Significance
A DNA double-strand break (DSB) can be repaired accurately by homologous recombination. The Mre11-Rad50-Nbs1 (MRN) complex is responsible for initiating homologous recombination by degrading 5′-ended DNA strand, where its activation by the Ctp1 cofactor plays a pivotal role. Here, by using purified fission yeast proteins, we show that two major elements comprise MRN activation. First, phosphorylation of Ctp1 promotes the physical interaction between MRN and Ctp1. Second, the C terminus of Ctp1 activates nucleolytic processing of DSB ends. In the latter case, a small peptide comprising only 15 amino acids from the Ctp1 C terminus is sufficient to activate MRN. Our results elucidate the core elements underlying MRN activation by Ctp1.
Keywords: Ctp1/CtIP, double-strand break repair, fission yeast, homologous recombination, Mre11-Rad50-Nbs1
Abstract
The Mre11-Rad50-Nbs1 complex (MRN) is important for repairing DNA double-strand breaks (DSBs) by homologous recombination (HR). The endonuclease activity of MRN is critical for resecting 5′-ended DNA strands at DSB ends, producing 3′-ended single-strand DNA, a prerequisite for HR. This endonuclease activity is stimulated by Ctp1, the Schizosaccharomyces pombe homolog of human CtIP. Here, with purified proteins, we show that Ctp1 phosphorylation stimulates MRN endonuclease activity by inducing the association of Ctp1 with Nbs1. The highly conserved extreme C terminus of Ctp1 is indispensable for MRN activation. Importantly, a polypeptide composed of the conserved 15 amino acids at the C terminus of Ctp1 (CT15) is sufficient to stimulate Mre11 endonuclease activity. Furthermore, the CT15 equivalent from CtIP can stimulate human MRE11 endonuclease activity, arguing for the generality of this stimulatory mechanism. Thus, we propose that Nbs1-mediated recruitment of CT15 plays a pivotal role in the activation of the Mre11 endonuclease by Ctp1/CtIP.
DNA double-strand breaks (DSBs) are potentially lethal lesions that threaten genomic integrity and cell viability. DSBs can occur spontaneously as a result of faulty DNA metabolism or by exposure to genotoxins. In eukaryotes, these DSBs have “dirty ends” that lack ligatable 3ʹ-hydroxyl/5ʹ-phosphate groups and are often firmly attached to proteins such as the Ku70-80 heterodimer and topoisomerases (1, 2). During meiosis, the topoisomerase-like Spo11 protein generates DSBs and remains covalently attached to the 5ʹ DNA ends (3). To enable further processing, these DSB ends must be converted to “clean” ends with 3ʹ-hydroxyl/5ʹ-phosphate groups properly exposed. This step is achieved by endonucleolytic cleavage, or clipping, by Mre11 (4–7).
In mammals, the MRE11, RAD50, and NBS1 complex (Mre11-Rad50-Nbs1 [MRN]), together with CtIP, is involved in the clipping reaction. MRE11 is the nuclease subunit that has both endonuclease and 3′-to-5′ exonuclease activities, but only the former is essential for clipping (4, 8–12). RAD50, a member of the Structural Maintenance of Chromosomes protein family, binds to MRE11 to form an (MRE11)2-(RAD50)2 ring structure (MR complex) (13–15). NBS1 binds to the MR complex via MRE11 to form the MRN complex (16). Homologs of CtIP include Ctp1 in Schizosaccharomyces pombe and Sae2 in Saccharomyces cerevisiae (17–19). Upon phosphorylation, these proteins physically interact with their cognate MRN complex via the N-terminal forkhead-associated domain of NBS1, leading to activation of the MRE11 endonucleolytic clipping activity (20–24). However, the mechanistic details underlying this activation have not yet been determined.
Through biochemical reconstitution using fission yeast proteins, we made three key findings regarding how Ctp1 activates MRN. First, MRN activation is mediated by Ctp1 phosphorylation, which promotes the direct association of Ctp1 with the Nbs1 subunit of MRN. Second, the highly conserved extreme C terminus of Ctp1 retains the ability to promote the endonuclease activity of MRN. Strikingly, a synthetic polypeptide comprising the 15 amino acids from the extreme C terminus of Ctp1 was sufficient for the full activation of MRN. Third, we verified the evolutionary significance of these findings by demonstrating that the conserved C-terminal polypeptide of CtIP can also stimulate the endonuclease activity of human MRN. Together, our results strongly suggest that the Ctp1-promoted MRN activation mechanism consists of at least two fundamentally separable elements: phosphorylation-induced Ctp1-MRN association and activation of MRN by the C-terminal peptide of Ctp1. Thus, recruitment of the Ctp1 C terminus to MRN is likely pivotal in this activation mechanism.
Results
Phosphorylation of Ctp1 by Casein Kinase II Promotes the Physical Interaction between MRN and Ctp1.
