Significance
The unique aspects of brain mitochondria composition and function are incompletely understood, and new approaches are required to understand their relationship. We addressed this challenge through a dual-process isolation procedure that yields both semipure brain mitochondria suitable for functional studies but not protein localization and ultrapure brain mitochondria that are suitable for determining protein localization but functionally compromised. We observed that contrary to the published literature, N-methyl-D-aspartate receptor, ryanodine receptor 1, glyceraldehyde 3-phosphate dehydrogenase, monocarboxylate transporter 1, excitatory amino acid transporter 1, and the L-type calcium channel Cav1.2α1 subunit are absent from brain mitochondria. Utilization of this dual-process brain mitochondria isolation technique will foster clarity on mechanisms of brain health and disease.
Keywords: mitochondria, channel, transporter, solute carrier, neuropsychiatric disease
Abstract
The brain requires continuously high energy production to maintain ion gradients and normal function. Mitochondria critically undergird brain energetics, and mitochondrial abnormalities feature prominently in neuropsychiatric disease. However, many unique aspects of brain mitochondria composition and function are poorly understood. Developing improved neuroprotective therapeutics thus requires more comprehensively understanding brain mitochondria, including accurately delineating protein composition and channel–transporter functional networks. However, obtaining pure mitochondria from the brain is especially challenging due to its distinctive lipid and cell structure properties. As a result, conflicting reports on protein localization to brain mitochondria abound. Here we illustrate this problem with the neuropsychiatric disease-associated L-type calcium channel Cav1.2α1 subunit previously observed in crude mitochondria. We applied a dual-process approach to obtain functionally intact versus compositionally pure brain mitochondria. One branch utilizes discontinuous density gradient centrifugation to isolate semipure mitochondria suitable for functional assays but unsuitable for protein localization because of endoplasmic reticulum (ER) contamination. The other branch utilizes self-forming density gradient ultracentrifugation to remove ER and yield ultrapure mitochondria that are suitable for investigating protein localization but functionally compromised. Through this process, we evaluated brain mitochondria protein content and observed the absence of Cav1.2α1 and other previously reported mitochondrial proteins, including the NMDA receptor, ryanodine receptor 1, monocarboxylate transporter 1, excitatory amino acid transporter 1, and glyceraldehyde 3-phosphate dehydrogenase. Conversely, we confirmed mitochondrial localization of several plasma membrane proteins previously reported to also localize to mitochondria. We expect this dual-process isolation procedure will enhance understanding of brain mitochondria in both health and disease.
Healthy brain function requires continuously high energy production to maintain neuronal integration, processing, and relaying of electrochemical signals. For example, after every action potential, neurons must rapidly restore their resting membrane potential and replenish spent neurotransmitter pools to regain responsiveness. This is tremendously energetically costly, yet neurons harbor only meager energy reserves (1). Furthermore, neurons metabolically depend upon glial synthesis of glutamate and GABA for coordinated neurotransmission (2). Neurons meet their resource challenges by importing large amounts of circulating fuels and neurotransmitter precursors through plasma membrane transporters and subsequently utilizing mitochondrial inner membrane solute transporters for further metabolism (3). As a result, neuronal electrochemical activity depends on coordination between trans- and intracellular channel–transporter functional networks comprising ion channels, pumps, and transporters of both plasma and mitochondrial membranes.
Disrupted channel–transporter networks have also been implicated and targeted in neuropsychiatric disease and brain injury (4–6). However, the functional organization of brain channel–transporter networks remains incompletely understood, especially with respect to mitochondrial components. Indeed, nearly half of mitochondrial transporters are currently orphan transporters with unknown substrate selectivity (6–8). Because even mild mitochondrial dysfunction may manifest as neuropsychiatric dysfunction (9), this knowledge deficit on mitochondrial transporters limits basic and therapeutic understanding of brain health. Mutations in many mitochondrial channels and transporters are known to drive neuropsychiatric diseases, such as inborn errors in the mitochondrial pyruvate carrier (MPC), glutamate carrier (GC1), and several additional mitochondrial carriers (10, 11). Moreover, deficiencies in neuronal energetics are also associated with Alzheimer’s disease, Parkinson’s disease, traumatic brain injury, stroke, depression, schizophrenia, and bipolar disorder (9, 12). Thus, rigorous delineation of the brain’s plasma membrane and mitochondrial channel–transporter functional networks is essential for developing new therapeutics for neuropsychiatric disease. Accurate determination of brain mitochondria protein content is critical to this endeavor.
Brain mitochondria isolation is uniquely challenging. For example, both the fatty myelin that insulates neurons and the synaptosomes produced from axonal arbor vesiculation readily fractionate with brain mitochondria (13). Accordingly, these contaminants must be removed for rigorous analysis of mitochondrial content. This challenge has been previously addressed with discontinuous density gradient centrifugation to remove synaptosome and myelin contaminants (14). However, removing endoplasmic reticulum (ER) membranes is more challenging (15). Indeed, removal of ER contamination from brain mitochondria preparations requires harsher isolation protocols, for which the trade-off in mitochondrial function has been poorly characterized (16). Thus, a critical challenge in brain research is developing mitochondria isolation approaches to obtain both functionally intact and compositionally pure brain mitochondria preparations. We encountered this challenge while investigating the subcellular localization of Cav1.2α1, the product of the prominent human neuropsychiatric risk gene CACNCA1C.
Cav1.2α1 is the pore-forming subunit of the predominant brain L-type voltage gated calcium channel (LTCC) (17). LTCCs enable activity-dependent calcium (Ca2+) transport into neurons for excitation–transcription coupling and long-term potentiation (18, 19). Cav1.2α1 has been linked to anxiety-like and depressive-like behavior, stress responses, addiction, recovery from posttraumatic stress, and survival of adult-born young hippocampal neurons (20–22). Cav1.2α1 also participates in channel–transporter functional networks between the plasma membrane and mitochondria, where its ablation curbs mitochondrial respiration and reactive oxygen species generation during excitotoxic injury (23, 24).
In addition to its canonical plasma membrane localization, a minor, yet persistent, subfraction of Cav1.2α1 has been observed in immunoblotted mitochondria (23). However, whether this is a biologically relevant, bona fide mitochondrial subfraction has remained undetermined. Because of our interest in the role of Cav1.2α1 in neuropsychiatric disease, we attempted to verify and study this phenomenon. We developed a dual-process mitochondrial isolation workflow that yields separate preparations of functional versus ultrapurified brain mitochondria, which enables powerful comparative tests of mitochondrial function and composition. This approach revealed that Cav1.2α1 is absent from brain mitochondria and has corresponding insignificant contributions to isolated brain mitochondria function. By then applying this dual-process mitochondrial isolation method to other disease-implicated and reportedly mitochondrially localized proteins, we revised the localization of several key brain channels and transporters.
Results
Cav1.2α1 in Semipure Brain Mitochondria Does Not Impact Mitochondrial Calcium Handling.
Plasma membrane Cav1.2 and its pore-forming subunit Cav1.2α1 have long been known to mediate depolarization-gated Ca2+ influx into neurons (17). Interestingly, residual Cav1.2α1 of uncertain biological relevance has been observed in Western blots of mitochondria purified by traditional methods (23). Because mitochondrial function is potently regulated by Ca2+ (25), we hypothesized that mitochondrial Cav1.2α1 might regulate mitochondrial Ca2+ homeostasis. Therefore, we tested the contribution of Cav1.2α1 to mitochondrial membrane potential, Ca2+ uptake, and Ca2+ capacitance. Crude mitochondria were isolated and further purified over a discontinuous Percoll density gradient to eliminate contaminating synaptosomes and myelin (14). Based on experiments described below, we designated this mitochondria preparation as “semipure.” Semipure mitochondria from Cacna1c brain-knockout (KO) and wild-type (WT) mice showed no significant differences in membrane potential, (SI Appendix, Fig. S1 A and B), Ca2+ uptake (SI Appendix, Fig. S1 C and D), or Ca2+ capacitance (SI Appendix, Fig. S1E).
Cav1.2α1 Enriches with the Outer Membrane Fraction of Semipure Mitochondria.
