Significance
Multisubunit motor nucleases such as RecBCD, AddAB, and AdnAB recognize, unwind, and resect DSB ends during homologous recombination in bacteria. Guided by recent cryo-EM structures of AdnAB bound to a forked duplex DNA, we conducted a thorough mutational analysis of the interface of the AdnB motor domain with the 3′ ssDNA tracking strand and the ssDNA–dsDNA junction. We thereby identified the tracking strand DNA contacts that, when severed, either completely decouple ATP hydrolysis from mechanical work or else markedly lower the efficiency of the helicase motor and hence its velocity during DSB resection. An analogy is drawn between the mechanism of chemomechanical coupling in AdnAB and that of an automobile clutch friction plate.
Keywords: helicase mechanism, DNA repair, Mycobacterium
Abstract
Mycobacterial AdnAB is a heterodimeric helicase–nuclease that initiates homologous recombination by resecting DNA double-strand breaks (DSBs). The N-terminal motor domain of the AdnB subunit hydrolyzes ATP to drive rapid and processive 3′ to 5′ translocation of AdnAB on the tracking DNA strand. ATP hydrolysis is mechanically productive when oscillating protein domain motions synchronized with the ATPase cycle propel the DNA tracking strand forward by a single-nucleotide step, in what is thought to entail a pawl-and-ratchet–like fashion. By gauging the effects of alanine mutations of the 16 amino acids at the AdnB–DNA interface on DNA-dependent ATP hydrolysis, DNA translocation, and DSB resection in ensemble and single-molecule assays, we gained key insights into which DNA contacts couple ATP hydrolysis to motor activity. The results implicate AdnB Trp325, which intercalates into the tracking strand and stacks on a nucleobase, as the singular essential constituent of the ratchet pawl, without which ATP hydrolysis on ssDNA is mechanically futile. Loss of Thr663 and Thr118 contacts with tracking strand phosphates and of His665 with a nucleobase drastically slows the AdnAB motor during DSB resection. Our findings for AdnAB prompt us to analogize its mechanism to that of an automobile clutch.
Multisubunit motor nucleases recognize, unwind, and resect DNA double-strand break (DSB) ends during homologous recombination in bacteria. Heterotrimeric Escherichia coli RecBCD, heterodimeric Bacillus subtilis AddAB, and heterodimeric Mycobacterium smegmatis AdnAB exemplify three distinct clades of end resection machines (1–15). AdnAB is thought to represent an ancestral state of the machine (11, 15). The mycobacterial AdnA and AdnB subunits are encoded in a two-gene operon that is transcriptionally up-regulated in response to DNA damage (16–18). AdnA and AdnB are paralogs composed of an N-terminal helicase motor domain and a C-terminal nuclease domain.
Ensemble biochemistry, mutagenesis, cryo-EM structures, and single-molecule studies of AdnAB (10–14) cohere into a working model for DSB resection whereby initial ATP-independent binding of AdnAB to a DSB end melts a short segment of the duplex to allow access of the 5′ terminal strand to the AdnA nuclease active site and access of the 3′ terminal strand to the AdnB motor. ATP hydrolysis catalyzed by the AdnB motor then drives translocation of tandem AdnB and AdnA motor domains in the 3′ to 5′ direction along the tracking strand, with the AdnB motor domain in the lead. Translocation propels the 3′ tracking strand along a sequential path through the AdnB and AdnA motor domains and then into the AdnB nuclease domain. ATPase-coupled DNA unwinding simultaneously threads the 5′ DNA strand through the AdnA nuclease domain.
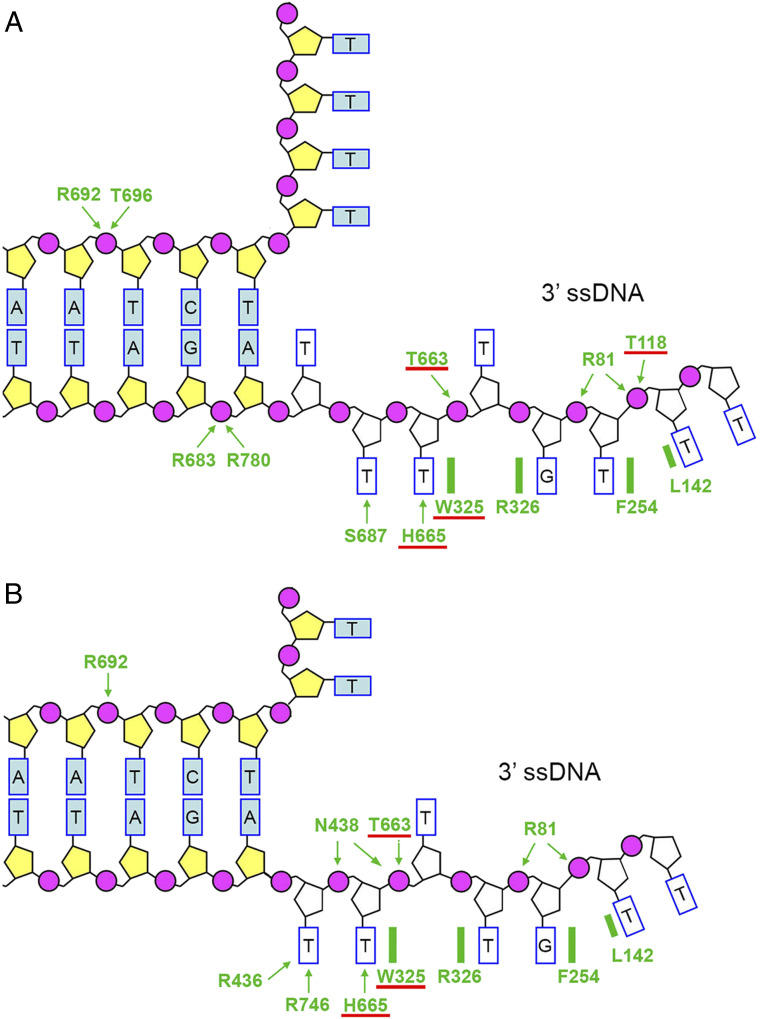
Cryo-EM structures of AdnAB bound to a duplex DNA with 5′ and 3′ tails revealed the path of the displaced 5′ strand and its interactions with the AdnA nuclease; the interface of the 3′ tracking strand with the AdnB motor; and an AdnA iron–sulfur cluster subdomain at the Y-junction that maintains the split trajectories of the unwound 5′ and 3′ strands (14). The AdnB–DNA interface, depicted in cartoon form in Fig. 1, entails an extensive network of amino acid side-chain contacts to the phosphates and nucleobases of a seven- to eight-nucleotide 3′ DNA tail and to two phosphates of the duplex segment at the single-strand (ss) DNA–double-strand (ds) DNA junction. The path of the 3′ tracking strand through the AdnB motor is punctuated by amino acids Trp325, Arg326, Phe254, and Leu142 that stack on and between the nucleobases to partition the 3′ ssDNA into discrete base-stacked segments.
Fig. 1.
AdnB motor domain interface with the fork junction and 3′ ssDNA tracking strand. (A) Cartoon summary of DNA contacts with protein side chains of the AdnB subunit in the DNA complex with the AdnA(D934A)–AdnB(D1014A) heterodimer. (B) Cartoon summary of DNA contacts with AdnB amino acid side chains in the DNA complex with the WT AdnAB heterodimer. Amino acids at the tracking strand interface that we show here are important for coupling ATP hydrolysis to mechanical work during DSB resection are underlined in red.
