Significance
Symbiotic dinitrogen (N2)-fixing trees fulfill a critical function in tropical forests by bringing in new nitrogen, yet it remains unclear how they overcome constraints by highly weathered, nutrient-poor tropical soils. We advance forest biogeochemistry and microbial ecology with the discovery from field trials in Panama that fast-growing N2-fixing trees in tropical forests exhibit accelerated mineral weathering and distinctive soil metagenomes that improve their access to inorganic nutrients in nutrient-poor soils. Furthermore, we show that N2-fixing trees exert similar effects on non-N2–fixing trees nearby thus having previously overlooked community-wide effects on tropical forest nutrient cycling. These results offer insights into the role of N2-fixing trees and their associated microbiomes in safeguarding the function of tropical forests within the global biosphere.
Keywords: mineral weathering, metagenomics, Acidobacteria, tropical forest, N2-fixing legume trees
Abstract
Legume trees form an abundant and functionally important component of tropical forests worldwide with N2-fixing symbioses linked to enhanced growth and recruitment in early secondary succession. However, it remains unclear how N2-fixers meet the high demands for inorganic nutrients imposed by rapid biomass accumulation on nutrient-poor tropical soils. Here, we show that N2-fixing trees in secondary Neotropical forests triggered twofold higher in situ weathering of fresh primary silicates compared to non-N2–fixing trees and induced locally enhanced nutrient cycling by the soil microbiome community. Shotgun metagenomic data from weathered minerals support the role of enhanced nitrogen and carbon cycling in increasing acidity and weathering. Metagenomic and marker gene analyses further revealed increased microbial potential beneath N2-fixers for anaerobic iron reduction, a process regulating the pool of phosphorus bound to iron-bearing soil minerals. We find that the Fe(III)-reducing gene pool in soil is dominated by acidophilic Acidobacteria, including a highly abundant genus of previously undescribed bacteria, Candidatus Acidoferrum, genus novus. The resulting dependence of the Fe-cycling gene pool to pH determines the high iron-reducing potential encoded in the metagenome of the more acidic soils of N2-fixers and their nonfixing neighbors. We infer that by promoting the activities of a specialized local microbiome through changes in soil pH and C:N ratios, N2-fixing trees can influence the wider biogeochemical functioning of tropical forest ecosystems in a manner that enhances their ability to assimilate and store atmospheric carbon.
The legume family is the most diverse angiosperm family in the Neotropics (1–3), with dinitrogen (N2)-fixing legume trees growing fast and supplying tropical forests with substantial quantities of nitrogen (N) during succession (4). This N2-fixing strategy requires that trees can access scarce sources of inorganic nutrients, including bioavailable forms of phosphorus (P) for metabolites and growth and molybdenum (Mo) for nitrogenase functioning (5–7) (the enzyme that catalyzes conversion of atmospheric N2 to bioavailable N).
However, in highly weathered tropical soils, in addition to their pools in organic matter (8, 9), large amounts of P and Mo are often occluded in an inorganic form in insoluble iron (Fe) and aluminum (Al)-bearing minerals (10, 11) and thus not available for immediate biological uptake. For example, both P and Mo are scarce in the oxisols and inceptisols that have developed from Mo-poor basalt bedrock in Panamanian tropical forests (7, 12) (SI Appendix, Table S1) and that contain P-adsorbing kaolinite, goethite, and hematite secondary minerals (SI Appendix, Fig. S1). Our own observations from these Panamanian forests show significant differences in the chemistry of soils beneath N2 fixing versus nonfixing trees, with significantly lower concentrations of nitric acid–extractable P, Fe, and Al and lower pH below N2-fixing trees (SI Appendix, Table S2). Moreover, a strong association between extracted P and Fe plus Al, but not between P and soil organic carbon, implies that soil P is significantly influenced by mineral dynamics within these tropical soils (SI Appendix, Fig. S2, P < 0.001 for Fe and Al and P > 0.10 for carbon, Pearson test).
These patterns raise the biogeochemical hypothesis that N2-fixing legume trees may strategically employ specific mechanisms to enhance mineral weathering (5, 13, 14), resulting in improved access to occluded inorganic mineral nutrients, enhanced N2-fixation, and enhanced carbon sequestration by forest biomass during succession. Here, we address this hypothesis by investigating 1) whether N2-fixing trees induce locally elevated rates of silicate mineral (olivine) weathering (compared to nonfixing trees), causing the depletion of elements critical to N2-fixation; 2) whether the altered soil beneath N2-fixing trees is linked to compositional and functional differences in the microbiomes and metagenomes associated with soil minerals; and 3) whether the presence of N2-fixers affects biogeochemical nutrient cycling in rooting zone soils beneath neighboring non-N2–fixing forest trees.
Results and Discussion
Enhanced Weathering Rates, Acidification, and Mo Leaching Beneath N2-Fixers.
We investigated these questions by undertaking a field study across floristically and biogeochemically well-characterized (4, 15–17) secondary tropical forests of Panama. The establishment of replicated experimental plots (0.1 ha each, n = 6) across a natural gradient in N2-fixing legume tree abundance (6 to 27% tree basal area [BA]) allowed us to evaluate whether N2-fixing legumes influence soil weathering rates and the soil microbiome to favor rapid growth. We further asked whether N2-fixing trees could affect soil weathering and microbiomes at scales of tree communities by influencing nearby (<5 m) compared to far away (>10 m) nonfixing trees. We determined in situ weathering by burying 504 mesh bags of crushed dunite, an olivine-rich (>90%) silicate rock (SI Appendix, Supplementary Note 3), for 8 months in the rooting zone (∼10 cm depth) beneath N2-fixing (5 species, n = 51 trees), nonfixing trees far from N2-fixers (NF-far) in legume-poor forests (5 species, n = 36 trees), and nonfixing trees near (NF-near) fixers in legume-rich forests (5 species, n = 39 trees). We used the primary ferromagnesian silicate olivine as a proxy for the dissolution of the secondary magnesium aluminosilicate mineral fractions that exist in these soils because although the dissolution of both linearly depends on pH, olivine weathers much more rapidly (18, 19).
