Significance
To accomplish the infection cycle, plant viruses usually employ various pathogenic factors to inhibit host antiviral defense. Revealing pathogenic mechanisms conserved among different viruses is essential for developing broad-spectrum disease control. Here, we show that a class of independently evolved transcriptional repressors is widely present in different plant viruses. Despite the diverse sequences and different evolutionary origins, these transcriptional repressors all modulate JA signaling by inhibiting the transcriptional activation of OsMYC transcription factors, disassociating the OsMED25-OsMYC complex, and cooperating with OsJAZ to improve their transcriptional repression activity. This facilitates viral infection and enhances vector feeding. Our findings reveal key functions of the transcriptional repressors in plant viruses and shed light on the general mechanism of viral pathogenicity and vector transmission.
Keywords: plant viruses, transcriptional repressor, jasmonic acid, antiviral defense, vector feeding
Abstract
Plant viruses employ diverse virulence strategies to achieve successful infection, but there are few known general strategies of viral pathogenicity and transmission used by widely different plant viruses. Here, we report a class of independently evolved virulence factors in different plant RNA viruses which possess active transcriptional repressor activity. Rice viruses in the genera Fijivirus, Tenuivirus, and Cytorhabdovirus all have transcriptional repressors that interact in plants with the key components of jasmonic acid (JA) signaling, namely mediator subunit OsMED25, OsJAZ proteins, and OsMYC transcription factors. These transcriptional repressors can directly disassociate the OsMED25-OsMYC complex, inhibit the transcriptional activation of OsMYC, and then combine with OsJAZ proteins to cooperatively attenuate the JA pathway in a way that benefits viral infection. At the same time, these transcriptional repressors efficiently enhanced feeding by the virus insect vectors by repressing JA signaling. Our findings reveal a common strategy in unrelated plant viruses in which viral transcriptional repressors hijack and repress the JA pathway in favor of both viral pathogenicity and vector transmission.
Plant viruses inflict serious damage to many crops throughout the world by hijacking the host plant’s cellular machinery for their replication and spreading inside the plant to achieve systemic infection (1). To establish successful infection, viruses that have little or no genetic relationship to one another may nevertheless employ similar pathogenicity strategies. For example, most plant viruses encode an RNA silencing suppressor to counter post-transcriptional gene silencing, which is a robust plant antiviral defense, but these suppressors are very diverse in sequence (2, 3). Thus, discovering functionally conserved viral proteins in different plant viruses is fundamental to understanding plant virology.
A number of very different RNA viruses are known to infect rice. Rice black-streaked dwarf virus (RBSDV) and Southern rice black-streaked dwarf virus (SRBSDV) are closely related members of the genus Fijivirus (family Reoviridae), with a genome of 10 segments of double-stranded RNA (dsRNA), and which are transmitted in a persistent propagative manner by the small brown planthopper (SBPH, Laodelphax striatellus) and the white-backed planthopper (WBPH, Sogatella furcifera), respectively (4). Rice stripe virus (RSV) is also transmitted by SBPH but is classified in the genus Tenuivirus (family Phenuiviridae) and has a single-stranded RNA (ssRNA) genome of four segments encoding seven proteins by an ambisense expression strategy (5). The recently identified virus rice stripe mosaic virus (RSMV) is a member of the genus Cytorhabdovirus (family Rhabdoviridae) and is naturally transmitted by the leafhopper Recilia dorsalis. RSMV has an undivided ssRNA genome encoding seven proteins on the complementary strand (6). These RNA viruses are widely distributed and cause holistic dwarfism, darkened leaves or stripes, chlorosis, and necrosis of rice plants, resulting in serious yield losses (7). There have been various studies on the function of individual viral proteins (8–11), but we recently reported that these viruses all employ diverse viral proteins to target an auxin response transcription factor and therefore facilitate infection (12), suggesting the presence of a conserved functional strategy among these otherwise unrelated RNA viruses.
Both plant growth and defense responses depend upon fine-tuning control of gene expression regulated by numerous transcriptional activators and repressors. Some plant pathogens also encode transcriptional regulators that assist their infection. An elegant and widely reported example is the phytopathogenic bacterium Xanthomonas, which secretes transcription activator-like (TAL) effectors into plant cells to specifically activate the expression of host genes. TAL effectors enter into the nucleus to induce disease or trigger resistance by binding to target promoters and activate gene expression (13). In plants, ∼10% of transcriptional regulators are repressors (14), including Aux/IAA proteins, Jasmonate ZIM-domain (JAZ) and Novel Interactor of JAZ (NINJA) proteins, and ethylene response factor-associated amphifilic repression (EAR) motif-containing repressors (15, 16). We recently showed that SRBSDV SP8 acts as a transcriptional repressor to interfere with OsARF17-mediated antiviral defense (12), but it is not known whether plant viruses commonly encode functionally conserved transcriptional repressors.
Plants have many defense strategies to counteract viruses and herbivore invasion (17), and several studies have shown the importance of phytohormone-mediated host immunity in combating virus infection, especially involving jasmonic acid (JA) (18–21). The general signal transduction processes following JA perception depend on basic helix–loop–helix (bHLH) transcription factors, of which MYC2 serves as a master regulator while MYC3 and MYC4 have overlapping functions and collectively trigger JA responses (22). Under normal conditions, a group of JAZ proteins physically interact with the NINJA or EAR motif to recruit TOPLESS (TPL) as a corepressor to directly suppress the transcriptional activity of MYC2 along with its homologs MYC3 and MYC4 (23). In response to the increase of JA-Ile, the JA receptor coronatine insensitive1 (COI1), a component of the SCF E3 ubiquitin ligase complex, specifically interacts with JAZ proteins for ubiquitination and degradation via the 26S protease system (24–27). This allows the released MYC2 to physically recruit MED25, a subunit of the mediator transcriptional coactivator complex, to activate the transcription of JA-responsive genes. In addition, the multifunctional MED25 physically interacts with, and coordinates the actions of, multiple regulators during different stages of transcriptional output in JA signaling (28–31). As a countermeasure, pathogens and herbivorous insects often employ their effector proteins to modulate the JA signaling pathway. For example, the effector MiSSP7 of the fungus Laccaria bicolor interacts with and protects PtJAZ6 protein from degradation to impair the JA pathway (32). In addition, it has been reported that the effector HARP1 secreted from the cotton bollworm also overcomes plant defense by interacting with JAZ repressors to restrain the COI1-mediated JAZ degradation, therefore blocking JA signaling (33). Although great progress has been made in studies of bacterial and fungal pathogens, it remains to be determined whether hijacking of JA signaling for pathogenicity is widely conserved among different plant viruses.
In this study, we show that SRBSDV, RBSDV, RSV, and RSMV, despite their diversity, all encode functionally conserved transcriptional repressors that directly inhibit the transcriptional activation of OsMYC3 by physical interaction with its conserved transcriptional activation domain (TAD) motif. These viral transcriptional repressors physically interact with OsMED25 and disrupt its association with OsMYC3. Meanwhile, these independently evolved transcriptional repressors are able to cooperate with host OsJAZ proteins, thereby constituting JA signaling repression complexes to cooperatively attenuate the JA pathway, conferring a benefit both to viral pathogenicity and to the feeding activity of their vectors.
Results
Identification of a Group of Independently Evolved Viral Transcriptional Repressors in Plant RNA Viruses.
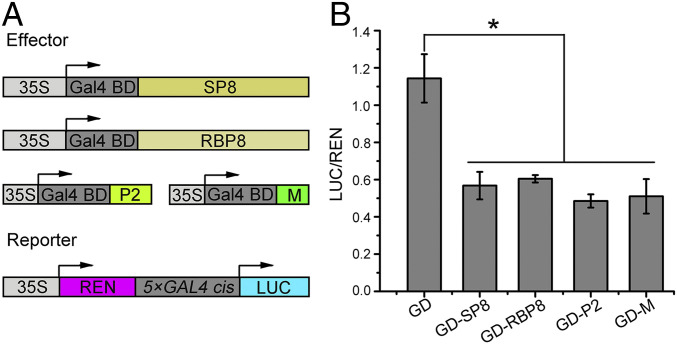
We recently showed that several different viral proteins (SRBSDV SP8, RBSDV P8, RSV P2, and RSMV M) from unrelated plant RNA viruses all target an auxin response transcription factor, but it is not clear whether these viral proteins are functionally conserved. Since RBSDV P8 protein exhibits intrinsic transcriptional repressor activity (34), we wondered whether the other viral proteins listed above also have this property. We first investigated the transcriptional repressor activity of SRBSDV SP8, a homolog of RBSDV P8, by using the GAL4 DNA-binding domain (GD) system, in which the GD specifically recognizes the 5×GAL4 cis element to activate the transcription of the luciferase (LUC) reporter gene (Fig. 1A). Compared with the empty effector GD, there was a striking decrease in LUC activity in the presence of GD-SP8 (Fig. 1B), suggesting that SP8 indeed acts as a transcriptional repressor. We then tested the transcriptional activity of the unrelated RSV P2 and RSMV M proteins (Fig. 1A) and found a dramatic decline of LUC activity in the presence of either GD-P2 or GD-M compared with the empty effector GD (Fig. 1B). Therefore, not only the dsRNA viruses (SRBSDV and RBSDV, Fijivirus) but also the single-stranded negative-sense RNA viruses (RSV, Tenuivirus and RSMV, Cytorhabdovirus) all encode a class of functionally conserved transcriptional repressors, although these repressors have very different sequences.