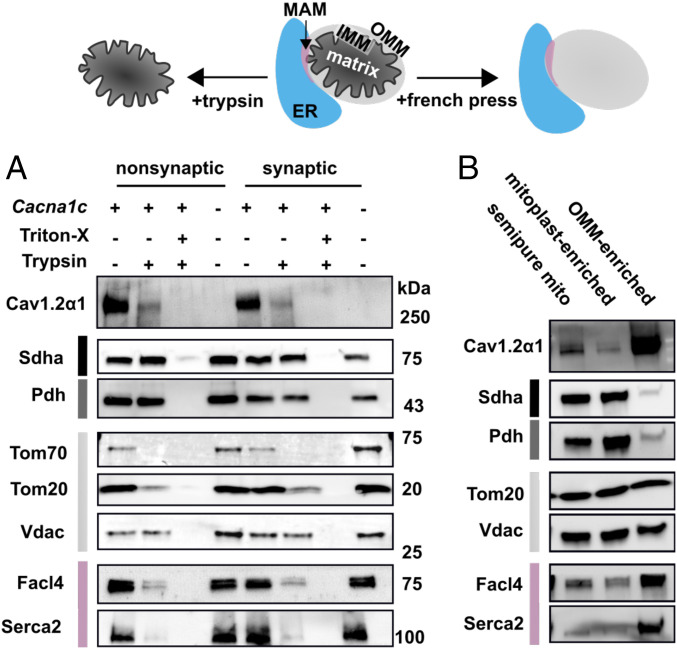
Because Cav1.2α1 KO did not overtly alter function of semipure isolated mitochondria, we next investigated whether a Cav1.2α1 mitochondrial subfraction was localized to the outer mitochondrial membrane (OMM). We reasoned that OMM localization would likely limit its ability to impact isolated mitochondrial function. Although the OMM is Ca2+-permeable, we considered that an OMM-localized Ca2+ channel could direct Ca2+ into mitochondria by concentrating supply to the mitochondrial Ca2+ uniporter (MCU) complex. Thus, semipure synaptic and nonsynaptic mitochondria from Cacna1c brain-KO and WT mice were digested with trypsin, with or without detergent, to expose inner mitochondrial membrane (IMM) and matrix proteins. Partial OMM digestion was then optimized by trypsin concentration (SI Appendix, Fig. S2A) and confirmed by IMM protein succinate dehydrogenase (Sdha) preservation (Fig. 1A). The voltage-dependent anion channel (Vdac), an integral OMM protein with minimal extra-OMM exposure, was also preserved (Fig. 1A). Conversely, the OMM proteins translocase of outer membrane (Tom) 20 and Tom70, which have larger extra-OMM globular domains expected to be more accessible to trypsin, were depleted (Fig. 1A) (26). Notably, digestion eliminated Cav1.2α1 (Fig. 1A), and the specificity of the antibody used to demonstrate this was confirmed in Cacna1c brain-KO brain tissue (Fig. 1A). Because the digest patterns between synaptic and nonsynaptic mitochondria were similar (Fig. 1A), nonsynaptic mitochondria were used for remaining isolations.
Fig. 1.
Cav1.2α1 fractionates with OMMs and MAMs in semipure mitochondria. (A) Representative immunoblots of submitochondrial fractions before and after trypsin digest in semipure mitochondria isolated from WT (+) and brain-Cacna1c brain-KO (−) mice. Subcellular compartments are color coded as ER (blue), MAM-enriched (pink), OMM (light gray), IMM (black), and mitochondrial matrix (dark gray). Trypsin was used to digest synaptic and nonsynaptic mitochondrially enriched fractions; 1% Triton-X was used as a control to solubilize all proteins, rendering them accessible to trypsin digest (n = 4, combined from three experiments). (B) Representative immunoblots from French press disrupted mitochondria on nondisrupted mitochondria (semipure mito), mitoplast-enriched fractions, and OMM-enriched cell fractions (n = 8, combined from three experiments).
After finding that OMM protein digestion from semipure mitochondria removed Cav1.2α1 (Fig. 1A), we next tested whether semipure OMM enrichment also increased Cav1.2α1. Semipure OMMs (OMM-enriched) were mechanically sheared from semipure mitochondria using a cell French press (Fig. 1B). As with trypsin digestion, Cav1.2α1 and the ER membrane markers fatty acyl-CoA synthetase 4 (Facl4) and sarcoplasmic reticulum/ER Ca2+-ATPase (Serca2) were depleted in IMM and mitoplast-enriched fractions (Fig. 1B) and enriched in OMM fractions (Fig. 1B). Replicate tryptic digest and French press fractionations are shown in SI Appendix, Fig. S2 B and C, respectively. Together, these data indicate that Cav1.2α1 associates with the crude OMM fraction from semipure mitochondria but do not distinguish whether this association is as a contaminant versus an OMM resident protein.
Immunoreactivity of Cav1.2α1 in the OMM Is an Artifact of ER Contamination.
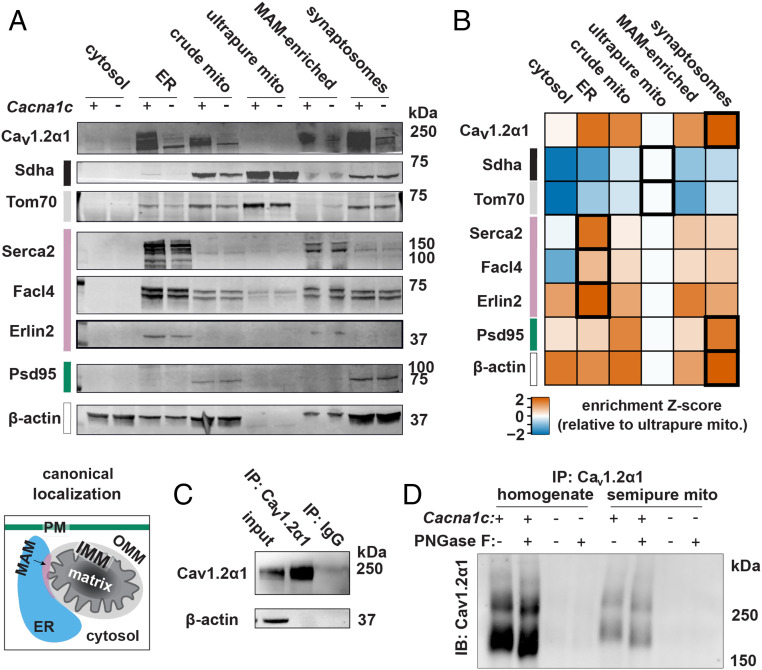
The OMM physically associates with functionally important mitochondria–ER contact structures called mitochondria-associated ER membranes (MAMs). To determine how much of the Cav1.2α1 signal originated from ER and MAM contaminants versus the OMM, we employed self-forming, continuous Percoll density gradient ultracentrifugation to investigate whether Cav1.2α1 would fractionate with ultrapure mitochondria or MAM-enriched ER fractions. We selected this approach because it separates functional MAMs from brain tissue (16), which prompted us to anticipate the generation of two mitochondrial fractions: 1) a higher-density ultrapure mitochondrial fraction and 2) a lower-density mixed fraction containing MAMs and mitochondria that could be separated by dilution and additional centrifugation. However, upon closer examination, we found that the latter fraction abundantly contained the synaptosome markers postsynaptic density protein 95 (Psd-95) and β-actin (Fig. 2 A and B). Thus, we designated this mixed mitochondrial and synaptosome fraction as “synaptosomes,” and we caution against MAM and mitochondrial localization assignments from this heterogeneous subcellular fraction. Fold enrichments for subcellular fraction-specific protein markers of cytosol, ER, crude mitochondria, ultrapure mitochondria, MAMs, and synaptosomes are shown in Fig. 2B, with replicate immunoblots in SI Appendix, Fig. S3A. Mitochondrial markers were further enriched by mitochondria ultrapurification (Fig. 2B). However, Cav1.2α1 signal was lost after ultrapurification, demonstrating that Cav1.2α1 is not mitochondrially localized. Conversely, Cav1.2α1 was retained alongside ER markers in the MAM-enriched fraction.
Fig. 2.
Cav1.2α1 in crude mitochondria is an artifact of Cav1.2α1 in the ER. (A) Representative immunoblots of brain mitochondria ultrapurification show depletion of Cav1.2α1 and MAM immunoreactivity from ultrapure mitochondria. Subcellular compartments are color coded as cytosol (white), PM (teal), ER (blue), MAM (pink), OMM (light gray), IMM (black), and mitochondrial matrix (dark gray) (n = 4, combined from three experiments). (B) Z score of marker enrichment relative levels in ultrapure mitochondria fraction during mitochondria ultrapurification. Highest enrichment fractions are boldly outlined. (C) Immunoprecipitation of Cav1.2α1 enriches Cav1.2α1 relative to IP input (Upper). (D) Representative immunoblots show bands from PNGase F treatment following Cav1.2α1 antibody immunoprecipitation from whole-cell homogenates and semipure mitochondria to evaluate for the presence of N-glycosylation protein modifications (n = 4, combined from two experiments).
To confirm that the Cav1.2α1 in semipure mitochondria was an ER contamination artifact, we next utilized the cellular phenomenon of N-glycosylation. Cav1.2α1, many other plasma membrane proteins, and a minimal fraction of ER resident proteins are N-glycosylated during processing in the ER (27). Conversely, N-glycosylation of mitochondrial proteins is extremely rare, with some uncertainty on whether the few reported observations are bona fide or artifactual (28). Therefore, we investigated whether Cav1.2α1 in semipure mitochondria was N-glycosylated. To cleave Cav1.2α1 N-glycosyl residues, we treated Cav1.2α1 immunoprecipitated from whole-cell and semipure mitochondria homogenates with PNGase F amidase, which cleaves internal glycoside bonds in oligosaccharides (Fig. 2 C and D). Immunoblotting control and PNGase F-treated Cav1.2α1 immunoprecipitates revealed the expected mobility shift induced by losing N-glycosylation (Fig. 2D). If Cav1.2α1 from the semipure mitochondria preparation was truly present in mitochondria, one would expect no mobility shift after PNGase F treatment, given the mitochondrial exclusion of N-glycosylated proteins. However, Cav1.2α1 from the semipure mitochondria showed the same PNGase F-dependent mobility shift as Cav1.2α1 from whole-cell homogenates (Fig. 2D), consistent with the presence of Cav1.2α1 in contaminating ER but not in mitochondria.
