Significance
Cellulose is the most abundant biopolymer on Earth and has many potential industrial applications, such as renewable energy and sustainable materials. Here we report the apo and UDP-glucose (UDP-Glc)–bound crystal structures of the catalytic domain of Arabidopsis thaliana CESA3. Our results offer a structural basis for how the substrate UDP-Glc and a metal ion, Mn2+, which is required for cellulose synthesis, are coordinated in plant CESAs. Furthermore, our structure reveals that CESAs may form homodimers through interactions between specific beta strands that likely aid in the early stages of CESA complex formation.
Keywords: cellulose synthase, UDP-glucose, plant cell wall, plant biology, structural biology
Abstract
Cellulose is synthesized by cellulose synthases (CESAs) from the glycosyltransferase GT-2 family. In plants, the CESAs form a six-lobed rosette-shaped CESA complex (CSC). Here we report crystal structures of the catalytic domain of Arabidopsis thaliana CESA3 (AtCESA3CatD) in both apo and uridine diphosphate (UDP)-glucose (UDP-Glc)–bound forms. AtCESA3CatD has an overall GT-A fold core domain sandwiched between a plant-conserved region (P-CR) and a class-specific region (C-SR). By superimposing the structure of AtCESA3CatD onto the bacterial cellulose synthase BcsA, we found that the coordination of the UDP-Glc differs, indicating different substrate coordination during cellulose synthesis in plants and bacteria. Moreover, structural analyses revealed that AtCESA3CatD can form a homodimer mainly via interactions between specific beta strands. We confirmed the importance of specific amino acids on these strands for homodimerization through yeast and in planta assays using point-mutated full-length AtCESA3. Our work provides molecular insights into how the substrate UDP-Glc is coordinated in the CESAs and how the CESAs might dimerize to eventually assemble into CSCs in plants.
Cellulose, a linear homopolymer of d-glucopyranose linked by β-1,4-glycosidic bonds, is the major structural component of the cell walls of plants, oomycetes, and algae and constitute the most abundant biopolymer on Earth (1). Cellulose is synthesized by cellulose synthases (CESAs) that belongs to the glycosyltransferase GT-2 superfamily (1, 2). In land plants, cellulose is produced at the plasma membrane by six-lobed rosette-shaped CESA complexes (CSCs) where each CESA is thought to synthesize one cellulose chain (3). The precise number of CESAs per CSC is unresolved but estimated to range between 18 and 36 (4–6).
Plants contain multiple cesa genes, with 10 found in the Arabidopsis genome (7). Of these, CESA1, CESA3, CESA6, and the CESA6-like CESAs (i.e., CESA2, CESA5, and CESA9) are involved in primary cell wall formation, whereas CESA4, CESA7, and CESA8 participate in secondary cell wall formation (8–12). These two types of CSCs form heterotrimeric complexes with a ratio of 1:1:1 (13, 14). The Arabidopsis CESAs share an overall sequence identity of ∼60% and have seven transmembrane helices (15). In plants, the catalytic domain (CatD) of the CESAs is located between the second and third transmembrane helices and contains a canonical D, D, D, QxxRW motif (1). While there are similarities between the plant CatD and its counterpart in bacterial cellulose synthases, the CatD is flanked by two plant-specific domains, the so-called plant-conserved region (P-CR) and class-specific region (C-SR) (16). These domains are proposed to have important functions in cellulose synthesis and CESA oligomerization (17).
The oligomerization of plant CESAs is thought to be important for the final CSC assembly, and multiple oligomeric states of CESAs, including homodimers, have been reported (18, 19). For example, immunoprecipitation assays using CESA7 fused to a dual His/STRP-tag demonstrated that CESA4, CESA7, and CESA8 could form independent homodimers, and it was hypothesized that the CESA homodimerization may contribute to early stages of CSC assembly. These homodimers might then be converted into CSC heterotrimeric configurations (19). This feature poses a marked difference from the bacterial cellulose synthase complex. However, how CESA homodimers are formed and how they function in cellulose synthesis are unknown.
To comprehend the mechanisms behind plant cellulose synthesis, it is essential to acquire structural information about plant CESAs. Indeed, the BcsA–BcsB complex structure from Rhodobacter greatly aided our understanding of the cellulose synthesis in bacteria (20). Nevertheless, there are many differences between bacterial and plant CESAs and the corresponding protein complexes. Extensive efforts have been undertaken to acquire plant CESA structural information, including homology modeling and small-angle X-ray scattering analyses (5, 6, 16, 21, 22). While these efforts have been important to form new hypotheses, they did not reveal significant insights into substrate coordination, cellulose chain extrusion, and complex assembly. Recently, a homotrimeric CESA8 structure from Populus tremula × tremuloides was resolved by cryogenic electron microscopy (cryo-EM), which offered significant new molecular understanding of cellulose microfibril biosynthesis and CESA coordination within the CSC (15). Here we report the crystal structures of Arabidopsis CESA3 CatD (AtCESA3CatD) in apo and uridine diphosphate (UDP)-glucose (UDP-Glc) bound forms and outline how the CatD might contribute to CESA homodimerization and substrate coordination.
Results
Crystal Structure of the Catalytic Domain of Arabidopsis CESA3.
After testing a number of Arabidopsis CESA3 cytosolic CatD constructs (see Materials and Methods), a construct spanning residues T317 to G810 from Arabidopsis CESA3 CatD (AtCESA3CatD) was expressed in Escherichia coli and subsequently purified (Fig. 1A). The corresponding crystal structure was resolved by the selenium single-wavelength anomalous diffraction (SAD) method in space group P212121 with two molecules in one asymmetric unit (SI Appendix, Table S1) (23). The crystal structure of AtCESA3CatD can be divided into three parts: the core catalytic domain, the P-CR domain, and the C-SR domain (Fig. 1B). The core catalytic domain, with the catalytic sites, is composed of a β-sheet with eight strands, flanked by two α-helices on one side and two α-helices with three β-strands on the other side. The N-terminal region of the core catalytic domain, composed of α1, α5, α6, and β1 to β5, aligns well with typical GT-2 GTs, e.g., the SpsA from Bacillus subtilis with a rmsd of 3.5 Å for 239 main chain atoms, indicating that the core GT domain structure is conserved (SI Appendix, Fig. S1A) (24). The remaining C-terminal region of the core catalytic domain includes the α9 helix and strands β6 to β12, which form two layers of β sheets (Fig. 1C). Here β6, β7, β9, and β12 are arranged in one layer that aligns parallelly with β1 to β4 and another layer that aligns parallelly with β8, β10, and β11. The proposed cellulose binding helix α9 inserts toward the catalytic region without forming a strong interaction with the rest of AtCESA3CatD, suggesting that it is flexible, at least in the absence of cellulose and transmembrane helices. Notably, we observed one disulfide bond formed between C618 and C782 in the β9 and β12 strands that perhaps contributes to catalytic domain stability (SI Appendix, Fig. S1B). Within AtCESA3CatD, the plant-specific domains, P-CR and C-SR, are clearly resolved and sandwich the core catalytic domain (Fig. 1B). The P-CR domain consists of α2 to α4 and β3 (Fig. 1D), similar to the previously reported individual P-CR segment (SI Appendix, Fig. S1C) (17). The C-SR (Fig. 1E), with two helices, α7 and α8, that are packed against α5 of the core catalytic domain, is enriched in positively charged amino acids, such as lysine and arginine (25).
