Significance
The potassium channel Kv1.3 in brain myeloid cells represents a promising therapeutic target for Alzheimer’s disease (AD). The patterns of expression and functional roles of Kv1.3 channels in brain myeloid subpopulations and the microglial-versus-peripheral myeloid origin of Kv1.3-expressing cells in AD remain unclear. Here, we show that in mice, Kv1.3 is selectively up-regulated in an Aβ-dependent manner by a subset of microglia-derived cells, expresses higher levels of proinflammatory genes, and confirms the presence of Kv1.3-expressing microglial subpopulations in human AD. Furthermore, blocking Kv1.3 in an AD model reduces Aβ neuropathology, increases synaptic protein expression, and skews the microglial transcriptome toward pro-phagocytic and protective phenotypes. Our findings strengthen the preclinical rationale for targeting microglial Kv1.3 channels for therapeutic immunomodulation in AD.
Keywords: microglia, Alzheimer’s disease, neurodegeneration, neuroinflammation, potassium channel
Abstract
Kv1.3 potassium channels, expressed by proinflammatory central nervous system mononuclear phagocytes (CNS-MPs), are promising therapeutic targets for modulating neuroinflammation in Alzheimer’s disease (AD). The molecular characteristics of Kv1.3-high CNS-MPs and their cellular origin from microglia or CNS-infiltrating monocytes are unclear. While Kv1.3 blockade reduces amyloid beta (Aβ) burden in mouse models, the downstream immune effects on molecular profiles of CNS-MPs remain unknown. We show that functional Kv1.3 channels are selectively expressed by a subset of CD11b+CD45+ CNS-MPs acutely isolated from an Aβ mouse model (5xFAD) as well as fresh postmortem human AD brain. Transcriptomic profiling of purified CD11b+Kv1.3+ CNS-MPs, CD11b+CD45int Kv1.3neg microglia, and peripheral monocytes from 5xFAD mice revealed that Kv1.3-high CNS-MPs highly express canonical microglial markers (Tmem119, P2ry12) and are distinct from peripheral Ly6chigh/Ly6clow monocytes. Unlike homeostatic microglia, Kv1.3-high CNS-MPs express relatively lower levels of homeostatic genes, higher levels of CD11c, and increased levels of glutamatergic transcripts, potentially representing phagocytic uptake of neuronal elements. Using irradiation bone marrow CD45.1/CD45.2 chimerism in 5xFAD mice, we show that Kv1.3+ CNS-MPs originate from microglia and not blood-derived monocytes. We show that Kv1.3 channels regulate membrane potential and early signaling events in microglia. Finally, in vivo blockade of Kv1.3 channels in 5xFAD mice by ShK-223 reduced Aβ burden, increased CD11c+ CNS-MPs, and expression of phagocytic genes while suppressing proinflammatory genes (IL1b). Our results confirm the microglial origin and identify unique molecular features of Kv1.3-expressing CNS-MPs. In addition, we provide evidence for CNS immunomodulation by Kv1.3 blockers in AD mouse models resulting in a prophagocytic phenotype.
Myeloid cells of the central nervous system (CNS) are composed of resident microglia and a smaller proportion of peripherally derived monocytes/macrophages that enter the CNS (1–4). These two distinct cell types are collectively referred to as CNS mononuclear phagocytes (CNS-MPs). CNS-MPs mediate immune responses (e.g., neuroinflammation), which play critical and causative roles in neurodegenerative disorders such as Alzheimer’s disease (AD). Nearly 30% of genetic risk factors of late-onset AD are immune genes that are expressed by CNS-MPs (5–8). Furthermore, modulation of immune genes such as Trem2 and Apoe in preclinical mouse models of AD pathology have confirmed the complex and seemingly opposing roles CNS-MPs play in neurodegeneration (9–11). These include proinflammatory and neurotoxic responses which are counterbalanced by antiinflammatory, prophagocytic, and neuroprotective responses (8, 12–14). The detrimental or protective effects of CNS-MP–mediated neuroinflammatory mechanisms are also highly context dependent and may be influenced by age and stage of neurodegeneration (3, 4, 15–18).
Bulk brain tissue and single-cell transcriptomic studies have provided molecular insights into microglial diversity and their role in AD pathogenesis (19, 20). These studies have consistently shown that homeostatic microglia seen in normal brain gradually transform to disease-/damage-associated microglia (DAM), which are characterized by high expression of Apoe, Axl, Cst7, Clec7a, and several other core DAM genes (19, 21). The transition from homeostatic to DAM appears to be partly dependent on Trem2 and Apoe and lies downstream of amyloid beta (Aβ) accumulation (22). DAM are typically found surrounding Aβ plaques, characterized by lipid inclusions and phagocytosed Aβ and typified by large cell bodies and retracted processes (19, 23). Whether DAM are protective, detrimental, or a dysfunctional group of brain myeloid cells is unclear; although, we have previously suggested the existence of proinflammatory and antiinflammatory DAM subprofiles (2).
While the majority of DAM are likely to originate from microglia, some contribution from the peripheral pool of blood-derived monocytes and macrophages is still a possibility. Within DAM, we recently identified a distinct proinflammatory subset of CNS-MPs that express genes such as Hif1α, Tlr2, and Il1b as well as the gene Kcna3, which encodes the Kv1.3 potassium channel (2). Elevated Kv1.3 channel expression has been confirmed in human postmortem AD brain by immunohistochemistry as well as by electrophysiological methods in acutely isolated CNS-MPs from mouse models of AD pathology (24, 25). Kv1.3 is a voltage-gated K+ channel that was first described in human T cells and suggested as a target for immunosuppression in 1984 (26). Following T cell receptor engagement, Kv1.3 provides the counterbalancing K+ efflux that is necessary to sustain calcium entry through calcium release-activated channels and to initiate downstream signaling and cytokine production (27, 28). Thus, while Kv1.3 was known to regulate membrane potential and calcium signaling in peripheral lymphocytes, for decades, its role in CNS-MPs was less clear. Pharmacologic inhibition of Kv1.3 channels is protective in mouse models of AD pathology by limiting Aβ accumulation potentially via increased clearance (2, 13, 25). In ischemic stroke mouse models, Kv1.3 channel expression by CNS-MPs increases by day 3 poststroke and inhibition of Kv1.3 channels limits infarct size (29, 30). Overall, inhibition of Kv1.3 channels represents a promising therapeutic strategy to specifically inhibit proinflammatory neuroinflammatory responses (25, 31).
The presence of Kv1.3 on freshly isolated CNS-MPs has currently only been demonstrated in mouse models of Aβ deposition (2, 25); however, the presence of Kv1.3-high CNS-MPs in models of tau pathology or, more importantly, in human AD brain has not been demonstrated. The distinct molecular features of Kv1.3-high CNS-MPs are also currently unknown because comparative molecular profiling of these cells has not been undertaken. It also remains unknown whether Kv1.3-high CNS-MPs in AD are derived from microglia or from peripheral monocytes/macrophages that enter the CNS. Addressing the cellular origin of Kv1.3-high CNS-MPs in AD is critical to better design and deliver Kv1.3 blockers to maximize treatment efficacy. Lastly, the ability of Kv1.3 channel blockers to skew neuroinflammatory responses and microglial profiles in mouse AD models is unknown.
In this study, we used flow cytometry to characterize Kv1.3 channel expression patterns in CNS-MPs isolated from Aβ and tau models of AD pathology, experimental allergic encephalomyelitis (EAE) models, and postmortem human AD brains. In AD models, we also undertook parallel transcriptomic profiling studies to contrast Kv1.3-high CNS-MPs with Kv1.3-negative (neg) microglia as well as with peripheral blood monocyte subsets. We used the irradiation bone marrow CD45.1/CD45.2 chimerism model (32) in an Aβ mouse model (5xFAD) to confirm whether Kv1.3-high CNS-MPs observed in 5xFAD brain originate from peripheral myeloid or from the brain-resident microglial compartment. Electrophysiological and signaling studies determined whether Kv1.3 channel function in microglia is coupled with regulation of membrane potential and early immune signaling events following activation. Lastly, we compared the transcriptomic profiles of microglia after long-term treatment of 5xFAD mice with a systemically delivered Kv1.3 blocker to investigate pharmacological targeting of Kv1.3 as a strategy to modulate neuroinflammation in AD.
Results
Kv1.3-High CNS-MPs Represent a Subset of CD11b+ CD45+ Myeloid Cells in Mouse AD Models as well as in Postmortem Human Brain.
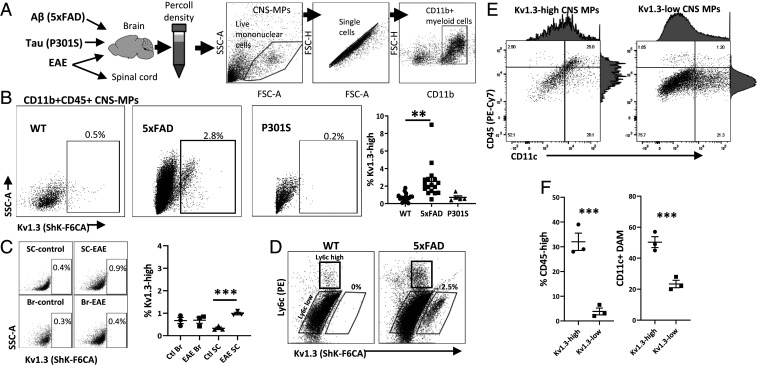
We recently found that voltage-gated Kv1.3 channels are expressed by CNS-MPs in human AD brain and in mouse models of AD pathology where they are coexpressed with proinflammatory genes such as Il1b, Cox2, and Hif1α (2). Furthermore, inhibition of Kv1.3 channels in AD models limits Aβ pathology (2, 25). We applied a validated flow cytometric assay of functional Kv1.3 channels expressed on the cell surface to rapidly phenotype acutely isolated CNS-MPs (Kv1.3-high CNS-MPs) from wild-type (WT), Aβ (5xFAD model), tau (P301S model), and EAE mouse brains (Fig. 1A) (2, 13, 33). We found that Kv1.3 channels are detectable in only ∼0.5 to 1% of WT CNS-MPs but are expressed in 3 to 5% of 5xFAD CNS-MPs (Fig. 1 B and C). In the P301S model, there were ∼0.2% of Kv1.3-high CNS-MPs isolated from 10-mo-old mice with established brain tau pathology (Fig. 1B) (34). Similarly, in the EAE mouse model, we observed minimal (∼0.4%) Kv1.3-high CNS-MPs in the brain and spinal cord as compared to WT mice, although a marginal increase in Kv1.3-high CNS-MPs was observed in EAE spinal cord (Fig. 1C). In 5xFAD brain, Kv1.3-high CNS-MPs were expressed (2.5%) specifically by a subset of CD45highLy6clow CNS-MPs (Fig. 1D). CD45highLy6chigh CNS-MPs, which represent inflammatory CNS-infiltrating monocytes/macrophages, did not express Kv1.3 channels detectable by flow cytometry (Fig. 1D). Within CD11b+CD45+ CNS-MPs in 5xFAD mice, a greater proportion of Kv1.3-high CNS-MPs were CD45high and CD11c+ (25%) as compared to Kv1.3-low CNS-MPs (Fig. 1 E and F), suggesting that Kv1.3-high CNS-MPs have greater similarities to DAM. Immunohistochemistry also confirmed the presence of Kv1.3 channel–positive CNS-MPs with activated morphology exclusively in 5xFAD brains as compared to their WT littermates (SI Appendix, Fig. S1). In contrast to CNS-MPs, peripheral splenic and blood monocytes (Ly6chigh or Ly6clow) did not express any measurable Kv1.3 channels in WT or 5xFAD mice (SI Appendix, Fig. S2). Overall, these mouse flow cytometry and immunohistochemistry studies show that Kv1.3-high expression is specific to CD11b+CD45highLy6clow CNS-MPs appears to be unique to the CNS with Aβ pathology. Kv1.3-high CNS-MPs may either represent Ly6clow microglia that up-regulate CD45 or CNS-infiltrating CD45high monocytes that down-regulate Ly6c. The fact that peripheral monocytes and resting microglia in WT mice do not express detectable Kv1.3 channel levels also indicates that unique factors in the CNS microenvironment of neurodegenerative disease may be responsible for Kv1.3 up-regulation.
