Significance
This work demonstrates that HK2-based assay can resolve a novel HK2high/CKneg CTC population with consistent genomic CNV but distinct transcriptome signatures compared to the CKpos counterpart in NSCLC patients. CK expression levels are found independent of cellular EMT status in these CTCs and may be related to distinct dissemination mechanisms in different types of body fluids. Selective association of CK subtypes in CTCs with patient EGFR mutation types may contribute to suboptimal EGFR inhibitor therapeutic efficacy in EGFRL858R mutant tumors, enabling prediction of patients with poor prognosis before therapy. More generally, HK2, as a metabolic function–associated marker, is likely to be useful in identifying CTCs from patients with a wide variety of cancers, independent of epithelial traits.
Keywords: hexokinase-2, circulating tumor cells, liquid biopsy, single-cell sequencing, non–small cell lung cancer
Abstract
Unlike other epithelial cancer types, circulating tumor cells (CTCs) are less frequently detected in the peripheral blood of non–small cell lung cancer (NSCLC) patients using epithelial marker–based detection approaches despite the aggressive nature of NSCLC. Here, we demonstrate hexokinase-2 (HK2) as a metabolic function–associated marker for the detection of CTCs. In 59 NSCLC patients bearing cytokeratin-positive (CKpos) primary tumors, HK2 enables resolving cytokeratin-negative (HK2high/CKneg) CTCs as a prevalent population in about half of the peripheral blood samples with positive CTC counts. However, HK2high/CKneg tumor cells are a minority population in pleural effusions and cerebrospinal fluids. Single-cell analysis shows that HK2high/CKneg CTCs exhibit smaller sizes but consistent copy number variation profiles compared with CKpos counterparts. Single-cell transcriptome profiling reveals that CK expression levels of CTCs are independent of their epithelial-to-mesenchymal transition (EMT) status, challenging the long-standing association between CK expression and EMT. HK2high/CKneg CTCs display metastasis and EGFR inhibitor resistance-related molecular signatures and are selectively enriched in patients with EGFRL858R driver oncogene mutation as opposed to EGFR19Del, which is more frequently found in patients with prevalent CKpos CTCs in the blood. Consistently, treatment-naïve patients with a larger number or proportion of HK2high/CKneg CTCs in the blood exhibit poor therapy response and shorter progression-free survival. Collectively, our approach resolves a more complete spectrum of CTCs in NSCLC that can potentially be exploited to identify patient prognosis before therapy.
Circulating tumor cells (CTCs) are known to spread from primary sites through blood circulation and other body fluids to seed distant metastases, the main cause of cancer-related deaths (1). The presence of CTCs correlates with increased metastatic propensity and burden; consequently, CTCs are widely considered as one of the most promising biomarkers for hematogenous metastases (2). Due to the extreme rarity of CTCs and the epithelial nature of many cancers, the epithelial cell adhesion molecule (EpCAM) and different members of the cytokeratin (CK) family are frequently employed for candidate CTC enrichment and identification prior to subsequent characterization (3–5). The EpCAM/CK-based detection strategy has been adopted by the most commonly used Food and Drug Administration–cleared CellSearch system for CTC enumeration, although this procedure has intrinsic limitations in detecting CTCs from nonepithelial malignancies (4).
While the CellSearch system provides reliable performance in breast, prostate, and colorectal cancers (3, 4), CTCs are much less frequently observed in peripheral blood of non–small cell lung cancer (NSCLC) patients when compared to other epithelial cancers using EpCAM/CK-based methods despite the highly aggressive nature of NSCLC (5–8). Previous studies reported CTC-positive rates among 21 to 40.8% of NSCLC patients using CellSearch-like systems, and frequently, no CTCs were detected in the blood for early-stage patients with localized disease (6, 9, 10). However, a significant portion of these early-stage patients with resectable tumors and few or no detectable CTCs develop metastatic relapse. These data suggest that significant populations of NSCLC CTCs that express low or no EpCAM or CK may exist but escape detection by these epithelial markers. While the introduction of mesenchymal markers such as N-Cadherin or Vimentin might help increase the coverage, these CKneg (or EpCAMneg) CTCs may not always have a concurrent expression of mesenchymal markers. Moreover, the presence of reactive mesenchymal stromal cells, tumor-derived circulating endothelial cells, and hematopoietic cells of mesenchymal origin compromise the specificity of these mesenchymal markers (3, 11–13). Ideally, to achieve a full spectrum for CTC detection in NSCLC, a marker that exploits a common feature of all cancer cells is desired.
Our approach to identifying and validating an NSCLC CTC marker arises from an aberrant function present in cancers of many different origins. A key hallmark of many cancers is the capacity to metabolize glucose at an elevated rate. This capacity has been exploited clinically by positron emission tomography (PET) for cancer diagnosis (14). The first enzymatic step of glycolysis, critical to the elevated glucose metabolism observed in most cancer cells, is the phosphorylation of glucose catalyzed by hexokinase (HK) (15). Four HK enzymes, HK1, HK2, HK3, and HK4 (glucokinase), have been identified in mammals. Glucokinase is expressed primarily in the liver. HK1 is expressed in the cells of many normal tissues. While HK2 is expressed in embryonic tissues and a few normal cells (muscle or adipose tissue), most normal cells express little or undetectable HK2. In contrast, HK2 is expressed at substantial levels in a wide range of cancers, including cancers of both epithelial and nonepithelial origins (15–18). The high level of HK2 expression and activity in glycolytic tumor cells revealed in PET imaging through the increased conversion of 18F-FDG to 18F-FDG-6P has been associated with poor overall survival in cancer patients (19). Given its selective expression in cancer cells and its relatively restricted expression in normal adult tissues, HK2 is a potential marker to discriminate highly glycolytic CTCs, independent of their expression levels of commonly used epithelial or mesenchymal markers, from other cell types in liquid biopsies.
Here, we investigate the utility of single-cell HK2 immunologic detection as a glycolytic activity–associated marker to identify CTCs in peripheral blood and other body fluids from lung adenocarcinoma (LUAD) patients. The HK2 marker, in combination with CK and CD45, allowed the detection of a greater spectrum of CTCs at a higher detection rate than traditional CellSearch-like systems relying on epithelial markers. HK2 analysis identified an HK2high/CKneg CTC population that is overlooked by the traditional EpCAM/CK-based detection strategy. This population was enriched in many peripheral blood samples but rarely detected in malignant pleural effusion (MPE) and cerebrospinal fluid (CSF). Physical and molecular features were compared at the single-cell level between CKpos and CKneg tumor cell populations, which revealed metastasis and therapy resistance–related transcriptome signatures in CKneg cells. In line with these findings, patients with prevalent CKneg CTCs were found to be associated with a specific EGFR mutation genotype and poor prognosis. More generally, the HK2 marker is likely to be useful in identifying CTCs from patients with a wide variety of cancers.
Results
Principle of HK2-based CTC Identification.
In contrast to normal tissues that primarily use HK1 for glycolysis, many tumors express elevated levels of HK2 (15–18). High levels of HK2 expression are observed in a wide range of cancer cell lines derived from tissues of different origins (Fig. 1A). HK2 expression, generally believed to be a driver of increased glycolysis in tumors (15), is evaluated here to detect CTCs in three types of liquid biopsy samples from lung cancer patients.
