Significance
The marine unicellular cyanobacterium Prochlorococcus is the most abundant photosynthetic organism on Earth. Members of this genus are classically thought to be adapted to high-oxygen and nutrient-poor ocean conditions, with a principle divergence between high-light and low-light ecotypes. We show that the most basal Prochlorococcus lineages are adapted to the low-oxygen, low-light, and high-nutrient conditions found in the dimly illuminated waters of anoxic marine zones. The most basal lineages have retained phycobilisomes as light-harvesting antennae—a characteristic of most other cyanobacteria—whose loss was thought to define all Prochlorococcus. As oxygenic photosynthesis drove ocean oxidation in the ancient Earth, oxygen appears to have played as much a role as light and nutrients in driving Prochlorococcus evolution.
Keywords: cyanobacteria, microbiology, genomics, oceanography, anoxia
Abstract
Marine picocyanobacteria of the genus Prochlorococcus are the most abundant photosynthetic organisms in the modern ocean, where they exert a profound influence on elemental cycling and energy flow. The use of transmembrane chlorophyll complexes instead of phycobilisomes as light-harvesting antennae is considered a defining attribute of Prochlorococcus. Its ecology and evolution are understood in terms of light, temperature, and nutrients. Here, we report single-cell genomic information on previously uncharacterized phylogenetic lineages of this genus from nutrient-rich anoxic waters of the eastern tropical North and South Pacific Ocean. The most basal lineages exhibit optical and genotypic properties of phycobilisome-containing cyanobacteria, indicating that the characteristic light-harvesting antenna of the group is not an ancestral attribute. Additionally, we found that all the indigenous lineages analyzed encode genes for pigment biosynthesis under oxygen-limited conditions, a trait shared with other freshwater and coastal marine cyanobacteria. Our findings thus suggest that Prochlorococcus diverged from other cyanobacteria under low-oxygen conditions before transitioning from phycobilisomes to transmembrane chlorophyll complexes and may have contributed to the oxidation of the ancient ocean.
The free-living, planktonic, unicellular cyanobacterium of the genus Prochlorococcus is the smallest and most abundant photosynthetic organism on Earth (1). Together with their closest phylogenetic relatives of the genus Synechococcus, they are collectively known as the marine picocyanobacteria (≤2 μm in cell diameter) and are significant contributors to primary production in the modern ocean (2). Distinguishing characteristics of Prochlorococcus, vis-a-vis other marine cyanobacteria, include an extremely small size (<1 μm in diameter) and the possession of transmembrane divinyl chlorophyll a and b complexes as their main photosynthetic light-harvesting antennae instead of the membrane-bound phycobilisomes that are present in most other cyanobacteria (1, 3). These features are readily discernible with flow cytometry and are widely utilized to selectively quantify Prochlorococcus in marine samples (1). The coding potential and diversity of Prochlorococcus have been studied in numerous phylogenetically distinct isolates and hundreds of single-cell–amplified genomes (SAGs) (4). Based on their physiology, genetic characteristics, and environmental niches, the genus is broadly divided into high-light (HL) and low-light (LL) adapted ecotypes (5). The current understanding is that light, temperature, and nutrients—including iron—play integral roles in the ecology and evolution of Prochlorococcus (6–10).
Prochlorococcus thrives in and is best known from sunlit and well-oxygenated, nutrient-poor, tropical and subtropical waters. However, Prochlorococcus is also the dominant photoautotroph in the nutrient-rich, oxygen-depleted waters of major anoxic marine zones (AMZs) in the Arabian Sea and the eastern tropical North (ETNP) and South Pacific (ETSP) (11, 12), where photosynthetic eukaryotes are not found. In these systems, Prochlorococcus can drive a cryptic oxygen cycle (13), by which aerobic metabolic processes can exist under apparent anoxic conditions due to the oxygen produced through oxygenic photosynthesis. Phylogenetic analysis of environmental sequences of the 16S to 23S ribosomal RNA (rRNA) internal transcribed spacer (ITS) region has revealed that the majority of the AMZ Prochlorococcus belong to two distinct phylogenetic lineages, originally termed LL V and LL VI (12) and renamed here as AMZ I and AMZ II. These lineages correspond to uncultured basal groups within the Prochlorococcus genus, for which there is no public genetic information except for their inferred capacity for nitrate utilization based on limited shotgun metagenomic sequencing data (14). Thus, further details on their coding potential and evidence for anaerobic or microaerobic metabolisms remain unconstrained for these AMZ lineages. Such information is important for understanding the origin and structure of the photosynthetic apparatus (15, 16), the evolution of marine pelagic picocyanobacteria (17), the functioning of ancient and modern AMZs (18), and the differential modes of metabolic coupling between Prochlorococcus and other AMZ microorganisms.
