Significance
The GII.4 genotype of human noroviruses accounts for the majority of gastroenteritis outbreaks worldwide. This predominance is characterized by the chronological emergence of new variants in response to immune pressure. We performed a comprehensive analysis on a large panel of GII.4 viruses to investigate the genetic determinants of antigenic diversification. Using immunoassays and neutralization data, we observed major changes to antigenic properties over the course of GII.4 evolution and increased intervariant cross-reactivity across contemporary viruses. Antigenic cartography and sequence analyses indicated a minimum number of coevolving amino acid changes on the structural protein necessary for the emergence of antigenically distinct variants. These insights could facilitate the monitoring and characterization of emerging GII.4 noroviruses and the development of cross-protective vaccines.
Keywords: norovirus, GII.4 noroviruses, pandemic, antigenic cartography, antigenic mapping
Abstract
Noroviruses are the predominant cause of acute gastroenteritis, with a single genotype (GII.4) responsible for the majority of infections. This prevalence is characterized by the periodic emergence of new variants that present substitutions at antigenic sites of the major structural protein (VP1), facilitating escape from herd immunity. Notably, the contribution of intravariant mutations to changes in antigenic properties is unknown. We performed a comprehensive antigenic analysis on a virus-like particle panel representing major chronological GII.4 variants to investigate diversification at the inter- and intravariant level. Immunoassays, neutralization data, and cartography analyses showed antigenic similarities between phylogenetically related variants, with major switches to antigenic properties observed over the evolution of GII.4 variants. Genetic analysis indicated that multiple coevolving amino acid changes—primarily at antigenic sites—are associated with the antigenic diversification of GII.4 variants. These data highlight complexities of the genetic determinants and provide a framework for the antigenic characterization of emerging GII.4 noroviruses.
Human noroviruses are a major worldwide cause of acute gastroenteritis. Symptoms typically manifest 12 to 48 h following contact with contaminated food or infected persons and include vomiting, diarrhea, cramps, and abdominal pain. Due to the nature of transmission, norovirus outbreaks are frequent in enclosed environments, such as schools, cruise ships, hospitals, nursing homes, and military bases (1–4), particularly during the winter season (5). Norovirus illness is self-limiting in healthy individuals, though continued asymptomatic viral shedding may persist for several weeks after recovery (6–8). In contrast, norovirus infections can be life-threatening to children, the elderly, malnourished, and immunocompromised individuals (9–11). Consequently, the majority of the estimated 200,000 deaths annually attributed to norovirus disproportionately occur in developing countries (10, 12). Unlike rotavirus, another major cause of gastroenteritis, there are currently no vaccines approved for norovirus.
Human noroviruses belong to the Caliciviridae family and present a ∼7.5 kb, single-stranded, positive-sense RNA genome organized into three open reading frames (ORFs). ORF1 encodes six nonstructural proteins involved in viral replication (NS1/2, NS3, NS4, NS5, NS6, and NS7), while ORF2 and ORF3 encode the major (VP1) and minor (VP2) capsid proteins, respectively. A mature norovirus virion is composed of 180 VP1 copies organized as an icosahedron with T = 3 symmetry (13). Heterologous expression of VP1 results in the self-assembly of virus-like particles (VLPs) that antigenically resemble wild-type virions (14). The VP1 protein is divided into two structurally defined domains: the shell (S) and the protruding (P), the latter of which is further organized into P1 and P2 subdomains (13). Studies have shown that key antigenic sites are mapped to the surface-exposed P2 subdomain (15–22). Strong interactions have also been identified between the P2 and cellular histo-blood group antigens (HBGA), which facilitate virus binding and infection on gut epithelial cells (23–25). Antibodies that block norovirus interaction with HBGA have been correlated with disease protection in human subjects (26), and are frequently employed as a surrogate of norovirus neutralization due to the strong correlation among the two assays (27, 28).
Noroviruses are organized into 10 genogroups (GI to GX) containing over 40 genotypes based on sequence differences of the VP1 structural protein (29). While over 30 of these genotypes (primarily within GI and GII) can infect humans, the GII.4 viruses have been responsible for over 70% of norovirus outbreaks worldwide for the past two decades (30). This sustained dominance is associated with the chronological emergence of new GII.4 variants, which persist for years until replacement by the next variant (Fig. 1A) (30). To date, six major variants have been reported on a pandemic scale—Grimsby 1995, Farmington Hills 2002, Hunter 2004, Den Haag 2006b, New Orleans 2009, and Sydney 2012—and four minor variants mostly associated with geographically restricted outbreaks: Sakai 2003, Yerseke 2006a, Osaka 2007, Apeldoorn 2007. The phylogenetic tree of GII.4 (Fig. 1B) shows the chronological relationship and diversification of these variants. The precise mechanism for GII.4 variant succession is poorly understood. The prevailing model posits an epochal evolution similar to that of H3N2 influenza viruses, wherein chronological succession is driven by selective pressures from herd immunity against older variants (31–33). Notably, the topology of the phylogenetic tree of GII.4 norovirus VP1 suggests that most successors are contemporary variants circulating at low levels, rather than directly evolved from the predominant virus (17, 34).
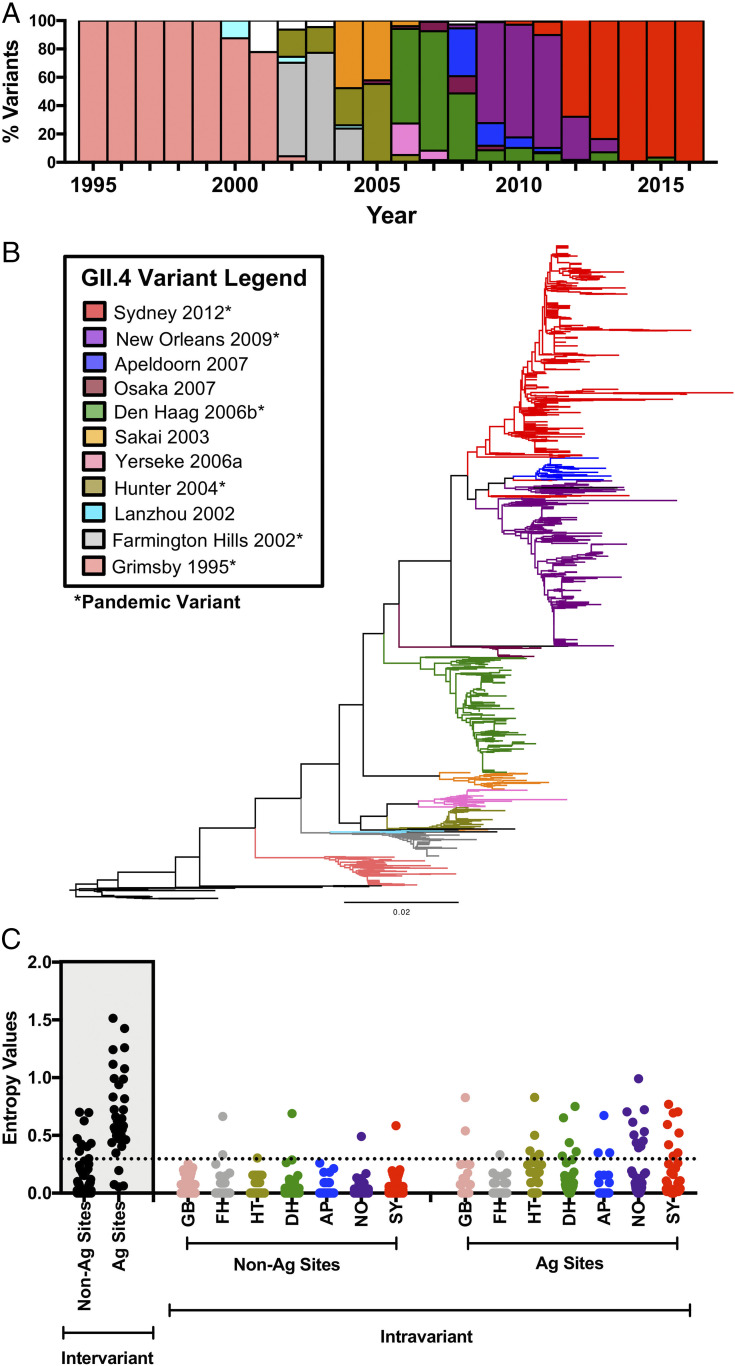
Fig. 1.