Accurate Localization in Brain Mitochondria Requires Ultrapurification to Remove ER.
After establishing that the immunoreactivity of Cav1.2α1 in semipure mitochondria was an artifact of ER contamination, we considered how the degree of mitochondrial purity affects apparent localization of other critical brain channel–transporter network proteins. Specifically, we tested proteins in three classes of reported localization: 1) plasma membrane, 2) mitochondria, and 3) dual-localized to the plasma membrane and mitochondria (Fig. 3A).
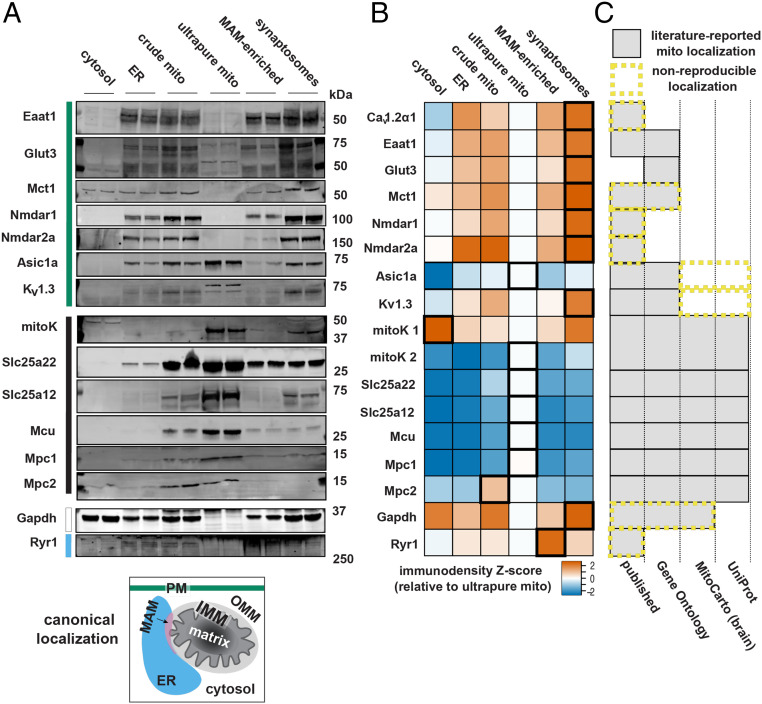
Fig. 3.
Plasma membrane proteins broadly contaminate semipure mitochondria. (A) Western blots show the presence or absence of solute carriers and ion channels with known membrane transporter functions in ultrapure brain mitochondria preparations isolated by continuous density gradient ultracentrifugation. The canonical localization of selected proteins is color coded to the left of the blots as cytosol (white), PM (teal), ER (blue), MAM (pink), OMM (light gray), IMM (black), and mitochondrial matrix (dark gray) (n = 4, combined from two experiments). (B) Z score of relative levels of marker enrichment in ultrapure mitochondria fraction. Highest enrichment fractions are outlined in bold (n = 2). (C) Comparative localization with published, Gene Ontology, MitoCarta, and Uniprot references with mitochondrial assignment shown by gray shading. Database-reported mitochondrial localization that was not reproduced in these experiments is outlined by yellow, dashed lines.
These three classes were distributed across four functional groups. The first functional group comprises ion channels functionally linked to Cav1.2α1 by involvement in Ca2+ handling: 1) the N-methyl-D-aspartate receptor subunits 1 and 2a (NR1 and NR2a), which together form an excitatory plasma membrane channel involved in neurodegenerative diseases (29, 30) previously reported in brain mitochondria (29); 2) the ryanodine receptor 1 (Ryr1), an ER Ca2+ channel that facilitates Ca2+-induced Ca2+ release (31, 32) also previously reported in brain mitochondria (33); and 3) the mitochondrial calcium uniporter (Mcu), the major mitochondrial Ca2+ importer, which is known to be involved in excitotoxic mitochondrial Ca2+ overload (34).
The second functional group comprises proteins involved in brain glucose metabolism: 1) glucose transporter 3 (Glut3), the primary neuronal plasma membrane glucose transporter implicated in Alzheimer’s disease (35, 36); 2) glyceraldehyde 3-phosphate dehydrogenase (Gapdh), a cytosolic glycolytic enzyme and apoptosis regulator in neurodegenerative disease that, while not a channel or transporter, has been reported in brain mitochondria (37, 38); 3) the mitochondrial pyruvate carrier subunits 1 and 2 (Mpc1 and Mpc2), which link glycolysis with mitochondrial metabolism and for which inactivating mutations result in severe neurodevelopmental impairments (11, 39, 40); 4) monocarboxylate transporter 1 (Mct1), a glial lactate and proton symporter that exports lactate to neurons and has been reported to localize in brain mitochondria (41, 42); and 5) the mitochondrial ATP-sensitive potassium channel (mitoK), a neuroprotective ATP-sensitive potassium channel that couples cellular energetics with excitability (43–45) and has also been reported in brain mitochondria (46, 47).
The third functional group comprises proteins mediating critical steps in the brain energy-producing and neurotransmission-supporting glutamate–glutamine cycle: 1) excitatory amino acid transporter 1 (Eaat1), an astrocytic glutamate/aspartate transporter necessary for glutamate uptake and reportedly localized in cardiac mitochondria (48, 49); 2) the mitochondrial aspartate/glutamate carrier (Slc25a12), an autism spectrum disorder and schizophrenia risk gene (50); and 3) the mitochondrial glutamate carrier (Slc25a22), implicated in lethal neonatal epilepsy (10).
The fourth and final functional group includes other ion channels that, like Cav1.2α1, have been reported to dual-localize to plasma and mitochondrial membranes: 1) acid-sensing ion channel isoform 1A (Asic1a), a proton-sensing cation channel with broad roles in learning and memory, pain sensation, and cell death (51–53), and 2) the voltage-gated potassium channel Kv1.3, a potassium channel regulating neuron membrane potential and excitability (54, 55). Of note, mitoK from the second group could also be classified into this fourth group as well (43).
All of the proteins listed above were localized in crude mitochondrial fractions from the brain (Fig. 3 A and B). As expected, Glut3 was absent from ultrapurified mitochondria. However, in contrast to previous reports of their mitochondrial localization, Eaat1, Mct1, Nmdar1, Nmdar2a, Gapdh, and Ryr1 were absent from ultrapurified mitochondria (Fig. 3 A and B) (Table 1). This implicates impurity of isolated mitochondria, most likely due to ER contamination, as the likely reason for the previous reports of their mitochondrial localization. By contrast, Asic1a, Kv1.3, and mitoK all retained localization in ultrapurified mitochondria.
Table 1.
Citation information for cellular localization of proteins previously reported as localizing in mitochondria but not observed in ultrapurified mitochondria here
| Protein | Localization | Tissue (species) | Method | Mitochondria purity | ||
| Main | Mitochondrial | Cell | Organ | |||
| Cav1.2α1 | Plasma membrane | Ref. 23 | HT22 (ms) | Brain (ms) | IB | Semipure |
| Eaat1 | Plasma membrane | Ref. 49 | — | Heart (ms) | IB, IF, fractionation | Crude |
| Mct1 | Plasma membrane | Ref. 41 | Neurons (rt) | Brain (rt) | IF, IB | Crude |
| Nmdar1 | Plasma membrane | Ref. 29 | — | Brain (rt) | IB, EM | Semipure |
| Gapdh | Cytoplasm | Ref. 37 and MitoCarta | HeLa (hu) | Liver (rt), brain (ms) | EM, IB, fractionation | Crude and ultrapure |
| Ryr1 | ER | Ref. 33 | Neurons (rt) | Brain (rt) | IB, IF, fractionation | Crude and semipure |
The main site of cellular localization and the reference(s) for reported mitochondrial localization are specified. Reported reference cell type, species (ms, mouse; rt, rat; gb, gerbil; hu, human), and localization method (IB, immunoblot; IF, immunofluorescence; EM, electron microscopy) are also indicated. Mitochondria purity was approximated to the designations used in this study.
