Significance
An outstanding challenge in GPCR pharmacology is quantifying the system-independent functional effects of ligand-receptor interactions. Current efforts to measure ligand efficacy at the level of the receptor (i.e., molecular efficacy) are limited by the requirement of extensive purification of receptor and G proteins to homogeneity. In this study, we present an accessible, scalable technology for the single point measurement of the molecular efficacy of GPCR ligands. Integrating this technology with insights from molecular dynamics simulations, we reveal that the transition of the G protein from an intermediate to a fully coupled interaction with the GPCR is a structural determinant of ligand molecular efficacy.
Keywords: G protein–coupled receptors, giant plasma membrane vesicles, FRET sensors, ligand efficacy, molecular dynamics simulations
Abstract
Classical pharmacological models have incorporated an “intrinsic efficacy” parameter to capture system-independent effects of G protein–coupled receptor (GPCR) ligands. However, the nonlinear serial amplification of downstream signaling limits quantitation of ligand intrinsic efficacy. A recent biophysical study has characterized a ligand “molecular efficacy” that quantifies the influence of ligand-dependent receptor conformation on G protein activation. Nonetheless, the structural translation of ligand molecular efficacy into G protein activation remains unclear and forms the focus of this study. We first establish a robust, accessible, and sensitive assay to probe GPCR interaction with G protein and the Gα C terminus (G-peptide), an established structural determinant of G protein selectivity. We circumvent the need for extensive purification protocols by the single-step incorporation of receptor and G protein elements into giant plasma membrane vesicles (GPMVs). We use previously established SPASM FRET sensors to control the stoichiometry and effective concentration of receptor–G protein interactions. We demonstrate that GPMV-incorporated sensors (v-SPASM sensors) provide enhanced dynamic range, expression-insensitive readout, and a reagent level assay that yields single point measurements of ligand molecular efficacy. Leveraging this technology, we establish the receptor–G-peptide interaction as a sufficient structural determinant of this receptor-level parameter. Combining v-SPASM measurements with molecular dynamics (MD) simulations, we elucidate a two-stage receptor activation mechanism, wherein receptor–G-peptide interactions in an intermediate orientation alter the receptor conformational landscape to facilitate engagement of a fully coupled orientation that tunes G protein activation.
G protein–coupled receptors (GPCRs) are a superfamily of over 800 integral membrane proteins that are a major target of modern drugs (1, 2). GPCRs trigger cellular signaling cascades through the ligand-dependent activation of G proteins. The C terminus (last 27 amino acids) of the G protein α-subunit (G-peptide) is one of the important structural elements in the selective G protein activation by GPCRs (3, 4). An activated GPCR engages the G-peptide to relay GDP exchange and consequent G protein activation (5). Swapping residues in the G-peptide is a long-established strategy to drive promiscuous GPCR–G protein coupling, attesting to its role in cognate G protein recognition (6, 7). Despite its established function in selective G protein activation, the role of the G-peptide in determining agonist efficacy remains poorly understood and forms the focus of this study.
Spectroscopic studies have highlighted dynamic conformational ensembles populated by the GPCR in response to ligand stimulation (8). In turn, single molecule Förster resonance energy transfer (FRET) studies of β2AR reveal that agonist binding causes a shift in the GPCR conformational ensemble, with corresponding changes in GDP–GTP exchange rates (9). The kinetics of conformational changes in the GPCR and consequent G protein activation were used to derive a “molecular efficacy” that quantifies the intrinsic effect that a ligand imposes directly onto its target GPCR. However, the structural basis of molecular efficacy and its association with the G-peptide requires further elucidation. Furthermore, high-resolution structures of GPCR–G protein complexes, stabilized by high efficacy agonists, reveal distinct orientations of the G-peptide bound to the receptor (10, 11). While these structures demonstrate that the transient GPCR–G protein interactions in cells are indeed dynamic, the relationship between these static snapshots and agonist molecular efficacy remains unclear.
Delineating the structural dynamics of GPCR–G-peptide interactions is limited by the requirement of high concentrations of receptors and G proteins purified to homogeneity. To circumvent this limitation, we have previously employed GPCR systematic protein affinity strength modulation (SPASM) sensors to gain insights into the GPCR–G protein interaction in live cells. GPCR SPASM sensors contain a GPCR and a G-peptide tethered through an ER/K linker flanked by FRET probes. The ER/K linker controls the stoichiometry and effective concentration of the intramolecular interaction in live cells. SPASM sensors are designed to detect changes in the affinity of interacting proteins/peptides (12, 13). GPCR SPASM sensors have been extensively utilized to monitor the interaction strength between GPCR and G-peptides and have dissected the structural elements that modulate these interactions (14–16). Using a combination of GPCR SPASM sensors and molecular dynamics (MD) simulations, we have successfully gained insights into the structural dynamics of GPCR–G-peptide interactions (14–16).
While GPCR SPASM sensors have been repeatedly used to probe the effects of full agonists (14–18), their narrow dynamic range, combined with the sensitivity of sensor measurements to live-cell expression levels, have limited reliable measurements with partial agonists. Here, we overcome these limitations by vesiculating GPCR SPASM sensors into giant plasma membrane vesicles (GPMVs) and use this system to probe into the structural and dynamic basis of agonist efficacy in GPCRs. GPMVs are large extracellular vesicles that can be generated from a variety of cell types via chemical vesiculation (19, 20). Isolated GPMVs that exhibit a size range of 0.1 to ∼15 μm are devoid of any detectable intracellular organelle signatures and are enriched in plasma membrane–associated proteins (21). GPMVs have been extensively used to study plasma membrane composition and architecture and provide a cell-free system to isolate plasma membrane–integrated proteins from the intracellular milieu. While GPMVs have been successfully utilized to study membrane proteins, such as receptors tyrosine kinases and ion channels (22, 23), their potential to investigate GPCR signaling remains unexplored.
In the current study, we demonstrate the functional incorporation of β2AR-Gαs-peptide SPASM sensors into GPMVs (v-β2AR-Spep). The v-β2AR-Spep sensors show over a twofold increase in ligand-binding capacity and a 3.5-fold increase in sensor dynamic range compared to live cells. Unlike live cells, v-β2AR-Spep readout is not sensitive to cellular expression levels and shows robust reproducibility compared to membranes (z’GPMV = 0.50 to ∼0.61 versus z’membrane = −0.22 to ∼−1.28). The v-β2AR-Spep sensors can be frozen or preserved on ice for at least 12 d without compromising sensor readout. We leveraged the combined benefits of v-β2AR-Spep over live cell and membrane assays to profile the nuanced changes in GPCR–G-peptide interactions with 16 adrenergic receptor ligands of varying efficacy. The FRET intensity changes in v-β2AR-Spep for various agonists do not correlate with classical pharmacological parameters, including ligand binding affinity, half maximal effective concentration (EC50), maximum response (Emax), and transducer ratio (τ). Instead, we find that the strength of the GPCR–G-peptide interaction shows perfect linear correlation between v-β2AR-Spep FRET sensor readout and both G protein activation and ligand molecular efficacy as reported using single molecule biophysical measurements (9). The partial agonists show reduced FRET intensity changes (ΔFRET) in v-β2AR-Spep compared to full agonists, revealing that the S-peptide interactions with partial-agonist–bound β2AR are weaker than with full-agonist–bound β2AR.
Probing the structural dynamics of the β2AR-Spep interaction using a combination of computational analysis and v-β2AR-Spep mutagenesis experiments reveals that β2AR-Spep interactions unique to an intermediate orientation of the S-peptide (11) are essential in proceeding toward the fully active orientation. Using GPMV SPASM sensors not only for β2AR but also for Gαs-coupled β1 adrenergic receptor (β1AR) and dopamine receptor D1R, we demonstrate the broader significance of the intermediate orientation for multiple Gs-coupled GPCRs. While the intermediate orientation is essential for the receptor–G-peptide interaction, ligand efficacy correlates with the strength of the interaction in the fully active orientation. Furthermore, our MD simulations demonstrate a significantly lower interaction strength and stability for the intermediate compared to the fully coupled orientation. This disparity leads to strong allosteric communication between the ligand and G-peptide–binding sites only in the presence of full agonists and the fully coupled orientation. Taken together, our study 1) establishes a cell-free technology platform to profile the molecular efficacy of GPCR ligands, 2) provides insights into the relationship between agonist efficacy and strength of GPCR–G-peptide interaction, and 3) provides a two-stage model that accentuates the importance of intermediate conformations that are necessary but not sufficient for full activation of the GPCR–G protein complex.
