Significance
Understanding microbial population dynamics during infection has important implications for disease management, transmission, and pathogen evolution. A quantitative analysis of microbial population structure requires the ability to track individual strains. We used a pool of individually barcoded strains to measure changes in Brucella abortus population structure during infection of bovine hosts via the ocular conjunctiva, a natural route of entry. Cattle exert a severe bottleneck on the bacterial population entering through the conjunctival mucosa such that individual cells have a 0.0001 probability of colonizing a local draining lymph node. The populations in lymph nodes, even on different sides of the same animal, are distinct and dominated by a small number of highly abundant, spatially distinct clones.
Keywords: brucellosis, bottleneck, TnSeq, bovine, infectious dose
Abstract
Cattle are natural hosts of the intracellular pathogen Brucella abortus, which inflicts a significant burden on the health and reproduction of these important livestock. The primary routes of infection in field settings have been described, but it is not known how the bovine host shapes the structure of B. abortus populations during infection. We utilized a library of uniquely barcoded B. abortus strains to temporally and spatially quantify population structure during colonization of cattle through a natural route of infection. Introducing 108 bacteria from this barcoded library to the conjunctival mucosa resulted in expected levels of local lymph node colonization at a 1-wk time point. We leveraged variance in strain abundance in the library to demonstrate that only 1 in 10,000 brucellae introduced at the site of infection reached a parotid lymph node. Thus, cattle restrict the overwhelming majority of B. abortus introduced via the ocular conjunctiva at this dose. Individual strains were spatially restricted within the host tissue, and the total B. abortus census was dominated by a small number of distinct strains in each lymph node. These results define a bottleneck that B. abortus must traverse to colonize local lymph nodes from the conjunctival mucosa. The data further support a model in which a small number of spatially isolated granulomas founded by unique strains are present at 1 wk postinfection. These experiments demonstrate the power of barcoded transposon tools to quantify infection bottlenecks and to define pathogen population structure in host tissues.
Brucella abortus commonly infects domesticated cattle (1) and is an etiologic agent of brucellosis, a global zoonotic disease that inflicts a significant burden on human and animal health and agricultural production (2). Our understanding of Brucella infection biology and host response has been greatly informed by in vitro studies of infection in tissue culture, or by in vivo modeling of brucellosis in rodents. Mouse models are particularly well developed and have proven useful in defining features of mammalian immune protection and in testing new vaccines and brucellosis therapies (3). However, unlike cattle, mice are not natural hosts of B. abortus. Moreover, the experimental routes of infection employed in laboratory studies of the mouse do not typically reflect natural routes of host entry that occur in bona fide mammalian hosts under field conditions. New studies of B. abortus using natural hosts infected through natural routes are therefore crucial as we work to advance understanding of the infection biology of this important group of pathogens.
Bacteriological analyses of infected cattle provide evidence that B. abortus entry through the ocular conjunctiva and upper respiratory passage is common under field conditions (4). Entry also occurs via mucosal tissue of the oral cavity, the respiratory tract, or the gastrointestinal tract (5–8). While invasion of the conjunctiva and other mucosal tissue induces an inflammatory response in the submucosa of cows (9), it is clear that some fraction of B. abortus 1) survive this inflammation after traversing the host barrier, 2) enter the lymphatic system, and 3) begin to colonize the parotid lymph node within 1 wk of infection (4, 9–11). The fraction of infecting B. abortus that eventually colonize lymph nodes though these natural routes of entry remains largely undefined.
Population bottlenecks are common during infection through natural routes as host barriers and immune functions serve to minimize pathogen entry and survival (12). Studies involving pools of strains with a limited number of unique tags have been utilized to assess bottlenecks during infection in model systems (e.g., refs. 13–16). Understanding the magnitude of bottlenecks is important in developing models of pathogen evolution and microbial adaptation to the host (17, 18). Knowledge of bottlenecks also greatly informs the design and interpretation of transposon-insertion sequencing (Tn-seq) studies aimed at discovering genes required for survival in the animal host (19). In this study, we aimed to define the magnitude of the bottleneck during the initial phase of B. abortus infection of its natural bovine host via a natural mucosal route of infection and to quantify B. abortus population structure in the host lymphatic tissue after infection.
We previously generated a highly diverse pool of ∼106 B. abortus transposon mutant strains (20, 21). A 20-base pair barcode present in each transposon mutant (22) provides a unique identification tag for each B. abortus strain and thus enables strain-level quantification of B. abortus populations. In this study, we used this diverse pool of individually tagged strains to study changes in B. abortus population structure upon infection through the conjunctival mucosa in cattle. Our study characterizes infection bottlenecks along the conjunctival route of entry, defines spatial distributions of clonally derived B. abortus strains in lymph nodes that drain the ocular and oral cavities, and demonstrates the feasibility and power of Tn-seq approaches to study B. abortus populations in a bovine infection model.
Results
Reproducible Infection of Calves via the Conjunctival Route with a B. abortus Tn-Himar Mutant Library.
We first sought to test whether our library of barcoded Tn-himar strains (21) infects cattle like wild-type B. abortus. Although pregnant animals present the most severe clinical effects of brucellosis, we chose young animals for this study of the early stage of infection. Cattle are routinely vaccinated with attenuated B. abortus at 4 to 12 mo of age and can remain infected with these strains for many weeks postvaccination (23). It is established that 3- to 4-mo-old calves are reproducibly infected after experimental conjunctival exposure to 5 × 108 colony-forming units (CFU) of B. abortus (10), so we administered 108 CFU from our barcoded B. abortus Tn-himar mutant library to the right and left ocular conjunctiva of six calves between 5 and 9 mo of age (Table 1). We chose to infect calves via the conjunctival mucosa because it is a natural route of host entry in the field (4) and is often used for vaccine challenge studies.
Table 1.