For rigorous verification of these results, we employed several antibody controls. Antibodies were selected for their previous validation against KO tissues [Mpc1 and Mpc2 (11) and Mcu and mitoK (43)] or for their specificity in our experiments using genetic KO tissue, siRNA-mediated KO in cultured cells, or blocking with manufacturer-provided peptides to detect nonspecific binding (Alomone Labs: Eaat1, Mct1, Kv1.3, and Slc25a12) (SI Appendix, Fig. S3 C–F). For Nmdar fractionation validation, three antibodies against different NR1 subunit epitopes, and one antibody against the NR2a subunit, were used to confirm lack of mitochondrial localization in ultrapurified fractions (SI Appendix, Fig. S3B). While the Nmdar antibody catalog numbers used in the original mitochondrial localization publication were not disclosed (29), we selected antibodies from the same vendor. Finally, Ryr1 antibody was selected for its vendor-demonstrated immunospecificity to differentiated versus undifferentiated myotubes, which respectively do and do not express Ryr1 (Product information sheet, Cell Signaling Technology, 2020, https://www.cellsignal.com/products/primary-antibodies/ryr1-d4e1-rabbit-mab/8153).
We next compared our protein localization results with three highly utilized protein localization databases: Gene Ontology, MitoCarta, and UniProt (Fig. 3B), each of which applies related but unique criteria for determination of mitochondrial localization. The Gene Ontology dataset uses vocabulary search terms to subcellularly localize gene products in literature and computational studies (56, 57). MitoCarta incorporates relative enrichment from crude to semipure preparations, computational, homology-based, and microscopy controls to assign mitochondrial localization in many tissues, including mouse brain (47, 58). The UniProt consortium combines automatic search term retrievals with expert manual annotations to confer protein localization (59). In comparison to ultrapure brain mitochondria here, the Gene Ontology dataset included false-positive mitochondrial localizations, such as Gapdh derived from MitoCarta (47, 58) (Fig. 3B). UniProt did not have false-positive mitochondrial localizations (59). However, compared to ultrapurified brain mitochondria, both the UniProt and brain MitoCarta datasets included false-negative mitochondrial localizations for dual-localized plasma membrane proteins Kv1.3 and Asic1a. Taken together, our findings show that ER contamination must be stringently removed to accurately evaluate brain mitochondria protein localization and that current protein localization databases do not fully account for dual-localized plasma membrane–mitochondrial proteins.
Different Mitochondrial Isolation Protocols Must Be Used to Determine Localization versus Function.
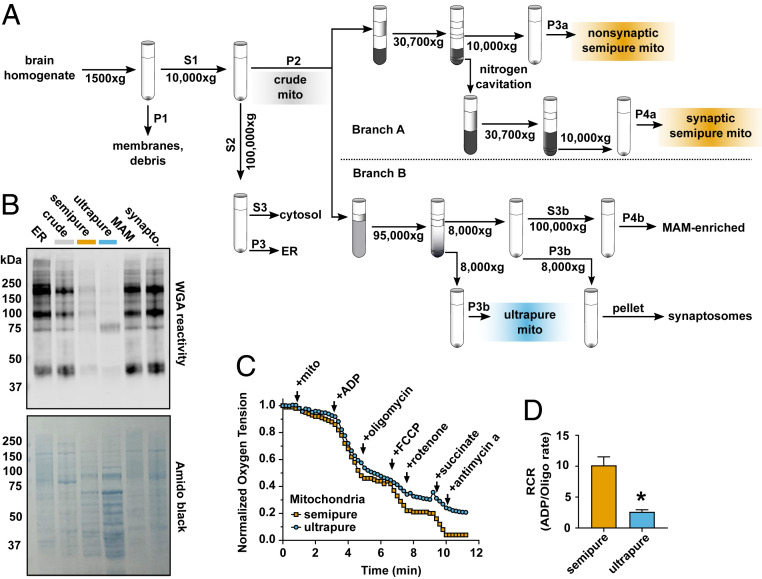
To broadly evaluate mitochondrial purity from crude, semipure, and ultrapure preparations (Fig. 4A), we examined mitochondrial fractions for total N-glycosyl content. For this, we used wheat germ agglutinin (WGA), a lectin that binds N-glycosyl adducts, in Western blots (Fig. 4A). We hypothesized that WGA reactivity would directly correlate with ER contamination and inversely correlate with mitochondrial purity. As expected, ER and crude mitochondrial fractions were rich in glycosylated proteins (Fig. 4B). While semipure mitochondria showed decreased N-glycosylated protein relative to the crude preparation, mitochondrial ultrapurification almost completely removed N-glycosylated protein (Fig. 4B).
Fig. 4.
Brain mitochondria ultrapurification removes ER contaminants but impairs function. (A) Schematic workflow for the dual-process brain mitochondria isolation protocol shows that after isolation of crude mitochondria (gray), branch A yields semipure mitochondria (orange) over a discontinuous Percoll gradient, and branch B yields ultrapure mitochondria (blue) over a self-forming continuous Percoll gradient. (B) Wheat germ agglutin (WGA) identification of N-glycosylated proteins in cellular subfractions shows that ER, crude mitochondria, semipure mitochondria, MAM, and synaptosomal brain cellular fractions are heavily glycosylated, while ultrapure brain mitochondria are relatively depleted of WGA immunoreactivity. Total protein quantification was visualized with amido black (n = 4 to 8, males and females, combined from three experiments). (C) Representative oxygen consumption trace from semipure brain mitochondria and ultrapure brain mitochondria. The change in oxygen saturation after manipulation of mitochondrial respiration was measured. (D) Brain mitochondrial RCR (ADP-stimulated/oligomycin-inhibited rates) shows that semipure brain mitochondria retained expected oxygen consumption, which is relatively depleted in ultrapure brain mitochondria (n = 4 to 8, males and females, combined from two experiments). *P = 0.01 by unpaired two-tailed Welch’s t test for unequal variances, Cohen’s d = 3.7.
Next, to test the consequence of mitochondrial ultrapurification on mitochondrial function, we performed oxygen consumption measurements with a Clark-type electrode (Fig. 4C). Specifically, we quantified the respiratory control ratio (RCR), a well-established measure of mitochondrial respiratory function that reveals mitochondrial ADP responsiveness and maximal respiratory capacity. Semipure mitochondria maintained RCRs similar to literature values (Fig. 4D) (60). Conversely, although ultrapurified mitochondria were able to respire, their RCR was significantly lower than semipure mitochondria. The ultrapurified mitochondria response to oligomycin and rotenone (Fig. 4C) suggests either that the process of ultrapurification damages mitochondria or that ER membranes or other nonmitochondrial material in semipure mitochondria enhance mitochondria respiration. These findings show that intact mitochondria can be ultrapurified to the point of minimal ER contamination but that the process is not suitable for functional assays. Thus, accurate functional and compositional analysis of isolated brain mitochondria is achieved through dual-process isolation.
Discussion
Investigation of mitochondrial transporter and plasma membrane channels is an active area of focus in the fields of basic brain function and neuropsychiatric disease (39). However, the extraordinarily complex functional interactions of brain transporters and channels in normal and disease conditions are poorly understood. Parsing these interactions is challenging and essential for fully understanding brain function and will likely require technically targeted approaches to better understand brain mitochondria composition versus function. While many reported brain mitochondrial isolation protocols produce functional mitochondria, they are highly contaminated with other organelles and thus insufficient for accurate determination of protein content. Conversely, more stringent isolations may damage mitochondria and impair functional analysis. Here we demonstrate an encompassing, dual-process isolation method that yields separate fractions of 1) analytical-grade, ultrapure mitochondria for protein localization assays and 2) respiration-competent, semipure mitochondria for functional assays.
Because semipure and ultrapure mitochondria are isolated with the same starting conditions and crude mitochondrial preparation in our dual-process approach, this approach enables powerful side-by-side enrichment comparisons that have not been possible with previous methods. Specifically, crude mitochondria purified by discontinuous Percoll gradient centrifugation, designated here as semipure, are functional but unsuitable for evaluating protein localization due to ER contamination. In parallel, subjecting the same crude mitochondria to more stringent purification through self-forming Percoll gradient ultracentrifugation produces ultrapure mitochondria freed of ER contamination. However, this ultrapurification method also impairs mitochondrial function.
We developed this dual-process protocol out of necessity when studying the neuropsychiatric disease-implicated channel subunit Cav1.2α1. After observing that Cav1.2α1 protein in semipure mitochondria does not significantly affect membrane potential, Ca2+ uptake, and Ca2+ capacitance, application of our dual-process mitochondrial isolation showed that the appearance of Cav1.2α1 in semipure mitochondria is due to ER contamination. Thus, the effects of Cav1.2α1 on mitochondrial function are likely limited to those downstream of its canonical role as a plasma membrane calcium channel (23, 24). We further applied ultrapurification to individually test mitochondrial localization of a panel of functionally linked brain channels and transporters by immunoblots and then in a broad, unbiased manner by WGA blots for total N-glycosylated protein content. These assays revealed that many well-characterized plasma membrane channels and transporters, as well as ER proteins, heavily contaminate even discontinuous Percoll gradient-purified semipure but not ultrapurified brain mitochondria. Our observations also support the emerging concept of dual-localized plasma membrane–mitochondrial membrane proteins by replicating the recently reported mitochondrial protein localization of Asic1a (51), Kv1.3 (54), and mitoK (43).