Results
Incorporation of Functional GPCR SPASM Sensors into GPMVs.
A prominent feature of receptor–G protein coupling is the interaction between the receptor and C-terminal α5 helix of the Gα subunit (G-peptide) (24, 25). We have previously utilized FRET-based SPASM sensors to measure the strength of the interaction between GPCRs and a native peptide encompassing the entire 27-amino-acid α5 helix of the G-peptide (16). Multiple studies have demonstrated the importance of the G-peptide in G protein selectivity and its structural role in receptor–G protein coupling (3, 6). Unlike the full G-peptide, G-peptide–based SPASM sensors are inherently independent of the heterogeneity of G protein nucleotide states in cells. We focused on the characterization of GPCR function in GPMVs with a SPASM sensor that probes the ligand-dependent interaction between the prototypical β2AR and the α5 helix of its cognate Gαs subunit (β2AR-Spep, Fig. 1A). GPMV formation in human embryonic kidney 293T (HEK293T) cells transiently expressing the β2AR sensors was induced with a buffer containing N-ethylmaleimide (NEM). GPMVs were separated from cells and debris using a two-step centrifugation protocol (Fig. 1B). Preparations of GPMVs using our optimized protocol average ∼4 μm in diameter and show sensor fluorescence localized to the membrane (Fig. 1C).
Fig. 1.
GPMVs incorporate GPCR SPASM sensors in their lipid bilayer. (A) Schematic of GPCR SPASM sensors (Right). The GPCR SPASM sensor used in this study comprises, from N to C terminus, GPCR (light gray), fluorescence acceptor (mCitrine), 10 nm ER/K linker (dark gray), fluorescence donor (mCerulean), and the C-terminal α5-helix peptide of a G-peptide (red). Protein domains are separated via a (GSG)4 linker to allow for rotational freedom between domains. Control sensors (No-pep) do not contain the C-terminal α5-helix peptide. A diagram of the sensors in equilibrium between dissociated (low FRET) and associated (high FRET) states is shown (Left). Unless stated otherwise, the GPCR utilized in the current study is the prototypical β2AR, and the C-terminal peptide is the α5-helix peptide of the cognate Gαs subunit. (B) Workflow for preparing and isolating GPMVs. (C) Representative fluorescence images of GPMVs expressing the β2AR-Spep sensors. (Scale bar, 5 µm.)
Saturation radioligand binding assays of GPMVs derived from HEK293T cells overexpressing wild-type (WT) β2AR demonstrate robust I125-(±)-cyanopindolol binding and competitive inhibition of radioligand by full agonist isoproterenol (Iso) (pKi = 5.68, SI Appendix, Fig. S1). To investigate ligand binding relative to traditional crude membrane preparations, we performed radioligand binding assays in matched membranes and GPMVs derived from the same batch of cells, transiently overexpressing β2AR-Spep. GPMVs exhibit a ∼2.5-fold increase in I125-(±)-cyanopindolol binding compared to crude membrane extracts (Fig. 2A). Furthermore, competitive inhibition of I125-(±)-cyanopindolol binding to GPMV samples via the adrenergic receptor agonist Iso displayed pKi values comparable with that of crude membrane extracts (Fig. 2 B and C). While binding affinity for the neutral antagonist I125-(±)-cyanopindolol is similar between GPMVs containing WT β2AR and β2AR-Spep, the latter shows significantly tighter binding to the full agonist Iso (∼40-fold). The Gα C terminus is known to enhance agonist binding affinity (26). We have previously reported ∼50-fold enhanced Iso binding in crude membranes for β2AR-Spep compared to a β2AR SPASM sensor lacking the G-peptide (27). Likewise, Liu et al. (11) observe a ∼45-fold enhanced Iso binding (IC50 high/IC50 low) to β2AR upon C-terminal fusion of the last 21 amino acids of the Gαs C terminus.
Fig. 2.
v-β2AR-Spep sensors are functional and responsive to agonist stimulations. (A) Binding of 125I-(±)-cyanopindolol to GPMVs or crude cell membrane extracts expressing 10 to 20 fmol β2AR-Spep sensors. (B) Competitive inhibition of 125I-(±)-cyanopindolol (Cya, 500 fM) binding to GPMVs or crude cell membrane extracts expressing 10 to 20 fmol β2AR-Spep sensors with increasing concentrations of Iso. (C) Table of binding and competitive inhibition parameters from A and B. (D) Change in ΔFRET (100 μM Iso treated, buffer control) following Iso stimulation in v-β2AR-No-pep and v-β2AR-Spep sensors. (E) Dose-dependent changes in ΔFRET in GPMVs expressing the β2AR-Spep sensor. Stimulation with increasing concentrations of Iso is shown in black. Stimulation with 100 μM Iso combined with increasing concentrations of ICI 118,551 (logIC50 = −6.18 ± 0.56) is shown in gray. Error bar denotes SEM. Data are derived from at least three independent experiments (at least three separate GPMV or membrane preparations). A Student’s t test was performed to evaluate significance between β2AR-Spep and β2AR-no-pep (−) control ΔFRET. ***P ≤ 0.001.
We next characterized sensor functionality in GPMVs relative to live cells. GPMV sensor measurements are denoted by prefix “v” (e.g., v-β2AR-Spep) to distinguish them from live cell measurements. The addition of Iso, an agonist for β2AR, stimulated an increase in FRET ratio (represented as ΔFRET) that was specific for the cognate S-peptide as compared to a no-peptide control (Fig. 2D and SI Appendix, Fig. S2). In parallel, two previously reported FRET sensors, β2AR-Gs (17) and β2AR–intracellular loop 3 (ICL3) (27) demonstrate comparable ΔFRET when vesiculated in GPMVs (SI Appendix, Fig. S3 A and B). This response was found to be dose dependent (black line in Fig. 2E) and subject to competitive inhibition via ICI 118,551, an β2AR-selective antagonist (gray line in Fig. 2E). Together, these results demonstrate the incorporation of functional GPCR SPASM sensors into GPMVs.
Characterization of Sensor Measurements in GPMVs.
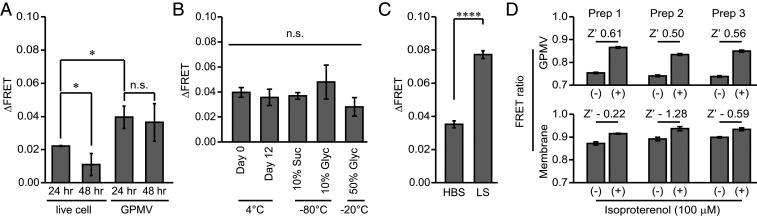
Live cell measurements require careful monitoring of expression levels and sensor localization (16, 17). Low levels of expression cause FRET spectra to be dominated by noise from scattering of the excited light by cells. Overexpression of receptors in live cells can lead to their progressive accumulation within intracellular compartments that may not be accessible to soluble extracellular ligands, resulting in the accumulation of sensors that are unresponsive to stimulation. On the contrary, GPMVs are vesiculated almost exclusively from the plasma membrane (21) and exhibit a generally uniform distribution of receptors across the vesicle lipid bilayer with little to no signs of internalization (Fig. 1C). Hence, we hypothesized that GPMVs should have a greater population of GPCRs that can access soluble ligands compared to live cells. We found that GPMVs expressing an N-terminally HA-tagged β2AR exhibit twofold greater labeling of the receptor with a fluorescently conjugated HA antibody as compared to live cells (SI Appendix, Fig. S4A). Furthermore, v-β2AR-Spep vesiculated at 24 h shows a twofold higher FRET response compared to live cell measurements (Fig. 3A). While longer expression times do enhance sensor expression in live cells (twofold increase from 24 to 48 h, SI Appendix, Fig. S4B), they result in substantial internal accumulation of sensor fluorescence with corresponding diminished sensor response (Fig. 3A). In contrast, similar levels of sensors are vesiculated into GPMVs prepared from cells expressing them for 24 and 48 h (SI Appendix, Fig. S4B). Accordingly, v-β2AR-Spep maintains its FRET response at 48 h compared to 24 h (Fig. 3A), resulting in an expression invariant twofold increase in sensor response compared to live cells.
