Chen et al. have studied how Caenorhabditis elegans copes with an acute heat stress that alters its development and induces the fragmentation of mitochondria. A DRP-1–dependent autophagy process takes place as part of the worm’s adaptive mechanism in response to heat stress. DRP-1 participates in the coordination between mitochondrial fission and autophagosome biogenesis.
Abstract
Acute heat stress (aHS) can induce strong developmental defects in Caenorhabditis elegans larva but not lethality or sterility. This stress results in transitory fragmentation of mitochondria, formation of aggregates in the matrix, and decrease of mitochondrial respiration. Moreover, active autophagic flux associated with mitophagy events enables the rebuilding of the mitochondrial network and developmental recovery, showing that the autophagic response is protective. This adaptation to aHS does not require Pink1/Parkin or the mitophagy receptors DCT-1/NIX and FUNDC1. We also find that mitochondria are a major site for autophagosome biogenesis in the epidermis in both standard and heat stress conditions. In addition, we report that the depletion of the dynamin-related protein 1 (DRP-1) affects autophagic processes and the adaptation to aHS. In drp-1 animals, the abnormal mitochondria tend to modify their shape upon aHS but are unable to achieve fragmentation. Autophagy is induced, but autophagosomes are abnormally elongated and clustered on mitochondria. Our data support a role for DRP-1 in coordinating mitochondrial fission and autophagosome biogenesis in stress conditions.
Introduction
Organisms are exposed to variations of the environmental temperature and should manage to preserve their cellular homeostasis in heat stress (HS) conditions. The HS response relies on the protective activity of the heat shock protein chaperones and the elimination of damage materials by macroautophagy, which are both activated by the transcription factor HSF1 (Desai et al., 2013; Dokladny et al., 2013). For instance, in Caenorhabditis elegans adults after a mild HS, HSF1 positively controls autophagy gene expression, which participates in a protective hormetic response resulting in lifespan extension (Kumsta et al., 2017). Organelles have evolved specific responses, aiming to maintain their proteostasis, in various stress conditions. Among them, the mitochondria can trigger the specific mitochondrial unfolded protein response (Haynes et al., 2013), an adaptive transcriptional pathway that promotes the recovery of mitochondrial function when it is stressed (Shpilka and Haynes, 2018). Moreover, a mitochondrial stress can stimulate the HS response during C. elegans aging (Labbadia et al., 2017). However, when dysfunctions are persistent and the mitochondrial unfolded protein response is overwhelmed, elimination of the mitochondria can occur by a selective macroautophagy process called mitophagy (Palikaras et al., 2018; Pickles et al., 2018). Mitophagy engulfs dysfunctional mitochondria in double-membrane vesicles, the autophagosomes, which fuse with the lysosome to allow their degradation. Mitophagy occurs at various basal levels, depending on the cell types, but is also induced in stress conditions, such as HS or oxidative stress (Lee et al., 2012).
Mitochondria are highly dynamic organelles that reorganize through fusion or fission events, which participate in the mitochondrial homeostasis and can modulate the establishment of contact sites with other cell organelles (Wu et al., 2018; Youle and van der Bliek, 2012). Among those, ER–mitochondria contacts could be a site of biogenesis of autophagosomes in yeast and mammalian cells (Hamasaki et al., 2013; Böckler and Westermann, 2014). Dynamin-related protein 1 (Drp1), a member of the dynamin family, is a key player in mitochondrial fission, a multistep process including the formation of an ER-mediated mitochondrial constriction site (Friedman et al., 2011; Labrousse et al., 1999; Lee et al., 2016). Drp1 oligomerization on the outer mitochondrial membrane (OMM) further increases the constriction, a prerequisite step to achieve membrane scission (Ji et al., 2015; Kraus and Ryan, 2017). Recent data highlighted a new function for Drp1 in the quality control of mitochondria via an interaction with the mitochondrial Zn2+ transporter Zip1 during mitophagy (Cho et al., 2019).
The nematode C. elegans, which develops and reproduces in a range of temperatures from 15°C to 25°C (Hirsh and Vanderslice, 1976), is a powerful model to study the effects of HS (Momma et al., 2017). For instance, adult animals submitted to 35°C or 37°C for 3–6 h present a mitochondrial fragmentation in the muscle cells (Momma et al., 2017). Adaptation to HS has been described mainly in adults, but the high heterogeneity of the HS (28°C to 37°C for 1–6 h or more) resulted in opposite effects, ranging from hormesis to lethality. A 1-h 36°C stress applied to young adult animals has been shown to trigger a hormetic stress response (Kumsta et al., 2017), while a 1.5-h 37°C stress on adults results in 40% lethality (Zevian and Yanowitz, 2014). Moreover, recent data on C. elegans adults have shown that, in muscle cells, a severe and prolonged HS induces a DCT-1–, PINK-1–, and PDR-1–dependent mitophagy (Palikaras et al., 2015).
Here we analyzed the contribution of autophagy to adaptation to stress during the development of C. elegans. We developed a novel paradigm to characterize the physiological effects of a sublethal acute HS (aHS) on mitochondrial homeostasis and development. aHS induces a rapid fragmentation of mitochondria, the formation of aggregates in the matrix, the decrease of mitochondrial respiration, and mitophagy events. Stressed animals first present a blockage of development, but an autophagic flux enables the rebuilding of the mitochondrial network and developmental recovery. We report that, in the epidermis, the autophagosome biogenesis occurs mainly on mitochondria upon HS but also in physiological conditions. Finally, we discovered that DRP-1 is important for the adaptation to aHS and participates in the coordination between mitochondrial fission and autophagy.
Results
aHS induces organelle modifications and developmental delay
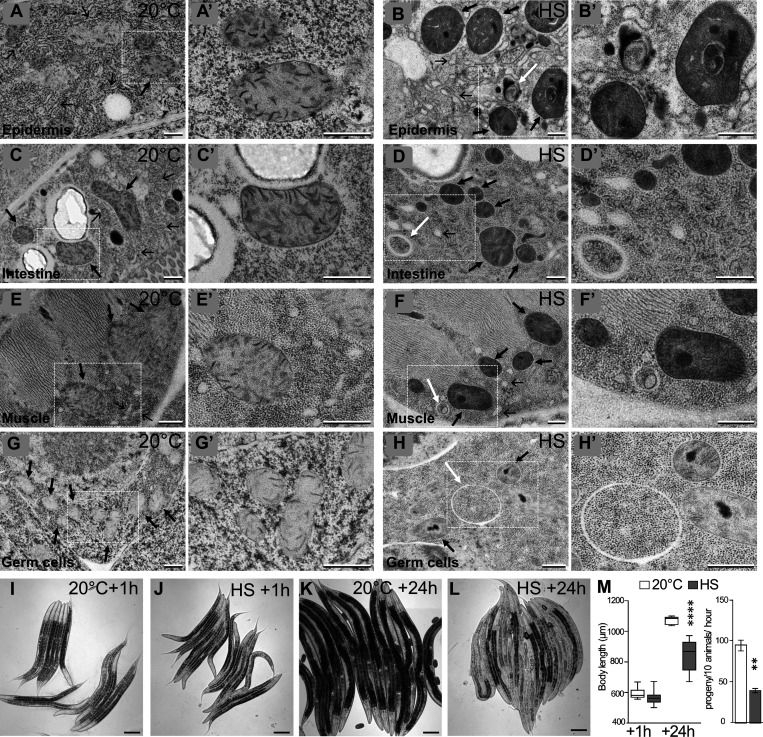
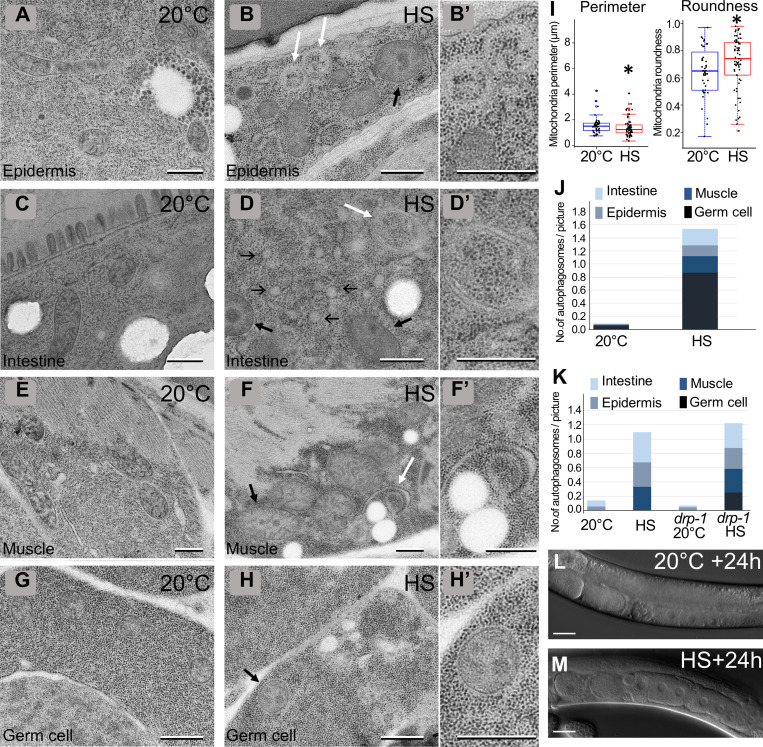
To analyze whether a sublethal HS could induce cellular damage, we submitted either young adults or fourth-stage larvae (L4) to 37°C during 60 min and performed transmission EM (TEM). The analysis of four tissues—the epidermis, the muscles, the intestine, and the germinal gonad—revealed three major subcellular alterations that were detected in both adults (Fig. 1) and L4 HS animals (Fig. S1) compared with control animals maintained at 20°C. First, the ER was fragmented and formed round, inflated vesicles compared with the tubular network in control animals (Fig. 1, A–H). Second, the appearance of mitochondria was modified; they looked darker, and the matrix contained irregular electron-dense structures, which could correspond to aggregates. Moreover, the measure of the area and the roundness of the mitochondrial sections in the epidermis indicates smaller but rounder mitochondria (Fig. S1). Third, a number of vesicles containing cytoplasmic materials and looking similar to autophagosomes (Manil-Ségalen et al., 2014) were present in stressed animals (Fig. 1, B, D, E, and F; quantification in Fig. S1). These data indicate that, independently of the stage of development, several tissues react similarly to HS, which alters both ER and mitochondrial morphology and triggers autophagy. It suggests that such a response to the HS is an intrinsic cellular property.
Figure 1.
Heat shock induces alterations of subcellular structures and a developmental delay. (A–H) TEM images of epidermis (A and B), intestine (C and D), muscle (E and F), and germ cells (G and H) in control (A, C, E, and G) and HS (B, D, F, and H). Animals were submitted to 37°C during 60 min followed by 60 min of recovery at 20°C. HS induces a fragmentation and inflation of the ER (thin black arrows), the formation of electron-dense structures within the matrix of mitochondria (thick black arrows), and the formation of autophagic vesicles (white arrows). (A′–H′) Magnifications of the boxed areas in A–H. (I–M) aHS in L4 animals results in a developmental delay but no lethality. Early L4 animals were submitted to 37°C during 60 min and analyzed after 1-h (I and J) or 24-h (K and L) recovery at 20°C. (M) Left: Boxplot showing the size of the animal. n = 27, 44, 8, 32; ****, P < 0.0001, t test. Right: Fertility was calculated after 24 h of recovery. Mean ± SEM from 4 independent experiments with 30 worms per condition. **, P < 0.005, paired t test. The scale bars are 0.5 µm (A–H, A′–H′) or 100 µm (I–M).
Figure S1.
Characterization of cellular defects after aHS (complementary to Fig. 1). (A–H) TEM images of epidermis (A and B), intestine (C and D), muscle (E and F), and germ cells (G and H) in control (A, C, E, and G) and HS (B, D, F, and H) L4. Thin black arrows, thick black arrows, and white arrows point to ER, mitochondria, and autophagosomes, respectively. (B′, D′, F′, and H′) Higher magnification of the area shown by white arrows in B, D, and F and a black arrow in G, respectively (I–K) Boxplots showing the quantification of the mitochondrial shape (I; *, P < 0.05, Kruskal-Wallis nonparametric test; n = 40, 65) and the numbers of autophagosomes in young adults (J; n = 156, 55) and L4 (K; n = 63, 90, 101, 94). (L and M) DIC pictures of the gonads of control (L) or aHS (M) animals. aHS results in a developmental delay but no sterility. The scale bars represent 0.5 µm (A–H, B′, D′, F′, and H′) or 20 µm (L and M).
We then analyzed whether the effects of HS on L4 could affect their survival, development, and fertility. After a 1-h recovery period at 20°C following the HS, the animals were alive, moving correctly, and presenting no obvious morphological difference from the non-HS controls (Fig. 1, I and J). 24 h later, there was no significant difference in survival between HS and control animals, but while control worms had reached adulthood and started laying eggs, HS animals were smaller and initiated egg laying with some delay (Fig. 1, K–M). Almost all HS animals became fertile in the next 24 h (Fig. 1 M and Fig. S1), and the progeny were viable. These data indicate that a 1-h 37°C HS in L4 animals results in a developmental delay without affecting the viability and fertility of the worms. The HS parameters, the cellular effects, and the developmental consequences on L4 animals indicate that these conditions correspond to a sublethal aHS and define a paradigm to analyze the mechanisms of adaptation. For that reason, the rest of this study concentrated on L4 to analyze the cellular mechanisms of response to aHS in C. elegans and focused on the mitochondrial phenotype and autophagy.