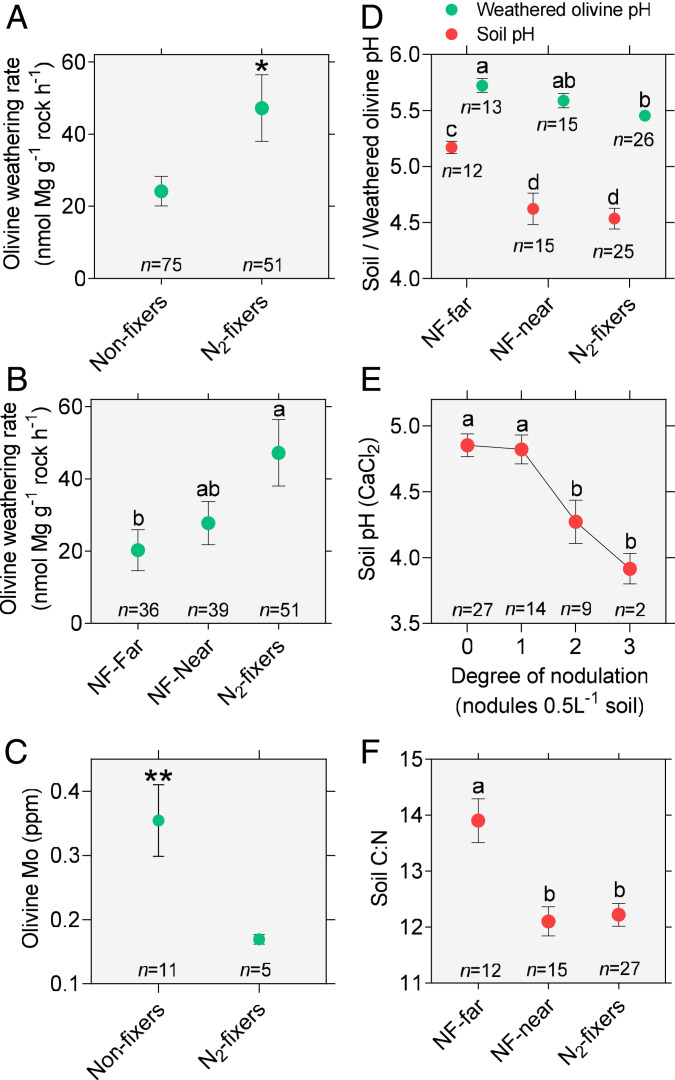
X-ray fluorescence (XRF) revealed that relative to the Mg concentration of fresh unweathered minerals (mean ± SE = 26.65 ± 0.11%) field-weathered olivine had lost significant amounts of Mg with olivine weathered beneath nonfixers exhibiting intermediate Mg concentration (mean ± SE = 26.22 ± 0.05%) and field-weathered olivine from beneath N2-fixers revealing the lowest concentration of Mg (mean ± SE = 25.97 ± 0.11%). Next, we utilized the olivine Mg:Si ratio, a parameter particularly sensitive to vegetation-driven dissolution (20), to calculate weathering rates (SI Appendix, Supplementary Note 3). We report that weathering in soil beneath the tropical N2-fixing legume trees occurred at a rate double that of nonfixers (Welch’s t test, P < 0.05) (Fig. 1A and SI Appendix, Supplementary Notes 2 and 3). This is consistent with an ∼64% increase in weathering by temperate forest nonlegume N2-fixing trees forming actinorhizal symbiosis (14, 21). Furthermore, the local N2-fixer effect on weathering extends to the neighboring tree community (Fig. 1B), with NF-near displaying weathering rates intermediate between N2-fixers and NF-far from fixers (Welch’s alternative to the traditional ANOVA P < 0.05, unpaired t test with Welch’s correction). The measured rate of olivine weathering was also linked to significant declines in soil pH (Spearman test, P < 0.01, SI Appendix, Fig. S5) and lower olivine pH after weathering, with conditions beneath N2-fixing trees and NF-near trees being notably more acidic than beneath NF-far trees (5, 22, 23) (Welch’s ANOVA, P < 0.0001 for both soil and olivine pH, Fig. 1D). In addition, complete digest and inductively coupled plasma mass spectroscopy (ICP-MS) analysis of weathered minerals (SI Appendix, Table S3 and Fig. S6) revealed significantly lower Mo levels in olivine from beneath N2-fixers compared to nonfixing trees (Welch’s t test, P < 0.01, Fig. 1C), highlighting the potential for N2-fixers to tap onto previously unavailable mineral sources vital for N2-fixation through enhanced weathering.
Fig. 1.
N2-fixing legume trees are linked to greater mineral weathering in tropical forest soils as well as lower soil C:N ratio and soil pH. (A) N2-fixers reveal doubled weathering rates compared to nonfixers (N2-fixers: mean = 47.21, SD = 65.89, nonfixers: mean = 24.18, SD = 35.76; two-tailed Welch’s t test, P = 0.026, Welch’s corrected t = 2.28, DFn = 70.14, Cohen’s d = 0.43). (B) NF-near N2-fixers show intermediate olivine weathering rates between NF-far and N2-fixers (N2-fixers: mean = 47.21, SD = 65.89, NF-near: mean = 27.79, SD = 37.41, NF-far: mean= 20.27, SD = 33.96; one-way classic ANOVA, P = 0.037, F = 3.40, DFd = 2, η2 = 0.052). (C) Mo content of olivine weathered in soils beneath N2-fixers is lower than that of nonfixing trees (N2-fixers: mean = 0.1692, SD = 0.0171, nonfixers: mean = 0.3546, SD = 0.1845; two-tailed Welch’s t test, P = 0.0076, Welch’s corrected t = 3.30, DFn = 10.37, Cohen’s d = 1.42). (D) Soil-weathered olivine pH is significantly lower beneath N2-fixers and NF-near in comparison to NF-far (N2-fixers: mean = 5.45, SD = 0.22, NF-near: mean = 5.59, SD = 0.25, NF-far: mean = 5.72, SD = 0.22; one-way classic ANOVA, P = 0.0002, F = 10.03, DFd = 2, η2 = 0.282) with similar pattern observed in soil pH (N2-fixers: mean = 4.54, SD = 0.46, NF-near: mean = 4.62, SD = 0.54, NF-far: mean = 5.17, SD = 0.19; one-way ANOVA, P = 0.0006, F = 8.73, DFd = 2, η2 = 0.263). (E) Degree of nodulation (number of nodules per 0.5 L soil) is associated to acidification of soil (0: mean = 4.85, SD = 0.45, 1: mean = 4.82, SD = 0.41, 2: mean = 4.27, SD = 0.49, 3: mean = 3.92, SD = 0.16; Welch’s ANOVA, P = 0.0027, W = 14.39, DFd = 6.654, ω2 = 0.790). (F) Soil C:N ratio is significantly lower beneath N2-fixers and NF-near relative to NF-far from fixers (N2-fixers: mean = 12.22, SD = 1.06, NF-near: mean = 12.10, SD = 1.01, NF-far: mean = 13.90, SD = 1.36; one-way classic ANOVA, P = 9.484e-05, F = 11.17, DFd = 2, η2 = 0.305). Multiple comparisons are carried out using Fisher’s least significant difference (LSD) tests or unpaired t tests with Welch’s correction (in the cases of Welch’s ANOVA). Error bars indicate SEM.
Increasing nodulation (number of nodules per 0.5 L soil) was associated with significantly lower soil pH (Welch’s ANOVA, P < 0.01) (Fig. 1E). Taken together, these results suggest that sites of active nodulation and N2 fixation promoted the acidification of soil [consistent with increased H+ exudation by nodules (24, 25) and N cycling related acidification (5)] which, in turn, enhances weathering and the release of critical elements. To further investigate this hypothesis, we compiled a total of 13 studies comparing soil pH between N2-fixing and -nonfixing trees (SI Appendix, Supplementary Note 1) in both planted (n = 7) and natural forest (n = 6) settings. These studies indicate a consistent trend of lower soil pH (mean = 0.25 pH units) under N2-fixing leguminous relative to nonfixing nonleguminous trees in both natural forest and planted agroforestry conditions (0.33 pH units lower pH under N2-fixers relative to nonfixers in our secondary forest plots, SI Appendix, Supplementary Note 1). This supports a causal relationship in which N2-fixing legume trees are drivers of acidification (“species effects”) as opposed to exhibiting a preference for acidic sites (“soil niche partitioning”). Moreover, the ratio of soil carbon to nitrogen (C:N hereafter) was significantly lower beneath N2-fixers than nonfixers and lower between nonfixers that were near versus far from individual fixers (Welch’s ANOVA, P < 0.01) (Fig. 1F), indicating an additional tree community effect of N2 fixers on soil chemistry in agreement with similar trends in temperate N2-fixing arboreal flora (26). Soil C:N ratios were also correlated with measured weathering rates (Spearman test, P < 0.01, SI Appendix, Fig. S5), identifying a potential link between weathering and the carbon and N cycles.
Microbial Function and Enhanced Silicate Weathering.
To analyze the role of the microbiome in our observed patterns of weathering rates, we constructed and sequenced 12 shotgun metagenome libraries of soil-weathered olivine mineral samples from beneath N2-fixers (n = 6), NF-near (n = 3), and NF-far (n = 3). Metagenomes provided information on the abundance of gene orthologs from 28 high level functional metabolic pathways [Level 1 in the Metagenomic Rapid Annotations using Subsystems Technology (MG-RAST) Subsystem database (27)]; these were analyzed using hierarchical clustering to objectively identify pathways associated with increased field-based weathering rates (Fig. 2A). A correlation heat-map and clustering analysis identified a single well-supported cluster of metabolic pathways (bootstrapping value > 75%) that linked enhanced weathering rates to a coordinated increase in gene abundance of metabolic pathways involved in microbial respiration, carbohydrate metabolism, and N cycling (Fig. 2A).
Fig. 2.