Conservation analysis of the major capsid protein of GII.4 noroviruses shows differential accumulation of amino acid changes at the inter- and intravariant level. (A) Year ranges of the chronological emergence, persistence and extinction of major GII.4 norovirus variants. VP1 sequences from 1,572 GII.4 viruses between the years 1995 and 2016 were used for this figure. Colored bars represent the major GII.4 variants within this time span with the same color scheme from B. Viruses that did not cluster with any of these variants are denoted in white. (B) Maximum-likelihood phylogenetic tree of major chronological GII.4 variants between the years 1974 and 2016. VP1 capsid amino acid sequences from 1,601 GII.4 noroviruses were used for tree construction. Colored branches represent antigenically distinct variants, while black branches denote viruses that did not cluster with any of these variants. Pandemic variants are marked with an asterisk. (C) Shannon entropy values were calculated to quantify amino acid changes accrued on nonantigenic (Non-Ag) or antigenic (Ag) residues of the VP1 capsid protein. These analyses were conducted at the intervariant (black points, shaded area) and intravariant (colored points) level. Variant names are abbreviated for convenience but retain the same color scheme from B. Residues with a Shannon entropy value ≤ 0.3 (dotted line) are considered conserved sites, while residues that exceed this score are considered variable (17).
Continuous selective pressures from herd immunity have resulted in accumulated amino acid substitutions in the GII.4 VP1 protein (17, 35). The majority of these substitutions are localized to at least five variable motifs (or antigenic sites A, C, D, E, and G) mapped to the P2 subdomain (SI Appendix, Fig. S1) (17). Mutations on these antigenic sites have been associated with variant emergence and changes in the antigenicity of GII.4 viruses (16, 17, 36). Contrary to the linear accumulation of amino acid changes at the intervariant level, the intravariant level presents a stochastic pattern of substitutions (17). The influence of those changes on intravariant antigenic diversification is poorly understood.
In this study we explore the factors involved in the antigenic diversification of GII.4 noroviruses at the inter- and intravariant level. A comprehensive HGBA blockade profile was generated for a large panel of GII.4 VLPs derived from archival viruses representing major and minor GII.4 variants. The antigenic relationship among the variants was validated with neutralization assays using the stem-cell–derived enteroid system for norovirus cultivation. Antigenic cartography of variant clustering was cross-analyzed against amino acid changes accrued to antigenic sites to establish the threshold for meaningful antigenic diversification.
Results
Intravariant Diversification of GII.4 Is Mostly Present at Antigenic Sites.
GII.4 norovirus evolution is characterized by an accrual of amino acid mutations at the P2 subdomain of the VP1 capsid protein (17) and chronological diversification in discrete variants (Fig. 1 A and B). A recent analysis of amino acid diversity (represented by Shannon entropy values) using VP1 sequences from 1,572 GII.4 viruses detected from 1995 to 2016 revealed that these changes are localized to a comparatively small number of residues that constitute key antigenic sites (17). Notably, while intravariant diversification was recorded, changes did not significantly accumulate during the circulation of variants (17). As a follow-up analysis, we calculated the Shannon entropy values of GII.4 noroviruses at the intravariant level (Fig. 1C). Results showed a higher number of changes at variable antigenic sites as compared to those from nonantigenic sites; however, these values were substantially lower than those observed at the intervariant level.
Differential Blockade Clustering between Archival and Contemporary GII.4 Variants.
An ongoing obstacle in the field of norovirus research is the lack of a traditional cell culture system for viral amplification and sera microneutralization analysis. This can be circumvented by the implementation of VLPs and HBGA blockade assays, which have been shown to correlate with neutralization assays (27, 28). To that end, we generated a panel of representative GII.4 norovirus VLPs encompassing the 10 variants from Grimsby 1995 to Sydney 2012 (SI Appendix, Table S1). The predominant virus sequence was selected as a representative for each respective variant. Additionally, up to two minor viruses were selected to account for the broad range of intravariant sequence diversity, particularly at antigenic sites. The list of VLPs and their amino acid differences at respective antigenic sites can be found in SI Appendix, Fig. S1.
Comprehensive HBGA blocking profiles were generated against the VLP library to explore the antigenic relationships between chronologically separated GII.4 variants. HBGA blockade titers, represented as half-maximal effective concentrations (EC50), were measured with hyperimmune mouse sera raised against VLPs from 10 viruses. These comprised six representative viruses from pandemic variants (GR_Grimsby 1995, FH_MD2004-3 2004, HT_Cumberland 2004, DH_DenHaag89 2006, NO_Virginia 2010, and SY_RockvilleD1 2012) and four representative viruses from minor variants (YE_Yerseke38 2006, SA_Sakai 2005, OS_Osaka 2007, and AP_Iwate4 2008) (Fig. 2). Robust blockade titers were observed between homologous sera and counterpart VLP interactions, and variable blockade titers were observed with heterologous sera (SI Appendix, Fig. S2). Interestingly, sera derived from archival GII.4 variants displayed minimal intervariant blockade reactivity, while broader cross-blockade was measured in variants that emerged after 2006 (i.e., Den Haag 2006b, Osaka 2007, Apeldoorn 2007, New Orleans 2009, and Sydney 2012) (Fig. 2A). Bidirectional cross-blockade was observed for variants within the contemporary cluster, but cross-blockade trends were, in general, unidirectional interactions against older variants (e.g., SY_RockvilleD1 2012 sera also blocked viruses in variants from New Orleans 2009 to Den Haag 2006b). Unidirectional blockade was observed on a more limited scale between Yerseke 2006a and the predominant Hunter 2004 virus. Rare examples of unidirectional blocking toward later emerged variants were also detected; however, these were the product of individual mice rather than a widespread trend. Unexpectedly, several of the more diverged intravariant representatives (e.g., HT_SG4091 2006 and SY_Arg12866 2014) showed only minimal interaction with sera derived from respective predominant variants. In particular, HT_SG4091 2006 did not present any cross-reactivity with the major Hunter 2004 virus (HT_Cumberland 2004) hyperimmune sera. Conversely, the HT_SG4091 2006-raised hyperimmune sera similarly failed to block any other VLPs from our panel, including the other two Hunter 2004 VLPs (SI Appendix, Fig. S3).
Fig. 2.
Comprehensive HBGA blockade assays reveal differential cross-reactivities between modern and archival GII.4 norovirus VLPs. (A, Left) Phylogenetic tree of representative GII.4 noroviruses, organized by amino acid sequence of the VP1 capsid protein. Colored branches represent variant clustering as indicated in Fig. 1. An asterisk (*) indicates viruses selected for generation of hyperimmune sera. (Right) Summary heat map of EC50 blockade titers of hyperimmune mouse sera against representative GII.4 VLPs. Rows are mapped to GII.4 VLPs on the phylogenetic tree. Columns denote hyperimmune mouse sera against one of the VLPs from their respective variant. Sera from four mice were employed as biological replicates for each sera and assayed as technical duplicates. Color gradient denotes EC50 blockade titers of each VLP:sera interaction. (B) Sera-specific, dose-dependent neutralization of Sydney 2012 virus 011617 in HEIs. Interactions with serial dilutions of mouse hyperimmune sera from A are color coded according to their respective variant. Data points indicated genome copies per microliter RNA of individual experimental replicates. Control measurements of 011617 as an input virus, 1 hpi, 3 dpi, and 3 dpi with sera raised against GII.17 VLP as non-GII.4 control, with 1 hpi set as the neutralization threshold (dashed line).