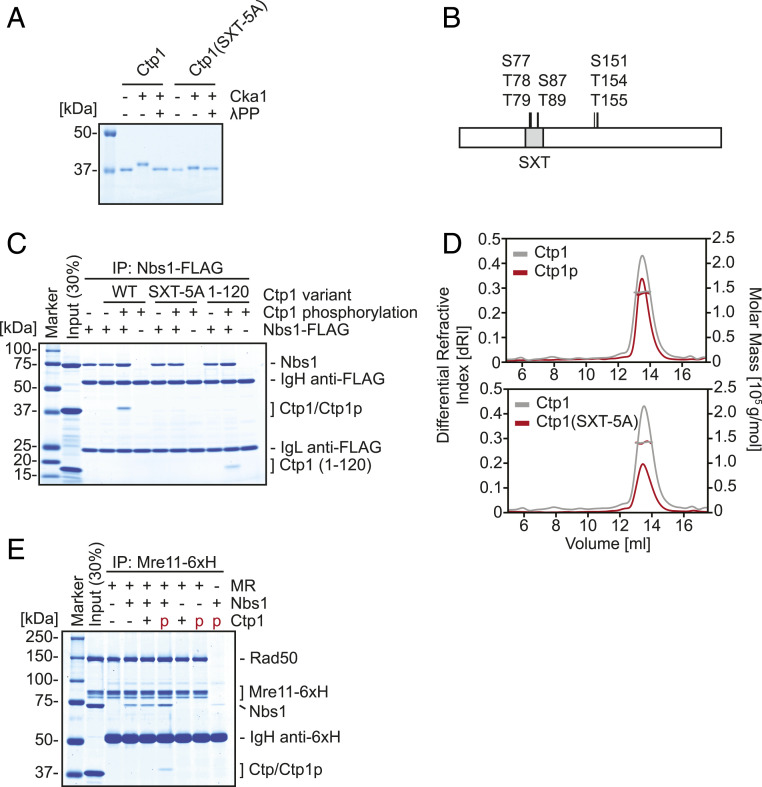
To analyze the interaction between MRN and Ctp1, we purified the S. pombe MR complex, Nbs1 and Ctp1 (SI Appendix, Fig. S1A). Previous studies suggested that casein kinase II (CK2) phosphorylates residues in the SXT domain of Ctp1 (Ser-77, Thr-78, Thr-79, Ser-87, and Thr-89) (17, 21, 25). Thus, the catalytic subunit of the S. pombe CK2 ortholog Cka1 was purified (SI Appendix, Fig. S1B) and incubated with Ctp1 in the presence of ATP. Upon Cka1 treatment, a mobility shift of Ctp1 was observed by sodium dodecyl sulfate/polyacrylamide gel electrophoresis (Fig. 1A). This shift was abolished upon λ-phosphatase treatment, suggesting that it was caused by phosphorylation of Ctp1. Notably, in a variant of Ctp1 where the five phosphorylatable residues of the SXT domain were all substituted for alanine (SXT-5A), the extent of the shift was greatly reduced, although a subtle mobility shift still remained (Fig. 1A). Further analysis of phosphorylated Ctp1 by mass spectrometry revealed additional phosphorylated residues in addition to those in the SXT domain (Ser-151, Thr-154, and Thr-155) (Fig. 1B and SI Appendix, Table S1).
Fig. 1.
Phosphorylation of the SXT site of Ctp1 by Cka1 recruits Ctp1 to the MRN complex. (A) In vitro phosphorylation of Ctp1 variants by the CK2 catalytic subunit Cka1. (B) Summary of phosphorylation sites of Ctp1 revealed by mass spectrometry. (C) Interaction of Ctp1 variants and Nbs1 examined by coimmunoprecipitation with Nbs1-FLAG. (D) Molecular masses of phosphorylated Ctp1 and SXT-5A mutant were evaluated by SEC-MALS. Estimated molecular mass: Ctp1 (unmodified) = 142.0 kDa; phosphorylated Ctp1 = 138.5 kDa; and SXT-5A = 141.6 kDa. Theoretical molecular mass of unmodified Ctp1 monomer is 33.1 kDa. (E) MRN-Ctp1 complex formation and its requirement, examined by coimmunoprecipitation with hexahistidine-tagged Mre11 (Mre11-6xH).
The forkhead-associated domain situated in the N-terminal region of Nbs1 has been shown to bind several phosphopeptides derived from the Ctp1 SXT site with micromolar order affinity (21). Pull-down experiments with FLAG-tagged Nbs1 demonstrated that full-length Ctp1 was pulled down by Nbs1 in a phosphorylation-dependent manner (Fig. 1C). Cka1-treated SXT-5A was not pulled down, whereas an N-terminal 120-amino-acid fragment of Ctp1 (1 to 120) that lacked all the phosphorylation residues except those within the SXT domain was pulled down by Nbs1-FLAG, demonstrating that Ctp1 binds to Nbs1 via the phosphorylated SXT site.
Ctp1 is known to form a tetramer (26). Thus, we asked if phosphorylation affects its oligomerization. Size-exclusion chromatography with multiangle light scattering (SEC-MALS) was employed to determine the molecular weight of Ctp1 variants. Similar to unphosphorylated wild-type (WT) Ctp1, the molecular weight of phosphorylated Ctp1 corresponded well to that of a tetramer (Fig. 1 D, Top). Similarly, the molecular weight of Ctp1(SXT-5A) also corresponded well to that of a tetramer without any appearance of extra SEC peaks which might indicate altered oligomeric status (Fig. 1 D, Bottom), arguing that five mutations within the SXT domain do not alter its oligomerization.
We next reconstituted the higher-order complex of MRN-Ctp1 (Fig. 1E). In the presence of Rad50 and Nbs1, hexahistidine-tagged Mre11 (Mre11-6×H) pulled down phosphorylated Ctp1, but not unphosphorylated Ctp1. Notably, Mre11-6×H did not pull down phosphorylated Ctp1 in the absence of Nbs1, indicating that incorporation of phosphorylated Ctp1 into the higher-order complex requires Nbs1.
Ctp1 Activates the Endonuclease Activity of MRN through Interaction with Nbs1.