The partitioning of the tracking strand into base-stacked segments is seen in two different cryo-EM structures of mycobacterial AdnAB in which the register of the nucleobases is shifted by a one-nucleotide step (Fig. 1 A and B). We speculated that this register shift mimics (in part) the changes occurring during the mechanical power stroke, in which protein subdomain movements within the AdnB motor, synchronized to the ATP hydrolytic cycle, drive passage of the nucleobases over the amino acid baffles in the 3′ to 5′ direction, and the baffles prevent backward sliding in a pawl-and-ratchet–like fashion (14). Consistent with this model, we found that double-alanine mutation of the Trp325–Arg326 dipeptide (a constituent of SF1 helicase motif III) decoupled DNA-dependent ATP hydrolysis from mechanical work by the AdnB motor.
In the present study, we conduct a comprehensive alanine scan of the 16 amino acids at the AdnB–DNA interface seen in the cryo-EM structures in order to gain insights into which DNA contacts are necessary to couple ATP hydrolysis to motor activity. The mutant AdnAB enzymes were surveyed for DNA-dependent ATP hydrolysis, DNA translocation, and DSB resection in ensemble and single-molecule assay formats. The results pinpoint the AdnB amino acid Trp325 as the singular essential constituent of the ratchet pawl, while establishing that Arg326, Phe254, and Leu142 are inessential. The results also demonstrate that loss of Thr663 and Thr118 contacts to the tracking strand phosphates and of His665 to a nucleobase drastically slow the AdnAB motor during DSB resection. We suggest that the AdnAB interface with the DNA tracking strand is analogous to an automotive clutch, whereby the ATPase engine can be uncoupled from AdnAB movement by mutations that affect the molecular equivalent of the clutch friction disk.
Results
Alanine Scan of the AdnB–DNA Interface (Round 1).
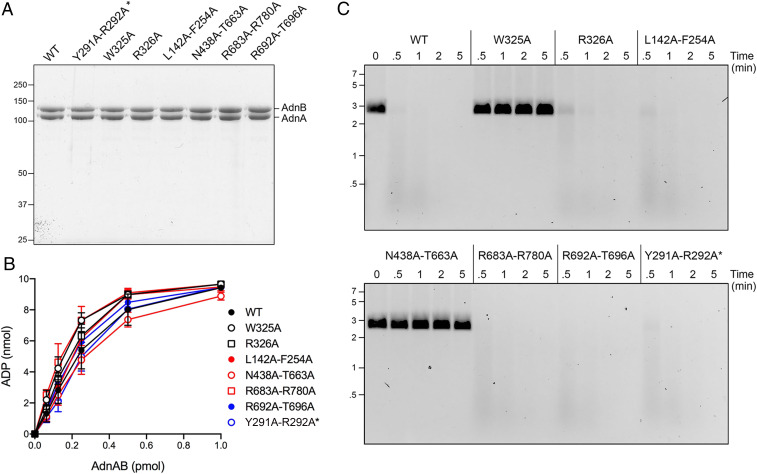
Having shown previously that a W325A–R326A double-mutation uncoupled ATP hydrolysis from translocation and DSB resection (14), we aimed to test the effects of single W325A and R326A mutations. We also interrogated other points of AdnB–DNA contact by simultaneously mutating pairs of amino acids that interact with the same, overlapping, or adjacent DNA moieties. These included: Leu142 and Phe254 that stack on vicinal nucleobases of the tracking strand; Asn438 and Thr663 that contact adjacent phosphates of the tracking strand; Arg683 and Arg780 that contact the same phosphate in the 3′ strand of the duplex; and Arg692 and Thr696 that contact the same phosphate in the 5′ strand of the duplex. Wild-type (WT) or mutant AdnAB heterodimers were produced in E. coli and purified from soluble bacterial extracts (Fig. 2A). Enzyme titrations showed that the specific activities of the mutants in ssDNA-dependent ATP hydrolysis were similar to that of WT AdnAB (Fig. 2B). Assay of the enzymes for their ability to catalyze ATP-dependent resection of linear duplex pUC19 plasmid DNA revealed that whereas the R326A, L142A–F254A, R683A–R780A, and R692A–T696A mutants digested the majority of input plasmid in 0.5 min, the W325A and N438A–T663A mutants elicited no apparent DNA resection during a 5-min reaction (Fig. 2C).
Fig. 2.
Effects of round 1 AdnAB mutations on DNA-dependent ATPase activity and DSB resection. (A) Aliquots (5 µg) of recombinant WT AdnAB and the indicated mutant heterodimers were analyzed by SDS-PAGE. All of the mutations are in the AdnB subunit, except for Y291A–R292A* in the AdnA subunit (distinguished by an asterisk next to the label). The Coomassie blue-stained gel is shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. The AdnA and AdnB polypeptides are indicated on the right. (B) ATPase reaction mixtures (10 µL) containing 20 mM Tris⋅HCl (pH 8.0), 2 mM MgCl2, 1 mM DTT, 1 mM (10 nmol) [α32P]ATP, 50 μM 44-mer ssDNA oligonucleotide, and AdnAB as specified were incubated for 10 min at 37 °C. The reactions were quenched by adding 2 μL of 5 M formic acid. Aliquots (2 μL) of the mixtures were applied to PEI–cellulose TLC plates, which were developed with 0.45 M ammonium sulfate. [α-32P]ATP and [α-32P]ADP were quantified by scanning the plates with a Typhoon FLA 7000 imager and processing the data with ImageQuant software. The extent of ATP hydrolysis is plotted as a function of input AdnAB. Each datum in the graph is the average of three independent titration experiments ± SEM. (C) DSB resection reaction mixtures (50 μL) containing 20 mM Tris⋅HCl (pH 8.0), 2 mM MgCl2, 1 mM ATP, 200 ng linear pUC19 DNA (cut with SmaI), and 2.5 pmol of WT or mutant AdnAB were incubated at 37 °C. Aliquots (10 µL) were withdrawn at the times specified, and the reactions were quenched immediately by adding 2.5 µL of 500 mM EDTA (pH 8.0). The reactions were supplemented with 5 μL of a solution containing 10 mM Tris⋅HCl (pH 7.6), 60% glycerol, 60 mM EDTA, and 0.15% Orange G and then analyzed by electrophoresis through a horizontal 0.8% agarose gel containing 0.5x TBE (45 mM Tris-borate, 1.2 mM EDTA). DNA was visualized under shortwave UV illumination after soaking the gel in 0.5x TBE containing 0.5 μg/mL ethidium bromide. The positions and sizes (kbp) of linear duplex DNA markers are indicated on the left.
As a measure of ssDNA translocation, we assayed the WT and mutant AdnAB enzymes for nuclease activity on a 5′ 32P-labeled 24-mer ssDNA substrate (SI Appendix, Fig. S1). The 5′ end of the ssDNA can access the AdnA nuclease from solution and thread into the active site without need for ATP hydrolysis (10–12), resulting in DNA cleavage by the AdnA nuclease at three principal sites to yield radiolabeled 5-mer, 6-mer, and 8-mer products (SI Appendix, Fig. S1). Each of the mutant AdnAB proteins was adept at ATP-independent cleavage by the AdnA nuclease (SI Appendix, Fig. S1). In the presence of ATP, WT AdnAB couples ATP hydrolysis to unidirectional translocation (3′ to 5′) on the 24-mer ssDNA and thereby “pumps” the 3′ end of the ssDNA through the tandem AdnB and AdnA motor domains and into the AdnB nuclease domain, resulting in incision at vicinal phosphodiesters to yield 5′ 32P-labeled 16-mer and 17-mer cleavage products (SI Appendix, Fig. S1). The salient findings were that the ATPase-active, DSB resection-defective W325A and N438A–T663A mutants could not translocate 3′ to 5′ on ssDNA when hydrolyzing ATP and thus could not pump the ssDNA into the AdnB nuclease (SI Appendix, Fig. S1). By contrast, the R326A, L142A–F254A, R683A–R780A, and R692A–T696A mutants recapitulated the WT ATP-dependent appearance of the AdnB nuclease cleavage products (SI Appendix, Fig. S1), signifying that they are active translocases, consistent with their activity in DSB end resection.