Calves utilized in the study
| Animal | Sex | Age at time of infection, mo | Days after infection at necropsy |
| 1 | Bull | 5 | 8 |
| 2 | Bull | 5 | 9 |
| 3 | Heifer | 9 | 9 |
| 4 | Heifer | 9 | 8 |
| 5 | Heifer | 5 | 8 |
| 6 | Heifer | 5 | 9 |
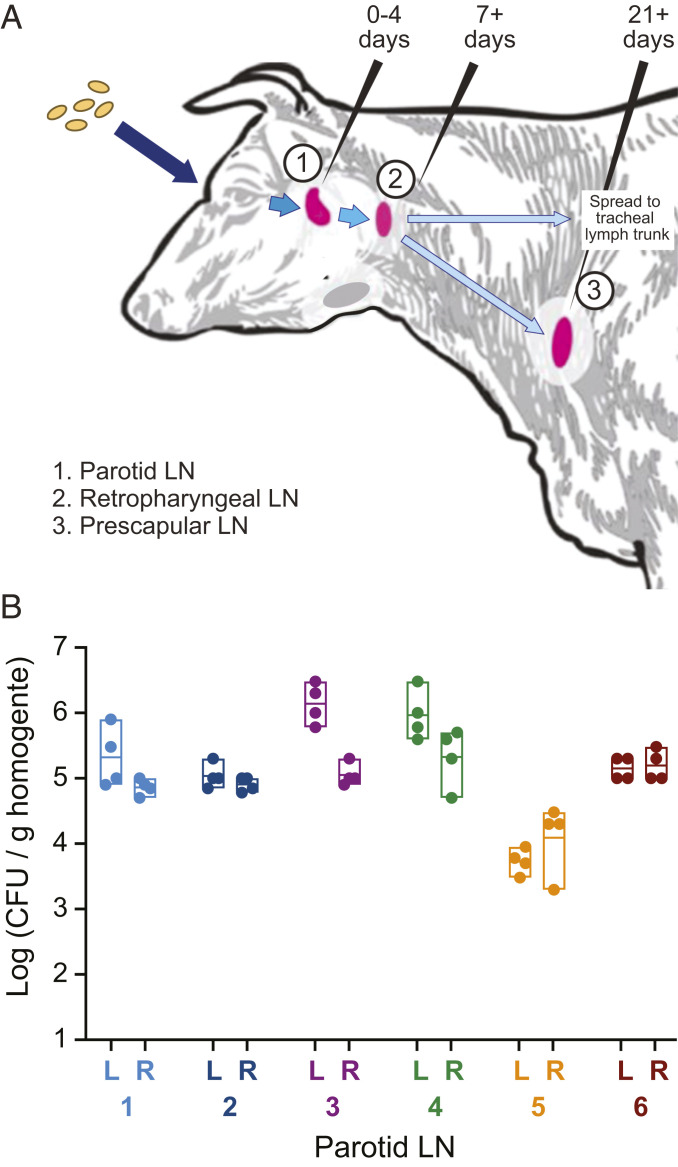
Infected animals were necropsied 8 or 9 d postinfection to assess B. abortus mutant populations in the parotid and retropharyngeal lymph nodes (Fig. 1A). Homogenates of the right and left parotid lymph nodes of each of the six calves yielded ∼105 CFU/g of tissue (Fig. 1B), which is consistent with infection studies using wild-type B. abortus (10). We further assessed colonization of the left retropharyngeal lymph nodes, which are distally draining compared to the parotid, and noted high variability in the extent of colonization between animals. Populations of B. abortus were isolated from retropharyngeal homogenates of only three of the six infected animals at our 8- to 9-d time point. This result is consistent with the work of Meador et al. (10), which demonstrated that B. abortus introduced via the conjunctival route is detected in the retropharyngeal lymph node between 7 and 21 d postinfection.
Fig. 1.
Colonization of bovine parotid lymph nodes by the barcoded Tn-himar B. abortus strains. (A) A barcoded pool of B. abortus Tn-himar strains (tan ovals) were inoculated into the left and right ocular conjunctiva of six calves. These bacterial cells disseminate to the lymph nodes (LN) of the head and neck (pink). B. abortus can be detected in (1) the parotid LN at 0 to 4 d postinfection, (2) the retropharyngeal LN at 7 to 21 d postinfection, and (3) the prescapular LN at time points at least 21 d after infection (10). (B) Expected levels of colonization were observed in the parotid LN 8 to 9 d postinfection. Parotid LN from the left (L) and right (R) sides of the infected animals were removed, quartered, and homogenized. Dots represent CFU per gram of tissue that were enumerated from each homogenized lymph node quadrant. Boxes indicate the minimum, maximum, and mean titer of the quadrants in each parotid LN of each animal (numbered 1 to 6).
Amplification and Sequencing of Barcodes from B. abortus Strains Isolated from Bovine Lymph Nodes.
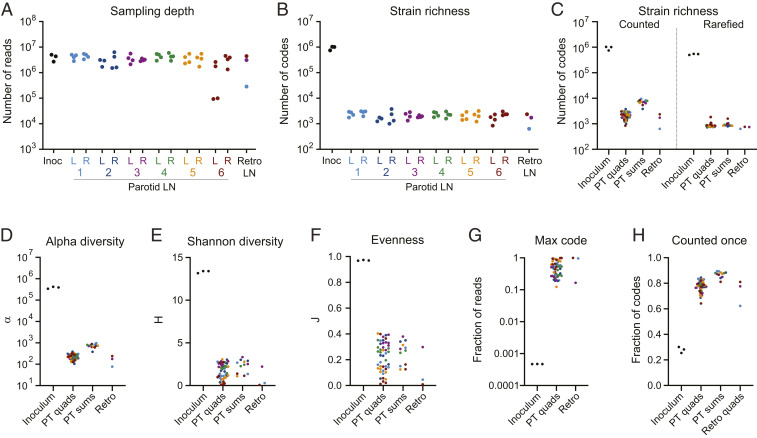
To assess the B. abortus Tn-strain distribution in host tissue, lymph nodes were dissected into quadrants and homogenized. Node homogenates were spread on TSA-SK agar (tryptic soy agar supplemented with 5% serum and 50 μg/mL kanamycin), the same medium used to outgrow the library for inoculation. Transposon barcodes were PCR-amplified from genomic DNA extracted from these plated samples, and barcode amplicons were sequenced and counted. The numbers of sequence reads and unique barcodes detected in each sample are detailed in Dataset S1. Using this approach, barcodes were reliably amplified, identified, and tallied across samples. Lymph node quadrants and inoculum samples yielded a similar number of total reads (2 to 5 × 106), indicating similar sampling depths for all samples (Fig. 2A).
Fig. 2.
Loss of strain diversity in lymph node samples. (A) Number of barcode sequencing reads from each sample. Each dot represents a sample from the inoculum (inoc), a parotid lymph node (LN) quadrant from the left (L) or right (R) side of each animal (numbered 1 to 6), or a colonized retropharyngeal lymph node (Retro LN). (B) Number of unique barcodes (i.e., strains) counted in each sample. Each dot represents a sample as in A. (C) Number of codes counted in each type of sample before (Left) and after (Right) rarefaction to normalize for differences in sequencing depth. (D–F) Fisher’s alpha diversity, α, Shannon diversity, H, and Shannon evenness, J, calculated for each sample. (G) Fraction of reads represented by the most abundant barcode in each sample. (H) Fraction of barcodes counted only once in each sample. PT quads: individual quadrants from parotid LNs. PT sums: the sum of barcode counts for the quadrants of each parotid LN. Retro; fragment from colonized retropharyngeal LN. Points are colored by animal as in Fig. 1.
The Tn-Himar Mutant Pool Contains a Diverse and Evenly Distributed Population of B. abortus Strains.