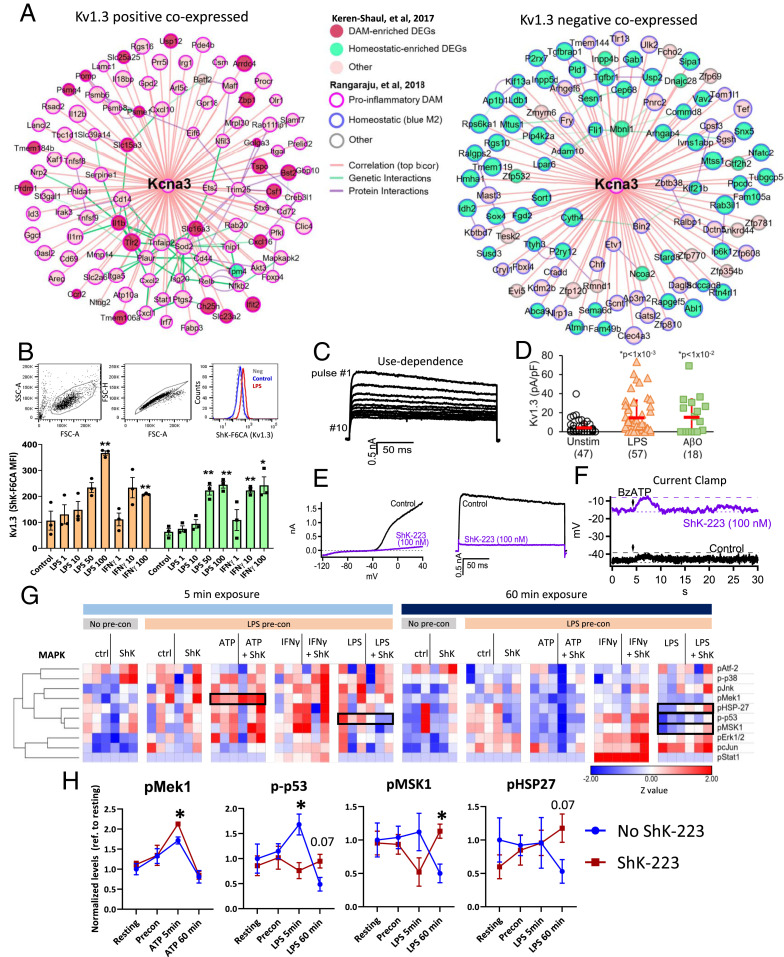
Fig. 1.
Kv1.3 channels are highly expressed in CNS-MPs in a mouse model of Aβ pathology. (A) Schematic of mouse models used to isolate CNS-MPs. This demonstrates a representative gating strategy for CNS-MPs. (B) Kv1.3 channel expression in CD11b+CD45+ CNS-MPs in WT (n = 8, age 6 to 8 mo), 5xFAD (n = 14, age 6 to 8 mo), and P301S (n = 4, age 10 mo). Quantification is shown on the right. (C) Kv1.3 channel expression in CD11b+CD45+ CNS-MPs from EAE brain (Br, n = 3) and spinal cord (SC, n = 3). Quantification is shown on the right. (D) WT and 5xFAD CD45highLy6chigh CNS-MPs (inflammatory CNS-infiltrating monocytes) did not express detectable Kv1.3 channels, while only 5xFAD CD45highLy6clow CNS-MPs expressed Kv1.3. (E and F) Within CD11b+CD45+ CNS-MPs in 5xFAD mice, a greater proportion of Kv1.3-high CNS-MPs were CD45high and CD11c+ (**P < 0.01, ***P < 0.001).
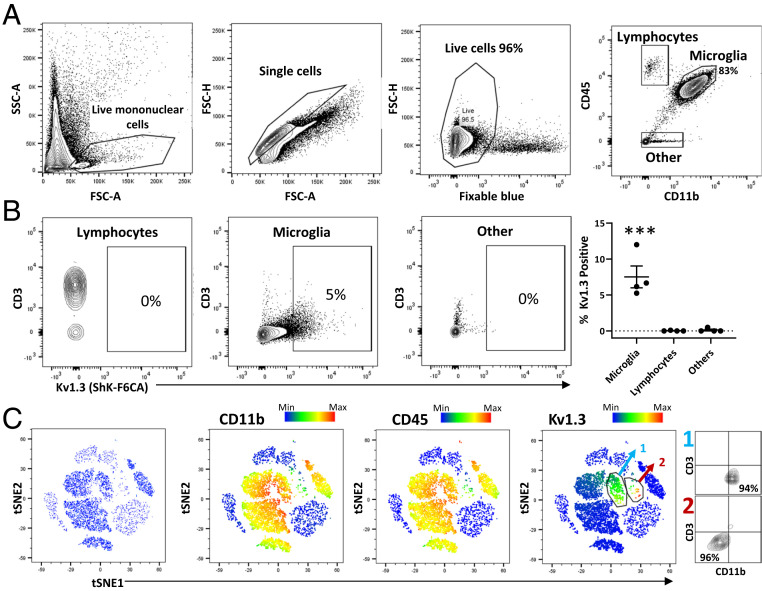
Previous studies have shown the presence of Kv1.3-high CNS-MPs in postmortem human brain by immunohistochemistry (24, 25); however, proof of Kv1.3 channel expression by flow cytometry is lacking due to limited access to fresh, unfrozen postmortem human brain tissue. Therefore, we acutely isolated CNS-MPs from fresh (3 to 12 h postmortem interval) postmortem human frontal cortex (gray matter) from four individuals: three cases with AD pathology and a fourth case with frontotemporal dementia with TDP-43 pathology (SI Appendix, Table S1). Viability of CNS-MPs was >95% in all samples (Fig. 2A). Within CD11b+CD45+ CNS-MPs, Kv1.3 channel expression was observed in ∼5 to 7% of cells (Fig. 2B, Microglia) while almost no Kv1.3 expression was detected in CNS lymphocytes (CD11b−CD45highCD3+ or CD3neg) or cells that were CD11bneg and CD45neg (Fig. 2B, P < 0.001). Using t-distributed stochastic neighbor embedding (tSNE) to visualize our data, we confirmed that the majority of Kv1.3-high cells were CD11b+CD45+ CNS-MPs, although a very small group of CD11bnegCD45neg CD3neg cells that highly expressed Kv1.3 were also detected in human brain (Fig. 2C). Although we did not have a nondisease human brain sample for comparison, our findings of Kv1.3 channel expression by subsets of human CNS-MPs closely mimic the observation in AD mouse models.
Fig. 2.
Kv1.3 channel expression by CNS-MPs in human neurodegeneration. (A) Flow cytometric gating strategy of acutely isolated brain mononuclear cells from fresh, unfrozen postmortem human frontal cortex gray matter (n = 4 cases with either AD or frontotemporal degeneration; see SI Appendix, Table S1). Within live single mononuclear cells, there were >80% CD11b+CD45intermediate microglia, small proportions of CD11bnegCD45high lymphocytes, and CD11bnegCD45neg other cell types. (B) Measurement of functional Kv1.3 expression using ShK-F6CA, comparing lymphocytes (predominantly CD3+ T cells), microglia, and other unlabeled cell populations in acutely isolated mononuclear cells (***P < 0.001). (C) tSNE analysis of flow cytometry data based on CD11b, CD45, CD3, Kv1.3, CD66b, and CD146 expression shows a dominant myeloid population. Kv1.3 expression was limited mostly to CD11b+CD45+ microglia (labeled population 1, light blue), while a small subset of Kv1.3-high cells of unknown lineage (labeled population 2, red) were also identified. Color scale bar represents relative expression (blue: low; red: high).
Kv1.3-High CNS-MPs in AD Models Closely Resemble CNS-Resident Microglia rather than Peripheral Monocytes, although with Unique Transcriptomic Features.