Metagenomics of the microbial community associated with weathered minerals in tropical forests link increased respiration, nitrogen, and carbohydrate metabolic potential of the microbial community to enhance weathering beneath N2-fixers. (A) Correlation heatmap matrix of gene abundance allocated to MG-RAST Subsystem-Level 1 (“High Level Metabolic”) pathways for the 12 sequenced metagenomes of reacted olivine at different weathering rates reveals that well-supported cluster (bootstrap value >70) coupling olivine weathering rates to respiration, N, and carbohydrate metabolism and virulence and defense response potential within the metagenome. The heatmap is constructed using R with Manhattan dissimilarity index, complete clustering method, and Pearson test correlations; values in italics indicate bootstrapping for each major node. High level pathways encompassing the cumulative abundance of all N metabolism (B and E) and respiration (C and F) genes and the Krebs (tricarboxylic acid) cycle (D and G) genes all correlate with weathering rates (Pearson test, *P < 0.05; respiration: Pearson correlation test r = 0.607, P = 0.036, F = 5.84, DFd =10, N metabolism: Pearson correlation test r = 0.606, P = 0.037, F = 5.79, DFd = 10, Krebs cycle: Pearson correlation test r = 0.619, P = 0.032, F = 6.20, DFd = 10) and reveal patterns of increase following the order NF-far < NF-near < N2-fixers. Gene abundance in B–G is normalized using the single copy gene rpoC (DNA-directed RNA polymerase beta' subunit) to account for number of sequenced genomes. Error bars indicate SEM.
Analysis of the metagenomes, normalized using the single copy marker gene rpoC to account for different number of covered genomes in a metagenomic sample (28), reinforced this result by identifying significant correlations between in situ olivine weathering beneath trees and the cumulative abundance of genes comprising entire high-level pathways for N metabolism, respiration, and the Krebs cycle (Pearson test, P < 0.05, Fig. 2 B–D). Normalized cumulative gene abundance for each process followed the same pattern: N2-fixers > NF-near > NF-far (Fig. 2 E–G). We interpret these results to indicate that below N2-fixing trees, microbes exhibit elevated activity linked to the potential for increased cycling of reduced carbon substrates and enhanced respiratory CO2 and acid production, which, in turn, promote mineral weathering. Additionally, the metagenomic N metabolism pathway response is consistent with increased inputs of symbiotically fixed N beneath N2-fixers, and the generation of acidity due to nitrification, potentially exacerbated by nitrate leaching (5, 22, 23). These findings support the suggestion that N2-fixing trees change soil biogeochemistry to enhance the weathering release of elements needed for N2 fixation as well as plant growth. This benefit also extended to the soil of nearby non-N2-fixing individuals in the tree community landscape.
Increased Potential for Anaerobic Metabolism under N2-Fixers: Implications for Inorganic P Dissolution.
Our findings of enhanced silicate weathering under N2-fixers linked to acidification and microbiome changes raised the question of whether the dissolution of native soil minerals, particularly those adsorbing growth-limiting P such as Fe oxides (goethite, hematite), would be similarly affected. Although the dissolution of native kaolinite (SI Appendix, Fig. S1) would be expected to exhibit the same linear relationship to pH as olivine (18, 19), Fe oxides and the subsequent release of inorganic P follow more complex dynamics with major effects of not just soil pH but also redox processes (29, 30).
Redox dissolution, brought up by low O2 availability, can frequently occur in soils of tropical forests and significantly impact inorganic P solubility (31). Anaerobic microsites generate low redox conditions in which microbes reduce ferric iron [Fe(III)] or sulfate (SO42-) by using them as final electron acceptors (32). Up-regulation of anaerobic reduction processes could, therefore, allow legumes to access inorganic P occluded in insoluble Fe(III)-bearing minerals in tropical soils by reducing the highly insoluble Fe(III)P to Fe(II) + P (Fe cycling) (10) and Fe(II)S + P (S cycling) (33, 34) as well as enable the release of sorbed Mo (35). Soil Fe(III) reduction rates are affected by both abiotic (precipitation, temperature, soil type and structure, and soil pH) and biotic (organic C content, microbial C mineralization rates, and abundance of Fe(III)-reducing bacteria) factors (29, 36–38).
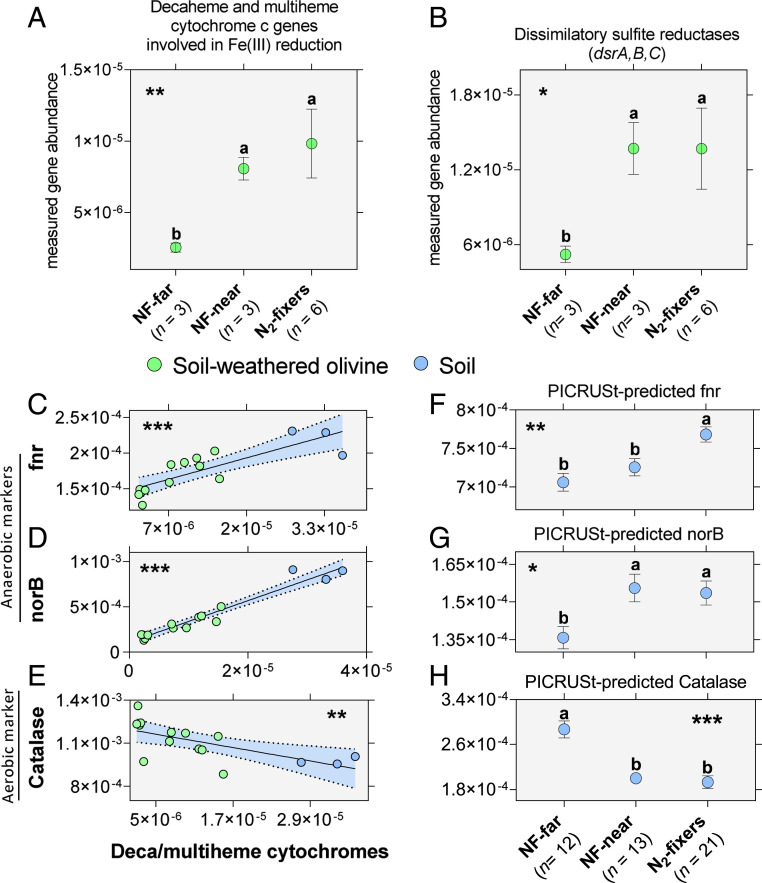
In our project, we have extracted, sequenced, and characterized a total of 15 belowground metagenomes (n = 12 from total DNA extracted from soil-weathered olivine and n = 3 from total DNA extracted from the rooting zone soil of N2-fixers, SI Appendix, Table S7). In olivine metagenomes, we recorded a higher relative abundance of Fe(III) reduction pathway genes, in particular respiratory decaheme and multiheme cytochrome c protein-coding genes, beneath N2-fixers and NF-near relative to NF-far as well as greater abundance of sulfite reductase genes (Welch’s ANOVA, P < 0.01, Fig. 3 A and B). Unfortunately, we have fewer rooting zone soil metagenomes preventing direct comparison of metagenomic Fe(III) reduction potential between soil-weathered olivine and soil samples.
Fig. 3.