Results and Discussion
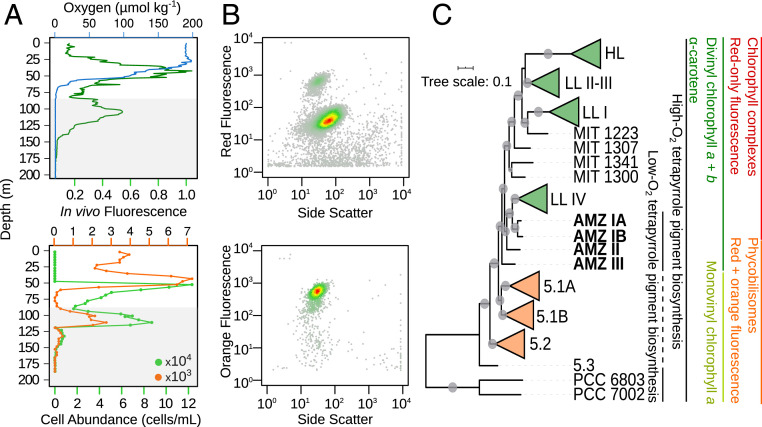
To explore the genetic makeup of these uncultured picocyanobacteria from AMZ waters, we analyzed 128 SAGs from water samples of the secondary fluorescence maximum in the eastern Pacific Ocean off northern Chile and Mexico (Fig. 1A, Table 1, and SI Appendix, Table S1). The geochemical characteristics of these AMZ waters (18) include undetectable levels of oxygen, no accumulation of sulfide or ammonium, the presence of significant levels of major nutrients, and, particularly of nitrite, a product of the anaerobic process of dissimilative nitrate reduction. Cells with optical characteristics typical of either Prochlorococcus (red fluorescence only) or Synechococcus (red and orange fluorescence) (Fig. 1B) were sorted for SAG generation and sequencing using previously described techniques (19). The AMZ SAG assemblies had an estimated completeness of up to 75%, an estimated genome size of ∼2.5 Mbp, and a guanine–cytocine content of ∼50% (SI Appendix, Table S1). These characteristics are more similar to those previously observed for the Prochlorococcus LL IV and marine Synechococcus lineages than for HL Prochlorococcus (6, 20).
Fig. 1.
Representative biogeochemical and flow-cytometry profiles and phylogenetic tree of unicellular cyanobacteria with Prochlorococcus from AMZs. (A) Dissolved oxygen concentration (blue), in vivo chlorophyll a fluorescence in relative units (dark green), and Synechococcus-like (orange) and Prochlorococcus-like (light green) cell abundance at Station 6 in the AMZ of the ETNP (Table 1). The gray area corresponds to depths with <0.5 μmol ⋅ kg−1 O2. (B) Fluorescence versus light scatter cytograms (in relative units) of cyanobacteria-like particles in the secondary deep-chlorophyll maximum. The upper panel shows particles with red fluorescence, a proxy for chlorophyll a-containing cells, and the lower panel shows particles with orange fluorescence, a proxy for phycoerythrin-containing cells. (C) Maximum-likelihood tree determined by phylogenetic inference using 49 concatenated ribosomal proteins: Prochlorococcus (collapsed genomes of recognized ecotypes in green), Synechococcus (collapsed genomes of marine subclusters in orange), and AMZ Prochlorococcus (in bold). PCC 7002 is the marine onshore Synechococcus sp. strain PCC 7002, and PCC 6803 is the freshwater Synechosystis sp. strain PCC 6803. The circles on each node represent support values higher than 50% (n = 1,000 iterations). Characteristic traits of the different cyanobacteria lineages examined are given in colored lines. The dashed line indicates that only a few members of the Synechococcus subcluster have that trait.
Table 1.
Sampling location and hydrographic characteristics of waters from which samples for single-cell genomics were collected
| Cruise | Station | Latitude (°N) | Longitude (°W) | Date (dd/mm/yy) | Depth (m) | Temperature (°C) | Salinity | CTD oxygen (µmol/kg) | Phosphate (µmol/kg) | Nitrate + nitrite (µmol/kg) | Prochlorococcus-like (104 cells/mL) | Synechococcus-like (104 cells/mL) |
| MV1015 | 1 | −20,083 | 70,800 | 19/11/10 | 53 | 12.874 | 34.731 | 3.3 | 2.72* | 17.85* | 1.04 | 0.37 |
| NH1315 | 6 | 18,920 | 104,891 | 19/07/13 | 100 | 13.995 | 34.781 | 1.5 | 2.52 | 20.44 | 60.8 | 4.3 |
Values from 50 m in separate cast.
Prochlorococcus SAGs affiliated with the uncharacterized AMZ I lineage were recovered with the traditional red-fluorescence-only cell sorting approach. However, SAGs affiliated with the previously uncharacterized AMZ II lineage were obtained only from cells that exhibited red and orange fluorescence, which is traditionally associated with Synechococcus. Moreover, we discovered a previously unknown Prochlorococcus lineage, termed here as AMZ III, among the cells with both red and orange fluorescence. According to phylogenetic analyses of individual and concatenated universal single-copy marker genes, AMZ I is a sister clade to LL IV and presents two subclades, termed here as AMZ IA and AMZ IB, with AMZ II occupying a basal position to the LL IV and AMZ I lineages (Fig. 1C and SI Appendix, Fig. S1). This tree topology is consistent with the established ITS phylogeny (8, 12). Furthermore, the newly discovered AMZ III lineage is located at the base of the Prochlorococcus radiation, thus representing the extant Prochlorococcus relatives closest to marine Synechococcus identified to date. The phylogenetic grouping of the AMZ Prochlorococcus SAGs was further corroborated by their genomic similarity (SI Appendix, Fig. S2).
