Significance
Pentapeptide-repeat proteins, such as MfpA and Qnr proteins, have an intriguing right-handed quadrilateral β-helical fold that gives rise to an elongated roughly cylindrical structure that resembles the shape of a double-stranded DNA helix. It has been speculated that these proteins, particularly those that interact with bacterial DNA gyrase, act as DNA mimics, competing with DNA to fulfil their functions. Until now there has been no direct evidence for this. Using enzymology and X-ray crystallography, we show the mycobacterial MfpA appears to act as a mimic of the transported (T) DNA segment during the gyrase supercoiling cycle, protecting the enzyme from fluoroquinolone antibiotics. These data suggest a mechanism to limit fluoroquinolone efficacy.
Keywords: fluoroquinolones, DNA gyrase, topoisomerase, pentapeptide-repeat proteins, tuberculosis
Abstract
DNA gyrase, a type II topoisomerase, introduces negative supercoils into DNA using ATP hydrolysis. The highly effective gyrase-targeted drugs, fluoroquinolones (FQs), interrupt gyrase by stabilizing a DNA-cleavage complex, a transient intermediate in the supercoiling cycle, leading to double-stranded DNA breaks. MfpA, a pentapeptide-repeat protein in mycobacteria, protects gyrase from FQs, but its molecular mechanism remains unknown. Here, we show that Mycobacterium smegmatis MfpA (MsMfpA) inhibits negative supercoiling by M. smegmatis gyrase (Msgyrase) in the absence of FQs, while in their presence, MsMfpA decreases FQ-induced DNA cleavage, protecting the enzyme from these drugs. MsMfpA stimulates the ATPase activity of Msgyrase by directly interacting with the ATPase domain (MsGyrB47), which was confirmed through X-ray crystallography of the MsMfpA–MsGyrB47 complex, and mutational analysis, demonstrating that MsMfpA mimics a T (transported) DNA segment. These data reveal the molecular mechanism whereby MfpA modulates the activity of gyrase and may provide a general molecular basis for the action of other pentapeptide-repeat proteins.
The bacterium Mycobacterium tuberculosis (Mtb) is the causative agent of tuberculosis (TB), which is one of the most serious global health problems, killing more than 1.5 million people every year (1). In an effort to address this problem, many compounds have been developed targeting crucial enzymes of M. tuberculosis, including gyrase (2).
Similar to Escherichia coli gyrase, mycobacterial gyrase is a type II topoisomerase with two GyrA and two GyrB subunits that assemble as an A2B2 complex (3). Gyrase binds to DNA (∼130 bp) forming a wrapped complex (4–7); three protein interfaces have been identified in this complex: the N (ATPase) gate, the DNA gate, and the C (exit) gate (3, 8). Gyrase catalyzes changes in the topology of DNA by promoting the passage of one DNA duplex (the T segment) via a transient break in a second double-stranded DNA segment (the G segment). As a consequence of the DNA wrap, the T segment and G segment are closely located on the same piece of DNA, distinguishing gyrase from other topoisomerases (9). Gyrase, which is an essential enzyme in all bacteria, is the only type II topoisomerase in mycobacteria, which lack DNA topoisomerase IV, a preferential decatenase, that is present in most other bacterial species (10–13). Consequently, mycobacterial gyrase has to fulfill the functions of supercoiling, relaxation, and decatenation, thus participating in both DNA replication and transcription.
Currently, there are many drugs and toxins targeting gyrase; among them, the broad-spectrum antibiotics, the quinolones, are thought to be the most effective (2, 3, 14, 15). Fluoroquinolones (FQs), which are derived from quinolones, have promising activity against TB, especially for multidrug-resistant and extensively drug-resistant TB (16). In bacteria, the FQs can interact with the cleaved DNA (G segment) and the GyrA and GyrB proteins to stabilize a cleavage complex and inhibit the religation of the cleaved DNA, potentially resulting in lethal double-strand DNA breaks in the genome (15, 17).
However, bacteria have survival strategies to defeat quinolone inhibition, such as pentapeptide-repeat proteins (PRPs) (18–20). Currently the origins, evolution, and mechanisms of PRPs are not clear. PRPs contain a repeat motif [S,T,A,V][D,N][L,F][S,T,R][G] that folds as a right-handed quadrilateral β-helix and adopts an elongated homodimeric quaternary structure with similar dimensions to a fragment of double-stranded DNA (18–20). It has been proposed that several bacterial PRPs including, Qnr (from Klebsiella pneumoniae, Proteus mirabilis, Salmonella enterica, Shigella flexneri, Aeromonas hydrophila, and Enterococcus faecalis), McbG (from E. coli), AlbG (Xanthomonas albilineans), and MfpA (from mycobacterial species) interact with gyrase and protect it from toxins (18, 20, 21).
In 2001, a PRP, MfpA (mycobacterial FQ resistance protein A), was first identified from the chromosome of Mycobacterium smegmatis by screening a genomic library (22), and was found to moderately increase the minimum inhibitory concentrations of M. smegmatis to FQs, but not to nalidixic acid (22). Subsequently, the structure and function of M. tuberculosis MfpA (MtbMfpA) was characterized using X-ray crystallography and gyrase assays (19). MtbMfpA, belongs to the PRP family, and resembles the molecular shape of B-DNA by forming an elongated twofold-symmetric homodimer through C-terminal interactions (19). Both MtbMfpA and MsMfpA (from M. smegmatis) are capable of inhibiting the supercoiling activity of gyrase, but unlike QnrB1, MtbMfpA has been reported to be unable to protect gyrase from FQs (21), although it has been proposed that this may require the presence of the MfpB protein (23). Based on these results, a possible working model was proposed for the action of MfpA as a G-segment mimic, binding at the DNA gate and inhibiting the supercoiling cycle of gyrase (19); it has also been suggested from modeling studies to mimic a T segment (24). However, to date, no direct experimental evidence supports these hypotheses.
Here we propose that MfpA regulates gyrase through acting as a T-segment mimic. Using M. smegmatis proteins, we find that MsMfpA inhibits the negative supercoiling activity of M. smegmatis gyrase (Msgyrase) in the absence of FQs and, additionally, protects Msgyrase from the FQs ciprofloxacin (CFX) and moxifloxacin (MFX) through decreasing FQ-induced cleavage. We also find that MsMfpA can stimulate ATP hydrolysis of Msgyrase through direct interaction with its ATPase domain (MsGyrB47). Additionally, we present crystal structures of MsMfpA and MsGyrB47 alone and of a MsMfpA–MsGyrB47 complex, strongly suggesting that MsMfpA interacts with MsGyrB47 like a T segment. Taking these data together, our study reveals MfpA as a T-segment mimic and suggests a regulatory mechanism of MfpA on DNA gyrase; similar results have also been obtained with QnrB1 (25).
Results
MsMfpA Inhibits the Supercoiling Activity of DNA Gyrase and Protects It from FQs.
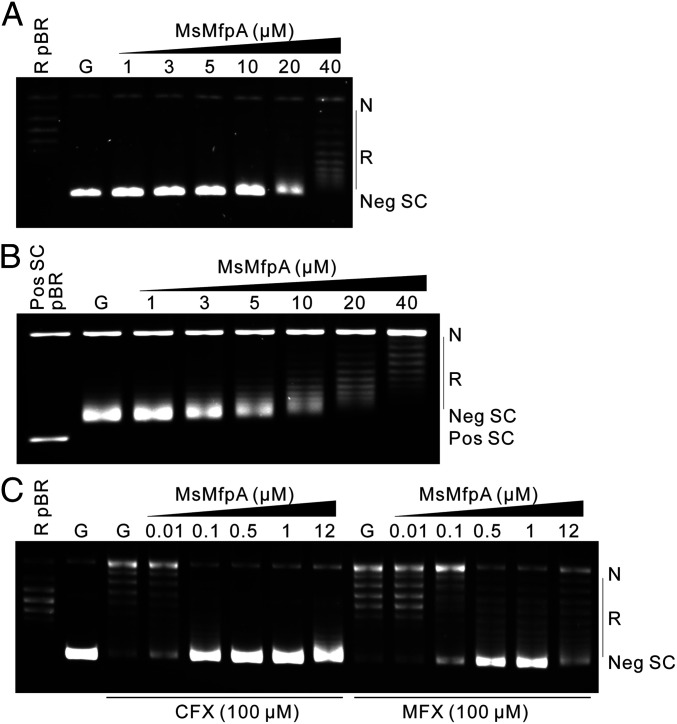
In order to investigate the action of MfpA on DNA gyrase, we initially purified MfpA and gyrase from M. tuberculosis. However, we found that MtbMfpA suffered from instability issues, so we switched species to M. smegmatis, which, given the high sequence identities between orthologous proteins (62% identity in this case), has frequently been used as a surrogate for M. tuberculosis. To determine if MsMfpA affects the actions of gyrase, we purified MsMfpA and His-tagged Msgyrase and performed gyrase assays. We titrated the supercoiling activity of Msgyrase and found that 2 nM enzyme was sufficient to achieve a high rate of negative supercoiling in 30 min, indicating that the enzyme had high activity (SI Appendix, Fig. S1A). Using this concentration of enzyme, we tested the effect of MsMfpA on Msgyrase in supercoiling assays. We found that 20 μM MsMfpA reduced the supercoiling activity and that 40 μM was capable of largely abolishing supercoiling (Fig. 1A): that is, inhibition appears to require a large excess of MfpA over gyrase. As previously reported, mycobacterial gyrase can catalyze the conversion of positively supercoiled DNA to negatively supercoiled DNA through ATP hydrolysis (10). In this reaction, the enzyme catalyzes both the relaxation of positive supercoils and the subsequent negative supercoiling of the relaxed DNA (SI Appendix, Fig. S1). Using positively supercoiled pBR322 as substrate and 3 nM Msgyrase, we found that MsMfpA could inhibit the negative supercoiling reaction, but did not appear to inhibit the relaxation of positively supercoiled DNA under these conditions (Fig. 1B). In addition, we examined MsMfpA in relaxation reactions using negatively supercoiled pBR322 without ATP, and found that it was unable to inhibit Msgyrase in this reaction (SI Appendix, Fig. S1 C and D). Based on these data, we suggest that MsMpfA may interact with Msgyrase to inhibit the ATP-dependent negative supercoiling reaction, without affecting the ATP-independent relaxation reaction. It also appears that ATP-dependent relaxation of positive supercoils is unaffected by MfpA.
