Significance
Monitoring orientation of biological molecules in living cells is a key approach to understanding the mechanisms of emergence/dissolution of molecular architectures. However, it has been challenging due to lack of general methodologies that can be used for living cells. We developed POLArIS, a genetically encoded and versatile probe for molecular orientation imaging. POLArIS is a recombinant binder rigidly connected to a fluorescent protein and can target biomolecules of interest by combining with phage display screening. Live imaging of starfish embryos expressing POLArIS that specifically binds to F-actin revealed that actin filaments radially extend in association with microtubule asters during mitosis. POLArIS can be applied in a variety of living specimens including whole bodies of or specific cell types in animals.
Keywords: fluorescence microscopy, fluorescence polarization, actin, starfish, mitosis
Abstract
Biomolecular assemblies govern the physiology of cells. Their function often depends on the changes in molecular arrangements of constituents, both in the positions and orientations. While recent advancements of fluorescence microscopy including super-resolution microscopy have enabled us to determine the positions of fluorophores with unprecedented accuracy, monitoring the orientation of fluorescently labeled molecules within living cells in real time is challenging. Fluorescence polarization microscopy (FPM) reports the orientation of emission dipoles and is therefore a promising solution. For imaging with FPM, target proteins need labeling with fluorescent probes in a sterically constrained manner, but because of difficulties in the rational three-dimensional design of protein connection, a universal method for constrained tagging with fluorophore was not available. Here, we report POLArIS, a genetically encoded and versatile probe for molecular orientation imaging. Instead of using a direct tagging approach, we used a recombinant binder connected to a fluorescent protein in a sterically constrained manner that can target specific biomolecules of interest by combining with phage display screening. As an initial test case, we developed POLArISact, which specifically binds to F-actin in living cells. We confirmed that the orientation of F-actin can be monitored by observing cells expressing POLArISact with FPM. In living starfish early embryos expressing POLArISact, we found actin filaments radially extending from centrosomes in association with microtubule asters during mitosis. By taking advantage of the genetically encoded nature, POLArIS can be used in a variety of living specimens, including whole bodies of developing embryos and animals, and also be expressed in a cell type/tissue specific manner.
Many biomolecules function as molecular assemblies such as protein complexes, nucleic acids, and lipid bilayers. Their function often depends on the changes in molecular arrangements of their constituents, for example, conformational changes during movement of molecular motors along cytoskeletons (1–3), rotation of F1-ATPases (4), and bending/extending of integrins (5, 6). Molecular orientation is the key information to study the relationship between such mutual changes of molecular arrangements and their function. Fluorescence microscopy has been widely used to study the structures and the dynamics of biomolecules in living cells, but monitoring the orientation of fluorescently labeled molecules in living cells is challenging even with the latest fluorescent microscopy including super-resolution approaches. Fluorophores, such as fluorescent proteins and organic compound dyes, emit fluorescence by radiating dipoles. The light emitted from a single dipole is fully polarized along the dipole axis. Therefore, when fluorescent labels are rigidly bound to molecules, the orientation of molecules can be monitored by analyzing the polarization state of the fluorescence. Fluorescence polarization microscopy (FPM) has been used for this approach (1–9) and is especially useful for detecting the presence of orderly assembled biomolecules and changes in their arrangements (10–12). In addition, FPM can report molecular orientations at a single-molecule level even in living cells when target molecules are labeled at proper density (3, 7). Super-resolution FPM methods have also been recently reported (13–16).
Despite such promising advantages and advancements in methodology, FPM has not been widely used in biomedical research because of the difficulties in the rotationally constrained labeling of target molecules with fluorescent proteins. To monitor the molecular orientation, a target molecule and a fluorescent protein must be rigidly connected so that the orientation of the target and that of the dipole of the fluorophore are sterically fixed. For many cases, intensive screening of the linkage was required for each target molecule since there was no universal method for constrained tagging due to difficulties in the rational three-dimensional design of protein connection. A novel, more versatile and easier approach is therefore imperative for expanding the application of FPM.
For this purpose, we have developed a versatile molecular orientation probe, named the Probe for Orientation and Localization Assessment, recognizing specific Intracellular Structures of interest (POLArIS). POLArIS is a high-affinity recombinant binder rigidly connected to a fluorescent protein. The recombinant binder can be screened by phage display to specifically target biomolecules of interests. Thus, POLArIS can tag biomolecules of interest with fluorescent proteins in a rotationally constrained manner. As a proof of principle, we developed a POLArIS that specifically binds to F-actin (POLArISact) and demonstrated that POLArISact reports the orientations of actin assembly in living cells. Moreover, by observing starfish embryos expressing POLArISact, we found an F-actin–based subcellular architecture that associates with the microtubule aster during mitosis, which had not been previously reported, demonstrating that POLArISact is useful for detecting ordered structures made of actin filaments in living cells.
Results
Constrained Tagging of a Recombinant Binder, Adhiron, with Circularly Permutated Superfolder Green Fluorescent Protein (GFP).
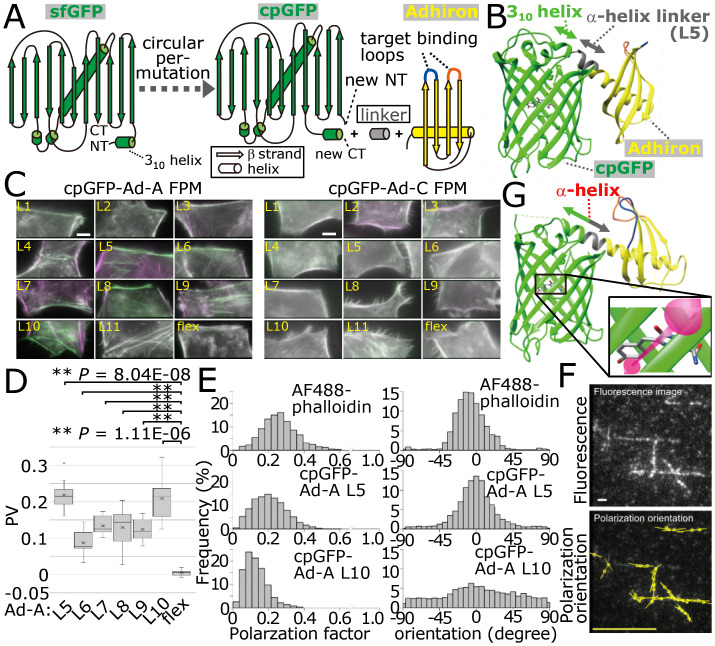
Most of the successful constrained tagging approaches used the direct connection of the C-terminal α-helices of target molecules to the N-terminal 310 helix of GFP (8–11). To develop POLArIS, we sought a recombinant binder protein that has an α-helix available for tagging with GFP. We found that Adhiron [the nomenclature was later changed to “affimer” (17)], a small protein (∼12 kDa) with a consensus sequence of plant-derived phytocystatins (18), has an α-helix at its N terminus lying on top of four anti-parallel β sheets (Fig. 1A). Two variable peptide regions are inserted between anti-parallel β sheets (Fig. 1A) to create binding sites for a molecule of interest. Phage display screening is generally used for selecting Adhirons that specifically bind to target molecules (19, 20). We chose specific three Adhirons (Adhiron-6, 14, and 24; for simplicity, we call them Ad-A, B, and C, respectively) that were reported to bind to F-actin in vitro as our initial test case.* The dissociation constant (Kd) of these Adhirons for F-actin was reported to be less than 0.5 μM, indicating that they bind to F-actin with a higher affinity than both Lifeact (Kd = 2.2 μM) (21) and UtrCH (Kd = 19 μM) (22). To connect the N-terminal α-helix of Adhiron to the 310 helix of GFP, we used a circularly permutated version of superfolder GFP (sfGFP) recently developed in our laboratory (Fig. 1A), which has the 310 helix at its C terminus and emits fluorescence as bright as the original sfGFP (9) (hereinafter simply called cpGFP). After the elimination of the end of N terminus that does not form α-helix, exposed terminal helices of Adhiron and cpGFP were connected with the EAAAK linker (L5). This linker was reported to form an α-helix (23). We made variations of Adhiron–cpGFP fusion constructs with extended α-helix linkers (L1-4), truncated α-helix linkers (L6-9), and constructs without any linkers (L10 and 11) (SI Appendix, Fig. S1 A and B). No serious steric hindrance (e.g., collision of main and/or side chains) between cpGFP and Adhiron connected by candidate linkers was presumed by UCSF Chimera (24) inspection for all fusion proteins (Fig. 1B shows an example for cpGFP-Ad-A L5).
Fig. 1.