To assess if the ATPase-active, translocation-defective mutants W325A and N438A–T663A were compromised in their affinity for ssDNA, we measured their ATPase activity, and that of WT AdnAB, as a function of ssDNA concentration (SI Appendix, Fig. S2). The extents of ATP hydrolysis by 25 nM WT and mutant AdnAB enzymes displayed a hyperbolic dependence on the input level of a 44-mer ssDNA oligonucleotide. Apparent Km values of 192, 236, and 511 nM were calculated for WT, W325A, and N438A–T663A AdnAB, respectively (SI Appendix, Fig. S2). Thus, the unwinding defect of the W325A mutant is not attributable to reduced affinity for the ssDNA activator of ATP hydrolysis. These results implicate the Trp325 nucleobase stacking interaction as a singular key constituent of a pawl in the molecular ratchet required for unidirectional movement of the AdnB motor on ssDNA in response to ATP hydrolysis. The N438A–T663A mutation, which eliminated two contacts to the tracking strand, elicited an ∼2.7-fold reduction in affinity for the ssDNA activator of ATP hydrolysis.
Is There a Corresponding Pawl in the AdnA Motor-like Domain?
The cryo-EM structures show that the AdnA motor-like domain binds AMPPNP•Mg2+ and has a seemingly intact ATPase active site, but it is not certain if AdnA is an active ATP phosphohydrolase (14). Ensemble biochemical studies showed that alanine mutation of AdnA Asp255—the motif II side chain that coordinates the metal cofactor necessary for ATP hydrolysis—did not exert a significant effect on the rate of plasmid DNA unwinding (12). Single-molecule DNA curtain assays showed no apparent effect of the AdnA–D255A mutation on the dynamics of DSB resection by AdnAB (14). Thus, it seems that ATP hydrolysis by the lagging AdnA motor-like domain is not contributing in a major way to the overall motor activity of the AdnAB complex. Whereas the tailed duplex ligand employed in the cryo-EM experiments had a 21-nucleotide 3′ tracking strand tail that should, in principle, be long enough to reach through the AdnA motor domain and into the AdnB nuclease domain, there was no observable density for the 3′ strand beyond its interface with the AdnB motor domain. Although the interface of the tracking strand with the AdnA subunit is undefined, alignment of the AdnA and AdnB primary structures indicates that several of the AdnB amino acids that contact the 3′ ssDNA tail are conserved in AdnA. Specifically, the AdnB Trp325–Arg326 motif is conserved as Tyr291–Arg292 in AdnA, thereby raising the prospect that Tyr291–Arg292 might comprise a pawl-like element within the AdnA subunit. To test this idea, we produced recombinant AdnAB with a Y291A–R292A double mutation in the AdnA subunit (Fig. 2A, denoted by an asterisk to indicate that AdnA is mutated). The Y291A–R292A enzyme retained DNA-dependent ATPase (Fig. 2B), DSB resection (Fig. 2C), and DNA translocation (SI Appendix, Fig. S1) activities similar to those of WT AdnAB. These results suggest that AdnA Tyr291–Arg292 is not functionally equivalent to its AdnB Trp325–Arg326 counterpart.
Alanine Scan of the AdnB–DNA Interface (Rounds 2 and 3).
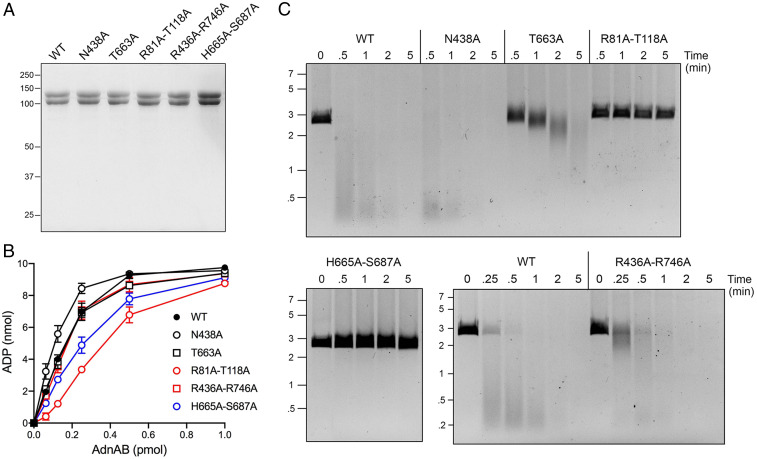
The N438A–T663A mutant identified in round 1 as uncoupling ATP hydrolysis from motor activity was reduced to its component N438A and T663A single mutations. We also mutated the remaining AdnB DNA contacts by pairs of alanine substitutions of: Arg81 and Thr118 that contact adjacent phosphates of the tracking strand; His665 and Ser687 that contact adjacent nucleobases of the tracking strand (Fig. 1A); and Arg436 and Arg746 that contact the same nucleobase on the tracking strand adjacent to the duplex junction (Fig. 1B). The recombinant AdnAB proteins were purified (Fig. 3A) and assayed for ssDNA-dependent ATPase activity (Fig. 3B). The ATPase specific activities of the mutants were similar (within a factor of two) to that of WT AdnAB (Fig. 3B). The temporal profile of ATP-dependent resection of linear duplex pUC19 plasmid DNA by the N438A mutant echoed that of the WT enzyme, whereby much of the input dsDNA was converted to fragments of ≤0.5 kbp within 30 s (Fig. 3C). By contrast, the T663A mutant was markedly sluggish in its rate of DSB resection; i.e., the bulk of the DNA was >2 kbp in length after 2 min and >1 kbp after 5 min (Fig. 3C). Thus, the T663A change appears to be the dominant effector of the uncoupling of resection seen for the N438A–T663A double mutant, thereby underscoring the importance of the Thr663 contact to the ssDNA phosphate of the flipped-out deoxynucleoside on which Trp325 stacks (Fig. 1 A and B). Simultaneous alanine mutations of Arg436 and Arg746 that contact one nucleobase at the ssDNA–dsDNA junction had a modest effect on the rate of DSB resection, whereby there was a significant population of intact and partially resected (∼2 kbp) plasmids visible at the 0.25- and 0.5-min time points, although the DNA was extensively resected by 1 min and thereafter (Fig. 3C). This pattern is suggestive of a lag in the initiation step of resection (discussed further below). Round 2 double mutants R81A–T118A and H665A–S687A were apparently unable to degrade pUC19 during a 5-min reaction, signifying that AdnB contacts to additional ssDNA phosphates on the leading edge of the 3′ tracking strand, and certain nucleobase contacts with the 3′ tracking strand adjacent to the ssDNA–dsDNA junction are important for chemomechanical coupling (Fig. 1 A and B).
Fig. 3.
