Significance
Protein folding and oligomeric complex assembly are essential for protein homeostasis. These processes are orchestrated by a network of host chaperones that interact with nascent and formed polypeptides. We investigated the mechanism by which a ubiquitous, essential, molecular chaperone, TRiC, folds an aggregation-prone protein subunit of a viral capsid. Structural studies, mass spectrometry, and in vitro folding and assembly experiments provided insights into the mechanism by which TRiC, in cooperation with a cochaperone, prefoldin, folds and assembles protein multimers. As the principles of protein folding and assembly are evolutionarily conserved, these findings likely point to common functions for TRiC and prefoldin in assembling a diversity of intracellular protein complexes.
Keywords: molecular chaperones, protein folding, virus assembly, TRiC, prefoldin
Abstract
Intracellular protein homeostasis is maintained by a network of chaperones that function to fold proteins into their native conformation. The eukaryotic TRiC chaperonin (TCP1-ring complex, also called CCT for cytosolic chaperonin containing TCP1) facilitates folding of a subset of proteins with folding constraints such as complex topologies. To better understand the mechanism of TRiC folding, we investigated the biogenesis of an obligate TRiC substrate, the reovirus σ3 capsid protein. We discovered that the σ3 protein interacts with a network of chaperones, including TRiC and prefoldin. Using a combination of cryoelectron microscopy, cross-linking mass spectrometry, and biochemical approaches, we establish functions for TRiC and prefoldin in folding σ3 and promoting its assembly into higher-order oligomers. These studies illuminate the molecular dynamics of σ3 folding and establish a biological function for TRiC in virus assembly. In addition, our findings provide structural and functional insight into the mechanism by which TRiC and prefoldin participate in the assembly of protein complexes.
Chaperones perform the essential function of folding proteins that cannot achieve a native conformation in an unassisted manner. The eukaryotic chaperonin TRiC (TCP1-ring complex, also called CCT for cytosolic chaperonin containing TCP1) mediates the adenosine triphosphate (ATP)-dependent folding of a subset (∼10%) of newly translated cytosolic proteins (1). TRiC substrates include the cytoskeletal proteins actin and tubulin (2, 3), tumor suppressor proteins (4, 5), and aggregation-prone proteins that accumulate in neurodegenerative diseases such as Huntington disease (6, 7). TRiC has a toroidal structure formed by two rings stacked back to back, each composed of eight paralogous subunits that form a barrel with a central cavity (8, 9). The central cavity functions as a chamber within which newly translated polypeptides are sequestered in a protected protein-folding environment (10, 11). ATP binding and hydrolysis trigger cyclic changes in the conformation of TRiC between an open form in the nucleotide-free state and a closed, folding-active conformation in the ATP-hydrolysis state (12, 13). Despite its quasisymmetry, there is a high degree of specialization within the eight subunits that form TRiC, with individual subunits differing in substrate-binding capacity (14), ATP-binding and hydrolytic activity (12, 15, 16), and surface hydrophobicity (8). The diversification of these subunits is thought to contribute to the capacity of TRiC to fold a wide array of substrates with complex topologies.
TRiC functions in concert with other molecular chaperones, including Hsp70 (17) and prefoldin (PFD) (18), to direct protein-folding pathways. Other chaperones, including ribosome-associated chaperones such as Hsp70, may function more promiscuously and earlier in the folding process to stabilize unfolded or partially folded proteins in conformations that can either fold without further assistance or be recognized by more specialized chaperones (19). PFD, a jellyfish-structured TRiC cochaperone (20), can also bind nascent polypeptides (18, 21). Hsp70 and PFD may direct the transfer of certain substrates to TRiC (17, 18). PFD can contribute directly to TRiC-mediated protein folding (22), but mechanisms by which cochaperones cooperate with TRiC to fold individual substrates are poorly defined.
As obligate intracellular pathogens, viruses replicate within host cells and use host chaperones to fold viral polypeptides. Proteins from diverse families of viruses have been identified as TRiC substrates (23–25). TRiC performs an essential folding function in the replication of mammalian orthoreoviruses (reoviruses) (26). Reoviruses are ubiquitous, infecting most mammals early in life, and have been linked to celiac disease in humans (27). Reoviruses have a proteinaceous outer capsid composed of μ1 and σ3, which coalesce in a 1:1 stoichiometric ratio to form heterohexamers. The σ3 component of the outer capsid is an obligate TRiC substrate (26). σ3 is aggregation prone (28) and exists in multiple oligomeric forms in the cell (26). The mechanism of TRiC/σ3 binding, folding, and release is unclear. In addition, the chaperones that cooperate with TRiC to fold and assemble σ3 into a complex with its binding partner, μ1, are unknown.
In this study, we identify a network of host chaperones that interact with reovirus σ3, including Hsp70, Hsp90, PFD, and TRiC. We establish a function for PFD in preventing σ3 aggregation and enhancing σ3 transfer to TRiC. Cryoelectron microscopy (cryo-EM) and cross-linking mass spectrometry (XL-MS) resolve the structure of σ3 within the TRiC folding chamber, revealing TRiC/σ3 interaction interfaces and the orientation of the σ3/μ1 oligomerization domain within TRiC. Biochemical studies establish a function for TRiC in assembling folded σ3 into a complex with μ1 through a process that is enhanced by PFD. Functional assays demonstrate that TRiC folds σ3 into its biologically active and infectious conformation. Together, this work provides mechanistic insight into the structure and function of TRiC and PFD in the folding and assembly of a heterooligomeric protein complex.
Results
Identification of a Chaperone Network Interacting with the Reovirus σ3 Major Outer-Capsid Protein.
To better understand the chaperones that fold and assemble protein oligomers, we investigated the biogenesis of the reovirus outer capsid, which comprises 200 heterohexamers each containing three copies of σ3 and μ1 (Fig. 1A). σ3 is a substrate of the TRiC chaperonin (26) and likely associates with TRiC cotranslationally or following nascent chain release (Fig. 1B). We hypothesized that an unbiased approach to identify host proteins that interact with σ3 would reveal additional chaperones, thus providing clues about the protein-folding networks responsible for the biogenesis of the outer capsid.
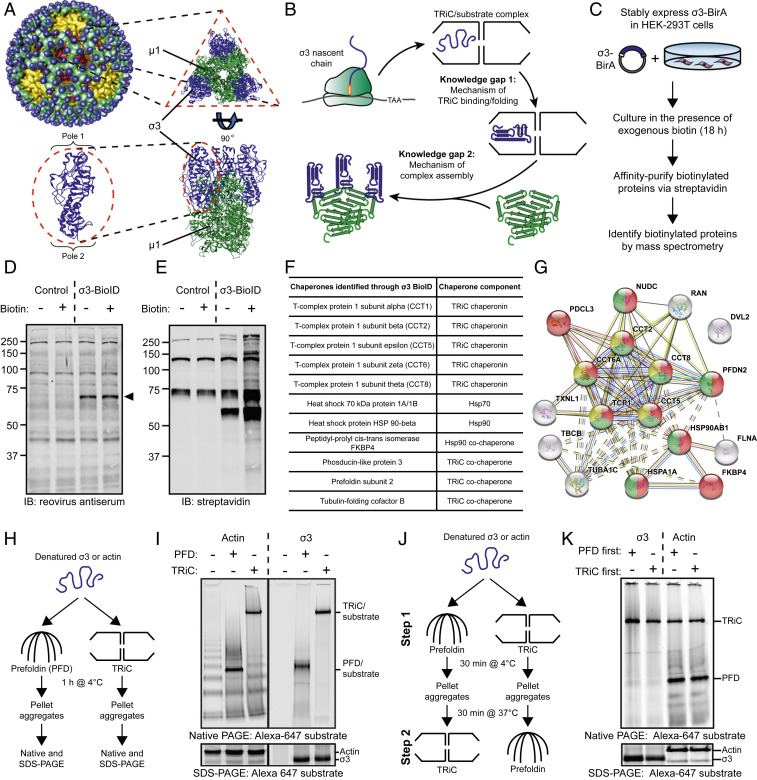
Fig. 1.
A chaperone network interacts with the reovirus σ3 major outer-capsid protein. (A) Structure of the reovirus particle (Upper Left) (44). Structure of the µ13σ33 outer-capsid heterohexamer and the σ3 monomer (Bottom Left); Protein Data Bank: 1JMU. (B) Model of the protein-folding pathway and knowledge gaps in the biogenesis of the reovirus outer capsid. (C) BioID workflow to identify host proteins that interact with the reovirus σ3 outer-capsid protein. (D and E) SDS-PAGE of control or σ3-BirA-expressing (σ3-BioID) HEK-293 cells incubated in the presence (+) or absence (−) of exogenous biotin for 18 h and immunoblotted with a reovirus-specific antiserum (D) or streptavidin (E). Arrowhead denotes σ3-BioID fusion protein. (F) Table of chaperones identified by σ3-BioID. (G) STRING analysis of host proteins identified by σ3-BioID. Yellow, TRiC subunits; red, protein-folding function; green, unfolded protein-binding function. (H) Workflow of PFD/TRiC/σ3-binding experiment. (I) Native- (Top) or SDS-PAGE (Bottom) of PFD/TRiC-binding experiment with σ3 or actin. (J) Workflow of two-step PFD/TRiC-binding experiment. (K) Native- (Top) or SDS-PAGE (Bottom) of two-step PFD/TRiC-binding experiment.