Mitochondrial function and morphology are transitorily altered upon aHS
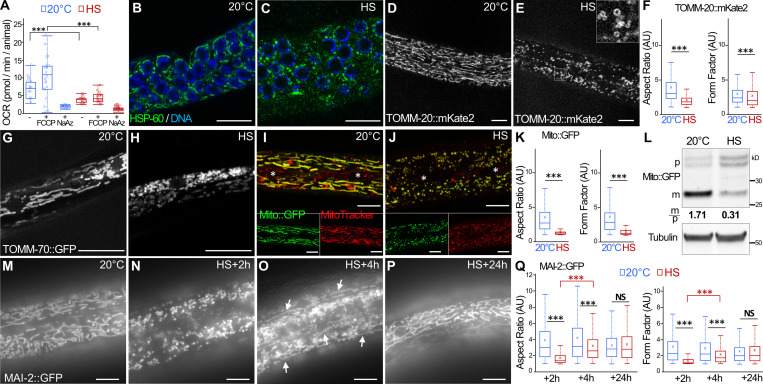
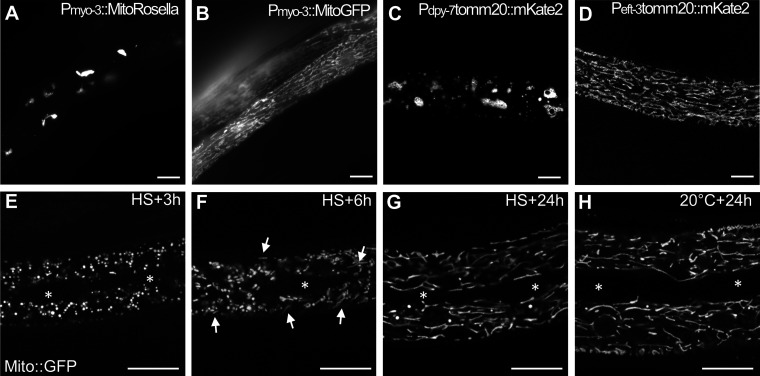
We next addressed whether the altered mitochondrial morphology and the accumulation of abnormal structures in the matrix observed by TEM correlate with mitochondrial dysfunction. We used the Seahorse technology to measure the basal and the maximal respiratory capacity in aHS and control animals. 1 h after the aHS, both the basal and maximal oxygen consumption of aHS animals were strongly reduced, indicating that the mitochondrial respiration was altered (Fig. 2 A). Because TEM is limited for analyzing the mitochondrial network integrity, we performed immunofluorescence or in vivo imaging using various mitochondrial markers expressed in the gonad, the muscle cells, the intestine, and the epidermis (Fig. 2 and Fig. S2). Among those, two transgenic strains displayed an abnormal mitochondrial network at 20°C (Fig. S2) and were not further used for the study. aHS results in the fragmentation of mitochondria in the gonad, muscles, and epidermis, indicating that the mitochondrial stress–induced phenotype observed by TEM is accompanied by a complete reorganization of the network. Visualization of mitochondria using matrix mitoGFP (Zhao et al., 2017), CMXRos dye, OMM TOMM-20::mKate2 (Ahier et al., 2018), and mitochondrial ATPase inhibitor 2 (MAI-2)::GFP (Fernández-Cárdenas et al., 2017) revealed that mitochondria in the epidermis are very strongly affected by aHS, forming numerous small and round vesicular structures instead of the long and reticulated network (Fig. 2, D, E, I, J, M, and N). Quantitative analyses of the length (aspect ratio) and the branching (form factor; Koopman et al., 2016; Marchi et al., 2017) confirmed the massive fragmentation of the mitochondria (Fig. 2, F and K).
Figure 2.
aHS induces transitory fragmentation and dysfunctions of mitochondria. (A) Oxygen consumption rate (OCR) in control and HS animals. FCCP, cyanide-4(trifluoromethoxy)phenylhydrazone; NaAz, sodium azide. Boxplot, triplicates 15 < n < 22; ***, P < 0.001, Wilcoxon test. (B and C) Confocal images of mitochondrial network in the gonad (anti-HSP60 staining) of control (B) and HS (C) animals. DNA of germ cells is stained with Hoechst (blue). (D–F) Confocal images of the mitochondria in the epidermis, visualized with mKate2 fused with the outer membrane localization domain of TOMM-20, in control (D) and HS (E) animals. Quantification of the mitochondrial shape descriptors aspect ratio and form factor using TOMM-20::mKate2 (F). Boxplot showing a total of 200–500 mitochondria analyzed from five animals. ***, P < 0.001, Wilcoxon test. (G and H) Confocal images of the mitochondria in the muscle visualized with GFP fused with the outer membrane localization domain of TOMM-70 in control (G) and HS (H) animals. (I–K) Colocalization of mito::GFP (green) and MitoTracker CMXRos (red) in the epidermis of control (I) and HS (J) animals. The white asterisks show the positions of the specialized seam cells of the epidermis that do not express Mito::GFP but are labeled with CMXRos. Quantification of the mitochondrial shape descriptors aspect ratio and form factor using Mito::GFP (K). Boxplot showing >300 mitochondria measured per animal from five animals. ***, P < 0.001, Wilcoxon test. (L) Western blot analysis of epidermal Mito::GFP. The quantification of the cytoplasmic precursor (p) and mitochondrial (m) forms shows a decrease of the GFP import after HS. (M–Q) Epifluorescence images of MAI-2::GFP showing the mitochondria in the epidermis in control (M) and HS after 2-h (N), 4-h (O), or 24-h (P) recovery. The rebuilding of the network is visible 4 h after HS by the presence of tubular mitochondria (white arrows). The quantification of the mitochondrial shape descriptors (aspect ratio and form factor) indicates a partial rebuilding after 4 h of recovery (Q). Boxplot, five animals were analyzed, and >500 mitochondria were measured per animal. ***, P < 0.001, Wilcoxon test. The scale bars are 10 µm.
Figure S2.
Specific reporters can affect mitochondrial network morphology (complementary to Fig. 2). (A–D) Live images of the mitochondria in the body wall muscles of the MitoRosella (A) or MitoGFP (B) and in the epidermis of TOMM-20::mKate2 under the control of an epidermal (C) or ubiquitous promoter (D). Both the type and the level of expression of the fluorescent reporter could affect the mitochondrial network at 20°C. (E–H) Confocal images of Mito::GFP expressed in the epidermis and showing the rebuilding of the mitochondrial network after aHS. The white asterisks show the position of the specialized seam cells of the epidermis that do not expressed Mito::GFP. White arrows in F point to tubular mitochondria. The scale bars represent 10 µm.
The amount of the epidermal mitoGFP was further quantified by Western blotting in aHS and control animals. In control animals, a 25–30-kD major band, corresponding to the mitochondrial matrix form after removal of the N-terminal presequence, was detected, as well as two weak bands corresponding to the precursor forms (Fig. 2 L). After aHS, the amount of mitochondrial GFP decreased and the amount of the precursor forms increased, suggesting that the mitochondrial import is somehow affected.
Despite the strong fragmentation of mitochondria after aHS, L4 were able to resume development and became fertile after 24 h (Fig. 1). Therefore, we analyzed the mitochondrial network 2, 4, and 24 h after aHS (Fig. 2, M–Q; and Fig. S2). After 4 h of recovery, some tubular mitochondria were already visible (Fig. 2 O), and in almost all animals studied, a reticulated mitochondrial network was rebuilt after 24 h (Fig. 2 P). Altogether, our data indicate that aHS triggers a rapid mitochondrial fragmentation with functional decrease, followed by a slow rebuilding and recovery of a normal network.
aHS induces an autophagic flux
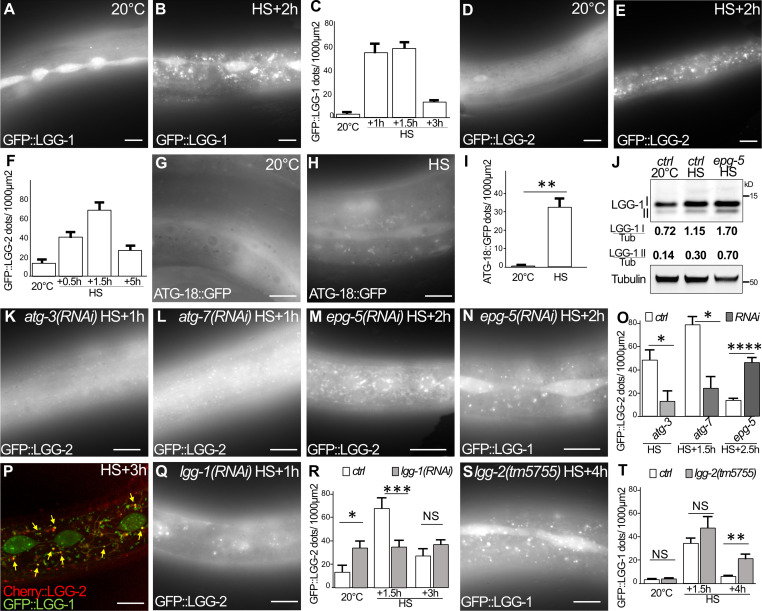
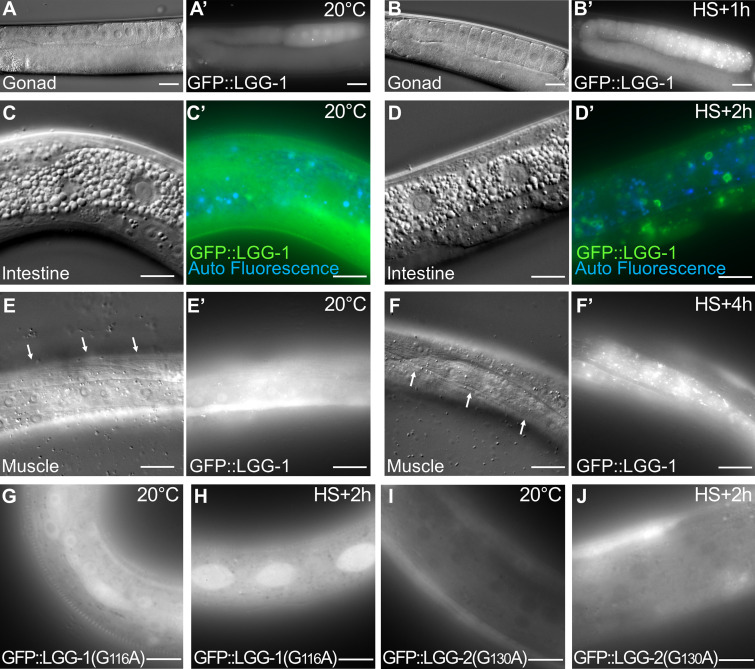
Our TEM analyses after aHS revealed the presence of autophagic vesicles in various tissues (Fig. 1 and Fig. S1). We next wanted to confirm that these structures are bona fide autophagosomes and addressed whether aHS triggers a functional autophagic flux (Fig. 3 and Fig. S3). Using GFP-tagged LGG-1, LGG-2, and ATG-18, the C. elegans homologues of GABARAP, LC3, and WIPI1/2, respectively (Alberti et al., 2010; Jenzer et al., 2014; Jia et al., 2007), we first examined the formation of puncta in the gonad, the intestine, the muscle, and the epidermis upon aHS and confirmed that aHS induces an increase in the number of autophagosomes in various tissues (Fig. S3) with a stronger response in the epidermis (Fig. 3, A–I). The ATG-18 dots, which correspond to initiation of autophagosomes, were detected during aHS, while the increase of LGG-1 and LGG-2 dots was maximum 90 to 120 min after aHS (Fig. 3, A–H). To exclude the possibility that the dots correspond to GFP aggregates induced by HS, we analyzed the mutated GFP::LGG-1(G116A) and GFP::LGG-2(G130A), which are unable to localize to autophagosomes (Alberti et al., 2010; Manil-Ségalen et al., 2014), and we observed no formation of puncta but a diffuse staining after HS (Fig. S3, G–J). Western blot analysis and quantification of the lipidated and nonlipidated forms of endogenous LGG-1 confirmed the induction of an autophagic flux (Fig. 3 J). The last evidence that aHS induces a functional autophagic flux was obtained by a genetic approach. Using mutants and RNAi that block either the formation (atg-3, atg-7, lgg-1) or the degradation (epg-5, lgg-2) of autophagosomes (Chen et al., 2017), we observed a decrease or an increase in the number of autophagosomes, respectively (Fig. 3, K–O).