Design of POLArIS and development of POLArISact. (A) Schematic of the circular permutation of sfGFP to create cpGFP, and fusion of cpGFP and Adhiron with a linker. NT and CT mean N terminus and C terminus, respectively. (B) Modeling of cpGFP-Adhiron (L5) structure with UCSF Chimera. (C) Two-axis FPM images of HeLa cells expressing cpGFP–Ad-A/C constructs. Horizontal and vertical components are shown in green and magenta, respectively. (Scale bar: 10 µm.) (D) PVs of indicated cpGFP–Ad-A constructs are shown by boxplots with mean values (cross marks). Statistics: Student’s t test with Šidák correction. **P < 0.01. n = 10. (E) In vitro evaluations of cpGFP–Ad-A L5/L10 by particle analysis with instantaneous FluoPolScope. Histograms of the polarization factor (Left) and polarization orientation (Right) for AF488-phalloidin (Top), cpGFP–Ad-A L5 (Middle), and L10 (Bottom). Orientation at 0° and ±90° indicates the polarization orientation of the particle is parallel and perpendicular, respectively, to actin filaments. (F) Representative images of instantaneous FluoPolScope analysis of cpGFP–Ad-A L5. (F, Top) Fluorescence image. (Scale bar: 1 µm.) (F, Bottom) Polarization orientations. The length of bars represents the polarization factor. Yellow bar: polarization factor = 0.5. (G) The crystal structure of POLArISact (T57S). Magenta two-way arrow: presumed orientation of the dipole of cpGFP (depicted according to ref. 70).
Screening and Characterization of POLArIS Candidates for F-Actin.
To test the binding selectivity of Adhirons to F-actin in mammalian cells, we made Adhirons connected with cpGFP via flexible linker (flex controls, SI Appendix, Fig. S1 A and B). As shown in SI Appendix, Fig. S1C, exogenously expressed flex controls for cpGFP–Ad-A and C were colocalized with F-actin labeled with phalloidin-Atto565 in fixed HeLa cells. In contrast, cpGFP–Ad-B flex showed significant background fluorescence in cytoplasm and nucleus (arrows in SI Appendix, Fig. S1C), and therefore we did not test Ad-B–based constructs for further analyses. Other cpGFP–Ad-A and C constructs (L1-11) showed colocalization with F-actin stained with phalloidin-Atto565 in HeLa cells, similarly to the cells expressing their corresponding flex controls (SI Appendix, Fig. S1D).
Next, we evaluated the anisotropy of the fluorescence for cpGFP–Ad-A/C constructs in living HeLa cells expressing each construct by fluorescence polarization analysis using two-axis FPM (Fig. 1C). In this FPM setup, fluorescent molecules are excited with isotropically polarized light-emitting diode illumination, and the emitted fluorescence is split into two orthogonal polarization orientations, namely 0° (horizontal) and 90° (vertical) (9). For the analysis, horizontal and vertical components are pseudocolored with green and magenta, respectively. The brightness of each pseudocolor of actin filaments represents the degree of the anisotropy of fluorescence. We found that six Ad-A–based constructs, namely L5-10, showed high brightness of green and magenta color on actin filaments running horizontally and vertically, respectively, in the image plane. The orientations of fluorescence polarization for these constructs were parallel to the axes of the actin filaments.
To quantitatively analyze the anisotropy of fluorescence of the constructs, we observed the anisotropy of fluorescence at the cortex of prometaphase-arrested HeLa M cells expressing each construct. We defined the polarization value, PV, for evaluation of the degree of anisotropy of fluorescence. In brief, the mean fluorescence intensity of a small rectangular region of interest enclosing vertical cortex in the cross-section of a mitotic cell in both vertical polarization and horizontal polarization images was measured and normalized, and the difference between the values were divided by the sum (see SI Appendix, SI Materials and Methods and SI Appendix, Fig. S2A for details). cpGFP–Ad-A L5 and 10 showed higher PV values (the absolute value of PV > 0.2) than those of other constructs (Fig. 1D and SI Appendix, Fig. S2B). These results were further confirmed by the analysis using the instantaneous FluoPolScope (7), which reports the absolute two-dimensional orientation of fluorescence polarization (SI Appendix, Fig. S2 C and D).
Next, we determined the absolute polarization orientation and the anisotropy of fluorescence (polarization factor) of single cpGFP–Ad-A L5 and L10 particles bound to F-actin in vitro. The polarization factor varies from zero (isotropic) to one (fully anisotropic) depending on the inclination angle of the dipole with respect to the focus plane and three-dimensional (3D) wobble of the probe [see a previous report (7) for more details]. The probes with a high polarization factor and a small variance in the polarization orientation histograms are preferable for studying the 3D architectural dynamics of actin filaments, such as the actin network in lamellipodia during the retrograde flow. Fluorescence polarization of cpGFP–Ad-A/C particles sparsely bound to actin filaments was analyzed using instantaneous FluoPolScope (Fig. 1 E and F). Alexa Fluor 488 Phalloidin (AF488-phalloidin), which is known to show high anisotropy of fluorescence when bound to F-actin filaments (7), was used for comparison. The polarization factor for cpGFP–Ad-A L5 was higher than that of L10, similarly to that of AF488-phalloidin. The distribution of the orientation histogram obtained for L5 was as narrow as that of AF488-phalloidin, while the orientation histogram for L10 showed broader distribution (Fig. 1E). We could observe the local orientation and the dynamics of the actin network during the retrograde flow in the lamellipodia of Xenopus tissue culture cells expressing cpGFP–Ad-A L5 (Movie S1). Thus, we found cpGFP–Ad-A L5 as the best orientation probe for monitoring the orientation of F-actin in vitro and in living cells and hereinafter call it POLArISact.
Structure Determination of POALrISact.
The structure of POLArISact was determined at 2.5-Å resolution by X-ray crystallography (Fig. 1G). We used the T57S mutant (residue 57 in cpGFP corresponds to residue 65 in the original avGFP) of POLArISact because we could not obtain the crystal of original POLArISact protein suitable for structure determination, possibly because of the peptide bond cleavage known as “backbone fragmentation” during chromophore formation or maturation (25) (for details, see SI Appendix, Fig. S3). No apparent difference was reported in the structure and orientation of chromophores of avGFP and S65T-avGFP (26), and therefore the T57S mutant is supposed to have the same structure as that of the original POLArISact.
Remarkably, the 310 helix at the C terminus of cpGFP was reorganized to an α-helix (Fig. 1 B and G). The new α-helix was continuously linked to the N-terminal α-helix of Ad-A to form a long α-helix, thereby achieving the constrained tagging. Ligand-binding loops (282SSVPHWWWT290 and 316RDPNMIFKI324 in Protein Data Bank ID: 7C03) and cpGFP were distantly located, and therefore linked cpGFP is unlikely to interfere with the binding of Ad-A to actin filaments. The longitudinal axis of the chromophore was positioned almost perpendicularly to the long axis of the connecting α-helix (Fig. 1 G, Inset).
POLArISact Preferentially Binds to F-Actin In Vitro and in Living Cells.
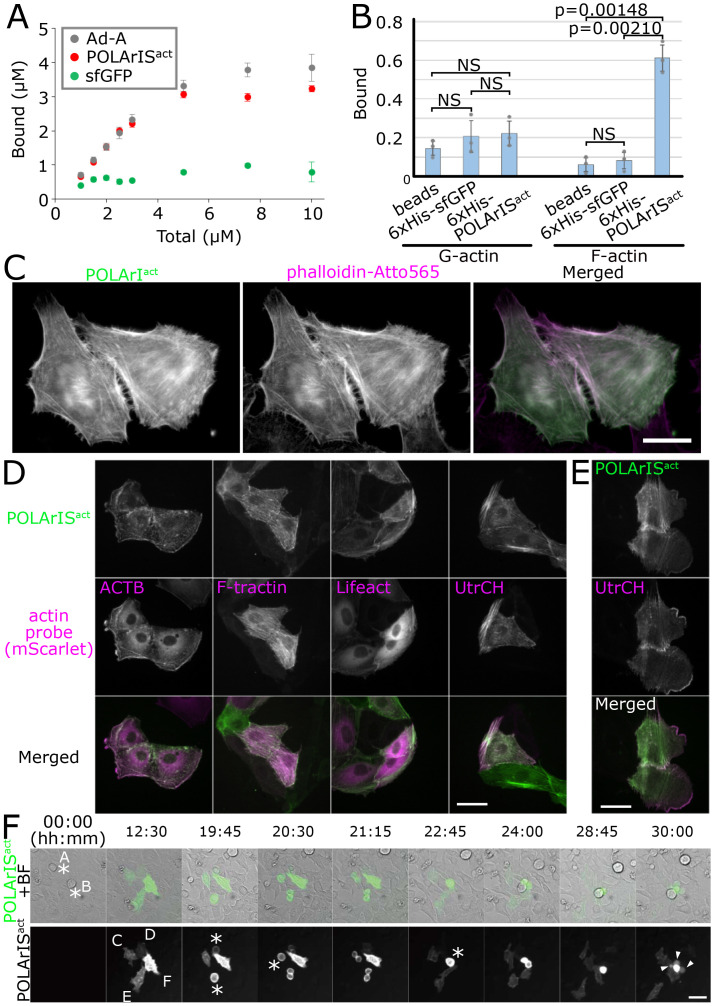
To investigate the binding property of POLArISact for F-actin, we performed the cosedimentation assay with F-actin. We found that POLArISact binds to F-actin with about 1:1 stoichiometry and with a dissociation constant (Kd) of 0.32 ± 0.18 µM (Fig. 2A and SI Appendix, Fig. S4A). Pull-down experiments indicated that POLArISact has a clear preference for F-actin to G-actin, as its binding to G-actin was undetectable in the assay (Fig. 2B and SI Appendix, Fig. S4B).
Fig. 2.