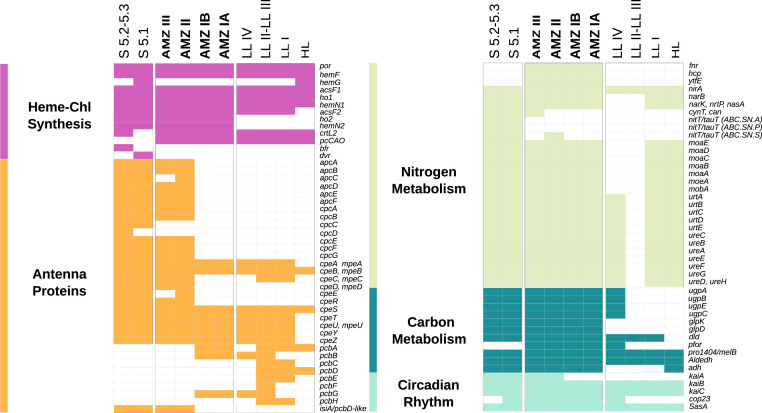
An analysis of the individual SAGs (Fig. 2 and SI Appendix, Fig. S3 and Tables S2 and S3) revealed that all three AMZ lineages encode genes for chlorophyll b synthase pcCao (21) and lycopene ε-cyclase crtL2 (22) but lack the gene dvr encoding for the divinyl chlorophyll a reductase (23). This coding potential contributes to the characteristic pigment traits of all Prochlorococcus, such as divinyl chlorophyll a (instead of the monovinyl form present in most of the other cyanobacteria, including marine Synechococcus), divinyl chlorophyll b, and α-carotene (3, 24). This genomic information is consistent with the abundance of these diagnostic pigments in AMZ waters of the Arabian Sea and eastern North Pacific (11) as well as of the eastern South Pacific (SI Appendix, Table S4). However, the AMZ II and AMZ III SAGs were also found to contain genes encoding for complete synthesis of phycobilisomes, coinciding with the orange fluorescence of these two lineages (Fig. 1B). Specifically, AMZ II and AMZ III SAGs encoded genes for the synthesis of the phycobiliproteins allophycocyanin, phycocyanin, and phycoerythrin (I and II), as well as the corresponding linkers, and lack the chlorophyll light-harvesting pcb genes typical of Prochlorococcus (Fig. 2 and SI Appendix, Fig. S4 and Table S3). Furthermore, AMZ II and AMZ III SAGs encoded a chlorophyll-binding protein of the iron-stress–induced (IsiA) type, which is typical of marine Synechococcus and is phylogenetically distant from the Prochlorococcus-typical pcb genes (25), including those in AMZ I SAGs (SI Appendix, Fig. S5). Likewise, we found the kaiA gene in AMZ II and AMZ III (Fig. 2 and SI Appendix, Table S3), a critical component of the circadian clock KaiABC system (26) that has not been found in Prochlorococcus before (27, 28). Thus, our results indicate that the replacement of phycobilisomes for divinyl chlorophyll a and b complexes and the loss of the kaiA gene occurred within the Prochlorococcus diversification and support previous observations regarding genetic remnants of such evolutionary events still being present in some Prochlorococcus lineages (28, 29). As shown above, these evolutionary steps appear to have occurred after Prochlorococcus acquired the capacity to synthesize divinyl chlorophyll b (Fig. 1C).
Fig. 2.
Presence–absence of selected genes in AMZ Prochlorococcus SAGs (see SI Appendix, Tables S2 and S3 for details).
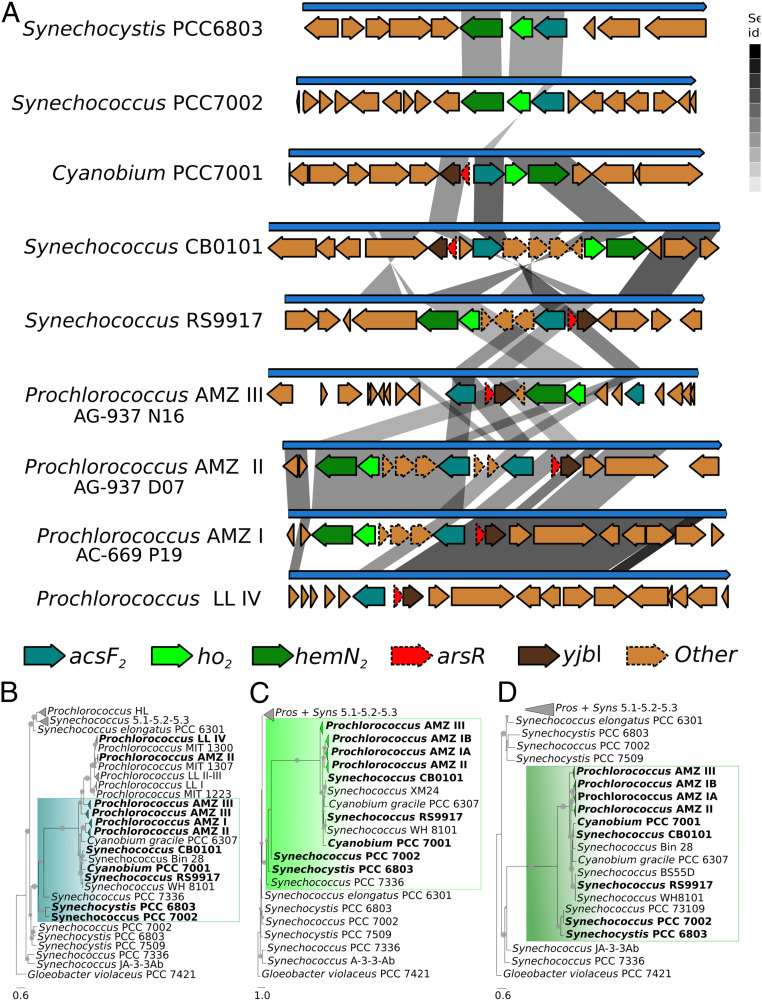
One of the major challenges modern cyanobacteria experience when living in anoxia and temporary darkness is the synthesis of tetrapyrrole pigments, as oxygen is required in several of their shared enzymatic steps. These biomolecules have important functions, including electron transfer, oxygen binding, and light absorption, with chlorophyll a being essential for photosynthesis (30). Certain cyanobacteria, however, have alternative or complementary enzymes for pigment biosynthesis that work under oxygen-limited conditions (31). In particular, three genes, hemN2, acsF2, and ho2, encode secondary versions of enzymes involved in heme, chlorophyll, and phycocyanobilin biosynthesis, respectively. The gene hemN2 encodes the oxygen-independent version of the enzyme that catalyzes the biosynthesis of protoporphyrinogen IX, the common precursor of Chl-a, hemes, and bilins; acsF2 encodes the cyclase that converts Mg-Proto monomethyl ester to divinyl protochlorophyllide during the synthesis of Chl-a; and