To verify the biological relevance of the observed blockade results, the neutralizing capabilities of hyperimmune mouse sera was tested in stem-cell derived human intestinal enteroid (HIE) cells. Sera from two representative mice for each virus used in Fig. 2A were tested against GII.4 011617 (GenBank accession no. MN782359), an infectious Sydney 2012 variant virus that showed replication in HIE cells and that differs in three amino acids on nonantigenic sites as compared with SY_RockvilleD1 2012 (27). With the exception of GR_Grimsby 1995 and HT_Cumberland 2004 sera, results showed a dose-dependent reduction in GII.4 011617 genome copies across all major variant sera (Fig. 2B). A stronger pattern of neutralization was observed from the SY_RockvilleD1 2012 sera, as well as sera raised against other viruses in the contemporary cluster, consistent with the observed HBGA blockade results. Partial neutralization was observed at high sera concentrations for the nonblocking pre-2006 viruses, which suggests the presence of additional antigenic sites and antibodies that are involved in neutralizing but not in blocking the HBGA binding. Additionally, no neutralization was observed from the sera raised against a different genotype (GII.17), supporting data that GII.4 variants are part of the same serotype (27). Overall, these results reinforce the biological relevance of the blockade data and downstream analyses of this study.
Antigenic Cartography Shows Intervariant Clustering between Contemporary GII.4 Viruses.
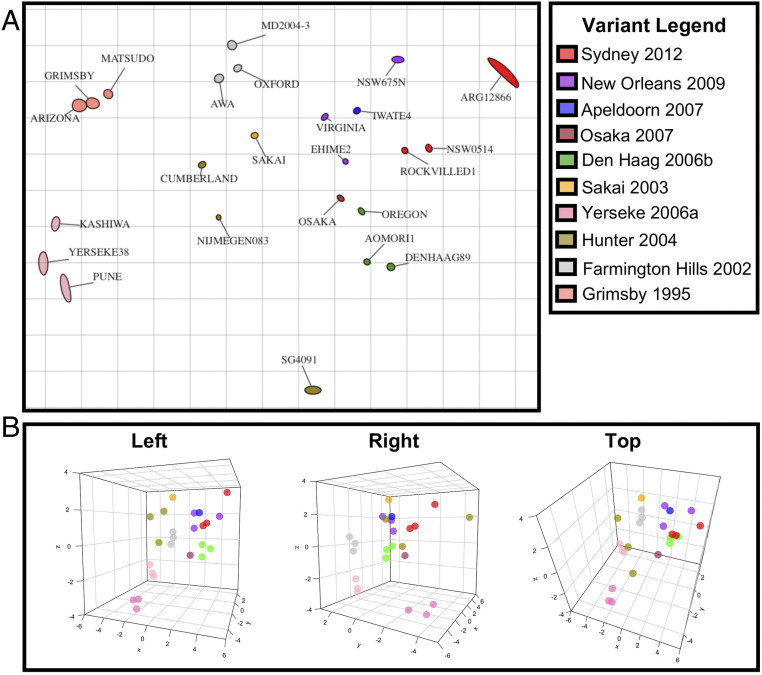
While summarizing HBGA blockade titers as a collection of one-to-one interactions between each respective antigen and sera provides important insights on the level of cross-reactivity among different viruses, it is unable to directly measure antigenic distances between the entire panel of tested viruses. In the field of influenza, this issue has been addressed by using multidimensional scaling analyses (antigenic cartography) (37). Thus, we employed antigenic cartography to further characterize the antigenic relationships of GII.4 viruses at the inter- and intravariant level. We constructed a two-dimensional (2D) projection that geometrically mapped EC50 blockade titers between antigen and sera as antigenic distance where one unit of antigenic distance corresponds to a twofold blockade difference (37). Consistent with the patterns observed in HBGA blocking assays, intravariant clustering was observed except for Hunter 2004 variant (Fig. 3A). Similar to the blockade titer analysis, Grimsby 1995, Farmington Hills 2002, Hunter 2004, and Yerseke 2006a—variants whose sera overwhelmingly blocked only homologous VLPs—were shown to each be antigenically distant from the other viruses when clustering was examined at 2D projections (Fig. 3A). As a notable exception, the virus HT_SG4091 2006 showed extreme antigenic separation from the other two Hunter viruses, consistent with the unique blockade profile (SI Appendix, Fig. S3). In contrast, closer intervariant clustering was observed between the contemporary viruses with cross-reactive blockade profiles (Fig. 2).
Fig. 3.
Antigenic cartography reveals clustering relationships of representative GII.4 VLPs. (A) Two-dimensional antigenic map of GII.4 norovirus VLPs derived from EC50 blockade titers against hyperimmune sera. Variants are color coded for parity with the phylogenetic tree in Fig. 1. Each unit of antigenic distance (grid lines) denote a twofold difference in HBGA blockade titers. Individual viruses are displayed as blobs with a stress parameter of 0.1. (B) Three-dimensional cartographic projection of GII.4 variant clustering. Cluster positions from key angles (left, right, top) are displayed for added context. One unit of antigenic distance denotes a twofold difference in HBGA blockade titers.
To verify that clustering among Den Haag 2006b, Osaka 2007, Apeldoorn 2007, New Orleans 2009, and Sydney 2012 variants was not an artifact of the 2D projection, antigen coordinates were reconfigured into three-dimensional (3D) projections (Fig. 3B). A fully animated 3D projection (displaying both antigens and sera) can be found in Movie S1. The results of the 2D projection were largely recapitulated in 3D space, though a 3D projection recapitulated the distance matrix generated from EC50 blockade titers better than 2D projection (regression analyses assessing table distance vs. map distance generated an adjusted R2 = 0.41 for 3D projection, R2 = 0.35 for 2D projection) (SI Appendix, Fig. S4). To investigate the degree to which minor variants influenced observed clustering trends, 2D and 3D antigenic cartography were conducted using only sera and VLPs from the major GII.4 variants (SI Appendix, Fig. S5). This resulted in refined clustering of viruses that emerged after 2006. Analyses of table vs. map distances were conducted for all tested projections, which demonstrated consistently higher R2 scores for 2D and 3D projections (adjusted R2 = 0.41 for 2D projection, and R2 = 0.52 for 3D projection) when compared with projections done using all the variants, illustrating how intermediate viruses influence major clustering trends and robustness of antigenic relationship projections.
Complex Relationship among Amino Acid Determinants That Lead to Antigenic Differences among GII.4 Viruses.
Previous studies identified that ∼87% of the VP1 sequence is highly conserved, while the majority of amino acid differences accrue at antigenic sites (17). Indeed, the phylogenetic organization of the representative GII.4 viruses was preserved even when only the antigenic sites were considered (Fig. 4A). In contrast, calculations using only the nonvariable VP1 regions (ΔAgs) disrupt the intervariant phylogenetic organization, reaffirming the importance of the antigenic sites for variant diversity. Interestingly, systematic omissions of single antigenic sites showed minimal changes to tree organization, suggesting that multiple sites play a role in diversification (SI Appendix, Fig. S6).
Fig. 4.
Phylogenetic organization of analyzed GII.4 viruses is largely influenced by residues present in antigenic sites. (A) Maximum-likelihood phylogenetic trees of representative GII.4 viruses calculated using the full VP1 amino acid sequence (full VP1), only the residues involved in antigenic sites (only Ags), or the full VP1 sequence with antigenic sites excluded (ΔAgs). (B) Heatmap of pairwise amino acid differences between representative virus pairs. Amino acid differences are displayed for full VP1, only Ags, and ΔAgs sequences. Organization of viruses in heatmaps followed those for full VP1 to facilitate direct comparisons. Color gradients correspond to the number of amino acid differences between virus pairs and have been normalized to the maximum differences recorded for each map.