Properties of POLArISact as an F-actin probe. (A) Plots of the mean values in cosedimentation assays with F-actin were shown for Ad-A (gray), sfGFP (green), or POLArISact (red) with error bars showing SDs. Data were from three independent experiments. (B) The mean values of the bound fraction in pull-down assays were shown for G- or F-actin with the error bars showing SDs. P values were calculated by applying Šidák multiple comparison correction to Student’s t test. Data were from three independent experiments. Gray circles indicate the values of individual experiments. (C) High-resolution images of fixed HeLa cells expressing POLArISact stained with phalloidin-Atto565. (Scale bar: 20 µm.) (D and E) Comparison of the localization of POLArISact with ACTB and other F-actin probes tagged with mScarlet in living LLC-PK1 cells. Compared with ACTB, F-tractin, and Lifeact, POLArISact showed a lower cytoplasmic signal (D). Localization of POLArISact was similar to that of UtrCH (D), with a relatively lower affinity to F-actin in lamellipoidia (E). (Scale bars: 50 µm.) (F) Live-cell time lapse-imaging of HeLa cells transfected with POLArISact plasmid. Cells A and B in mitosis at time 0 (soon after transfection) divided into cells C and D and E and F, respectively, and then expressed POLArISact (time 12:30). The order of expression level was: F > E > C > D. While cells C, D, and E divided successfully, cell F failed to divide and died forming blebs (time 30:00). Asterisks indicate cells in mitosis; arrowheads indicate blebs. (Scale bar: 50 µm.)
The intracellular localization of POLArISact was reinvestigated by costaining with phalloidin-Atto565 in fixed cells using a high-magnification objective lens instead of the low-magnification lens we had used for the screening (SI Appendix, Fig. S1D). The striking resemblance between their localization (Fig. 2C) proved the specific binding of POLArISact to F-actin in cells.
Next, we expressed POLArISact in living LLC-PK1 cells and compared its localization with exogenously expressed actin (ACTB), Lifeact, UtrCH, or F-tractin. The distribution of POLArISact was most similar to that of UtrCH (Fig. 2D). POLArISact showed a clearly lower cytoplasmic/nuclear background than ACTB, Lifeact, or F-tractin (Fig. 2D). As far as we have tested, POLArISact labels all F-actin structures that can be labeled by other F-actin probes, except F-actin networks in lamellipodia with a moderate affinity (Fig. 2E). These results demonstrate that POLArISact can be used as an excellent F-actin marker in living cells. It is worth noting that all cell lines we tested, including HeLa, HEK293, LLC-PK1, and Jurkat, grew as normally as the cells without expressing POLArISact unless POLArISact was overexpressed (Fig. 2F shows an example of HeLa).
Fluorescence Polarization Analysis of F-Actin in Living Starfish Oocytes by Using POLArISact.
We observed the fluorescence polarization of POLArISact in living starfish oocytes to test if the probe reveals assembly dynamics of actin in the physiological condition of living cells. Drastic and dynamic changes in F-actin organization have been reported during maturation and fertilization of starfish oocytes (27–32). We expressed POLArISact in starfish oocytes by microinjection of messenger RNA (mRNA) and monitored their early development processes with two-axis FPM.
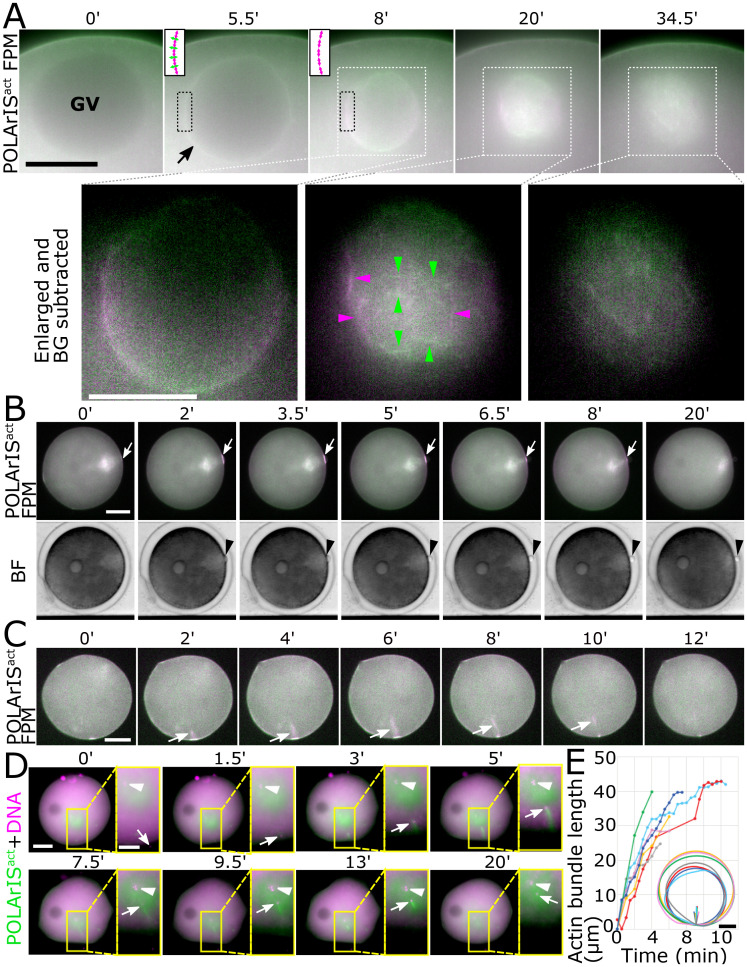
First, we focused on the F-actin dynamics during the breakdown of the germinal vesicle (GV) (Fig. 3A and Movie S2). Before GV breakdown, almost no fluorescence of POLArISact was observed on the GV membrane (Fig. 3A, 0 min). GV breakdown started ∼30 min after 1-methyladenine treatment. POLArISact-labeled F-actin abruptly appeared on the GV membrane with no apparent fluorescence polarization (5.5 min). When the GV started to shrink, we observed the fluorescence polarization signal of POLArISact as shown in the pseudocolor images of the GV membrane with green color in top/bottom regions and magenta in right/left (8 min). The transient accumulation of F-actin on the nucleoplasmic side of the GV membrane was previously observed during the initial phase of the GV breakdown called F-actin shell (27, 31). F-actin shell was reported to have two components: actin filaments running parallel to the GV membrane, forming a “base” of the shell, and filopodia-like spikes that protrude perpendicularly to the GV membrane from the base (31). Actin filaments running in the optical plane mainly contribute to the ensemble fluorescence polarization detected by FPM. Thus, the fluorescence of POLArISact bound to actin filaments of the base is supposed to have the polarization orientation parallel to the GV membrane, while the orientation of fluorescence polarization from spikes is supposed to be vertical to the GV membrane (schematics in insets in the top row, 5.5 and 8 min). The apparent absence of fluorescence polarization (5.5 min) during the initial phase of the GV breakdown might be caused by the cancellation of fluorescence polarization originated from POLArISact bound to actin filaments in the F-actin shell (inset, 5.5 min), as the base and spikes were observed as distinct structures only with high-resolution imaging such as confocal laser scanning microscopy (CLSM) (27). The green color in top/bottom regions of GV and magenta in right/left (8 min) indicates the strong fluorescence polarization of the structures, suggesting that basal network is the dominant component of F-actin network stained with POLArISact (inset, 8 min). This is consistent with the previous reports (27, 31) showing that the actin spikes were transient structures.
Fig. 3.
FPM observation of GV breakdown and PB extrusion and fertilization of starfish oocytes expressing POLArISact. (A) Time-lapse images of two-axis FPM of starfish oocytes expressing POLArISact during GV breakdown. Time 0 is just before the breakdown, ∼30 min after 1-MA addition. Regions marked by white dashed line boxes are magnified in the Bottom (enlarged). An arrow indicates the “F-actin shell”. Arrowheads in Bottom row indicate actin filaments with clear polarization in the meshwork (green: horizontal, magenta: vertical). Two-way arrows in insets in the top row schematically indicate supposed orientations of fluorescence polarization of POLArISact bound to local F-actin structures (magenta, the base of the shell; green, spikes) in regions marked by black dashed line boxes. In Bottom, the background signal was subtracted for the better presentation of actin meshwork. (B) Time-lapse images of starfish oocytes expressing POLArISact during the PB extrusion with two-axis FPM (Top) and bright-field view (Bottom). Arrowheads indicate the PB and arrows indicate contractile ring-like F-actin, respectively. (C) Time-lapse two-axis FPM images of a starfish oocyte expressing POLArISact during fertilization. Arrows indicate actin bundles associated with fertilization. (Scale bars: 25 µm (A), 50 µm (B and C).) (D) Time-lapse fluorescence microscopy images of starfish oocyte expressing POLArISact (green) with Hoechst staining (magenta) during fertilization. Areas enclosed by yellow boxes in the Left are magnified in the Right. In some Right, the background signal was subtracted for better visibility. After fertilization (1.5 min), sperm DNA (arrows) was positioned at the tip of the actin bundle until it stopped moving (3 to 13 min). The actin bundle was gradually depolymerized (9.5 to 13 min) and finally disappeared (20 min). Interestingly, after the extension of the actin bundle, the sperm DNA and the egg DNA (arrowheads) were closely apposed (9.5 to 20 min), as if the sperm DNA was conveyed to the egg DNA by the extension of the actin bundle. (Scale bars: 50 µm (Left), 25 µm (Right).) (E) Plots of the length of actin bundles over time in different colors for each oocyte (n = 8). Actin bundles typically extended perpendicularly to the cell surface and reached about 40 μm deep. The schematic shows the trajectory of the actin bundle in each egg and the outline of the same egg, with the same color as that of the plot graph. (Scale bar: 50 µm.)