Figure 3.
aHS induces an autophagic flux in the epidermis. (A–F) Autophagosomes are visualized with GFP::LGG-1 (A and B) or GFP::LGG-2 (D and E) in control (A and D) and HS (B and E) animals and quantified (mean ± SEM; n = 10, 8, 11, 11 in C; n = 13, 10, 10, 8 in F). (G–I) The initiation sites of autophagosomes are visualized with ATG-18::GFP after 30-min treatment in control (G) and HS (H) animals and quantified (I; mean ± SEM, n = 7, 6; **, P < 0.005, Wilcoxon test). (J) Western blot analysis using anti–LGG-1 antibody to quantify the cleaved (I) and lipidated (II) forms. aHS induces an increase of the autophagic flux with a further accumulation of lipidated LGG-1 in epg-5(RNAi) animals where maturation of autophagosomes is altered. (K–O) The autophagic flux induced by HS was analyzed by imaging of GFP::LGG-2 or GFP::LGG-1 autophagosomes in the autophagy-deficient animals atg-3 (K), atg-7 (L), and epg-5 (M and N) and quantified (O; mean ± SEM, n = 5, 5, 4, 5, 7, 11; *, P < 0.05; ****, P < 0.0001, Wilcoxon test). (P–T) LGG-1 acts upstream of LGG-2 during aHS autophagic flux. Three types of autophagosomes are detected after HS: LGG-1 only (green), LGG-2 only (red), and double positive (yellow arrows; P). The depletion of lgg-1 (Q and R) decreases the number of LGG-2 autophagosomes, while the depletion of lgg-2 (S and T) increases the number of LGG-1 autophagosomes. Mean ± SEM; n = 13, 10, 10, 10, 8, 8; *, P < 0.05; **, P < 0.005; ***, P < 0.0005, two-way ANOVA. The scale bars are 10 µm.
Figure S3.
aHS induces autophagy in various tissues (complementary to Fig. 3). (A–F) DIC pictures and the corresponding fluorescence images (A′–F′) of GFP::LGG-1 in animals at 20°C (A, C, and E) and after aHS (B, D, and F). The increase of LGG-1 puncta is shown in the germ cells (A′ and B′), the intestine (C′ and D′), and the body wall muscles (white arrows in E and F) of aHS animals. Note that in the intestine, the GFP::LGG-1 stains large vesicular structures. (G–J) Images of the epidermis showing that the nonlipidated GFP::LGG-1(G116A) (G and H) and GFP::LGG-2(G130A) (I and J) do not form puncta after HS (H and J). The scale bars represent 20 µm (A and B) or 10 µm (C–J).
We reported previously that the localization of LGG-1 and LGG-2 defines three populations of autophagosomes in C. elegans embryo (Manil-Ségalen et al., 2014). The in vivo analysis of mCherry::LGG-2 (Jenzer et al., 2019) and GFP::LGG-1 in the epidermis revealed that aHS induces three types of autophagosomes: LGG-1 only, LGG-2 only, and double positive (Fig. 3 P). Moreover, we observed that the depletion of LGG-1 partially decreases the number of LGG-2 dots (Fig. 3 Q and R), while the depletion of LGG-2 rather increases the number of LGG-1 dots (Fig. 3, S and T). These observations suggest that the sequential functions of LGG-1 and LGG-2, previously described in the early embryo (Manil-Ségalen et al., 2014), could be conserved in stress-induced autophagy.
Autophagy protects larvae from aHS and facilitates mitochondrial rebuilding
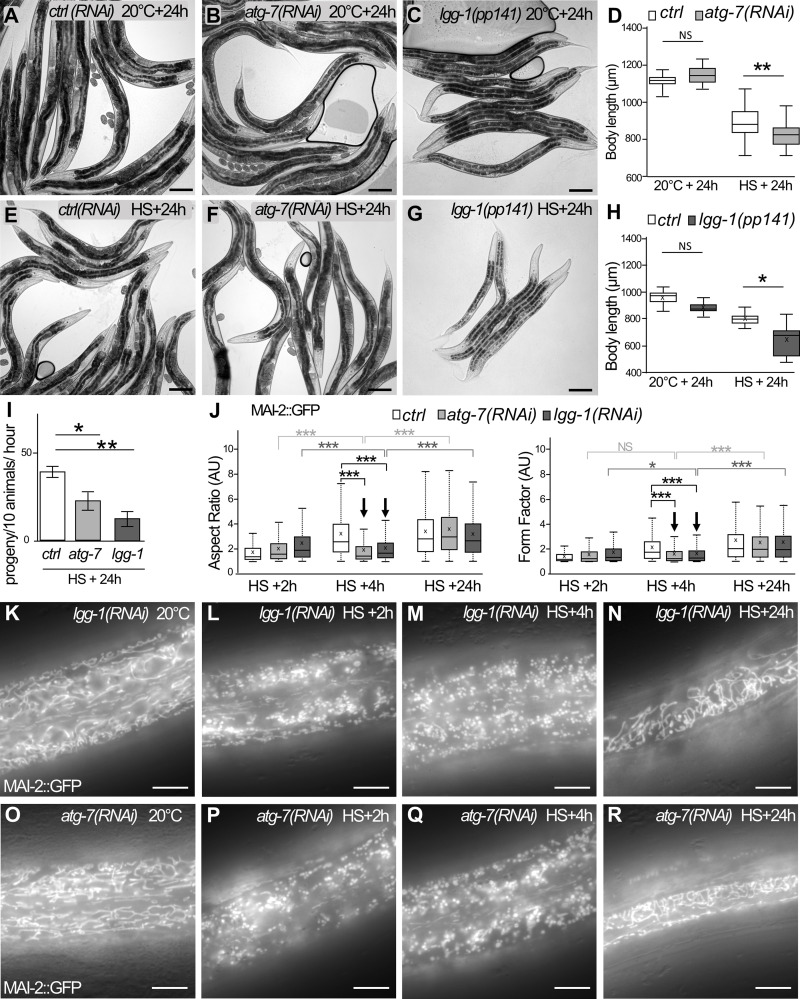
We next asked whether the induction of autophagic flux upon aHS could be involved in the adaptive response and the worm development recovery. We compared the survival and the size after aHS of autophagy mutants with wild-type animals (Fig. 4, A–F). 3 h after HS, the lgg-1, atg-3, or atg-7 animals did not look different from control animals, suggesting that the autophagic flux is not essential for the immediate survival of animals upon aHS. However, 24 h after aHS, atg-7(RNAi) or lgg-1(RNAi) animals were smaller than wild-type animals (Fig. 4, D–H). Moreover, autophagy-deficient animals started to lay eggs later than the controls (Fig. 4 I), indicating that the aHS-induced developmental delay was further increased. These data demonstrate that the autophagic flux is protective for the larvae and important for developmental recovery after aHS.
Figure 4.
Autophagy is protective and facilitates mitochondrial rebuilding after aHS. (A–I) The blockage of the autophagic flux exacerbates the developmental phenotypes induced by aHS. ctrl(RNAi) (A and E), atg-7(RNAi) (B and F), or lgg-1(pp141) (C and G) animals were maintained at 20°C (A–C) or submitted to aHS (E–G) and analyzed after 24-h recovery. The body length of the autophagy-defective animals is decreased 24 h after aHS (D, boxplot; n = 65, 72, 78, 61; **, P < 0.005, t test; H; boxplot; n = 9, 8, 8, 9; **, P < 0.001, Kruskal-Wallis test). The number of progeny of atg-7 and lgg-1 animals is decreased (I; mean ± SEM from 4 independent experiments, >10 animals; *, P < 0.05; **, P < 0.005, t test). (J–R) Blocking autophagy affects the mitochondrial rebuilding in the epidermis. Boxplot of the mitochondrial shape descriptors aspect ratio and form factor using MAI-2::GFP (J). 4 h after aHS, the rebuilding of the network is less efficient in lgg-1(RNAi) and atg-7(RNAi) animals. Three to five animals were analyzed, and >500 mitochondria were measured per animal. ***, P < 0.0001, t test. (K–R) Epifluorescence images of MAI-2::GFP showing the mitochondrial network in the epidermis at 20°C (K and O) and after HS with 2-h (L and P), 4-h (M and Q), or 24-h (N and R) recovery. 4 h after aHS, almost all mitochondria are still fragmented in lgg-1(RNAi) and atg-7(RNAi) animals (compare with Fig. 2 O showing wild-type animals). The scale bars are 100 µm (A–G) or 10 µm (K–R).
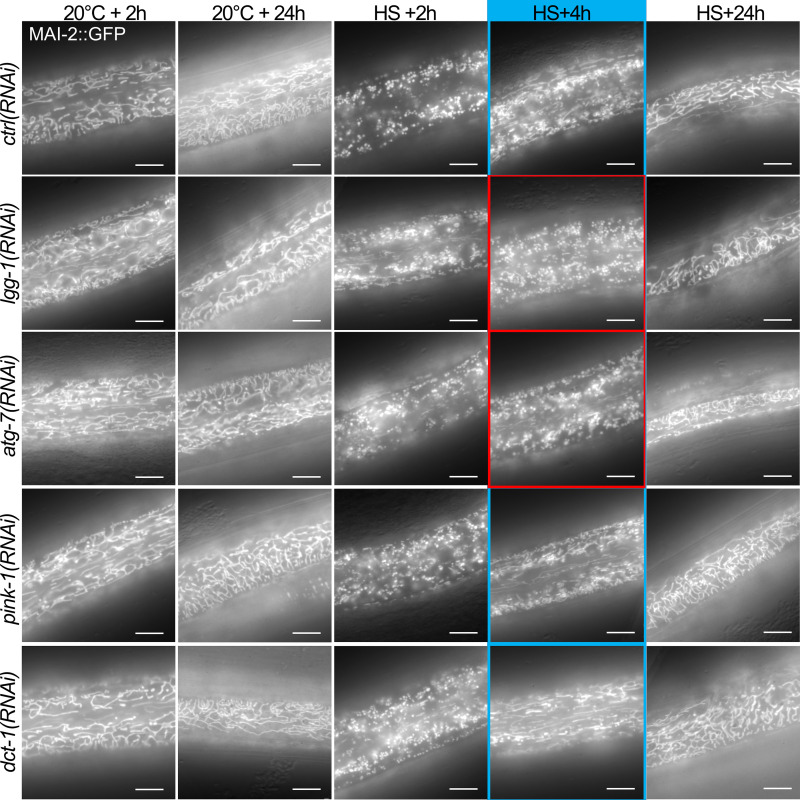
We then investigated whether the autophagic flux is involved in the rebuilding of the mitochondrial network in the epidermis after aHS (Fig. 4, J–R; and Fig. S4). The quantification of mitochondrial rebuilding was performed 2 h, 4 h, and 24 h after aHS in control and autophagy-deficient animals (lgg-1, atg-7). lgg-1(RNAi) and atg-7(RNAi) animals presented no alteration of the mitochondrial network at 20°C. However, a delay in the rebuilding was observed in these animals 4 h after aHS (Fig. 4, J, M, and Q). 24 h after aHS, most of the lgg-1(RNAi) and atg-7(RNAi) animals showed a recovery of the tubular mitochondrial network (Fig. 4, J, N, and R). Our data demonstrate that autophagy is important for the dynamic of mitochondrial rebuilding after aHS-induced fragmentation.
Figure S4.
Blocking autophagy but not mitophagy delays the rebuilding of the mitochondrial network after aHS (complementary to Figs. 4 and 5). Epifluorescence pictures of MAI-2::GFP illustrating the rebuilding of the mitochondrial network in the epidermis after aHS. At 20°C, the depletion of autophagy or mitophagy genes does not result in a strong modification of the mitochondrial network. In lgg-1–, atg-7–, pink-1–, and dct-1–depleted animals, the aHS results in the strong fragmentation of the mitochondria, but after 4 h of recovery, atg-7(RNAi) and lgg-1(RNAi) animals display a less efficient rebuilding. The blue- and red-framed images show regions of the hypodermis with or without tubular mitochondria, respectively. The scale bars are 10 µm.
