Significance
Cellular membranes take on an elaborate set of highly curved and bent shapes which are essential to diverse cellular functions from endocytosis to cell division. The prevailing view has been that membrane bending is driven by proteins with curved shapes, which assemble at the membrane surface to form solid scaffolds. In contrast, here we show that proteins which form liquid-like assemblies on membranes are also potent drivers of bending. These “liquid scaffolds” apply compressive stress to the membrane surface, generating a diverse and dynamic family of membrane shapes. These data, which come at a time when liquid-like protein assemblies are being identified throughout the cell, suggest that protein liquids may play an important role in shaping cellular membranes.
Keywords: membrane biophysics, membrane curvature, protein phase separation
Abstract
Membrane bending is a ubiquitous cellular process that is required for membrane traffic, cell motility, organelle biogenesis, and cell division. Proteins that bind to membranes using specific structural features, such as wedge-like amphipathic helices and crescent-shaped scaffolds, are thought to be the primary drivers of membrane bending. However, many membrane-binding proteins have substantial regions of intrinsic disorder which lack a stable three-dimensional structure. Interestingly, many of these disordered domains have recently been found to form networks stabilized by weak, multivalent contacts, leading to assembly of protein liquid phases on membrane surfaces. Here we ask how membrane-associated protein liquids impact membrane curvature. We find that protein phase separation on the surfaces of synthetic and cell-derived membrane vesicles creates a substantial compressive stress in the plane of the membrane. This stress drives the membrane to bend inward, creating protein-lined membrane tubules. A simple mechanical model of this process accurately predicts the experimentally measured relationship between the rigidity of the membrane and the diameter of the membrane tubules. Discovery of this mechanism, which may be relevant to a broad range of cellular protrusions, illustrates that membrane remodeling is not exclusive to structured scaffolds but can also be driven by the rapidly emerging class of liquid-like protein networks that assemble at membranes.
From endocytic buds (1) to needle-like filopodial protrusions (2), curved membrane surfaces play critical roles in many cellular processes (3). The energetic cost of creating these highly curved surfaces is considerable, such that spontaneous membrane fluctuations are insufficient to establish and stabilize the shapes of cellular membranes (4). Instead, work during the past two decades has revealed that interactions between proteins and lipids drive membrane curvature (5). Multiple physical mechanisms underlie the ability of proteins to shape membrane surfaces. These include amphipathic helices that insert like wedges into one leaflet of the membrane, creating an interleaflet area mismatch that drives curvature (6). Alternatively, proteins with inherently curved membrane binding domains such as BAR domains, dynamin, and ESCRTs act as scaffolds that can stabilize curved membrane geometries (7, 8). While each of these mechanisms relies on structured protein domains, we have recently reported that intrinsically disordered proteins, which lack a stable three-dimensional structure, can also be potent drivers of membrane bending (9, 10). Specifically, when noninteracting disordered domains are crowded together in cellular structures, steric repulsion among them drives the membrane to buckle outward, taking on a curved shape.
Interestingly, rather than repelling one another, many disordered proteins have recently been found to assemble together via weak, multivalent interactions, forming networks that have the physical properties of liquids (11). Notably, recent studies have suggested that liquid–liquid phase separation of membrane-bound proteins plays an important role in diverse cellular processes including nucleation of actin filaments (12), immunological signaling (13), and assembly of virions (14).
How might liquid–liquid phase separation of proteins at membrane surfaces impact membrane curvature? To address this question, we examined phase separation of the N-terminal low-complexity domain of fused in sarcoma, FUS LC, on the surfaces of synthetic and cell-derived membrane vesicles. FUS LC was chosen as a model protein for this study because it is among the most thoroughly characterized examples of a domain that undergoes liquid–liquid protein phase separation in solution (15). Here, we assemble FUS LC on membrane surfaces using an N-terminal histidine tag (16) that binds strongly to lipids with Ni-NTA headgroups. As FUS LC accumulated at the membrane surface, we observed protein phase separation in the two-dimensional plane of the membrane followed by spontaneous inward bending of the membrane, such that protein-lined tubules were created. Similar tubules were observed with two other domains implicated in liquid–liquid phase separation, the low-complexity domain of hnRNPA2 (17) and the RGG domain of LAF-1 (18). Interestingly, the tubules had undulating morphologies, similar to a string of pearls. This phenomenon is associated with an area mismatch between the two membrane leaflets (19, 20), suggesting that protein phase separation pulls lipids toward one another, creating a net compressive stress on one side of the membrane. In line with this hypothesis, a continuum mechanics model, built on the standard Helfrich framework, recreated the tubule morphology when a compressive stress was imposed using spontaneous curvature on the outer membrane surface. Further, the model predicted that tubule diameter should increase with increasing membrane rigidity and increasing rigidity ratio, trends confirmed by our experiments. Collectively, these findings suggest that protein phase separation on membrane surfaces generates considerable stresses that can drive the spontaneous assembly of membrane buds and tubules with physiologically relevant dimensions.
Results
Protein Phase Separation on Membranes Drives Assembly of Protein-Lined Tubules.
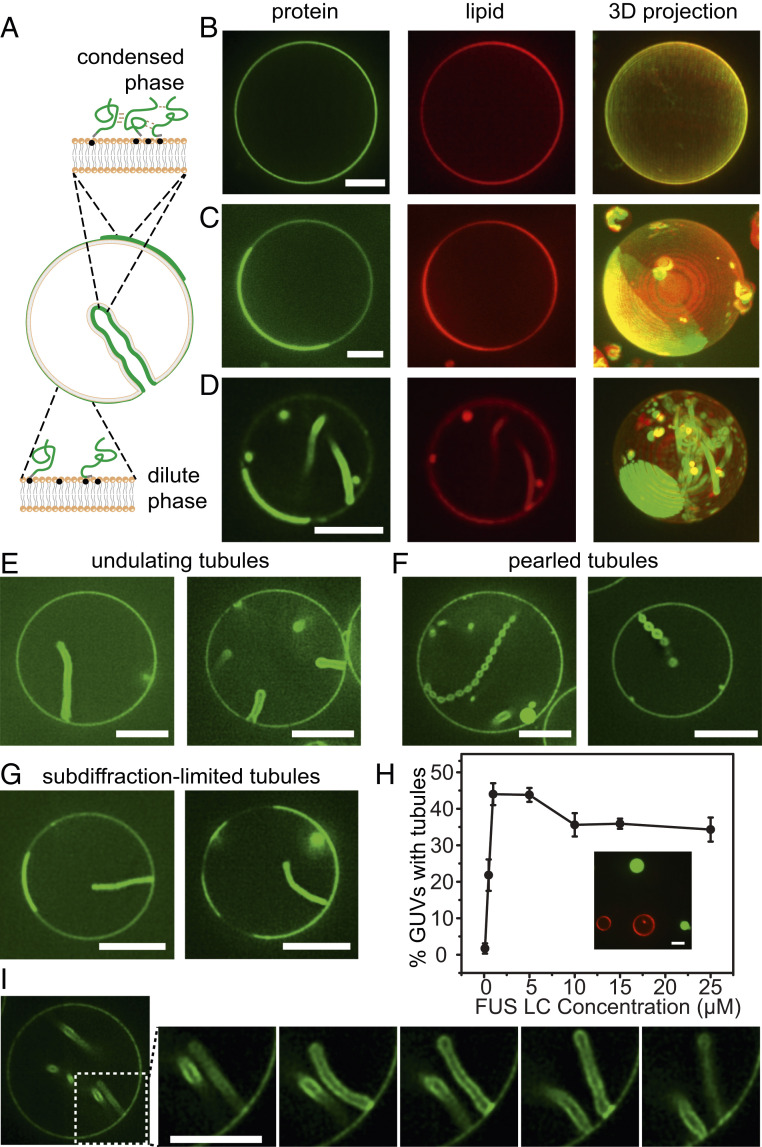
To examine the impact of protein phase separation on membrane surfaces, we combined an N-terminal 6 histidine-tagged version of FUS LC, his-FUS LC, with giant unilamellar vesicles consisting of 93 mol% POPC, 5 mol% Ni-NTA, 2 mol% DP-EG10 biotin for coverslip tethering, and 0.1 mol% Texas Red-DHPE for visualization (Fig. 1A). The protein was labeled at the N terminus with an N-hydroxysuccinimide (NHS)-reactive dye, Atto 488 for visualization, as described under Materials and Methods. Samples were imaged using multichannel, high-magnification spinning disk confocal microscopy. When a protein concentration of 0.5 μM was applied to the vesicles, a relatively dim, uniform signal from the protein was observed at the membrane surface (Fig. 1B). In contrast, when the protein concentration was increased to 1 μM, more intense regions of fluorescence in the protein channel were observed around the vesicle periphery (Fig. 1C). Three-dimensional reconstruction of image stacks revealed that these bright regions formed hemispherical domains on the vesicle surfaces which were surrounded by dimmer regions (Fig. 1C, protein panel).