Fig. 3.
GPMV-incorporated (v-) β2AR-Spep sensors exhibit enhanced stability, response, and reproducibility over live cells and membrane preparations. (A) Change in ΔFRET following Iso stimulation of live cells or GPMVs prepared from HEK293T cells expressing the β2AR-Spep sensor. In comparison with live cells, GPMVs show enhanced ΔFRET that does not diminish when prepared from HEK293T cells with increased expression time. (B) GPMV sensor readout (100 µM Iso stimulation) is stable over 12 d on ice or following storage at −80 °C (10% sucrose or 10% glycerol) and −20 °C (50% glycerol). (C) ΔFRET comparison between v-β2AR-Spep sensors in HBS or LS. (D) Iso-stimulated (+) and nonstimulated (−) ΔFRET measurements from three independent GPMVs and membrane preparations. Error bar denotes SEM. Data are derived from at least three independent experiments (at least three separate GPMV, membrane, or cell preparations). A Student’s t test was performed to evaluate significance between indicated conditions. *P ≤ 0.05; ****P ≤ 0.0001. n.s., nonsignificant.
Unlike purified receptor or crude membrane preparations, live cells are not suitable for long-term storage on ice or freezing conditions. However, GPMVs containing β2AR-Spep retained sensitivity to agonist stimulation for 12 d either on ice with 50% glycerol at −20 °C or following freeze–thaw at −80 °C with 10% sucrose or glycerol (Fig. 3B). Additionally, live cells require specific buffer and osmolarity to maintain cell integrity. In contrast, because GPMVs are a cell-free system, they should be more resilient to changes in buffer conditions. We report that lower salt concentrations (25 mM KCl) increased v-β2AR-Spep response 2.2-fold (Fig. 3C). Taken together, the inherently higher sensor response in GPMVs, combined with the increase in dynamic range in lower salt concentrations, provides a 3.5-fold higher response compared to live cells. To leverage this higher dynamic range, v-β2AR-Spep measurements were performed in low-salt buffer (LS) from this point onward unless otherwise noted.
GPMVs versus Crude Membrane Preparations.
While crude membrane extracts are commonly used for radioligand binding and G protein activation measurements using 35S-GTPγS, the inherent heterogeneity in the composition and architecture of these membranes is likely to impact the reproducibility of sensor measurements across distinct preparations. By comparison, GPMVs are vesiculated exclusively from the plasma membrane and generally exhibit a homogenous spherical architecture with reproducible size distributions across preparations. To facilitate a direct comparison between GPMV and membrane sensor measurements, they were both prepared from a single batch of β2AR-Spep HEK293T cells. Three such independent preparations of GPMV and membrane were used to assess the reproducibility of sensor measurements (Fig. 3D). The change in sensor response relative to the SD of the measurement was used to obtain a Z′ for both GPMV and membrane (Fig. 3D). GPMVs exhibit significantly higher statistical effect size (Z′ ≥ 0.5) when compared to crude membrane extracts (Z′ < 0). Thus, in comparison with live cells and crude membrane extracts, GPMVs provide a stable and reliable reagent for β2AR SPASM sensor measurements.
Improved Accessibility of GPMV Sensor Measurements.
To improve the accessibility of GPMV-incorporated sensors for the measurement of G protein activation, we evaluated their utility in standard fluorescence plate reader and luminescence formats (SI Appendix, Fig. S5). A modified FRET measurement protocol was used to directly measure the FRET efficiency of GPMV sensors (see Materials and Methods). Changes in FRET efficiency correlated linearly with measurements performed individually on a more sensitive fluorometer, albeit requiring a three- to fourfold higher solution concentration of GPMVs (SI Appendix, Fig. S5B). To reduce the GPMV concentration necessary for FRET measurements in a screening format, we replaced the FRET donor and acceptor fluorophores with the two subunits of a nano-luciferase complementation reporter (28). The change in luminescence intensity reported by the nano-luciferase sensor correlated linearly with the change in ΔFRET measured using a fluorometer (SI Appendix, Fig. S5C). Importantly, the nano-luciferase sensor readout requires significantly lower amounts of GPMVs compared to the fluorometer and fluorescence plate reader measurements using the FRET sensors (SI Appendix, Table S2). Furthermore, we observe enhanced statistical effect size (Z′ = 0.84) with the nano-luciferase sensor. Hence, the nano-luciferase sensor embedded in GPMVs provides an accessible and scalable technology to profile the G protein activation rates by different ligands, with potential implications for high throughput screening.
Alternative Method of GPMV Preparation.
GPMVs incorporating sensors for two other Gs-coupled receptors (v-β1AR-Spep and v-D1R-Spep) report robust FRET responses (SI Appendix, Fig. S3A). However, a previously reported Gi-coupled sensor (α2AR-Ipep) and a Gq-coupled sensor (V1AR-Qpep) did not report a FRET response in GPMVs despite a previously documented robust responses in live cells (14). The traditional method to vesiculate cells to form GPMVs utilizes NEM that covalently modifies cysteine residues in proteins (29). While our data suggest that NEM treatment does not adversely impact β2AR functionality, we sought to address concerns for other receptors and Gα C-terminal peptides that might be affected by cysteine modifications [notably, the cysteine residue in Gαi that is the primary target of pertussis-toxin treatment (30)]. Hence, we used an alternate approach that triggers vesiculation using a bicine buffer that should not directly react with cellular proteins (31). GPMVs prepared via bicine buffer maintain sensor functionality for the Gs-, Gi-, and Gq-coupled receptors (SI Appendix, Fig. S3B). These findings demonstrate that GPCR sensors incorporated into GPMVs are a versatile technology to probe GPCR function.
The IC50 values for ICI 118,551 are lower than those observed in our previous measurements using the identical FRET sensors in live cells (16). Our data suggest that ICI 118,511 binds weakly to β2AR in GPMVs. While the molecular basis for this discrepancy is unknown, the use of NEM in GPMV vesiculation could selectively compromise ligand binding. To examine the potential influence of NEM, we examined competitive inhibition of Iso (100 μM) with ICI 118,551 in bicine buffer–prepared vesicles (SI Appendix, Fig. S3C). Both methods yield similar logIC50 values (NEM—6.18 ± 0.56; bicine buffer—6.37 ± 0.54), suggesting NEM alone is not the source of the discrepancy between GPMV and cellular measurements. Additionally, ΔFRET measurements correlate linearly between v-β2AR-Spep sensor prepared via either NEM or bicine (SI Appendix, Fig. S3D). Together, these observations indicate that potential cystine modifications via NEM do not impact the relative effects of ligands on the v-β2AR-Spep sensor.
Sensor Measurements Correlate with G Protein Activation and Molecular Efficacy.
We have previously reported that β2AR SPASM sensors probe ligand and receptor-dependent G protein selectivity by measuring the strength of the interaction between the receptor and the α5-helix of distinct Gα subunits (16). However, because of a limited dynamic range of the sensor response and sensitivity to sensor expression levels in live cells (16), a reliable comparison of sensor measurements for agonists of varying efficacy was not feasible. Here, we leveraged the benefits of GPMVs outlined earlier to compare sensor measurements to known metrics of receptor pharmacology (32), including binding affinity (KD), downstream signaling (EC50 and Emax), and efficacy ratio (KD/EC50 = τ−1, SI Appendix, Eq. S1) (33). For saturating concentrations of 13 ligands, β2AR sensor readout does not show any correlation with previously reported KD, EC50, efficacy ratio, or Emax (Fig. 4 A–E) (33). In contrast with these downstream measures of ligand efficacy, β2AR sensor readout in GPMVs correlates linearly (R2 = 0.99) with previously reported G protein activation (9) using receptor and G protein purified to homogeneity (Fig. 5A and SI Appendix, Table S1). Recently, Gregorio et al. (9) used extensive single-molecule biophysical measurements of ligand-dependent changes in β2AR conformation for different kinetic states in the receptor–G protein engagement cycle to measure the ligand molecular efficacy, which denotes the effects of the ligand at the level of the receptor. In addition to G protein activation, the v-β2AR-Spep also correlates linearly with molecular efficacy (Fig. 5B) (9). Interestingly, v-β2AR-Gs displays a molecular efficacy–independent small but significant increase in FRET signal.
