Abstract
SARS-CoV-2 genetic material is detectable in the faeces of a considerable part of COVID-19 cases and hence, in municipal wastewater. This fact was confirmed early during the spread of the COVID-19 pandemic and prompted several studies that proposed monitoring its incidence by wastewater. This paper studies the fate of SARS-CoV-2 genetic material in wastewater treatment plants using RT-qPCR with a two-fold goal: i) to check its presence in the water effluent and in the produced sludge and ii) based on the understanding of the virus particles fate, to identify the most suitable spots for detecting the incidence of COVID-19 and monitor its evolution. On the grounds of the affinity of enveloped virus towards biosolids, we hypothesized that the sludge line acts as a concentrator of SARS-CoV-2 genetic material. Sampling several spots in primary, secondary and sludge treatment at the Ourense (Spain) WWTP in 5 different days showed that, in effect, most of SARS-CoV-2 particles cannot be detected in the water effluent as they are retained by the sludge line. We identified the sludge thickener as a suitable spot for detecting SARS-CoV-2 particles thanks to its higher solids concentration (more virus particles) and longer residence time (less sensitive to dilution caused by precipitation). These findings could be useful to develop a suitable strategy for early warning of COVID-19 incidence based on WWTP monitoring.
Keywords: SARS-CoV-2, Covid-19, Wastewater, Wastewater treatment plant, Incidence monitoring, Sludge
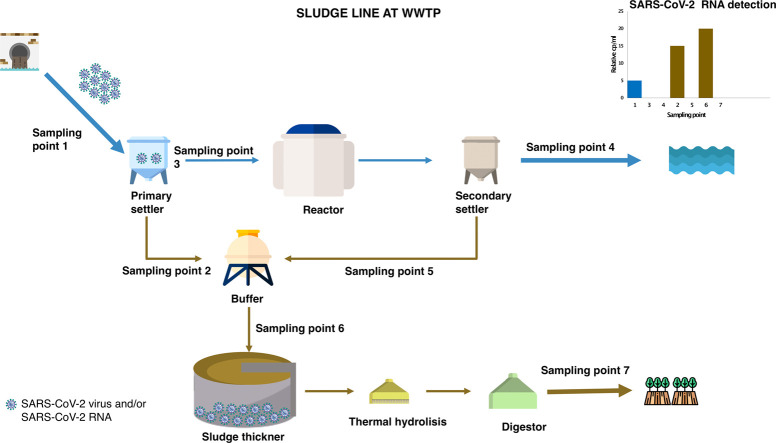
Graphical abstract
1. Introduction
Municipal wastewater can be a vector for the spread of viral diseases, especially viruses that are transmitted through the faecal-oral route. There are numerous reports on detection of viruses in wastewater treatment plants, including Norovirus, Sapovirus, Hepatitis A virus, Adenovirus, Poliovirus or Enterovirus among others (Ehlers et al., 2005; Sassi et al., 2018; Symonds et al., 2016; Taboada-Santos et al., 2020). Current knowledge regarding the behaviour of SARS-CoV-2 in wastewater is very limited, although its RNA has been detected in faeces of symptomatic (Holshue et al., 2020; Wölfel et al., 2020) and asymptomatic individuals (Park et al., 2020; Tang et al., 2020), and in municipal wastewater in different countries, starting in the Netherlands (Medema et al., 2020). Accounts of SARS-CoV-2 in wastewater quickly followed in several countries, e.g. Spain (Randazzo et al., 2020a, Randazzo et al., 2020b), Italy (La Rosa et al., 2020), USA (Sherchan et al., 2020; Wu et al., 2020), Japan (Haramoto et al., 2020) and Turkey (Kocamemi et al., 2020a) among others. Although it is assumed that the enveloped viruses are not excreted in high concentrations and that their survival in water is limited, there is little experimental evidence to confirm these hypotheses in wastewater.
Municipal wastewater constitutes a complex matrix which includes suspended solid materials, colloidal and dissolved biodegradable organic matter, nutrients, pathogens, etc. In wastewater treatment plants (WWTP) most of solids are separated from the water to the so-called sludge line. Many WWTPs have a first stage of solid separation (primary settler) and a secondary settler where the activated sludge is separated from the clarified water. Finally, the two types of sludge (primary and secondary) are concentrated in the thickener from where they are sent to the sludge treatment unit.