Subsequently to the disappearance of the F-actin shell, a meshwork-like F-actin network (30, 32) was observed with fluorescence polarization of POLArISact (Fig. 3A, 20 min, arrowheads). The fluorescence signal was gradually weakened with time (34.5 min).
Following the process of GV breakdown, the polar body (PB) was extruded (Fig. 3B and Movie S3). POLArISact signal began to be enriched on the cortex near the animal pole (Fig. 3B, 0 to 2 min, arrow). The PB appeared at the animal pole immediately after the F-actin accumulation (2 min, arrowhead). POLArISact signal rapidly decreased while half of the PB appeared on the membrane (5 to 6.5 min) and disappeared as the PB extrusion completed (8 to 20 min). In Fig. 3B, the fluorescence polarization signal of POLArISact at the PB extrusion site was clearly observed as a magenta color by two-axis FPM, indicating that actin filaments are aligned parallel to the division plane. This is consistent with the proposed model of the contractile ring formation during the PB extrusion (28).
Next, we monitored the F-actin dynamics associated with fertilization (Fig. 3 C–E). We found that a bright POLArISact signal appeared on the cell surface where sperm entered (Fig. 3C, 0 min). From there, an actin-based structure grew into the egg (2 to 8 min), similarly to the previous observations associated with fertilization (29). The strong fluorescence polarization was detected with FPM, indicating that this structure was composed of bundles of well-aligned actin filaments. The tip of the bundle, typically reaching about 40 μm from the surface membrane (Fig. 3E), had been connected to the sperm nucleus in the cytoplasm until the nucleus stopped moving in the cytoplasm (Fig. 3D and Movie S4), which was reminiscent of actin-based motilities of bacterial pathogens (33). From these observations, we confirmed that our two-axis FPM setup using POLArISact can distinguish F-actin assemblies from randomly oriented F-actin accumulations in living starfish eggs and zygotes.
Formation of Radially Extended Actin Filaments during Mitosis in Zygotes and Blastomeres of Early Starfish Embryos.
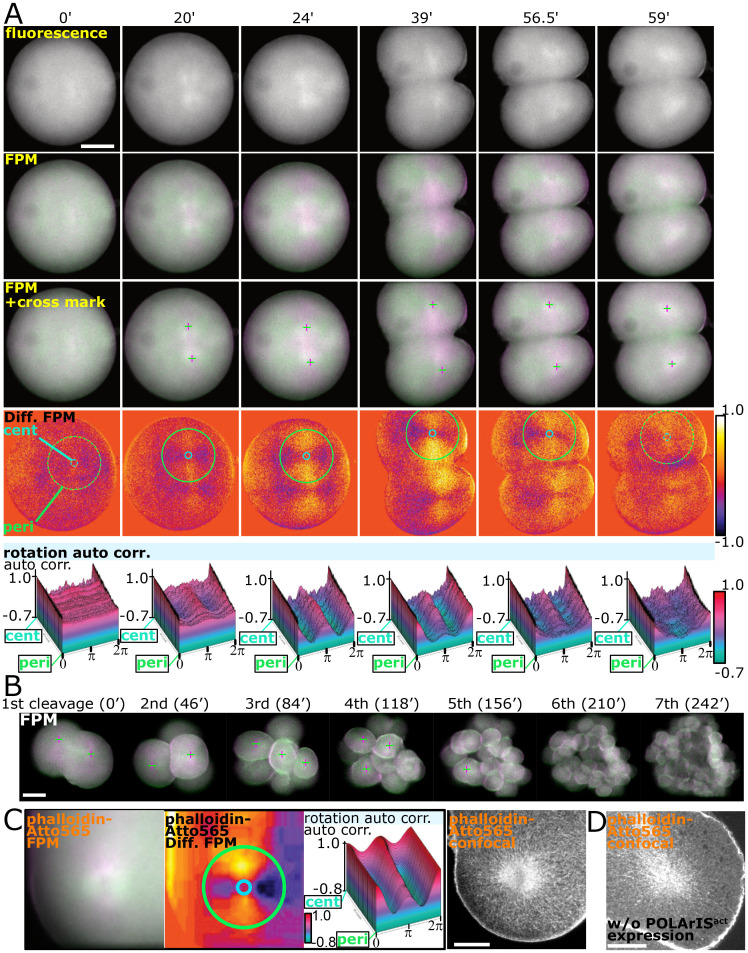
We next observed F-actin dynamics during early development of starfish embryos. Unexpectedly, we found the strong fluorescence polarization of POLArISact observed as the green (horizontal)/magenta (vertical) cross patterns appeared in pseudocolor images during the first cleavages (Fig. 4A, second and third rows, and Movies S5 and S6). The presence of the cross patterns was further examined by using the differential images of horizontal/vertical polarization (Fig. 4A, fourth row). Two small cross patterns appeared near the center of the zygote (Fig. 4A, 20 min) and rapidly expanded and reached the cell surface before the ingression occurred (24 min). The cross patterns persisted during the cleavage (39 to 56.5 min) and disappeared when the cleavage finished (59 min). We also observed similar structures in the blastomeres repeatedly in cleavages after the two-cell stage (Fig. 4B and Movies S7 and S8). Of note, the cross pattern also appeared using UG3, the probe of F-actin for FPM that we recently reported (SI Appendix, Fig. S9 B and C). However, the pattern was not distinct, probably due to UG3’s low fluorescence polarization (9).
Fig. 4.
Organization and dynamics of FLARE during starfish embryo cleavages. (A) Time-lapse images during the first cleavage of a starfish embryo expressing POLArISact with fluorescence microscopy (the top row) and two-axis FPM (green/magenta in second and third rows and differential in the fourth row). Cross marks in the third row indicate the centers of the green/magenta cross patterns. The bottom row shows the 3D plots of the rotational autocorrelation analyses of green/magenta cross patterns. Light blue circles (annotated as cent for center) and light green circles (annotated as peri for peripheral) in differential images indicate circles of minimum and maximum diameters used in the autocorrelation analyses. Z-axis shows the normalized autocorrelation. (Scale bar: 50 µm.) (B) Time-lapse two-axis green/magenta FPM observation of a starfish embryo expressing POLArISact from the first to the seventh cleavage. FLARE could not be recognized after the fifth cleavage because of the out-of-focus fluorescence from blastomeres that were overlapped with in-focus blastomeres in the z-axis direction. Time 0 was set arbitrarily to the frame in which FLAREs were clearly visible during the first cleavage. (Scale bar: 50 µm.) (C) Green/magenta (Left), differential (Middle Left, with circles of min/max diameters used in autocorrelation analyses), 3D plot of rotational autocorrelation analysis (Middle Right) of two-axis FPM, and a CLSM (Right) image of phalloidin-Atto565 staining of a glyoxal-fixed starfish embryo expressing POLArISact in the second cleavage. Images of FPM and CLSM were taken from the same region with almost the same z-axis planes of the same embryo. (Scale bar: 25 µm.) (D) CLSM image of phalloidin-Atto565 staining in a noninjected embryo during the third cleavage. Regardless of POLArISact expression, actin filaments of FLARE were observed in fixed starfish embryos. (Scale bar: 25 µm.)
For the quantitative analysis of the changes in the local distribution of the cross patterns, we deduced the center of the cross pattern from the differential polarization image by image analysis, set concentric circular shells around the deduced center of the cross pattern, and plotted the autocorrelation as the circular shell is rotated around the center (see SI Appendix, SI Materials and Methods for details). A cross pattern should result in high autocorrelation when rotated by π (peak) and low autocorrelation when rotated by π/2 or 3π/2 (troughs). The amplitude (the difference between the peak and the troughs) becomes higher when the cross pattern is more evident. The analyses clearly showed the dynamic appearance and disappearance of cross patterns during cleavages (Fig. 4A, bottom row, indicated with “auto-corr”).
The observation above implies the appearance and disappearance of actin filaments radially aligned from two centers in a wide 3D range during the cleavages. To confirm the presence of radially aligned actin filaments, we observed fixed embryos with conventional CLSM. As fixatives based on paraformaldehyde (PFA) or PFA in combination with glutaraldehyde (GA) (SI Appendix, SI Materials and Methods) failed to preserve these F-actin structures in dividing starfish embryos (SI Appendix, Fig. S5), we used the recently reported glyoxal fixation (34). Although fluorescence of POLArISact was diminished by glyoxal fixation, two-axis FPM observation of phalloidin-Atto565 staining in a fixed POLArISact-expressing embryo showed the characteristic cross pattern as we had observed in live starfish embryos with POLArISact (Fig. 4C, Left three panels). Using CLSM, radially extending actin filaments were certainly visible in the same embryo (Fig. 4C, Right). The observation of the similar radial distribution of F-actin in embryos without injection of POLArISact mRNA (Fig. 4D) excluded the possibility of artifacts caused by the expression of POLArISact.
