Significance
Lipid-enveloped viruses such as flaviviruses and coronaviruses underlie numerous human illnesses. Strategies which target this shared membrane feature may be useful as broad-spectrum antiviral agents. The mosquito protein AEG12 disrupts lipid membranes via delivery of a fatty acid in exchange for diacylphospholipids. This mechanism allows the mosquito to digest red blood cells after a meal and disrupts lipid coat viruses as part of the mosquito immune response to infection. It may be possible to engineer this strategy for human therapeutics.
Keywords: antiviral, membrane disruption, cytolysis, insect digestion, insect immune response
Abstract
The mosquito protein AEG12 is up-regulated in response to blood meals and flavivirus infection though its function remained elusive. Here, we determine the three-dimensional structure of AEG12 and describe the binding specificity of acyl-chain ligands within its large central hydrophobic cavity. We show that AEG12 displays hemolytic and cytolytic activity by selectively delivering unsaturated fatty acid cargoes into phosphatidylcholine-rich lipid bilayers. This property of AEG12 also enables it to inhibit replication of enveloped viruses such as Dengue and Zika viruses at low micromolar concentrations. Weaker inhibition was observed against more distantly related coronaviruses and lentivirus, while no inhibition was observed against the nonenveloped virus adeno-associated virus. Together, our results uncover the mechanistic understanding of AEG12 function and provide the necessary implications for its use as a broad-spectrum therapeutic against cellular and viral targets.
Mosquito-borne flaviviruses such as Dengue and Zika represent a major public health risk; 96 million new cases of Dengue infection are reported every year (1), while the Zika infection can cause neurological sequleae and microencephaly (2). Vaccines have had limited success in preventing human flavivirus infections, demonstrating the need for new therapeutic approaches (2). Like humans, mosquitos mount a robust anti-viral immune response against flaviviruses. For example, the mosquito protein AEG12 and its homologs have been shown to be up-regulated in Aedes aegypti in response to Zika virus, West Nile virus, dengue, and Yellow Fever virus infection via JAK-STAT pathways (3–7). AEG12 homologs are also up-regulated upon infection by the parasite Brugia malayi (8), suggesting a role against a wide range of viral- and cell-based targets. Previous sequence analysis of AEG12 revealed it to be a member of the major allergy (MA) protein family, so named because the prototypical protein for this family (Bla g 1) is a major human allergen (9). While many insects contain MA proteins, the family is significantly expanded in Aedes (∼20) and Culex (∼8) mosquitoes; both are major vectors of arboviruses (9, 10). Elucidating the molecular basis for this antiviral activity may provide valuable insight into the development of novel flavivirus and broad-spectrum therapeutic strategies.
Curiously, AEG12 was initially described not in the context of flavivirus infection but digestion. Here, AEG12 proteins were found to be highly up-regulated in the midgut of female A. aegypti and subsequently localized to the microvilli following a blood feed but not a sugar feed (9, 11, 12). A similar expression pattern was observed in the AcG12 homolog from Anopheles cilicifacies, suggesting a conserved function across mosquito species (13). In cockroaches, suppression of Bla g 1 expression significantly hindered digestion, resulting in starvation despite adequate food intake suggesting a broader role for MA domain proteins in breaking down and absorbing nutrients from both blood and nonblood food materials (14, 15). However, the function and mechanistic roles of AEG12 proteins remains unknown.
The structure of Bla g 1 consists of two consecutive units, each containing five helices (α1 to α5) arranged in a pentagon with the sixth helix (α6) stacking on top. The two pentagonal structures come together to enclose an exceptionally large hydrophobic cavity of ∼3,800 Å3 that can accommodate up to eight fatty acid ligands (16, 17). Bla g 1 isolated from its natural allergen source was found to contain a mixture of palmitate, oleate, and stearate fatty acids (nMix), the presence of which significantly enhanced Bla g 1 thermostability (16, 17). We hypothesize that these lipid cargoes play a key role in the biological function of both Bla g 1 and AEG12 in digestion and antiviral activity. Indeed, previous studies showed that free fatty acids have viricidal effects against enveloped viruses because of their ability to permeabilize and disrupt their lipid membranes (18–21). Additionally, both free fatty acids and fatty acid–protein conjugates have been shown to destabilize and lyse mammalian cells membranes (22, 23). The ability of AEG12 to mobilize and deliver a range of fatty acid cargoes could provide a common mechanism through which it fulfills both putative functions.
Here, we demonstrate that AEG12 adopts a similar structure as Bla g 1, enabling it to bind a range of fatty acid ligands within its central hydrophobic cavity. Selective delivery of these fatty acid cargoes into phosphatidylcholine (PC) lipid bilayers results in membrane destabilization and cytotoxicity against eukaryotic cells consistent with a role in erythrocyte digestion. Similarly, AEG12 displayed antiviral activity against a range of enveloped, but not nonenveloped viruses suggesting that both putative functions of AEG12 share a common mechanism of action centered on lipid exchange resulting in membrane destabilization.
Results
Structure of AEG12 Suggests Potential Role in Membrane Remodeling.
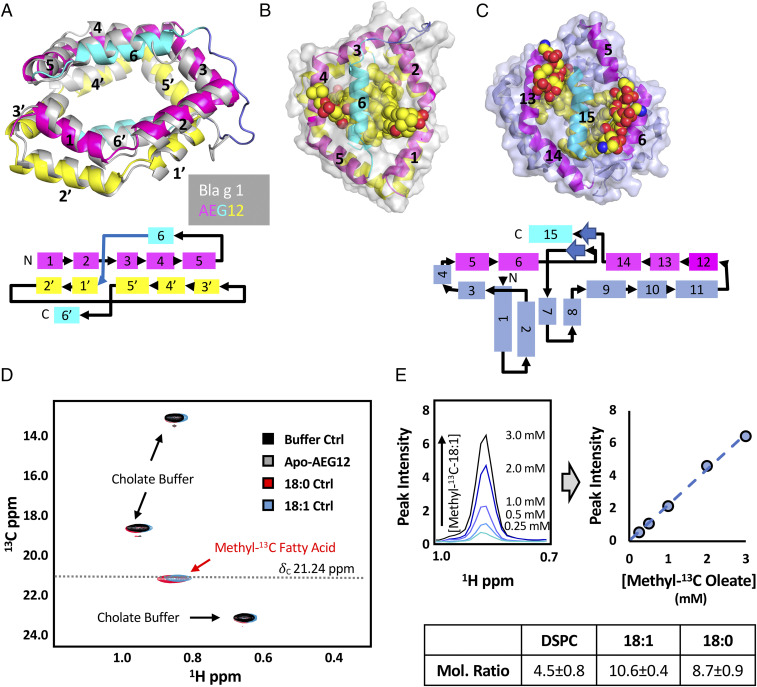
Based on sequence analysis, it is predicted that AEG12 will adopt an MA-like fold (9). To gain insight into AEG12 structure, function, and mechanism, we determined its structure to 1.95 Å resolution (Fig. 1A). AEG12 aligns closely with Bla g 1 (1.5 Å rmsd over 147 C-α atoms) and encloses a similarly sized hydrophobic cavity (17). Unlike Bla g 1, no additional electron density was observed corresponding to ligands bound within this cavity. Molecular modeling suggests that four distearoylphosphatidylcholine (DSPC) phospholipids, representing a total of eight saturated acyl chains can readily pack into the central cavity without significant structural rearrangements relative to the solved X-ray crystal structure (Fig. 1B). Removing the acyl chains resulted in the collapse of helix 6, causing it to lie in plane with the other helices of AEG12 (SI Appendix, Fig. S1). This theoretical stoichiometry could be exceeded with unsaturated acyl chain ligands, with AEG12 able to accommodate up to 12 C18:1 oleate molecules with no increase in protein backbone rmsd (SI Appendix, Fig. S1) over the course of an extended (500 ns) simulation time. To validate this conjecture, samples of AEG12 were loaded with DSPC, methyl-13C–labeled oleate, or methyl-13C–labeled stearate, and the bound ligands quantified using 1D 31P or 2D 1H-13C-NMR (Fig. 1D). The resulting mole ratios of 4.5 ± 0.8, 10.6 ± 0.4, and 8.7 ± 0.9 for DSPC, oleate, and stearate, respectively, compared favorably with both the simulations described above and the experimentally determined values for Bla g 1 (17). Previous studies indicate that Materazzi, a Drosophila homolog of Bla g 1 and AEG12, functions as a lipid-binding protein in vivo, suggesting that this large binding capacity may represent a common feature across the MA domain protein family and may contribute to their biological functionality (24).
Fig. 1.
