Significance
Diseases associated with amyloid formation affect millions of people worldwide. Great progress has been made to understand the process of amyloid formation in test tube reactions, but it has so far remained unclear if the same biophysical principles also dominate in living cells and organisms. Here, we use the nematode C. elegans to demonstrate that we can apply kinetic modeling approaches to quantitatively analyze the process of protein aggregation in a living animal. The mathematical models that we develop are not restricted to this system and are applicable to other disease-associated proteins and animal models. The ability to quantitatively understand aggregation mechanisms in living organisms will be critical to advance our understanding of human disease and to design therapeutic strategies.
Keywords: protein aggregation, amyloid, chemical kinetics, polyglutamine, C. elegans
Abstract
Protein aggregation is associated with a wide range of degenerative human diseases with devastating consequences, as exemplified by Alzheimer’s, Parkinson’s, and Huntington’s diseases. In vitro kinetic studies have provided a mechanistic understanding of the aggregation process at the molecular level. However, it has so far remained largely unclear to what extent the biophysical principles of amyloid formation learned in vitro translate to the complex environment of living organisms. Here, we take advantage of the unique properties of a Caenorhabditis elegans model expressing a fluorescently tagged polyglutamine (polyQ) protein, which aggregates into discrete micrometer-sized inclusions that can be directly visualized in real time. We provide a quantitative analysis of protein aggregation in this system and show that the data are described by a molecular model where stochastic nucleation occurs independently in each cell, followed by rapid aggregate growth. Global fitting of the image-based aggregation kinetics reveals a nucleation rate corresponding to 0.01 h−1 per cell at 1 mM intracellular protein concentration, and shows that the intrinsic molecular stochasticity of nucleation accounts for a significant fraction of the observed animal-to-animal variation. Our results highlight how independent, stochastic nucleation events in individual cells control the overall progression of polyQ aggregation in a living animal. The key finding that the biophysical principles associated with protein aggregation in small volumes remain the governing factors, even in the complex environment of a living organism, will be critical for the interpretation of in vivo data from a wide range of protein aggregation diseases.
Protein aggregation is a pathological hallmark of a wide range of neurodegenerative and systemic diseases (1, 2). A mechanistic understanding of the pathways of amyloid formation has been obtained in vitro by approaches of chemical kinetics, providing fundamental insights into the microscopic aggregation steps of disease-associated proteins and peptides, most notably the amyloid-β peptide associated with Alzheimer’s disease (AD) (3). These studies have been invaluable to the design of small molecules that reduce the generation of potentially toxic oligomeric species (4, 5), to understand the action of molecular chaperones in inhibiting protein aggregation (6, 7), and to rationalize the efficacy of antibodies in clinical trials for AD (8). This framework has been extended to include stochastic and spatial effects that control kinetics in small volumes (femtoliter-picoliter range) (9–11), with the premise of being applicable to protein aggregation in living cells.
However, cells and organisms have evolved intricate protein homeostasis pathways to ensure correct protein folding and to suppress misfolding and aggregation (12, 13), and it has not yet been established whether a chemical kinetics approach is sufficient to describe protein aggregation in vivo. The fundamental question is whether the complex nature of the cellular and organismal environment induces a major remodeling of the aggregation network as studied in vitro, or whether the same biophysical principles remain dominant.
The nematode Caenorhabditis elegans provides the high level of control and the tools necessary to perform a quantitative kinetic analysis and determine the mechanisms governing protein aggregation in a living animal. C. elegans has a well-defined anatomy, and the animals within a population are genetically identical. Perhaps the most beneficial feature of this animal model system is its optical transparency, allowing the aggregation of a fluorescently labeled protein to be directly visualized. Specifically, we take advantage of the C. elegans muscle tissue that corresponds to 95 physiologically identical cells, and propose that these postmitotic cells can be quantitatively modeled as individual “test tubes” in which the deposition of expanded polyQ takes place by a mechanism of nucleated aggregation.
Results
Establishing a Framework for In Vivo Protein Aggregation Kinetics.