The biotinylation identification (BioID) proximity-based labeling approach was used to identify σ3-binding proteins. The BirA biotin ligase was fused to the C terminus of reovirus σ3 (σ3-BioID), enabling biotinylation of interacting proteins within a ∼10- to 15-nm radius (29) (Fig. 1C). This fusion protein was expressed in HEK-293 cells. To assess σ3-BioID biotinylating activity, cell lysates were harvested, resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and probed with reovirus antiserum (Fig. 1D) or streptavidin (Fig. 1E). Compared with untransfected cells, multiple additional biotinylated protein species were detected in cells expressing σ3-BioID. Affinity purification and mass spectrometry of biotinylated proteins in cells expressing σ3-BioID identified 26 unique proteins across the three independent experimental replicates (SI Appendix, Fig. S1A) [biotinylated proteins expressed in control HEK-293 cells and naturally biotinylated carboxylases (30) were excluded]. Seventeen proteins were identified in at least two of the three replicates and included in subsequent analysis (SI Appendix, Table S1). Eleven of 17 σ3-interacting proteins are molecular chaperones, including TRiC, Hsp70, Hsp90, and PFD (Fig. 1F). Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) analysis of these proteins revealed an enrichment of chaperones with known protein–protein interactions (Fig. 1G), including subunits of the TRiC chaperonin (CCT1,2,5,6,8), heat shock proteins 70 and 90 (HSPA1A and HSP90AB1), and TRiC cochaperones (PDCL, PDCL3, and PFDN2).
The TRiC cochaperone PFD is thought to promote delivery of unfolded substrates to TRiC, thereby increasing the efficiency of the protein-folding process. To determine whether PFD enhances TRiC/σ3-binding efficiency, we reconstituted these chaperone interactions with purified components. Chemically denatured σ3 was fluorescently labeled and incubated alone, with PFD, or with TRiC (Fig. 1H). Actin, a canonical PFD and TRiC substrate, was used as a control. Following a 1 h incubation, insoluble aggregates were pelleted, and the soluble supernatant was resolved by native- and SDS-PAGE. Incubation of denatured actin with PFD or TRiC produced defined protein species that migrated at the anticipated molecular weight of PFD-actin and TRiC-actin complexes (Fig. 1I), whereas incubation of actin alone in buffer resulted in multiple aggregated species. Incubation of σ3 with TRiC yielded a protein species migrating at the molecular weight of a TRiC-substrate complex. Incubation of σ3 in the presence of PFD produced a protein species migrating at the anticipated molecular weight for a PFD-substrate complex. However, incubation of denatured σ3 alone resulted in loss of soluble protein detectable by native- or SDS-PAGE, suggesting that a majority of denatured σ3 forms aggregates when incubated in the absence of TRiC or PFD.
To determine whether PFD promotes σ3 association with TRiC, we conducted a two-step binding experiment in which either actin or σ3 was incubated with PFD or TRiC for 30 min, subsequently incubated with the reciprocal chaperone, and resolved by native-PAGE (Fig. 1J). The proportion of actin bound to PFD and TRiC was equivalent in both conditions (Fig. 1K). In contrast, σ3 migrated primarily as a TRiC-bound species regardless of the chaperone incubation order. In addition, the proportion of TRiC-bound σ3 was ∼50% higher when σ3 was first incubated with PFD and followed by TRiC (SI Appendix, Fig. S1B). To assess whether ternary complexes of TRiC, σ3, and PFD form in vitro, σ3 was incubated alone, with TRiC, or with TRiC and PFD and resolved by native-PAGE. Incubation of σ3 with TRiC and PFD together yielded a higher molecular weight complex, reflecting a TRiC/σ3/PFD ternary complex (SI Appendix, Fig. S1C). Together, these data indicate that the binding equilibrium of TRiC/σ3/PFD favors the transfer of σ3 between PFD and TRiC through a ternary TRiC/σ3/PFD intermediate complex.
Next, we examined whether PFD functions during reovirus replication in vivo. Immunofluorescence microscopy revealed a punctate cytoplasmic staining pattern of PFD in uninfected cells, whereas in reovirus-infected cells, PFD redistributed to the periphery of viral factories (SI Appendix, Fig. S2 A and B), similar to the pattern observed with TRiC (26). Reovirus replication yield was reduced 10-fold following siRNA-mediated PFD knockdown in human brain microvascular endothelial cells (HBMECs) (SI Appendix, Fig. S2C). Finally, σ3 protein abundance was substantially reduced in reovirus-infected cells in the setting of PFD knockdown (SI Appendix, Fig. S2D). Collectively, these biochemical and biological data establish a function for PFD in σ3 biogenesis and viral replication.
Cryoelectron Microscopy Reveals σ3 in a Near-Native Conformation in the Closed TRiC Folding Chamber.
To define the structural basis of TRiC-mediated substrate binding and folding, TRiC/σ3 complexes were purified (SI Appendix, Fig. S3), and the structure was determined by single-particle cryo-EM. TRiC can adopt an open, apo-conformation and a closed, ATP-induced folding-active state (31). Purified TRiC/σ3 was incubated in the presence of the ATP-hydrolysis transition state analog ATP/AlFx to yield the closed conformation and visualized by cryo-EM. Images of vitrified TRiC/σ3 revealed monodispersed particles (Fig. 2A), and two-dimensional (2D) class averages included TRiC in front, side, and tilt orientations with an extra density present in one of the two TRiC rings corresponding to the bound substrate (Fig. 2B, arrows). Initial classification revealed subpopulations of TRiC in open and closed conformations. We were unable to discern defined substrate density, such as α-helices, in the open TRiC state, suggesting that σ3 is largely disordered and highly flexible before TRiC cycling.
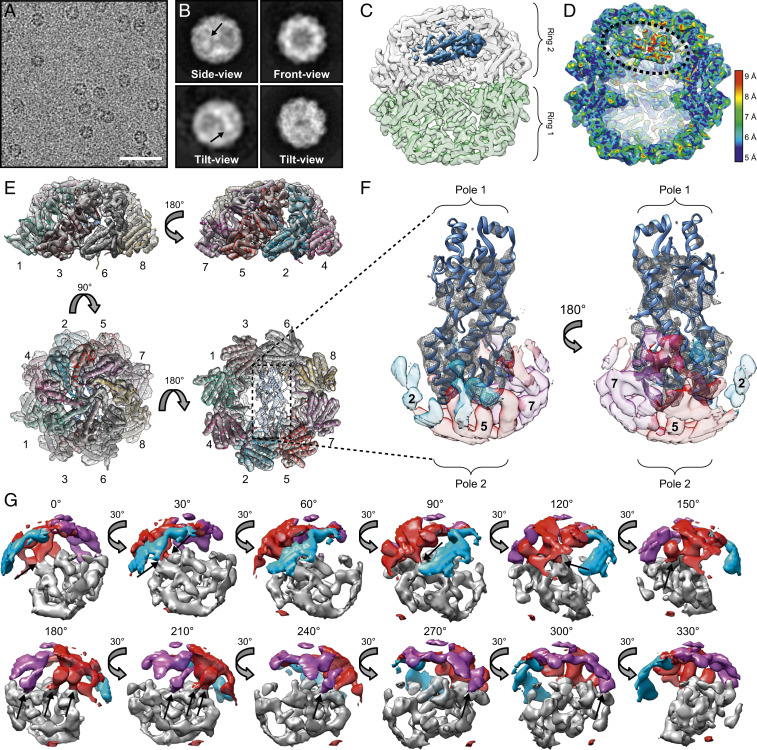
Fig. 2.
Cryoelectron microscopy reveals σ3 in a near-native conformation in the closed TRiC folding chamber. (A) Electron micrograph of ice-embedded TRiC/σ3 complex. (Scale bar: 50 nm.) (B) Selected 2D reference-free averages showing top, side, and tilt views of TRiC/σ3. Arrows indicate extra density. (C) Single model refinement of closed TRiC/σ3 complex with diffuse densities of σ3 shown in blue. A slight cut-through view of the top TRiC ring (white) and bottom TRiC ring (green) are displayed so that σ3 density within the chamber can be visualized. (D) A cut-through view of the local resolution map of the TRiC/σ3 complex. Dotted line encircles σ3 density. (E) Single molecule refinement of TRiC/σ3 overlaying secondary structure of CCT1 to CCT8 subunits shown in different colors from a single TRiC ring containing the σ3 density. (F) Single molecule of σ3 with ribbon tracing (Protein Data Bank: 1FN9) and overlying density mesh with transparent surface density maps of CCT2 (light blue), CCT5 (red), and CCT7 (purple). (G) Surface density focused on σ3 pole 2 (gray), CCT2 (blue), CCT5 (red), and CCT7 (purple) rotated 330°. Arrows indicate overlapping σ3/CCT density.