To explore the relationship between amino acid diversity and antigenic differences, we generated heatmaps of pairwise amino acid differences among all viruses tested in this study (Fig. 4B). Results indicated minimal amino acid differences within intervariant boundaries within the context of both whole VP1 sequences and antigenic sites. Surrounding intervariant amino acid differences, particularly on the antigenic sites, showed a major grouping between Osaka 2007, Den Haag 2006b, Apeldoorn 2007, New Orleans 2009, and Sydney 2012, and a second clustering for Hunter 2004 and Yerseke 2006a variants, suggesting a genetic basis for the observed cross-reactivity in these clusters. In contrast, no clear pattern was observed when amino acid differences were analyzed in the context of ΔAgs, further demonstrating the importance of antigenic sites.
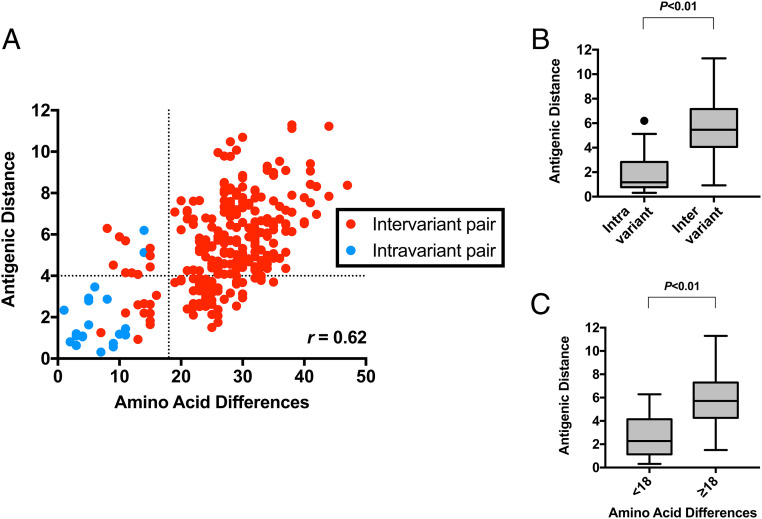
To empirically assess the minimal amino acid differences that could result in antigenic differences, we plotted the correlation of pairwise amino acid differences with the antigenic distance matrix derived from the 3D antigenic cartography map (Fig. 5A). Overall, the amino acid differences in VP1 and antigenic distance were moderately correlated, as assessed by correlation coefficient r = 0.62. The resulting data indicated that a minimum of 18-amino acid changes to the VP1 sequence is required to reach an antigenic distance that exceeds that of intravariant pairings (antigenic distance ≥4; i.e., ≥16-fold blockade difference) (Fig. 5 B and C). As antigenic cartography incorporated multiple heterologous blocking pairs under the detection limit (EC50 titer <50), the robustness of antigenic distance was assessed by replotting it against original EC50 titers (SI Appendix, Fig. S7A). Results observed a strong association between EC50 titers of <50 and antigenic distances of ≥4, suggesting parity between analytical methods. While the correlation between amino acid differences and antigenic distance was observed for the majority of intervariant pairings that exceeded 18-amino acid differences (n = 183 of a total 234), roughly 22% of those pairings (n = 51) failed to achieve significant antigenic distance. Conversely, 10 intervariant pairings were observed to reach an antigenic distance of ≥4 with fewer than 18-amino acid differences. These results suggest that the formation of antigenically distinct variants is either borne from mutations at key antigenic sites, several of them changing at once, or both.
Fig. 5.
Pairwise analysis of inter- and intravariant differences indicated that a minimum of 18-amino acid changes in the VP1 sequence are required for significant antigenic differences. (A) Scatterplot analysis that compares the antigenic distance and amino acid differences on the full VP1 sequences for representative GII.4 VLP pairs. Antigenic distance and amino acid difference data were derived from Figs. 3B and 4B, respectively. Virus (represented by VLPs) pairs are color coded to show intervariant (red) and intravariant (blue) pairs. The vertical dotted line indicates the amino acid difference = 18, while the horizontal dotted line indicates where antigenic distance = 4. (B) Statistical analyses of antigenic distances between intra- and intervariant pairings. P < 0.01 in unpaired t test. (C) Statistical analyses of antigenic distances between VLP pairs at <18 or ≥18 amino acid differences, respectively. P < 0.01 in unpaired t test.
Coevolution of Key Residues Mapped to Antigenic Sites Results in Antigenic Differences among GII.4 Viruses.
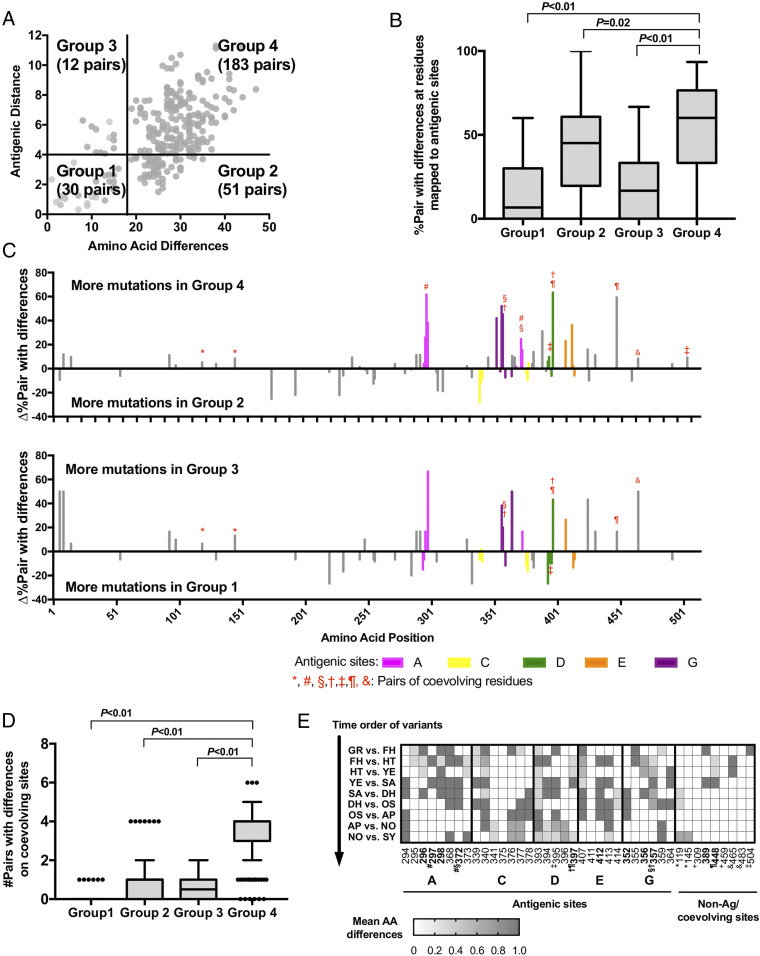
To explore whether differences of mutational patterns could explain discrepancies among genetic and antigenic differences at intra- and intervariant levels, virus pairings were divided into quadrants based on their positions on the scatterplot (Fig. 6A): group 1, antigenic distance < 4 and amino acid differences < 18 (30 pairs); group 2, antigenic distance < 4 and amino acid differences ≥ 18 (51 pairs); group 3, antigenic distance ≥ 4 and amino acid differences < 18 (12 pairs); group 4, antigenic distance ≥ 4 and amino acid differences ≥ 18 (183 pairs). A significantly higher frequency of mutations on antigenic sites was observed on virus pairs from group 4 compared to other groups (P ≤ 0.02 in one-way ANOVA with post hoc Dunnett’s multiple-comparison test) (Fig. 6B). We then compared the excess of mutations at each residue between groups 4 and 2 and groups 3 and 1, respectively (Fig. 6C). Virus pairs from group 4 showed an excess of amino acid mutations at antigenic sites compared to group 2. Notably, eight pairs of residues coevolved during the diversification of GII.4 viruses (Table 1), with five of them located on the antigenic sites and showed excess of mutations on group 4, indicating their positive association with antigenic differences (Fig. 6C). However, five residues that map to antigenic site C (339, 340, 341, 376, and 377) were in mutational excess on group 2 viruses when compared to those from group 4. Two residues (389 and 448) that did not map to any corroborated antigenic sites presented a mutational excess of >20% on group 4; the residue 389 mapped close to the HBGA binding pocket and the residue 448 coevolved with residue 397 from antigenic site D, suggesting a role in the emergence of GII.4 variants. Evidence of coevolving residues at antigenic sites was similarly observed when comparing groups 3 and 1, raising a possible explanation for how significant antigenic distance was attained with minimal amino acid differences.
