and their main effects are reported in Table 3. A GEE with binomial variance and logit link including these predictors was then used to model the log odds of C-peptide detection as follows:
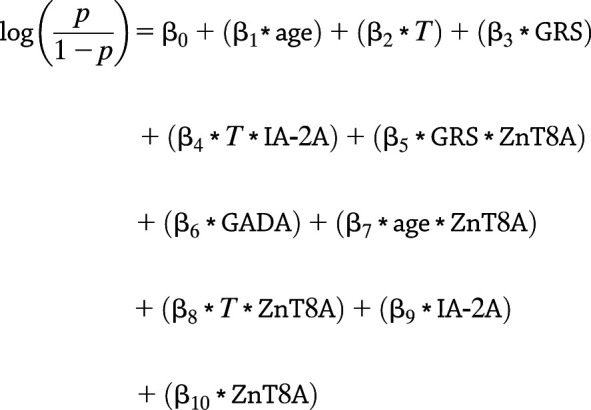 |
where p is the probability of detecting C-peptide, T is disease duration in years, and the intercept (β0) is −0.251. The remaining β-coefficients are given in exponentiated form (as ORs) (Table 3).
Table 3.
Summary of repeated train-test split results for selection of the most significant predictors of C-peptide detection
| Probability of inclusion | Coefficient from model | OR† | P | |
|---|---|---|---|---|
| Onset age | 1.00 | β1 | 1.110 | <0.0001 |
| Duration | 1.00 | β2 | 0.701 | <0.001 |
| GRS | 0.80 | β3 | 0.748 | 0.061 |
| Duration * IA–2A | 0.72 | β4 | 0.896 | 0.175 |
| GRS * ZnT8A | 0.65 | β5 | 1.380 | 0.057 |
| GADA | 0.58 | β6 | 1.230 | 0.128 |
| Onset age * ZnT8A | 0.52 | β7 | 1.080 | <0.001 |
| Duration * ZnT8A | 0.51 | β8 | 0.908 | 0.190 |
| IA–2A | 0.47 | β9 | 1.680 | 0.041 |
| ZnT8A | 0.39 | β10 | 0.625 | 0.198 |
Disease duration; onset age; GRS; titers for IA–2A, ZnT8A, and GADA; and all two-way interaction effects of predictor variables were tested for inclusion in the C-peptide model simultaneously with penalized logistic regression with repeated 10-fold cross validation for feature selection. The data set was repeatedly partitioned in half for training and testing, and the probability of each predictor (including all two-way interaction effects) being in the model was calculated over 1,000 iterations. All coefficients with at least 50% probability of inclusion and their main effects are reported. Interaction effects are denoted as two variables separated by an asterisk. The glmnet, version 3.0, package in R was used for penalized logistic regression and feature selection.
Regression coefficients (βi) are reported in exponentiated form as ORs, such that OR = eβ. The coefficient values correspond to the β values from the overall model formula.
Because the GEE method is based on quasi-likelihood theory, Akaike information criterion was not directly applicable for model selection. Instead we used quasi-likelihood information criterion (QIC), a modification of Akaike information criterion (27), to compare the two models (QIC results summarized in Supplementary Table 1). The calculation of QIC considers how well the model fits the data while favoring simpler models via a penalization term for the number of variables included (27). Statistical analyses were performed with R 3.6.1. Models were fit with use of the geeglm function in the geepack version 1.2 R package (28–30). When comparing the odds of detecting C-peptide between percentiles, a difference in estimated detection probability < 0.01 was considered to represent similar odds. All tests were two sided, with P < 0.05 considered significant. All CIs were estimated with the bootstrap method by sampling of subjects with replacement (sampling was performed 1,000 times).
Data and Resource Availability
The data sets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request. No applicable resources were generated or analyzed during the current study.
Results
Disease Duration, Age at Onset, GRS, and Their Interactions Affect Probability of Detection of C-Peptide in Individuals With Type 1 Diabetes
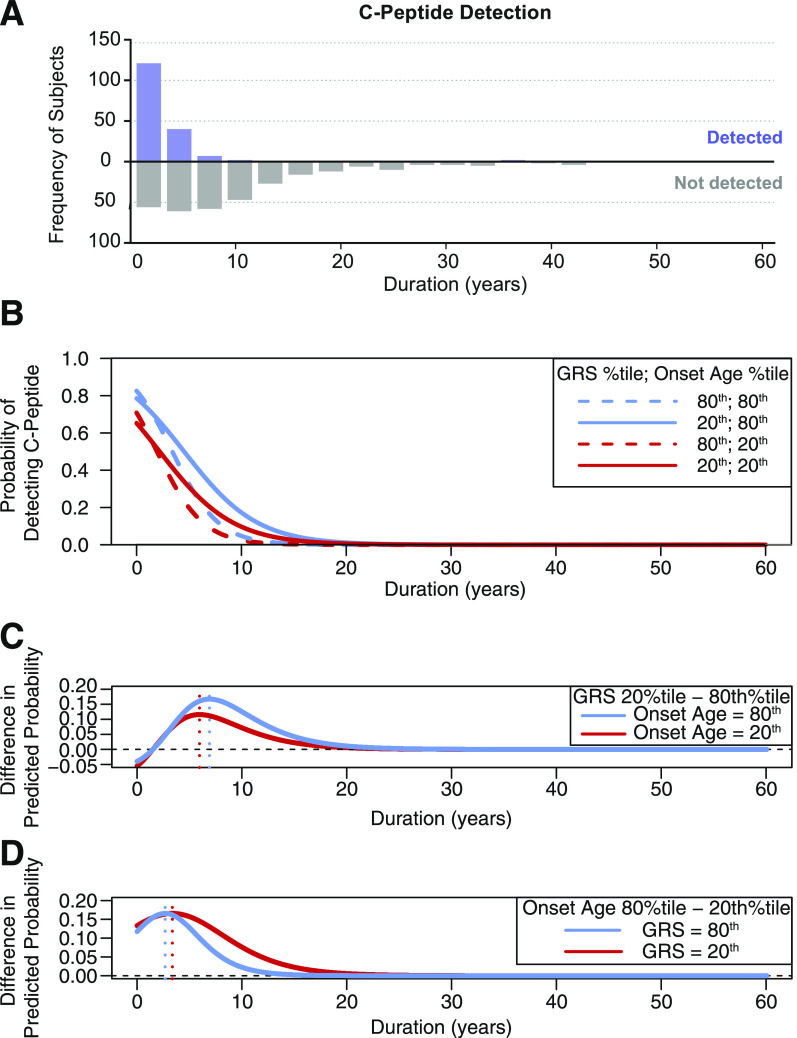
As expected, C-peptide detection decreased with increasing disease duration (Fig. 1A). We modeled the effect of type 1 diabetes duration, age at onset, and GRS on C-peptide detection using GEE (model 1). As expected, disease duration was negatively associated with odds of detecting C-peptide: at the mean GRS (0.278), each additional duration year decreased the odds of C-peptide detection (OR 0.697 [95% CI 0.59, 0.82], P < 0.0001) (Table 2). Older onset age was associated with increased odds of C-peptide detection (OR 1.070 [95% CI 1.01, 1.14], P = 0.033). We found a significant interaction effect between disease duration and GRS on C-peptide detection odds (OR 0.895 [95% CI 0.84, 0.96], P < 0.001). At increasing disease durations and fixed onset age, subjects at the 80th GRS quantile had substantially lower odds of C-peptide detection compared with subjects at the 20th GRS quantile (Fig. 1B). Although younger age at onset (Fig. 1B, red curves) was generally associated with lower odds of C-peptide detection compared with older age at onset (Fig. 1B, blue curves), onset age did not affect the rate of decline in the odds of C-peptide detection over time. In contrast, higher GRS (Fig. 1B, dashed curves) was associated with a more rapid decline in the odds of C-peptide detection compared with lower GRS (Fig. 1B, solid curves). Sex was not a significant predictor of C-peptide detection (P = 0.443) and was therefore not included in the model.