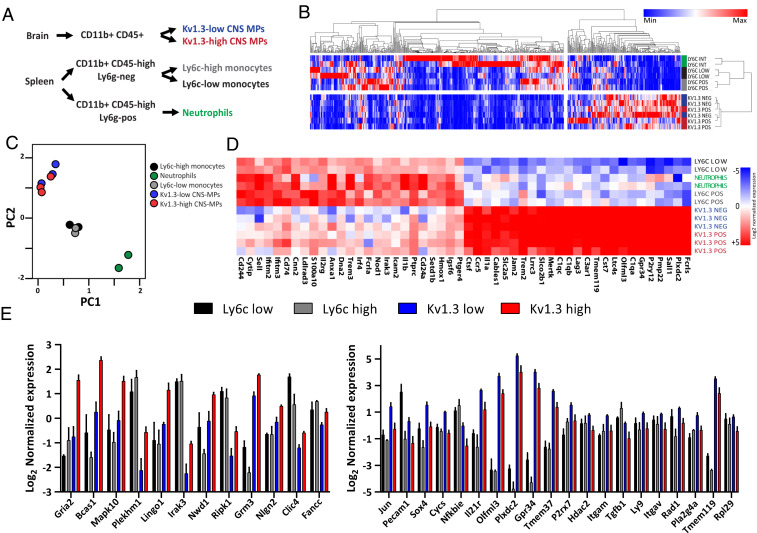
Since flow cytometric immunophenotyping cannot definitively confirm the microglial or monocyte-derived origin of Kv1.3-high CNS-MPs in AD models, we undertook transcriptomic phenotyping of Kv1.3-high and Kv1.3-low CNS-MPs and compared these to peripheral Ly6chigh and Ly6clow monocytes from 7 to 8 mo old 5xFAD mice in which Kv1.3 channel expression is apparent (Fig. 3A). We sorted ∼3,000 Kv1.3-high and 15,000 Kv1.3-low CNS-MPs per mouse brain as well as 5,000 Ly6chigh splenic monocytes, 4,000 Ly6clow splenic monocytes, and Ly6g+Ly6cintermediate neutrophils from 5xFAD mice. We preamplified using equal amount of RNA per sample and performed neuroinflammatory transcriptomic profiling by NanoString (770 neuroinflammatory gene panel) as described earlier (35). Of 770 genes, 617 were included for analysis (Dataset S1), and hierarchical clustering analysis showed that both CNS-MP populations (Kv1.3-high and Kv1.3-neg) were markedly different from peripheral monocytes, while the segregation between Kv1.3-high and Kv1.3-neg and between Ly6chigh and Ly6clow monocytes was less apparent (Fig. 3B). Principal component analysis (PCA) of expression data showed that nearly 65% of variance in the data were explained by two principal components (PCs), of which PC1 explained 43% while PC2 explained 22% of variance (Fig. 3C). PCA showed three distinct cell clusters: 1) CNS-MPs (Kv1.3-high and Kv1.3-neg), 2) peripheral splenocytes (Ly6chigh and Ly6clow), and 3) neutrophils (Fig. 3C). Kv1.3-neg CNS-MPs, which are primarily CD11b+CD45intLy6clow microglia, and Kv1.3-high CNS-MPs shared high-level expression of several canonical microglial genes, such as P2yr12, Tmem119, and Olfml3, indicating microglia-like molecular features (Fig. 3D). Kv1.3-high and Kv1.3-low CNS-MPs also highly expressed genes such as Trem2 and Cst7, indicative of DAM in 5xFAD mice (Fig. 3D). Overall, these unbiased analyses show that Kv1.3-high CNS-MPs resemble microglia and may originate from CNS-resident microglia rather than peripheral monocytes. However, it also remains possible that peripheral monocytes that take up residence in the CNS over a long period of time adopt unique CNS-specific profiles, making them transcriptomically indistinguishable from microglia.
Fig. 3.
Kv1.3-high CNS-MPs resemble CNS-resident microglia rather than peripheral monocytes in the 5xFAD model. (A) Experimental workflow for NanoString transcriptomic phenotyping of Kv1.3-high and Kv1.3-low CNS-MPs, peripheral Ly6chigh and Ly6clow monocytes, and neutrophils isolated from 5xFAD mice (n = 3, age 7 to 8 mo, females). (B) Hierarchical clustering differentiated peripheral Ly6chigh and Ly6clow monocytes from Kv1.3-high and Kv1.3-low CNS-MPs, although differences within the monocyte and CNS-MPs populations were less apparent. (C) PCA revealed a clear separation of three cell populations: CNS-MPs (Kv1.3-high and -low), peripheral splenocytes (Ly6c-high and -low), and neutrophils. (D) Heatmap displaying the transcriptomic differences between peripheral Ly6chigh and Ly6clow monocytes, neutrophils, and Kv1.3-high and Kv1.3-low CNS-MPs. Color scale bar represents low (blue) to high (red) log2-transformed normalized gene expression. (E) Log2-transformed normalized expression of top 20 down-regulated genes in Kv1.3-high CNS-MPs and 12 up-regulated genes in Kv1.3-high CNS-MPs.
Despite the molecular similarities between Kv1.3-high CNS-MPs and Kv1.3-neg microglia, we also identified unique molecular signatures of Kv1.3-high CNS-MPs in 5xFAD mice (48 differentially expressed genes). Differential expression analysis showed higher expression of 12 genes and lower expression of 36 genes in Kv1.3-high CNS-MPs as compared to Kv1.3-low microglia (Fig. 3E and SI Appendix, Fig. S3A). Genes up-regulated in Kv1.3-high CNS-MPs included Gria2 (GluA2), Bcas1, Mapk10 (Jnk3), Plekhm1, Lingo1, Irak3, and Clic4. Interestingly, Clic4 was previously reported to be highly coexpressed with the Kv1.3 gene in proinflammatory DAM in AD models, supporting the overall validity of our findings (36). Gene set enrichment analysis (GSEA) (Dataset S2) showed enrichment of glutamatergic synaptic transmission, serine/threonine protein kinase signaling, and ATP binding (SI Appendix, Fig. S3C) in Kv1.3-high CNS-MPs. Genes with lower expression in Kv1.3-high CNS-MPs as compared to Kv1.3-low CNS-MPs included Jun, Pecam1, Sox4, and Cycs as well as known homeostatic microglial genes Olfml3, P2rx7, Itgam (CD11b), and Tmem119. GSEA showed down-regulation of transmembrane transporter activity, cell development, mitochondrial outer membrane proteins, and ribosomal protein-encoding genes in Kv1.3-high CNS-MPs as compared to Kv1.3-low CNS-MPs (SI Appendix, Fig. S3D).
We contrasted transcriptomic signatures of Kv1.3-high and Kv1.3-neg CNS-MPs with homeostatic and DAM gene signatures identified by previous single-cell RNA sequencing (RNAseq) studies (19). Genes highly expressed in Kv1.3-low CNS-MPs were selectively enriched for homeostatic microglial markers (SI Appendix, Fig. S3E). We also contrasted our results with microglial transcriptomic coexpression modules that we previously identified (2). In this coexpression network analysis of microglial transcriptomes, 19 distinct modules of genes were identified, which were then classified as those associated with AD pathology (homeostatic, proinflammatory DAM, and antiinflammatory DAM) and those not associated with AD pathology. We found that Kv1.3-low CNS-MPs were enriched for homeostatic genes while Kv1.3-high CNS-MPs appeared to be enriched for proinflammatory DAM genes (SI Appendix, Fig. S3F). Accordingly, several genes up-regulated in Kv1.3-high CNS-MPs are also key members of the proinflammatory DAM module, including Kcna3 (Kv1.3), Clic4, and Irak3.
Of the genes uniquely up-regulated in Kv1.3-high CNS-MPs, we characterized the expression of Gria2 (GluA2) in WT and 5xFAD mouse brains using immunofluorescence staining. In WT mouse brain, Gria2 protein was expressed primarily by neurons, both in the cell body as well as axons, but not by microglia (SI Appendix, Fig. S4A). Gria2 expression in 5xFAD brain was increased specifically in swollen axons in the white matter (SI Appendix, Fig. S4A, arrows) and minimal in plaque-associated Iba1+-activated microglia (SI Appendix, Fig. S4B, arrows). In human AD postmortem brain, compared to control brain, we observed increased Gria2 immunoreactivity in axons compared to control brain and minimal immunoreactivity in glia (SI Appendix, Fig. S4C). We stained 5xFAD mouse brains for Vglut1, another glutamatergic target, and observed Vglut1 immunofluorescence at proximity to Iba1+ microglia and axons (SI Appendix, Fig. S4D), similar to Gria2 staining patterns. This suggests that the higher expression of low-level glutamatergic transcripts in Kv1.3-high CNS-MPs may reflect phagocytic uptake of neuronal elements (37, 38) rather than proteins translated within CNS-MPs.
Overall, Kv1.3-high CNS-MPs, despite their similarities to Kv1.3-low microglia, also significantly differed from Kv1.3-low microglia due to increased representation of glutamatergic and proinflammatory DAM transcripts and decreased expression of homeostatic microglial genes.
Confirmation of Microglial Origin of Kv1.3-Positive CNS Myeloid Cells in Mouse AD Models.
Our transcriptomic findings show that Kv1.3-high CNS-MPs more closely resemble microglia than monocytes. CD45 and Ly6c expression is insufficient to assign microglia or monocytic origin of CNS-MPs in the brain. Transcriptomic similarities between microglia and Kv1.3-high CNS-MPs also do not definitely confirm cellular origin, as residence of peripherally derived cells in the brain itself could possibly induce a microglia-like phenotype in this population. Thus, we created bone marrow chimeras using total body irradiation and transfer of bone marrow from WT and 5xFAD mice into the reciprocal mouse strains, leveraging the resistance of microglia but not peripheral myeloid cells to myeloablative doses of irradiation (1, 39, 40).
First, we irradiated 3- to 4-mo-old WT and 5xFAD mice (n = 5/group) and then euthanized the mice 5 d after irradiation. We observed equal weight loss in WT and 5xFAD mice and no mortality prior to euthanasia. Peripheral blood monocytes and splenocytes (lymphocytes, neutrophils, and monocytes) were decreased >95% in irradiated WT and 5xFAD mice compared to nonirradiated mice (SI Appendix, Fig. S5 A and B). In the brain, mononuclear cells were unchanged between all animals (SI Appendix, Fig. S5 C, Top). Importantly, there were equal proportions of CD11b+CD45int microglia and an observable decrease in CD11b+CD45high infiltrating macrophages as well as CD45int lymphocytes in irradiated WT and 5xFAD mice compared to nonirradiated mice (SI Appendix, Fig. S5 C, Bottom). These studies confirmed comparable radioresistance of microglia and equal susceptibility of peripheral leukocytes to irradiation in both WT and 5xFAD mice.