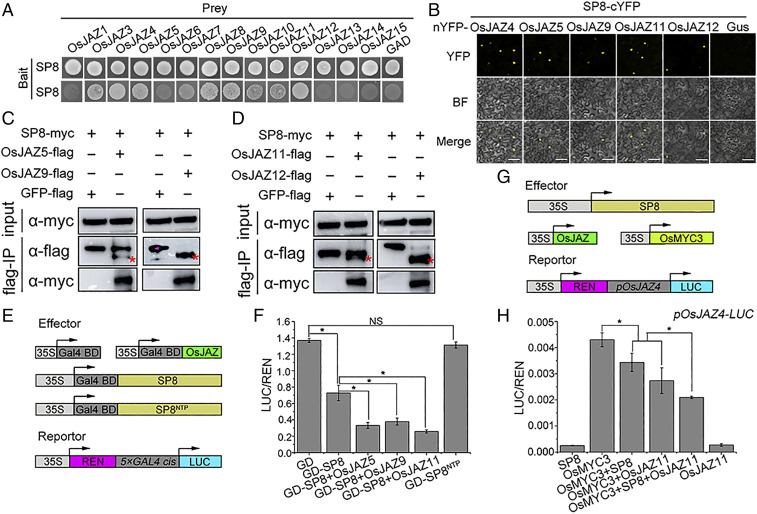
Fig. 1.
Identification of a group of independently evolved viral transcriptional repressors in plant RNA viruses. (A) Diagrams of the reporter and a series of effectors (SRBSDV SP8, RBSDV P8, RSV P2, and RSMV M) used in the GD system. The reporter gene LUC is driven by a concatenated (5×) GAL4 promoter, and specifically recognized GD effectors are driven by the CaMV 35S promoter. REN, renilla luciferase, an internal control; LUC, firefly luciferase. (B) Relative LUC activities measured in N. benthamiana cells. The 5×GAL4::LUC reporter was cotransformed with the indicated GD effectors into N. benthamiana leaves and measured at 48 hpi. * indicates a significant difference between samples analyzed by ANOVA at P ≤ 0.05 by Fisher's least significant difference tests.
SP8 Interacts with OsMYC3.
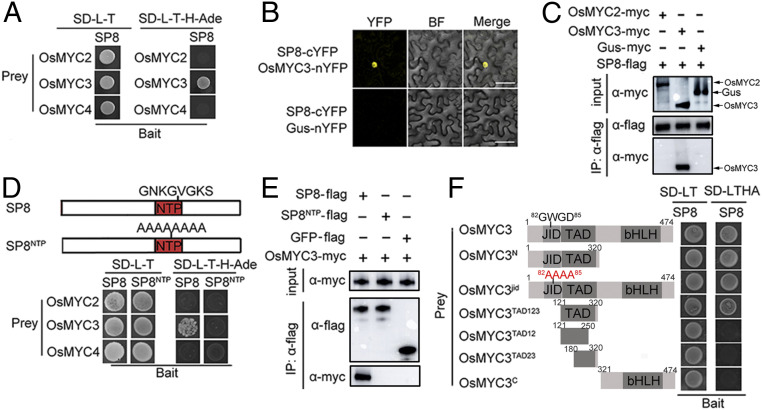
To identify the biological roles of these viral transcriptional repressors, we initially used SRBSDV SP8 as bait to screen a rice complementary DNA (cDNA) library by yeast two-hybrid (Y2H) assay. Preliminary analysis identified the rice gene LOC_Os01g50940, which encodes a bHLH transcription factor OsMYC3, as one of the potential interaction partners of SP8. To validate the interaction between OsMYC3 and SP8, the full-length sequence of OsMYC3 fused with the GAL4 activation domain (AD) and the SP8 protein fused with the GAL4 DNA-binding domain (BD) were coexpressed in yeast cells. As expected, the Y2H results showed that SP8 specifically interacted with OsMYC3 but not with the OsMYC3 homologs OsMYC2 and OsMYC4 (Fig. 2 A and B). Bimolecular fluorescence complementation (BiFC) assays verified the relationship between OsMYC3 and SP8 because strong fluorescence was observed in the nucleus when SP8-cYFP and OsMYC3-nYFP were coinfiltrated in Nicotiana benthamiana leaves but not in the negative control (Fig. 2C). This association was further confirmed by coimmunoprecipitation (Co-IP) experiments. The SP8-flag was expressed together with OsMYC3-myc or OsMYC2-myc in N. benthamiana leaves, and the total proteins were harvested 48 hours post inoculation (hpi) for incubation with FLAG beads. OsMYC3-myc, but not OsMYC2-myc, was coimmunoprecipitated by SP8-flag as detected by the c-myc antibody (Fig. 2D). Similar results from luciferase complementation imaging (LCI) assays confirmed that SP8 only specifically interacts with OsMYC3 in planta (SI Appendix, Fig. S1 A and B). Additionally, our experiments found that OsMYC3 also interacted with RBSDV P8 protein (SI Appendix, Fig. S2), a homolog of SRBSDV SP8, indicating that the viral protein-OsMYC3 interaction was conserved in members of the genus Fijivirus.
Fig. 2.
SP8 interacts with OsMYC3. (A) Interaction of SP8 with OsMYC3, but not with the OsMYC3 homologs OsMYC2 and OsMYC4, in the Y2H system. The full-length sequences of OsMYC2, OsMYC3, and OsMYC4 were amplified into pGADT7 (AD) and SP8 protein fused with pGBKT7 (BD) vector were cotransformed into the yeast strain AH109. Positive yeast transformants were selected on SD-L-T-H-Ade plates at 30 °C. Photos were taken after 3 days. (B) BiFC assays for potential interaction between SP8 and OsMYC3. SP8-cYFP was transiently coexpressed with OsMYC3-nYFP in N. benthamiana leaves and confocal imaged at 48 hpi. Agro-infiltration with SP8-cYFP and Gus-nYFP vector served as a negative control. (Scale bar, 20 μm.) (C) BiFC assays for potential interaction between SP8 and OsMYC3. SP8-cYFP was transiently coexpressed with OsMYC3-nYFP in N. benthamiana leaves and confocal imaged at 48 hpi. Agro-infiltration with SP8-cYFP and Gus-nYFP vector served as a negative control. (Scale bar, 20 μm.) (D) Co-IP assays to analyze the interactions among SP8 with OsMYC2 and OsMYC3 in vivo. Total proteins were incubated with FLAG beads and detected using anti-myc or anti-flag antibody. The sample coexpressing SP8-flag and GUS-myc was the negative control. (E) Co-IP assay confirms the interaction between OsMYC3 and the conserved NTP domain of SP8 in planta. SP8NTP-myc and OsMYC3-flag were coexpressed in N. benthamiana leaves, and total protein was incubated with FLAG beads and detected by anti-myc and anti-flag antibodies. Coinfiltration of SP8-myc and OsMYC3-flag was the positive control, while SP8NTP-myc cotransformed with flag-GFP served as a negative control. (F) Schematic representation of the deleted variations of OsMYC3 used in the Y2H assay. JID, JAZ interaction domain; TAD, transcriptional activation domain; bHLH, the basic helix–loop–helix domain responsible for homo-/hetero-dimerization. The right panel shows that OsMYC3TAD, but not OsMYC3jid or OsMYC3C, interacts with SP8.
MYC2 is a master transcription factor in the JA pathway, while heterodimer of the related bHlH TF MYC3 with MYC2 act redundantly in activating the JA response in the well-studied Arabidopsis (22, 35, 36). In rice, OsMYC2 is known to play important roles in JA signaling (37), but the functions of OsMYC3 in the JA pathway are currently largely unclear. To investigate whether OsMYC3 is involved in the JA pathway, the interactions between OsMYC3 and OsJAZ proteins were initially examined by Y2H assays. The majority of OsJAZ proteins were able to tightly combine with OsMYC3 via the conserved JAZ interaction domain (JID) motif (SI Appendix, Fig. S3 A and C). This conclusion was confirmed by BiFC assays where the reconstituted YFP signals occurred specifically in the nucleus as a result of the physical interaction between OsMYC3 and OsJAZ11 (SI Appendix, Fig. S3B). Similar results were obtained using OsJAZ5 or OsJAZ9 (SI Appendix, Fig. S3B), confirming that OsMYC3, like OsMYC2, functions in association with a subset of OsJAZ proteins. To understand the role of OsMYC3 in the JA pathway, we constructed CRISPR/Cas9 mutant Osmyc3 lines (SI Appendix, Fig. S4A) in the background of rice cultivar Zhonghua11 (ZH11) and then treated the mutants with 0.1 μM and 1 μM methyl jasmonate (MeJA) for 7 days under hydroponic conditions. Intriguingly, the root lengths of ZH11 controls were distinctly reduced in response to MeJA treatment, whereas the repression effect was much smaller in Osmyc3 mutant lines (SI Appendix, Fig. S3 D and E). Further qRT-PCR analysis revealed that, in wild-type ZH11 seedlings, MeJA treatment greatly induced many genes, including OsLOX1, OsLOX2, OsAOC (involved in JA biosynthesis) and OsJAmyb, OsJAZ8, OsJAZ9 (involved in JA signaling transduction), but this induction was significantly compromised in Osmyc3 mutant lines (SI Appendix, Fig. S3 F–K). Together, we conclude that OsMYC3 indeed plays an important role in the JA signaling pathway.
Functional Domains Required for the Interaction between SP8 Protein and OsMYC3 Transcription Factor.
Analysis of the SP8 sequence revealed that it contained a conserved nucleoside triphosphate (NTP)-binding motif in the middle (38). To define the OsMYC3-interacting domains, we constructed a truncated mutant SP8NTP to test the interaction with OsMYC3 (Fig. 2D, Upper). The OsMYC3-SP8 interaction was completely abolished in yeast cells when this conserved NTP-binding motif “GNKGVGKS” was equally substituted with Ala (Fig. 2D, Bottom). Similarly, co-IP assays in leaves of N. benthamiana showed that SP8-myc was successfully coimmunoprecipitated by OsMYC3-flag, while the SP8NTP mutant failed to interact with OsMYC3 (Fig. 2E). These results therefore indicate that the NTP domain of SP8 is essential for the interaction with OsMYC3.