Images of σ3 complexed with TRiC in the closed state were processed using a single class refinement procedure to yield a 4.5 Å cryo-EM map of TRiC containing internal density (SI Appendix, Fig. S4A). Focused classification of the additional density revealed a large population (47%) that contained structural features of σ3 (e.g., α-helices) and was of appropriate size (SI Appendix, Fig. S4B). A second round of classification using this population (class 1) revealed a subclass that closely matched the crystal structure of σ3 at a resolution of 6.2 Å (Fig. 2 C and D and SI Appendix, Fig. S4C class 1A). An additional large subpopulation contained a less-defined density in the central chamber in a similar orientation to σ3; this density may represent a folding intermediate, although we cannot confidently ascribe this intermediate to σ3 (SI Appendix, Fig. S4C class 1B).
The cryo-EM map of TRiC containing σ3 in a near-native conformation was of sufficient resolution to allow identification of individual TRiC subunits based on differences in the density maps of helix H8 of CCT1 to 8 (Fig. 2E and SI Appendix, Fig. S4D and Movie S2). Additionally, many σ3 helical densities were resolved at 5 to 7 Å (Movies S1 and S2). Strikingly, the substrate density in this TRiC/σ3 map corresponded to a near-native conformation of σ3 with well-defined identifiable secondary structure (Fig. 2F). Thus, we could identify a majority of the μ1-binding domain (pole 2) and a long α-helix that links pole 2 with pole 1. Interestingly, the normally solvent-exposed pole 1 of σ3 did not display well-defined density in the cryo-EM map, suggesting disorder or mobility in this region of the molecule. Closer examination of the well-resolved pole 2 of σ3 revealed close proximity to areas of density corresponding to CCT5 and CCT7, suggestive of TRiC-substrate contact points (Fig. 2G and Movie S3).
σ3 Bridges Two TRiC Hemispheres with Its Oligomerization Domain Oriented Toward the Center of the Folding Chamber.
The TRiC/σ3 cryo-EM map was used to build a molecular model of the TRiC/σ3 structure (Fig. 3A). In this structure, σ3 adopts an orientation inside the TRiC folding chamber stretching across two hemispheres of a single ring (Fig. 3B). An internal view of σ3 within the folding chamber suggests each of the poles of the elongated σ3 structure engages in subunit-specific interactions within the TRiC chamber (Fig. 3 C–E). Pole 2 of σ3 appears to engage the internal surface of CCT5 and CCT7 (Fig. 3 E, Top), while density corresponding to pole 1 lies in proximity to internal surfaces of CCT1, CCT3, and CCT6 (Fig. 3 E, Bottom). The CCT1/3/6 and CCT5/7 interaction surfaces (Fig. 3 E, Insets) and the corresponding surface on each pole of σ3 are enriched in hydrophilic residues (Fig. 3 F and G), pointing to a mostly polar TRiC-substrate interface in the closed chamber.
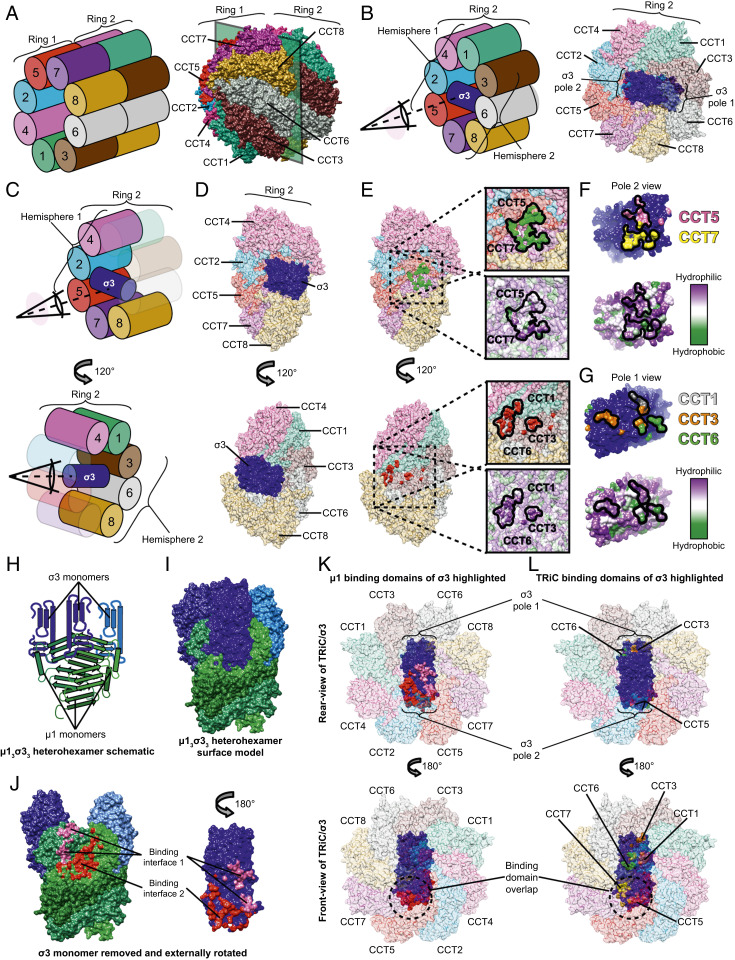
Fig. 3.
σ3 bridges two TRiC hemispheres with its oligomerization domain oriented toward the center of the folding chamber. (A) Schematic (Left) and structural model (Right) of the complete TRiC/σ3 complex in an oblique orientation. (B) Schematic (Left) and structural model (Right) of TRiC/σ3 complex with ring 1 omitted, allowing visualization of σ3. (C and D) Schematic (C) and structural model (D) of σ3 bound to TRiC. Top images omit CCT1,3,6 and Bottom images omit CCT2,5,7. (E) Identical to D except with σ3 removed, revealing the interaction interface. In the Top image, surface residues of CCT5 and CCT7 in proximity to σ3 are colored green. In the Bottom image, surface residues of CCT1,3,6 in proximity to σ3 are colored red. TRiC surface hydrophobicity maps are shown in expanded views with the interaction interface encircled. (F and G) σ3 protein rotated 180° relative to its position in D (Top) image (F) or D (Bottom) image (G). σ3 residues in proximity to CCT5 and CCT7 are colored pink and yellow, respectively. Residues in proximity to CCT1,3,6 are colored gray, orange, and green, respectively. Surface hydrophobicity plots are shown below. (H and I) Schematic (H) and surface (I) representation of µ13σ33 heterohexamer (Protein Data Bank: 1JMU). (J) µ13σ33 heterohexamer with the proximal σ3 monomer removed and externally rotated. Surface residues forming contacts in the μ1/σ3 interface are colored pink and red. (K and L) Rear view (Top) and front view (Bottom) of TRiC/σ3 complex with μ1 (K) or TRiC (L) interaction surfaces highlighted.
σ3 is a heterohexamer subunit, polymerizing with μ1 to form reovirus capsid assemblies (Fig. 3 H and I). To define the orientation of the σ3 polymerization domain within TRiC/σ3, the μ1 interaction interface was mapped onto σ3. In its quaternary state, σ3 forms an interaction interface with two separate molecules of μ1 that involves a majority of σ3 pole 2 (Fig. 3J). This dual μ1 interaction interface was mapped onto TRiC/σ3 to identify its orientation within the folding chamber. Front (Fig. 3 K, Bottom) and rear views (Fig. 3 K, Top) of σ3 within a single TRiC ring revealed that the μ1-binding interface is oriented internally, facing the central equatorial domains and away from the TRiC opening. This orientation maximizes the distance between the μ1-binding interface and the canonical TRiC substrate exit channel. Side-by-side comparison with TRiC/σ3 interaction surfaces demonstrated significant overlap between μ1 and CCT5/7-binding interfaces (Fig. 3 K and L circles). Together, this structural information indicates that the oligomerization domain of σ3 faces the most central region of the chaperonin.
Cross-Linking Mass Spectrometry Reveals TRiC/σ3 Contacts in Different States of ATP Cycling.
We used XL-MS to identify TRiC/σ3 contacts in open and closed TRiC states. Purified TRiC/σ3 was cross-linked with disuccinimidyl suberate (DSS) without ATP, leaving TRiC in an open, folding-incompetent conformation, or in the presence of ATP/AlFx to lock TRiC in a closed state (Fig. 4A). Native- and SDS-PAGE confirmed the expected changes in electrophoretic mobility associated with cross-linking and ATP/AlFx treatment (Fig. 4B).
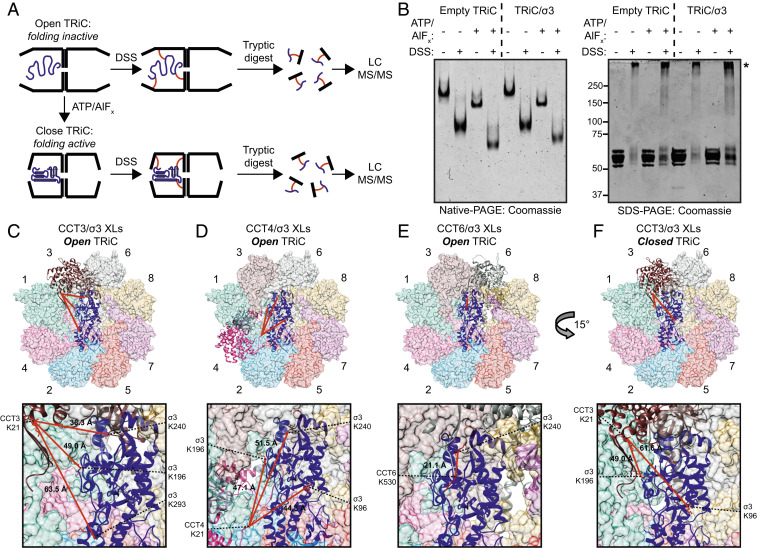
Fig. 4.