By contrast, we found that previous reports for mitochondrial localization of Mct1 (41), Ryr1 (31), Nmdar (29), and Gapdh (37) are false-positives almost certainly due to ER contamination in crude and semipure mitochondria. In addition, Eaat1, reportedly localized to cardiac mitochondria (49), did not localize to brain mitochondria. Previously, Nmdar and Gapdh were reported to localize to mitochondria based on immunoelectron microscopy (29, 37). However, antibody negative controls were not used, and immunoreactivity was also observed in the ER. Thus, hypothesized roles of these proteins, especially the more highly referenced Nmdar (61–64) and Mct1 (65, 66), in disease-related mitochondrial function require reevaluation.
We also compared our mitochondria ultrapurification results to established proteome databases. The MitoCarta2.0 database is rooted in mitochondrial proteomics with orthogonal verification measures (47, 58). This currently field-standard database has rigorously screened and identified mitochondrial proteins based on relative enrichment from crude to semipure preparations with robust microscopy, computational, and homology-based controls. Mitochondrial assignments by MitoCarta matched our localization findings in ultrapure mitochondria in all cases except Gapdh and the dual-localized proteins Asic1a and Kv1.3. Thus, the careful enrichment corrections used by MitoCarta may be overly conservative in the case of proteins that are dual-localized at the plasma and mitochondrial membranes. The dual-localized proteins that we tested were found in mitochondrial subfractions at lower relative quantities than in other cellular compartments (SI Appendix, Fig. S3G). Furthermore, we found that the minor isoform of mitoK localized to mitochondria (43, 59). This underscores the important and likely limitation whereby minor isoforms that localize to mitochondria could be erroneously excluded as noise in peptide-based mass-spectrometry proteomics, which could fail to observe peptides distinguishing minor splice isoforms localized to mitochondria (67). Taken together, our data show that although canonical mitochondrial proteins further enrich after ER and plasma membrane removal, dual-localized proteins may partially deenrich with a resident mitochondrial subfraction persisting through mitochondrial ultrapurification. Thus, accurately testing for dual-localization of plasma membrane proteins to mitochondria requires mitochondrial ultrapurification.
Last, we consider limitations of this work and potential future directions. We acknowledge that ultrapure mitochondria does not equate to perfectly pure mitochondria. Nonetheless, mitochondria ultrapurification by ultracentrifugation over a self-forming, continuous Percoll gradient broadly distinguished between mitochondrial resident versus contaminating proteins with far greater accuracy compared to the discontinuous Percoll gradient separation utilized by semipurification. However, it did not decipher whether protein contaminants were captured while being trafficked through the secretory pathway or whether they were resident ER or MAM proteins. Indeed, while proteins trafficked through the secretory pathway, such as Cav1.2α1, commonly bear N-linked glycosylation, and this modification is generally barred from mitochondrial localization, resident ER and MAM-enriched ER proteins have been reported be N-glycosylated in limited instances (68, 69). Future MAM research will clarify the role of N-glycosylation and important channel–transport networks between mitochondria, ER, and the plasma membrane (15). It will also be important to pursue technical advances enabling isolation of functionally intact ultrapure mitochondria. This will likely need to be differentially optimized depending on the tissue and physiologic condition under examination. For example, here we excluded calcium chelators from experimental buffers but not the initial isolation buffer, which enables calcium modulation as a variable during tests of mitochondrial function (70). However, in other circumstances of excessive calcium or sensitized mitochondria, the confounding factor of the mitochondrial permeability transition pore activation could come into play and require addition of a calcium chelator to buffers used for testing mitochondrial function (71). Future advances to this method may include development of new, designer buffer systems combined with genetically encoded organellar tags (72) to enhance mitochondrial purification. We also note that potential ER and MAM contributions to isolated mitochondria function cannot be dismissed and acknowledge the intriguing possibility that removing ER and MAMs might impair isolated mitochondria function. Finally, because we conducted our experiments in normal mouse brain, localization results reported here cannot account for nonbrain tissue or disease-induced changes in the mitochondrial proteome.
Decades of neuroscience research have demonstrated the profound importance of brain metabolism on function and that numerous neuropsychiatric conditions feature mitochondrial aberrations. Indeed, a full understanding of brain mitochondria would undoubtedly facilitate the development of new neurotherapeutics. The increased accessibility of powerful technologies such as metabolomics and proteomics provides abundant opportunity to decipher fundamental relationships between mitochondrial composition and brain metabolism and function. However, despite the impressive capacities of these next-generation technologies, basic research techniques like subcellular fractionation remain essential for accurate interpretation of their results. Thus, dual-process isolation of brain mitochondria reported here can be used in the future to more accurately evaluate both functional and compositional disease-specific changes and undergird the use of evermore powerful analytical technologies. This simple and rigorous approach is sufficiently stringent and sensitive to detect likely false-positive and false-negative mitochondrial localizations that have been reported in the literature. We expect that application of this dual-process method of brain mitochondria purification will robustly advance understanding of basic brain function and neuropsychiatric health and disease.
Materials and Methods
Animals.
Animal procedures were performed in accordance with University of Iowa AAALAC accredited institutional animal care and use committee (IACUC) regulations. Group-housed adult male and female Cacna1c brain-KO mice and WT littermates were used. Cacna1c brain-KO mice were generated by crossing homozygous Cacna1c floxed mice (73) with mice expressing Cre recombinase under control of a transgenic Nestin promoter. In this mouse line, Cacna1c is broadly lost throughout the brain, thereby minimizing artifacts from nontargeted cell populations during brain mitochondrial isolation. Animals were euthanized for experiments between Zeitgeber time ZT3 and ZT9.
Semipure Brain Mitochondria Isolation by Discontinuous Percoll Gradient Separation.
Crude mitochondria were isolated from adult mouse brain by homogenization and centrifugation. Isolated crude mitochondria were then further purified by Percoll discontinuous gradient centrifugation into semipure synaptic and semipure nonsynaptic mitochondria fractions, similar to as previously described with modifications (14). Whole brains were rapidly excised and minced in 10 mL of mitochondria isolation buffer (MIB; 225 mM mannitol, 75 mM sucrose, 2 mM K2HPO4, 5 mM Hepes, 1 mM EGTA, 0.1% fatty acid free bovine serum albumin, pH 7.2). Importantly, we selected an isolation buffer pH at the lower side of published protocols to recapitulate cytosolic pH. The additional inclusion of phosphate is also expected to improve pH buffering via the endogenous mitochondrial phosphate exchange system. After mincing, MIB was decanted and replaced with fresh MIB at a 1:10 dilution of brain mass to buffer volume, followed by homogenization with eight passes of a Teflon-on-glass Potter–Elvehjem homogenizer at 400 rpm. These and the remaining differential centrifugation steps were performed in round-bottom, 50-mL centrifuge tubes. Homogenates were diluted 1:1 in MIB and then were centrifuged at 1,500 × g for 5 min to pellet nuclei, plasma membranes, and nonlysed cells with a Beckman JA-25.50 fixed angle rotor. The resulting supernatant was then transferred to new tubes and centrifuged at 10,000 × g for 10 min in a Beckman JA-25.50 fixed angle rotor to pellet a crude mitochondrial fraction. Of note, this slower centrifugation step compared to the previously published speed of 21,200 × g decreases visible red blood cell debris within the crude mitochondrial pellet and is compatible with the adapted and merged ultrapurification arm described in the next section. The supernatant from this 10,000 × g spin contains bulk ER and cytosolic proteins and was ultracentrifuged at 100,000 × g for 1 h in 13.2-mL ultracentrifuge tubes in a Beckman SW-41 Ti swinging-bucket rotor for 1 h to pellet ER proteins, separating cytosolic proteins into the supernatant.
The crude mitochondrial pellet from the 10,000 × g spin noted above contains mitochondria and contaminating synaptosomes, myelin, ER, and MAMs. This pellet was gently resuspended with a loose-fitting, 2-mL glass-on-glass hand homogenizer in 3.5 mL of 15% Percoll in MIB (vol/vol). All Percoll solutions were made using a 100% Percoll stock which contains all MIB components at the same concentrations by adding dry mannitol, sucrose, and K2HPO4 to pure Percoll liquid and adding 100× concentrated EGTA and Hepes stock solutions (pH 7.2), which minimally alter Percoll concentration. Discontinuous Percoll layers were prepared in 10-mL, round-bottom ultracentrifuge tubes by underlaying 1.5 mL of 40% Percoll in MIB beneath 3.5 mL 24% Percoll in MIB. The 15% Percoll mixture containing suspended crude mitochondria was then layered on the top of the gradient. The resulting discontinuous gradient tubes were nested into 50-mL ultracentrifuge tubes and then separated at 30,700 × g for 8 min in a Beckman JA-25.50 fixed angle rotor. This rotor was used for the remainder of semipure mitochondria isolation. Synaptosomes were collected between the 15 and 24% Percoll interfaces, and nonsynaptic mitochondria were collected from the 24 and 40% Percoll interface. Synaptosomes were diluted to a final Percoll concentration of 12% (1:1 dilution of synaptosomes in MIB) and lysed under 1,500 psi nitrogen cavitation for 15 min with stirring. The resulting mixture of ruptured synaptosomes and freed mitochondria was layered over 4 mL of 24% Percoll in MIB, forming a discontinuous gradient and then centrifuged at 30,700 × g for 10 min in 10-mL, round-bottom tubes, again nested into 50-mL ultracentrifuge tubes. This step separates mitochondria, which form the higher density band, from synaptosomal debris above the 24% Percoll layer. Both synaptosomal and nonsynaptosomal mitochondria were resuspended and washed with 10 mL MIB, then pelleted at 10,000 × g for 10 min in 50-mL, round-bottom tubes. This wash step was repeated once more, and the resulting semipure mitochondria pellets were kept on ice for functional and protein analyses. All steps were performed on ice with centrifugation at 4 °C.