Fig. 1.
The effect of MsMfpA on the activities of Msgyrase. (A) MsMfpA (at the concentrations indicated) was used in negative supercoiling reactions with 2 nM Msgyrase; relaxed pBR322 as a control is shown in the first lane. (B) MsMfpA was used in reactions with positively supercoiled pBR322 (being converted to negatively supercoiled pBR322) with 3 nM Msgyrase; positively supercoiled pBR322 as a control is shown in the first lane. (C) Protective effects of MsMfpA on 5 nM Msgyrase in supercoiling reactions in the presence of 100 μM CFX or MFX; relaxed pBR322 is shown in the first lane. G: Msgyrase; N: nicked DNA; Neg SC: negatively supercoiled DNA; Pos SC: positively supercoiled DNA; Pos SC pBR: positive supercoiled pBR322; R: relaxed DNA; R pBR: relaxed pBR322.
Previously, the pentapeptide protein QnrB1 was found to protect DNA gyrase against CFX (26–28). To test the action of MsMfpA on Msgyrase in the presence of FQs, supercoiling assays with CFX or MFX were performed using 5 nM (Fig. 1C) Msgyrase. We found that 0.1 to 0.5 μM MsMfpA was enough to protect Msgyrase against CFX (100 μM), while 0.5 to 1 μM of MsMfpA was required against MFX (100 μM). To confirm the protective effects, gyrase assays with positively supercoiled pBR322 were performed using 10 nM Msgyrase in the presence of ATP (SI Appendix, Fig. S1 E and F). The result shows that, consistent with the negative supercoiling reactions, MsMfpA protects Msgyrase against both CFX and MFX during both the relaxation of positive supercoils and supercoiling of relaxed DNA. These data suggest that, similar to QnrB1, MsMfpA may protect Msgyrase from FQs through interacting with the enzyme.
To confirm whether MsMfpB can help MsMfpA to protect gyrase, supercoiling assays were performed using 2 nM gyrase in the presence of 1 mM GTP (SI Appendix, Fig. S1G). Without CFX, 2 nM gyrase is able to catalyze the supercoiling of relaxed DNA; no significant change was observed after adding increasing concentrations of MsMfpB. With 100 μM CFX in the assay, the supercoiling activity was dramatically inhibited and 0.5 μM MsMfpA was able to partially rescue gyrase from CFX. Under this condition, no significant increase of the protective effect was observed after adding MsMfpB into the assays. These results suggest that MsMfpB may not play a role in the protection of gyrase from FQs.
MsMfpA Inhibits FQ-Induced Cleavage in an ATP-Dependent Manner.
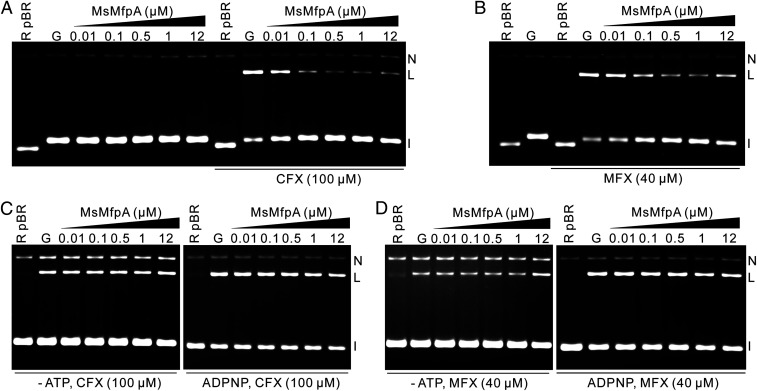
FQs stabilize the DNA gyrase–DNA cleavage complex, which is an essential intermediate in the supercoiling cycle (29, 30). To ascertain whether MsMfpA affects FQ-induced cleavage, we performed cleavage assays with relaxed pBR322 using 20 nM Msgyrase in the presence of ATP. We found that relatively low concentrations (∼0.01 µM) of MsMpfA were capable of reducing the CFX (100 µM)-induced cleavage (Fig. 2A). As a control, cleavage assays without CFX showed that MsMfpA was unable to cleave DNA on its own (Fig. 2A). To test if MsMfpB can help MsMfpA to inhibit CFX-induced cleavage, cleavage assays performed in the presence of 1 mM GTP and 0.1 µM MsMfpA showed that MsMfpB was unable to increase the inhibitory effect by MsMfpA (SI Appendix, Fig. S2A). All the data with MsMfpB suggest that MsMfpB may not help MsMfpA to protect gyrase from FQs, so we did not carry out any further investigations with this protein.
Fig. 2.
FQ-induced cleavage is inhibited by MsMfpA in an ATP-dependent manner. Effect of MsMspA on (A) CFX- or (B) MFX-induced cleavage complexes with Msgyrase. (A) As a control, cleavage assays with Msgyrase were performed in the absence of FQs and the presence of MsMfpA. The inhibitory effects of MfpA on CFX- (C) or MFX- (D) induced cleavage reactions were abolished in the absence of ATP or in the presence of ADPNP. I: intact DNA; L: linear DNA; N: nicked DNA. Note that the gel is run in the presence of ethidium bromide.
We examined MsMfpA in cleavage assays with MFX (40 µM) and found that MsMfpA was also able to reduce MFX-induced cleavage (Fig. 2B), although to a lesser extent. To further explore the inhibitory effects, we tested MsMfpA in cleavage assays with positively supercoiled pBR322 in the presence of ATP and found that MsMfpA was capable of reducing CFX- or MFX-induced cleavage (SI Appendix, Fig. S2B). Based on these findings, we suggest that MsMfpA can inhibit FQ-induced cleavage to protect the activities of Msgyrase against FQs.
Cleavage of DNA by gyrase is an ATP-independent reaction (31, 32), and FQs can stabilize the cleavage complex in the absence of ATP. To test if MsMfpA could still work on Msgyrase without ATP, cleavage assays were performed in the presence of CFX without ATP or with ADPNP (5′-adenylyl-β,γ-imidodiphosphate). The data show that MsMfpA was unable to protect Msgyrase under these conditions (Fig. 2C). Similar to cleavage assays with CFX, MsMfpA lost the protective effects on Msgyrase without ATP or with ADPNP in the presence of MFX (Fig. 2D). To confirm the crucial role of ATP, we performed cleavage assays with CFX or MFX using negatively and positively supercoiled pBR322 in the absence or presence of ATP. All the results showed that MsMfpA had protective effects on Msgyrase only with ATP (SI Appendix, Fig. S2 C–F). These data strongly suggest that the hydrolysis of ATP is necessary for MsMfpA to protect Msgyrase from FQs.
MsMfpA May Facilitate the Religation of DNA from FQ-Induced Cleavage Complex.
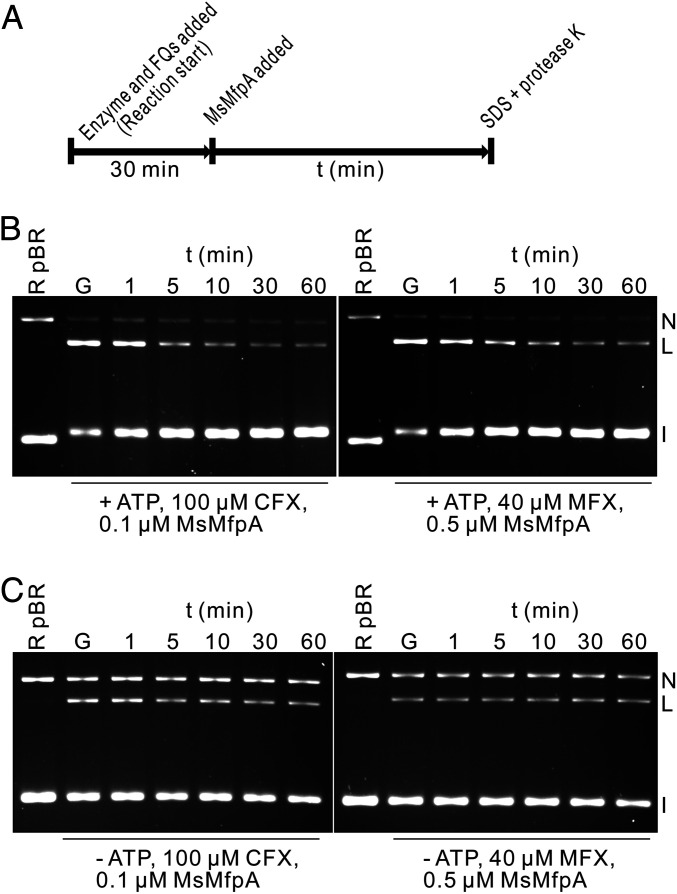
To test if MsMfpA could dissociate FQ-stabilized cleavage complexes, time-course cleavage assays were performed using MsMfpA and Msgyrase. In these assays, cleavage complexes were initially obtained by adding enzyme and CFX or MFX, before MsMfpA was added into the reactions (Fig. 3A). As shown in Fig. 3B, MsMfpA can decrease the amount of linear DNA induced by CFX or MFX in the presence of ATP. The time-course cleavage assays were also performed without ATP. As anticipated, in the absence of ATP, the protective effects of MsMfpA were abolished (Fig. 3C). These results suggest that MsMfpA may disrupt the stabilized cleavage complexes when ATP is hydrolyzed by gyrase.