Fig. 4.
ΔFRET measurements from v-β2AR-Spep sensors are not representative of classic pharmacological parameters. (A) List of the adrenoreceptor ligands utilized in this study. (B–E) ΔFRET measurements of v-β2AR-Spep sensors stimulated with 13 ligands listed in A were plotted against previously documented pharmacological measurements that determine classical ligand efficacy (33). Ligand-induced ΔFRET in LS does not correlate with (B) KD (R2 = 0.01), (C) EC50 ([3H]-cAMP accumulation; R2 = 0.31), (D) efficacy ratio (KD/EC50; R2 = 0.26), or (E) Emax ([3H]-cAMP accumulation; R2 = 0.59). Error bar denotes SEM. Data are derived from at least three independent experiments (at least three separate GPMV preparations).
Fig. 5.
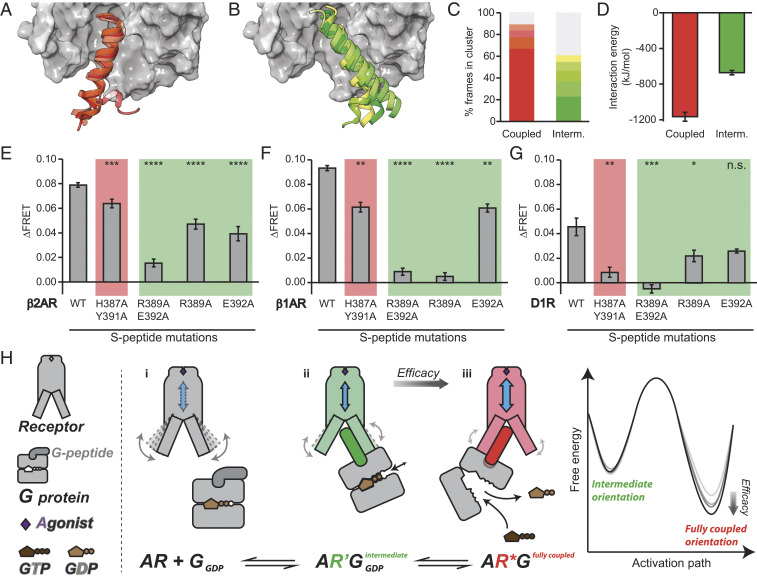
ΔFRET measurements from v-β2AR-Spep sensors correlate with molecular efficacy and β2AR-Spep interaction energy in the fully coupled orientation. (A) List of the adrenoreceptor ligands presented in B–G. (B) Change in ΔFRET for v-β2AR-Spep sensors compared to previously reported GTP turnover for purified recombinant β2AR combined with Gs (9). LS is in black (see Materials and Methods). HBS buffer is in gray (see Materials and Methods). (C) ΔFRET for v-β2AR-Spep sensors compared to previously documented molecular efficacies of indicated ligands (9). Ligand-induced ΔFRET correlates strongly with molecular efficacy and GTP turnover (R2 values indicated). (D) Fully active state structures of β2AR (Protein Data Bank [PDB]: 3SN6) (25) and AA2AR (PDB: 5G53) (34) are shown with receptor in gray surface representation (β2AR) and Gα-α5 helices shown as ribbons (residues 369 to 394 in maroon and residues 368 to 394 in pink). Hot spot residues H387, Y391, and E392 are shown as lines and R389 as balls and sticks. In the AA2AR structure, only Cα and Cβ atoms of residue R389 could be resolved and no atoms from D368. Relative rmsd between peptide orientations in two structures was 2.0 Å. (E) The intermediate state structure of β2AR and Gαs (PDB: 6E67) (11) is shown with crystallized Gαs peptide residues 381 to 394 as a green ribbon. (F and G) Intermolecular β2AR-Spep interaction energy was calculated over S-peptide residues 368 to 394 based on MD simulations of the fully coupled (F) and intermediate (G) states. MD data points were derived from the last 100 ns of five or eight replicates (as noted in SI Appendix, Table S3). ΔFRET data are derived from at least three independent experiments (at least three separate GPMV preparations). Error bar denotes SEM.
Sensor Measurements Correlate with Interaction Energy in the Fully Coupled Orientation.
Structural studies of β2AR report two distinct orientations of the C terminus peptide of the Gs protein when crystallized with the either peptide (14 amino acids) (11) or the full Gs protein (25) (Fig. 5 D and E). The first orientation, widely regarded as the fully active conformation, has been observed in both Class A GPCR–GS complex structures that have been reported to date (Fig. 5D) (25, 34). The second orientation, conjectured as an intermediate state, was identified in an intramolecularly fused β2AR-Spep chimeric construct (Fig. 5E) (11). To probe the molecular details of the β2AR-Spep interaction, we performed MD simulations starting with the S-peptide in either orientation and bound to agonists with varying efficacy. We find that average interaction energy of the S-peptide with the β2AR with different agonists bound, derived from MD simulations in the fully coupled orientation, correlated well with the ΔFRET measurements from v-β2AR-Spep sensors. (Fig. 5F, R2 = 0.87). In contrast, the average interaction energy in the intermediate orientation was similar regardless of agonist efficacy (Fig. 5G, R2 = 0.06).
Relative Stability and Interaction Energies of the Fully Coupled and Intermediate Orientations.
All-atom MD simulations of the Iso-bound β2AR-Spep complex revealed that the fully coupled orientation is significantly more stable than the intermediate orientation (Fig. 6 A–C). Clustering of the S-peptide conformations showed there are more conformation clusters in the intermediate orientation, indicating greater flexibility compared to the fully coupled orientation (Fig. 6 A–C and SI Appendix, Fig. S6). This observation is true for all of the full and partial agonists studied here (SI Appendix, Fig. S7A). The MD simulations starting from the fully coupled orientation are dominated by a single highly occupied cluster, which contained 67% of all the snapshots from MD simulations in comparison to 23% of the snapshots for the intermediate orientation (Fig. 6C). The total number of conformation clusters in the MD simulations starting from the intermediate orientation was fourfold higher than that started from the fully coupled orientation (SI Appendix, Fig. S6). The rmsd in the coordinates of the S-peptide is higher in the intermediate conformation of β2AR with both full and partial agonists (SI Appendix, Fig. S7B), and the number of sustained residue interactions of the S-peptide with the β2AR is higher in the fully active state compared to the intermediate state (SI Appendix, Fig. S7C). Collectively, these observations demonstrate that the β2AR-bound S-peptide exhibits significantly greater flexibility in the intermediate orientation compared to the fully coupled orientation. The enhanced conformational flexibility is also reflected in the near twofold-weaker β2AR-Spep interaction energy when compared to the fully coupled orientation (Fig. 6D). Using the computational method Allosteer (35), we calculated the allosteric communication pipeline from the extracellular site to the G protein coupling sites for Iso and clenbuterol in the fully coupled versus intermediate states. As seen from SI Appendix, Fig. S8, the allosteric communication is stronger in the Iso-bound fully active state compared to intermediate state (SI Appendix, Fig. S8 A and C). For the partial agonist clenbuterol, the allosteric communication is more dispersed in the fully active state and is weaker in the intermediate state. Taken together, these results show that the weakening of the allosteric communication to the G protein coupling site in the intermediate state for any agonist leads to no correlation of the S-peptide interaction energy to the ligand efficacy.
Fig. 6.