Sequence analysis showed that OsMYC3 displays the typical characteristics of a member of the bHLH transcription factor family. In addition to a basic bHLH domain in the C-terminal portion, it contains a putative TAD and a conserved JID in the N-terminal region (SI Appendix, Fig. S4B). We therefore initially divided OsMYC3 into N-terminal and C-terminal regions for Y2H analysis, finding that only the yeast transformant carrying AD-OsMYC3N and BD-SP8 grew well on SD-L-T-H-Ade selective medium (Fig. 2F), indicating that the N-terminal of OsMYC3 is vital for the interaction with SP8. To further determine whether the JID or TAD domains mediated the OsMYC3-SP8 interaction, we carried out JID site-direct mutagenesis. The mutant OsMYC3jid (in which the conserved JID motif “GWGD” was replaced by “AAAA”) still interacted with SP8 in yeast cells, suggesting that the JID motif was not responsible for the interaction with SP8. Interestingly, the deletion variant OsMYC3TAD123 (amino acid 121 to 320) was competent for interaction with SP8, rather than the deletions OsMYC3TAD12 (aa 121 to 250), OsMYC3TAD23 (aa 180 to 320), or OsMYC3C (aa 321 to 474), in which the TAD domain was partly destroyed (Fig. 2F). Collectively, these results suggest that the functional TAD domain of OsMYC3 is necessary and sufficient for maintaining the interaction with SP8.
SP8 Suppresses the Transcriptional Activation of OsMYC3 to Attenuate JA Signaling.
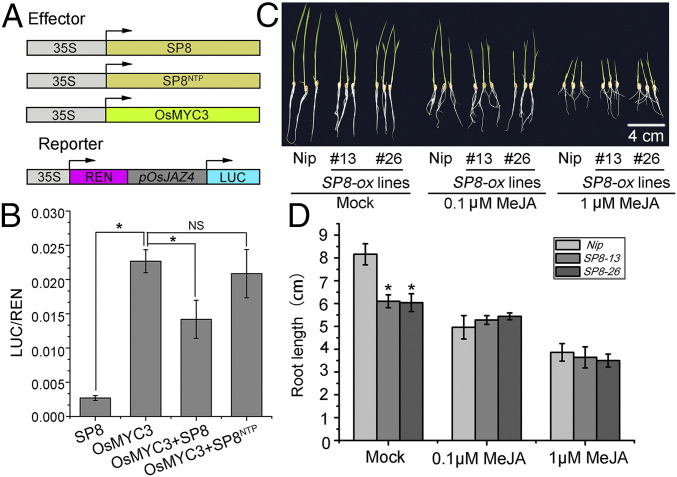
Since SP8 specifically associates with the TAD domain of OsMYC3, which is known to be responsible for transcriptional activation, it seemed likely that SP8 has a role in fine-tuning of the transcriptional activation of OsMYC3. Promoter analysis revealed that the cis elements targeted by the OsMYC3 transcription factor were enriched in the promoter region OsJAZ4 (SI Appendix, Fig. S4C), and we therefore fused the promoter of OsJAZ4 with a firefly luciferase (LUC) vector to construct the reporter pOsJAZ4::LUC (Fig. 3A). In this dual-luciferase transient transcriptional activity assay, the expression level of pOsJAZ4::LUC was significantly enhanced by OsMYC3, but this activation was obviously hindered in the presence of SP8, whereas SP8NTP displayed no detectable repressive effect (Fig. 3B). This therefore shows that SP8 directly suppresses the transcriptional activation of OsMYC3.
Fig. 3.
SP8 suppresses the transcriptional activation of OsMYC3 to attenuate JA signaling. (A) Scheme of the reporter and multiple effectors employed in dual-luciferase transient transcriptional activity assay. The reporter gene LUC was driven by the OsJAZ4 promoter and the effectors shown by the CaMV 35S promoter. REN, renilla luciferase, an internal control; LUC, firefly luciferase. (B) SP8 represses the transcription activity of OsMYC3 on OsJAZ4 promoter. The pOsJAZ4::LUC reporter was cotransformed with the effectors shown into N. benthamiana leaves and measured at 48 hpi. Relative luciferase activities were analyzed by the ratio of LUC/REN, error bars represent SD (n = 6). NS: no significance. (C) Phenotypes of SP8-ox lines (SP8-13 and SP8-26) grown on rice nutrient solution containing 0.1 μM or 1 μM MeJA for 7 days. The root lengths of SP8-ox seedlings were mildly inhibited by MeJA compared with the Nip control. Abbreviation: Nip., Nipponbare, the wild-type rice variety. (D) Quantification of the relative root lengths showing that SP8-ox lines were more insensitive to JA-inhibitory root growth than the control Nip. Error bars represent SD, * in B and D indicates a significant difference between samples analyzed by ANOVA at P ≤ 0.05 in Fisher's least significant difference tests.
To determine the biological significance of SP8 in the JA response, we next generated SP8-ox transgenic lines and identified two homozygous lines with strong expression of SP8, named SP8-13 and SP8-26. We then tested the JA sensitivity of these lines in a root growth assay. In the absence of MeJA, SP8-ox lines exhibited a slightly stunted phenotype (Fig. 3C), consistent with the stunting symptoms of SRBSDV. However, in the presence of 0.1 μM MeJA, the inhibitory effect of root growth was significantly enhanced in the Nipponbare (Nip) background rice seedlings but only slightly in SP8-13 and SP8-26 transgenic plants. There was a similar effect from 1 μM MeJA treatment (Fig. 3 C and D). Further qRT-PCR analysis revealed that most genes involved in the JA pathway, including OsLOX1, OsLOX2, OsAOC involved in JA biosynthesis and OsJAmyb, OsJAZ8, OsJAZ9 involved in JA signaling transduction, were highly induced in Nip seedlings in response to MeJA treatment, whereas this induction was significantly compromised in SP8-13 and SP8-26 plants (SI Appendix, Fig. S5). Together, these observations imply that expression of SP8 protein aggravates the repressive effect on OsMYC3, consistent with the fact that SP8-ox lines were less sensitive to OsMYC3-mediated JA-inhibitory root growth.
SP8 Disturbs the Association between OsMED25 and OsMYC3.
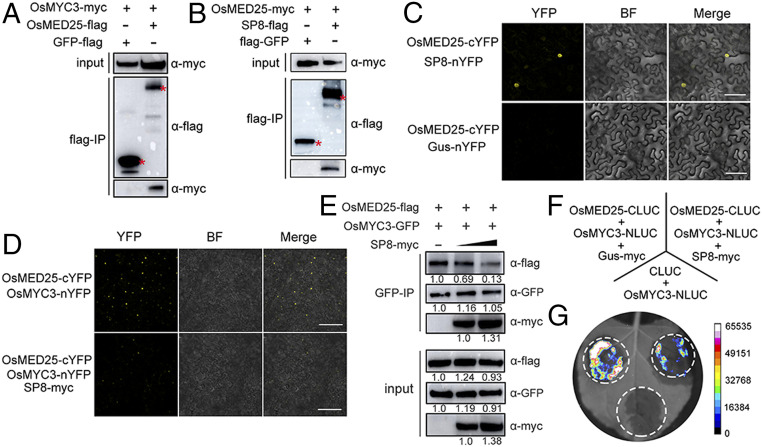
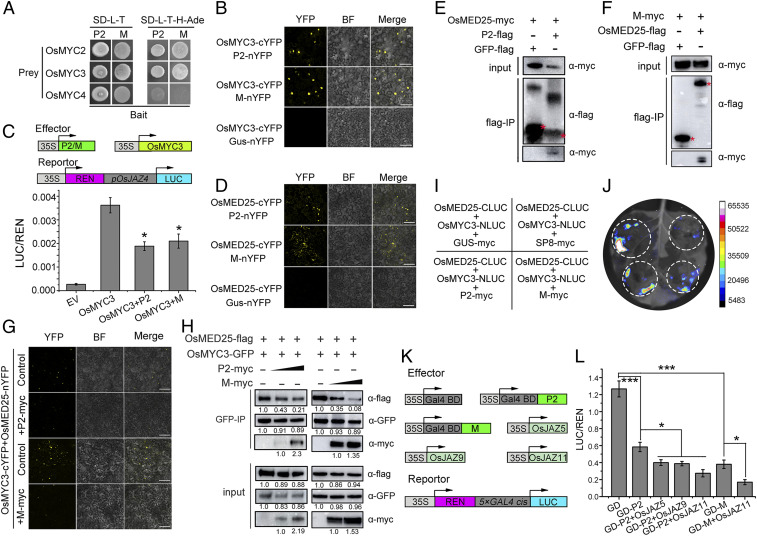
Previous studies have shown that the mediator subunit MED25 functionally interacts with the TAD domain of MYC2, which acts as an integrative hub to coordinate the MYC2-directed transcriptional program (30, 39). More recently, a small group of JA-inducible bHLH proteins, MYC2-TARGETED BHLH1 (MTB1), MTB2, and MTB3, have been identified to antagonistically affect the function of the MYC2-MED25 transcriptional activation complex and therefore negatively regulate JA-mediated transcriptional responses (29). We therefore wondered whether SP8 also causes alterations in the JA signaling pathway by disturbing the association between OsMED25 and OsMYC3. Co-IP assays first showed that OsMED25 fusion protein interacted not only with OsMYC3 but also with SP8 protein in leaves of N. benthamiana, whereas there was no interaction with the negative control green fluorescent protein (GFP)-flag (Fig. 4 A and B). In addition, BiFC assays demonstrated the nuclear localization of OsMED25-cYFP/OsMYC3-nYFP and OsMED25-cYFP/SP8-nYFP (Fig. 4 C and D), further providing direct evidence that they indeed interact with each other in vivo. Importantly, Y2H assays showed that OsMED25 interacted with the same TAD motif of OsMYC3 as SP8 (Fig. 2F and SI Appendix, Fig. S6), indicating that OsMED25 and SP8 may competitively bind to OsMYC3.