Implications of improved metagenomic potential for Fe and S reduction and anaerobic metabolism to release of Fe-bound P—a benefit N2-fixers may also pass to neighboring nonfixing trees. (A) Significant enrichments in Fe reduction gene orthologs (cumulative gene abundance including decaheme and multiheme cytochrome c genes) are observed in the soil mineral metagenomes of N2-fixers and neighboring nonfixers near them (N2-fixers: mean = 9.83e-6, SD = 5.91 to 6, n = 6, NF-near: mean = 8.06e-6, SD = 1.38e-6, n = 3, NF-far: mean = 2.54e-6, SD = 5.41e-7, n = 3, Welch’s ANOVA, **P = 0.0047, W = 21.44, DFd = 4.58, ω2 = 0.844). (B) Sulfite reduction genes (cumulative abundance of sulfite reductases dsr A, B, and C) are signficantly enriched in the soil mineral metagenomes of N2-fixes and nonfixers near fixers relative to nonfixers far from fixers (N2-fixers: mean = 1.369e-5, SD = 7.97e-6, n = 6, NF-near: mean = 1.370e-5, SD = 3.6e-6, n = 3, NF-far: mean = 5.21e-6, SD = 1.15e-6, n = 3, Welch’s ANOVA, *P = 0.0286, W = 8.90, DFd = 4.39, ω2 = 0.681). (C) The anaerobic marker fnr correlates positively with deca/multiheme cytochrome c genes involved in Fe(III) reduction (Pearson correlation test r = 0.828, ***P = 0.0001, F = 28.33, DFd = 13). (D) The anaerobic marker norB correlates positively with deca/multiheme cytochrome c genes (Pearson correlation test r = 0.971, ***P = 2e-9, F = 213.3, DFd = 13). (E) The aerobic marker catalase correlates negatively with deca/multiheme cytochrome c genes (Pearson correlation test r = - 0.660, **P = 0.0074, F = 10.02, DFd = 13). (F) The PICRUSt-predicted fnr gene abundance is significantly greater in soil metagenomes of N2-fixers and NF-near relative to NF-far trees (N2-fixers: mean = 7.68e-4, SD = 4.54e-5, n = 21; NF-Near: mean = 7.26e-4, SD = 4.08e-5, n = 13; NF-Far: mean = 7.06e-4, SD = 4.00e-5, n = 12; Welch’s ANOVA **P = 0.0012, W = 8.881, DFd = 25.78, ω2 = 0.354). (G) The PICRUSt-predicted norB gene abundance is significantly greater in soil metagenomes of N2-fixers and NF-near relative to NF-far trees (N2-fixers: mean = 1.54e-4, SD = 2.20e-5, n = 21, NF-Near: mean = 1.56e-4, SD = 1.99e-5, n = 13, NF-Far: mean = 1.36e-4, SD = 1.52e-5, n = 12; Welch’s ANOVA *P = 0.012, W = 5.278, DFd = 26.62, ω2 = 0.224). (H) The PICRUSt-predicted catalase (CAT) gene abundance is significantly lower in soil metagenomes of N2-fixers and NF-near relative to NF-far trees (N2-fixers: mean = 1.93e-4, SD = 5.09e-5, n = 21, NF-Near: mean = 2.00e-4, SD = 2.82e-5, n = 13, NF-Far: mean = 2.87e-4, SD = 5.23e-5, n = 12; Welch’s ANOVA ***P = 6.22e-5, W = 14.636, DFd = 24.96, ω2 = 0.494). Multiple comparisons are carried out using unpaired t tests with Welch’s correction. Error bars reveal SEM.
To circumvent this limitation and to establish whether our observations from soil-weathered olivine hold in a more general way for soils, we evaluated microbiome structure and function in the rooting zone soils beneath our three categories of trees using Next-Generation 16S ribosomal RNA (rRNA) sequencing for prokaryotes. Analysis of genus-level 16S rRNA data revealed significant differences in soil microbiome composition, with the microbial communities under N2-fixers and NF-near trees exhibiting substantial overlap but differing from those of NF-far (Permutational multivariate analysis, P < 0.001, SI Appendix, Fig. S8).
To provide further insight, we also carried out 16S rRNA-informed metagenomic reconstruction of soil metagenomes using the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) platform (39). Kyoto Encyclopedia of Genes and Genomes (KEGG)-based PICRUSt annotations typically do not generate Fe(III) reduction gene assignments per se. However, anaerobic lifestyle markers such as fumarate and nitrate reduction regulatory protein (fnr) (40) and nitric oxide reductase (norB) (41) gene abundances and aerobic lifestyle markers such as catalase gene abundance (42), which are available in PICRUSt data, can be used as reliable proxies for Fe(III) reduction gene abundance because they 1) share strong positive and negative correlations, respectively (Pearson tests P < 0.001, P < 0.001, and P < 0.01, correspondingly, Fig. 3 C–E), and 2) are also functionally associated. Furthermore, KEGG-based PICRUSt predicted fnr, norB, and catalase significantly correlated in abundance with that of measured fnr, norB, and catalase in our 15 shotgun metagenomic samples (Pearson tests P < 0.001 and P < 0.01, SI Appendix, Fig. S9)—confirming that PICRUSt-based predictions for those three genes are reliable and directly comparable with shotgun metagenomic data. In PICRUSt-predicted soil metagenomes of N2-fixers, fnr is of significantly greater abundance than in those of NF-far and NF-near, the latter occupying an intermediary position (Fig. 3F, Welch’s ANOVA, P < 0.01) and norB is of significantly greater abundance in metagenomes of N2-fixers and NF-near relative to NF-far trees (Fig. 3G, Welch’s ANOVA, P < 0.01). PICRUSt-based predicted abundance values for catalase follow the opposite pattern with NF-far exhibiting significantly greater abundance than both NF-near and N2-fixing legume trees (Fig. 3H, Welch’s ANOVA, P < 0.001). Therefore, PICRUSt predictions based on 16S rRNA abundance indicate increased anaerobic metagenomic potential in rooting zone soils of N2-fixing legume trees and their nearby nonfixing neighbors compared to nonfixers far from legumes in legume-poor forests. This further supports the hypothesis that the legume soil microbiome facilitates Fe(III) reduction and access to occluded P and may help explain reduced soil P beneath N2-fixers (SI Appendix, Table S2). To explore this further, we evaluated the mechanisms driving increased metagenomic Fe(III) reduction potential in soils of N2-fixers and NF-near relative to NF-far.
Soil pH Is a Major Determinant of Metagenomic Fe(III) Reduction Potential.
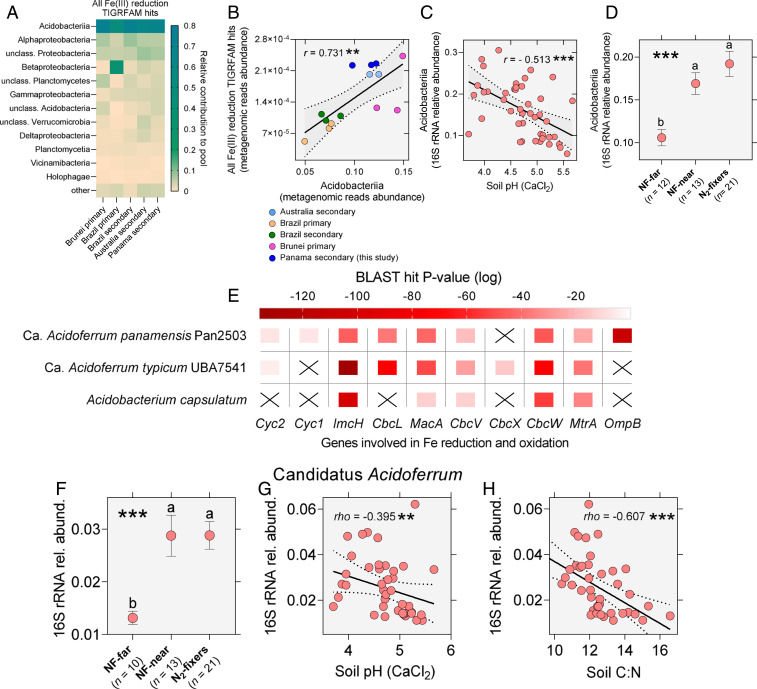
The dissolution of Fe(III) oxides and the subsequent liberation of its adsorbed inorganic P are thermodynamically favored at low pH as the forward reactions consume protons (43, 44). Consequently, many Fe(III)-reducing bacteria are acidophilic and thrive in acidic environments (43, 45, 46). To investigate the link between pH and Fe(III)-reducing potential of the soil community in tropical forest systems, we performed searches using the domain-specific Basic Local Alignment Search Tools (BLASTs) of our three soil metagenomes against the TIGRFAM domain database. Our soil metagenomes contained genes mapping to all three domains implicated in Fe reduction processes including OmcA/MtrC, DmsE/MtrA, and MtrB/PioB (TIGRFAM03507-3509) domains. Taxonomic profiling revealed that the Acidobacteriia dominated all pools, with ∼66% of hits matching to members of this class (Fig. 4A). Acidobacteriia are a class of predominantly acidophilic bacteria within the phylum Acidobacteria and their dominance of Fe(III) reduction gene pools in our tropical soils supports the Fe(III) reduction potential—pH link that we have described.
Fig. 4.