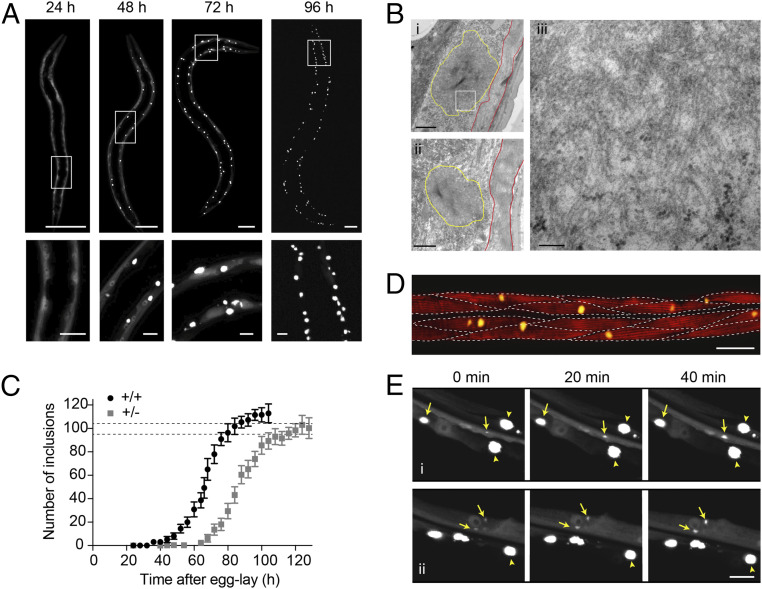
PolyQ-containing proteins have been studied extensively in vitro and in vivo, and expansions of polyQ were predicted (14) and have been observed to form cross-β fibrils in vitro (15, 16) and to accumulate into insoluble inclusions in polyQ diseases in vivo (15, 17, 18). Most polyQ diseases have a pathogenic threshold at an expansion of around 35 to 40 glutamine residues (19), and this threshold is conserved across experimental systems including C. elegans (20). When expressed in C. elegans muscle cells, a Q40 protein C-terminally tagged with yellow fluorescent protein (Q40-YFP, SI Appendix, Materials and Methods) forms intracellular inclusions that have a relatively uniform size and shape (Fig. 1A and SI Appendix, Fig. S1) (20). Visualized by transmission electron microscopy (TEM), the polyQ inclusions contain fibrillar material with widths on the order of 10 nm (Fig. 1B and SI Appendix, Fig. S2). The appearance of immobile amyloid-like inclusions is consistent with fluorescence lifetime imaging (FLIM) (21) and fluorescence recovery after photobleaching (FRAP) (20) experiments carried out previously. Protein aggregation kinetics can thus be monitored in living animals using the visualization of intracellular Q40-YFP inclusions as a direct measure of the total aggregate amount. The appearance of inclusions over time is reminiscent of the kinetics of amyloid formation observed in vitro, displaying a rapid increase from zero toward a plateau (Fig. 1C).
Fig. 1.
Q40-YFP expressed in C. elegans body wall muscle cells displays concentration-dependent, amyloid-like aggregation kinetics with each cell acquiring one inclusion on average. (A) Confocal images of C. elegans strain AM141, progressively displaying bright inclusions of a relatively uniform size and shape in the body wall muscle cells. Lower panels are magnifications of the boxed areas in the Upper panels. (Scale bars: Upper, 50 µm; Lower, 10 µm.) (B) Transmission electron micrographs of embedded sections of 96-h-old animals showing the subcellular localization of the inclusions (yellow outlines) below the muscle sarcomere (red outlines). Right shows a higher magnification image corresponding to the boxed region in the Upper Left panel, displaying a meshwork of fibrils with a typical width on the order of 10 nm. (Scale bars: Left, 1 µm; Right, 100 nm.) (C) Average number of inclusions per animal over time for age-synchronized populations of C. elegans expressing Q40-YFP in body wall muscle cells for homozygous (+/+) and heterozygous (+/−) animals. The plateau value for the number of inclusions approximates the number of cells in which the protein is expressed, indicated by the dashed lines at 95 for the body wall muscle cells and 104 for the total number of muscle cells in which Q40-YFP is observed. n = 12 to 20 animals for each timepoint; error bars indicate the SD. (D) Confocal image of an animal stained with phalloidin around the midpoint of aggregation (66 h) to reveal the muscle filaments. Dashed lines indicate the approximate boundaries between muscle cells, revealing that some have acquired an inclusion by this time, whereas in other cells visible aggregation has not yet taken place. (Scale bar, 20 µm.) (E) Panels i and ii: close-up confocal images of the muscle tissue in strain AM141 followed for 40 min at around the midpoint of aggregation (64 h) (SI Appendix, Fig. S1B). Yellow arrows point to inclusions that are observed to grow during this time, whereas arrowheads indicate mature inclusions that do not change in size. Note that diffuse signal of soluble Q40-YFP is depleted around the mature inclusions. (Scale bar, 10 µm.)
Also analogous to in vitro reactions, inclusion formation in vivo is concentration dependent, as evidenced by the comparison of homozygous and heterozygous Q40-YFP animals (Fig. 1C), the latter having 50% of the Q40-YFP gene copies and exhibiting 50% of the fluorescence intensity prior to the onset of aggregation (SI Appendix, Fig. S3). Despite the different kinetic profiles for the animals expressing the two Q40-YFP protein concentrations, the plateau value for the number of inclusions per animal is very similar (∼112 for homozygous versus ∼100 for heterozygous animals) (Fig. 1C). This number of aggregates corresponds closely to the number of cells in which Q40-YFP is expressed (dashed lines in Fig. 1C). The unc-54 promoter predominantly directs expression to the 95 body wall muscle cells, and in addition strain AM141 displays fluorescence in the eight vulval muscle cells and the anal depressor cell, leading to a total of 104 cells in which Q40-YFP is expressed and aggregation can occur. This result reveals that, on average, each cell acquires one inclusion over the course of the aggregation process. The aggregation curve can therefore be interpreted as the cumulative number of cells in which visible inclusion formation has taken place. Indeed, at the midpoint of the aggregation reaction we find that the majority of cells have either zero or one inclusion, whereas only rarely a cell contains two inclusions (Fig. 1D).