Structure of AEG12 suggests lipid-binding functionality. (A) Crystal structure of AEG12 overlaid on that of Bla g 1 (gray) (Protien Data Bank [PDB] ID: 4JRB). AEG12 color scheme corresponds to the topographical diagram shown below. (B) Surface view of AEG12 modeled with four phospholipids with spherical rendering. (C) X-ray structure of poxvirus VMAP with phospholipids (PDB ID: 6BR9). The pentagonal ring and vertically displaced capping helix colored in pink and cyan, respectively as in A and B. Helices which have no parallel in AEG12 are shown in slate. (D) Representative 1H-13C-HMQC spectra of 5% cholate NMR buffer alone (black) and in the presence of methyl-13C-stearate (red), methyl-13C-oleate (blue), or Apo-AEG12 (gray). Peaks corresponding to the cholate buffer and the methyl-13C–labeled fatty acids are indicated. A representative standard curve generated using the 13C-methyl-fatty acid peak (δC 21.24 ppm) is shown in E. Stearate:AEG12 and Oleate:AEG12 mol ratios calculated using this method are shown in the table (E, Bottom). DSPC binding stoichiometries were obtained using 31P-NMR as described elsewhere (17). Uncertainty (±) values reported here and in all subsequent tables represent the SD obtained from at least three independent trials from at least two biological replicates unless otherwise specified.
Curiously, it was noted that the C-terminal domain of the poxvirus viral membrane associated protein (VMAP) A6 has a similar architecture as Bla g 1/AEG12, consisting of similar sized planar arrangements of amphipathic helices capped by a similar C-terminal helix (Fig. 1C) (25). Crystal structures of A6 revealed electron density corresponding to phospholipids bound into the hydrophobic cavity with head groups protruding on either side of the C-terminal helix consistent with the proposed mode of binding in AEG12/Bla g 1. In a biological context, VMAPs have been shown to facilitate viral budding through membrane remodeling (25, 26). Based on these findings, we hypothesize that AEG12 could potentially play a similar function in destabilizing lipid bilayers, a process which may be facilitated by its ability to bind and deliver fatty acid cargoes within its hydrophobic cavity.
AEG12 Displays Hemolytic Activity.
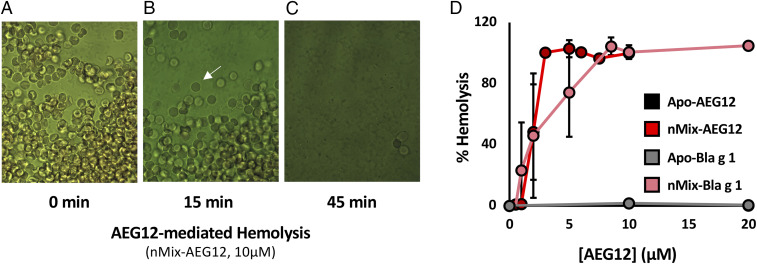
Lysis of erythrocytes represents the initial stage of digestion following a blood meal; a process which could be facilitated by the lipid binding and membrane destabilizing potential of AEG12. To explore this hypothesis, human erythrocytes were treated with AEG12. No hemolytic activity was observed for Apo-AEG12, suggesting that fatty acid ligands may be required for its biological function. Since the natural ligands of AEG12 have not been characterized, nMix—a mixture of palmitate, oleate, and stearate fatty acids mirroring the natural ligands of Bla g 1—was employed as a surrogate. nMix-loaded AEG12 (nMix-AEG12) produced a visible change in erythrocyte morphology, transitioning from the typical biconcave shape to a slightly expanded sphere. This was accompanied by the release of hemoglobin, which was manifested by the increased opacity of the background media in Fig. 2 A–C indicative of hemolytic activity. Plotting hemolysis as a function of AEG12 concentration (Fig. 2D) yields a 50% cytotoxicity (CC50) value of ∼2 μM for nMix-AEG12. Neither Apo-AEG12 nor AEG12 loaded entirely with saturated fatty acids (C12, C14, C16, C20) showed any detectable activity (<3% lysis) at the conditions tested, suggesting that lysis is dependent on the presence of specific fatty acid cargoes. Apo- and nMix-Bla g 1 yielded similar CC50 values as its Aedes counterpart suggesting that ligand-dependent lytic activity may be a common property shared by other insect MA proteins and may underlie their biological function.
Fig. 2.
AEG12 displays ligand-dependent hemolytic activity. Light microscopy images taken at 20× of human erythrocytes (50% hematocrit) incubated with 10 μM nMix-AEG12 for 0 (A), 15 (B), and 45 (C) min showing the loss of cellular structure (white arrow) and the subsequent release of hemoglobin, as represented by the loss of contrast between the cell interior and the extracellular environment. The amount of released hemoglobin could be measured spectroscopically to provide a quantitative measure of hemolysis. (D) Percent hemolysis of erythrocytes after a 60-min incubation with nMix-AEG12 or nMix-Bla g 1. Note that percent hemolysis is normalized with the negative control (PBS) and positive control (2% Triton 100) representing 0 and 100%, respectively. Note that the data points for Apo-AEG12 and Apo-Bla g 1 overlap, as neither displayed any detectable hemolysis. Error bars reported here and in all subsequent figures represent the SD obtained from at least three independent trials from at least two biological replicates unless otherwise specified.
AEG12 Functions as a Broad-Spectrum Lytic Agent.
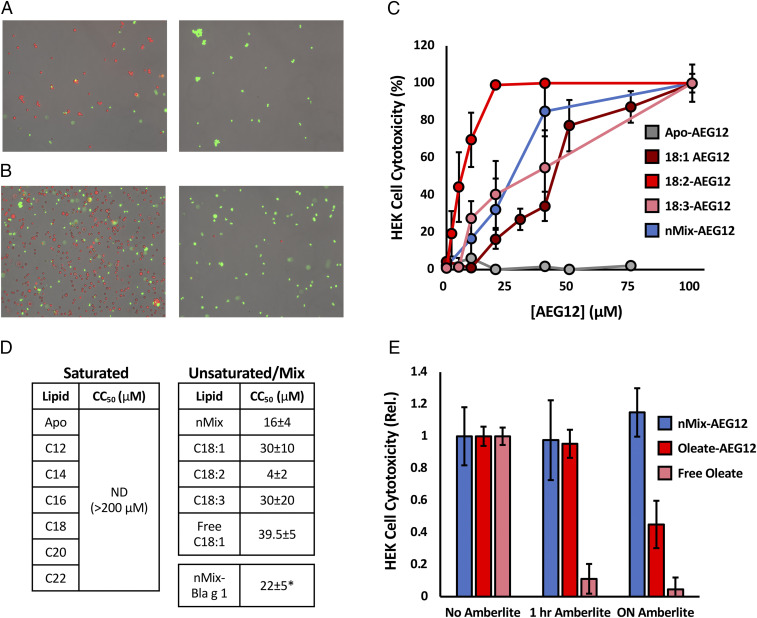
The structure and lipid binding properties of AEG12 suggest that hemolytic activity is mediated through interactions with the plasma membrane, rather than a specific molecular target or receptor. Based on this mechanism, one would expect robust cytotoxic activity across a wide range of cell types and species. Using the AO/PI (acridine-orange/propidium iodine) stain system it was found that both nMix-Bla g 1 and nMix-AEG12 could lyse human embryonic kidney (HEK) cells (Fig. 3 A and B) with CC50 values in the μM range (Fig. 3 C and D), supporting this conjecture. Loading AEG12 entirely with unsaturated or polyunsaturated fatty acids yielded similar levels of cytotoxicity. As with hemolysis, neither Apo-AEG12 nor AEG12 loaded with entirely saturated fatty acid ligands showed any significant lytic activity, supporting a shared mode of action between these two biological functions. Similar cytotoxic activity was observed against other eukaryotic cells in suspension including monkey (Vero), insect (SF9), and even yeast (Saccharomyces cerevisiae) despite the presence of a cell wall around the latter, consistent with its proposed function as a broad-spectrum lytic agent targeting biological membranes (SI Appendix, Fig. S3). Curiously no lytic activity was observed against Escherichia coli, suggesting that AEG12 may be able to differentiate between prokaryotic and eukaryotic cell membranes and direct its cytotoxic activity accordingly.
Fig. 3.
AEG12 cytotoxic activity in mammalian cells is restricted to unsaturated fatty acid ligands. (A) Microscopy images obtained at 10× for HEK cells treated with PBS (Left) or 2% Triton 100 (Right) representing negative and positive controls, respectively. Viable (live) and dead cells are identified using the AO/PI stain system and are colored red and green, respectively. (B) Cells treated with 40 μM Apo-AEG12 (Left) or nMix-AEG12 (Right) and imaged as in A. HEK cell cytotoxicity, defined as the proportion of dead cells to total cells, following treatment with varying concentrations of AEG12 shown in C. 50% cytotoxicity (CC50) values for AEG12 loaded with various saturated and unsaturated cargoes are shown in D. ND denotes fatty acid cargoes for which no detectable lysis was observed at any of the concentrations tested. (E) Decrease in cytotoxicity of nMix-AEG12 (blue), Oleate-AEG12 (red), or free oleate alone (pink) following 1 h and overnight incubation with Amberlite resin relative to an untreated (no Amberlite) control. * denotes values obtained from two independent trials.
AEG12 Demonstrates Broad-Spectrum Inhibition of Enveloped Viruses.