The distribution of barcodes in the B. abortus mutant pool (i.e., the inoculum) reflects a diverse and evenly distributed set of unique strains. We identified 0.7 to 1.0 × 106 barcodes in each of three inoculum aliquots, which collectively represent 1.5 × 106 unique codes (SI Appendix, Fig. S5). We previously mapped 5 × 105 barcodes to distinct positions in the B. abortus genome with high confidence (21), and in this present study we captured most of the previously mapped barcodes in the inoculum (SI Appendix, Fig. S5). Within each inoculum sample, barcode abundances were evenly distributed with a small tail of more abundant codes (SI Appendix, Fig. S6). These samples contained few codes represented by 100 or more reads and, accordingly, our filtering approach outlined in Materials and Methods and SI Appendix, Fig. S2 removed only 0.2% of reads and 0.7% of the codes from these samples (SI Appendix, Fig. S3 and Dataset S1). The apparent strain diversity of the inoculum samples may therefore be modestly inflated by sequencing errors, which are expected in ∼1% of the barcode reads (24–26) (and examples in SI Appendix, Fig. S1).
B. abortus Strain Diversity in Lymph Nodes Is Highly Reduced Relative to the Inoculum.
We next used our barcode sequence datasets to assess changes in B. abortus population structure when B. abortus enters the host via the conjunctival route. Though we administered 108 CFU of a library containing at least 106 unique strains to the right and left ocular conjunctiva of each calf, fewer than 104 unique barcodes were detected in the parotid lymph nodes on the right and left sides of the animal (Fig. 2C). Each lymph node quadrant contained an average of 2 × 103 unique barcodes (Fig. 2 B and C). To determine if differences in apparent strain diversity between host tissue samples were due to variability in sequencing depth (SI Appendix, Fig. S3), we randomly selected 106 reads from each sample for barcode analysis. This rarefaction approach to normalize for differences in sampling depth shows that a nearly uniform number of strains were present in each lymph node (Fig. 2C). We conclude that the number of unique strains that colonize each parotid lymph node is similar between animals. These data provide evidence for a strong restriction of strains between the conjunctival mucosa and the parotid lymph node at this infectious dose. Specifically, we observe a loss of at least two orders of magnitude in strain diversity between the site of infection and the parotid lymph nodes. This population restriction is corroborated by marked decreases in population diversity metrics, including Fisher’s alpha diversity (α) and Shannon diversity (H), in the lymph nodes relative to the inoculum (Fig. 2 D and E). These widely used diversity indices take into account both the richness (i.e., the number of different strains present) and their relative abundances (27, 28). Richer and more evenly distributed populations exhibit higher diversity index scores, while populations with fewer and more dominant strains have lower scores.
Strain Abundance Distribution Is Skewed in Host Tissue.
The highly diverse B. abortus population in the inoculum has an even strain (i.e., barcode) abundance distribution (SI Appendix, Fig. S6) and a Shannon’s evenness index (J) near 1 (Fig. 2F). In contrast, the distribution of strains in the B. abortus populations isolated from bovine lymph nodes is highly skewed: Small numbers of strains were highly abundant—counted on the order of 106 times—and the majority of codes in these tissues were rare (SI Appendix, Fig. S6). This skew is reflected in low Shannon evenness values for host tissues (Fig. 2F). The most-abundant strain in each lymph node accounted for 12 to 99% of total sequence reads (Fig. 2F and Dataset S1), and in most lymph node samples 80% of strains were counted only once (Fig. 2G). In comparison, the most abundant strain in the inoculum accounted for only 0.047% of total reads. A positive relationship between abundance in the inoculum and the likelihood that a strain will colonize the host is expected. Indeed, strains that occupy the most extreme end of the abundance tail of the inoculum are more likely to be observed in the host.
Abundant strains in technical replicates from a subset of lymph node fragments are highly correlated (SI Appendix, Fig. S4). This lends confidence to our conclusion that such strains are truly abundant in the lymph node and are not an artifact of sample processing or jackpot effects during PCR barcode amplification. Variation between replicates for less-abundant codes reflects the limits of sampling low-abundance members of a community using this method. The profound skew in the strain abundance distribution that we observe in lymph node samples provides evidence that a very limited number of strains that reach the lymph node are hyperabundant during the early stages of infection.
Populations of the Most Abundant Strains in Each Lymph Node Are Distinct.
The observed skew in the strain abundance distribution in vivo raised the question of whether the most-abundant B. abortus strains were unique to each lymph node or the same across nodes. Hyperabundance of distinct sets of strains across animals (and lymph nodes) is expected if the governing processes are stochastic. On the other hand, if certain strains have a genetic propensity to hyperproliferate, we expect to see a shared set of highly abundant strains between animals (and lymph nodes). To evaluate these possibilities, we extracted the set of barcodes that comprised at least 2% of the sequencing reads in any lymph node sample. We then evaluated the relative abundance of these 160 barcodes in every sample. We observe a distinct set of highly abundant strains in each lymph node (Fig. 3). There was no overlap in hyperabundant strains between animals and, in most cases, no overlap in hyperabundant strains between the right and left parotid lymph nodes within an animal. Furthermore, in many cases, abundant strains were distinct even between quadrants of an individual lymph node. This provides evidence that strains that establish residence and proliferate in the lymph node are, to some extent, spatially restricted within this tissue. However, there are cases in which abundant strains are shared among dissected quadrants of an individual lymph node. Thus, it is likely that highly proliferating strains eventually spread through the node. Indeed, the two parotid lymph nodes exhibiting the most homogeneity in abundant strains across the four quadrants (the left parotid lymph nodes of calves 3 and 4) also had the highest bacterial loads (Figs. 1 and 3). We conclude that at early stages of infection B. abortus populations are dominated by a small, random set of strains.
Fig. 3.
Distinct sets of hyperabundant strains are observed in each lymph node. Heat map of the abundance of 160 barcodes that represent more than 2% of reads in at least one sample. Each row is a sample, each column represents a hyperabundant barcode (i.e., strain), and gray scale intensity indicates fractional abundance of each barcode in each sample. Empty squares indicate barcodes that were not detected in a sample. Horizontal lines separate samples from each animal. Note that the PCR amplification of barcodes from calf 4, left parotid, quadrant A was contaminated with template from calf 1, left parotid, quadrant B. This signal is detected in the abundance data from the calf 4 Left_A sample.
