Wood et al. use EM to visualize cytonemes, the specialized filopodia that mediate cell–cell signaling. Cytonemes of Drosophila cells have regions defined by oscillating diameters and contents that include ER, mitochondria, and ribosomes. Cytonemes contact target cells by penetrating into invaginations of target cell membranes.
Abstract
Cytonemes are specialized filopodia that mediate paracrine signaling in Drosophila and other animals. Studies using fluorescence confocal microscopy (CM) established their general paths, cell targets, and essential roles in signaling. To investigate details unresolvable by CM, we used high-pressure freezing and EM to visualize cytoneme structures, paths, contents, and contacts. We observed cytonemes previously seen by CM in the Drosophila wing imaginal disc system, including disc, tracheal air sac primordium (ASP), and myoblast cytonemes, and identified cytonemes extending into invaginations of target cells, and cytonemes connecting ASP cells and connecting myoblasts. Diameters of cytoneme shafts vary between repeating wide (206 ± 51.8 nm) and thin (55.9 ± 16.2 nm) segments. Actin, ribosomes, and membranous compartments are present throughout; rough ER and mitochondria are in wider proximal sections. These results reveal novel structural features of filopodia and provide a basis for understanding cytoneme cell biology and function.
Introduction
Cytonemes are thin, actin-based filopodia that distribute morphogen signaling proteins in developing tissues (Kornberg, 2014a). They directly link signal-producing and signal-receiving cells in the Drosophila wing imaginal disc (Chen et al., 2017; González-Méndez et al., 2017), abdominal histoblast nest (Bischoff et al., 2013), and gonadal stem cell niche (Rojas-Ríos et al., 2012), as well as in zebrafish neural plate (Stanganello et al., 2015; Mattes et al., 2018), chick limb bud (Sanders et al., 2013), and human embryonic stem cells (Junyent et al., 2020). Signaling by morphogen proteins, such as Hedgehog (Hh), decapentaplegic (Dpp; a bone morphogenic protein [BMP] family member), wingless (Wg/Wnt), and branchless (Bnl; a fibroblast growth factor family member) is dependent on cytonemes and the contacts they make with target cells (Kornberg, 2017). Despite the prevalence and critical roles of cytonemes, their ephemeral nature and wispy structure have limited their physical characterization.
The contacts cytonemes make with target cells share both structural and functional attributes with neuronal synapses. The contacts are <40 nm, close enough for complementary fragments of GFP at the surface of presynaptic and postsynaptic cells to reconstitute (Roy et al., 2014; Huang et al., 2019). The contacts are constituted with proteins that also function at neuronal synapses, are excitatory, and are glutamatergic (Huang et al., 2019; Roy et al., 2014); they are sites where signals exchange between transmitting and recipient cells; and they are essential for signaling (Mattes et al., 2018; Roy et al., 2014; González-Méndez et al., 2017). Further parallels include the growth cone filopodia of extending axons, which involve signaling proteins and receptors and involve signaling molecules such as Wg/Wnts, Hhs, and BMPs (Zou and Lyuksyutova, 2007). These parallels between neuronal synapses and cytoneme contacts encouraged us to ask whether ultrastructural analysis might reveal further similarities.
EM studies of several types of membrane extensions have been reported. These include studies of thin, actin-based extensions, such as transzonal projections (Baena and Terasaki, 2019), tunneling nanotubes (TNTs; Rustom et al., 2004; Sartori-Rupp et al., 2019; Alarcon-Martinez et al., 2020), and axonal growth cone filopodia (Bastiani and Goodman, 1984). Serial transmission EM (TEM) analysis of grasshopper neuron growth cones identified filopodia inserting deep into invaginations of target cell membranes (Bastiani and Goodman, 1984). These burrowing extensions induce formation of coated pits and vesicles in the cell membranes. In mouse ovarian follicles, transzonal projections of granulosa cells contact the oocyte through gap junctions, and they have been proposed to facilitate adherens junction–mediated paracrine signaling to drive cumulus cell differentiation (Baena and Terasaki, 2019). Other granulosa cells not in contact with the oocyte project filopodia that end by inserting into invaginations of neighboring granulosa cells without membrane fusion.
The TNT term refers to a diverse set of cell protrusions that have been characterized almost exclusively in adherent cultured cells (Dupont et al., 2018). Like cytonemes, they are actin-rich cytoplasmic cell–cell connections, and because they have been distinguished mostly by their lack of adherence to matrix or plastic growth substrates (Dupont et al., 2018; McCoy-Simandle et al., 2016), it is not clear if they are a type of filopodia distinct from cytonemes (Yamashita et al., 2018). EM analysis of TNTs in cultured neuronal cell lines reveals individual TNTs extending in parallel bundles held together by N-cadherin linkages. TNTs have outward bulging sections with mitochondria and other membranous compartments. TNTs were originally defined as open-ended, but close-ended TNTs that burrow into invaginations of target cells have also been observed (Sartori-Rupp et al., 2019). A scanning electron microscope study of pericytes surrounding adult mouse retinal capillaries identified individual TNTs with gap junctions at closed-ended foot-like processes (Alarcon-Martinez et al., 2020). Membrane nanotubes have also been characterized in virus-infected T cells. In this context, viral transfer to uninfected cells is mediated by membrane nanotubes and is receptor and contact dependent (Sowinski et al., 2008).
Another type of cellular extension involved in signaling is the vertebrate primary cilium, which localizes specific receptors and signal transduction proteins (Anvarian et al., 2019; Kornberg, 2014b). Although the receptor and lipid composition of primary cilia membranes are specialized and regulated, primary cilia have no ribosomes (Rosenbaum and Child, 1967), and their protein constituents selectively enter through structures in the transition zone of the basal body. EM studies of cilia have identified their highly organized ultrastructure and functionally and structurally distinct regions (Fisch and Dupuis-Williams, 2011).
For this paper, we refer to filopodia that connect directly to another cell as cytonemes and refer to others as filopodia. Using high-pressure freezing (HPF) and EM, we examined the structures of filopodia and cytonemes in the Drosophila wing disc and analyzed cytonemes and their contact points to determine whether they share features with other types of signaling membrane extensions and synapses. Compared with chemical fixation, HPF better preserves tissue morphology (Studer et al., 1992), providing images of organelles and membranes that have smoother, more regular appearance (Kaneko and Walther, 1995; Royer and Kinnamon, 1996), denser and more homogenous cytoplasmic matrices (Kaneko and Walther, 1995), and better preservation of physiological extracellular space (Korogod et al., 2015). Following HPF and freeze substitution (FS), we used both serial scanning EM (SEM) and TEM to track filopodia that extend from three different cell types and visualized their connections to target cells. Serial SEM of automated tape-collected sections allowed for relatively seamless imaging across a large depth of tissues, while TEM provided higher resolution images. We found that filopodia are present in greater numbers and diversity than confocal microscopy (CM) had revealed and that they have distinctive shapes characterized by alternating thin and wide sections that had not been recognized by CM studies. Cytonemes and their contact points contained ribosomes, although there was heterogeneity in amount. Membranous compartments of different sizes and shapes were present along the length of filopodia, while mitochondria and rough ER were largely confined to wider areas of membrane extensions proximal to the projecting cell. Cytonemes connecting air sac primordium (ASP) cells to each other and cytonemes connecting myoblasts to each other were identified in addition to previously studied cytoneme populations.
Results
ASP and myoblast filopodia project toward multiple cell types
Cytoneme lengths, orientations, and dynamics have been studied in the wing disc and other systems using membrane-targeted fluorescent markers, and the role of cytonemes in signaling has been analyzed genetically and with fluorescently tagged ligands and receptors (Kornberg, 2014a). Bipartite GFP reconstitution experiments show that cytonemes make close contact with target cells. Yet, the structure and dimensions of cytonemes and the nature of the cellular contacts have not been resolved because their thin diameter is below the limits of light microscopy and they are not well preserved by chemical fixation. To overcome these issues, we used HPF to preserve the wing disc rapidly before chemical fixation and serial SEM and TEM to image the fine structures of cytonemes, the cells from which they project and to which they connect.
The wing disc is a flattened epithelial sac with a layer of columnar epithelial cells on one side that includes the primordia for the wing blade and the dorsal exoskeleton of the adult thorax (notum; Fig. 1 A). The notum primordium has several deep folds, and its basal side is attached to the cells of the tracheal ASP, myoblasts, and ECM. The ASP is a tubular extension with ∼120 tracheal epithelial cells that extends posteriorly across the wing disc from the transverse connective tracheal branch. It is a single-cell–layered epithelial tube with a thin proximal stalk and bulbous distal end. Myoblasts proliferate to cover most of the notum primordium during the third instar, and some lie between the disc and ASP.
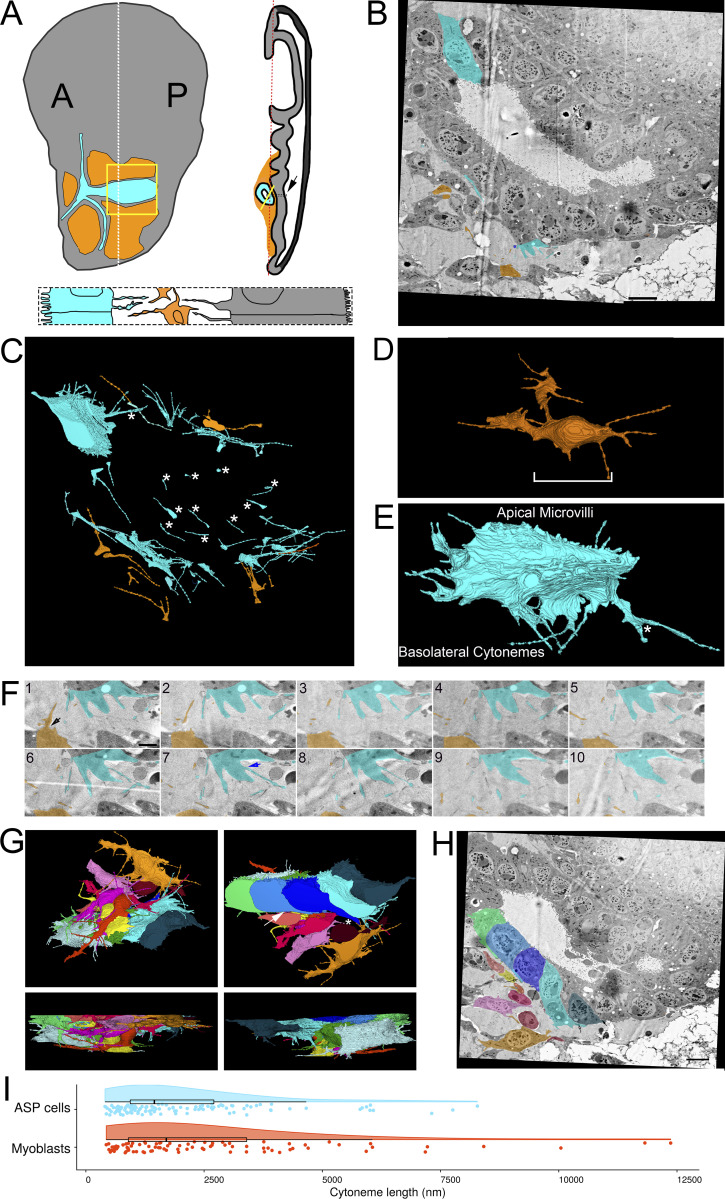
Figure 1.
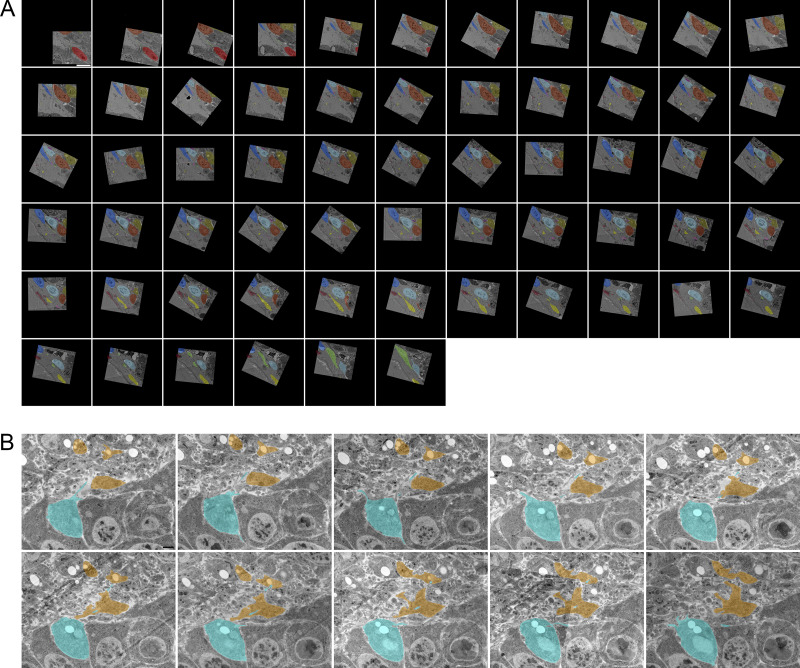
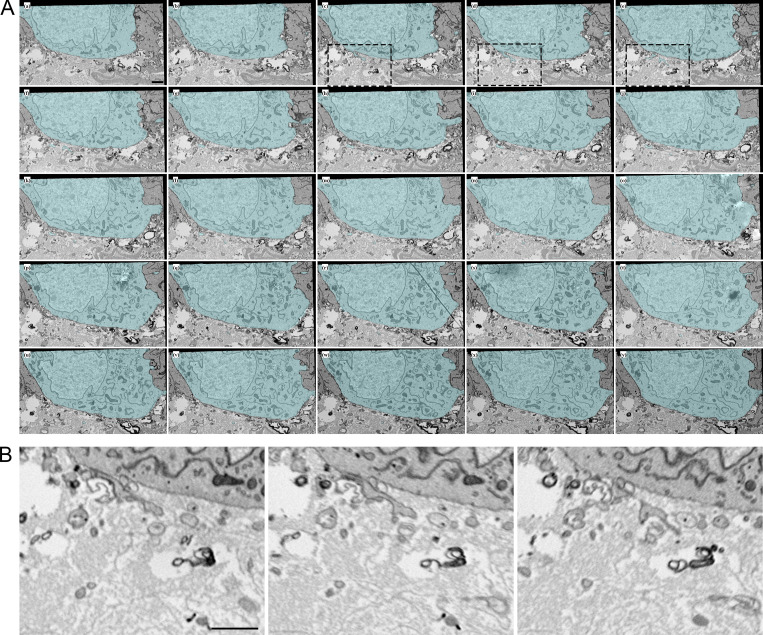
Filopodia project from ASP epithelial cells and myoblasts. (A) Illustration depicting the Drosophila wing imaginal disc (gray), ASP (cyan), and myoblasts (orange) from frontal (left) and sagittal (right) perspectives. The dashed lines indicate the position of the sagittal (white) and frontal (red) sections. The solid yellow lines indicate the area shown in B. An enlargement (bottom) of the black box in the sagittal view depicts the microvilli on the apical surfaces, and filopodia on the basal surfaces, of both ASP and disc epithelial cells. The intervening myoblasts project filopodia in multiple directions. A and P indicated the anterior and posterior regions of the disc. (B) One section of 172 serial SEM images showing the ASP surrounded by myoblasts. Disc epithelial cells are visible in the top-right corner. Each image was segmented to highlight a portion of filopodia projecting from ASP (cyan) and myoblasts (orange). (C) Segmentation was used to create a 3D reconstruction of the filopodia projecting from ASP cells (cyan) and myoblasts (orange). One reconstructed ASP cell is included in the 3D rendering. Asterisks indicate filopodia projecting in the direction of cells of the same type. (D) A reconstructed myoblast projecting filopodia in multiple directions. The main cell body, marked along its length with a white bracket, is 15.4 × 5.9 × 2.4 µm (measured at the widest points). (E) A reconstructed ASP cell with microvilli on its apical surface and filopodia projecting from its basolateral surfaces. The asterisk indicates a filopodia, which bifurcates. The cell body, not including filopodia, is 12.2 × 5.09 × 4.68 µm (measured at the widest points). (F) An enlarged region from segmented serial SEM images showing filopodia projecting from an ASP cell and a myoblast in the direction of the other cell type. The black arrow (section 1) indicates a filopodia extending directly from the cell body, while the blue arrow (section 7) indicates a filopodia extending from a thick membrane extension. (G) Top, bottom, and side views of eight myoblasts (warm colors) and seven ASP cells (cool colors) reconstructed in 3D. The white asterisk indicates an example of extracellular space, and the white arrow indicates an area where myoblast and ASP cell membranes are apposed. (H) SEM image with segmentation of myoblasts and ASP cells corresponding to those in (G). The arrows point to thick extensions that extend from almost all ASP cells toward the ASP tip. (I) A raincloud plot of the lengths of filopodia extended by the 3D-reconstructed ASP cells and myoblasts. Data points for 113 ASP and 89 myoblast cytonemes are shown with probability distribution functions and box-and-whisker plots for each group. Scale bars: 5 µm (B), 500 nm (F), and 5 µm (H).