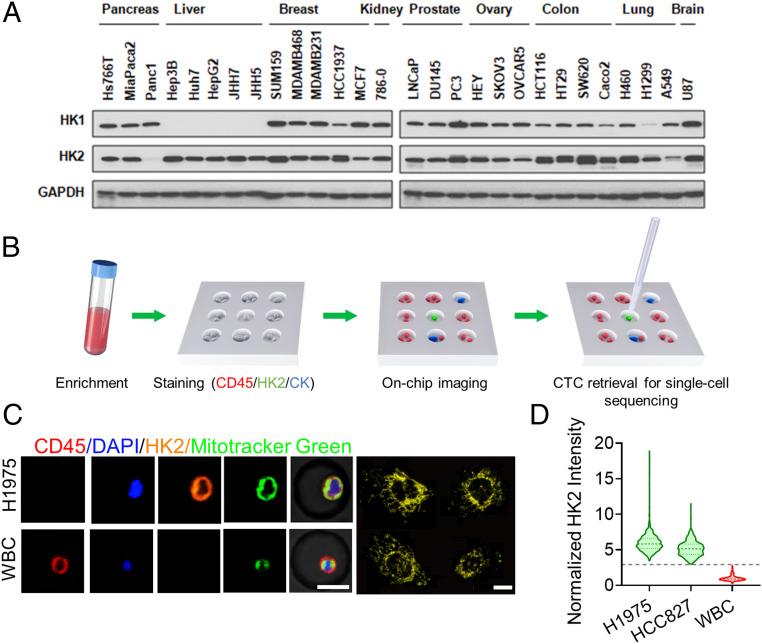
Fig. 1.
Overall strategy and technological platform validation. (A) HK2 overexpression in a wide range of cancer cell lines. (B) The working flow of CTC enrichment, identification, and single-cell manipulation. A PDMS microwell chip was used with each well 30 μm in diameter and 20 μm in height. (C, Left) Colocalization of HK2 and MitoTracker Green in H1975 cells. White blood cells (WBC) are used as a negative control for HK2 staining. (C, Right) Fluorescence image of HK2-stained H1975 cells at 60× magnification. HK2 staining exhibits a fragmented morphology, with many spheroid-shaped staining, or a reticulated morphology, very similar to the morphology of mitochondria. (Scale bar: 15 μm.) (D) Violin plots of HK2 intensity of H1975 and HCC827 cells that were normalized to the mean of white blood cells (n = 591, 433, and 1,027, respectively). The dashed and dotted lines of each violin plot denote the median and first and third quartiles, respectively. The gray dashed line indicates the average HK2 intensity of leukocytes plus five SDs.
The overall strategy of the HK2-based CTC identification assay is illustrated in Fig. 1B. Briefly, a RosetteSep CTC Enrichment Mixture in combination with density gradient centrifugation is used to remove red blood cells and platelets followed by depletion of CD45pos cells. Enriched cells are recovered and applied into a polydimethylsiloxane (PDMS) microwell chip with tens of thousands of addressable microwells for on-chip cell fixation, permeabilization, and immunostaining of HK2, pan-CK (CK7/8), CD45, and 4′,6-diamidino-2-phenylindole (DAPI), followed by washing and imaging. CK7 and CK8 are highly expressed cytokeratins in NSCLC and are commonly used in the cytopathological analysis of LUAD tumors. To avoid cell loss during on-chip operations, a porous membrane is used to seal the chip after cell loading and prior to immunostaining. Putative CTCs including CD45neg/HK2high/CKpos, CD45neg/HK2high/CKneg, and CD45neg/HK2low/CKpos cells are identified based on the calculated fluorescence cutoffs. While the CD45neg/HK2low/CKneg cell population may contain some CTCs with potentially low glycolytic activities, there also exist in this population confounding cell types that include CD45neg immune cells (e.g., plasma cells), circulating endothelial cells, mesothelial cells (in MPE), and dying CTCs (SI Appendix, Fig. S1). Consequently, they are excluded from our analysis. All the putative CTCs are then retrieved individually by a motorized micromanipulator, based on recorded addresses, for single-cell sequencing to determine the malignancy of each cell (Fig. 1B).
As a proof-of-concept demonstration, we spiked tumor cells from two representative LUAD cell lines known to express substantial levels of HK2 into 5 mL of healthy donor blood and then followed with CTC enrichment and the on-chip immunostaining protocol described above. Successive dilutions of the spiked H1975 and HCC827 cells indicated consistent 55 to 65% recovery rates for 15 or more spiked cells (SI Appendix, Figs. S2 and S3), close to the recovery rates obtained by other commercial systems (CellSearch, VTX-1, etc.) using the same cell lines (20–22). We found that HK2 staining colocalized with mitochondrial staining (Fig. 1C) (15). The average HK2 fluorescence signals of the spiked H1975 and HCC827 cells are approximately fivefold greater than those of leukocytes (Fig. 1D). A cutoff line defined by the average HK2 signal plus five times the SD of CD45pos leukocytes can segregate all leukocytes from spiked cancer cells (Fig. 1D). In subsequent analyses of biological fluids from LUAD patients, we designated levels greater or smaller than this cutoff value to be HK2high or HK2low. Additionally, leukocytes are normally deemed CK7/8neg (23); consequently, their average background signal of CK plus five times the SD can cover the signal range of almost all leukocytes and was set as the cutoff for gating CKneg from CKpos cells (Fig. 2A).
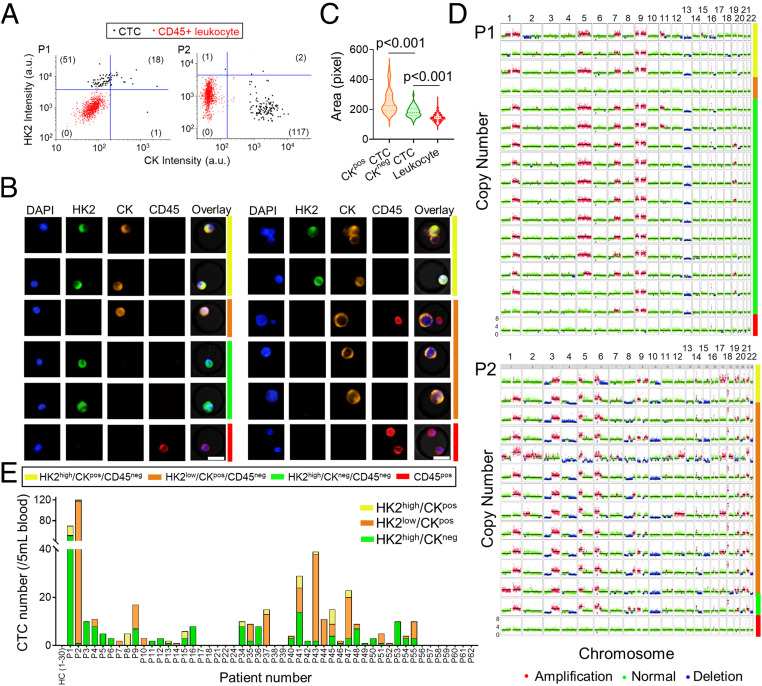
Fig. 2.
Identification and characterization of CTCs in the blood samples of LUAD patients. (A) Scatter plots report HK2 and CK levels of CTCs and CD45pos leukocytes in the blood samples from P1 and P2. HK2high and CKpos cells are gated out by five SDs above the mean of HK2 or CK levels of CD45pos leukocytes. Numbers of HK2high/CKneg/CD45neg, HK2low/CKpos/CD45neg, HK2high/CKpos/CD45neg, and HK2low/CKneg/CD45neg CTC subsets are displayed in the figure. (B) Representative fluorescence images of CTCs from P1 (Left) and P2 (Right). Images are color coded by respective bars to the right with cell type annotations below. (C) Comparison of cell sizes among CKpos CTCs, CKneg CTCs, and leukocytes from P1 and P2 (n = 138, 52, and 2,045, respectively). The dashed and dotted lines of each violin plot denote the median and first and third quartiles, respectively. (D) Single-cell CNV profiles across the autosomes of randomly selected CTCs and leukocytes from P1 and P2, highlighting their genome-wide similarity independent of CK expression. The CTC subtypes and leukocytes are color coded to the right in the same way as B. (E) CTC counts and classification of 30 healthy donors and 50 LUAD patients whose blood samples were analyzed in this study. No CTC was detected in any of the 30 healthy donors.
HK2high/CKneg CTCs as a Prevalent Phenotype in About Half of Peripheral Blood Samples of LUAD Patients.