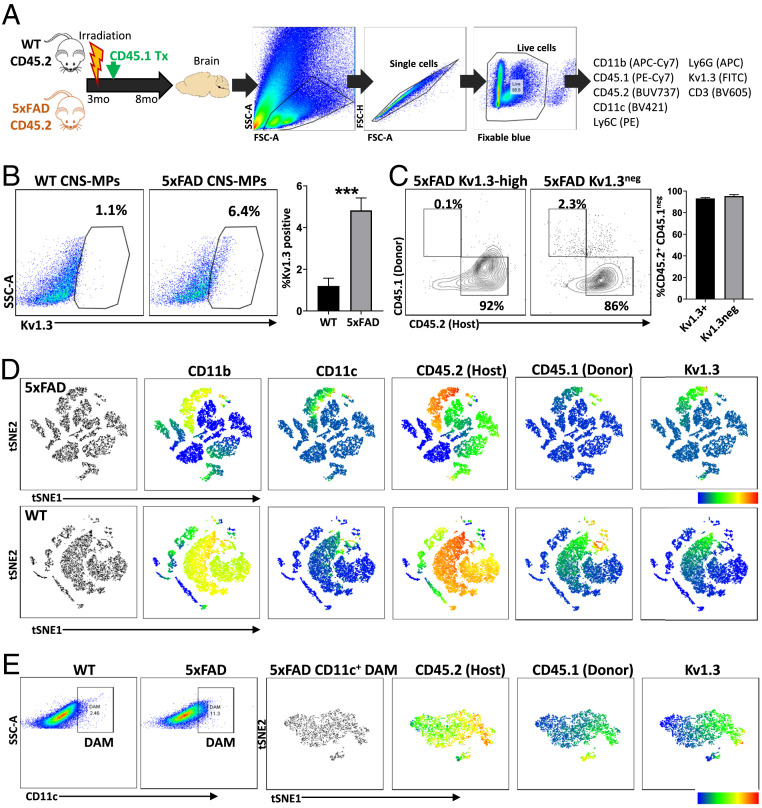
Next, 3-mo-old CD45.2+ WT and 5xFAD mice received total body irradiation, and the hematopoietic system was reconstituted with CD45.1+ WT donor bone marrow cells (Fig. 4A). At day 30 posttransplant, we confirmed >95% of blood monocytes, and blood lymphocytes were CD45.1+ while only a small fraction of blood monocytes and lymphocytes expressed CD45.2 (SI Appendix, Fig. S6A). We maintained these mice till 8 mo of age, at which time we expect Kv1.3 CNS-MPs to appear in the brain myeloid compartment in 5xFAD mice. At 8 mo, the majority of splenic monocytes and lymphocytes were CD45.1 positive, indicating stable maintenance of peripheral immune chimerism (SI Appendix, Fig. S6B). In the brain, majority (95%) of CNS-MPs were CD45.2 positive (host-derived) (Fig. 4A). Like nonirradiated mice, we observed that 4 to 5% CD11b+CD45int CNS-MPs were Kv1.3 high in irradiated, chimeric 5xFAD mice while <1% of CNS-MPs in chimeric WT mice were Kv1.3 high (Fig. 4B, P < 0.005). The burden of Aβ pathology and plaque-associated microglial response were equal in both irradiated/chimeric and nonirradiated age-matched 5xFAD mice (SI Appendix, Fig. S7). In the chimeric 5xFAD mice, over 95% Kv1.3-high CNS-MPs were CD45.2 positive (host/microglia) while only 3 to 4% of Kv1.3-high CNS-MPs expressed CD45.1 (donor/peripheral monocytes), confirming that Kv1.3-high CNS-MPs are primarily of microglial origin (Fig. 4C).
Fig. 4.
Confirmation of microglial origin of Kv1.3-positive CNS-MPs in 5xFAD mouse model. (A) Experimental workflow of irradiation bone marrow chimerism and flow cytometry experiments in WT (n = 5) and 5xFAD (n = 5) mice. The 3-mo-old CD45.2+ WT and 5xFAD mice received whole-body irradiation and bone marrow transplant with CD45.1+ donor cells. At 8 mo of age, CNS-MPs isolated from brain were analyzed by flow cytometry (antibody panel: CD11b, CD45.1, CD45.2, CD11c, CD3, Ly6c, live/dead indicator, and fluorescent Kv1.3 channel blocker, ShkF6CA). (B) Kv1.3 channel expression in CNS-MPs was significantly higher in 5xFAD than WT mice (***P < 0.001). (C) Expression of CD45.2 and CD45.1 in Kv1.3-positive and -negative CNS-MPs confirmed microglial origin Kv1.3-positive CNS-MPs. (D) tSNE-based visualization of flow cytometry data of CNS-MPs from 5xFAD and WT mouse brain postirradiation and bone marrow transplantation confirm microglial origin of Kv1.3-positive CNS-MPs. (E) Within CD11c+ DAM from CD45.1/CD45.2 chimeric 5xFAD mice, tSNE shows Kv1.3 expression primarily by CD45.2+ microglia but not CD45.1 CNS-MPs.
Using unbiased tSNE-based visualization of data (Fig. 4D), we observed almost no Kv1.3 expression in lymphocytic and pan-negative CNS mononuclear cells, indicating that in 5xFAD adult mouse brain, nearly the entire pool of Kv1.3-positive CNS-MPs is a subset of microglia (Fig. 4D). We also observed increased CD11c+ DAM in 5xFAD mice compared to WT mice (Fig. 4E, 11% versus 2 to 3%, P < 0.05), and the proportion of CD11c+ DAM in irradiated/chimeric 5xFAD mice at 8 mo was similar to that observed in nonirradiated 5xFAD mice. Additionally, tSNE of the CD11c+ DAM in irradiated 5xFAD mice revealed a subset of Kv1.3-high CNS-MPs that only expressed CD45.2 but not CD45.1, further supporting their microglial origin (Fig. 4E).
Kv1.3 Is Highly Coexpressed with Proinflammatory Microglial Profiles and Lies Downstream of the Trem2 Immune Checkpoint.
To delineate potential cellular mechanisms regulated by Kv1.3 channels in proinflammatory DAM, we analyzed existing transcriptomic data from mouse microglia to identify genes that are most strongly positively and negatively correlated with Kcna3 expression. The top 50 genes positively correlated with Kv1.3 included proinflammatory genes (Il1b, Tlr2, Ifit2, Ptgs2, and Stat1) as well as DAM genes (Csf1, Tspo, and Slc16a3) based on reference single-cell RNAseq data from mouse microglia (Fig. 5A). Conversely, genes negatively correlated with Kcna3 included canonical microglial genes (e.g., P2ry12) primarily associated with homeostatic phenotypes (Fig. 5B). GSEA showed that Kv1.3-positively correlated genes were enriched terms related to immune response, nitric oxide biosynthesis, response to infections, regulation of Tnf production and cell migration, and pathways involved in response to lipopolysaccharide (LPS), IL1 signaling, NFκB pathway, and regulation of type 1 interferon production (SI Appendix, Fig. S8, Top). Kv1.3-negatively correlated genes were enriched in gene ontology terms, including small GTPase-mediated signal transduction, endocytosis, and regulation of apoptosis (SI Appendix, Fig. S8, Bottom).
Fig. 5.
Kv1.3 is coexpressed with proinflammatory DAM genes and regulates membrane potential and early MAPK signaling in microglia. (A) Representation of top 50 genes most highly positively (Left) and negatively (Right) coexpressed with Kcna3 (Kv1.3) in mouse microglia. Data were derived from previously published transcriptomic analyses of microglia from in vivo (WT, 5xFAD, and APP/PS1) and in vitro models (control, LPS, IL4). Strength of correlations with Kcna3 is inversely proportional to distance from Kcna3 (hub). Known genetic and protein—protein interactions are shown. (B) Flow cytometry-based measurement of cell surface functional Kv1.3 channels expressed by MMC microglial cells when stimulated by LPS or IFNγ (n = 3/group; **P < 0.01). (C) Whole-cell patch-clamp recordings of outward rectifying K+ currents of LPS-activated MMCs depolarized to 40 mV for 200-ms duration. The characteristic use-dependent inactivation of Kv1.3 is shown (pulsed every second). (D) Comparison of Kv1.3 current density (pA/pF) in unstimulated or LPS activated MMCs. (E) Blockade of Kv1.3 currents in MMCs by ShK-223 (100 nM), resulting in minimal residual K+ current. (Left) Ramp currents. (Right) Voltage-activated outward Kv1.3 current following depolarization to +40 mV. (F) Current clamp traces showing resting membrane potential of LPS-stimulated MMCs in the absence (black) or presence (purple) of ShK-223 (100 nM). BzATP was added at 5 s (arrow) of the recording to induce P2X-mediated calcium entry and membrane depolarization, evidenced by a transient increase in resting membrane potential (RMP). (G) Phosphoprotein measurements by Luminex analysis of MMC cell lysates, shown as a heatmap. MMCs with preconditioned (pre-con) with LPS to increase Kv1.3 channel expression and then exposed to either control, ATP, IFNg, or LPD for 5 min or 60 min with or without the addition of ShK-223. Luminex data was normalized to no-preconditioned (no pre-con) MMCs for each timepoint and then Z-standardized. Pairwise comparisons (without vs. with ShK-223) with P < 0.1 are highlighted (n = 3/condition). (H) Quantitative analysis of Luminex data highlighting MAPK phospho-analytes that showed significant effects of ShK-223 treatment (*P < 0.05).
Since microglia transition from homeostatic to DAM states via a Trem2-dependent checkpoint, we also asked whether Kv1.3 lies upstream or downstream of this Trem2 checkpoint. We interrogated existing microglial transcriptomic datasets from WT, Trem2-KO, and 5xFAD as well as 5xFAD–Trem2KO mouse brain (41) and found that up-regulation of Kv1.3 in 5xFAD microglia was suppressed in 5xFAD/Trem2KO mice while Apoe was only partly suppressed by loss of Trem2 (SI Appendix, Fig. S9A). We then performed flow cytometric studies on acutely isolated microglia from 7- to 8-mo-old WT and 5xFAD mice and found that cell-surface Trem2 expression was indeed increased in 5xFAD microglia as compared to WT microglia and that Kv1.3-high CNS-MPs also expressed significantly higher Trem2 as compared to Kv1.3-neg CNS-MPs (SI Appendix, Fig. S9 B and C). Overall, these data suggest that Kv1.3 channel expression is highly coexpressed with proinflammatory DAM genes and lies downstream of the Trem2 checkpoint (SI Appendix, Fig. S9D).
Kv1.3 Channels Regulate Microglial Membrane Potential and MAPK Signaling in Response to Inflammatory Stimuli.