Cytonemes extending from the various cells of the wing disc are distinguished by the cell types they connect and by the signaling proteins they transmit. Disc cell cytonemes connect to other wing disc cells, to ASP cells, or to closely apposed myoblasts (Huang and Kornberg, 2015). These cytonemes transport Hh and Dpp between disc epithelial cells; Hh, Dpp, and Bnl from disc to ASP cells; Hh and Wg between disc cells and myoblasts; and Delta between myoblasts and ASP cells.
To examine cytonemes at high resolution, we dissected wing discs gently to preserve physical associations with the ASP and myoblasts. The specimens were placed in a planchet with a cryoprotectant solution and within 1 min were subjected to HPF. Frozen tissues were processed by FS, embedding, and sectioning and were examined by SEM and TEM. Filopodia were observed extending from cells throughout the samples; some connected to other cells. We do not know if the free-ended extensions may be cytonemes that were in the process of extending to a target or had disconnected from a target cell or, alternatively, if they may be filopodia with another function.
We tracked filopodia extending from ASP cells, myoblasts, and disc epithelial cells (Fig. 1 A). To examine filopodia extending from the ASP cells and myoblasts, 172 serial sections 60 nm thick (total depth 10.32 μm) were collected, imaged by serial SEM (Fig. 1 B and Fig. S1), and aligned. Representative filopodia and cells were segmented and reconstructed in 3D to determine directions of projection (Fig. 1 C). Whereas myoblasts are not polarized cells and filopodia project from all sides (Fig. 1 D and Video 1), filopodia of the polarized epithelial ASP cells project only from lateral and basal surfaces and only in the direction of myoblasts (Fig. 1, E and F; and Video 1). ASP cells also have microvilli on their apical surface (Fig. 1 E and Video 1). Some of the myoblast filopodia project in the direction of the ASP (Fig. 1, C and E). Filopodia were also observed projecting between cells of the same type (Fig. 1 C, asterisks). In some instances, a single filopodium projects from a cell before bifurcating (Fig. 1 E, asterisk). Some filopodia project directly from the cell body (Fig. 1 F, section 1 arrow), while others project from thicker membrane extensions (Fig. 1 F, section 7 arrow). Thick membrane extensions were present as protrusions from the main cell body, having wide proximal sections with distal sections that sometimes branched off into one or more thin filopodia. Thick extensions had widths typically between 300 and 600 nm. Frequently, thin filopodia branched off from thick extensions at a distinct point, but in some cases, a thick extension gradually narrowed to thin filopodia without a distinct transition point.
Figure S1.
ASP cells and myoblast project filopodia and cytonemes, which connect with target cells. Serial SEM images, including section 27 shown in Fig. 1 B. A portion of ASP cytonemes and one cell (cyan) and myoblast cytonemes (orange) are shown segmented. Image resolution has been scaled by 50%. Scale bar: 5 µm.
Video 1.
Complete 3D reconstructions of an ASP cell and a myoblast from the segmented serial SEM stack. The thick membrane extension that projects distally from the basal surface of the ASP cell and narrows to a cytoneme is labeled with an arrow in some frames. The apical side projecting microvilli and the basal side projecting cytonemes are also labeled in some frames. The cells are the same as those found in Fig. 1, D and E, but the colors correspond to how they are displayed in Fig. 1, G and H. Playback speed, 15 frames per second.
Reconstruction of seven ASP cells and eight nearby myoblasts (Fig. 1 G) revealed regions with extracellular space as well as closely apposed surfaces (Fig. 1 H). 113 ASP and 89 myoblast filopodia extend from these 15 cells; although most are <7 μm, the longest are 8.28 μm (ASP) and 12.4 μm (myoblast; Fig. 1 I). 20 filopodia extend beyond the imaged area, and their complete length could not be measured. The serial SEM stack, which was ∼58 × 58 × 10 μm, included filopodia projecting between the ASP and surrounding myoblasts. Previous CM studies reported that cells at the tip of the ASP extend cytonemes up to 40 μm (Roy et al., 2014), but the imaged area did not include tip ASP cells in their entirety and, therefore, precluded finding the longest cytonemes between ASP and disc cells.
Although ASP cells project thick extensions and filopodia in many directions, a thick extension that tracks distally along the basal surface is a consistent ASP cell feature. Most ASP cells, including both cells that were and were not reconstructed in 3D, have a substantial portion of their basal surface within the imaged area. Of those cells, 93% project a thick membrane extension (Fig. 1 H, arrows; and Video 1) that tracks distally along the surface of the neighboring cell. 98% of those thick extensions narrow to filopodia, of which the majority (79%) continue to project distally. The myoblasts project filopodia in multiple directions, both toward and away from the ASP. Using serial SEM, we visualized previously identified populations of ASP and myoblast filopodia that extend between these cell types (Huang and Kornberg, 2015); we also identified filopodia that project in the direction of cells of the same type.
Filopodia extend from basal and lateral surfaces of disc epithelial cells
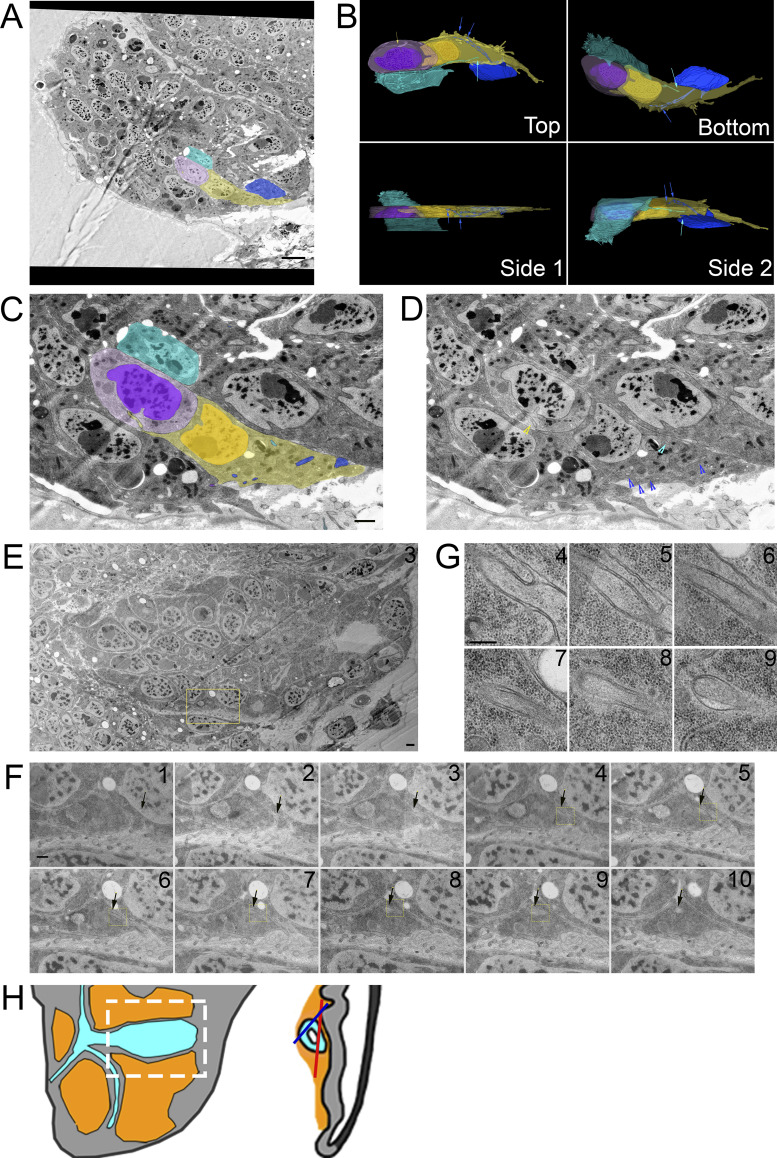
We next visualized filopodia extending from the disc where basolateral cytonemes have been previously analyzed by CM (Callejo et al., 2011). We identified filopodia projecting from the basal side of disc epithelial cells that appear similar in size and shape to those of the ASP and myoblasts (Fig. 2, A and B). We also aligned, segmented, and reconstructed in 3D a series of 10 sections at the basal side of a fold region (Fig. 2, C and D). Filopodia extend along the fold (i.e., aligned with the anterior/posterior axis), but there are no filopodia crossing the fold perpendicular to the basal surfaces. Thicker membrane extensions were also present, and some of these membrane extensions transitioned from thick to thin within the stack (Fig. 2 D, pink).
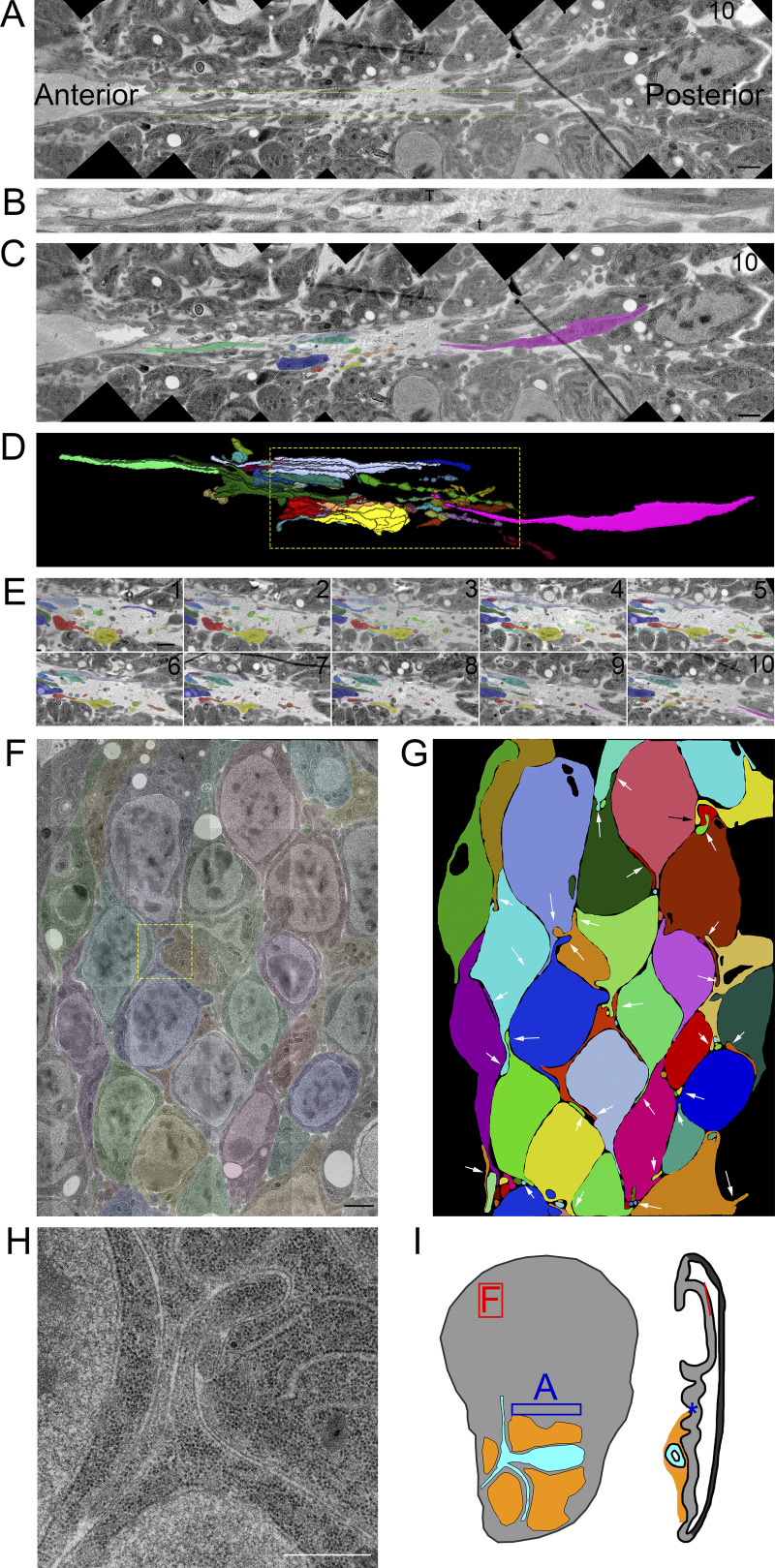
Figure 2.
Filopodia extend from basal and lateral surfaces of disc cells. (A) One montaged image (see Materials and Methods) of a 10-section TEM series showing the basal side of the disc in a fold region (asterisk on disc diagram in Fig. 2 I). Thick membrane extensions and thin filopodia extend along the fold. (B) An enlargement of the dashed green box in A with a mix of thick membrane extensions and thin filopodia with constrictions and varicosities. (C) Segmentation of individual membrane extensions and filopodia. The section number is labeled in the top-right corner. (D) 3D reconstruction of segmentation showing thick membrane extensions, thin filopodia, and a thick membrane extension that narrows to a filopodia (pink). The reconstructed volume containing the extensions is 21.6 × 2.7 × 0.7 µm. (E) Each of the 10 segmented sections used to produce the boxed region of D. (F) One section of an eight-section TEM series of the wing pouch, which shows filopodia extending from the lateral surfaces of disc cells. The montage of original images has been overlayed with transparent segmentation of individual cells. (G) Segmentation of the cells shown in F with arrows pointing to the lateral filopodia. (H) Enlargement of the yellow boxed area in panel F shown without transparent segmentation. (I) Disc diagram representing the areas imaged and shown in A (blue box and asterisk) and F (red box and line). Scale bars: 1 µm (A, C, and F) and 500 nm (H).