B. abortus Strain Populations within Individual Lymph Nodes Are Spatially Heterogeneous.
To more comprehensively assess B. abortus population structure in bovine lymph tissue after infection, we compared the full set of strains (i.e., barcodes) present in each lymph node and across lymph node quadrants. We utilized two different metrics for pairwise population comparisons between each sample. Jaccard dissimilarity is a metric that compares the presence or absence of population members (29). Bray–Curtis dissimilarity is a related metric that also takes into account abundance of population members (30). When considering presence/absence of barcodes in each population, we observe high levels of dissimilarity between all host tissue samples, even samples dissected from the same lymph node (Fig. 4A). When abundance is considered, we again observe high levels of dissimilarity between populations of B. abortus strains from different lymph nodes in the same or different animals. However, the Bray–Curtis metric reveals moderate similarity between dissected quadrants of individual lymph nodes (Fig. 4B). Lymph nodes in which the populations in each quadrant exhibit the lowest Bray–Curtis dissimilarity (i.e., greatest similarity) tend to share more hyperabundant strains between quadrants (Figs. 3 and 4B). The largely distinct sets of strains colonizing each animal, each lymph node, and each region of a lymph node are consistent with a significant population bottleneck leading to random subsets of strains in each tissue. In addition, the spatial heterogeneity within tissues suggests that most strains in lymph nodes are spatially restricted and do not spread evenly throughout the lymph node at early infection time points. This is consistent with granuloma formation in the lymph nodes, which is a feature typical of host tissue infected with Brucella (31).
Fig. 4.
B. abortus strain populations are similar within a lymph node but different between animals. Heat map of (A) Jaccard and (B) Bray–Curtis dissimilarity values for each pair of samples (1 = dissimilar, 0 = similar). For the inoculum (inoc), each individual replicate (A–C) and the sum of the three replicates (total) were compared. Horizontal lines separate samples from each animal. Samples are colored as in Fig. 1. As in Fig. 3, Bray–Curtis dissimilarity scores between calf 4 Left_A and calf1 Left quadrants reflect contaminating PCR signal.
Populations in Retropharyngeal Lymph Nodes Reflect the Parotid Lymph Node in the Same Animal.
In the three cases for which we obtained population data from retropharyngeal lymph nodes, there was a significant degree of similarity between the retropharyngeal node and its associated (draining) parotid lymph node. Specifically, the highly abundant codes in the retropharyngeal lymph node populations were shared with the highly abundant codes in the parotid lymph node from the same side of the animal (Fig. 3), and Bray–Curtis metrics indicate population level similarity between retropharyngeal lymph nodes and the parotid lymph node from the same side of the animal (Fig. 4B). Thus, a strain that establishes residence and proliferates in a parotid lymph node is more likely to establish residence in directly connected nodes.
Strain-Level Probability of Parotid Lymph Node Colonization via the Conjunctival Route.
Experimentally defined populations of B. abortus in the inoculum and in the lymph nodes of bovine hosts provide the opportunity to assess the probability of host infection at an individual strain level. There are many host immune mechanisms that function to restrict microbial entry and proliferation in the host. Some strains present in the inoculum will successfully traverse these immune obstacles and establish a colony in the host tissue, while others will be eliminated from the host. There is evidence that host entry and colonization is shaped by stochastic factors (32). However, infection is also influenced by the genetic properties of the particular strain (33). Other Tn mutagenesis studies involving Brucella infection in tissue culture models (34–37) or in animal models (38–41) indicate that a small fraction of mutants are attenuated. Similarly, only a small fraction of genes are required for survival of Mycobacterium spp. in mammalian hosts (42, 43). Thus, it is reasonable to predict that most (>90%) transposon insertions that are tolerated in axenic culture will be neutral or near neutral during infection.
If a Tn-himar barcode insertion at a particular chromosomal site is neutral with respect to infection, then infection can be thought of as a simple “yes–no” event that may be modeled as a binomial sampling process. In such a model, the expectation of observing a neutral barcode in the host should follow a binomial sampling distribution. Specifically, the success rate of individual strains (i.e., barcodes) in colonizing a lymph node is a function of 1) the probability that a single cell will successfully infect and 2) the abundance of each strain in the inoculum. In a binomial framework, the number of cells of a particular strain in the inoculum can be considered the number of infection trials for that strain.
Following this model, we empirically determined the probability that any single cell will colonize by binning the strains in the inoculum by abundance and surveying the fraction of strains in each bin that are not detected in each parotid lymph node (i.e., that are lost during infection). As expected, more abundant strains in the inoculum are more likely to be detected in a lymph node. In addition, all lymph nodes across all animals showed strikingly similar rates of strain loss in each abundance bin, demonstrating that this frequency-dependent pattern of infection is general. Furthermore, the relationship between relative abundance of strains in the inoculum and the likelihood of observing those strains in lymph nodes follows a binomial probability (Fig. 5). We compared the experimentally determined relationship between strain abundance in the inoculum and detection frequency in the lymph nodes to theoretical expectations of binomial sampling with different probabilities of success. Based on this analysis, we estimate the probability that any individual cell in the inoculum will successfully colonize a parotid lymph node via the ocular conjunctiva to be 0.0001. Furthermore, fitting the experimentally observed patterns of strain loss versus colonization to a log-logistic dose–response model (44) yields a median infectious dose (ID50) corresponding to fractional (inoculum) abundance of 10−4.4, or ∼3,500 CFU (SI Appendix, Fig. S7).
Fig. 5.
Lymph node colonization by individual B. abortus strains is a function of abundance in the inoculum and follows a binomial probability. (A) Barcodes (i.e., unique strains) were binned by fractional abundance in the inoculum. The number of barcodes in each bin is plotted where abundance corresponds to the center of the bin. (B) Fraction of strains not detected in parotid lymph node samples (i.e., lost during infection) are plotted as a function of barcode abundance in the inoculum. The presence or absence of each barcode was recorded for each parotid lymph node. Mean (± SD) fraction of codes not detected in each bin is plotted in black (n = 12 parotid lymph nodes). Gray lines represent expected fractions of codes not detected assuming a binomial sampling at different probabilities (P). To simulate a binomial sampling, the fractional strain abundance in the inoculum was converted to an approximate number of strains in the inoculum (fractional abundance * 108 CFU in inoculum; gray axis label) and used as the number of trials.
Discussion
Tn-seq studies are most often aimed at high-throughput assessment of gene function in a particular environment (45). We constructed a barcoded pool of B. abortus Tn-himar mutants and used this population to quantify bovine colonization through a natural route of infection at the individual strain level. This work demonstrates the feasibility of applying Tn-seq approaches to study how infection shapes B. abortus populations in a large animal (bovine) system.
The barcoded Tn mutant collection used in this study is highly diverse (∼106 unique strains; Fig. 2 and SI Appendix, Fig. S5). We reproducibly recovered strains from this pool from two lymphatic sites and we were able to reliably amplify B. abortus strain barcodes from this host tissue. The extreme stochastic loss of strain diversity between the conjunctival infection site and lymph nodes, and the corresponding skew in B. abortus strain abundances in vivo precluded a global analysis of the function(s) of individual genes during infection. Nonetheless, we were able to leverage the barcoded nature of our Tn-himar pool to quantify population-level features of B. abortus infection biology, shedding light on an infection bottleneck and the spatial distribution of B. abortus clones in colonized tissue. Tn-seq studies aimed at defining the roles of individual B. abortus genes in bovine infection will require a less-diverse infecting pool and are in development. Given the observed bottleneck and the number of CFU in the infecting inoculum, we estimate that a mutant pool containing approximately 1,000 strains, each represented by 104 to 105 clones, would be suitable to assess individual gene function in the context of this infection model.