Cross-linking mass spectrometry reveals changes in the conformational mobility of the σ3 substrate in different states of TRiC cycling. (A) Workflow of the XL-MS approach. (B) Coomassie-stained native- (Left) or SDS- (Right) PAGE of TRiC/σ3 incubated with the conditions shown. Asterisk denotes cross-linked complexes. (C–F) TRiC/σ3 intermolecular cross-links identified in the open (C–E) and closed (F) conformations mapped onto the TRiC/σ3 structural model. Expanded views are shown below. Lysine residues participating in cross-links are colored red, and Cα–Cα cross-linking distance is listed.
The presence of intermolecular cross-links was determined by mass spectrometry. Analysis of DSS-treated TRiC/σ3 incubated in the absence of ATP/AlFx revealed 61 unique interprotein cross-links between TRiC subunits (SI Appendix, Table S2) and seven unique cross-links between TRiC subunits and σ3. TRiC/σ3 cross-links observed between surface-exposed lysines of σ3 and CCT3, CCT4, and CCT6 were mapped onto the cryo-EM structure of TRiC/σ3 in the closed state (Fig. 4 C–E). All TRiC cross-links with σ3 mapped to TRiC subunit tail domains, which are disordered and highly flexible. CCT3 K21 cross-linked with K196, K240, and K293 of σ3 with Cα–Cα distances of 49 Å, 38 Å, and 64 Å, respectively. Similar cross-linking patterns were observed with CCT4/σ3 and CCT6/σ3. The theoretical maximum distance of cross-linked lysine side chains and the spacer of DSS is <23 Å. The long TRiC/σ3 cross-linking distances in the open TRiC conformation suggest a large degree of conformational mobility and flexibility in either TRiC subunit tail domains, σ3, or both. This conclusion is consistent with the high mobility of the N- and C-terminal tails of TRiC, which are not well resolved in most chaperonin structures (12, 32), and supported by the observation that σ3 K240 cross-linked with tail domains of three different TRiC subunits. The restriction of TRiC/σ3 cross-links in the open state to CCT3, CCT4, and CCT6 indicates that even before TRiC cycling, unfolded σ3 has limited conformational mobility.
XL-MS of TRiC/σ3 in a closed, folding competent state identified 56 unique interprotein cross-links between TRiC subunits (SI Appendix, Table S3). Only two cross-links between TRiC and σ3 were identified (K21 of CCT3 and K96 and K196 of σ3) (Fig. 4F). We hypothesized that the reduced number of TRiC/σ3 contacts in this TRiC state reflects decreased σ3 mobility in its structured, folded conformation. It is unlikely that the reduction in TRiC/σ3 cross-links in the closed state is due to decreased DSS accessibility to the internal folding chamber, as the number of internal TRiC inter- and intrasubunit cross-links did not change substantially in ATP/AlFx-treated TRiC. A higher number of redundant TRiC/σ3 cross-linked peptide pairs was observed in the open state (fifteen) compared to the closed state (two). These data were used to build a model of TRiC subunit/σ3 cross-links reflecting σ3 mobility restrictions in the open and closed TRiC states (SI Appendix, Fig. S5).
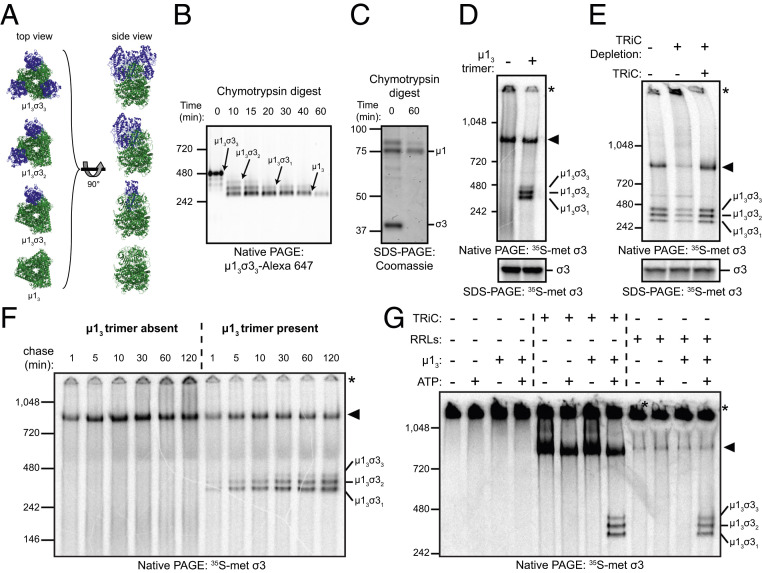
The TRiC Chaperonin Catalyzes σ3 Folding and Assembly onto the µ13 Trimer Base.
To determine whether TRiC/σ3 engages in μ13σ33 assembly, we prepared μ13 trimers from μ13σ33 heterohexamers (SI Appendix, Fig. S6). Three molecules of µ1 intertwine to form a pedestal that is capped by three finger-like projections of σ3 during reovirus assembly (Fig. 5A). Digestion of purified μ13σ33 with chymotrypsin removed σ3 and yielded the μ13 trimer, which migrated as a ∼250-kDa complex by native-PAGE (Fig. 5B). SDS-PAGE of the digested μ13σ33 heterohexamer demonstrated that this complex contains µ1 and lacks σ3 (Fig. 5C).
Fig. 5.
The TRiC chaperonin catalyzes the folding and assembly of σ3 onto the μ13 trimer base. (A) Ribbon tracings of μ1/σ3 assemblies (Protein Data Bank: 1JMU). σ3 monomers are colored blue, and the µ13 trimer is colored green. (B) Native-PAGE of µ13σ33 digested for the intervals shown with chymotrypsin. (C) Coomassie-stained SDS-PAGE of purified µ13σ33 digested for the intervals shown. (D) Native- (Top) and SDS- (Bottom) PAGE of σ3 in vitro translation reactions incubated in the presence or absence of the µ13 trimer. (E) Native- (Top) or SDS- (Bottom) PAGE of σ3 in vitro translation reactions conducted in mock-depleted (−) or TRiC-depleted (+) RRLs with or without addition of purified TRiC (final concentration: 0.2 μM). (F) Native-PAGE of σ3 translated in RRLs for 2 min with [35S]methionine and incubated with cold methionine for the intervals shown in the presence or absence of µ13. (G) Native-PAGE of denatured [35S]methionine-labeled σ3 incubated with the conditions shown.
To assess whether newly translated σ3 assembles onto μ13 trimers, σ3 was translated in rabbit reticulocyte lysates (RRLs) in the presence or absence of purified μ13 and resolved by native-PAGE. In the absence of μ13, in vitro translated σ3 migrated in a ∼800-kDa complex with TRiC or as a high-molecular-weight aggregate (Fig. 5D). When translated in the presence of μ13, σ3 formed soluble μ1/σ3 assemblies. Cotranslation of σ3 with μ13 also limited the accumulation of high-molecular-weight σ3 aggregates, suggesting that the presence of μ13 limits σ3 misfolding and aggregation.
To determine whether TRiC is required for efficient μ1/σ3 assembly during σ3 translation, σ3 was translated in TRiC-depleted RRLs in the presence of μ13. TRiC-associated σ3 and μ1/σ3 assemblies were substantially reduced and aggregated σ3 was significantly increased in TRiC-depleted RRLs (Fig. 5E and SI Appendix, Fig. S7 A–C). Reconstitution of RRLs with purified TRiC restored TRiC/σ3 binding, reduced σ3 aggregation, and increased the abundance of μ1/σ3 assemblies to levels observed using untreated RRLs, indicating that TRiC is required for efficient σ3 assembly into complexes with μ1.
To define the kinetics of μ1/σ3 assembly, σ3 was translated in RRLs supplemented with [35S]methionine in the presence or absence of μ13, incubated with cold methionine, and resolved by native-PAGE. In the absence of μ13, σ3 accumulated as a TRiC-bound species and a high-molecular-weight aggregate (Fig. 5F). In the presence of μ13, σ3 rapidly formed μ1/σ3 assemblies with a majority of the assembly reaction completed by 30 min (SI Appendix, Fig. S7D). Aggregated σ3 increased over time in reactions conducted without μ13 (SI Appendix, Fig. S7E), whereas levels of aggregated σ3 remained unchanged over the same interval in the presence of μ13. The kinetics of TRiC/σ3 binding appeared similar in both reaction conditions, with a rapid accumulation of TRiC/σ3 peaking at 10 to 30 min and gradually declining (SI Appendix, Fig. S7F). Together, these data indicate that σ3 rapidly forms soluble μ1/σ3 assemblies when synthesized in the presence of μ13.