Having confirmed elevated HK2 levels in multiple cell lines including LUAD lines, we applied this method to interrogate peripheral blood samples from a cohort of 50 treatment-naïve stage III/IV LUAD patients (SI Appendix, Tables S1 and S2). Putative CTCs were detected through a combination of HK2, pan-CK (CK7/8), DAPI, and CD45 staining based on the marker criteria discussed above, followed by validation with single-cell sequencing of genome-wide copy number variation (CNV) profiles. Although all the patients were bearing CKpos primary tumors, two typical CTC subtypes were identified in the blood samples. For Patient 1 (P1), a total of 70 putative CTCs were identified in 5 mL of blood, including 51 HK2high/CKneg/CD45neg cells, 18 HK2high/CKpos/CD45neg cells, and 1 HK2low/CKpos/CD45neg cell (Fig. 2 A and B and SI Appendix, Table S1). These putative CTCs were all DAPI positive, and ∼73% of them exhibited a CKneg subtype with high HK2 staining. In contrast, for P2, a total of 120 putative CTCs were identified in 5 mL of blood, including 117 HK2low/CKpos/CD45neg cells, 2 HK2high/CKpos/CD45neg cells, and 1 HK2high/CKneg/CD45neg cell (Fig. 2 A and B and SI Appendix, Table S1). Around 99% of detected CTCs exhibited a CKpos subtype, and 97.5% showed low HK2 staining. Remarkably, the sizes of CKpos CTCs were statistically larger than those of CKneg CTCs in P1 and P2 blood samples (Fig. 2C).
Single-cell CNV analysis allowed us to survey the entire genomic landscape and determine the malignancy (i.e., the cancer cell genotype) of the putative CTCs at single-cell precision based on chromosomal gains and losses of each cell. The CNV profiles of 16 randomly selected CTCs of P1 displayed largely reproducible copy number alterations across the genome, distinct from leukocytes (Fig. 2D). The copy number gain in chromosome 9 and loss in chromosome 13, in particular, were recurrent across nearly all the putative CTCs sequenced. Such reproducible global CNV profiles were shared in 100% of sequenced HK2high cells regardless of their CK levels, confirming the tumor origin of these cells. Likewise, all sequenced putative CTCs from P2 displayed reproducible gain and loss in CNV patterns, independent of CK or HK2 levels (Fig. 2D). Single-cell sequencing results from six additional patients also showed highly reproducible CNV patterns across all the sequenced putative CTCs within a patient, attesting the malignancy of these cells (SI Appendix, Fig. S4). Taken together, these results validate the high specificity of this marker combination (i.e., CD45neg/HK2high/CKpos/neg and CD45neg/HK2low/CKpos) in detecting CTCs in LUAD patient blood samples.
Among the 50 LUAD patients, CTCs were detected in 72% (≥1 CTC in 5 mL blood) or 66% (≥2 CTCs in 5 mL blood) of patients based on our markers (Fig. 2E and SI Appendix, Table S1). A CTC number ≥5 in 5 mL of blood was detected in ∼44% of patients. In contrast, only 21 to 54.4% of NSCLC patients were detected to have ≥1 CTC (or 21 to 40.8% for ≥2 CTCs) in 7.5 mL blood using EpCAM/CK-based approaches in previous studies (SI Appendix, Table S3) (6, 9, 10, 24–27). A CTC number ≥5 in 7.5 mL of blood was detected in only 9 to 19.2% of patients in those studies (6, 10, 27). In addition, our assay showed a clear reproducibility in CTC enumeration with a near unity Pearson correlation coefficient (CC = 0.995, P < 0.0001) between measurements of duplicate blood draws across seven NSCLC patients (SI Appendix, Figs. S5 and S6). In line with the high assay specificity verified by single-cell CNV analysis, no CTCs were detected in any of the blood samples acquired from 30 healthy donors (Fig. 2E and SI Appendix, Table S4). Interestingly, while the CNV profiles of CTCs from the same patient displayed a high consistency, clear heterogeneous CNV patterns were observed in CTCs from different patients (SI Appendix, Fig. S7). For patients with positive CTC counts, HK2high/CKneg CTCs were detected in ∼81% of these patients and were the prevalent phenotype (>50% of total CTCs in a patient) in 47% of them (Fig. 2E and SI Appendix, Table S1). Around 31% of the patients with positive CTC counts had only HK2high/CKneg CTCs detected in their blood samples. These cells were not accessible to EpCAM/CK-based detection approaches. Consequently, the use of the glycolytic activity–associated HK2 assay can achieve a higher CTC detection sensitivity and a broader CTC spectrum than the traditional EpCAM/CK-based approach used by CellSearch-like systems, which was further confirmed by a side-by-side comparison between these two approaches across seven NSCLC patients (SI Appendix, Figs. S8 and S9). These results prompted us to investigate whether other types of liquid biopsies from LUAD patients, such as MPE and CSF, contain a significant portion of CKneg disseminated tumor cells.
HK2high/CKneg Tumor Cells as a Minority Phenotype in MPE and CSF Samples from LUAD Patients.
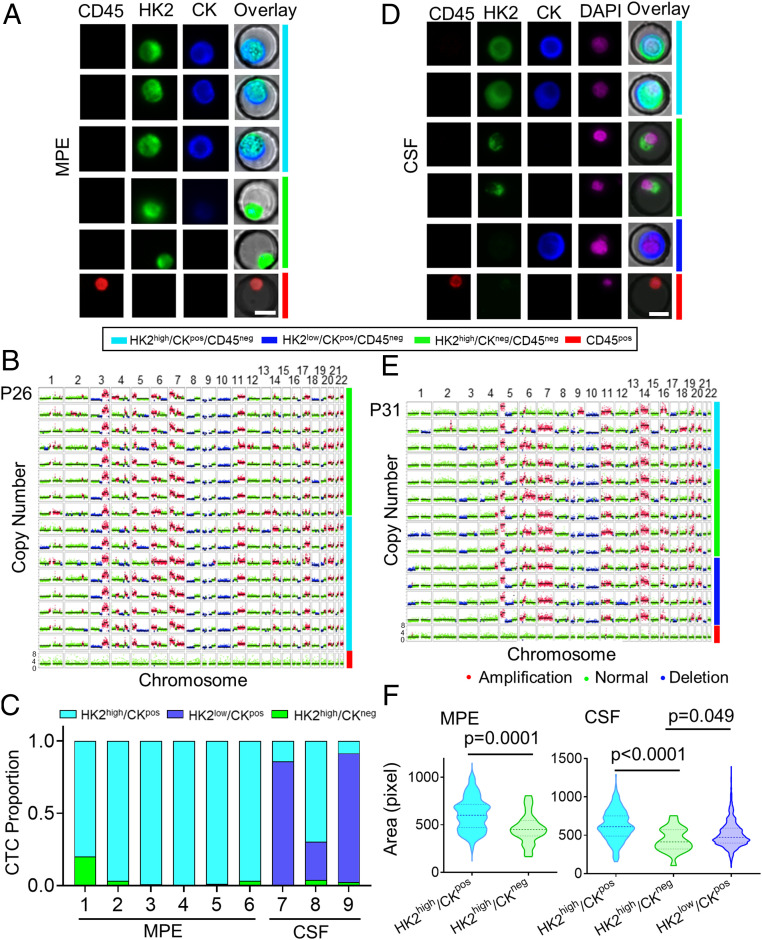
CKneg tumor cells were found not only in peripheral blood but also in other types of liquid biopsies, such as MPE and CSF, of LUAD patients. For lung cancer patients, disseminated tumor cells present in pleural effusion are a common manifestation and denote an advanced stage of disease with metastasis (28). In 10 mL of the pleural effusion sample from P26, we identified a total of 367 putative tumor cells that can be classified into two categories, including 355 HK2high/CKpos cells and 12 HK2high/CKneg cells, based on their CK levels. (Fig. 3A and SI Appendix, Table S3). We did not assess HK2low/CKpos/CD45neg cells because, in contrast to blood samples, there are many detached mesothelial cells that also express CK7/8 but have low HK2 levels in pleural effusion samples, confounding the tumor cell identification in those cells. Single-cell sequencing demonstrated reproducible CNV profiles across all the putative tumor cells sequenced, regardless of their CK expression levels, confirming that they are indeed tumor cells (Fig. 3B). Similar CNV profile consistency was also observed among disseminated tumor cells in the MPE samples of P25 and P27, while to a slightly lesser degree (SI Appendix, Figs. S10 and S11). Unlike blood samples, across the six MPE samples analyzed from LUAD patients, we found that HK2high/CKneg tumor cells constituted a minority population in the pleural effusions, ranging from 0.2 to 20%; HK2high/CKpos tumor cells are the majority population (Fig. 3C and SI Appendix, Table S5).