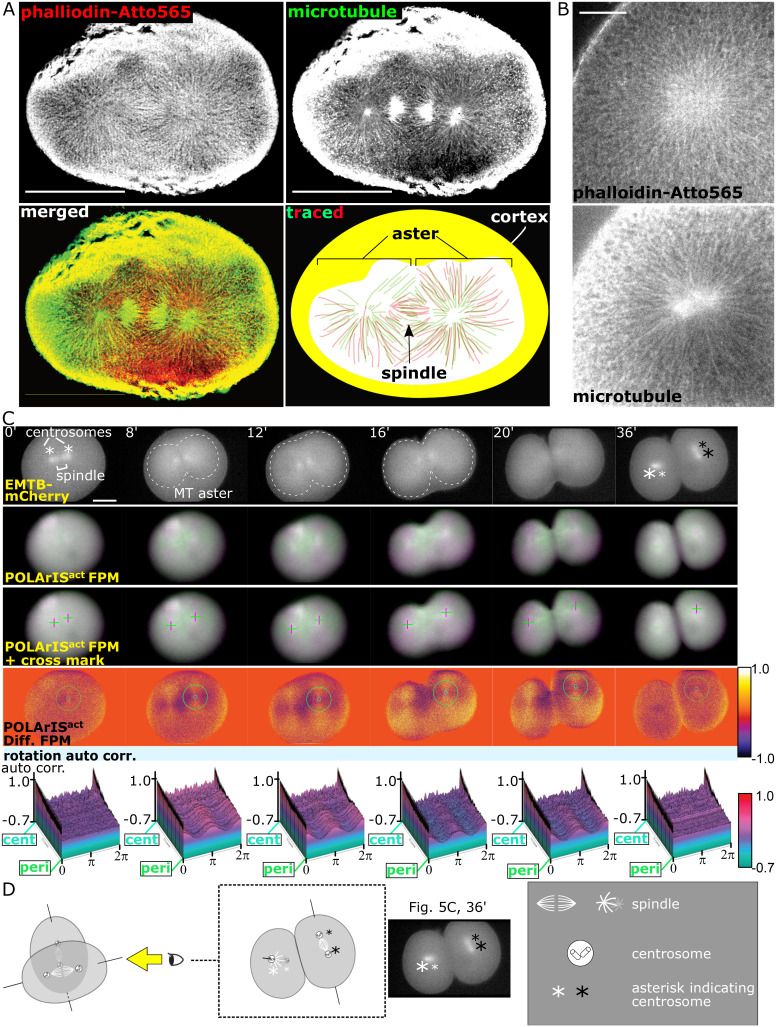
We next compared the distribution of F-actin and microtubules in a fixed starfish embryo expressing POLArISact during the first cleavage (Fig. 5A). CLSM observation revealed numerous actin filaments extended from two points. Staining microtubules with α-tubulin antibody indicated that these two points were centrosomes. Similar distributions of F-actin and microtubules were also observed during the third cleavage (Fig. 5B). Compared with microtubule-based asters, F-actin staining seemed to have a more intricate that is, “fluffy” texture. From these observations, we named this structure as FLARE (FLuffy And Radial actin-aster associated with mitosis in Embryo). To our best knowledge, no report has demonstrated the presence of this structure made of F-actin in vivo.
Fig. 5.
Comparisons of the distribution and dynamics of FLARE and microtubule asters. (A and B) CLSM images of F-actin (phalloidin-Atto565) and microtubules (anti-α-tubulin staining) in glyoxal-fixed starfish embryos expressing POLArISact in the first (A) and the third (B) cleavages. In the schematic, filaments of actin (red) and microtubule (green) were manually traced from microscopic pictures (A, Bottom Right). (Scale bars: 100 µm (A), 20 µm (B).) (C) Time-lapse green/magenta (second and third rows) and differential (fourth row, with circles of min/max diameters in autocorrelation analyses) two-axis FPM images of the first cleavage of a starfish embryo expressing POLArISact and EMTB-mCherry. The top row shows vertical polarization images of EMTB-mCherry. Cross marks (third row) indicate centers of green/magenta cross patterns. The bottom row shows 3D plots of autocorrelation analyses of FLARE. Centrosomes are shown by asterisks (see D for details). Contours of microtubule asters are shown by dashed lines. In POLArISact images, the background signal was subtracted for a better presentation of cross patterns. (Scale bar: 50 µm.) (D) Schematic of the arrangement of spindles and centrosomes in the panel of C, 36 min. Larger asterisks indicate centrosomes nearer the objective lens. Asterisks with the same color indicate centrosomes in the same blastomere.
Dynamics of FLARE Is Tightly Associated with that of Microtubule Aster.
We next carried out live-cell observation of FLARE with microtubules visualized with mCherry-tagged Enconsin microtubule-binding domain (EMTB-mCherry) (35) (Fig. 5C and Movies S9 and S10). With EMTB-mCherry, microtubule asters were observed as widespread staining of mCherry rather than radially aligned microtubules (Fig. 5C, contours of asters were indicated by white dashed lines). FLARE and microtubule aster appeared at the same time, expanded at a similar speed, and disappeared simultaneously. Before cleavage initiation, EMTB-mCherry fluorescence was concentrated around two points corresponding to centrosomes (indicated by asterisks, see Fig. 5D for details). EMTB-mCherry fluorescence was also detected between centrosomes, which corresponds to a mitotic spindle. Very weak and tiny cross patterns of FLARE were observed around centrosomes (Fig. 5C, 0 min). Concentrated fluorescence of EMTB-mCherry gradually became faint and spread as the cross patterns of FLARE expanded (8 min). The front of the expanding EMTB-mCherry fluorescence and the cross pattern of FLARE reached cell surface at 12 min and stayed during the cleavage (16 to 20 min). When the division finished, mCherry fluorescence was localized only at two centers, indicating that astral microtubules were depolymerized (36 min). Similarly, the cross patterns of FLARE became very weak and small at this time point. These synchronized dynamics of microtubules and F-actin were also observed in the later cleavages of the same embryo and all observed embryos coexpressing POLArISact and EMTB-mCherry that we observed (SI Appendix, Fig. S6).
Dynamics and Maintenance of FLARE Are Microtubule Dependent.
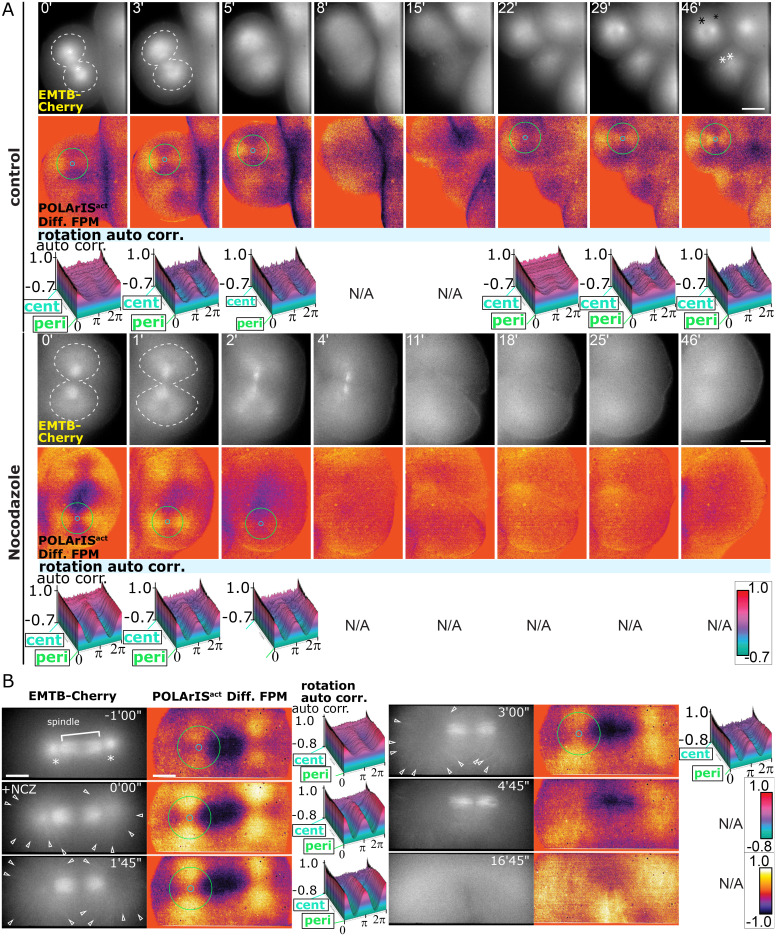
As described above, FLARE is very similar to microtubule asters in their dynamics and the distribution. To test possible interactions between FLARE and microtubule aster, we carried out pharmacological experiments. First, we used cytochalasin D, the actin polymerization inhibitor (SI Appendix, Fig. S7). Treatment of a starfish embryo with cytochalasin D resulted in the loss of FLARE formation, while microtubule aster still showed the repeated dis- and reappearance, indicating that microtubule aster dynamics is not dependent on FLARE.