To uncouple translation from folding and determine whether TRiC is sufficient to assemble σ3 onto μ13, we used purified denatured 35S-labeled σ3 as a substrate for in vitro folding reactions. σ3 was incubated alone, with TRiC, with μ13, or with TRiC and μ13 together. As an additional control, denatured σ3 was diluted into RRLs, which contain TRiC as well as additional chaperones. Samples were untreated or supplemented with ATP for 1 h and resolved by native-PAGE. In the absence of TRiC, σ3 exclusively formed aggregates (Fig. 5G). Soluble μ1/σ3 assemblies were detected only when reactions contained μ13, ATP, and either TRiC or RRLs. These data indicate that TRiC and ATP are necessary for μ1/σ3 assembly.
μ13 may promote the release of σ3 from TRiC by preventing rebinding after ATP-induced release or, alternatively, there may be a direct handover of σ3 from TRiC to μ13. Visualization of TRiC/σ3/μ13 assembly reactions by transmission electron microscopy (TEM) revealed a subpopulation of TRiC particles complexed with μ13 (SI Appendix, Fig. S8 A–D), suggesting that ternary TRiC/σ3/μ13 complexes function as an intermediate during σ3 assembly onto μ1.
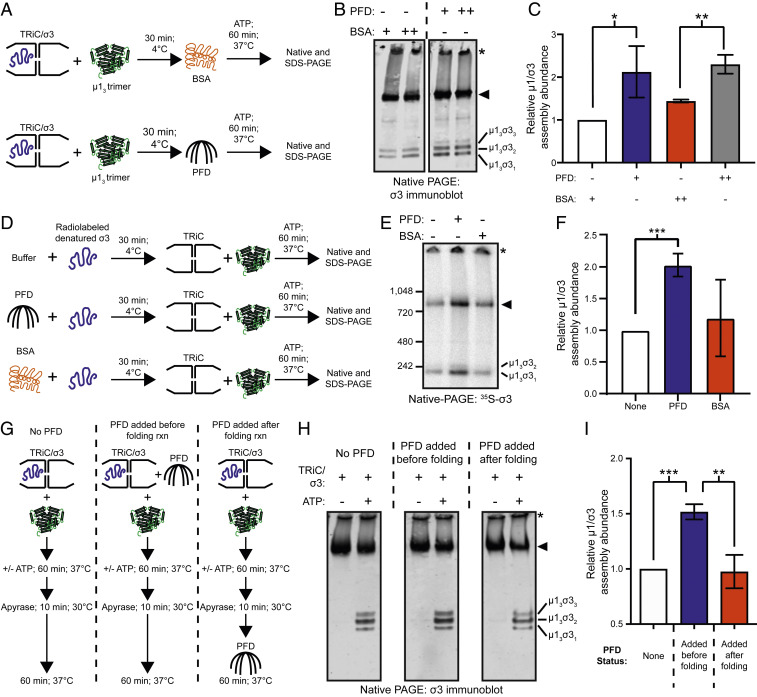
Prefoldin Enhances the Efficiency of TRiC-Mediated μ13σ33 Assembly.
We hypothesized that PFD cooperates with TRiC to maximize μ1/σ3 assembly efficiency. To test this hypothesis, we conducted in vitro assembly assays using purified TRiC/σ3, PFD, and μ13 (Fig. 6A). μ13 was added to TRiC/σ3, and reactions were supplemented with PFD or bovine serum albumin (BSA) as a control. ATP was added to initiate TRiC folding, and reactions were incubated for 1 h, followed by SDS- or native-PAGE and Coomassie staining (SI Appendix, Fig. S9 A and B) or immunoblotting to identify σ3-containing protein assemblies (Fig. 6B). Addition of PFD to reactions resulted in a twofold increase in μ1/σ3 assembly abundance (Fig. 6 B and C). This experimental approach was repeated using radiolabeled denatured σ3 as a substrate. Preincubation of denatured σ3 with PFD alone or PFD and TRiC resulted in a similar twofold increase in μ1/σ3 assembly abundance relative to BSA preincubation (Fig. 6 D–F and SI Appendix, Fig. S9 C–E), indicating that PFD enhances μ1/σ3 assembly efficiency.
Fig. 6.
Prefoldin enhances the efficiency of TRiC-mediated µ13σ33 heterohexamer assembly. (A) Workflow for µ13σ33 heterohexamer assembly assay. (B) Native-immunoblot of µ13σ33 assembly reactions conducted in the presence of BSA or PFD. +,++, 1:1 or 4:1 PFD/BSA:TRiC molar ratio. (C) Quantification of μ1/σ3 assembly abundance from B. (D) Workflow for assembly assay with denatured 35S-labeled σ3 and buffer, PFD, or BSA preincubation. (E) Native-PAGE of µ13σ33 assembly reactions conducted with buffer, PFD, or BSA. (F) Quantification of μ1/σ3 assembly abundance from E. (G) Workflow for PFD time-of-addition µ13σ33 heterohexamer assembly assay. (H) Native-immunoblot of µ13σ33 assembly reactions using independent PFD conditions. (I) Quantification of μ1/σ3 assembly abundance from H. For all quantifications, data are plotted as mean ± SD for three independent replicates (*P < 0.05; **P < 0.01; ***P < 0.001; unpaired two-tailed t test). Asterisk corresponds to aggregated σ3 and the arrowhead corresponds to TRiC/σ3.
We considered two possible mechanisms by which PFD might augment TRiC-mediated μ13σ33 assembly. PFD could directly enhance TRiC-folding activity, thereby increasing the amount of folded σ3 produced during ATP-dependent TRiC cycling. Alternatively, PFD could transfer folded σ3 from TRiC to μ13. To distinguish between these possibilities, TRiC-mediated folding and assembly reactions were uncoupled by conducting time-of-addition experiments with PFD (Fig. 6G). TRiC/σ3 was incubated with PFD at different intervals, followed by the addition of apyrase to terminate folding reactions to determine the contribution of PFD to assembly after halting TRiC cycling. Reactions were resolved by SDS- and native-PAGE and Coomassie stained (SI Appendix, Fig. S9 F and G) or immunoblotted with a σ3-specific monoclonal antibody (Fig. 6H). PFD presence during ATP-driven folding resulted in a ∼1.5-fold increase in μ1/σ3 assembly abundance relative to reactions lacking PFD (Fig. 6I). PFD addition after the termination of TRiC folding had no significant effect on μ1/σ3 assembly abundance relative to reactions lacking PFD. These data support a function for PFD in enhancing μ1/σ3 oligomer assembly through a cooperative mechanism that occurs during TRiC cycling.
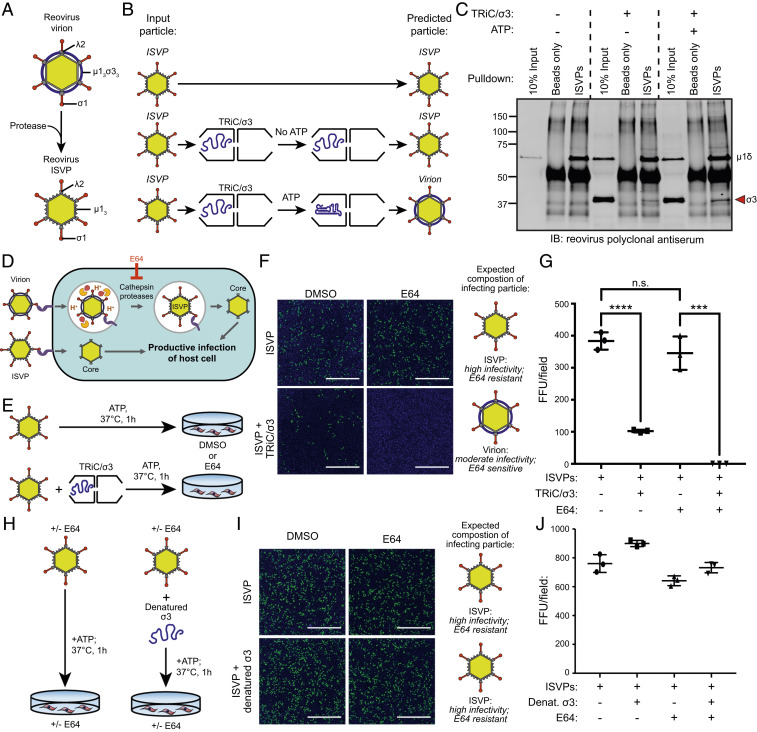
TRiC Folds σ3 Into Its Biologically Active, Infectious Conformation.