Fig. 3.
MsMfpA promotes the religation of DNA. (A) Outline of the experiment. Msgyrase and CFX or MFX were added into the reaction to obtain the cleavage complexes, then 0.1 or 0.5 μM MsMfpA was then added into the reaction. Samples were taken at different times and mixed with SDS and proteinase K to release the linear DNA product. (B and C) The action of MsMfpA in time-course cleavage assays with 100 μM CFX or 40 μM MFX in the presence (B) or absence (C) of ATP. G: gyrase.
MsMfpA Stimulates the ATPase Activity of Mycobacterial DNA Gyrase by Directly Interacting with the ATPase Domain.
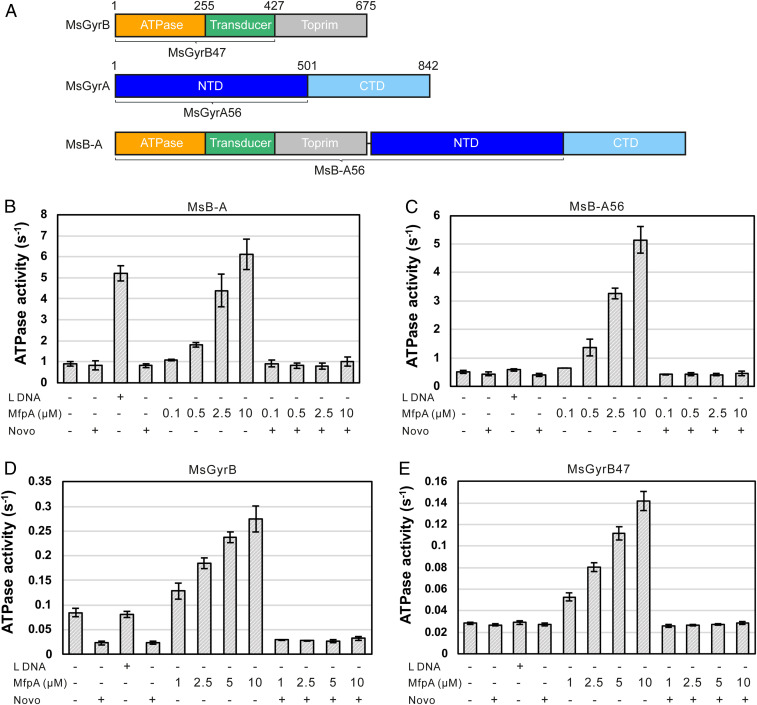
As shown above, ATP is required for MsMfpA to inhibit FQ-induced cleavage and protect Msgyrase against FQs. This observation led us to hypothesize that MsMfpA might affect the hydrolysis of ATP by Msgyrase. To test this hypothesis, we performed ATPase assays using 100 nM gyrase full-length fusion protein (MsB-A), in which the two subunits are linked by a Lys residue (Fig. 4A). The resultant protein performs in supercoiling and cleavage assays at a level comparable to wild-type Msgyrase (A2B2) (SI Appendix, Fig. S3 A and B). Linear DNA, as a positive control, was able to significantly activate the ATPase activity of MsB-A (Fig. 4B), which has been observed previously (12). When novobiocin, an inhibitor of gyrase that binds at the ATPase active site (33), was added, the ATPase activity was essentially abolished (Fig. 4B). Addition of MsMfpA dramatically increased the hydrolysis of ATP by MsB-A (Fig. 4B); addition of novobiocin abolished this activity (Fig. 4B), suggesting that MsMfpA specifically stimulates ATP hydrolysis by MsB-A.
Fig. 4.
The hydrolysis of ATP by Msgyrase is activated by MsMfpA. (A) Architecture of M. smegmatis DNA gyrase and the B–A full-length fusion. Gyrase domains are colored as follows: GyrB-ATPase subdomain, orange; GyrB-transducer subdomain, green; GyrB-Toprim subdomain, silver; GyrA-NTD, blue; GyrA-CTD, light blue. CTD, C-terminal domain; NTD, N-terminal domain; Toprim, topoisomerase primase domain. ATPase activity assays were carried out using MsB-A (B), MsB-A56 (C), MsGyrB (D), and MsGyrB7 (E) with linear DNA or different concentrations of MsMfpA in the absence or presence of 100 μM novobiocin. Linear DNA was used as a control in the assays; 100 μM novobiocin was used as a specific inhibitor of ATPase activity. L DNA: linear pBR322; Lys: lysine linker; Novo: novobiocin.
To determine the binding site of MsMfpA on MsB-A, the C-terminal domain of MsGyrA, responsible for inducing the DNA wrap, was removed to generate a His-tagged fusion protein MsB-A56. Cleavage assays, performed using 20 nM MsB-A56, showed that MsMfpA was able to reduce the FQ-induced cleavage activity of MsB-A56, in the presence of ATP (SI Appendix, Fig. S3C). This suggests that MsMfpA interacts with MsB-A56 to inhibit the formation of the stabilized cleavage complexes. In ATPase assays, the activity of MsB-A56, which is unable to wrap DNA, could not be activated by linear DNA (Fig. 4C), but MsMfpA was still capable of stimulating the ATPase activity of MsB-A56 (Fig. 4C). Again, the stimulation was inhibited to baseline by novobiocin (Fig. 4C). All these results suggest that MsMfpA interacts with MsB-A56 and stimulates its ATPase activity to protect it against FQs.
To determine if MsMfpA could directly interact with the MsGyrB subunit, we examined the activity of MsMfpA on 3 μM His-tagged MsGyrB in ATPase assays. A low ATPase activity of MsGyrB, which is inhibited to baseline by novobiocin, was observed (Fig. 4D). Unlike linear DNA, MsMfpA was still able to stimulate ATP hydrolysis, which was inhibited by novobiocin (Fig. 4D), suggesting that the interaction of MfpA with GyrB differs from that of DNA. To test the activity of MsMfpA on the ATPase domain, we made a plasmid expressing MsGyrB47, the ATPase (N-terminal) domain of M. smegmatis GyrB. No ATPase activity for this domain was observed using 5 μM protein (Fig. 4E), suggesting that removing the C-terminal domain of MsGyrB further decreased the ATPase activity. The ATPase activity of MsGyrB47, like that of MsGyrB, was significantly activated by MsMfpA, but not by DNA (Fig. 4E). DNA stimulation of the ATPase activity of the isolated ATPase domain has previously been observed with yeast topo II and human topo IIα (34, 35).
To investigate if MsMfpA and DNA compete in the stimulation of ATP hydrolysis, we did ATPase assays in the presence of linear DNA, MsMfpA, or both (SI Appendix, Fig. S3D). The results showed that either linear DNA or MsMfpA was able to stimulate the ATPase activity. But when both linear DNA and MsMfpA were added into the reactions, the concentration-dependent activation by MsMfpA disappeared, while the stimulation by linear DNA could be observed. These results indicate that there is a competition between MsMfpA and linear DNA in binding to gyrase.
Structure of MsMfpA–MsGyrB47 Complex.
We used X-ray crystallography to further understand the regulatory mechanism of MsMfpA on Msgyrase. First, we solved the structure of MsMfpA alone to 1.77-Å resolution. As shown in SI Appendix, Fig. S4A, MsMfpA, like MtbMfpA (19), forms an elongated homodimer through the interaction of its C termini and folds as a right-handed β-helix resembling the dimensions of B-form DNA. As reported previously, a T segment can be captured by the ATPase domains of Streptococcus pneumoniae topo IV to form a complex in the presence of ADPNP and Mg2+ (36) (SI Appendix, Fig. S4F). Based on these data, we hypothesized that MsMfpA, like a T segment, could be captured by MsGyrB47 in the presence of Mg2+ and ADPNP. We then attempted cocrystallization of MsMfpA and MsGyrB47, but only obtained crystals of the MsGyrB47 dimer with ADPNP and solved this structure to 1.56-Å resolution (SI Appendix, Fig. S4D).