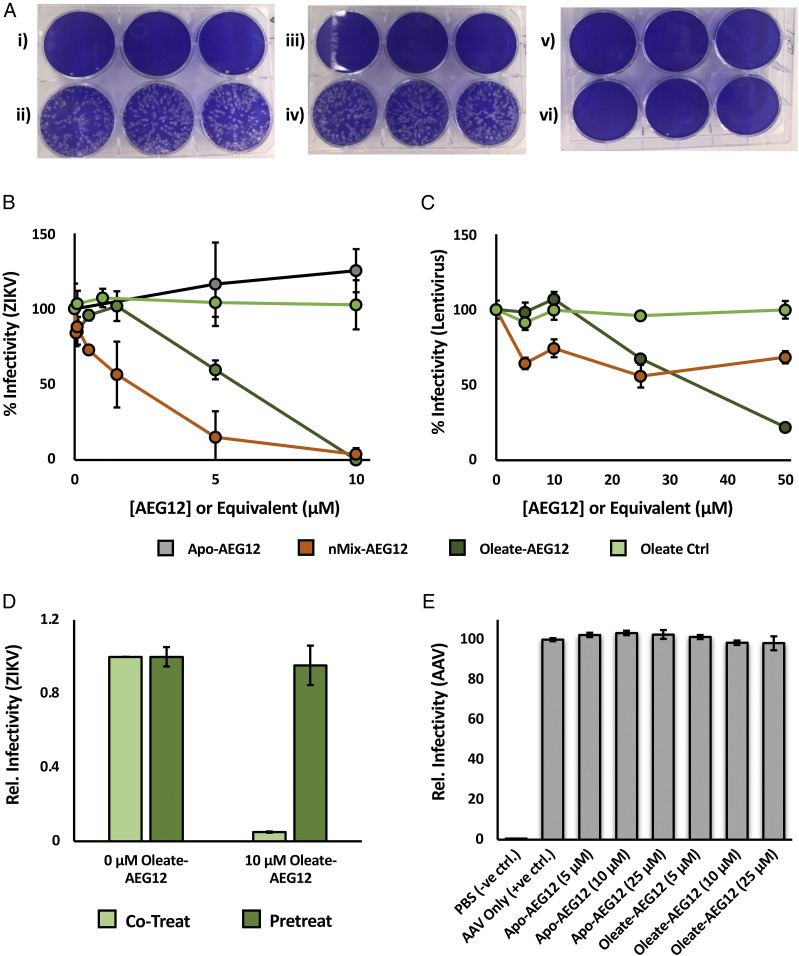
Previous data have suggested that AEG12 is up-regulated in response to flavivirus infection (4–6). The genetic material of flaviviruses is encapsulated within a lipid bilayer derived from host cell membranes, potentially making them vulnerable to the lytic activity of AEG12 and giving rise to the latter’s proposed antiviral activity. To test this hypothesis, samples of Zika virus (ZIKV Paraiba 2015) were exposed to sublytic concentrations of Apo- or nMix-AEG12 for 30 min at 37 °C and then applied to monkey Vero cells. We noted that AEG12 itself did not have any effect on Vero cell viability, potentially due to their adherent nature, or the presence of Ca2+ in the cell media as discussed in the following section. However, nMix-AEG12 significantly reduced infectivity against Vero cells while the Apo-AEG12 had no noticeable effect (Fig. 4 and SI Appendix, Fig. S4). While Oleate-AEG12 yielded similar antiviral activity as its nMix-loaded counterpart, free Oleate alone produced no effect suggesting that AEG12 may play a more active role in the lipid delivery process against viral targets (Fig. 4). To determine whether AEG12 was acting upon the viral particles or the Vero cell membranes, Vero cells were pretreated with Oleate-AEG12, washed, and subsequently infected with ZIKV (Fig. 4). Pretreatment produced no noticeable effect on infectivity, indicating that the antiviral activity of AEG12 is due to its action on the viral particle rather than perturbations to the host cell membrane. Similar levels of antiviral activity were observed against another ZIKV strain (ZIKV H/PF 2013) and the closely related flavivirus dengue (DENV), with IC50 values <10 μM in all cases (SI Appendix, Figs. S4 and S5). More distantly related enveloped viruses such as human α-coronavirus (HCoV 229e) and lentivirus were also inhibited, albeit with ∼10-fold reduced efficacy (Fig. 4 and SI Appendix, Fig. S5). Neither Apo- nor Oleate-AEG12 was able to inhibit infection by the nonenveloped adeno-associated virus (AAV) at concentrations up to 25 μM (Fig. 4). These results suggest that the cargo-dependent antiviral activity of AEG12 is directed against the lipid bilayer of enveloped viruses, mirroring the destabilizing effect observed against eukaryotic cells.
Fig. 4.
AEG12 inhibits Zika virus, lentivirus infection. (A) Representative plaque assay plate showing the formation of Zika virus (ZIKV Paraiba 2015)-induced plaques on confluent Vero cells (stained blue). Rows i) and ii) depict plates treated with no virus, or untreated virus, representing the negative and positive controls, respectively. Treating the virus with 10 μM Apo-AEG12 for 30 min did not significantly reduce infectivity (iv), while treatment with 10 μM nMix-AEG12 completely inactivated the virus (vi). Treatment of the cells with identical concentrations of Apo-AEG12 or nMix-AEG12 alone did not significantly hinder proliferation in the absence of virus, as shown in iii) and v), respectively. A plot of the relative number of viral plaques relative to AEG12 concentration yields an inhibition curve (B) from which approximate IC50 values can be obtained. Note that the concentration of free oleate employed in the oleate control is eight times that of AEG12 in order to account for the observed 8:1 binding stoichiometry. The inhibition curve of AEG12 against the enveloped lentivirus is shown in C. Preincubating target cells with Oleate-AEG12 prior to infection by ZIKV (pretreat) yields no protective effect (D). Infectivity of ZIKV incubated with the equivalent concentration of Oleate-AEG12 as described in B (cotreat) shown for comparison. (E) Infectivity of AAV is undiminished after treatment with Apo- and Oleate-AEG12.
AEG12 as a Delivery Vehicle for Unsaturated Fatty Acids.
Incorporation of free unsaturated and polyunsaturated fatty acids has been shown to destabilize phospholipid membranes, contributing to the destabilization of both eukaryotic cells and enveloped virions (19–23), while their saturated counterparts have a much more muted effect (27, 28). This mirrors the ligand selectivity of AEG12 hemolysis and cytotoxicity, suggesting a mode of action involving the delivery and incorporation of fatty acid ligands into the plasma membranes of targeted cells. However, the cytolytic activity of free fatty acids also raises questions concerning the necessity of AEG12 within its biological context. Indeed, a positive control in which HEK cells were incubated with free oleate alone yielded a CC50 of 39.5 μM—equivalent to 4.0 μM Oleate-AEG12 after accounting for the 10:1 binding stoichiometry of the latter, with similar results reported for SF9 cells (SI Appendix, Fig. S3). It should be noted that free fatty acids are difficult to solubilize and transport within a biological system (29, 30). This is best illustrated by the inability to obtain aqueous solutions of the nMix ligands at the concentrations required to carry out our cytotoxicity assays in the absence of AEG12. In this regard, the low solubility of AEG12’s natural ligands further highlights the potential utility of AEG12 in a biological context. A more practical consequence of the phenomenon is the necessity to employ free oleate alone as a positive control both within the assay presented here and throughout the remainder of this manuscript, with Oleate-AEG12 representing the ligand-bound equivalent because of its similar cytotoxicity profile and functional properties as nMix-AEG12 under most conditions.
Within a biological setting, the low solubility of AEG12’s natural ligands ensures that they are quickly removed from the aqueous environment either through partitioning into biological membranes or binding to various scavenger proteins, further hindering their utility. Sequestering these ligands within AEG12 thus provides a convenient way of maintaining a large pool of mobile fatty acids in the aqueous phase while simultaneously protecting it from scavenger proteins. To illustrate this principle, samples of nMix-AEG12, Oleate-AEG12, and free oleate were incubated with Amberlite—an ion exchange resin which binds and sequesters detergents and fatty acids. Amberlite treatment did not significantly alter the cytotoxicity or thermostability of nMix-AEG12 relative to an untreated (no Amberlite) control (Fig. 3E). Oleate-AEG12 showed a modest decrease in cytotoxicity only after prolonged (overnight) incubation, potentially reflecting a greater propensity for Oleate to dissociate from the AEG12 complex because of its higher solubility relative to the nMix ligands as discussed above. In contrast, a 1-h incubation was sufficient to completely eliminate the cytotoxic activity of free Oleate (Fig. 3E). These results indicate that cytolytic activity is mediated solely through ligands bound within the AEG12 hydrophobic cavity, rather than residual free fatty acids which are carried through the purification process. These ligands are securely retained with limited exchange between the bound and free forms, protecting them from competing fatty acid binding proteins or partitioning into nontargeted lipid structures. Thus, while free fatty acids are capable of lysing cells in the absence of AEG12, the latter likely acts as a delivery vehicle, mobilizing insoluble and difficult to transport fatty acids and controlling their delivery to target cell membranes for lysis within a biological setting.