Next, to examine if the microtubule aster plays a role in the formation and/or the maintenance of FLARE, we treated starfish embryos with microtubule polymerization inhibitor nocodazole (NCZ) at different time points. When NCZ was added to the embryos immediately after microtubule asters started to extend, both microtubule asters and FLARE stopped expanding (Fig. 6A, NCZ, 0 to 1 min). EMTB-mCherry fluorescence and FLARE polarization gradually disappeared (2 to 11 min), and the embryo failed to complete the cleavage (18 to 46 min). Thereafter, cleavages did not occur, and neither the microtubule aster nor FLARE reappeared in the same embryo. These observations indicate that the expansion of the microtubule aster is a prerequisite for the FLARE formation. Next, we treated an embryo with NCZ after microtubules sufficiently extended throughout the cytoplasm (Fig. 6B and Movie S11). In this case, microtubule aster was depolymerized outwardly from its center, and the fluorescence polarization of FLARE disappeared simultaneously (Fig. 6B, 1′45″ to 4′45″), indicating that the maintenance of FLARE requires the integrity of astral microtubules. These results demonstrate that both the formation and the maintenance of FLARE depend on the dynamics of microtubules and the integrity of the microtubule aster.
Fig. 6.
FLARE dynamics is disrupted by inhibiting polymerization of microtubules. (A and B) Time-lapse two-axis FPM observation of dimethyl sulfoxide (control) or nocodazole-treated embryos expressing POLArISact and EMTB-mCherry in the fourth (A) and the third cleavages (B). (A) Top rows show vertical polarization images of EMTB-mCherry (asterisks indicate centrosomes, dashed lines indicate contours of microtubule asters), middle rows show differential FPM images of POLArISact with circles of min/max diameters used in autocorrelation analyses, and bottom rows show 3D plots of autocorrelation analyses of FLARE. N/A means that no center of the cross pattern was identified in our search algorithm. The drug was added at time 0. (Scale bar: 50 µm.) (B) Left columns show vertical polarization images of EMTB-mCherry (asterisks indicate centrosomes), middle columns show differential FPM images with circles of min/max diameters used in autocorrelation analyses, and right columns show 3D plots of autocorrelation analyses of FLARE structures. N/A means that no center of the cross pattern was identified in our search algorithm. The drug was added after at least one astral microtubule reached the cell surface (time 0). A silicone immersion 40× objective lens and a 1.5× intermediate magnification lens were used to visualize microtubule filaments (arrowheads). (Scale bar: 25 µm.)
FLARE Formation Is Not Arp2/3 Complex Dependent.
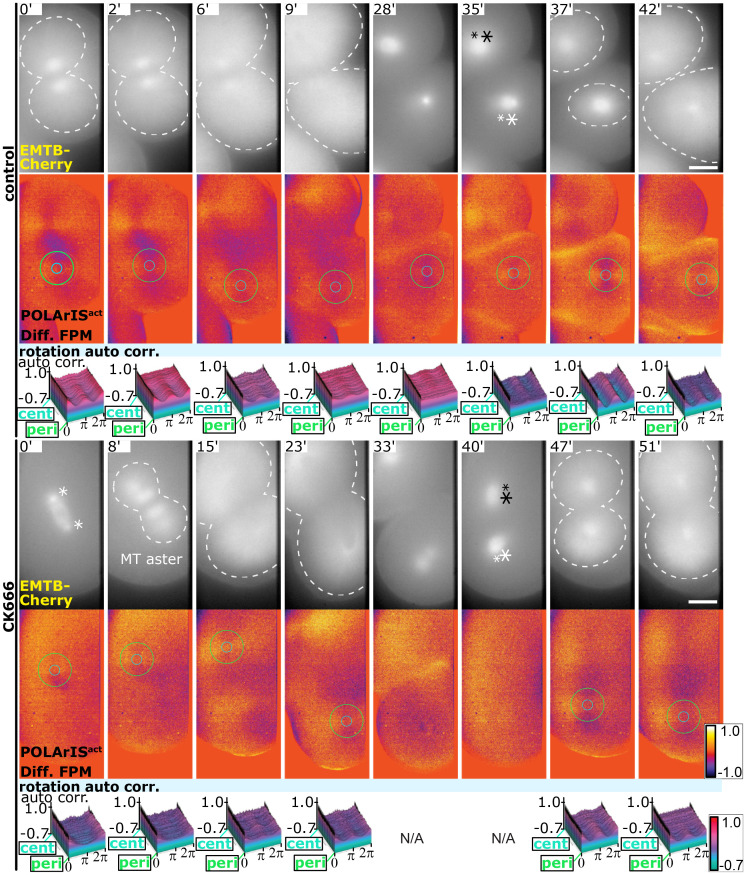
Arp2/3 protein complex and formin family proteins are known as major actin nucleation factors (36, 37). We attempted to determine which factor is responsible for the formation of FLARE by pharmacological inhibition of the activity of Arp2/3 complex and formins with CK666 (38) and SMIFH2 (39), respectively. Since SMIFH2 turned out to be impermeable to fertilization envelopes (SI Appendix, Fig. S8A), we microinjected SMIFH2 under the elevated fertilization envelopes and perfused SMIFH2 directly to embryos (SI Appendix, Fig. S8B). We unexpectedly found that not only actin but also microtubules lost their dynamics (10 to 30 min), and embryos failed to divide (40 to 90 min) in the presence of SMIFH2 (SI Appendix, Fig. S8B). This implies that SMIFH2 might have nonspecific side effects in starfish embryos in addition to the specific inhibition of formin, so we could not determine if formin family proteins are involved in the formation of the FLARE in our preparations.
When embryos were treated with CK666 immediately after the first cleavage (Fig. 7), subsequent cleavages did not occur, but microtubule aster showed repeated disappearance and reappearance, confirming that CK666 specifically inhibited Arp2/3-dependent actin nucleation and thereby prevented embryos from dividing without affecting microtubule dynamics. FLARE also appeared and disappeared repeatedly, indicating that Arp2/3 is not essential for the FLARE formation.
Fig. 7.
FLARE formation and dynamics are independent of the actin nucleation by Arp2/3. Time-lapse two-axis FPM observations of dimethyl sulfoxide (control) or CK666-treated starfish embryos expressing POLArISact and EMTB-mCherry. Drugs were added immediately after the first cleavage completed (time 0). Top rows show vertical polarization images of EMTB-mCherry (asterisks indicate centrosomes, dashed lines indicate contours of microtubule asters), middle rows show differential FPM images of POLArISact with circles of min/max diameters used in autocorrelation analyses, and bottom rows show 3D plots of autocorrelation analyses of FLARE. N/A means that no center of the cross pattern was identified in our search algorithm. (Scale bar: 25 µm.)
Discussion
Our previous studies on the cytoskeletal dynamics (40, 41) have led us to focus on fluorescence polarization techniques, as the polarized fluorescence microscopy might detect the initial phase of polymerization even when the polymerized cytoskeletal protein molecules cannot be detected as filaments. Through the years of our trials to use FPM to analyze the dynamics of cytoskeletal proteins, we noticed that the biggest challenge was the difficulty in tagging each protein of interest with a fluorescent protein in a rotationally constrained manner. Designing more versatile and affordable methods were required for the constrained tagging of any biomolecules of interests with fluorescent labels. We have achieved this task by using a small recombinant binder protein Adhiron/affimer as a scaffold to connect target molecules with circularly permutated sfGFP. As described in this report, we established a versatile tool, POLArIS, taking advantage of Adhiron/affimer that can be screened through phage display for any specific target molecules of interest. There are many Adhirons/affimers reported, targeting a variety of specific biomolecules (19, 20, 42–48) from laboratories, and the production service is commercially available. The two-axis fluorescence polarization imaging is a robust system for mapping fluorescence anisotropy with the aid of commonly used software for image analysis (NIS-Elements Advanced Research from Nikon and ImageJ). We expect that POLArIS will help a broader range of biomedical researchers to use fluorescence polarization techniques without any difficulties in the constrained tagging not only for proteins but also for nucleic acids or lipids.
Here, by using POLArISact as the first POLArIS example of POLArIS series, we show that POLArISact can report the orientation and localization of F-actin in living cells. The observations of starfish eggs and embryos expressing POLArISact revealed a previously unidentified highly ordered F-actin–based architecture, FLARE. We did not find such ordered structures with conventional fluorescence microscopy, but with POLArISact, radially orientated actin bundles extending from centrosomes were visible by their polarized fluorescence. This is reminiscent of Inoué’s first visualization of mitotic spindle dynamics in living cells using a polarizing microscope in the late 1940s, which could not have been achieved with conventional microscopic techniques for living cells (49, 50). While the association of actin filaments with spindle microtubules have been reported in several mammalian oocytes (51–53) and Xenopus embryonic epithelial cells (54, 55), to our best knowledge, the presence of radially aligned actin filaments that extend toward the cortex in association with growing astral microtubules in living cells has not been reported previously. Besides the use of a fluorescence polarization technique, one of the most plausible reasons that FLARE has been left unidentified until today was the effect of fixatives. We found that glyoxal is one of the best fixatives for actin filaments in FLARE. Indeed, immunostaining of glyoxal-fixed neurons with anti-β-actin antibody showed a significantly brighter staining than that of PFA-fixed neurons (34). Recently, in Xenopus egg extract, the formation of radially aligned actin filaments around centrosomes were observed (56). It would be interesting to see if actin filaments form FLARE-like structures in eggs/embryos of Xenopus and other species during cleavages in vivo.