Fig. 1.
Protein phase separation on membranes drives assembly of protein-lined tubules. (A) Pictorial representation of his-FUS LC liquid–liquid protein phase separation on GUV membranes and inward tubule formation. Green lines represent FUS LC proteins. Gray domains indicated 6×histidine tags, and the black dots indicate Ni-NTA lipids. (B–G) Representative superresolution images of GUVs incubated with 0.5 μM (B) and 1 μM atto-488–labeled his-FUS LC (C–G) in 25 mM Hepes, 150 mM NaCl buffer, pH 7.4. (B–D) Representative confocal images (lipid and protein channels) and corresponding maximum intensity projects of GUVs incubated with his-FUS LC. Some GUVs are covered uniformly by the protein (B), while others display 2D liquid–liquid phase separation (C), which is frequently correlated with the formation of lipid tubules (D). (E–G) Three kinds of membrane tubule structures were observed: undulating tubules (E), tubules consisting of a string of pearls (F), and subdiffraction limited tubules, the structure of which cannot be clearly resolved (G). GUV membrane composition: 93 mol% POPC, 5 mol% Ni-NTA, 2 mol% DP-EG10 biotin, and 0.1 mol% Texas Red-DHPE. (H) The fraction of GUVs displaying inward tubules as a function of his-FUS LC concentration. Data represent mean ± SD, n = 3 independent experiments, and n > 100 GUVs were acquired in each replicate. When the addition of his-FUS LC was greater than 5 μM, protein droplets were observed in the surrounding medium (Inset in H). (I) Confocal image series illustrating dynamic fluctuations in tubule shape. (Scale bars, 5 μm.)
The appearance of these vesicles is remarkably similar to vesicles undergoing phase separation into two coexisting lipid phases (21, 22). In particular, the protein-rich regions in Fig. 1 C and D have smooth, rounded boundaries, suggesting that they enclose an easily deformable liquid (21). However, the membrane composition used in the present study consisted entirely of unsaturated lipids with melting temperatures well below room temperature, such that phase separation of the underlying lipid membrane was not expected. Furthermore, a control protein that is not involved in protein phase separation, histidine-tagged green fluorescent protein (GFP), covered the surfaces of these vesicles uniformly (SI Appendix, Fig. S1). These results suggest that the variations in intensity in the his-FUS LC protein channel did not arise from lipid heterogeneity. Instead, the FUS LC protein appeared to organize on the two-dimensional membrane surface into protein-rich and protein-poor phases. Notably, the head-labeled lipid probe, Texas Red-DHPE, was slightly brighter within the protein-rich regions, likely owing to affinity between the aromatic fluorophore on the lipid headgroup and the FUS LC domain which is enriched in aromatic tyrosine residues (15). However, photophysical effects of FUS LC on Texas Red could also play a role. To separate the lipid label from FUS LC, we also conducted experiments with a tail group–labeled lipid, Texas Red-ceramide. Here, enrichment of the labeled lipid in the protein-rich regions was lost, further suggesting that lipid phase separation does not occur in these vesicles (SI Appendix, Fig. S2).
A few minutes after the addition of his-FUS LC, we observed that many of the vesicles developed lipid tubules spontaneously. These tubules originated at the surfaces of the vesicles and protruded into the vesicle lumen, such that they were lined by the his-FUS LC protein (Fig. 1D). Some of the tubules had an undulating, wavy appearance (Fig. 1 D and E) while others formed a series of tight spheres, resembling a string of pearls (Fig. 1F). Still others were so slender that their morphology could not be precisely determined (Fig. 1G). In some instances, tubules remain associated with protein-rich membrane domains (Fig. 1G), while in other cases, the domains appear to have been consumed, transforming fully into tubules (Fig. 1F). Vesicles were incubated with proteins and given time to come approximately into equilibrium before imaging began. While experiments were performed under isosmotic conditions, the impact of tubule formation on membrane tension was not precisely mapped.
Tubules were observed more frequently as the concentration of his-FUS LC increased (Fig. 1H and SI Appendix, Table S1). Specifically, less than 2% of vesicles formed lipid tubules in the presence of 0.1 μM FUS LC, while 22% and 44% formed tubules after addition of 0.5 μM and 1 μM FUS LC, respectively. However, for protein concentrations above 1 μM, the fraction of vesicles with tubules reached a plateau, likely owing to the appearance of three-dimensional protein droplets in the surrounding solution, which did not appear to be membrane associated (Fig. 1H, Inset). These droplets likely compete with the membrane surface for protein molecules, limiting the further accumulation of protein on the membrane surface.
Importantly, dynamic changes were observed in the morphology of the tubules over time, suggesting that the protein layer on the membrane surface remained highly deformable rather than assembling into a rigid scaffold (Fig. 1I and Movie S1). Additionally, domains of the protein-depleted phase had rapidly fluctuating boundaries and diffused randomly within the protein-enriched phase, observations which further demonstrate the fluid-like nature of the protein-rich phase (Movie S2). To further quantify the relationship between protein concentration and tubule formation, we next varied the strength of protein–protein and protein–membrane interactions and observed the impact on the membrane tubules.
Assembly of Lipid Tubules Depends on the Strength of Protein–Protein and Protein–Membrane Interactions.
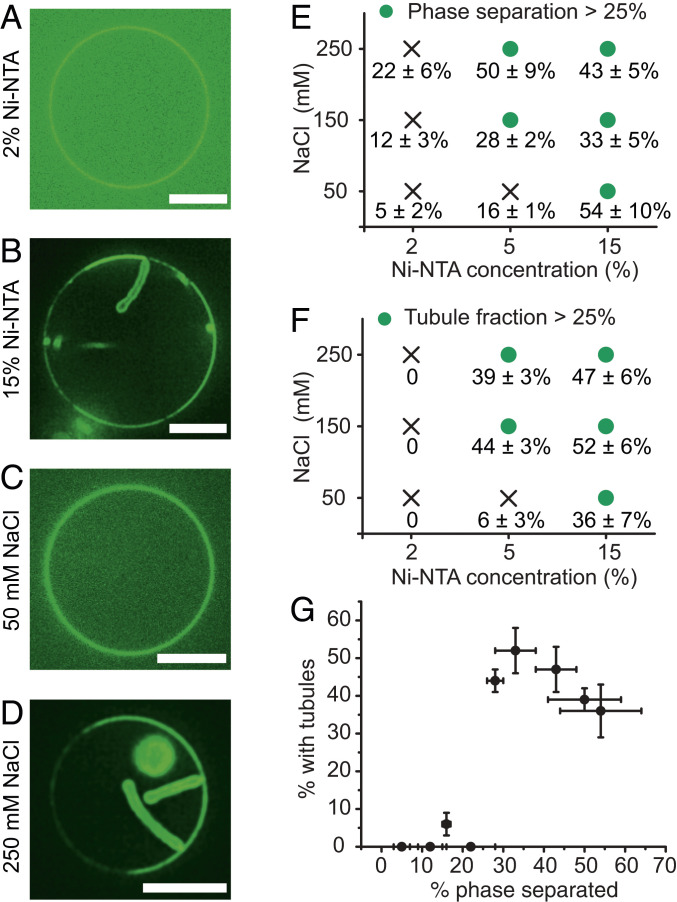
The membrane tubules in Fig. 1 appear to emerge from the protein-rich regions of the membrane surface, suggesting that they rely on self-association of membrane-bound his-FUS LC molecules. We would expect that the ability of these proteins to come together on membrane surfaces depends on both the extent of protein–membrane binding and the strength of protein–protein interactions. Therefore, the assembly of membrane tubules likely depends upon these parameters. To vary the extent of protein–membrane binding, we varied the concentration of Ni-NTA-DOGS lipids in the membrane vesicles. To vary the strength of interactions between his-FUS LC proteins, we varied the concentration of sodium chloride in the solution. This approach is based on published studies showing that the saturation concentration for liquid–liquid phase separation of FUS LC decreases as sodium chloride concentration increases, resulting in enhanced phase separation (16).