AEG12-Mediated Delivery of Unsaturated Fatty Acids Destabilizes Biological Membranes.
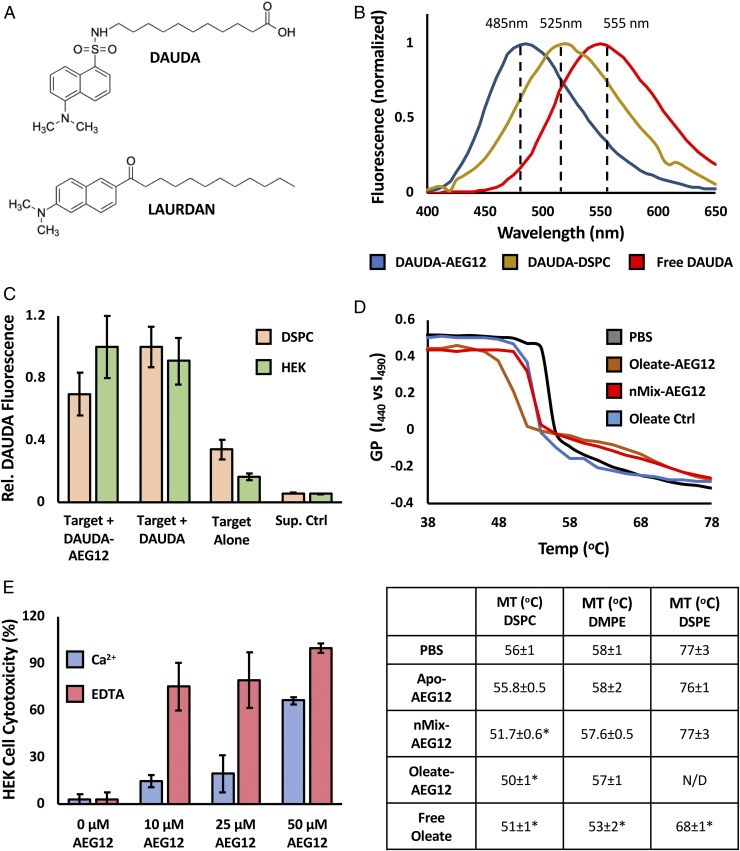
The ability of AEG12 to deliver and incorporate its fatty acid cargoes into biological membranes was further assessed using the fluorescent fatty acid analog 11-(Dansylamino)undecanoic acid (DAUDA), the structure of which is shown in Fig. 5A. Because of its lipophilic properties, DAUDA can be readily loaded into the hydrophobic cavity of AEG12 or incorporated into model membrane systems: in this case DSPC vesicles. The latter was chosen as a model membrane system due to its ability to replicate the 16 to 18 carbon average acyl chain lengths found in mammalian membrane phospholipids, while having a phase transition temperature that was above ambient room temperature. The fluorescence spectra of the DAUDA-AEG12 complex shows a characteristic blue shift (∼495 nm) relative to both equivalent DAUDA-DSPC complex (∼520 nm) and free DAUDA (∼550 nm), providing a convenient mechanism to differentiate the three species (Fig. 5B). Incubating DAUDA-AEG12 with unlabeled DSPC vesicles resulted in the formation of the DAUDA-DSPC complex, as evidence by an increase in fluorescence at 520 nm that persists even after several wash steps to remove any residual DAUDA-AEG12 complex (Fig. 5C). A similar transfer was observed in HEK cells treated with the DAUDA-AEG12 (Fig. 5C). This indicates that that fatty acids and other hydrophobic cargoes can be efficiently transferred from the AEG12 hydrophobic cavity and incorporated into biological membranes.
Fig. 5.
Cargo delivery by AEG12 selectively disrupts PC bilayers. (A) Chemical structure of DAUDA and LAURDAN fluorophores. (B) Normalized fluorescence intensity of DAUDA complexed with AEG12 (blue), DSPC (yellow), or in aqueous solution (Red) showing the effect of local environment on fluorescence maximum. (C) Transfer of DAUDA into DSPC vesicles (orange) or HEK Cells (green) bilayer targets following incubation with DAUDA-AEG12 or DAUDA alone followed by a three washes with PBS. Equivalent values obtained for a PBS-treated negative control (target alone) and supernatant (Sup. Ctrl) from the DAUDA-AEG12 sample shown for reference. (D) Representative melting curves showing the temperature dependence of LAURDAN GP incorporated into DSPC bilayer vesicles following incubation with 28 μM Apo-nMix, or Oleate-AEG12, or the equivalent concentration (224 μM) of free oleate. GP values were calculated from the LAURDAN emission intensity at 440 and 490 nm as discussed in the methods section. The temperature at the inflection point represents the phase transition temperature, values of which are summarized below for DSPC and DSPE vesicles treated with 28 μM or 40 μM AEG12, respectively. Conditions which differ significantly from the PBS control as determined via Student’s t test assuming equal variance are denoted with *. (E) Cytotoxicity of Oleate-AEG12 against HEK Cells in the presence of 1.3 mM Ca2+ (blue) or 2.5 mM EDTA (red) showing the inverse dependence of cell lysis on extracellular calcium and by extension the cell membrane repair pathway.
Delivery of unsaturated fatty acids into phospholipid bilayers by AEG12 via this mechanism is expected to decrease membrane stability, giving rise to the observed cytotoxic activity. This can be assessed through the use of a 6-Dodecanoyl-N,N-dimethyl-2-naphthylamine (LAURDAN) fluorescent probe (Fig. 5A), whose fluorescent properties or generalized polarization (GP) is dependent on the phase behavior of the model membrane system into which it is inserted (31). Monitoring LAURDAN GP over a range of temperatures produces a melting curve, the inflection point of which represents the liquid-solid phase transition temperature (MT) of the targeted bilayer system as shown in Fig. 5D. Incubating DSPC vesicles with 28 μM nMix-AEG12, Oleate-AEG12, or the equivalent concentration of free oleate (accounting for the 8:1 oleate–AEG12 binding stoichiometry) yielded a significant decrease in the MT of DSPC vesicles relative to a negative control incubated with phosphate-buffered saline (PBS) alone. Addition of Apo-AEG12 had no observable effect, consistent with the proposed mode of action. Phosphatidylethanolamine lipids of equivalent acyl chain length (distearoylphosphatidylethanolamine, DSPE) or MT (dimyristoylphosphatidylethanolamine) as DSPC were similarly destabilized by free oleic acid. However, nMix-AEG12 failed to yield any observable effect even at higher (40 μM) concentrations suggesting that AEG12 cargo delivery is dependent on the target membrane composition.
Mammalian cell membrane repair is strongly dependent on Ca2+, the removal of which has been shown to enhance the cytotoxicity of other known membrane-disrupting protein fatty acid complexes (32). Removal of Ca2+ thus provides an elegant experiment to confirm the proposed model for AEG12 activity. Indeed, the cytolytic activity of Oleate-AEG12 against HEK cells was significantly inhibited by the presence of 1.3 mM Ca2+, while the addition of 2.5 mM ethylenediaminetetraacetic acid (EDTA) yielded the opposite effect (Fig. 5E), consistent with the interpretation that AEG12 damages cell membranes through delivery of its fatty acid cargo.
Ligand Binding Preferences Provide Thermodynamic Basis for AEG12 Cytotoxicity and Selectivity.