Kv1.3 is a voltage-gated K+ channel that hyperpolarizes the cell membrane to allow sustained calcium influx in T cells in response to T cell receptor engagement and activating stimuli. Cytoplasmic domains of Kv1.3 are also known to interact with proteins such as Kvβ2 and scaffolding proteins involved in mitogen-activated protein kinase (MAPK) signaling, potentially linking Kv1.3 channel function with signaling pathways (42). In LPS-stimulated microglia that express high levels of Kv1.3 channels, Kv1.3 regulates membrane potential and calcium entry from the plasma membrane (43) and potentially downstream signaling cascades which may explain the transcriptomic effects observed in vivo following Kv1.3 channel blockade in AD mouse models. We used the MMC (mouse microglial cell) line, which is more representative of primary mouse microglia than the BV2 cell line, to investigate the importance of Kv1.3 channels in regulating microglial membrane potential and signaling mechanisms (44, 45). At baseline, MMCs did not express detectable Kv1.3 channels on the surface as measured by both flow cytometric detection by ShK-F6CA and patch-clamp electrophysiology. Following LPS or interferon gamma (IFN-γ) stimulation for 48 to 72 h, we observed a dose-dependent increase in surface Kv1.3 channel expression measured by flow cytometry (Fig. 5B) and patch-clamp (Fig. 5C). Patch-clamp studies of MMCs stimulated by Aβ42 oligomer (100 nM) also showed increased Kv1.3 currents, similar to LPS-stimulated MMCs (Fig. 5D) (46). Kv1.3 currents in MMCs demonstrated typical features, including use-dependent inactivation and blockade by ShK-223 (100 nM) (Fig. 5C). Following blockade of Kv1.3 channels, we observed minimal residual K currents, suggesting that Kv1.3 is the dominant K channel expressed by activated MMCs (Fig. 5E) (47). Kv1.3 channel blockade by ShK-223 also resulted in membrane depolarization confirming the dependence on Kv1.3 channels for maintenance of resting membrane potential (Fig. 5F). These complimentary studies confirmed that Kv1.3 channels regulate membrane potential in proinflammatory microglia as recently published by a group using primary neonatal microglia (43).
To assess whether Kv1.3 channels regulate shifts in membrane potential when microglia face acute immune stimuli, we prestimulated MMC with LPS for 48h to up-regulate Kv1.3 channel expression and then exposed the cells to the purinergic agonists ATP and BzATP (Fig. 5F). These were selected because BzATP activates P2X channels (e.g., P2 × 4 and P2 × 7), resulting in calcium (Ca2+) entry and membrane depolarization, which should subsequently activate Kv1.3 channels. P2 × 4 is also up-regulated by DAM in AD models where it is coexpressed with Kv1.3 (2, 30), raising the possibility that Kv1.3 and P2X-mediated Ca2+ entry mechanisms are functionally coupled. BzATP transiently induced a small membrane depolarization in LPS-stimulated MMCs while ShK-223 mediated Kv1.3 blockade prior to BzATP exposure amplified this transient membrane depolarization (Fig. 5F). This confirms that Kv1.3 channel function minimizes membrane depolarization induced by calcium entry via P2X in microglia. Collectively, these in vitro studies align with recent observations made in primary microglia regarding the importance of Kv1.3 channels in regulating microglial membrane potential and their coupling with Ca2+ entry mechanism in LPS-activated conditions (43).
We next asked whether Kv1.3 channels regulate signaling cascades when microglia are activated by immune stimuli. We first primed MMCs with LPS (100 ng/mL) for 48 h to up-regulate Kv1.3 channels and then exposed Kv1.3-expressing MMCs to high dose LPS (1,000 ng/mL), IFN-γ (1,000 ng/mL), and ATP (200 µM) to activate both P2X channels for 5 min and 60 min with or without ShK-223 (100 nM) pretreatment. Cell lysates were analyzed by Luminex to measure phosphoproteins involved in MAPK signaling pathways, including ERK, p38, and Jnk cascades, as well as mTOR/Akt pathways, which are known to play important roles in microglia-mediated neuroinflammatory responses in AD (35, 48). LPS preconditioning increased levels of pErk1/2, pStat1, and pcJun in MMCs, and ShK-223 pretreatment did not impact phospho-MAPK levels in the absence of stimuli (Fig. 5G). Of the three stimuli at 5 min postexposure timepoints (ATP, IFN-γ, or LPS), ATP exposure increased p-p53 levels although ShK-223 pretreatment increased pMek1 and p-p53 levels in ATP-stimulated microglia (Fig. 5G). IFN-γ increased pStat1 levels at 5 min while ShK-223 pretreatment did not impact pStat1 levels although ShK-223 pretreatment increased pMek1 levels. LPS exposure increased pJnk, pStat1, and pcJun levels, and ShK-223 pretreatment further increased pJnk along with p-p38 levels but reduced p-p53 levels. At 60 min postexposure, ATP decreased pErk1/2 and pMSK1 levels while ShK-223 pretreatment had no effect. IFN-γ increased pStat1 levels without any effect of ShK-223. LPS exposure increased pcJun levels and decreased pMSK1 and p-p53 levels, and ShK-223 pretreatment increased p-p53 and pMSK1 levels (Fig. 5 G and H). While LPS preconditioning for 48 h increased Akt, GSK3β, PTEN, and mTOR levels, additional acute stimulation with ATP, IFN-γ, or LPS with or without ShK-223 pretreatment did not affect levels of analytes in the Akt/mTOR signaling panel (SI Appendix, Fig. S10). Overall, these signaling studies provide evidence for regulation of early MAPK but not Akt/mTOR signaling events by Kv1.3 channels in proinflammatory microglia.
Kv1.3 Blockade Reduces Aβ Pathology, Increases CD11c+ DAM, and Impacts Neuroinflammatory Profiles of Microglia in Mouse AD Models.
Prior studies have shown that blockade of Kv1.3 channels by the highly Kv1.3-selective 35-amino acid peptide ShK-223 decreased Aβ plaque burden in the 5xFAD brain and increased Aβ phagocytosis by microglia derived from ShK-223–treated mice (2). Previous coexpression network analysis studies also predicted that Kv1.3 blockade should inhibit proinflammatory and potentially augment antiinflammatory and prophagocytic gene expression (2). However, direct evidence for immunomodulatory effects of Kv1.3 blockers on microglial profiles in AD models is lacking. We treated 3-mo-old 5xFAD mice with ShK-223 or phosphate-buffered saline (PBS) for a duration of 3 mo, after which mice were euthanized, and whole-brain and fluorescence-activated cell sorting (FACS)-purified CD11b+CD45+ microglia were used for transcriptomics and immunohistochemical studies of fixed brain (Fig. 6A).
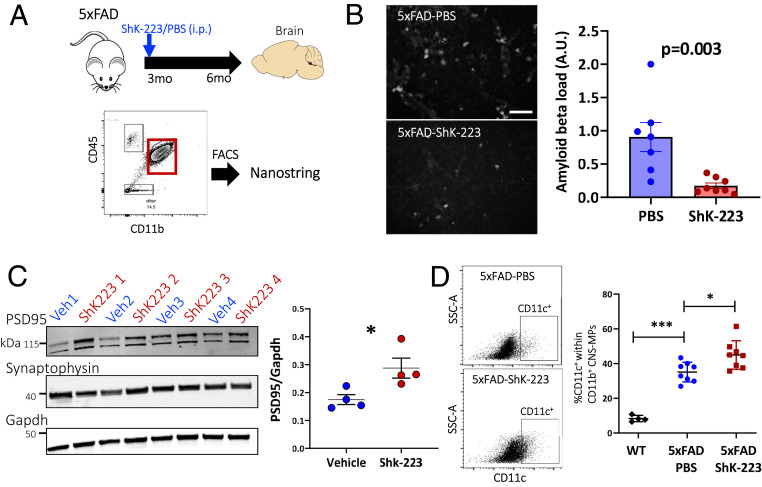
Fig. 6.
Kv1.3 channel blockade reduces Aβ pathology, increases synaptic markers, and increases CD11c+ DAM. (A) Experimental workflow outlining treatment of 3-mo-old 5xFAD mice with ShK-223 (n = 4) or PBS (n = 4) through intraperitoneal injection until 6 mo of age and FACS of CD11b+CD45+ CNS-MPs for NanoString-based neuroinflammatory gene profiling. (B) Representative images of 5xFAD–PBS (n = 4) and 5xFAD–ShK-223 (n = 4) frontal cortex used for quantitative analysis of Aβ plaque load shown in adjacent histogram show a significant decrease in Aβ plaque load in 5xFAD–ShK-223 cortex. A total of 4 to 5 images (10× magnification) were used for quantitative analysis of Aβ plaque burden (white accumulations) using ImageJ. (C) Western blot analysis of brain lysates comparing PSD95 and Synaptophysin protein levels in PBS- and ShK-223–treated groups. The housekeeping protein was Gapdh. Quantitative analysis is shown on the right. (D) Increased CD11c+ cells within CD11b+ CNS-MPs in ShK-223–treated mice as compared to PBS-treated 5xFAD and untreated WT mice. *P < 0.05, ***P < 0.001.
Consistent with prior studies (2), we found that Aβ plaque load was significantly lower in ShK-223–treated 5xFAD mice (Fig. 6B, P = 0.003). We also found that postsynaptic density protein PSD95 expression was higher in the brains of ShK-223–treated 5xFAD mice as compared to vehicle-treated mice while pan-neuronal synaptophysin was unchanged, suggesting a protective effect of ShK-223 on synaptic health (Fig. 6C) (49). Transcriptomic analyses of the whole-brain messenger RNA (mRNA) (NanoString glial profiling panel) also showed concordant increased expression of neuronal synaptic and carbohydrate metabolic genes (SI Appendix, Fig. S11 and Dataset S3). Flow cytometric analyses of acutely isolated CNS-MPs showed no differences in microglial numbers but showed a significantly increased CD11c+ microglia in ShK-223–treated mice compared to PBS-treated mice (Fig. 6D, P < 0.05), indicating increased presence of a DAM phenotype.
We next performed NanoString neuroinflammatory profiling (551 genes) of FACS-purified CD11c+CD45+ microglia (Dataset S4). PCA differentiated ShK-223–treated from PBS-treated 5xFAD mice (Fig. 7A) and 80 genes were differentially expressed (P < 0.05) with 57 up-regulated and 23 down-regulated genes following ShK-223 treatment (Fig. 7B). Of these, nine genes were up-regulated and 18 genes were down-regulated by at least 1.5-fold (SI Appendix, Fig. S12 A and B). GSEA showed that ShK-223 treatment increased expression of genes involved in signal transduction (Tgfbr1, Pak1, and Ikbkb), antigen presentation and processing (Fcgr1, Fcgr3, and Ifi30), surface receptors (Cxcr1, C3ar1, and Ccr2), microglial phagocytosis (including C1qc, Fc receptors, and Itgam), and ribosomal translational machinery and transmembrane proteins (Fig. 7 C–F, SI Appendix, Fig. S12C, and Dataset S5). ShK-223 treatment also suppressed genes involved in proinflammatory signaling (IL1b), endocytosis (Dock1, Lsr, and Pld2), and lipid metabolism (Pik3r5) (Fig. 7 C and F and SI Appendix, Fig. S12D). We identified miR-155, a well-known regulator of proinflammatory responses, and ATF1 as potential upstream regulators of genes increased by ShK-223 treatment. Conversely, EBF1 was predicted to be an upstream regulator of genes decreased by ShK-223 treatment (Fig. 7 D and E).