Characterizing a Bottleneck in a Natural Route of Infection.
We administered 108 B. abortus Tn-himar mutants to six calves via the conjunctival route. This is 10 to 100 times lower than typical B. abortus immunization doses administered to cattle (46–48) but similar to challenge strain doses in vaccine or experimental infection studies (4, 10, 47). Thus, the number of strains delivered to calves in our study is relevant when considering the population biology of B. abortus in experimental infection contexts. In a field setting, it is reasonable to expect that cattle contacting Brucella via abortions in the field can be exposed to 108 cells (or more) given that aborted fetal tissue can contain 1013 CFU/g (49).
Our dose is significantly higher than the reported B. abortus infectious dose of ∼500 CFU in mice (50, 51) and guinea pigs (52), when administered via aerosol. The magnitude of strain loss that we observe between the conjunctiva and the site of initial lymph node colonization (Fig. 2) suggests that the number of B. abortus cells required to ensure reliable infection from the ocular conjunctiva is much larger than 500 CFU. Indeed, controlled cattle vaccination studies (53, 54) have shown that only 78% of unvaccinated animals are infected at a dose of 3.7 × 105 CFU via the conjunctival route, while 89% are infected at a dose of 7.4 × 105 CFU. Doses of 1.5 × 107 to 1 × 108 CFU typically result in 100% infection in experimental conjunctival infections (54). From these data, an infection “bottleneck” along the conjunctival route is inferred.
Several groups have presented thorough discussions of the nature of infection bottlenecks from both experimental and theoretical perspectives (12, 55, 56). These reviews and others highlight ways in which host restriction mechanisms shape bottlenecks which, in turn, influence the population biology and evolutionary dynamics of microbial pathogens (57). Given the difference in B. abortus population size between the inoculum (108) and the lymph node (105 to 106), a bottleneck in this path is evident. However, one cannot accurately gauge the magnitude of a bottleneck from cell count alone because population size is a function of both the bottleneck itself and subsequent microbial growth that occurs between the time of colonization and the time of sampling. By uniquely tagging individual strains in the population and leveraging the distribution of strain frequencies in our tagged pool we were able assess the magnitude of the B. abortus population bottleneck via the conjunctival route of infection.
At an inoculum of 108 CFU, each strain in the initial pool (diversity of ∼106) was represented on average 100 times in the infecting dose. If we assume that each barcode in a lymph node reflects a single colonization event, our data indicate each node was colonized by an average of 7 × 103 individuals from a pool of 108 CFU in the inoculum (Fig. 2 and Dataset S1). Thus, only 1 in 14,000 strains present in the inoculum are detected in vivo 8 to 9 d postinfection; this could be viewed as an infection bottleneck of 1:14,000. This rough estimate likely undercounts colonization events because highly abundant strains in the inoculum are expected to successfully colonize more than once. As such, we extended our analysis of the probability of host colonization taking advantage of the fact that the abundance of strains in the inoculum is not completely uniform. This permitted us to assess the probability of colonization of the parotid lymph nodes as a function of strain abundance in the infecting pool. Specifically, we considered colonization at the individual strain level as a binary event, that is, whether a strain barcode was present or absent in a node. Plotting the probability of observing a particular barcode—in any node, across all six animals—against the fractional abundance of barcodes in the infecting pool yielded a binomial-like probability curve, which is well modeled by a single-cell colonization probability of 0.0001 across the vast majority of strain abundances. From our analyses, we conclude that the infection bottleneck via the conjunctiva at a dose of 108 CFU is 1:10,000, while the ID50 is 3,500 CFU.
Multiple models can explain the bottleneck we observe. At one extreme, it could be the case that 99.99% of strains in the Tn-himar pool contain insertions in genes required to colonize the host. These strains would therefore be lost by the time we isolated B. abortus from host tissue 8 to 9 d after infection. Certainly, our pool of Tn-himar strains is expected to contain mutants that have reduced fitness in the bovine host environment. However, previous in vitro and in vivo studies with less complex Brucella spp. mutant pools (34–41) are not consistent with a model in which such a large fraction of mutants are attenuated in infection. In a second model, the host exerts selective pressure on all members of the infecting population irrespective of strain genotype, and only 1:10,000 cells randomly colonize the host. In reality, aspects of both models influence the infection bottleneck we observe. Indiscriminate strain killing by host defenses is the dominant mechanism that accounts for loss of strain diversity in this experiment.
Brucella Tn-Seq Studies of Natural Host via Natural Routes.
This study demonstrates that barcoded Tn-seq studies provide a means to quantify changes in the population structure of B. abortus in a bovine host, infected via a natural route. Given the success of the approach outlined here, one can envision using similar approaches for studying B. abortus transmission in suitably controlled field settings. We have also recently constructed a barcoded Brucella ovis mutant library (58), which may be similarly useful for assessing the population biology of this ovine pathogen during infection/transmission.
Materials and Methods
Bacterial Strains and Select Agent Considerations.
All experiments using live B. abortus 2308 were performed in biosafety level 3 facilities according to select agent regulations at the US Department of Agriculture (USDA) National Animal Disease Center. B. abortus Tn-himar strains were cultivated on TSA-SK at 37 °C with 5% CO2.
Preparation of B. abortus Transposon Library.
The B. abortus Tn-Himar library was previously generated and mapped (21) using a barcoded transposon mutagenesis strategy developed by Wetmore et al. (22). In this study, a 1-mL aliquot of the mutant library containing ∼2.3 × 109 CFU was thawed and spread on a 150-mm TSA-SK plate. After 2 d of growth at 37 °C in the presence of 5% CO2, cells were scraped from the plate, resuspended in phosphate-buffered saline (PBS), and diluted to a concentration of 2 × 109 CFU/mL in PBS. This mixture of strains served as the inoculum for all six animals. Three 500-μL aliquots of this mixture were saved as representatives of the inoculum.
Animal Care and Welfare.
Holstein calves between 5 and 9 mo of age (Table 1) housed on the grounds of the National Animal Disease Center in Ames, IA were transferred to an agriculture biosafety level 3 (Ag-BSL-3) containment facility, where they were housed for the duration of the study. All animal care was performed under the guidance of the associated institutional animal care and use committee protocols at the National Animal Disease Center.
Infection of Cattle with B. abortus Transposon Library.