ho2 encodes a heme oxygenase involved in bilin synthesis. These genes are clustered together in the genomes of the freshwater cyanobacterium Synechocystis PCC 6803 and the euryhaline coastal marine cyanobacterium Synechococcus PCC 7002. In both isolates, such genes are highly expressed under oxygen-limited conditions but not under fully oxic conditions (32, 33). We identified the gene cluster acsF2-ho2-hemN2 in SAGs of the three AMZ lineages and—with a similar genomic configuration—also in marine Synechococcus sp. strains RS9917, WH 8101, and CB0101 (Figs. 2 and 3A). This gene cluster has not been previously reported in any of the prior studies of Prochlorococcus and marine Synechococcus (31). Phylogenetic analyses revealed that the gene encoding the AcsF2 cyclase branches closest to homologs in the LL Prochlorococcus and Synechococcus strains RS9917, WH 8101, and CB0101 but not with homologs in other marine Synechococcus nor with those in HL Prochlorococcus ecotypes (Fig. 3B). The cluster-forming ho2 and hemN2 genes branch in a similar way as acsF2, except that no homologs from LL Prochlorococcus are grouped closely with them (Fig. 3 C and D).
Fig. 3.
Synteny and phylogeny of three genes involved in tetrapyrrole biosynthesis under oxygen-limited conditions. (A) Synteny of three orthologous genes involved in chlorophyll (acsF2), phycocyanobilin (ho2), and heme (hemN2) biosynthesis across selected cyanobacterial lineages. The gene arsR encodes a putative transcriptional regulator, and yjbL encodes an unknown protein. Maximum-likelihood phylogenetic trees of (B) acsF-, (C) ho-, and (D) hemN-predicted amino acid sequences. The colored boxes mark the gene versions that appear organized in a cluster and should work under oxygen-limited conditions; lineages appearing in A are in bold. Conserved residues for several functions are given in SI Appendix, Fig. S6. Filled circles on each node correspond to support values higher than 50% (n = 1,000 iterations).
The phylogeny and conserved gene arrangement shared between freshwater and marine cyanobacteria suggests that Prochlorococcus may have evolved before the ocean became fully oxygenated. In this scenario, ho2 and hemN2 would have been lost as Prochlorococcus adapted to an oxic ocean, but acsF2 would have been retained in the LL lineages (Fig. 3 A and B). The alternative scenario that the low-oxygen pigment biosynthesis trait was acquired after Prochlorococcus evolved in an oxic ocean cannot be totally discarded. However, a detailed analysis of contigs containing the genes for pigment biosynthesis (as well as those for the light-harvesting antenna) using previously described approaches (14, 34, 35) did not provide any evidence of recent horizontal gene transfer or of hallmark viral signatures. If Prochlorococcus evolved in an oxygen-depleted ocean before it could adapt to fully oxygenated oligotrophic surface waters, the replacement of phycobilisomes for transmembrane divinyl chlorophyll complexes may have been driven by the low levels and narrow spectral characteristics of the light available at depth (36). Moreover, continuous changes in the light field at depth due to variable optical conditions of surface waters or due to internal waves could have allowed for the coexistence of lineages with different light-harvesting antennae, as is currently observed. However, in the proposed scenario, only those Prochlorococcus with chlorophyll complexes would have been able to colonize the high-oxygen surface waters.