Fig. 7.
Kv1.3 blockade by ShK-223 in 5xFAD mice skews neuroinflammatory profiles of microglia toward a protective phenotype. (A) PCA of gene expression data based on the first three PCs shows three mouse groups (red: 5xFAD–ShK-223, blue: 5xFAD–PBS, and black/gray: 9- to 10-mo-old 5xFAD mice). ShK-223–treated mice were most distinct from the other mouse groups. (B) Heatmap displaying the transcriptomic differences (551 genes) between CD11c+CD45+ CNS-MPs from PBS-treated and ShK-223–treated 5xFAD mice. (C) Heatmap representation of GSEA showing overrepresented gene ontology terms in genes up-regulated and down-regulated following ShK-223 treatment. (D) Transcriptional factors predicted to lie upstream of genes up-regulated and down-regulated by ShK-223 treatment. (E) MicroRNAs (miRNA) predicted to regulate the expression of genes up-regulated (Right, green) and down-regulated (Left, purple) by ShK-223 treatment. The x-axis represents the enrichment z-score. (F) ShK-223–treated 5xFAD mouse microglia showed up-regulation of genes encoding transmembrane proteins (Left) and down-regulation of genes involved in the Il1 signaling (Right).
We also explored sex differences between 5xFAD mice and identified 28 DEGs. Male 5xFAD microglia expressed higher levels of Pilra (a receptor for herpes simplex virus type 1 [HSV1] and complement 4a) and C3 while female microglia expressed higher levels of proinflammatory genes such as Cxcl10, Ccl4, Ccl2, Ccl3, and IL1a (Dataset S4). These findings support greater proinflammatory activation of microglia in females compared to males, consistent with known increased Aβ plaque pathology and neuroinflammation in AD models and in human AD (50–52). There was no overlap between DEGs in response to ShK-223 and DEGs based on sex, suggesting that the neuroinflammatory effects of Kv1.3 blockade are independent of sex. Overall, these in vivo and transcriptomic studies provide evidence for the ability of a Kv1.3 channel blocker (ShK-223) to modulate neuroinflammatory profiles of microglia toward a more phagocytic state with increased expression of translational machinery, transmembrane receptors, and protective gene signatures while decreasing proinflammatory IL1-related signaling (Fig. 7F).
Discussion
Selective inhibition of proinflammatory CNS-MP responses has emerged as a promising therapeutic approach for disease modification in neurodegenerative diseases, ischemic stroke, and neuroinflammatory disorders (25, 29, 30, 33, 53, 54). Among potential therapeutic targets, the voltage-gated potassium channel Kv1.3 has been identified as a regulator of proinflammatory CNS-MP responses (25, 29, 30, 55), and in mouse models of AD pathology and ischemic stroke, selective Kv1.3 channel blockers have shown pathological and behavioral efficacy (2, 25, 29). Electrophysiological and immunohistochemical studies have demonstrated increased functional Kv1.3 channel expression by CNS-MPs in AD (2, 25), although specific subtypes of CNS-MPs that up-regulate Kv1.3 have not been characterized in mouse models of AD pathology or in human AD brain. To better define the peripheral monocyte-versus-microglial origin of Kv1.3-high CNS-MPs in neurodegenerative diseases, molecular phenotyping and chimerism strategies are necessary. While the presence of Kv1.3-high CNS-MPs has been shown in mouse models of Aβ deposition (2, 25), the presence Kv1.3-high CNS-MPs in tau pathology models or other models of chronic neuroinflammation has also not been confirmed. Furthermore, the ability of Kv1.3 channel blockers to skew neuroinflammatory responses and microglial profiles in mouse AD models was unknown.
Transcriptomic and proteomic studies fail to detect Kv1.3 at high levels due to the low abundance of Kv1.3 transcript and protein in microglia, despite its critical functional roles. Thus, patch-clamp has been the gold standard for electrophysiological characterization of functional Kv1.3 channel expression; although, the limited sampling of this method cannot easily identify differences in Kv1.3 level expression by cell subpopulations (56, 57). In this study, we used a validated flow cytometry approach to detect functional, cell-surface Kv1.3 channels in CNS-MP subsets from mouse models of Aβ pathology (5xFAD), tau pathology (P301S), and EAE as well as from fresh unfrozen human AD brain. This flow cytometric assay leverages the ability of ShK-F6CA (33), a fluorescent-tagged peptide blocker of Kv1.3 channels, to bind cell-surface functional Kv1.3 channels with a detection limit of more than 500 to 600 Kv1.3 channels per cell (33, 58). Thus, ShK-F6CA is an excellent Kv1.3-selective probe to easily measure functional cell-surface Kv1.3 channels and detect immune cells with high Kv1.3 channel expression, allowing phenotyping of large populations and subsets of CNS-MPs acutely isolated from the adult brain including fresh human postmortem brain (2, 30, 58). Using this approach, we confirmed that Kv1.3 channel expression is restricted to a subset of CD45lowLy6clow CNS-MPs in 5xFAD mice but not in P301S or EAE mice. This pattern of Kv1.3 expression is similar to that observed in Kv1.3-high CNS-MPs following transient middle cerebral artery occlusion (30). We also found that Kv1.3-high CNS-MPs in 5xFAD mice express relatively higher levels of CD45 as well as CD11c, a DAM marker, suggesting that Kv1.3-high CNS-MPs may have DAM-like properties. Importantly, we also show that Kv1.3 expression is restricted to a subpopulation of CNS-MPs in human AD brain with patterns of expression that resemble the 5xFAD mouse model. Since we found Kv1.3-high CNS-MPs in Aβ but not tau mouse models and also in human AD which exhibit both in Aβ and tau pathologies, we suspect that up-regulation of Kv1.3 expression might be driven primarily by progressive Aβ accumulation. Indeed, Aβ42 fibrils in prior studies (25) and Aβ42 oligomers as shown in this study up-regulate Kv1.3 channels in primary microglia.
To directly contrast the molecular features of Kv1.3-high and Kv1.3-low CNS-MPs with peripheral myeloid cells, we sorted Kv1.3-high and Kv1.3-low CNS-MPs along with peripheral monocyte Ly6chigh and Ly6clow subsets from 5xFAD mice. At the transcriptomic level, Kv1.3-high CNS-MPs more closely resemble resident microglia via shared expression of canonical microglial markers such as Tmem119, Cx3cr1, and P2ry12, albeit at relatively lower levels. Despite these similarities, we observed some differences between Kv1.3-high and Kv1.3-low CNS-MPs in 5xFAD mice. The relatively lower homeostatic microglial gene expression along with higher expression of proinflammatory DAM genes in Kv1.3-high CNS-MPs suggest that Kv1.3-high CNS-MPs may represent a proinflammatory subset of DAM. This conclusion is further supported by prior coexpression network analyses of microglial transcriptomes which showed that Kv1.3 (gene Kcna3) is a member of a proinflammatory gene module in AD models and is coexpressed with genes such as Hif1a, IL1b, Ptgs2, Tlr2, and Irak4 while being highly anticorrelated with homeostatic genes such as Tmem119, Olfml3, and P2ry12 (2). Another unique feature of Kv1.3-high CNS-MPs is their higher levels of glutamatergic transcripts such as Gria2. Given this overall low relative abundance of glutamatergic mRNAs in Kv1.3-high CNS-MPs and our observed lack of Gria2 protein expression by CNS-MPs in 5xFAD brain, we suspect that the presence of these glutamatergic and synaptic transcripts in Kv1.3-high CNS-MPs may be indicative of phagocytosed neuronal and synaptodendritic components (59) rather than de novo transcription and translation in CNS-MPs. Whether Kv1.3-high CNS-MPs exhibit higher synaptic phagocytic activity needs to be clarified in future studies.
Kv1.3 blockers have displayed promising treatment effects in mouse models of AD pathology and other neurologic diseases (2, 25, 29). Given this, it is imperative to determine the cellular origin of Kv1.3-high CNS-MPs as this may impact future therapeutic approaches to inhibit their cellular function more definitively. If peripheral monocytes give rise to Kv1.3-high CNS-MPs, this would raise the possibility for their modulation before entering the CNS. Alternatively, their microglial origin would indicate that factors specific to the CNS mediate Kv1.3 up-regulation and would indirectly imply CNS bioavailability of ShK toxin–based peptide blockers of Kv1.3 channels (2, 25, 29). Our transcriptomic findings indicate that Kv1.3-high CNS-MPs closely resemble resident microglia in AD models; however, these data do not confirm the microglial origin of Kv1.3-high CNS-MPs because blood monocyte–derived CNS-MPs also resemble microglia at the transcriptomic level after residing in the CNS over a long period of time. Previous irradiation bone marrow chimeric studies showed that blood-derived myeloid cells adopted microglial morphology and microglia-like transcriptomic states three months after total body irradiation and bone marrow transplantation while retaining some distinctions from microglia (60). It is also possible that microglia, in response to progressive neuropathology, adopt features that resemble peripheral myeloid cells. Furthermore, our flow cytometry data show that Kv1.3-high CNS-MPs are CD45high but are Ly6cneg/low, providing conflicting evidence regarding their origin from microglia (CD45int/low and Ly6clow) or from CNS-infiltrating brain macrophages/monocytes (CD45high Ly6chigh). To definitively clarify the cellular origin of Kv1.3-high CNS-MPs in AD models, we used the irradiation bone marrow CD45.1/CD45.2 chimerism model in 5xFAD mice and show that more than 90% of Kv1.3-high CNS-MPs are derived from microglia and not from bone marrow–derived CNS-infiltrating monocytes/macrophages. While this does not exclude the possibility of some contribution of peripheral monocytes to the Kv1.3-high CNS-MP pool in the AD brain, the majority of Kv1.3-high CNS-MPs are derived from microglia. Since peripheral monocyte populations in 5xFAD mice express nearly no Kv1.3 channels and Kv1.3-high CNS-MPs are a subset of microglia, the observed therapeutic effects of Kv1.3 blockers in mouse AD models must be mediated via direct CNS effects.