Pollutants of hydrophobic nature are mostly retained in primary or secondary sludge (Prado et al., 2014), a phenomenon described for some viruses already many decades ago (Wellings et al., 1976). It is known that enveloped viruses, due to the presence of a lipid bilayer surrounding the protein capsid, have a different affinity to non-enveloped viruses, with a greater tendency to adsorb to solid and/or colloidal particles (Ye et al., 2016). This was experimentally proved to occur for two enveloped viruses: Murine coronavirus MHV (murine hepatitis virus) and Pseudomonas phage ϕ6 (Ye et al., 2016). Therefore, most probably, SARS-CoV-2 and particles thereof are indeed hydrophobic and, accordingly, they would be associated to the solids and/or colloidal material. Yet, most of the current literature concerning SARS-CoV-2 or its genetic material in wastewater deals with their presence in the water phase and very little attention has been paid to their fate in the sludge line, with the exception of the work carried out by Peccia et al. (2020) and Kocamemi et al. (2020).
Another aspect of concern for water boards and utilities is the potential transmission of SARS-CoV-2 in WWTP and their effluents. Actually, what is known so far about the transmission of SARS-CoV-2 is not particularly worrisome, given that WWTP operation is already intended to avoid the transmission of potential pathogens present in wastewater, although only a fraction of WWTP count with tertiary treatment enabling effective virus removal. Being sludge and the water effluents the main outflows from a WWTP, it is important to ascertain whether SARS-CoV-2 can be detected in these streams. However, whether the absence or detection of SARS-CoV-2 genetic material can lead to any conclusion on the infectivity of water or sludge remains to be ascertained.
Hence, this manuscript pays special attention to the sludge in WWTPs. The sludge treatment is very heterogeneous in WWTPs as large plants often feature anaerobic digestion treatment, usually at moderate temperatures (35–40 °C) and long residence times (10–20 days), which would help to inactivate the possible viral load. Furthermore, thermal hydrolysis or similar thermal treatment of sludge is increasingly common in large WWTPs and provides effective inactivation of viral inputs. In contrast, in smaller plants sludge can receive a mere heat drying treatment before being shipped to an authorized manager, or even just centrifugation to reduce its water content.
The goal of this manuscript is two-fold: first, to shed some light on the fate of SARS-CoV-2 in WWTPs by examining the detection of its genetic material along the water and sludge lines, a topic which needs to be further investigated (Foladori et al., 2020) and second to check whether the hydrophobic nature of SARS-CoV-2 make the sludge line, and particularly the thickened sludge, as a suitable spot for monitoring its incidence in the WWTP catchment area.
2. Materials and methods
2.1. Wastewater and sludge samples
Wastewater and sludge samples were taken from Ourense WWTP in north-western Spain (characteristics and sampling points in Table 1 ). The 250 mL samples were taken twice a week from April 6 to April 21, 2020, kept at 4 °C before being sent in less than 24 h to the Universidade de Santiago de Compostela (USC) facilities to be concentrated.
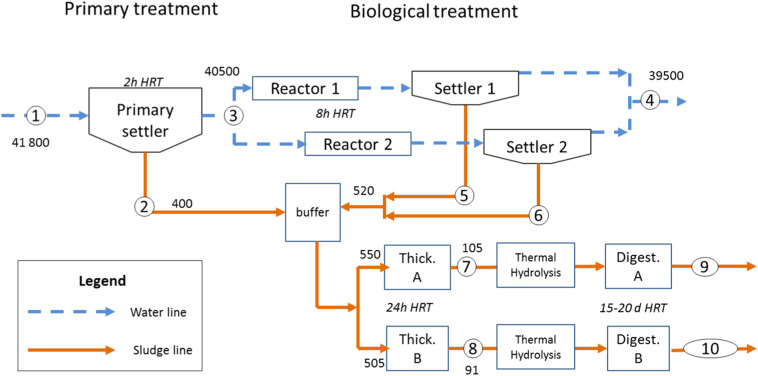
Fig. 1.
Simplified description of Ourense WWTP layout and sampling points (circled numbers corresponding to Table 2 labels) in the water and sludge line. Measured flowrates (in m3/d) during a representative dry weather period (here taken as 21st April) are shown next to the corresponding streams and nominal residence times are next to the corresponding unit.
Table 1.
Wastewater treatment plant characteristics and sampling points for wastewater and sludge.
| Ourense WWTP characteristics | |
|---|---|
| Nominal size | 350,000 population equivalent |
| Pretreatment | Grit and sand separator, oil and grease removal |
| Wastewater treatment | Primary settler Biological SBR for COD and N removal Chemical removal of P Microfiltration of secondary effluent |
| Sludge treatment | Gravity thickening and centrifuge, thermal hydrolysis, anaerobic digestion |
| Sampling points water line | Wastewater after grit removal; outflow primary settler; outflow secondary treatment |
| Sampling points sludge line | Primary sludge, secondary, thickened mixed sludge, digested sludge |
For the water line, 24-h composite samples were taken and characterised following standard methods (APHA, 2017) in terms of pH, conductivity, total and volatile suspended solids (Standard Methods 2540-D), chemical oxygen demand (spectrophotometry, Standard Methods 5220-D), ammonium (spectrophotometry, Standard Methods 4500-F), nitrate (kit equivalent to DIN 38405-9 (DIN, 2011)), total nitrogen (kit equivalent to ISO 11905-, (International Organization for Standardization, 1997)) and total phosphorus (spectrophotometry, Standard Methods 4500-P). For the sludge lines, point samples were taken the pH and total and volatile suspended solids were measured.