Ultrapure Brain Mitochondria Isolation by Continuous Self-Forming Percoll Gradient Separation.
To remove ER and MAM proteins more stringently than semipurification via discontinuous Percoll gradient purification, crude mouse brain mitochondria were ultrapurified by ultracentrifugation through a self-forming, continuous Percoll gradient, similar to as previously described with modifications (16, 74). These modifications enabled assembly of semipurification and ultrapurification into two arms of our dual-process mitochondrial isolation, with both products stemming from the same crude mitochondrial isolation. As noted above in the previous section, we decreased the spin speed for crude mitochondrial isolation compared to that used previously for mitochondrial semipurification (14). We modified ultrapurification by utilizing MIB, as defined in the previous section, which contains sucrose and K2HPO4 in addition to the Hepes, mannitol, EGTA, and BSA previously used (16, 74). We expect that K2HPO4 provides additional buffering capacity compared to Hepes alone. We also redesignated a mixed mitochondrial fraction to a mixed synaptosomal fraction as described here and in Results.
Specifically, crude mitochondria from one mouse brain were generated as described for semipurification in the preceding section as the second pellet of sequential 1,500 × g and 10,000 × g spins, which were then resuspended in 1 mL MIB and layered over 10 mL of 30% Percoll (vol/vol in MIB) in 13.2-mL ultracentrifuge tubes and centrifuged in a Beckman SW 41 Ti swinging-bucket rotor at 95,000 × g for 30 min. During this centrifugation, the high spin speed causes the colloidal silica Percoll particles sediment to form a continuous gradient. The resulting highest density band was aspirated, resuspended in a 10× volume of MIB, and pelleted at 8,000 × g for 10 min in a Beckman JA-25.50 fixed angle rotor to obtain a mitochondrial fraction substantially deenriched from ER compared to semipure mitochondria. The pellet was resuspended in a small amount of buffer by gentle agitation, washed again by dilution in a 10× volume of MIB, and repelleted at 8,000 × g for 10 min. This pellet was designated ultrapure mitochondria and set aside on ice for experiments. Lower-density bands produced from the continuous gradient ultracentrifugation were aspirated, resuspended in a 10× volume of MIB, and centrifuged at 10,000 × g for 10 min in a Beckman JA-25.50 fixed angle rotor. The pellet was resuspended in a small amount of buffer by gentle agitation, washed again by dilution in a 10× volume of MIB, and repelleted at 8,000 × g for 10 min. Compared to the previous literature on isolating this fraction by a similar process, we redesignated this pellet as a mixed synaptosomal fraction. The resulting supernatant was centrifuged at 100,000 × g for 1 h in a Beckman SW 41 Ti swinging-bucket rotor to pellet MAM fractions. All steps were performed on ice with centrifuging at 4 °C.
Isolated Brain Mitochondria Respiratory Coupling.
Mitochondria were suspended to 0.5 mg/mL in respiration buffer (120 mM KCl, 5 mM Hepes, 5 mM K2HPO4, pH 7.2) containing 5 mM glutamate and malate. Basal oxygen consumption rate was recorded with a Clark-type oxygen sensitive electrode under a 37 °C water jacket with constant stirring. State III respiration was stimulated by adding 100 μM ADP. Next, state IV respiration was measured by adding 1 μM oligomycin to inhibit ATP synthase. Maximum uncoupled respiration was elicited by adding 0.2 μM FCCP. Complex I activity was inhibited with 1 μM rotenone. Complex II activity was stimulated with 5 mM succinate. Finally, 1 μM antimycin A was added to inhibit electron transfer at complex III. Measurement was continued after each addition until a stable respiration rate was obtained. RCR under glutamate and malate was determined by calculating the slope of oxygen consumption over time and dividing this rate from state III by state IV plus oligomycin as shown in the following equation:
Isolated Mitochondrial Membrane Potential.
Mitochondrial membrane potential in isolated semipure brain mitochondria was determined using ratiometric red and green fluorescence imaging after staining with JC-1 dye following kit protocol (Sigma Aldrich). Briefly, 0.5 μg of nonsynaptic mitochondria were loaded with Ca2+-free JC-1 staining solution energized with ATP, ADP, and succinate in a 96-well plate. In this assay, the negative membrane potential of mitochondria leads to the aggregation of the positively charged JC-1 dye in the matrix, where aggregation induces red fluorescence. Valinomycin was added to mitochondria as a negative control for mitochondrial membrane potential. Ratios of aggregated vs. nonaggregated JC-1 dye (525/590 and 490/530 excitation/emission, respectively) were measured on a fluorimeter every 30 s for 4 h at room temperature. Maximum JC-1 red to green ratio was quantified.
Isolated Mitochondrial Calcium Capacitance.
Isolated semipure nonsynaptic mitochondrial Ca2+ uptake and capacitance were measured using the fluorescent calcium indicator Calcium Green 5N. Briefly, 100 µg of mitochondrial protein was suspended in respiration solution (150 mM KCl, 5 mM Hepes, 2 mM K2HPO4, 5 mM glutamate, 5 mM malate, pH 7.2) with cell membrane impermeable Calcium Green 5N and then loaded into 96-well plates. Treatment solutions were added, and basal fluorescence was recorded for three minutes at 506/531 excitation/emission. Ca2+ uptake rate was measured by titrating CaCl2 at 50 μM additions into the mitochondrial suspension, with 5 to 10 min of recording mitochondrial Ca2+ uptake after each addition. Ca2+ capacitance was defined as the Ca2+ addition after which there was no further loss of fluorescent signal. Ruthenium red (10 μM) was used as a control for inhibited mitochondrial Ca2+ uptake. Mitochondria-free wells were used to generate a standard curve to convert Calcium Green 5N fluorescence to nanomoles (nmol) of extramitochondrial Ca2+. The rate of Ca2+ uptake was determined by the rate constant of the change in nmol of Ca2+ over time. Ca2+ capacitance was determined by the maximum amount of CaCl2 addition that mitochondria were able to buffer, indicated by a negative slope.
Brain Mitochondria Subfractionation.
Nonsynaptic mitochondria-enriched fractions were collected as described above and then resuspended at 1.5 mg/mL in MIB. Mitochondria were loaded into a precooled mini-French press cell and extruded at a rate of 1 drop per 4 s at 2,000 psi. Mitoplast-enriched fractions were collected by pelleting at 12,000 × g in a Beckman JA-25.50 fixed angle rotor. The resulting supernatant was ultracentrifuged at 59,000 × g in 10-mL ultracentrifuge tubes in a Beckman Type 50.2 Ti fixed-angle rotor, and the pellet was collected as the outer membrane-enriched fraction.
Synaptic and nonsynaptic mitochondria-enriched fractions were each collected by approaches described above and subjected to trypsin digest. Briefly, 100 µg of mitochondrial protein was suspended in several concentrations of trypsin and incubated at room temperature for 25 min. Triton-X served as a control for complete membrane solubilization that rendered all proteins accessible to trypsin.
N-Glycosylation Cleavage.
Nonsynaptic brain mitochondria were collected as described above. Cav1.2α1 was immunoprecipitated from nonsynaptic mitochondria and whole-cell homogenates. Proteins were incubated in lysis buffer (20 mM Hepes, 150 mM NaCl, 2 mM EDTA, 1% n-dodecyl-beta-maltoside, pH 7.4) with protease inhibitors. Samples were washed in lysis buffer without N-dodecyl-beta-maltoside and then sonicated three times for 10 s at 60% amplitude (Branson Digital Sonifier SFX 150). Insoluble proteins were pelleted at 10,000 × g. The resulting supernatant was precleared with protein A/G agarose beads for 30 min with rotation. Beads were spun out of solution, and the resulting supernatant was treated for 1 h with 2 μg Cav1.2α1 antibodies (a gift from Amy Lee, University of Iowa, Iowa City, IA). Protein A/G agarose was then added for two more hours of rotating, and bead immunoprecipitate complexes were washed and pelleted. Immunoprecipitated proteins were treated with PNGase F following New England Biolabs protocols to cleave N-glycosylation sites. Briefly, 20 μg protein was denatured at 100 °C for 10 min and then cooled on ice. Nonidet P-40 and GlycoBuffer #2 were added to final concentrations of 10% and 1×, respectively. PNGase F was added at a 1:20 dilution, and the sample was then gently mixed and incubated at 37 °C for 1 h. Shift in band size as an effect of PNGase activity was measured via Western blot.