Fig. 6.
Coevolving changes on antigenic sites determine antigenic diversity. (A) Scatterplot from Fig. 5 organized into quadrants. Group 1: antigenic distance < 4, amino acid differences < 18. Group 2: antigenic distance < 4, amino acid differences ≥ 18. Group 3: antigenic distance ≥ 4, amino acid differences < 18. Group 4: antigenic distance ≥ 4, amino acid differences ≥ 18. The number of virus (represented by VLPs) pairs in each quadrant are displayed. (B) Statistical analysis of virus pairs in each quadrant with a mutation on each residue of the antigenic sites, compared with percent pair in group 4 and others; P ≤ 0.02 in one-way ANOVA with post hoc Dunnett’s multiple-comparison test. (C) Comparison of excess percent accumulation and sequence position of amino acid changes between groups 4 and 2 and groups 3 and 1. Mutated residues are color coded by respective antigenic sites. Instances of coevolved residues are denoted with shared red symbols. (D) Statistical analysis of accrued coevolved residues at antigenic sites from each group, compared with those in group 4 and others; P < 0.01 in one-way ANOVA with post hoc Dunnett’s multiple-comparison test. (E) Mean number of amino acid differences on individual residues from antigenic sites and coevolving sites between viruses from two consecutive variants. Key residues which showed >20% excess mutations in group 4 against group 2 are highlighted in bold and coevolved residues are marked with shared symbols as shown in C.
Table 1.
Coevolving amino acid residues on VP1 protein
Based on these results, we hypothesized GII.4 antigenic diversity is driven by simultaneous changes to multiple antigenic sites. Indeed, when we compared the number of coevolving pairs for each group of viruses, those from group 4 showed the highest number of coevolving sites (median: 4; P < 0.01 Dunnett’s multiple-comparison test) (Fig. 6D), with no overlap on the 25th to 75th percentiles when compared with any of the other three groups of virus pairs. Finally, we sorted out the timeline of those key amino acid mutations on the antigenic sites and calculated the mean of amino acid differences between consecutive variant pairs (Fig. 6E). While several residues (e.g., 340, 368, 393, 395, 407, 413) changed constantly, variant transitions presented mutations at different residues. Particularly, the coevolving residues from group 4 (Fig. 6A) were variable among pre-2006 variants but remained mostly conserved among contemporary variants, thus supporting the major role of those residues on the antigenic diversification of GII.4 viruses and the cross-reactivity observed for the contemporary variants.
Discussion
GII.4 noroviruses are the predominant of all viral gastroenteritis outbreaks worldwide and therefore a major target for vaccine development efforts. Indeed, the two vaccines under clinical trials include GII.4 antigenic components in their formulations (38, 39). The ever-changing nature of GII.4 noroviruses poses a challenge to effective vaccine development, so a better understanding of their antigenic diversification is needed.
Antigenic differences among chronologically emerged GII.4 variants have been demonstrated using monoclonal antibodies and polyclonal sera (15–20, 33, 36), and have been attributed to changes in five major variable antigenic sites (A, C, D, E, G) that map on the outermost domain (P2) of the major capsid protein. Differences at those antigenic sites can also be detected at intravariant levels but minimal effort has been made to dissect their role in antigenic diversification. To better understand the antigenic differences at inter- and intravariant level, we performed a comprehensive antigenic analysis with the largest panel of GII.4 VLPs developed to date. We found that, despite presenting differences at the major antigenic sites, there is strong antigenic clustering among viruses within the same variant. Thus, while small changes could lead to antigenic differences (40), those might not be enough to antigenically separate viruses from the same variant when compared with all other viruses (or variants) in global antigenic analyses.
Phylogenetic separation of viruses does not necessarily reflect underlying antigenic diversity (41–43). Thus, large-scale antigenic profiling coupled with multidimensional scaling analyses are indispensable tools for exploring relationships of viruses (37, 41, 44, 45). It has been demonstrated that phylogenetically distinct H3N2 influenza viruses contain numerous underlying antigenic differences (37). In contrast, dengue viruses present cross-reactive epitopes that preclude full separation at the antigenic level despite being traditionally separated as serotypes and four discrete phylogenetic clusters (41). Our analyses revealed antigenic and phylogenetic relationships among GII.4 variants. While GII.4 variants are phylogenetically discrete, basal phylogenetic clustering results in antigenic similarities. These similarities were validated by the recently developed neutralization assay using infectious virus and the HIE cell culture system (46). While not all antigenic similarities were explained by the phylogenetic tree of whole VP1, sequence similarity of antigenic sites explained the observed antigenic clustering.
Sequence information to infer antigenic properties and epidemiology of viruses has been extremely helpful to select the viruses incorporated into seasonal influenza vaccines. Thus, numerous efforts have been made to predict antigenicity from genetic sequences (47–50). Predictions of influenza H3N2 virus at a viral population level revealed that a subset of all substitutions results in major antigenic changes (48). Specifically, mutations on only 7 of 131 residues forming the antigenic sites were responsible for major antigenic changes and repeatedly involved in emergence of new H3N2 variants (51). Moreover, experimental data showed that minimal mutations in influenza hemagglutinin could produce antigenic changes that escape immunity elicited by polyclonal sera, suggesting that most protective immunity targets a single immunodominant epitope (52). In contrast to the influenza viruses, our analyses show that multiple amino acid changes—primarily at antigenic sites—are necessary for the emergence of an antigenically distinct GII.4 virus. Major antigenic differences are associated with synchronous changes occurring on four different antigenic sites (A, D, E, G), with antigenic site C exerting minimal influence on the antigenic diversification of GII.4 noroviruses. Antigenic sites A and G have been shown to be immunodominant (17, 36), while antigenic sites D and E have been shown to be involved in antigenic differences and regulating HBGA binding (53, 54). Thus, simultaneous mutations at immunodominant sites seem to be needed for major shifts in the antigenic properties of GII.4 noroviruses.
Importantly, a subset of noroviruses presenting a large number of amino acid mutations were antigenically similar, while another subset of viruses presenting only few amino acid mutations were antigenically distinct. This suggests that the chemical properties (52), epistatic interactions (55, 56), and physical locations of the residues making up the antigenic sites play a significant role on the antigenic topology of GII.4 noroviruses, warranting further research. Our previous study revealed key residues (352, 355, 357, 368, and 378) involved in the evolution and emergence of new variants (17). Three of them mapped on the antigenic site G (352, 355, 357), and residues 352 and 357 showed an excess of mutations that lead to antigenic differences (Fig. 6). The other two residues (368 and 378), albeit showing excess of mutations on group 4 (<5% more than group 2) and being associated with changes in blockade activity of specific monoclonal antibodies (17, 36), were not strongly associated with antigenic differences measured with polyclonal sera. Amino acid changes that drive the emergence of new variants could not always alter the antigenicity of those variants. Rather, there could be mutations required for the adaptation or the transition to the next variant without resulting in major changes on the antigenicity. Notably, in this study we found that key residues associated with the major antigenic changes mostly mutated during the transitions between pre-2006 variants, while other residues from the same antigenic sites mutated only among contemporary variants without resulting in major changes in antigenicity. Thus, it seems that noroviruses explore new mutations on key residues to escape from immune pressure, but are also constantly exploring new sequence spaces outside those key residues that could provide new properties and epidemic potential.