All animal work was conducted in the BSL-3 Agricultural Suite at the USDA National Animal Disease Center, Agricultural Research Service according to USDA-approved protocols. Animals were acclimated to the biosafety containment barn for 1 wk prior to infection. Fifty microliters of the B. abortus mutant library prepared as described above was inoculated into the conjunctiva of each eye of each calf. Eight or nine days postinfection, the calves were humanely euthanized and the parotid and retropharyngeal lymph nodes were collected. The lymph nodes were immediately placed at 4 °C. Parotid lymph nodes were processed the day of collection. Retropharyngeal lymph nodes were processed the following day.
Harvesting B. abortus Transposon Strains from Lymph Nodes.
Parotid lymph nodes were divided into four equal sections. Each section was weighed and then homogenized in 2 mL PBS in gentleMACS M tubes and a gentleMACS Octo Dissociator (Miltenyi Biotec) using the RNA_1 setting. Homogenates were gently mixed with detergent (0.1% Triton X-100) and incubated at room temperature for ∼10 min to lyse the eukaryotic cell membranes and allow tissue debris to settle. Two hundred microliters of the homogenate were serially diluted to enumerate CFU on TSA-SK plates. One milliliter of each homogenate was evenly spread on a 150-mm TSA-SK plate to recover B. abortus strains. Plates were incubated at 37 °C in the presence of 5% CO2. After 72 h, the bacterial lawn was scraped from the plate and resuspended in PBS and cell pellets were stored at −20 °C.
Due to limitations in sample processing throughput and the expectation that retropharyngeal lymph nodes may not be colonized at the time of harvest (10), only the left retropharyngeal lymph node from each animal was processed. These were divided into two sections and homogenized as above. One milliliter of each homogenate was spread on a 150-mm TSA-SK plate to recover bacteria and assess colonization. After 72 h of incubation, zero to a few Brucella-like colonies were observed from three animals; thousands to a lawn of Brucella-like colonies were observed from three animals. Bacteria were scraped from the later three samples and processed with the parotid lymph node samples.
B. abortus Genomic DNA Preparation.
Genomic DNA was recovered from stored cell pellets using the Ultradeep Microbiome Prep Kit (Molzym), omitting the initial steps for removal of host DNA. Briefly, ∼20 µL in volume of each bacterial pellet (derived from lymph node scrapings) was resuspended in kit Buffer RL. Cell pellets stored from the inoculum were also processed in parallel. Bacterial cells were lysed with addition of kit BugLysis buffer at 37 °C, lysate was treated with proteinase K at 56 °C, and genomic DNA was purified using kit spin columns following a guanidine isothiocyanate treatment step. An additional ethanol precipitation was conducted on the eluates, and final pellets were each dissolved in 100 µL of ultrapure water. DNA samples were stored at −20 °C or colder prior to PCR analysis.
PCR Barcode Amplification and Sequencing.
Transposon barcodes were amplified as described by Wetmore et al. (22). Briefly, 20-μL PCR reactions contained 0.2 to 1.0 μg of genomic DNA, 0.5 μM of each primer [Barseq_P1 as the universal forward primer and an indexed Barseq_P2 primer unique to each sample (22)], and Q5 hotstart polymerase with GC enhancer (New England Biolabs). Reactions were cycled as follows: 98 °C for 4 min, 25 cycles of 98 °C, 55 °C, and 72 °C for 30 s at each temperature, then 72 °C for 5 min. Amplification was confirmed by resolving 2 μL of each reaction in a 2% agarose gel. Reactions were pooled (5 μL each) and the pooled products were cleaned using GeneJet PCR purification kit (Thermo Fisher). Amplified barcodes were sequenced using an Illumina HiSeq 4000 with Illumina TruSeq primers.
Barcode Counting, a Filtering Approach to Minimize Spurious Inflation of Barcode Diversity.
Our analysis workflow is schematized in SI Appendix, Fig. S2 and described as follows. The sequencing reads were processed with the custom Perl script MultiCodes.pl (22) to generate lists of the number of times each barcode was observed in each sample. To assess the fitness consequences of gene disruptions by barcoded transposons, the analysis pipeline developed by Wetmore et al. (22) analyzes only codes that are mapped with high confidence to positions in the genome, and the pipeline to map barcodes to specific positions in the genome is stringent (22). An advantage of this approach is that it effectively minimizes the effect of sequencing errors, as codes in experimental samples that are not identical to a mapped code are ignored. However, in the analysis presented herein, we aimed to use barcodes to assess the diversity and structure of the entire population and the unmapped codes constitute a sizeable fraction of the population (SI Appendix, Fig. S5). Illumina sequencing errors occur at a frequency of 10−3 per base (24–26); thus, given a 20-base pair barcode, one expects ∼1 to 2% of barcode sequence reads to contain an error. Such errors inflate the apparent diversity of barcodes in a sampled population. In an effort to more accurately define B. abortus population distributions in host tissue, we developed the following approach to minimize the influence of sequencing errors on our analysis.
There are multiple approaches to reduce error in sequence-based population measurements (59). In our case, the B. abortus Tn mutant library is highly diverse and contains many true rare codes (21). Thus, approaches based on removing codes under a threshold of coverage were inappropriate. We therefore applied two selective approaches to reduce inflation of barcode diversity due to sequence error. First, we systematically removed codes that were “off-by-one” from any code that was counted ≥100 times. Based on error rates, we expect that codes sequenced more than 100 times are likely to be sequenced erroneously at least once. The most abundant codes in our host datasets were counted on the order of 106 times in select lymph node samples (Dataset S1). For these extremely abundant codes, we detected all 60 possible codes arising from single site errors at an observed rate of 0.5 to 1 × 10−3 per site, consistent with expectations (see SI Appendix, Fig. S1 for examples). When summed, the reads corresponding to these “off-by-one” codes total 1 to 2% of the reads of the abundant parent code. Applying this filtering approach resulted in the removal of a small fraction (1.4 ± 0.3%) of total sequencing reads, and 24 ± 4% of codes from each lymph node sample. As a second step, we removed all codes from our lymph node samples that were not detected in at least one of the inoculum replicates. Together, these two filtering steps removed 80.3 ± 9.7% of codes, which represented only 1.7 ± 0.3% of the total reads from the lymph node samples (SI Appendix, Fig. S3 and Dataset S1). Importantly, the total number of reads filtered using this approach is consistent with the expected number of erroneous reads based on known Illumina error rates. After filtering, revised lists of barcode counts were generated for each sample. Barcode counts for each lymph node were generated by totaling the reads from the respective quadrant samples.
Population Analysis of Barcodes.
To generate the proportional Euler graphs to visualize overlap in barcode sets we used the R package eulerr (60). The R package vegan (61) was used for population diversity metrics including Fisher alpha diversity, Shannon diversity (H), and evenness (J). The vegan function rarefy was used to assess diversity in a randomly selected set of 106 sequence reads. Dissimilarity between samples was calculated with the vegan function vegdist. Analysis of the hyperabundant barcodes involved first identifying barcodes that comprised at least 2% of the reads in any sample; 160 barcodes met this threshold. We then extracted the fractional abundance of this set of 160 codes in all samples.