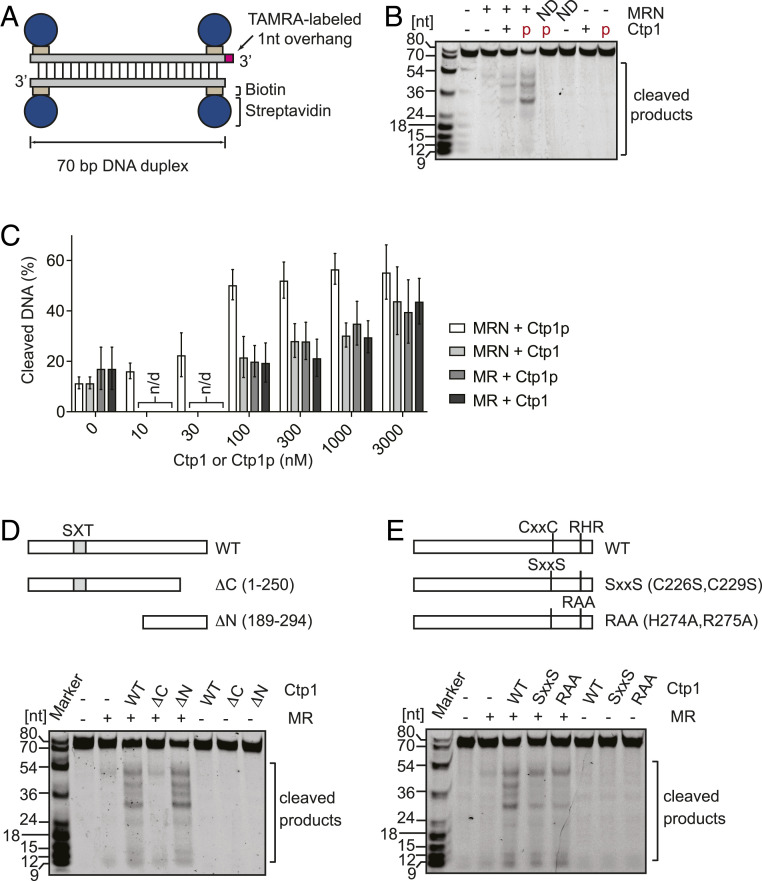
To test the endonucleolytic activity of the MRN-Ctp1 complex, we employed a fluorescently labeled 70-bp DNA duplex with biotin-streptavidin bound to the ends of each strand (Fig. 2A) (27). Addition of phosphorylated Ctp1 (100 nM monomers) to the reaction containing an equimolar amount of MRN subunits and double-stranded DNA (dsDNA) ends caused active cleavage of the streptavidin-bound substrate, whereas very little cleavage was seen with MRN alone (Fig. 2B). In the absence of MRN, Ctp1 did not display any endonuclease activity. The MRN complex containing a nuclease-deficient Mre11 (H134S; ND in Fig. 2B) did not yield any cleavage products, even in the presence of phosphorylated Ctp1, indicating that the endonucleolytic cut was performed by the Mre11 active site. Similar to previous reports with human and S. cerevisiae proteins (9, 28), both Mn2+ and Mg2+ are required for phosphorylated Ctp1-dependent stimulation of MRN (SI Appendix, Fig. S2A). ATP stimulated the phosphorylated Ctp1-dependent endonuclease of MRN, but ADP, AMP-PNP, and ATPγS did not (SI Appendix, Fig. S2B), suggesting that ATP hydrolysis by the ABC-type ATPase of Rad50 is required for this stimulation (9, 11, 29, 30).
Fig. 2.
Stimulation of Mre11 endonuclease activity by Ctp1 is enhanced by Cka1 phosphorylation in the presence of Nbs1. (A) Substrate used for assaying Mre11 endonuclease activity. (B) MRN endonuclease activity was assayed with phosphorylated (red “p”) or unphosphorylated Ctp1. ND, an MRN complex containing nuclease-deficient Mre11H134S. (C) Titration of unphosphorylated Ctp1 (Ctp1) and phosphorylated Ctp1 (Ctp1p) in the endonuclease assay. Error bars, SD; n = 3. n/d, not determined. A representative gel image is shown in SI Appendix, Fig. S3. (D) Mre11 endonuclease activity in assays involving the Mre11-Rad50 complex and full-length or truncated Ctp1. (E) The effect of substitutions in the CxxC and RHR motifs on MRN endonuclease stimulation. Ctp1 (WT, truncated, or mutant proteins) is at 3 µM in D and E.
Notably, unphosphorylated Ctp1 stimulated the endonuclease activity of Mre11, albeit to a lesser extent than phosphorylated Ctp1 (Fig. 2B). Titration experiments (Fig. 2C and SI Appendix, Fig. S3) demonstrated that near-maximal levels of DNA cleavage were achieved at an equimolar concentration to MRN (100 nM) with phosphorylated Ctp1, while unphosphorylated Ctp1 did not fully stimulate the endonuclease activity, even at a 30-times higher concentration. These results suggest that the phosphorylation of Ctp1, most likely at the SXT site, stimulates MRN endonuclease activity through enhanced interaction of Ctp1 with MRN. Unlike the endonuclease activity, neither the exonuclease nor the ATPase activity of MRN was stimulated by phosphorylated Ctp1 (SI Appendix, Fig. S4).
Because higher-order MRN-Ctp1 complex formation requires Nbs1, we assessed the influence of Nbs1 on endonuclease stimulation (Fig. 2C and SI Appendix, Fig. S3). In the absence of Nbs1, the stimulation conferred by Ctp1 was independent of phosphorylation and comparable in magnitude to that seen with unphosphorylated Ctp1 in the presence of Nbs1. Ctp1(SXT-5A) also stimulated MR endonuclease to a similar level as unphosphorylated Ctp1 (SI Appendix, Fig. S5A). These results argue that Nbs1 is required only for the endonuclease stimulation by phosphorylated Ctp1, and also raises the possibility that a region(s) other than the phosphorylated SXT domain might contribute to this activity.
The C Terminus of Ctp1 Is Critical for Stimulating the Endonuclease Activity of MRN.