Holding the concentration of his-FUS LC constant at 1 μM, we mapped the prevalence of two-dimensional protein phase separation and lipid tubules as a function of both NaCl concentration (50 mM to 250 mM) and the concentration of Ni-NTA-DOGS lipids (2 to 15 mol%) (Fig. 2 A–D). We observed that increasing either parameter led to an increase in both the fraction of vesicles exhibiting phase separation (Fig. 2E and SI Appendix, Table S2) and the fraction of vesicles exhibiting lipid tubules (Fig. 2F and SI Appendix, Table S3). Plotting the fraction of phase separated vesicles versus the fraction of vesicles with lipid tubules reveals a sharp transition to strong tubule formation when ∼25% or more of the vesicles display phase separation (Fig. 2G, Pearson’s correlation coefficient, 0.8). Additionally, the brightness of the protein-rich phases and the tubules were each three to four times greater than the brightness of the protein-depleted phases (SI Appendix, Fig. S3), further suggesting that tubules emerged from the protein-rich phase, though the accuracy may be limited by the errors associated with comparing the intensity of membrane surfaces with varying curvatures (23). Some tubules clearly emerge from protein-rich regions (Fig. 1G Right, Fig. 2 B and D), while the majority appear to consume the protein-rich regions from which they formed (Fig. 1 E and G Left), as summarized in SI Appendix, Fig. S4. Importantly, most protein-rich phases appear to consist of a single layer of protein bound to the membrane surface, based on quantitative fluorescence analysis (SI Appendix, Fig. S5) and poor recruitment of FUS LC proteins lacking a histidine tag to bare membranes and membranes covered by histidine-tagged FUS LC (SI Appendix, Figs. S6 and S7). Notably, lipid phase boundaries have previously been observed to drive membrane budding (22). However, we almost exclusively observe tubules, which have substantially higher curvature than a bud of equal surface area would have, suggesting that phase boundaries are not the primary driver of membrane curvature in the present study.
Fig. 2.
Protein phase separation and tubule formation depend on the concentration of membrane-bound proteins and the strength of protein–protein interactions. (A) Representative confocal images of FUS LC bound to GUVs (composition: 96 mol% POPC, 2 mol% Ni-NTA, 2 mol% DP-EG10-biotin, and 0.1 mol% Texas Red-DHPE) containing 2 mol% Ni-NTA, and (B) GUVs (composition: 83 mol% POPC, 15 mol% Ni-NTA, 2 mol% DP-EG10-biotin, and 0.1 mol% Texas Red-DHPE) containing 15% Ni-NTA. GUVs were incubated with 1 μM Atto-488–labeled his-FUS LC in 25 mM Hepes, 150 mM NaCl pH 7.4 buffer. (C and D) Representative images of GUVs (93 mol% POPC, 5 mol% Ni-NTA, 2 mol% DP-EG10 biotin, and 0.1 mol% Texas Red-DHPE made in 560 mOsmo glucose solution) incubated with 1 μM atto-488–labeled FUS LC in 25 mM Hepes pH 7.4 buffer containing (C) 50 mM and (D) 250 mM NaCl, respectively. Glucose was added to the buffers accordingly to maintain osmotic pressure balance. (Scale bars, 5 μm.) (E and F) Percentage of GUVs displaying (E) protein phase separation and (F) inward lipid tubules as a function of Ni-NTA content and NaCl concentration. Green dots indicate fractions exceeding 25%. (G) Percentage of all GUVs that formed inward tubules as a function of percentage of GUVs with phase separation. Here, the Pearson’s correlation coefficient between phase separation and tubule formation was 0.8. Data are shown as mean value ± SD. n > 100 GUVs were analyzed cumulatively from three independent replicates for each condition. Approximately 18 ± 1% for vesicles containing 15% Ni-NTA and exposed to 1 μM FUS LC displayed both phase-separated regions and membrane tubules.
Collectively, these results demonstrate that formation of protein-lined lipid tubules is strongly correlated with phase separation of his-FUS LC on membrane surfaces. However, it remains unclear why phase separation on membrane surfaces drives the membrane to bend inward toward the lumen of the vesicle. In order to understand this phenomenon, we developed a continuum mechanical model of membrane bending in the presence of protein phase separation.
A Continuum Mechanics Model Predicts Tubule Shape and Dependence of Tubule Diameter on Membrane Bending Rigidity.
The morphologies of the tubules that we have observed can provide insights into the mechanism by which protein phase separation drives membrane bending. Some tubules consist of a well-defined “string of pearls” in which spherical shapes are separated by thin necks (Fig. 1F). Other tubules have an undulating morphology in which the “pearls” are less well defined, with some tubules being nearly cylindrical (Figs. 1E and 2 B and D). This set of shapes—pearls, undulations, and cylinders—can be classified as Delaunay surfaces (24), which have a constant, nonzero mean curvature (Fig. 3A). Unduloids are surfaces of revolution of an elliptic catenary (24, 25). With small changes in geometric parameters, a range of unduloid surfaces can be constructed (24) (Fig. 3A). More importantly, Delaunay surfaces, particularly unduloids and their variants, are known to minimize the Helfrich energy for membrane bending (25). The radius and shape of the unduloids depends on a single dimensionless parameter , in which is the membrane tension, is the bending modulus, and is the spontaneous curvature. When , the membrane resembles a cylinder and for , the membrane resembles an unduloid (Fig. 3A).
Fig. 3.
Mechanical model of undulating and pearled tubule formation. (A) Unduloid-like shapes solution for Helfrich energy minimization at different values of nondimensional parameter, . For α ∼0.75, the membrane takes on a cylindrical shape (purple line); for α > 0.75, the unduloid becomes a sphere similar to a string of pearls (gray line). (B) Schematic depiction of membrane tubule formation due to the compressive stresses applied by liquid–liquid phase separation on the membrane. On a flat membrane, the density of protein segments decreases with increasing distance from the membrane surface, such that x is greater than x’. Therefore, if the membrane remains flat, there will be an increasing number of unsatisfied potential protein–protein interactions as distance from the surface increases. These unsatisfied interactions create a driving force for membrane bending, which increases the density of protein segments at a distance from the membrane (x’ < x), leading to more overlap among the proteins and stronger protein–protein interactions. (C) Schematic of the axisymmetric simulations depicting the simulation domain and the boundary conditions. The yellow region represents the bare membrane, and the green region is the area coated by the proteins. The dashed lines indicate the cap of the tubule, assumed to have a constant curvature. The inset shows the spontaneous curvature distribution along the tubule region used to model the membrane shape. (D) Undulating tubules minimize the membrane bending energy as the spontaneous curvature increases for uniform bending rigidity of the membrane (κ = 80 kBT). (E) Percentage of change in the tubule diameter ((D−Dκ = 25 kBT)/Dκ = 25 kBT) as a function of the bending rigidity for three different values of spontaneous curvature. The dashed lines show a square root dependence on the bending modulus by fitting to the curve (), where for the gray line, A = 5.4, B = −26.44; for the pink line, A = 2.71, B = −12.9; and for the blue line, A = 1.53 and B = −7.4. (F) Pearled tubules minimize the bending energy of the membrane for heterogeneous membrane rigidity (κratio= κprotein-domain/κbare membrane), C0 = 3.5 μm−1. (G) Percentage of change in the tubule diameter ((D − Dκ = 25 kBT)/Dκ = 25 kBT) as a function of the bending rigidity for three different values of spontaneous curvature for κratio = 20. The dashed lines are the fitted curve (), where for the gray line, A = 10.98, B = −51.31; for the pink line, A = 4.22, B = −19.58; and for the blue line, A = 3.1 and B = −13.
Tubules with unduloid-like morphologies are known to arise when there is an area mismatch between the inner and outer leaflets of the lipid bilayer, such that the membrane has a finite spontaneous curvature (26). For example, addition of lipids (19), polymers (20), and proteins (27) to the surfaces of membrane vesicles have each been shown to produce such tubules. However, in these cases, the tubules protruded outward from the membrane surfaces, as would be expected when the area of the outer leaflet exceeds that of the inner leaflet. In contrast, we observe tubules that protrude inward from the membrane surface, suggesting that protein phase separation reduces the area of the outer leaflet relative to that of the inner leaflet (Fig. 3B).