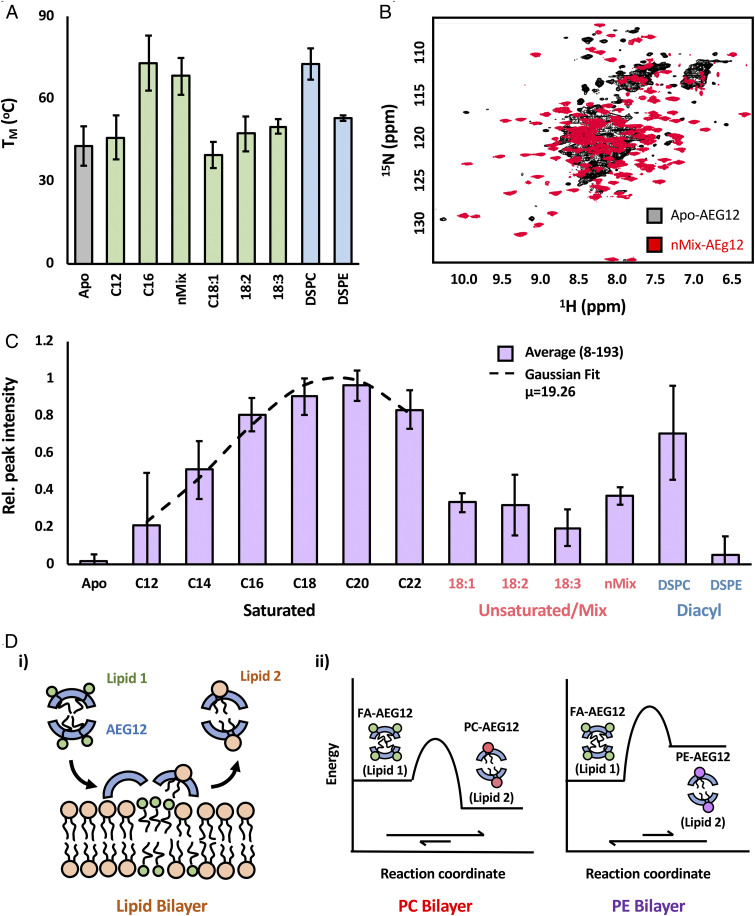
Given the cargo dependence of cytotoxicity, it is prudent to examine the interactions between AEG12 and its fatty acid ligands in a more systematic manner. Previous work has identified a mixture of palmitate (16:0), stearate (18:0), and oleate (18:1) fatty acids as the natural ligands for Bla g 1 when purified from its natural allergen source (16, 17). The slow binding and dissociation of hydrophobic ligands from AEG12 (Fig. 3E) suggests that this process is under kinetic rather than thermodynamic control, hindering efforts to identify the natural ligand of AEG12 based on equilibrium KD measurements. The ΔG of ligand binding results in a change in melting temperature of the target protein (ΔTm). This thermal shift thus provides a convenient, if indirect method of comparing binding affinities (33), as evidenced by the ∼20 °C increase in Tm observed for Bla g 1 when bound to its nMix natural ligands or their saturated equivalents (16:0 to 18:0) (17). Binding of nMix ligands did not significantly perturb the overall alpha-helical content of AEG12 when assessed using circular dichroism (SI Appendix, Fig. S1), in line with the molecular modeling described above. Monitoring the temperature-dependent loss of secondary structure reveals a significant thermal shift upon binding of both saturated fatty acids and nMix ligands (Fig. 6A and SI Appendix, Fig. S1), with the latter yielding maximal enhancement. These results suggest that AEG12 broadly mirrors the binding preference of its cockroach homolog, with the nMix lipids representing a close approximation of the natural ligands for both MA proteins. Loading AEG12 with entirely mono- (18:1) and polyunsaturated (18:2 and 18:3) ligands yielded lower Tm values than their saturated counterparts, suggesting a lower affinity for these ligands. Curiously, ΔTm values were significantly reduced under acidic conditions, with no significant difference observed between 18:1, DSPC, and nMix-loaded AEG12 (SI Appendix, Fig. S1), potentially providing a mechanism through which AEG12 ligands could be exchanged within endosomal vesicles prior to secretion into the basic (pH 7.5 to 8) postblood meal midgut environment (34, 35).
Fig. 6.
Thermodynamics of AEG12 ligand binding provides potential basis for cytotoxic selectivity. (A) Effect of ligand binding on AEG12 thermostability as assessed using circular dichroism. Representative CD spectra and melting curves shown in SI Appendix, Fig. S2. (B) 1H-15N-HSQC spectra of perdeuterated 15N-labeled AEG12. Apo-AEG12 (black) shows significant peak broadening indicative of intermediate (μs-ms) exchange dynamics. Loading AEG12 with long chain (nMix) fatty acids (red) significantly reduces peak broadening. 15N-HSQC peak intensity of 15N-labeled AEG12 loaded with a range of saturated and unsaturated fatty acids or diacyl chain ligands is shown in C. Relative peak intensities for AEG12 loaded with various hydrophobic ligands. Purple bars represent the average values across all peaks which could be confidently assigned and quantified, while the associated error bars represent the SD about this mean value. Data obtained from the saturated fatty acids could be fit to a Gaussian curve (dashed line) relative to acyl chain length, with a maximum peak intensity occurring at 19. Complete peak intensity values for all residues can be found in SI Appendix, Fig. S2. (D) Proposed model for AEG12-mediated cargo delivery. (i) General model illustrating the transfer of initial cargo (green) from AEG12 (blue) to the target bilayer. This is accompanied by the abstraction of phospholipids (orange) from said bilayer to occupy the otherwise empty AEG12 hydrophobic cavity. Cargo transfer is dependent on the relative stability of AEG12 in complex with Lipid 1 and Lipid 2, as illustrated by the free energy diagrams depicting the transfer of FA cargoes to PC and PE bilayers based on this model (ii).
To gain further insight into the structural response to ligand binding, 15N labeled AEG12 was prepared using the same range of fatty acid cargoes and analyzed using solution-state NMR. 1H-15N-heteronuclear single quantum coherence (HSQC) spectra of AEG12 either in its Apo form or loaded with shorter chain (e.g., C12) fatty acids yields predominantly broad, poorly defined peaks with low intensity consistent with elevated levels of conformational exchange or dynamics (Fig. 6B). Addition of longer-chain cargoes enhanced both peak intensity and dispersion indicating a single globular conformation from which the assignments could be determined (Fig. 6B and SI Appendix, Fig. S2). The intensity of the amide backbone resonances could be fit to a Gaussian function with respect to fatty acid chain length with 19 carbons representing the theoretical optimum (Fig. 6C and SI Appendix, Fig. S2). Unsaturated fatty acids failed to yield similar enhancements to conformational stability, suggesting that their kinked acyl chains are not accommodated in the same way unless accompanied by saturated counterparts. The effect was generally observed across all the residues for which there were dispersed peaks that could be confidently assessed. These results broadly mirror those reported for the thermostability assays, suggesting that ligand selectivity is mediated through global effects on the conformational stability of the entire protein consistent with its shell-like structure in which nearly all the residues lie in close proximity to the ligand binding cavity.
In addition to fatty acids, Bla g 1 can also bind diacyl chain ligands, where it displays a strong preference for PC lipids at the expense of phosphatidylethanolamine (PE) (16, 17). To determine whether AEG12 shares a similar selectivity, samples were subjected to the ligand-loading process in the presence of a 1:1 ratio of DSPC and DSPE. The resulting sample showed a significant enrichment of PC lipids assessed by 31P-NMR (SI Appendix, Fig. S6) indicative of preferential binding. Additionally, loading AEG12 with DSPC phospholipids yielded TM and NMR peak intensity values broadly consistent with its saturated fatty acid equivalents, while DSPE yielded a more modest enhancement (Fig. 6C) suggesting that binding of the latter is energetically unfavorable relative to the former (Fig. 6A). These results suggest that AEG12 broadly mirrors headgroup binding preferences of its cockroach counterpart and provide thermodynamic basis for this selectivity.
Based on these findings, we propose a model in which AEG12 acts as a delivery vehicle, mobilizing and transporting unsaturated and nMix fatty acid ligands to the membranes of the targeted cell. The unsaturated or nMix fatty acid ligands are then exchanged with diacyl chain ligands from the target membrane. The enhanced thermostability of the PC-AEG12 complex relative to nMix-AEG12 or its various unsaturated fatty acid equivalents illustrated in Fig. 6C makes this process thermodynamically favorable when targeting PC-based membranes. Conversely, the comparatively low thermostability of both Apo- and phosphatidylethanolamine-AEG12 hinders ligand delivery to PE-based membranes (Fig. 6D).
To assess whether this exchange process occurs, nMix-AEG12 was incubated with DSPC or DSPE vesicles, and the resulting mixture was analyzed using size-exclusion chromatography (SEC). Neither lipid yielded a significant shift in the elution volume relative to an AEG12 control (SI Appendix, Fig. S6), suggesting that AEG12 remains in the free form and does not strongly associate with lipid vesicles; a conjecture supported by cosedimentation assays (SI Appendix, Fig. S6). Mass spectrometry (MS) analysis of the isolated AEG12 revealed the presence of DSPC but not DSPE (SI Appendix, Fig. S6), suggesting that at least some of the initial nMix ligands are being exchanged for PC but not PE lipids. To further confirm this exchange process, DSPC vesicles were incubated with DAUDA for 1 h. The resulting DAUDA-labeled vesicles were washed three times to remove unbound fluorophore and incubated with Apo- or nMix-AEG12. A significant increase in fluorescence was observed in the soluble fraction with an emission maximum at ∼480 nm consistent with the formation of a free DAUDA-AEG12 complex via lipid exchange (SI Appendix, Fig. S6). These results, combined with the thermostability data presented above, provide an elegant mechanism through which the cytotoxicity of AEG12 can be targeted toward specific cells based on the relative thermodynamic stability of AEG12 in the ligand-bound and cargo-exchanged states (Fig. 6D)
Discussion
In this work, we demonstrated that the lytic activity of AEG12 is dependent on its ability to bind and exchange fatty acids with cell membranes. The fact that both AEG12 and Bla g 1 displayed maximal lytic activity when loaded with nMix, the natural ligands identified for Bla g 1, further supports cell lysis as a natural biological activity for MA domain proteins. While the ability of free fatty acids to destabilize membranes and lyse cells in vitro is well documented (22, 23), the difficulty in solubilizing and mobilizing large amounts of fatty acids makes this approach challenging to implement within a biological system. As an additional consideration, absorption of free fatty acids within the insect midgut is a highly efficient process aided in part by a large number of fatty acid-binding proteins, making it difficult to maintain a large pool of free fatty acids in solution (36). While MA domain proteins such as AEG12 are not strictly necessary for cell lysis in vitro, the biological utility of these proteins stems from their ability to package large numbers of fatty acids in a convenient, easy to transport molecule that can protect them from the external environment while allowing for their rapid secretion in response to a blood meal. The resilience of the AEG12-ligand complex also provides an elegant mechanism through which cargo delivery and thus lytic activity can be restricted to specific cellular targets as evidenced by the apparent selectivity of AEG12 against PC but not PE membranes potentially contributing to its inability to lyse prokaryotic cells, something that is not possible with free fatty acids alone.