The observation that most muscle cells acquire a single polyQ inclusion suggests a rate-limiting nucleation event that initiates the aggregation process. For simple polyQ stretches, nucleation has been proposed to arise from a conformational change of a single molecule (22, 23). However, oligomeric species formed by polyQ-containing proteins have also been observed in several model systems including C. elegans (24), and have been suggested to act as precursors on the pathway toward fibril formation (25–27). Irrespective of the molecular nature of the nucleation event in our model, this step is required to initiate the formation of visible inclusions through fibril elongation and secondary processes that together amplify the fibril mass. In particular, branching of fibrils or secondary nucleation with slow off-rates of the newly formed nuclei can result in three-dimensional growth of the inclusion. Consistent with this framework, the growth of polyQ inclusions is experimentally observable in the cells of live animals and proceeds at a rapid radial expansion rate of ∼0.5 µm/h (Fig. 1E). Since mature inclusions typically have a diameter of 3 to 5 µm, the obtained rate reveals that the inclusions can reach their maximum size on a timescale of several hours. Diffuse signal corresponding to soluble Q40-YFP is no longer observed surrounding mature inclusions (Fig. 1E), indicating that inclusion growth is limited by the available amount of protein in each cell. Whereas we cannot exclude the possibility that newly synthesized protein continues to be incorporated into the inclusion, translation is probably limited as a consequence of the mounted stress response (28). Supporting this notion, polysomes have been shown to decline in Q40-YFP animals (29).
Nucleation Occurs Stochastically in Individual Muscle Cells.
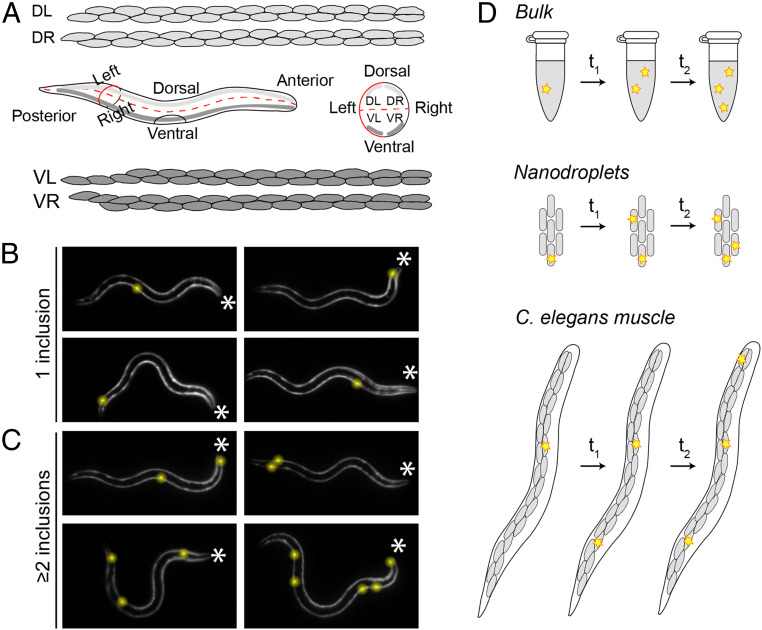
The increase over time of the number of cells in which an inclusion has formed (Fig. 1 C and D) implies that the initiation of protein aggregation occurs at different moments in time in individual cells. One possible explanation for this observation could be biological variation that makes some cells more prone to undergo aggregation than others. However, aside from the vulval muscle cells and the anal depressor muscle, the 95 C. elegans body wall muscle cells (Fig. 2A) are highly similar in their lineage relationship, anatomical function, and gene expression profiles (30, 31). Consistent with the cells being physiologically identical and having equal aggregation propensities, the first inclusion observed for each of a set of animals appeared randomly among the cells along the posterior–anterior axis (Fig. 2B and SI Appendix, Fig. S4).
Fig. 2.
Q40-YFP aggregation occurs stochastically in individual muscle cells. (A) Illustration showing the distribution of the 95 body wall muscle cells in adult C. elegans. Four bundles of muscle cells run along the posterior–anterior axis. The animals typically crawl on their left or right side on solid media, resulting in a superposition of the two dorsal and the two ventral bundles (Middle Left). A schematic view from the anterior side shows the localization of the four bundles in the different quadrants (Middle Right). Cell shapes were drawn based on http://wormatlas.org. (B) The 32-h-old animals display their first inclusion at any position along the posterior–anterior axis. The asterisks mark the anterior of the animals; inclusions are highlighted by yellow spheres for better visualization. (C) The 32-h-old animals with two or more inclusions support the notion that nucleation is initiated stochastically in individual cells, in the absence of a pattern of spatial propagation from the first to subsequent inclusions. (D) Cartoon showing the occurrence of aggregation events (yellow stars) in bulk, nanodroplets, and in C. elegans muscle cells. In a typical test tube reaction (Top), all aggregation events take place within the same continuous volume. In nanodroplets (Middle) and C. elegans muscle tissue (Bottom, only one bundle of muscle cells is shown for clarity), the total volume is divided over multiple small volumes, in which the probability of nucleation is low. As a consequence, aggregation will be initiated in different droplets or cells at different points in time. Three aggregation events are shown in each system for clarity, but in reality the number of nucleation events in a given period of time is proportional to the size of the reaction vessel.