One of the striking findings in our analyses is that blockade interactions of VLPs and sera were not symmetrical between variants. In general, newer variants could produce cross-reactive antibodies against previous or contemporary viruses, but rarely vice versa. This unidirectional cross-reactivity suggests that immune responses elicited against the newer strains might suppress circulation of variants that preceded the current dominant strain. This observation is not universal for GII.4 noroviruses, as shown by the cross-blocking antibodies presented by New Orleans 2009-immunized animals to viruses from the next variant, Sydney 2012. Notably, viruses from Sydney 2012 presented a new viral RNA polymerase (GII.P31) that replaced the polymerase type (GII.P4) that predominated for over two decades (57). Thus, virus dominance might be multifactorial, including not only the antigenic changes on the VP1 but also other properties, like transmissibility, replication levels, receptor or HBGA attachment affinity (33, 56, 58–60). Changes in different proteins or domains, such as the viral RNA polymerase, might be involved in the emergence and predominance of noroviruses (30).
A potential limitation to this study is that hyperimmune sera used in blockade experiments was raised from naïve mice. In contrast, humans can be subject to multiple norovirus infections over the course of their lifetime. Indeed, broad blockade antibody responses were reported in vaccinated adults (61), suggesting that previous infections influence subsequent immune responses (4, 8). Additional studies are required to determine what role norovirus exposure history plays in the antigenic discrimination of the different GII.4 variants. Nevertheless, our study provides an important framework on the intravariant diversification and the amino acid changes that could produce antigenically distinct viruses. The reagents developed in this study and the information provided could facilitate the monitoring of newly emerging GII.4 viruses and the development of cross-protective vaccines.
Materials and Methods
Genetic Analyses of GII.4 Norovirus.
A total of 1,601 sequences from GII.4 norovirus detected from 1974 to 2016 were included in the analyses (17), and the nomenclature used for the GII.4 variants was adopted as previously indicated (35). Sequences and phylogenetic analyses were done using MEGA v7 (62) and visualized in FigTree (tree.bio.ed.ac.uk/software/figtree/). The Camberwell MD145-12 1987 virus was used to root the tree. Intravariant and intervariant Shannon entropy values were calculated using the Shannon Entropy-One tool as implemented in Los Alamos National Laboratory (www.hiv.lanl.gov/), and described previously (17). VP1 sequences from 1,572 GII.4 viruses circulating between the years 1995 to 2016 were used for Shannon entropy calculation. Analysis of amino acid diversification was plotted in GraphPad Prism v7.
Coevolving amino acid residues on VP1 protein were estimated using Bayesian Graphical Models for coevolving sites (BGM) (63) implemented in HyPhy package v2.22 (64) and Coevolution Analysis using Protein Sequences (CAPS) v2 (65). Pairs of residues supported in both methods (i.e., edge support > 0.95 in BGM and bootstrap > 0.80 in CAPS2) were assigned as coevolving residues in this study. To reduce the data size for the analyses, we used a randomly subsampled dataset with a maximum of 30 viruses per variant (total n = 308), as indicated previously (17).
Selection of Representative GII.4 Norovirus and VLP Production.
From those 1,601 viral sequences, 24 viruses within GII.4 variants Grimsby 1995, Farmington Hills 2002, Sakai 2003, Hunter 2004, Yerseke 2006a, Den Haag 2006b, Osaka 2007, Apeldoorn 2007, New Orleans 2009, and Sydney 2012 were selected for VLP and hyperimmune sera development. Three viruses for each of the pandemic variants and the Yerseke 2006a variant were chosen to account for sequence differences at the intravariant level. One representative virus was chosen for minor variants Sakai 2003, Osaka 2007, and Apeldoorn 2007. VLPs production was done as described previously (8, 18). Briefly, the VP1-encoding sequences from the representative GII.4 noroviruses (synthesized by GenScript) were cloned into pFastBac1 vectors at the SalI and NotI restriction sites. Alternatively, a synthesized P domain was cloned into pFatBac1-VP1 vectors containing same amino acid sequence of S domain using the PspXI and NotI restriction sites (18). VP1 inserts were transferred into baculovirus expression vectors via MAX Efficiency DH10Bac Competent cells (Thermo Fisher Scientific). Bacmids were transfected into Sf9 insect cells (ATCC) and passaged two to three times. Bacmid identity was confirmed by Sanger sequencing. Assembled VLPs were subsequently purified through a cesium chloride gradient and identity confirmed by Western blot with norovirus VP1 specific monoclonal antibodies (66). Structural integrity for each VLPs was validated with electron microscopy.
Hyperimmune Sera Production.
Murine hyperimmune sera against GR_Grimsby 1995, FH_MD2004-3 2004, HT_Cumberland 2004, HT_SG4091 2006, YE_Yerseke38 2006, DH_DenHaag89 2006, AP_Iwate4 2008, NO_Virginia 2010, and SY_RockvilleD1 2012 GII.4 noroviruses were produced following a schedule of three VLP + alum immunizations spaced 4 wk apart. Terminal bleed was done on week 10. Animal protocols were approved by the Food and Drug Administration Institutional Animal Care and Use Committee (protocol no. 2018-41).
HBGA Blocking Assays.
HBGA blocking assays were used to test the reactivity of GII.4 norovirus VLPs against VLP-derived hyperimmune sera, as previously described (17). Sera from four different mice were collected for each norovirus VLP. Briefly, polyvinyl 96 well “U”-bottom microtiter plates (ThermoFisher) were coated (overnight at 4 °C) with saliva and blocked for 1 h at room temperature with 5% blocking buffer (Bio-Rad). Twofold serial dilutions of mouse hyperimmune sera were plated in duplicate and incubated with representative VLPs at 1 μg/mL for 1 h at 37 °C. Plates were washed four times. Pooled sera from guinea pigs immunized against FH_MD2004-3 2004 and SY_RockvilleD1 2012 VLPs were used as primary detection antibodies against bound VLPs (17, 18). Antibody detection of VLP binding was visualized using goat anti-guinea pig IgG conjugated with horseradish peroxidase and 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS; SeraCare). The deepening blue-green color of the reaction was used to estimate an optical density endpoint at 405 nm (OD 405) around 1.3 to 1.5, whereupon the reaction was quenched with an equal volume of 1% sodium dodecyl sulfate (SDS; Bio-Rad). The OD 405 of sera dilutions was measured with a SPECTROstarnano plate reader (BMG Labtech). For quality control purposes, OD 405 curves were normalized with the average negative control wells used as a lower bound (OD 405 = 0.2). EC50 measurements were calculated from nonlinear regression of normalized OD 405 curves using GraphPad Prism v7. VLP blocking by hyperimmune sera was evaluated through OD 405 quantification, as well as EC50 of sera dilutions. Sera that did not block at the lowest dilution were assigned an EC50 titer of below one-half the detection limit of the assay (<50). Summary heat maps of hyperimmune sera reactivity against representative GII.4 VLPs were constructed via GraphPad Prism v7.
HIE Culture, Neutralization, and RT-PCR.