Effects of round 2 AdnAB mutations on DNA-dependent ATPase and DSB resection. (A) Aliquots (5 µg) of recombinant WT AdnAB and the indicated AdnB mutant heterodimers were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. (B) ATPase reaction mixtures (10 µL) containing 20 mM Tris⋅HCl (pH 8.0), 2 mM MgCl2, 1 mM DTT, 1 mM (10 nmol) [α32P]ATP, 50 μM 85-mer ssDNA oligonucleotide, and AdnAB as specified were incubated for 10 min at 37 °C. The extent of ATP hydrolysis is plotted as a function of input AdnAB. Each datum in the graph is the average of three independent titration experiments ± SEM. (C) DSB resection reaction mixtures (50 μL) containing 20 mM Tris⋅HCl (pH 8.0), 2 mM MgCl2, 1 mM ATP, 200 ng linear pUC19 DNA (cut with SmaI), and 2.5 pmol of WT or mutant AdnAB were incubated at 37 °C. Aliquots (10 µL) were withdrawn and quenched at the times specified. The products were analyzed by agarose gel electrophoresis and staining with ethidium bromide. The positions and sizes (kbp) of linear duplex DNA markers are indicated on the left.
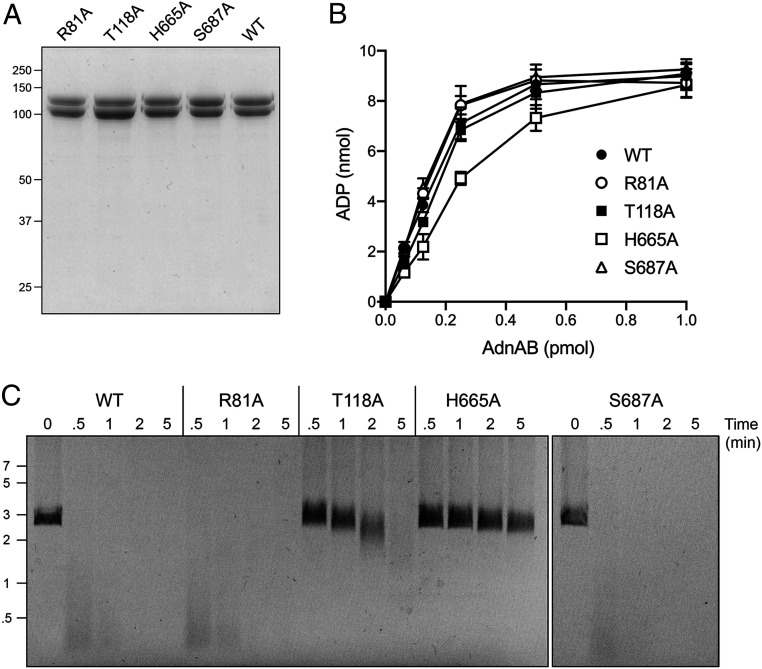
To follow up on these results, we performed a third round of alanine scanning, entailing reduction of the resection-dead double mutants to their component single mutations: R81A, T118A, H665A, and S687A. The recombinant AdnAB mutants were purified (Fig. 4A) and titrated for ssDNA-dependent ATPase activity (Fig. 4B). The ATPase specific activities of the mutants were similar (within a factor of two) to that of WT AdnAB (Fig. 4B). The temporal profiles of pUC19 end resection by the R81A and S687A mutants were similar to that of the WT enzyme; i.e., the input 2.7 kbp dsDNA was converted to fragments of ≤0.5 kbp within 30 s (Fig. 4C). Thus, the contacts of the Arg81 with two phosphates at the leading edge of the ssDNA tracking strand and of Ser687 with a nucleobase proximal to the ssDNA–dsDNA junction are not important per se for motor activity during end resection. By contrast, the T118A mutant AdnAB was markedly slower at resecting the linear plasmid (most of the DNA was still >2 kbp after 2 min and >1 kbp after 5 min), and the H665A mutant was even weaker (all of the DNA was >2 kbp after 5 min). We surmise that the contact between Thr118 and one of the tracking strand phosphates (also contacted by Arg81) is important per se for end resection. Note that the resection defect of the R81A–T118A double mutant was more severe than that of the T118A single mutant. We conclude that His665, via its interactions with the second unpaired nucleobase at the ssDNA–dsDNA junction (the same base on which the essential Trp325 side-chain stacks), is critical per se for AdnAB’s vigorous resection activity.
Fig. 4.
AdnB Thr118 and His665 are important for DNA end resection. (A) Aliquots (5 µg) of recombinant WT AdnAB and the indicated mutant heterodimers were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. (B) ATPase reaction mixtures (10 µL) containing 20 mM Tris⋅HCl (pH 8.0), 2 mM MgCl2, 1 mM DTT, 1 mM (10 nmol) [α32P]ATP, 50 μM 85-mer ssDNA oligonucleotide, and AdnAB as specified were incubated for 10 min at 37 °C. The extent of ATP hydrolysis is plotted as a function of input AdnAB. Each datum in the graph is the average of three independent titration experiments ± SEM. (C) DSB resection reaction mixtures (50 μL) containing 20 mM Tris⋅HCl (pH 8.0), 2 mM MgCl2, 1 mM ATP, 200 ng linear pUC19 DNA (cut with SmaI), and 2.5 pmol of WT or mutant AdnAB were incubated at 37 °C. Aliquots (10 µL) were withdrawn and quenched at the times specified. The products were analyzed by agarose gel electrophoresis and staining with ethidium bromide. The positions and sizes (kbp) of linear duplex DNA markers are indicated on the left.
Arg436 and Arg746 Mutations Affect the Initiation Step of DSB Resection.
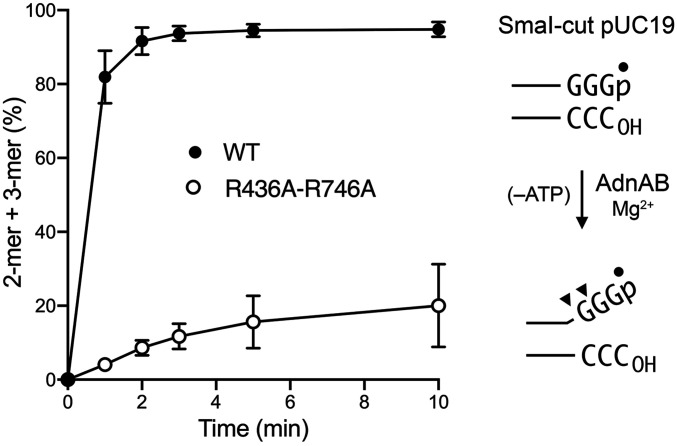
We showed previously that ATP-independent binding of AdnAB to a blunt DSB end of Sma1-cut pUC19 melts a short segment of the duplex and allows access of the 5′ strand to the AdnA nuclease active site, which cleaves the melted 5′ strand two and three nucleotides from the 5′ end to liberate 32P-labeled dinucleotide and trinucleotide products (10) (Fig. 5). With the insight from the AdnAB cryo-EM structures that the AdnA nuclease cleaves the displaced 5′ strand two nucleotides from the ssDNA–dsDNA junction, it was surmised that the ATP-independent end-melting step unpairs four or five nucleotides at the blunt DSB end, which would suffice for the 3′ strand to be engaged by the essential Trp325, Thr663, and His665 amino acids of the AdnB motor. As noted in the preceding section, the apparent short lag in the decay of the full-length pUC19 plasmid during resection by the R436A–R746A mutant raised the prospect of a delay or defect in end melting. As shown in Fig. 5, whereas reaction of WT AdnAB with 5′ 32P-labeled Sma1-cut pUC19 DNA in the absence of ATP resulted in rapid and efficient release of 5′ 32P-labeled dinucleotide and trinucleotide products of AdnA cleavage, the R436A–R746A mutant did so 20-fold more slowly and with lower yield. We infer that the contacts of Arg436 and Arg746 to the first unpaired nucleotide of the tracking strand at the ssDNA–dsDNA junction (Fig. 1) are important to promote and stabilize the melted end conformation that precedes the onset of ATP-dependent motor activity.