The Fe cycling gene pool in tropical forest soils is dominated by acidophilic Acidobacteriia. Characterization of the MAGs of Ca. Acidoferrum panamensis, genus novus, species novum, and Ca. Acidoferrum typicum, species novum (part of the newly proposed order Ca. Acidoferrales, ordo novus) compared to the known Fe(III) reducer Acidobacterium capsulatum reveals that this new group of uncultured acidobacteriia may be the most numerous Fe cyclers in the tropical soil microbiome. (A) Members of class Acidobacteriia (phylum Acidobacteria) dominate the pools of TIGRFAM03507-03509 domain hits involved in Fe(III) reduction across tropical forest soils. (B) The MGX-based relative abundance of metagenomic reads mapping to Acidobacteria exhibit a strong positive correlation with the summed metagenomic relative abundance of MtrC/OmcA, DmsE/MtrA, and MtrB/PioB domain hits (Pearson correlation test r = 0.731, **P = 0.0030, F = 13.77, DFd = 12). (C) The QIIME 16S rRNA-based relative abundance of Acidobacteriia exhibits a negative correlation with soil pH (Pearson correlation test r = − 0.513, ***P = 0.0004, F = 15.00, DFd = 42). (D) The QIIME 16S rRNA-based relative abundance of Acidobacteriia is significantly greater in soils beneath N2-fixing legume trees and nonfixing trees near N2-fixers compared to soils of nonfixing trees far from N2-fixers (N2-fixers: mean = 0.0788, SD = 0.0668, n = 21, NF-Near: mean = 0.0867, SD = 0.0456, n = 13, NF-Far: mean = 0.0562, SD = 0.0328, n = 12; one-way classic ANOVA, ***P = 0.0003, F = 9.788, DFd = 43, η2 = 0.313). (E) Heatmap revealing the BLAST P values (log-normalized) of hits against a set of selected Fe cycling proteins (refer to the Main Text for more details). (F) The IDTAXA classifier 16S rRNA hits against the GTDB (set at default parameters) reveal that the relative abundance of Ca. Acidoferrum is significantly greater in soil microbiomes of N2-fixers and nonfixers near N2-fixers relative to nonfixers far from N2-fixers (N2-fixers: mean = 0.0288, SD = 0.0120, n = 21, NF-Near: mean = 0.0288, SD = 0.0141, n = 13, NF-Far: mean = 0.0131, SD = 0.0040, n = 10; Welch’s ANOVA, ***P = 1.25e-05, W = 18.82, DFd = 23.83, ω2 = 0.571). (G) The relative abundance of Ca. Acidoferrum negatively correlates with soil pH (Spearman correlation test, rho = - 0.395, **P = 0.0096, DFd = 40). (H) The relative abundance of Ca. Acidoferrum negatively correlates with soil C:N (Spearman correlation test, rho = − 0.607, ***P < 0.0001, DFd = 41). Multiple comparisons are carried out using Post-Hoc Fisher LSD tests (classic ANOVA) or unpaired t tests with Welch’s correction (Welch’s ANOVA). Error bars show SEM.
To test if the dominance of Acidobacteriia over Fe(III)-reducing gene pools holds up for different tropical forest systems, we repeated the analysis for a group of publicly available metagenomes from tropical forests sites including Amazon, Brunei, and Australia (SI Appendix, Table S8). We found that Acidobacteriia similarly dominated the soil pools of Fe(III)-reducing TIGRFAM hits in all tested tropical ecosystems (Fig. 4A). Furthermore, the metagenomic abundance of Acidobacteriia exhibited a strong positive correlation with the total abundance of Fe(III)-reducing domain hits (TIGRFAM03507 + TIGRFAM03508 + TIGRFAM03509) when combining data from across all sites (Fig. 4B, Pearson test, P < 0.01), confirming that the more abundant the Acidobacteriia as a component of the microbiome, the greater the Fe(III)-reducing potential encoded by the metagenome.
Within our site in Panama, the abundance of Acidobacteriia in the soil microbial 16S rRNA libraries negatively correlated with measured soil pH (Fig. 4C, Pearson test, P < 0.001). Acidobacteriia were also significantly more abundant in soils beneath N2-fixers and nonfixers near fixers than in soils of nonfixers far from fixers (Fig. 4D, ANOVA, P < 0.001). Together, these findings corroborate the active involvement of acidophilic Acidobacteriia in the processes of Fe(III) reduction and resulting inorganic P release in tropical soils. This also provides a mechanistic link explaining the increased abundance of Fe(III) reduction genes (Fig. 3A) in the significantly more acidic soils of N2-fixing legume trees and their neighboring nonfixers (Fig. 1D).
To investigate the identity of the Fe cycling Acidobacteriia at our site, we performed genome assembly on our soil samples and recovered several metagenome-assembled genomes (MAGs), one of which (Pan2503), was assigned to the Acidobacteriia. Genome-wide analyses against the Genome Taxonomy Database (GTDB) indicated that MAG Pan2503 is closely related to the previously isolated Acidobacteriial MAG of UBA7541. The MAGs of Pan2503 and UBA7541 and the genome of Acidobacterium capsulatum—their most closely related, cultured known Fe(III)-reducing agent (47)—were all searched for Fe cycling gene homologs. The two MAGs contained more Fe cycling gene homologs (SI Appendix, Supplementary Note 6 and Fig. S12) than are found in the genome of the confirmed Fe(III) reducer A. capsulatum (Fig. 4E). These findings support the active involvement of these uncultured Acidobacteriia in the Fe cycle. Here, we propose renaming the generically named order UBA7541 to Candidatus (Ca.) Acidoferrales, ordo novus, and genus UBA7541 to Ca. Acidoferrum, genus novus in order to acknowledge their Fe cycling potential. Additionally, we propose that MAG Pan2503, isolated from our total soil metagenomes, is named Ca. Acidoferrum panamensis, species novum, while MAG UBA7541 is renamed to Ca. Acidoferrum typicum, combinatio nova (refer to SI Appendix, Supplementary Note 8 for detailed descriptions).
Taxonomic annotation of the raw 16S rRNA soil microbiome sequences against the GTDB 16S database in the IDTAXA classifier (48) reveals that the newly discovered candidate genus Ca. Acidoferrum occupies a substantial proportion of the soil microbial community at our site. Its relative abundance, similar to that of Acidobacteriia as a whole, is ∼twofold greater in the soil microbiomes under N2-fixing trees and their near nonfixing neighbors than under nonfixers far from fixers (Fig. 4F, Welch’s ANOVA, P < 0.001). Its abundance also negatively correlates with soil pH and soil C:N ratio (Fig. 4 G and H), indicating that the genus, as many Acidobacteriia, reveals acidophilic traits and is affected by the C:N ratio of soil organic matter and acquired organic substrates (49). As the third most abundant genus in the soil microbiome, members of Ca. Acidoferrum are likely representing the most prominent Fe cycling lineage in these tropical soils.
Together, our findings indicate that differences in soil pH and soil C:N can select for uncultured Fe cycling Acidobacteriia that would account for the observed greater abundance of Fe(III)-reducing genes and Fe cycling potential beneath N2-fixers and their tree neighbors.
Legume-Mediated Soil pH and C:N and the Soil Microbiome.
We tested if preference for sites of generally lower soil redox in N2-fixing legumes could be responsible for the greater Fe cycling potential of their soil metagenomes. We found that the percent BA of N2-fixing trees in sites across the Agua Salud Dynamic Forest Plot Network did not differ between paired up-slope and down-slope transects that offer different soil redox conditions (SI Appendix, Supplementary Note 7 and Fig. S13). In contrast, our data indicate that the increased potential for Fe cycling in soils under N2-fixing legume trees and their nonfixing neighbors in the forest community is contributable and correlates to lower soil pH and C:N ratio, favoring increases in Fe(III)-reducing acidophiles, such as the hereby described Ca. Acidoferrum (Fig. 4).