Autophagosomes form on mitochondria, but selective mitophagy is dispensable for adaptation to aHS
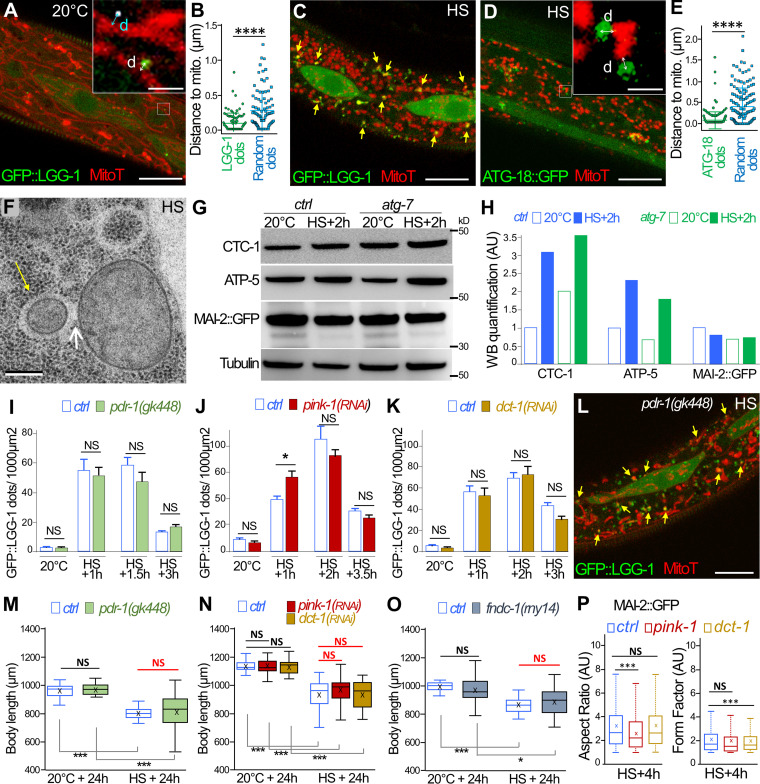
The correlation between the induction of an autophagic flux and the mitochondrial rebuilding defect prompted us to investigate mitophagy after aHS. In the epidermis of control animals, the analysis of GFP::LGG-1 and MitoTracker showed no colocalization between mitochondria and autophagosomes (Fig. 5 A). However, measurement of the minimal distance between autophagosomes and mitochondria revealed a nonrandom distribution and a close association of the two organelles (Fig. 5, A and B). In aHS animals, numerous autophagosomes were found in close proximity with mitochondria, and a fraction of them colocalized with fragmented mitochondria, suggestive of mitophagy events (Fig. 5 C). The measure of the distance between ATG-18::GFP and mitochondria (Fig. 5, D and E) confirmed that, upon aHS, the autophagosome biogenesis is initiated in very close proximity to mitochondria. The presence of small mitochondria within vesicular structures was also observed several times by TEM (Fig. 5 F). We also used Western blot analysis to monitor the level of mitochondrial degradation induced upon aHS (Fig. 5, G and H). 2 h after aHS, the quantification of the mitochondrial encoded cytochrome c oxidase subunit 1, CTC-1/COX1, and the nuclear encoded ATP synthase subunit, ATP-5, indicated an increase of the mitochondrial proteins in both control or atg-7 mutant animals. These results suggest that aHS induces the degradation of some mitochondria by autophagy but also an increase of the mitochondrial mass.
Figure 5.
Mitochondria are the main site of autophagosome biogenesis in the epidermis, but mitophagy is not essential for adaptation and mitochondrial rebuilding after aHS. (A–E) Colocalization analysis of autophagosomes and mitochondria. Confocal images of GFP::LGG-1 (green) and MitoTracker (red) in the epidermis of a control animal (A). Insets in A and D show the minimal distance (d, indicated by the bidirectional arrows) between autophagosome and mitochondria, which is compared with random dots (white puncta). (B) Horizontal bars represent mean and SEM, n = 89, from 9 animals; ****, P < 0.0001, t test with Welch correction. (C) In an HS animal, a fraction of the fragmented mitochondria are colocalizing with autophagosomes (yellow arrows). (D) Confocal images of ATG-18::GFP (green) and MitoTracker (red) in the epidermis of an aHS animal showing that the initiation of the autophagosome occurs in close proximity with mitochondria (insets) compared with random dots (mean and SEM in E, n = 151, from 9 animals; ****, P < 0.0001, t test with Welch correction). (F) TEM images after aHS showing mitochondria within a vesicle (yellow arrow) in contact (white arrow) with nonengulfed mitochondria. (G and H) Western blot (WB) analysis of total protein extracts of control and atg-7 animals at 20°C or after aHS plus 2 h of recovery (G). (H) Quantification of the mitochondrial proteins CTC-1/Cox1, ATP-5, and MAI-2::GFP in a whole animal shows that aHS does not induce a massive degradation of mitochondria. The signal was normalized using tubulin, and, for each mitochondrial protein, the control at 20°C was arbitrarily set at 1. (I–K) The depletion of pdr-1 (I; n = 10, 10, 8, 8, 11, 12, 11, 15), pink-1 (J; n = 6, 7, 18, 12, 7, 11, 11, 12), and dct-1 (K; n = 6, 9, 21, 15, 17, 16, 19, 17) does not modify the number of LGG-1 autophagosomes formed after HS. Mean ± SEM; *, P < 0.05, two-way ANOVA. (L) A single confocal image of GFP::LGG-1 (green) and MitoTracker (red) shows that colocalization events are present in pdr-1(gk448) mutant animals (yellow arrows). (M–P) The blockage of the mitophagy pathways does not exacerbate the developmental phenotypes induced by aHS. ctrl, pdr-1(gk448), pink-1(RNAi; N), dct-1(RNAi; N), and fundc1(rny14; O) animals were maintained at 20°C or submitted to aHS, and their body size was measured after 24 h of recovery. Boxplots (n > 30); *, P < 0.01; ***, P < 0.0001, Wilcoxon test. (P) Quantification of the mitochondrial shape descriptors aspect ratio and form factor using MAI-2::GFP. 4 h after aHS, the rebuilding of the network is not strongly altered in pink-1(RNAi) or dct-1(RNAi) animals (corresponding pictures in Fig. S4). Three to five animals were analyzed, and >500 mitochondria were measured per animal. ***, P < 0.0001, t test. The scale bars represent 10 µm (A, C, D, and L), 1 µm (insets in A and D), or 0.2 µm (F).
Next, to analyze whether mitophagy is involved in the adaptation to aHS, we depleted the PINK/PARKIN or Nix or FUN14 domain containing 1 (FUNDC1) pathways, which are involved in various selective mitophagy processes in C. elegans (Leboutet et al., 2020). Using mutants or RNAi for pink-1, pdr-1, dct-1, and fndc-1, we characterized the formation of autophagosomes, the size of the animals, and the mitochondrial rebuilding after aHS (Fig. 5, I–P; and Fig. S4). The number of LGG-1 autophagosomes did not decrease in pdr-1 mutant animals or in pink-1(RNAi) or dct-1(RNAi) (Fig. 5, I–K), and colocalization events between autophagosomes and mitochondria were still present (Fig. 5 L). Moreover, the depletion of pdr-1, pink-1, dct-1, or fndc-1 did not affect the size of the worm 24 h after aHS (Fig. 5, M–O), contrarily to atg-7 or lgg-1 (Fig. 4). Finally, the mitochondrial network rebuilding was not strongly affected in pink-1(RNAi) and dct-1(RNAi) animals (Fig. 5 P and Fig. S4), as observed in lgg-1(RNAi) and atg-7(RNAi) animals (Fig. 4). These data indicate that pdr-1, pink-1, dct-1, or fndc-1 is not essential for the adaptation to aHS and suggest that part of the fragmented mitochondria could be addressed to autophagosomes either by an unidentified mitophagy pathway or through bulk autophagy.
DRP-1 is necessary for aHS-induced mitochondrial fragmentation
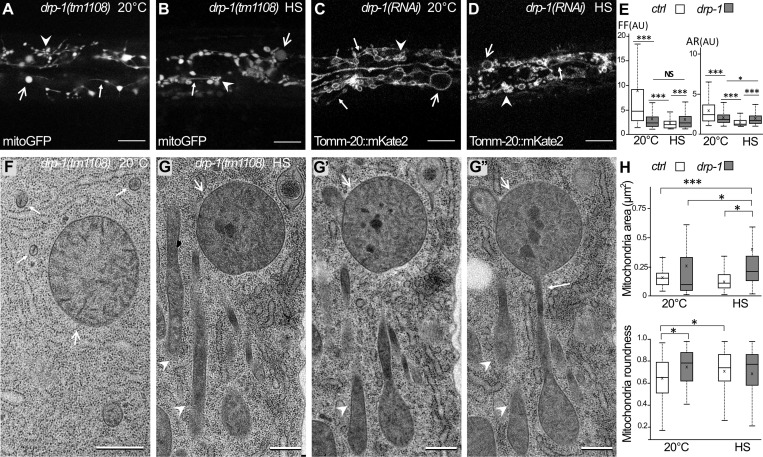
Our results indicate that autophagy is involved in mitochondrial network recovery after fragmentation. To address whether the mitochondrial fragmentation is a prerequisite for autophagy induction, we analyzed a null mutant of drp-1, the C. elegans homologue of Drp1, which affects mitochondrial fission (Breckenridge et al., 2008; Labrousse et al., 1999). DRP-1 function has been studied mainly in the muscle cells of C. elegans (Labrousse et al., 1999; Scholtes et al., 2018; Shen et al., 2014) but is not well characterized in the epidermis. The qualitative and quantitative analyses of the morphology of the mitochondria in drp-1 animals were performed using mitoGFP and TOMM-20::mKate2 (Fig. 6, A–E). In the epidermis, at 20°C, the mitochondrial network of drp-1 animals showed no regular tubular structures but three categories of abnormal mitochondria: the enlarged globular, the filament-like, and the clustered (Fig. 6 A; and Fig. 2 I for control). The OMM labeling showed that the three types of altered mitochondria remain connected (Fig. 6 C; and Fig. 2 D for control). These data confirmed that, similarly to muscle cells, maintenance of mitochondrial morphology in the epidermis is dependent on DRP-1 fission activity. After aHS, the mitochondrial network of drp-1 animals did not form small, rounded vesicles, as observed in wild-type animals, indicating that the HS-induced fragmentation is DRP-1 dependent (Fig. 6, B and D; and Fig. 2, E and J, for controls). However, both the qualitative and quantitative analyses indicated that the mitochondrial network of drp-1 animals was modified after aHS (Fig. 6 E). The clustering of mitochondria was more obvious, and the filament-like was more difficult to observe. To characterize further the phenotypes of mitochondria, we performed TEM analysis (Fig. 6, F–H). At 20°C, mitochondrial sections were very round but highly variable in size (Fig. 6 G; quantification in Fig. 6 F), confirming the in vivo analysis. After aHS, the presence of dark aggregates in the matrix was observed (Fig. 6 H), similarly to wild-type animals, but the mitochondria displayed a very heterogeneous shape and formed a connected network (Fig. 6 G; quantification in Fig. 6 H). Together, our results indicate that, in the absence of DRP-1, heat-stressed mitochondria tend to modify their shape but are unsuccessful in achieving fission.
Figure 6.
HS-induced mitochondrial fragmentation is dependent on DRP-1. (A–D) Confocal images of the mitochondria in the epidermis of drp-1 animals visualized with the matrix-located mitoGFP (A and B) or outer membrane TOMM-20::mKate2 (C and D) at 20°C (A and C) or after HS (B and D). Abnormally shaped mitochondria are classified as enlarged (large arrow), clustered (arrowhead), and filament-like (small arrow). For comparison with wild type, see Fig. 2, D, E, I, and J. (E) Boxplots of the mitochondrial shape descriptors form factor (FF) and aspect ratio (AR) using mitoGFP. Seven or eight animals were analyzed, and >700 mitochondria were measured per animal. ***, P < 0.001, Wilcoxon test. (F–G″) EM pictures of mitochondria in the epidermis of drp-1 animals at 20°C (F) or after HS (G). G, G′, and G″ are serial sections of stress mitochondria containing dark occlusions and displaying enlarged (large arrows), clustered (arrowhead), and filament-like (small arrow) shapes. For comparison with wild type, see Fig. 1, A and B. (H) Measurements of the area and the roundness of mitochondria in the epidermis were performed on EM pictures. Boxplots; n = 40, 65, 37, 51; *, P < 0.05; ***, P < 0.0005, Kruskal-Wallis test. The scale bars are 10 µm (A–D) or 0.5 µm (F and G).