Metagenomic fragment recruitment (SI Appendix, Fig. S7) showed that AMZ I, II, and III lineages are found only in oxygen-depleted oceanic waters. Moreover, AMZ I outnumbers II and III, which is consistent with the flow cytometry data (11, 12) indicating that red-only fluorescent cells dominate in AMZ waters. The presence of indigenous lineages with a specific gene cluster for pigment synthesis under oxygen-limited conditions (Fig. 3) is thus consistent with the designation of Prochlorococcus “AMZ” ecotypes. Moreover, these ecotypes, which inhabit nutrient-rich waters, have a complete gene repertoire for the use of ammonium, urea, nitrite, and nitrate (Fig. 2 and SI Appendix, Table S3) as nitrogen sources. On the other hand, according to the analysis of public metatranscriptomic data (SI Appendix, Fig. S8), at least some of the photosynthesis genes discussed above are being actively transcribed in AMZs. However, targeted gene expression studies over the light–dark periods will be required to address this issue fully.
Other genotypic properties that appear to be specific to AMZ ecotypes and that may confer them adaptations to an anoxic environment include the presence of a hybrid-cluster protein (hcp) gene and cognate fnr regulator of the anaerobic Cpr/Fnr family as well as a gene ytfE encoding an iron-sulfur cluster repair protein YtfE (Fig. 2 and SI Appendix, Table S3). These three genes are in the same genomic neighborhood in AMZ I and AMZ II, but ytfE is in a different location in AMZ III. The HCPs and YtfE are associated with anaerobic nitrosative stress (e.g., ref. 37), while HCPs are also essential in protein S-nitrosylation by nitric oxide (38), a fundamental redox-based cellular signaling mechanism. Additionally, AMZ Prochlorococcus SAGs encode enzymes involved in glycerol oxidation (glpK and glpD) and alcohol fermentation (adh) (Fig. 2). These additional features indicate that AMZ Prochlorococcus may be capable of supplementing their carbon and energy requirements through the use of organic compounds, as previously suggested for other lineages (39, 40), to survive under dark anoxic conditions.
Marine planktonic picocyanobacteria likely emerged in the Neoproterozoic 1,000 to 542 Mya (17), roughly at the time of the oxygenation of the deep ocean (41). We have shown that extant lineages inhabiting AMZs are basal to the Prochlorococcus genus, can have phycobilisomes as light-harvesting antenna, and carry ancestral adaptations to low-oxygen conditions. These findings suggest that Prochlorococcus emerged in low-oxygen environments and may have contributed to sustaining aerobic metabolisms and providing a significant carbon source through oxygenic photosynthesis, in the ancient, anoxic upper ocean, as they do today (13). The evolutionary history of Prochlorococcus thus appears intimately related to the availability of oxygen in the ocean.
Materials and Methods
Sampling and Hydrographic Data Collection.
Sampling was carried out during Cruise MV1015 (DOI: 10.7284/903808), the “Biogeochemical Gradients: Role in Arranging Planktonic Assemblages (BiG RAPA) Expedition” (https://hahana.soest.hawaii.edu/cmoreserver/cruises/big_rapa/plan.htm), at Station 1 in the ETSP and during Cruise NH1315 (DOI: 10.7284/903869) at Station 6 in the ETNP (Table 1). For Cruise MV1015, hydrographic data and seawater samples were collected with a rosette/conductivity-temperature-depth (CTD) system, consisting of Niskin bottles and a CTD profiler (Sea-Bird SBE 911plus) with dissolved-oxygen, fluorometer, and transmissometer sensors. For Cruise NH1315, hydrographic data and seawater samples were collected with a pump profiling system, similar to the one previously used in AMZ waters (13).
Picocyanobacteria Cell Count by Flow Cytometry.
For each site, duplicate samples of 1 mL raw seawater were fixed in a final concentration of 1% (vol/vol) glutaraldehyde at room temperature, in the dark, for 10 min and then fast frozen in liquid nitrogen and stored at −80 °C until analysis. In the laboratory, samples were thawed on ice and passed through a 60 μm mesh net, and cell abundance was determined with a FACSCalibur flow cytometer (Becton Dickinson) by using red and orange fluorescence as proxies for cells containing chlorophyll a and phycobiliproteins, respectively (42).
AMZ Prochlorococcus Single-Cell Sample Collection.
For the MV1015 (ETSP) cruise, water samples for single-cell sorting were collected from the Niskin bottles and prefiltered through a 60 μm mesh-size net. Then, ∼4,000 cells of Prochlorococcus-like cells were sorted on board into 1 mL of sterile glycerol–Tris-EDTA (TE) buffer (43) with an InFlux flow cytometer (BD Biosciences). Sorting was performed gating on putative Prochlorococcus cells in the red emission channel (excited by a 488 nm laser), using forward scattered light as a proxy for particle size (42). These presorted Prochlorococcus were cryopreserved in liquid nitrogen and then stored at −80 °C. For the NH1410 (ETNP) cruise, water samples were collected from the Pump Profiling System, filtered through a 60 μm mesh size, and stored in glycerol–TE buffer at −80 °C until cell sorting.
SAG Sequencing, Assembly, Gene Prediction, and Annotation.