The actin filaments that extend from centrosomes toward cell cortex in mitosis may imply an actin-based physical connection between the cell cortex and centrosomes and/or the mitotic spindle. The mechanism of cytokinesis initiation, for example, how the cleavage plane is determined and how furrow ingression is induced, has been actively studied and is still one of the fundamental questions remain to be solved in cell biology. This is especially enigmatic in large cells such as eggs, since the mitotic spindle, which is believed to provide positional cues to induce contractile formation and furrowing, is far away from the cell cortex. While astral microtubules have been thought to have central roles in these pathways (57–59), roles of F-actin are currently known to be only at the cell cortex (60). Since the existence of actin filaments associated with astral microtubules has not been recognized until now, it would be fascinating to test if FLARE is involved in the signal transduction in cytokinesis initiation and/or other processes related to cytokinesis.
The mechanism of the FLARE formation is currently unknown. Centrosomes isolated from Jurkat cells formed radial F-actin array in vitro (61), which resembles FLARE, and actin filaments were found to be localized around centrosomes in HeLa and Jurkat cells (62, 63). Moreover, radially extended actin filaments from centrosomes were observed in HeLa cells during the forced exit of mitosis after prometaphase arrest with S-trityl-l-cysteine (62). Actin nucleation activity of centrosomes in these examples requires Arp2/3 complex. However, we found that FLARE formation was not affected by Arp2/3 inhibition. Thus, it is likely that FLARE is intrinsically different from these F-actin related structures around centrosomes in cultured human cells.
Some of fluorescently labeled phalloidin probes (7) and SiR-actin (64) can be used to monitor the fluorescence polarization of F-actin. Their live-cell applications are limited to specific biological preparations where these organic compounds can be delivered to cytoplasm (for example, isolated cells and monolayered culture cells). Fluorescently labeled phalloidin has been commonly used as an F-actin probe, but its strong F-actin stabilizing effect is not suitable for physiological studies in living cells where the activity of actin assembly/disassembly is essential. The successful application of POLArISact to study the dynamics of the actin cytoskeleton during early development of starfish eggs and embryos strongly suggests that POLArISact is a promising alternative to fluorescent phalloidin for F-actin studies in living cells.
We compared POLArISact with other widely used F-actin probes to find its excellence. In fixed cells, the localization of POLArISact was indistinguishable from that of phalloidin staining, which is currently thought to be the most reliable marker for F-actin. When observed in living cells, the cytoplasmic background signal from POLArISact was apparently lower than that of Lifeact or F-tractin and as low as that of UtrCH. We recently reported UtrCH-based F-actin probes for FPM, namely UG3 and UG7, can be used for orientation monitoring of F-actin with FPM (9). Similarly to POLArISact, UG3 and UG7 are genetically encoded and the orientations of their fluorescence polarization are both parallel to the actin filament, but their polarization factor in living cells (<0.1) (9) was substantially lower than that of POLArISact (∼0.3, SI Appendix, Fig. S2C). We expressed UG3 in starfish oocytes and found that the fluorescence polarization on the actin meshwork upon GV breakdown was substantially weaker than that of POLArISact (SI Appendix, Fig. S9A). In addition, it was very difficult to recognize FLARE with FPM during cleavages by using UG3 (SI Appendix, Fig. S9 B and C), probably due to UG3’s low fluorescence polarization. Together, these results demonstrate that at present, POLArISact is the best tool for monitoring F-actin dynamics with FPM.
Using recent super-resolution techniques such as stimulated emission depletion (STED) and localization microscopy techniques (e.g., photoactivated localization microscopy/stochastic optical reconstruction microscopy), we can observe single actin filaments, and therefore their orientations are observed even in a dense actin network. However, the time resolution of STED is supposed to be sometimes too low to follow the dynamic changes in actin organization when observing large objects, for example, whole cross sections of starfish eggs. Phototoxicity and photobleaching caused by STED imaging will also be problematic for long-period time-lapse imaging. The time resolution of localization microscopy techniques is low, and single-molecule imaging of this kind is very difficult when observing deep structures in large specimens. Therefore, we consider that POLArISact is useful for orientation imaging of actin filaments in many situations.
Since polarization is one of the most fundamental properties of light, POLArIS is expected to open a new avenue to various fluorescent spectroscopic applications such as, but not limited to, high throughput screening (65), super-resolution (13–16), resonance energy transfer (66, 67), and fluorescence correlation (68, 69). By taking advantage of the genetically encoded nature, POLArIS has the potential of supporting broader applications of fluorescence polarization techniques to a variety of cell types, species, and many types of specimens such as developing embryos and whole bodies of animals. Furthermore, POLArIS can be expressed in a cell type/tissue-specific manner when combined with specific promoters.
Materials and Methods
Experimental materials and method details for cell culture, starfish egg preparation and drug treatment, plasmid construction, protein purification, crystallization and structure determination, structural modeling, pull-down assay, cosedimentation assay, plasmid introduction into cells, fixation and staining of cultured cells and starfish eggs/embryos, in vitro F-actin labeling, fluorescence microscopy, FPM, and data analysis are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to N. Nakai (TMDU) and H.T. Miyazaki (National Institute for Materials Science) for helpful discussion, O. Hoshi (TMDU) for continuous encouragement, and M. Taguchi (TMDU) for secretarial and experimental assistance. We deeply thank H. Iwasaki (Gunma University) for the suggestion of making the initial opportunity of the collaborative research using starfish oocytes, R. Nitta and M. Aoki (RIKEN) for discussion and initial trial for the crystallization of POLArISact, K. Hanada and M. Inoue (RIKEN) for plasmid preparation for the expression of POLArISact protein in bacterial cells, and T. M. Watanabe (RIKEN) for discussion and initial assistance for FPM construction. We are grateful to the late Dr. Shinya Inoué, the father of polarized light microscopy, for his great support and his pioneering work of polarized fluorescence in green fluorescent protein crystals. For funding, we are grateful to the Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research (KAKENHI) (C) and (B) (Grants 18K06819 to K. Sato, 17K07405 to K.C., and 17H04013 and 26293038 to S.T.); Fostering Joint International Research (B) (Grant 18KK0222 to S.T.); JSPS Bilateral Joint Research Projects (Open Partnership Joint Research Projects) (Grant 1006285 to S.T.); JSPS Home-Returning Researcher Development Research (Grant 18K19962 to T.T.); Takeda Science Foundation (to K.C.); NIH (Grant R01 GM100160 to T.T.); Marine Biological Laboratory Telfer Fund (to T.T.); and Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research) from AMED (Grant JP19am0101082 to M.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
*A. Lopata et al., Biophysical Society 60th Annual Meeting, February 27–March 2, 2016, Los Angeles, CA.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019071118/-/DCSupplemental.
Data Availability
All data are included in the manuscript and supporting information. Plasmids for POLArISact expression, cpGFP-Ad-A flex expression, and mRNA production of POLArISact are available from Addgene (164971 at https://www.addgene.org/164971/, 165044 at https://www.addgene.org/165044/, and 164970 at https://www.addgene.org/164970/, respectively). The ImageJ plugin for FLARE analysis can be found in Gitlab at https://gitlab.com/mkwnana/s4firp.
Change History
May 31, 2022: Figures 1–7 have been updated.