We might expect such a reduction in area if attractive interactions between his-FUS LC peptides generates compressive forces at the membrane surface. How might these compressive forces arise? As an intrinsically disordered domain, FUS LC behaves more like a polymer than like a structured protein domain (15). When polymers are tethered to surfaces, the density of polymer segments decreases substantially as the distance from the surface increases (28) (Fig. 3B). If the membrane were to remain flat, this reduced density of segments would result in a reduction in interactions between the amino acids within FUS LC, as the distance from the membrane surface increased. These unsatisfied interactions create a driving force for membrane curvature. Specifically, if the membrane bends, such that protein-lined buds and tubules are formed, the density of protein segments will increase with increasing density from the membrane surface, such that some portion of the unsatisfied interactions can now be satisfied (i.e., x > x’ in Fig. 3B). Another perspective on this potential mechanism comes from the work of Lipowsky (29) and Sung (30) who have examined the impact of adsorption of polymers on membrane surfaces. The assembly of FUS LC proteins at the membrane surface is analogous to adsorption of a polymer because the network of protein–protein interactions creates a macromolecular condensate that adheres to the membrane at multiple points. Using thermodynamic arguments in combination with the principles of membrane mechanics, these authors showed that when a polymer adsorbs strongly at multiple points to the membrane surface, the membrane will bend in order to maximize contact with the polymer. This bending causes the membrane to form protein-lined structures that effectively engulf the polymer (SI Appendix, Fig. S8). A detailed physical argument and derivation can be found in SI Appendix which shows how the adsorption of FUS LC condensates to the membrane surface could generate compressive stresses that act as an effective spontaneous curvature, driving membrane bending to form protein-lined membrane tubules with diameters that are consistent with our experimental results.
To examine the set of membrane shapes created by this mechanism, we used the Helfrich model with spontaneous curvature to simulate a compressive stress being applied to one leaflet of a lipid bilayer (31). The area difference between the two leaflets was modeled using a locally specified spontaneous curvature for simplicity in simulations (Fig. 3C). See SI Appendix for detailed model assumptions, derivations, and the relationship between the spontaneous curvature and the stresses in the bilayer (SI Appendix, Tables S5 and S6). The spontaneous curvature effectively represents the stresses due to the area difference between the two leaflets (32). The governing equations were solved in an axisymmetric parametrization for ease of computation to demonstrate the principles underlying the formation of undulating and pearled tubules.
We first simulated a domain of fixed area and homogeneous bending rigidity that included the protein-enriched phase and the surrounding protein-depleted phase. Our results showed that increasing the spontaneous curvature in the protein-rich phase resulted in the formation of undulating tubules (Fig. 3D) similar to those observed in experiments (Fig. 1). Furthermore, the simulations predicted that the tubule diameter would increase linearly in proportion to the square root of the bending modulus (Fig. 3E). The bending energy corresponding to the formation of the undulating and pearled tubules is shown in SI Appendix, Fig. S9. Notably, similar morphologies will arise anytime the membrane has a sufficient isotropic spontaneous curvature (19, 20).
It is likely that the protein-enriched phase has an increased bending rigidity compared to the protein-depleted phase, owing to the higher density of protein contacts. Therefore, we next asked if the ratio of bending rigidities in the attached protein layer and the underlying membrane layer could impact the shapes of the tubules. We defined the ratio of bending rigidities, , and varied the ratio in the range of 1 to 20, in which denotes uniform bending rigidity. With increasing , we observed that the tubules took on a more clearly defined pearled morphology (Fig. 3 F and G and SI Appendix, Fig. S10) similar to those observed in some of our experiments (Fig. 1F). We next sought to test these predictions.
Tubule Diameter Varies with Membrane Bending Rigidity and Salt Concentration.
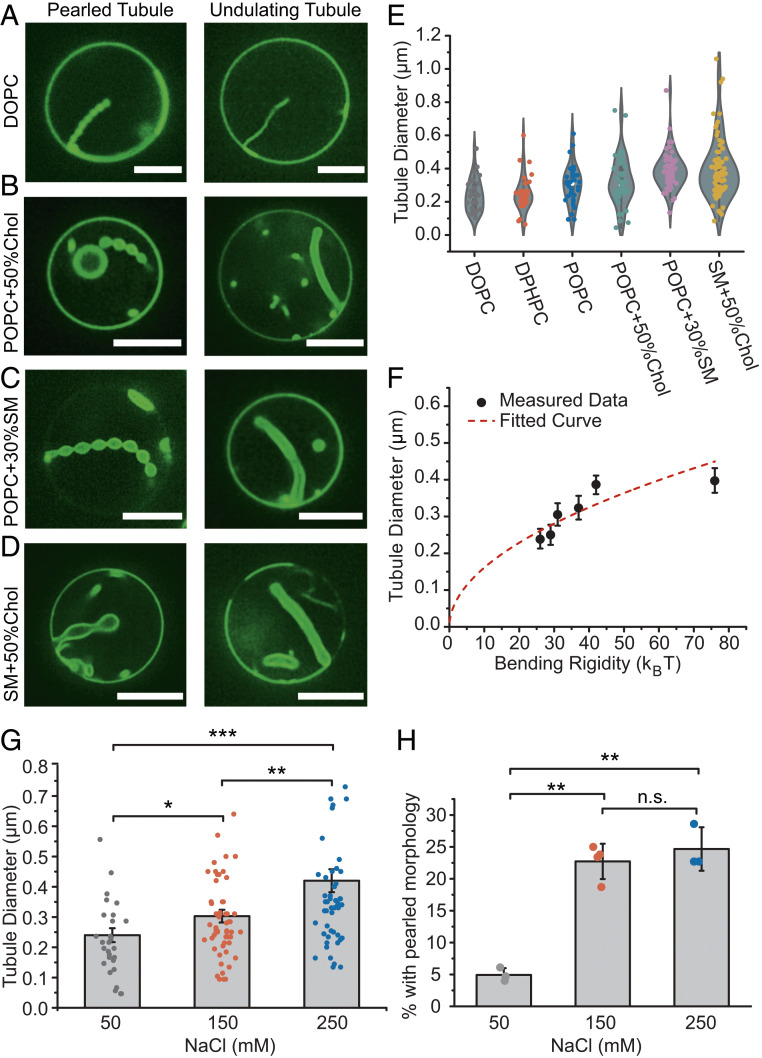
The continuum model predicted that the radii of the tubules should increase in proportion to the square root of the membrane bending rigidity. To examine this prediction, we measured the diameters of the resolvable lipid tubules formed by assembly of his-FUS LC on membrane surfaces as a function of membrane bending rigidity (Fig. 4 A–E and SI Appendix, Table S4). Here the bending rigidity of vesicles having each membrane composition was estimated based on published values for similar compositions, as noted in SI Appendix, Table S1. As membrane bending rigidity was increased from ∼20 kBT to ∼76 kBT, through variations in membrane lipid composition, we observed a substantial increase in membrane tubule diameter from 240 ± 100 nm (SD) to 400 ± 190 nm (SD) (Fig. 4E). For each lipid composition, tubules with both pearled and undulating morphologies were observed (Fig. 4 A–D). Notably, the exact lipid composition and membrane tension likely vary somewhat between vesicles within the same preparation. These differences likely contribute to the variability of tubule diameter, frequency, and morphology. Nonetheless, the data were reasonably well fit by a curve in which tubule diameter was proportional to the square root of bending rigidity, in agreement with the predictions of the simulation (compare Figs. 4F and3 E and G). Here, optical reassignment during spinning disk confocal microscopy, followed by deconvolution, was used to increase the optical resolution to better than 150 nm (33).
Fig. 4.
Tubule diameter varies with membrane bending rigidity and salt concentration. (A–F) Six groups of GUVs with different compositions (listed in SI Appendix, Table S4 and SI Appendix, Materials and Methods) were used to vary membrane bending rigidity. GUVs were incubated with 1 μM atto-488–labeled his-FUS LC in 25 mM Hepes, 150 mM NaCl, pH 7.4 buffer. (A–D) Representative superresolution confocal images tubules with pearled (Left) and undulating morphologies (Right), from GUVs consisting primarily of (A) DOPC, (B) POPC + 50% Chol, (C) POPC + 30% SM, and (D) SM + 50% Chol. (Scale bars, 5 μm.) (E) Violin plot showing the measured tubule diameter distribution for tubules formed using each GUV composition. (F) GUV tubule diameter as a function of membrane bending rigidity. Data points from left to right represent DOPC, DPHPC, POPC, POPC + 50% Chol, POPC + 30% SM, and SM + 50% Chol, respectively. Data are displayed as mean ± SE from at least 60 tubules per composition, gathered during three independent experiments. The measured tubule diameters increase roughly as the square root of membrane bending rigidity (red dash line, R2 = 0.64). (G) Bar chart displaying average tubule diameter under different NaCl concentrations. GUVs (composition: 83 mol% POPC, 15 mol% Ni-NTA, 2 mol% DP-EG10-biotin, and 0.1% Texas Red-DHPE) were incubated with 1 μM atto-488–labeled his-FUS LC in 25 mM Hepes, pH 7.4 buffer with corresponding NaCl concentration under iso-osmotic conditions. Error bars correspond to SE. Each point is a mean value of diameters measured at three positions along the same tubule. n > 100 GUVs were acquired cumulatively from three independent replicates for each condition. (H) Fraction of tubules that displayed a pearled morphology as a function of NaCl concentration. Data are displayed as mean ± SD from three independent experiments (n = 3) on separate preparations of vesicles, with cumulatively n > 100 vesicles categorized. Brackets show statistically significant comparisons using an unpaired, two-tailed Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, and n.s. indicates a difference that was not statistically significant.