Probability.
To assess abundance of each code in the inoculum, the counts for all three inoculum replicates were summed. The total counts for each barcode were normalized by the total number of reads in all inoculum replicates. These fractional abundance values were then log10-transformed. Barcodes were then binned by log10(fractional abundance). Most bins were 0.1 log unit in width. However, the bins for the most abundant codes were wider due to the scarcity of highly abundant codes. Next, we assessed the fraction of codes in each bin that were or were not detected in the summary list of barcodes for each parotid lymph node. Then for each bin, we averaged the fraction not detected in all 12 lymph nodes.
To generate the probability curves we used the binomial distribution function [p(x) = choose(n,x) Px(1 − P)(n−x)] to calculate the probability (p) of colonization occurring exactly zero times (x = 0) with different probabilities (P) of colonization over a range of number of trials (n) using the pbinom function in R. Here the number of trials corresponds to abundance in the inoculum. Specifically, n was calculated by multiplying fractional abundance in the inoculum by the size of the inoculum (108).
To estimate the infectious dose at which 50% of lymph nodes would be colonized (ID50), the observed mean fraction of strains not detected in lymph nodes was fit to a log-logistic model with three parameters using the drm function in the drc package in R (44). In this model, f(x) = d/(1+exp(b(log(x) − log(e)))), where d is the upper limit, b corresponds to the steepness of the curve, and e indicates the dose (i.e., fractional abundance) corresponding to a 0.5 probability of success in colonization.
Supplementary Material
Acknowledgments
We acknowledge the efforts of the project technicians, especially Darl Pringle, the animal care and veterinary staff, and the biosafety staff at the National Animal Disease Center. We thank David Hershey, members of the S.C. laboratory, Andrew Olive, and Ashley Shade for helpful discussions. This work was funded by in part by NIH grants R01AI107159 and R35GM131762.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023500118/-/DCSupplemental.
Data Availability
Raw barcode amplicon sequence data have been deposited in the NCBI sequence read archive (SRA) under BioSample accession SAMN10145474, BioProject accession PRJNA604627 (62). All data files and R analytical scripts/code used in this manuscript have been deposited in GitHub (https://github.com/amfiebig/Brucella_barcodes).
References
- 1.Godfroid J., Brucella spp. at the wildlife-livestock interface: An evolutionary trajectory through a livestock-to-wildlife “host jump”? Vet. Sci. 5, 81 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hull N. C., Schumaker B. A., Comparisons of brucellosis between human and veterinary medicine. Infect. Ecol. Epidemiol. 8, 1500846 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grilló M. J., Blasco J. M., Gorvel J. P., Moriyón I., Moreno E., What have we learned from brucellosis in the mouse model? Vet. Res. (Faisalabad) 43, 29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plommet M., “Brucellosis in domestic animals” in Bovine Brucellosis: An International Symposium, Crawford R. P., Hidalgo R. J., Eds. (Texas A&M University Press, 1977), pp. 116–134. [Google Scholar]
- 5.Gorvel J. P., Brucella: A Mr “Hide” converted into Dr Jekyll. Microbes Infect. 10, 1010–1013 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Paixão T. A., et al., Establishment of systemic Brucella melitensis infection through the digestive tract requires urease, the type IV secretion system, and lipopolysaccharide O antigen. Infect. Immun. 77, 4197–4208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossetti C. A., et al., Systems biology analysis of Brucella infected Peyer’s patch reveals rapid invasion with modest transient perturbations of the host transcriptome. PLoS One 8, e81719 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xavier M. N., Paixão T. A., Poester F. P., Lage A. P., Santos R. L., Pathological, immunohistochemical and bacteriological study of tissues and milk of cows and fetuses experimentally infected with Brucella abortus. J. Comp. Pathol. 140, 149–157 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Enright F. M., “The pathogenesis and pathobiology of Brucella infection in domestic animals” in Animal Brucellosis, Nielsen K., Duncan J. R., Eds. (CRC Press, Boca Raton, FL, 1990), pp. 301–320. [Google Scholar]
- 10.Meador V. P., Warner D. P., Deyoe B. L., Distribution of Brucella abortus organisms in calves after conjunctival exposure. Am. J. Vet. Res. 49, 2015–2017 (1988). [PubMed] [Google Scholar]
- 11.Payne J. M., The pathogenesis of experimental brucellosis in the pregnant cow. J. Pathol. Bacteriol. 78, 447–463 (1959). [DOI] [PubMed] [Google Scholar]
- 12.Abel S., Abel zur Wiesch P., Davis B. M., Waldor M. K., Analysis of bottlenecks in experimental models of infection. PLoS Pathog. 11, e1004823 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abel S., et al., Sequence tag-based analysis of microbial population dynamics. Nat. Methods 12, 223–226 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin C. J., et al., Digitally barcoding Mycobacterium tuberculosis reveals in vivo infection dynamics in the Macaque model of tuberculosis. MBio 8, e00312–e00317 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Troy E. B., et al., Understanding barriers to Borrelia burgdorferi dissemination during infection using massively parallel sequencing. Infect. Immun. 81, 2347–2357 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rego R. O., Bestor A., Stefka J., Rosa P. A., Population bottlenecks during the infectious cycle of the Lyme disease spirochete Borrelia burgdorferi. PLoS One 9, e101009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bacigalupe R., Tormo-Mas M. A., Penadés J. R., Fitzgerald J. R., A multihost bacterial pathogen overcomes continuous population bottlenecks to adapt to new host species. Sci. Adv. 5, eaax0063 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCrone J. T., Lauring A. S., Genetic bottlenecks in intraspecies virus transmission. Curr. Opin. Virol. 28, 20–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahmutovic A., Abel Zur Wiesch P., Abel S., Selection or drift: The population biology underlying transposon insertion sequencing experiments. Comput. Struct. Biotechnol. J. 18, 791–804 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrou J., et al., Brucella periplasmic protein EipB is a molecular determinant of cell envelope integrity and virulence. J. Bacteriol. 201, e00134–e00119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrou J., et al., Periplasmic protein EipA determines envelope stress resistance and virulence in Brucella abortus. Mol. Microbiol. 111, 637–661 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wetmore K. M., et al., Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. MBio 6, e00306–e00315 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheville N. F., et al., Effects of age at vaccination on efficacy of Brucella abortus strain RB51 to protect cattle against brucellosis. Am. J. Vet. Res. 57, 1153–1156 (1996). [PubMed] [Google Scholar]
- 24.Pfeiffer F., et al., Systematic evaluation of error rates and causes in short samples in next-generation sequencing. Sci. Rep. 8, 10950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fox E. J., Reid-Bayliss K. S., Emond M. J., Loeb L. A., Accuracy of next generation sequencing platforms. Next Gener. Seq. Appl. 1, 1000106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodwin S., McPherson J. D., McCombie W. R., Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17, 333–351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisher R. A., Corbet A. S., Williams C. B., The relation between the number of species and the number of individuals in a random sample of an animal population. J. Anim. Ecol. 12, 42–58 (1943). [Google Scholar]