The β2AR/Gαs interaction in the intermediate orientation is weak but crucial. (A and B) The entire 4 μs MD ensembles of the fully coupled (A) and intermediate (B) orientations bound to Iso were aligned on the transmembrane helix backbone and rmsd clustered based on the backbone atoms of residues 377 to 389 (1.5 Å cutoff). The centroid of S-peptide clusters with greater than 5% occupancy are shown as ribbons with β2AR shown as surface representation in gray. Displayed residues are 372 to 394; unstructured residues 368 to 371 were removed for clarity. (C) Relative population of clusters (number of frames in each cluster divided by the total number of simulated frames, 50,000). The topmost populated clusters with greater than 5% occupancy are colored in red (fully coupled) or green (intermediate), following the color scheme in A and B. The remaining clusters are colored in gray. (D) Intermolecular β2AR-Spep interaction energy was calculated over S-peptide residues 368 to 394 based on MD. Iso was the bound ligand for both fully coupled and intermediate datasets. Each data point was derived from the last 100 ns of eight replicates. (E–G) Change in ΔFRET following Iso stimulation in GPMVs expressing β2AR (E), β1AR (F), and D1R (G) S-peptide sensors of WT or mutants previously reported from crystal structures as unique for the fully coupled (3SN6, red background) or intermediate (6E67, green background) orientations. (H) A schematic of the proposed model for the engagement and activation of G proteins by an agonist-stimulated GPCR. (i) Agonist binding enhances receptor conformational dynamics; (ii) G protein initially engages receptor with G-peptide in an intermediate orientation (green G-peptide) that in turn alters the receptor conformational landscape (highlighted in green); and (iii) G protein engages receptor with G-peptide in a fully coupled orientation (red G-peptide) with strong allosteric coupling with the agonist-binding site (blue arrow denotes allosteric coupling; receptor conformation is highlighted in red). Free energy diagram depicts ligand molecular efficacy being tuned by the strength and stability of the interaction in the fully coupled orientation. Error bar denotes SEM. Data are derived from at least three independent experiments (at least three separate GPMV preparations). A Student’s t test was performed to evaluate significance between WT and mutant conditions. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.000. n.s., nonsignificant.
A Single Hot Spot Selective for the Intermediate Orientation Is Essential for β2AR-Spep Interaction.
The comparison of crystal structures displaying the two orientations in β2AR previously suggested that Gαs residues R389 and E392 are sites of receptor–peptide interaction unique to the intermediate orientation (11). Furthermore, the mutation of R389 and E392 together (R389A/E392A) was reported to have a negative impact on G protein activation, as measured by TM6 displacement, GDP release, and Iso affinity (11). Hence, we pursued mutagenesis of R389 and E392 to examine whether this intermediate orientation is sampled in v-β2AR-Spep sensor measurements. The R389A/E392A double mutant reduced FRET measurements by 80% (Fig. 6E). However, an examination of MD simulations revealed that while R389 contacts β2AR exclusively in the intermediate orientation, E392 additionally contributes to the β2AR-Spep interaction in the fully coupled orientation. To avoid overinterpretation of a potential dual role of E392, we next examined the effects of single mutations at R389 and E392 (R389A and E392A, Fig. 6E). R389A and E392A mutants decreased measured ΔFRET by 40% and 50% relative to WT control. Thus, despite the instability and weaker binding in the intermediate orientation, selectively destabilizing this orientation substantially diminished the overall β2AR-Spep interaction. In contrast, mutagenesis of two interaction sites reported as unique to the fully active conformation (H387A/Y391A double mutant) (11) only reduced sensor ΔFRET by 19% relative to WT. While these interaction sites are unique to the fully active conformations in both the crystal structure and MD simulations, it must be noted that the S-peptide in this orientation makes numerous contacts with β2AR (25). Hence, despite a modest effect of the H387A/E392A double mutant, our site-directed mutagenesis results do not yield definitive insights into the contribution of the fully active conformation. Introduction of additional mutations is complicated by the potential disruption of secondary structural contacts that could impact both orientations. To expand on the significance of the intermediate orientation interaction hot spot R389, we examined the effects of these S-peptide mutations on the β1 adrenergic receptor (v-β1AR-Spep) and the dopamine 1 receptor (v-D1R-Spep). Both the R389A/E392A double mutation and R389A single mutation induced significant decrease in ΔFRET readout relative to WT, attesting to the broader significance of these interaction hot spots in the ensemble receptor–S-peptide interaction.
Discussion
GPCR signaling involves the serial amplification of ligand–receptor interactions to downstream second messenger responses (1). High-efficacy ligands can saturate the amplification cascade, leading to nonlinear downstream responses that do not correlate with receptor-level effects. Hence, classical pharmacological models have incorporated an “intrinsic efficacy” to capture system-independent effects of GPCR ligands (36, 37). Nonetheless, intrinsic efficacy has remained elusive in the absence of quantitative receptor-level information. Recently, an extensive single molecule biophysical study has characterized a ligand “molecular efficacy” parameter that captures the influence of ligand-dependent receptor conformation on G protein activation. The structural translation of ligand molecular efficacy into G protein activation remains unclear and forms the focus of this study. To bridge this knowledge gap, we first establish a robust, accessible, and sensitive assay to probe GPCR interaction with G protein and the Gα C terminus (G-peptide), an established structural determinant of G protein selectivity. We circumvent the need for extensive purification protocols by the single-step incorporation of receptor and G protein elements into GPMVs. We use previously established SPASM sensors to control the stoichiometry and effective concentration of receptor–G protein interactions. Taken together, GPMV-incorporated SPASM sensors (v-SPASM sensors) provide enhanced dynamic range, expression-insensitive readout, and a reagent-level assay to profile ligand effects at the level of the receptor. Leveraging this technology, we find that receptor–G-peptide and not receptor–G protein interactions for the prototypical β2AR show a near perfect correlation with ligand molecular efficacy. Thus, we establish the receptor–G-peptide interaction as a sufficient structural determinant of ligand molecular efficacy. The Gs-peptide (S-peptide) is known to interact with β2AR in two distinct orientations, previously termed intermediate and fully coupled (11, 25). MD simulations show that β2AR-Spep interaction energies in the fully coupled but not intermediate orientations show near perfect correlation with sensor measurements, refining interactions in the fully coupled orientations as the structural basis for molecular efficacy. MD simulations further demonstrate that unlike the fully coupled orientation, the intermediate orientation is a weak, unstable interaction that does promote allosteric coupling with the ligand-binding site. Paradoxically, we find that destabilization of a single intermediate orientation “hot spot” residue is sufficient to substantially disrupt the GPCR–G-peptide interaction in three distinct Gs-coupled receptors (β2AR, β1AR, and D1R). These observations suggest a two-stage receptor activation mechanism, wherein interactions in the intermediate orientation alter the receptor conformational landscape to facilitate engagement of the fully coupled orientation and G protein activation.
Downstream activity in GPCR signaling is quantified via classical pharmacological parameters, including EC50, Emax, and efficacy ratio (KD/EC50) (33). The inverse of efficacy ratio, EC50/KD, is a measure of the fractional receptor occupancy necessary to drive half the maximal downstream response (Emax for full agonist) (SI Appendix, Eq. S1). The majority of adrenergic ligands utilized in this study have a high efficacy ratio (KD/EC50 > 10, Fig. 4D) and consequently yield near saturating downstream responses comparable to the full agonist Iso (Fig. 4E). Therefore, the Emax of downstream responses for high-efficacy ligands does not differentiate between the underlying rates of G protein activation. EC50, and consequently efficacy ratio measurements, derive from second messenger responses that encapsulate ligand-dependent effects on multiple pathways that converge on a downstream effector, such as adenylyl cyclase. Consequently, ligand effects at the level of the receptor do not necessarily correlate with classical pharmacological parameters. In contrast, GTP turnover and molecular efficacy measurements are typically derived from cell-free, highly purified systems (9). Therein, the observed GTP turnover and molecular efficacy are a direct consequence of the biochemical and biophysical processes occurring at the level of the GPCR–G protein complex and are not convolved by nonlinear signaling amplifications after G protein activation. Likewise, the v-β2AR-Spep sensors derive their FRET measurements from the direct interaction between β2AR and S-peptide in an equilibrated environment. Accordingly, v-β2AR-Spep sensor measurements show near perfect correlation with previously reported measurements of G protein activation and molecular efficacy (Fig. 5 B and C) rather than classical pharmacological parameters (Fig. 4 A–E). Taken together with the enhanced sensor dynamic range compared to live cell measurements, extended shelf life and stability of readout (Fig. 3), and utility in standard plate reader fluorescence/luminescence formats (SI Appendix, Fig. S5), our data support v-SPASM sensors as an accessible technology to characterize ligand efficacy at the level of the receptor.