In order to examine the relationship between Ctp1 oligomerization and MR endonuclease activation, two mutant proteins that affect oligomer formation differently were employed. Ctp1(15-294) forms dimers whereas Ctp1(189-294) exists as a monomer (SI Appendix, Fig. S5B). In both cases, stimulation of MR was comparable to that by unphosphorylated Ctp1, indicating that tetramerization per se is not essential for MR activation. Strikingly, while the N-terminal truncation (i.e., Ctp1[189-294]) stimulated MR endonuclease activity similarly to the full-length Ctp1, the C-terminal truncation (residues 1 to 250) had little stimulatory effect (Fig. 2D). This strongly argues that the C-terminal region of Ctp1, from residue 189 onward and lacking the SXT domain, has a role in the activation of the MR endonuclease that is independent of Nbs1 (17, 18, 31, 32).
Next, the CxxC and RHR motifs commonly found in the C-terminal region of CtIP-family proteins (17, 18) were investigated by mutating CxxC to SxxS or RHR to RAA. These mutant proteins still stimulated MRN endonuclease activity, but at a lower level than wild-type Ctp1 (Fig. 2E), indicating that these two motifs in the C-terminal region are relevant to, but not essential for, endonuclease stimulation.
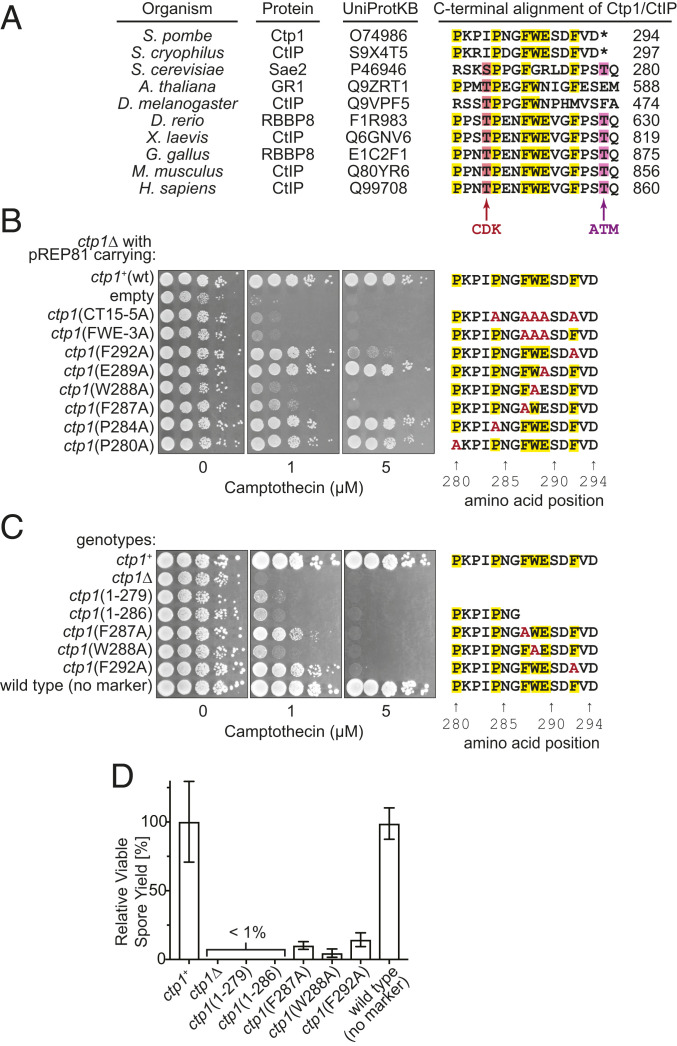
Next, we turned our attention to a C-terminal 15-amino-acid sequence (CT15 hereafter), which is relatively conserved in eukaryotes (Fig. 3A). We assessed the importance of this sequence by a multicopy plasmid-based complementation test using the ctp1∆ mutant and DNA damage sensitivity to camptothecin (CPT). CPT inhibits topoisomerase I, resulting in the accumulation of reaction intermediates with topoisomerase I covalently attached to DNA ends, which require MRN to be removed (2, 33). Overexpression of two full-length Ctp1 variants with multiple conserved residues replaced with alanine—CT15-5A (P284A, F287A, W288A, E289A, F292A) and FWE-3A (F287A, W288A, E289A)—were unable to complement the DNA damage sensitivity of the ctp1∆ strain (Fig. 3B). Furthermore, ctp1∆ cells expressing Ctp1 variants with a single-residue substitution—F287A, W288A, or F292A—showed a severe defect in DNA repair (Fig. 3B). Strains with the corresponding mutations at the native chromosomal locus also showed similar DNA damage sensitivity (Fig. 3C).
Fig. 3.
The ability of Ctp1 to promote Mre11 activity in vivo is located within a conserved 15-amino-acid sequence at the C terminus (CT15). (A) Sequence alignment of the CtIP/Ctp1 C terminus. Thr-847 and Thr-859 in human CtIP are phosphorylation sites for CDK and ATM, respectively. (B) Complementation of ctp1Δ camptothecin sensitivity with plasmids expressing Ctp1 variants. (C) Camptothecin sensitivity of strains with mutations in CT15 at the endogenous ctp1 locus. Serially diluted cells were spotted onto Yeast Extract with Supplements plates containing camptothecin. (D) Relative viable spore yield of various ctp1 mutants. Error bars, SD; n = 3.
Sporulation requires the removal of covalently attached Rec12 (Spo11) from DSB ends by the MRN endonuclease (31, 34). The viable spore yield of strains containing a short C-terminal truncation, ctp1(1-279) and ctp1(1-286), was as severely reduced as the ctp1∆ mutant (Fig. 3D). Mutations at the three critical amino acids identified above (F287A, W288A, or F292A; Fig. 3B) also showed a substantial reduction in viable spore yield.