Human jejunal enteroids (J2) were kindly provided by Mary Estes, Baylor College of Medicine, Houston, Texas. HIE cells were grown as previously described (27, 46). Briefly, HIE cells were maintained as 3D cultures in Human IntestiCult Organoid Growth Medium (Components A and B; Stem Cell Technologies). For neutralization experiments, HIE cells were plated as monolayers on 96-well plates. Monolayers were differentiated for 5 to 6 d until confluent in 1:1 of Component A and CMGF- media (Advanced Dulbecco Modified Eagle Medium [DMEM/F12] enriched with 1% GlutaMAX, 1% 1M Hepes, and 1% penicillin/streptomycin, ThermoFisher Scientific) (67). The monolayers were incubated in this differentiation media + 0.5 mM of GCDCA for 2 to 3 d prior to infection/neutralization. A Sydney 2012 variant virus, GII.4 011617, was filtered from 10% stool suspensions and was used for all neutralization experiments (27). The use of human stool was approved by the FDA Institutional Review Board (protocol number CBER IRB 16-069B). For neutralization, a 1:200 dilution of the virus was incubated with serial dilutions of mouse hyperimmune sera for 1 h. HIE cells were infected with 100 μL of virus or virus-sera mixes (+0.5 mM GCDCA) and incubated at 37 °C to allow for virus adsorption. Following washes, the monolayers were overlaid with fresh differentiation media (+0.5 mM GCDCA) and incubated at 37 °C for 3 d. At 1 h postinfection (hpi) and 3 d postinfection (dpi), the cells were repeatedly thawed and frozen to release intracellular viruses. Viral RNA was extracted from whole-cell content using the MagMAX-96 Viral RNA Isolation Kit (ThermoFisher Scientific). Real-time RT-PCR was performed using MGB GII primers and probe (68) with the following conditions: 50 °C (5 min), 95 °C (20 s), followed by 45 cycles of 95 °C (15 s) and 60 °C (1 min). A full-length GII.4 RNA transcript was used as the standard curve (69). The results were analyzed with CFX Maestro software and compiled in GraphPad Prism v7. The neutralization threshold was defined as the number of genome copies after 1 hpi. Thus, viruses that were neutralized presented values at or below the threshold, while viruses that were able to replicate had values above the threshold.
Antigenic Cartography.
The previously calculated EC50 measurements of all representative VLPs against all hyperimmune sera were used as the foundation of the antigenic cartography. EC50 titers below the detection limit were set to <50. Table distance Dij, which was the distance between VLP i and serum j derived from the corresponding EC50 titer, was calculated in ACMACS as described in Smith et al. (37). The table distance was then projected on the 2D map via ACMACS antigenic cartography algorithm (https://acmacs-web.antigenic-cartography.org) (37). The confidence area of positions of the antigens was estimated with stress parameter 0.1, and shown as blobs in the 2D map. The 3D cartographic projections were generated using ACMACS, and antigenic distance (i.e., map distance) and coordinates data were separately downloaded for the following mutational analyses. The 3D map was drawn using plot3D package in R v3.6.0 (70).
Correlation of Antigenic and Amino Acid Distances.
Antigenic distance among GII.4 VLPs was calculated based on the 3D antigenic cartography map generated from the blocking EC50 values. The number of amino acid differences on the whole VP1 and on key residues on antigenic and/or coevolving sites were calculated using amino acid sequences from GII.4 VLPs using R v3.6.0 and ape package. Data were merged and organized by using R v3.6.0, and association between antigenic distance and number of amino acid differences was plotted as a scatter plot or Tukey’s boxplot using GraphPad Prism v7. The Pearson correlation coefficient (r) between antigenic distance and amino acid differences from GII.4 VLPs was calculated using Mantel method implemented in vegan package in R v3.6.0.
Pairs of the VLPs were divided into four groups based on their antigenic and amino acid differences: group 1, antigenic distance < 4 and amino acid differences < 18; group 2, antigenic distance < 4 and amino acid differences ≥ 18; group 3, antigenic distance ≥ 4 and amino acid differences <18; and group 4, antigenic distance ≥ 4 and amino acid differences ≥ 18. Amino acid mutations were profiled on every single residue from each of the pairs using R v3.6.0. Ratio (%) of pairs with mutations in the groups were then calculated at each single residue and visualized using GraphPad Prism v7. Statistical significance was assessed using t test, one-way ANOVA, and Dunnett’s multiple-comparison test implemented in GraphPad Prism v7.
Supplementary Material
Acknowledgments
We sincerely thank the animal technicians and veterinarians at the Food and Drug Administration for help with the animal immunization protocol and Yamei Gao for the electron microscopy images technical support. Financial support for this work was provided by the Food and Drug Administration intramural funds Program Z01 BK 04012 LHV (to G.I.P.). K.T. and C.J.L. were recipients of a Center for Biologics Evaluation and Research/Food and Drug Administration-sponsored Oak Ridge Institute for Science and Education fellowship.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. D.J.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015874118/-/DCSupplemental.
Data Availability
Raw EC50 data are available as Dataset S1. Virus-like particles and animal sera are available from the authors upon request.
References
- 1.Isakbaeva E. T., et al., Norovirus transmission on cruise ship. Emerg. Infect. Dis. 11, 154–158 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kambhampati A., Koopmans M., Lopman B. A., Burden of norovirus in healthcare facilities and strategies for outbreak control. J. Hosp. Infect. 89, 296–301 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lew J. F., Kapikian A. Z., Jiang X., Estes M. K., Green K. Y., Molecular characterization and expression of the capsid protein of a Norwalk-like virus recovered from a Desert Shield troop with gastroenteritis. Virology 200, 319–325 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Karangwa C. K., et al., Sequential gastroenteritis outbreaks in a single year caused by norovirus genotypes GII.2 and GII.6 in an institutional setting. Open Forum Infect. Dis. 4, ofx236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed S. M., Lopman B. A., Levy K., A systematic review and meta-analysis of the global seasonality of norovirus. PLoS One 8, e75922 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saito M.et al.; Norovirus Working Group in Peru , Multiple norovirus infections in a birth cohort in a Peruvian Periurban community. Clin. Infect. Dis. 58, 483–491 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atmar R. L., et al., Norwalk virus shedding after experimental human infection. Emerg. Infect. Dis. 14, 1553–1557 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parra G. I., Green K. Y., Sequential gastroenteritis episodes caused by 2 norovirus genotypes. Emerg. Infect. Dis. 20, 1016–1018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bok K., Green K. Y., Norovirus gastroenteritis in immunocompromised patients. N. Engl. J. Med. 367, 2126–2132 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel M. M., et al., Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 14, 1224–1231 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardemil C. V., Parashar U. D., Hall A. J., Norovirus infection in older adults: Epidemiology, risk factors, and opportunities for prevention and control. Infect. Dis. Clin. North Am. 31, 839–870 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pires S. M., et al., Aetiology-specific estimates of the global and regional incidence and mortality of diarrhoeal diseases commonly transmitted through food. PLoS One 10, e0142927 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prasad B. V., et al., X-ray crystallographic structure of the Norwalk virus capsid. Science 286, 287–290 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Jiang X., Wang M., Graham D. Y., Estes M. K., Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66, 6527–6532 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Debbink K., Donaldson E. F., Lindesmith L. C., Baric R. S., Genetic mapping of a highly variable norovirus GII.4 blockade epitope: Potential role in escape from human herd immunity. J. Virol. 86, 1214–1226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindesmith L. C., et al., Immunogenetic mechanisms driving norovirus GII.4 antigenic variation. PLoS Pathog. 8, e1002705 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tohma K., Lepore C. J., Gao Y., Ford-Siltz L. A., Parra G. I., Population genomics of GII.4 noroviruses reveal complex diversification and new antigenic sites involved in the emergence of pandemic strains. mBio 10, e02202-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parra G. I., et al., Multiple antigenic sites are involved in blocking the interaction of GII.4 norovirus capsid with ABH histo-blood group antigens. J. Virol. 86, 7414–7426 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen D. J., Gray J. J., Gallimore C. I., Xerry J., Iturriza-Gómara M., Analysis of amino acid variation in the P2 domain of the GII-4 norovirus VP1 protein reveals putative variant-specific epitopes. PLoS One 3, e1485 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koromyslova A. D., Morozov V. A., Hefele L., Hansman G. S., Human norovirus neutralized by a monoclonal antibody targeting the histo-blood group Antigen pocket. J. Virol. 