2.2. Sample processing
Water samples were concentrated by ultrafiltration. To do so, 100 mL were first gently centrifuged to remove large particles at 4600 ×g during 30 min to avoid blocking the filtration membrane. Supernatants were concentrated by filtration using Amicon 15 mL 10 K centrifugal devices to a 1 mL sample. Then, 10 mL of phosphate buffer saline pH 7.4 (PBS, with composition 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 and 1.8 mM KH2PO4) were added and the sample was concentrated again to 1 mL.
Samples taken from the sludge lines were concentrated by precipitation with polyethylene glycol (PEG) according to Hjelmsø et al. (2017). Then, 1:8 (v/v) of Glycine buffer (0.05 M glycine, 3% beef extract) was added to 50 mL of sludge, incubated for 2 h at 4 °C to detach viruses bound to organic material. Samples were then centrifuged at 8000 ×g during 30 min and filtered through a 0.45 μm polyethersulfone (PES) membrane to remove eukaryotic and prokaryotic cells. Then, viruses were precipitated by adding 1:5 (v/v) of PEG 8000 (80 g/L) and NaCl (17.5 g/L) during an overnight shaking (150 rpm) at 4 °C and a centrifugation at 13,000 ×g during 90 min. Samples were then resuspended in PBS buffer pH 7.4 and stored at −80 °C for further analysis.
Concentration control was performed with bacteriophage MS2, by inoculating each sample with 250 μL the virus (5.5 × 106 plaque forming units/mL) before starting the concentration process.
2.3. RNA extraction and RT-qPCR detection
RNA extraction and RT-qPCR was carried out at the department of Microbiology of Complexo Hospitalario Universitario de Vigo. Nucleic acid extraction from both water and sludge concentrated samples was performed using MicrolabStarlet IVD using the STARMag 96 × 4 Universal Cartridge Kit (Seegene, Seoul, South Korea) according to manufacturer specifications.
Viral RNA was detected and quantified by a one-step multiplex RT-qPCR Allplex system™ 2019-nCoV (Seegene, Seoul, South Korea). The assay is designed to detect RNA-dependent polymerase (RdRP) gene and nucleocapsid (N) gene, both specific to SARS-CoV-2, and a region conserved in the E gene of the structural protein envelope for the detection of pan-Sarbecoviruses including SARS-CoV-2. The test uses internal RNA control for sample preparation and control of the PCR amplification process. For the RT-PCR, the CFX96 system was used ™ (Bio-Rad Laboratories, Hercules, CA, USA). The analysis of the results was performed using the specific Seegene viewer software 2019-nCoV. Following the kit manufacturer instruction, amplification and extraction efficiency is evaluated by the internal RNA control Ct, which should be in the 25–30 range (indicating inhibition otherwise).
In parallel, a SARS-CoV-2 EDX standard (Bio-Rad, Hercules, CA) containing synthetic RNA transcripts of five SARS-CoV-2 genetic targets (genes E, N, ORF1ab, RdRp and S) of known concentration was used to establish a linear regression curve and obtain the concentration in copies/mL.
2.4. RNA quantification
RNA was quantified by an external standard curve built as follows. The Allplex™2019-nCoV (STARlet) assay was run in triplicate using 2 × 104 DNA copies/mL and 2-serial dilutions of SARS-CoV-2 standard at starting concentrations of 2 × 105 DNA copies/mL to 31.25 DNA copies/mL. Additional dilutions were tested yielding 40, 20 and 2 DNA copies/mL. The sensitivity of the assay was considered as the lowest concentration of SARS-CoV-2 giving a positive result for any target. The EDX SARS-CoV-2 Standard (Exact Diagnostics, TX, USA) containing 200,000 copies/mL of synthetic RNA transcripts from five gene targets (E, N, ORF1ab, RdRP and S Genes of SARS-CoV-2) was used. The limit of detection resulted in 4 genome copies/reaction for Allplex™2019-nCoV assay after nucleic acid extraction with STARlet system (Hamilton (US). The gene target N was preferably used for quantification and the gene target RdRP to confirm detection and specificity. To continuously assure the correct performance of the quantification, in each PCR plate several controls are included, namely, a positive control (that coincides with one of the values of the standard), a negative control (non-template control) and endogenous controls provided by the RNA extraction kit.