A second possible hypothesis for the different aggregation onset across cells is that nucleation is first randomly triggered in one cell, after which aggregation spreads to neighboring cells. The spreading of aggregation throughout the brain by means of cell-to-cell transfer of aggregate seeds has been proposed to contribute to the pathology of several neurodegenerative diseases, including Huntington’s (32, 33). However, we do not find evidence for a spatial correlation between the first and subsequent inclusions (Fig. 2C and SI Appendix, Fig. S4B), suggesting that Q40-YFP aggregation is triggered by independent nucleation events in the corresponding cells in C. elegans. The absence of direct aggregate transfer between adjacent cells is supported by a recently published observation that Q40-RFP expressed in C. elegans body wall muscle does not spread to neighboring tissues (34), and is further verified below using kinetic analysis.
These observations motivate the question whether the inclusions form at different times in each cell because of the intrinsic stochasticity of the nucleation process in small volumes (Fig. 2D). In bulk in vitro experiments, nucleation events occur with high frequency throughout the continuous, microliter-sized reaction volume. In contrast, these events become very rare when one considers individual picoliter-sized volumes, such as that of a cell (9–11). The intrinsic nucleation properties of the molecules remain the same when a given reaction volume is divided into multiple small volumes, but one cannot predict in which of the small volumes a given nucleation event will occur. In the C. elegans muscle tissue, aggregate nuclei will thus appear randomly distributed in a cell-by-cell fashion over time (Fig. 2D). The probability of a second nucleation event occurring within the same cell is low, especially given the rapid speed of inclusion growth which quickly depletes the surrounding soluble protein (Fig. 1E), hence decreasing the probability of another nucleation event.
In the absence of aggregate spreading, the question remains what causes the sigmoidal shape of the aggregation curve (Fig. 1C). In strain AM141, inclusion formation begins during larval stages and proceeds as the animal is growing. We found that the marked increase in muscle cell volume during development (∼50-fold from embryo to adult, SI Appendix, Fig. S5A) dominates the shape of the aggregation curve, given that the probability of nucleation is expected to scale linearly with cell volume at equal concentration (SI Appendix, Fig. S5).
Altogether, our observations are in agreement with a model in which Q40-YFP aggregation in C. elegans body wall muscle cells is governed by stochastic nucleation in individual cells, followed by inclusion growth.
Quantitative Analysis of In Vivo Protein Aggregation Kinetics.
To consolidate these findings of stochastic nucleation and inclusion growth in a quantitative manner, we turned toward the application of chemical kinetics. Kinetic analysis has transformed the mechanistic understanding of protein aggregation in vitro and allows macroscopic measurements of aggregation processes to be connected with their microscopic mechanisms. Global fitting of in vitro data depends on the availability of a range of protein concentrations (35). Moreover, under in vitro conditions, the volume within which the aggregation reaction takes place is typically constant throughout the aggregation time course. To address these issues, we generated a new series of four C. elegans strains (designated A–D) that express different amounts of Q40-YFP under the control of the unc-54 promoter. The reduced protein concentrations compared to strain AM141 restrict nucleation during developmental stages, and instead the new lines show an onset of inclusion formation at early adulthood, when the muscle cells are close to reaching their maximum volume (SI Appendix, Fig. S5A). In the following kinetic analysis, we therefore do not consider the developmental stage and define the first day of adulthood as t = 0.
To monitor the aggregation time course for all four strains in parallel, we employed a high-throughput confocal imaging platform with semiautomated image analysis (Fig. 3 and SI Appendix, Figs. S6 and S7 and Materials and Methods). This method yields numbers that are comparable to those obtained by manual inclusion counting (SI Appendix, Fig. S6E), but has the advantage of being unbiased and more quantitative. The intracellular protein concentrations were quantified from Western blot analysis, yielding 1 mM, 0.7 mM, 0.5 mM, and 0.3 mM for strains A–D, respectively (SI Appendix, Fig. S8 and Materials and Methods). These values were proportional to the fluorescence intensities for the four strains (SI Appendix, Fig. S8D). We monitored the animals from day 1 up to day 8 of adulthood, during which fluorescence intensities remained approximately constant (SI Appendix, Fig. S7B), and as expected, strains expressing higher protein concentrations exhibited faster kinetics (Fig. 3 and SI Appendix, Fig. S7A).
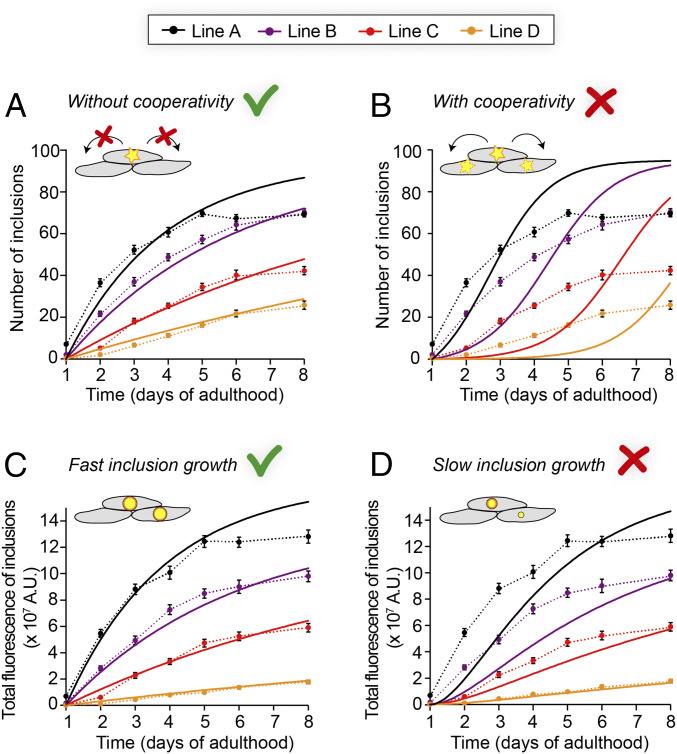
Fig. 3.