Fig. 3.
Identification and characterization of disseminated tumor cells in the MPE and CSF samples of LUAD patients. (A) Representative fluorescence images of tumor cells from an MPE sample of P26. Images are color coded by respective bars to the right with cell type annotations below. (B) Single-cell CNV profiles of randomly selected tumor cells and leukocytes from the MPE sample of P26. The cell subtypes are color coded to the right in the same way as A. (C) Tumor cell subtype proportions across all the MPE and CSF samples. (D) Representative fluorescence images of tumor cells from a CSF sample of P31. Images are color coded by respective bars to the right with cell type annotations below. (E) Single-cell CNV profiles of randomly selected tumor cells and leukocytes from the CSF sample of P31. The cell subtypes are color coded to the right in the same way as D. (F) Comparison of cell sizes between HK2high/CKpos and HK2high/CKneg tumor cells in the MPE sample (Left, n = 100 and 25, respectively) and HK2high/CKpos, HK2high/CKneg, and HK2low/CKpos tumor cells in the CSF sample (Right, n = 258, 22, and 1,498, respectively). The dashed and dotted lines of each violin plot denote the median and first and third quartiles, respectively.
HK2high/CKneg tumor cells were also detected in the CSF samples of LUAD patients. In 1.5 mL of CSF from P32, a total of 202 HK2high/CKpos cells, 11 HK2high/CKneg cells, and 77 HK2low/CKpos cells were identified (Fig. 3D and SI Appendix, Table S6). Similar to MPE samples, both the CKpos and CKneg phenotypes exhibited consistent CNV profiles at the single-cell level (Fig. 3E). Across the three CSF samples measured, HK2high/CKneg tumor cells constituted a minority population in the CSF, ranging from 0.44 to 3.8% (Fig. 3C and SI Appendix, Table S6). Similar to the CTCs found in blood samples, morphological evaluation of disseminated tumor cells in MPE and CSF samples revealed larger sizes of CKpos cells than CKneg cells, which may be attributed to the loss of cytokeratins of the CKneg cells (Fig. 3F)
The Connection between CKneg CTCs and Epithelial–Mesenchymal Transition.
Different members of the CK family are common epithelial markers used in the cytopathological analysis. Reduced CK expression in cancer cells is often associated with loss of epithelial signatures and epithelial-to-mesenchymal transition (EMT)—a process that has been implicated in the intravasation of CTCs into blood vessels (29). To verify whether the CKneg CTCs acquired mesenchymal features and thus became more invasive, a comparison of transcriptome signatures between CKpos and CKneg CTCs across a statistical number of single cells is desired. However, the extreme rarity of CTCs in blood samples challenges such analysis. In contrast, MPE samples normally contain larger numbers of disseminated tumor cells that are better suited to single-cell transcriptome analysis. In NSCLC, cancer cells from primary lesions infiltrate the pleura and spread to the pleural space, resulting in an overproduction of pleural fluid and disrupted adsorption. This forms a collection of disseminated tumor cell–containing fluid in the pleural cavity (28). Similar to CTCs in the bloodstream, previous reports have indicated that many tumor cells present in MPEs have EMT signatures and high propensities for seeding distant metastases (30–32). Therefore, transcriptome analysis of tumor cells in MPE samples may be informative for discerning the unique molecular features associated with CKneg CTCs.
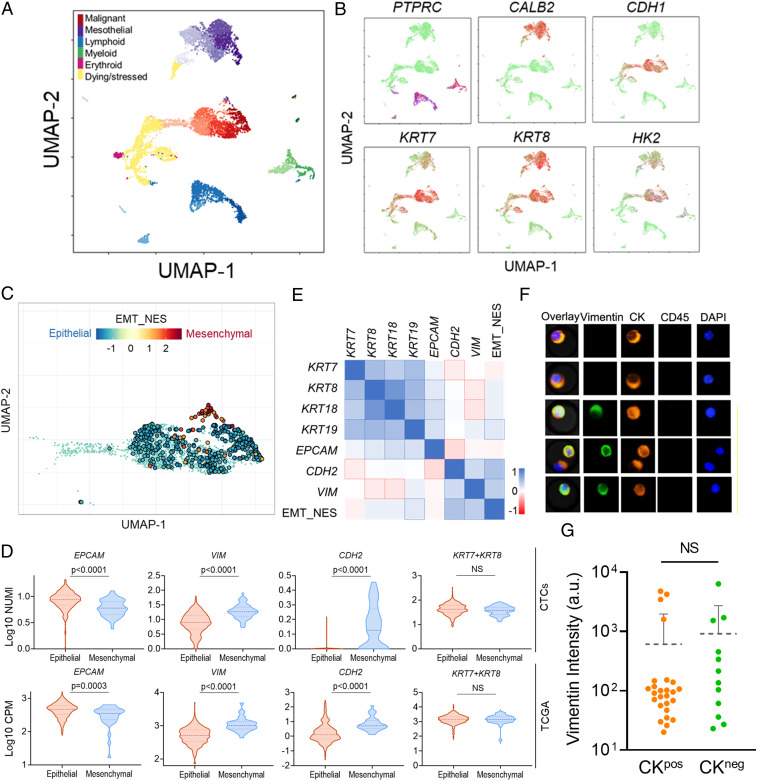
We performed single-cell RNA sequencing of CKneg and CKpos tumor cells in an MPE sample from a LUAD patient, using 10X Genomics. A dimension reduction of the single-cell data using the Uniform Manifold Approximation and Projection (UMAP) algorithm clearly segregated the immune cells, mesothelial cells, and putative tumor cells into different clusters, confirmed by the respective marker expression profiles (Fig. 4 A and B) (33). Specifically, the immune cell populations (blue and green clusters) showed clear CD45 expression. The cells in the upper purple cluster exhibited high calretinin expression and loss of E-cadherin—a marker combination that is specific to reactive mesothelial cells in cytological specimens (Fig. 4B) (34, 35). In contrast, most cells in the central red cluster display high E-cadherin and CK7/8 expression without CD45 and mesothelial marker calretinin, suggesting a high likelihood of malignant cells (Fig. 4 A and B). Dying and stressed cells, as identified by the yellow cluster, exhibited extensive mitochondrial gene contamination and were removed from the subsequent analyses (Fig. 4A and SI Appendix, Fig. S12 and Supplementary Materials and Methods).
Fig. 4.
Single-cell transcriptome analysis of an MPE sample from a LUAD patient. (A) UMAP clustering of different cell types in the MPE sample. Different cell types are color coded and separated at the different regions in the two-dimensional space. The subclusters in each cell type cluster are labeled by the respective color with varying lightness. (B) Expression of selected markers across different cell types visualized in the two-dimensional UMAP space. Green color denotes low expression, and red color denotes high expression. (C) Enrichment of EMT features of all the tumor cells shown in the two-dimensional UMAP space. The normalized enrichment scores (NES) are color coded, and cells with statistically significant enrichment scores (FDR < 0.05) are highlighted by enlarged circles. (D) Comparison of expression levels of several EMT-related genes between epithelial and mesenchymal tumor cells (Top, n = 426 and 58, respectively) and LUAD patient samples curated in TCGA (Bottom, n = 349 and 32, respectively). The dashed and dotted lines of each violin plot denote the median and first and third quartiles, respectively. (E) Spearman correlations between relevant CK family genes and EMT-related genes. Correlation coefficients are color coded, and statistically significant correlations (Bonferroni corrected P < 0.05) are highlighted by outlines. (F) Representative fluorescence images showing the existence of both CKpos/Vimentinpos and CKpos/Vimentinneg CTCs in patient blood samples. (G) Comparison of vimentin levels between CKpos and CKneg CTCs isolated from blood samples of two LUAD patients (P43 and P53). The dashed line indicates the average intensity of each group with an error bar denoting SD. NS: not significant.