The triplicate external standard curve for target N led to the following equation: ln(copies/mL) = −0.7121 (0.044) Ct + 31.913 (1.54) where bracketed values are the standard errors of the slope and intercept. Confidence intervals of single sample quantifications were estimated by error propagation from the calibration curve error. From the calibration curve, we estimated a limit of quantification of 25 copies/mL in the concentrated sample which corresponds to 1.0 copy/mL in preconcentrated raw wastewater.
3. Results
3.1. Wastewater physicochemical characterisation
The characterisation of the sample in terms of chemical oxygen demand (COD), suspended solids and, for water samples, total nitrogen is shown in Table 2 . It can be seen how the influent has a very variable composition, which is most probably caused by precipitation events.
Table 2.
Physicochemical characterisation of samples (full characterisation in Supplementary materials). Number in brackets after sampling spot corresponds to Fig. 1. NT = total nitrogen, TSS = total suspended solids, VSS = volatile suspended solids. NA = Non-available sample.
| Sampling spot | 6-Apr | 7-Apr | 14-Apr | 16-Apr | 21-Apr | |
|---|---|---|---|---|---|---|
| Influent flowrate | m3/d | 72,778 | 41,235 | 54,349 | 54,398 | 41,876 |
| Influent (1) | COD (mgCOD/L) | 1211 | 164 | 613 | 535 | 387 |
| TSS (mg/L) | 990 | 100 | 280 | 350 | 230 | |
| TN (mgN/L) | 56 | 21 | 52 | 38 | 37 | |
| Outflow primary (3) | COD (mgCOD/L) | 101 | 53 | 212 | 160 | 59 |
| TSS (mg/L) | 51 | 75 | 128 | 107 | 33 | |
| TN (mgN/L) | 35 | 26 | 41 | 38 | 30 | |
| Treated effluent(4) | COD (mgCOD/L) | 17 | 13 | 23 | 19 | 13 |
| TSS (mg/L) | 2.5 | 6.0 | 8.6 | 3.0 | 1.4 | |
| TN (mgN/L | 8.5 | 8.0 | 9.8 | 11 | 6.7 | |
| Primary sludge (2) | TSS (g/L) | 5.54 | 1.50 | 11.4 | 14.5 | 9.33 |
| VSS (g/L) | 2.93 | 1.10 | 8.89 | 10.2 | 6.22 | |
| Biological sludge A (5) | TSS (g/L) | 6.15 | 6.87 | 9.00 | 5.43 | 7.63 |
| VSS (g/L) | 4.29 | 5.12 | 7.00 | 4.05 | 5.60 | |
| Biological sludge B (6) | TSS (g/L) | 6.34 | 5.64 | 6.68 | 5.67 | 6.31 |
| VSS (g/L) | 4.29 | 4.52 | 4.33 | 4.12 | 4.59 | |
| Thickened sludge A (7) | TSS (g/L) | 28.2 | 27.3 | 26.1 | 20.8 | NA |
| VSS (g/L) | 20.8 | 20.1 | 20.2 | 15.1 | NA | |
| Thickened sludge B (8) | TSS (g/L) | 28.7 | 16.9 | 25.1 | 19.2 | 17.5 |
| VSS (g/L) | 20.6 | 19.8 | 19.1 | 13.8 | 10.5 | |
| Digested sludge A (9) | TSS (g/L) | 43.8 | 41.4 | 41.4 | 44.0 | 43.7 |
| VSS (g/L) | 23.8 | 21.7 | 22.2 | 23.6 | 23.6 | |
| Digested sludge B (10) | TSS (g/L) | 40.8 | 38.7 | 37.9 | 38.5 | 41.2 |
| VSS (g/L) | 23.0 | 21.1 | 20.2 | 21.5 | 22.5 |
Indeed, the influence of rain on sewerage streams is a hurdle to the use of the WWTP influent as an epidemiological indicator if influent dilution prevents the virus particles concentration from being compared on a day-to-day basis. However, the sludge streams tend to have a more steady content of biosolids as it is related to the mass flow of solids and COD entering the WWTP, which mostly depends on the population served by the WWTP. Properly operated settlers would lead to sludge streams with steady solid contents despite incoming rain in sewerage. Among the different sludge streams in a WWTP, solid concentration in thickeners seems to be much more stable than the characteristics of the primary influent thanks to characteristics of the thickening stage and to the comparably larger retention time.