Determining the mechanisms of Q40-YFP aggregation in vivo by global fitting of aggregation time courses at multiple protein concentrations. (A) Global fit (solid lines) of the average number of inclusions per animal, assuming a constant nucleation rate over time and no cooperativity. The two global free parameters are the nucleation rate and the reaction order. (B) Global fit (solid lines) of the same dataset shown in A, but using a model that forces significant cooperativity. Cooperativity is not spatially restricted in this model and can occur between any of the cells. The two global free parameters are the nucleation rate and the reaction order. (C) Prediction of the total aggregate amount based on the fit in A and assuming fast inclusion growth on a timescale of hours (solid lines), compared to the observed integrated fluorescence over the inclusions per animal. (D) Prediction as in C, but assuming slow inclusion growth on a timescale of days. Data are representative of two independent experiments. The dashed lines connecting the datapoints in A–D are to guide the eye, and error bars indicate the SEM. n = 20 animals per strain and timepoint.
In order to define the Q40-YFP aggregation reaction in C. elegans muscle cells by means of the underlying nucleation rate constant and the corresponding reaction order, we developed a model of stochastic nucleation with explicit dependence on the protein concentration (see SI Appendix, Materials and Methods). In this model, we assume that the time between nucleation and the appearance of an observable aggregate is short relative to the timescale of the measurement, which is supported by the rapid growth of detectable aggregates (Fig. 1E), and that the aggregation behavior is deterministic once a nucleus has formed. Furthermore, we chose to fix the plateau value at 95 following our earlier observation that, on average, each body wall muscle cell acquires one inclusion. We first fitted the numbers of inclusions for the four C. elegans lines to a model that assumes a constant nucleation rate over time (Fig. 3A), and extracted a reaction order of 1.6 with respect to the protein concentration and a nucleation rate constant of 6 × 10−13 M−0.6 s−1. At an intracellular protein concentration of 1 mM, as in line A, this rate constant corresponds to a nucleation rate of 7 × 10−18 M s−1, or 0.01 molecules h−1 per cell. The reaction order of 1.6 implies a significant dependence of the nucleation rate on the protein concentration and suggests that a nucleus size of less than 2 is unlikely. The fits to this model generally represent the experimental data well, with the exception of the last timepoints in the highest expressing line A animals. It is unclear why the number of inclusions ceases to increase beyond day 5 of adulthood for line A, and we speculate that it may be due to positive selection for animals that remain healthy, or due to the increased proteasomal activity that has been observed in aging C. elegans (36), leading to degradation of the remaining soluble protein.
As noted above, there is currently no evidence to support direct transfer of aggregated species between neighboring cells in C. elegans polyQ models (Fig. 2C and SI Appendix, Fig. S4B) (34). Communication through the regulation of proteostasis mechanisms, however, is not spatially restricted and can occur between all cells in a tissue, and even between different tissues within an organism (37). This phenomenon could lead to cooperativity in the aggregation behavior, even in the absence of a spatial correlation in the appearance of aggregates. To test if such a phenomenon plays a significant role, we extended our mathematical model to include a cooperative mechanism by which the presence of cells with an inclusion increases the aggregation probability in other cells. The predicted kinetic profiles display initial upwards curvature and do not reproduce the data well (Fig. 3B), leading us to conclude that Q40-YFP aggregation in the C. elegans body wall muscle cells occurs independently in individual cells with a constant nucleation rate.
The image analysis furthermore allowed us to extract measures for the aggregate mass per inclusion and per animal, based on the integrated fluorescence intensity of the thresholded inclusions. We observed that the average total fluorescence per inclusion converges to a plateau value for each strain (SI Appendix, Fig. S9A), and that these values are approximately proportional to the protein concentration (SI Appendix, Fig. S9B). This finding is in line with our previous notion that, in each cell, the inclusion grows until all diffuse, soluble protein is depleted (Fig. 1 A and E). Using the average maximum fluorescence for the inclusions in each strain combined with the fits obtained from the number of aggregates, we predicted the development of the total fluorescence intensity of the inclusions per animal under different assumptions about the timescale of inclusion growth. Assuming rapid growth (reaching the maximum inclusion size within ∼4 h), the predictions closely match the experimentally obtained values as determined by the integrated fluorescence over the inclusions per animal (Fig. 3C). Predictions for slow growth (inclusions reaching their maximum size in ∼3 d), on the other hand, are not in agreement with the experimental data (Fig. 3D).