DRP-1 is involved in autophagosome formation and adaptation to aHS
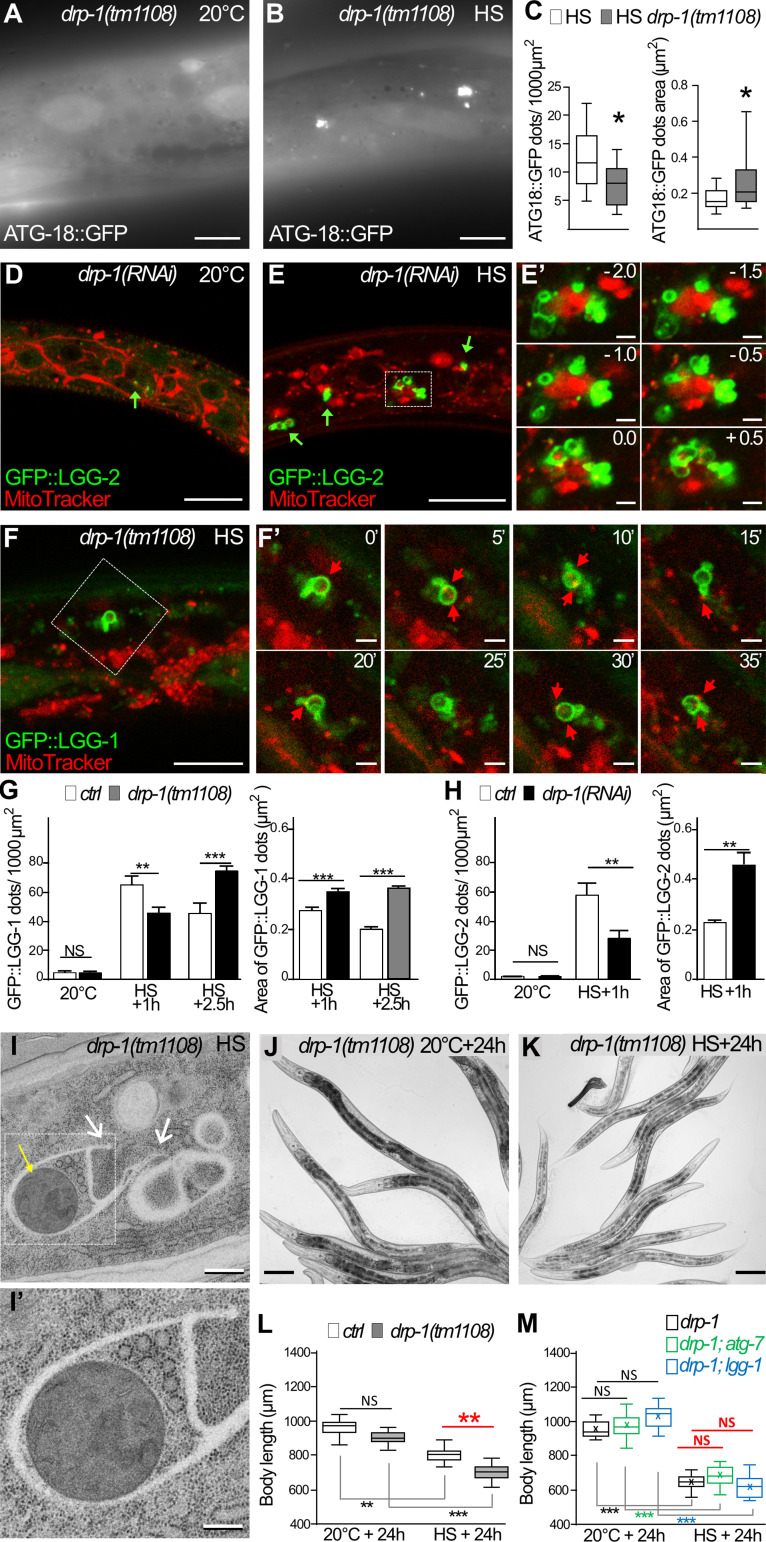
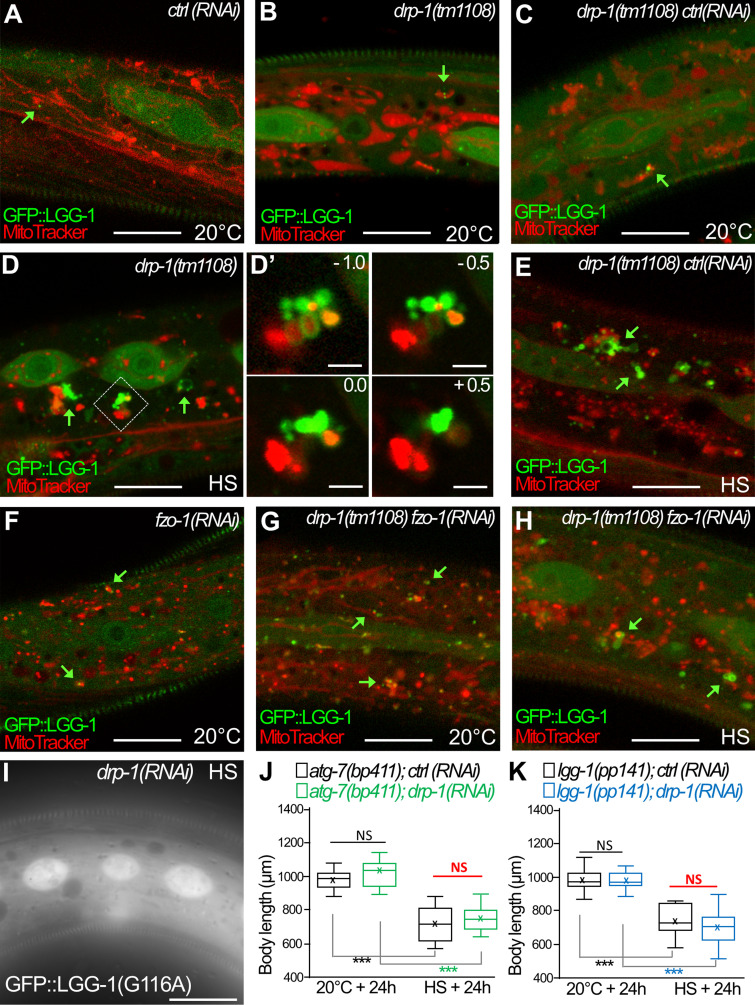
We then analyzed whether autophagy was induced upon aHS in the absence of fission of the mitochondria. The localization of ATG-18::GFP, GFP::LGG-1, and GFP::LGG-2 was monitored in drp-1 animals at 20°C and after aHS (Fig. 7 and Fig. S5). At 20°C, the three autophagy markers presented a distribution pattern similar to that in wild-type animals (Fig. 7, A, C, D, G, and H; compare with Figs. 3 and 5 for controls). This indicated that, in basal conditions, DRP-1 is dispensable for the formation of autophagosomes, and its depletion does not induce autophagy. However, the distribution and appearance of the three autophagy markers were very different after aHS of drp-1 animals (Fig. 7, B, E, and F). The autophagic markers formed groups of vesicle-like structures in close proximity to or even intermingled with mitochondria. Quantifications of the number and size of the dots suggested that, in the absence of DRP-1, the aHS induces a clustering of autophagic structures (Fig. 7, C, G, and H). The analysis of the nonlipidated GFP::LGG-1(G116A) indicates that the LGG-1 clusters are not protein aggregates (Fig. S5). To analyze the dynamics of these autophagic structures, time-lapse movies were acquired 30 min after aHS (Fig. 7, F and F′). Among the 31 LGG-1–positive structures that were visualized for 45–130 min, 74.2% persisted during the whole recording. The slight changes in their morphology and their long persistence support the notion that these structures are dysfunctional autophagosomes. EM in drp-1 animals after aHS further confirmed the formation of irregular membranous structures that engulfed mitochondria and extended unclosed arms (Fig. 7 I). These data show that DRP-1 is dispensable for the induction of autophagy, but in its absence, the biogenesis of autophagosomes is aberrant and the completion or closure of autophagosomes is not correctly achieved.
Figure 7.
DRP-1 is necessary for functional autophagy after aHS. (A–C) The initiation sites of autophagosomes are visualized with ATG-18::GFP in the epidermis of drp-1 animals at 20°C (A) and after HS (B). Upon HS, the initiation of autophagy is triggered in drp-1 animals, but ATG-18 puncta are clustered (compare with Fig. 3, G and H, for a wild-type control). (C) Boxplots showing the number and the area of puncta (n = 13, 14; *, P < 0.05, Wilcoxon test). (D and E) Confocal images of mitochondria stained with MitoTracker (red) and autophagosomes visualized with GFP::LGG-2 (green) in the epidermis of drp-1 animals at 20°C (D) and after aHS (E). Insets in E′ are a 0.5-µm Z-series corresponding to the dotted square in E. After aHS, the drp-1–depleted animals display abnormal autophagosomes (green arrows) that clustered and intermingled with mitochondria. (F) Confocal time-lapse images of mitochondria stained with MitoTracker (red) and autophagosomes visualized with GFP::LGG-1 (green) in the epidermis of drp-1(tm1108) mutant after aHS. F′ shows time lapse images, corresponding to the dotted square in F, captured every 5 mins. Despite changes in their morphology, the abnormal autophagosomes are persistent. (G and H) Quantification of the number and the area of LGG-1– and LGG-2–positive structures in drp-1 animals at 20°C or after aHS (G, mean ± SEM, n = 19, 16, 12, 11, 10, 10, 12, 11, 10, 10, two-way ANOVA; H, mean ± SEM, Wilcoxon test, n = 10, 4, 9, 10, 11, 10; **,P < 0.005; ***, P < 0.0005). (I) EM picture of mitochondria (yellow arrow) and cytoplasmic material engulfed in membranous structures in the epidermis of a drp-1 animal after HS. Membranous nonclosed extensions are visible (white arrows). I′ is a zoom of the dotted square in I. (J–L) The depletion of DRP-1 exacerbates the developmental phenotypes induced by aHS. ctrl (see Fig. 1) and drp-1(tm1108) (J and K) animals were maintained at 20°C or submitted to aHS, and their body size was analyzed after 24-h recovery (L; boxplot, n = 9, 13, 8, 13; **, P < 0.001, Kruskal-Wallis test). (M) Blocking autophagy in drp-1 mutants does not further increase the developmental delay after aHS. ctrl(RNAI), atg-7(RNAi), or lgg-1(RNAi) were applied to drp-1(tm1108) animals at 20°C or submitted to aHS, and the size of the animals was measured after 24 h (boxplot, n > 10, Kruskal-Wallis test). The scale bars are 10 µm (A–F), 1 µm (E′), 2 µm (F′), 0.5 µm (I), 0.5 µm (I′) and 100 µm (J and K).
Figure S5.
The clustering of autophagosomes at mitochondria is maintained in drp-1;fzo-1 animals after aHS (complementary to Fig. 7). (A–H) Confocal images of autophagosomes (GFP::LGG-1; green) and mitochondria (MitoTracker; red) in the epidermis of animals at 20°C (A–C, F, and G) and after aHS (D, E, and H). Green arrows point to autophagosomes in contact with mitochondria. In drp-1 animals (A–D), the aHS induces an accumulation of autophagic structures intermingled with mitochondria. Insets in D′ are 0.5-µm Z-series corresponding to the dotted square in D. The depletion of FZO-1 induces a fragmentation of the mitochondrial network in control animals (F) but partially restores a tubular mitochondrial network in drp-1(tm1108) animals (G). In drp-1(tm1108);fzo-1(RNAi) animals, upon aHS, the GFP::LGG-1 forms large clusters in contact with mitochondria. (I) The nonlipidated GFP::LGG-1(G116A) does not form puncta or clusters after HS in drp-1 animals. (J and K) The depletion of DRP-1 does not further increase the developmental delay of autophagy mutants (atg-7, lgg-1) induced by aHS. Single mutants with ctrl(RNAi) or dpr-1(RNAi) were maintained at 20°C or submitted to aHS and measured after 24-h recovery (boxplots, n > 30; ***, P < 0.0001, Kruskal-Wallis test). The scale bars represent 10 µm or 2 µm (D′).
The functional implication of DRP-1 in adaptation to aHS was confirmed by measuring the size of drp-1 mutant animals after 24-h recovery (Fig. 7 I). drp-1 animals presented an increase of the developmental delay similar to that of the autophagy mutants lgg-1 and atg-7. Moreover, the depletion of both drp-1 and lgg-1 or atg-7 resulted in a defect of similar size with no additive effect (Fig. 7 M and Fig. S5). Altogether, these data indicate that DRP-1 is required in the autophagy response and adaptation to aHS. The deregulation of autophagy observed in drp-1 mutants suggests a functional link between mitochondrial fission and autophagosome biogenesis.
Autophagosomes form at mitochondrial fission sites upon aHS
The clustering of autophagosomes observed in drp-1 animals upon aHS could be an indirect consequence of the large, misshapen mitochondria. However, a careful examination of drp-1 mutants revealed that abnormal autophagosomes can be formed on small mitochondria and not only on mitochondrial blebs (Fig. 7 E and Fig. S5). To assess this possibility, we submitted the drp-1 mutants to an RNAi against fzo-1, the worm homologue of human mitofusin, implicated in the mitochondrial fusion (Fig. S5). The depletion of fzo-1 results in the accumulation of fragmented mitochondria in the epidermis of animals grown at 20°C and partially restores some tubular mitochondria in the drp-1 mutant animals (Fig. S5 G). Upon aHS, the formation of mitochondrial clustered autophagosomes was detected in the epidermis of drp-1;fzo-1 depleted animals (Fig. S5 H). These data indicate that the formation of autophagosome clusters in drp-1 animals is not a consequence of the inability of elongating autophagosomes to engulf the mitochondria due to their bigger size.