To determine whether TRiC-bound σ3 assembles onto reovirus particles, we took advantage of a unique aspect of reovirus biology to develop an in vitro assembly assay. During cell entry, the reovirus outer capsid is cleaved by endosomal cathepsins, forming intermediates called infectious subvirion particles (ISVPs) (Fig. 7A). ISVPs lack σ3 and retain a cleaved form of μ1 (μ1δ) that can reform a complex with σ3 to yield particles that are structurally and functionally indistinguishable from mature reovirus virions (33, 34). To assess whether σ3 transfers from TRiC to viral particles, ISVPs were incubated alone or with TRiC/σ3 in the presence or absence of ATP (Fig. 7B). Immunoprecipitation of ISVPs incubated without TRiC/σ3 yielded particles that exclusively contained μ1δ (Fig. 7C). Particles immunoprecipitated from assembly reactions that contained ISVPs and TRiC/σ3 in the absence of ATP yielded μ1δ and a small amount of σ3, likely reflecting the transfer of a minority population of preexisting folded σ3. Immunoprecipitation of ISVPs from TRiC/σ3/ISVP assembly reactions incubated with ATP yielded a substantial σ3 band, reflecting the efficient transfer of σ3 from TRiC to ISVPs.
Fig. 7.
TRiC/σ3 recoats ISVPs producing biologically active mature reovirus particles. (A) Schematic of reovirus virion and ISVP resulting from proteolytic disassembly of virions. (B) Workflow of ISVP recoating experiment. (C) Immunoblot of immunoprecipitated ISVPs incubated in the presence or absence of TRiC/σ3. (D) Schematic illustrating the reovirus virion and ISVP entry pathways and inhibition of virion disassembly by E64. (E) Workflow to assess ISVP infectivity when incubated with TRiC/σ3. (F) Immunofluorescence images of HeLa cells cultured in DMSO or E64 (200 μM) and inoculated with ISVPs (Top) or ISVPs incubated with TRiC/σ3 (Bottom) and stained with DAPI (blue) and a reovirus-specific antiserum (green). (Scale bar: 1 mm.) (G) Quantification of fluorescent focus units (FFUs) per imaging field from the experiment in F. (H) Workflow to assess the infectivity of ISVPs incubated with denatured σ3. (I) Immunofluorescence images of HeLa cells incubated with DMSO or E64 and inoculated with ISVPs (Top) or ISVPs incubated with denatured σ3 (Bottom) and stained with DAPI (blue) and a reovirus-specific antiserum (green). (Scale bar: 1 mm.) (J) Quantification of FFUs per imaging field from the experiment in I. For quantifications, results shown are the mean ± SD of a single experiment conducted in triplicate and are representative of three independent experiments (***P < 0.001; ****P < 0.0001; unpaired two-tailed t test; n.s., not significant).
To determine whether TRiC folds σ3 into a biologically active conformation, we took advantage of another unique aspect of reovirus biology. Unlike mature virions that undergo endosomal processing (Fig. 7 D, Top), ISVPs access the cytoplasm by directly penetrating the cell membrane (Fig. 7 D, Bottom) (35–37). Infection by virions can be blocked by cysteine protease inhibitors, which neutralize endosomal proteases (38). ISVPs bypass endosomes and are unaffected by protease inhibition. However, ISVPs recoated with purified σ3 behave phenotypically like virions and require endosomal processing to infect cells.
ISVPs were incubated in the presence or absence of purified TRiC/σ3 and ATP and inoculated onto HeLa cells treated with E64 (cysteine protease inhibitor) or dimethyl sulfoxide (DMSO) (Fig. 7E). ISVPs incubated in the absence of TRiC/σ3 infected HeLa cells efficiently in the presence or absence of E64 (Fig. 7 F and G). E64 completely blocked infection by ISVPs previously incubated with TRiC/σ3 and ATP (Fig. 7 F, Bottom Right), similar to the infection pattern observed when the identical experiment was conducted with purified reovirus virions (SI Appendix, Fig. S10 A–C). These data indicate that TRiC folds and assembles σ3 onto ISVPs. As a control, we tested whether incubation of ISVPs with denatured σ3 produced the changes in particle infectivity consistent with native recoating (Fig. 7H). The infection pattern of ISVPs incubated in the presence or absence of denatured σ3 was unchanged with E64 treatment (Fig. 7 I and J), indicating that denatured σ3 cannot recoat ISVPs. Together, these findings indicate that TRiC folds and transfers biologically active σ3 to ISVPs, thereby forming mature reovirus particles.
Discussion
In this study, we discovered a network of host chaperones that interact with σ3, including TRiC, PFD, Hsp70, and Hsp90. PFD maintains σ3 solubility and promotes efficient substrate transfer to TRiC. Once bound to TRiC, σ3 has a large degree of conformational flexibility. After TRiC cycles to its closed state, σ3 adopts a native structure with its oligomerization domain oriented internally. TRiC and PFD cooperate to maximize the efficiency of σ3 assembly into a complex with μ1. This folding pathway produces a biologically active form of σ3 that assembles to yield mature virions. This work illuminates a chaperone network that directs the folding and assembly of a complex heterooligomeric protein.
Cryo-EM of TRiC/σ3 demonstrates an intact chaperone–substrate complex with TRiC in a closed, folding-competent state. This post-ATP-hydrolysis conformation of TRiC contains folded σ3 with identifiable secondary structures. σ3 orients perpendicularly across a single TRiC ring and contacts two hemispheres. Although TRiC contains two rings, TRiC did not complex with more than one σ3 molecule, suggesting negative cooperativity in substrate binding, consistent with a previous study (39). The position of σ3 in the closed TRiC conformation differs from that of other TRiC-bound substrates, such as the G protein β-subunit bound to apo-TRiC, found associated with apical domains (40), or the mTOR subunit mLST8, which appears to bind at the interring equator in the closed TRiC state (41). In the closed conformation, the normally solvent-exposed region of σ3 (pole 1) is proximal to CCT3 and 6 (Fig. 3 C–F) with the μ1-binding domain (pole 2) oriented toward the opposite side of the chamber in proximity to CCT5. In this position, μ1 may compete with CCT5 for σ3 binding. The orientation of the σ3 oligomerization domain toward the TRiC equator is also intriguing. Amino- and carboxyl-terminal tails of TRiC subunits are present in this region and could help stabilize this sticky domain of σ3. Somewhat surprisingly, the clearest density for the σ3 structure lies in the μ1-binding domain (Fig. 2E, pole 2), whereas the domain that is solvent exposed in the final assembled state (pole 1) has less clear density in the three-dimensional (3D) reconstruction. The strong density for the μ1-binding domain indicates that this region is well structured and bound in a stable interface with TRiC, whereas the lack of density for the solvent-exposed region could be due to disorder in the fold or flexibility in the positioning relative to TRiC. Therefore, TRiC appears to stabilize the oligomerization domain of σ3, which may contribute to its essential function in mitigating σ3 aggregation.
The inability to resolve secondary structure elements of σ3 in the cryo-EM analysis of the open TRiC conformation suggests that the form of σ3 bound to TRiC before ATP cycling is largely unstructured and mobile. This conclusion is supported by XL-MS data demonstrating multiple cross-links between individual σ3 amino acids and the tail domains of different TRiC subunits (Fig. 4 C–E). In the closed TRiC chamber, σ3 appears in a near-native state, and the limited number of TRiC/σ3 XL-MS cross-links indicates limited substrate mobility. These data provide only snapshots of the σ3 conformational cycles, and intermediates that occur during σ3 folding are still unclear. Individual TRiC subunits hydrolyze ATP at different rates, and these subunits are arranged in high ATP-affinity and low ATP-affinity hemispheres (15). In its TRiC-bound state, σ3 spans these two hemispheres with pole 1 and pole 2 in proximity to the low- and high-affinity hemispheres, respectively. Repeated binding and release of pole 2 from the high-affinity hemisphere combined with infrequent release of pole 1 from the low-affinity hemisphere may contribute to σ3 folding. Insight into a potential folding intermediate comes from cryo-EM subpopulation 1B (SI Appendix, Fig. S4C), which reveals a structure with an orientation similar to that of folded σ3 without discernible σ3 secondary structure. This subclass also lacks density at pole 2 of σ3, which is observed interacting with CCT5 in the TRiC/σ3 cryo-EM map. This structure could represent a folding intermediate along the pathway to native σ3 or a terminally misfolded form of σ3 bound to TRiC. The lack of density at pole 2 suggests that this region has more folding constraints than the rest of the σ3 molecule.
Once folded, σ3 must assemble into a larger structure with μ1. Many TRiC substrates are subunits of higher-order multiprotein assemblies, including VHL (4), Cdc20 (42), and G protein β (40). Although TRiC functions in folding subunits of oligomers, it is unknown whether TRiC directly coordinates oligomer assembly. In folding assays with σ3 and TRiC, σ3 remains associated with TRiC after many ATP cycles (Fig. 5 D–G). While we cannot exclude σ3 release and rebinding in the absence of μ13, assembly of μ1/σ3 oligomers is dependent on both μ13 and ATP, suggesting the occurrence of direct substrate handoff. Consistent with this model, TEM of assembly reactions containing TRiC/σ3 and μ13 reveal subpopulations of TRiC/σ3 and μ13 in ternary complexes (SI Appendix, Fig. S8). Due to the orientation of the μ1-binding domain of σ3 toward the TRiC equator, transfer to μ1 could occur by release through an alternate 40-Å opening between apical domains of TRiC subunits CCT1 and CCT4 in the open conformation (22). Direct transfer via this mechanism would minimize exposure of the aggregation-prone σ3 oligomerization domain, limiting the formation of nonproductive folding intermediates and maximizing oligomer assembly.