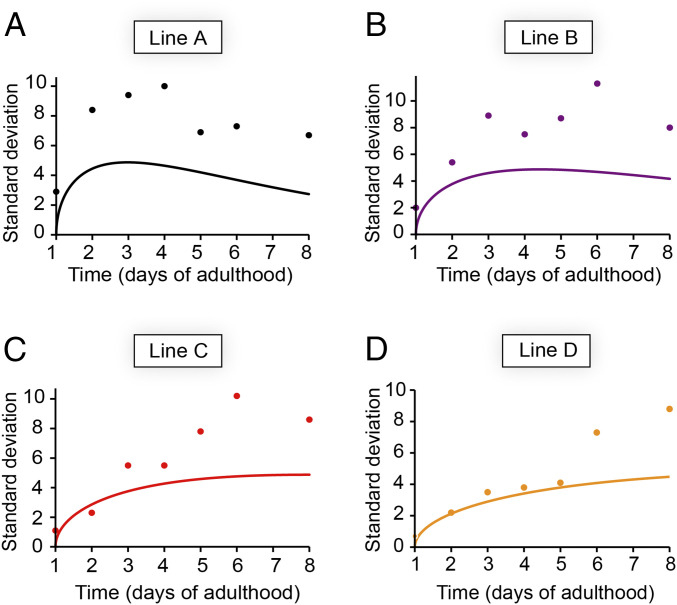
The stochastic nature of nucleation in small volumes not only leads to cell-to-cell differences within a single animal, but may also play a role in animal-to-animal variation given the finite number of cells within each animal. Interanimal differences in the number of inclusions were previously attributed primarily to biological variation (20). However, simulations for a 95-cell system, based solely on our fits of the data in Fig. 3A, show that stochasticity causes a considerable SD in the number of inclusions in a population of animals (Fig. 4). We find that the predictions are in remarkable agreement with the experimental values obtained for the lowest expressing strain D up to day 5 of adulthood (Fig. 4D) and present a lower boundary for the experimental values at later timepoints and in higher expressing strains (Fig. 4 A–C), for which measurement errors and biological variation presumably make additional contributions.
Fig. 4.
The stochasticity of nucleation sets a lower boundary for the SD of the inclusion numbers in a population of animals. (A–D) Prediction (solid line) and experimental datapoints of the SDs for (A) line A, (B), line B, (C) line C, (D) line D. The predictions are based on the nucleation rate for each strain as determined from global fitting as shown in Fig. 3A. n = 20 animals for each strain and timepoint; data are representative of two independent experiments.
Discussion
In this study, we present a kinetic model for polyQ aggregation in the muscle cells of living C. elegans. The first key finding emerging from our work is that Q40-YFP aggregation in each C. elegans muscle cell is governed by stochastic nucleation, followed by growth of the inclusion on a timescale that is fast compared to that of nucleation (Figs. 1E and 3 C and D). Notably, typically one inclusion per cell is observed in cell culture (38–40), in mouse models of polyQ diseases (18, 41, 42), and in the brains of juvenile Huntington’s disease patients (17), consistent with a rate-limiting nucleation event followed by fast inclusion growth as observed in this study.
The second key outcome is the ability to quantify the nucleation rate constant and reaction order from global fitting of in vivo kinetic data (Fig. 3A), allowing for a comparative analysis across experimental systems. The nucleation step for Q40-YFP aggregation in the C. elegans body wall muscle cells shows a significant dependence on the protein concentration, with a reaction order of ∼1.6. This value is similar to that found in previous work on several polyQ proteins both in vitro (43) and in cell culture (44), raising the possibility of a conserved nucleation mechanism. The timescale of nucleation as demonstrated in cultured cells is comparable to that in C. elegans, but the intracellular protein concentrations in these cells (low micromolar) were approximately two orders of magnitude lower than for C. elegans (44). Therefore, the rate constants of the reaction differ significantly, being orders of magnitude higher in the cells. Our modeling approach can be applied in future studies to clarify whether this discrepancy is due to the use of different polyQ constructs, the difference between the respective cell types, between isolated cells and a living organism, or a combination of these factors. Including cellular processes such as aggregate clearance may also resolve some of the differences in the effective rate constants between experimental systems.
Our third key finding is that nucleation occurs independently in each body wall muscle cell, i.e., the extent to which aggregation in one cell increases aggregation in other cells is negligible (Fig. 3 A and B). The observation that each cell operates essentially as an individual entity in which nucleation occurs stochastically is consistent with cell culture data (44), but is remarkable in the context of a multicellular animal. Our results suggest that spreading of Q40-YFP aggregates either does not occur or does not contribute substantially to the appearance of new inclusions (Figs. 2C and 3B), which is in contrast to observations for a yeast prion protein (45) and for α-synuclein (34) expressed in C. elegans. The collapse in proteostasis that is thought to underlie age-associated protein aggregation (36, 46–48), on the other hand, would also lead to cooperative aggregation kinetics. Possibly, the protein concentrations in our C. elegans models are associated with nucleation rates that are sufficiently high to dominate over cooperative effects, such as those mediated by proteostasis. The concentration of a polyQ protein such as huntingtin in the human brain, however, is about four orders of magnitude lower (49). Considering the nucleation rate and reaction order that we find in C. elegans, and using a cell volume of 4 pL, we calculate that nucleation would only occur in about 0.5% of cells in the human brain over a period of 40 y. However, a fraction of ∼20 to 30% of neurons has been reported to contain inclusions in patient material (17), suggesting that aggregation kinetics in the human brain are faster than extrapolated from our results obtained in C. elegans. Moreover, no inclusions were found in a presymptomatic individual that carried the disease allele (17), suggesting that protein aggregation is only initiated in midlife when symptoms appear. A significantly cooperative model may therefore be more appropriate to describe the progression of human polyQ aggregation around the pathogenic threshold. In addition, disease-specific flanking regions likely modulate the aggregation kinetics and potentially the ability to spread, and it will be interesting to extend our approach to different disease proteins.