The v-SPASM sensors provide insights into the structural mechanisms that dictate molecular efficacy. Ligand molecular efficacy for GPCRs was first quantified by single-molecule kinetics of β2AR conformation (9). Extensive biophysical measurements in this study revealed that molecular efficacy is determined by the rate of an inactive to active conformational transition in the GPCR coupled with a diminished G protein affinity for GDP, indicative of its activation. However, ligand molecular efficacy did not substantially impact the kinetics of the GPCR–G protein interaction, which were limited by an efficacy-independent G protein on-rate and efficacy-independent G protein off-rate in either GDP- or GTP-bound states. These findings suggest that sensors that probe a ligand-dependent GPCR–G protein association are less likely to differentiate between varying efficacy agonists. Accordingly, we find that v-β2AR-Gs sensors report a subtle but ligand efficacy–invariant increase in interaction strength (SI Appendix, Fig. S9). In contrast, our v-β2AR-Spep sensor measurements show near perfect correlation with molecular efficacy (Fig. 5C). The G-peptide is a well-established determinant of GPCR–G protein coupling specificity and a prominent structural element in all reported GPCR–G protein structures (3, 6, 7, 25, 34, 38–40). Hence, our findings suggest that ligand molecular efficacy is sufficiently determined by the strength of the β2AR-Spep interaction. Interactions with the G-peptide in the fully coupled orientation are, in turn, relayed through the G protein core-to-trigger nucleotide exchange and consequent G protein activation (5).
It has been demonstrated via two different crystal structures that β2AR can engage the S-peptide in distinct intermediate or fully coupled orientations (11, 25). Our SPASM sensors with the free G-peptide are designed to probe the ensemble of interaction states between GPCR and the peptide. Accordingly, the dynamics of the β2AR-Spep engagement in distinct orientations is veiled within our ensemble FRET measurements. Hence, MD simulations were employed to gain structural insights into the two S-peptide orientations. In simulations with a high-efficacy full agonist, the G-peptide interaction in the fully coupled orientation demonstrates tight binding characterized by a number of intermolecular contacts that yield a relatively high binding energy and limit conformational flexibility (Fig. 6 A–D and SI Appendix, Figs. S6 and S7). Additionally, the stability of the G-peptide in the fully coupled orientation is reinforced by strong allosteric communication with the ligand-binding site (SI Appendix, Fig. S8). Reducing ligand efficacy weakens the G-peptide interaction in the fully coupled orientation and dampens the allosteric communication between ligand and G-peptide–binding sites (Fig. 5F and SI Appendix, Fig. S8). Importantly, the interface binding energy displayed a linear correlation with v-β2AR-Spep sensor measurements, G protein activation rates, and molecular efficacy (Fig. 5). In contrast, agonists were unable to sustain a stable interaction between receptor and G-peptide in the intermediate orientation, as witnessed by peptide conformational flexibility (Fig. 6 B and C and SI Appendix, Fig. S7) and lower interaction energies that are invariant with ligand efficacy (Fig. 5G). Taken together, these observations suggest that high efficacy agonists form a stable ternary complex only when the G-peptide interacts in the fully coupled orientation. In turn, the strength of the interaction in the fully coupled orientation determines G protein activation and, consequently, the efficacy of signal transduction at the level of the receptor.
With the discovery of the intermediate orientation, it has been proposed that β2AR must initially engage the Gs protein in the intermediate orientation, as key residues involved in the fully coupled interaction are embedded within the G protein (11). However, our v-β2AR-Spep sensors specifically utilize the isolated G-peptide, wherein all residues are solvent exposed. Thus, the β2AR in our sensors has consistent access to hot spot residues required for both the intermediate and fully coupled interactions with the S-peptide. Nonetheless, MD simulations suggest that the intermediate orientation is unlikely to be populated in the context of the S-peptide in v-β2AR-Spep sensors given that receptor interactions with the G-peptide in the intermediate orientation are significantly weaker and unstable compared to the fully coupled orientation. Interestingly, we observed that mutagenesis of an intermediate orientation–selective hot spot (R389 in Gαs) is sufficient to significantly disrupt the receptor–G-peptide interaction, leading to a substantial (40%) decline in our v-β2AR-Spep measurements (Fig. 6E). These contrasting observations are consistent with a two-stage interaction of β2AR with the S-peptide, with an initial engagement in the intermediate orientation. Unlike the previously proposed mechanism (11), which allocates the intermediate orientation as an obligatory prerequisite due to embedment of fully coupled orientation residues within the G protein, our findings suggest that regardless of the accessibility of key residues involved in the two orientations, the intermediate orientation is a transient prerequisite to the formation of a stable fully coupled state. The requirement of the intermediate orientation in G protein coupling is also evident in previously reported effects on ligand binding. C-terminal fusion of the last 21 amino acids of Gαs to β2AR enhanced agonist binding affinity, consistent with ternary complex formation (9). Mutagenesis of a single intermediate orientation hot spot in the fused Gs peptide (R389A) significantly weakened agonist affinity, further attesting to the role of the intermediate orientation in the formation of the fully coupled state (9). The structural mechanisms that position the intermediate orientation as a prerequisite for the fully coupled state in the isolated G-peptide remain unclear. We have previously shown that the receptor occupies a dynamic conformational landscape, wherein transient, latent G protein binding cavities facilitate interactions with distinct G proteins (15). While these previously modeled cavities were observed using the fully coupled orientation as the template, and thus are distinct from the intermediate orientation, our observations demonstrate that insertion of either the cognate or noncognate G-peptide into receptor intracellular cavities imposes a dynamic conformational heterogeneity onto the receptors (11, 15, 25). Furthermore, previous studies have clearly established that receptor–G protein interactions can influence receptor conformation (41, 42). Hence, we speculate that interactions in the intermediate orientation not only prime the G protein for activation (11) but also influence the receptor conformational landscape, priming the receptor for the fully coupled orientation.
In conclusion, we propose a two-stage receptor activation mechanism that advances a structural mechanism for ligand molecular efficacy for G protein activation (Fig. 6H and SI Appendix, Fig. S10). Regardless of ligand efficacy, the receptor initially engages the GDP-bound G protein in the intermediate orientation. Receptor binding to the G protein releases the Gα C terminus to expose key residues required for the formation of the fully coupled state (11). In parallel, engagement of the Gα C terminus in the intermediate orientation alters the receptor conformational landscape (9, 15, 27, 41–43), priming an interaction with the G protein in the fully coupled orientation. The transition of the G protein to fully coupled orientation enhances allosteric communication with the ligand-binding site and significantly enhances nucleotide exchange, stimulating G protein activation. Consequently, ligand molecular efficacy is tuned by the strength and stability of the interaction in the fully coupled orientation (Fig. 5 C and F). These findings are in agreement with a recent study on the Neurotensin receptor 1-Gi1 complex, wherein a transition from an intermediate to fully active state is proposed to enhance nucleotide exchange and, consequently, G protein activation (39). Taken together, our mechanistic insights reveal structural roles for the distinct intermediate and fully coupled orientations in the GPCR–G protein activation cycle.
Materials and Methods
More detailed descriptions of all methods utilized in the current study are available in SI Appendix, Supplementary Methods.
Experimental Methods.
Reagents.
(−)-Iso (+)-Bitartrate salt, clenbuterol hydrochloride, ICI 118,551 hydrochloride, fenoterol hydrobromide, metaproterenol hydrosulfide, (−)-norepinephrine, R(−)-denopamine, ritodrine hydrochloride, bamethane sulfate, terbutaline hemisulfate, and dobutamine hydrochloride were purchased from Sigma Aldrich. Epinephrine hydrochloride, carazolol, and formoterol hemifumarate were purchased from Cayman Chemical. Salbutamol hemisulfate and alprenolol hydrochloride were purchased from Tocris.
Molecular cloning.
The construction of β2AR-Spep has been previously described (16). All constructs were cloned into the PCDNA5/FRT vector (Thermo Fisher). FRET-based GPCR SPASM sensors comprise, from N to C terminus, GPCR, fluorescence acceptor (mCitrine), 10 nm ER/K linker, fluorescence donor (mCerulean), and the last 27 C-terminal residues of a WT or mutant G-peptide, each cloned between unique restriction sites. Protein domains are separated via a (GSG)4 linker to secure rotational freedom between domains. The luciferase versions of our sensors substitute fluorescent proteins with the large and small fractions of the split nano-luciferase protein. The intramolecular β2AR-ICL3 FRET sensor was engineered based on previous designs (44, 45).
Cell culture.