Filopodia also extend from basolateral sides of disc epithelial cells. In contrast to the filopodia that extend from the ASP, myoblasts, and basal side of the disc, basolateral disc filopodia are tightly confined by narrow extracellular spaces (Fig. 2, F–H; and Fig. S2). The paths of these filopodia are defined by the contours of the cells and adjacent filopodia.
Figure S2.
Filopodia extend laterally from wing disc epithelial cells. Serial TEM images, including section 1 shown partially and segmented in Fig. 2, F and G. 36 individual TEM images were montaged for each section. Images are in serial order from left to right and top to bottom. Scale bar: 1 µm.
Cytonemes extend into invaginations of target cells
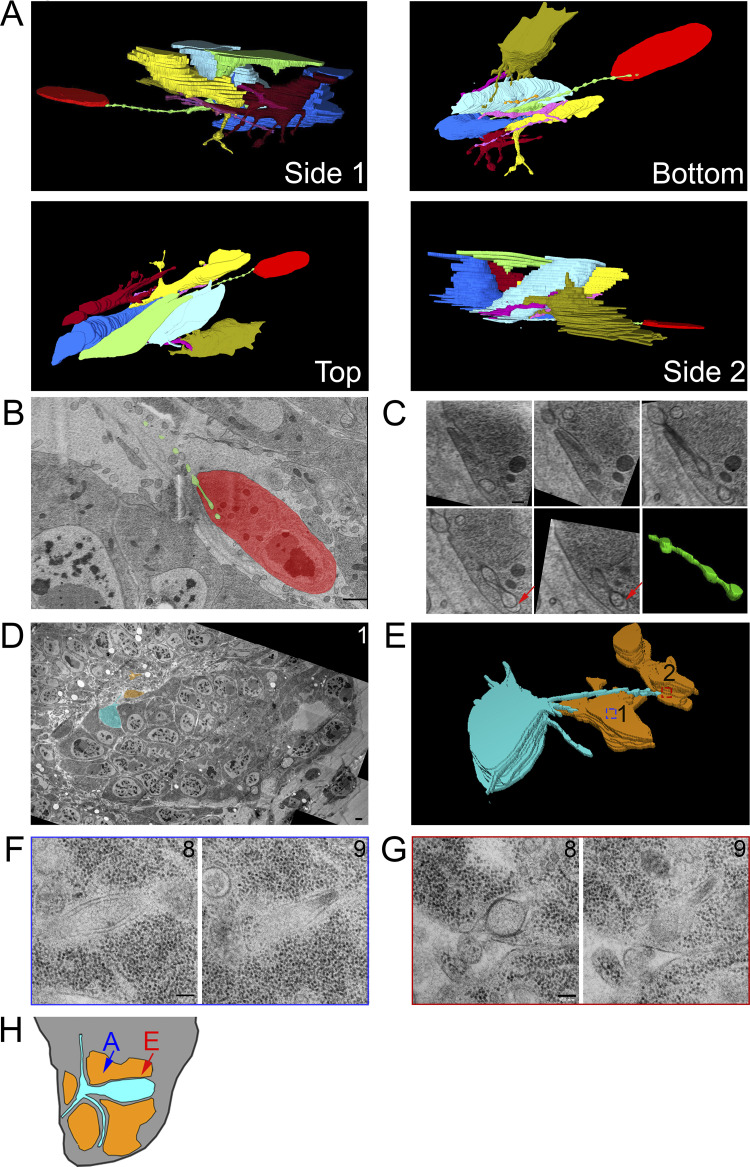
To investigate how cytonemes physically interact with target cells, we used SEM and TEM to track cytonemes projecting from their origins to cells they contact. 3D reconstruction of a serial SEM stack (Fig. S3 A) revealed a 13-μm-long cytoneme that extends from a myoblast (green) past several myoblasts and contacts a more distant myoblast (Fig. 3 A, red). This cytoneme terminates in a cavity that envelops the cytoneme with target cell membrane (Fig. 3 B). The diameter of the cytoneme shaft varies several times along the length of the cytoneme. These undulations are spaced at regular intervals both in the proximal regions far from the target cell and in distal regions that are inside the target cell cavity where the target cell membrane closely mirrors the shape of the undulating shaft. At the end of the cytoneme, there is a gap of as much as 82 nm between the cytoneme tip and the cell membrane (Fig. 3 C). The locations of cytoneme and myoblasts are indicated in the diagram in Fig. 3 H (blue arrow).
Figure S3.
Myoblasts are connected by cytonemes to other myoblasts and ASP cells. Serial SEM images, including section 1 shown in Fig. 3 A. (A) Individual myoblasts and their cytonemes were segmented in different colors. The segmentation was used to produce the 3D rendering in Fig. 3, A and C. (B) Serial TEM images showing the area segmented to produce the 3D rendering in Fig. 3 E. The series is a partial view of larger montages used to create each section. A full montage of section 1 is shown in Fig. 3 D. Images are in serial order from left to right and top to bottom. Scale bar: 5 µm (A), 1 µm (B).
Figure 3.
Myoblast and ASP cytonemes end in membrane invaginations of myoblasts. (A) Four perspectives of a 3D reconstruction of myoblasts and their cytonemes from 61 SEM images spanning a depth of 6.4 µm. The cytoneme extended by the green cell end in an invagination of the red cell membrane. (B) A segmented section showing the ending of the green cytoneme in a membrane invagination of the red cell. The blue arrow on the disc diagram (Fig. 3 H) shows the area imaged. (C) Higher magnification SEM images of the five serial sections containing the portion of the green cytoneme inside the red cell invagination. The sixth panel contains a 3D reconstruction of the part of the cytoneme inside the invagination, showing that it retains the same shape and structure when connected to the target cell. (D) One segmented TEM image montage highlighting an ASP cell (cyan) with two cytonemes that connect to two different myoblasts (orange). (E) 3D reconstruction of 10 segmented serial TEM images showing ASP cytonemes in contact with and invaginating into two myoblasts (labeled 1 and 2). The blue and red boxes mark the XY position of the end of each cytoneme and the area imaged at higher magnification. (F and G) High-magnification images showing the last two sections of the cytonemes in contact with myoblast #1 (F) and myoblast #2 (G). (H) A disc diagram indicating the positions of the segmented myoblasts shown in A (blue) and E (red). Scale bars: 1 µm (B), 200 nm (C), 1 µm (F), and 100 nm (G).
Images of ASP cells from serial TEM (Fig. S3 B and Fig. 3 D) and 3D reconstruction (Fig. 3 E) reveal similar structures. The ASP cell highlighted (cyan) in these panels extends cytonemes that project toward two myoblasts (orange). The cytoneme extending to myoblast #1 is 13.1 μm in total length, of which 2.2 μm is inside the myoblast. The cytoneme extending to myoblast #2 is 5.4 μm in total length, of which 0.5 μm is inside the myoblast. A higher magnification view of the two sections containing the most distal portions of these cytonemes has cytoneme termini surrounded at close proximity by myoblast membranes (Fig. 3, F and G). An area of high density is present at the point of contact between the membrane of myoblast #2 and the cytoneme (Fig. 3 G, section 8). In contrast to the rounded ending of the burrowing cytoneme in Fig. 3 C, the ends of both cytonemes in this section are narrow (section 9) relative to the more proximal portions (section 8). The location of these two myoblasts are indicated in the diagram in Fig. 3 H (red arrow).
Previous work identified cytonemes connecting disc epithelial cells to each other (Huang and Kornberg, 2015; González-Méndez et al., 2020), and our SEM and TEM analysis also identified cytonemes that extend between ASP cells. Partial 3D reconstruction of four neighboring ASP cells (Fig. 4 A) from serial SEM images reveals cytonemes that extend from two cells (blue and cyan) that terminate within membrane invaginations of another (yellow). The blue cell cytoneme bifurcates into two branches within the yellow cell, and both branches extend further into the yellow cell (Fig. 4 B). The yellow cell extends a cytoneme that terminates within membrane invaginations of the purple cell (Fig. 4, B–D).
Figure 4.
ASP cytonemes connect to neighboring ASP cells in membrane invaginations. (A) Four neighboring ASP cells visualized by serial SEM were segmented. The cyan cell was segmented throughout the entire stack, while the other cells were partially segmented to allow for visualization of their cytoneme connections in 3D. (B) Four perspectives of the 3D-reconstructed ASP cells showing cytonemes from the blue and cyan cell ending within an invagination of the yellow cell, while the yellow cell projects a cytoneme ending within the purple cell. The reconstructed volume containing the cells is 22.6 × 9.4 × 6.2 µm. (C and D) One segmented (C) and nonsegmented (D) section showing cytonemes inside membrane invaginations of neighboring cells. Arrowheads point to the cytonemes of corresponding color. (E) A TEM image montage of the ASP and surrounding myoblasts (location represented by white box and red line shown on disc diagrams in Fig. 4 H) shows a cytoneme from one ASP cell inside an invagination a neighboring cell. (F and G) A series of 10 image montages (enlargements of the boxed region of E) shows the cytoneme and its path (marked by arrows) within the membrane of the target cell. Boxed regions show the area imaged at higher magnification of sections 4–9 shown in G. (H) Both A (blue) and E (red) are images of the ASP and surrounding myoblasts (white box). The sagittal perspective shows the depth and angle of each section. Scale bars: 5 µm (A), 500 nm (C), 1 µm (E), 500 nm (F), and 200 nm (G).
We also examined inter-ASP cell and inter-disc cytonemes by TEM. An example of an inter-ASP cytoneme and its connection to a neighboring cell is shown at high magnification in the series of micrographs in Fig. 4, E and F. This cytoneme–cell connection is similar to the connections of inter-myoblast cytonemes (Fig. 3, A–C) and ASP to myoblast cytonemes (Fig. 3, D–G). The cytoneme and cell membranes are closely apposed, except for a gap at the end of the cytoneme that is 45 nm wide at its largest point (Fig. 4 G, section 9). Disc cytonemes, like ASP and myoblast cytonemes, also extend into membrane invaginations of target cells. Fig. 5 A shows a cytoneme originating at the basolateral surface of a disc cell (cyan) and terminating within an invagination near the apical surface of a neighboring disc cell. This cytoneme bifurcates into two branches that both terminate within the target cell (Fig. 5 B). A small uniform space separates the cytoneme and cell membrane throughout the length of their apposition. A cytoneme–cell connection with similar topology present near the basal side of the disc is shown in Fig. 5 C. This disc cell cytoneme terminates within a membrane invagination near the basal surface of its neighbor (Fig. 5 C). In this instance, the gap between the cytoneme and cell membranes is also small and uniform.
Figure 5.
Disc cytonemes extend into invaginations near the apical and basal surfaces. (A) A series of five TEM image montages showing a disc epithelial cell (cyan) projecting a cytoneme that ends in a membrane invagination of a neighboring disc cell. The region imaged is shown as an asterisk on the disc diagram. Part of the target cell’s apical surface and microvilli are seen in the upper left corner. (B) The yellow box represents the area montaged at higher magnification where the path of the cytoneme in the invagination is seen. Inside the invagination, the cytoneme branches off into two terminal varicosities. (C) A TEM image shows a disc epithelial cell (yellow) projecting a cytoneme from its lateral surface, which ends in an invagination of the membrane of a neighboring disc cell near its basal surface. The area imaged is in the fold indicated on the disc diagram. Progressive enlargements of the boxed regions are shown from left to right. The middle two images are the same with and without segmentation of the cytoneme-projecting cell. (D) TEM images of four separate endings of cytonemes within invaginations of target cell membranes, showing the variability in the size of the space between the end of the cytoneme and target cell membranes. Scale bars: 500 nm (A and B), 2 µm (C panel 1), 500 nm (C panel 4), and 100 nm (D).
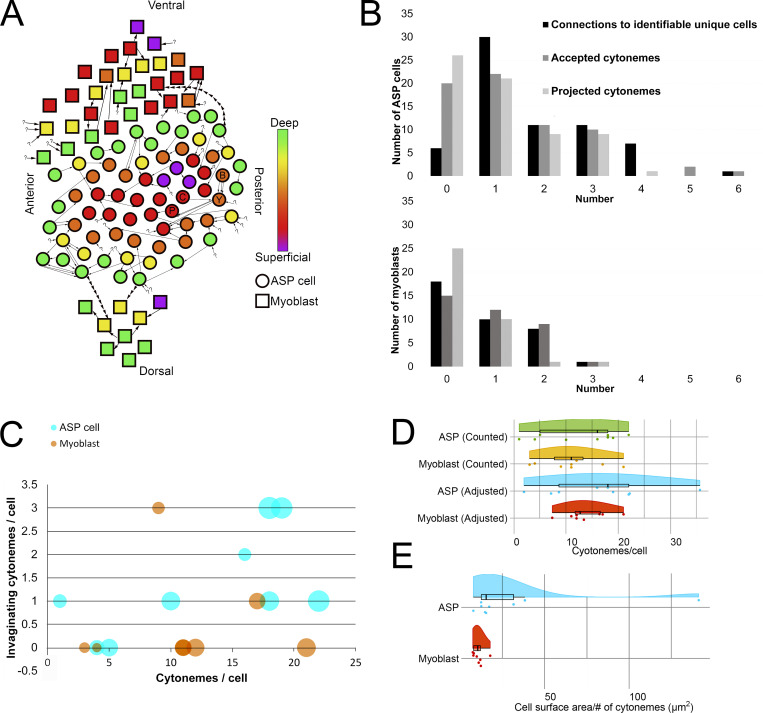
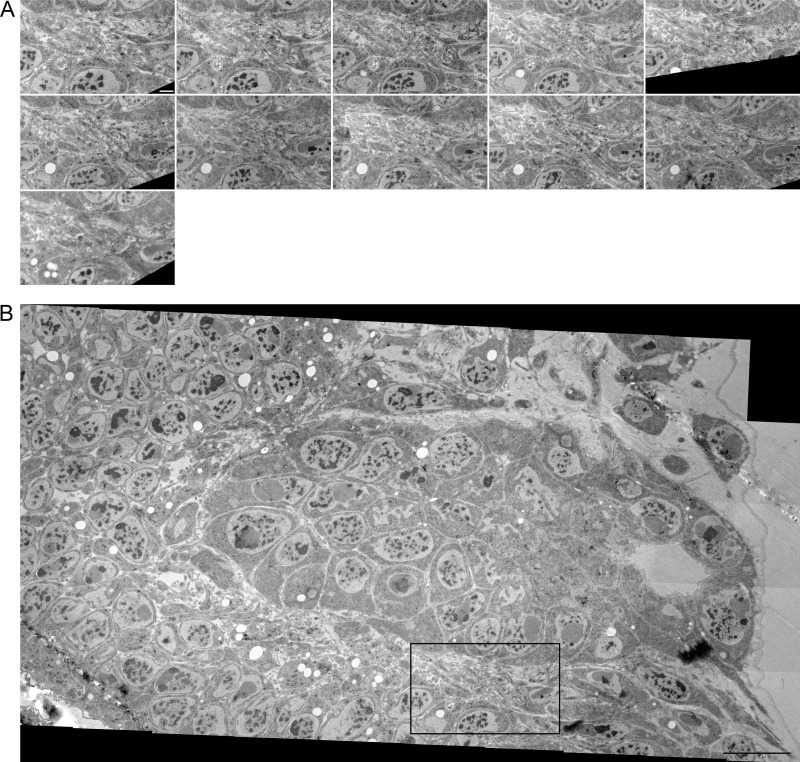
To obtain a more complete understanding of the prevalence and variations of inter-ASP cell and inter-myoblast cytonemes, the 66 ASP cells and 37 myoblasts visible in the SEM stack (Fig. 1, Fig. 4, and Fig. S1) were examined, and each cytoneme was tracked from its terminus inside an invagination to its originating cell body. 33 of 123 total invaginating cytonemes could not be traced back to a source cell either because they extended outside the imaged area or the images lacked sufficient resolution. A rough 2D map of the ASP and surrounding myoblasts was created by placing each cell in an approximate XY position and assigning a color corresponding to the Z position within the stack (Fig. 6 A). Four pairs of ASP cells and one pair of myoblasts reciprocally project cytonemes to each other. Five pairs of ASP cells and one pair of myoblasts connect by more than one cytoneme projected by one cell in the pair. Many cells both project and receive cytonemes, but some only project (14 ASP cells and 4 myoblasts) or receive (20 ASP cells and 14 myoblasts). Only three ASP cells project cytonemes that connect to myoblasts (Fig. 6 A). By excluding connections made by received cytonemes that could not be traced back to a source cell and by counting repeated connections (multiple cytonemes between the same two cells) as one connection, the number of identifiable unique connections was counted for ASP cells and myoblasts (Fig. 6 B). The cell with the most unique identifiable connections was an ASP cell linked to five other ASP cells and one myoblast. 60 of the 66 ASP cells connect by cytonemes to another identifiable cell; 19 of 37 myoblasts link to another identifiable cell either by the presence of a cytoneme from another cell in an invagination or by extending one (Fig. 6 B). ASP cells receive 90 cytonemes, and myoblasts receive 33. ASP cells project 70 cytonemes that end in the invagination of another cell; myoblasts project 15 (Fig. 6 B). These results reveal that very few cells lack cytoneme-mediated cell–cell contacts, that inter-ASP cell cytoneme networks connect ASP cells, and that the area of cell–cell contact is increased with invaginations that juxtapose cytoneme and cell membranes.