We then directly compared C-terminal mutants with those where CxxC or RHR are mutated (i.e., C226S, C229S and H274A, R275A, respectively). ctp1∆ cells overexpressing these Ctp1 variants showed similar CPT sensitivity to two CT15 mutants, FWE-3A and W288A, which themselves resemble the null mutant (SI Appendix, Fig. S6A). Strains carrying these mutations at the ctp1+ locus also showed severe CPT sensitivity comparable to the null mutant, except that, at 0.5 μM of CPT, the H274A, R275A mutant showed slightly milder sensitivity than other mutants (SI Appendix, Fig. S6B). The levels of DNA damage sensitivity seen in the CxxC and RHR motif mutants are essentially in line with those reported previously (18, 26, 31). This observation, together with the data showing that purified CxxC and RHR-mutated proteins still retain the ability to stimulate MR endonuclease activity, suggests the involvement of CxxC and RHR motifs in enhancing the effect of CT15.
Collectively, these results support the idea that CT15 and its three conserved amino acids are critical for the role of MRN in the repair of DNA damage during vegetative growth and the removal of DSB-associated Rec12 during sporulation.
Conserved 15-Amino-Acid Peptide from the C Terminus of Ctp1 Is Sufficient for Stimulating the Endonuclease Activity of MRN.
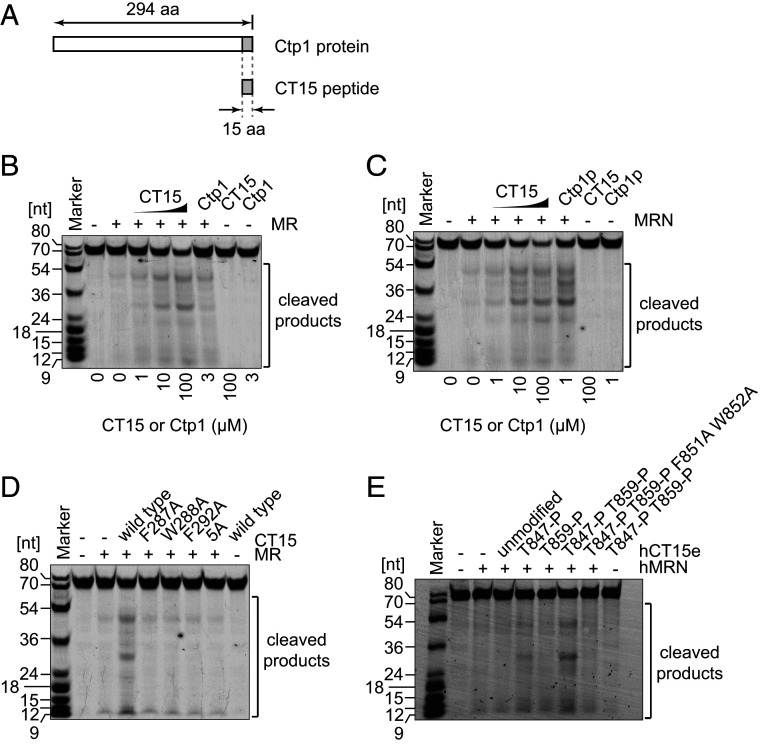
Our results suggested that CT15 might be the functional element responsible for the stimulation of MR endonuclease activity. We therefore introduced a synthetic CT15 peptide into an endonuclease assay containing the MR complex, but lacking Nbs1 (Fig. 4A). Strikingly, the CT15 peptide stimulated the endonuclease activity of MR similarly to the unphosphorylated, full-length Ctp1 (Fig. 4B). Moreover, stimulation of MR at higher concentrations of the CT15 peptide (100 µM) was comparable to the maximal levels achieved with the MRN complex and phosphorylated full-length Ctp1 (Ctp1p in Fig. 4C). Importantly, CT15 stimulated both MRN and MR to a similar level (compare Fig. 4B and Fig. 4C), suggesting that a high dose of CT15 bypassed the requirement for the Nbs1-mediated phosphorylation-dependent Ctp1 recruitment mechanism. CT15 synthetic peptide variants, each with one of the three critical aromatic residues replaced by alanine (F287A, W288A, or F292A), were severely defective in MR stimulation (Fig. 4D). As synthetic CT15 itself has no endonuclease activity (Fig. 4 B–D), these results strongly suggest that the CT15 peptide itself is practically sufficient to function as an activator for the MR endonuclease.
Fig. 4.
A conserved 15-amino-acid peptide (CT15) is sufficient to stimulate MRN endonuclease activity. (A) Location of CT15 in reference to the full length Ctp1. (B) Endonuclease activity of MR was assayed in the presence of CT15 peptides. (C) Same as B except that MRN was used. (D) Same as B but CT15 variants mutated at conserved amino acids were used at 100 µM. (E) A synthetic peptide of the human CT15 counterpart (hCT15e) stimulates hMRN endonuclease activity when its CDK and ATM target sites (Thr-847 and Thr-859, respectively) are phosphorylated. In this assay, hMRN was introduced at 100 nM with respect to the concentration of RAD50 as a monomer. The peptides were used at 50 µM, and potassium chloride, instead of sodium chloride, was used at 100 mM.
We considered the possibility that the function of CT15 might be evolutionarily conserved. Interestingly, alignment of CT15 sites revealed that T847 and T859 in human CtIP, which are phosphorylated by CDK and ATM, respectively, are located within, or one amino acid outside of, the aligned site (Fig. 3A). Mutation of these residues to alanine has been shown to severely compromise the ability of CtIP to stimulate MRN (22, 23, 35, 36); we therefore speculated that they might directly regulate the CT15 peptide. The CT15 site in human CtIP was extended to a total of 19 amino acids to encompass both phosphorylation sites (human CT15 extended, hCT15e), and several peptides were synthesized (wild type and mutant variants).