A second prediction from our simulation is that the tubule diameter should increase as the rigidity of the protein-rich phase increases while the rigidity of the underlying membrane is held constant. To test this prediction, we examined the impact of sodium chloride concentration on tubule diameter. Increasing sodium chloride concentration has been previously shown to increase the strength of interactions between FUS LC molecules in condensed phases (15). Therefore, we inferred that his-FUS LC might assemble into a more rigid protein layer at high salt concentration.
As the sodium chloride concentration increased from 50 mM to 250 mM, we observed an increase in tubule diameter of ∼75%, from 240 ± 120 nm to 420 ± 280 nm (SD), in qualitative agreement with simulation results (compare Fig. 4G with Fig. 3G). Similarly, increasing either the concentration of histidine-binding lipids or the concentration of his-FUS LC in solution drove a significant increase in tubule diameter (SI Appendix, Fig. S11). One interpretation is that these perturbations result in a greater density of proteins being recruited to the membrane surface, resulting in a more rigid protein layer, which is predicted by the model to increase tubule diameter (Fig. 3G).
Further, the incidence of tightly pearled tubules increased significantly as NaCl concentration increased from 50 mM to 250 mM, in agreement with simulations (Fig. 4H). A second means of increasing the ratio of protein to lipid rigidity is to decrease the rigidity of the lipids, which similarly resulted in an increase in the fraction of pearled tubules (SI Appendix, Fig. S12). Notably, phase separation and formation of protein-lined tubules increased with increasing glutamine content of the FUS LC domain (SI Appendix, Fig. S13) consistent with previous work on the importance of glutamine to phase separation of FUS LC into three-dimensional droplets (16). Collectively, these data suggest that protein phase separation applies a compressive stress to the membrane surface, resulting in assembly of protein tubules directed inward from the membrane surface.
Notably, tubules formed by protein phase separation generally have larger diameters in comparison to tubules formed by rigid protein scaffolds such as BAR domains (34). Additionally, the pearled and unduloid morphologies of tubules formed by phase separation are in contrast to those of tubules formed by rigid scaffolds, which generally have cylindrical morphologies of constant diameter (34). Our simulation generates cylindrical morphologies only when we assume that the protein applies an anisotropic (deviatoric) curvature, as is the case for BAR domain scaffolds (SI Appendix, Fig. S14). Therefore, the pearled and undulating morphologies of tubules observed in the current work suggest that protein phase separation drives membrane bending through a physical mechanism that is distinct from the mechanisms that rigid scaffolds use to deform membranes. However, we acknowledge that several unknowns, including the strength of protein–protein and protein–membrane interactions, the rigidity of the protein layer, and the magnitude of the compressive stresses produced by these interactions, limit our ability to describe this phenomenon in mechanistic detail at present.
Membrane Bending by Protein Phase Separation Is a General Phenomenon that Can Be Driven by Diverse Protein Domains.
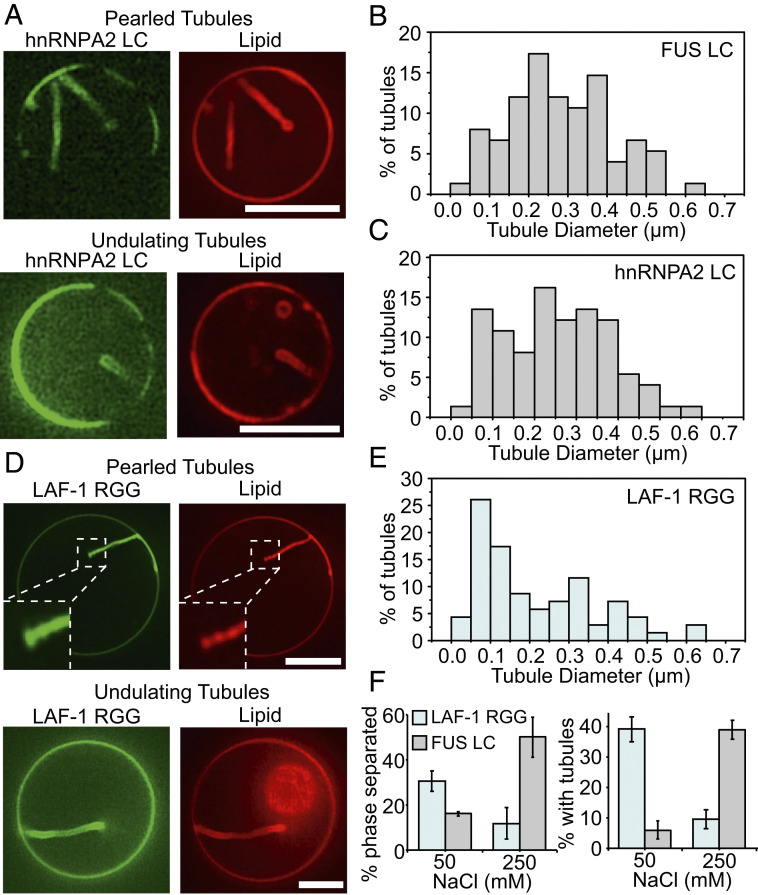
The model we have developed does not take into account the specific amino acid sequence of the FUS LC domain or the particular types of molecular interactions that drive the protein to phase separate. Instead, we have described tubule formation as a general process that could arise whenever protein phase separation occurs at the membrane surface. Therefore, we next asked whether the ability to drive lipid tubule formation is specific to FUS LC or whether it is a general property of membrane-bound domains that undergo liquid–liquid phase separation. To address this question, we evaluated two additional domains known to be involved in liquid–liquid phase separation, the low-complexity domain of hnRNPA2 (hnRNPA2 LC), a protein involved in RNA processing and transport granule formation (17), and the RGG domain of LAF-1 (LAF-1 RGG), a DDX3 RNA helicase found in Caenorhabditis elegans P granules (18). Both proteins contained N-terminal histidine tags, which we used to bring them to the membrane surface, as we did with FUS LC (Fig. 5).
Fig. 5.
hnRNPA2 LC and Laf-1 RGG domains drive formation of inwardly directed membrane tubules with similar morphologies to those formed by FUS LC. (A) his-hnRNPA2 LC at a concentration of 1 μM drove formation of inwardly directed tubules with pearled and undulating morphologies when introduced to GUVs consisting of 83 mol% POPC, 15 mol% Ni-NTA, 2 mol% DP-EG10 biotin, and 0.1 mol% Texas Red-DHPE. (B) Distribution of tubule diameters formed upon exposure of GUVs to his-FUS LC, 75 total tubules. (C) Distribution of tubule diameters formed upon exposure of GUVs to his-hnRNPA2 LC, 75 total tubules. (D) his-Laf-1 RGG at a concentration of 1 μM drove formation of inwardly directed tubules with pearled and undulating morphologies when introduced to GUVs of the same composition as in A. (E) Distribution of tubule diameters formed upon exposure of GUVs to his-Laf-1 RGG, 70 total tubules. (F) The fraction of vesicles exhibiting two-dimensional protein phase separation and tubule formation by his-Laf-1 RGG decreased with increasing salt concentration. This is the opposite trend of that observed for vesicles exposed to his-FUS LC (data repeated from Fig. 2 E and F, for comparison). Error bars represent the SD of three trials, with cumulatively n > 300 GUVs analyzed. (Scale bar in A and D, 5 μm.)