In Vitro Gene Knockdown.
HT22 mouse hippocampal (Sigma Aldrich, SCC129), C2C12 mouse myoblast (ATCC, CTRL-1772), and NIH/3T3 mouse fibroblast (ATCC, CTRL-1658) cells were grown in High Glucose DMEM supplemented with 10% Fetal Bovine Serum, 1xGlutaMax, and 1% Penicillin-Streptomycin. Cells were cultured at 37 °C with 21% oxygen and 5% carbon dioxide. Cells were reverse transfected at 50 to 70% confluency with Lipofectamine RNAiMax reagent following kit instructions and using two distinct mouse siRNA pairs (generated from Integrated DNA Technology libraries; SI Appendix, Table S1) per gene. Transfected cells were harvested 72 h after treatment by direct lysis in Laemmli’s buffer before moving to Western blotting.
Western Immunoblotting.
Protein concentrations were quantified with bicinchoninic acid (BCA) assay. Protein concentrations were standardized, and samples were denatured for 10 min in Laemmli’s buffer either at 95 °C or at 65 °C, with the lower temperature used for hydrophobic, transmembrane proteins. Proteins were separated on 10% Tris-glycine eXtended gels (BioRad) for mitochondrial purification studies and on 5% SDS-PAGE gels for PNGase assays. Proteins were then transferred with BioRad’s Trans-Blot Turbo System onto 0.45-μm low-fluorescence PVDF membranes. Membranes were blocked with 5% milk in TBS (50 mM Tris, 150 mM NaCl) for 1 h at room temperature and then incubated with primary antibodies in 5% TBS (50 mM Tris, 150 mM NaCl, 0.05% Tween-20) at 4 °C. Antibodies included Anti-Cav1.2 (Alomone Labs, AGP-001), Gapdh (Santa Cruz, sc-365062), Sdha (Cell Signaling, 11998), Tom20 (Cell Signaling, 42406), Facl4 (ThermoFisher, PA5-27137), Tom70 (Santa Cruz Biotechnology, sc-390545, a gift from Stefan Strack, University of Iowa, Iowa City, IA), Pdh E1α (Cell Signaling, 3205), Vdac (Cell Signaling, 4661), Serca2 (Cell Signaling, 4388), Erlin2 (Cell Signaling, 2959), Psd95 (Cell Signaling, 2507), β-actin (Santa Cruz, sc-47778), Eaat1 (Alomone, AGC-021), Glut3 (Millipore, AB1344), Mct1 (Alomone, AMT-011), Asic1 (biorbyt, orb13257), Kv1.3 (Alomone, APC-002), Ryr1 (Cell Signaling, 8153), mitoK (Sigma, HPA010980), Slc25a12 (Alomone, ANT-112), Slc25a22 (Proteintech, 25402-1-AP), Mcu (Sigma, HPA016480, a gift from Yuriy Usachev, University of Iowa), Mpc1 (Cell Signaling, 14462S), and Mpc2 (Cell Signaling, 46141S). Complementary antibody panels against the Nmda receptor complex (Nmdar) were selected for their unique epitopes shown in SI Appendix, Table S2.
For some antibodies as indicated in the text, preabsorption controls were run on mirror immunoblots using blocking peptides from Alomone Labs following vendor protocols. Glycosylation was detected with wheat germ agglutin (WGA) (Biotium, 29029-1), which has an affinity toward N-acetylglucosamine and sialic acid, with or without overnight PVDF membrane treatment in 55 mM NaOH overnight after transfer and before blocking, to hydrolyze O-glycosylation. Immunoreactivity was detected using fluorescently labeled secondary antibodies visualized on a Li-COR Odyssey CLx system.
Data Analysis and Statistics.
Data processing and normalization were performed in Excel. Relative immunodensity enrichment on immunoblots was quantified in ImageJ and log normalized. Relative enrichment for each individual organelle marker was determined by calculating mean deviation of each cellular compartment’s signal from the respective crude mitochondrial signal. Z scores were then calculated for each marker’s fractional distribution by dividing individual mean deviation scores by the SD of the marker in all fractions. Replicates were averaged and represented by heat map using Heatmapper (75). Statistical analysis was performed using GraphPad Prism. Normality was in respirometry, and calcium flux measurements were assessed by Shapiro–Wilk test for normal distributions. Genotype or treatment comparisons were analyzed by Student’s t test, and genotype versus treatment or time conditions were analyzed using two-way ANOVA with Bonferroni-corrected P values used for multiple comparisons. Statistical significance threshold was set to P = 0.05. Effect size was calculated with Cohen’s d. Data are represented as mean ± SEM.
Supplementary Material
Acknowledgments
We thank Adam Rauckhorst for technical training and helpful discussion. We thank Stefan Strack, Yuriy Usachev, and Aislinn Williams for providing manuscript feedback. We thank Eric Schon and Delfina Larrea for generously discussing and providing insight on MAM purification. We thank Amy Lee and Jussara Fernandes Hagen for helpful discussion on immunoprecipitation. This work was supported by NIH Grant F31 1NS106773-01 (to M.F.N.), NIH NIDDK Grant R01 DK104998 (to E.B.T.), NIH NINDS Grant R01 NS084190 (to A.A.P.), NIH RO1DA029122 (to A.M.R.), and NSF Research Fellowship Grant 1048957 (to M.F.N.). Grant P30CA086862 to George Weiner contributed to the support of core facilities utilized for this research. A.A.P. was supported by the Brockman Foundation; he also received gracious support from the Elizabeth Ring Mather and William Gwinn Mather Fund, the S. Livingston Samuel Mather Trust, the G. R. Lincoln Family Foundation, the Leonard Krieger Fund of the Cleveland Foundation, the Maxine and Lester Stoller Parkinson’s Research Fund, and Gordon and Evie Safran. We thank the University of Iowa Fraternal Order of Eagles Diabetes Research Center, the Iowa Neuroscience Institute, and the Louis Stokes VA Medical Center for resources and facilities.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019046118/-/DCSupplemental.
Data Availability
All study data are included in the article and SI Appendix.
References
- 1.Attwell D., Laughlin S. B., An energy budget for signaling in the grey matter of the brain. J. Cereb. Blood Flow Metab. 21, 1133–1145 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Hertz L., The glutamate-glutamine (GABA) cycle: Importance of late postnatal development and potential reciprocal interactions between biosynthesis and degradation. Front. Endocrinol. (Lausanne) 4, 59 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rolfe D. F., Brown G. C., Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol. Rev. 77, 731–758 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Mantegazza M., Curia G., Biagini G., Ragsdale D. S., Avoli M., Voltage-gated sodium channels as therapeutic targets in epilepsy and other neurological disorders. Lancet Neurol. 9, 413–424 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Zamponi G. W., Targeting voltage-gated calcium channels in neurological and psychiatric diseases. Nat. Rev. Drug Discov. 15, 19–34 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Palmieri F., Monné M., Discoveries, metabolic roles and diseases of mitochondrial carriers: A review. Biochim. Biophys. Acta 1863, 2362–2378 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Taylor E. B., Functional properties of the mitochondrial carrier system. Trends Cell Biol. 27, 633–644 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunningham C. N., Rutter J., 20,000 picometers under the OMM: Diving into the vastness of mitochondrial metabolite transport. EMBO Rep. 21, e50071 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pei L., Wallace D. C., Mitochondrial etiology of neuropsychiatric disorders. Biol. Psychiatry 83, 722–730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molinari F., et al., Impaired mitochondrial glutamate transport in autosomal recessive neonatal myoclonic epilepsy. Am. J. Hum. Genet. 76, 334–339 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oonthonpan L., Rauckhorst A. J., Gray L. R., Boutron A. C., Taylor E. B., Two human patient mitochondrial pyruvate carrier mutations reveal distinct molecular mechanisms of dysfunction. JCI Insight 5, e126132 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schapira A. H. V., Olanow C. W., Greenamyre J. T., Bezard E., Slowing of neurodegeneration in Parkinson’s disease and Huntington’s disease: Future therapeutic perspectives. Lancet 384, 545–555 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Whittaker V. P., Michaelson I. A., Kirkland R. J. A., The separation of synaptic vesicles from nerve-ending particles (‘synaptosomes’). Biochem. J. 90, 293–303 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kristian T., Isolation of mitochondria from the CNS. Curr. Protoc. Neurosci. 52, 7.22.21–27.22.12 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Csordás G., et al., Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 174, 915–921 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Area-Gomez E., et al., Presenilins are enriched in endoplasmic reticulum membranes associated with mitochondria. Am. J. Pathol. 175, 1810–1816 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hell J. W., et al., Identification and differential subcellular localization of the neuronal class C and class D L-type calcium channel alpha 1 subunits. J. Cell Biol. 123, 949–962 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simms B. A., Zamponi G. W., Neuronal voltage-gated calcium channels: Structure, function, and dysfunction. Neuron 82, 24–45 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Sridharan P. S., Lu Y., Rice R. C., Pieper A. A., Rajadhyaksha A. M., Loss of Cav1.2 channels impairs hippocampal theta burst stimulation-induced long-term potentiation. Channels (Austin) 14, 287–293 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee A. S., et al., The neuropsychiatric disease-associated gene Cacna1c; mediates survival of young hippocampal neurons. eneuro 3, ENEURO.0006-0016.2016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bavley C. C., et al., Dopamine D1R-neuron cacna1c deficiency: A new model of extinction therapy-resistant post-traumatic stress. Mol. Psychiatry (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabir Z. D., Martínez-Rivera A., Rajadhyaksha A. M., From gene to behavior: L-type calcium channel mechanisms underlying neuropsychiatric symptoms. Neurotherapeutics 14, 588–613 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michels S., et al., Downregulation of the psychiatric susceptibility gene Cacna1c promotes mitochondrial resilience to oxidative stress in neuronal cells. Cell Death Discov. 4, 54 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hotka M., et al., L-type Ca2+ channel-mediated Ca2+ influx adjusts neuronal mitochondrial function to physiological and pathophysiological conditions. Sci. Signal. 13, eaaw6923 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schanne F. A., Kane A. B., Young E. E., Farber J. L., Calcium dependence of toxic cell death: A final common pathway. Science 206, 700–702 (1979). [DOI] [PubMed] [Google Scholar]