While we found that microglia, not monocytes, represent the cellular origin of Kv1.3-high CNS-MPs in mouse AD models, higher levels of Kv1.3 mRNA have been reported in human monocytes as compared to postmortem human microglia in the aging brain (2, 24). One explanation could be that Kv1.3 mRNA levels do not correlate well with protein-level or functional-channel expression. Another possibility is that Kv1.3 expression patterns are different in mouse and human brain such that Kv1.3-high CNS-MPs in mouse models are microglia derived, while in human AD, they are monocyte derived. The third possibility is that accumulating AD pathology activates a unique transcriptomic program in microglia which up-regulates Kv1.3 expression. In support of this idea, several genes that are not highly expressed by homeostatic microglia in the brain are up-regulated following build-up of Aβ pathology. Here, we also show that Kv1.3 channel up-regulation lies downstream of the Trem2 immune checkpoint which is critical for microglial state transition from homeostatic to DAM profiles in response to Aβ accumulation (22). While the expression level of Kv1.3 in postmortem microglia from human AD as compared to controls is unknown, our flow cytometric data show a discrete population of Kv1.3-high CNS-MPs in human AD cases and confirm lack of Kv1.3 expression by infiltrating macrophages and lymphocytes in the brain. Future transcriptomic and proteomic characterization of Kv1.3-high CNS-MPs from human brain will determine whether our findings are valid in human AD. Our work also provides mechanistic insights into the role of Kv1.3 channels in regulating membrane potential and stimulus-induced early cellular signaling events in microglia. Using the MMC line, we show that Kv1.3 channels regulate membrane potential of microglia when activated by proinflammatory stimuli. When microglia were exposed to BzATP to activate calcium entry via P2X channels, we found that Kv1.3 blockade accentuated BzATP-induced membrane depolarization, suggesting that K efflux via Kv1.3 helps maintain the membrane potential in a hyperpolarized state. This observed functional coupling between P2X and Kv1.3 agrees with recent work in primary microglia (43). In addition to regulating membrane potential and calcium influx, Kv1.3 channels are known to interact with signaling proteins (e.g., MAPKs), raising the possibility that Kv1.3 function may be functionally coupled with early signaling events. Consistent with this idea, we found that Kv1.3 blockade suppressed LPS-induced p53 phosphorylation and increased MEK/ERK activation without impacting Akt/PI3k/mTOR signaling. While demonstrating the ability of Kv1.3 channels to fine tune early MAPK signaling events, further investigation is warranted to understand the molecular basis and consequences of this observed functional coupling between Kv1.3 channel function and MAPK signaling.
From a translational perspective, our in vivo studies with ShK-223 in 5xFAD mice validate prior studies demonstrating reduction in Aβ plaque pathology in 5xFAD mice and APP/PS1 mice via Kv1.3 channel blockade by peptidergic (ShK analogs) and small molecules (PAP1) (2, 25). This validation is important as it strengthens the preclinical rationale to pursue Kv1.3 channel blockade as a human AD-relevant therapeutic target. We additionally show that reduction in Aβ plaque burden by ShK-223 treatment is associated with increased postsynaptic protein PSD95 expression with concordantly increased synaptic gene expression in 5xFAD brain, consistent with an overall neuroprotective effect of Kv1.3 blockade. We also present evidence for cell-surface Kv1.3 channel expression by a subpopulation of acutely isolated CNS-MPs with microglial properties from fresh postmortem human AD brain, confirming immunohistochemistry studies that showed Kv1.3 channel protein expression in CNS-MPs in AD brain (24). Since a Kv1.3 channel–blocking peptide belonging to the ShK analog family (ShK-186 or Dalazatide) has already completed early-phase human studies of safety for systemic autoimmunity (61), our preclinical findings provide a compelling rationale to develop Kv1.3 blockers as potential therapeutics for human AD.
The CNS harbors multiple subfamilies of Kv1 channels expressed across various cell types. Targeting microglial Kv1.3 channels with high specificity could assist us in modulating their neuroinflammatory effects in AD. Therefore, in our studies, we used ShK-223 because of its higher selectivity for Kv1.3 over other Kv1 channels (expressed in neurons) and its superior stability and suitability as a therapeutic peptide. Future pharmacokinetics and pharmacodynamics studies of ShK-223 and related analogs are now needed to facilitate translation of our findings. In addition to neuropathological effects of ShK-223 in 5xFAD mice, we also show that Kv1.3 blockade can impact the molecular profile of microglia by increasing surface expression of CD11c and increased expression of several genes encoding surface receptors (Cx3cr1, Mertk, Itgam, and Tgfbr1) that are highly expressed by homeostatic microglia and required for their ability to sense the CNS microenvironment (62). ShK-223 treatment also increased expression of phagocytic genes that are part of the Tyrobp causal network of AD (8) and up-regulated protective genes (Tnfsf13b, Itgb5, and Pi3kcg) that are downstream of miR-155 (63) while suppressing proinflammatory genes such as Il1b and others involved in Il1-related signaling. The ability of ShK-223 to modulate the immune profile of microglia is an important proof of concept that Kv1.3 blockers can inhibit proinflammatory microglial responses in vivo and augment potentially protective responses (2). For example, ShK-223–treated 5xFAD mouse microglia showed higher expression of Ccr2 and lower expression of Hdac2. Ccr2 is a chemokine receptor expressed by CNS-MPs involved in chemotaxis to sites of inflammation. In AD mouse models, loss of Ccr2 accelerated disease progression and Aβ accumulation while impairing microglial accumulation and Aβ clearance, suggesting that increased Ccr2 by Kv1.3 blockade is a protective and pro-Aβ clearance effect of ShK-223 (64). Conversely, we found that Hdac2 expression was significantly reduced in ShK-223–treated 5xFAD microglia. Interestingly, Hdac2 loss in mouse AD models resulted in increased microglial phagocytosis of Aβ and decreased neuropathology (65), suggesting that lower Hdac2 in ShK-223–treated mice is also a beneficial effect of Kv1.3 blockade. Another gene up-regulated by ShK-223 was Tmem173, which encodes the protein STING that mediates type 1 interferon responses that reduce microglia-mediated neuroinflammation (66). Collectively, our observations based on whole-brain and microglia-specific transcriptomics and histological studies support the idea that the microglial immune profile induced by Kv1.3 blockade is protective.
A limitation of our transcriptomics studies is the relatively narrow scope of the NanoString neuroinflammatory panel, which is restricted to ∼700 genes; however, these are highly representative of inflammatory, homeostatic, and DAM genes as well as risk genes associated with late-onset AD. Future studies will assess global and single-cell transcriptomic along with proteomic changes in microglia and whole brain in conjunction with cytokine profiling in brain, plasma, and cerebrospinal fluid to identify the neuroinflammatory effects and their corresponding biofluid biomarkers of therapeutic efficacy and target engagement. Future studies using conditional Kv1.3 deletion in microglia are also necessary to better define the role of Kv1.3 channels in regulating microglial function in AD pathology since low-level Kv1.3 channel expression below the detection limit of our flow cytometric assay has been reported in other brain cell types.
In conclusion, our results demonstrate that microglia, but not infiltrating macrophages, express Kv1.3 channels in AD in an Aβ- rather than a tau-dependent manner. Kv1.3-high microglia have a unique transcriptomic signature that distinguishes them from Kv1.3-negative microglia, and long-term Kv1.3 blockade in AD mouse models inhibits proinflammatory gene expression while promoting a prophagocytic signature in microglia. These results strengthen the preclinical rationale for targeting microglial Kv1.3 channels as a therapeutic approach for AD.
Methods
A detailed list of all reagents used for our experiments is provided in SI Appendix, Table S2. Experimental details on animals, acute isolation of adult mouse CNS-MPs (2, 13, 67), PBMCs and splenocytes and human CNS-MPs, irradiation bone marrow chimerism (32), long-term in vivo Kv1.3 channel blockade (2, 13, 68), flow cytometry studies (2, 13, 67) and fluorescence-activated cell sorting, RNA extraction and Nanostring analyses (41, 67, 69), whole-cell patch-clamp studies (43, 47), immunostaining and image analysis, Western blotting, Luminex studies, mouse microglia cultures, Netscreen analysis (2), and statistical considerations are described in detail in SI Appendix, SI Methods.
Ethics Approval and Consent to Participate.
Approval from the Emory University IACUC was obtained prior to all animal-related studies (IACUC protocol number 300123). Human postmortem brain samples were obtained at time of brain autopsy for experiments within the scope of an IRB-approved study at Emory University.