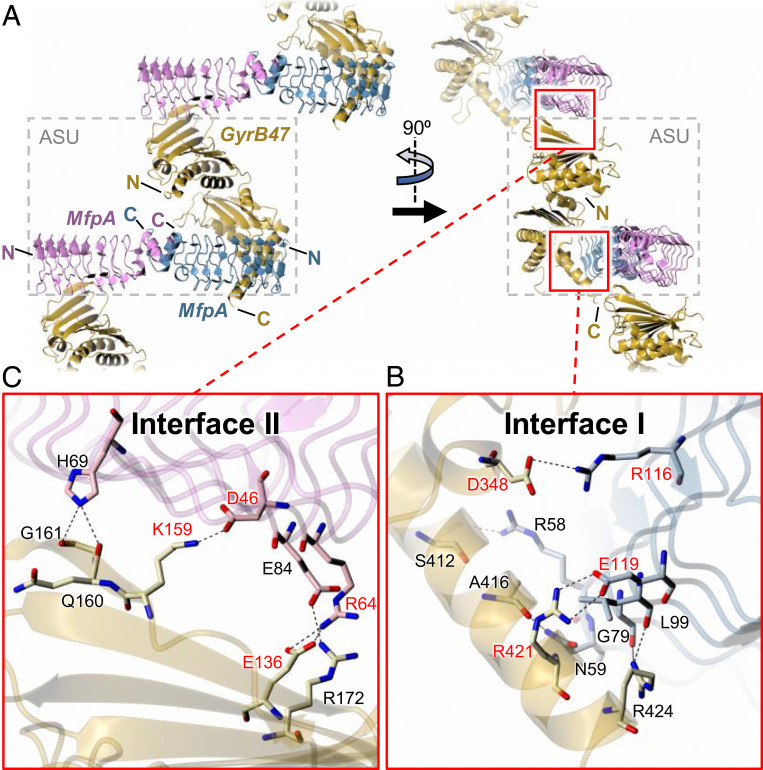
After extensive screening, we obtained crystals of a MsMfpA–MsGyrB47 complex and collected X-ray data to 2.2-Å resolution. To our surprise, the structure revealed an asymmetric unit comprised of an MsMfpA homodimer and a single MsGyrB47 subunit (Fig. 5 and SI Appendix, Fig. S4B). Even through the application of crystallographic symmetry, it was not possible to generate a recognizable MsGyrB47 dimer. The structure of the MsMfpA dimer was very similar to that determined in isolation (rmsd values in the range 0.43 to 0.55 Å for pairwise subunit comparisons and 0.94 Å for the dimer:dimer comparison), while compared with the subunit in the MsGyrB47-only structure, there was a slight hinge bending motion of around 20° at the junction between the ATPase subdomain and the transducer subdomain in the MsGyrB47 domain in the complex with MsMfpA (SI Appendix, Figs. S5 and S6). As a result, superposition of the whole subunits gave a comparatively large rmsd value of 2.80 Å, while separate comparisons of the corresponding ATPase and transducer subdomains gave smaller rmsd values of 0.83 and 1.59 Å, respectively. This conformational change causes a loop from the transducer domain bearing Gln370 and Lys372 (the “QK loop”), which would otherwise coordinate the γ-phosphate of ATP, to move away from the active site (SI Appendix, Fig. S5). Coupled with the lack of potential interactions with Tyr12 from the N-terminal arm of an opposing MsGyrB47 subunit, we would not expect this structure to bind nucleotide, even if it were present. Within the crystal lattice, there are two distinct interfaces between the MsMfpA dimer and the MsGyrB subunit (Fig. 5), which are named interface I, involving the C-terminal end of one MsGyrB47 subunit with one subunit of the MsMfpA dimer (with an interface area of ∼700 Å2) (Fig. 5), and interface II, involving the N-terminal end of a symmetry-related MsGyrB47 subunit with the other subunit of MsMfpA (with an interface area of ∼410 Å2) (Fig. 5). These interactions generate a zig-zag arrangement of the components that pervades the crystal. We next sought to determine which, if any, of these interfaces were biologically relevant.
Fig. 5.
Structure of MfpA-MsGyrB47 complex. The Top panels (A) show orthogonal views of the crystal packing, which reveals a 2:1 MsMfpA:MsGyrB47 stoichiometry with two potentially significant interfaces between MsMfpA and MsGyrB47 that together create a zig-zag arrangement of the molecules. Interface I lies within the asymmetric unit (ASU; delineated by dashed gray box) and involves the C-terminal region of MsGyrB47 interacting with one half of the MsMfpA dimer. Interface II lies at the junction of two neighboring ASUs and involves the N-terminal region of MsGyrB47 interacting with the other half of the MsMfpA dimer. These interfaces are shown in detail in the two Insets, (B and C), below. Within each, there are both hydrogen bonds and salt bridges. We sought to probe the importance of these interfaces by disruption of two salt bridges in each interface through site-directed mutagenesis. The residues that were mutated are labeled in red. For clarity, in the Lower panels, the backbone traces are depicted as semitransparent ribbons.
The Interactions on Interface I Are Essential for MsMfpA to Protect Mycobacterial DNA Gyrase.
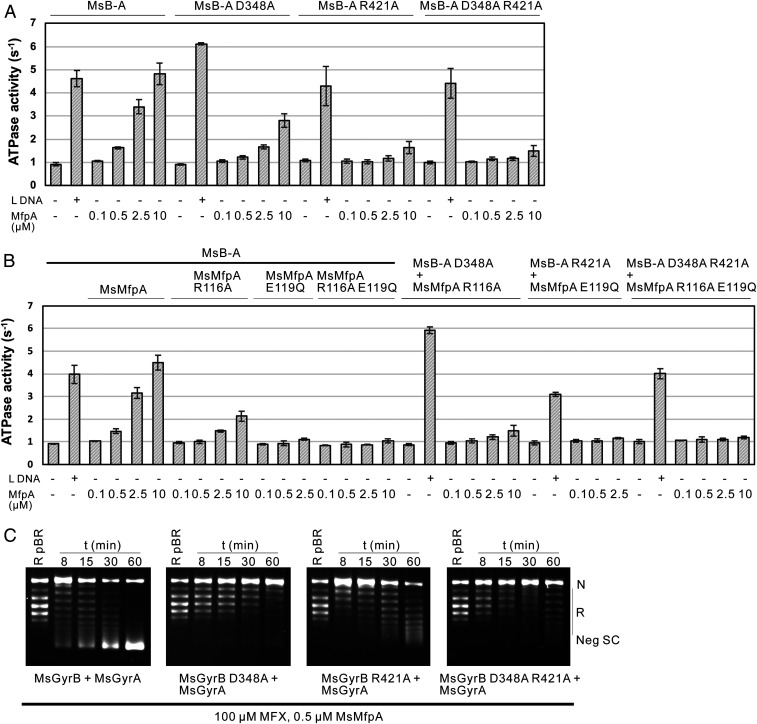
To test the importance of interface I in the protection of Msgyrase by MsMfpA, we disrupted two salt bridges within the interface that link MsGyrB47 residues Asp348 and Arg421 to MsMfpA residues Arg116 and Glu119, respectively. We first mutated these residues to Ala on MsGyrB47 to obtain MsGyrB47 (D348A), MsGyrB47 (R421A), and MsGyrB47 (D348A, R421A) and performed ATPase assays using wild-type MsMfpA and these mutated proteins. Compared with wild-type MsGyrB47, the ATPase stimulation of MsGyrB47 (D348A) by MsMfpA was decreased about twofold, while the stimulation of both MsGyrB47 (R421A) and MsGyrB47 (D348A, R421A) was almost abolished (SI Appendix, Fig. S8A). These data suggest that the interactions of interface I may be important for MsMfpA to stimulate Msgyrase ATPase activity.
To verify these results, we constructed plasmids to express the gyrase fusion proteins MsB-A (D348A), MsB-A (R421A), and MsB-A (D348A, R421A). These mutants, which showed similar supercoiling activities with the wild-type protein (SI Appendix, Fig. S8B), were used in ATPase assays. We found that, similar to wild-type MsB-A, the hydrolysis of ATP by these mutants could be significantly activated by linear DNA (Fig. 6A). But unlike linear DNA, the effects of MsMfpA on these mutants, consistent with the results of MsGyrB47 mutants, were significantly decreased (Fig. 6A). These results suggest that interface I could be required for MsMfpA protection Msgyrase.
Fig. 6.
The interactions on the interface I are required for MsMfpA to stimulate the hydrolysis of ATP and protect gyrase from FQs. The stimulation of ATP hydrolysis by MsB-A or MsB-A mutants by MsMfpA or MsMfpA mutants (A and B). (C) Time-course supercoiling assays were performed with MsMfpA, Msgyrase, and Msgyrase mutants. The wild-type MsMfpA protected Msgyrase from 100 μM MFX, but not Msgyrase (D348A), Msgyrase (R421A), and Msgyrase (D348A, R421A).
To further confirm the results, we made MsMfpA mutants—MsMfpA (R116A), MsMfpA (E119Q) (we found that E119A was not stable, as evidenced by protein precipitation), and MsMfpA (R116A, E119Q)—and tested the activities of these proteins on wild-type MsB-A and its mutants in ATPase assays. As shown in Fig. 6B, the stimulation of wild-type MsB-A or MsB-A mutants by MsMfpA mutants was dramatically decreased; some precipitate was observed in the assays with 10 μM MsMfpA (E119Q). All of these findings indicate that the residues on the interface I are necessary for MsMfpA to interact with MsGyrB47 and stimulate the hydrolysis of ATP.