Fig. 1.
Crystal structure of the Arabidopsis CESA3 catalytic domain (AtCESA3CatD). (A) Domain organization of Arabidopsis AtCESA3. TM: transmembrane helix. GT domain: glycosyltransferase domain. The domain boundary of the GT domain is indicated by residue numbering. The zinc finger domain and TM helices are colored blue and red, respectively. The GT domain is colored as in B. (B) The crystal structure of AtCESA3CatD is shown as a cartoon. The P-CR domain, C-SR domain, and core GT domain of AtCESA3CatD are colored wheat, yellow, and salmon, respectively. Secondary structure elements are labeled numerically. The unmodeled region is connected by dashed lines. (C–E) Detailed view of the C terminus of the core GT domain (C), P-CR domain (D), and C-SR domain (E).
AtCESA3CatD Forms a Homodimer via Conserved Residues along the Periphery of the Core Catalytic Domain.
Plant CESA oligomerization is thought to be important for the CSC assembly (19). We found that two molecules of AtCESA3CatD within an asymmetric unit form a homodimer (Fig. 2A), with a total buried surface area of ∼711.3 Å2 as calculated by PISA (26). The dimer interface mainly involves the amino acids residing in β6 and α9 (Fig. 2A). β6 from both protomers form an antiparallel β-sheet, establishing tight interactions (mainly via hydrogen bonds between O and N atoms in the main chains of Q571, V572, C573, Y574) (Fig. 2B). These amino acids are highly conserved among the Arabidopsis CESA proteins, implying a common dimerization mode (SI Appendix, Fig. S2). By contrast, we observed only weak interaction between the two α9 helices. In line with these crystallographic data, purified AtCESA3CatD was observed as a homodimer in a solution based on analytical gel filtration calibration (Fig. 2C). In addition, we used small-angle X-ray scattering (SAXS) profiles of AtCESA3CatD (at 2 and 5 mg/mL) to further study the oligomeric state of AtCESA3CatD in solution (SI Appendix, Fig. S3A). The corresponding profiles overlapped nicely, and we used the scattering profile from the highest concentration (5 mg/mL) for further data analysis due to its superior signal-to-noise ratio. The Guinier plot at low angles appeared linear with no indication of protein aggregation (SI Appendix, Fig. S3A). The derived radius of gyration (Rg) for AtCESA3CatD from the Guinier approximation and the distance distribution function [P(r)] were 3.96 ± 0.15 and 4.0 ± 0.06 nm, respectively (SI Appendix, Table S2). The maximum particle dimension (Dmax) of AtCESA3CatD was 12.2 nm. Based on the Porod volume determined from the scattering pattern, the molecular mass of AtCESA3CatD in solution was calculated to be 106 ± 11 kDa, indicating that AtCESA3CatD forms a dimer under these conditions. This was further confirmed by overlapping the theoretical scattering curve derived from the monomeric or dimeric crystallographic structure of AtCESA3CatD with the experimental data, in which the dimeric crystallographic structure fitted well to the experimental curve, with a χ2 value of 1.47 (SI Appendix, Fig. S3B)
Fig. 2.
AtCESA3CatD forms a homodimer via conserved amino acid pairs. (A) Crystal structure of two paired AtCESA3CatD molecules resolved in an asymmetric unit. The interacting segments are displayed in salmon. The P-CR and C-SR domains are colored wheat and yellow, respectively. (B) Details of the homodimerization mechanism of AtCESA3CatD dimer formation. β6 of AtCESA3CatD is shown as sticks and a cartoon with amino acids labeled, with N, O, and C atoms colored blue, red, and salmon, respectively. Hydrogen bonds are indicated as dashed lines. Note that the prime (′) indicates the second protomer in the homodimer. (C) Analytical size exclusion chromatography of AtCESA3CatD wild type and point mutants. The elution volumes for the four constructs are labeled with the same color as that of construct. The native AtCESA3317−792 (residues 317 to 792) demonstrates two peaks, corresponding to a dimer (left peak) and a monomer (right peak). (D) Yeast split-ubiquitin pairwise result of wild-type AtCESA3 and its mutants (AtCESA3Q571P, AtCESA3C573P, and AtCESA3Q571P:C573P) on SD/-Leu-Trp-His-Ade when 5 mM 3-AT was added to the medium. R1.1 is the positive control with PTSU2-APP as the bait and PNubG-Fe65 as the prey; R1.2 is the negative control with empty plasmid PBT3-N as the bait and PPR3-N as the prey. R2.1 to R2.4: AtCESA3, AtCESA3Q571P, AtCESA3C573P, and AtCESA3Q571P:C573P fused to Cub bait vector. R3.1 to R3.4: AtCESA3, AtCESA3Q571P, AtCESA3C573P, and AtCESA3Q571P:C573P fused to NubG prey vector. R4.1 to R4.4: AtCESA3, AtCESA3Q571P, AtCESA3C573P, and AtCESA3Q571P:C573P fused to Cub bait vector and NubG prey vector, respectively. (E) BiFC analyses in epidermal tobacco leaf cells with indicated constructs. Note the punctate localization of the BiFC signal in wild-type CESA3, consistent with Golgi localized proteins. (Scale bar, 100 µm.)
To further investigate the dimerization, we generated truncated AtCESA3CatD constructs (AtCESA3317−792) by removing the amino acids from A793 to G810 where the α9 resides in the crystal structure. AtCESA3317−792 can still exist as a dimer despite the monomeric state observed (Fig. 2C). Next, residues Q571 and C573 in AtCESA3CatD were mutated separately to proline to disrupt the dimer interface because proline lacks backbone amide protons, thus blocking the formation of hydrogen bonds observed in the homodimer structure (Fig. 2B) (27). Surprisingly, the effect of the two mutations, Q571P and C573P, on dimer formation is distinct in solution, where only the C573P mutation completely abolished homodimer formation of AtCESA3CatD, whereas Q571P had minor effects. We also carried out the circular dichroism (CD) spectroscopy measurement of the native and mutated proteins AtCESA3CatD, AtCESA3CatDQ571P, and AtCESA3CatDC571P to demonstrate that there was no evidence of structural misfolding of the two proline mutants (SI Appendix, Fig. S4A). Together, our data indicated that β6 is important for CatD homodimerization, whereas α9 provides minor contributions.