Figure 1.
A: The y-axis indicates the number of individuals with (upper histogram, purple bars) and without (lower histogram, gray bars) detectable C-peptide in the cohort. B–D: The odds of C-peptide detection were compared for the 20th (GRS = 0.260) and the 80th (GRS = 0.301) GRS quantiles within our cohort for comparison of lower versus higher genetic load. For comparison of younger versus older onset age, odds of C-peptide detection were compared for the 20th (6.0 years) and the 80th (15.7 years) onset age quantiles. B: Solid curves show the predicted probability of C-peptide detection according to disease duration when GRS is at the 20th percentile and onset age is set to either the 20th percentile (red solid curve) or the 80th percentile (blue solid curve). Dashed curves show the predicted probability of C-peptide detection according to disease duration when GRS is at the 80th percentile and onset age is set to either the 20th percentile (red dashed curve) or the 80th percentile (blue dashed curve). C: The difference in predicted probability of detectable C-peptide between the 20th and 80th GRS percentiles was plotted according to disease duration when onset age is at either the 20th percentile (red curve) or the 80th percentile (blue curve). The vertical dotted lines mark the duration at which the difference in predicted probability between the 20th and 80th GRS percentiles is the greatest (6.0 years when onset age is at the 20th percentile [red dotted vertical line] and 6.9 years when onset age is at the 80th percentile (blue dotted vertical line]). D: The difference in predicted probability of detectable C-peptide between the 20th and 80th onset age percentiles was plotted according to disease duration when GRS is at either the 20th percentile (red curve) or the 80th percentile (blue curve). The vertical dotted lines mark the duration at which the difference in predicted probability between the 20th and 80th onset age percentiles is the greatest (3.4 years when GRS is at the 20th percentile [red dotted vertical line] and 2.7 years when GRS is at the 80th percentile [blue dotted vertical line]). %tile, percentile.
At the median onset age (10.2 years), GRS had the greatest effect on the odds of C-peptide detection at 6.3 years’ disease duration (95% CI 1.14, 8.14), while longer disease durations (≥19.1 years [95% CI 10.99, 25.16]) yielded similar odds of C-peptide detection regardless of GRS. For comparison of the effect of low versus high GRS on C-peptide detection, the difference in predicted probability of detectable C-peptide between the 20th and 80th GRS percentiles was plotted according to disease duration with onset age set to either the 20th or the 80th percentile (Fig. 1C). When age at onset was at the 20th percentile (6.0 years), GRS had the greatest effect on C-peptide detection odds at 6.0 years’ disease duration (95% CI 0.33, 7.54), with lower GRS corresponding to higher odds of C-peptide detection (Fig. 1C, red curve). When age at onset was set to the 80th percentile (15.7 years), GRS had the greatest effect on C-peptide detection odds at 6.9 years’ duration (95% CI 1.98, 9.09) (Fig. 1C, blue curve).
At the median GRS (0.280), onset age had the largest effect on odds of C-peptide detection at 3.0 years’ disease duration (95% CI 2.42, 3.57), while longer disease durations (≥14.4 years [95% CI 10.39, 20.22]) yielded similar odds of C-peptide detection regardless of onset age. For comparison of the effect of older versus younger age at onset on C-peptide detection, the difference in predicted probability of detectable C-peptide between the 20th and 80th age at onset percentiles was plotted according to disease duration with GRS set to either the 20th or 80th percentile (Fig. 1D). When GRS was at the 20th percentile (0.260), age at onset had the greatest effect on C-peptide detection odds at 3.4 years’ duration (95% CI 2.72, 4.32), with younger age at onset corresponding to lower odds of C-peptide detection (Fig. 1D, red curve). When GRS was set to the 80th percentile (0.301), age at onset had the greatest effect on C-peptide detection odds at 2.7 years’ duration (95% CI 1.80, 3.22), with younger age at onset corresponding to lower odds of C-peptide detection (Fig. 1D, blue curve).