Single-cell sorting, whole-genome amplification, 16S rRNA real-time PCR screening, and PCR product sequencing were performed at the Bigelow Laboratory for Ocean Sciences’ Single Cell Genomics Center (SCGC, http://scgc.bigelow.org/), as described previously (19). For the ETSP SAGs, an Illumina standard shotgun library was constructed, and samples were sequenced utilizing the HiSeq 2000 platform at Canada’s Michael Smith Genome Sciences Centre. Artifactual sequences were filtered off the raw data and draft SAGs were assembled using SPAdes 3.5. SAG contigs of >1 kbp were decontaminated using ProDeGe (44), and sequences with lengths between >200 bp and <2 kbp were considered whenever they had >80% identity (>75% alignment coverage) with Prochlorococcus reference genomes (SI Appendix, Table S1). Quality filtered sequences of <200 bp were discarded. The decontaminated SAG sequences were uploaded for gene prediction and annotation to the Integrated Microbial Genomes & Microbiomes (IMG/MER) system (45). Genome completeness was estimated by the presence of 114 single-copy conserved bacterial phylogenetic markers among the predicted protein-coding genes of each SAG (46). The ETNP SAGs were sequenced at SCGC as described previously (19). Gene predictions, functional annotation, manual curation, and pathway reconstruction for all individual SAGs were performed within the IMG/MER platform (https://img.jgi.doe.gov) (45). The presence of genes of interest was determined by their functional annotation, according to their assigned Kyoto Encyclopedia of Genes and Genomes ID. Genomic synteny (Fig. 3 and SI Appendix, Figs. S3 and S4) were plotted with Trebol (https://inf.imo-chile.cl). Sequence identity between genomic sequences was computed using BLASTn with a bit-score cutoff of 50. The contigs were checked for evidence of recent horizontal gene transfer as in ref. 14 and for viral signatures using VirSorter (34) and VirFinder (35).
Retrieval of AMZ SAGs Phylogenetic Marker Sequences.
Prochlorococcus ITS sequences were retrieved from reference genomes (SI Appendix, Table S1) and utilized as BLASTn queries to retrieve those from the Prochlorococcus AMZ SAGs. Then, transfer RNA genes were removed and the identity was determined for all ITS sequences that ended with >600 bp. The same approach was adopted with the rbcL gene, encoding the RuBisCO large subunit and the petB gene, a component of the cytochrome b6f complex. Average nucleotide identity (ANI) analysis (SI Appendix, Fig. S2) was performed using FastANI (47).
Identification and Conserved Residues of Cluster-Forming Tetrapyrrole Biosynthesis Genes.
The gene cluster acsF2-ho2-hemN2, associated with pigment biosynthesis under oxygen-limited conditions (31), was identified in individual Prochlorococcus AMZ SAGs within IMG/MER. Conserved residues in AcsF2, HO2, and HemN2 (SI Appendix, Fig. S6) were visually detected and manually annotated according to information provided in the literature (48–50). Though alignments were also analyzed with PROSITE (https://prosite.expasy.org), no additional relevant domains were found.
Phylogenetic and Phylogenomic Analyses.
Predicted amino acid sequences were aligned using MAFFT (51) version 7.271. Maximum-likelihood phylogenetic and phylogenomic inferences were performed using IQ-TREE (52) version 1.6.12. The models of sequence evolution were selected with ModelFinder (53) according to the Bayesian information criterion. The best fit models for single-gene phylogenies are given in the respective figure legends. Branch support values were computed using the Shimodaira–Hasegawa approximate likelihood ratio test within IQ-TREE. For phylogenomic reconstructions, concatenated amino acid–aligned sequences of 49 ribosomal proteins (Fig. 1C) and 137 single-copy genes (SI Appendix, Fig. S1C) were used (54). The best fit model for the tree in Fig. 1C was LG+R10, and LG+I+G4 was the best fit model for the tree in SI Appendix, Fig. S1C.
Fragment Recruitment.
The recruitment of sequence fragments from global ocean metagenomes (SI Appendix, Table S5) was performed against the contigs associated with the AMZ Prochlorococcus ecotypes obtained in the ETNP and ETSP. Fragment recruitment onto Prochlorococcus LL genomes was included for comparison (SI Appendix, Fig. S7). Details about the procedure can be found in SI Appendix.
Transcript Recruitment.
The recruitment of transcripts encoding pigment-biosynthesis and photosynthetic proteins from public AMZ metatranscriptomes (SI Appendix, Table S5) was carried out against the AMZ SAGs and Prochlorococcus and marine Synechococcus genomes (SI Appendix, Fig. S8). The details about the procedure can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank the chief scientists Dan Repeta (MV1015) and Frank J. Stewart (NH1315) and the captains, crews, and scientific support personnel of the R/V Melville and the R/V New Horizon. We also thank Gadiel Alarcón and Montserrat Aldunate for assistance in sample collection, María Lorena González for cell sorting during the BiG RAPA Expedition, Cristian Venegas for assistance in flow cytometry analysis, Marcela Montoya for help with the SAG analyses, and the SCGC staff. This work was supported by grants from the National Agency for Research and Development of Chile (Grants ICN12_019-IMO, 1130784, 1161483, and 3180724) and from the Agouron Institute to O.U. Support for this research was also provided by the US NSF (Grants OCE-1335810 and OIA-1826734 to R.S.) and the Tula Foundation, the G. Unger Vetlesen and Ambrose Monell Foundations, the Natural Sciences and Engineering Research Council of Canada, and Compute/Calcul Canada to S.J.H. A.M.P. received postdoctoral support through the Tula Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025638118/-/DCSupplemental.