Fig. 5.
Arg436 and Arg746 mutation affects the initiation step of DSB resection. Binding of AdnAB to a blunt DSB end (Sma1-cut pUC19 DNA) in the absence of ATP leads to local melting of the duplex terminus that allows ingress of the 5′ strand to the AdnA nuclease, which incises the 5′ strand at sites denoted by arrowheads in the right panel. Reaction mixtures (70 µL) containing 20 mM Tris⋅HCl (pH 8.0), 1 mM DTT, 2 mM MgCl2, 1,400 ng (798 fmol) 5′ 32P-labeled SmaI-cut pUC19, and 10.7 pmol WT AdnAB or R436A–R746A mutant were incubated at 37 °C. Aliquots (10 µL) were withdrawn at the times specified and quenched by adding 10 µL of 90% formamide, 50 mM EDTA. The samples were heated for 5 min at 100 °C, and the reaction products were analyzed by electrophoresis through a 40-cm 24% polyacrylamide gel containing 7 M urea, 45 mM Tris-borate, 1.25 mM EDTA. The formation of labeled 5′-pGGG and 5'-pGG cleavage products (expressed as the percent of total labeled DNA) is plotted as a function of reaction time. Each datum in the graph is the average of two independent experiments; error bars denote the range of values.
Single-Molecule DNA Curtain Analysis of DSB Resection Velocity by AdnAB Mutants.
We recently applied single-molecule DNA curtain methods (19, 20) to study the dynamics of DSB resection by AdnAB (14). The curtain consists of a parallel array of bacteriophage λ dsDNA molecules tethered at one end to a lipid bilayer surface and aligned along a metallic barrier oriented perpendicular to the direction of liquid flow (19, 20). The DNAs are stained with the fluorescent dye YOYO-1 and visualized by total internal refection fluorescence microscopy. Engagement of the free DNA ends by AdnAB in the presence of ATP and magnesium results in a time-dependent shortening of the fluorescent linear DNAs. DNA shortening is taken to be synonymous with helicase unwinding and concomitant DSB resection. Our previous DNA curtain experiments with WT AdnAB revealed that: 1) the enzyme is highly processive, but prone to spontaneous pausing; 2) the velocity of the AdnAB motor speeds up, while the frequency and duration of pausing declines, with increasing ATP concentration in the range of 25–100 µM; and 3) the velocity of DSB resection slows after the enzyme experiences a spontaneous pause (14). Here we conducted single-molecule analyses of DSB resection by selected AdnAB mutants that were characterized above via ensemble assay methods. The salient mechanistic questions that are particularly suited to DNA curtain studies concern whether the effects of AdnB mutations on end resection detected in ensemble assays are the result of decreased rate of AdnB movement, decreased processivity, enhanced pausing, or a combination thereof.
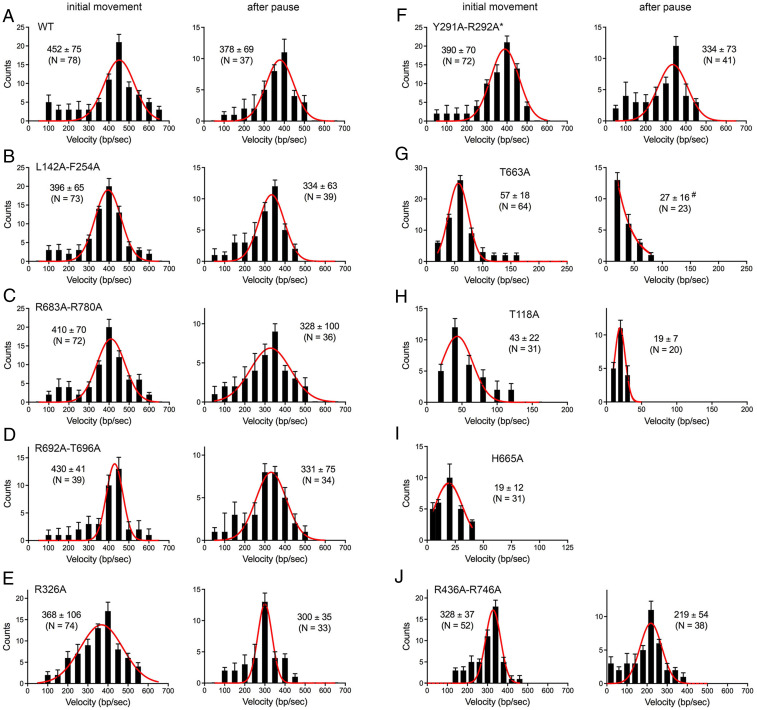
DNA curtain resection experiments were performed in the presence of 100 µM ATP. The W325A and N438A–T663A mutants that were inert in pUC19 DNA resection in the ensemble assay format were incapable of resecting λ DNA in the curtain assay. The distributions of velocities of λ DNA shortening by WT AdnAB and other mutants during individual resection tracts are shown in Fig. 6. The data were plotted as histograms and fit to Gaussian distributions (red curves) in Fig. 6. Scatter plots of the velocities are shown in Fig. 7A. For each AdnAB protein in Figs. 6 and 7A, we plotted separately: 1) the distribution of velocities at which AdnAB resects from the free end of the phage DNA; and 2) for those events entailing pausing and resumption of resection after a transient pause, the velocity of movement after a pause. As noted previously, the rate of WT AdnAB resection determined from the Gaussian fits was slower after a pause (378 ± 69 bp/s) than before a pause or in the absence of a pause (452 ± 75 bp/s) (Fig. 6A). This postpause slowing of the AdnAB motor was maintained in each of the AdnAB mutants (with the exception of H665A, for which no postpause resection events were observed), irrespective of whether the motor domain mutations had an effect on initial resection velocity (Fig. 6 B–J). The differences in initial resection versus postpause resection velocities were statistically significant in each case (Fig. 7A).
Fig. 6.
Single-molecule DNA curtain analysis of DSB resection velocity by AdnAB mutants. Velocity distributions of individual resection tracts are shown for WT AdnAB (A) and the indicated AdnAB mutants (B–J). For each AdnAB protein, we plotted separately the distribution of velocities at which AdnAB resected from the original end of the phage DNA (initial movement, graphs at left in each panel) and, for those events entailing pausing and resumption of resection after a transient pause, the velocity of movement after a pause (graph at right in each panel). Black error bars represent 95% confidence intervals calculated from bootstrap analysis. The solid red lines represent Gaussian fits to the data. The number of events scored (N) and mean velocities (± SD) derived from the Gaussian fit are shown in each graph (except for G, where the postpause mean velocity, denoted by #, was calculated from the scatter plot data shown in Fig. 7A).