- 26.Hiller S., et al., Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 321, 1206–1210 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lazniewska J., Weiss N., Glycosylation of voltage-gated calcium channels in health and disease. Biochim. Biophys. Acta Biomembr. 1859, 662–668 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Gupta R., Brunak S., Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput., 310–322 (2002). [PubMed] [Google Scholar]
- 29.Korde A. S., Maragos W. F., Identification of an N-methyl-D-aspartate receptor in isolated nervous system mitochondria. J. Biol. Chem. 287, 35192–35200 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaul M., Garden G. A., Lipton S. A., Pathways to neuronal injury and apoptosis in HIV-associated dementia. Nature 410, 988–994 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Beutner G., et al., Type 1 ryanodine receptor in cardiac mitochondria: Transducer of excitation-metabolism coupling. Biochim. Biophys. Acta 1717, 1–10 (2005). [DOI] [PubMed] [Google Scholar]
- 32.McPherson P. S., et al., The brain ryanodine receptor: A caffeine-sensitive calcium release channel. Neuron 7, 17–25 (1991). [DOI] [PubMed] [Google Scholar]
- 33.Jakob R., et al., Molecular and functional identification of a mitochondrial ryanodine receptor in neurons. Neurosci. Lett. 575, 7–12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu J., et al., Mitochondrial calcium uniporter Mcu controls excitotoxicity and is transcriptionally repressed by neuroprotective nuclear calcium signals. Nat. Commun. 4, 2034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simpson I. A., Chundu K. R., Davies-Hill T., Honer W. G., Davies P., Decreased concentrations of GLUT1 and GLUT3 glucose transporters in the brains of patients with Alzheimer’s disease. Ann. Neurol. 35, 546–551 (1994). [DOI] [PubMed] [Google Scholar]
- 36.Liu Y., Liu F., Iqbal K., Grundke-Iqbal I., Gong C.-X., Decreased glucose transporters correlate to abnormal hyperphosphorylation of tau in Alzheimer disease. FEBS Lett. 582, 359–364 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarze A., et al., GAPDH, a novel regulator of the pro-apoptotic mitochondrial membrane permeabilization. Oncogene 26, 2606–2620 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Hara M. R., et al., S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 7, 665–674 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Bricker D. K., et al., A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science 337, 96–100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herzig S., et al., Identification and functional expression of the mitochondrial pyruvate carrier. Science 337, 93–96 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Hashimoto T., Hussien R., Cho H.-S., Kaufer D., Brooks G. A., Evidence for the mitochondrial lactate oxidation complex in rat neurons: Demonstration of an essential component of brain lactate shuttles. PLoS One 3, e2915 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fünfschilling U., et al., Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature 485, 517–521 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paggio A., et al., Identification of an ATP-sensitive potassium channel in mitochondria. Nature 572, 609–613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inoue I., Nagase H., Kishi K., Higuti T., ATP-sensitive K+ channel in the mitochondrial inner membrane. Nature 352, 244–247 (1991). [DOI] [PubMed] [Google Scholar]
- 45.Paucek P., et al., Reconstitution and partial purification of the glibenclamide-sensitive, ATP-dependent K+ channel from rat liver and beef heart mitochondria. J. Biol. Chem. 267, 26062–26069 (1992). [PubMed] [Google Scholar]
- 46.Bajgar R., Seetharaman S., Kowaltowski A. J., Garlid K. D., Paucek P., Identification and properties of a novel intracellular (mitochondrial) ATP-sensitive potassium channel in brain. J. Biol. Chem. 276, 33369–33374 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Calvo S. E., Clauser K. R., Mootha V. K., MitoCarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44, D1251–D1257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shank R. P., Bennett G. S., Freytag S. O., Campbell G. L., Pyruvate carboxylase: An astrocyte-specific enzyme implicated in the replenishment of amino acid neurotransmitter pools. Brain Res. 329, 364–367 (1985). [DOI] [PubMed] [Google Scholar]
- 49.Ralphe J. C., Segar J. L., Schutte B. C., Scholz T. D., Localization and function of the brain excitatory amino acid transporter type 1 in cardiac mitochondria. J. Mol. Cell. Cardiol. 37, 33–41 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Haziza S., et al., Fluorescent nanodiamond tracking reveals intraneuronal transport abnormalities induced by brain-disease-related genetic risk factors. Nat. Nanotechnol. 12, 322–328 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Wang Y. Z., et al., Intracellular ASIC1a regulates mitochondrial permeability transition-dependent neuronal death. Cell Death Differ. 20, 1359–1369 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wemmie J. A., et al., The acid-activated ion channel ASIC contributes to synaptic plasticity, learning, and memory. Neuron 34, 463–477 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Xiong Z.-G., et al., Neuroprotection in ischemia: Blocking calcium-permeable acid-sensing ion channels. Cell 118, 687–698 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Szabò I., et al., A novel potassium channel in lymphocyte mitochondria. J. Biol. Chem. 280, 12790–12798 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Fadool D. A., et al., Kv1.3 channel gene-targeted deletion produces “Super-Smeller Mice” with altered glomeruli, interacting scaffolding proteins, and biophysics. Neuron 41, 389–404 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ashburner M.et al.; The Gene Ontology Consortium , Gene ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.The Gene Ontology Consortium , The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 47, D330–D338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pagliarini D. J., et al., A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.UniProt Consortium , UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brand M. D., Nicholls D. G., Assessing mitochondrial dysfunction in cells. Biochem. J. 435, 297–312 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lobysheva N. V., et al., Glutamate induces H2O2 synthesis in nonsynaptic brain mitochondria. Free Radic. Biol. Med. 65, 428–435 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Shevtsova E. F., et al., (2017) Mitochondrial permeability transition pore as a suitable target for neuroprotective agents against Alzheimer's disease. CNS Neurol. Disord. Drug Targets 16, 677–685. [DOI] [PubMed] [Google Scholar]
- 63.Fecher C., et al., Cell-type-specific profiling of brain mitochondria reveals functional and molecular diversity. Nat. Neurosci. 22, 1731–1742 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Szabo I., Zoratti M., Mitochondrial channels: Ion fluxes and more. Physiol. Rev. 94, 519–608 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Brooks G. A., Cell-cell and intracellular lactate shuttles. J. Physiol. 587, 5591–5600 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Verkhratsky A., Nedergaard M., Physiology of Astroglia. Physiol. Rev. 98, 239–389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stastna M., Van Eyk J. E., Analysis of protein isoforms: Can we do it better? Proteomics 12, 2937–2948 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stroud D. A., et al., Composition and topology of the endoplasmic reticulum-mitochondria encounter structure. J. Mol. Biol. 413, 743–750 (2011). [DOI] [PubMed] [Google Scholar]
- 69.Kornmann B., et al., An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477–481 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pan X., et al., The physiological role of mitochondrial calcium revealed by mice lacking the mitochondrial calcium uniporter. Nat. Cell Biol. 15, 1464–1472 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brookes P. S., Yoon Y., Robotham J. L., Anders M. W., Sheu S.-S., Calcium, ATP, and ROS: A mitochondrial love-hate triangle. Am. J. Physiol. Cell Physiol. 287, C817–C833 (2004). [DOI] [PubMed] [Google Scholar]
- 72.Bayraktar E. C., et al., MITO-Tag Mice enable rapid isolation and multimodal profiling of mitochondria from specific cell types in vivo. Proc. Natl. Acad. Sci. U.S.A. 116, 303–312 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moosmang S., et al., Role of hippocampal Cav1.2 Ca2+ channels in NMDA receptor-independent synaptic plasticity and spatial memory. J. Neurosci. 25, 9883–9892 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vance J. E., Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265, 7248–7256 (1990). [PubMed] [Google Scholar]
- 75.Babicki S., et al., Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 44, W147–W153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.