Remarkably, we found that the hCT15e peptide with both T847 and T859 phosphorylated stimulated the endonuclease activity of human MRN (hMRN; Fig. 4E). By contrast, weak stimulation of hMRN was observed for hCT15e with only T847 phosphorylated, while stimulation was barely seen when unphosphorylated peptide, or a peptide phosphorylated only at T859, were employed. Importantly, alanine substitution of conserved residues corresponding to F851 and W852 resulted in a loss of stimulation even when both T847 and T859 were phosphorylated. These results demonstrate that the hCT15e peptide also retains the capacity to stimulate human MRN endonuclease activity, but only when conserved CDK and ATM target sites are phosphorylated.
Discussion
Through biochemical reconstitution, we demonstrate that phosphorylation of Ctp1 within the SXT domain by casein-kinase II promotes robust binding of Ctp1 to Nbs1, stimulating the endonuclease activity of MRN. What is central to this stimulation mechanism is the extreme C terminus of Ctp1 designated as CT15, which contains only 15 amino acids and can fully stimulate the endonuclease activity of MR. Furthermore, the human counterpart of CT15 can also stimulate human MRN as long as two associated residues, T847 and T859, are phosphorylated. We propose that the core of MRN endonuclease stimulation is provided by CT15 whose effectiveness is promoted by the phosphorylation-dependent recruitment of Ctp1 to MRN.
Our current understanding of the role of Ctp1/Sae2/CtIP in DNA end resection primarily stems from the in vivo and in vitro analysis of loss-of-function mutants. In this paper, we take a synthetic approach and demonstrate that the MR complex is stimulated by the CT15 peptide. This observation narrows down the MR activation domain from the loosely defined C terminus to this particularly small peptide element, shedding light on the interpretation of previously described Ctp1 mutants. The peak of MR activity stimulated by the CT15 peptide was comparable to that of MRN stimulated by CK2-phosphorylated Ctp1. However, while phosphorylated Ctp1 could achieve this level of stimulation at approximately stoichiometric concentrations, a vast excess of CT15 was required to potentiate MR to a similar degree. Based on this, we posit that the phosphorylation-dependent recruitment of Ctp1 to MRN up-regulates the activity of the CT15 peptide by 100- to 1,000-fold. A flexible intrinsically disordered region within Ctp1 connects the CT15 site in the extreme C terminus to the N-terminally located SXT domain. Phosphorylation of the SXT domain promotes its binding to the N-terminal FHA domain of Nbs1. The FHA domain of Nbs1 is connected to the C-terminally located Mre11-interacting domain via a flexible loop. Taken together, these topological observations suggest that the CT15 site is flexibly tethered by Nbs1 at the phosphorylated SXT domain and thus accessible to the area within a radius of ∼300 Å from the Mre11 dimer–dimer interface (21, 26). Sequestering CT15 in close proximity to MR is expected to result in a local concentration increase that likely compensates for the suboptimal activity of CT15. The 300-Å tethering range would allow CT15 to potentially interact with any part of Mre11 including the catalytic site, the head domain of Rad50, and even part of the coiled-coil regions as well as any bound DNA.
We are considering the possibility that CT15 binding to MR induces/stabilizes a critical conformational change leading to efficient cleavage of end-blocked DNA. Recently, the structural homolog of the MR complex from bacteria, SbcCD, was shown to adopt a drastic conformational change (“cutting state”) upon sensing a DNA break (13). SbcCD also exhibits endonuclease activity on DNA end-blocked substrates, but, unlike MR, this activity appears to be inherent to SbcCD since neither Nbs1 nor Ctp1 has been identified in prokaryotes. A structural element named the fastener loop of SbcD (Mre11) interacts with the outer β-sheet of SbcC (Rad50) in the cutting state of the complex, and this binding surface corresponds to the cluster of Rad50S mutations within S. cerevisiae. Additionally, interaction of the phosphorylated C terminus of Sae2 with Rad50 is abolished upon introduction of a rad50S mutation (K81I) found in the same cluster (37). These observations raise the possibility that CT15 might promote the association of Mre11 and Rad50 through the fastener loop interface, possibly even bridging the interaction between Rad50 and Mre11, which would work in favor of forming/maintaining the cutting-state conformation of the MR complex similar to SbcCD (38, 39).
It is also possible that CT15 promotes MR endonuclease through DNA binding. The core FWE motif within the CT15 site conforms to the mismatch recognition FxE motif found within MSH6 of the MutSα complex (40, 41). The “x” in the FxE motif is usually an aromatic residue, with human MSH6 harboring FYE and S. cerevisiae Msh6 containing FFE. However, unlike the FWE motif, the conserved FxE motif is located within a β-strand situated in a structurally well-defined environment of MutS where it contacts a mispaired base (with support from multiple other residues). If there is any functional similarity within the FXE motifs of Ctp1/CtIP and MSH6, it is likely achieved in conjunction with structural support from another well-structured protein like Mre11 or Rad50.
Ctp1 and its human homolog CtIP form a tetramer (26, 42). Mutations that impair tetramer formation in CtIP (9) and Sae2 (37) did not fully abolish its capacity to activate MR; a residual stimulation was still seen in both cases. Similarly, in our assay, stimulation of MR by Ctp1 was seen regardless of its oligomeric status, indicating tetramerization per se is not essential for MR activation (SI Appendix, Fig. S5B). This, however, does not exclude the involvement of tetramerization in MR activation; impairment of tetramerization leads to a reduction in MR activation and DNA repair deficiency overall (26, 43). It is reasonable, for example, to assume that the recruitment of a tetramer containing four CT15 activators, mediated through the interaction between Nbs1 and phosphorylated Ctp1, would stimulate MR more than the recruitment of a single Ctp1 molecule by elevating the local concentration of CT15. Tetramerization of Ctp1 is necessary for bridging DNA molecules in the absence of MRN (26, 43). Thus, it is also possible that some form of DNA bridging acts in favor of enhancing the overall efficiency of MR activation.