3.2. Presence of SARS-CoV-2 genetic material in water and sludge
A total of 15 samples of water and 35 samples of sludge collected from April 6 to April 21 (2020) were tested for the presence of SARS-CoV-2 RNA. All samples were positive for our internal control, bacteriophage MS2, although with variable efficiency (36.0 ± 15.4% for wastewater and 32.1 ± 15.8% for sludge). Such a variation has been described before (Petterson et al., 2015; Silva-Sales et al., 2020), and it is probably caused by the physicochemical complexity and variability of sewage samples during the different sampling days.
The interpretation of results in Table A1 (Supplementary material) was based on considering a positive sample when the cycle threshold took place below cycle 40, for either RNA-dependent polymerase (RdRP) and nucleocapsid (N) SARS-CoV-2 specific genes. The analysis was repeated to confirm the quantification in these cases. This strategy is coincident with the results reported by Hur et al. (2020) who found, in average, higher Ct values for N gene than for RdRp. The kit used in this work was also evaluated by Sung et al. (2020) who suggest that the possible detection of only one of the RdRP or N genes is caused either by the matrix of the sample or a consequence of the lower limit of detection inherent to multiplex analyses. In samples where only gene E (characteristic of pan-Sarbecoviruses) was detected, the analysis was repeated for confirmation, usually leading to a confirmed negative result.
For most entries in Table A1 (Supplementary material), both specific genes were detected, suggesting that non-specific amplification is unlikely although full confirmation would require sequencing of the amplified fragments.
3.3. Quantification of SARS-CoV-2 particles in water and sludge line WWTP
Following the presence of SARS-CoV-2 genetic material along the WWTP allows inferring its fate in the different processes. For that purpose, samples were quantified using commercial standards, as explained in Section 2, leading to the results shown in Table 3 .
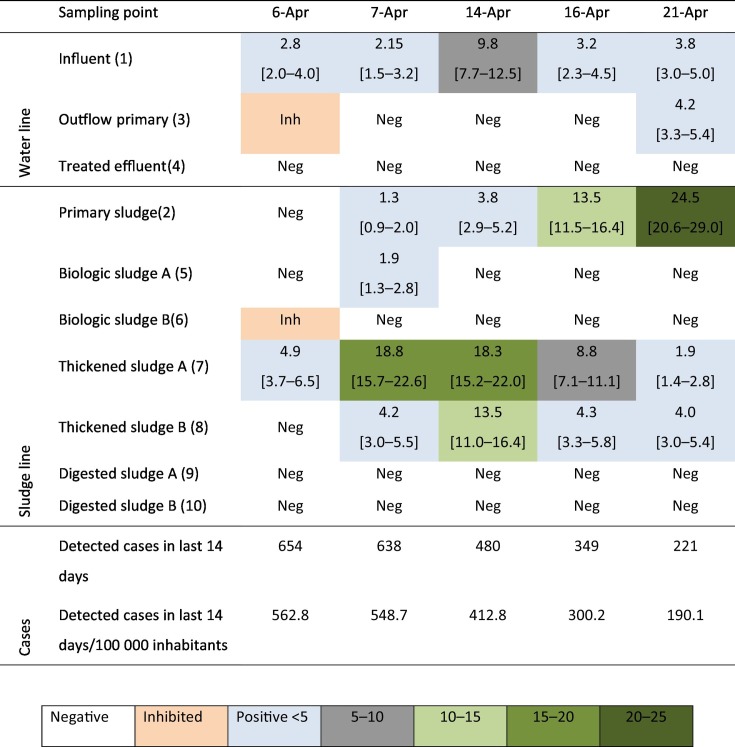
Table 3.
Quantification of SARS-CoV-2 genetic material at several WWTP sampling spots. (Pos = positive; Neg = negative; Inh = inhibited). All values in copies/mL, α = 0.1 confidence interval in square brackets.
4. Discussion
The level of SARS-CoV-2 RNA detected in the influent was low in general, at most up to 9 copies/mL (cp/mL) rising to more than 20 cp/mL in some sludge samples. The titers in raw wastewater are in the low range compared to other results currently reported. For instance, titers were reported from 2 to 20 cp/mL in Istanbul (Turkey) area WWTPs (Kocamemi et al., 2020a), 5–20 cp/mL in a WWTP in Massachussets (Wu et al., 2020) increasing to 100–1000 cp/mL in Murcia region, Spain (Randazzo et al., 2020b) and 50–3000 cp/mL in Paris, France (Wurtzer et al., 2020). Foladori et al. (2020), in probably the first review of the work rapidly being reported on SARS-CoV-2 content in wastewater, point out to very different levels of loadings in WWTP, with a range of 20 copies/L to 3·106 copies/L. According to these authors, many factors are identified as responsible for this large range, including the occurrence of rainwater in combined sewers, the high variability of the viral load in faeces (up to 4 orders of magnitude) and methodological differences in sampling, concentration and quantification. Such a large variability in the detected virus loadings suggests that a widely standardised methodology would be required to obtain more comparable figures. Still, it is likely that SARS-CoV-2 surveillance should be based on time-comparison of measurements from the same location and that comparisons between different sites will be inaccurate.