To determine whether these residues play roles in the protection of Msgyrase, we generated plasmids to coexpress MsGyrB mutants and wild-type MsGyrA to obtain Msgyrase (GyrB D348A), Msgyrase (GyrB R421A), and Msgyrase (GyrB D348A, R421A). All the mutants showed high negative supercoiling activity (SI Appendix, Fig. S8C). We tested MsMfpA on Msgyrase and its mutants in time-course supercoiling assays and found that MsMfpA was able to protect the wild-type enzyme against 100 μM CFX, but not the mutant enzymes (SI Appendix, Fig. S8D). We then performed the assays with 100 μM MFX. Similar to the results with CFX, the protective effects to the mutant enzymes by MsMfpA were significantly decreased (Fig. 6C). To further confirm these results, time-course supercoiling assays were performed using MsMfpA and gyrase mutants. As shown in SI Appendix, Fig. S8E, MsMfpA mutants lost the protection on the wild-type enzyme (SI Appendix, Fig. S8 E, Left) as well as the mutant enzymes (SI Appendix, Fig. S8 E, Right). These data indicate that the interactions of interface I play an essential role in MsMfpA-dependent protection of Msgyrase from FQs.
The Interactions on Interface II Are Not Required for MsMfpA to Protect Mycobacterial DNA Gyrase.
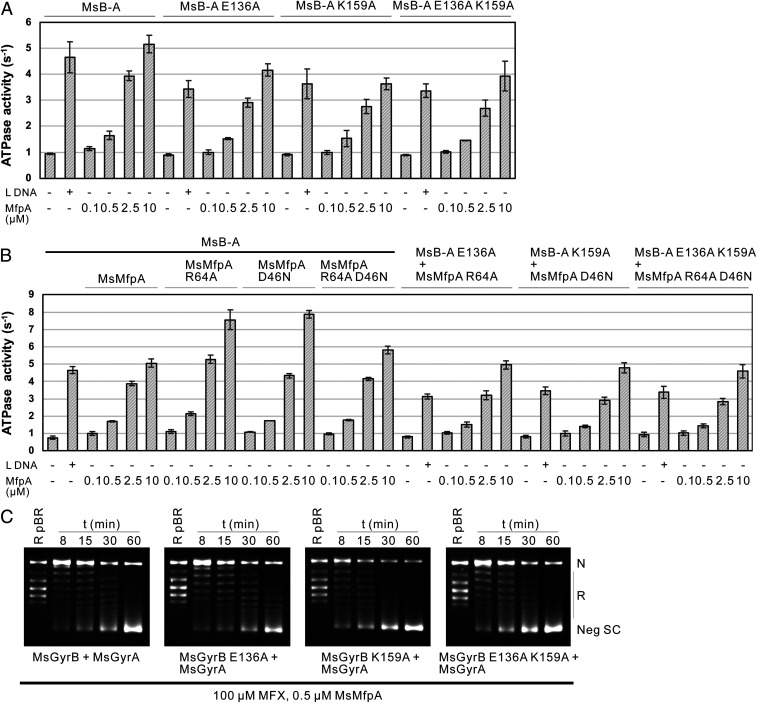
To test the importance of interface II, again we disrupted two salt bridges within the interface, this time linking MsGyrB47 residues Glu136 and Lys159 to MsMfpA residues Arg64 and Asp46, respectively. We made mutants on MsB-A to generate MsB-A (E136A), MsB-A (K159A), and MsB-A (E136A, K159A) and tested MsMfpA on wild-type and mutated enzymes in ATPase assays, and found that, similar to linear DNA, MsMfpA was able to stimulate the ATPase activity of all these enzymes (Fig. 7A).
Fig. 7.
The interactions on the interface II are not necessary for MsMfpA to stimulate the hydrolysis of ATP and protect gyrase from FQs. ATPase assays were performed using MsMfpA, MsMfpA mutants, MsB-A, and MsB-A mutants (A and B). (C) Time-course supercoiling assays were carried out using MsMfpA, Msgyrase, and Msgyrase mutants. MsMfpA could protect Msgyrase (Upper and Right) and its mutants (Msgyrase (E136A) (Upper, Right), Msgyrase (K159A) (Lower, Left), and Msgyrase (E136A, K159A) (Lower, Right) against 100 μM MFX.
To verify the results, the plasmids expressing MsMfpA (R64A), MsMfpA (D46N) (we found that D46A was insoluble), and MsMfpA (R64A, D46N) were constructed. ATPase assays performed using these mutants showed that all mutated MsMfpA proteins were able to stimulate ATP hydrolysis of the wild-type MsB-A, as well as the mutated enzymes (Fig. 7B). Among these data, hyperstimulations were observed in the assays with MsMfpA (R64A) and MsMfpA (D46N) on the wild-type MsB-A, suggesting that these mutations may change the properties of MsMfpA. All the results of ATPase assays suggest that the interactions of interface II are not required for the protective effects of MsMfpA on Msgyrase.
To test if these interactions are important for MsMfpA to protect Msgyrase, we also constructed plasmids coexpressing MsGyrB mutants and wild-type MsGyrA to generate Msgyrase (GyrB E136A), Msgyrase (GyrB K159A), and Msgyrase (GyrB E136A, K159A). Time-course supercoiling assays performed using these mutated enzymes showed that MsMfpA still had protective activity on these mutants (Fig. 7C), suggesting that these interactions are not vital for MsMfpA to protect Msgyrase against FQs. To confirm these results, we performed the assays with mutated MsMfpA and enzymes. Unexpectedly, the protective activity on Msgyrase (SI Appendix, Fig. S9, Left) and its mutants (SI Appendix, Fig. S9, Right) by MsMfpA mutants were obviously decreased. This suggests that the properties of MsMfpA may be affected by mutating these sites.
Taking the results of both the ATPase assays and time-course assays together, we find that the interactions on interface II are not essential for MsMfpA to stimulate ATP hydrolysis and protect Msgyrase from FQs. Therefore, it seems likely that interface II is simply a crystal contact and has no biological relevance; however, it is feasible that this interface might have a significant role in vivo.
Discussion
Effect of MfpA on the Topoisomerase Reactions of DNA Gyrase.
MfpA has been described as a regulator for mycobacterial DNA gyrase that can both inhibit the enzyme as well as provide protection from FQs, although there is controversy regarding the latter point (21, 23). In this study, using biochemical assays and X-ray crystallography, we analyzed the actions of MfpA on gyrase using M. smegmatis proteins. Gyrase catalyzes the hydrolysis of ATP to introduce negative supercoils through the passage of a DNA T segment through a G segment. We found that a large excess of MfpA (∼>10,000-fold) was needed to inhibit supercoiling under these conditions (Fig. 1). However, under the same conditions, MfpA was apparently unable to inhibit the ATP-dependent relaxation of positive supercoils (Fig. 1), which is essentially the same (reduction in DNA linking number). These results can be interpreted as MfpA “competing” with the DNA T segment for binding to gyrase. DNA wrapping around gyrase leads to the efficient presentation of a T segment to the DNA gate (5) and it is perhaps not surprising that high concentrations of MfpA are required to compete with the T segment. ATP-dependent relaxation of positively supercoiled DNA by gyrase is highly efficient (37) and this reaction would present a significant barrier to competition by MfpA; a positively supercoiled DNA substrate is thought to have a coupling efficiency (i.e., probability of T-segment capture) of ∼100% (37). Conversely, ATP-independent relaxation of negatively supercoiled DNA was unaffected by MfpA (SI Appendix, Fig. S1). This reaction is thought to proceed via entrance of the T segment through the “exit” gate (38) (i.e., binding of MfpA to the ATPase domains would not be expected to greatly affect this reaction). If MfpA acted as a G-segment mimic, we would have expected both the relaxation and supercoiling reactions to be affected. Taken together with other data in this paper, we have essentially disproved the idea that MfpA acts as a G-segment mimic (19) and proved the notion, suggested previously (24), that it is a T-segment mimic, consistent with recent results on QnrB1 (25).
Effect of MfpA on Quinolone Action.
FQs, such as CFX and MFX, can inhibit the gyrase-catalyzed DNA supercoiling reaction (Fig. 1) (2, 3). We found that far less of an excess of MfpA is required to relieve supercoiling inhibition by FQs (∼100-fold) than is required to inhibit gyrase-catalyzed supercoiling (>10,000-fold) (Fig. 1). It is well-known that quinolones can interact with the gyrase–DNA complex without DNA wrapping, including DNA segments as short as 20 bp (39–41). Therefore, the requirement for DNA wrapping in the presence of FQs is reduced and could potentially indicate that MfpA has a higher probability of outcompeting the T segment.
Under appropriate conditions, FQs can stabilize the gyrase–DNA cleavage complex leading to the generation of linear DNA from a closed-circular substrate (Fig. 2). In the presence of ATP, MfpA can inhibit the FQ-induced gyrase cleavage reaction. In these reactions, the ratio of MfpA over gyrase was lower than above (∼25-fold), although complete inhibition of cleavage was not observed. We suggest that the effect of MfpA on the gyrase supercoiling and cleavage reactions can be rationalized by it essentially behaving as a T-segment mimic. In the case of supercoiling, MfpA has to outcompete the T-segment in binding to the ATP-operated clamp of GyrB; this is progressively more difficult when the DNA is positively supercoiled. In the presence of a quinolone, the binding of MfpA, acting as a T-segment mimic, forces the DNA gate (i.e., the protein–protein interface) open. Under these conditions, the quinolone dissociates, due to the disruption of its binding site allowing gyrase to religate the DNA. This idea is discussed in more detail below.