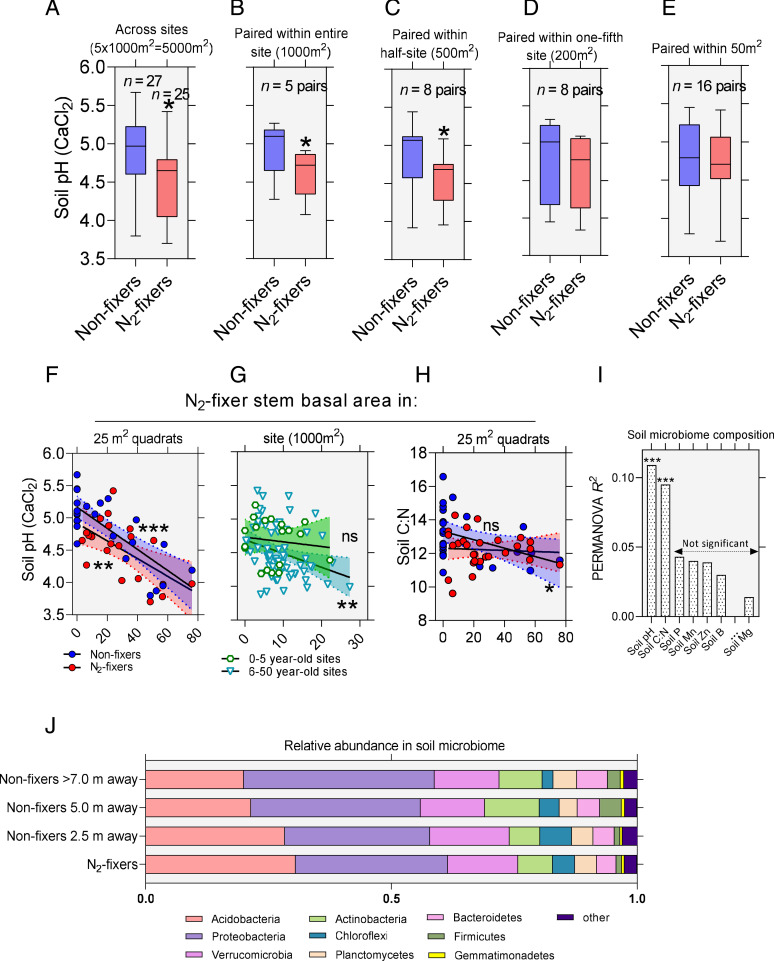
Replicated measurements of under-crown soil pH beneath N2-fixers and nonfixers across our five forest sites reveal significantly more acidic soils under N2-fixers (unpaired two-tailed t test, P < 0.05, Fig. 5A). To establish if such changes are driven by fixers and to exclude the effects of between and within site differences in soil chemistry, we paired the mean for each of the two tree groups for different area sizes (scales) and carried out paired two-tailed t tests. We found that the significant differences in soil pH between N2-fixers and nonfixers hold up at the 1,000 m2 as well as the 500 m2 scale but are less pronounced at 200 m2 scale and completely disappear at the 50 m2 scale (Fig. 5 B–E) as a result of nonfixers and N2-fixers being in a fully neighboring state, growing close to each other. However, even at finer scales N2-fixers are still linked to differences in soil pH. For example, the stem BA of N2-fixers at 25 m2 quadrat level (5 × 5 m) is found to significantly correlate (Pearson test, P < 0.001) with measured soil pH (Fig. 5F). Three independent lines of evidence presented in this work support that the presence and abundance of N2-fixing legume trees are causal to these changes in pH rather than the opposite: 1) nodulation, a process known to be negatively affected by pH (50) but also to stimulate H+ exudation from root systems (24, 51), correlates positively with soil pH (Fig. 1E); 2) our meta-analysis of before and after measurements of soil pH pre- and postplanting of N2-fixing trees in forestry plantations discussed above (SI Appendix, Supplementary Note 1); and 3) the observation that site-level BA of N2-fixing trees does not correlate with site pH at the onset of secondary succession (0 to 5-y-old sites) but correlates with pH later on during succession (6- to 50-y-old forests, Fig. 5G).
Fig. 5.
N2-fixing legumes modify the soil microbiome through changes in soil pH and C:N ratio. (A) Under-crown sampling from our five sites reveals that N2-fixers have on average more acidic topsoil than nonfixers (unpaired two-tailed t test, P = 0.017, t = 2.48, DFn = 26, Cohen’s d = 0.688). (B–E) Pairing of N2-fixing and nonfixing trees at different area sizes (scales) demonstrate that N2-fixers have significantly lower under-crown soil pH than nonfixers when paired at 1,000 m2 or 500 m2 scales with the effect fading at the 250 m2 and near absent at the 50 m2 scale (paired two-tailed t tests, *P < 0.05 comparing site means). (F) The BA of N2-fixers at the 25 m2 scale significantly correlates with soil pH for both fixers and nonfixers (Pearson test, ***P < 0.0001, r = −0.78 for nonfixers and Pearson test, **P = 0.0042, r = -0.60 for N2-fixers); (G) The BA of N2-fixers at the 1,000 m2 scale significantly correlates with soil pH for 6 to 50 y old forests (Pearson test, **P = 0.0098, r = −0.30, n = 72) but not for 0- to 5-y-old forests (Pearson test, ns P = 0.615, r = −0.12, n = 19). This is consistent with fixer effects on soil pH associated with their successional growth and symbiotic N2-fixation in the course of forest secondary succession rather than initial species filtering at the onset of secondary successional processes. (H) The BA of N2-fixers at the 25 m2 significantly correlates with soil C:N under the crown of nonfixers (Pearson test, *P = 0.0297, r = −0.42), but not fixers (Pearson test, ns P = 0.766, r = −0.06), suggesting that the presence of fixers instigates changes in organic matter composition beneath nonfixers that is dependent on the percent BA occupied by N2-fixers locally. (I) PERMANOVA R2 highlights soil pH and C:N as the major covariates of soil microbiome composition (non-metric multidimensional scaling analysis with Manhattan dissimilarity index of species-level operational taxonomic units [OTUs]). Soil elemental concentrations are based on nitric acid digests of soil powders. (J) Analysis of nonfixer soil microbiomes at different distances away from N2-fixers (2.5 m, n = 11, 5.0 m, n = 11, and >7.0 m, n = 3) and N2-fixers (n = 21) reveals stepwise changes in the relative abundance of major prokaryotic phyla (distance effect on the microbiomes of nonfixing trees: PERMANOVA P = 0.014, R2 = 0.152; Manhattan distance matrix based on phylum-level OTUs). A detailed account of methodology behind the pairing at different scales can be found in Dataset S1.
The lower C:N litter generated by actively nodulating N2-fixers can instigate changes in underlying soil organic matter and particularly its C:N ratios (26). Consequently, it can be anticipated that when nonfixers grow in neighborhoods rich in fixers, they will exhibit a decline in soil C:N ratios owing generation and redistribution of litter by nearby legume trees. We find evidence for this in the correlation between under-crown soil C:N measurements in nonfixers and fixer stem BA at the 25 m2 scale (Fig. 5H). At the level of microbial nutrition, differences in C:N ratios will translate into selection for disparate C substrates and metabolic pathways that would influence the microbiome structure.
Further analysis revealed that among parameters of soil chemistry, soil pH and soil C:N ratios (both heavily influenced by symbiotic N2-fixation, as discussed above) significantly covaried with soil microbial composition, whereas the nitric acid-extractable elemental concentration of P, Ca, K, Mg, Al, and Fe in the soil did not (Fig. 5I and SI Appendix, Table S6). Previously, we established that the soil microbiomes of N2-fixers and NF-near cluster together and separately from those of NF-far (SI Appendix, Fig. S8). As the sampled population of NF-near (transects four and five) and NF-far trees (transects one, two, and three) are drawn from different sites at different frequencies, this may act as a confounding factor in the observed site effects. To circumvent this, we compared the soil microbiomes under nonfixers as a function of their physical distance (in meters) to the nearest N2-fixing tree. We found that, not unlike the grouping of nonfixers in NF-far and NF-near, the actual physical distance to N2-fixers significantly impacted the soil microbiome composition of nonfixers (permutational multivariate ANOVA [PERMANOVA], P < 0.05) in a stepwise fashion with soils under nonfixers closest to N2-fixers (2.5 m away) resembling the microbiomes of N2-fixers the most and those farthest (>7.0 m away), the least (Fig. 5J).
Although our study concerns N2-fixers in young tropical forests, the general effects of symbiotic N2-fixers on soil pH and C:N ratios have also been recorded from different natural and agricultural ecosystems (21, 26, 52–54). Such effects on soil chemistry may lead to similar changes in different systems, their soil microbiomes and associated biogeochemical cycling, including increases in acidophilic Fe cyclers such as the newly described members of Ca. Acidoferrum, enhanced mineral weathering, and consequent effects on the short- and long-term carbon cycles (5).