In conclusion, we propose that the same biophysical principles that govern protein aggregation in small volumes in vitro, namely spatial confinement and stochastic nucleation, are the dominant driving forces controlling polyQ aggregation in the C. elegans muscle tissue. We anticipate that our modeling approach will be applicable to analyze the features of amyloid-like protein aggregation in a wide range of biological systems, including human tissues affected by protein misfolding diseases.
Materials and Methods
A detailed description of the experimental methods and the mathematical models can be found in SI Appendix. C. elegans were maintained on nematode growth media (NGM) seeded with Escherichia coli OP50 at 20 °C using standard methods (50). Age-synchronized populations were generated by performing a timed egg lay of 1 to 2 h, and adult animals were transferred daily to separate them from their offspring. C. elegans strains were generated by plasmid microinjection followed by genomic integration using ultraviolet (UV) irradiation. The resulting strains were backcrossed with wild-type N2 animals five times. Manual inclusion counting was performed using a fluorescence stereomicroscope (Leica MZ16FA) at 115× magnification, while the animals freely crawled on seeded NGM plates. For each timepoint, 12 to 20 animals were counted from the same age-synchronized pool. For semiautomated inclusion counting, 20 animals for each strain and timepoint were picked into M9 buffer in a 384-well plate, and confocal imaging was performed on the ImageXpress high-content imaging system (Molecular Devices) equipped with a 20× objective. Confocal imaging of live and phalloidin-stained animals was performed on a Leica SP8 microscope. TEM on sections of resin-embedded animals was performed on a JEOL 1230 GEM.
Supplementary Material
Acknowledgments
We thank the Biological Imaging Facility (RRID: SCR017767), High-Throughput Analysis Lab, BioCryo Facility, the Keck Biophysics Facility, and the Lackner laboratory at Northwestern University for instrument use and technical assistance. We are grateful to Renée Brielmann for C. elegans microinjections and to Charlene Wilke for freeze substitution and assistance with TEM. This work was supported by grants from the NIH (National Institute on Aging R56AG059579, R37AG026647, RF1AG057296, and P01AG054407) and the Daniel F. and Ada L. Rice Foundation to R.I.M.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021888118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Knowles T. P. J., Vendruscolo M., Dobson C. M., The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15, 384–396 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Chiti F., Dobson C. M., Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu. Rev. Biochem. 86, 27–68 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Cohen S. I. A., et al., Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl. Acad. Sci. U.S.A. 110, 9758–9763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Habchi J., et al., Systematic development of small molecules to inhibit specific microscopic steps of Aβ42 aggregation in Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 114, E200–E208 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Habchi J., et al., An anticancer drug suppresses the primary nucleation reaction that initiates the production of the toxic Aβ42 aggregates linked with Alzheimer’s disease. Sci. Adv. 2, e1501244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen S. I. A., et al., A molecular chaperone breaks the catalytic cycle that generates toxic Aβ oligomers. Nat. Struct. Mol. Biol. 22, 207–213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arosio P., et al., Kinetic analysis reveals the diversity of microscopic mechanisms through which molecular chaperones suppress amyloid formation. Nat. Commun. 7, 10948 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linse S., et al., Kinetic fingerprints differentiate the mechanisms of action of anti-Aβ antibodies. Nat. Struct. Mol. Biol. 27, 1125–1133 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Knowles T. P. J., et al., Observation of spatial propagation of amyloid assembly from single nuclei. Proc. Natl. Acad. Sci. U.S.A. 108, 14746–14751 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michaels T. C. T., et al., Fluctuations in the kinetics of linear protein self-assembly. Phys. Rev. Lett. 116, 258103 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Michaels T. C. T., Dear A. J., Knowles T. P. J., Stochastic calculus of protein filament formation under spatial confinement. New J. Phys. 20, 055007 (2018). [Google Scholar]
- 12.Labbadia J., Morimoto R. I., The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84, 435–464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sala A. J., Bott L. C., Morimoto R. I., Shaping proteostasis at the cellular, tissue, and organismal level. J. Cell Biol. 216, 1231–1241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perutz M. F., Johnson T., Suzuki M., Finch J. T., Glutamine repeats as polar zippers: Their possible role in inherited neurodegenerative diseases. Proc. Natl. Acad. Sci. U.S.A. 91, 5355–5358 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scherzinger E., et al., Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90, 549–558 (1997). [DOI] [PubMed] [Google Scholar]
- 16.Chen S., Berthelier V., Hamilton J. B., O’Nuallain B., Wetzel R., Amyloid-like features of polyglutamine aggregates and their assembly kinetics. Biochemistry 41, 7391–7399 (2002). [DOI] [PubMed] [Google Scholar]