The CT15 equivalent in CtIP, which we identified based on sequence homology to S. pombe CT15, is highly conserved within higher eukaryotes. Interestingly, two of the critically important phosphorylation sites in human CtIP, T847, and T859 are located within and adjacent to the CT15 site, which prompted us to expand the CT15 site to include them—generating the hCT15e peptide—and test the phosphorylated versions of this peptide. Notably, mutations of these residues to alanine have been shown to severely impact the effect of CtIP related to MRN endonuclease activity both in vivo and in vitro (22, 35, 36, 44). Here, we show that phosphorylation of T847 and T859 together serve the role of reconstituting the active form of the CT15 cofactor necessary for the stimulation of hMRN endonuclease activity. The peptide, when doubly phosphorylated, showed the most robust MRN stimulation. Mutation of the critical aromatic residues within the CT15 site resulted in a loss of stimulation even in the double-phosphorylated peptide, suggesting that the phosphorylation of these sites directly regulates the activity of CT15 in CtIP.
Defining the conserved CT15 site in the Ctp1/Sae2/CtIP ortholog family shifts the focus from the already recognized CXXC and RHR motifs and clearly defines the critical site for MRN activation. This is important for our interpretation of the previously reported mutant phenotypes. It is likely that the Seckel and Jawad syndromes are in fact at least in part caused by the loss of CT15 functionality as both reported truncations lack this site (45). Furthermore, reducing the size of an MRN activator to that of a peptide opens the possibility of practical uses. Modification of the CT15 peptide could yield a stronger activator or a potential inhibitor of the MRN complex. Activation or inhibition of the MRN complex could be particularly useful in gene editing downstream from CRISPR-Cas9 or other targeted nucleases as it could shift the bias between nonhomologous end joining and homologous recombination repair. Alternatively, CT15-derived peptides could prove to be useful as cancer therapeutics or radiotherapy sensitizers (46, 47).
Materials and Methods
Endonuclease Assay.
Standard endonuclease assays were performed in 4 µL of reaction mixture containing 150 mM NaCl, 20 mM Tris⋅HCl, pH 7.5, 10% glycerol, 1 mM dithiothreitol, 0.5 mM MnCl2, 5 mM MgCl2, 50 nM DNA, 2 mM ATP, 1 µM streptavidin, 50 nM dsDNA, 50 nM (Mre11)2(Rad50)2, 100 nM Nbs1, and the indicated amounts of Ctp1. Reactions were incubated for 1 h at 30 °C and then stopped by addition of 1 µL stop reagent (1:1:1 mixture of proteinase K [Takara], 0.5 M ethylenediaminetetraacetic acid [EDTA], pH 8.0, and 10% sodium dodecyl sulfate) and by incubation at 50 °C for 20 min. Four volumes of formamide loading buffer (20 mM EDTA, 0.05 mg/mL Orange G, and 96% formamide) were added to the samples, which were incubated at 98 °C for 7 min and then loaded on 13.3% polyacrylamide gels containing 8 M urea. DNA substrate and products attached to TAMRA were visualized with a Typhoon FLA 9500 imager (GE Healthcare). Band quantification was performed with ImageQuant TL v8.1.0.0 (GE Healthcare). Briefly, for each lane, total and uncut DNA band signals were quantified; background based on unoccupied corresponding areas of the gel was subtracted; and the amount of degraded DNA was expressed as the percentage of the total lane signal, based on the initial uncut-band signal.
Additional materials and methods are in SI Appendix.
Supplementary Material
Acknowledgments
We thank Yumiko Kurokawa for help with protein purification and members of the H.I. laboratory for discussion and encouragement. This study was supported in part by the Japan Society for the Promotion of Science, Grants-in-Aid for Scientific Research on Innovative Areas (15H059749 to H.I.), for Scientific Research (A) (18H03985 to H.I.), for Young Scientists (B) (17K15061 to B.A.), for Scientific Research (B) (18H02371 to H.T. and 19H03160 to Y.M.), for Early-Career Scientists (19K16039 to K.I. and 20K15713 to B.A.), and by NIH Grant R35 CA241801 (to P.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016287118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information. Requests for reagents or further information should be directed to H.T. (htsubouchi@bio.titech.ac.jp), P.S. (sungp@uthscsa.edu), or H.I. (hiwasaki@bio.titech.ac.jp).