Incorporating GADA, IA–2A, and ZnT8A Titers Improves Model Fit
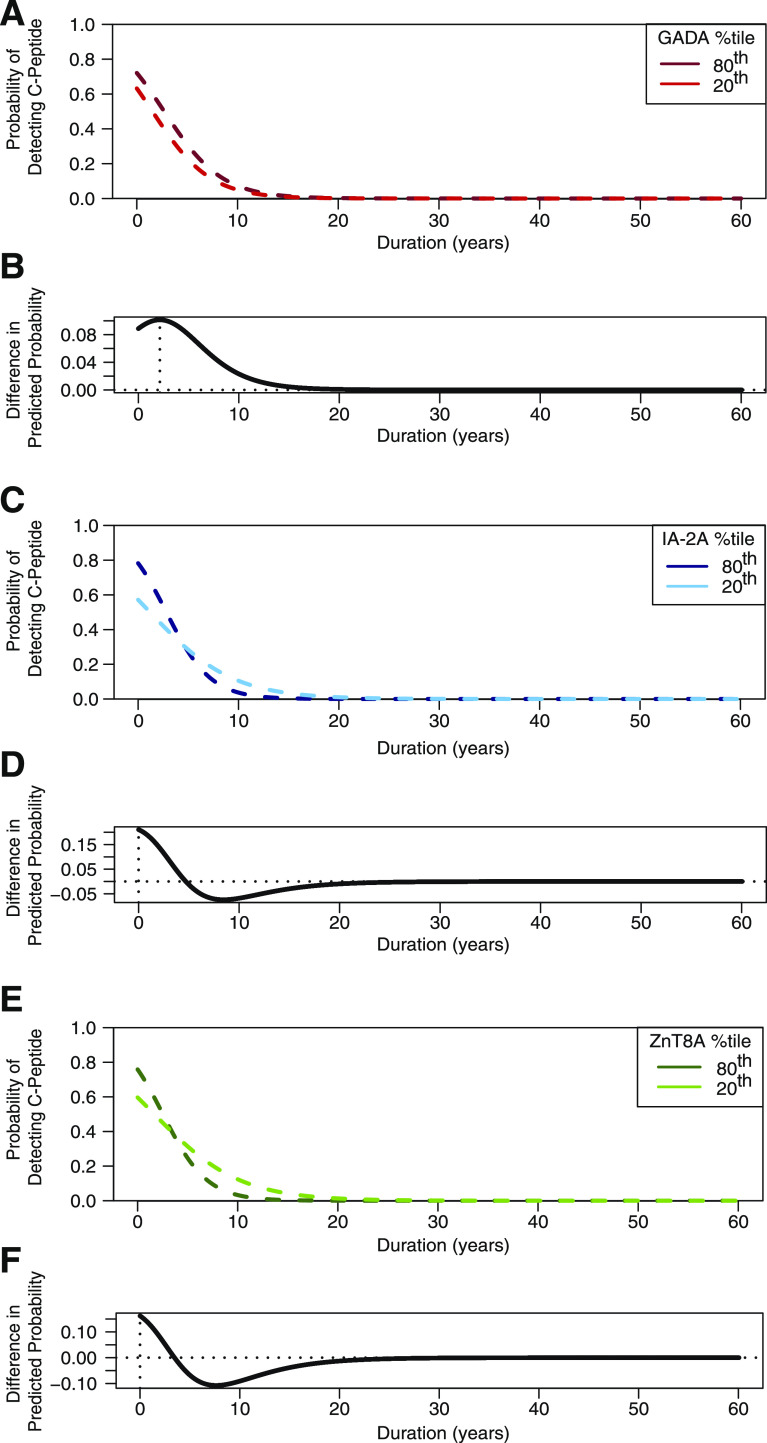
We next examined whether autoantibody titers and their interactions with disease duration, onset age, or GRS had an effect on C-peptide detection odds using logistic regression with a lasso penalty (model 2 [results summarized in Table 3]). For model 2, the most significant predictors of C-peptide detection were onset age and disease duration (each with 100% probability of model inclusion). As with model 1, older age at onset was associated with greater odds of C-peptide detection (OR 1.110, P < 0.0001) and longer duration was associated with decreased odds of C-peptide detection (OR 0.701, P < 0.001). GRS was the third most informative predictor in model 2, with 80% probability of inclusion. Higher GRS was associated with reduced odds of C-peptide detection (OR 0.748, P = 0.061). The predictive capacity of IA–2A and ZnT8A for C-peptide detection was dependent on their interactions with other variables (summarized in Table 3). In contrast, GADA titer was individually predictive of C-peptide detection, with higher titers associating with increased odds for C-peptide detection (OR 1.230, P = 0.128) (Fig. 2A), and had 58% probability of model inclusion. Although, overall, autoantibodies were less informative for C-peptide detection compared with onset age, duration, and GRS, their incorporation improved model fit compared with model 1 (Supplementary Table 1).
Figure 2.
Significant effect of GADA, IA–2A, and ZnT8A levels on the probability of detecting C-peptide. A, C, and E: Dashed lines show the fitted predicted probability of C-peptide detection (y-axis) according to type 1 diabetes duration. GRS is set to the median. B, D, and F: Difference in predicted probability between the 80th and 20th autoantibody titer percentiles was plotted according to disease duration. The vertical dotted line marks the duration at which the difference in predicted probability between the 80th and 20th autoantibody percentiles was the greatest. A: Fitted predicted probability of C-peptide detection for subjects at the 80th GADA titer percentile (dark-red dashed line) and at the 20th GADA titer percentile (light-red dashed line). B: Difference in predicted probability between the 80th and 20th GADA titer percentiles according to disease duration. The vertical dotted line marks the duration at which the difference in predicted probability between the 80th and 20th GADA percentiles is the greatest (2.1 years’ duration). C: Fitted predicted probability of C-peptide detection for subjects at the 80th IA–2A titer percentile (dark-blue dashed line) and at the 20th IA–2A titer percentile (light-blue dashed line). D: Difference in predicted probability between the 80th and 20th IA–2A titer percentiles according to disease duration. The vertical dotted line marks the duration at which the difference in predicted probability between the 80th and 20th IA–2A titer percentiles is the greatest (0 years’ duration). E: Fitted predicted probability of C-peptide detection for subjects at the 80th ZnT8A titer percentile (dark-green dashed line) and at the 20th ZnT8A titer percentile (light-green dashed line). F: Difference in predicted probability between the 80th and 20th ZnT8A titer percentiles according to disease duration. The vertical dotted line marks the duration at which the difference in predicted probability between the 80th and 20th ZnT8A titer percentiles is the greatest (0 years’ duration). %tile, percentile.
GADA titer had the largest effect on odds of C-peptide detection at disease duration of 2.1 years (95% CI 1.27, 2.97), while longer disease durations (≥12.6 years [95% CI 6.22, 17.87]) yielded similar odds of C-peptide detection regardless of GADA titer (Fig. 2B). While IA–2A and ZnT8A titers were not individually predictive of C-peptide detection, both interacted significantly with type 1 diabetes duration to predict C-peptide detection odds. At disease durations <4 years, higher IA–2A titers were associated with a higher probability of C-peptide presence followed by a more rapid decline in the probability of C-peptide detection compared with lower levels of IA–2A (Fig. 2C). This effect was most pronounced at disease onset (duration 0 years [95% CI 0, 6.55]) (Fig. 2D). Similarly, higher ZnT8A titers were associated with a higher probability of C-peptide detection close to disease onset (i.e., <2 years’ duration) followed by a more rapid decline in the probability of C-peptide detection compared with lower ZnT8A titers (Fig. 2E). As with IA–2A, the effect of ZnT8A titer on the probability of C-peptide detection was most pronounced at disease onset (duration 0 years [95% CI 0, 7.67]) (Fig. 2F).