References
- 1.Ohmachi M., et al., Fluorescence microscopy for simultaneous observation of 3D orientation and movement and its application to quantum rod-tagged myosin V. Proc. Natl. Acad. Sci. U.S.A. 109, 5294–5298 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forkey J. N., Quinlan M. E., Shaw M. A., Corrie J. E. T., Goldman Y. E., Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization. Nature 422, 399–404 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Sosa H., Peterman E. J. G., Moerner W. E., Goldstein L. S. B., ADP-induced rocking of the kinesin motor domain revealed by single-molecule fluorescence polarization microscopy. Nat. Struct. Biol. 8, 540–544 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Nishizaka T., et al., Chemomechanical coupling in F1-ATPase revealed by simultaneous observation of nucleotide kinetics and rotation. Nat. Struct. Mol. Biol. 11, 142–148 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Swaminathan V., et al., Actin retrograde flow actively aligns and orients ligand-engaged integrins in focal adhesions. Proc. Natl. Acad. Sci. U.S.A. 114, 10648–10653 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nordenfelt P., et al., Direction of actin flow dictates integrin LFA-1 orientation during leukocyte migration. Nat. Commun. 8, 2047 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mehta S. B., et al., Dissection of molecular assembly dynamics by tracking orientation and position of single molecules in live cells. Proc. Natl. Acad. Sci. U.S.A. 113, E6352–E6361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kampmann M., Atkinson C. E., Mattheyses A. L., Simon S. M., Mapping the orientation of nuclear pore proteins in living cells with polarized fluorescence microscopy. Nat. Struct. Mol. Biol. 18, 643–649 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakai N., et al., Genetically encoded orientation probes for F-actin for fluorescence polarization microscopy. Microscopy (Oxf.) 68, 359–368 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vrabioiu A. M., Mitchison T. J., Structural insights into yeast septin organization from polarized fluorescence microscopy. Nature 443, 466–469 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Vrabioiu A. M., Mitchison T. J., Symmetry of septin hourglass and ring structures. J. Mol. Biol. 372, 37–49 (2007). [DOI] [PubMed] [Google Scholar]
- 12.DeMay B. S., et al., Septin filaments exhibit a dynamic, paired organization that is conserved from yeast to mammals. J. Cell Biol. 193, 1065–1081 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhanghao K., et al., Super-resolution dipole orientation mapping via polarization demodulation. Light Sci. Appl. 5, e16166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valades Cruz C. A., et al., Quantitative nanoscale imaging of orientational order in biological filaments by polarized superresolution microscopy. Proc. Natl. Acad. Sci. U.S.A. 113, E820–E828 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Backer A. S., Lee M. Y., Moerner W. E., Enhanced DNA imaging using super-resolution microscopy and simultaneous single-molecule orientation measurements. Optica 3, 3–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhanghao K., et al., Super-resolution imaging of fluorescent dipoles via polarized structured illumination microscopy. Nat. Commun. 10, 4694 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bedford R., et al., Alternative reagents to antibodies in imaging applications. Biophys. Rev. 9, 299–308 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiede C., et al., Adhiron: A stable and versatile peptide display scaffold for molecular recognition applications. Protein Eng. Des. Sel. 27, 145–155 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes D. J., et al., Generation of specific inhibitors of SUMO-1- and SUMO-2/3-mediated protein-protein interactions using Affimer (Adhiron) technology. Sci. Signal. 10, eaaj2005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopata A., et al., Affimer proteins for F-actin: Novel affinity reagents that label F-actin in live and fixed cells. Sci. Rep. 8, 6572 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riedl J., et al., Lifeact: A versatile marker to visualize F-actin. Nat. Methods 5, 605–607 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winder S. J., et al., Utrophin actin binding domain: Analysis of actin binding and cellular targeting. J. Cell Sci. 108, 63–71 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Arai R., Ueda H., Kitayama A., Kamiya N., Nagamune T., Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 14, 529–532 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Pettersen E. F., et al., UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Barondeau D. P., Kassmann C. J., Tainer J. A., Getzoff E. D., Understanding GFP posttranslational chemistry: Structures of designed variants that achieve backbone fragmentation, hydrolysis, and decarboxylation. J. Am. Chem. Soc. 128, 4685–4693 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Brejc K., et al., Structural basis for dual excitation and photoisomerization of the Aequorea victoria green fluorescent protein. Proc. Natl. Acad. Sci. U.S.A. 94, 2306–2311 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mori M., et al., An Arp2/3 nucleated F-actin shell fragments nuclear membranes at nuclear envelope breakdown in starfish oocytes. Curr. Biol. 24, 1421–1428 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Hamaguchi Y., Numata T., K Satoh S., Quantitative analysis of cortical actin filaments during polar body formation in starfish oocytes. Cell Struct. Funct. 32, 29–40 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Santella L., Limatola N., Vasilev F., Chun J. T., Maturation and fertilization of echinoderm eggs: Role of actin cytoskeleton dynamics. Biochem. Biophys. Res. Commun. 506, 361–371 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Mori M., et al., Intracellular transport by an anchored homogeneously contracting F-actin meshwork. Curr. Biol. 21, 606–611 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Wesolowska N., et al., Actin assembly ruptures the nuclear envelope by prying the lamina away from nuclear pores and nuclear membranes in starfish oocytes. eLife 9, e49774 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lénárt P., et al., A contractile nuclear actin network drives chromosome congression in oocytes. Nature 436, 812–818 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Lamason R. L., Welch M. D., Actin-based motility and cell-to-cell spread of bacterial pathogens. Curr. Opin. Microbiol. 35, 48–57 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richter K. N., et al., Glyoxal as an alternative fixative to formaldehyde in immunostaining and super-resolution microscopy. EMBO J. 37, 139–159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller A. L., Bement W. M., Regulation of cytokinesis by Rho GTPase flux. Nat. Cell Biol. 11, 71–77 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amann K. J., Pollard T. D., The Arp2/3 complex nucleates actin filament branches from the sides of pre-existing filaments. Nat. Cell Biol. 3, 306–310 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Pruyne D., et al., Role of formins in actin assembly: Nucleation and barbed-end association. Science 297, 612–615 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Nolen B. J., et al., Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature 460, 1031–1034 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rizvi S. A., et al., Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol. 16, 1158–1168 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terada S., Nakata T., Peterson A. C., Hirokawa N., Visualization of slow axonal transport in vivo. Science 273, 784–788 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Terada S., Kinjo M., Hirokawa N., Oligomeric tubulin in large transporting complex is transported via kinesin in squid giant axons. Cell 103, 141–155 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Kyle H. F., et al., Exploration of the HIF-1α/p300 interface using peptide and Adhiron phage display technologies. Mol. Biosyst. 11, 2738–2749 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Tiede C., et al., Affimer proteins are versatile and renewable affinity reagents. eLife 6, e24903 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kearney K. J., et al., Affimer proteins as a tool to modulate fibrinolysis, stabilize the blood clot, and reduce bleeding complications. Blood 133, 1233–1244 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hesketh E. L., et al., Affimer reagents as tools in diagnosing plant virus diseases. Sci. Rep. 9, 7524 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klont F., Hadderingh M., Horvatovich P., Ten Hacken N. H. T., Bischoff R., Affimers as an alternative to antibodies in an affinity LC-MS assay for quantification of the soluble receptor of advanced glycation end-products (sRAGE) in human serum. J. Proteome Res. 17, 2892–2899 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson J. I., et al., Affimer proteins inhibit immune complex binding to FcγRIIIa with high specificity through competitive and allosteric modes of action. Proc. Natl. Acad. Sci. U.S.A. 115, E72–E81 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michel M. A., Swatek K. N., Hospenthal M. K., Komander D., Ubiquitin linkage-specific affimers reveal insights into K6-linked ubiquitin signaling. Mol. Cell 68, 233–246.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inoué S., [Polarization optical studies of the mitotic spindle. I. The demonstration of spindle fibers in living cells]. Chromosoma 5, 487–500 (1953). [DOI] [PubMed] [Google Scholar]
- 50.Inoué S., Cell division and the mitotic spindle. J. Cell Biol. 91, 131s–147s (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Azoury J., et al., Spindle positioning in mouse oocytes relies on a dynamic meshwork of actin filaments. Curr. Biol. 18, 1514–1519 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Schuh M., Ellenberg J., A new model for asymmetric spindle positioning in mouse oocytes. Curr. Biol. 18, 1986–1992 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Mogessie B., Schuh M., Actin protects mammalian eggs against chromosome segregation errors. Science 357, eaal1647 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Woolner S., O’Brien L. L., Wiese C., Bement W. M., Myosin-10 and actin filaments are essential for mitotic spindle function. J. Cell Biol. 182, 77–88 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kita A. M., et al., Spindle-F-actin interactions in mitotic spindles in an intact vertebrate epithelium. Mol. Biol. Cell 30, 1645–1654 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Field C. M., Pelletier J. F., Mitchison T. J., Disassembly of actin and keratin networks by Aurora B kinase at the midplane of cleaving Xenopus laevis eggs. Curr. Biol. 29, 1999–2008.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rappaport R., Experiments concerning the cleavage stimulus in sand dollar eggs. J. Exp. Zool. 148, 81–89 (1961). [DOI] [PubMed] [Google Scholar]
- 58.Field C. M., Groen A. C., Nguyen P. A., Mitchison T. J., Spindle-to-cortex communication in cleaving, polyspermic Xenopus eggs. Mol. Biol. Cell 26, 3628–3640 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.von Dassow G., Concurrent cues for cytokinetic furrow induction in animal cells. Trends Cell Biol. 19, 165–173 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Woolner S., Bement W. M., Unconventional myosins acting unconventionally. Trends Cell Biol. 19, 245–252 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Farina F., et al., The centrosome is an actin-organizing centre. Nat. Cell Biol. 18, 65–75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farina F., et al., Local actin nucleation tunes centrosomal microtubule nucleation during passage through mitosis. EMBO J. 38, e99843 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Inoue D., et al., Actin filaments regulate microtubule growth at the centrosome. EMBO J. 38, e99630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spira F., et al., Cytokinesis in vertebrate cells initiates by contraction of an equatorial actomyosin network composed of randomly oriented filaments. eLife 6, e30867 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang X., Aulabaugh A., Application of fluorescence polarization in HTS assays. Methods Mol. Biol. 1439, 115–130 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Mattheyses A. L., Hoppe A. D., Axelrod D., Polarized fluorescence resonance energy transfer microscopy. Biophys. J. 87, 2787–2797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ojha N., Rainey K. H., Patterson G. H., Imaging of fluorescence anisotropy during photoswitching provides a simple readout for protein self-association. Nat. Commun. 11, 21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamamoto J., et al., Rotational diffusion measurements using polarization-dependent fluorescence correlation spectroscopy based on superconducting nanowire single-photon detector. Opt. Express 23, 32633–32642 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Oura M., et al., Polarization-dependent fluorescence correlation spectroscopy for studying structural properties of proteins in living cell. Sci. Rep. 6, 31091 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rosell F. I., Boxer S. G., Polarized absorption spectra of green fluorescent protein single crystals: Transition dipole moment directions. Biochemistry 42, 177–183 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the manuscript and supporting information. Plasmids for POLArISact expression, cpGFP-Ad-A flex expression, and mRNA production of POLArISact are available from Addgene (164971 at https://www.addgene.org/164971/, 165044 at https://www.addgene.org/165044/, and 164970 at https://www.addgene.org/164970/, respectively). The ImageJ plugin for FLARE analysis can be found in Gitlab at https://gitlab.com/mkwnana/s4firp.