Conclusions
Overall, our results indicate that the interaction between N2-fixers and their soil microbial community provides trees within the forest community with improved access to mineral resources that can help meet their high nutrient demands for fast growth and high rates of carbon accumulation. Our metagenomics reveal that N2-fixing trees stimulate weathering through their effects on soil pH, C and N cycling, and enrichment of genes of specific classes of metabolic pathways that link microbial energy metabolism with inorganic mineral nutrient cycling. These gene enrichments are consistent with 1) acidification of the immediate soil matrix by enhanced C mineralization, respiratory CO2 evolution, and carbonic acid production; 2) pH-dependent selection for increased Fe(III)-reducing potential of the microbiome resulting in enhanced reductive dissolution capacity; 3) the acid dissolution of minerals by fermentative and Krebs cycle acid products; and 4) generation of excess H+ by enhanced NO3− leaching as a consequence of greater N inputs and nitrification as well as nodulation. Finally, N2-fixing trees have broad effects on their surrounding forest community, including on weathering rates and soil physicochemical factors, that influence the metagenomes and microbiomes in the surrounding soil. Consequently, increased prominence of acidophilic Fe reducers in soils of N2-fixers, here exemplified by the newly described Ca. Acidoferrum, would in turn enable enhancements in Fe cycling and release inorganic P to support forest growth. Our findings highlight the previously unrealized central role of fast growing N2-fixing legume trees and their soil microbiomes in tropical forest nutrient cycling to support forest production and carbon sequestration.
Methods and Materials
The text below represents only a concise version of the full methods and materials available in SI Appendix, Supplementary Note 8.
The field work and mineral deposition in mesh bags (a total of 504 bags each containing 4 g crushed olivine and placed at each site of beneath 126 individual trees) were carried out in six 0.1 ha transects of secondary tropical forest located in the Agua Salud Secondary Forest Dynamics Network in the Panama Canal Area. Soils in the area are classified as P-poor deep oxisols and inceptisols. Mineralogically, the soils are dominated by kaolinite, quartz, goethite, and hematite (SI Appendix, Supplementary Note 1). The six chosen sites (transects) were of different N2-fixer BA ranging from 6 to 27% total tree stem BA. Soil pH (0.01 M CaCl2), 3 M nitric acid digests, and subsequent inductively coupled plasma optical emission spectrometry (ICP-OES) analyses were performed at the Smithsonian Tropical Research Institute (STRI) Soil Lab. XRF of weathered and fresh minerals and hydrofluoric acid digests of minerals was carried out at the University of Sheffield and ICP-MS was performed by the Environmental Science Soil Lab at the University of Nottingham (SI Appendix, Supplementary Notes 2 and 3). Total DNA was extracted from 0.50 g of each reacted olivine sample pool prior to further homogenization to powder for XRF analyses and from 0.25 g rooting zone topsoil using the MoBio PowerSoil DNA isolation kit according to the manufacturers’ protocols. Amplicon library preparation and MiSeq sequencing were carried out at the STRI Marine and Molecular Lab following procedures outlined in Werner (55).
Shotgun metagenome library preparation and HiSeq sequencing were carried out at the Genomics Institute, University of Edinburgh, using the Nextera XT DNA Library Prep Kit and following manufacturer’s protocols with sequencing carried out at 1 run of HiSeq 4000 sequencer (2 × 150 bp PE). Taxonomy of the rooting zone soil microbial community based on amplicon data were obtained through QIIME1 against the GreenGene database as well as through the IDTAXA Classifier against the GTDB. Shotgun metagenomes were submitted to and annotated by the MG-RAST pipeline using default submission settings. Functional protein domain analyses of the shotgun metagenomes as well as taxonomic assignment of metagenomic reads was achieved using the MGX platform (56). Construction of MAGs was carried out using the following pipeline: MEGAHIT (57), MaxBin2.0 (58), CheckM (59), KBase in-built RAST, and Galaxy-embedded National Center for Biotechnology Information (NCBI) BLAST + blastx function (60). The latter was used for searching the genomes for Fe cycling gene hits against an author’s curated database of Fe cycling genes and their amino acid sequences obtained from several Fe cycling model microorganisms (Dataset S3). Phylogenomic analyses based on average amino acid identity were carried out by uploading the MAG genomes of Pan2503 and UBA7541 in amino sequence FASTA format together with the genomes of representatives of all known acidobacterial classes and orders (downloaded from the NCBI website at https://www.ncbi.nlm.nih.gov/) to EDGAR2.3 (61) and using in-built analytical pipelines therein. Shotgun metagenomes from this work and the MAG of Ca. Acidoferrum panamensis Pan2503 are publicly available on MG-RAST (https://www.mg-rast.org/mgmain.html?mgpage=project&project=mgp80870) and NCBI servers (https://www.ncbi.nlm.nih.gov/assembly/GCA_013450225.1), respectively.
Supplementary Material
Acknowledgments
We gratefully acknowledge funding of D.Z.E. through a European Research Council (ERC) Advanced grant to D.J.B. (CDREG, 2993) and the Leverhulme Trust through a Leverhulme Research Centre award (RC-2015-019). S.A.B. acknowledges funding from the Princeton University Carbon Mitigation Initiative, Natural Environment Research Council Grants NE/M019497/1 and NE/N012542/1, British Council Grant #275556724, and the Leverhulme Trust. This work is a contribution of the Agua Salud Project, a collaboration between the STRI, the Panama Canal Authority, and the Ministry of the Environment of Panama. The Agua Salud Secondary Forest Dynamics network was set up with support from HSBC, and core site operations were supported by Stanley Motta, Small World Institute Fund, the Hoch family, and NSF (Grant EAR-1360391 to J.S.H) during the study period. Agua Salud is part of the Smithsonian Institution Forest Global Earth Observatory.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022241118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.ter Steege H., et al., Continental-scale patterns of canopy tree composition and function across Amazonia. Nature 443, 444–447 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Hedin L. O., Brookshire E. N. J., Menge D. N. L., Barron A. R., The nitrogen paradox in tropical forest ecosystems. Annu. Rev. Ecol. Evol. Syst. 40, 613–635 (2009). [Google Scholar]
- 3.Sprent J. I., Ardley J., James E. K., Biogeography of nodulated legumes and their nitrogen-fixing symbionts. New Phytol. 215, 40–56 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Batterman S. A., et al., Key role of symbiotic dinitrogen fixation in tropical forest secondary succession. Nature 502, 224–227 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Epihov D. Z., et al., N2-fixing tropical legume evolution: A contributor to enhanced weathering through the Cenozoic? Proc. Biol. Sci. 284, 20170370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macleod K. C., Holland P. L., Recent developments in the homogeneous reduction of dinitrogen by molybdenum and iron. Nat. Chem. 5, 559–565 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barron A. R., et al., Molybdenum limitation of asymbiotic nitrogen fixation in tropical forest soils. Nat. Geosci. 2, 42–45 (2008). [Google Scholar]
- 8.Turner B. L., Engelbrecht B. M. J., Soil organic phosphorus in lowland tropical rain forests. Biogeochemistry 103, 297–315 (2011). [Google Scholar]
- 9.Wichard T., Mishra B., Myneni S. C. B., Bellenger J.-P., Kraepiel A. M. L., Storage and bioavailability of molybdenum in soils increased by organic matter complexation. Nat. Geosci. 2, 625–629 (2009). [Google Scholar]
- 10.Chacon N., Silver W. L., Dubinsky E. A., Cusack D. F., Iron reduction and soil phosphorus solubilization in humid tropical forests soils: The roles of labile carbon pools and an electron shuttle compound. Biogeochemistry 78, 67–84 (2006). [Google Scholar]
- 11.Goldberg S., Forster H. S., Godfrey C. L., Molybdenum adsorption on oxides, clay minerals, and soils. Soil Sci. Soc. Am. J. 60, 425–432 (1996). [Google Scholar]