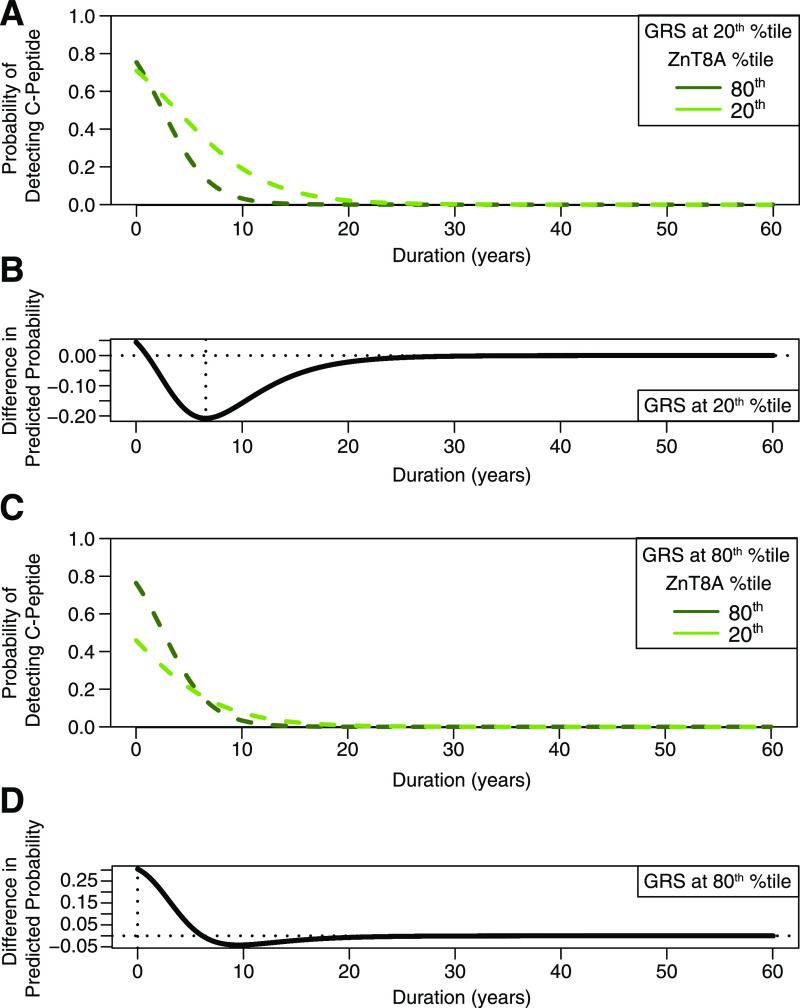
ZnT8A was the only autoantibody that had an interactive effect with GRS: at lower GRS values, the probability of C-peptide detection declined more rapidly when ZnT8A titer was high compared with when ZnT8A titer was low (Fig. 3A). At the 20th GRS percentile, ZnT8A titer had the greatest effect on the odds of C-peptide detection at 6.6 years’ duration (95% CI 0, 7.99) (Fig. 3B). In contrast, when GRS was high, the probability of detecting C-peptide declined with increased disease duration independent of ZnT8A titer (Fig. 3C). At the 80th GRS percentile, ZnT8A titer had the greatest effect on the odds of C-peptide detection at disease onset (duration 0 years [95% CI 0, 5.07]) (Fig. 3D).
Figure 3.
Interaction effect of ZnT8A titer, GRS, and type 1 diabetes duration on the probability of detecting C-peptide. For illustration of the effect of GRS on the association between ZnT8A titer and detectable C-peptide, the predicted probability of C-peptide detection was plotted with GRS set to the 20th percentile (A and B) and the 80th percentile (C and D) according to disease duration. A and C: Dashed lines show the fitted predicted probability of C-peptide detection (y-axis) at the 80th ZnT8A titer percentile (dark-green dashed line) and at the 20th ZnT8A titer percentile (light-green dashed line). B and D: Difference in predicted probability of C-peptide detection between the 80th and 20th ZnT8A titer percentiles according to disease duration. The vertical dotted line marks the duration at which the difference in predicted probability between the 80th and 20th ZnT8A titer percentiles is the greatest. A: Interaction effect between ZnT8A and type 1 diabetes duration when GRS is set to the 20th percentile. At the 20th GRS percentile, the probability of C-peptide detection declined more rapidly when ZnT8A titer was high (dark-green dashed line) compared with when ZnT8A titer was low (light-green dashed line). B: At the 20th GRS percentile, the difference in predicted probability between the 80th and 20th ZnT8A titer percentiles was the greatest at 6.6 years’ duration. C: Interaction effect between ZnT8A and type 1 diabetes duration when GRS is set to the 80th percentile. At the 80th GRS percentile, the probability of C-peptide detection declined with increased disease duration, regardless of ZnT8A titer. D: At the 80th GRS percentile, the difference in predicted probability between the 80th and 20th ZnT8A titer percentiles was the greatest at disease onset (i.e., 0 years). %tile, percentile.
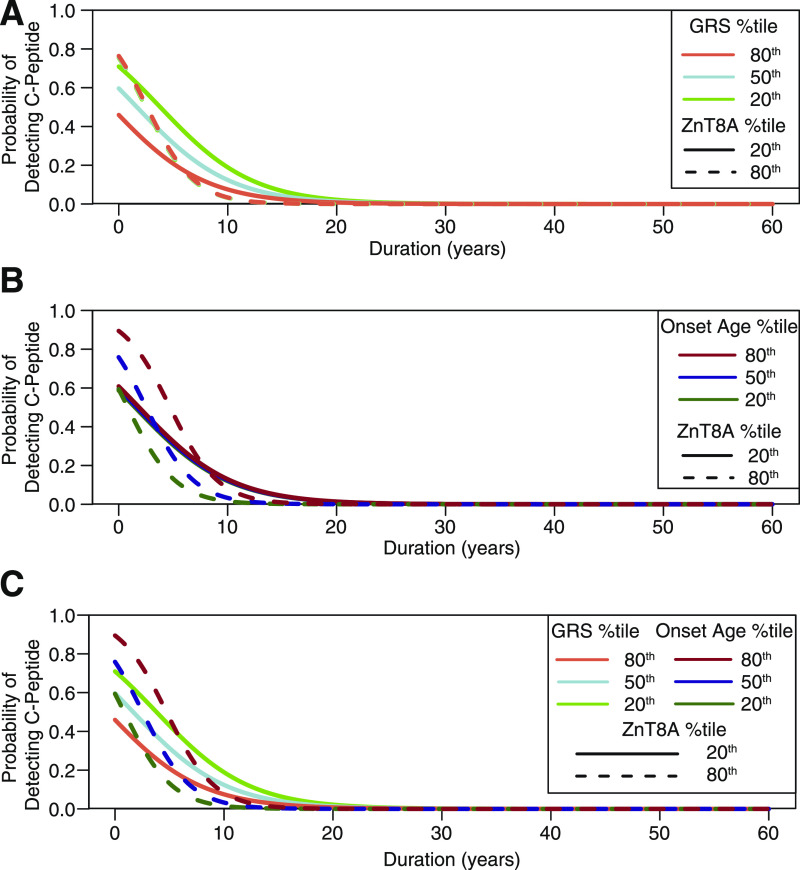
The effects of GRS and age at onset on the probability of C-peptide detection varied as a function of ZnT8A titer (Fig. 4). The effect of GRS on C-peptide detection probability was more pronounced when ZnT8A titer was low, whereas GRS had almost no effect on C-peptide detection when ZnT8A titer was high (Fig. 4A). Specifically, when ZnT8A titer was at the 20th percentile, the maximum difference in C-peptide detection probability between high and low GRS was 0.26 (at 1.6 years’ duration, 95% CI 0, 3.49). However, when ZnT8A titer was at the 80th percentile, the greatest difference in C-peptide detection probability between high and low GRS was only 0.014 (at 2.5 years’ duration, 95% CI 1.69, 3.27). In contrast with that of GRS, the effect of age at onset on C-peptide detection probability was more pronounced when ZnT8A titer was high and was diminished when ZnT8A titer was low (Fig. 4B). Specifically, when ZnT8A titer was at the 80th percentile, the maximum difference in C-peptide detection probability between younger (6.0 years) and older (15.7 years) onset age was 0.41 at 2.8 years’ duration (95% CI 2.02, 3.64). However, when ZnT8A titer was at the 20th percentile, the greatest difference in C-peptide detection probability between younger and older onset age was only 0.022 (at 1.7 years’ duration [95% CI 0, 3.57]). Overall, high ZnT8A titer was associated with a more rapid decline in the probability of C-peptide detection compared with low ZnT8A titer, and this association was modulated by age at type 1 diabetes onset (Fig. 4B and C, dashed curves). Low ZnT8A titer was associated with a more gradual decline in probability of C-peptide detection, and this association was modulated by GRS (Fig. 4A and C, solid curves).