PFD associates with σ3 in vitro and in vivo and enhances σ3 loading onto TRiC. PFD could function as a sink for excess σ3 when the TRiC folding system is overwhelmed due to the abundance of newly translated σ3 in reovirus-infected cells. This idea is supported by the capacity of PFD to maintain σ3 solubility in the absence of TRiC for subsequent transfer upon TRiC availability. PFD also increases the efficiency of μ1/σ3 assembly from the σ3-TRiC complex, although the mechanism governing this effect is unknown. PFD could redirect nonnative conformers of σ3 back to TRiC to allow for additional rounds of TRiC cycling, thus maximizing the likelihood of σ3 achieving its native state. PFD also modifies the electrostatic and hydrophobic environment in the TRiC folding chamber (22). These PFD-induced modifications in the biochemical environment experienced by σ3 during TRiC cycling could reduce the conformational freedom allowable to σ3 and enhance the kinetics of σ3 folding. Studies of actin folding indicate that PFD acts directly on TRiC-actin complexes to maximize productive folding (22). A similar synergy between TRiC and PFD could prevent the premature release of nonnative, aggregation-prone σ3, thereby maximizing assembly efficiency. Since the addition of PFD to assembly reactions after the termination of TRiC folding does not enhance μ1/σ3 assembly, it is less likely that PFD functions as a shuttle to transfer folded σ3 onto μ13 trimers. Together, these data point to a dynamic function of PFD in assisting substrate loading onto TRiC and maximizing the kinetics of folding and assembly.
Our study illuminates details of TRiC-substrate interactions, provides insight into the folding mechanism of TRiC, and establishes a function for TRiC in directing the assembly of a heterooligomeric protein complex. Furthermore, these data provide a clear function for PFD in assisting TRiC in folding aggregation-prone substrates. Finally, biological approaches with viable virus provide in vivo evidence of the in vitro folding and assembly activity of TRiC. Together, these findings contribute to an understanding of chaperone-assisted oligomer folding and assembly, a process fundamental to protein homeostasis and dysfunctional in many age-related diseases.
Materials and Methods
PFD- and TRiC-Binding Assays.
PFD and TRiC were diluted to 4 μM or 0.2 μM, respectively, in folding buffer (30 mM Hepes pH 7.4, 100 mM KCl, 5 mM MgCl2, 10% glycerol) supplemented with dithiothreitol (DTT) to a final concentration of 1 mM. Purified chemically denatured Alexa-647 labeled σ3 or actin was diluted 1:100 into PFD or TRiC to achieve a final σ3/actin concentration of ∼0.4 μM and incubated at 4 °C for 60 min. Protein aggregates were pelleted by centrifugation at 21,000 × g at 4 °C for 10 min. Clarified supernatants were resolved by native- and SDS-PAGE, and fluorescent gels were imaged using a Typhoon scanner.
PFD and TRiC Substrate Transfer Assays.
PFD and TRiC were diluted to 4 μM or 0.2 μM, respectively, in folding buffer supplemented with DTT to a final concentration of 1 mM. Purified chemically denatured Alexa-647 labeled σ3 or actin was diluted 1:100 into PFD or TRiC to achieve a final σ3/actin concentration of ∼0.4 μM and incubated at 4 °C for 30 min. Protein aggregates were pelleted by centrifugation at 21,000 × g at 4 °C for 10 min. Clarified supernatants were transferred to new microfuge tubes, supplemented with the reciprocal chaperone (TRiC to a final concentration of 0.2 μM or PFD to a final concentration of 4 μM), and incubated at 37 °C for 30 min. Reactions were resolved by native- and SDS-PAGE, and fluorescent gels were imaged using a Typhoon scanner.
μ13σ33 Assembly Assays.
Assembly of in vitro translated σ3.
Reovirus σ3 was translated in vitro using the TNT coupled rabbit reticulocyte lysate (RRL) system (Promega, L4610) according to the manufacturer’s instructions (26). A pcDNA3.1+ template encoding the reovirus T1L S4 gene (σ3 protein) was transcribed and translated in RRLs supplemented with RNasin Plus RNase Inhibitor (Promega, N2611) and [35S]-methionine (Perkin-Elmer, NEG709A500UC) for radiolabeling. Where indicated, σ3 was translated in RRLs that were TRiC immuno-depleted (26). Reactions were supplemented with the μ13 trimer to a final concentration of 23 nM where indicated. For assembly kinetics, reactions were chased with cold methionine added to a final concentration of 2 mM. Translation reactions were terminated by fourfold dilution in stop buffer (20 mM Hepes-KOH pH 7.4, 100 mM potassium acetate, 5 mM magnesium acetate, 5 mM ethylenediaminetetraacetic acid, 2 mM methionine, 1 mM DTT, 2 mM puromycin). Reactions were resolved by blue native- or SDS-PAGE and imaged using a Perkin-Elmer Cyclone Phosphor System Scanner.
Assembly of denatured 35S-labeled σ3.
Assembly of denatured 35S-labeled σ3 was conducted in folding buffer supplemented with 1 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP). TRiC and the μ13 trimer were diluted to final concentrations of 0.2 μM and ∼23 nM, respectively. Assembly reactions were supplemented with 5 μL of RRLs where indicated. RRLs were ATP depleted by pretreatment with apyrase (New England Biolabs, M0398) (0.04 U per 20 μL RRLs) at 37 °C for 30 min. Denatured 35S-labeled σ3 purified from Escherichia coli was diluted 1:100 into reactions and then immediately supplemented with ATP to a final concentration of 10 mM. Reactions were incubated at 37 °C for 60 min, resolved by native- or SDS-PAGE, and imaged using a phosphorimager.
PFD/TRiC/σ3/μ13 Assembly Assays.
Purified TRiC/σ3 and μ13 were preincubated together at 4 °C for 30 min at final concentrations of 0.6 μM and 86 nM, respectively, in folding buffer supplemented with DTT to a final concentration of 1 mM. For initial PFD experiments, PFD (or BSA as a control) was added to reactions at a final concentration of 0.6 μM or 2.4 μM. Folding was initiated by the addition of ATP to a final concentration of 10 mM, and samples were incubated at 37 °C for 90 min. For PFD time-of-addition assays, folding reactions were conducted with the following modifications: PFD was added to TRiC/σ3 to achieve a 1:1 PFD:TRiC molar ratio (0.6 μM each) before the addition of ATP or after the termination of folding by the addition of apyrase (10 U/mL). For μ13 time-of-addition assays, folding reactions were conducted with the following modifications: μ13 (86 nM final concentration) was preincubated with TRiC/σ3 at 4 °C for 30 min or added immediately before the initiation of folding with ATP or after the termination of folding by the addition of apyrase (10 U/mL). All assembly reactions were resolved by native- or SDS-PAGE, Coomassie-stained, and immunoblotted with a σ3-specific monoclonal antibody.
TRiC/σ3/ISVP Recoating Assays and Immunoprecipitation.
Assembly reactions were conducted in folding buffer containing 1 mM TCEP with 109 ISVPs per reaction. TRiC/σ3 was added to a final concentration of 2.3 μM to achieve an estimated 50:1 ratio of TRiC-bound σ3 molecules to σ3-binding sites in ISVPs (assuming 50% occupancy of TRiC with σ3). ATP was added to indicated reactions at a final concentration of 10 mM, and reactions were incubated at 37 °C for 60 min. Assembly reactions using denatured σ3 were conducted identically, except chemically denatured σ3 was diluted 1:100 into folding buffer containing 109 ISVPs to achieve a final σ3 concentration of 0.84 μM. Recoated ISVPs were further processed by immunoprecipitation or tested for infectivity.
To immunoprecipitate ISVPs, 5 μg of a mouse μ1-specific monoclonal antibody (43) was added to recoating reactions and incubated at 4 °C for 60 min with rotation. Samples were added to Protein G Dynabeads (Thermo Fisher Scientific, 10004D) and incubated at 4 °C for 60 min. Beads were washed five times with 400 μL of folding buffer, resuspended in sample buffer, resolved by SDS-PAGE, and transferred to nitrocellulose. Immunoblotting was conducted using a reovirus-specific polyclonal antiserum and membranes were imaged with the Odyssey CLx Imaging System (LI-COR).
Detailed information about cell and virus growth conditions, ISVP infectivity and E64 bypass assays, recombinant DNA, σ3-BioID, protein purification, cross-linking mass spectrometry, confocal microscopy, cryo-EM/TEM, and statistical analyses can be found in SI Appendix.
For a summary of materials and resources used, see SI Appendix, Table S4.
Supplementary Material
Acknowledgments
This work was supported in part by Public Health Service Awards AI032539 (to T.S.D.), AI122563 (to J.J.K.), GM007347 (to J.J.K.), TR000445 (to J.J.K. via the Vanderbilt Institute for Clinical and Translational Research), GM074074 (to J.F.), GM103124 (to D.G.), and P41GM103832, P01NS092525, and S10OD021600 (to W.C.); European Research Council AdvG 233226 and AdvG 670821 (to R.A.); and Robert Welch Foundation Grant Q1279 (to B.V.V.P). σ3-BioID sample processing and analysis was conducted at the University of Pittsburgh Biomedical Mass Spectrometry Center with the help of Pamela Cantrell and Xuemei Zeng. We thank Stephen Harrison and Tobias Herrmann for sharing reagents and methods to prepare the μ13σ33 heterohexamer and the μ13 trimer.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018127118/-/DCSupplemental.