- 17.DiFiglia M., et al., Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277, 1990–1993 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Davies S. W., et al., Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90, 537–548 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Lieberman A. P., Shakkottai V. G., Albin R. L., Polyglutamine repeats in neurodegenerative diseases. Annu. Rev. Pathol. 14, 1–27 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morley J. F., Brignull H. R., Weyers J. J., Morimoto R. I., The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 99, 10417–10422 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laine R. F., et al., Fast fluorescence lifetime imaging reveals the aggregation processes of α-synuclein and polyglutamine in aging Caenorhabditis elegans. ACS Chem. Biol. 14, 1628–1636 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S., Ferrone F. A., Wetzel R., Huntington’s disease age-of-onset linked to polyglutamine aggregation nucleation. Proc. Natl. Acad. Sci. U.S.A. 99, 11884–11889 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kar K., Jayaraman M., Sahoo B., Kodali R., Wetzel R., Critical nucleus size for disease-related polyglutamine aggregation is repeat-length dependent. Nat. Struct. Mol. Biol. 18, 328–336 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beam M., Silva M. C., Morimoto R. I., Dynamic imaging by fluorescence correlation spectroscopy identifies diverse populations of polyglutamine oligomers formed in vivo. J. Biol. Chem. 287, 26136–26145 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olshina M. A., et al., Tracking mutant huntingtin aggregation kinetics in cells reveals three major populations that include an invariant oligomer pool. J. Biol. Chem. 285, 21807–21816 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legleiter J., et al., Mutant huntingtin fragments form oligomers in a polyglutamine length-dependent manner in vitro and in vivo. J. Biol. Chem. 285, 14777–14790 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ossato G., et al., A two-step path to inclusion formation of huntingtin peptides revealed by number and brightness analysis. Biophys. J. 98, 3078–3085 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satyal S. H., et al., Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 97, 5750–5755 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirstein-Miles J., Scior A., Deuerling E., Morimoto R. I., The nascent polypeptide-associated complex is a key regulator of proteostasis. EMBO J. 32, 1451–1468 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gieseler K., Qadota H., Benian G. M., Development, structure, and maintenance of C. elegans body wall muscle. WormBook 2017, 1–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao J., et al., Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeon I., et al., Human-to-mouse prion-like propagation of mutant huntingtin protein. Acta Neuropathol. 132, 577–592 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masnata M., et al., Demonstration of prion-like properties of mutant huntingtin fibrils in both in vitro and in vivo paradigms. Acta Neuropathol. 137, 981–1001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandhof C. A., et al., Reducing INS-IGF1 signaling protects against non-cell autonomous vesicle rupture caused by SNCA spreading. Autophagy 16, 878–899 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meisl G., et al., Molecular mechanisms of protein aggregation from global fitting of kinetic models. Nat. Protoc. 11, 252–272 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Walther D. M., et al., Widespread proteome remodeling and aggregation in aging C. elegans. Cell 161, 919–932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morimoto R. I., Cell-nonautonomous regulation of proteostasis in aging and disease. Cold Spring Harb. Perspect. Biol. 12, a034074 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S., Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Ramdzan Y. M., et al., Tracking protein aggregation and mislocalization in cells with flow cytometry. Nat. Methods 9, 467–470 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Hageman J., et al., A DNAJB chaperone subfamily with HDAC-dependent activities suppresses toxic protein aggregation. Mol. Cell 37, 355–369 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Bichelmeier U., et al., Nuclear localization of ataxin-3 is required for the manifestation of symptoms in SCA3: In vivo evidence. J. Neurosci. 27, 7418–7428 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goti D., et al., A mutant ataxin-3 putative-cleavage fragment in brains of Machado-Joseph disease patients and transgenic mice is cytotoxic above a critical concentration. J. Neurosci. 24, 10266–10279 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kakkar V., et al., The S/T-rich motif in the DNAJB6 chaperone delays polyglutamine aggregation and the onset of disease in a mouse model. Mol. Cell 62, 272–283 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Colby D. W., Cassady J. P., Lin G. C., Ingram V. M., Wittrup K. D., Stochastic kinetics of intracellular huntingtin aggregate formation. Nat. Chem. Biol. 2, 319–323 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Nussbaum-Krammer C. I., Park K.-W., Li L., Melki R., Morimoto R. I., Spreading of a prion domain from cell-to-cell by vesicular transport in Caenorhabditis elegans. PLoS Genet. 9, e1003351 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ben-Zvi A., Miller E. A., Morimoto R. I., Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc. Natl. Acad. Sci. U.S.A. 106, 14914–14919 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.David D. C., et al., Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 8, e1000450 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Labbadia J., Morimoto R. I., Repression of the heat shock response is a programmed event at the onset of reproduction. Mol. Cell 59, 639–650 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Macdonald D., et al., Quantification assays for total and polyglutamine-expanded huntingtin proteins. PLoS One 9, e96854 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brenner S., The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.