When FQ-induced gyrase cleavage reactions were carried out in the absence of ATP, MfpA did not affect the reaction (Fig. 2), consistent with our previous results on the supercoiling and relaxation reactions, and the notion that MfpA interacts with the ATPase domain of GyrB. Gyrase modulates DNA topology, the precise activity depending on the starting substrate (i.e., relaxed, negatively supercoiled or positively supercoiled DNA). In the presence or absence of ATP, FQs are able to poison all these reactions by stabilizing cleavage complexes, while MfpA is capable of inhibiting the formation of cleavage complexes only in the presence of ATP (Fig. 2 and SI Appendix, Fig. S2), suggesting that ATP hydrolysis by gyrase is required in order for MfpA to protect gyrase from FQs.
In both the inhibition and cleavage experiments, more MfpA was required in the case of reactions involving MFX as compared to those involving CFX. This difference likely reflects the higher potency of MFX toward mycobacterial gyrases (12), which indicates a higher affinity of this FQ for the gyrase–DNA complex.
Promotion of the Gyrase ATPase Activity by MfpA.
We found that MfpA, like DNA (12), can stimulate the ATPase reaction of Msgyrase (Fig. 4). This result is consistent with the proposal that MfpA, along with other PRPs, behaves as a DNA mimic (20). By investigating a range of Msgyrase constructs, we showed that MfpA can activate the ATPase reaction of the minimal protein domain required for ATPase activity (GyrB47). Interestingly the GyrB47 ATPase activity could not be activated by DNA; this is consistent with observations made with other gyrases [e.g., E. coli (42)], but contrasts with data on the corresponding domains of topo II, where both the human enzyme (topo IIα) and yeast topo II show DNA-stimulated ATP hydrolysis (34, 35). The fact that the ATPase activity of MsGyrB47 can be activated by MfpA but not by DNA suggests that it might interact with this domain in a different manner to DNA. However, exactly how MfpA stimulates ATP hydrolysis and how ATP hydrolysis correlates with FQ rescue need further investigation. Nonetheless, our other data suggest that MfpA can compete with the T segment, and overall that it behaves as a T-segment mimic.
Structure of the MfpA–GyrB Complex.
We solved the structure of MsMfpA (SI Appendix, Fig. S4A) and found that it was very similar to that of MfpA proteins from other species (19, 20) and other PRPs, such as Qnr (28, 43, 44). Considering their consistent structure and biological function, we hypothesize that other gyrase-targeted PRPs in bacteria act as T-segment mimics to stimulate ATP hydrolysis and protect gyrase. Our initial efforts to crystallize the MfpA–GyrB complex led to a structure of the N-terminal domain of MsGyrB (MsGyrB47) alone with ADPNP bound (SI Appendix, Fig. S4D); this structure closely resembles those from other bacterial species (45–47). In the presence of ADPNP and Mg2+, MsGyrB47 shows a dimeric clamp-closed structure with a “hole” between the two monomers, which has been proposed to accommodate the T-segment DNA (47, 48).
Under other conditions, and in the absence of nucleotide, we obtained a structure of the MsMfpA–MsGyrB47 complex (Fig. 5 and SI Appendix, Fig. S4B). These structures can be compared with that of the ATPase domain of S. pneumoniae topoisomerase IV with a captured T-segment, which forms a complex in the presence of ADPNP (36) (SI Appendix, Fig. S4F). In this case, the ATPase domain is in a dimeric clamp-closed conformation with a 14-bp DNA segment bound in the cavity formed between the two subunits (47). A similar cavity is present in the MsGyrB47 homodimer, although the clamp adopts a slightly more closed conformation (SI Appendix, Fig. S4D). In the case of the MfpA–GyrB–N terminal domain (NTD) complex, a monomer of the ATPase domain is bound to two MfpA dimers forming two interfaces. Attempts to recapitulate a recognizable GyrB47 dimer within the context of this complex resulted in severe clashes of the second GyrB47 subunit with MfpA (SI Appendix, Fig. S4C). When viewed in the “standard” GyrB orientation (SI Appendix, Fig. S4E), it is clear that the relationship between MsMfpA and GyrB47 is entirely different from that expected for T-segment DNA (SI Appendix, Fig. S4F), since the long axes of MfpA and DNA, respectively, are orthogonal in this comparison. Subsequent mutagenesis experiments showed that only mutations at interface I (at the C terminus of MsGyrB47) significantly affect the actions of MsMfpA on Msgyrase, suggesting that the interactions at interface II could simply be crystal-packing interactions.
Nevertheless, the 2:1 MsMfpA:MsGyrB47 stoichiometry we observe remains counter-intuitive, although we cannot rule out the possibility that steric clashes in the somewhat artificial environment of the crystal lattice could be preventing the formation of the expected 2:2 complex. With this in mind, we sought to generate a hypothetical 2:2 complex, which is essentially a chimera of the crystal structures of the MsGyrB47 homodimer and the MsMfpA–MsGyrB47 2:1 complex (SI Appendix, Fig. S6). This model is twofold symmetric and retains the observed interactions between the pair of ATPase subdomains in the former structure, as well as the observed interactions between the transducer domain of MsGyrB47 and MsMfpA in the latter structure (SI Appendix, Fig. S7). As a result, the relationship between the ATPase subdomains and the transducer domains is strikingly different from any known homologous structure. Nevertheless, this extreme conformation approximates to an extrapolation of the hinge-bending motion we observe for MsGyrB47 in the comparison between the homodimer and 2:1 complex crystal structures (SI Appendix, Figs. S5 and S6). This model has led us to speculate that the binding of MsMfpA may promote the opening of the DNA gate, bringing about the dissociation of the bound FQs. Moreover, a mode of MfpA interaction with gyrase that is distinct from that of a T segment could explain why PRPs can rescue FQ-stabilized cleavage complexes, while the T segment cannot.
In the crystal structure of the 2:1 complex, the hinge bending motion pulls the QK loop away from the nucleotide binding site, and thus likely represents a conformational state that is incapable of hydrolyzing ATP because, by analogy with the E. coli enzyme, Lys372 plays a critical role in stabilizing the transition state (49). This would also apply in the hypothetical 2:2 model with the more extreme bending. We therefore speculate that these models reflect the situation after ATP hydrolysis has occurred. Indeed, the withdrawal of the QK loop would promote phosphate release. Separation of the ATPase subdomains (i.e., opening of the GyrB clamp), might be necessary to eject the ADP.
A Model of MfpA Action on Gyrase.
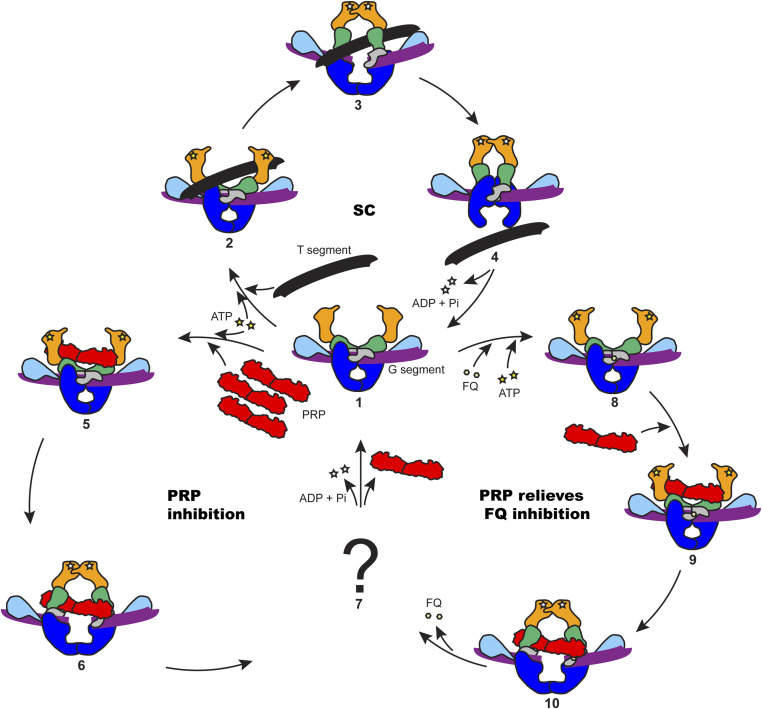
In this study, we detailed the molecular actions of MfpA in the regulation of DNA gyrase using M. smegmatis proteins and proposed a possible working model for PRPs (Fig. 8). Without FQs, ATP binding to GyrB drives the closing of the GyrB clamp to capture a T segment. Under these conditions, excess PRP is required to compete with the T segment and interact with the ATPase domains of GyrB. The binding of PRP to the ATPase domains competes with the T segment and activates the hydrolysis of ATP, and the supercoiling activity of gyrase is abolished as T segments can no longer be transported. In the presence of FQs, the gyrase–DNA cleavage complex can be stabilized by FQ binding, and the reaction cycle is stalled with a closed DNA gate and a cleaved G segment. Under these conditions, PRPs can interact with the ATPase domains of GyrB. This interaction stimulates ATP hydrolysis, resulting in a conformational change of the ATPase domains that drives the opening of the blocked DNA gate, and expelling the FQ molecules, allowing religation of the G segment. Meanwhile, the PRP is released from gyrase and can potentially go on to relieve another poisoned enzyme. In the absence of further FQ binding, gyrase can then continue with the normal supercoiling reaction cycle. Further work will be needed to substantiate this model.