To corroborate the importance of the β6 strand in CESA dimerization in vivo, we used full-length wild-type and mutated AtCESA3 constructs (AtCESA3, AtCESA3Q571P, AtCESA3C573P, and AtCESA3Q571P:C573P) in split-ubiquitin yeast two-hybrid assays. The yeast split-ubiquitin pairwise result of wild-type AtCESA3 revealed strong growth on SD/-Leu-Trp-His-Ade when either 5 or 10 mM 3-AT was added to the medium (Fig. 2D and SI Appendix, Fig. S5 A and B). By contrast, AtCESA3Q571P displayed only slight interaction with itself (grew on 5 mM 3-AT but not on 10 mM 3-AT); AtCESA3C573P and AtCESA3Q571P:C573P did not result in any detected interactions in our assays (Fig. 2D and SI Appendix, Fig. S5 A and B). In addition, we generated similar full-length wild-type and mutated AtCESA3 constructs for bimolecular fluorescence complementation (BiFC) experiments in planta. We observed a clear BiFC signal from the wild-type AtCESA3 that appeared as puncta in transiently transformed tobacco leaf cells (Fig. 2E and SI Appendix, Fig. S5 C–M), consistent with Golgi localization of the CESAs prior to their secretion to the plasma membrane (28). By contrast, the mutants AtCESA3Q571P, AtCESA3C573P, and AtCESA3Q571P:C573P did not show any BiFC signal (Fig. 2E). To ensure that the introduced point mutations did not interfere with the folding of the protein and thus the fluorescent signal, we also generated fluorescently (YFP) tagged versions of the wild-type and mutated AtCESA3 and again transiently transformed the constructs into tobacco. These constructs all displayed fluorescence reminiscent of the Golgi apparatus (SI Appendix, Fig. S5 C–F). Taken together, our results indicate that the amino acids Q571 and C573 in β6 contribute to the dimerization of plant CESAs in vitro and in vivo, corroborating that β6 is important for homodimer formation.
UDP-Glc and Manganese Are Coordinated by Conserved Amino Acids at the Core of AtCESA3CatD.
CESAs are thought to utilize UDP-Glc as their substrate. We therefore aimed to determine the crystal structure of UDP-Glc–bound AtCESA3. We managed to obtain the UDP-Glc–bound structure using the construct AtCESA3317−792, which lacks the canonical QxxRW binding motif, as well as the apo structure of AtCESA3317−792 at 2.05 and 2.35 Å resolution, respectively. We superimposed the apo structure of AtCESA3317−792 and AtCESA3CatD on UDP-Glc–bound AtCESA3317−792, which demonstrated that they were almost structurally identical overall (Fig. 3A and SI Appendix, Fig. S1D). The exception was that residues K349, K520, and K521 were stabilized upon UDP-Glc binding (SI Appendix, Fig. S1 E and F). We were unable to show cocrystallization with cellobiose (a glucose disaccharide), indicating that larger fragments of CESA are needed to coordinate cellobiose.
Fig. 3.
UDP-Glc is coordinated via multiple amino acids and Mn2+ in the core catalytic grove of AtCESA3317−792. (A) Crystal structure of AtCESA3CatD (colored salmon) is superimposed onto the UDP-Glc–bound crystal structure of AtCESA3317−792 (colored lime). The UDP-Glc is shown as sticks, with C, O, N, and P atoms colored lime, red, blue, and orange, respectively (the same color scheme is used in all panels). The loop with catalytic residue D765 is outlined by a blue dashed ellipse. The putative glucan-binding region is indicated by a gray dashed ellipse. (B) Unbiased difference Fourier (Fo-Fc) omit electron density map of Mn2+ and UDP-Glc. The map, represented as green mesh, is contoured at 2.2σ. The bound Mn2+ is shown as violet spheres. (C) The binding of UDP-Glc and Mn2+ to AtCESA3317−792. The water molecules (w1, w2, w3) are shown as red spheres. The residues participating in the binding are labeled and shown as sticks. (D) Detailed coordination of Mn2+. The interacting residues D545 and K521 are shown, and interaction distances are labeled (in Å).
In our structure, UDP-Glc is clearly contained in a binding pocket formed by the β1 strand, the loop connecting β1 and α1, and the α5 helix. In contrast to the relatively poor unbiased difference Fourier (Fo-Fc) map of the glucose unit of UDP-Glc, the nucleotide uridine fitted well to its Fo-Fc map, indicating that the glucose unit is relatively flexible in our structure and is poorly coordinated (Fig. 3 A and B). We therefore paid particular attention to the nucleotide uridine of UDP-Glc, which projects deep inside the pocket of the CatD to establish extensive interactions (Fig. 3C). Here residue D379 forms a hydrogen bond with uridine N3 of UDP-Glc. In addition, residue D379 interacts with the side chain of R510 through a salt bridge. Four residues, D379, R510, K512, and H518, coordinate the binding of one water molecule, w1, which is in direct contact with uridine O4. This coordinated interaction may, perhaps, contribute to the exclusion of adenine and guanine as nucleotides but allow uracil due to size restrictions. Furthermore, residue S343 forms bilateral interactions with uridine O2 and ribose. Ribose is further bound via E350, while residues V345 and K520 sandwich the nucleobase uracil through hydrophobic interaction (like a clamp). The positively charged residues K349 and K520 form salt bridges with the α- and β-phosphates of UDP-Glc (Fig. 3C). For the catalytic activity of GTs, a divalent cation (such as a magnesium or manganese ion) is required (1, 4). Interestingly, we observed a manganese ion (Mn2+) in the catalytic site (Fig. 3D). The bound Mn2+ was supported by an X-ray fluorescence scan (SI Appendix, Fig. S4B). The Mn2+ ion is coordinated by the oxygen atom in the carboxylic acid group of D545, the amino group of the K521 side chain, the hydroxyl group (O3) in ribose of UDP-Glc, and two well-resolved water molecules (w2 and w3) (Fig. 3D).
Comparison of Plant and Bacterial Cellulose Synthases Indicates Some Conservation but also Structural Divergence.