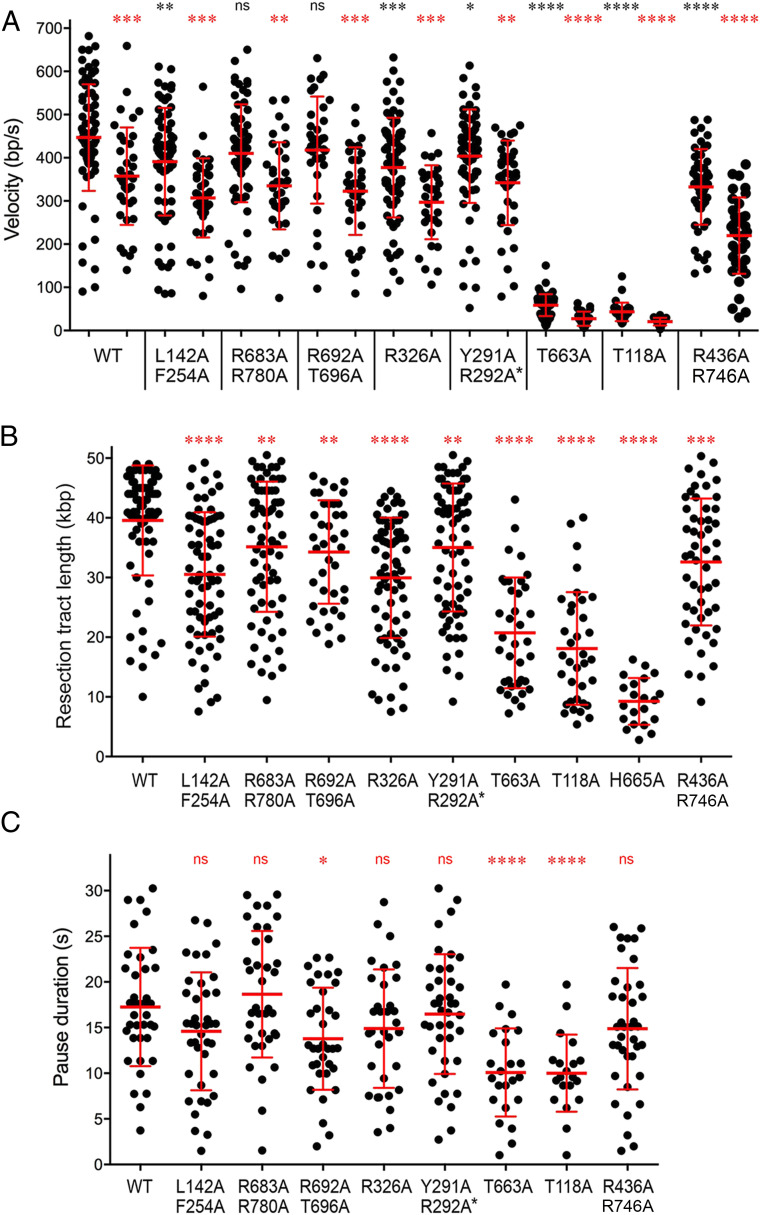
Fig. 7.
Summary of AdnAB mutational effects on single-molecule DNA resection. (A) Scatter plots of resection velocities during initial movement and after pausing. For each AdnAB protein specified below the x axis, the initial movement velocities are plotted on the left, and the postpausing velocities are plotted on the right. Red lines indicate the mean ± SD. Black asterisks above each initial movement scatter plot summarize the P values of unpaired t tests of mutant versus WT initial movement velocities. Red asterisks above each postpause scatter plot summarize the P values of unpaired t tests of postpause versus initial movement for each AdnAB enzyme. (B) Resection tract length. Scatter plots of total DNA resection tract lengths. The observation period was 20–25 min for slow-velocity mutants T663A, T118A, and H665A. All other AdnAB proteins were observed for 5–7 min. Red lines indicate the mean ± SD. Red asterisks above each mutant scatter plot summarize the P values (unpaired t test of mutant versus WT). (C) Pause duration. Scatter plots of pause times between initial movement and resumption of resection. Red lines indicate the mean ± SD. Red asterisks above each mutant scatter plot summarize the P values (unpaired t test of mutant versus WT). P value symbols: ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant.
Focusing on the initial resection tracts, we found that several AdnB motor mutants of AdnAB, which had little or no discernible effect on pUC19 DSB resection, were capable of sustaining resection velocities similar to (81–95% of the Gaussian mean value) that of WT AdnAB, as follows: R692A–T696A (430 ± 41 bp/s); R683–R780 (410 ± 70 bp/s); L142A–F254A (396 ± 65 bp/s); R326A (368 ± 106 bp/s). The AdnA motor domain mutation Y291A–R292A also had little effect on initial resection velocity (390 ± 70 bp/s). By contrast, the AdnB motor mutations H665A (19 ± 12 bp/s), T118A (43 ± 22 bp/s), and T663A (57 ± 18 bp/s) resulted in drastic reductions in initial resection velocity, by 8- to 24-fold vis-à-vis WT AdnAB, which could account for the kinetic defects seen for these mutations in the ensemble pUC19 resection assays. The R436A–R746A enzyme, which was suggested based on ensemble assays to have a delayed onset of pUC19 DSB resection, resected the λ DNA curtain at a velocity of 328 ± 37 bp/s (73% of WT velocity). Note that the curtain assays, as implemented here, are not able to visualize the AdnAB enzyme (which is not fluorescently labeled) and hence cannot gauge the duration of the initiation step of the DSB resection reaction, which precedes the onset of DNA shortening.
Mutational Effects on DSB Resection Tract Length.
The DNA curtain reaction endpoint, at which no further DNA shortening is observed, is presumed to reflect dissociation of AdnAB from the DNA substrate. The lengths (in kbp) of individual DNAs that were resected at the endpoint by WT and mutant AdnAB enzymes are plotted in Fig. 7B. The mean length of DNA resected by WT AdnAB was 39.6 ± 9.2 kbp. The “resection-competent” mutants displayed slightly lower resection track lengths (statistically significant), as follows: R692A–T696A (34.3 ± 10.1 kbp); R683–R780 (35.2 ± 10.9 kbp); L142A–F254A (30.5 ± 10.4 kbp); R326A (30.0 ± 10.1 kbp); R436A–R746A (32.6 ± 9.2 kbp); Y291–R292A (35.0 ± 10.7 kbp). For events entailing a pause and resumption of DNA resection, the distributions of individual pause durations are plotted in Fig. 7C. There was little or no statistically significant difference between the WT pause durations (17.3 ± 6.5 s) and those of the resection-competent mutants, as follows: R692A–T696A (13.8 ± 5.6 s); R683–R780 (18.7 ± 6.9 s); L142A–F254A (14.6 ± 6.5 s); R326A (14.9 ± 6.5 s); R436A–R746A (14.9 ± 6.6 s); Y291–R292A (16.5 ± 6.6 s). By contrast, the resection tract lengths of the three “slow-velocity” mutants during an extended 20- to 25-min observation period (sufficient to establish an endpoint of the resection reaction) were curtailed significantly, reflected in shorter mean resection lengths for H665A (9.3 ± 3.9 kbp), T118A (18.1 ± 9.4 kbp), and T663A (20.7 ± 9.3 kbp) (Fig. 7B). The slow-velocity T118A and T663A enzymes also displayed distinctive behavior with respect to their observed pausing events (lasting 10.0 ± 4.2 s for T118A and 10.8 ± 4.8 s for T663A), which were significantly shorter than those of WT AdnAB (Fig. 7C). It is conceivable that the T118A and T663A enzymes are prone to dissociate prematurely from the DNA upon pausing and are therefore less likely to resume resection after longer pause intervals.