Data Availability
Model coordinates and density maps from the cryo-EM study have been deposited to the Protein Data Bank and the Electron Microscopy Data Bank: closed TRiC particles before classification [4.5 Å map] (PDB ID 7LUM, EMD-23522) and Class 1A TRiC/σ3 [6.2 Å map] (PDB ID 7LUP, EMD-23526). Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the Proteomics Identifications Database partner repository (identifier: PXD019751). All other data related to this paper are within the main text or the SI Appendix.
References
- 1.Yam A. Y., et al., Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat. Struct. Mol. Biol. 15, 1255–1262 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao Y., Thomas J. O., Chow R. L., Lee G. H., Cowan N. J., A cytoplasmic chaperonin that catalyzes beta-actin folding. Cell 69, 1043–1050 (1992). [DOI] [PubMed] [Google Scholar]
- 3.Gao Y., Vainberg I. E., Chow R. L., Cowan N. J., Two cofactors and cytoplasmic chaperonin are required for the folding of alpha- and beta-tubulin. Mol. Cell. Biol. 13, 2478–2485 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feldman D. E., Thulasiraman V., Ferreyra R. G., Frydman J., Formation of the VHL-elongin BC tumor suppressor complex is mediated by the chaperonin TRiC. Mol. Cell 4, 1051–1061 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Trinidad A. G., et al., Interaction of p53 with the CCT complex promotes protein folding and wild-type p53 activity. Mol. Cell 50, 805–817 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tam S., et al., The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat. Struct. Mol. Biol. 16, 1279–1285 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behrends C., et al., Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol. Cell 23, 887–897 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Leitner A., et al., The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure 20, 814–825 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zang Y., et al., Development of a yeast internal-subunit eGFP labeling strategy and its application in subunit identification in eukaryotic group II chaperonin TRiC/CCT. Sci. Rep. 8, 2374 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thulasiraman V., Yang C. F., Frydman J., In vivo newly translated polypeptides are sequestered in a protected folding environment. EMBO J. 18, 85–95 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gestaut D., Limatola A., Joachimiak L., Frydman J., The ATP-powered gymnastics of TRiC/CCT: An asymmetric protein folding machine with a symmetric origin story. Curr. Opin. Struct. Biol. 55, 50–58 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zang Y., et al., Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM. Nat. Struct. Mol. Biol. 23, 1083–1091 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Douglas N. R., et al., Dual action of ATP hydrolysis couples lid closure to substrate release into the group II chaperonin chamber. Cell 144, 240–252 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joachimiak L. A., Walzthoeni T., Liu C. W., Aebersold R., Frydman J., The structural basis of substrate recognition by the eukaryotic chaperonin TRiC/CCT. Cell 159, 1042–1055 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reissmann S., et al., A gradient of ATP affinities generates an asymmetric power stroke driving the chaperonin TRIC/CCT folding cycle. Cell Rep. 2, 866–877 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amit M., et al., Equivalent mutations in the eight subunits of the chaperonin CCT produce dramatically different cellular and gene expression phenotypes. J. Mol. Biol. 401, 532–543 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Melville M. W., McClellan A. J., Meyer A. S., Darveau A., Frydman J., The Hsp70 and TRiC/CCT chaperone systems cooperate in vivo to assemble the von Hippel-Lindau tumor suppressor complex. Mol. Cell. Biol. 23, 3141–3151 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vainberg I. E., et al., Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell 93, 863–873 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Balchin D., Hayer-Hartl M., Hartl F. U., In vivo aspects of protein folding and quality control. Science 353, aac4354 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Siegert R., Leroux M. R., Scheufler C., Hartl F. U., Moarefi I., Structure of the molecular chaperone prefoldin: Unique interaction of multiple coiled coil tentacles with unfolded proteins. Cell 103, 621–632 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Martín-Benito J., et al., Structure of eukaryotic prefoldin and of its complexes with unfolded actin and the cytosolic chaperonin CCT. EMBO J. 21, 6377–6386 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gestaut D., et al., The chaperonin TRiC/CCT associates with prefoldin through a conserved electrostatic interface essential for cellular proteostasis. Cell 177, 751–765.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kashuba E., Pokrovskaja K., Klein G., Szekely L., Epstein-Barr virus-encoded nuclear protein EBNA-3 interacts with the epsilon-subunit of the T-complex protein 1 chaperonin complex. J. Hum. Virol. 2, 33–37 (1999). [PubMed] [Google Scholar]
- 24.Hong S., et al., Type D retrovirus Gag polyprotein interacts with the cytosolic chaperonin TRiC. J. Virol. 75, 2526–2534 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue Y., et al., Chaperonin TRiC/CCT participates in replication of hepatitis C virus genome via interaction with the viral NS5B protein. Virology 410, 38–47 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Knowlton J. J., et al., The TRiC chaperonin controls reovirus replication through outer-capsid folding. Nat. Microbiol. 3, 481–493 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouziat R., et al., Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science 356, 44–50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huismans H., Joklik W. K., Reovirus-coded polypeptides in infected cells: Isolation of two native monomeric polypeptides with affinity for single-stranded and double-stranded RNA, respectively. Virology 70, 411–424 (1976). [DOI] [PubMed] [Google Scholar]
- 29.Kim D. I., et al., Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl. Acad. Sci. U.S.A. 111, E2453–E2461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roux K. J., Kim D. I., Burke B., May D. G., BioID: A screen for protein-protein interactions. Curr. Protoc. Protein Sci. 91, 19.23.11–19.23.15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer A. S., et al., Closing the folding chamber of the eukaryotic chaperonin requires the transition state of ATP hydrolysis. Cell 113, 369–381 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Cong Y., et al., 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc. Natl. Acad. Sci. U.S.A. 107, 4967–4972 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jané-Valbuena J., et al., Reovirus virion-like particles obtained by recoating infectious subvirion particles with baculovirus-expressed σ3 protein: An approach for analyzing σ3 functions during virus entry. J. Virol. 73, 2963–2973 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandran K., et al., In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins μ1 and σ3. J. Virol. 73, 3941–3950 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chandran K., Farsetta D. L., Nibert M. L., Strategy for nonenveloped virus entry: A hydrophobic conformer of the reovirus membrane penetration protein micro 1 mediates membrane disruption. J. Virol. 76, 9920–9933 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nibert M. L., Odegard A. L., Agosto M. A., Chandran K., Schiff L. A., Putative autocleavage of reovirus μ1 protein in concert with outer-capsid disassembly and activation for membrane permeabilization. J. Mol. Biol. 345, 461–474 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Tosteson M. T., Nibert M. L., Fields B. N., Ion channels induced in lipid bilayers by subvirion particles of the nonenveloped mammalian reoviruses. Proc. Natl. Acad. Sci. U.S.A. 90, 10549–10552 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baer G. S., Dermody T. S., Mutations in reovirus outer-capsid protein σ3 selected during persistent infections of L cells confer resistance to protease inhibitor E64. J. Virol. 71, 4921–4928 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobrzynski J. K., Sternlicht M. L., Peng I., Farr G. W., Sternlicht H., Evidence that beta-tubulin induces a conformation change in the cytosolic chaperonin which stabilizes binding: Implications for the mechanism of action. Biochemistry 39, 3988–4003 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Plimpton R. L., et al., Structures of the Gβ-CCT and PhLP1-Gβ-CCT complexes reveal a mechanism for G-protein β-subunit folding and Gβγ dimer assembly. Proc. Natl. Acad. Sci. U.S.A. 112, 2413–2418 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuéllar J., et al., Structural and functional analysis of the role of the chaperonin CCT in mTOR complex assembly. Nat. Commun. 10, 2865 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camasses A., Bogdanova A., Shevchenko A., Zachariae W., The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol. Cell 12, 87–100 (2003). [DOI] [PubMed] [Google Scholar]
- 43.H. W. Virgin, 4th, Mann M. A., Fields B. N., Tyler K. L., Monoclonal antibodies to reovirus reveal structure/function relationships between capsid proteins and genetics of susceptibility to antibody action. J. Virol. 65, 6772–6781 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nason E. L., et al., A monoclonal antibody specific for reovirus outer-capsid protein σ3 inhibits σ1-mediated hemagglutination by steric hindrance. J. Virol. 75, 6625–6634 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Model coordinates and density maps from the cryo-EM study have been deposited to the Protein Data Bank and the Electron Microscopy Data Bank: closed TRiC particles before classification [4.5 Å map] (PDB ID 7LUM, EMD-23522) and Class 1A TRiC/σ3 [6.2 Å map] (PDB ID 7LUP, EMD-23526). Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the Proteomics Identifications Database partner repository (identifier: PXD019751). All other data related to this paper are within the main text or the SI Appendix.