Data Availability
Single-cell Amplified Genome data are available in Integrated Microbial Genomes, http://img.jgi.doe.gov/cgi-bin/w/main.cgi (accession nos. 2626541501, 2626541503–2626541507, 2626541509–2626541537, 2716884926, 2716884927, 2718217657, 2718217658, 2724679665, 2724679666, 2724679668, 2724679669, 2724679671, 2724679674–2724679676, 2724679678–2724679681, 2724679684, 2724679692, 2724679753, 2724679758, 2724679760, 2724679773, 2724679775, 2818991481–2818991488, 2818991490, 2818991493, 2818991494, 2818991497–2818991503, 2818991506–2818991511, 2818991513–2818991516, 2818991519–2818991530, 2818991533, 2818991535, 2818991536, 2818991539–2818991542, 2818991544–2818991547, 2818991549, 2818991551, 2818991553, 2818991555, 2818991557–2818991559, 2818991561, 2818991563–2818991568, 2818991570–2818991573, and 2818991575). All other study data are included in the article and/or SI Appendix.
References
- 1.Chisholm S. W., et al., A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature 334, 340–343 (1988). [Google Scholar]
- 2.Flombaum P., et al., Present and future global distributions of the marine cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. U.S.A. 110, 9824–9829 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chisholm S. W., et al., Prochlorococcus marinus nov. gen. nov. sp.: An oxyphototrophic marine prokaryote containing divinyl chlorophyll a and b. Arch. Microbiol. 157, 297–300 (1992). [Google Scholar]
- 4.Pachiadaki M. G., et al., Charting the complexity of the marine microbiome through single-cell genomics. Cell 179, 1623–1635.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moore L. R., Rocap G., Chisholm S. W., Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393, 464–467 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Kettler G. C., et al., Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 3, e231 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Partensky F., Garczarek L., Prochlorococcus: Advantages and limits of minimalism. Annu. Rev. Mar. Sci. 2, 305–331 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Biller S. J., Berube P. M., Lindell D., Chisholm S. W., Prochlorococcus: The structure and function of collective diversity. Nat. Rev. Microbiol. 13, 13–27 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Martiny J. B. H., Jones S. E., Lennon J. T., Martiny A. C., Microbiomes in light of traits: A phylogenetic perspective. Science 350, aac9323 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Braakman R., Follows M. J., Chisholm S. W., Metabolic evolution and the self-organization of ecosystems. Proc. Natl. Acad. Sci. U.S.A. 114, E3091–E3100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goericke R., Olson R., Shalapyonok A., A novel niche for Prochlorococcus sp. in low-light suboxic environments in the Arabian Sea and the eastern tropical North Pacific. Deep Sea Res. Part I Oceanogr. Res. Pap. 47, 1183–1205 (2000). [Google Scholar]
- 12.Lavin P., González B., Santibáñez J. F., Scanlan D. J., Ulloa O., Novel lineages of Prochlorococcus thrive within the oxygen minimum zone of the eastern tropical South Pacific. Environ. Microbiol. Rep. 2, 728–738 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Robledo E., et al., Cryptic oxygen cycling in anoxic marine zones. Proc. Natl. Acad. Sci. U.S.A. 114, 8319–8324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Astorga-Eló M., Ramírez-Flandes S., DeLong E. F., Ulloa O., Genomic potential for nitrogen assimilation in uncultivated members of Prochlorococcus from an anoxic marine zone. ISME J. 9, 1264–1267 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bibby T. S., Mary I., Nield J., Partensky F., Barber J., Low-light-adapted Prochlorococcus species possess specific antennae for each photosystem. Nature 424, 1051–1054 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Ting C. S., Rocap G., King J., Chisholm S. W., Cyanobacterial photosynthesis in the oceans: The origins and significance of divergent light-harvesting strategies. Trends Microbiol. 10, 134–142 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Sánchez-Baracaldo P., Origin of marine planktonic cyanobacteria. Sci. Rep. 5, 17418 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ulloa O., Canfield D. E., DeLong E. F., Letelier R. M., Stewart F. J., Microbial oceanography of anoxic oxygen minimum zones. Proc. Natl. Acad. Sci. U.S.A. 109, 15996–16003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stepanauskas R., et al., Improved genome recovery and integrated cell-size analyses of individual uncultured microbial cells and viral particles. Nat. Commun. 8, 84 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dufresne A., et al., Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 9, R90 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satoh S., Tanaka A., Identification of chlorophyllide a oxygenase in the Prochlorococcus genome by a comparative genomic approach. Plant Cell Physiol. 47, 1622–1629 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Stickforth P., Steiger S., Hess W. R., Sandmann G., A novel type of lycopene ε-cyclase in the marine cyanobacterium Prochlorococcus marinus MED4. Arch. Microbiol. 179, 409–415 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Nagata N., Tanaka R., Satoh S., Tanaka A., Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell 17, 233–240 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goericke R., Repeta D. J., The pigments of Prochlorococcus marinus: The presence of divinyl chlorophyll a and b in a marine procaryote. Limnol. Oceanogr. 37, 425–433 (1992). [Google Scholar]