- 12.Wurzburger N., Bellenger J. P., Kraepiel A. M. L., Hedin L. O., Molybdenum and phosphorus interact to constrain asymbiotic nitrogen fixation in tropical forests. PLoS One 7, e33710 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nasto M. K., et al. , Interactions among nitrogen fixation and soil phosphorus acquisition strategies in lowland tropical rain forests. Ecol. Lett. 17, 1282–1289 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Perakis S. S., Pett-Ridge J. C., Nitrogen-fixing red alder trees tap rock-derived nutrients. Proc. Natl. Acad. Sci. U.S.A. 116, 5009–5014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Breugel M., et al., Succession of ephemeral secondary forests and their limited role for the conservation of floristic diversity in a human-modified tropical landscape. PLoS One 8, e82433 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogden F. L., Crouch T. D., Stallard R. F., Hall J. S., Effect of land cover and use on dry season river runoff, runoff efficiency, and peak storm runoff in the seasonal tropics of Central Panama. Water Resour. Res. 49, 8443–8462 (2013). [Google Scholar]
- 17.Lai H. R., Hall J. S., Batterman S. A., Turner B. L., van Breugel M., Nitrogen fixer abundance has no effect on biomass recovery during tropical secondary forest succession. J. Ecol. 106, 1415–1427 (2018). [Google Scholar]
- 18.Carroll-Webb S. A., Walther J. V., A surface complex reaction model for the pH-dependence of corundum and kaolinite dissolution rates. Geochim. Cosmochim. Acta 52, 2609–2623 (1988). [Google Scholar]
- 19.Oelkers E. H., Declercq J., Saldi G. D., Gislason S. R., Schott J., Olivine dissolution rates: A critical review. Chem. Geol. 500, 1–19 (2018). [Google Scholar]
- 20.Hausrath E. M., et al., Short- and long-term olivine weathering in Svalbard: Implications for Mars. Astrobiology 8, 1079–1092 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Homann P. S., van Miegroet H., Cole D. W., Wolfe G. V., Cation distribution, cycling, and removal from mineral soil in Douglas-fir and red alder forests. Biogeochemistry 16, 121–150 (1992). [Google Scholar]
- 22.Tang C., Unkovich M. J., Bowden J. W., Factors affecting soil acidification under legumes. III. Acid production by N -fixing legumes as influenced by nitrate supply. New Phytol. 143, 513–521 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Van Miegroet H., Cole D. W., The impact of nitrification on soil acidification and cation leaching in a red alder ecosystem. J. Environ. Qual. 13, 586–590 (1984). [Google Scholar]
- 24.Ding X., et al., Synergistic interactions between Glomus mosseae and Bradyrhizobium japonicum in enhancing proton release from nodules and hyphae. Mycorrhiza 22, 51–58 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Blossfeld S., Schreiber C. M., Liebsch G., Kuhn A. J., Hinsinger P., Quantitative imaging of rhizosphere pH and CO2 dynamics with planar optodes. Ann. Bot. 112, 267–276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cools N., Vesterdal L., De Vos B., Vanguelova E., Hansen K., Tree species is the major factor explaining C:N ratios in European forest soils. For. Ecol. Manage. 311, 3–16 (2014). [Google Scholar]
- 27.Meyer F., et al., The metagenomics RAST server–A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aserse A. A., Räsänen L. A., Aseffa F., Hailemariam A., Lindström K., Diversity of sporadic symbionts and nonsymbiotic endophytic bacteria isolated from nodules of woody, shrub, and food legumes in Ethiopia. Appl. Microbiol. Biotechnol. 97, 10117–10134 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Patrick W. H. Jr, Gotoh S., Williams B. G., Strengite dissolution in flooded soils and sediments. Science 179, 564–565 (1973). [DOI] [PubMed] [Google Scholar]
- 30.Dakora F. D., Phillips D. A., Root exudates as mediators of mineral acquisition in low-nutrient environments. Plant Soil 245, 35–47 (2002). [Google Scholar]
- 31.Chacon N., Flores S., Gonzalez A., Implications of iron solubilization on soil phosphorus release in seasonally flooded forests of the lower Orinoco River, Venezuela. Soil Biol. Biochem. 38, 1494–1499 (2006). [Google Scholar]
- 32.Tiedje J., Sexstone A. J., Parkin T. B., Revsbech N. P., Shelton D. R., Anaerobic processes in soil. Plant Soil 76, 197–212 (1984). [Google Scholar]
- 33.Chi R., Xiao C., Gao H., Bioleaching of phosphorus from rock phosphate containing pyrites by Acidithiobacillus ferrooxidans. Miner. Eng. 19, 979–981 (2006). [Google Scholar]
- 34.Hamilton W. A., Sulphate-reducing bacteria and anaerobic corrosion. Annu. Rev. Microbiol. 39, 195–217 (1985). [DOI] [PubMed] [Google Scholar]
- 35.Smedley P. L., Kinniburgh D. G., Molybdenum in natural waters: A review of occurrence, distributions and controls. Appl. Geochem. 84, 387–432 (2017). [Google Scholar]
- 36.Brzezińska M., Stępniewska Z., Stępniewski W., Soil oxygen status and dehydrogenase activity. Soil Biol. Biochem. 30, 1783–1790 (1998). [Google Scholar]
- 37.Flessa H., Beese F., Effects of sugarbeet residues on soil redox potential and nitrous oxide emission. Soil Sci. Soc. Am. J. 59, 1044–1051 (1995). [Google Scholar]
- 38.Keiluweit M., Gee K., Denney A., Fendorf S., Anoxic microsites in upland soils dominantly controlled by clay content. Soil Biol. Biochem. 118, 42–50 (2018). [Google Scholar]
- 39.Langille M. G. I., et al., Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cruz-García C., et al., Fnr (EtrA) acts as a fine-tuning regulator of anaerobic metabolism in Shewanella oneidensis MR-1. BMC Microbiol. 11, 64 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Householder T. C., Fozo E. M., Cardinale J. A., Clark V. L., Gonococcal nitric oxide reductase is encoded by a single gene, norB, which is required for anaerobic growth and is induced by nitric oxide. Infect. Immun. 68, 5241–5246 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lipson D. A., et al., Metagenomic insights into anaerobic metabolism along an Arctic peat soil profile. PLoS One 8, e64659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu S., et al., Ecophysiology of Fe-cycling bacteria in acidic sediments. Appl. Environ. Microbiol. 76, 8174–8183 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams B. G., Patrick Jun W. H., Effect of Eh and pH on the dissolution of strengite. Nature 234, 16–17 (1971). [Google Scholar]
- 45.Johnson D. B., Kanao T., Hedrich S., Redox transformations of iron at extremely low pH: Fundamental and applied aspects. Front. Microbiol. 3, 96 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coupland K., Johnson D. B., Evidence that the potential for dissimilatory ferric iron reduction is widespread among acidophilic heterotrophic bacteria. FEMS Microbiol. Lett. 279, 30–35 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Blöthe M., et al., pH gradient-induced heterogeneity of Fe(III)-reducing microorganisms in coal mining-associated lake sediments. Appl. Environ. Microbiol. 74, 1019–1029 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murali A., Bhargava A., Wright E. S., IDTAXA: A novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 6, 140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foesel B. U., et al., Determinants of Acidobacteria activity inferred from the relative abundances of 16S rRNA transcripts in German grassland and forest soils. Environ. Microbiol. 16, 658–675 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Bala A., Murphy P. J., Osunde A. O., Giller K. E., Nodulation of tree legumes and the ecology of their native rhizobial populations in tropical soils. Appl. Soil Ecol. 22, 211–223 (2003). [Google Scholar]
- 51.Sas L., Rengel Z., Tang C., Excess cation uptake, and extrusion of protons and organic acid anions by Lupinus albus under phosphorus deficiency. Plant Sci. 160, 1191–1198 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Bolan N. S., Hedley M. J., White R. E., Processes of soil acidification during nitrogen cycling with emphasis on legume based pastures. Plant Soil 134, 53–63 (1991). [Google Scholar]
- 53.Leary J. K., Hue N. V., Singleton P. W., Borthakur D., The major features of an infestation by the invasive weed legume gorse (Ulex europaeus) on volcanic soils in Hawaii. Biol. Fertil. Soils 42, 215–223 (2006). [Google Scholar]
- 54.Berthold D., Vor T., Beese F., Effects of cultivating black locust (Robinia pseudoacacia L. ) on soil chemical properties in Hungary. Forstarchiv (Hann.) 80, 307–313 (2009). [Google Scholar]
- 55.Werner J. J., Zhou D., Caporaso J. G., Knight R., Angenent L. T., Comparison of Illumina paired-end and single-direction sequencing for microbial 16S rRNA gene amplicon surveys. ISME J. 6, 1273–1276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jaenicke S., et al., Flexible metagenome analysis using the MGX framework. Microbiome 6, 76 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li D., Liu C.-M., Luo R., Sadakane K., Lam T.-W., MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Wu Y.-W., Simmons B. A., Singer S. W., MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W., CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Afgan E., et al., The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537–W544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blom J., et al., EDGAR 2.0: An enhanced software platform for comparative gene content analyses. Nucleic Acids Res. 44, W22–W28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.