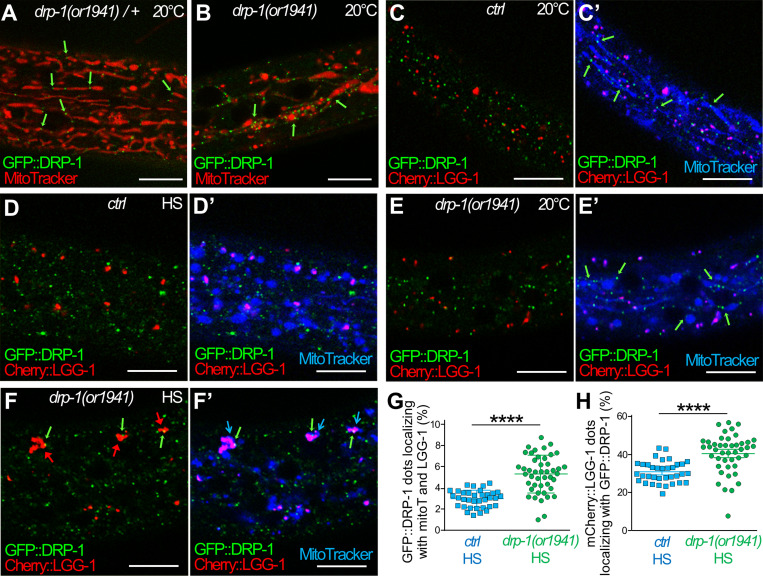
Because DRP-1 oligomerizes at mitochondrial fission sites, we next studied whether the formation of an autophagosome induced by aHS occurs at such sites (Fig. 8). To circumvent the difficulty of analyzing the rapid and transient fission process, we used a particular CRISPR-engineered allele of drp-1(or1941) (Lowry et al., 2015). drp-1(or1941) is an in-frame GFP insertion that results in a GFP::DRP-1 that correctly localizes at mitochondrial fission sites (constrictions and tips in heterozygous animal in Fig. 8 A) but is not able to achieve fission (abnormal mitochondrial network in homozygous animals in Fig. 8 B; Montecinos-Franjola et al., 2020). Therefore, drp-1(or1941), which phenocopies a drp-1 mutant, was used to visualize the presumptive mitochondrial fission sites and was compared with a wild-type drp-1 as a positive control. The colocalization of GFP::DRP-1 with mitochondria (MitoTracker) was used to identify fission sites and analyzed together with autophagosomes (mCherry::LGG-1) in the epidermis of animals at 20°C or after aHS. The fission sites were visible in control animals at 20°C, where GFP::DRP-1 formed puncta associated with tubular mitochondria (Fig. 8 C′). After aHS, GFP::DRP-1 puncta were present and generally not associated with fragmented mitochondria (Fig. 8 D′), but on rare occasions, a codistribution of GFP::DRP-1 with mCherry::LGG-1 and mitochondria was observed (quantification in Fig. 8 G). In drp-1(or1941) at 20°C, GFP::DRP-1 formed puncta, in contact with blebs of mitochondria (Fig. 8, B and E′), suggestive of abortive fission sites. After aHS of drp-1(or1941) animals, clusters of autophagosomes associated with mitochondria were observed (Fig. 8 F′) recapitulating the drp-1 loss-of-function phenotype. The number of GFP::DRP-1 dots associated with both mCherry::LGG-1 and mitochondria was increased but remained low. However, the fraction of mCherry::LGG-1 puncta positive for GFP::DRP-1 was increased to 41% (Fig. 8 H). Moreover, 68% of the large autophagic clusters formed in drp-1(or1941) mutants presented a GFP::DRP-1 puncta (arrows in Fig. 8 F). Together, these results support the hypothesis that, upon HS, autophagosomes are formed at mitochondrial fission sites and remain associated with presumptive fission sites in a defective drp-1 background. Altogether, our data suggest that DRP-1 could be involved in the coordination between mitochondrial fission and autophagosome biogenesis during aHS response.
Figure 8.
The dysfunction of DRP-1 during aHS induces the accumulation of autophagosomes at mitochondrial fission sites. (A and B) Confocal images of GFP::DRP-1 (green) and MitoTracker (red) in the epidermis of heterozygous control (A) and homozygous drp-1(or1941) mutants (B). drp-1(or1941) is an in-frame insertion of GFP at the drp-1 locus that results in an abnormal mitochondrial network (blebs in B), indicating that GFP::DRP-1 is correctly localized (green arrows) but is not functional. This particular GFP allows visualizing the presumptive mitochondrial fission points in a context-phenocopying drp-1 mutant (B). (C) Another transgenic GFP::DRP-1 strain where the drp-1 locus wild type is used as a positive control. (C–F) Confocal live imaging of the epidermis of GFP::DRP-1 in control (C and D) and drp-1(or1941) mutant animals (E and F) at 20°C (C and E) and after HS (D and F). The mitochondria are stained with MitoTracker DeepRed (blue), and autophagosomes are visualized with mCherry::LGG-1 (red). C′, D′, E′, and F′ show three colors merging. (C) In control animals at 20°C, the GFP::DRP-1 puncta are associated to tubular mitochondria but not to mCherry::LGG-1 dots (C′). (D) Upon aHS, most of the GFP::DRP-1 puncta are not localizing with mitochondria (B) or with mCherry::LGG-1 dots. (E) In drp-1(or1941) mutant at 20°C, the presumptive mitochondrial fission points (GFP puncta) are associated with blebs or filamentous mitochondria but not with mCherry::LGG-1 dots (E′). (F) After aHS of drp-1(or1941) mutants, the clustering of autophagosomes (red arrows pointing to mCherry::LGG-1) is associated with presumptive mitochondrial fission points (green and blue arrows in F′). (G) Quantification of the colocalization of GFP::DRP-1 puncta with MitoTracker (mitoT) and mCherry::LGG-1 in the epidermis of control and drp-1(or1941) animals after aHS (mean and SEM, n = 35, 44; ****, P < 0.0001, t test with Welch correction). (H) Quantification of the colocalization of mCherry::LGG-1 puncta with GFP::DRP-1 in the epidermis of control and drp-1(or1941) animals after aHS (mean and SEM, n = 35, 44; ****, P < 0.0001, t test with Welch correction). The scale bars are 10 µm.
Discussion
Stress response pathways help the organism to adapt to and survive changes in environmental conditions. Autophagy is one of the mechanisms used by the cell to resist and adapt to stress conditions. We have characterized a new aHS paradigm for studying the interplay between autophagy and mitochondrial homeostasis. Our data indicate that aHS modifies mitochondrial function, induces a DRP-1–dependent fission of the mitochondrial network, and triggers an autophagic flux with mitophagy events.
The aHS (1 h at 37°C) during the early fourth larval stage induces a marked developmental delay but no lethality or sterility, confirming previous analyses at 35°C or 37°C (Jovic et al., 2017; Zevian and Yanowitz, 2014). When young adults are submitted to a similar HS, no obvious anatomical phenotype has been reported, and worms display a hormetic response and increased lifespan (Kumsta et al., 2017). This indicates that, during development, animals are more sensitive to a short but elevated heat than in adulthood. The adaptation to HS is a priority and may be detrimental to the ongoing developmental program. Thus, a transcriptomic analysis revealed a very global shift in expression dynamics during the first 3–4 h of HS and identified cuticle synthesis among the major developmental processes that are disrupted (Jovic et al., 2017). Interestingly, our study revealed a massive HS response in the epidermis, the main tissue involved in cuticle synthesis during larval stages (Page and Johnstone, 2007).
Our data show that mitochondria are rapidly fragmented upon aHS and present a decrease in oxygen consumption. Moreover, the TEM analysis revealed the characteristic presence of dark occlusions in the matrix of heat-stressed mitochondria, which could correspond to protein aggregates due to temperature-induced misfolding of proteins. Such inclusions have not been observed in the cytosol or in the inflated ER, suggesting that they reflect some specificity of the mitochondria. Noticeably, similar inclusions have been described in sperm-derived mitochondria before their elimination by mitophagy (Wang et al., 2016; Zhou et al., 2016; Al Rawi et al., 2011; Sato and Sato, 2011). A similar phenotype has also been reported in a fraction of mitochondria in C. elegans mutants for the fusion genes eat-3/OPA1 and fzo-1/mfn1 (Byrne et al., 2019), supporting a link between dark aggregate formation and mitochondrial morphological defects.
After aHS, the fragmented mitochondria recover a tubular shape, demonstrating the reversibility of the stressed phenotype. Our data demonstrate that autophagy is important for the mitochondrial rebuilding and for the developmental delay recovery. Studies on adult worm muscles after various stresses suggest a balanced response between PINK-1/PDR-1–dependent mitophagy and mitochondrial biogenesis (Palikaras et al., 2015), but they did not document the rebuilding of the network. After aHS, we observed a strong autophagy response in the epidermis but a weaker one in the muscles, confirming a previous report (Kumsta et al., 2017). We did not detect an obvious implication of the PINK/PARKIN or Nix or FUNDC1 pathways, which are involved in selective mitophagy in C. elegans (Leboutet et al., 2020). One possibility is that some mitochondria are eliminated through bulk autophagy, but the hypothesis of a redundancy or a different mitophagy pathway cannot be excluded.
Our study provides the first evidence that, in both basal and aHS-induced autophagy, the autophagosomes are formed close to mitochondria in C. elegans epidermis. In yeast and mammal cells, ER–mitochondria contact zones are a major site for autophagosome biogenesis in stress conditions (Hamasaki et al., 2013; Böckler and Westermann, 2014). One can suppose that a similar mechanism occurs in C. elegans at specific contact sites between the mitochondria and ER.
Studies in Drp1 mouse models have shown that the mitochondrial network morphology is highly variable between tissues (Kageyama et al., 2014; Wakabayashi et al., 2009), which could be linked to differences in regard to mitochondrial needs such as ATP production. In C. elegans, the mitochondrial phenotype of the drp-1 mutant has been studied mainly in muscles and neurons, but it has not been characterized in the epidermis. Our in vivo and TEM analyses confirm that, in the absence of DRP-1, the mitochondria form a highly connected network composed of blebs and thin tubules. This could result from the initial constriction steps for mitochondrial fission involving ER contacts (Elgass et al., 2015) and actin filament (Friedman et al., 2011; Korobova et al., 2013; Kraus and Ryan, 2017) but the impossibility of processing further in the absence of DRP-1. The modification of the mitochondrial morphology of drp-1 animals after HS suggests that mitochondria are initiating an unsuccessful fission process.
The autophagy/mitophagy response to aHS is affected in drp-1 mutant worms, which could be due to impaired mitochondrial fission or a quality control defect. The selective elimination of mitochondria by mitophagy was initially linked to mitochondrial fission (Youle and van der Bliek, 2012). Several reports have shown that blocking fission or increasing fusion reduces the mitophagy (Gomes et al., 2011; Rambold et al., 2011; Twig et al., 2008). Fission could allow a “stress test,” permitting mitochondrial screening and further triggering the complete depolarization followed by mitophagy when a mitochondrion is subfunctional. A recent report proposed that the coordination of mitochondrial fission and control quality involves the interaction of Drp1 with the Zn2+ transporter Zip1 (Cho et al., 2019). However, an alternative process has been proposed with a local mitochondrial removal mediated by the extending isolation membrane/phagophore during mitophagy (Yamashita et al., 2016). This study performed in hypoxia or deferiprone-induced mitophagy conditions showed that most of the autophagosomes are formed on or near mitochondria and concluded that Drp1 is not essential for this process, despite a decrease in mitophagy efficiency. Other studies in cultured cells and yeast also suggest that Drp1/Dnm1-dependent fission is not required for mitophagy (Burman et al., 2017; Murakawa et al., 2015; Mendl et al., 2011). Our data support a DRP-1–dependent mechanism of formation of autophagosomes, following aHS in C. elegans, in contact with mitochondria, and they open the possibility that DRP-1 plays a role in bulk autophagy. It is possible that the mechanisms of formation of autophagosomes vary depending on the types of stress and cells used. In the epidermis after aHS, the clustering of autophagosomes is observed on both small and large mitochondria and is not abolished in drp-1;fzo-1–depleted animals, suggesting that this is not an indirect effect due to a mitochondrial size limitation. Moreover, our data suggest that mitochondrial fission could regulate the autophagic flux and support the hypothesis that DRP-1 is implicated in the coordination between mitochondrial fission and autophagosome biogenesis. It is tempting to speculate that, in the absence of drp-1, the contact sites between ER and mitochondria are maintained for a longer time because mitochondrial fission is not achieved. If these contact sites are involved in autophagosome biogenesis, a prolonged initiation-elongation of isolation membrane could explain the clustering observed upon aHS.
Materials and methods
C. elegans culture and strains
Nematode strains were grown on nematode growth media (NGM: 1.5 g NaCl [Sigma-Aldrich, 60142], 1.5 g bactopeptone [Becton-Dickinson, 211677], 0.5 ml of 5 mg/ml cholesterol [Sigma-Aldrich, C8667], 10 g bacto agar [Becton-Dickinson, 214010], add 500 ml of H2O. Autoclave the NGM medium and supplement it with 500 µl of 1 M CaCl2 [Sigma-Aldrich, C3306], 500 µl of 1 M MgSO4 [Sigma-Aldrich, M5921], 10 ml of 1 M KH2PO4 [Sigma-Aldrich, P5655], 1,650 µl of 1 M K2HPO4 [Sigma-Aldrich, 60356]) and fed with Escherichia coli strain OP50 (Caenorhabditis Genetic Center). The C. elegans Bristol N2 strain was used as a wild-type strain. Genotypes of all the strains used in this study are listed in Table S1.
For HS, adult worms were allowed to lay eggs for 1–2 h at 20°C on NGM plates and then removed. NGM plates were maintained at 20°C until the progeny reached early L4 stage and were submitted to 37°C for 60 min in an incubator (Binder). For egg-laying analysis, 10 worms were placed on NGM-OP50 plates for 1 h at 20°C, then removed, and the number of eggs was immediately scored. Each score was calculated in triplicate and reproduced four times independently.
For MitoTracker staining, L1–L2 worms were transferred to NGM agar plates containing 3.7 µM of Red CMXRos or DeepRed MitoTracker (Molecular Probes, Invitrogen) and incubated for overnight in the dark. Worms were transferred for 1 h into a normal NGM plate for lowering background staining.