References
- 1.Wu D., Topper L. M., Wilson T. E., Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics 178, 1237–1249 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartsuiker E., Neale M. J., Carr A. M., Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol. Cell 33, 117–123 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keeney S., Giroux C. N., Kleckner N., Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Shibata A., et al., DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell 53, 7–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Z., Chung W.-H. H., Shim E. Y., Lee S. E., Ira G., Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134, 981–994 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia V., Phelps S. E. L., Gray S., Neale M. J., Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 479, 241–244 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langerak P., Mejia-Ramirez E., Limbo O., Russell P., Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks. PLoS Genet. 7, e1002271 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Syed A., Tainer J. A., The MRE11-RAD50-NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu. Rev. Biochem. 87, 263–294 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anand R., Ranjha L., Cannavo E., Cejka P., Phosphorylated CtIP functions as a Co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell 64, 940–950 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Deshpande R. A., Lee J. H., Arora S., Paull T. T., Nbs1 converts the human Mre11/rad50 nuclease complex into an endo/exonuclease Machine specific for protein-DNA adducts. Mol. Cell 64, 593–606 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Wang W., Daley J. M., Kwon Y., Krasner D. S., Sung P., Plasticity of the Mre11-Rad50-Xrs2-Sae2 nuclease ensemble in the processing of DNA-bound obstacles. Genes Dev. 31, 2331–2336 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reginato G., Cannavo E., Cejka P., Physiological protein blocks direct the Mre11-Rad50-Xrs2 and Sae2 nuclease complex to initiate DNA end resection. Genes Dev. 31, 2325–2330 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Käshammer L., et al., Mechanism of DNA end sensing and processing by the Mre11-rad50 complex. Mol. Cell 76, 382–394.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Tatebe H., et al., Rad50 zinc hook functions as a constitutive dimerization module interchangeable with SMC hinge. Nat. Commun. 11, 370 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y., et al., ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex. EMBO J. 35, 743–758 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schiller C. B., et al., Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nat. Struct. Mol. Biol. 19, 693–700 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akamatsu Y., et al., Molecular characterization of the role of the Schizosaccharomyces pombe nip1+/ctp1+ gene in DNA double-strand break repair in association with the Mre11-Rad50-Nbs1 complex. Mol. Cell. Biol. 28, 3639–3651 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Limbo O., et al., Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell 28, 134–146 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKee A. H. Z., Kleckner N., A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics 146, 797–816 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sartori A. A., et al., Human CtIP promotes DNA end resection. Nature 450, 509–514 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams R. S., et al., Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell 139, 87–99 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H., et al., The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 9, e1003277 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anand R., et al., NBS1 promotes the endonuclease activity of the MRE11-RAD50 complex by sensing CtIP phosphorylation. EMBO J. 38, 1–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cannavo E., Reginato G., Cejka P., Stepwise 5′ DNA end-specific resection of DNA breaks by the Mre11-Rad50-Xrs2 and Sae2 nuclease ensemble. Proc. Natl. Acad. Sci. U.S.A. 116, 5505–5513 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dodson G. E., Limbo O., Nieto D., Russell P., Phosphorylation-regulated binding of Ctp1 to Nbs1 is critical for repair of DNA double-strand breaks. Cell Cycle 9, 1516–1522 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andres S. N., et al., Tetrameric Ctp1 coordinates DNA binding and DNA bridging in DNA double-strand-break repair. Nat. Struct. Mol. Biol. 22, 158–166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anand R., Pinto C., Cejka P., Methods to study DNA end resection I: Recombinant protein purification. Methods Enzymol. 600, 25–66 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Cannavo E., Cejka P., Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature 514, 122–125 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Deshpande R. A., Lee J.-H., Paull T. T., Rad50 ATPase activity is regulated by DNA ends and requires coordination of both active sites. Nucleic Acids Res. 45, 5255–5268 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prasada Rao H. B. D., et al., A SUMO-ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination. Science 355, 403–407 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma L., Milman N., Nambiar M., Smith G. R., Two separable functions of Ctp1 in the early steps of meiotic DNA double-strand break repair. Nucleic Acids Res. 43, 7349–7359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen K. L., Russell P., Ctp1-dependent clipping and resection of DNA double-strand breaks by Mre11 endonuclease complex are not genetically separable. Nucleic Acids Res. 44, 8241–8249 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pommier Y., et al., Repair of topoisomerase I-mediated DNA damage. Prog. Nucleic Acid Res. Mol. Biol. 81, 179–229 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartsuiker E., et al., Ctp1CtIP and Rad32Mre11 nuclease activity are required for Rec12Spo11 removal, but Rec12Spo11 removal is dispensable for other MRN-dependent meiotic functions. Mol. Cell. Biol. 29, 1671–1681 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huertas P., Jackson S. P., Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 284, 9558–9565 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deshpande R. A., et al., DNA-dependent protein kinase promotes DNA end processing by MRN and CtIP. Sci. Adv. 6, eaay0922 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cannavo E., et al., Regulatory control of DNA end resection by Sae2 phosphorylation. Nat. Commun. 9, 4016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Connelly J. C., de Leau E. S., Leach D. R. F., Nucleolytic processing of a protein-bound DNA end by the E. coli SbcCD (MR) complex. DNA Repair (Amst.) 2, 795–807 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Saathoff J. H., Käshammer L., Lammens K., Byrne R. T., Hopfner K. P., The bacterial Mre11-Rad50 homolog SbcCD cleaves opposing strands of DNA by two chemically distinct nuclease reactions. Nucleic Acids Res. 46, 11303–11314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato N., et al., Sensing and processing of DNA interstrand crosslinks by the mismatch repair pathway. Cell Rep. 21, 1375–1385 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warren J. J., et al., Structure of the human MutSalpha DNA lesion recognition complex. Mol. Cell 26, 579–592 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Davies O. R., et al., CtIP tetramer assembly is required for DNA-end resection and repair. Nat. Struct. Mol. Biol. 22, 150–157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andres S. N., Li Z. M., Erie D. A., Williams R. S., Ctp1 protein-DNA filaments promote DNA bridging and DNA double-strand break repair. J. Biol. Chem. 294, 3312–3320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huertas P., Cortés-Ledesma F., Sartori A. A., Aguilera A., Jackson S. P., CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455, 689–692 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qvist P., et al., CtIP mutations cause Seckel and Jawad syndromes. PLoS Genet. 7, e1002310 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dupré A., et al., A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat. Chem. Biol. 4, 119–125 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kavitha C. V., Choudhary B., Raghavan S. C., Muniyappa K., Differential regulation of MRN (Mre11-Rad50-Nbs1) complex subunits and telomerase activity in cancer cells. Biochem. Biophys. Res. Commun. 399, 575–580 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information. Requests for reagents or further information should be directed to H.T. (htsubouchi@bio.titech.ac.jp), P.S. (sungp@uthscsa.edu), or H.I. (hiwasaki@bio.titech.ac.jp).