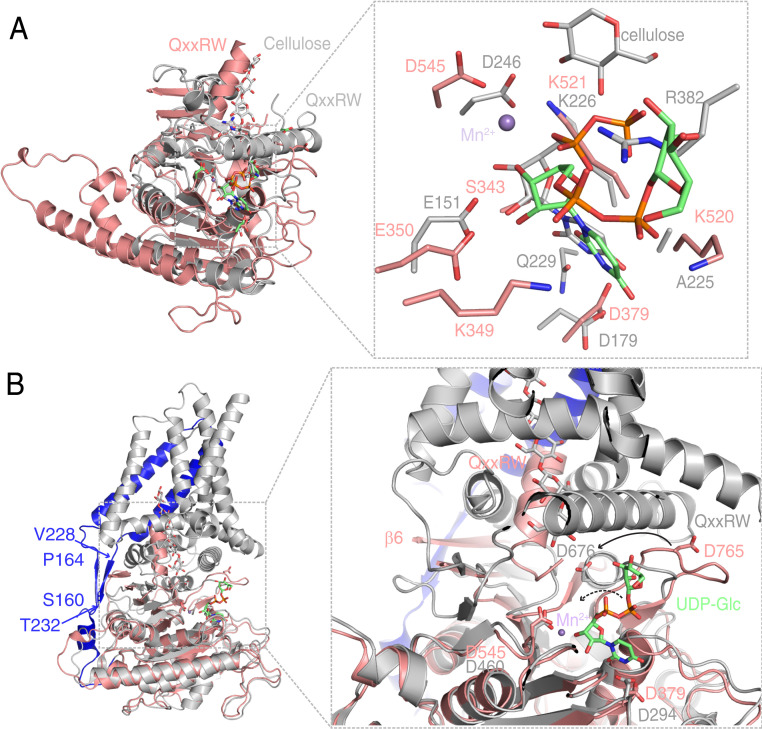
The catalytic mechanism of bacterial cellulose synthesis is well characterized through structural studies of the BcsA–BcsB complex (20, 29, 30). For structural comparison, we aligned the structure of the bacterial BcsA (residues 127 to 401) and the UDP-Glc–bound AtCESA3317−792 using the DALI server (31), with a rmsd of ∼2.7 Å for all main chain atoms, indicating some variations in the structures (Fig. 4A). Particular attention was given to the canonical D, D, D, and QxxRW motifs, D379, D545, D765, and Q803xxR806W807 for AtCESA3 as well as D179, D246, D343, and Q379xxR382W383 for the BcsA, respectively (Fig. 4A). The first two aspartic acids (D379 and D545 in AtCESA3, D179 and D246 in BcsA) interact with the uridine group of UDP-Glc and coordinate the metal ion in both AtCESA3 and BcsA (20) and thus aligned well. However, we observed two major structural differences for the third aspartic acid and the QxxRW motif between AtCESA3 and BcsA. In bacteria, the third D, D343, located in the “finger helix,” serves as the general base to attack the C4 hydroxyl group of the acceptor cellulose chain. Furthermore, the QxxRW motif in BscA helps stabilize the phosphate group of UDP-Glc by R382, as well as the cellulose chain by Q379 and W383. By contrast, the third D, D765, in AtCESA3 resides in a loop connecting β10 and β11, and the corresponding residues, which were observed to form the finger helix in BcsA, were modeled as the β11 strand in our crystal structure. The QxxRW motif, which is located in helix α9 in AtCESA3CatD, here points toward the catalytic pocket, possibly because of its flexibility and lack of stabilization of transmembrane helices (Fig. 4 A and B). In addition, there are some differences in residues that coordinate UDP-Glc. In AtCESA3, residues S343 and E350 form direct interactions with the ribose of UDP-Glc, while Q229 and E151 bind to the counterpart in BcsA. Moreover, K349 and K520 in AtCESA3 form salt bridges with the phosphate group in UDP-Glc, while it is the QxxRW motif in BcsA that aids in coordinating the phosphate group (Fig. 4A). Taken together, the UDP-Glc binding in plant CESAs differs from that in bacteria. Unlike BcsA, which is activated by its C-terminal PilZ domain through binding to c-di-GMP, it remains unclear whether plant CESAs are required to be activated and, if so, how they are activated.
Fig. 4.
Structural comparisons of AtCESA3CatD to BcsA and PttCESA8 reveal different mechanisms in coordinating UDP-Glc. (A) Structural comparison of AtCESA3CatD (salmon) to the BcsA catalytic domain (PDB ID code 4HG6; gray). An enlarged view of the catalytic sites involving UDP-Glc and metal ion binding in AtCESA3 and BcsA is shown on the Right. The bound cellulose and UDP-Glc are shown as sticks. For cellulose, C and O atoms are colored gray and red; for UDP-Glc in BcsA, C, O, N, and P atoms are colored gray, red, blue, and orange, respectively. (B) Structural alignment of AtCESA3CatD and full-length PttCESA8 (PDB ID code 6WLB). The PttCESA8 is colored gray, with its N-terminal region (E157-Q249) prior to the GT domain in blue. The two β-strands connecting the first two transmembrane helices to cytosolic domains are indicated by the starting and ending amino acids, respectively. A detailed view of catalytic sites and the glucan chain channel of AtCESA3CatD and PttCESA8 is shown on the Right. The conserved residues that bind UDP-Glc are show as sticks, and the bound Mn2+ is shown as spheres. The cellulose chain in PttCESA8 is shown as sticks, with C and O atoms colored gray and red. The conformational change in the catalytic residue D765 in AtCESA3CatD compared with its equivalent residue D676 in PttCESA8 is indicated by the solid arrow. A proposed change in UDP-Glc in AtCESA3CatD, following the same change direction as that of D765, is indicated by the dashed arrow. β6 in AtCESA3CatD and canonical QxxRW motif in both structures are labeled. Structural comparison also clearly demonstrates a large conformational change in the helix comprising the QxxRW motif, implying that it is dynamic and flexible.
The homotrimeric cryo-EM structure of the full-length Poplus CESA8 (PttCESA8) was published recently (21), and structural comparison revealed that, overall, our crystal structure of AtCESA3CatD aligned well with that of PttCESA8 (Fig. 4B and Movie S1). The comparison also suggested that the catalytic residue D765 in our structure likely needs to be relocated to its active conformation (D676 in PttCESA8), i.e., positioned between UDP-Glc and cellulose chain sites upon CSC formation (Fig. 4B). Notably, the glucose unit of UDP-Glc in AtCESA3317−792 is in close proximity (∼10 Å) to the nonreducing end of the bound cellulose in the PttCESA8 structure. A proposed change in UDP-Glc, mainly the glucose and phosphate groups, would take place following the same change direction as that of D765, but note that the nucleoside uridine positioning and its binding pocket remain unchanged, consistent with its well-ordered electron density map (Figs. 3B and 4B and Movie S1). The incoming UDP-Glc and nascent glucan chain therefore nicely come together to connect the catalytic site and extruding cellulose channel to explain the catalytic mechanism of cellulose synthesis (Fig. 4B). In addition, the P-CR domain aligns well with the PttCESA8 structure and also with the previously solved rice P-CR domain structure (SI Appendix, Fig. S1C). By contrast, both AtCESA3 and PttCESA8 revealed only parts of the C-SR domain, indicating that the majority of it is, perhaps, intrinsically disordered or flexible.