HEK293T-Flp-In (hereafter HEK293T) cells were cultured in Dulbecco's Modified Eagle medium (DMEM) (Thermo Fisher) containing 4.5 g/L d-glucose, with 10% fetal bovine serum (FBS) (Millipore) (vol/vol), and 1% l-glutamine, 20 mM Hepes, pH 7.5. Cells were maintained in a humidified atmosphere containing 5% CO2 at 37 °C. Transfected cells were grown to at least 95% confluence prior to harvesting for GPMV induction and isolation.
Transfection.
Constructs described in this study were transiently transfected in HEK293T cells via an optimized protocol utilizing polyethylenimine (PEI, linear; molecular weight (MW), 25,000; PolySciences). A transfection mixture containing 40 µg sensor DNA and 120 µg PEI mixed together in 1 mL Opti-minimal essential medium media (Thermo Fisher) was prepared and incubated for 30 min at 25 °C. The media containing the transfection reagents were exchanged with 20 mL fresh medium after 4 h of incubation at 37 °C.
GPMV preparation and purification.
Transfected HEK293T cells were harvested after 24 to 48 h of expression time and spun down (300 × g, 5 min), and the cell pellet was resuspended in 10 mL GPMV buffer twice. The GPMV buffer contains 10 mM Hepes pH 7.5, 150 mM NaCl, and 2 mM CaCl2. A total of 2 mM NEM was supplemented after the second resuspension step, prior to incubation at 28 °C. After 2 h of incubation at 28 °C, the samples underwent three centrifugation steps to pellet and discard cells and debris. The final supernatant was then centrifuged (3,220 × g, 40 min) and resuspended twice to pellet the GPMVs and remove residual NEM, which were resuspended in 2 mL buffer of choice.
FRET experiment and data collection.
Experiments were carried out in Hepes-buffered saline (HBS) buffer (20 mM Hepes pH 7.4, 5 mM KCl, 145 mM NaCl, 2 mM CaCl2, and 1 mM MgCl2) or low-salt ΔFRET buffer (20 mM Hepes pH 7.4, 25 mM KCl, and 5 mM MgCl2). Ligands were prepared in the same buffers as the GPCR SPASM sensors, supplemented with 1 mM ascorbic acid. GPCR SPASM sensors were treated with the indicated ligands (100 μM unless stated otherwise) and incubated for 5 min. The emission spectra of the ligand-stimulated GPCR sensors were read on the fluorimeter (excitation 430 nm, bandpass 8 nm, emission 450 to 600 nm, and bandpass 4 nm) or the plate reader (excitation 435 and 485 nm, emission 500 to 550 nm, and slit width 10 nm). GPCR sensors treated with buffer supplemented with 1 mM ascorbic acid were used as the baseline for which the changes in ΔFRET or FRET efficiency were measured. The ΔFRET was calculated as 525 nm emission/475 nm emission.
Membrane preparation.
Crude membranes were prepared as described in detail previously (27). Briefly, HEK293T cells expressing β2AR-Spep SPASM sensors were treated with a hypotonic buffer and mechanically disrupted in the presence of protease inhibitors, and crude membranes were separated from intact cells, nuclei, and debris by a low-speed spin (1,000 × g, 2 min, 4 °C). Membranes were harvested by ultracentrifugation at 135,000 × g, 25 min, 4 °C. Harvested membranes were washed in buffer (20 mM Hepes, pH 7.4, 0.5 mM EDTA, 50 mM NaCl, 10 mM MgCl2, and 1 mM DTT) and resuspended for storage in the same buffer, containing 12% sucrose (weight/volume).
Radiolabeled antagonist binding assay.
GPMVs and membranes containing expressed β2AR-Spep sensors (7 × 104 mCerulean cps, ∼2 nM) were resuspended in HBS supplemented with 10 mg/mL bovine serum albumin and 1 mM ascorbic acid and sonicated briefly. Increasing concentrations of (±)-[125I] iodocyanopindolol (0 to 800 pM) (PerkinElmer, catalog number NEX189) were added to each sample. Nonspecific (±)-[125I] iodocyanopindolol binding was assessed by repeating the same assay conditions listed above in the presence of 1 mM alprenolol (Tocris, catalog number 2806). Reactions were equilibrated on ice for 90 min. Equilibrated reactions were passed through PEI-treated GF/C filters (GE Healthcare, catalog number 1822) via vacuum manifold and washed three times with 5 mL ice-cold Tris-buffered saline (50 mM Tris pH 7.4, 150 mM NaCl). Filters were air dried, and filter-bound [125I] was measured by automatic gamma counting (PerkinElmer, Wallac Wizard-2 2470).
Competition binding assay.
GPMV and membranes containing expressed β2AR-Spep sensors (3 × 105 mCerulean cps, ∼10 nM) were resuspended following the same procedure as the radiolabeled antagonist binding assay. Saturating concentrations of (±)-[125I] iodocyanopindolol (500 pM) and increasing concentrations of Iso were added to the resuspended membrane samples. Nonspecific (±)-[125I] iodocyanopindolol binding was assessed by measuring the signal from 500 pM or 1 nM (±)-[125I] iodocyanopindolol in the presence of 10 mM alprenolol. Maximum (±)-[125I] iodocyanopindolol signal was assessed using 500 pM or 1nM (±)-[125I] iodocyanopindolol without competing unlabeled ligand. Reactions were equilibrated, washed, and measured following the same procedure listed above for the radioligand antagonist binding assay.
Fluorescence imaging.
Samples were prepared via flowing in GPMVs expressing the β2AR-spep sensor into a flow chamber constructed using a glass slide, a nitrocellulose coated 22 × 22 glass coverslip, and two strips of double-stick tape. The flow chamber containing the GPMVs were incubated at 4 °C for 24 h with the coverslip facing downward to allow GPMVs to sediment near the coverslip surface. All images were acquired on a Nikon A1Rsi laser scanning confocal microscope (University Imaging Center). Images were acquired using a 60× oil immersion objective (Nikon). Images were prepared using Fiji (46).
Computational Methods.
Modeling of fully active β2AR-Spep-agonist complex structures.
SI Appendix, Table S3 summarizes the starting structures and other modeling conditions. The procedure we used for generating the starting structure of the fully active β2AR-Spep complex has been described previously (14).
Modeling of the intermediate state of β2AR-Spep-Iso structure.
The crystal structure of β2AR fused to a short C-terminal peptide of Gαs (residues 381 to 394) is posited to be an intermediate state of β2AR bound to Gαs that precedes the fully active receptor state (11). The crystal construct contained four thermostabilizing mutations and two mutations to form a disulfide bond between the Gαs (381 to 394) (L394C) and β2AR (A226C). To make the 6E67 model for this study as close as possible to WT β2AR and to our fully active state model, all six mutations were reverted to WT, the disulfide bond was removed, and the β2AR-Spep fusion protein was separated into individual chains (Maestro Version 11.1.012, Release 2017-1, Schrödinger, LLC). Disulfide bonds found in the fully active state (C106–C191 and C184–C190) were included in the model.
β2AR-GαSpep interaction energy calculation.
The interaction energy between β2AR and S-peptide was calculated using the Gromacs “energy” program taken from the total nonbond energy from short-range (i.e., within 12 Å) van der Waals and Coulombic forces.
Contact persistence.
Contacts between β2AR and S-peptide were determined using the online server Get Contacts (47) based on the full 4 μs trajectory. Calculated contacts included hydrogen bonds, salt bridges, pi-cation, pi-stacking, T-stacking, hydrophobic, and van der Waals, as defined on the Get Contacts home page. A persistent contact was defined as a β2AR residue (sidechain or backbone) that maintained an interaction with an S-peptide residue for greater than 40% of the entire simulation.
Supplementary Material
Acknowledgments
This research was funded by the NIH (R35-GM126940 to S.S. and R01-GM117923 to N.V. and S.S.) and American Heart Association (18PRE34080117 to K.K.). We thank Daniel Hilger for sharing published data on GTP turnover.
Footnotes
Competing interest statement: The University of Minnesota currently has a published patent application on this technology (US Patent application number 16/769,852; Inventors: K.K. and S.S.).