Similar to FUS LC, hnRNPA2 LC is a prion-like domain composed primarily of polar and aromatic residues. It contains relatively few aliphatic residues and is depleted in charged residues (17). Both FUS LC and hnRNPA2 LC have an increased propensity to undergo liquid–liquid phase separation as the ionic strength of the surrounding medium increases (15, 17). Based on these similarities, we might expect hnRNAPA2 LC and FUS LC to have similar interactions at the membrane surface and therefore to behave similarly in our assays. As expected, when hnRNPA2 LC was added to giant vesicles at a concentration of 1 μM, inwardly directed lipid tubules with undulating and pearled morphologies were observed (Fig. 5A). Further, the distribution of tubule diameters was similar between hnRNPA2 LC and FUS LC (Fig. 5 B and C).
In contrast to hnRNPA2 LC and FUS LC, LAF-1 RGG is dense in charged residues such as arginine and aspartic acid (18). In this way, increasing the ionic strength of the surrounding medium opposes liquid–liquid phase separation of LAF-1 RGG (18), suggesting that the dominant driving force for liquid–liquid phase separation is electrostatic attraction between oppositely charged residues. To examine the impact of these differences on the formation of membrane tubules, we added 1 μM of LAF-1 RGG to giant vesicles. Interestingly, we observed inwardly directed lipid tubules, which were similar to those formed by FUS LC and hnRNPA2 LC (Fig. 5D). The diameters of tubules formed by the three proteins covered approximately the same range, though tubules formed by LAF-1 RGG had a somewhat smaller average diameter (Fig. 5E). Importantly, the fraction of giant vesicles that displayed lipid tubules upon exposure to LAF-1 RGG decreased with increasing salt concentration. This trend, which is the opposite of what we observed for FUS LC (Fig. 5F), is expected owing to the ability of high ionic strength solutions to screen the electrostatic interactions that support liquid–liquid phase separation of LAF-1 RGG (18). Notably, changes in the salt concentration may also impact membrane bending rigidity (35). However, the opposite impact of increased salt concentration in FUS LC and LAF-1 RGG experiments indicates that the dominant effect of salt concentration is on protein phase separation, not on membrane bending rigidity. Additionally, the diameter of lipid tubules formed by exposure to LAF-1 RGG increased with increasing membrane bending rigidity (SI Appendix, Fig. S15) while the fraction of pearled tubules decreased (SI Appendix, Fig. S12B), in agreement with our findings for tubules formed by exposure to FUS LC (Fig. 4F and SI Appendix, Fig. S12A).
Collectively, these results demonstrate that the ability of liquid–liquid phase separation at membrane surfaces to drive inward membrane protrusions is a general phenomenon that is not dependent on the specific molecular interactions that drive each protein to phase separate. Instead, liquid–liquid phase separation itself, rather than a particular pattern of electrostatic or hydrophobic interactions between proteins and lipids, appears to be responsible for generating the compressive stress that drives membrane deformation.
Notably, none of the proteins examined in this work are expected to insert into the membrane surface. Indeed, FUS LC did not bind measurably to membranes when its histidine tag was cleaved (SI Appendix, Fig. S6), suggesting that insertion into the membrane is very weak, if it exists. However, proteins that insert into membranes are common and often result in outward membrane bending (36, 37). Such insertions, if coupled to domains that drive liquid–liquid phase separation, could work against the impact of liquid–liquid phase separation on membrane curvature.
Protein Phase Separation Drives Tubule Formation from Cell-Derived Membranes.
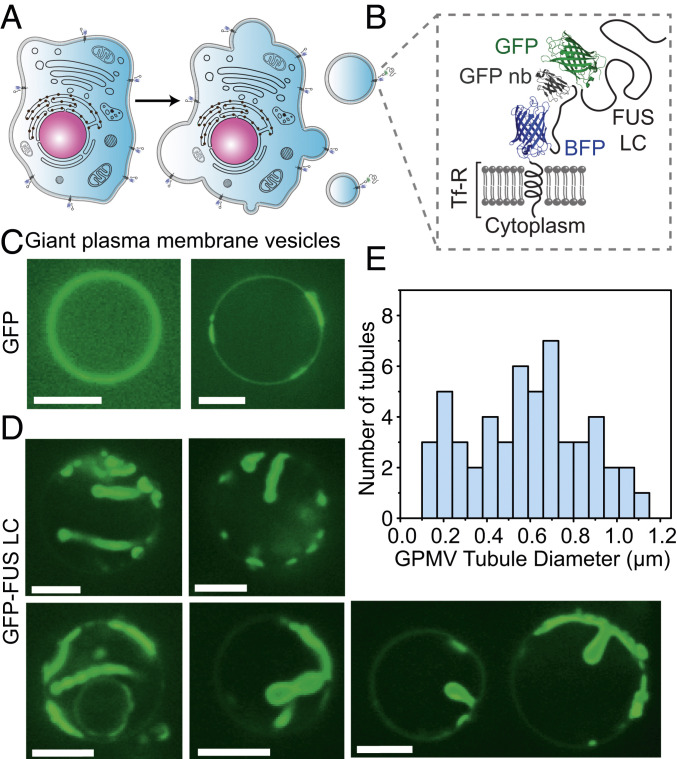
We next asked whether protein phase separation at membrane surfaces is sufficient to drive remodeling of cellular membranes. To address this question, we derived membrane vesicles from the plasma membranes of mammalian retinal pigmented epithelial (RPE) cells. To facilitate binding of FUS LC to the surfaces of these vesicles, we engineered the donor cells to express a chimeric transmembrane protein that consisted of the transmembrane domain of the transferrin receptor, fused to an extracellular blue fluorescing protein (BFP) domain for visualization. This chimera displayed a nanobody against GFP on the cell surface (Fig. 6 A and B). Membrane blebs extracted from these cells also displayed the nanobody on their surfaces, which facilitated the recruitment of GFP-tagged proteins (Fig. 6 A and B). Adding soluble GFP domains to the solution surrounding these blebs resulted in GFP being strongly concentrated at the bleb surfaces (Fig. 6C). Notably, the GFP signal appeared to separate into brighter and dimmer regions on the surfaces of some of the blebs. This separation within blebs has been observed previously (38) and is thought to arise from lipid phase separation in which the transferrin receptor transmembrane domain is known to prefer the liquid disordered membrane phase (39).
Fig. 6.
Protein phase separation can drive tubule formation from cell-derived membranes. (A) Cartoon showing extraction of GPMVs from donor RPE cells. (B) Schematic of the architecture of the membrane receptor and ligand protein. GFP-FUS LC is recruited to the GPMV membrane by binding to a GFP nanobody displayed on the cell surface. (C) Confocal images of GPMVs incubated with 2 μM GFP and (D) GFP-FUS LC in buffer containing 10 mM Hepes, 2 mM CaCl2, 150 mM NaCl, pH 7.4. (Scale bar, 5 μm.) (E) Distribution of diameters of tubules formed from GPMVs. n = 50 tubules measured.
When a GFP-tagged version of FUS LC, GFP-FUS LC (40), was introduced to blebs taken from the same donor cells, the GFP signal was similarly concentrated at the bleb surfaces (Fig. 6D). However, the surfaces of blebs exposed to GFP-FUS LC did not remain flat. Instead, regions of the bleb surfaces with intense GFP signal bent inward, creating protein-lined membrane buds and tubules. Many of the tubules had pearl-like and undulating morphologies, similar to tubules formed by exposure of synthetic vesicles to his-FUS LC (compare Figs. 1 and 6D). The diameter of the tubules ranged broadly from 150 nm to more than 1 μm (Fig. 6E). Here, the average tubule diameter, 570 ± 260 nm (SD), was somewhat larger than that of tubules formed from synthetic membranes. This difference could arise from the enhanced bending rigidity of cell-derived membranes which contain a high density of transmembrane proteins. Alternatively, the GFP-FUS LC protein, which has been observed to form gel-like assemblies in solution (40, 41), may increase the rigidity of the protein layer. Nonetheless, the range of curvatures observed in these cell-derived vesicles encompasses that of many cellular structures including filopodia, dendritic spines, phagosomes, and many organelles (3). These results demonstrate that liquid–liquid phase separation of membrane-bound proteins is sufficient to deform complex, cell-derived membranes. Additionally, because these experiments use an antibody–antigen interaction to bring FUS LC to the membrane surface, rather than a histidine tag, these results show that histidine–lipid interactions are not required for membrane bending by liquid–liquid phase separation. Building on these findings, future work could examine the ability of protein phase separation to drive membrane remodeling in live cells, using either natural or engineered proteins.