To further verify that the cells in the red cluster are indeed tumor derived, we employed an expectation-maximization algorithm to infer the CNV of all viable cells using single-cell transcriptome data as input (SI Appendix, Supplementary Materials and Methods) (36). The putative tumor cells showed distinct CNV patterns compared to normal cells. A k-mean clustering of the inferred single-cell CNV profiles clearly separated all the putative tumor cells in the red cluster from the nonmalignant mesothelial and immune cells in the other clusters, with no exception (SI Appendix, Fig. S13). Compared with the baseline CNV profile of normal cells, disseminated tumor cells showed drastically different CNV profiles across many chromosomal arms, including clear amplification in chromosomes 5p and 14q as well as deletions in chromosomes 13q and 17p. These results confirmed the tumor origin of these putative cells identified by the UMAP algorithm. We further assessed the expression levels of HK2, CK7, and CK8 across single cells. A large number of tumor cells showed significantly higher expression levels of HK2, CK7, and CK8 than immune cells (SI Appendix, Fig. S14A). These results are consistent with the immunostaining results of the MPE samples, where the majority of the disseminated tumor cells are CKpos.
To interrogate whether the CKneg tumor cells are those undergoing EMT and acquiring mesenchymal phenotype, we defined a quantitative EMT score using the hallmark EMT gene set that contains 200 EMT-defining genes curated in the molecular signature database (MSigDB, Broad Institute) (37). The location of each single tumor cell in the EMT spectrum was quantified by the respective enrichment score obtained in the gene set enrichment analysis with respect to an average reference tumor cell (SI Appendix, Supplementary Materials and Methods). Positive enrichment scores denote acquisition of mesenchymal signature, and negative scores represent epithelial phenotype. A total of 426 cells and 58 cells were identified to have statistically significant epithelial and mesenchymal signatures (false discovery rate [FDR] < 0.05), respectively, with many other cells sitting in the intermediate states of the EMT spectrum (Fig. 4C).
In line with the classifications described above, mesenchymal cells with high EMT scores show statistically lower EPCAM expression as well as higher VIM and CDH2 expression than their epithelial counterparts (Fig. 4D). However, to our surprise, the difference of CK7/8 (KRT7 + KRT8) expression levels between mesenchymal and epithelial cells is marginal (Fig. 4D). This lack of a clear difference in the expression of these markers also holds true for the other two types of highly expressed cytokeratin, KRT18 and KRT19, in LUAD cells (SI Appendix, Fig. S14B). The indistinguishable CK expression levels between epithelial and mesenchymal tumor cells are consistent with the results that only weak or insignificant correlations were observed between various types of CK expressions and EMT scores (Fig. 4E). To test whether this observation is also present in CTCs circulating in the blood, we used vimentin as a mesenchymal marker and assessed the difference of vimentin levels between CKpos and CKneg CTCs collected from patient blood samples. In line with the results obtained from the MPE sample, no statistically significant difference of vimentin levels was identified between CKpos and CKneg CTCs using immunostaining (Fig. 4 F and G). We found both CKpos/VIMpos and CKneg/VIMneg CTCs in patient blood samples, supporting that CK expression levels are not correlated with the EMT status of CTCs.
These unexpected results between the CK expression and cellular EMT status in CTCs prompted us to further interrogate whether differential expression of CK7/8 between mesenchymal and epithelial phenotypes exists in patient tumor tissues. We evaluated the gene expression data of LUAD patient samples curated in The Tumor Genome Atlas (TCGA) database, which includes 533 samples from primary tumor tissues and 2 from recurrent tumors. To classify their phenotypes in the EMT spectrum, we calculated the EMT scores using the analysis we employed for the MPE sample above (SI Appendix, Supplementary Materials and Methods). Consistently, no statistically significant difference was identified in CK7/8 (KRT7 + KRT8) expression levels between samples with epithelial signatures and those with mesenchymal signatures (FDR < 0.05), while significant differences were observed in the expression levels of other EMT markers, including EPCAM, VIM, and CDH2 (Fig. 4D). Taken together, these results suggest that the expression levels of CK7/8/18/19—commonly used epithelial markers in cytopathological staining of LUAD—cannot effectively distinguish epithelial and mesenchymal CTC phenotypes in LUAD. Therefore, CKpos and CKneg CTCs cannot be simply attributed to epithelial and mesenchymal phenotypes, respectively.
Elevated Metastasis and Drug Resistance–Related Features in the CKlow Tumor Cell Population.
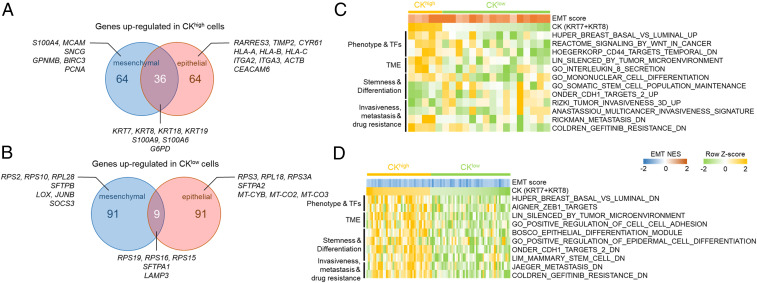
The distribution of CK7/8 expression in epithelial and mesenchymal tumor cell populations is largely overlapping (Fig. 4D). Both phenotypic populations have a few outlier tumor cells with high (or low) CK expression levels and a large number of cells with intermediate CK levels. To interrogate the differential transcriptome profiles between CK-outlier cells in each phenotype, we first ranked all the tumor cells within a phenotype based on their CK7/8 expression levels. We then denoted the cells within the top 10% and bottom 10% of CK7/8 expression levels across all the disseminated tumor cells as CKhigh and CKlow, respectively, and identified differentially expressed genes (DEGs) between them (SI Appendix, Supplementary Materials and Methods).
A survey of the top 100 DEGs up-regulated in CKhigh cells revealed a number of cytokeratins and S100 family proteins shared by two phenotypic populations (Fig. 5A and SI Appendix, Fig. S15). CKhigh cells displayed elevated γ-synuclein (SNCG) compared to CKlow counterparts in the mesenchymal population. Increased expression of γ-synuclein has been implicated in the pathogenesis of NSCLC by promoting cell survival and proliferation (38). Consistently, increased expression of proliferating cell nuclear antigen (PCNA) was also observed in CKhigh cells in the mesenchymal population. In contrast, CKhigh cells in the epithelial population were found to have increased expression in genes encoding integrin subunits (ITGA2, ITGA3, etc.) and MHC-I (HLA-A, HLA-B, HLA-C). For CKlow cells, the top 100 up-regulated DEGs included many genes encoding 40S or 60S ribosome protein components (RPS2, RPL18, etc.) as well as pulmonary surfactant-associated proteins (SFTPA1, SFTPB, etc.), which are either unique to a single phenotypic population or shared between two populations (Fig. 5B and SI Appendix, Fig. S14). Overexpression of genes encoding ribosome protein components in CTCs has recently been shown to contribute to tumor metastasis (39). Consistently, in CKlow cells of the mesenchymal population, we also found increased expression of LOX and JUNB, which have been implicated in enhanced invasiveness and tumor metastasis in lung cancer (40–42).
Fig. 5.
Molecular signatures associated with CKhigh and CKlow cells. (A) Venn diagram showing the top 100 DEGs between CKhigh and CKlow tumor cells up-regulated in CKhigh cells shared between mesenchymal and epithelial populations as well as specific to each population. (B) Venn diagram showing the top 100 DEGs between CKhigh and CKlow tumor cells up-regulated in CKlow cells shared between mesenchymal and epithelial populations as well as specific to each population. (C and D) Enrichment scores (row z-score) of CKhigh and CKlow tumor cells in the (C) mesenchymal and (D) epithelial populations for selected gene sets analyzed by GSVA. The normalized enrichment scores of EMT for those cells are color coded on top of the GSVA enrichment score map.