- 25.Garczarek L., Hess W. R., Holtzendorff J., van der Staay G. W. M., Partensky F., Multiplication of antenna genes as a major adaptation to low light in a marine prokaryote. Proc. Natl. Acad. Sci. U.S.A. 97, 4098–4101 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishiura M., et al., Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281, 1519–1523 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Rocap G., et al., Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424, 1042–1047 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Holtzendorff J., et al., Genome streamlining results in loss of robustness of the circadian clock in the marine cyanobacterium Prochlorococcus marinus PCC 9511. J. Biol. Rhythms 23, 187–199 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Hess W. R., et al., Coexistence of phycoerythrin and a chlorophyll a/b antenna in a marine prokaryote. Proc. Natl. Acad. Sci. U.S.A. 93, 11126–11130 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chew A. G., Bryant D. A., Chlorophyll biosynthesis in bacteria: The origins of structural and functional diversity. Annu. Rev. Microbiol. 61, 113–129 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Fujita Y., Tsujimoto R., Aoki R., Evolutionary aspects and regulation of tetrapyrrole biosynthesis in cyanobacteria under aerobic and anaerobic Environments. Life (Basel) 5, 1172–1203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ludwig M., Bryant D. A., Transcription profiling of the model cyanobacterium Synechococcus sp. strain PCC 7002 by Next-Gen (SOLiD) sequencing of cDNA. Front. Microbiol. 2, 41 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aoki R., Takeda T., Omata T., Ihara K., Fujita Y., MarR-type transcriptional regulator ChlR activates expression of tetrapyrrole biosynthesis genes in response to low-oxygen conditions in cyanobacteria. J. Biol. Chem. 287, 13500–13507 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roux S., Enault F., Hurwitz B. L., Sullivan M. B., VirSorter: Mining viral signal from microbial genomic data. PeerJ 3, e985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren J., Ahlgren N. A., Lu Y. Y., Fuhrman J. A., Sun F., VirFinder: A novel k-mer based tool for identifying viral sequences from assembled metagenomic data. Microbiome 5, 69 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holtrop T., et al., Vibrational modes of water predict spectral niches for photosynthesis in lakes and oceans. Nat. Ecol. Evol., 5, 55–66 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Balasiny B., et al., Release of nitric oxide by the Escherichia coli YtfE (RIC) protein and its reduction by the hybrid cluster protein in an integrated pathway to minimize cytoplasmic nitrosative stress. Microbiology (Reading) 164, 563–575 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Seth D., et al., A multiplex enzymatic machinery for cellular protein S-nitrosylation. Mol. Cell 69, 451–464.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biller S. J., Coe A., Roggensack S. E., Chisholm S. W., Heterotroph interactions alter Prochlorococcus transcriptome dynamics during extended periods of darkness. mSystems 3, e00040–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muñoz-Marín M. C., et al., Mixotrophy in marine picocyanobacteria: Use of organic compounds by Prochlorococcus and Synechococcus. ISME J. 14, 1065–1073 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lyons T. W., Reinhard C. T., Planavsky N. J., The rise of oxygen in Earth’s early ocean and atmosphere. Nature 506, 307–315 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Petersen T. W., Brent Harrison C., Horner D. N., van den Engh G., Flow cytometric characterization of marine microbes. Methods 57, 350–358 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Rinke C., et al., Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Tennessen K., et al., ProDeGe: A computational protocol for fully automated decontamination of genomes. ISME J. 10, 269–272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Markowitz V. M., et al., IMG ER: A system for microbial genome annotation expert review and curation. Bioinformatics 25, 2271–2278 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W., CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jain C., Rodriguez-R L. M., Phillippy A. M., Konstantinidis K. T., Aluru S., High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 5114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen G. E., Canniffe D. P., Hunter C. N., Three classes of oxygen-dependent cyclase involved in chlorophyll and bacteriochlorophyll biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 114, 6280–6285 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muñoz-Sánchez J., Chánez-Cárdenas M. E., A review on hemeoxygenase-2: Focus on cellular protection and oxygen response. Oxid. Med. Cell. Longev. 2014, 604981 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Layer G., Moser J., Heinz D. W., Jahn D., Schubert W.-D., Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of radical SAM enzymes. EMBO J. 22, 6214–6224 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen L.-T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S., ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Campbell J. H., et al., UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. Proc. Natl. Acad. Sci. U.S.A. 110, 5540–5545 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell Amplified Genome data are available in Integrated Microbial Genomes, http://img.jgi.doe.gov/cgi-bin/w/main.cgi (accession nos. 2626541501, 2626541503–2626541507, 2626541509–2626541537, 2716884926, 2716884927, 2718217657, 2718217658, 2724679665, 2724679666, 2724679668, 2724679669, 2724679671, 2724679674–2724679676, 2724679678–2724679681, 2724679684, 2724679692, 2724679753, 2724679758, 2724679760, 2724679773, 2724679775, 2818991481–2818991488, 2818991490, 2818991493, 2818991494, 2818991497–2818991503, 2818991506–2818991511, 2818991513–2818991516, 2818991519–2818991530, 2818991533, 2818991535, 2818991536, 2818991539–2818991542, 2818991544–2818991547, 2818991549, 2818991551, 2818991553, 2818991555, 2818991557–2818991559, 2818991561, 2818991563–2818991568, 2818991570–2818991573, and 2818991575). All other study data are included in the article and/or SI Appendix.