Fig. 4.
SP8 disturbs the association between OsMED25 and OsMYC3. (A) Co-IP assay analyzing the interaction between OsMED25 and OsMYC3 in vivo. Total proteins were extracted from N. benthamiana leaves infiltrated with OsMED25-flag and OsMYC3-myc and then incubated with FLAG beads and detected using c-myc antibody. The sample coexpressing GFP-flag and OsMED25-myc is a negative control. (B) Co-IP assay analyzing the interaction between OsMED25 and SP8 in vivo. Total proteins were extracted from N. benthamiana leaves infiltrated with OsMED25-myc and GFP-flag and then incubated with FLAG beads and detected using c-myc antibody. The sample coexpressing GFP-flag and OsMED25-myc is a negative control. (C) BiFC assays to detect interaction between OsMED25 and SP8. OsMED25-cYFP was transiently coexpressed with SP8-nYFP in N. benthamiana leaves and confocal imaged at 48 hpi. Agro-infiltration with OsMED25-cYFP and Gus-nYFP vector served as a negative control. (Scale bar, 50 μm.) (D) SP8 disturbs the OsMED25-OsMYC3 association. Fusion proteins were transiently expressed in leaves of N. benthamiana and observed by confocal microscopy. The YFP signals were reduced in the presence of SP8. (Scale bar, 50 μm.) (E) Interaction between OsMYC3 and OsMED25 was weakened by viral protein SP8. OsMED25-flag combined with OsMYC3-GFP and increasing amounts of SP8-myc were coincubated in leaves of N. benthamiana. The immunoprecipitated fractions were probed with anti-flag, anti-myc, and anti-GFP antibodies. Intrinsic protein levels were evaluated by input in the lower panel. (F) Scheme of LCI assays for coexpression in leaves of N. benthamiana. (G) Results from LCI assays showing that SP8 protein impaired the interaction between OsMYC3 and OsMED25.
We therefore next tested whether SP8 protein disturbs the direct association between OsMED25 and OsMYC3. As shown in Fig. 4D, the reconstituted YFP signal was observed when OsMED25-cYFP and OsMYC3-nYFP were coexpressed in leaves of N. benthamiana, but this fluorescence was much less in the presence of SP8-myc. Quantitative fluorescence signals confirmed that SP8 does indeed disturb the OsMED25-OsMYC3 interaction, and immunoblot analysis of sampling points suggested that all the fusion proteins were successfully expressed in leaves of N. benthamiana (SI Appendix, Fig. S7). Consistent with this idea, we further used a protein competition Co-IP assay to test whether SP8 affects the association of OsMYC3 and OsMED25. Immunoblot analysis showed that both SP8-myc and OsMED25-myc were coimmunoprecipitated by OsMYC3-flag (Fig. 4E). Notably, the ability of OsMED25 and OsMYC3 to associate was markedly and progressively reduced in the presence of increasing amounts of SP8 protein (Fig. 4E). In additional LCI assays, coexpression of OsMYC3 fused with the N-terminal half of LUC and OsMED25 fused with the C-terminal half of LUC in N. benthamiana leaves produced intense luminescence, but this luminescence was much less in the presence of SP8-myc (Fig. 4 F and G). Collectively, these results suggest that SP8 protein indeed competes with OsMED25 for binding to the TAD domain of OsMYC3 and therefore interrupts the OsMED25-OsMYC3 interaction.
SP8 Associates with OsJAZ Proteins to Synergistically Attenuate JA Signaling.
Because MYC-regulated transcriptional activity largely depends on its interaction with endogenous JAZ repressors, it seemed possible that SP8 interacts with OsJAZ proteins and therefore affects downstream transcriptional activation of OsMYC3. A Y2H screen showed that SP8 strongly interacted with OsJAZ4, OsJAZ5, OsJAZ9, OsJAZ11, and OsJAZ12 (Fig. 5A), whereas these interactions were completely abolished in OsJAZ mutants where the “TIFY” motif was changed to Ala (SI Appendix, Fig. S8), supporting the view that these OsJAZ proteins specifically interacted with SP8. In N. benthamiana leaves, both BiFC and Co-IP experiments strongly confirmed that SP8 interacts with OsJAZ proteins in planta (Fig. 5 B–D). We also obtained similar results when RBSDV P8 was combined with OsJAZ5, OsJAZ9, and OsJAZ12 in yeast cells (SI Appendix, Fig. S9). These observations were at first surprising, suggesting that SP8 protein indeed associates with the OsJAZ-OsMYC3 complex, and this formation of a ternary complex organized by viral transcriptional repressors may occur commonly, at least in the genus Fijivirus.
Fig. 5.
SP8 associates with OsJAZ proteins synergistically to attenuate JA signaling. (A) SP8 interacts with many OsJAZ family proteins in Y2H. BD-SP8 was cotransformed with AD-OsJAZs proteins (OsJAZ1 to OsJAZ15, except OsJAZ2) into yeast strain AH109, and positive transformants were selected on SD-l-T-H-Ade plates at 30 °C. Photos were taken after 3 days. (B) BiFC assays confirming the interaction between SP8 and OsJAZs proteins. SP8-cYFP was agro-infiltrated with OsJAZ4/5/9/11/12-nYFP into N. benthamiana leaves and confocal imaged at 48 hpi. The expression pair SP8-cYFP and Gus-nYFP served as a negative control. (Scale bar, 50 μm.) (C and D) Co-IP analysis of SP8 and OsJAZs proteins including OsJAZ5, OsJAZ9, OsJAZ11, and OsJAZ12 in vivo. Total proteins in C and D were purified by FLAG beads and probed with anti-myc antibody. (E) Diagrams of the reporter and a range of effectors used in the GD system. The reporter gene LUC was driven by the 5×GAL4 promoter and specifically recognized GD effectors by the CaMV 35S promoter. REN, renilla luciferase, an internal control; LUC, firefly luciferase. (F) Overexpression of OsJAZs proteins facilitates the transcriptional repressor activity of SP8. The 5×GAL4::LUC reporter was cotransformed with the indicated GD effectors into N. benthamiana leaves and measured at 48 hpi. (G) Schematic representation of the pOsJAZ4::LUC reporter and various effectors in dual-luciferase transient transcriptional activity assay. (H) SP8 interacts with OsJAZs to synergistically suppress the transcriptional activation of OsMYC3. The pOsJAZ4::LUC reporter was cotransformed with the indicated effectors into N. benthamiana leaves and measured at 48 hpi. Relative luciferase activities in F and H were analyzed by the ratio LUC/REN. Error bars represent SD (n = 6); * indicates a significant difference between samples analyzed by ANOVA at P ≤ 0.05 by Fisher's least significant difference tests.
Next, we investigated whether SP8 disturbed the OsJAZ-OsMYC3 association as it had done for the OsMED25-OsMYC3 complex. Because several OsJAZ proteins were involved in interaction with SP8, we initially chose OsJAZ9 for BiFC experiments, finding that the reconstituted YFP signals appeared in the nucleus when OsMYC3-cYFP and OsJAZ9-nYFP were coexpressed in leaves of N. benthamiana and that this fluorescence was not significantly different in the presence of SP8 protein (SI Appendix, Fig. S10, Upper). Further BiFC experiments showed that the SP8 protein does not affect the OsJAZ11-OsMYC3 and OsJAZ12-OsMYC3 interactions (SI Appendix, Fig. S10, Middle and Bottom). Together, these observations demonstrate a negligible effect of SP8 protein on the interaction between OsJAZ and OsMYC3.
Finally, we hence wondered whether the SP8-OsJAZ interaction directly affected the function of SP8. Mutant analysis showed that SP8NTP apparently failed to interact with OsJAZ proteins (SI Appendix, Fig. S11 A and B). To further explore the detailed domain of SP8 interacting with OsJAZs, we then constructed several different NTP-truncated SP8 mutants including SP8N1 (aa 1 to 337, without NTP domain), SP8N2 (aa 1 to 396, with NTP domain), SP8C1 (aa 338 to 592, with NTP domain), and SP8C2 (aa 397 to 592, without NTP domain). Interestingly, both SP8C1 and SP8C2 interacted with a set of OsJAZ proteins, although SP8C2 lacked the NTP motif. However, the interaction was completely abolished in the C-terminal deletion versions SP8N1 and SP8N2, despite the inclusion of functional NTP residues in SP8N2 (SI Appendix, Fig. S11 C and D). These results indicated that the C-terminal of SP8 was involved in association with OsJAZ proteins rather than the NTP domain. It therefore seems that NTP may be more necessary for the enzymatic function or tertiary structure of SP8. On the other hand, it was noteworthy that all the partial fragments of SP8 protein consistently failed to interact with OsMYC3 (SI Appendix, Fig. S11 C and D). It seems possible, therefore, that the domains of SP8 responsible for interacting with OsMYC3 and OsJAZs are independent.