Apart from the structural observations above, we also noted some differences between the structures of AtCESA3 and PttCESA8 (SI Appendix, Fig. S4C). First, the residues spanning D565 to G617 in AtCESA3 were built as Q571-V575 (β6), N594-D599 (β7), and V612-G615 (β8) (residues from Q576 to N592 and N601 to Q609 were not modeled because of the lack of clear electron density). However, the counterpart in PttCESA8 (D480 to G532) was modeled as a helix with long loops. Second, the residues spanning G768 to M772 were modeled as β11 and T777 to C782 as β12 in AtCESA3, while the counterpart residues in PttCESA8 (G671 to C691) were modeled as the finger helix and R689 to Y692 as the β-strand but occupying β7, rather than β12, as observed in our structure. Such modeling discrepancies may be due to stabilization of the structure by transmembrane helices and the flexibility of certain parts of the protein or combinations of these factors. Nevertheless, taken together, these two structures (one with substrate, one with a nascent cellulose chain) offer detailed insights into cellulose synthesis.
Discussion
Cellulose is of immense importance to a range of industrial applications. Mechanistic insights into how cellulose is produced is critical to engineer cellulose synthesis. We have solved the crystal structure of the AtCESA3 catalytic domain and revealed how it coordinates its cognate substrate UDP-Glc together with the metal ion Mn2+ and outlined how it is involved in homodimerization.
A comparison of AtCESA3’s structure with that of BcsA from bacteria revealed some similarities but also remarkable differences in substrate binding. Glycosyltransferase GT-2 family members catalyze glycosidic bond formation with the aid of a divalent cation (Mn2+ or Mg2+), which is usually coordinated by the second aspartic acid residue in the canonical D, D, D, and QxxRW motif and two phosphates from a nucleoside phosphate, such as UDP-Glc (1, 2). Consistently, the second D (D545) in AtCESA3 coordinates Mn2+ (Fig. 4A). However, local conformational changes of UDP-Glc, especially the glucose and phosphate groups, would here be required to coordinate Mn2 (indicated by the dashed arrow in Fig. 4B). Interestingly, such conformational changes are in line with the potential movement of the catalytic residue D765 and its loop in the AtCESA3 to become aligned with their counterparts (D676) in PttCESA8 (Fig. 4B and Movie S1). The finger helix in BcsA comprising the TED motif is very important for cellulose elongation and translocation (20, 29, 30). As mentioned before, the equivalent regions of the finger helix in our structure and PttCESA8 were modeled as the β-strand and α-helix, respectively. This structural difference is likely the result of the lack of stabilization by amphipathic and transmembrane helices in our structure. Nevertheless, the difference demonstrates the flexibility and structural plasticity of this region. Given the conserved TED motif linking to this region, its flexibility and structural plasticity would be important for its function, particularly for glycosyl transfer and nascent glucan chain translocation. Indeed, the movement and destabilization of the finger helix were proposed previously as it functions in glycosyl transfer and glucan chain translocation (29).
AtCESA3CatD exists in a dimeric state in solution, revealed by both analytical size exclusion chromatography and SAXS. The dimer formation involves β−β-strand backbone interaction. Mutation of C573 to P573 (AtCESA3C573P) abolished the AtCESA3 dimer formation in vitro and in vivo, which further validates the importance of the site for the dimerization. Our CD spectrum measurements of purified AtCESA3CatD and its proline mutants indicate that the purified proline mutation proteins have a secondary structure similar to the wild type. The full-length AtCESA3 and its proline mutants do move through the endomembrane system to the Golgi when tagged with full-length YFP in BiFC experiments, indicating the proline mutation does not introduce folding defects; otherwise, it would be targeted for degradation (SI Appendix, Fig. S5). Although our in vivo assays cannot distinguish the AtCESA3 oligomerizations between dimers and trimers, it was revealed that the P-CR region is involved in CSC trimer formation from the poplar CESA8 structure, so the proline mutations would have little influence on the trimer interface based on the poplar CESA8 structure. Analysis of the β6 sequences of Arabidopsis CESAs shows that the corresponding position of Q571 is favored for lysine or at least polar residues. The position of V572 is hydrophobic, with uncharged residues (V, L, I, and C), and C573 is conserved among all AtCESAs (SI Appendix, Fig. S2). The amino acid conservation implies a common interaction mode between different AtCESAs to form homo- or heterodimers. Notably, homodimerization of CESA4, CESA7, and CESA8 was observed using the irregular xylem (irx) mutants (irx1, irx3, and irx5 with mutations in the three CESA genes involved in secondary wall synthesis, respectively), and the authors speculated that CESA homodimerization perhaps precludes CSC formation in the absence of the complete set of secondary wall CESAs (19). It is therefore plausible that homodimerization occurs prior to the assembly of CSC, perhaps to prohibit premature CSC formation (19, 21). Notably, structural alignment between AtCESA3CatD and that of the PttCESA8 showed that the β6 that we propose partakes in CESA dimerization is partly obscured by two β-strands, formed by S160 to P164 and V228 to T232 in the full-length PttCESA8 homotrimer structure (Fig. 4B). This makes it unlikely that the β6 residues would contribute to the CSC assembly and complex maintenance. However, given the flexibility of the CatD, which is actually rather isolated from the transmembrane domains and the two β-strands (Movie S1), it is plausible that structural shifts of the two β-strands and the CatD in the context of the transmembrane domain may expose β6 for dimer formation prior to CSC assembly. Nevertheless, how such homodimerization of full-length CESAs contributes to CSC trimer formation and assembly requires further investigation.
