Abstract
Background
Cytochrome P450 (CYP) genes are necessary for the production or metabolism of fetal sex hormones during pregnancy. The second-to-fourth digit ratio (2D: 4D) is formed in the early stage of human fetal development and considered an indicator reflecting prenatal sex steroids levels. We explored the association between 2D: 4D and single-nucleotide polymorphisms (SNPs) of CYP.
Material/Methods
Correlation analysis between 2D: 4D and 8 SNPs, rs2687133 (CPY3A7), rs7173655 (CYP11A1), rs1004467, rs17115149, and rs2486758 (CYP17A1), and rs4646, rs2255192, rs4275794 (CYP19A1), was performed using data from 426 female and 412 male Chinese university students. SNP genotyping was conducted using PCR. Digit lengths were photographed and measured by image processing software.
Results
rs2486758 (CYP17A1) correlated with left hand 2D: 4D in men (P=0.026), and rs1004467 (CYP17A1) correlated with right hand 2D: 4D in men (P=0.008) and the whole population (P=0.032). In men, allele G rs1004467 decreased right hand 2D: 4D, while allele C of rs2486758 increased left hand 2D: 4D. In women, left hand 2D: 4D was higher in genotypes with allele A of SNP rs4646 (CYP19A1) under the dominant genetic model; female DR-L was higher in genotypes with allele T of rs17115149 (CYP11A1). SNPs rs2687133 (CYP3A7) and rs1004467 (CYP17A1) were significantly correlated with right hand 2D: 4D (P=0.0107).
Conclusions
SNPs rs1004467 and rs2486758 of CYP17A1 are significant in the relationship between 2D: 4D and CYP gene polymorphisms under different conditions. SNP interactions between CYP genes probably impact 2D: 4D. The correlation between 2D: 4D and some sex hormone-related diseases may be due to the effect of CYP variants on the 2 phenotypes.
Keywords: Fetal Development; Genetics; Gonadal Steroid Hormones; Polymorphism, Single Nucleotide
Background
The ratio of the second-to-fourth digit length (2D: 4D) in human hands is a sex dimorphism trait that tends to be higher in females than in males and is believed to reflect prenatal sex steroid levels [1]. This ratio is possibly determined by the balance of prenatal testosterone and estrogen levels in a narrow window during the fetal period and remains stable after birth [2]. Thus, 2D: 4D is commonly used as a noninvasive retrospective index of prenatal exposure to sex steroids, especially androgen and estrogen. Studies have shown that 2D: 4D is strongly associated with health conditions and diseases of sex dimorphisms or those affected by sex hormones. Therefore, 2D: 4D might be an indicator of human conditions such as body composition [3], personality traits [4], and physical performance [5], and of disease susceptibilities, including cancer [6], congenital adrenal hyperplasia [7], infertility [8], and mental disorders [9].
Genetic studies on 2D: 4D in twins have shown that 2D: 4D is highly heritable (about 80%) [10]. Gene polymorphisms and the 2D: 4D ratio are significant in studies on factors that can affect the upper limbs [11]. Therefore, 3 genome-wide association studies (GWAS) have been conducted to identify the single-nucleotide polymorphisms (SNPs) associated with 2D: 4D. In 2010, Medland et al studied a sample of 1382 twins and their siblings from Australia, and the SNP rs314277 of gene LIN28B was found to be strongly associated with 2D: 4D [12]. In 2013, Lawrance-Owen et al conducted a 2D: 4D GWAS in the UK and found that the SMOC1 polymorphism was correlated with 2D: 4D [13]. In 2018, Warrington et al performed the largest 2D: 4D genome-wide association meta-analysis, in which 11 loci were identified (GLIS1, rs4927012; EFNA1, rs11581730; LDAH, rs340600; OLA1, rs12474669; FLI1, rs10790969; HOXD11/12, rs847158; GLI3, rs77640775; SMOC1, rs2332175; SALL1, rs6499762; TOX3, rs1080014; and SALL3, rs4799176), and were believed to explain only 3.8% of the 2D: 4D discrepancy [14]. It is worth noting that none of these SNPs were located in the sex chromosome, and an Asian population was not involved in the studies. Later, other 2D: 4D correlation studies targeting hormone-related gene polymorphisms were conducted, with the majority mainly focusing on estrogen and androgen receptor genes [15,16]. However, 2D: 4D-associated SNPs of enzyme-related genes involved in hormone synthesizing and metabolism have been less frequently studied.