Figure 4.
ZnT8A titer interacts with GRS and type 1 diabetes onset age to predict odds of C-peptide detection. A: The probability of C-peptide detection is plotted according to disease duration when GRS is set to the 80th (orange curves), 50th (blue curves), or 20th (green curves) percentile. Solid and dashed curves represent the probability of C-peptide detection when ZnT8A titer is at the 20th and 80th percentiles, respectively. The probability of C-peptide detection was sensitive to changes in GRS when ZnT8A titer was low (solid lines) but not when ZnT8A titer was high (dashed lines, which appear superimposed). B: The probability of C-peptide detection is plotted according to disease duration when onset age is set to the 80th (red curves), 50th (blue curves), or 20th (green curves) percentile. Solid and dashed curves represent the probability of C-peptide detection when ZnT8A titer is at the 20th and 80th percentiles, respectively. The probability of C-peptide detection was sensitive to changes in onset age when ZnT8A titer was high (dashed lines) but not when ZnT8A titer was low (solid lines, which appear superimposed). C: The probability of C-peptide detection is plotted according to disease duration with respect to ZnT8A titer, GRS, and age at disease onset. Dark dashed curves represent the probability of C-peptide detection when ZnT8A titer is high and age at onset is high (dashed dark-red curve), medium (dashed dark-blue curve), and low (dashed dark-green curve). Light solid curves represent the probability of C-peptide detection when ZnT8A titer is low and GRS is high (light-red/orange curve), medium (light-blue curve), and low (light-green curve). %tile, percentile.
QIC was then used for comparisons between model 1 (initial model without autoantibody titers) (Table 2) and model 2 (prediction model based on variable inclusion frequencies) (Table 3). Despite a penalty for the higher number of variables, model 2 (QIC = 334.6) was found to have a better fit versus model 1 (QIC = 359.2) (Supplementary Table 1).
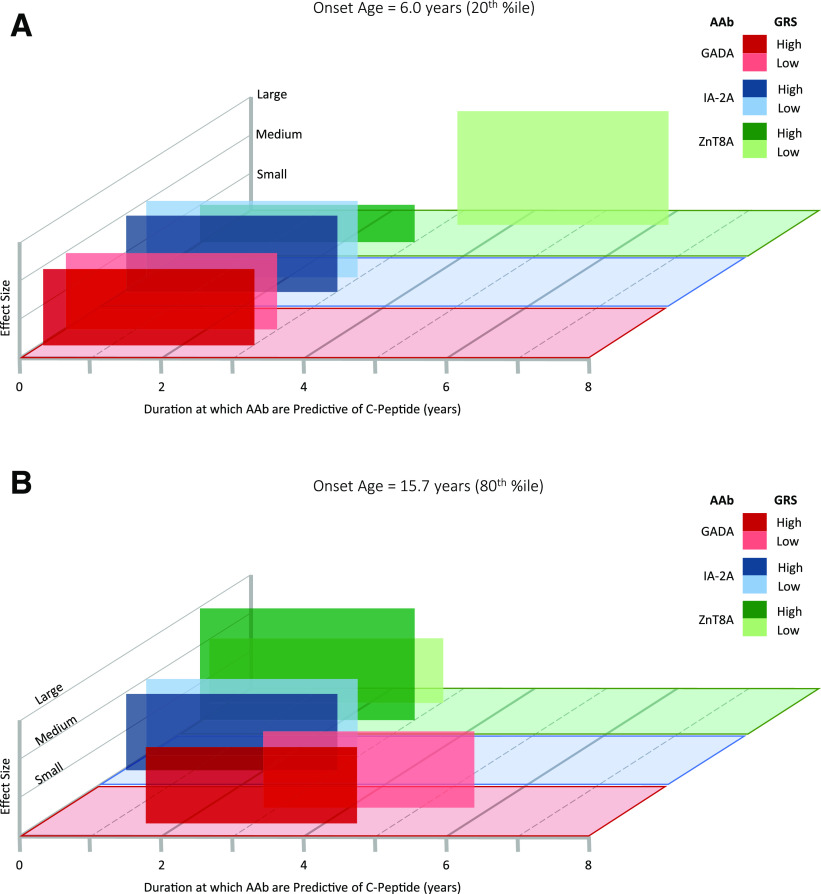
Overall, the most informative factors for C-peptide detection odds in our final model were onset age, duration, and GRS. However, inclusion of GADA, IA–2A, and ZnT8A titers improved model fit compared with model 1, which only included onset age, duration, and GRS (Supplementary Table 1). Both age at diagnosis and GRS influenced the duration at which each autoantibody was most informative (i.e., the duration at which the autoantibody effect on C-peptide was >95th percentile across all duration times) for C-peptide detection odds (Fig. 5). Specifically, when onset age was at the 20th percentile (6.0 years), both GADA and IA–2A levels were most informative for C-peptide detection from disease onset to 3 years’ duration, regardless of GRS (Fig. 5A). When GRS was high, ZnT8A titer was also most informative for C-peptide detection from 0 to 3 years’ duration, although the magnitude of this effect was small. In contrast, when GRS was low, ZnT8A titer was most informative for C-peptide detection at 3.2–6.2 years’ duration (Fig. 5A). When onset age was at the 80th percentile (15.7 years), both IA–2A and ZnT8A levels were most informative for C-peptide detection from disease onset to 3 years’ duration, regardless of GRS (Fig. 5B). GRS affected the duration at which GADA had the greatest effect on C-peptide detection odds: when GRS was high, GADA was most informative at 1.5–4.5 years’ duration. When GRS was low, GADA was most informative for C-peptide detection at 2.8–5.8 years’ duration (Fig. 5B).