Figure 6.
Intratissue cytonemes predominate in the ASP epithelium and myoblast population. (A) A map of the approximate position of 66 ASP cells (circles) and 37 myoblasts (squares) contained within the serial SEM stack and their connections to other cells. The color indicates the Z depth of each cell within the stack. Arrows begin at the cell from which a cytoneme originates and point toward the cell a cytoneme ends within. Question marks indicate invaginated cytonemes that could not be tracked back to a cell body. Positional labels (e.g., anterior) refer to the direction of the underlying disc compartments. The four cells segmented in Fig. 4, A–C, are indicated on the map with letters corresponding to their segmentation color (B, blue; Y, yellow; C, cyan; P, purple). (B) For each ASP cell (top) or myoblast (bottom), the number of connections (either sent or received invaginating cytonemes) to other verifiably unique cells, the number of invaginating cytonemes accepted, and the number of invaginating cytonemes projected to other cells were determined and graphed as a histogram. (C) For nine 3D-reconstructed ASP cells (cyan) and eight 3D-reconstructed myoblasts (orange), the number of total cytonemes was compared with the number of those cytonemes that made contact and invaginated another cell. The area of the dot representing each cell scales with the surface area of the reconstructed cell. (D) The number of cytonemes counted for each reconstructed cell along with the number of expected cytonemes if all cells were contained fully within the stack are shown as individual data points and summarized as box plots with the probability density function. The adjustment factor is based on the surface area of each cell compared with the surface area of a completely reconstructed cell of the same type. (E) The cell surface area per cytoneme for each reconstructed cell.
The ASP and myoblast cell map (Fig. 6 A) shows only cytonemes that terminate in a target cell invagination. To estimate the percentage of the total filopodia population that cell-to-cell cytonemes represent, nine typical ASP cells and eight typical myoblasts were reconstructed in 3D, and the total number of filopodia was compared with the number of invaginating cytonemes for each cell (Fig. 6 C). The percentage of cytonemes varies between cell types (20.07% ± 30.8% for ASP cells and 4.9% ± 10.9% for myoblasts). Because most of the reconstructed cells have some portion outside of the imaged area, either in the XY or in the Z direction, the number of cytonemes counted for those cells is likely incomplete. To adjust for this, the surface area of a complete cell for each type was divided by the surface area of each incomplete cell, and this factor was used to estimate the total number of filopodia: 16.71 ± 9.43 for ASP cells and 13.76 ± 3.95 for myoblasts (Fig. 5 D). The surface areas of the two complete myoblasts are 124 and 129 μm2. Two other myoblasts with portions outside the imaged area have surface areas larger than the complete myoblasts (145 and 165 μm2), and the filopodia number for these cells was not adjusted. The complete ASP cell has a surface area of 272 μm2, which was used as the basis to estimate the cytoneme number for partial ASP cells. One nearly complete ASP cell has a surface area slightly higher (274 μm2) and was not adjusted for cytoneme number. The counted and adjusted numbers of filopodia are shown in raincloud plots (Allen et al., 2019). The cell surface area that corresponded to projection of one filopodium tended to be smaller for myoblasts (10.6 ± 3.17 μm2) than ASP cells (16.7 ± 9.43 μm2; Fig. 6 E), indicating that filopodia are denser on myoblasts, even though the total number is similar to that of ASP cells.
Although the presence of cytonemes extending into invaginations of target cells is consistent for all three cell types, some aspects are variable. Whereas a space separates all the cytoneme:cell membrane juxtapositions, in some areas, the space is better described as a gap (Fig. 6 D and Fig. S4). Both the size and the shape of these gaps varies, but the differences are not characteristics of the particular cell type that either projects or receives a cytoneme. Invaginating cytonemes with the same morphologies are present in all 10 discs examined by EM.
Figure S4.
Cytonemes end in membrane invaginations of target cells. (A–D) High magnification views of the cytonemes shown in Fig. 5 D panel 1 (A), panel 2 (B), panel 3 (C), and panel 4 (D) of cells. Arrows point to the leading edge of the cytoneme in each section. Images are in serial order from left to right and top to bottom. Scale bars: 100 nm.
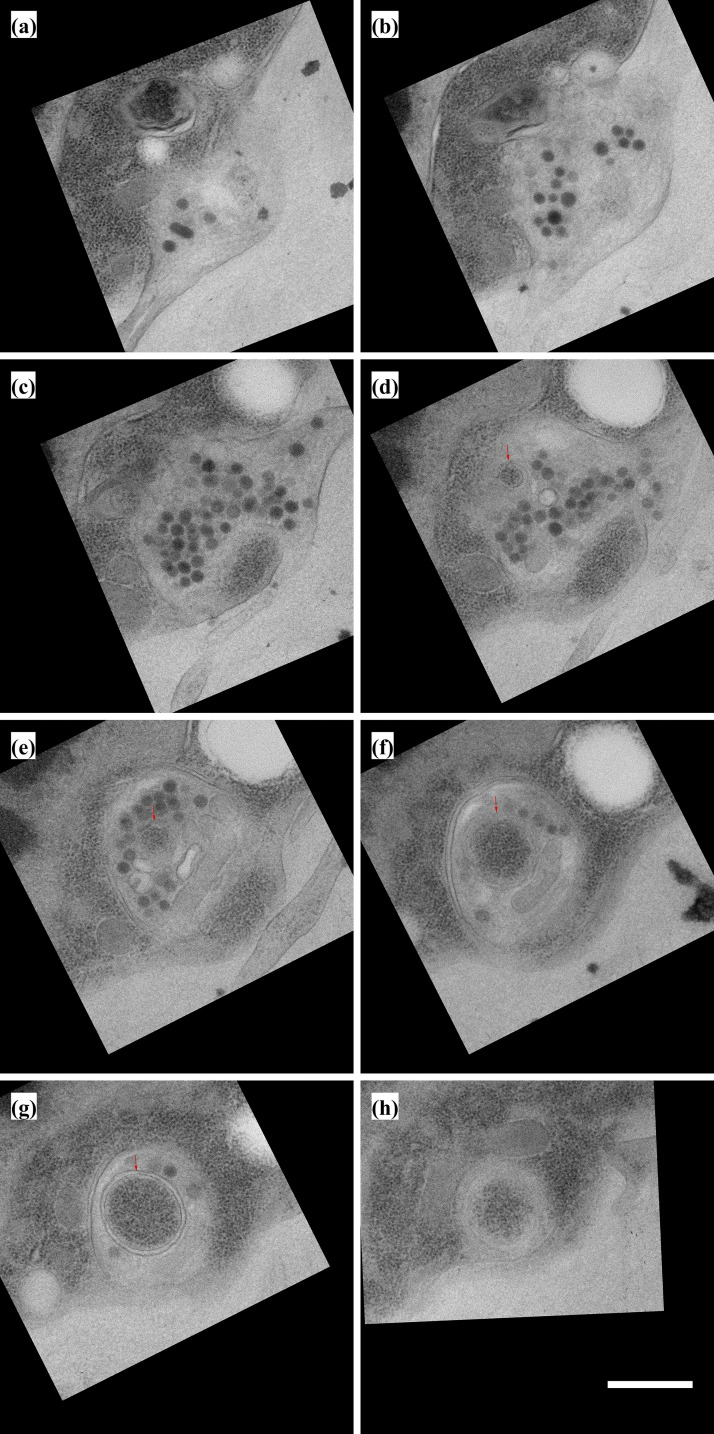
Periodic variations in shaft diameter create distinct varicosities and constrictions
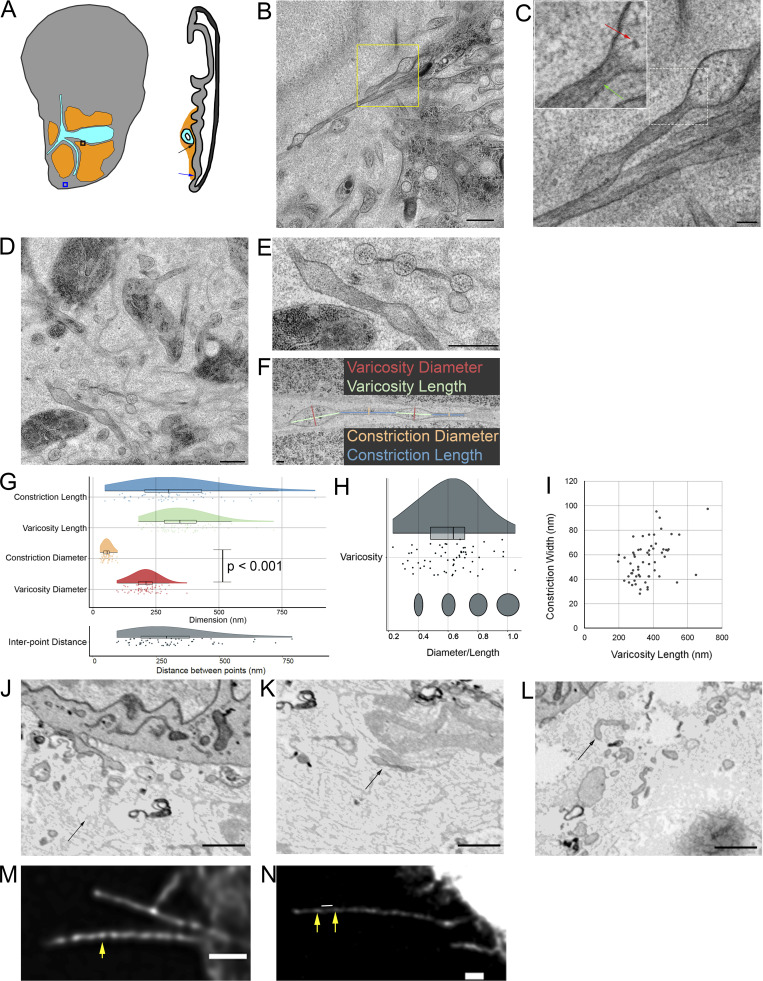
Based on the general understanding of filopodia structure, we expected to find that filopodia in our preparations would have shafts with a uniform and consistent diameter. Instead, we observed that with the exception of short (<1 μm) extensions, all filopodia emanating from ASP, myoblast, and disc cells have diameters that oscillate between narrow constrictions and well-defined wider sections (varicosities; Fig. 7, A and B). The constrictions have prominent actin filaments and little cytosol (Fig. 7 C, green arrow), and the varicosities have more cytosol, including ribosomes (Fig. 7 C, red arrow). No microtubules were identified in filopodia, consistent with previous CM experiments. Although the presence of repeating sequences of constriction/varicosity/constriction/varicosity is a common feature of the cytonemes we observed, the shape, size, and spacing is variable (Fig. 7, D and E; and Fig. S5). The number of varicosities per cytoneme depended on varicosity and constriction length and the total length of the cytoneme, with longer cytonemes generally having more varicosities. Cytoneme termini also vary, having widths that are as narrow as the constrictions or an expanded, bulbous structure, similar to varicosities. Bulbous termini are present in filopodia that are entirely extracellular as well as cytonemes that burrow into membrane invaginations. The EM images do not distinguish differences in composition between the bulbous termini and shaft varicosities (Fig. 5 D). The morphology of filopodia was consistent across all 10 discs examined by EM.
Figure 7.
Filopodia diameters vary between regions of varicosities and constrictions. (A) Boxes and arrows indicate the imaged area shown in B (blue) and D (black). (B) TEM image of two exemplar filopodia with regions of varicosities and constrictions extended by disc epithelial cells (blue arrow in A). (C) Higher magnification view of filopodia corresponding to dashed box in B. The inset is an enlargement of the area shown in the white dashed box. Ribosomes (red arrow) are visible in a varicosity, while actin filaments (green arrow) are visible in the constriction. (D) TEM image of filopodia among myoblasts (black arrow in A). (E) Enlargement of two filopodia from D showing the variance in size and shape of varicosities and constrictions. (F) The width and length of 64 varicosities and 60 constrictions of cytonemes projected by the ASP and myoblasts were measured as shown. (G and H) Raincloud plots of (G) filopodia dimensions measured from EM images (top) and compared with a two-sided t test, constriction lengths (n = 56) measured from filopodia of T-TRAP–expressing myoblasts (shown in N), and (H) ratio between width and length of varicosities (dark gray) and constrictions (light gray) measured from EM images. (I) Plot of constriction width versus varicosity length showing a moderate linear correlation (ρ = 0.42). (J) SEM image of a chemically fixed filopodium extending (arrow) from a disc epithelial cell with wide and narrow sections. (K) SEM image of a portion of a chemically fixed filopodium (arrow) with two varicosities separated by a constriction. (L) SEM image of a chemically fixed filopodium that does not show signs of having wide or narrow regions. (M and N) Deconvoluted CM images of filopodia projecting from live ASP cells expressing the membrane marker CD2-GFP (M) and a live myoblast expressing T-TRAP (N). Yellow arrows point to examples of points along the filopodia with greater diameter. The white line shown in N is an example of the measured distance between two points with greater diameter (shown in the fluorescence plot in G). Scale bars: 500 nm (B), 100 nm (C), 500 nm (D and E), 100 nm (F), and 1 µm (M and N).