Discussion
Here, we demonstrate that protein phase separation at membrane surfaces can drive the assembly of protein-lined membrane tubules of physiologically relevant dimensions. This mechanism appears to be physically distinct from membrane bending by solid scaffolds which include the rigid, tubular assemblies of BAR domains, dynamin, and shiga toxin as well as the cage-like geometries of protein coats formed by clathrin, COPII, and many viral capsids (7, 42–44). In contrast, we show that a family of model proteins that form liquid-like assemblies can drive the formation of membrane tubules with dynamic cylindrical and unduloid morphologies (Fig. 1 and Movie S1). These results illustrate that increasing the spontaneous curvature of a membrane, which is the fundamental requirement for membrane bending (4, 45–48), is not exclusive to structured scaffolds but can also arise from liquid-like protein interactions that generate stresses at membrane surfaces. Using this liquid scaffolding mechanism, cytosolic proteins that phase separate at membrane surfaces could contribute to outward membrane protrusions such as filopodia, dendritic spines, viral buds, and cilia. In contrast, proteins and receptors that assemble into liquid scaffolds on the outer cell surface could contribute to structures that bud into the cell, such as endocytic vesicles.
The inward tubule formation observed here in response to liquid–liquid phase separation is in direct contrast to the outwardly protruding tubules generated by repulsive interactions among self-avoiding disordered domains found in endocytic proteins (9, 10). These two sets of observations can be understood as two extremes of the same mechanism. Specifically, the membrane protein composite can be thought as two layers of a two-dimensional fluid, one layer consisting of lipids and the other consisting of proteins. Many studies have shown that lipid bilayers can only be stretched or compressed by a few percent (49, 50). In contrast, the protein layer is capable of dramatic changes in density. When self-avoiding domains become crowded on the membrane surface, they push each other apart. As the protein layer expands, the nearly inextensible lipid bilayer is forced to bend outward. In contrast, when self-interacting proteins undergo liquid–liquid phase separation on the membrane surface, the protein layer contracts, forcing the nearly incompressible lipid bilayer to bend inward. Similar behavior has been observed in simplified models of biological tissues such as intestine (51) and brain (52), in which tissues fold owing to the differential compressibility of adjacent two-dimensional layers (53), suggesting a common mechanism in soft matter. While structured protein scaffolds are known to induce anisotropic spontaneous curvature, liquid-like scaffolds arising from assembly of disordered proteins are likely to induce isotropic spontaneous curvature. Notably, the formation of tubules and pearls due to anisotropic protein curvatures have been studied extensively using mechanical models (54–57).
What advantage might a liquid scaffold offer for membrane remodeling? We speculate that the lower energy barriers to assembly and disassembly associated with a liquid may allow the membrane greater freedom to deform into a variety of shapes and dimensions rather than the more narrowly defined set of geometries observed for most structured scaffolds. Indeed, many curved membrane structures, from cytoskeletal protrusions (58) to the endoplasmic reticulum (59), are known to have heterogenous and dynamic morphologies. In particular, the unduloid morphology reported here has been observed in the endosomal networks of plants (60). In light of the ongoing discovery of liquid-like behavior in many membrane-bound protein networks (61), the ability of protein phase separation to shape membranes has the potential to impact membrane-associated processes throughout the cell.
Materials and Methods
Reagents.
1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPHPC), 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC), Sphingomyelin (Brain, Porcine), cholesterol, and 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (nickel salt) were purchased from Avanti Polar Lipids, Inc. NaCl, Tris hydrochloride (TrisHCl), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes), neutravidin, Texas Red-DHPE, isopropyl-β-D-thiogalactopyranoside (IPTG), β-mercaptoethanol (β-ME), BODIPY TR Ceramide, and Triton X-100 were obtained from Thermo Fisher Scientific. 2-(N-Morpholino)ethanesulfonic acid hydrate, 4-Morpholineethanesulfonic acid, 3-(Cyclohexylamino)-1-propanesulfonic acid, Urea, NaH2PO4, Na2HPO4, Na3PO4, sodium tetraborate, Ethylenediaminetetraacetic acid (EDTA), phenylmethanesulfonyl fluoride, EDTA-free protease inhibitor tablets, imidazole, poly-L-lysine, ATTO-594 NHS-ester, and ATTO-488 NHS-ester were purchased from Sigma-Aldrich. Dipalmitoyl-decaethylene glycol-biotin (DP-EG10-biotin) was kindly provided by Darryl Sasaki from Sandia National Laboratories (62). Amine reactive polyethylene glycol (mPEG-Succinimidyl Valerate MW 5000) and PEG-biotin (Biotin-PEG SVA, MW 5000) were purchased from Laysan Bio, Inc. Fetal bovine serum, trypsin, penicillin, streptomycin, L-glutamine, phosphate-buffered saline, Ham’s F-12, Ham’s F-12 without phenol red, Dulbecco’s modified Eagle medium (DMEM) and DMEM without phenol red were purchased from GE Healthcare. N-ethylmalemide was purchased from Bio Basic. All reagents were used without further purification.
Plasmids.
The pRP1B FUS 1-163 plasmid for FUS LC (residues 1 to 163) protein incorporating a Tobacco Etch Virus nuclear inclusion-a endopeptidase (TEV)-cleavable N-terminal hexahistidine tag was a gift from Nicolas Fawzi Laboratory, Brown University (16). This plasmid is available from AddGene (https://www.addgene.org/127192/). The plasmid for expression of GFP-FUS LC (residues 2 to 214) was generously provided by the laboratory of Steven McKnight at the University of Texas Southwestern Medical Center (40). The pRSET vector coding for the nondimerizable hexa-his-tagged eGFP (hisGFP A206K) was kindly shared by Dr. Adam Arkin (University of California, Berkeley). The plasmids for QQ4xSS#1 and S12xQ are available from AddGene (https://www.addgene.org/127194/ and https://www.addgene.org/127193/, respectively). The plasmids for hnRNPA2 LC and LAF-1 RGG domain were also obtained from AddGene (https://www.addgene.org/98657/ (Fawzi laboratories) and www.addgene.org/124929/ (Good, Hammer, and Schuster laboratories), respectively).The plasmid for mammalian expression of TfR-∆ecto-BFP-HA-GFPnb was generated by inserting a hemagglutinin (HA)-tag into the TfR-∆ecto-BFP-GFPnb plasmid previously described (63). First, the GFPnb sequence from this original plasmid was amplified by PCR using primers that introduced the HA-tag sequence. The amplified HA-GFPnb sequence was then restriction cloned back into the original plasmid using NotI sites. All constructs were confirmed by DNA sequencing. Notably, the HA-tag was included for screening purposes and did not play a functional role in the present study.
Online Materials and Methods.
SI Appendix, Materials and Methods includes cell culture and production of stable cell line, protein expression and purification, protein labeling, gian unilamellar vesicle (GUV) preparation, GUV tethering, GUV fluorescence imaging, single molecule calibration, giant plasma membrane vesicles (GPMVs), GPMV imaging, statistical analysis, and model development.
Supplementary Material
Acknowledgments
This research was supported by the NIH through Grants R01GM120549 and R35GM139531 (J.C.S.), R01GM132106 (P.R.), R01GM118530 (N.L.F.), R01NS116176 (N.L.F.), F32GM133138 (K.J.D.), NSF 1845734 (N.L.F.), the Human Frontier Science Program RGP0045/2018 (N.L.F.), and by the Welch Foundation F-2047 (J.C.S.). The plasmid DNA for GFP FUS LC was provided by the laboratory of S. McKnight, University of Texas Southwestern Medical Center.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2017435118/-/DCSupplemental.
Data Availability
All processed study data are available in the Supporting Information and main text. Owing to the size of the image files, the raw image data are not available in a public repository. However, all raw and processed data are available upon request to the corresponding authors.