Figure 5.
Duration at which autoantibodies (AAb) are most informative for C-peptide detection. Model representation illustrating the duration at which each autoantibody is most informative for C-peptide detection odds when age at disease onset is set to either the 20th percentile (6.0 years [A]) or the 80th percentile (15.7 years [B]). Boxes span the disease duration (x-axis) at which the autoantibody effect on C-peptide is >95th percentile across all duration times. The y-axis represents the effect size of each autoantibody on C-peptide detection odds. For illustration of how GRS modulates the duration at which autoantibodies are most informative in the model, each autoantibody is shown for when GRS is set to the 80th percentile and the 20th percentile. A: Duration at which GADA, IA–2A, and ZnT8A levels are informative for C-peptide detection odds when age at disease onset is set to the 20th percentile (6.0 years). At this younger onset age, GRS did not affect the duration interval at which GADA (dark- and light-red boxes) or IA–2A (dark- and light-blue boxes) were informative for C-peptide detection odds. GRS did affect the duration interval at which ZnT8A were informative for C-peptide detection odds: when GRS was high, ZnT8A titer was most informative for C-peptide detection odds from 0 to 3 years’ duration (dark-green box). When GRS was low, ZnT8A titer was most informative for C-peptide detection odds at 3.2–6.2 years’ duration (light-green box). B: Duration at which GADA, IA–2A, and ZnT8A levels are informative for C-peptide detection odds when age at disease onset is set to the 80th percentile (15.7 years). At this older onset age, GRS did not affect the duration interval at which IA–2A (dark- and light-blue boxes) or ZnT8A (dark- and light-green boxes) are informative for C-peptide detection odds. GRS did affect the duration interval at which GADA titer was most informative for C-peptide detection odds: when GRS was high, GADA titer was most informative at 1.5–4.5 years’ duration (dark-red box). When GRS was low, GADA titer was most informative for C-peptide detection odds at 2.8–5.8 years’ duration (light-red box). %tile, percentile.
Discussion
Within our cohort, the odds of detecting C-peptide following type 1 diabetes onset was best predicted by a combined model with incorporation of disease duration, age at disease onset, GRS, and titers for GADA, IA–2A, and ZnT8A. In accordance with previous studies, we found longer disease duration and younger age at onset to be associated with lower odds of C-peptide detection in subjects with type 1 diabetes (16,31,32).
An interaction effect between GRS and disease duration was found such that higher overall genetic load for type 1 diabetes was associated with a more rapid decline in the odds of C-peptide detection. High-risk alleles from the HLA class I (A*24) and class II (DR3 and DR4) loci have been associated with younger age at onset (33), and risk conferred by these alleles comprises a large component of GRS (22). Because younger age at onset has been associated with more rapid decline in residual β-cell function (13), higher GRS was expected to also result in reduced odds of C-peptide detection through its intrinsic link with onset age. Yet, in the current study, GRS and age at onset had different effects on C-peptide detection: younger onset age resulted in a downward shift of the predicted probability curve of C-peptide detection compared with older onset age, whereas higher GRS resulted in a steeper decline in C-peptide detection odds compared with lower GRS (Fig. 1B). Thus, although related through high-risk HLA associations, GRS and onset age exert differing effects on the odds of C-peptide detection over time.
GADA titer was directly related to the odds of C-peptide detection, and because GADA did not have any interaction effects with other variables, it may offer unfettered utility as a biomarker. In accordance with our findings, Ziegler and Bonifacio recently addressed the observation that GAD65 autoimmunity is associated with slower progression to clinical type 1 diabetes, speculating that GADA may be reflective of β-cell–protective mechanisms (34). Longitudinal studies are required to explore whether these purported mechanisms (or the enduring effects thereof) could extend beyond clinical onset to prolong the preservation of β-cell function.
The effects of IA–2A and ZnT8A titers on C-peptide detection were dependent on their interactions with either disease duration (IA–2A) or disease duration, GRS, and onset age (ZnT8A). Our results suggest that, following diagnosis, subjects with low or undetectable titers of IA–2A and ZnT8A may experience a slower decline in functional β-cell mass compared with subjects with high titers and, thus, a longer period during which they could benefit from clinical interventions aimed at preserving β-cell function. This is in agreement with previously reported findings that IA–2A and ZnT8A tend to emerge and peak closer to type 1 diabetes onset, typically following insulin autoantibodies and/or GADA seroconversion (35). Therefore, the presence of IA–2A and ZnT8A is thought to reflect a more advanced stage of disease and/or β-cell destruction. In support of this notion, Juusola et al. (36) found that among children and adolescents, ZnT8A presence at the time of diagnosis was associated with reduced C-peptide levels at 12, 18, and 24 months postonset compared with levels in ZnT8A-negative subjects.
ZnT8A was unique in that its relationship to C-peptide detection odds depended on its interaction with not only disease duration but also GRS and onset age. When ZnT8A titer was low, its effect on C-peptide detection odds was more sensitive to change in GRS and less sensitive to change in age at onset. When ZnT8A titer was high, its effect was more sensitive to change in age at onset versus GRS. Because ZnT8 was more recently identified as a type 1 diabetes autoantigen (37), fewer studies have longitudinal data on ZnT8A’s association with residual β-cell function as compared with GADA and IA–2A (38,39). Notwithstanding, associations have been uncovered of ZnT8A levels, HLA alleles, and age at disease onset (36,40,41). Although these associations are supported by our model, more studies are needed to untangle the complexities of these interaction effects on residual β-cell function.