Figure S5.
Cytonemes have distinct regions of varicosities and constrictions. Serial TEM images, including section 3 shown partially inFig. 6, D and E. The series is a partial view of larger montages used to create each section. Images are in serial order from left to right and top to bottom. Scale bar: 1 µm.
To quantify the shapes of constrictions and varicosities, we imaged filopodia that project from ASP cells and myoblasts by serial SEM and analyzed 64 varicosities and 60 constrictions (Fig. 7 F). Diameters were measured at the midpoint of each varicosity and constriction; because of their shape, the midpoint diameter of a varicosity is equivalent to the maximum diameter. Varicosity (356 ± 105 nm) and constriction (331 ± 171 nm) lengths are similar, and both are varied (Fig. 7 G). The period length (the length between midpoints of successive varicosities) was 716.3 ± 206.6 nm. Varicosity diameters (206 ± 51.8 nm) are almost four times larger than constriction diameters (55.9 ± 16.2 nm). The diameter-to-length ratio of varicosities is a measure of variability in varicosity shape (Fig. 7 H). Most varicosities are ovoid, although some are more round (i.e., diameter/length equal to 1) and some more elongated. The relationship between varicosity length and constriction width is moderately positive (Fig. 7 I), but no other relationship between varicosity or constriction width, length, or shape was found.
To determine whether chemical fixation also preserved the structure of the filopodia in HPF-derived sections, we examined sections of chemically fixed wing discs. Chemical fixation followed by reduced osmium-thiocarbohydrazide–reduced osmium staining and serial SEM examination revealed cellular and organellar membranes with good preservation and demarcation (Fig. S6). Although filopodia and filopodia membranes were also preserved (Fig. 7 J and Fig. S6), regions of varicosities and constrictions are not well defined (compare Fig. 1 F and Fig. 7, J–L).
Figure S6.
Varicosities and constrictions are less well preserved in chemically fixed samples. (A) Segmented serial SEM images, including section 4 (d) shown in Fig. 7 J. Boxed regions are shown enlarged without segmentation. Scale bars: 1 µm.
Using CM, we examined filopodia morphology of live ASP cells expressing CD2-GFP, which localizes to the plasma membrane. The widths of the filopodia vary along their lengths (Fig. 7 M), consistent with EM images. Bright points with wider widths (yellow arrow) are separated by dimmer, narrow sections, presumably marking the varicosities and constrictions seen by EM.
TEM images show that varicosities generally have a higher density of ribosomes than constrictions. To determine if filopodial varicosities can be identified in preparations of live cells, we expressed a GFP-tagged version of ribosomal protein Rpl10 (tandemly tagged ribosomal affinity purification [T-TRAP]; Zhang et al., 2016). T-TRAP localizes throughout the cell, including filopodia (Fig. 7 N). Filopodia have wider, brighter T-TRAP points (Fig. 7 N, yellow arrows) and intervening sections that appear narrower and dimmer. T-TRAP fluorescence distinguished these features more clearly than CD2-GFP, presumably because ribosomal density varies significantly between varicosities and constrictions, whereas CD2-GFP is evenly distributed throughout the membranes of both. We used T-TRAP CM images to measure the distance (Fig. 7 N, white line) between the ends of adjacent wide, bright points along filopodia (Fig. 7 N, yellow arrows). Although measurement of interpoint distances was less precise than measurement of constriction lengths in EM images, their distributions (Fig. 7 G) and averages (306 ± 154 nm for CM versus 331 ± 171 nm for EM) are similar.
Large membrane extensions contain cellular organelles
Although some filopodia extend directly from cells as thin projections, many cells project wider membrane extensions that narrow gradually into thin filopodia (Fig. 8 A, red arrow). Within these wide membrane extension regions, mitochondria, rough ER, and membranous compartments are visible (Fig. 8, A and B; and Fig. S7). We did not observe mitochondria or rough ER in the thinner filopodia segments. This result is consistent with confocal images of filopodia that extend from ASPs that overexpress either mito-GFP (and membrane-bound mCherry; Fig. 8 C) or RFP tagged with KDEL (lysine-asparate-glutamate-leucine), an ER localization signal (and membrane-bound GFP; Fig. 8 D). Both the mitochondria and the ER markers are prevalent in the wider regions of membrane extensions that are proximal to the cell body but not in thinner regions of membrane extensions. ASP membrane extensions do not all have the same proportions of mitochondria and ER markers.
Figure 8.
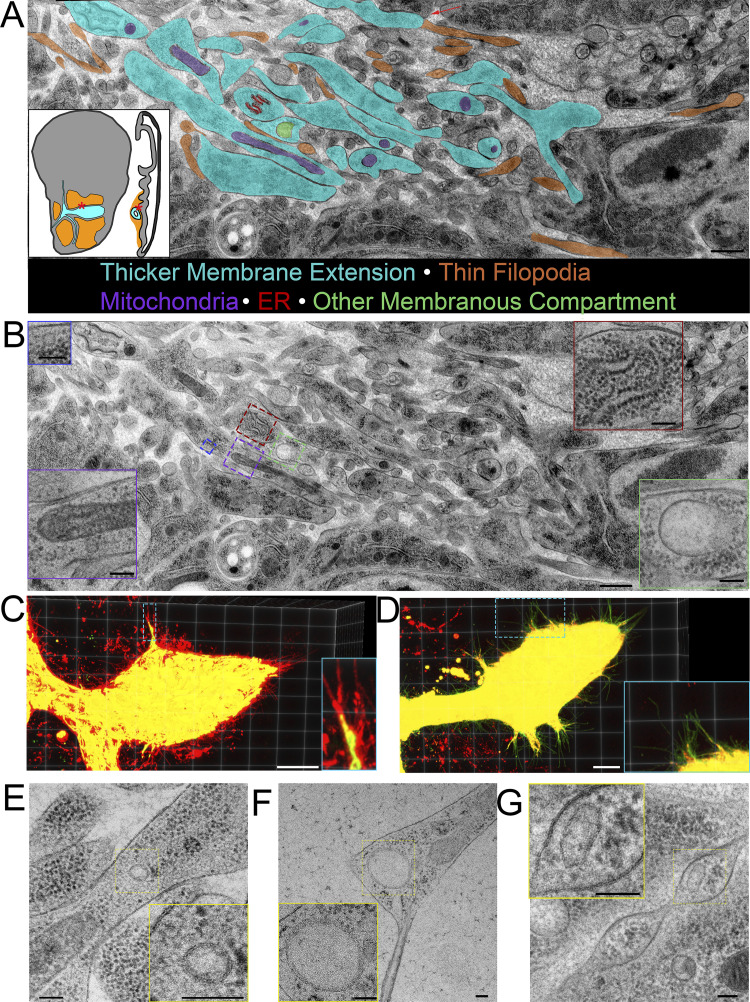
Thick membrane extensions contain mitochondria, ER, and membranous compartments. (A) Partially segmented TEM image montage of thick membrane extensions (cyan) and thin filopodia (orange) running between ASP and myoblasts (red asterisks on disc diagrams). Some cells project thick membrane extensions that narrow into thin cytonemes (red arrow). The distinction between thick extensions and thin filopodia was made based on 10 serial sections of the region, not only the section shown. Thick membrane extensions contain mitochondria (purple), rough ER (red), and other membranous compartments of different sizes (green). Membrane extensions and filopodia that were in cross section were not segmented. (B) TEM image shown in A without segmentation. Insets show enlarged examples of mitochondria (purple border), rough ER (red border), and other membranous compartments (green and blue borders). (C and D) 3D projection of confocal image stack of ASP expressing CD8-mCherry and mito-GFP (C) and CD8-GFP and KDEL-RFP (D). (E–G) TEM images of a small membranous compartment inside a thick area of a membrane extension (E), a larger membranous compartment within a thick portion of a membrane extension and bifurcation into thin extensions (F), and a membranous compartment inside a varicosity of a cytoneme (G). Scale bars: 500 nm (A and B), 100 nm (B, inset), 20 µm (C and D), and 100 nm (E–G).
Figure S7.
Membrane extensions contain organelles. (A) Serial TEM images, including section 4 shown in Fig. 7, A and B. (B) A large montage of TEM images (corresponding to section 4) showing the ASP and surrounding myoblasts. The box represents the region shown in A. Scale bars: 1 µm (A) and 5 µm (B).
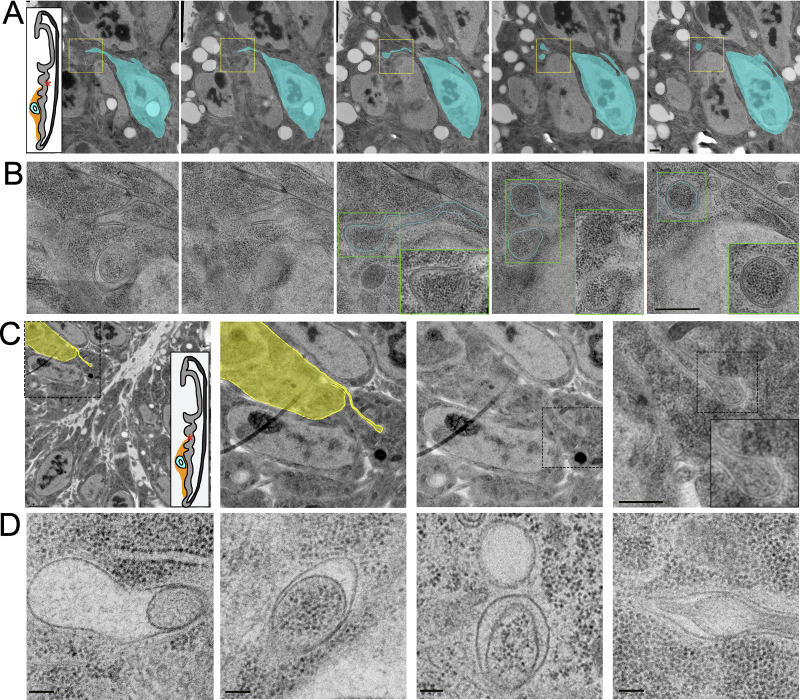
Round or oval membranous compartments are also present in many wide membrane extensions (Fig. 8, E and F). These membranous compartments range widely in size from <50 nm to nearly 800 nm. Membranous compartments also varied in shape, with some being circular (Fig. 8, E and F) and others ovoid (Fig. 7 G). Membranous compartments in thin parts of the extensions are infrequent (Fig. 7 G).
Synaptic vesicles are present at immature neuron–myoblast synapses
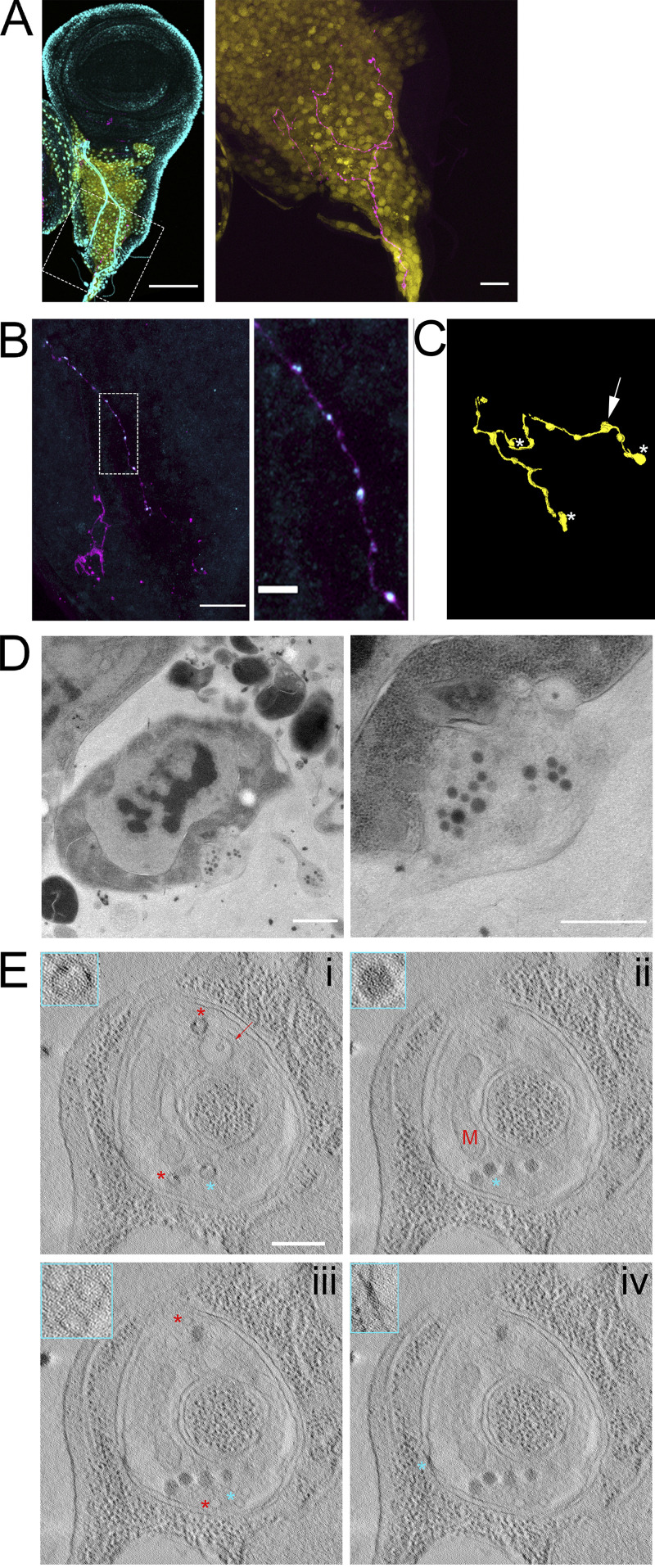
Although membranous compartments similar in size to synaptic vesicles were infrequent in cytonemes or in the cytosol of the target cell surrounding cytoneme termini, we did observe synaptic vesicles concentrated at neuronal synapses in the sections of wing discs. Although there are no previous reports of synapsing neurons in third instar wing discs, we discovered an axon that synapses with multiple myoblasts. This axon was observed only in preparations that minimized physical disruption; it was identified by staining with α-HRP antibody in >40 discs (Fig. 9, A and B) and was identified in 1 disc by TEM (Fig. 9, C–E). HRP staining highlights the axon that extends along the disc stalk and among the myoblasts and branches at several points (Fig. 9 A). Varicosities are present along the axon (Fig. 9 A). Immunostaining confirmed the presence of synaptotagmin within the neuron; staining was especially enriched at some varicosities (Fig. 9 B).
Figure 9.