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2017201118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Pierce K. L., Premont R. T., Lefkowitz R. J., Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 3, 639–650 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Hauser A. S., Attwood M. M., Rask-Andersen M., Schiöth H. B., Gloriam D. E., Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 16, 829–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamm H. E., et al., Site of G protein binding to rhodopsin mapped with synthetic peptides from the alpha subunit. Science 241, 832–835 (1988). [DOI] [PubMed] [Google Scholar]
- 4.Oldham W. M., Hamm H. E., Heterotrimeric G protein activation by G-protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 9, 60–71 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Kaya A. I., et al., A conserved phenylalanine as a relay between the α5 helix and the GDP binding region of heterotrimeric Gi protein α subunit. J. Biol. Chem. 289, 24475–24487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conklin B. R., Farfel Z., Lustig K. D., Julius D., Bourne H. R., Substitution of three amino acids switches receptor specificity of Gq alpha to that of Gi alpha. Nature 363, 274–276 (1993). [DOI] [PubMed] [Google Scholar]
- 7.Conklin B. R., et al., Carboxyl-terminal mutations of Gq alpha and Gs alpha that alter the fidelity of receptor activation. Mol. Pharmacol. 50, 885–890 (1996). [PubMed] [Google Scholar]
- 8.Nygaard R., et al., The dynamic process of β(2)-adrenergic receptor activation. Cell 152, 532–542 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gregorio G. G., et al., Single-molecule analysis of ligand efficacy in β2AR-G-protein activation. Nature 547, 68–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du Y., et al., Assembly of a GPCR-G protein complex. Cell 177, 1232–1242.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X., et al., Structural insights into the process of GPCR-G protein complex formation. Cell 177, 1243–1251.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sivaramakrishnan S., Spudich J. A., Systematic control of protein interaction using a modular ER/K α-helix linker. Proc. Natl. Acad. Sci. U.S.A. 108, 20467–20472 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swanson C. J., Sivaramakrishnan S., Harnessing the unique structural properties of isolated α-helices. J. Biol. Chem. 289, 25460–25467 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Semack A., Sandhu M., Malik R. U., Vaidehi N., Sivaramakrishnan S., Structural elements in the Gαs and Gβq C termini that mediate selective G protein-coupled receptor (GPCR) signaling. J. Biol. Chem. 291, 17929–17940 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandhu M., et al., Conformational plasticity of the intracellular cavity of GPCR-G-protein complexes leads to G-protein promiscuity and selectivity. Proc. Natl. Acad. Sci. U.S.A. 116, 11956–11965 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malik R. U., et al., Detection of G protein-selective G protein-coupled receptor (GPCR) conformations in live cells. J. Biol. Chem. 288, 17167–17178 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malik R. U., Dysthe M., Ritt M., Sunahara R. K., Sivaramakrishnan S., ER/K linked GPCR-G protein fusions systematically modulate second messenger response in cells. Sci. Rep. 7, 7749 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Touma A. M., Malik R. U., Gupte T., Sivaramakrishnan S., Allosteric modulation of adenosine A1 and cannabinoid 1 receptor signaling by G-peptides. Pharmacol. Res. Perspect. 8, e00673 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levental K. R., Levental I., Giant plasma membrane vesicles: Models for understanding membrane organization. Curr. Top. Membr. 75, 25–57 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Scott R. E., Plasma membrane vesiculation: A new technique for isolation of plasma membranes. Science 194, 743–745 (1976). [DOI] [PubMed] [Google Scholar]
- 21.Scott R. E., Perkins R. G., Zschunke M. A., Hoerl B. J., Maercklein P. B., Plasma membrane vesiculation in 3T3 and SV3T3 cells. I. Morphological and biochemical characterization. J. Cell. Sci. 35, 229–243 (1979). [DOI] [PubMed] [Google Scholar]
- 22.Sarabipour S., Hristova K., Effect of the achondroplasia mutation on FGFR3 dimerization and FGFR3 structural response to fgf1 and fgf2: A quantitative FRET study in osmotically derived plasma membrane vesicles. Biochim. Biophys. Acta 1858, 1436–1442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bricogne C., et al., TMEM16F activation by Ca2+ triggers plasma membrane expansion and directs PD-1 trafficking. Sci. Rep. 9, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung K. Y., et al., Conformational changes in the G protein Gs induced by the β2 adrenergic receptor. Nature 477, 611–615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasmussen S. G. F., et al., Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasenick M. M., Watanabe M., Lazarevic M. B., Hatta S., Hamm H. E., Synthetic peptides as probes for G protein function. Carboxyl-terminal G α s peptides mimic Gs and evoke high affinity agonist binding to β-adrenergic receptors. J. Biol. Chem. 269, 21519–21525 (1994). [PubMed] [Google Scholar]
- 27.Gupte T. M., Malik R. U., Sommese R. F., Ritt M., Sivaramakrishnan S., Priming GPCR signaling through the synergistic effect of two G proteins. Proc. Natl. Acad. Sci. U.S.A. 114, 3756–3761 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dixon A. S., et al., NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 11, 400–408 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Sezgin E., et al., Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nat. Protoc. 7, 1042–1051 (2012). [DOI] [PubMed] [Google Scholar]
- 30.West R. E. Jr, Moss J., Vaughan M., Liu T., Liu T. Y., Pertussis toxin-catalyzed ADP-ribosylation of transducin. Cysteine 347 is the ADP-ribose acceptor site. J. Biol. Chem. 260, 14428–14430 (1985). [PubMed] [Google Scholar]
- 31.Del Piccolo N., Placone J., He L., Agudelo S. C., Hristova K., Production of plasma membrane vesicles with chloride salts and their utility as a cell membrane mimetic for biophysical characterization of membrane protein interactions. Anal. Chem. 84, 8650–8655 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neubig R. R., Spedding M., Kenakin T., Christopoulos A.; International Union of Pharmacology Committee on Receptor Nomenclature and Drug Classification , International union of pharmacology committee on receptor nomenclature and drug classification. XXXVIII. Update on terms and symbols in quantitative pharmacology. Pharmacol. Rev. 55, 597–606 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Baker J. G., The selectivity of β-adrenoceptor agonists at human β1-, β2- and β3-adrenoceptors. Br. J. Pharmacol. 160, 1048–1061 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carpenter B., Nehmé R., Warne T., Leslie A. G. W., Tate C. G., Erratum: Structure of the adenosine A2A receptor bound to an engineered G protein. Nature 538, 542 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Bhattacharya S., Vaidehi N., Differences in allosteric communication pipelines in the inactive and active states of a GPCR. Biophys. J. 107, 422–434 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephenson R. P., A modification of receptor theory. Br. J. Pharmacol. Chemother. 11, 379–393 (1956). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenakin T. P., A Pharmacology Primer: Techniques for More Effective and Strategic Drug Discovery (Elsevier Inc., ed. 4, 2014). [Google Scholar]
- 38.Koehl A., et al., Structure of the µ-opioid receptor-Gi protein complex. Nature 558, 547–552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato H. E., et al., Conformational transitions of a neurotensin receptor 1-Gi1 complex. Nature 572, 80–85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim K., et al., Structure of a hallucinogen-activated Gq-coupled 5-HT2A serotonin receptor. Cell 182, 1574–1588.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao X. J., et al., The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc. Natl. Acad. Sci. U.S.A. 106, 9501–9506 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee S., Nivedha A. K., Tate C. G., Vaidehi N., Dynamic role of the G protein in stabilizing the active state of the adenosine A2A receptor. Structure 27, 703–712.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupte T. M., Ritt M., Dysthe M., Malik R. U., Sivaramakrishnan S., Minute-scale persistence of a GPCR conformation state triggered by non-cognate G protein interactions primes signaling. Nat. Commun. 10, 4836 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakanishi J., Takarada T., Yunoki S., Kikuchi Y., Maeda M., FRET-based monitoring of conformational change of the beta2 adrenergic receptor in living cells. Biochem. Biophys. Res. Commun. 343, 1191–1196 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Reiner S., Ambrosio M., Hoffmann C., Lohse M. J., Differential signaling of the endogenous agonists at the β2-adrenergic receptor. J. Biol. Chem. 285, 36188–36198 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schindelin J., et al., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venkatakrishnan A. J., et al., Uncovering patterns of atomic interactions in static and dynamic structures of proteins. bioRxiv [Preprint] (2019). 10.1101/840694 (13 November 2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.