References
- 1.Kirchhausen T., Owen D., Harrison S. C., Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harb. Perspect. Biol. 6, a016725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mattila P. K., Lappalainen P., Filopodia: Molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 9, 446–454 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Hurley J. H., Boura E., Carlson L.-A., Różycki B., Membrane budding. Cell 143, 875–887 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zimmerberg J., Kozlov M. M., How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 7, 9–19 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Stachowiak J. C., Brodsky F. M., Miller E. A., A cost-benefit analysis of the physical mechanisms of membrane curvature. Nat. Cell Biol. 15, 1019–1027 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drin G., Antonny B., Amphipathic helices and membrane curvature. FEBS Lett. 584, 1840–1847 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Daumke O., Roux A., Haucke V., BAR domain scaffolds in dynamin-mediated membrane fission. Cell 156, 882–892 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Christ L., Raiborg C., Wenzel E. M., Campsteijn C., Stenmark H., Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem. Sci. 42, 42–56 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Busch D. J., et al., Intrinsically disordered proteins drive membrane curvature. Nat. Commun. 6, 7875 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snead W. T., et al., BAR scaffolds drive membrane fission by crowding disordered domains. J. Cell Biol. 218, 664–682 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brangwynne C. P., Tompa P., Pappu R. V., Polymer physics of intracellular phase transitions. Nat. Phys. 11, 899 (2015). [Google Scholar]
- 12.Banjade S., Rosen M. K., Phase transitions of multivalent proteins can promote clustering of membrane receptors. eLife 3, e04123 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang W. Y. C., et al., A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science 363, 1098–1103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alenquer M., et al., Influenza A virus ribonucleoproteins form liquid organelles at endoplasmic reticulum exit sites. Nat. Commun. 10, 1629 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burke K. A., Janke A. M., Rhine C. L., Fawzi N. L., Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell 60, 231–241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murthy A. C., et al., Molecular interactions underlying liquid-liquid phase separation of the FUS low-complexity domain. Nat. Struct. Mol. Biol. 26, 637–648 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryan V. H., et al., Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol Cell 69, 465–479.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elbaum-Garfinkle S., et al., The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U.S.A. 112, 7189–7194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanborn J., Oglecka K., Kraut R. S., Parikh A. N., Transient pearling and vesiculation of membrane tubes under osmotic gradients. Faraday Discuss. 161, 167–176, discussion 273–303 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Tsafrir I., et al., Pearling instabilities of membrane tubes with anchored polymers. Phys. Rev. Lett. 86, 1138–1141 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Veatch S. L., Keller S. L., Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophys. J. 85, 3074–3083 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baumgart T., Hess S. T., Webb W. W., Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature 425, 821–824 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Domanov Y. A., et al., Mobility in geometrically confined membranes. Proc. Natl. Acad. Sci. U.S.A. 108, 12605–12610 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreyszig E., Differential Geometry, Mathematical Expositions (University of Toronto Press, Toronto, 1959), pp. xiv + 352. [Google Scholar]
- 25.Naito H., Okuda M., Zhong-can O. Y., New solutions to the Helfrich variation problem for the shapes of lipid bilayer vesicles: Beyond Delaunay’s surfaces. Phys. Rev. Lett. 74, 4345–4348 (1995). [DOI] [PubMed] [Google Scholar]
- 26.Bar-Ziv R., Moses E., Instability and “pearling” states produced in tubular membranes by competition of curvature and tension. Phys. Rev. Lett. 73, 1392–1395 (1994). [DOI] [PubMed] [Google Scholar]
- 27.Snead W. T., et al., Membrane fission by protein crowding. Proc. Natl. Acad. Sci. U.S.A. 114, E3258–E3267 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dimarzio E. A., McCrackin F. L., One-dimensional model of polymer adsorption. J. Chem. Phys. 43, 539–547 (1965). [Google Scholar]
- 29.Hiergeist C., Indrani V. A., Lipowsky R., Membranes with anchored polymers at the adsorption transition. Europhys. Lett. 36, 491–496 (1996). [Google Scholar]
- 30.Kim Y. W., Sung W., Membrane curvature induced by polymer adsorption. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 63, 041910 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Helfrich W., Elastic properties of lipid bilayers–Theory and possible experiments. Z. Naturforsch. C 28, 693–703 (1973). [DOI] [PubMed] [Google Scholar]
- 32.Wiese W., Harbich W., Helfrich W., Budding of lipid bilayer vesicles and flat membranes. J. Phys. Condens. Matter 4, 1647–1657 (1992). [Google Scholar]
- 33.Hayashi S., Resolution doubling using confocal microscopy via analogy with structured illumination microscopy. Jpn. J. Appl. Phys. 55, 082501 (2016). [Google Scholar]
- 34.Frost A., Unger V. M., De Camilli P., The BAR domain superfamily: Membrane-molding macromolecules. Cell 137, 191–196 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helfrich W., Prost J., Intrinsic bending force in anisotropic membranes made of chiral molecules. Phys. Rev. A Gen. Phys. 38, 3065–3068 (1988). [DOI] [PubMed] [Google Scholar]
- 36.Janosi L., Li Z., Hancock J. F., Gorfe A. A., Organization, dynamics, and segregation of Ras nanoclusters in membrane domains. Proc. Natl. Acad. Sci. U.S.A. 109, 8097–8102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ford M. G., et al., Curvature of clathrin-coated pits driven by epsin. Nature 419, 361–366 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Lorent J. H., et al., Structural determinants and functional consequences of protein affinity for membrane rafts. Nat. Commun. 8, 1219 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harder T., Scheiffele P., Verkade P., Simons K., Lipid domain structure of the plasma membrane revealed by patching of membrane components. J. Cell Biol. 141, 929–942 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato M., et al., Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murray D. T., et al., Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell 171, 615–627.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faini M., Beck R., Wieland F. T., Briggs J. A., Vesicle coats: Structure, function, and general principles of assembly. Trends Cell Biol. 23, 279–288 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Luque D., Castón J. R., Cryo-electron microscopy for the study of virus assembly. Nat. Chem. Biol. 16, 231–239 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Römer W., et al., Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature 450, 670–675 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Campelo F., Hernández-Machado A., Model for curvature-driven pearling instability in membranes. Phys. Rev. Lett. 99, 088101 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Callan-Jones A., Bassereau P., Curvature-driven membrane lipid and protein distribution. Curr. Opin. Solid State Mater. Sci. 17, 143–150 (2013). [Google Scholar]
- 47.Fošnarič M., et al., Theoretical study of vesicle shapes driven by coupling curved proteins and active cytoskeletal forces. Soft Matter 15, 5319–5330 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Alimohamadi H., Rangamani P., Modeling membrane curvature generation due to membrane−protein interactions. Biomolecules 8, 120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans E. A., Waugh R., Melnik L., Elastic area compressibility modulus of red cell membrane. Biophys. J. 16, 585–595 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rawicz W., Olbrich K. C., McIntosh T., Needham D., Evans E., Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys. J. 79, 328–339 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang S., Walton K. D., Gumucio D. L., Signals and forces shaping organogenesis of the small intestine. Curr. Top. Dev. Biol. 132, 31–65 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tallinen T., et al., On the growth and form of cortical convolutions. Nat. Phys. 12, 588–593 (2016). [Google Scholar]
- 53.Hughes A. J., et al., Engineered tissue folding by mechanical compaction of the mesenchyme. Dev. Cell 44, 165–178.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kralj-Iglic V., et al., Amphiphile-induced tubular budding of the bilayer membrane. Eur. Biophys. J. 34, 1066–1070 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Iglic A., Babnik B., Gimsa U., Kralj-Iglic V., On the role of membrane anisotropy in the beading transition of undulated tubular membrane structures. J. Phys. Math. Gen. 38, 8527–8536 (2005). [Google Scholar]
- 56.Iglic A., Kralj-Iglic V., Majhenc J., Cylindrical shapes of closed lipid bilayer structures correspond to an extreme area difference between the two monolayers of the bilayer. J. Biomech. 32, 1343–1347 (1999). [DOI] [PubMed] [Google Scholar]
- 57.Sens P., Turner M. S., Theoretical model for the formation of caveolae and similar membrane invaginations. Biophys. J. 86, 2049–2057 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ridley A. J., Life at the leading edge. Cell 145, 1012–1022 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Pendin D., McNew J. A., Daga A., Balancing ER dynamics: Shaping, bending, severing, and mending membranes. Curr. Opin. Cell Biol. 23, 435–442 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buono R. A., et al., ESCRT-mediated vesicle concatenation in plant endosomes. J. Cell Biol. 216, 2167–2177 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Snead W. T., Gladfelter A. S., The control centers of biomolecular phase separation: How membrane surfaces, PTMs, and active processes regulate condensation. Mol. Cell 76, 295–305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Momin N., et al., Designing lipids for selective partitioning into liquid ordered membrane domains. Soft Matter 11, 3241–3250 (2015). [DOI] [PubMed] [Google Scholar]
- 63.DeGroot A. C. M., et al., Entropic control of receptor recycling using engineering ligands. Biophys. J. 114, 1377–1388 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All processed study data are available in the Supporting Information and main text. Owing to the size of the image files, the raw image data are not available in a public repository. However, all raw and processed data are available upon request to the corresponding authors.