Synaptic vesicles are preserved in a neuron synapsing with wing disc myoblasts. (A) A fixed wing and leg disc from an L3 larva expressing GFP (yellow) under the control of 1151-Gal4 labeled with DAPI (cyan) and HRP-Alexa Fluor 647. The neuron (magenta) can be seen among the myoblast layer (yellow) of the wing disc. The right panel is shown without DAPI signal. (B) A portion of a fixed disc stained for HRP (magenta) and synaptotagmin (cyan). Confocal images are shown as Z projections. (C) A portion of the myoblast region was imaged by TEM in 40 serial 100-nm sections. The montaged and aligned images were used to segment and 3D reconstruct the neuron. The location of four identified synapses along the neuron are indicated by an arrow, corresponding to the synapse shown in D, or by asterisks. (D) A TEM image showing a section of one of the observed neuron–myoblast synapses. At higher magnification (right), vesicles are apparent. (E) Images from a tomographic tilt series containing the synapse shown in D. Tomography revealed vesicles with electron-dense cargo appearing as puncta on the inner edge of vesicles (i, asterisks), a membranous compartment containing a smaller vesicle (i, arrow), a mitochondrion (ii, M), dense core vesicles (ii, asterisk), clear synaptic vesicles (iii), and electron-dense junctions (iv) between the neuron and myoblast membranes. The insets outlined in cyan show enlarged portions of the image that correspond to cyan asterisks, while red asterisks indicate other examples of the feature shown in the inset. Scale bars: 100 µm (A, left), 20 µm (A, right, and B, right), 1 µm (D, left), and 500 nm (D, right, and E).
The myoblast region with the axon was analyzed in 40 100-nm sections by TEM, and images were used to segment and reconstruct a portion of the neuron in 3D (Fig. 9 C). Four synapses between the neuron and myoblasts were identified (locations marked in Fig. 3 C) and were imaged at higher magnification. Synaptic vesicles were found clustered at synapses (Fig. 9 D) and along the length of the axon, especially at varicosities. A tomographic tilt series of one of the four synapses (Fig. 9 D and Fig. S8) revealed vesicles with different contents (Fig. 9 E), including vesicles with cargo that appeared as dense puncta (Fig. 9 Ei, asterisks), vesicles contained within a larger membranous compartment (Fig. 9 Ei, arrow), dense core vesicles (Fig. 9 Eii), and clear vesicles (Fig. 9 Eiii). An ∼50-nm cleft separates the neuronal and myoblast membranes except at electron-dense junctions (Fig. 9 Eiv). Mitochondria (Fig. 9 Eii) were also observed at synapses. The synapsing portion of the neuron is a spheroid varicosity positioned within a concave pocket of the myoblast membrane (Fig. S8). A portion of the myoblast membrane is enveloped in an invagination of the neuron membrane (Fig. S8, arrows). The features of these synapses suggest that they are not fully mature, as they lack the characteristic T-bar structure of mature Drosophila synapses. The presence of vesicles at neuronal, but not cytoneme synapses in our samples shows that vesicles are preserved but that cytoneme synapses do not have the same type of standing reservoir of vesicles. This suggests that vesicles at cytoneme synapses are much fewer and/or transient than at neuronal synapses.
Figure S8.
The myoblast membrane is partially enveloped by an invagination of the neuronal membrane.Aligned serial TEM images of the synapse shown in Fig. 9, D and E. The section shown in Fig. 9 D corresponds to b. Arrows indicate the portion of the myoblast that is enveloped by an invagination of the neuron. Scale bar: 500 nm.
Discussion
Imaging filopodia has historically been limited by technical impediments. The small diameter of filopodia, which has been approximated at ∼200 nm, limits detection by standard transmitted light microscopy, and because their optical properties are similar to cell bodies, they are essentially invisible where they extend over cells. They are also preserved poorly by chemical fixatives (Ramírez-Weber and Kornberg, 1999). Recent technical innovations, however, now make it possible to see and investigate filopodia in ways that have not previously been possible. Contemporary confocal microscopes have better spatial resolution and more sensitive detectors, filopodia can be brightly marked with fluorescent proteins, and there are efficient methods for mosaic expression of fluorescent, genetically encoded proteins. With these tools, light-emitting filopodia can be detected where they extend over nonfluorescent cells.
The Drosophila wing disc system has many attributes that make it favorable for viewing cytonemes. Its three most abundant cell types—disc epithelia, trachea epithelia, and mesenchymal myoblasts—have low autofluorescence. It is relatively flat and has few cell layers. It can be isolated and mounted quickly without fixation for microscope viewing such that cytonemes extending across epithelia lie in a small number of optical planes. In addition, there are many ways to make genetic mosaics, and signaling systems that communicate within and between the tissues are well characterized. Studies performed during the past 20 years have revealed many novel features of cytonemes in the wing disc system.
Cytonemes are abundant in the wing disc, and many are specialized for signaling with components such as receptors, transporters, and channels. As dedicated signaling organelles, they are designated cytonemes, and as imaging and detection methods have improved since cytonemes were discovered, the inventory of number and variety of cytonemes has increased substantially. Cytonemes have been detected extending from every cell type in the disc system. They are dynamic, vary in length to >100 μm, make synaptic contacts that are essential for exchanging signaling proteins and for signaling, and are regulated such that their numbers correlate with signaling level. Nevertheless, although much has been learned, the limitations of fluorescence microscopy leave many questions unresolved. We do not know, for instance, how many cytonemes a particular cell makes, the precise architecture of the actin-based extensions, the macromolecular contents, or the fine structure of the connections they make with target cells. To address these questions, this study imaged filopodia and cytoneme connections using HPF, serial SEM, and TEM to better resolve the ultrastructure and contents of these specialized filopodia. Our findings not only provide novel insights into these essential signaling organelles but, in addition, have important general implications for the nature of filopodia and the process of synaptogenesis.
By optimally preserving cells in the near-native state with HPF and analyzing cell morphologies at nanometer resolution, we determined the inventory of filopodia that extend from individual epithelial and mesenchymal cells at an instant in time. Every cell we analyzed has at least one filopodia, with ASP cells and myoblasts averaging 17 and 14, respectively. The cell with the highest number is an ASP cell with 22. ASP cytonemes are exclusively basal and basolateral and have varied orientation. The basal surface of most ASP cells projects at least one thick membrane extension that transitions to a thin filopodia that tracks distally along the basal surface of its distal neighbor cell. Myoblasts lack apparent apical/basal polarity but have an expanded, oblong medial region from which both thick projections and filopodia extend. All prominent projections extend radially in the plane of the disc surface and without apparent polarization with respect to anteroposterior or dorsoventral disc axes or the proximodistal ASP axis. In this landscape of epithelia and mesenchyme, we conclude that projections extend the reach of every cell.
Fluorescence microscopy identified cytonemes that extend between anterior compartment disc cells and posterior compartment disc cells (Chen et al., 2017; González-Méndez et al., 2017), between ASP and disc cells (Sato and Kornberg, 2002; Roy et al., 2011), and between myoblasts and ASP cells (Huang and Kornberg, 2015). However, cytonemes connecting cells of the same type in the myoblast layer or ASP had not been previously investigated. Our EM analysis reveals that whereas most filopodia do not contact another cell, when they do, it is most commonly with cells of the same type.
Based on our previous studies and studies of others (Huang et al., 2019; Junyent et al., 2020), we expected that connections between cytonemes and target cells might share structural features with neuronal synapses. Neuronal synapses are characterized by specializations of the pre- and postsynaptic compartments, including the postsynaptic density (Kennedy, 2000), a cleft between the two membranes containing electron-dense complexes (Zuber et al., 2005), and clusters of vesicles in the presynaptic active zone (Harris and Weinberg, 2012). The presence and precise characteristics of these specializations vary with the stage and size of the synapse. During synaptogenesis in insects, the first contact between synaptic partners is initiated by growth cone filopodia that burrow into their target (Bastiani and Goodman, 1984), and in vertebrate immature synapses, electron densities and vesicle-rich active zones are absent (Stelzner et al., 1973; Altman, 1971; Bunge et al., 1967; Glees and Sheppard, 1964; Fröhlich and Meinertzhagen, 1982). The cytoneme connections we imaged burrow into target cell invaginations and do not have the electron densities and vesicles that are characteristic of mature neuronal synapses. There are regions of high density between the cytoneme and target cell membranes (Fig. 3 G and Fig. 4 I) that are similar to specializations that have been observed in developing house fly brains (Fröhlich and Meinertzhagen, 1982). These cytoneme connections appear to share features with the early, maturing neuronal synapses, which acquire structural components stepwise (Fröhlich and Meinertzhagen, 1982). The lack of a standing pool of vesicles at cytoneme synapses does not invalidate evidence that vesicles are important for cytoneme signaling (Huang et al., 2019; González-Méndez et al., 2020); rather, it suggests that cytoneme vesicles might be few and transient.
Structures with cardinal features of neuronal synapses were identified in our EM preparations. Although synapsing neurons have been identified in the wing disc system during the pupal but not larval stage (Jan et al., 1985), the use of collagenase before fixation in this previous study may have resulted in the loss of the neuron we identified that contacts myoblasts in third instar discs. In addition, this neuron is thin, and the increased brightness of fluorophores and increased sensitivity of detectors available today may also explain why it had not yet been detected. The soma of this neuron is outside of the wing disc, but its axon appears to extend into the hypodermal stalk of the wing disc and through the myoblast layer. The connections between the neuron and myoblast have synaptic vesicles, mitochondria, and electron-dense junctions that are consistent with synaptic function, although they lack the T-bars that are characteristic of mature Drosophila synapses.
The cytoneme–cell connections we detected penetrate several microns into cell invaginations, a feature shared with signaling cellular projections, such as those extended of granulosa cells of the mouse ovarian follicle (Baena and Terasaki, 2019) and growth cone filopodia of the grasshopper embryo (Bastiani and Goodman, 1984). Membrane invagination by closed-ended TNTs has also been observed (Sartori-Rupp et al., 2019), and invaginating structures are common at mammalian synapses (Petralia et al., 2015, 2018). Membrane protrusions of T cells end in invaginations of dendritic cells and are characteristic of the immunological synapse (Leithner et al., 2021). Sponges elongate processes that make invaginating contacts with other cells, although they do not have neurons (Petralia et al., 2015).
Penetrating cytonemes are mirrored closely by the cell membrane except at the end where a gap is frequent. This gap ranges in size and shape, perhaps representing different stages of maturing cytoneme–cell connections. Growth cone filopodia that selectively invaginate the membranes of other growth cones and neuron cell bodies have a space along both the sides and the tip of the filopodia that increases during maturation (Bastiani and Goodman, 1984). It is possible that the cytonemes we observed with large amounts of space around the terminus represent a more advanced stage of cytoneme–cell interaction.
In both extracellular space and invaginations, cytoneme shafts have distinct regions of wide-diameter varicosities and narrow-diameter constrictions. Although filopodia have been examined by EM before, to our knowledge, this is the first study to examine filopodia in HPF tissue by serial EM. HPF filopodia have been visualized in cross-section before, but the shape along the length of filopodia was not assessed (Lak et al., 2015). HPF-preserved membrane protrusions of human mammary epithelial cells imaged via focused ion beam SEM have bulges and uneven thicknesses (Jorgens et al., 2017), although they are more irregular than the varicosities we observed and do not have a repeating pattern.
Filopodia imaged after chemical fixation typically have more uniform diameter and less smooth membranes compared with the structures observed here. Chemical fixation is known to cause shrinkage of tissues and distortion of cellular and organellar membranes (McDonald, 2014) and might be responsible for this difference. Alternatively, other types of filopodia may have more ordered interiors and different cytoskeletal proteins that confer better stabilization during chemical processing. In this study, HPF and FS, the gold standard for EM processing (McDonald, 2014), were employed to cryoimmobilized cells and tissues in order to effectively fix and protect ultrastructural components through the processing of the tissues into acetone and resin.
The varicosity/constriction filopodia architecture has features reminiscent of en passant synapses, which have variably spaced varicosities at synaptic sites (Giachello et al., 2012; Shepherd et al., 2002). TEM analysis of human intraretinal ganglion axons has identified varicosities with very similar shapes as cytoneme varicosities, although the axon diameter is >10 times that of cytonemes (Wang et al., 2003). And although neuronal varicosities store synaptic vesicles and other synaptic machinery, most filopodia varicosities have high ribosomal content, and few contain vesicles or membranous compartments. It is unclear whether varicosities we observed in filopodia might be related to the periodic pearling seen in some membrane protrusions (Caballero et al., 2019; Heinrich et al., 2014). Bulges have been observed in TNTs by SEM (Scholkmann et al., 2018), but their contents, including vesicles and other membranous compartments, distinguish them from filopodia varicosities.
Although some filopodia extend directly from cells, others extend from thicker membrane projections where mitochondria, rough ER, and membrane-bound compartments are common. These organelles are extremely rare in filopodia. Ribosomes, in contrast, are abundant and widely disseminated in filopodia and are often at the same density as the cellular cytosol. This suggests that thick membrane projections and filopodia are both cytosolic extensions of the cell, with organellar exclusion occurring based on size rather than selective filtering. In contrast, primary cilia filter content and have no ribosomes.
Materials and methods
HPF
Prior to dissection, planchets (Ted Pella) were coated with 1-hexadecene (Sigma-Aldrich) and thoroughly dried. Wing imaginal discs were dissected in either PBS or Grace’s Insect Medium (Thermo Fisher Scientific) from wandering third instar larva. Using forceps, the bottom third of the larva was removed. One tine of a forceps was inserted into the mouth, and the larva was turned inside out along the tine using the other forceps. Fat bodies, intestines, and dorsal trunks of the trachea were removed carefully. The wing disc was pulled away from the carcass and transferred via capillary action between forceps to a drop of PBS or medium on a staging dish. 20% BSA solution made with either PBS or Grace’s Insect Medium was used as cryoprotectant. A 0.1-mm “A” planchet was partially filled with BSA solution. Approximately 100 μl of BSA solution was added to the PBS or medium drop containing the disc. The disc was transferred serially to two drops of BSA solution before being transferred to the planchet. The level of BSA solution in the planchet was adjusted to a slight convex meniscus with forceps. The planchet was transferred to the specimen holder, and the flat side of “B” planchet was placed on top of the specimen-containing planchet. The sample holder was transferred to the BAL-TEC HPM-010 high-pressure freezer and frozen. This process was performed as quickly as possible in order to reduce the time from dissection to freezing and exposure of the disc to BSA solution. The entire process from dissection to freezing took between 3 and 5 min, and the disc was exposed to BSA for <1 min.
FS
FS was done according to the super-quick FS protocol (McDonald and Webb, 2011). After HPF, planchets were separated, and the sample-containing sides were placed in cryotubes containing acetone-based media with 4% OsO4, 0.1% uranyl acetate, 2% MeOH, and 5% H2O. No more than three samples were placed in any one tube. The media had been prefrozen and stored in liquid nitrogen (LN2). At all times after freezing and before FS, samples were kept at LN2 temperature by performing transfers in LN2 or LN2 vapor, although care was taken to not introduce LN2 into the cryotube before FS. Prior to FS, an aluminum block with slots for 13-mm tubes was cooled with LN2 in a rectangular foam ice bucket. Cryotubes containing the samples and FS media were transferred into one of the end rows of the LN2-filled block. The temperature was monitored by wrapping a temperature probe around a tube filled with acetone, which was then placed in the block. To begin FS, the block was turned on its side within the ice bucket so that the row with the tubes was closest to the bottom of the bucket and so that the tube caps were facing one wall of the ice bucket. The LN2 was poured out of the ice bucket, and the uncovered bucket was placed on a shaker and agitated at 100 rpm. The bucket was shaken until the temperature of the tubes reached 2–4°C. The FS process took between 2.5 and 3 h.