- 28.Shannon C. E., The mathematical theory of communication. 1963. MD Comput. 14, 306–317 (1997). [PubMed] [Google Scholar]
- 29.Jaccard P., Contribution au problème de l’immigration post‐glaciaire de la flore alpine. Bull. Soc. Vaud. Sci. Nat. 36, 87–130 (1900). [Google Scholar]
- 30.Bray J. R., Curtis J. T., An ordination of the upland forest communities of Southern Wisconsin. Ecol. Monogr. 27, 325–349 (1957). [Google Scholar]
- 31.Poester F. P., Samartino L. E., Santos R. L., Pathogenesis and pathobiology of brucellosis in livestock. Rev. Sci. Tech. 32, 105–115 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Duneau D., et al., Stochastic variation in the initial phase of bacterial infection predicts the probability of survival in D. melanogaster. eLife 6, e28298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delrue R. M., Lestrate P., Tibor A., Letesson J. J., De Bolle X., Brucella pathogenesis, genes identified from random large-scale screens. FEMS Microbiol. Lett. 231, 1–12 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Kim D. H., et al., Identification of genes contributing to the intracellular replication of Brucella abortus within HeLa and RAW 264.7 cells. Vet. Microbiol. 158, 322–328 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Kim S., et al., Isolation and characterization of mini-Tn5Km2 insertion mutants of Brucella abortus deficient in internalization and intracellular growth in HeLa cells. Infect. Immun. 71, 3020–3027 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sternon J. F., et al., Transposon sequencing of Brucella abortus uncovers essential genes for growth in vitro and inside Macrophages. Infect. Immun. 86, e00312–e00318 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Q., Pei J., Turse C., Ficht T. A., Mariner mutagenesis of Brucella melitensis reveals genes with previously uncharacterized roles in virulence and survival. BMC Microbiol. 6, 102 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lestrate P., et al., Identification and characterization of in vivo attenuated mutants of Brucella melitensis. Mol. Microbiol. 38, 543–551 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Lestrate P., et al., Attenuated signature-tagged mutagenesis mutants of Brucella melitensis identified during the acute phase of infection in mice. Infect. Immun. 71, 7053–7060 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian M., et al., Identification of novel genes essential for Brucella abortus to establish infection by signature-tagged mutagenesis. Vet. Microbiol. 230, 130–137 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Zygmunt M. S., Hagius S. D., Walker J. V., Elzer P. H., Identification of Brucella melitensis 16M genes required for bacterial survival in the caprine host. Microbes Infect. 8, 2849–2854 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Mendum T. A., et al., Transposon libraries identify novel Mycobacterium bovis BCG genes involved in the dynamic interactions required for BCG to persist during in vivo passage in cattle. BMC Genomics 20, 431 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sassetti C. M., Rubin E. J., Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U.S.A. 100, 12989–12994 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ritz C., Baty F., Streibig J. C., Gerhard D., R., Dose-response analysis using R. PLoS One 10, e0146021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cain A. K., et al., A decade of advances in transposon-insertion sequencing. Nat. Rev. Genet. 21, 526–540 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dorneles E. M., Sriranganathan N., Lage A. P., Recent advances in Brucella abortus vaccines. Vet. Res. (Faisalabad) 46, 76 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plommet M., Plommet A.-M., Vaccination against bovine brucellosis with a low dose of strain 19 administered by the conjunctival route. Ann. Rech. Vet. 7, 1–8 (1976). [PubMed] [Google Scholar]
- 48.Yang X., et al., Progress in Brucella vaccine development. Front. Biol. (Beijing) 8, 60–77 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alexander B., Schnurrenberger P. R., Brown R. R., Numbers of Brucella abortus in the placenta, umbilicus and fetal fluid of two naturally infected cows. Vet. Rec. 108, 500 (1981). [DOI] [PubMed] [Google Scholar]
- 50.Kahl-McDonagh M. M., Arenas-Gamboa A. M., Ficht T. A., Aerosol infection of BALB/c mice with Brucella melitensis and Brucella abortus and protective efficacy against aerosol challenge. Infect. Immun. 75, 4923–4932 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olsen S. C., Waters W. R., Stoffregen W. S., An aerosolized Brucella spp. challenge model for laboratory animals. Zoonoses Public Health 54, 281–285 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Teske S. S., et al., Animal and human dose-response models for Brucella species. Risk Anal. 31, 1576–1596 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Manthei C. A., “Brucellosis in cattle” in Brucellosis: A Symposium Under the Joint Auspices of National Institutes of Health of the Public Health Service, Federal Security Agency, United States Dept. of Agriculture [and] National Research Council, September 22-23, 1949 (American Association for the Advancement of Science, Bethesda, MD, 1950), pp. 172–187. [Google Scholar]
- 54.Manthei C. A., Brucellosis. Application of research to bovine brucellosis control and eradication programs. J. Dairy Sci. 51, 1115–1120 (1968). [DOI] [PubMed] [Google Scholar]
- 55.Johnson K. E. E., Ghedin E., Quantifying between-host transmission in influenza virus infections. Cold Spring Harb. Perspect. Med. 10, a038422 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zwart M. P., Elena S. F., Matters of size: Genetic bottlenecks in virus infection and their potential impact on evolution. Annu. Rev. Virol. 2, 161–179 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Moxon R., Kussell E., The impact of bottlenecks on microbial survival, adaptation, and phenotypic switching in host-pathogen interactions. Evolution 71, 2803–2816 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Varesio L. M., Willett J. W., Fiebig A., Crosson S., A carbonic anhydrase pseudogene Sensitizes select Brucella lineages to low CO2 tension. J. Bacteriol. 201, e00509–e00519 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar R. C., Flyvbjerg H., Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31, 3476–3482 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Larsson J.. eulerr: Area-proportional Euler and Venn diagrams with ellipses. R package version 6.1.0. https://cran.r-project.org/package=eulerr. Accessed 20 August 2020.
- 61.Oksanen J., et al., vegan: Community ecology package. R package version 2.5-6. https://CRAN.R-project.org/package=vegan. Accessed 10 January 2020.
- 62.Fiebig A., Crosson S., Strain-level assessment of bovine colonization by Brucella abortus. NCBI BioProject. https://www.ncbi.nlm.nih.gov/bioproject/6046275. Deposited 3 February 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw barcode amplicon sequence data have been deposited in the NCBI sequence read archive (SRA) under BioSample accession SAMN10145474, BioProject accession PRJNA604627 (62). All data files and R analytical scripts/code used in this manuscript have been deposited in GitHub (https://github.com/amfiebig/Brucella_barcodes).