4.1. Fate of SARS-CoV-2 particles in the WWTP
SARS-CoV-2 RNA was systematically detected in the influent to the primary settler (up to 9 cp/mL) but not in the secondary treatment effluent. Regarding SARS-CoV-2 transmission, the effluent is probably safe for reuse and discharge to water bodies, as other studies have also reported (Randazzo et al., 2020b; Rimoldi et al., 2020) given the absence of SARS-CoV-2 genetic material in the effluent although infectivity tests were not carried out. Another potential mechanism of transmission of airborne viruses is the production of aerosol in secondary treatment, particularly if aeration is provided by horizontal rotors or surface turbines (Gotkowska-Płachta et al., 2013; Sánchez-Monedero et al., 2008). Given the rare occurrence of SARS-CoV-2 RNA in the inflow to the secondary treatment, the potential of dispersion by aerosols created during aeration is low.
As for the sludge line, it appears that SARS-CoV-2 RNA is mainly retained at the primary settler (up to 24 cp/mL) and only detected in one occasion in the biological sludge, which suggest that, as hypothesized, the virus particles have a higher affinity for the sludge. On the 5th sampling day (April 21st) it can be seen, that virus RNA is detected in the outflow of the primary settler. This is consistent with a partition between the water and the sludge line and would correspond with the high loading detected in the primary sludge (24 cp/mL).
From these results it is concluded that the virus particles are mostly diverted to the sludge line although RNA degradation may also contribute to their absence in the water line beyond the primary settler. Interestingly, the virus RNA concentration increases in the thickeners which have a longer retention time, (approximately 24 h) and a higher solid content. In the interpretation of results from Table 3, it must be understood that a WWTP should not be seen as a “plug-flow” but as a complex system with many recirculation streams to the plant inlet. As an example, it is seen that the virus RNA detected in the thickeners on the 5th sampling day is lower than in the remaining streams and in contrast with the very high loading of the primary sludge. However, the thickeners integrate the incoming concentration of the previous 24 h and therefore are not a complete correspondence to the primary sludge of the same day. Interestingly, the solid concentration in the thickened sludge is significantly lower on April 21st compared to the rest of days (less than 30% of the solid concentration of the remaining days), which can be related to maintenance and washing and may explain the low virus RNA loadings.
No genetic material was detected in the digested sludge, which is surely related both to the severe temperature undergone during thermal hydrolysis and to the long residence time in the anaerobic digesters. Therefore, the results confirm the safety of the sludge after thermal treatment and anaerobic digestion. However, in smaller WWTPs is only treated by volume reduction methods with no thermal treatment, the safety of sludge disposal remains to be verified.
4.2. Primary and/or thickened sludge as indicators of incidence
It is seen that the concentration of SARS-CoV-2 genetic material is systematically higher in some sludge sampling spots (particularly at primary sludge and thickened sludge) compared to the influent samples (Table 3). This result confirms one of the hypotheses of this work, namely that the affinity of virus particles for biosolids would divert the genetic material of SARS-CoV-2 towards the sludge line. Peccia et al. (2020) also pointed out at the sludge line as suitable for indication or detection of SARS-CoV-2 in the population. Although the number of samples analysed in this work is limited and replication in other WWTPs is required, this finding suggests that detecting COVID-19 incidence in the population in the sludge might have a higher sensitivity than in the wastewater… In this WWTP, the primary settler and the sludge thickeners would act in effect as “concentrators” of SARS-CoV-2 genetic material. Furthermore, the retention times in sludge thickeners (~24 h) are usually higher than in primary settlers (~1–2 h). In small WWTPs with limitations of staff or instrumental (in particular flow-proportional automatic samplers), these spots could replace or complement the influent as the preferred spot for sampling.
This higher retention time results in dampening the potential variations of SARS-CoV-2 particles in wastewater in an even more effective way than taking composite samples. Such a buffering is not helpful when the phenomenon to be monitored has fast dynamics, but in the case of COVID-19 population incidence, the desired monitoring dynamics would be in the order of days, making both the thickened and the primary sludge, suitable sampling spots even if used for early detection of outbreaks (Orive et al., 2020). Further investigation will be needed to establish the validity of this work in smaller WWTPs in absence of a primary settler, where all the sludge would be collected after aerobic biological treatment with larger solid retention times.