Discussion
Chemomechanical coupling by ssDNA translocating helicases is driven by iterative protein domain motions synchronized to the steps of the ATP hydrolytic cycle. ATP hydrolysis is mechanically productive when the oscillating protein domain motions drive the DNA tracking strand forward in one direction, typically by a single-nucleotide step (21). For AdnAB, we envisioned, based on the cryo-EM structures, that the step motion of the tracking strand through the AdnB motor entails the flipping of nucleobases between amino acid baffles that demarcate the base-stacked segments of the tracking strand (Fig. 1). Such baffles would act in a pawl-and-ratchet–like fashion to prevent backward sliding of the AdnB motor on the DNA. Disabling the enzymic pawl could, in principle, preserve DNA-dependent ATPase activity but render it futile, or at least less efficient, with respect to mechanical work, whereby efficiency is defined as the probability of ATP hydrolysis triggering irreversible forward motion of the helicase on the tracking strand. Of the four AdnB baffle amino acids, we find that Trp325 is uniquely essential for DNA translocation and DSB resection. Trp325 makes a π stack on the tracking strand nucleobase 5′ of the flipped-out nucleoside (Fig. 1). Its neighbor Arg326, which makes a cation–π stack on the nucleobase 3′ of the flipped-out nucleoside, is not essential, although its replacement by alanine does slow the rate of DSB resection by ∼19% compared to WT AdnAB. A noteworthy finding in the present study is that Leu142 and Phe254 that intercalate into the tracking strand further downstream do not appear to be important for motor activity. Thus, Trp325 is the singular essential constituent of the tracking strand-intercalating ratchet pawl. Trp325 could disfavor backsliding in either of two ways: via the stabilizing effect of its π stack over the nucleobase; or by promoting the flipping of the adjacent nucleoside.
A motif III Trp is present in bacterial helicases PcrA and UvrD, wherein the Trp indole stacks on a nucleobase of the 3′ tracking strand (22, 23). Mutating the Trp to Ala in PcrA did not affect kcat for ATP hydrolysis at supersaturating concentrations of ssDNA cofactor, but it did result in a 200-fold decrement in the affinity of the mutant PcrA for the ssDNA cofactor, and it interdicted PcrA unwinding of a 3′-tailed duplex DNA (22). By contrast, the motif III W325A mutation in the AdnB subunit of AdnAB did not affect AdnAB’s affinity for the ssDNA activator of ATP hydrolysis. An expanded scan of the ssDNA interface of PcrA revealed additional mutations that uncoupled ATP hydrolysis from duplex unwinding, as gauged by ensemble biochemical assays (24).
Of the 16 amino acids that comprise the AdnB–DNA interface, only three others are overtly important for chemomechanical coupling: Thr118, Thr663, and His665. The T118A, T663A, and H665A mutants retained vigorous ssDNA-dependent ATPase activity but displayed feeble DSB resection activity in ensemble assays, the basis of which was clarified by single-molecule DNA curtain assays. These mutants resected DNA with drastically reduced velocity vis-à-vis WT AdnAB. We surmise that loss of Thr118, Thr663, and His665 contacts to the tracking strand lowers the microscopic efficiency of the AdnB motor, such that a significant fraction of the ATP hydrolytic events are unproductive with respect to translocation, resulting in a monotonously slow motor. An alternative scenario, wherein the slow ensemble resection activity of the mutants reflects more frequent pausing without affecting translocation velocity, is convincingly ruled out by the DNA curtain studies. Indeed, the fraction of DNA curtain events that entailed a pause and resumption of resection by T118A (17/41; 41%) and T663A (21/58; 36%) was similar to, but no greater than, that of WT AdnAB (37/78; 47%). His665 makes a hydrogen bond to the thymine nucleobase on which Trp325 is stacked and a van der Waals contact to the thymidine deoxyribose. Thr663 makes a hydrogen bond to the 3′-phosphate of the nucleotide on which Trp325 stacks (Fig. 1). Thus, three of the four key enablers of chemomechanical coupling are engaged in contacts to a single nucleotide of the tracking strand. The fourth residue, Thr118, coordinates a tracking strand phosphate three nucleotides distal to that contacted by Thr663 (Fig. 1A).
Our present findings anent AdnAB prompt us to analogize its mechanism to that of an automobile clutch. A clutch is the mechanical device that transfers all power from the engine to the transmission of a motor vehicle. On the molecular scale, the engine corresponds to the ATPase module of AdnB as it resides on and is activated by the tracking DNA single strand. The ATPase cycle entails oscillating movements of the RecA-like domains (21). Because the velocity of DSB resection by WT AdnAB in the DNA curtain assay is quite similar to the kcat for ssDNA-dependent ATP hydrolysis by a nuclease-inactive AdnAB in ensemble assays (415 s−1) (11), we surmise that 1 bp is unwound by WT AdnAB per ATP hydrolyzed and that the AdnAB motor is highly efficient. The various atomic contacts to the tracking strand that we find are important for translocation velocity comprise the analog of the clutch friction plate. These contacts couple the output of the ATP hydrolysis cycle to forward movement of the DNA strand through the AdnB motor domain during the power stroke of ATP hydrolysis, after which they are reset, and offset, by a one-nucleotide interval of the tracking strand. An interfacial mutation like W325A that fully uncouples the ATP hydrolysis from forward movement has the effect of disengaging the clutch entirely so that the engine is running in neutral. The threonine and histidine mutations that slow the velocity of DNA resection without slowing the ssDNA-dependent ATPase engine can be thought of as making the clutch slippery and hence less efficient (i.e., prone to pop in and out of gear on the millisecond timescale). It is reasonable to ask whether there is evidence for clutch-like behavior in the WT AdnAB complex, and indeed we think there is, insofar as our DNA curtain analysis showed that WT AdnAB is prone to pause transiently and then resume movement during processive resection of λ DNA (14). We envision that this may reflect “normal” disengagement of the clutch so that WT AdnAB experiences episodes of mechanically unproductive ATP hydrolysis as it idles on the tracking strand.
Our results here also shed light on the issue of how much DNA interaction is needed to activate the engine and to get the motor “in gear” to initiate duplex unwinding and resection. Previous studies addressed the effect of ssDNA length on the steady-state parameters for ATP hydrolysis by a nuclease-inactive AdnAB. Whereas the apparent Km for ssDNA decreased progressively as the ssDNA cofactor was lengthened from 12 to 44 nucleotides, there was no difference between a 12-mer and a 44-mer with respect to the apparent kcat (11). A six-nucleotide ssDNA was ineffective in triggering ATP hydrolysis. It is likely that the ssDNA length required to achieve ATP hydrolysis approximates the seven- to eight-nucleotide tracking strand segment visualized in the cryo-EM structures. The behavior of the R436A–R746A mutant reported here highlights the particular contributions of interactions with the first unpaired tracking strand nucleobase at the ssDNA–dsDNA junction during the ATP-independent initiation phase of DSB resection (entailing melting of the terminal duplex upon AdnAB binding) but less so during the subsequent ATP-dependent phase of processive helicase activity, in which the velocity of the R436A–R746A mutant was only 27% slower than WT AdnAB. The available data suggest that the ATP-independent melting of a blunt DSB suffices to direct a five-nucleotide 3′ ssDNA tail into the AdnB motor domain, which would satisfy many of the enzymic contacts needed to get the engine in gear with respect to translocation. It is conceivable that several early cycles of ATP hydrolysis then drive the tracking strand forward to establish contact with Thr118 before a processive and efficiently coupled state of the motor is established.
Translocation by the AdnB motor is thought to thread the 3′ tracking strand along a path through the AdnA motor domain and then into the AdnB nuclease domain that resects the 3′ strand (14). Because the AdnB and AdnA motor domains are homologous, we queried whether the AdnA equivalent of the essential AdnB base-stacking pawl plays an important role in DSB resection. Our finding that the AdnA Y291A–R292A mutant retained vigorous resection activity in ensemble and single-molecule assay formats argues against the existence of an analogous pawl within the AdnA motor-like domain. At present, there is no direct evidence that the AdnA subunit has auxiliary motor activity or that AdnA hydrolyzes ATP during DSB resection, notwithstanding that the AdnA subunit can bind ATP and has a seemingly intact set of phosphohydrolase motifs in its ATP-binding site (14). It is conceivable that AdnA provides a passive conduit for the tracking strand en route to the AdnB nuclease.