RNA-mediated interference
RNAi was delivered by feeding (Kamath and Ahringer, 2003; Timmons and Fire, 1998). L4 animals or embryos were raised onto 1 mM IPTG-containing NGM plates seeded with bacteria E. coli HT115[DE3] carrying the empty vector L4440 (pPD129.36) as a control or the bacterial clones from the J. Ahringer library, Open Biosystem (lgg-1: C32D5.9, lgg-2: ZK593.6, atg-7: M7.5, epg-5: C56C10.12, pink-1: EEED8.9: dct-1: C14F5.1, drp-1: T12E12,4).
Light microscopy imaging
Epifluorescence images were captured on an AxioImagerM2 microscope (Zeiss) with 10×/0.3 NA EC Plan Neofluar or 100×/1.4 NA plan apochromat differential interference contrast (DIC) oil objectives using ZEN acquisition software (Zeiss) coupled to a camera (AxioCam506mono). Worms were immobilized by 40 mM sodium azide (in M9) and mounted on 2% agarose pads. Considering the effect of sodium azide on mitochondrial respiration, the observation duration after mounting was limited within 10 min.
Confocal images were captured with LAS-X software on a confocal Leica TCS SP8 DMI6000 microscope using an HC plan apochromat 63×/1.4 NA oil objective, with serial z sections of 0.3–1 µm. Images were analyzed using the ImageJ Cell Counter plugin (http://imagej.nih.gov/ij). The time of recovery after HS is indicative and could vary to a maximum of 30 min due to mounting and acquisition processes.
To perform time-lapse microscopy experiments, worms were grown in the dark on plates containing MitoTracker. Then, L4 worms were transferred for 1 h onto a normal plate for lowering background staining and then submitted to 1 h at 37°C. After 40 min of recovery at 20°C, the animals were immobilized in 30% (wt/vol) Pluronic F-127 gel (Sigma-Aldrich) containing 30-µm polystyrene microbeads. Confocal images were captured with serial z sections of 0.8 µm every 5 min (3% of 488-nm laser intensity, gain 600, binning 2 × 2; 10% of 638-nm laser intensity, gain 850, binning 2 × 2). Image analyses were performed with ImageJ or Fiji software.
Gonads were dissected in 1.25% PFA-PBS and fixed for 30 min in 4% PFA-PBS. After 30 min of saturation in PBS containing 4% BSA (Sigma-Aldrich; A7030) and 0.1% Triton X-100 (Sigma-Aldrich; T9284), samples were incubated overnight with 60-kD heat shock protein (HSP60) monoclonal antibodies (1:5; Developmental Studies Hybridoma Bank, University of Iowa). After incubation with anti-mouse antibodies Alexa Fluor 488 (Molecular Probes; 1:500, A28175) and Hoechst (Molecular Probes; 1:500), gonads were mounted in 1,4-diazabicyclo[2.2.2]octane (Sigma-Aldrich).
Analyses of mitochondrial shape
The aspect ratio and the form factor, which are indicators of the mitochondrial length and branching, respectively, have been quantified using ImageJ software. Briefly, the images are convolved using the matrix developed by Koopman (Koopman et al., 2016; Marchi et al., 2017), then a threshold is applied to isolate mitochondria from background. In the case of drp-1 mutant, the convolution was not applied, because it generated artificial objects. Shape descriptors of the mitochondria were calculated by implementing the Analyze Particles tool with a minimum filter of 15. For EM pictures, the outlines of mitochondria were manually drawn using freehand selections in ImageJ, then area, perimeter, and shape descriptors (roundness) were measured.
Statistics
All statistical analyses were performed using either GraphPad Prism or R software (https://www.r-project.org/). The Shapiro-Wilk test was used to evaluate the normal distribution of the values, and the Hartley Fmax test was used for similar variance analysis. Data derived from different genetic backgrounds were compared by two-sided Student’s t test, ANOVA, Kruskal-Wallis test, or Wilcoxon-Mann-Whitney test. Fisher’s exact test was used for nominal variables. Boxplot representations indicate the first (Q1/25th percentile), median (Q2/50th percentile), and third (Q3/75th percentile) quartiles, and whiskers are a maximum of 1.5 times the interquartile range (excluding any outliers). The cross X indicates the mean.
EM
L4 larvae were transferred to M9 20% BSA (Sigma-Aldrich; A7030) on 200-µm-deep flat carriers (Leica Biosystems), followed by cryoimmobilization in the EMPACT-2 high-pressure freezer apparatus (Leica Microsystems) as described previously (Largeau and Legouis, 2019). Cryosubstitution was performed using an automated freeze-substitution system (AFS2) with an integrated binocular lens and incubating chamber (Leica Microsystems) with acetone. Blocks were infiltrated with 100% Epon and embedded in fresh Epon (Agar Scientific; R1165). Ultrathin sections of 80 nm were cut on a ultramicrotome (Leica Microsystems; EM UC7) and collected on a formvar- and carbon-coated copper slot grid (LFG; FCF-2010-CU-50). Sections were contrasted with 0.05% oolong tea extract for 30 min and 0.08 M lead citrate (Sigma-Aldrich; 15326) for 8 min. Sections were observed with a Jeol 1400 TEM instrument at 120 kV, and images were acquired with a Gatan 11-megapixel SC1000 Orius charge-coupled device camera.
Oxygen consumption measurements
Synchronized young L4 larvae were recovered in M9 buffer (Brenner, 1974) and washed three times in the following solution: 60 mg MgSO4–7H2O, 60 mg CaSO4–2H2O, 4 mg KCl per liter of double distilled water (Luz et al., 2015) and separated by 2-min centrifugation at 100 g at 21°C. Worms were transferred in Seahorse XF plate wells in a final 180-µl volume, and six measurements were performed with a setting corresponding to 2 min mixing, 30 s waiting, and 2 min measuring. 15 mM carbonyl cyanide-4(trifluoromethoxy)phenylhydrazone (Sigma; C2920) in 0.17% DMSO was used to determine maximal respiration, and 40 mM sodium azide was used to inhibit mitochondrial respiration. The number of animals (15–30 per well) was determined to normalize the oxygen consumption rate calculated using Wave software (Agilent). Experiments were performed in triplicates and repeated independently.
Western blot analysis
Total protein extracts were prepared from a synchronized population of L4 larvae. The worms were collected after centrifugation and then mixed with the lysis buffer (PBS–Triton 2%) containing glass beads. They were then denatured using the Precellys 24 machine at 6,000 rpm with incubation for ∼5 min twice to cool the sample. The protein extracts are then centrifuged at 15,000 rpm and separated on a NuPAGE 4–12% Bis-Tris gel (Life Technologies; NP0321BOX). The nonspecific sites are then blocked after the incubation for 1 h with TBS–Tween 1× (Tris Base NaCl, Tween 20) 5% (w/v) milk. Blots were probed with antibodies against GFP (1:1,000, mouse, 11814460001; Sigma-Aldrich), LGG-1 (1:200; rabbit), tubulin (1:2,000, 078K4763; Sigma-Aldrich), ATP-5 (1:1,000, ab14705; Abcam), and mitochondrially encoded cytochrome c oxidase I (1:2,000, ab14748; Abcam) and revealed using HRP-conjugated antibodies (anti-rabbit, 1:1,000, W401B, and anti-mouse, 1:5,000, W4021; both from Promega) and SuperSignal Pico Chemiluminescent Substrate (Thermo Fisher Scientific; 34579). Signals were revealed on an LAS-3000 photoimager (Fuji) and quantified with ImageJ.
Online supplemental material
Fig. S1 (complementary to Fig. 1) characterizes cellular defects after aHS. Fig. S2 (complementary to Fig. 2) illustrates how specific reporters can affect mitochondrial network morphology. Fig. S3 (complementary to Fig. 3) shows that aHS induces autophagy in various tissues. Fig. S4 (complementary to Figs. 4 and 5) shows that blocking autophagy but not mitophagy delays the rebuilding of the mitochondrial network after aHS. Fig. S5 (complementary to Fig. 7) shows that the clustering of autophagosomes at mitochondria is maintained in drp-1;fzo-1 animals after aHS. Table S1 lists strains used in the study.
Supplementary Material
lists C. elegans strains used in the study.
Acknowledgments
This paper is dedicated to the memory of our colleague and friend Agnès Delahodde.
For providing reagents, the authors thank Hong Zhang (National Laboratory of Biomacromolecules, Beijing, China), Chonglin Yang (Center for Life Science, Yunnan University, China), Nektarios Tavernarakis (Institute of Molecular Biology and Biotechnology, Heraklion, Greece), Steven Zuryn (The University of Queensland, Brisbane, Australia), Rosa Navarro (Universidad Nacional Autónoma de México, Ciudad de México, México), Ming Li (School of Life Sciences, Yunnan University, China), Sylvie Hermann-Ledenmat (I2BC, Gif-sur-Yvette, France), and Raynald Cossard (I2BC, Gif-sur-Yvette, France), and the Caenorhabditis Genetic Center, which is funded by the National Institutes of Health, National Center for Research Resources. We are grateful to Legouis laboratory members for helpful discussions.
This work was supported by the Agence Nationale de la Recherche (project EAT; ANR-12-BSV2-018), the Association pour la Recherche sur le Cancer (SFI20111203826), and the Ligue Contre le Cancer (M29506). Y. Chen received a fellowship from the China Scholarship Council, and R. Leboutet received a fellowship from the Fondation pour la Recherche Médicale. The present work has benefited from the core facilities of Imagerie-Gif (http://www.i2bc.paris-saclay.fr), a member of Infrastructures en Biologie Santé et Agronomie (http://www.ibisa.net), supported by France BioImaging (ANR-10-INBS-04-01) and the Labex Saclay Plant Science (ANR-11-IDEX-0003-02). The monoclonal antibody HSP60 was obtained from the Developmental Studies Hybridoma Bank, created by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health and maintained at the Department of Biology, University of Iowa, Iowa City, IA.
The authors declare no competing financial interests.
Author contributions: Y. Chen: methodology, investigation, validation, formal analysis, visualization. R. Leboutet: methodology, investigation, validation, visualization. C. Largeau: methodology, investigation, validation, formal analysis, visualization. S. Zentout and C. Lefebvre: investigation, validation, formal analysis, visualization. A. Delahodde: conceptualization, supervision, resources. E. Culetto: methodology, investigation, validation, formal analysis, supervision, writing – original draft. R. Legouis: conceptualization, project administration, supervision, funding acquisition, visualization, writing – original draft. All authors: review and editing.