To resolve the molecular programs associated with CKhigh and CKlow cells in both epithelial and mesenchymal populations, we performed gene set variation analysis (GSVA) across all the CK-outlier tumor cells in each phenotype (43). We interrogated the enrichment profiles of selected gene sets that displayed statistically significant differences between CKhigh and CKlow cells (Fig. 5 C and D). Among these gene sets, CKlow cells showed enrichment in stem cell features and target genes in response to CDH1 knockdown in both mesenchymal and epithelial populations (Onder_CDH1_targets_2) (44). These cells also displayed signatures associated with metastasis and EGFR tyrosine kinase inhibitor (TKI) resistance, as evidenced by the enrichment profiles in several relevant gene sets. For example, most CKlow cells in both phenotypic populations have negative enrichment scores and consequently reduced expression levels in the metastasis-related gene sets that include genes down-regulated in metastatic tumors (Fig. 5 C and D) (45). Similarly, a majority of CKlow cells in both populations also displayed negative enrichment scores in a set of genes down-regulated in EGFR-TKI–resistant tumors (Fig. 5 C and D). Taken together, both the DEG and GSVA analyses suggest that CKlow cells exhibit metastasis and EGFR-TKI resistance–related molecular signatures compared with their CKhigh counterparts regardless of their EMT status.
LUAD Patients with Prevalent HK2high/CKneg CTCs Associated with Poor Therapy Responses.
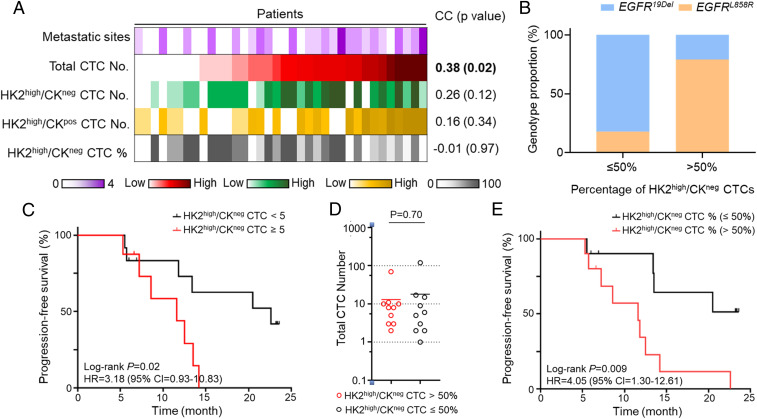
The elevated metastasis and drug resistance–related transcriptome signatures of disseminated tumor cells with low CK expression inspired us to investigate the difference in clinical outcomes between LUAD patients with different numbers and ratios of HK2high/CKneg CTCs. The 36 LUAD patients with positive CTC counts in the blood can be classified into two subgroups based on the percentages of HK2high/CKneg CTCs, including 17 patients with prevalent HK2high/CKneg CTC populations (>50%) and the remaining 19 patients with HK2high/CKneg CTCs ≤50% (Fig. 2E and SI Appendix, Table S1). In line with previous reports, we found a statistically significant correlation between the total CTC numbers and numbers of metastatic sites across patients (Fig. 6A and SI Appendix, Table S1) (46). Consistent with enriched metastasis-related signatures in CKlow CTCs, the correlation of HK2high/CKneg CTC numbers and numbers of metastatic sites is clearly higher than that of HK2high/CKpos CTC number, while the P value did not reach the significance threshold of 0.05, presumably due to our small sample size. (Fig. 6A).
Fig. 6.
Clinical implications of LUAD patients with HK2high/CKneg CTCs. (A) Heatmap showing the relationship between numbers of metastatic sites and numbers (or percentages) of various CTC subtypes across patients with positive CTC counts in the blood (patient n = 36). The z-scores of the CTC numbers across these patients were color coded and displayed from low to high. Spearman correlation coefficients between the CTC numbers (or percentages) and numbers of metastatic sites and respective P values were shown to the right. (B) EGFR mutation subtype proportions for EGFR-mutant patients with different HK2high/CKneg CTC percentages (patient n = 25). (C) Kaplan–Meier curves showing PFS of the treatment-naïve LUAD patients with positive CTC counts in blood segregated by the number of HK2high/CKneg CTCs (patient n = 20). The log-rank P value and hazard ratio are indicated. (D) Comparison of total CTC numbers between two patient groups defined by the percentage of HK2high/CKneg CTCs in blood. The solid line indicates the average total CTC number of each group. No statistically significant difference was found between the total CTC numbers of the two groups. (E) Kaplan–Meier curves showing PFS of the treatment-naïve LUAD patients with positive CTC counts segregated by the percentage of HK2high/CKneg CTCs in blood (patient n = 20). The log-rank P value and hazard ratio are indicated.
All of the 36 patients with positive CTC counts in the blood were treatment naïve at the time of blood collection for CTC analysis, including 25 patients with EGFR-TKI sensitizing mutations (12 EGFR19Del, 13 EGFRL858R) (SI Appendix, Table S1). Surprisingly, we found a preferential enrichment of EGFRL858R mutation subtype in EGFR-TKI–sensitive patients with prevalent HK2high/CKneg CTCs in the blood (Fig. 6B). By contrast, the EGFR19Del subtype was enriched in the remaining EGFR-TKI–sensitive patients with a low percentage of HK2high/CKneg CTCs in the blood (Fig. 6B). Among these patients, 25 patients with EGFR-sensitizing mutations received the first-generation EGFR-TKI treatment as the first-line therapy after blood draw, and the remaining patients without targetable mutations received chemotherapy or chemotherapy + antiprogrammed cell death 1 as the first-line therapy (except for P51, who received ALK inhibitor). In line with enriched drug resistance signatures of CKneg CTCs, within the 20 patients with progression-free survival (PFS) data on record (SI Appendix, Table S1), patients with a higher number of HK2high/CKneg CTCs (HK2high/CKneg CTCs ≥ 5) in blood showed unfavorable therapy responses as evidenced by shorter PFS compared with those with HK2high/CKneg CTCs < 5 (Fig. 6C). Consistent with our expectation, the number of HK2high/CKneg CTCs provided a better prediction of patient PFS than the total CTC number in our cohort (SI Appendix, Fig. S16). In addition to the absolute CTC number, the percentage of HK2high/CKneg CTCs was also found to be predictive for patient therapy response when the two groups of patients have comparable total CTC numbers on average (Fig. 6 D and E). Specifically, patients with prevalent HK2high/CKneg CTCs (>50%) in their blood showed much shorter PFS compared with those with a low percentage of HK2high/CKneg CTCs (Fig. 6E), while the total CTC numbers of these two patient populations were statistically indistinguishable (Fig. 6D). These results suggested an unfavorable prognosis of CKneg CTCs compared with CKpos counterparts and may partially explain the relatively suboptimal therapy responses of EGFRL858R-mutant patients to EGFR-TKI reported by previous studies (47–51).
Discussion
Cancer cells often elevate glucose metabolism to fuel their uncontrolled growth. Exploiting this altered metabolism allowed us to develop metabolic activity-based methods for rapid identification of CTCs from liquid biopsy samples for both epithelial and nonepithelial malignancies (52). In this study, we evaluated a key enzyme in glucose metabolism—HK2—as a surrogate for metabolic activity-based CTC detection. HK2 catalyzes glucose phosphorylation, the first enzymatic step in glycolysis, and is expressed at easily detectable levels in many cancer cells. The use of HK2 as a marker allows us to analyze fixed blood cells preserved in TransFix/ethylenediaminetetraacetic acid (EDTA) vacuum blood collection tubes within 3 d of blood collection, without significant signal loss. This approach relieves the time constraint and pressure of sample delivery, processing, and metabolic analysis and thus significantly improves the adaptability of our assay. Single-cell sequencing confirmed the malignancy of HK2-identified putative CTCs. Nearly all the randomly selected HK2high cells from different types of liquid biopsy samples were confirmed to be malignant by single-cell genome-wide CNV analysis (Fig. 2D and 3 B and E and SI Appendix, Figs. S3 and S4), attesting to the high specificity of HK2 as a glycolytic activity–associated marker in identifying CTCs present in a high background of leukocytes and other confounding cell types.