5. Conclusions
The affinity of SARS-CoV-2 by biosolids was seen to govern to a large extent its fate in WWTPs by being associated to sludge streams. As a consequence, SARS-CoV-2 genetic material was not detected in the WWTP effluent, indicating its probable safety which could be confirmed with infectivity tests. The combined treatment of thermal hydrolysis and anaerobic digestion also prevented the detection of SARS-CoV-2 in sludge leaving the plant. The primary sludge and mostly the thickened sludge showed higher and steadier concentrations, which suggests that COVID-19 incidence could be monitored in the sludge line, possibly in addition to the raw wastewater sampled in the influent. Longer residence times and higher solid concentrations in sludge thickeners would make it a robust sampling spot, which merits being further investigated.
CRediT authorship contribution statement
Sabela Balboa: Methodology, Investigation, Sample processing, Data analysis, Writing - Original Draft.
Miguel Mauricio-Iglesias: Conceptualization, Data Analysis, Writing - Original Draft.
Santiago Rodriguez: Resources, Writing - Review & Editing.
Lucía Martínez-Lamas: Investigation, Sample processing, Data analysis, Writing - Original Draft.
Francisco J. Vasallo: Investigation, Sample processing, Supervision; Writing - Review & Editing.
Benito Regueiro: Supervision; Writing - Review & Editing.
Juan M. Lema: Conceptualization, Funding acquisition; Supervision; Writing - Review & Editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Sabela Balboa, Miguel Mauricio-Iglesias and Juan M. Lema belong the following programs co-funded by ERDF (EU): the CRETUS Strategic Partnership (ED431E 2018/01) and the Galician Competitive Research Group (ED431C2017/029).
Editor: Damia Barcelo
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2021.145268.
Appendix A. Supplementary data
Supplementary material
References
- APHA . American Public Health Association; American Water Works Association, Water Environment Federation: 2017. Standard Methods for the Examination of Water and Wastewater. [Google Scholar]
- DIN, 2011. 38405-9:2011-09 German Standard Methods for Examination of Water, Waste Water and Sludge - Anions (Group D) - Part 9: Spectrometric Determination of Nitrate (D 9).
- Ehlers M.M., Grabow W.O.K., Pavlov D.N. Detection of enteroviruses in untreated and treated drinking water supplies in South Africa. Water Res. 2005;39:2253–2258. doi: 10.1016/j.watres.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743:140444. doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotkowska-Płachta A., Filipkowska Z., Korzeniewska E., Janczukowicz W., Dixon B., Gołaś I., Szwalgin D. Airborne microorganisms emitted from wastewater treatment plant treating domestic wastewater and meat processing industry wastes. CLEAN – Soil, Air, Water. 2013;41:429–436. doi: 10.1002/clen.201100466. [DOI] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737:140405. doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmsø M.H., Hellmér M., Fernandez-Cassi X., Timoneda N., Lukjancenko O., Seidel M., Elsässer D., Aarestrup F.M., Löfström C., Bofill-Mas S., Abril J.F., Girones R., Schultz A.C. Evaluation of methods for the concentration and extraction of viruses from sewage in the context of metagenomic sequencing. PLoS One. 2017;12:1–17. doi: 10.1371/journal.pone.0170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L.A., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur K.H., Park K., Lim Y., Jeong Y.S., Sung H., Kim M.N. Evaluation of four commercial kits for SARS-CoV-2 real-time reverse-transcription polymerase chain reaction approved by emergency-use-authorization in Korea. Front. Med. 2020 doi: 10.3389/fmed.2020.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Organization for Standardization, 1997. ISO 11905-1:1997 Water Quality—Determination of Nitrogen—Part 1: Method Using Oxidative Digestion With Peroxodisulfate.