CESA mutations greatly aided in understanding cellulose synthesis and generally result in dwarf plants with reduced growth and cellulose content (32). Our crystal structures of AtCESA3CatD and the UDP-Glc–bound AtCESA3317−792 offer a wealth of detailed information to help us understand how these mutations affect CESA function and CSC formation. The cesa mutants that have missense mutations in the catalytic domain, with corresponding cellulose defects, are summarized in SI Appendix, Table S3 and shown in SI Appendix, Fig. S4D (2). Mutation sites (P578, D588, and G604) cannot be modeled due to the lack of electron density, indicating they may reside in flexible loops. S377F, A533V, and G617E may disrupt the internal structure of the GT domain, as such mutations yield an enlarged side chain that could cause steric clashes with internal structures. Notably, A522V, G615S, S761L, E764K, and D765N are in close vicinity of the catalytic region, of which D765 is a catalytic residue. In the context of our structure, A522V and G615S may occlude the glucose transfer by altering the catalytic area; S761L and E764K possibly change the electrostatic environment of the catalytic area, which could affect catalysis; D765 is a catalytic residue; and D765N mutation may thus abolish glucose transfer. Hence, based on our structure, these mutations likely affect UDP-Glc binding and catalysis. Consistent with our crystal structure analysis, a number of these important mutations were nicely mapped in computational models of the plant CESAs in the past (16). Remarkably, R437 and H773 are both at the protein surface but still lead to a reduction in root growth and cellulose content when mutated (33, 34). These phenotypes are, however, relatively mild and are likely not due to changes in enzyme activity but possibly affect the CSC assembly and secretion.
In summary, our crystal structures of the apo and UDP-Glc–bound forms of the AtCESA3 CatD reveal how plant CESAs bind to their substrate UDP-Glc. The structure of AtCESA3CatD can form a homodimer that might contribute initial support for the CESAs to become assembled into CSCs. Our data thus outline important information about how cellulose is produced in plant cells and how the CESAs might assemble to functional CSCs.
Materials and Methods
Cloning.
Secondary structure prediction was performed based on the full-length AtCESA3, and subsequently, the catalytic domain was mapped between approximately amino acid 310 and 830. Based on this information, a number of truncations were designed for soluble expression tests. We managed to express the soluble protein of the catalytic domain spanning amino acids 317 to 792, which was the only soluble construct. Because the QXXRW motif is important for the GT family function, we included the predicted QXXRW helix in an extended construct (AtCESA3CatD). AtCESA3317−792 and AtCESA3CatD were cloned into a derivative of the pET28a (+) vector using primers listed in SI Appendix, Table S4 with tobacco etch virus (TEV) protease cleavable N-terminal hexahistidine tag and C-terminal hexahistidine tag, respectively. The constructs with an internal loop (631 to 701) substituted with a GSGSG linker and mutants (AtCESA3CatDQ571P and AtCESA3CatDC573P) were generated by quick-change mutagenesis and verified by sequencing. The full-length coding sequence of AtCESA3 was cloned in pENTR/D-TOPO using primers listed in SI Appendix, Table S4. Point mutations were induced via site-directed mutagenesis using the primers outlined in SI Appendix, Table S4 to generate entry clones for AtCESA3Q571P, AtCESA3C573P, and AtCESA3Q571P:C573P. To generate the full-length translational fusions, an LR reaction was performed between the aforementioned entry clones and the destination vector pEARLYGATE104 that contains the constitutive promoter 35S driving the expressions of YFP upstream of the Gateway site. To generate constructs for bimolecular fluorescence complementation, AtCESA3, AtCESA3Q571P, AtCESA3C573P, and AtCESA3Q571P:C573P were amplified using primers listed in SI Appendix, Table S4 and cloned into pAMON and pSUR (35) using the NEB NEBuilder HiFi DNA Assembly system. All constructs were sequence verified and transformed into Agrobacterium. To generate constructs for the split-ubiquitin yeast two-hybrid system, AtCESA3, AtCESA3Q571P, AtCESA3C573P, and AtCESA3Q571P:C573P were amplified with primers listed in SI Appendix, Table S4 and cloned into PBT3-N and PPR3-N using restriction enzyme digestion and ligation. All constructs were sequence verified and transformed into a NMY51 yeast cell.
Protein Expression and Purification.
The sequenced plasmids expressing the AtCESA3 catalytic domains were transformed into E. coli BL21 (DE3). Bacteria were induced by 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 16 °C for 16 h for protein expression and harvested by centrifugation at 4,000 × g for 10 min. The pellet was resuspended in buffer (25 mM HEPES pH 7.5, 150 mM NaCl), lysed by an LM20 microfludizor, and clarified by centrifugation at 20,000 rpm (JA- 25.5, Beckman) at 4 °C for 1 h. The supernatant was collected and incubated with 5 mL Ni-nitrilotriacetic acid (NTA) beads (Biobasic), and the eluted fractions were pooled. For purification of N-terminal cleavable hexahistidine tag protein (AtCESA3317−792), the protein was dialyzed overnight against buffer (25 mM HEPES pH 7.5, 150 mM NaCl, 5 mM β-mercaptoethanol [β-ME]) by adding TEV protease to remove the hexahistidine tag. For the C-terminal uncleavable hexahistidine tagged protein (AtCESA3317−810 and its mutants), the proteins were further purified by a Hitrap Q HP column (GE Healthcare). All the proteins were further purified by loading to a HiLoad Superdex 200 16/60 (GE Healthcare) which was preequilibrated with buffer (25 mM HEPES pH 7.5, 150 mM NaCl, 5 mM β-ME). The eluted protein fractions were pooled, concentrated to 10 mg/mL, and stored at −80 °C for later use after snap freezing in liquid nitrogen.
Analytical Size Exclusion Chromatography.
The Superdex 200 increase 10/300 GL column (GE Healthcare) preequilibrated with buffer (25 mM HEPES pH 7.5, 150 mM NaCl, 5 mM β-ME) was used for analytical size exclusion chromatography following the operation manual. Briefly, 100 μL AtCESA3 proteins (AtCESA3317−792, AtCESA3CatD, AtCESA3CatDQ571P, and AtCESA3CatDC573P) were loaded onto the column at a concentration of 2 mg/mL. All size exclusion chromatography profiles were exported and analyzed by Unicorn software.
Circular Dichroism Spectra Measurement of AtCESA3CatD.
AtCESA3 wild type, AtCESA3CatDQ571P, and AtCESA3CatDQ571P were buffer exchanged to 25 mM HEPES pH 7.5, 150 mM NaCl, and 5 mM β-ME. Two hundred microliters of 0.5 mg/mL protein samples were loaded to a 0.1 mm quartz cuvette. The CD spectrum was recorded by Chirascan from 196 to 260 nm with 0.5 nm per step and plotted by Excel.
SAXS Data Collection and Analysis.