The use of HK2 as a tumor cell marker permits us to reveal a HK2high/CKneg CTC population which is not accessible to and is normally overlooked by current epithelial marker-based CTC detection methods. Although new markers (e.g., oncofetal chondroitin sulfate) or a combination of multiple markers have been developed to capture multiple phenotypes of CTCs (53, 54), these methods are still dependent on CK expression and thereby inherently biased toward identifying CKpos CTCs. In liquid biopsy samples, the existence of CKneg malignant cells are not easily identifiable because many benign cell types with low or undetectable CK expression levels confound such examination. Sequencing all the CKneg cells in a liquid biopsy sample at the single-cell level to identify CKneg CTCs is neither practical nor cost effective. Additionally, our results indicate that CK expression levels are largely independent of cellular EMT status in LUAD. Both epithelial and mesenchymal populations may contain cells with high (or low) CK expression (Fig. 4D). Therefore, the inclusion of mesenchymal markers, such as vimentin and N-cadherin, in the detection panel would not necessarily identify CKneg CTCs in the epithelial population. HK2 and CD45 costaining can distinguish CTCs from a large number of immune cells and other confounding cell types in liquid biopsy samples and allows for identification of both CKpos and CKneg CTCs.
Although all the patients were bearing CKpos primary tumors in this study (SI Appendix, Tables S1, S5, and S6), HK2high/CKneg CTCs were found to be a prevalent CTC subtype in 47% of blood samples from 36 patients with detectable CTCs (Fig. 2E). Overall, 11 of these 36 patients had only CKneg CTCs detected, and standard EpCAM/CK-based CTC detection methods are unable to detect these cells. This observation partially explains why CTCs are less frequently observed in peripheral blood of NSCLC patients compared with other epithelial cancers when detected by CellSearch-like systems. Some patients have predominantly HK2low/CKpos CTC population in their blood, as exemplified by P2, which may be attributed to a quiescent or stressed cell state with reduced glycolysis and HK2 expression (54). Our approach does not detect those CKneg CTCs with low HK2 expression in blood because of contaminating circulating cells of normal tissue or blood origin. Nevertheless, we detected the presence of CTCs in 72% stage III/IV NSCLC patients with five or more CTCs in 5 mL blood samples from 44% of them, including four stage IIIB patients with localized disease (SI Appendix, Table S1). This level of sensitivity is clearly better than that of previous reports, where CTCs were detected in 7.5 mL blood from 21 to 54.4% of NSCLC patients using CellSearch-like approaches without sequencing confirmation of the tumor origin of detected cells (SI Appendix, Table S3) (6, 9, 10, 24–27). The increased sensitivity for CTC detection in LUAD patients is mainly attributed to the introduction of the glycolytic activity–associated HK2 marker that permits identification of CKneg CTCs inaccessible to EpCAM/CK-based detection strategies.
This study characterized disseminated tumor cells from multiple liquid biopsy forms of the same tumor type. HK2high/CKneg cells were a prevalent CTC subtype in 47% of blood samples with positive CTC counts but were rarely detected in MPE and CSF samples that contain prevalent CKpos tumor cells. The differences in CK subpopulations across different types of liquid biopsy may be the consequences of different microenvironments of these liquid biopsy samples and/or distinct mechanisms of dissemination between CKpos and CKneg cells. Given that the primary tumor lesions of all the patients are CKpos, the enrichment of CKneg CTCs in peripheral blood samples implicates that more CTCs in the blood tend to transition to a CKneg subtype, acquire a more metastatic signature, and, consequently, seed distant organs. The selective enrichment of the EGFRL858R mutation subtype in patients with prevalent CKneg CTCs in blood suggests a high propensity of CKpos-to-CKneg transition driven by L858R mutation.
In all types of liquid biopsy samples, CKneg tumor cells have smaller sizes than CKpos cells, possibly due to the reduced expression of intermediate filaments (Figs. 2C and 3F). However, their CNV profiles resemble those of CKpos tumor cells, suggesting that CK expression levels are likely to be epigenetically regulated rather than reflecting the existence of genetically distinct clones (Figs. 2D and 3 B and E and SI Appendix, Figs. S4, S10, and S11) (55). Single-cell transcriptome analysis of the disseminated tumor cells from an MPE sample failed to identify correlations between CK expression and cellular EMT status. (Fig. 4 D–G). However, caution needs to be taken when extrapolating these results to CTCs in blood samples. We therefore performed independent validation using immunostaining assays. The results indeed pointed to indistinguishable vimentin levels between CKpos and CKneg CTCs in blood samples (Fig. 4G), echoing the uncorrelated relationship between CK expression and cellular EMT status found in analyzing the MPE sample and TCGA data (Fig. 4D). Our results are also supported by an earlier report showing the existence of Vimentinpos mesenchymal subpopulation in CKpos CTCs of NSCLC patients (10).
In addition, disseminated tumor cells with low CK expression were found to be enriched in metastasis and EGFR-TKI resistance–related transcriptional signatures compared with those with high CK expression. Consistently, we observed a shorter PFS in patients with a higher number or percentage of CKneg CTCs in the blood (Fig. 6 C–E). The selective enrichment of EGFRL858R (or EGFR19Del) mutation subtype in patients with a high (or low) percentage of CKneg CTCs is consistent with previous findings that exon 19 deletions are associated with longer PFS and/or overall survival compared with L858R mutation after first-line EGFR-TKIs for advanced NSCLC (Fig. 6B) (47–51). However, the mechanistic difference that underlies therapeutic efficacies and survival benefits between these two EGFR genotypes is still unclear. The differential prevalence of CKneg CTCs in these two EGFR genotypes revealed in our study may help explain the suboptimal therapeutic efficacy and survival benefits of EGFRL858R-mutant tumors. The mechanistic understanding of phenotypic transitions between CKpos and CKneg CTCs as well as the clinical implications of CKneg CTCs as a prognostic marker or a drug target in LUAD patients warrant further investigation in large patient cohorts.
Materials and Methods
Peripheral blood and pleural effusion samples were obtained from LUAD patients in Shanghai Chest Hospital with written informed consent. CSF samples were obtained from LUAD patients in Huashan Hospital with written informed consent. All patients were treatment naïve and at stage III or IV. A total of 50 patients who contributed peripheral blood samples were enrolled from June 2018 to December 2018 and from April 2020 to November 2020 and followed until November 2020. The clinical study was approved by the institutional ethics review committees at the Shanghai Chest Hospital and Huashan Hospital and was performed according to the Declaration of Helsinki Principles. Please refer to SI Appendix, Supplementary Materials and Methods for cell lines and reagents used, microwell chip fabrication, and protocols for CTC identification, sequencing, and data analysis.
Supplementary Material
Acknowledgments
We thank the following agencies and foundations for support: National Natural Science Foundation of China Grants 21775103 (to Q.S.), 81701852 (to L.Y.), and 81672272 (to S.L.); Andy Hill CARE Fund (to W.W.); Washington Research Foundation Technology Development Grant (to W.W.); National Key R&D Program 2016YFC1303300 (to S.L.); the Science and Technology Innovation Program of Shanghai (to S.L.); and Shanghai Chest Hospital Project of Collaborative Innovation Grant YJXT20190209 (to S.L.).
Footnotes
Competing interest statement: A patent has been filed in China by Shanghai General Hospital.
This article is a PNAS Direct Submission. C.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2012228118/-/DCSupplemental.
Data Availability
The single-cell sequencing data reported in this paper have been deposited in the ArrayExpress database (accession no. E-MTAB-8767) (56).