Since the active transcriptional repression activity of SP8 completely depends on a functional NTP domain (Fig. 3B), we speculated that OsJAZ proteins might affect the transcriptional repressor activity of SP8. To test this hypothesis, we conducted dual-luciferase transient transcriptional activity assays. Interestingly, the transcriptional levels of 5×GAL4::LUC were more severely suppressed when OsJAZ proteins were coexpressed with GD-SP8 in contrast with the single GD-SP8 effector (Fig. 5 E and F). As a consequence, the SP8 regulated transcriptional activation of OsMYC3 was markedly repressed when OsJAZ proteins were also infiltrated into N. benthamiana. Our results also showed that OsJAZs have no obvious effect on accumulation of SP8 protein (Fig. 5 G and H and SI Appendix, Fig. S12 A–D). We next tested whether SP8 directly affects the stability of the various OsJAZ proteins. When coexpressed with SP8-myc, there were no significant differences in GFP fluorescence intensity (measured by laser scanning confocal microscopy at 36 hpi) compared to the GUS-myc controls for any of the GFP-tagged OsJAZ proteins (SI Appendix, Fig. S13 A–D), and immunoblot assays also showed that the accumulation levels of OsJAZs-GFP did not seem to be affected by SP8 at 36 hpi (SI Appendix, Fig. S13 E–H). These results support the hypothesis that SP8 associates with OsJAZ proteins to cooperatively repress the OsMYC3-mediated transcriptional program.
Manipulation of the JA Pathway by Viral Transcriptional Repressor Is a Conserved Strategy of Different Plant Viruses.
To explore whether this action of hijacking JA signaling was conserved among very different rice viruses, we performed Y2H assays to test for potential interactions between the viral proteins and JA signaling components. The active viral transcriptional repressors RSV P2 and RSMV M proteins both interacted with OsMYC2 and OsMYC3 but not with OsMYC4 (Fig. 6A). Homology and phylogenetic analysis revealed that OsMYC2 and OsMYC3 were closely related, while OsMYC4 was not (SI Appendix, Fig. S14). In BiFC assays, a strong fluorescence was observed in the nucleus when OsMYC3-cYFP was expressed with either P2-nYFP or M-nYFP in leaves of N. benthamiana, while there was no YFP signal in the negative control (Fig. 6B). We then mapped the key regions required for interaction, finding that the TAD domain of OsMYC3 was indispensable for the association with RSV P2 and RSMV M proteins (SI Appendix, Fig. S15), implying that the P2 and M proteins might also regulate the transcription of OsMYC3. Dual-luciferase transient transcriptional activity assays then showed that the transcriptional activity of OsMYC3 was strikingly reduced when the P2 or M effectors were expressed in plant cells but not when P2 or M were expressed together with the empty GD effector, excluding the possibility of nonspecific binding (Fig. 6C). Additionally, we also found that OsMYC3 directly interacted with OsMYC2 (SI Appendix, Fig. S16 A–D) and cooperatively promoted the transcriptional activity of OsMYC2, but its enhancement of the LUC activity was substantially suppressed when P2 protein was expressed (SI Appendix, Fig. S16 E and F). Taken together, these findings indicate that the P2 and M proteins efficiently repress the transcriptional activity of OsMYC3.
Fig. 6.
Manipulation of the JA pathway by viral transcriptional repressors is conserved in plant viruses. (A) Y2H assays showing the interactions of viral proteins RSV P2 and RSMV M protein with OsMYC transcription factors. Viral proteins were fused with BD while OsMYC transcription factors (OsMYC2, OsMYC3, and OsMYC4) were cloned into AD yeast vectors. All transformants were selected on SD-L-T-H-Ade plates at 30 °C and photographed after 3 days. (B) BiFC assays confirming the interactions of RSV P2 and RSMV M protein with OsMYC3. OsMYC3 and viral proteins P2/M were respectively cloned into cYFP and nYFP vectors and then agro-infiltrated into N. benthamiana leaves together. The samples were imaged by confocal microscopy at 48 hpi. (Scale bar, 50 μm.) (C) Schematic diagrams of the pOsJAZ4::LUC reporter and various effectors. REN, renilla luciferase, an internal control; LUC, firefly luciferase. The reporter was coinfiltrated with OsMYC3 and viral proteins P2/M into N. benthamiana leaves and measured at 48 hpi, while the sample with pOsJAZ4::LUC empty vector was the negative control. Relative luciferase activities were analyzed by the LUC/REN ratio. Error bars represent SD (n = 6); * indicates a significant difference between samples analyzed by ANOVA and evaluated at P ≤ 0.05 by Fisher's least significant difference tests. (D) BiFC assays confirming the interactions of RSV P2 and RSMV M protein with OsMED25. OsMED25 and viral proteins P2/M were respectively cloned into cYFP and nYFP vectors and then agro-infiltrated into N. benthamiana leaves together. The samples were imaged by confocal microscopy at 48 hpi. (Scale bar, 50 μm.) (E) Co-IP assays to examine the interactions between OsMED25 and RSV P2 in vivo. Total proteins were extracted from N. benthamiana leaves coexpressing OsMED25-myc and P2-flag, the supernatant was precipitated with FLAG beads, and the associated proteins were verified using anti-myc antibody. The sample expressing OsMED25-myc and GFP-flag served as a negative control. (F) Co-IP assays to examine the interactions between OsMED25 and RSMV M protein in vivo. Total proteins were extracted from N. benthamiana leaves coexpressing OsMED25-flag and M-myc, the supernatant was precipitated with FLAG beads, and the associated proteins were verified using anti-myc antibody. The sample expressing M-myc and GFP-flag served as a negative control. (G) Viral proteins P2 and M disturb the association of OsMED25-OsMYC3. Fusion proteins were transiently expressed in leaves of N. benthamiana and observed by confocal microscopy. The YFP signals were reduced in the presence of P2 or M protein. (Scale bar, 100 μm.) (H) Interaction between OsMYC3 and OsMED25 was weakened by viral proteins RSV P2 and RSMV M. OsMED25-flag combined with OsMYC3-GFP and increasing amounts of P2/M-myc were coincubated in leaves of N. benthamiana. The immunoprecipitated fractions were detected by anti-flag, anti-myc, and anti-GFP antibody, respectively. Intrinsic protein levels were evaluated by input in the lower panel. (I) Scheme of LCI assays for coexpression in leaves of N. benthamiana. (J) Results from LCI assays showing that RSV P2 and RSMV M proteins impaired the interaction between OsMYC3 and OsMED25. SP8 acts as a positive control. (K) Schematic diagrams of the reporter and a series of effectors employed in the GD system. The reporter gene LUC driven by the 5×GAL4 promoter specifically recognized GD effectors driven by the CaMV 35S promoter. REN, renilla luciferase, an internal control; LUC, firefly luciferase. (L) RSV P2 and RSMV M protein have transcriptional repressor activity and are aggravated by OsJAZs proteins. The 5×GAL4::LUC reporter was coinfiltrated with the GD-P2/M effectors alone or together with OsJAZs proteins into N. benthamiana leaves and measured at 48 hpi. Data were analyzed by the LUC/REN ratio. Error bars represent SD (n = 6); * indicates a significant difference between samples at P ≤ 0.05 by Fisher's least significant difference tests.
BiFC experiments then demonstrated that the viral transcriptional repressors P2 and M interacted with OsMED25 since YFP fluorescence was observed in the combinations OsMED25-cYFP/P2-nYFP and OsMED25-cYFP/M-nYFP (Fig. 6D). These observations were further confirmed by Co-IP experiments, finding that OsMED25 fusion proteins were both coimmunoprecipitated by P2 and M but that there was no interaction with the negative GFP-flag (Fig. 6 E and F). Together, these results indicate that both P2 and M interact with OsMED25. The effect of P2 or M proteins on the OsMED25-OsMYC3 interaction was further investigated by BiFC assays. In contrast to the control, the fluorescence signal of OsMED25-cYFP/OsMYC3-nYFP was significantly decreased in the presence of P2 or M protein (Fig. 6G and SI Appendix, Fig. S17). To test whether this disruption of the OsMED25-OsMYC3 interaction by viral transcriptional repressors occurs generally in all the viruses being studied, we further designed protein competition Co-IP assays in leaves of N. benthamiana. As shown in Fig. 6H, immunoblot analysis demonstrated that OsMYC3 could combine with OsMED25 and viral proteins (RSV P2 and RSMV M) but that the association between OsMED25 and OsMYC3 was markedly reduced in the presence of increasing amounts of P2 or M. In addition, LCI assays showed that coexpression of OsMYC3-NLUC and OsMED25-CLUC in N. benthamiana leaves produced intense luminescence, but this luminescence was much less in the presence of P2-myc or M-myc (Fig. 6 I and J). Together, these results therefore support the conclusion that disruption of the OsMED25-OsMYC3 interaction by viral transcriptional repressors is a conserved strategy of the different plant viruses studied.
Several OsJAZ proteins were identified as interaction partners of both RSV P2 and RSMV M protein in yeast cells (SI Appendix, Fig. S18A), and these interactions were again confirmed by BiFC assays in vivo (SI Appendix, Fig. S18B). BiFC assays also showed that neither P2 nor M protein affected the OsJAZ11-OsMYC3 interaction (SI Appendix, Fig. S18 C and D). Together, the evidence demonstrates that these new active viral transcriptional repressors, RSV P2 and RSMV M protein, resemble the fijivirus P8 proteins by interacting with the key components of JA signaling and suggest the existence of a P2/M-OsMYC-OsJAZ ternary complex. We therefore examined whether the transcriptional repressor activity of P2 and M proteins could be affected by OsJAZ proteins. As shown in the dual-luciferase transient transcriptional activity assays, when OsJAZ proteins were present, the transcription level of LUC was much lower compared with the single GD-P2 or GD-M effector (Fig. 6 K and L), indicating that P2 and M protein prefer to associate with OsJAZ proteins to enhance their repression activity. In addition, confocal observation and immunoblot analysis jointly proved that, like SP8, RSV P2 and RSMV M proteins do not alter the stability of OsJAZ proteins (SI Appendix, Fig. S19). Taken together, these results reveal a conserved strategy among very diverse plant RNA viruses in which independently evolved viral transcriptional repressors manipulate the JA signaling pathway.