Through targeting the plasma membrane, AEG12 is able to lyse a broad spectrum of different cells types, even accounting for the selectivity described above. While this may seem redundant given the specific association of AEG12 with blood feeding in mosquitoes, its utility becomes apparent when we consider the biological context of other MA domain proteins such as Bla g 1. Here, the omnivorous nature of cockroaches and other insects would favor such a general-purpose digestive function. Indeed, knocking down Bla g 1 expression was found to significantly hinder digestion and uptake of food pellets composed of a variety of plant- and meat-based materials consistent with its action as a broad-spectrum digestive agent (15). Conversely, the prominent role of midgut bacteria in nutrient uptake and digestion would provide evolutionary pressure against lysis of PE membranes consistent with the observations presented herein (37). The importance of MA domain proteins in digestion is further underscored by the fact that invertebrates lack a gall bladder to dissolve lipids. As such, they must rely on other strategies for dissolving membranes from food and extracting lipids from their diet (38). Phospholipases produced in the midgut hydrolyze triacylglycerides, potentially facilitating cytolysis and lipid absorption in insects (39, 40). As early as 1975 it was noticed that phospholipase activity could be enhanced by the addition of free oleate (41). The ability of AEG12 and other MA domain proteins to deliver such ligands into biological membranes could thus contribute to digestion not only through directly lysing ingested cells but also through increasing the ability of phospholipases to access substrates which would normally be buried within the lipid leaflet.
Besides being related to a blood feed, the gene for AEG12 has been identified in several studies as being up-regulated in response to flavivirus infections (3–6). The experiments presented here show that AEG12 does indeed possess antiviral properties, which are mediated by a similar lipid membrane-disrupting mechanism as cytolysis. The elevated activity against flaviviruses such as ZIKV and DENV may reflect the fact that their viral envelopes are derived from the endoplasmic reticulum of host cells rather than the plasma membrane as is the case for Lentiviruses, resulting in a higher proportion of PC lipids (42). Such a correlation would mirror the dependence of cytotoxic activity on target membrane composition, further supporting a shared mode of action between the two putative functions of AEG12. However, the more moderate activity against HCoV’s, which also bud from the endoplasmic reticulum, suggests the presence of additional determinants of antiviral specificity which warrant further investigation. Previous studies have demonstrated that free unsaturated fatty acids can neutralize enveloped viruses through membrane destabilization (20, 21). More recently, it has been shown that phospholipases are able to neutralize flaviviruses and other enveloped viruses through inducing similar perturbations, demonstrating that disruption of the viral lipid envelope is a viable mechanism for achieving antiviral activity despite the presence of a protective protein capsid (43, 44). Taken together, the results presented herein describe an elegant model in which the ability of AEG12 to destabilize lipid bilayers enables it to serve a dual function: breaking down erythrocytes following a blood meal while simultaneously neutralizing enveloped virus particles which may have been ingested. In mosquitoes, ingestion of a blood meal has the counterintuitive effect of decreasing the activity of several immune pathways, further underscoring the “gatekeeper” functionality of AEG12 during this period of increased vulnerability (45). Indeed, one might speculate that the expansion of the MA family in Aedes and Culex (9), the only major insect vectors for arboviruses, was due to evolutionary pressure favoring this secondary antiviral function. While the AEG12 content within the Aedes midgut have yet to be quantified in terms of physical units, studies carried out on the cockroach equivalent report ∼2 μmol/kg in the excreted frass, suggesting that micromolar concentrations of Bla g 1 are feasible in the midgut (14, 16). Similar concentrations of AEG12 would be sufficient to support both of its putative biological functions based on the CC50 and IC50 values reported in this work (11). It should also be noted that the quantification studies were carried out in the absence of specific induction or stimulation of Bla g 1. Given the significant up-regulation of AEG12 following a blood meal, even higher concentrations might be achievable within the mosquito midgut.
The simplicity of AEG12’s antiviral and cytolytic activity could also provide the basis for the development of human therapeutics that deliver fatty acid cargoes to specific targets, be they malignant cells or virion particles, based on their molecular profile. Recently, it was discovered that α-lactalbumin (aLa) can form large (up to 850 kDa) macromolecular complexes with fatty acids such as oleic acid. These complexes, or liprotides, consist of a micellular core of fatty acids surrounded by partially unfolded aLa’s (46). Liprotides are able to bind and transfer their cargoes into biological membranes, destabilizing them and ultimately contributing to cell lysis (32). Thus, while the architecture of these liprotides differs significantly from AEG12, their overall mechanism of action remains the same. The toxicity and selectivity of liprotides or AEG12 could potentially be enhanced through conjugation with IgG antibodies. Indeed, similar lipoprotein complexes formed from oleic acid and erythrocyte-specific IgGs significantly enhanced hemolytic activity, highlighting the potential of such antibody-targeted lipid delivery systems (47). However, because of their large size and heterogeneous nature, the fatty acid cargoes from some of these disordered lipid–protein complexes have been observed to undergo rapid exchange between free and bound form, limiting their therapeutic utility (48). In this regard, the small size and homogeneous nature of AEG12 coupled with the limited ability of its bound ligands to exchange with the aqueous environment, provides a compelling basis for the development of therapeutic agents with both targeted cytotoxic and antiviral properties; a task which is facilitated by the well-defined three-dimensional (3D) structure and systematic study of fatty-acid ligand binding presented in this work. Finally, the broad spectrum of AEG12’s mechanism of action, as demonstrated by the range of antiviral activities reported in this work, means that therapeutic derivatives can be quickly designed and implemented against new and emerging threats such as SARS-CoV-2 (COVID-19).
Materials and Methods
Protein Expression, Purification, and X-ray Crystallography.
The AEG12 gene representing a single insect MA domain repeat was expressed as a Glutathione S-Transferase (GST) fusion. The resulting protein was purified from crude lysate using affinity chromatography. The GST tag was cleaved, and the resulting AEG12 was stripped of its endogenously bound ligands using reverse-phase high-performance liquid chromotography and refolded either in the Apo-form or in the presence of the desired cargo as described previously for Bla g 1 (16, 17, 49). Amberlite-treated samples were generated by incubated nMix-AEG12, Oleate-AEG12, or free Oleate with 30 mg Amberlite XAD-2 Resin (Sigma) per milligram of fatty acid. The soluble fraction was recovered and showed no significant AEG12 absorption onto the resin beads. For X-ray crystallography studies, AEG12 was expressed as a maltose-binding protein fusion and purified using affinity chromatography and gel filtration. Crystals were obtained via sitting vapor diffusion. Full expression and crystallization details are available in SI Appendix, Supplemental Materials and Methods and Table S1.
Computational Studies.
Molecular Dynamics simulations were carried out as described previously for Bla g 1 (17), with additional details provided in SI Appendix, Supplemental Materials and Methods. In brief, the X-ray structure of AEG12 was used to generate an initial model, to which fatty acids or phospholipids were manually added and energy minimized using Amber (50). The resulting Apo- and Ligand-loaded models were solvated in a water box with counterions. Following proper equilibrations, molecular dynamics trajectories were calculated using Amber.16 over the span of 120 ns (DSPC) or 500 ns (Oleate) (50).
Circular Dichroism.
Circular dichroism spectra and thermodenaturation curves were collected on 0.5 μM AEG12 in Tris buffer (100 mM ⋅ Tris pH 7.5, 50 mM ⋅ NaCl) or 10 mM citrate buffer (pH 5.8) using Jasco J-815 circular dichroism (CD) spectropolarimeter as described previously (17). Uncertainty values for all data presented in this work represent the SD obtained from at least three independent trials unless otherwise specified. Statistically significant differences were identified using a Student’s t test assuming equal variance.
NMR.
Backbone amide assignments (SI Appendix, Fig. S2) were determined using perdeuterated 13C,15N-labeled nMix-AEG12 using an 800 MHz Agilent DD2 spectrometer equipped with a cryogenically cooled HCN probe. Standard triple-resonance experiments were analyzed using NMRPipe (51) and NMRViewJ (52) and were used to establish connectivity. HSQC spectra described in Fig. 6 and SI Appendix, Fig. S2 were obtained using uniformly 15N-labeled AEG12 with a similarly equipped 600 MHz Agilent DD2 spectrometer. Binding stoichiometries were obtained using samples of AEG12 loaded with either DSPC, methyl-13C–stearate, or methyl-13C–oleate (Cambridge Isotope Laboratories) diluted 1:1 with cholate buffer (100 mM ⋅ Tris pH 8.0, 100 mM ⋅ NaCl, 10% wt/vol cholate). Phospholipid concentrations were quantified using 1D 31P-NMR spectra acquired using a 600 MHz spectrometer equipped with a Varian 5-mm Z-gradient probe as described previously (17), while 13C-fatty acid concentrations were assessed using 1H-13C SOFAST heteronuclear multi-quantum coherence (HMQC) spectra obtained with the 600 MHz Agilent HCN system described above (53).