Fig. 8.
Proposed model for PRPs in the protection of DNA gyrase. Gyrase domains are colored as in Fig. 4. (1) Gyrase binds to a G segment, the N gate is open. (2) ATP is bound to GyrB and a T segment is presented to the ATPase domains of GyrB; the G segment is cleaved. (3) The presented T segment is captured by the GyrB clamp; ATP hydrolysis promotes the rotation of GyrB to open the DNA gate. (4) The T segment passes through the DNA gate and goes out through C (exit) gate; ADP and phosphate are released from GyrB. (5) Excess PRP is able to access the ATPase domains. (6) PRP is captured by the GyrB clamp and stimulates ATP hydrolysis, changing the conformation of GyrB to open the DNA gate. (7) The PRP is released in an unknown manner. (8) FQ stabilizes the gyrase–DNA cleavage complex. (9) PRP binds to the poisoned complex. (10) PRP activates the hydrolysis of ATP and changes the conformation of GyrB to open the DNA gate and dissociate the bound quinolones from the cleavage complex; the FQ is released from the complex. The supercoiling cycle (SC) contains 1, 2, 3, and 4. The PRP inhibition cycle contains 1, 5, 6, and 7. The PRP relieves FQ inhibition cycle contains 1, 8, 9, 10, and 7.
Overall, we have shown that MfpA binds to the GyrB subunit of gyrase such that it interferes with the binding of the T segment and stimulates the ATPase activity; at high concentration, it can inhibit gyrase-catalyzed DNA supercoiling. The MfpA–GyrB interaction relieves gyrase from FQ inhibition and can reverse quinolone-induced gyrase-catalyzed DNA cleavage. Given that quinolones are not natural products and FQs have only been in clinical use for ∼35 y, it seems unlikely that MfpA’s principal function is FQ protection. Indeed, the reduced level of quinolone susceptibility conferred by MfpA is relatively modest (22). Given that MfpA “mimics” DNA suggests that it might have a regulatory role in the bacterial cell (20), either specific to gyrase or encompassing other DNA-binding proteins. The similarity in protein structure of other PRPs (e.g., Qnr proteins) suggests that other members of this family will have similar roles. Our description of the molecular basis of MfpA–GyrB interaction could be utilized in the design of inhibitors with potential as novel antibiotics.
Materials and Methods
Cloning and Protein Purification.
Cloning and protein purification methods are described in SI Appendix.
Gyrase Assays.
Supercoiling, relaxation, DNA cleavage, and ATPase assays were carried out as described for M. tuberculosis gyrase (50, 51) with minor modifications, as described in SI Appendix.
Crystallization, Data Collection, Structure Determination, and Refinement.
Crystallization experiments were performed using sitting-drop vapor diffusion in 96-well plates, as described in SI Appendix. X-ray data were recorded at the Diamond Light Source; data collection statistics are summarized in Table 1. Full details are given in SI Appendix.
Table 1.
Summary of X-ray data and model parameters
| Protein | MsMfpA | MsGyrB47 | MsMfpA-GyrB47 complex | |
| Data collection | ||||
| Diamond Light Source beamline | I04 | I04 | I03 | |
| Wavelength (Å) | 0.9795 | 0.9795 | 0.9763 | |
| Detector | Pilatus3 6M | Eiger2 XE 16M | Eiger2 XE 16M | |
| Resolution range (Å) | 66.28–1.77 (1.81–1.77) | 65.37–1.56 (1.59–1.56) | 75.20–2.20 (2.26–2.20) | |
| Space group | P21 | P6122 | C2 | |
| Cell parameters (Å°) | a = 31.01, b = 85.73, c = 66.34, β = 92.41 | a = b = 76.99, c = 261.48 | a = 150.55, b = 59.38, c = 141.22, β = 119.71 | |
| Total measured intensities | 213,565 (8,389) | 2,578,084 (127,674) | 380,771 (29,306) | |
| Unique reflections | 33,772 (1,901) | 66,509 (3,231) | 55,380 (4,481) | |
| Multiplicity | 6.3 (4.4) | 38.8 (39.5) | 6.9 (6.5) | |
| Mean I/σ(I) | 8.9 (0.9) | 16.7 (1.9) | 14.4 (0.8) | |
| Completeness (%) | 99.8 (97.1) | 100.0 (100.0) | 100.0 (100.0) | |
| Rmerge* | 0.097 (1.517) | 0.141 (2.486) | 0.048 (1.857) | |
| Rmeas† | 0.106 (1.729) | 0.143 (2.518) | 0.052 (2.022) | |
| CC1/2‡ | 0.999 (0.507) | 1.000 (0.845) | 0.996 (0.608) | |
| Wilson B value (Å2) | 32.5 | 19.9 | 68.8 | |
| Refinement | ||||
| Resolution range (Å) | 52.49–1.77 (1.82–1.77) | 64.69–1.56 (1.60–1.56) | 75.32–2.20 (2.26–2.20) | |
| Reflections: working/free§ | 32,049/1,697 (2,344/114) | 63,082/3,287 (4,566/247) | 52,549/2,825 (3,845/181) | |
| Rwork/Rfree¶ | 0.208/0.234 (0.392/0.368) | 0.147/0.194 (0.251/0.253) | 0.208/0.251 (0.371/0.379) | |
| Ramachandran plot: favored/allowed/disallowed# (%) | 98.4/1.6/0.0 | 97.7/2.3/0 | 97.6/2.4/0 | |
| Rms bond distance deviation (Å) | 0.008 | 0.007 | 0.007 | |
| Rms bond angle deviation (°) | 1.50 | 1.37 | 1.44 | |
| No. of protein residues by chain [residue no. ranges] | A: 180 [10–189]; B: 181 [10–190] | A: 394 [0–213,246–425] | MfpA-A: 183 [9–191]; MfpA-B: 181 [9–189]; GyrB47-C: 345 [34–104,124–214,244–426] | |
| No. of water/other molecules∥ | 160/3 | 296/11 | 86/3 | |
| Mean B factors: protein/water/other∥ (Å2) | 35.5/39.4/47.0 | 24.7/36.4/25.2 | 77.9/67.2/91.9 | |
| PDB accession code | 6ZT4 | 6ZT3 | 6ZT5 | |
Each dataset was acquired from a single crystal. Values in parentheses are for the outer resolution shell.
Rmerge = ∑hkl ∑i |Ii(hkl) − ⟨I(hkl)⟩|/∑hkl ∑iIi(hkl).
Rmeas = ∑hkl [N/(N − 1)]1/2 × ∑i |Ii(hkl) − ⟨I(hkl)⟩|/∑hkl ∑iIi(hkl), where Ii(hkl) is the ith observation of reflection hkl, ⟨I(hkl)⟩ is the weighted average intensity for all observations i of reflection hkl and N is the number of observations of reflection hkl.
CC1/2 is the correlation coefficient between symmetry equivalent intensities from random halves of the dataset.
The dataset was split into “working” and “free” sets consisting of 95% and 5% of the data respectively. The free set was not used for refinement.
The R-factors Rwork and Rfree are calculated as follows: R = ∑(| Fobs - Fcalc |)/∑| Fobs |, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively.
As calculated using MolProbity.
“Other” refers to ligands (including buffer components or cryoprotectant molecules) and ions.
Supplementary Material
Acknowledgments
We thank Shannon McKie, Dmitry Ghilarov, and Jonathan Heddle for comments on the manuscript and Dmitry Ghilarov and Jonathan Heddle for sharing results prior to publication. This work was supported by the Centre of Excellence for Plant and Microbial Science, established between the John Innes Centre and the Chinese Academy of Sciences and funded by the UK Biotechnology and Biological Sciences Research Council (BBSRC) and the Chinese Academy of Sciences; an Investigator Award from the Wellcome Trust (110072/Z/15/Z to A.M.); and by BBSRC Institute Strategic Programme Grant BB/P012523/1. This work was also supported by grants from the Ministry of Science and Technology of China (2018YFC1603900, 2017YFA0505901 to K.M.), National Natural Science Foundation of China (31970136, 31670137 to K.M.) and International Joint Research Project of the Institute of Medical Science, University of Tokyo (Extension-2019-K3006 to K.M.). The Diamond Light Source is acknowledged for access to beamlines I03 and I04 under proposal MX18565.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016705118/-/DCSupplemental.
Data Availability
Crystal-structure data have been deposited in the Protein Data Bank (PDB ID codes 6ZT3, 6ZT4, and 6ZT5). All other study data are presented within the paper and SI Appendix.