In our model, age at diagnosis and GRS influenced the effect size and duration at which the autoantibodies were most indicative of C-peptide detection, and these factors had differing effects depending on autoantibody specificity, as summarized in Fig. 5. The duration at which GADA exerted its peak effect was sensitive to changes in GRS when onset age was older (∼16 years) (Fig. 5B, red). In contrast, GRS modulated the duration at which ZnT8A were most effective in the model but only when onset age was younger (∼6 years) (Fig. 5A, green). IA–2A’s effect was uniquely resilient to changes in onset age and GRS (Fig. 5A and B, blue). Notably, onset age and GRS influenced the magnitude of the effect of ZnT8A but not GADA or IA–2A. Hence, in terms of a clinical application, our model suggests that for individuals diagnosed with type 1 diabetes at a young age, GADA and IA–2A levels are most informative for C-peptide detection odds during the first 3 years following diagnosis, but thereafter, ZnT8A become the predominant marker, specifically among subjects with lower GRS. In older-onset subjects, IA–2A and ZnT8A levels are most informative within 3 years of diagnosis, while GADA are most informative slightly later, particularly among subjects with low GRS.
One limitation of our study is that our cohort is largely cross-sectional, and autoantibodies are known to fluctuate temporally (42). Since exogenous insulin therapy may increase autoantibody titers, insulin autoantibodies were excluded from this study. We aim to investigate the contribution of insulin autoantibodies to C-peptide detection odds in a longitudinal study that includes samples collected at diagnosis. Another caveat is the use of random serum C-peptide. Stimulated C-peptide tests, although more accurate in assessing β-cell function, are invasive, costly, and time-consuming. On the other hand, implementing fasting restrictions on patients would be difficult, and assuring compliance would not be feasible. Random C-peptide has been shown to correlate with mixed-meal tolerance test outcomes (43) and, hence, was our most practical option. Additionally, the GRS applied here was found to be less robust for assessment of risk in non-Caucasian populations (22); thus, our analysis was restricted to subjects with genetically imputed European ancestry. Efforts to generate population-specific GRS models are underway (44), and incorporation of such GRS into our C-peptide model would theoretically allow for modeling among ethnically diverse cohorts.
Collectively, our data support a model wherein age at onset, GRS, and islet autoantibody titers can be used to project the loss of C-peptide over the course of type 1 diabetes duration. Although it requires validation in an independent longitudinal cohort, we envisage that this model, based on underlying genetic composition and minimally invasive serological markers, may be used to project C-peptide trajectory and thereby inform clinical trial inclusion criteria for a precision medicine approach.
Article Information
Acknowledgments. The authors thank Ezio Bonifacio (Dresden University of Technology) for valuable feedback and constructive comments.
Funding. These studies were supported by funding from the National Institutes of Health (P01 AI042288) and JDRF (1-SRA-2019-764-A-N and 2-PDF-2016-207-A-N) and endowments from the American Diabetes Association, the McJunkin Family Charitable Foundation, and the Jeffrey Keene Family Professorship.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. M.D.W. researched the data and wrote the manuscript. R.B. analyzed the data and wrote the manuscript. D.J.P., C.R.G., and K.M.M. researched the data and reviewed and edited the manuscript. A.L.P., A.M., and M.J.H. contributed to discussion and reviewed and edited the manuscript. S.C. analyzed the data and reviewed and edited the manuscript. D.A.S., T.M.B., and M.A.A. contributed to discussion and reviewed and edited the manuscript. C.H.W. conceived of the study and wrote the manuscript. C.H.W. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.13516808.
References
- 1.Insel RA, Dunne JL, Atkinson MA, et al. Staging presymptomatic type 1 diabetes: a scientific statement of JDRF, the Endocrine Society, and the American Diabetes Association. Diabetes Care 2015;38:1964–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winkler C, Krumsiek J, Buettner F, et al. Feature ranking of type 1 diabetes susceptibility genes improves prediction of type 1 diabetes. Diabetologia 2014;57:2521–2529 [DOI] [PubMed] [Google Scholar]
- 3.Oram RA, Patel K, Hill A, et al. A type 1 diabetes genetic risk score can aid discrimination between type 1 and type 2 diabetes in young adults. Diabetes Care 2016;39:337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel KA, Oram RA, Flanagan SE, et al. Type 1 diabetes genetic risk score: a novel tool to discriminate monogenic and type 1 diabetes. Diabetes 2016;65:2094–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Redondo MJ, Geyer S, Steck AK, et al.; Type 1 Diabetes TrialNet Study Group . A type 1 diabetes genetic risk score predicts progression of islet autoimmunity and development of type 1 diabetes in individuals at risk. Diabetes Care 2018;41:1887–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmer JP, Fleming GA, Greenbaum CJ, et al. C-peptide is the appropriate outcome measure for type 1 diabetes clinical trials to preserve β-cell function: report of an ADA workshop, 21–22 October 2001. Diabetes 2004;53:250–264 [DOI] [PubMed] [Google Scholar]
- 7.Steffes MW, Sibley S, Jackson M, Thomas W. β-Cell function and the development of diabetes-related complications in the diabetes control and complications trial. Diabetes Care 2003;26:832–836 [DOI] [PubMed] [Google Scholar]
- 8.Oram RA, Jones AG, Besser REJ, et al. The majority of patients with long-duration type 1 diabetes are insulin microsecretors and have functioning beta cells. Diabetologia 2014;57:187–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shields BM, McDonald TJ, Oram R, et al.; TIGI Consortium . C-peptide decline in type 1 diabetes has two phases: an initial exponential fall and a subsequent stable phase. Diabetes Care 2018;41:1486–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu MG, Keenan HA, Shah HS, et al. Residual β cell function and monogenic variants in long-duration type 1 diabetes patients. J Clin Invest 2019;129:3252–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wasserfall C, Nick HS, Campbell-Thompson M, et al. Persistence of pancreatic insulin mRNA expression and proinsulin protein in type 1 diabetes pancreata. Cell Metab 2017;26:568–575.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damond N, Engler S, Zanotelli VRT, et al. A map of human type 1 diabetes progression by imaging mass cytometry. Cell Metab 2019;29:755–768.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmer JP. C-peptide in the natural history of type 1 diabetes. Diabetes Metab Res Rev 2009;25:325–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komulainen J, Knip M, Lounamaa R, et al.; The Childhood Diabetes in Finland Study Group . Poor beta-cell function after the clinical manifestation of type 1 diabetes in children initially positive for islet cell specific autoantibodies. Diabet Med 1997;14:532–537 [DOI] [PubMed] [Google Scholar]