Cell Lysis.
Detailed protocols for the culturing and preparation of cells employed in these assays are provided in SI Appendix, Supplemental Materials and Methods. To measure hemolysis, erythrocytes were incubated with either AEG12, PBS (negative control), or 2% triton X (positive control) at 37 °C for 1 h. The resulting mixture was then pelleted at 600× g for 10 min. The OD490 of the supernatant was measured and used to calculate % hemolysis as described previously (54). Still images depicted in Fig. 2 were obtained using a Zeiss AxioObserver Z1 fluorescence microscope (Carl Zeiss Incorporated) with a metal halide light source at 20× magnification. HEK-293, Vero, and SF9 cells were similarly treated with AEG12. The resulting mixture was diluted 1:1 with AO/PI stain (Nexcelom), and cell viability was assessed as per manufacturer recommended protocols using either the Zeiss system described above at 10× magnification or a Cellometer Auto 2000 automatic cell counter (Nexcelom). In both cases, eight images were acquired per sample to minimize inconsistency. Yeast cells were similarly treated with AEG12, PBS (negative control), or 3% vol/vol Zymolyase (positive control), washed two times with PBS, serially diluted, and plated onto yeast peptone dextrose adenine media agar plates. The plates were incubated at 30 °C for 48 h and then colonies were counted to determine survival. E. coli BL21 DE3 cells were incubated with AEG12 or free oleic acid for 1 h at 37 °C. A serial dilution was plated on Luria broth (LB)+kanamycin plates, which were incubated overnight at 37 °C prior to colony counting. The resulting cell viability data were fit to an exponential decay function versus AEG12 concentration to obtain CC50 values.
DAUDA Fluorescence Transfer.
Phospholipids (Avanti Polar Lipids) were resuspended as 1.0 μm unilamellar vesicles using an Avanti Mini-Extruder as per manufacturer’s directions. To assess cargo transfer from AEG12 into biological membranes, AEG12 was loaded with DAUDA ( Sigma) and incubated with either DSPC vesicles (5 mg/mL) or HEK-293 cells (1 × 106 cells or mL) for 1 h at 37 °C. Cells or vesicles were recovered through centrifugation, rinsed three times with PBS, and diluted 10 times . DAUDA fluorophore transfer to the pelleted vesicles was assessed using a Fluoromax-Plus-CP Spectrophotometer (Horiba) at an excitation wavelength of 340 nm and an emission window of 400 to 650 nm; spectra obtained from the wash steps confirmed removal of unbound fluorophore. Positive and negative controls were generated through treating the vesicles/cells with the equivalent concentration of free DAUDA or PBS, respectively.
To assess the reverse transfer process, DAUDA was incorporated into DSPC vesicles by incubating vesicles (5 mg/mL) with 40 μM free DAUDA for 1 h at 37 °C. Vesicles were washed three times with PBS, and removal of free DAUDA was confirmed as discussed above. DAUDA-DSPC vesicles were incubated with 25 μM Apo- or nMix-AEG12 at 37 °C. Samples were removed at 15, 30, and 60 min, diluted 10 times in PBS, and centrifuged to remove the DAUDA-DSPC vesicles. AEG12-bound DAUDA remaining in the supernatant was assessed as described above.
Vesicle Binding.
A total of 25 μM nMix-AEG12 were incubated with/without 10 mg/mL DSPC or DSPE vesicles in PBS for 1 h at 37 °C prior to SEC (S75, GE). Fractions were collected and analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and matrix-assisted laser desorption/ionization time of flight mass spectrometry to confirm the presence of AEG12 and phospholipid, respectively. Details on the latter are available in SI Appendix, Supplemental Materials and Methods.
For cosedimentation assays, 5 mg/mL DSPC vesicles were incubated with 25 μM Apo- or nMix-AEG12 for 1 h. Vesicles were pelleted and resuspended to match the original starting volume. Samples of the supernatant and resuspended pellet were analyzed using SDS-PAGE. SDS-PAGE band intensities were normalized to a control sample representing the total protein in both the vesicle and soluble fractions obtained prior to centrifugation.
Vesicle Destabilization.
Lipid vesicles (3.70 mg/mL) were incubated with 25 μM LAURDAN (Sigma) and the appropriate concentration of AEG12 or oleic acid at 37 °C for 1 h. Samples were diluted 10 times prior to spectroscopic analysis using a Fluoromax-Plus-CP Spectrophotometer (Horiba). Samples were heated from 25 to 88 °C at a rate of 2 °C/min. Emission spectra (400 to 500 nm) were collected at 2-degree increments using an excitation wavelength of 355 nm and a data pitch of 10 nm. Fluorescence intensity at 440 and 490 nm were used to calculate GP values as described by Parasassi et al. (31) which could then be fit to a two-state Boltzmann curve to obtain phase-transition temperatures.
Viral Assays.
A full description of the viral strains and culturing employed in this study, along with detailed methodology, is available in SI Appendix, Supplemental Materials and Methods. In brief, flavivirus (ZIKV, DENV) and Coronavirus (HCoV) were incubated with varying concentrations of AEG12 for 1 h at 37 °C. The resulting mixture was applied to a confluent monolayer of Vero (ZIKV, DENV) or MRC-5 (HCoV) cells for 1 h. Positive and negative controls were prepared using untreated virus, or PBS/media alone. Cells were washed, fixed with methyl cellulose containing media, and incubated for 2 to 6 d at room temperature. Viral infectivity was assessed through assessing the formation of viral plaques (ZIKV, HCoV) or immunostaining (DENV).
To assess AAV and lentivirus infectivity, green fluorescent protein (GFP)-modified strains were treated with AEG12 for 30 min at 37 °C and applied to a confluent monolayer of Vero cells for an additional hour. Cells were washed with media and incubated for 48 h. The proportion of cells expressing GFP viral proteins was determined by flow cytometry (BD LSRFortessa) and used as a measure of infectivity. Infectivity values were normalized to the positive and negative controls and fit to an exponential decay function relative to AEG12 concentration to obtain IC50 values.
Supplementary Material
Acknowledgments
We thank Dr. Tom Kirby, Scott Gabel, and Dr. Robert London for their help and assistance throughout this work, along with Dr. Bob Petrovich and Lori Edwards for providing some of the cell lines and instrumentation employed in this study and for their assistance in generating the AEG12 constructs employed in this study. We thank Dr. Jason Williams and Andrea Adams for assistance with the mass spectrometry. We thank Dr. Jeff Tucker, Erica Scappini, and Dr. Agnes Janoshazi for their assistance with microscopy and fluorescent imaging; Dr. Carl Bortner and Maria Sifre for assistance with the flow cytometry studies; and Dr. Natasha Degtyareva for assistance with the yeast cell cytotoxicity assays. Finally, we would like to thank Drs. Scott Williams and Traci Hall for their critical reading of this manuscript. We kindly acknowledge Dr. Aravinda de Silva for his support in generating DENV and ZIKV FRNT data. This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences, Z01-ES102906 (G.A.M.), ZIC ES102506-09 (N.M.), 1ZIA-ES102645 (L.C.P.), and Z01-ES043010 (L. Perera). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019251118/-/DCSupplemental.
Data Availability
Structure data have been deposited in the RCSB Protein Data Bank (6xrx).