Supplementary Material
Acknowledgments
We are grateful to those who donated their brains for research through the Goizueta ADRC and to Dr. Michael Pennington for synthesis of ShK-223. This work was supported by Goizueta ADRC (P50AG025688), Alzheimer’s Association (#37102 to S. Rangaraju), National Institute of Neurological Disorders and Stroke (NINDS)/National Institute on Aging (K08-NS099474-1 and R01 NS114130-01A1 to S. Rangaraju; R01AG053960, R01AG061800, RF1AG057471, RF1AG057470, RF1AG062181, and R01AG057339 to N.T.S.; F32AG064862 to S. Rayaprolu; 1R01NS087142 to D.J.K.; and R01NS100294 to H.W.), and the Accelerating Medicines Partnership for AD (U01AG061357 to N.T.S. and A.I.L.). This study was also supported in part by the Emory Integrated Genomics Core and Emory Flow Cytometry Core, which are subsidized by the Emory University School of Medicine and are among the Emory Integrated Core Facilities. This research project was supported in part by the Neuropathology Core of the Emory Neuroscience NINDS Core Facilities grant (P30NS055077) and the Georgia Clinical and Translational Science Alliance of the NIH (UL1TR002378). The content is solely the responsibility of the authors and does not necessarily reflect the official views of the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013545118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Ransohoff R. M., Perry V. H., Microglial physiology: Unique stimuli, specialized responses. Annu. Rev. Immunol. 27, 119–145 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Rangaraju S., et al., Identification and therapeutic modulation of a pro-inflammatory subset of disease-associated-microglia in Alzheimer’s disease. Mol. Neurodegener. 13, 24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prinz M., Priller J., Microglia and brain macrophages in the molecular age: From origin to neuropsychiatric disease. Nat. Rev. Neurosci. 15, 300–312 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Perry V. H., Nicoll J. A., Holmes C., Microglia in neurodegenerative disease. Nat. Rev. Neurol. 6, 193–201 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Villegas-Llerena C., Phillips A., Garcia-Reitboeck P., Hardy J., Pocock J. M., Microglial genes regulating neuroinflammation in the progression of Alzheimer’s disease. Curr. Opin. Neurobiol. 36, 74–81 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Colonna M., Wang Y., TREM2 variants: New keys to decipher Alzheimer disease pathogenesis. Nat. Rev. Neurosci. 17, 201–207 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Jonsson T., et al., Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 368, 107–116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang B., et al., Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153, 707–720 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang T., et al., Upregulation of TREM2 ameliorates neuropathology and rescues spatial cognitive impairment in a transgenic mouse model of Alzheimer’s disease. Neuropsychopharmacology 39, 2949–2962 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynch J. R., Morgan D., Mance J., Matthew W. D., Laskowitz D. T., Apolipoprotein E modulates glial activation and the endogenous central nervous system inflammatory response. J. Neuroimmunol. 114, 107–113 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Zhao N., Liu C.-C., Qiao W., Bu G., Apolipoprotein E, receptors, and modulation of Alzheimer’s disease. Biol. Psychiatry 83, 347–357 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lambert J. C.et al.; European Alzheimer’s Disease Initiative (EADI); Genetic and Environmental Risk in Alzheimer’s Disease; Alzheimer’s Disease Genetic Consortium; Cohorts for Heart and Aging Research in Genomic Epidemiology , Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45, 1452–1458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rangaraju S., et al., A systems pharmacology-based approach to identify novel Kv1.3 channel-dependent mechanisms in microglial activation. J. Neuroinflammation 14, 128 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Umoh M. E., et al., A proteomic network approach across the ALS-FTD disease spectrum resolves clinical phenotypes and genetic vulnerability in human brain. EMBO Mol. Med. 10, 48–62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crotti A., Ransohoff R. M., Microglial physiology and pathophysiology: Insights from genome-wide transcriptional profiling. Immunity 44, 505–515 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Sarlus H., Heneka M. T., Microglia in Alzheimer’s disease. J. Clin. Invest. 127, 3240–3249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trotta T., et al., Microglia-derived extracellular vesicles in Alzheimer’s disease: A double-edged sword. Biochem. Pharmacol. 148, 184–192 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Asai H., et al., Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat. Neurosci. 18, 1584–1593 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keren-Shaul H., et al., A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290.e17 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Mathys H., et al., Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep. 21, 366–380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deczkowska A., et al., Disease-associated microglia: A universal immune sensor of neurodegeneration. Cell 173, 1073–1081 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Krasemann S., et al., The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamphuis W., Kooijman L., Schetters S., Orre M., Hol E. M., Transcriptional profiling of CD11c-positive microglia accumulating around amyloid plaques in a mouse model for Alzheimer’s disease. Biochim. Biophys. Acta 1862, 1847–1860 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Rangaraju S., Gearing M., Jin L. W., Levey A., Potassium channel Kv1.3 is highly expressed by microglia in human Alzheimer’s disease. J. Alzheimers Dis. 44, 797–808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maezawa I., et al., Kv1.3 inhibition as a potential microglia-targeted therapy for Alzheimer’s disease: Preclinical proof of concept. Brain 141, 596–612 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeCoursey T. E., Chandy K. G., Gupta S., Cahalan M. D., Voltage-gated K+ channels in human T lymphocytes: A role in mitogenesis? Nature 307, 465–468 (1984). [DOI] [PubMed] [Google Scholar]
- 27.Negulescu P. A., Shastri N., Cahalan M. D., Intracellular calcium dependence of gene expression in single T lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 91, 2873–2877 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feske S., Wulff H., Skolnik E. Y., Ion channels in innate and adaptive immunity. Annu. Rev. Immunol. 33, 291–353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y. J., Nguyen H. M., Maezawa I., Jin L. W., Wulff H., Inhibition of the potassium channel Kv1.3 reduces infarction and inflammation in ischemic stroke. Ann. Clin. Transl. Neurol. 5, 147–161 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao T., et al., Temporal profiling of Kv1.3 channel expression in brain mononuclear phagocytes following ischemic stroke. J. Neuroinflammation 16, 116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rangaraju S., Chi V., Pennington M. W., Chandy K. G., Kv1.3 potassium channels as a therapeutic target in multiple sclerosis. Expert Opin. Ther. Targets 13, 909–924 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Li J. M., et al., Absence of vasoactive intestinal peptide expression in hematopoietic cells enhances Th1 polarization and antiviral immunity in mice. J. Immunol. 187, 1057–1065 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beeton C., et al., A novel fluorescent toxin to detect and investigate Kv1.3 channel up-regulation in chronically activated T lymphocytes. J. Biol. Chem. 278, 9928–9937 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Yoshiyama Y., et al., Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron 53, 337–351 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Chen M. J., et al., Microglial ERK signaling is a critical regulator of pro-inflammatory immune responses in Alzheimer’s disease. bioRxiv [Preprint] (2019). 10.1101/798215 (Accessed 10 October 2019). [DOI]
- 36.Rangaraju S., et al., Quantitative proteomics of acutely-isolated mouse microglia identifies novel immune Alzheimer’s disease-related proteins. Mol. Neurodegener. 13, 34 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji K., Akgul G., Wollmuth L. P., Tsirka S. E., Microglia actively regulate the number of functional synapses. PLoS One 8, e56293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bie B., Wu J., Foss J. F., Naguib M., Activation of mGluR1 mediates C1q-dependent microglial phagocytosis of glutamatergic synapses in Alzheimer’s rodent models. Mol. Neurobiol. 56, 5568–5585 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shemer A., et al., Engrafted parenchymal brain macrophages differ from microglia in transcriptome, chromatin landscape and response to challenge. Nat. Commun. 9, 5206 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Menzel F, et al., Impact of X-irradiation on microglia. Glia 66, 15–33 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Wang Y., et al., TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell 160, 1061–1071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pérez-García M. T., Cidad P., López-López J. R., The secret life of ion channels: Kv1.3 potassium channels and proliferation. Am. J. Physiol. Cell Physiol. 314, C27–C42 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Nguyen H. M., et al., Biophysical basis for Kv1.3 regulation of membrane potential changes induced by P2X4-mediated calcium entry in microglia. Glia 68, 2377–2394 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarkar S., et al., Kv1.3 modulates neuroinflammation and neurodegeneration in Parkinson’s disease. J. Clin. Invest. 130, 4195–4212 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarkar S., et al., Characterization and comparative analysis of a new mouse microglial cell model for studying neuroinflammatory mechanisms during neurotoxic insults. Neurotoxicology 67, 129–140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stine W. B., Jungbauer L., Yu C., LaDu M. J., Preparing synthetic Aβ in different aggregation states. Methods Mol. Biol. 670, 13–32 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen H. M., et al., Differential Kv1.3, KCa3.1, and Kir2.1 expression in “classically” and “alternatively” activated microglia. Glia 65, 106–121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ulland T. K., et al., TREM2 maintains microglial metabolic fitness in Alzheimer’s disease. Cell 170, 649–663.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu D. S., et al., APOE4 enhances age-dependent decline in cognitive function by down-regulating an NMDA receptor pathway in EFAD-Tg mice. Mol. Neurodegener. 10, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sala Frigerio C., et al., The major risk factors for Alzheimer’s disease: Age, sex, and genes modulate the microglia response to Aβ plaques. Cell Rep. 27, 1293–1306.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanamsagar R., Bilbo S. D., Sex differences in neurodevelopmental and neurodegenerative disorders: Focus on microglial function and neuroinflammation during development. J. Steroid Biochem. Mol. Biol. 160, 127–133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferretti M. T., et al., Sex and gender differences in Alzheimer’s disease: Current challenges and implications for clinical practice. Eur. J. Neurol. 27, 928–943 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Bittner S., Meuth S. G., Targeting ion channels for the treatment of autoimmune neuroinflammation. Ther. Adv. Neurol. Disord. 6, 322–336 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bozic I., et al., Voltage gated potassium channel Kv1.3 is upregulated on activated astrocytes in experimental autoimmune encephalomyelitis. Neurochem. Res. 43, 1020–1034 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Di Lucente J., Nguyen H. M., Wulff H., Jin L. W., Maezawa I., The voltage-gated potassium channel Kv1.3 is required for microglial pro-inflammatory activation in vivo. Glia 66, 1881–1895 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kruskal P. B., Jiang Z., Gao T., Lieber C. M., Beyond the patch clamp: Nanotechnologies for intracellular recording. Neuron 86, 21–24 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Jouhanneau J.-S., Poulet J. F. A., Multiple two-photon targeted whole-cell patch-clamp recordings from monosynaptically connected neurons in vivo. Front. Synaptic Neurosci. 11, 15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Norton R. S., Pennington M. W., Wulff H., Potassium channel blockade by the sea anemone toxin ShK for the treatment of multiple sclerosis and other autoimmune diseases. Curr. Med. Chem. 11, 3041–3052 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Rajendran L., Paolicelli R. C., Microglia-mediated synapse loss in Alzheimer’s disease. J. Neurosci. 38, 2911–2919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cronk J. C., et al., Peripherally derived macrophages can engraft the brain independent of irradiation and maintain an identity distinct from microglia. J. Exp. Med. 215, 1627–1647 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tarcha E. J., et al., Safety and pharmacodynamics of dalazatide, a Kv1.3 channel inhibitor, in the treatment of plaque psoriasis: A randomized phase 1b trial. PLoS One 12, e0180762 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hickman S. E., et al., The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 16, 1896–1905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kou X., Chen D., Chen N., The regulation of microRNAs in Alzheimer’s disease. Front. Neurol. 11, 288 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.El Khoury J., et al., Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat. Med. 13, 432–438 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Datta M., et al., Histone deacetylases 1 and 2 regulate microglia function during development, homeostasis, and neurodegeneration in a context-dependent manner. Immunity 48, 514–529.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Mathur V., et al., Activation of the STING-dependent type I interferon response reduces microglial reactivity and neuroinflammation. Neuron 96, 1290–1302.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Min Y. G., et al., B cell immunophenotyping and transcriptional profiles of memory B cells in patients with Myasthenia gravis. Exp. Neurobiol. 28, 720–726 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rangaraju S., et al., Differential phagocytic properties of CD45low microglia and CD45high brain mononuclear phagocytes-activation and age-related effects. Front. Immunol. 9, 405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emig D., et al., AltAnalyze and DomainGraph: Analyzing and visualizing exon expression data. Nucleic Acids Res. 38, W755–W762 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.