Methods
Recombinant AdnAB Proteins.
WT and mutant AdnAB heterodimers were produced in E. coli by coexpression of His10–Smt3-tagged AdnA and untagged AdnB subunits as described previously (10) and purified from soluble extracts (14) by sequential nickel–agarose affinity chromatography, cleavage of the His10Smt3 tag by treatment with the Smt3-specific protease Ulp1, separation of tag-free AdnAB from His10Smt3 by passage over a second nickel–NTA agarose column, and a final Superdex-200 gel filtration step. The protein concentrations were determined by using the BioRad dye reagent with bovine serum albumin (BSA) as the standard.
Single-Molecule DNA Curtain Assays.
All experiments were performed with a custom-built prism-type total internal reflection fluorescence microscope (Nikon) equipped with a 488-nm laser (Coherent Sapphire, 200 mM) and two Andor iXon EMCCD cameras (25). Flow cells and single-tethered dsDNA curtains were prepared as described previously (26) using bacteriophage λ DNA (48.5 kbp; NEB Cat No. N3011S) that was biotinylated at one end and anchored to a lipid bilayer through a biotin–streptavidin linkage. All single-molecule experiments were performed at 37 °C in resection buffer (20 mM Tris⋅HCl, pH 8.0, 1 mM DTT, 2 mM MgCl2, 0.1 mM ATP, 0.2 mg/mL BSA). DNA was aligned along chromium barriers under a constant buffer flow at 0.15 mL/min and stained with 0.1 mM YOYO-1 for 1 min. Free YOYO-1 was flushed out of the flow cell at 0.5 mL/min for 2 min. End resection was then immediately initiated by the injection of 5 nM AdnAB through a 150-µL sample loop at 0.15 mL/min, and unbound AdnAB was flushed from the sample chamber. End resection was then visualized by the gradual reduction of the length of the fluorescently stained DNA molecules (14). Image acquisition was initiated immediately prior to the protein injections and continued for the durations of the resection assays, which were typically 5–7 min for WT AdnAB and the resection-competent mutants but were 20–25 min for determination of resection tract lengths for the slow-velocity mutants T663A, T118A, and H665A. Images were acquired at 1 frame per 0.5 s or 2 s with 0.1 s integration time, and the illumination laser was shuttered between each image to minimize photo-bleaching. For measurement of resection tract lengths for the slow-velocity mutants T663A, T118A, and H665A, the images were acquired at 1 frame per 10 s.
Optical Microscopy Image Processing and Data Analysis.
Raw TIFF images were imported as image stacks to ImageJ. Images were corrected for drift using the StackReg function in ImageI. Kymographs were then generated from the corrected image stacks by defining a 1-pixel-wide region of interest encompassing individual dsDNA molecules, and these kymographs were used for analysis of AdnAB resection tract length and velocity as described (26). AdnAB end resection events from the kymographs were fitted with a linear function to calculate AdnAB translocation velocity for each individual molecule. AdnAB translocation velocities were then obtained from Gaussian fits of the distributions of observed velocities.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants R35-GM126945 and R01-AI64693 (to S.S.) and R01-CA236606 and R35-GM118026 (to E.C.G.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023955118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Dillingham M. S., Kowalczykowski S. C., RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72, 642–671 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singleton M. R., Dillingham M. S., Gaudier M., Kowalczykowski S. C., Wigley D. B., Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 432, 187–193 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Wilkinson M., Chaban Y., Wigley D. B., Mechanism for nuclease regulation in RecBCD. eLife 5, e18277 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng K., Wilkinson M., Chaban Y., Wigley D. B., A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair. Nat. Struct. Mol. Biol. 27, 71–77 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chédin F., Kowalczykowski S. C., A novel family of regulated helicases/nucleases from gram-positive bacteria: Insights into the initiation of DNA recombination. Mol. Microbiol. 43, 823–834 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Yeeles J. T., Dillingham M. S., A dual-nuclease mechanism for DNA break processing by AddAB-type helicase-nucleases. J. Mol. Biol. 371, 66–78 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Yeeles J. T., Gwynn E. J., Webb M. R., Dillingham M. S., The AddAB helicase-nuclease catalyses rapid and processive DNA unwinding using a single Superfamily 1A motor domain. Nucleic Acids Res. 39, 2271–2285 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saikrishnan K., et al., Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex. EMBO J. 31, 1568–1578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krajewski W. W., et al., Structural basis for translocation by AddAB helicase-nuclease and its arrest at χ sites. Nature 508, 416–419 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinha K. M., Unciuleac M. C., Glickman M. S., Shuman S., AdnAB: A new DSB-resecting motor-nuclease from mycobacteria. Genes Dev. 23, 1423–1437 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unciuleac M. C., Shuman S., Characterization of the mycobacterial AdnAB DNA motor provides insights into the evolution of bacterial motor-nuclease machines. J. Biol. Chem. 285, 2632–2641 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Unciuleac M. C., Shuman S., Double strand break unwinding and resection by the mycobacterial helicase-nuclease AdnAB in the presence of single strand DNA-binding protein (SSB). J. Biol. Chem. 285, 34319–34329 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta R., Unciuleac M. C., Shuman S., Glickman M. S., Homologous recombination mediated by the mycobacterial AdnAB helicase without end resection by the AdnAB nucleases. Nucleic Acids Res. 45, 762–774 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia N., et al., Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection. Proc. Natl. Acad. Sci. U.S.A. 116, 24507–24516 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wigley D. B., Bacterial DNA repair: Recent insights into the mechanism of RecBCD, AddAB and AdnAB. Nat. Rev. Microbiol. 11, 9–13 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Rand L., et al., The majority of inducible DNA repair genes in Mycobacterium tuberculosis are induced independently of RecA. Mol. Microbiol. 50, 1031–1042 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Namouchi A., et al., The Mycobacterium tuberculosis transcriptional landscape under genotoxic stress. BMC Genomics 17, 791 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Müller A. U., Imkamp F., Weber-Ban E., The mycobacterial LexA/RecA-independent DNA damage response is controlled by PafBC and the Pup-proteasome system. Cell Rep. 23, 3551–3564 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Collins B. E., Ye L. F., Duzdevich D., Greene E. C., DNA curtains: Novel tools for imaging protein-nucleic acid interactions at the single-molecule level. Methods Cell Biol. 123, 217–234 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Finkelstein I. J., Visnapuu M. L., Greene E. C., Single-molecule imaging reveals mechanisms of protein disruption by a DNA translocase. Nature 468, 983–987 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang W., Lessons learned from UvrD helicase: Mechanism for directional movement. Annu. Rev. Biophys. 39, 367–385 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dillingham M. S., Soultanas P., Wigley D. B., Site-directed mutagenesis of motif III in PcrA helicase reveals a role in coupling ATP hydrolysis to strand separation. Nucleic Acids Res. 27, 3310–3317 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J. Y., Yang W., UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 127, 1349–1360 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dillingham M. S., Soultanas P., Wiley P., Webb M. R., Wigley D. B., Defining the roles of individual residues in the single-stranded DNA binding site of PcrA helicase. Proc. Natl. Acad. Sci. U.S.A. 98, 8381–8387 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Tullio L., Kaniecki K., Greene E. C., Single-stranded DNA curtains for studying the Srs2 helicase using total internal reflection fluorescence microscopy. Methods Enzymol. 600, 407–437 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue C., et al., Regulatory control of Sgs1 and Dna2 during eukaryotic DNA end resection. Proc. Natl. Acad. Sci. U.S.A. 116, 6091–6100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.