References
- 1.Wolf M. C., et al., A broad-spectrum antiviral targeting entry of enveloped viruses. Proc. Natl. Acad. Sci. U.S.A. 107, 3157–3162 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins M. H., Metz S. W., Progress and works in progress: Update on flavivirus vaccine development. Clin. Ther. 39, 1519–1536 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Sim S., et al., Transcriptomic profiling of diverse Aedes aegypti strains reveals increased basal-level immune activation in dengue virus-refractory populations and identifies novel virus-vector molecular interactions. PLoS Negl. Trop. Dis. 7, e2295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Etebari K., et al., Global transcriptome analysis of Aedes aegypti mosquitoes in response to zika virus infection. mSphere 2, e00456-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saldaña M. A., et al., Zika virus alters the microRNA expression profile and elicits an RNAi response in Aedes aegypti mosquitoes. PLoS Negl. Trop. Dis. 11, e0005760 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colpitts T. M., et al., Alterations in the Aedes aegypti transcriptome during infection with West Nile, dengue and yellow fever viruses. PLoS Pathog. 7, e1002189 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Souza-Neto J. A., Sim S., Dimopoulos G., An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc. Natl. Acad. Sci. U.S.A. 106, 17841–17846 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Juneja P., et al., Exome and transcriptome sequencing of Aedes aegypti identifies a locus that confers resistance to Brugia malayi and alters the immune response. PLoS Pathog. 11, e1004765 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Randall T. A., Perera L., London R. E., Mueller G. A., Genomic, RNAseq, and molecular modeling evidence suggests that the major allergen domain in insects evolved from a homodimeric origin. Genome Biol. Evol. 5, 2344–2358 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nanfack Minkeu F., Vernick K. D., A systematic review of the natural virome of anopheles mosquitoes. Viruses 10, 1–21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shao L., Devenport M., Fujioka H., Ghosh A., Jacobs-Lorena M., Identification and characterization of a novel peritrophic matrix protein, Ae-Aper50, and the microvillar membrane protein, AEG12, from the mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 35, 947–959 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Bonizzoni M., et al., Strain variation in the transcriptome of the dengue fever vector, aedes aegypti. G3 (Bethesda) 2, 103–114 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miglani D., Identification, characterization and analysis of expression of midgut specific G12 gene of Anopheles culicifacies (Diptera: Culicidae). Int. J. Biotechnol. Biochem. 13, 285–299 (2017). [Google Scholar]
- 14.Gore J. C., Schal C., Gene expression and tissue distribution of the major human allergen Bla g 1 in the German cockroach, Blattella germanica L. (Dictyoptera: Blattellidae). J. Med. Entomol. 41, 953–960 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Suazo A., Gore C., Schal C., RNA interference-mediated knock-down of Bla g 1 in the German cockroach, Blattella germanica L., implicates this allergen-encoding gene in digestion and nutrient absorption. Insect Mol. Biol. 18, 727–736 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Mueller G. A., et al., The novel structure of the cockroach allergen Bla g 1 has implications for allergenicity and exposure assessment. J. Allergy Clin. Immunol. 132, 1420–1426 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foo A. C. Y., et al., Hydrophobic ligands influence the structure, stability, and processing of the major cockroach allergen Bla g 1. Sci. Rep. 9, 18294 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohn A., Gitelman J., Inbar M., Interaction of polyunsaturated fatty acids with animal cells and enveloped viruses. Antimicrob. Agents Chemother. 18, 962–968 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohn A., Gitelman J., Inbar M., Unsaturated free fatty acids inactivate animal enveloped viruses. Arch. Virol. 66, 301–307 (1980). [DOI] [PubMed] [Google Scholar]
- 20.Thormar H., Isaacs C. E., Brown H. R., Barshatzky M. R., Pessolano T., Inactivation of enveloped viruses and killing of cells by fatty acids and monoglycerides. Antimicrob. Agents Chemother. 31, 27–31 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hilmarsson H., Larusson L. V., Thormar H., Virucidal effect of lipids on visna virus, a lentivirus related to HIV. Arch. Virol. 151, 1217–1224 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Rumsby M. G., Sparrow S., Little C., The oleic acid-induced shape transformation of human erythrocytes [proceedings]. Biochem. Soc. Trans. 7, 922–924 (1979). [DOI] [PubMed] [Google Scholar]
- 23.Hoque M., Dave S., Gupta P., Saleemuddin M., Oleic acid may be the key contributor in the BAMLET-induced erythrocyte hemolysis and tumoricidal action. PLoS One 8, e68390 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X., Rommelaere S., Kondo S., Lemaitre B., Renal purge of hemolymphatic lipids prevents the accumulation of ROS-induced inflammatory oxidized lipids and protects Drosophila from tissue damage. Immunity 52, 374–387.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Pathak P. K., et al., Structure of a lipid-bound viral membrane assembly protein reveals a modality for enclosing the lipid bilayer. Proc. Natl. Acad. Sci. U.S.A. 115, 7028–7032 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krijnse Locker J., Chlanda P., Sachsenheimer T., Brügger B., Poxvirus membrane biogenesis: Rupture not disruption. Cell. Microbiol. 15, 190–199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ibarguren M., López D. J., Escribá P. V., The effect of natural and synthetic fatty acids on membrane structure, microdomain organization, cellular functions and human health. Biochim. Biophys. Acta 1838, 1518–1528 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Onuki Y., Morishita M., Chiba Y., Tokiwa S., Takayama K., Docosahexaenoic acid and eicosapentaenoic acid induce changes in the physical properties of a lipid bilayer model membrane. Chem. Pharm. Bull. (Tokyo) 54, 68–71 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Ralston A. W., Hoerr C. W., The solubilities of the normal saturated fatty acids. J. Org. Chem. 7, 546–555 (1942). [DOI] [PubMed] [Google Scholar]
- 30.Robb I. D., Determination of the aqueous solubility of fatty acids and alcohols. Aust. J. Chem. 19, 2281–2284 (1966). [Google Scholar]
- 31.Parasassi T., Krasnowska E. K., Bagatolli L., Gratton E., Laurdan and prodan as polarity-sensitive fluorescent membrane probes. J. Fluoresc. 8, 365–373 (1998). [Google Scholar]
- 32.Frislev H. S., Boye T. L., Nylandsted J., Otzen D., Liprotides kill cancer cells by disrupting the plasma membrane. Sci. Rep. 7, 15129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall J., A simple model for determining affinity from irreversible thermal shifts. Protein Sci. 28, 1880–1887 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Billker O., Miller A. J., Sinden R. E., Determination of mosquito bloodmeal pH in situ by ion-selective microelectrode measurement: Implications for the regulation of malarial gametogenesis. Parasitology 120, 547–551 (2000). [DOI] [PubMed] [Google Scholar]
- 35.del Pilar Corena M., et al., Carbonic anhydrase in the adult mosquito midgut. J. Exp. Biol. 208, 3263–3273 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Arrese E. L., et al., Lipid storage and mobilization in insects: Current status and future directions. Insect Biochem. Mol. Biol. 31, 7–17 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Gaio Ade. O., et al., Contribution of midgut bacteria to blood digestion and egg production in aedes aegypti (diptera: culicidae) (L.). Parasit. Vectors 4, 105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Canavoso L. E., Jouni Z. E., Karnas K. J., Pennington J. E., Wells M. A., Fat metabolism in insects. Annu. Rev. Nutr. 21, 23–46 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Terra W. R., Ferreira C., “Biochemistry and molecular biology of digestion” in Insect Molecular Biology and Biochemistry, L. I. Gilbert, Ed. (Academic Press, 2012), pp. 365–418. [Google Scholar]
- 40.Stanley D., The non-venom insect phospholipases A2. Biochim. Biophys. Acta 1761, 1383–1390 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Lachowiez R. M., Hammoud N. M., Teibel J. L., Dix J. A., Phospholipase activation, free fatty acids and the proton permeability of a biological membrane. FEBS Lett. 234, 195–198 (1988). [DOI] [PubMed] [Google Scholar]
- 42.Casares D., Escribá P. V., Rosselló C. A., Membrane lipid composition: Effect on membrane and organelle structure, function and compartmentalization and therapeutic avenues. Int. J. Mol. Sci. 20, 2167 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller V. D., et al., Phospholipase A2 isolated from the venom of Crotalus durissus terrificus inactivates dengue virus and other enveloped viruses by disrupting the viral envelope. PLoS One 9, e112351 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen M., et al., Broad-spectrum antiviral agents: Secreted phospholipase A2 targets viral envelope lipid bilayers derived from the endoplasmic reticulum membrane. Sci. Rep. 7, 15931 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu Y., et al., Blood meal acquisition enhances arbovirus replication in mosquitoes through activation of the GABAergic system. Nat. Commun. 8, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frislev H. S., Jessen C. M., Oliveira C. L. P., Pedersen J. S., Otzen D. E., Liprotides made of α-lactalbumin and cis fatty acids form core-shell and multi-layer structures with a common membrane-targeting mechanism. Biochim. Biophys. Acta 1864, 847–859 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Hoque M., Gupta J., Saleemuddin M., Augmenting the cytotoxicity of oleic acid-protein complexes: Potential of target-specific antibodies. Biochimie 137, 139–146 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Nielsen S. B., et al., The interaction of equine lysozyme:oleic acid complexes with lipid membranes suggests a cargo off-loading mechanism. J. Mol. Biol. 398, 351–361 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Foo A. C. Y., Thompson P. M., Mueller G. A., Removal and replacement of endogenous ligands from lipid-bound proteins and allergens. J. Vis. Exp. 168, e61780 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Case D. D., et al., AMBER (University of California, San Francisco, 2016).
- 51.Delaglio F., et al., NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 52.Johnson B. A., Blevins R. A., NMR view: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 (1994). [DOI] [PubMed] [Google Scholar]
- 53.Schanda P., Kupĉe E., Brutscher B., SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J. Biomol. NMR 33, 199–211 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Evans B. C., et al., Ex vivo red blood cell hemolysis assay for the evaluation of pH-responsive endosomolytic agents for cytosolic delivery of biomacromolecular drugs. J. Vis. Exp. 50166, e50166 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structure data have been deposited in the RCSB Protein Data Bank (6xrx).