- 15.Decochez K, Keymeulen B, Somers G, et al.; Belgian Diabetes Registry . Use of an islet cell antibody assay to identify type 1 diabetic patients with rapid decrease in C-peptide levels after clinical onset: Belgian Diabetes Registry. Diabetes Care 2000;23:1072–1078 [DOI] [PubMed] [Google Scholar]
- 16.Mortensen HB, Swift PGF, Holl RW, et al.; Hvidoere Study Group on Childhood Diabetes . Multinational study in children and adolescents with newly diagnosed type 1 diabetes: association of age, ketoacidosis, HLA status, and autoantibodies on residual beta-cell function and glycemic control 12 months after diagnosis. Pediatr Diabetes 2010;11:218–226 [DOI] [PubMed] [Google Scholar]
- 17.Wenzlau JM, Walter M, Gardner TJ, et al. Kinetics of the post-onset decline in zinc transporter 8 autoantibodies in type 1 diabetic human subjects. J Clin Endocrinol Metab 2010;95:4712–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrone A, Spoletini M, Zampetti S, et al.; Immunotherapy Diabetes (IMDIAB) Group . The PTPN22 1858T gene variant in type 1 diabetes is associated with reduced residual β-cell function and worse metabolic control. Diabetes Care 2008;31:1214–1218 [DOI] [PubMed] [Google Scholar]
- 19.Fløyel T, Brorsson C, Nielsen LB, et al. CTSH regulates β-cell function and disease progression in newly diagnosed type 1 diabetes patients. Proc Natl Acad Sci U S A 2014;111:10305–10310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roshandel D, Gubitosi-Klug R, Bull SB, et al.; DCCT/EDIC Research Group . Meta-genome-wide association studies identify a locus on chromosome 1 and multiple variants in the MHC region for serum C-peptide in type 1 diabetes. Diabetologia 2018;61:1098–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKeigue PM, Spiliopoulou A, McGurnaghan S, et al. Persistent C-peptide secretion in type 1 diabetes and its relationship to the genetic architecture of diabetes. BMC Med 2019;17:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perry DJ, Wasserfall CH, Oram RA, et al. Application of a genetic risk score to racially diverse type 1 diabetes populations demonstrates the need for diversity in risk-modeling. Sci Rep 2018;8:4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wasserfall C, Montgomery E, Yu L, et al. Validation of a rapid type 1 diabetes autoantibody screening assay for community-based screening of organ donors to identify subjects at increased risk for the disease. Clin Exp Immunol 2016;185:33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cortes A, Brown MA. Promise and pitfalls of the ImmunoChip. Arthritis Res Ther 2011;13:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Onengut-Gumuscu S, Chen WM, Burren O, et al.; Type 1 Diabetes Genetics Consortium . Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet 2015;47:381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das S, Forer L, Schönherr S, et al. Next-generation genotype imputation service and methods. Nat Genet 2016;48:1284–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan W. Akaike’s information criterion in generalized estimating equations. Biometrics 2001;57:120–125 [DOI] [PubMed] [Google Scholar]
- 28.Yan J. geepack: yet another package for generalized estimating equations. R News 2002;2/3:12–14 [Google Scholar]
- 29.Yan J, Fine J. Estimating equations for association structures. Stat Med 2004;23:859–874; discussion 875–877, 879–880 [DOI] [PubMed] [Google Scholar]
- 30.Halekoh U, Højsgaard S, Yan J. The R package geepack for generalized estimating equations. J Stat Softw 2006;15:1–11 [Google Scholar]
- 31.Greenbaum CJ, Beam CA, Boulware D, et al.; Type 1 Diabetes TrialNet Study Group . Fall in C-peptide during first 2 years from diagnosis: evidence of at least two distinct phases from composite Type 1 Diabetes TrialNet data. Diabetes 2012;61:2066–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barker A, Lauria A, Schloot N, et al. Age-dependent decline of β-cell function in type 1 diabetes after diagnosis: a multi-centre longitudinal study. Diabetes Obes Metab 2014;16:262–267 [DOI] [PubMed] [Google Scholar]
- 33.Noble JA, Valdes AM. Genetics of the HLA region in the prediction of type 1 diabetes. Curr Diab Rep 2011;11:533–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ziegler A-G, Bonifacio E. Why is the presence of autoantibodies against GAD associated with a relatively slow progression to clinical diabetes? Diabetologia 2020;63:1665–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziegler A-G, Nepom GT. Prediction and pathogenesis in type 1 diabetes. Immunity 2010;32:468–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Juusola M, Parkkola A, Härkönen T, et al.; Childhood Diabetes in Finland Study Group . Positivity for zinc transporter 8 autoantibodies at diagnosis is subsequently associated with reduced β-cell function and higher exogenous insulin requirement in children and adolescents with type 1 diabetes. Diabetes Care 2016;39:118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wenzlau JM, Juhl K, Yu L, et al. The cation efflux transporter ZnT8 (Slc30A8) is a major autoantigen in human type 1 diabetes. Proc Natl Acad Sci U S A 2007;104:17040–17045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petersen JS, Dyrberg T, Karlsen AE, et al.; The Canadian-European Randomized Control Trial Group . Glutamic acid decarboxylase (GAD65) autoantibodies in prediction of beta-cell function and remission in recent-onset IDDM after cyclosporin treatment. Diabetes 1994;43:1291–1296 [DOI] [PubMed] [Google Scholar]
- 39.Lan MS, Wasserfall C, Maclaren NK, Notkins AL. IA-2, a transmembrane protein of the protein tyrosine phosphatase family, is a major autoantigen in insulin-dependent diabetes mellitus. Proc Natl Acad Sci U S A 1996;93:6367–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wenzlau JM, Frisch LM, Hutton JC, Fain PR, Davidson HW. Changes in zinc transporter 8 autoantibodies following type 1 diabetes onset: the Type 1 Diabetes Genetics Consortium Autoantibody Workshop. Diabetes Care 2015;38(Suppl. 2):S14–S20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salonen KM, Ryhänen S, Härkönen T, Ilonen J, Knip M; Finnish Pediatric Diabetes Register . Autoantibodies against zinc transporter 8 are related to age, metabolic state and HLA DR genotype in children with newly diagnosed type 1 diabetes. Diabetes Metab Res Rev 2013;29:646–654 [DOI] [PubMed] [Google Scholar]
- 42.Decochez K, Tits J, Coolens JL, et al. High frequency of persisting or increasing islet-specific autoantibody levels after diagnosis of type 1 diabetes presenting before 40 years of age: the Belgian Diabetes Registry. Diabetes Care 2000;23:838–844 [DOI] [PubMed] [Google Scholar]
- 43.Hope SV, Knight BA, Shields BM, Hattersley AT, McDonald TJ, Jones AG. Random non-fasting C-peptide: bringing robust assessment of endogenous insulin secretion to the clinic. Diabet Med 2016;33:1554–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Onengut-Gumuscu S, Chen W-M, Robertson CC, et al.; SEARCH for Diabetes in Youth; Type 1 Diabetes Genetics Consortium . Type 1 diabetes risk in African-ancestry participants and utility of an ancestry-specific genetic risk score. Diabetes Care 2019;42:406–415 [DOI] [PMC free article] [PubMed] [Google Scholar]