References
- 1.WHO , Global Tuberculosis Report (WHO, Geneva, Switzerland, 2019). [Google Scholar]
- 2.Nagaraja V., Godbole A. A., Henderson S. R., Maxwell A., DNA topoisomerase I and DNA gyrase as targets for TB therapy. Drug Discov. Today 22, 510–518 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Bush N. G., Evans-Roberts K., Maxwell A., DNA topoisomerases. Ecosal Plus 6, 10.1128/ecosalplus.ESP-0010-2014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisher L. M., Mizuuchi K., O’Dea M. H., Ohmori H., Gellert M., Site-specific interaction of DNA gyrase with DNA. Proc. Natl. Acad. Sci. U.S.A. 78, 4165–4169 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heddle J. G., Mitelheiser S., Maxwell A., Thomson N. H., Nucleotide binding to DNA gyrase causes loss of DNA wrap. J. Mol. Biol. 337, 597–610 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Kirkegaard K., Wang J. C., Mapping the topography of DNA wrapped around gyrase by nucleolytic and chemical probing of complexes of unique DNA sequences. Cell 23, 721–729 (1981). [DOI] [PubMed] [Google Scholar]
- 7.Morrison A., Cozzarelli N. R., Contacts between DNA gyrase and its binding site on DNA: Features of symmetry and asymmetry revealed by protection from nucleases. Proc. Natl. Acad. Sci. U.S.A. 78, 1416–1420 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roca J., Wang J. C., DNA transport by a type II DNA topoisomerase: Evidence in favor of a two-gate mechanism. Cell 77, 609–616 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Kampranis S. C., Maxwell A., Conversion of DNA gyrase into a conventional type II topoisomerase. Proc. Natl. Acad. Sci. U.S.A. 93, 14416–14421 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aubry A., Fisher L. M., Jarlier V., Cambau E., First functional characterization of a singly expressed bacterial type II topoisomerase: The enzyme from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 348, 158–165 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Cole S. T., et al., Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Manjunatha U. H., et al., Functional characterisation of mycobacterial DNA gyrase: An efficient decatenase. Nucleic Acids Res. 30, 2144–2153 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zechiedrich E. L., Cozzarelli N. R., Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes Dev. 9, 2859–2869 (1995). [DOI] [PubMed] [Google Scholar]
- 14.Maxwell A., Bush N. G., Germe T., McKie S. J., “Non-quinolone topoisomerase inhibitors” in Antimicrobial Resistance and Implications for the 21st Century, Fong I. W., Shlaes D., Drlica K., Eds. (Springer, Switzerland, 2018), pp. 593–618. [Google Scholar]
- 15.Collin F., Karkare S., Maxwell A., Exploiting bacterial DNA gyrase as a drug target: Current state and perspectives. Appl. Microbiol. Biotechnol. 92, 479–497 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillespie S. H., The role of moxifloxacin in tuberculosis therapy. Eur. Respir. Rev. 25, 19–28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drlica K., et al., Quinolones: Action and resistance updated. Curr. Top. Med. Chem. 9, 981–998 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collin F., Maxwell A., The microbial toxin microcin B17: Prospects for the development of new antibacterial agents. J. Mol. Biol. 431, 3400–3426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hegde S. S., et al., A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science 308, 1480–1483 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Vetting M. W., et al., Pentapeptide repeat proteins. Biochemistry 45, 1–10 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mérens A., et al., The pentapeptide repeat proteins MfpAMt and QnrB4 exhibit opposite effects on DNA gyrase catalytic reactions and on the ternary gyrase-DNA-quinolone complex. J. Bacteriol. 191, 1587–1594 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montero C., Mateu G., Rodriguez R., Takiff H., Intrinsic resistance of Mycobacterium smegmatis to fluoroquinolones may be influenced by new pentapeptide protein MfpA. Antimicrob. Agents Chemother. 45, 3387–3392 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao J., et al., Mycobacterium fluoroquinolone resistance protein B, a novel small GTPase, is involved in the regulation of DNA gyrase and drug resistance. Nucleic Acids Res. 41, 2370–2381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shah S., Heddle J. G., Squaring up to DNA: Pentapeptide repeat proteins and DNA mimicry. Appl. Microbiol. Biotechnol. 98, 9545–9560 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Mazurek Ł., et al., Pentapeptide repeat proteins QnrB1 and AlbG require ATP hydrolysis to rejuvenate poisoned gyrase complexes. bioRxiv:10.1101/2020.09.24.310243 (26 September 2020). [DOI] [PMC free article] [PubMed]
- 26.Briales A., et al., In vitro effect of qnrA1, qnrB1, and qnrS1 genes on fluoroquinolone activity against isogenic Escherichia coli isolates with mutations in gyrA and parC. Antimicrob. Agents Chemother. 55, 1266–1269 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacoby G. A., et al., qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 50, 1178–1182 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vetting M. W., et al., Structure of QnrB1, a plasmid-mediated fluoroquinolone resistance factor. J. Biol. Chem. 286, 25265–25273 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown P. O., Cozzarelli N. R., A sign inversion mechanism for enzymatic supercoiling of DNA. Science 206, 1081–1083 (1979). [DOI] [PubMed] [Google Scholar]
- 30.Mizuuchi K., Fisher L. M., O’Dea M. H., Gellert M., DNA gyrase action involves the introduction of transient double-strand breaks into DNA. Proc. Natl. Acad. Sci. U.S.A. 77, 1847–1851 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gellert M., Mizuuchi K., O’Dea M. H., Itoh T., Tomizawa J. I., Nalidixic acid resistance: A second genetic character involved in DNA gyrase activity. Proc. Natl. Acad. Sci. U.S.A. 74, 4772–4776 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugino A., Peebles C. L., Kreuzer K. N., Cozzarelli N. R., Mechanism of action of nalidixic acid: Purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. U.S.A. 74, 4767–4771 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis R. J., et al., The nature of inhibition of DNA gyrase by the coumarins and the cyclothialidines revealed by X-ray crystallography. EMBO J. 15, 1412–1420 (1996). [PMC free article] [PubMed] [Google Scholar]
- 34.Campbell S., Maxwell A., The ATP-operated clamp of human DNA topoisomerase IIalpha: Hyperstimulation of ATPase by “piggy-back” binding. J. Mol. Biol. 320, 171–188 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Olland S., Wang J. C., Catalysis of ATP hydrolysis by two NH(2)-terminal fragments of yeast DNA topoisomerase II. J. Biol. Chem. 274, 21688–21694 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Laponogov I., et al., Trapping of the transport-segment DNA by the ATPase domains of a type II topoisomerase. Nat. Commun. 9, 2579 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bates A. D., O’Dea M. H., Gellert M., Energy coupling in Escherichia coli DNA gyrase: The relationship between nucleotide binding, strand passage, and DNA supercoiling. Biochemistry 35, 1408–1416 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Williams N. L., Maxwell A., Probing the two-gate mechanism of DNA gyrase using cysteine cross-linking. Biochemistry 38, 13502–13511 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Cove M. E., Tingey A. P., Maxwell A., DNA gyrase can cleave short DNA fragments in the presence of quinolone drugs. Nucleic Acids Res. 25, 2716–2722 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gmünder H., Kuratli K., Keck W., In the presence of subunit A inhibitors DNA gyrase cleaves DNA fragments as short as 20 bp at specific sites. Nucleic Acids Res. 25, 604–611 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Higgins N. P., Cozzarelli N. R., The binding of gyrase to DNA: Analysis by retention by nitrocellulose filters. Nucleic Acids Res. 10, 6833–6847 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ali J. A., Jackson A. P., Howells A. J., Maxwell A., The 43-kilodalton N-terminal fragment of the DNA gyrase B protein hydrolyzes ATP and binds coumarin drugs. Biochemistry 32, 2717–2724 (1993). [DOI] [PubMed] [Google Scholar]
- 43.Hegde S. S., Vetting M. W., Mitchenall L. A., Maxwell A., Blanchard J. S., Structural and biochemical analysis of the pentapeptide repeat protein EfsQnr, a potent DNA gyrase inhibitor. Antimicrob. Agents Chemother. 55, 110–117 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiong X., Bromley E. H., Oelschlaeger P., Woolfson D. N., Spencer J., Structural insights into quinolone antibiotic resistance mediated by pentapeptide repeat proteins: Conserved surface loops direct the activity of a Qnr protein from a gram-negative bacterium. Nucleic Acids Res. 39, 3917–3927 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brino L., et al., Dimerization of Escherichia coli DNA-gyrase B provides a structural mechanism for activating the ATPase catalytic center. J. Biol. Chem. 275, 9468–9475 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Stanger F. V., Dehio C., Schirmer T., Structure of the N-terminal gyrase B fragment in complex with ADP⋅Pi reveals rigid-body motion induced by ATP hydrolysis. PLoS One 9, e107289 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wigley D. B., Davies G. J., Dodson E. J., Maxwell A., Dodson G., Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature 351, 624–629 (1991). [DOI] [PubMed] [Google Scholar]
- 48.Tingey A. P., Maxwell A., Probing the role of the ATP-operated clamp in the strand-passage reaction of DNA gyrase. Nucleic Acids Res. 24, 4868–4873 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith A. B., Maxwell A., A strand-passage conformation of DNA gyrase is required to allow the bacterial toxin, CcdB, to access its binding site. Nucleic Acids Res. 34, 4667–4676 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karkare S., et al., The naphthoquinone diospyrin is an inhibitor of DNA gyrase with a novel mechanism of action. J. Biol. Chem. 288, 5149–5156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karkare S., Yousafzai F., Mitchenall L. A., Maxwell A., The role of Ca2+ in the activity of Mycobacterium tuberculosis DNA gyrase. Nucleic Acids Res. 40, 9774–9787 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystal-structure data have been deposited in the Protein Data Bank (PDB ID codes 6ZT3, 6ZT4, and 6ZT5). All other study data are presented within the paper and SI Appendix.