Resin infiltration
Samples were washed five times with glass distilled acetone (Electron Microscopy Sciences). During the last wash, the tubes were placed on a rocker for 5 min. The samples were gradually infiltrated with epon/araldite (EA) resin through progressive steps (25%, 50%, 75%) of EA/acetone mixture (McDonald and Müller-Reichert, 2002). EA resin consisted of 6.2 g Eponate 12, 4.4 g Araldite 502, and 12.2 g dodecenyl succinic anhydride. 1 ml of EA/acetone mixture was pipetted into the tube after removal of the previous step. Using a PELCO BioWave Pro (Ted Pella), uncapped tubes were placed in direct contact with the cold spot and microwaved under vacuum at 150 mW for 1 min. For the 25% and 50% EA/acetone steps, tubes were rotated for 1 h at room temperature, after which they were briefly centrifuged to ensure that the samples were at the bottom of the tube. Some samples came out of the planchets on their own during rotation, the ones that did not were visualized using a dissecting microscope and removed using a needle tip to carefully trace around the border between the sample and planchet and gently lift. 75% EA/acetone was added to the samples, which were then microwaved and rotated overnight. The next day, the resin mixture was replaced with 100% EA and rotated for 1 h. The samples were placed in a new tube containing accelerated EA (same recipe as above with 0.8 ml benzyldimethylamine added) and rotated for 3 h with exchanges of fresh EA every hour. Between each exchange, samples were briefly centrifuged down. Samples were then flat embedded between two Teflon-coated slides, with Parafilm used as a spacer (Müller-Reichert et al., 2003), and baked at 60°C for 48 h.
Chemical fixation
In preparation for EM, larvae were dissected in cold cacodylate buffer and fixed with 2% glutaraldehyde (EM grade) in 0.1 M sodium cacodylate buffer (pH 7.4) at room temperature. Samples were stored in the 2% glutaraldehyde solution until reduced osmium-thiocarbohydrazide–reduced osmium staining was ready to be performed (>24 h). As previously described (Terasaki et al., 2013), the samples were postfixed for 1 h with a 1:1 mixture of 4% aqueous osmium tetroxide and 3% potassium ferrocyanide in 0.2 M cacodylate buffer at pH 7.4. The samples were then rinsed with distilled water and treated with 1% aqueous thiocarbohydrazide for 20 min. Following a rinse with distilled water, samples were treated with 2% aqueous osmium tetroxide for 30 min and rinsed again with distilled water. Overnight at 4°C, samples were treated with 1% aqueous uranyl acetate. Following a distilled water rinse, samples were treated with 0.066% lead acetate for 30 min and again rinsed with water. Samples were dehydrated with a series of increasing ethanol solutions and infiltrated with epoxy resin (Poly/Bed; Polysciences).
Section preparation
For TEM, a Reichert-Jung Ultracut E microtome was used to cut 70- or 100-nm-thick sections with a diamond knife (Ultra 45°; DiATOME), which were collected onto formvar film–coated copper/rhodium slot grids. Sections were poststained with 2% uranyl acetate and Reynold’s lead citrate.
For SEM, 60-nm sections were cut using a diamond knife (Ultra 45°, DiATOME) and collected using an automated tape-collecting ultramicrotome. The tape was transferred to silicon wafers and carbon coated (Baena et al., 2019).
EM
TEM was performed using a Tecnai 12 microscope (FEI) operating at 120 kV. Images were acquired with Gatan Microscopy Suite version 3 software and a Gatan Ultrascan 2000 or Gatan Rio camera. Montage imaging was performed using SerialEM (Mastronarde, 2005). Custom software was used to extract individual image files from the montage imaging stack and to produce text files that simplified their import into TrakEM2. Pixel sizes for TEM images range between 0.52 and 3.54 nm/pixel.
Tomography was performed on a Tecnai 20 electron microscope (FEI). A ±60° single-tilt series was performed, with images acquired every 1°. IMOD software was used to reconstruct the tilt series (Kremer et al., 1996).
Serial SEM imaging was performed using two microscopes: a Zeiss Sigma field-emission scanning electron microscope in backscatter mode at 8 keV (Fig. 1 and Fig. 6) and an FEI Verios field-emission scanning electron microscope in backscatter mode at 5 keV (Fig. 3). The manufactures’ software was used to acquire images on both microscopes. Original pixel size for SEM images shown in Fig. 1 and Fig. 4 was 5 nm/pixel and 4.5 nm/pixel for SEM images shown in Fig. 7. Resolution for both image sets was reduced to 33% of the original image size (i.e., 15 nm/pixel and 13.5 nm/pixel) for ease of alignment. Pixel size is 6.77 nm/pixel for Fig. 3 B and 0.886 nm/pixel for Fig. 3 C.
The TrakEM2 module (Cardona et al., 2012) of FIJI ImageJ (Schindelin et al., 2012) was used to reconstruct montage data sets, align image stacks from serial sections for both SEM and TEM, segment images, and produce 3D reconstructions of segmentation. Images acquired on the Zeiss Sigma scanning electron microscope were prealigned using the Linear Stack Alignment with SIFT ImageJ plugin before being imported into the TrakEM2 module.
ASP and myoblast connection map
For the cytoneme connection map shown in Fig. 6, ASP cells and myoblasts within the 172-image serial SEM stack (the image stack also used in Fig. 1 and Fig. 4, A–D) were assigned a number to track their connections. Each cell was examined to identify all cytonemes that invaginated with its membrane, and when possible, the cytonemes were tracked back to the originating cell. The rough XY positioning of each cell was mapped, and each cell was assigned to one of five Z depth colors determined by the placement of its nucleus within the stack. Green cells have the majority of their nuclei within sections 1–40 (deepest sections), yellow cell nuclei correspond to sections 41–80, orange cell nuclei correspond to sections 81–120, red cell nuclei correspond to sections 121–160, and purple cell nuclei correspond to sections 161–172. Arrows mark the direction of the connection only (originating at the filopodia sending cell and pointing toward the cytoneme receiving cell) and do not indicate information about the length or path of the cytoneme.
Filopodia dimensions
Filopodia lengths were estimated from nine ASP cells and eight myoblasts reconstructed in 3D. Coordinates of points at the beginning and end of each cytoneme were obtained and lengths estimated, with the assumption that they were straight between the beginning and end points, using the distance formula: AB = √(x2 − x1)2 + (y2 − y1)2 + (z2 − z1)2.
Varicosities and constrictions were defined by the change in diameter along their lengths. Constrictions were relatively uniform in diameter, while varicosities had smoothly increasing then decreasing diameters along their lengths. The beginning and ends of varicosities were defined as the point at which the diameter began increasing from the preceding constriction (beginning of varicosity) and the point at which the diameter decreased to the diameter of the next constriction (end of varicosity). Varicosity and constriction diameters were measured at their midpoints.
CM
Live imaging
Wing imaginal discs were dissected as described above in Grace’s Insect Medium. Discs were mounted by placing the disc in a drop of medium on a coverslip with the ASP side down. Medium was slowly pipetted from the drop until the level was such that the surface tension had just begun to compress the disc against the coverslip. The coverslip was inverted onto a concavity slide with small amounts of silicone vacuum grease applied to the area surrounding the slide depression. Imaging was performed at room temperature. Tissue was imaged using 488 and 560 wavelength lasers, an inverted laser scanning confocal microscope (Olympus FV-3000), a 60× (NA 1.35; Fig. 7) or 40× oil objective (NA 1.3; Fig. 8), and GaAsp photomultiplier tube detectors. Images were acquired with Olympus FV31S-SW software. Images were 1,024 × 1,024 pixels, except for the CD2-GFP images represented in Fig. 7 N, which were 2,048 × 2,048 pixels. Line averaging from two scans and 3× or 4× zoom was used to acquire CM images in Fig. 7. Images were acquired as Z-stacks, with step sizes of 0.3 μm (Fig. 7) or 0.4 μm (Fig. 8). Fig. 7 images were deconvoluted using Olympus cellSens software with three iterations of an advanced maximum likelihood estimation algorithm. Fig. 8 image stacks were 3D rendered with the ImageJ plugin ClearVolume (Royer et al., 2015). Pixel sizes were 198 nm/pixel (Fig. 8), 26 nm/pixel (Fig. 7 M), and 69 nm/pixel (Fig. 7 N).
Constriction lengths were measured in filopodia from T-TRAP–expressing myoblasts. High expression of T-TRAP in myoblasts appeared to affect cell viability, as rounded cells and cell debris were present within the myoblast layer. Constriction lengths were measured only in filopodia extending from myoblasts with a healthy appearance and normal morphology. FIJI ImageJ was used to measure the length between the ends of adjacent bright points.
Immunohistochemistry
The posterior third of the larva was removed, the cuticle was inverted, and fat bodies were removed. Carcasses were fixed in 4% PFA for 25 min. After three washes of PBS, 0.2% Triton X-100 in PBS (TBST) was used to permeabilize cells for 10 min followed by blocking with Roche blocking reagent diluted 1:5 in TBST for 30 min. Antibodies were diluted in blocking reagent and incubated overnight at 4°C. After three 10-min TBST washes, secondary antibody goat anti-mouse Alexa Fluor 555 (1:500, A-21422; Invitrogen) or Alexa 405 conjugated phalloidin (1:400, A-30104; Invitrogen) was diluted in blocking reagent and incubated with the carcasses for 45 min at room temperature. After three 10-min TBST washes, the wing discs were detached from the trachea and gently separated from the carcass. Two strips of double-sided tape were used to provide space between the slide and coverslip. Discs were mounted in VECTASHIELD mounting reagent (Vector Laboratories). Confocal images are shown as maximum Z projections of multiple confocal slices. Pixel sizes were 1.24 μm/pixel (Fig. 9 A, left), 414 nm/pixel (Fig. 9 A, right), and 621 nm/pixel (Fig. 9 B).
Primary antibodies
Alexa Fluor 647–conjugated goat anti-HRP (1:100, 123-605-021; Jackson ImmunoResearch) and mouse anti-synaptotagmin (1:5, 3H2 2D7; Developmental Studies Hybridoma Bank).
Fly lines
For CM, btl-Gal4 (Roy et al., 2014), 1151-Gal4 from K. VijayRaghavan (National Center for Biological Sciences Tata Institute of Fundamental Research, Bangalore, Karnataka, India; Roy and VijayRaghavan, 1997), btl-LHG, lexO-CD2-GFP (Roy et al., 2014), T-TRAP (Zhang et al., 2016), UAS-mcd8:GFP, UAS-mcd8:mCherry, UAS-RFP:KDEL (#30910; Bloomington), UAS-mito:GFP (#8443; Bloomington), and UAS-GFP/Tm6B. For EM, w1118.
Sample number and statistical analysis
EM
HPF discs from 10 larvae were examined by EM. All samples had the same filopodia and cytoneme synapse morphology. Images shown in figures are representative of all discs examined. Chemically fixed discs from four larvae were examined for filopodia morphology.
CM
Nine (KDEL-RFP) and 10 (mito-GFP) ASP were examined for ER and mitochondria localization. Cytonemes projecting from four ASP (btl-LHG, lexO-CD2-GFP) and the myoblast layers of seven discs were photographed, and images were processed by deconvolution; filopodia morphology was consistent with the filopodia shown in Fig. 7, M and N. 56 constrictions were measured across 16 cytonemes to determine interpoint distance from deconvoluted images of T-TRAP–expressing myoblasts. More than 40 discs were examined with HRP staining.
Constriction and varicosity widths were compared with a Student’s t test. When reported in the text, average and SD are listed.
Online supplemental material
Serial section images are shown in Figs. S1, S2, S3, S4, and S5 and Figs. S7 and S8. Fig. S6 shows segmented serial SEM images. Video 1 shows 360° rotations of the 3D-reconstructed cells shown in Fig. 1, D and E.
Supplementary Material
Acknowledgments
We thank Reena Zalpuri and Dr. Kent McDonald at the University of California, Berkeley, Electron Microscope Laboratory for training and advice in HPF and TEM techniques. We thank Dr. Richard Fetter for advice, sharing of custom software, and help with tomography data acquisition.
This work was supported by National Institutes of Health grants 5F32HL147624 (B.M. Wood) and R35GM122548 (T.B. Kornberg).
The authors declare no competing financial interests.
Author contributions: B.M. Wood contributed to the conceptualization, formal analysis, funding acquisition, investigation, project administration, visualization, and writing of the original draft. V. Baena, H. Huang, and D.M. Jorgens contributed to the investigation and writing, review, and editing. M. Terasaki contributed to the investigation, resources, and writing, review, and editing. T.B. Kornberg contributed to the conceptualization, funding acquisition, resources, supervision, and writing, review, and editing.