OsMYC3 Actively Regulates Broad-Spectrum Antiviral Defense against the dsRNA Virus SRBSDV and the ssRNA Virus RSV.
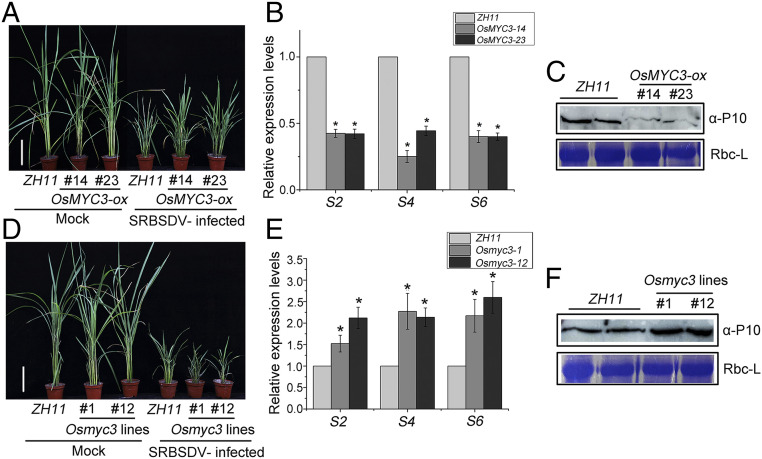
The ability of viral transcriptional repressors to conservatively repress the transcriptional activation of OsMYC3 prompted us to explore the potential effect of the JA signaling pathway on viral infection. We first investigated the impact of OsMYC3 on dsRNA virus (SRBSDV) infection by inoculating transgenic rice plants from the T2 generation: two homozygous OsMYC3-ox lines, named OsMYC3-14 and OsMYC3-23, and CRISPR/Cas9 mutants Osmyc3-1 and Osmyc3-12. About 30 days after inoculation, most plants were distinctly stunted but slightly more so in OsMYC3-14 and OsMYC3-23 overexpression lines than in the ZH11 controls (Fig. 7A). The amounts of viral RNA (three different SRBSDV genomic RNA segments S2, S4, and S6) assessed by qRT-PCR and the amount of the viral outer capsid protein P10 detected by Western blot were all less in OsMYC3-ox plants than in ZH11 controls (Fig. 7 B and C). Conversely, Osmyc3-1 and Osmyc3-12 mutants were more severely dwarfed by SRBSDV (Fig. 7D) and had greater accumulations of virus both in RNA and protein levels relative to ZH11 controls (Fig. 7 E and F). Thus, OsMYC3-ox lines were more resistant, whereas the Osmyc3 mutants were more susceptible to SRBSDV infection. Together, these results suggest that OsMYC3 plays crucial roles in rice defense against SRBSDV.
Fig. 7.
OsMYC3 positively regulates rice resistance to SRBSDV. (A and D) Symptoms in OsMYC3-ox lines (OsMYC3-14 and OsMYC3-23) (A) and Osmyc3 mutants (Osmyc3-1 and Osmyc3-12) (D) following mock-inoculation or SRBSDV infection. The diseased rice plants were verified by RT-PCR and photographs were taken at 30 dpi. (Scale bar, 10 cm.) (B and E) qRT-PCR results showing the relative expression of viral RNA (three different RBSDV genomic RNA segments S2, S4, and S6) in SRBSDV-infected OsMYC3-ox lines (B) and Osmyc3 mutants (E) at 30 dpi. OsUBQ5 was used as the internal reference gene and data were compared with ZH11 background from three biological replicates in a one-way ANOVA and evaluated at P ≤ 0.05 by Fisher's least significant difference tests. Abbreviation: S2, Segment 2; S4, Segment 4; S6, Segment 6. (C and F) Western blot to assess the accumulation of SRBSDV P10 in SRBSDV-infected OsMYC3-ox lines (C) and Osmyc3 mutants (F) compared with ZH11 at 30 dpi. Total proteins were extracted from SRBSDV-infected transgenic rice leaves and examined by anti-P10 antibody.
In similar experiments using the ssRNA virus RSV, typical symptoms of chlorosis, striping, and necrosis began to appear in leaves at about 20 days post inoculation (dpi). Symptoms were milder in the overexpression lines OsMYC3-14 and OsMYC3-23, whereas the area of necrosis on Osmyc3-1 and Osmyc3-12 plants tended to be more severe than that on ZH11 wild-type plants (SI Appendix, Fig. S20 A and B). Furthermore, qRT-PCR analysis showed that the transcription level of RSV coat protein (CP) was approximately fourfold lower in OsMYC3-ox lines and about twofold higher in Osmyc3 mutants than in the ZH11 controls at 30 dpi (SI Appendix, Fig. S20 C and D). Consistently, the accumulation of RSV CP protein detected by Western blot was lower in OsMYC3-14 and OsMYC3-23 but higher in Osmyc3-1 and Osmyc3-12 than in ZH11 controls (SI Appendix, Fig. S20E). These results indicate that OsMYC3 positively regulates rice defense against RSV. Thus, OsMYC3 shows broad-spectrum antiviral defense against the dsRNA virus SRBSDV and the single-stranded negative-sense RNA virus RSV.
Blocking of JA Signaling Promotes Vectors Feeding.
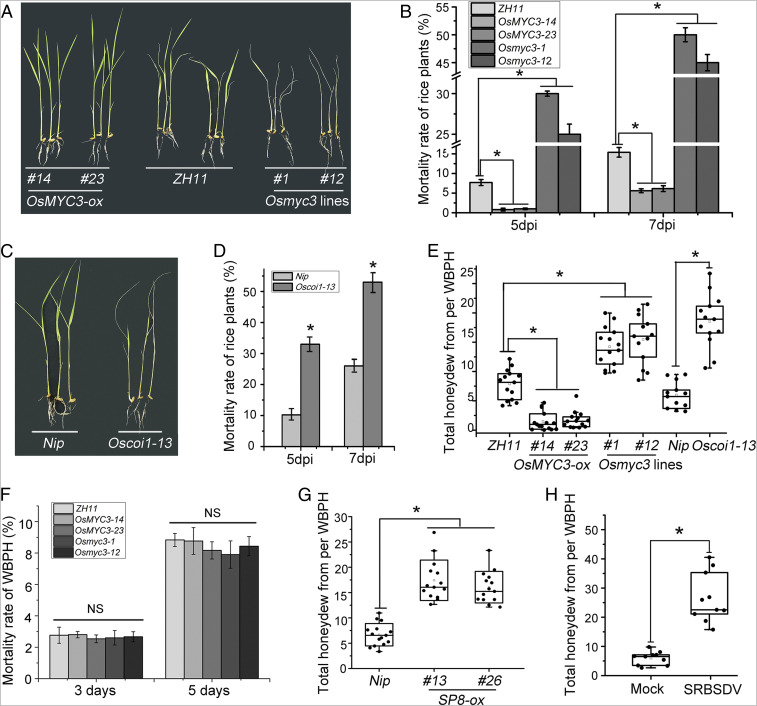
It has previously been shown that the JA pathway plays a central role in response to insect attack (40), and it is generally accepted that JA is effective against chewing herbivores, while salicylic acid (SA) is involved in plant defense against phloem-sucking insects (33, 41–43). However, these results have been from dicotyledonous plants, and the effects of JA on resistance to the planthoppers (WBPH and SBPH) are not known, despite the fact that these two phloem-feeding insects also seriously threaten rice production by transmitting viruses. The JA-insensitive mutants (Osmyc3-1 and Osmyc3-12) and overexpression lines (OsMYC3-14 and OsMYC3-23) were therefore exposed to a WBPH colony for 3, 5, or 7 days. There were obvious differences between these genotypes in their tolerance of WBPH: Osmyc3-1 and Osmyc3-12 mutants tended to be more susceptible with wilting and even death while the survival rates of OsMYC3-14 and OsMYC3-23 overexpression lines were consistently greater than that of the ZH11 control (Fig. 8 A and B). Similar results were obtained using Oscoi1-13, a JA receptor OsCOI1 mutant (Fig. 8 C and D), and also when plants were exposed to SBPH (SI Appendix, Fig. S21 A–D). These results indicate that suppression of JA signaling results in decreased resistance to the phloem-feeding insects WBPH and SBPH.
Fig. 8.
Blocking of JA signaling promotes vector feeding. (A) Phenotypes of OsMYC3-ox, Osmyc3 mutants, and ZH11 plants 7 days after infestation with five adult WBPHs per seedling. (B) Mortality rate of OsMYC3-ox, Osmyc3 mutants, and ZH11 plants infested by adult WBPHs. For each genotype, ∼20 seedlings were tested, and the mortality was counted at 5 dpi and 7 dpi. Data were analyzed by one-way ANOVA and evaluated at P ≤ 0.05 by Fisher's least significant difference tests. (C) Phenotypes of Oscoi1-13 mutants and Nip plants 7 days after infestation with five adult WBPHs per rice seedling. (D) Mortality rate of Oscoi1-13 mutants and Nip plants infested by adult WBPHs. For each genotype, ∼20 seedlings were tested, and the mortality was counted at 5 dpi and 7 dpi. Data were analyzed by one-way ANOVA and evaluated at P ≤ 0.05 by Fisher's least significant difference tests. (E) Total honeydew secreted from each adult WBPH individually fed on OsMYC3-ox, Osmyc3 mutants, Oscoi1-13 mutants, and Nip or ZH11 wild-type plants for about 36 hours. Data were analyzed by one-way ANOVA and evaluated at P ≤ 0.05 by Fisher's least significant difference tests. (F) Mortality rates of WBPH. For each genotype, ∼30 seedlings were tested with four virus-free WBPHs each, and the mortality was counted at 3 dpi and 5 dpi. (G) Total honeydew secreted from each adult WBPH individually fed on SP8-ox transgenic rice plants or Nip controls for about 36 hours. Data were analyzed by ANOVA and evaluated at P ≤ 0.05 by Fisher's least significant difference tests. (H) Total honeydew secreted from each adult WBPH individually fed on healthy and SRBSDV-infected rice plants for about 36 hours. Data were analyzed by ANOVA and evaluated at P ≤ 0.05 by Fisher's least significant difference tests.