References
- Ahier, A., Dai C.-Y., Tweedie A., Bezawork-Geleta A., Kirmes I., and Zuryn S.. 2018. Affinity purification of cell-specific mitochondria from whole animals resolves patterns of genetic mosaicism. Nat. Cell Biol. 20:352–360. 10.1038/s41556-017-0023-x [DOI] [PubMed] [Google Scholar]
- Al Rawi, S., Louvet-Vallée S., Djeddi A., Sachse M., Culetto E., Hajjar C., Boyd L., Legouis R., and Galy V.. 2011. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 334:1144–1147. 10.1126/science.1211878 [DOI] [PubMed] [Google Scholar]
- Alberti, A., Michelet X., Djeddi A., and Legouis R.. 2010. The autophagosomal protein LGG-2 acts synergistically with LGG-1 in dauer formation and longevity in C. elegans. Autophagy. 6:622–633. 10.4161/auto.6.5.12252 [DOI] [PubMed] [Google Scholar]
- Böckler, S., and Westermann B.. 2014. Mitochondrial ER contacts are crucial for mitophagy in yeast. Dev. Cell. 28:450–458. 10.1016/j.devcel.2014.01.012 [DOI] [PubMed] [Google Scholar]
- Breckenridge, D.G., Kang B.-H., Kokel D., Mitani S., Staehelin L.A., and Xue D.. 2008. Caenorhabditis elegans drp-1 and fis-2 regulate distinct cell-death execution pathways downstream of ced-3 and independent of ced-9. Mol. Cell. 31:586–597. 10.1016/j.molcel.2008.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. 10.1093/genetics/77.1.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burman, J.L., Pickles S., Wang C., Sekine S., Vargas J.N.S., Zhang Z., Youle A.M., Nezich C.L., Wu X., Hammer J.A., et al. 2017. Mitochondrial fission facilitates the selective mitophagy of protein aggregates. J. Cell Biol. 216:3231–3247. 10.1083/jcb.201612106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, J.J., Soh M.S., Chandhok G., Vijayaraghavan T., Teoh J.-S., Crawford S., Cobham A.E., Yapa N.M.B., Mirth C.K., and Neumann B.. 2019. Disruption of mitochondrial dynamics affects behaviour and lifespan in Caenorhabditis elegans. Cell. Mol. Life Sci. 76:1967–1985. 10.1007/s00018-019-03024-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Scarcelli V., and Legouis R.. 2017. Approaches for studying autophagy in Caenorhabditis elegans. Cells. 6:27. 10.3390/cells6030027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H.M., Ryu J.R., Jo Y., Seo T.W., Choi Y.N., Kim J.H., Chung J.M., Cho B., Kang H.C., Yu S.-W., et al. 2019. Drp1-Zip1 interaction regulates mitochondrial quality surveillance system. Mol. Cell. 73:364–376.e8. 10.1016/j.molcel.2018.11.009 [DOI] [PubMed] [Google Scholar]
- Desai, S., Liu Z., Yao J., Patel N., Chen J., Wu Y., Ahn E.E.-Y., Fodstad O., and Tan M.. 2013. Heat shock factor 1 (HSF1) controls chemoresistance and autophagy through transcriptional regulation of autophagy-related protein 7 (ATG7). J. Biol. Chem. 288:9165–9176. 10.1074/jbc.M112.422071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokladny, K., Zuhl M.N., Mandell M., Bhattacharya D., Schneider S., Deretic V., and Moseley P.L.. 2013. Regulatory coordination between two major intracellular homeostatic systems: heat shock response and autophagy. J. Biol. Chem. 288:14959–14972. 10.1074/jbc.M113.462408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgass, K.D., Smith E.A., LeGros M.A., Larabell C.A., and Ryan M.T.. 2015. Analysis of ER-mitochondria contacts using correlative fluorescence microscopy and soft X-ray tomography of mammalian cells. J. Cell Sci. 128:2795–2804. 10.1242/jcs.169136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Cárdenas, L.P., Villanueva-Chimal E., Salinas L.S., José-Nuñez C., Tuena de Gómez Puyou M., and Navarro R.E.. 2017. Caenorhabditis elegans ATPase inhibitor factor 1 (IF1) MAI-2 preserves the mitochondrial membrane potential (Δψm) and is important to induce germ cell apoptosis. PLoS One. 12:e0181984. 10.1371/journal.pone.0181984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., and Voeltz G.K.. 2011. ER tubules mark sites of mitochondrial division. Science. 334:358–362. 10.1126/science.1207385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes, L.C., Di Benedetto G., and Scorrano L.. 2011. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 13:589–598. 10.1038/ncb2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki, M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., et al. 2013. Autophagosomes form at ER-mitochondria contact sites. Nature. 495:389–393. 10.1038/nature11910 [DOI] [PubMed] [Google Scholar]
- Haynes, C.M., Fiorese C.J., and Lin Y.-F.. 2013. Evaluating and responding to mitochondrial dysfunction: the mitochondrial unfolded-protein response and beyond. Trends Cell Biol. 23:311–318. 10.1016/j.tcb.2013.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh, D., and Vanderslice R.. 1976. Temperature-sensitive developmental mutants of Caenorhabditis elegans. Dev. Biol. 49:220–235. 10.1016/0012-1606(76)90268-2 [DOI] [PubMed] [Google Scholar]
- Jenzer, C., Manil-Ségalen M., Lefebvre C., Largeau C., Glatigny A., and Legouis R.. 2014. Human GABARAP can restore autophagosome biogenesis in a C. elegans lgg-1 mutant. Autophagy. 10:1868–1872. 10.4161/auto.29745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenzer, C., Simionato E., Largeau C., Scarcelli V., Lefebvre C., and Legouis R.. 2019. Autophagy mediates phosphatidylserine exposure and phagosome degradation during apoptosis through specific functions of GABARAP/LGG-1 and LC3/LGG-2. Autophagy. 15:228–241. 10.1080/15548627.2018.1512452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, W.K., Hatch A.L., Merrill R.A., Strack S., and Higgs H.N.. 2015. Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. eLife. 4:e11553. 10.7554/eLife.11553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, K., Hart A.C., and Levine B.. 2007. Autophagy genes protect against disease caused by polyglutamine expansion proteins in Caenorhabditis elegans. Autophagy. 3:21–25. 10.4161/auto.3528 [DOI] [PubMed] [Google Scholar]
- Jovic, K., Sterken M.G., Grilli J., Bevers R.P.J., Rodriguez M., Riksen J.A.G., Allesina S., Kammenga J.E., and Snoek L.B.. 2017. Temporal dynamics of gene expression in heat-stressed Caenorhabditis elegans. PLoS One. 12:e0189445. 10.1371/journal.pone.0189445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama, Y., Hoshijima M., Seo K., Bedja D., Sysa-Shah P., Andrabi S.A., Chen W., Höke A., Dawson V.L., Dawson T.M., et al. 2014. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 33:2798–2813. 10.15252/embj.201488658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R.S., and Ahringer J.. 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 30:313–321. 10.1016/S1046-2023(03)00050-1 [DOI] [PubMed] [Google Scholar]
- Koopman, M., Michels H., Dancy B.M., Kamble R., Mouchiroud L., Auwerx J., Nollen E.A.A., and Houtkooper R.H.. 2016. A screening-based platform for the assessment of cellular respiration in Caenorhabditis elegans. Nat. Protoc. 11:1798–1816. 10.1038/nprot.2016.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova, F., Ramabhadran V., and Higgs H.N.. 2013. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 339:464–467. 10.1126/science.1228360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus, F., and Ryan M.T.. 2017. The constriction and scission machineries involved in mitochondrial fission. J. Cell Sci. 130:2953–2960. 10.1242/jcs.199562 [DOI] [PubMed] [Google Scholar]
- Kumsta, C., Chang J.T., Schmalz J., and Hansen M.. 2017. Hormetic heat stress and HSF-1 induce autophagy to improve survival and proteostasis in C. elegans. Nat. Commun. 8:14337. 10.1038/ncomms14337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia, J., Brielmann R.M., Neto M.F., Lin Y.-F., Haynes C.M., and Morimoto R.I.. 2017. Mitochondrial stress restores the heat shock response and prevents proteostasis collapse during aging. Cell Rep. 21:1481–1494. 10.1016/j.celrep.2017.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrousse, A.M., Zappaterra M.D., Rube D.A., and van der Bliek A.M.. 1999. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell. 4:815–826. 10.1016/S1097-2765(00)80391-3 [DOI] [PubMed] [Google Scholar]
- Largeau, C., and Legouis R.. 2019. Correlative light and electron microscopy to analyze LC3 proteins in Caenorhabditis elegans embryo. Methods Mol. Biol. 1880:281–293. 10.1007/978-1-4939-8873-0_18 [DOI] [PubMed] [Google Scholar]
- Leboutet, R., Chen Y., Legouis R., and Culetto E.. 2020. Mitophagy during development and stress in C. elegans. Mech. Ageing Dev. 189:111266. 10.1016/j.mad.2020.111266 [DOI] [PubMed] [Google Scholar]
- Lee, J., Giordano S., and Zhang J.. 2012. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem. J. 441:523–540. 10.1042/BJ20111451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.E., Westrate L.M., Wu H., Page C., and Voeltz G.K.. 2016. Multiple dynamin family members collaborate to drive mitochondrial division. Nature. 540:139–143. 10.1038/nature20555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry, J., Yochem J., Chuang C.-H., Sugioka K., Connolly A.A., and Bowerman B.. 2015. High-throughput cloning of temperature-sensitive Caenorhabditis elegans mutants with adult syncytial germline membrane architecture defects. G3 (Bethesda). 5:2241–2255. 10.1534/g3.115.021451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz, A.L., Rooney J.P., Kubik L.L., Gonzalez C.P., Song D.H., and Meyer J.N.. 2015. Mitochondrial morphology and fundamental parameters of the mitochondrial respiratory chain are altered in Caenorhabditis elegans strains deficient in mitochondrial dynamics and homeostasis processes. PLoS One. 10:e0130940. 10.1371/journal.pone.0130940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manil-Ségalen, M., Lefebvre C., Jenzer C., Trichet M., Boulogne C., Satiat-Jeunemaitre B., and Legouis R.. 2014. The C. elegans LC3 acts downstream of GABARAP to degrade autophagosomes by interacting with the HOPS subunit VPS39. Dev. Cell. 28:43–55. 10.1016/j.devcel.2013.11.022 [DOI] [PubMed] [Google Scholar]
- Marchi, S., Bonora M., Patergnani S., Giorgi C., and Pinton P.. 2017. Methods to assess mitochondrial morphology in mammalian cells mounting autophagic or mitophagic responses. Methods Enzymol. 588:171–186. 10.1016/bs.mie.2016.09.080 [DOI] [PubMed] [Google Scholar]
- Mendl, N., Occhipinti A., Müller M., Wild P., Dikic I., and Reichert A.S.. 2011. Mitophagy in yeast is independent of mitochondrial fission and requires the stress response gene WHI2. J. Cell Sci. 124:1339–1350. 10.1242/jcs.076406 [DOI] [PubMed] [Google Scholar]
- Momma, K., Homma T., Isaka R., Sudevan S., and Higashitani A.. 2017. Heat-induced calcium leakage causes mitochondrial damage in Caenorhabditis elegans body-wall muscles. Genetics. 206:1985–1994. 10.1534/genetics.117.202747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecinos-Franjola, F., Bauer B.L., Mears J.A., and Ramachandran R.. 2020. GFP fluorescence tagging alters dynamin-related protein 1 oligomerization dynamics and creates disassembly-refractory puncta to mediate mitochondrial fission. Sci. Rep. 10:14777. 10.1038/s41598-020-71655-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakawa, T., Yamaguchi O., Hashimoto A., Hikoso S., Takeda T., Oka T., Yasui H., Ueda H., Akazawa Y., Nakayama H., et al. 2015. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat. Commun. 6:7527. 10.1038/ncomms8527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, A.P., and Johnstone I.L.. 2007. The cuticle (March 19, 2007). WormBook, ed. The C. Elegans Research Community, WormBook, http://www.wormbook.org. 10.1895/wormbook.1.138.1 [DOI]
- Palikaras, K., Lionaki E., and Tavernarakis N.. 2015. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature. 521:525–528. 10.1038/nature14300 [DOI] [PubMed] [Google Scholar]
- Palikaras, K., Lionaki E., and Tavernarakis N.. 2018. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 20:1013–1022. 10.1038/s41556-018-0176-2 [DOI] [PubMed] [Google Scholar]
- Pickles, S., Vigié P., and Youle R.J.. 2018. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 28:R170–R185. 10.1016/j.cub.2018.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold, A.S., Kostelecky B., Elia N., and Lippincott-Schwartz J.. 2011. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. USA. 108:10190–10195. 10.1073/pnas.1107402108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M., and Sato K.. 2011. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 334:1141–1144. 10.1126/science.1210333 [DOI] [PubMed] [Google Scholar]
- Scholtes, C., Bellemin S., Martin E., Carre-Pierrat M., Mollereau B., Gieseler K., and Walter L.. 2018. DRP-1-mediated apoptosis induces muscle degeneration in dystrophin mutants. Sci. Rep. 8:7354. 10.1038/s41598-018-25727-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Q., Yamano K., Head B.P., Kawajiri S., Cheung J.T.M., Wang C., Cho J.-H., Hattori N., Youle R.J., and van der Bliek A.M.. 2014. Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Mol. Biol. Cell. 25:145–159. 10.1091/mbc.e13-09-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpilka, T., and Haynes C.M.. 2018. The mitochondrial UPR: mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell Biol. 19:109–120. 10.1038/nrm.2017.110 [DOI] [PubMed] [Google Scholar]
- Timmons, L., and Fire A.. 1998. Specific interference by ingested dsRNA. Nature. 395:854. 10.1038/27579 [DOI] [PubMed] [Google Scholar]
- Twig, G., Elorza A., Molina A.J.A., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., et al. 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27:433–446. 10.1038/sj.emboj.7601963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi, J., Zhang Z., Wakabayashi N., Tamura Y., Fukaya M., Kensler T.W., Iijima M., and Sesaki H.. 2009. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J. Cell Biol. 186:805–816. 10.1083/jcb.200903065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Zhang Y., Chen L., Liang Q., Yin X.-M., Miao L., Kang B.-H., and Xue D.. 2016. Kinetics and specificity of paternal mitochondrial elimination in Caenorhabditis elegans. Nat. Commun. 7:12569. 10.1038/ncomms12569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., Carvalho P., and Voeltz G.K.. 2018. Here, there, and everywhere: the importance of ER membrane contact sites. Science. 361:eaan5835. 10.1126/science.aan5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita, S.-I., Jin X., Furukawa K., Hamasaki M., Nezu A., Otera H., Saigusa T., Yoshimori T., Sakai Y., Mihara K., et al. 2016. Mitochondrial division occurs concurrently with autophagosome formation but independently of Drp1 during mitophagy. J. Cell Biol. 215:649–665. 10.1083/jcb.201605093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle, R.J., and van der Bliek A.M.. 2012. Mitochondrial fission, fusion, and stress. Science. 337:1062–1065. 10.1126/science.1219855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zevian, S.C., and Yanowitz J.L.. 2014. Methodological considerations for heat shock of the nematode Caenorhabditis elegans. Methods. 68:450–457. 10.1016/j.ymeth.2014.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y.G., Chen Y., Miao G., Zhao H., Qu W., Li D., Wang Z., Liu N., Li L., Chen S., et al. 2017. The ER-localized transmembrane protein EPG-3/VMP1 regulates SERCA activity to control ER-isolation membrane contacts for autophagosome formation. Mol. Cell. 67:974–989.e6. 10.1016/j.molcel.2017.08.005 [DOI] [PubMed] [Google Scholar]
- Zhou, Q., Li H., Li H., Nakagawa A., Lin J.L.J., Lee E.-S., Harry B.L., Skeen-Gaar R.R., Suehiro Y., William D., et al. 2016. Mitochondrial endonuclease G mediates breakdown of paternal mitochondria upon fertilization. Science. 353:394–399. 10.1126/science.aaf4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists C. elegans strains used in the study.