- Kocamemi B.A., Kurt H., Sait A., Sarac F., Saatci A.M., Pakdemirli B. 2020. SARS-CoV-2 Detection in Istanbul Wastewater Treatment Plant Sludges. medRxiv 2020.05.12.20099358. [DOI] [Google Scholar]
- Kocamemi, B.A., Kurt, H., Hacıoglu, S., Yaralı, C., Saatci, A.M., Pakdemirli, B., Affiliations, 2020a. First Data-set on SARS-CoV-2 Detection for Istanbul Wastewaters in Turkey. medRxiv. doi: 10.1101/2020.05.03.20089417 [DOI]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Orive G., Lertxundi U., Barcelo D. Early SARS-CoV-2 outbreak detection by sewage-based epidemiology. Sci. Total Environ. 2020;732:139298. doi: 10.1016/j.scitotenv.2020.139298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Lee C.-W., Park D.-I., Woo H.-Y., Cheong H.S., Shin H.C., Ahn K., Kwon M.-J., Joo E.-J. Detection of SARS-CoV-2 in fecal samples from patients with asymptomatic and mild COVID-19 in Korea. Clin. Gastroenterol. Hepatol. 2020 doi: 10.1016/j.cgh.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petterson S., Grøndahl-Rosado R., Nilsen V., Myrmel M., Robertson L.J. Variability in the recovery of a virus concentration procedure in water: implications for QMRA. Water Res. 2015;87:79–86. doi: 10.1016/j.watres.2015.09.006. [DOI] [PubMed] [Google Scholar]
- Prado T., Gaspar A.M.C., Miagostovich M.P. Detection of enteric viruses in activated sludge by feasible concentration methods. Braz. J. Microbiol. 2014;45:343–349. doi: 10.1590/s1517-83822014000100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Cuevas-Ferrando E., Sanjuán R., Domingo-Calap P., Sánchez G., Sanjuan R., Domingo-Calap P., Sanchez G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. Int. J. Hyg. Environ. Health. 2020;230 doi: 10.1016/j.ijheh.2020.113621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Ferrando E.C., Simon P., Allende A., Sanchez G. SARS-CoV-2 RNA titers in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C., Moja L., Gismondo M.R., Salerno F. Presence and vitality of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744 doi: 10.1101/2020.05.01.20086009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Monedero M.A., Aguilar M.I., Fenoll R., Roig A. Effect of the aeration system on the levels of airborne microorganisms generated at wastewater treatment plants. Water Res. 2008;42:3739–3744. doi: 10.1016/j.watres.2008.06.028. [DOI] [PubMed] [Google Scholar]
- Sassi H.P., Tuttle K.D., Betancourt W.Q., Kitajima M., Gerba C.P. Persistence of viruses by qPCR downstream of three effluent-dominated rivers in the Western United States. Food Environ. Virol. 2018;10:297–304. doi: 10.1007/s12560-018-9343-7. [DOI] [PubMed] [Google Scholar]
- Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana. USA. Sci. Total Environ. 2020;743:104743. doi: 10.1016/j.phrs.2020.104743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Sales M., Martínez-Puchol S., Gonzales-Gustavson E., Hundesa A., Gironès R. High prevalence of rotavirus a in raw sewage samples from Northeast Spain. Viruses. 2020;12:318. doi: 10.3390/v12030318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H., Han M.G., Yoo C.K., Lee S.W., Chung Y.S., Park J.S., Kim M.N., Lee H., Hong K.H., Seong M.W., Lee K.H., Chun S., Lee W.G., Kwon G.C., Min W.K. Nationwide external quality assessment of SARS-CoV-2 molecular testing. South Korea. Emerg. Infect. Dis. 2020 doi: 10.3201/EID2610.202551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds E.M., Sinigalliano C., Gidley M., Ahmed W., McQuaig-Ulrich S.M., Breitbart M. Faecal pollution along the southeastern coast of Florida and insight into the use of pepper mild mottle virus as an indicator. J. Appl. Microbiol. 2016;121:1469–1481. doi: 10.1111/jam.13252. [DOI] [PubMed] [Google Scholar]
- Taboada-Santos A., Rivadulla E., Paredes L., Carballa M., Romalde J., Lema J.M. Comprehensive comparison of chemically enhanced primary treatment and high-rate activated sludge in novel wastewater treatment plant configurations. Water Res. 2020;169:115258. doi: 10.1016/j.watres.2019.115258. [DOI] [PubMed] [Google Scholar]
- Tang A., Tong Z., Wang H., Dai Y., Li K., Liu J., Wu W., Yuan C., Yu M., Li P., Yan J. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child. China. Emerg. Infect. Dis. 2020 doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellings F.M., Lewis A.L., Mountain C.W. Demonstration of solids-associated virus in wastewater and sludge. Appl. Environ. Microbiol. 1976;31:354–358. doi: 10.1128/aem.31.3.354-358.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Gu X., Lee W.L., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Moniz K., Erickson T., Chai P., Thompson J., Alm E. 2020. SARS-CoV-2 Titers in Wastewater Are Higher Than Expected From Clinically Confirmed Cases. medRxiv 2020.04.05.20051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.-M., Moulin L. 2020. Time Course Quantitative Detection of SARS-CoV-2 in Parisian Wastewaters Correlates With COVID-19 Confirmed Cases. medRxiv 2020.04.12.20062679. [DOI] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material