Molecular responses of plants to phloem-feeding insects are strongly correlated with the feeding and the degree of tissue damage at the feeding site. The efficiency of insect feeding is therefore believed to be a very important factor in plant resistance. Accordingly, we measured the honeydew excretion produced within 36 hours by each adult WBPH as this is widely accepted as a good indicator of insect feeding (44–46). The production of honeydew was about 7.88 ± 2.54 mg on ZH11 background rice seedlings, substantially more than that on OsMYC3-14 (1.59 ± 1.48 mg) or OsMYC3-23 (1.65 ± 1.45 mg). In contrast, the amounts of honeydew on Osmyc3-1 (14.24 ± 3.36 mg) and Osmyc3-12 (15.09 ± 4.27 mg) were statistically greater than on the ZH11 control (Fig. 8E) and that improved ingestion potentially enhanced virion acquisition and transmission without any significant effect on the motility of the vector insects (Fig. 8F). There was also a higher secretion of honeydew on the JA signaling mutant Oscoi1-13 (17.97 ± 4.94 mg) than on Nip plants (5.99 ± 2.28 mg) (Fig. 8E). Together with similar data from SBPH (SI Appendix, Fig. S21 E and F), these results demonstrate that interfering with the JA signaling pathway enhances insect vector feeding, resulting in a rapid breakdown of resistance against WBPH and SBPH.
To further confirm the potential effects on insect resistance when the virus proteins directly block JA signaling through transcriptional repression of OsMYC3, we measured honeydew excretion on rice expressing the individual viral proteins. Compared with the Nip control (6.67 ± 2.15 mg), there was a significant increase in honeydew excretions on the lines SP8-ox (treated with WBPH) and RSVP2-ox (treated with SBPH) to respectively 17.49 ± 2.14 mg and 14.13 ± 3.26 mg (Fig. 8G and SI Appendix, Fig. S21G). Consistent with these observations, infection by SRBSDV and RSV also induced an approximately fourfold greater accumulation of honeydew from their vector insects (Fig. 8H and SI Appendix, Fig. S21H), providing definitive evidence that this viral hijacking of the JA pathway enhances the feeding of the vectors, which is likely to be significant for virus transmission and pathogenicity. Thus, our results show that by repressing the JA signaling pathway, viral transcriptional repressors alter plant immunity in ways that enhance the viruses themselves and also the feeding activity of their vectors.
Discussion
Plants have evolved a complex innate immune system to protect themselves against pathogen invasion, and defensive phytohormones are a part of this resistance strategy (47, 48). To counteract this response, pathogens have developed various methods to manipulate hormone pathways for their own advantage. Some pathogens employ their effectors to target the various components of the SA and JA signaling pathways to achieve colonization. In bacteria, it seems likely that the AvrPtoB effector inhibits SA-mediated basal immunity by directly interacting with the SA master regulator NPR1 (49), while other effectors, for example, HopZ1a and HopX1, impair the SA pathway by activating JA signaling (50, 51). There is increasing evidence that viral proteins can also directly regulate plant hormone homeostasis during virus infections. In rice dwarf virus (RDV), Pns11 interacts with S-adenosyl-l-methionine synthetase to promote ethylene biosynthesis (52), whereas RDV P2 binds OsIAA10 and reprograms auxin signaling to enhance virus infection (53). However, little work has yet explored how viruses directly manipulate JA signaling. In a previous report, we showed that JA-mediated antiviral defense plays an essential role in rice resistance to RBSDV (18), but the precise mechanism remains poorly understood. More recently, we found that several different viral proteins (SRBSDV SP8, RBSDV P8, RSV P2, and RSMV M) from very diverse plant RNA viruses target an auxin response transcription factor, making the plants more susceptible to viruses (12), but it will need further research to investigate whether this consequence of OsARF17-mediated antiviral defense is relevant to the induction of JA signaling. In this study, we further report a strategy in both double-stranded and single-stranded negative RNA viruses. Viral proteins disturb the direct association of OsMED25 and OsMYC and interact with OsJAZ proteins synergistically interfering with the OsMYC transcription factors, therefore alleviating the JA-mediated defense response. It is noteworthy that these viral proteins are active transcriptional repressors. Taking SRBSDV as an example, the viral protein SP8 directly associates with OsMYC3 through the TAD and efficiently represses its transcriptional activation. SP8 physically interacts with OsMED25 and competitively binds OsMYC3, thereby resulting in the disassociation of the OsMED25-OsMYC3 complex. Furthermore, OsJAZs also interact with SP8 protein, and this interaction enhances the transcriptional repressor activity of SP8. Thus, we propose that SP8 associated with OsJAZ proteins and OsMYC3 constituting a sophisticated complex modulates JA signaling that benefits virus infection. Similar scenarios were observed with RBSDV (P8 protein), RSV (P2 protein), and RSMV (M protein), suggesting that hijacking of JA signaling may be conserved among these very diverse viruses.
In recent decades, one of the best-studied antiviral defense mechanisms has been RNA silencing (3, 54). Viruses have evolved diverse mechanisms to avoid silencing mainly via viral suppressors of RNA silencing (VSRs). VSRs generally suppress host antiviral defense by directly targeting the key components of the RNA silencing machinery. VSRs have been found in almost all plant virus genera, and their sequences have no obvious similarity. For example, geminivirus βC1 (ssDNA genome) and TSWV NSs (ssRNA genome) have both been recognized as VSRs (55). In this study, we have identified a type of active viral transcriptional repressor common to unrelated viruses which directly suppresses host defense by hijacking the key components of the JA signaling pathway. Like VSRs, these viral transcriptional repressors have no clear sequence similarities. SRBSDV SP8 protein is a core capsid protein of 591 amino acids, while RSV P2 has only 199 amino acids, and no sequence similarity or common conserved protein domain can be identified. As shown in Fig. 2D, SRBSDV SP8 protein, but not RSV P2, contains a conserved NTP domain. Importantly, loss of function of NTP inactivated the SP8 protein and therefore irreversibly impaired its global interactions in planta (SI Appendix, Fig. S11 C and D), indicating that the NTP domain was required in the case of the SP8 activity but clearly not for other transcriptional repressors. Thus, we predict that these repressors may perhaps share some similarity in protein tertiary structure, which will be elucidated via protein crystal structure in the future. Moreover, SP8 specifically interacted with OsMYC3, whereas RSV P2 and RSMV M could associate with both OsMYC2 and OsMYC3. These results suggest that these repressors may act in different ways but with a similar result of suppressing the JA pathway.
Manipulation of the host defense pathway by pathogens generally benefits their own infection. We here particularly studied viruses which are naturally transmitted by vector insects, JA signaling has received most attention in connection with resistance against insects. In defending themselves against chewing insects, plants respond to both insect-derived signals and physical damage associated with insect feeding, triggering a PTI-like defense mechanism by activating the JA signaling pathway. However, there are also antagonistic roles played by the JA pathway in affecting host resistance to chewing and phloem-feeding insects in Arabidopsis (17, 56). For example, a whitefly salivary protein, Bt56, was reported to directly elicit the SA-signaling pathway by interacting with a tobacco class II KNOTTED 1-like homeobox (KNOX) transcription factor (NTH202), which likely leads to down-regulation of the JA defense pathway, thereby decreasing resistance to the whitefly (57). Both cucumber mosaic virus (Cucumovirus) 2b protein and tomato yellow leaf curl China virus (Begomovirus) βC1 were involved in JA pathway to achieve host manipulation to vectors in dicotyledon (58, 59). Here, our results provide insights into phloem-based defense in monocotyledonous plants. It is particularly interesting that manipulation of JA signaling by the viral transcriptional repressor alters resistance to the phloem-feeding vectors by promoting their feeding activity, surely enhancing virus transmission but with a negligible effect on the motility of the insects (Fig. 8F and SI Appendix, Fig. S21F). Vector resistance in JA-insensitive mutant Osmyc3 lines was almost completely impaired, while excessive OsMYC3 significantly enhanced host resistance against WBPH and SBPH. In agreement with this phenomenon, plants expressing individual viral transcriptional repressors or virus-infected plants all had a clearly suppressed OsMYC3-mediated JA response, conferring a benefit to vector feeding (Fig. 8 G and H, and SI Appendix, Fig. S21 G and H). Together, our results reveal that a variety of viruses commonly utilize their viral transcriptional repressor to directly modulate key components of JA signaling to overcome host defense against phloem-sucking vectors, thereby facilitating viral pathogenicity and spread (SI Appendix, Fig. S22).
In conclusion, this report describes a group of viral transcriptional repressors present widely in unrelated plant viruses. These hijack JA signaling to benefit both virus infection and vector feeding. This is a novel mechanism in itself, but the results also expand our knowledge of the interplay between viral pathogenicity, vector behavior, and host defense, opening the way for biological approaches to effectively manage vector-mediated virus diseases.