93, e02174-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Loben Sels J. M., Green K. Y., The antigenic topology of norovirus as defined by B and T cell epitope mapping: Implications for universal vaccines and therapeutics. Viruses 11, E432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mallory M. L., Lindesmith L. C., Graham R. L., Baric R. S., GII.4 human norovirus: Surveying the antigenic landscape. Viruses 11, E177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao S., et al., Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 81, 5949–5957 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marionneau S., et al., Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 122, 1967–1977 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haga K., et al., Genetic manipulation of human intestinal enteroids demonstrates the necessity of a functional fucosyltransferase 2 gene for secretor-dependent human norovirus infection. mBio 11, e00251-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atmar R. L., et al., Serological correlates of protection against a GII.4 norovirus. Clin. Vaccine Immunol. 22, 923–929 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ford-Siltz L. A., Wales S., Tohma K., Gao Y., Parra G. I., Genotype-specific neutralization of norovirus is mediated by antibodies against the protruding domain of the major capsid protein. J. Infect. Dis., 10.1093/infdis/jiaa116 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Atmar R. L., et al., Comparison of microneutralization and histo-blood group Antigen-blocking assays for functional norovirus antibody detection. J. Infect. Dis. 221, 739–743 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chhabra P., et al., Updated classification of norovirus genogroups and genotypes. J. Gen. Virol. 100, 1393–1406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parra G. I., Emergence of norovirus strains: A tale of two genes. Virus Evol. 5, vez048 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siebenga J. J., et al., Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J. Virol. 81, 9932–9941 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koelle K., Cobey S., Grenfell B., Pascual M., Epochal evolution shapes the phylodynamics of interpandemic influenza A (H3N2) in humans. Science 314, 1898–1903 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Lindesmith L. C., et al., Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 5, e31 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruis C., et al., Preadaptation of pandemic GII.4 noroviruses in unsampled virus reservoirs years before emergence. Virus Evol. 6, veaa067 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parra G. I., et al., Static and evolving norovirus genotypes: Implications for epidemiology and immunity. PLoS Pathog. 13, e1006136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindesmith L. C., et al., Emergence of a norovirus GII.4 strain correlates with changes in evolving blockade epitopes. J. Virol. 87, 2803–2813 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith D. J., et al., Mapping the antigenic and genetic evolution of influenza virus. Science 305, 371–376 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Bernstein D. I., et al., Norovirus vaccine against experimental human GII.4 virus illness: A challenge study in healthy adults. J. Infect. Dis. 211, 870–878 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaxart, Safety & Immunogenicity Study of Ad5 Based Oral Norovirus Vaccines (VXA-NVV-103), ClinicalTrials.gov: NCT03897309 (2019). https://clinicaltrials.gov/ct2/show/NCT03897309. Accessed 17 July 2020.
- 40.Lindesmith L. C., et al., Antigenic characterization of a novel recombinant GII.P16-GII.4 Sydney norovirus strain with minor sequence variation leading to antibody escape. J. Infect. Dis. 217, 1145–1152 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katzelnick L. C., et al., Dengue viruses cluster antigenically but not as discrete serotypes. Science 349, 1338–1343 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubin S. A., et al., Recent mumps outbreaks in vaccinated populations: No evidence of immune escape. J. Virol. 86, 615–620 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trento A., et al., Conservation of G-protein epitopes in respiratory syncytial virus (group A) despite broad genetic diversity: Is antibody selection involved in virus evolution? J. Virol. 89, 7776–7785 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang S. W., et al., Mapping enterovirus A71 antigenic determinants from viral evolution. J. Virol. 89, 11500–11506 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Y., et al., Limited antigenic diversity in contemporary H7 avian-origin influenza A viruses from North America. Sci. Rep. 6, 20688 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ettayebi K., et al., Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du X., et al., Mapping of H3N2 influenza antigenic evolution in China reveals a strategy for vaccine strain recommendation. Nat. Commun. 3, 709 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Neher R. A., Bedford T., Daniels R. S., Russell C. A., Shraiman B. I., Prediction, dynamics, and visualization of antigenic phenotypes of seasonal influenza viruses. Proc. Natl. Acad. Sci. U.S.A. 113, E1701–E1709 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao Y., et al., Predicting influenza antigenicity from hemagglutintin sequence data based on a joint random forest method. Sci. Rep. 7, 1545 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun H., et al., Using sequence data to infer the antigenicity of influenza virus. mBio 4, e00230-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koel B. F., et al., Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 342, 976–979 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Lee J. M., et al., Mapping person-to-person variation in viral mutations that escape polyclonal serum targeting influenza hemagglutinin. eLife 8, e49324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindesmith L. C., et al., Human norovirus epitope D plasticity allows escape from antibody immunity without loss of capacity for binding cellular ligands. J. Virol. 93, e01813–e01818 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lindesmith L. C., et al., Monoclonal antibody-based antigenic mapping of norovirus GII.4-2002. J. Virol. 86, 873–883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kryazhimskiy S., Dushoff J., Bazykin G. A., Plotkin J. B., Prevalence of epistasis in the evolution of influenza A surface proteins. PLoS Genet. 7, e1001301 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hensley S. E., et al., Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science 326, 734–736 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eden J. S., Tanaka M. M., Boni M. F., Rawlinson W. D., White P. A., Recombination within the pandemic norovirus GII.4 lineage. J. Virol. 87, 6270–6282 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jin M., et al., Characterization of the new GII.17 norovirus variant that emerged recently as the predominant strain in China. J. Gen. Virol. 97, 2620–2632 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheung S. K. C., et al., Higher viral load of emerging norovirus GII.P16-GII.2 than pandemic GII.4 and epidemic GII.17, Hong Kong, China. Emerg. Infect. Dis. 25, 119–122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bull R. A., Eden J. S., Rawlinson W. D., White P. A., Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog. 6, e1000831 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lindesmith L. C., et al., Broad blockade antibody responses in human volunteers after immunization with a multivalent norovirus VLP candidate vaccine: Immunological analyses from a phase I clinical trial. PLoS Med. 12, e1001807 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar S., Stecher G., Tamura K., MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poon A. F., Lewis F. I., Pond S. L., Frost S. D., An evolutionary-network model reveals stratified interactions in the V3 loop of the HIV-1 envelope. PLoS Comput. Biol. 3, e231 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pond S. L., Frost S. D., Muse S. V., HyPhy: Hypothesis testing using phylogenies. Bioinformatics 21, 676–679 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Fares M. A., McNally D., CAPS: Coevolution analysis using protein sequences. Bioinformatics 22, 2821–2822 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Parra G. I., et al., Identification of a broadly cross-reactive epitope in the inner shell of the norovirus capsid. PLoS One 8, e67592 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zou W. Y., et al., Human intestinal enteroids: New models to study gastrointestinal virus infections. Methods Mol. Biol. 1576, 229–247 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bok K., et al., Evolutionary dynamics of GII.4 noroviruses over a 34-year period. J. Virol. 83, 11890–11901 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu C., Wales S. Q., Mammel M. K., Hida K., Kulka M., Optimizing a custom tiling microarray for low input detection and identification of unamplified virus targets. J. Virol. Methods 234, 54–64 (2016). [DOI] [PubMed] [Google Scholar]
- 70.R Core Team , R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2019). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw EC50 data are available as Dataset S1. Virus-like particles and animal sera are available from the authors upon request.