References
- 1.Chaffer C. L., Weinberg R. A., A perspective on cancer cell metastasis. Science 331, 1559–1564 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Mohme M., Riethdorf S., Pantel K., Circulating and disseminated tumour cells–Mechanisms of immune surveillance and escape. Nat. Rev. Clin. Oncol. 14, 155–167 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Stoecklein N. H., Fischer J. C., Niederacher D., Terstappen L. W., Challenges for CTC-based liquid biopsies: Low CTC frequency and diagnostic leukapheresis as a potential solution. Expert Rev. Mol. Diagn. 16, 147–164 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Mohan S., Chemi F., Brady G., Challenges and unanswered questions for the next decade of circulating tumour cell research in lung cancer. Transl. Lung Cancer Res. 6, 454–472 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamilton G., Rath B., Detection of circulating tumor cells in non-small cell lung cancer. J. Thorac. Dis. 8, 1024–1028 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krebs M. G., et al., Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J. Clin. Oncol. 29, 1556–1563 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Crosbie P. A., et al., Circulating tumor cells detected in the tumor-draining pulmonary vein are associated with disease recurrence after surgical resection of NSCLC. J. Thorac. Oncol. 11, 1793–1797 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chemi F.et al.; TRACERx Consortium , Pulmonary venous circulating tumor cell dissemination before tumor resection and disease relapse. Nat. Med. 25, 1534–1539 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanssen A., Loges S., Pantel K., Wikman H., Detection of circulating tumor cells in non-small cell lung cancer. Front. Oncol. 5, 207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindsay C. R., et al., A prospective examination of circulating tumor cell profiles in non-small-cell lung cancer molecular subgroups. Ann. Oncol. 28, 1523–1531 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Schehr J. L., et al., High specificity in circulating tumor cell identification is required for accurate evaluation of programmed death-ligand 1. PLoS One 11, e0159397 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cima I., et al., Tumor-derived circulating endothelial cell clusters in colorectal cancer. Sci. Transl. Med. 8, 345ra89 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Plaks V., Koopman C. D., Werb Z., Cancer. Circulating tumor cells. Science 341, 1186–1188 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallamini A., Zwarthoed C., Borra A., Positron emission tomography (PET) in oncology. Cancers (Basel) 6, 1821–1889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathupala S. P., Ko Y. H., Pedersen P. L., Hexokinase II: cancer’s double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene 25, 4777–4786 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patra K. C., et al., Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell 24, 213–228 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L., et al., Hexokinase 2-mediated Warburg effect is required for PTEN- and p53-deficiency-driven prostate cancer growth. Cell Rep. 8, 1461–1474 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolf A., et al., Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J. Exp. Med. 208, 313–326 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathupala S. P., Rempel A., Pedersen P. L., Glucose catabolism in cancer cells: Identification and characterization of a marked activation response of the type II hexokinase gene to hypoxic conditions. J. Biol. Chem. 276, 43407–43412 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Kondo Y., et al., KRAS mutation analysis of single circulating tumor cells from patients with metastatic colorectal cancer. BMC Cancer 17, 311 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H. E., et al., Detection of EGFR mutations in cfDNA and CTCs, and comparison to tumor tissue in non-small-cell-lung-cancer (NSCLC) patients. Front. Oncol. 10, 572895 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yap Y. S., et al., Detection and prognostic relevance of circulating tumour cells (CTCs) in Asian breast cancers using a label-free microfluidic platform. PLoS One 14, e0221305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moll R., Divo M., Langbein L., The human keratins: Biology and pathology. Histochem. Cell Biol. 129, 705–733 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofman V., et al., Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: Comparison of the efficacy of the CellSearch Assay™ and the isolation by size of epithelial tumor cell method. Int. J. Cancer 129, 1651–1660 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Tanaka F., et al., Circulating tumor cell as a diagnostic marker in primary lung cancer. Clin. Cancer Res. 15, 6980–6986 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Hanssen A., et al., Characterization of different CTC subpopulations in non-small cell lung cancer. Sci. Rep. 6, 28010 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allard W. J., et al., Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 10, 6897–6904 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Feller-Kopman D., Light R., Pleural disease. N. Engl. J. Med. 378, 740–751 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Zeisberg M., Neilson E. G., Biomarkers for epithelial-mesenchymal transitions. J. Clin. Invest. 119, 1429–1437 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giarnieri E., et al., EMT markers in lung adenocarcinoma pleural effusion spheroid cells. J. Cell. Physiol. 228, 1720–1726 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Yin T., et al., Malignant pleural effusion and ascites induce epithelial-mesenchymal transition and cancer stem-like cell properties via the vascular endothelial growth factor (VEGF)/Phosphatidylinositol 3-kinase (PI3K)/Akt/Mechanistic target of rapamycin (mTOR) pathway. J. Biol. Chem. 291, 26750–26761 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen S. F., et al., Pulmonary adenocarcinoma in malignant pleural effusion enriches cancer stem cell properties during metastatic cascade. PLoS One 8, e54659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Becht E., et al., Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Chhieng D. C., Yee H., Cangiarella J. F., Symmans W. F., Cohen J. M., Use of E-cadherin and CD44 aids in the differentiation between reactive mesothelial cells and carcinoma cells in pelvic washings. Cancer 90, 299–306 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Kachali C., Eltoum I., Horton D., Chhieng D. C., Use of mesothelin as a marker for mesothelial cells in cytologic specimens. Semin. Diagn. Pathol. 23, 20–24 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Müller S., Cho A., Liu S. J., Lim D. A., Diaz A., CONICS integrates scRNA-seq with DNA sequencing to map gene expression to tumor sub-clones. Bioinformatics 34, 3217–3219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian A., et al., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma Z., et al., Gamma-synuclein binds to AKT and promotes cancer cell survival and proliferation. Tumour Biol. 37, 14999–15005 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Ebright R. Y., et al., Deregulation of ribosomal protein expression and translation promotes breast cancer metastasis. Science 367, 1468–1473 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kallergi G., et al., The prognostic value of JUNB-positive CTCs in metastatic breast cancer: From bioinformatics to phenotypic characterization. Breast Cancer Res. 21, 86 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng D. H., et al., ZEB1 induces LOXL2-mediated collagen stabilization and deposition in the extracellular matrix to drive lung cancer invasion and metastasis. Oncogene 36, 1925–1938 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao Y., et al., LKB1 inhibits lung cancer progression through lysyl oxidase and extracellular matrix remodeling. Proc. Natl. Acad. Sci. U.S.A. 107, 18892–18897 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hänzelmann S., Castelo R., Guinney J., GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Onder T. T., et al., Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 68, 3645–3654 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Jaeger J., et al., Gene expression signatures for tumor progression, tumor subtype, and tumor thickness in laser-microdissected melanoma tissues. Clin. Cancer Res. 13, 806–815 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Castro-Giner F., Aceto N., Tracking cancer progression: From circulating tumor cells to metastasis. Genome Med. 12, 31 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jackman D. M., et al., Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin. Cancer Res. 12, 3908–3914 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Mok T. S., et al., Improvement in overall survival in a randomized study that compared dacomitinib with gefitinib in patients with advanced non-small-cell lung cancer and EGFR-activating mutations. J. Clin. Oncol. 36, 2244–2250 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Soria J. C.et al.; FLAURA Investigators , Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N. Engl. J. Med. 378, 113–125 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Yang J. C., et al., Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-lung 3 and LUX-lung 6): Analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol. 16, 141–151 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y., et al., Patients with exon 19 deletion were associated with longer progression-free survival compared to those with L858R mutation after first-line EGFR-TKIs for advanced non-small cell lung cancer: A meta-analysis. PLoS One 9, e107161 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Z., et al., Liquid biopsy-based single-cell metabolic phenotyping of lung cancer patients for informative diagnostics. Nat. Commun. 10, 3856 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Agerbæk M. O., et al., The VAR2CSA malaria protein efficiently retrieves circulating tumor cells in an EpCAM-independent manner. Nat. Commun. 9, 3279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu M., et al., Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339, 580–584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flavahan W. A., Gaskell E., Bernstein B. E., Epigenetic plasticity and the hallmarks of cancer. Science 357, eaal2380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi Q., Hexokinase 2 detects cytokeratin negative circulating tumor cells in lung adenocarcinoma. ArrayExpress. https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-8767/. Deposited 17 January 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The single-cell sequencing data reported in this paper have been deposited in the ArrayExpress database (accession no. E-MTAB-8767) (56).