Materials and Methods
The transgenic plants expressing viral proteins, SP8-ox and RSVP2-ox, and some mutants of genes involved in the JA pathway, like Oscoi1, were created in the Nipponbare (Oryza sativa L. cv. Japonica, Nip) cultivar background. Rice seedlings overexpressing OsMYC3 and the Osmyc3 mutant plants used Zhonghua 11 (ZH11) seedlings as the background. Details of the experimental methods including screening the cDNA library, Y2H assay, agro-infection assays in N. benthamiana, BiFC, Co-IP, LCI, generation of transgenic plants, root growth inhibition assay, dual-luciferase transient transcriptional activity assay, virus inoculation assays, RNA extraction, qRT-PCR, Western blot analysis, and honeydew measurements are provided in SI Appendix, Materials and Methods. The primers used in this study are listed in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
We thank Professor Zuhua He (Shanghai Institute for Biological Sciences, Chinese Academy of Sciences, China) for the coi1-13 mutant, Professor Qingyun Bu (Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, China) for OsMED25 constructs, Professor Sheng Yang He (Michigan State University) for his important suggestions, and Mike Adams for critically reading and improving the manuscript. This work was funded by China National Funds for Excellent Young Scientists (32022072), the National Key Research and Development Plan (2016YFD0200804), National Natural Science Foundation of China (32001888), Zhejiang Provincial Natural Science Foundation (Q21C140013), and Ningbo Science and Technology Innovation 2025 Major Project (2019B10004). This work was sponsored by the K.C. Wong Magna Fund in Ningbo University.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016673118/-/DCSupplemental.
Data Availability
All of the materials and data that were used or generated in this study are described and available in the text and SI Appendix.
References
- 1.Wang A., Dissecting the molecular network of virus-plant interactions: The complex roles of host factors. Annu. Rev. Phytopathol. 53, 45–66 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Baulcombe D. C., How virus resistance provided a mechanistic foundation for RNA silencing. Plant Cell 31, 1395–1396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Z., et al., Jasmonate signaling enhances RNA silencing and antiviral defense in rice. Cell Host Microbe 28, 89–103.e8 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Wei T., Li Y., Rice reoviruses in insect vectors. Annu. Rev. Phytopathol. 54, 99–120 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Feng M., et al., Rescue of tomato spotted wilt virus entirely from complementary DNA clones. Proc. Natl. Acad. Sci. U.S.A. 117, 1181–1190 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang X., et al., Rice stripe mosaic virus, a novel Cytorhabdovirus infecting rice via leafhopper transmission. Front. Microbiol. 7, 2140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou G., Xu D., Xu D., Zhang M., Southern rice black-streaked dwarf virus: A white-backed planthopper-transmitted fijivirus threatening rice production in Asia. Front. Microbiol. 4, 270 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H., et al., Rice black-streaked dwarf virus P10 acts as either a synergistic or antagonistic determinant during superinfection with related or unrelated virus. Mol. Plant Pathol. 20, 641–655 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu G., et al., Tenuivirus utilizes its glycoprotein as a helper component to overcome insect midgut barriers for its circulative and propagative transmission. PLoS Pathog. 15, e1007655 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He L., et al., Rice black-streaked dwarf virus-encoded P5-1 regulates the ubiquitination activity of SCF E3 ligases and inhibits jasmonate signaling to benefit its infection in rice. New Phytol. 225, 896–912 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Y., et al., Rice stripe tenuivirus nonstructural protein 3 hijacks the 26S proteasome of the small brown planthopper via direct interaction with regulatory particle non-ATPase subunit 3. J. Virol. 89, 4296–4310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H., et al., Distinct modes of manipulation of rice auxin response factor OsARF17 by different plant RNA viruses for infection. Proc. Natl. Acad. Sci. U.S.A. 117, 9112–9121 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Römer P., et al., Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. New Phytol. 187, 1048–1057 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Ikeda M., Ohme-Takagi M., A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol. 50, 970–975 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Tiwari S. B., Hagen G., Guilfoyle T. J., Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16, 533–543 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazan K., Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci. 11, 109–112 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Jones J. D. G., Dangl J. L., The plant immune system. Nature 444, 323–329 (2006). [DOI] [PubMed] [Google Scholar]
- 18.He Y., et al., Jasmonic acid-mediated defense suppresses brassinosteroid-mediated susceptibility to Rice black streaked dwarf virus infection in rice. New Phytol. 214, 388–399 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Xie K., et al., Abscisic acid negatively modulates plant defence against rice black-streaked dwarf virus infection by suppressing the jasmonate pathway and regulating reactive oxygen species levels in rice. Plant Cell Environ. 41, 2504–2514 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Zhang H., et al., Suppression of auxin signalling promotes rice susceptibility to Rice black streaked dwarf virus infection. Mol. Plant Pathol. 20, 1093–1104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Y., et al., The OsGSK2 kinase integrates brassinosteroid and jasmonic acid signaling by interacting with OsJAZ4. Plant Cell 32, 2806–2822 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng Z., et al., The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol. Plant 4, 279–288 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Pauwels L., et al., NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thines B., et al., JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Pauwels L., Goossens A., The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant Cell 23, 3089–3100 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kazan K., Manners J. M., JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 17, 22–31 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Zhang F., et al., Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 525, 269–273 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhai Q., Deng L., Li C., Mediator subunit MED25: At the nexus of jasmonate signaling. Curr. Opin. Plant Biol. 57, 78–86 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Liu Y., et al., MYC2 regulates the termination of jasmonate signaling via an autoregulatory negative feedback loop. Plant Cell 31, 106–127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.You Y., Zhai Q., An C., Li C., LEUNIG_HOMOLOG mediates MYC2-dependent transcriptional activation in cooperation with the coactivators HAC1 and MED25. Plant Cell 31, 2187–2205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An C., et al., Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin. Proc. Natl. Acad. Sci. U.S.A. 114, E8930–E8939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plett J. M., et al., Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc. Natl. Acad. Sci. U.S.A. 111, 8299–8304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C.-Y., et al., An effector from cotton bollworm oral secretion impairs host plant defense signaling. Proc. Natl. Acad. Sci. U.S.A. 116, 14331–14338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H., Wei C., Zhong Y., Li Y., Rice black-streaked dwarf virus minor core protein P8 is a nuclear dimeric protein and represses transcription in tobacco protoplasts. FEBS Lett. 581, 2534–2540 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Kazan K., Manners J. M., MYC2: The master in action. Mol. Plant 6, 686–703 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Fernández-Calvo P., et al., The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogawa S., et al., OsMYC2, an essential factor for JA-inductive sakuranetin production in rice, interacts with MYC2-like proteins that enhance its transactivation ability. Sci. Rep. 7, 40175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nibert M. L., Kim J., Conserved sequence motifs for nucleoside triphosphate binding unique to turreted reoviridae members and coltiviruses. J. Virol. 78, 5528–5530 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen R., et al., The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24, 2898–2916 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Howe G. A., Jander G., Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59, 41–66 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Chung H. S., et al., Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 146, 952–964 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du B., et al., Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc. Natl. Acad. Sci. U.S.A. 106, 22163–22168 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y., et al., Map-based cloning and characterization of BPH29, a B3 domain-containing recessive gene conferring brown planthopper resistance in rice. J. Exp. Bot. 66, 6035–6045 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghaffar M. B., Pritchard J., Ford-Lloyd B., Brown planthopper (N. lugens Stal) feeding behaviour on rice germplasm as an indicator of resistance. PLoS One 6, e22137 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y., et al., A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat. Biotechnol. 33, 301–305 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Lu H. P., et al., Resistance of rice to insect pests mediated by suppression of serotonin biosynthesis. Nat. Plants 4, 338–344 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Yang L., et al., Pseudomonas syringae type III effector HopBB1 promotes host transcriptional repressor degradation to regulate phytohormone responses and virulence. Cell Host Microbe 21, 156–168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pieterse C. M. J., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S. C., Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Chen H., et al., A bacterial type III effector targets the master regulator of salicylic acid signaling, NPR1, to subvert plant immunity. Cell Host Microbe 22, 777–788.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Gimenez-Ibanez S., et al., The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol. 12, e1001792 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Macho A. P., Guevara C. M., Tornero P., Ruiz-Albert J., Beuzón C. R., The Pseudomonas syringae effector protein HopZ1a suppresses effector-triggered immunity. New Phytol. 187, 1018–1033 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Zhao S., et al., A viral protein promotes host SAMS1 activity and ethylene production for the benefit of virus infection. eLife 6, e27529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin L., et al., Rice dwarf virus P2 protein hijacks auxin signaling by directly targeting the rice OsIAA10 protein, enhancing viral infection and disease development. PLoS Pathog. 12, e1005847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo Z., Li Y., Ding S. W., Small RNA-based antimicrobial immunity. Nat. Rev. Immunol. 19, 31–44 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Burgyán J., Havelda Z., Viral suppressors of RNA silencing. Trends Plant Sci. 16, 265–272 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Jiang Y., Zhang C.-X., Chen R., He S. Y., Challenging battles of plants with phloem-feeding insects and prokaryotic pathogens. Proc. Natl. Acad. Sci. U.S.A. 116, 23390–23397 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu H.-X., et al., A salivary effector enables whitefly to feed on host plants by eliciting salicylic acid-signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 116, 490–495 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu D., et al., Viral effector protein manipulates host hormone signaling to attract insect vectors. Cell Res. 27, 402–415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li R., et al., Virulence factors of geminivirus interact with MYC2 to subvert plant resistance and promote vector performance. Plant Cell 26, 4991–5008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the materials and data that were used or generated in this study are described and available in the text and SI Appendix.