References
- Alarcon-Martinez, L., Villafranca-Baughman D., Quintero H., Kacerovsky J.B., Dotigny F., Murai K.K., Prat A., Drapeau P., and Di Polo A.. 2020. Interpericyte tunnelling nanotubes regulate neurovascular coupling. Nature. 585:91–95. 10.1038/s41586-020-2589-x [DOI] [PubMed] [Google Scholar]
- Allen, M., Poggiali D., Whitaker K., Marshall T.R., and Kievit R.A.. 2019. Raincloud plots: a multi-platform tool for robust data visualization. Wellcome Open Res. 4:63. 10.12688/wellcomeopenres.15191.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman, J. 1971. Coated vesicles and synaptogenesis. A developmental study in the cerebellar cortex of the rat. Brain Res. 30:311–322. 10.1016/0006-8993(71)90081-3 [DOI] [PubMed] [Google Scholar]
- Anvarian, Z., Mykytyn K., Mukhopadhyay S., Pedersen L.B., and Christensen S.T.. 2019. Cellular signalling by primary cilia in development, organ function and disease. Nat. Rev. Nephrol. 15:199–219. 10.1038/s41581-019-0116-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena, V., and Terasaki M.. 2019. Three-dimensional organization of transzonal projections and other cytoplasmic extensions in the mouse ovarian follicle. Sci. Rep. 9:1262. 10.1038/s41598-018-37766-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena, V., Schalek R.L., Lichtman J.W., and Terasaki M.. 2019. Serial-section electron microscopy using automated tape-collecting ultramicrotome (ATUM). Methods Cell Biol. 152:41–67. 10.1016/bs.mcb.2019.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiani, M.J., and Goodman C.S.. 1984. Neuronal growth cones: specific interactions mediated by filopodial insertion and induction of coated vesicles. Proc. Natl. Acad. Sci. USA. 81:1849–1853. 10.1073/pnas.81.6.1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, M., Gradilla A.-C., Seijo I., Andrés G., Rodríguez-Navas C., González-Méndez L., and Guerrero I.. 2013. Cytonemes are required for the establishment of a normal Hedgehog morphogen gradient in Drosophila epithelia. Nat. Cell Biol. 15:1269–1281. 10.1038/ncb2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunge, M.B., Bunge R.P., and Peterson E.R.. 1967. The onset of synapse formation in spinal cord cultures as studied by electron microscopy. Brain Res. 6:728–749. 10.1016/0006-8993(67)90129-1 [DOI] [PubMed] [Google Scholar]
- Caballero, D., Pinto I.M., Rubinstein B.Y., and Samitier J.. 2019. Protrusion membrane pearling emerges during 3D cell division. Phys. Biol. 16:066009. 10.1088/1478-3975/ab4549 [DOI] [PubMed] [Google Scholar]
- Callejo, A., Bilioni A., Mollica E., Gorfinkiel N., Andrés G., Ibáñez C., Torroja C., Doglio L., Sierra J., and Guerrero I.. 2011. Dispatched mediates Hedgehog basolateral release to form the long-range morphogenetic gradient in the Drosophila wing disk epithelium. Proc. Natl. Acad. Sci. USA. 108:12591–12598. 10.1073/pnas.1106881108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona, A., Saalfeld S., Schindelin J., Arganda-Carreras I., Preibisch S., Longair M., Tomancak P., Hartenstein V., and Douglas R.J.. 2012. TrakEM2 software for neural circuit reconstruction. PLoS One. 7:e38011. 10.1371/journal.pone.0038011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W., Huang H., Hatori R., and Kornberg T.B.. 2017. Essential basal cytonemes take up Hedgehog in the Drosophila wing imaginal disc. Development. 144:3134–3144. 10.1242/dev.149856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont, M., Souriant S., Lugo-Villarino G., Maridonneau-Parini I., and Vérollet C.. 2018. Tunneling nanotubes: intimate communication between myeloid cells. Front. Immunol. 9:43. 10.3389/fimmu.2018.00043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch, C., and Dupuis-Williams P.. 2011. Ultrastructure of cilia and flagella - back to the future! Biol. Cell. 103:249–270. 10.1042/BC20100139 [DOI] [PubMed] [Google Scholar]
- Fröhlich, A., and Meinertzhagen I.A.. 1982. Synaptogenesis in the first optic neuropile of the fly’s visual system. J. Neurocytol. 11:159–180. 10.1007/BF01258010 [DOI] [PubMed] [Google Scholar]
- Giachello, C.N.G., Montarolo P.G., and Ghirardi M.. 2012. Synaptic functions of invertebrate varicosities: what molecular mechanisms lie beneath. Neural Plast. 2012:670821. 10.1155/2012/670821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glees, P., and Sheppard B.L.. 1964. Electron microscopical studies of the synapse in the developing chick spinal cord. Z. Zellforsch. Mikrosk. Anat. 62:356–362. 10.1007/BF00339285 [DOI] [PubMed] [Google Scholar]
- González-Méndez, L., Seijo-Barandiarán I., and Guerrero I.. 2017. Cytoneme-mediated cell-cell contacts for Hedgehog reception. eLife. 6:e24045. 10.7554/eLife.24045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Méndez, L., Gradilla A.C., Sánchez-Hernández D., González E., Aguirre-Tamaral A., Jiménez-Jiménez C., Guerra M., Aguilar G., Andrés G., Falcón-Pérez J.M., and Guerrero I.. 2020. Polarized sorting of Patched enables cytoneme-mediated Hedgehog reception in the Drosophila wing disc. EMBO J. 39:e103629. 10.15252/embj.2019103629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, K.M., and Weinberg R.J.. 2012. Ultrastructure of synapses in the mammalian brain. Cold Spring Harb. Perspect. Biol. 4:a005587. 10.1101/cshperspect.a005587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich, D., Ecke M., Jasnin M., Engel U., and Gerisch G.. 2014. Reversible membrane pearling in live cells upon destruction of the actin cortex. Biophys. J. 106:1079–1091. 10.1016/j.bpj.2013.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H., and Kornberg T.B.. 2015. Myoblast cytonemes mediate Wg signaling from the wing imaginal disc and Delta-Notch signaling to the air sac primordium. eLife. 4:e06114. 10.7554/eLife.06114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H., Liu S., and Kornberg T.B.. 2019. Glutamate signaling at cytoneme synapses. Science. 363:948–955. 10.1126/science.aat5053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan, Y.N., Ghysen A., Christoph I., Barbel S., and Jan L.Y.. 1985. Formation of neuronal pathways in the imaginal discs of Drosophila melanogaster. J. Neurosci. 5:2453–2464. 10.1523/JNEUROSCI.05-09-02453.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgens, D.M., Inman J.L., Wojcik M., Robertson C., Palsdottir H., Tsai W.-T., Huang H., Bruni-Cardoso A., López C.S., Bissell M.J., et al. 2017. Deep nuclear invaginations are linked to cytoskeletal filaments - integrated bioimaging of epithelial cells in 3D culture. J. Cell Sci. 130:177–189. 10.1242/jcs.190967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junyent, S., Garcin C.L., Szczerkowski J.L.A., Trieu T.-J., Reeves J., and Habib S.J.. 2020. Specialized cytonemes induce self-organization of stem cells. Proc. Natl. Acad. Sci. USA. 117:7236–7244. 10.1073/pnas.1920837117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko, Y., and Walther P.. 1995. Comparison of ultrastructure of germinating pea leaves prepared by high-pressure freezing-freeze substitution and conventional chemical fixation. J. Electron Microsc. (Tokyo). 44:104–109. 10.1093/oxfordjournals.jmicro.a051149 [DOI] [PubMed] [Google Scholar]
- Kennedy, M.B. 2000. Signal-processing machines at the postsynaptic density. Science. 290:750–754. 10.1126/science.290.5492.750 [DOI] [PubMed] [Google Scholar]
- Kornberg, T.B. 2014a. Cytonemes and the dispersion of morphogens. Wiley Interdiscip. Rev. Dev. Biol. 3:445–463. 10.1002/wdev.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg, T.B. 2014b. The contrasting roles of primary cilia and cytonemes in Hh signaling. Dev. Biol. 394:1–5. 10.1016/j.ydbio.2014.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg, T.B. 2017. Distributing signaling proteins in space and time: the province of cytonemes. Curr. Opin. Genet. Dev. 45:22–27. 10.1016/j.gde.2017.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korogod, N., Petersen C.C., and Knott G.W.. 2015. Ultrastructural analysis of adult mouse neocortex comparing aldehyde perfusion with cryo fixation. eLife. 4:e05793. 10.7554/eLife.05793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer, J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Lak, B., Yushin V.V., Slos D., Claeys M., Decraemer W., and Bert W.. 2015. High-pressure freezing and freeze-substitution fixation reveal the ultrastructure of immature and mature spermatozoa of the plant-parasitic nematode Trichodorus similis (Nematoda; Triplonchida; Trichodoridae). Micron. 77:25–31. 10.1016/j.micron.2015.05.012 [DOI] [PubMed] [Google Scholar]
- Leithner, A., Altenburger L.M., Hauschild R., Assen F.P., Rottner K., Stradal T.E.B., Diz-Muñoz A., Stein J.V., and Sixt M.. 2021. Dendritic cell actin dynamics control contact duration and priming efficiency at the immunological synapse. J. Cell Biol. 220:e202006081. 10.1083/jcb.202006081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde, D.N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51. 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Mattes, B., Dang Y., Greicius G., Kaufmann L.T., Prunsche B., Rosenbauer J., Stegmaier J., Mikut R., Özbek S., Nienhaus G.U., et al. 2018. Wnt/PCP controls spreading of Wnt/β-catenin signals by cytonemes in vertebrates. eLife. 7:e36953. 10.7554/eLife.36953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy-Simandle, K., Hanna S.J., and Cox D.. 2016. Exosomes and nanotubes: Control of immune cell communication. Int. J. Biochem. Cell Biol. 71:44–54. 10.1016/j.biocel.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, K.L. 2014. Out with the old and in with the new: rapid specimen preparation procedures for electron microscopy of sectioned biological material. Protoplasma. 251:429–448. 10.1007/s00709-013-0575-y [DOI] [PubMed] [Google Scholar]
- McDonald, K., and Müller-Reichert T.. 2002. Cryomethods for thin section electron microscopy. Methods Enzymol. 351:96–123. 10.1016/S0076-6879(02)51843-7 [DOI] [PubMed] [Google Scholar]
- McDonald, K.L., and Webb R.I.. 2011. Freeze substitution in 3 hours or less. J. Microsc. 243:227–233. 10.1111/j.1365-2818.2011.03526.x [DOI] [PubMed] [Google Scholar]
- Müller-Reichert, T., Hohenberg H., O’Toole E.T., and McDonald K.. 2003. Cryoimmobilization and three-dimensional visualization of C. elegans ultrastructure. J. Microsc. 212:71–80. 10.1046/j.1365-2818.2003.01250.x [DOI] [PubMed] [Google Scholar]
- Petralia, R.S., Wang Y.-X., Mattson M.P., and Yao P.J.. 2015. Structure, distribution, and function of neuronal/synaptic spinules and related invaginating projections. Neuromolecular Med. 17:211–240. 10.1007/s12017-015-8358-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia, R.S., Wang Y.X., Mattson M.P., and Yao P.J.. 2018. Invaginating structures in mammalian synapses. Front. Synaptic Neurosci. 10:4. 10.3389/fnsyn.2018.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Weber, F.-A., and Kornberg T.B.. 1999. Cytonemes: cellular processes that project to the principal signaling center in Drosophila imaginal discs. Cell. 97:599–607. 10.1016/S0092-8674(00)80771-0 [DOI] [PubMed] [Google Scholar]
- Rojas-Ríos, P., Guerrero I., and González-Reyes A.. 2012. Cytoneme-mediated delivery of hedgehog regulates the expression of bone morphogenetic proteins to maintain germline stem cells in Drosophila. PLoS Biol. 10:e1001298. 10.1371/journal.pbio.1001298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum, J.L., and Child F.M.. 1967. Flagellar regeneration in protozoan flagellates. J. Cell Biol. 34:345–364. 10.1083/jcb.34.1.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S., and VijayRaghavan K.. 1997. Homeotic genes and the regulation of myoblast migration, fusion, and fibre-specific gene expression during adult myogenesis in Drosophila. Development. 124:3333–3341. [DOI] [PubMed] [Google Scholar]
- Roy, S., Hsiung F., and Kornberg T.B.. 2011. Specificity of Drosophila cytonemes for distinct signaling pathways. Science. 332:354–358. 10.1126/science.1198949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S., Huang H., Liu S., and Kornberg T.B.. 2014. Cytoneme-mediated contact-dependent transport of the Drosophila decapentaplegic signaling protein. Science. 343:1244624. 10.1126/science.1244624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer, S.M., and Kinnamon J.C.. 1996. Comparison of high-pressure freezing/freeze substitution and chemical fixation of catfish barbel taste buds. Microsc. Res. Tech. 35:385–412. [DOI] [PubMed] [Google Scholar]
- Royer, L.A., Weigert M., Günther U., Maghelli N., Jug F., Sbalzarini I.F., and Myers E.W.. 2015. ClearVolume: open-source live 3D visualization for light-sheet microscopy. Nat. Methods. 12:480–481. 10.1038/nmeth.3372 [DOI] [PubMed] [Google Scholar]
- Rustom, A., Saffrich R., Markovic I., Walther P., and Gerdes H.-H.. 2004. Nanotubular highways for intercellular organelle transport. Science. 303:1007–1010. 10.1126/science.1093133 [DOI] [PubMed] [Google Scholar]
- Sanders, T.A., Llagostera E., and Barna M.. 2013. Specialized filopodia direct long-range transport of SHH during vertebrate tissue patterning. Nature. 497:628–632. 10.1038/nature12157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori-Rupp, A., Cordero Cervantes D., Pepe A., Gousset K., Delage E., Corroyer-Dulmont S., Schmitt C., Krijnse-Locker J., and Zurzolo C.. 2019. Correlative cryo-electron microscopy reveals the structure of TNTs in neuronal cells. Nat. Commun. 10:342. 10.1038/s41467-018-08178-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M., and Kornberg T.B.. 2002. FGF is an essential mitogen and chemoattractant for the air sacs of the drosophila tracheal system. Dev. Cell. 3:195–207. 10.1016/S1534-5807(02)00202-2 [DOI] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholkmann, F., Seidel D., Funk R.H.W., and Roehlecke C.. 2018. Ultrastructural features (bulges) of membrane nanotubes between cone-like photoreceptor cells: an investigation employing scanning electron microscopy. Matters (Zur.). 10.19185/matters.201802000011 [DOI] [Google Scholar]
- Shepherd, G.M.G., Raastad M., and Andersen P.. 2002. General and variable features of varicosity spacing along unmyelinated axons in the hippocampus and cerebellum. Proc. Natl. Acad. Sci. USA. 99:6340–6345. 10.1073/pnas.052151299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowinski, S., Jolly C., Berninghausen O., Purbhoo M.A., Chauveau A., Köhler K., Oddos S., Eissmann P., Brodsky F.M., Hopkins C., et al. 2008. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat. Cell Biol. 10:211–219. 10.1038/ncb1682 [DOI] [PubMed] [Google Scholar]
- Stanganello, E., Hagemann A.I.H., Mattes B., Sinner C., Meyen D., Weber S., Schug A., Raz E., and Scholpp S.. 2015. Filopodia-based Wnt transport during vertebrate tissue patterning. Nat. Commun. 6:5846. 10.1038/ncomms6846 [DOI] [PubMed] [Google Scholar]
- Stelzner, D.J., Martin A.H., and Scott G.L.. 1973. Early stages of synaptogenesis in the cervical spinal cord of the chick embryo. Z. Zellforsch. Mikrosk. Anat. 138:475–488. 10.1007/BF00572291 [DOI] [PubMed] [Google Scholar]
- Studer, D., Hennecke H., and Müller M.. 1992. High-pressure freezing of soybean nodules leads to an improved preservation of ultrastructure. Planta. 188:155–163. 10.1007/BF00216809 [DOI] [PubMed] [Google Scholar]
- Terasaki, M., Shemesh T., Kasthuri N., Klemm R.W., Schalek R., Hayworth K.J., Hand A.R., Yankova M., Huber G., Lichtman J.W., et al. 2013. Stacked endoplasmic reticulum sheets are connected by helicoidal membrane motifs. Cell. 154:285–296. 10.1016/j.cell.2013.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Dong J., Cull G., Fortune B., and Cioffi G.A.. 2003. Varicosities of intraretinal ganglion cell axons in human and nonhuman primates. Invest. Ophthalmol. Vis. Sci. 44:2–9. 10.1167/iovs.02-0333 [DOI] [PubMed] [Google Scholar]
- Yamashita, Y.M., Inaba M., and Buszczak M.. 2018. Specialized intercellular communications via cytonemes and nanotubes. Annu. Rev. Cell Dev. Biol. 34:59–84. 10.1146/annurev-cellbio-100617-062932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K.X., Tan L., Pellegrini M., Zipursky S.L., and McEwen J.M.. 2016. Rapid changes in the translatome during the conversion of growth cones to synaptic terminals. Cell Rep. 14:1258–1271. 10.1016/j.celrep.2015.12.102 [DOI] [PubMed] [Google Scholar]
- Zou, Y., and Lyuksyutova A.I.. 2007. Morphogens as conserved axon guidance cues. Curr. Opin. Neurobiol. 17:22–28. 10.1016/j.conb.2007.01.006 [DOI] [PubMed] [Google Scholar]
- Zuber, B., Nikonenko I., Klauser P., Muller D., and Dubochet J.. 2005. The mammalian central nervous synaptic cleft contains a high density of periodically organized complexes. Proc. Natl. Acad. Sci. USA. 102:19192–19197. 10.1073/pnas.0509527102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.