SAXS data for AtCESA3CatD were collected with a Xenocs Nano-inXider SAXS instrument equipped with a microfocus sealed-tube X-ray source (Cu, 30 W, 40 μm focus) and a Dectris Pilatus 3 hybrid pixel detector. The X-rays are filtered through the two-dimensional single-reflection multilayer optics and collimated by a three-pinhole system. The sample-to-detector distance was set at 0.94 m, and the sample chamber and X-ray paths were evacuated prior to usage. This setup covers a range of momentum transfer of 0.08 < q < 4 nm−1 [q = 4π sin(θ)/λ, where 2θ is the scattering angle]. SAXS experiments of AtCESA3CatD were carried out at room temperature in the buffer composed of 25 mM HEPES pH 7.5, 150 mM NaCl, and 5 mM β-ME using the low-noise flow cell. The protein concentrations used were 2 and 5 mg/mL The data were collected for 60 min, and for each measurement a total of six frames at 10 min intervals were recorded. The scattered X-rays detected by a two-dimensional area detector were converted to one-dimensional scattering using the built-in SAXS software (Xenocs).
All the data processing steps were performed using the program package PRIMUS (36, 37). The scattering of the buffer was subtracted from the data. The experimental data obtained were analyzed for aggregation using a Guinier plot (38). The forward scattering I(0) and the radius of gyration Rg were computed using the Guinier approximation, which assumes that at very small angles (q < 1.3/Rg), the intensity is represented as I(q) = I(0) × exp[−(qRg)2/3] (38). These parameters were also computed from the extended scattering patterns using the indirect transform package GNOM (39), which provides the distance distribution function P(r) and hence the maximal particle dimension Dmax and the radius of gyration Rg. The hydrated volume Vp, which was used to estimate the molecular mass of the protein, was computed using the Porod invariant. The theoretical scattering profiles were generated from the crystallographic structure of AtCESA3CatD and evaluated against the experimental scattering profile using CRYSOL (40).
Crystallization and Structure Determination.
The protein samples were concentrated to 10 mg/mL by a 10 kDa MWCO concentrator (Millipore). Intelli-Plate 96-3 sitting drop crystallization plates (Hampton Research) were used for initial screening, which was set up by Mosquito (SPTLabtech) with three drop ratios (100:50, 75:75, 50:100/nL) using a commercial screening kit (Crystal screening, Index, Salt Rx, PEG Ion, PEG Rx) from Hampton Research. The kits were then stored in RockImager (Formulatrix) for imaging and tracking. The native and Se-Met derivative AtCESA3317−792 crystals were crystalized in 100 mM 2-(N-morpholino)ethanesulfonic acid (MES) pH 6.0, 5% polyethylene glycol (PEG) 3350, and 100 mM MnCl2. The UDP and UDP-Glc at a concentration of 10 mM were mixed with AtCESA3317−792 prior to crystallization to get the ligand-bound structure of AtCESA3. AtCESA3CatD was crystallized in 100 mM MES pH 6.5, 2% PEG 3350, and 200 mM potassium sodium tartrate. All the crystals were cryoprotected by adding 25% glycerol to the mother liquor. SAD datasets for Se-Met–labeled AtCESA3317−792 were collected at an inflection wavelength of 0.9795 Å at the Swiss Light Source. The native datasets of AtCESA3317−792 and AtCESA3CatD were collected from the Australian Light Source and the Taiwan Light Source. All the datasets were processed using XDS (41). The Phenix AutoSol program was used for phasing, and 20 selenium atoms were found in the substructure solution (figure of merit: 0.6). The models were built and refined using Phenix and Coot (42, 43). The resultant structure served as a template to solve both AtCESA3317−792 and AtCESA3CatD structures by molecular replacement. All the structure-related figures were generated by PyMol (44). The movie comparing AtCESA3CatD with PttCESA8 structure was generated by Chimera (45).
Split-Ubiquitin Analyses in Yeast Cells.
Split-ubiquitin yeast two-hybrid analysis was performed using a DUALmembrane starter kit (Dualsystems Biotech) following the manufacturer’s instructions. The yeast strain NMY51 was transformed with both bait and prey plasmids, and positive colonies were selected on synthetic-defined (SD) medium lacking leucine and tryptophan (SD/-Leu-Trp). Protein–protein interactions were detected after 12 d growth on SD medium additionally lacking adenine and histidine (SD/-Leu-Trp-His-Ade) and supplemented with 5 or 10 mM 3-aminotriazole (3-AT) to prevent the slight leakiness of the HIS3 gene. The bait PTSU2-APP and prey PNubG-Fe65 plasmids were used as a positive control, and the PBT3-N and PPR3-N plasmids were used as negative controls in the DUALmembrane protein functional assay.
Bifluorescent Complementation Analyses in Epidermal Tobacco Leaf Cells.
BiFC assays were carried out as previously described (46), except that the concentration of bacteria was increased to optical density (OD)600 0.2. Leaves coinfiltrated with Agrobacterium strains carrying 35S::CFP-N7 (47) and P19 (48) as well as the constructs of interest were examined for fluorescence 3 d postinfiltration and imaged as previously described (49) on an inverted Nikon Ti-E microscope body equipped with a CSU-W1 spinning disk head (Yokogawa). Sequential detection occurred using a 100× oil-immersion objective (Apo TIRF, numerical aperture [NA] 1.49) and a deep-cooled iXon Ultra 888 EM-CCD (Andor Technology). All BiFC combinations were imaged under the same conditions.
Supplementary Material
Acknowledgments
We thank beamline scientists at National Synchrotron Radiation Research Center (NSRRC, Taiwan), the Swiss Light Source, and Australian Synchrotron for their technical assistance in data collection. We also thank Dr. Liew Chong Wai from the NTU Institute of Structural Biology for assistance in data collection. We would like to acknowledge the Facility for Analysis, Characterization, Testing and Simulation, Nanyang Technological University, Singapore, for use of X-ray facilities. This work was supported by Tier II Grant MOE2019-T2-2-099 from the Ministry of Education of Singapore (to Y.-G.G.), Australian Research Council Future Fellowship and Discovery Project Grants DP190101941 and FT160100218 (to S.P.), Villum Investigator Grant Project ID 25915 (to S.P.), Australian Academy of Science Thomas Davies Research Grant (to E.R.L.), and Novo Nordisk Laureate Grant NNF19OC0056076 (to S.P.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2024015118/-/DCSupplemental.
Data Availability
Atomic coordinates and structure factors for AtCESA3317−792, UDP-Glc–bound AtCESA3317−792, and AtCESA3CatD have been deposited in the Protein Data Bank (PDB) with PDB ID codes 7CK1, 7CK2, and 7CK3, respectively.
References
- 1.Lairson L. L., Henrissat B., Davies G. J., Withers S. G., Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555 (2008). [DOI] [PubMed] [Google Scholar]
- 2.McFarlane H. E., Döring A., Persson S., The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 65, 69–94 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Mueller S. C., Brown R. M. Jr., Evidence for an intramembrane component associated with a cellulose microfibril-synthesizing complex in higher plants. J. Cell Biol. 84, 315–326 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purushotham P., et al., A single heterologously expressed plant cellulose synthase isoform is sufficient for cellulose microfibril formation in vitro. Proc. Natl. Acad. Sci. U.S.A. 113, 11360–11365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vandavasi V. G., et al., A structural study of CESA1 catalytic domain of Arabidopsis cellulose synthesis complex: Evidence for CESA trimers. Plant Physiol. 170, 123–135 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nixon B. T., et al., Comparative structural and computational analysis supports eighteen cellulose synthases in the plant cellulose synthesis complex. Sci. Rep. 6, 28696 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richmond T. A., Somerville C. R., The cellulose synthase superfamily. Plant Physiol. 124, 495–498 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arioli T., et al., Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Scheible W. R., Eshed R., Richmond T., Delmer D., Somerville C., Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis Ixr1 mutants. Proc. Natl. Acad. Sci. U.S.A. 98, 10079–10084 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Persson S., et al., Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 15566–15571 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner S. R., Somerville C. R., Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9, 689–701 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor N. G., Howells R. M., Huttly A. K., Vickers K., Turner S. R., Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. U.S.A. 100, 1450–1455 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill J. L. Jr., Hammudi M. B., Tien M., The Arabidopsis cellulose synthase complex: A proposed hexamer of CESA trimers in an equimolar stoichiometry. Plant Cell 26, 4834–4842 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonneau M., Desprez T., Guillot A., Vernhettes S., Höfte H., Catalytic subunit stoichiometry within the cellulose synthase complex. Plant Physiol. 166, 1709–1712 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purushotham P., Ho R., Zimmer J., Architecture of a catalytically active homotrimeric plant cellulose synthase complex. Science 369, 1089–1094 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Sethaphong L., et al., Tertiary model of a plant cellulose synthase. Proc. Natl. Acad. Sci. U.S.A. 110, 7512–7517 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rushton P. S., et al., Rice cellulose synthaseA8 plant-conserved region is a coiled-coil at the catalytic core entrance. Plant Physiol. 173, 482–494 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurek I., Kawagoe Y., Jacob-Wilk D., Doblin M., Delmer D., Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proc. Natl. Acad. Sci. U.S.A. 99, 11109–11114 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atanassov I. I. II, Pittman J. K., Turner S. R., Elucidating the mechanisms of assembly and subunit interaction of the cellulose synthase complex of Arabidopsis secondary cell walls. J. Biol. Chem. 284, 3833–3841 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Morgan J. L., Strumillo J., Zimmer J., Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493, 181–186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olek A. T., et al., The structure of the catalytic domain of a plant cellulose synthase and its assembly into dimers. Plant Cell 26, 2996–3009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scavuzzo-Duggan T. R., et al., Cellulose synthase ‘class specific regions’ are intrinsically disordered and functionally undifferentiated. J. Integr. Plant Biol. 60, 481–497 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Hendrickson W. A., Anomalous diffraction in crystallographic phase evaluation. Q. Rev. Biophys. 47, 49–93 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tarbouriech N., Charnock S. J., Davies G. J., Three-dimensional structures of the Mn and Mg dTDP complexes of the family GT-2 glycosyltransferase SpsA: A comparison with related NDP-sugar glycosyltransferases. J. Mol. Biol. 314, 655–661 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Baker J. A., Wong W. C., Eisenhaber B., Warwicker J., Eisenhaber F., Charged residues next to transmembrane regions revisited: “Positive-inside rule” is complemented by the “negative inside depletion/outside enrichment rule.” BMC Biol. 15, 66 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Joseph P. R., et al., Proline substitution of dimer interface β-strand residues as a strategy for the design of functional monomeric proteins. Biophys. J. 105, 1491–1501 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paredez A. R., Somerville C. R., Ehrhardt D. W., Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312, 1491–1495 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Morgan J. L., et al., Observing cellulose biosynthesis and membrane translocation in crystallo. Nature 531, 329–334 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan J. L., McNamara J. T., Zimmer J., Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat. Struct. Mol. Biol. 21, 489–496 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holm L., Benchmarking fold detection by DaliLite v.5. Bioinformatics 35, 5326–5327 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Williamson R. E., et al., Morphology of rsw1, a cellulose-deficient mutant of Arabidopsis thaliana. Protoplasma 215, 116–127 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Zhong R., Morrison W. H. III, Freshour G. D., Hahn M. G., Ye Z. H., Expression of a mutant form of cellulose synthase AtCesA7 causes dominant negative effect on cellulose biosynthesis. Plant Physiol. 132, 786–795 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bosca S., et al., Interactions between MUR10/CesA7-dependent secondary cellulose biosynthesis and primary cell wall structure. Plant Physiol. 142, 1353–1363 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J. E., Lampugnani E. R., Bacic A., Golz J. F., SEUSS and SEUSS-LIKE 2 coordinate auxin distribution and KNOXI activity during embryogenesis. Plant J. 80, 122–135 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Konarev P. V., Petoukhov M. V., Volkov V. V., Svergun D. I., ATSAS 2.1, a program package for small-angle scattering data analysis. J. Appl. Cryst. 39, 277–286 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I., PRIMUS: A windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 36, 1277–1282 (2003). [Google Scholar]
- 38.Guinier A., Diffraction of X-rays of very small angles-application to the study of unltramicroscopic phenomenon. Ann. Phys. (Paris) 12, 161–237 (1939). [Google Scholar]
- 39.Svergun D. I., Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 25, 495–503 (1992). [Google Scholar]
- 40.Svergun D., Barberato C., Koch M. H. J., CRYSOL–A program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 28, 768–773 (1995). [Google Scholar]
- 41.Kabsch W., Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams P. D., et al., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeLano W. L., PyMOL: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 40, 82–92 (2002). [Google Scholar]
- 45.Pettersen E. F., et al., UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Sánchez-Rodríguez C., et al., The cellulose synthases are cargo of the TPLATE adaptor complex. Mol. Plant 11, 346–349 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Quon T., Lampugnani E. R., Smyth D. R., PETAL LOSS and ROXY1 interact to limit growth within and between sepals but to promote petal initiation in Arabidopsis thaliana. Front. Plant Sci. 8, 152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garabagi F., Gilbert E., Loos A., McLean M. D., Hall J. C., Utility of the P19 suppressor of gene-silencing protein for production of therapeutic antibodies in Nicotiana expression hosts. Plant Biotechnol. J. 10, 1118–1128 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Schneider R., et al., Two complementary mechanisms underpin cell wall patterning during xylem vessel development. Plant Cell 29, 2433–2449 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for AtCESA3317−792, UDP-Glc–bound AtCESA3317−792, and AtCESA3CatD have been deposited in the Protein Data Bank (PDB) with PDB ID codes 7CK1, 7CK2, and 7CK3, respectively.