In humans, cytochrome P450s (CYPs) constitute one of the most abundant and diverse enzyme superfamilies, whose members catalyze the oxidative transformation of many organic substrates [17]. The function of CYP genes is critical for many metabolic processes, particularly for sex steroid conversion [18]. The altered expression of CYP genes involved in steroid metabolism may affect sex hormone levels and further increase the risk of some sex-related diseases, such as breast cancer [19], prostate cancer [20], infertility [21], polycystic ovary syndrome [22], and congenital adrenal hyperplasia [23]. During pregnancy, fetal sex hormones affect embryo development in many ways. The expression of specific CYP genes in the fetal liver, gonad, and placenta is indispensable across the development to synthesize sex hormones including progesterone, estrogen, and androgen [24]. First, CYP11A1, an enzyme initiating the steroidogenesis to converse cholesterol to pregnenolone, is pivotal for fetal survival in the early phase of pregnancy. Also, about 30% of the estrogen produced during pregnancy is yielded from the pregnenolone conversion by placental CYP17A1. In humans, dehydroepiandrosterone (DHEA) and DHEA sulfate produced by the fetal adrenal glands mainly undergo 16-hydroxylation by CYP3A7 in the liver, and are then metabolized by the placental enzyme aromatase (CYP19A1) [25]. Additionally, CYP19A1 is a rate-limiting enzyme involved in catalyzing the conversion of androgen to estrogen [26]. There is growing evidence to support that the analysis of sex hormone balance in a developing fetus might be helpful for predicting adult diseases during the lifespan [27].
In particular, some SNPs of CYP genes may affect the sex steroids, leading to physiological or pathological changes in humans. The C allele of rs45446698 in CYP3A7 causes the overexpression of the fetal CYP3A7 gene in adults and further impacts the levels of circulating endogenous sex hormones [28]. The SNP rs17115149 (C>A) of the CYP17A1 gene was strongly correlated with testosterone levels in men with infertility [29]. The SNPs in the CYP11A1 and CYP17 genes are significantly associated with testosterone plasma concentrations of patients with polycystic ovary syndrome, and SNPs rs4441215 and rs936306 of the CYP19A1 gene are associated with serum estradiol levels [30]. Further, SNPs in the CYP genes CYP3A7, CYP11A1, CYP17A1, and CYP19A1 have been shown to be associated with malignancy and androgenic enzyme expression in the fetal liver [31,32]. Therefore, it is proposed that CYP SNPs may have effects on prenatal sex hormones.
Herein, we conducted a study on 4 CYP genes involved in fetal or placental sex steroid synthesis and metabolism, namely CYP3A7, CYP11A1, CYP17A1, and CYP19A1. The association between these CYP gene polymorphisms and 2D: 4D were evaluated in a population of Chinese university students. Our study might elucidate the underlying mechanism of genetic influence on the formation of 2D: 4D. Also, the study might indirectly support 2D: 4D as a useful predictor for many hormone-related diseases, especially for those with significant sex differences in prevalence, incidence, and clinical manifestation.
Material and Methods
Study Subjects
The participants were 426 women aged 19.6±1.3 years and 412 men aged 19.2±1.2 years. All participants were recruited from the Ningxia Medical University in Yinchuan city of the Ningxia Hui Autonomous Region in northwestern China. All participants were physically and mentally healthy. The study design was approved by the Ningxia Medical University Ethics Committee (No.2019-057). Informed written consent was obtained from each participant. All the study samples are stored securely. The DNA samples, research data, and any information that can be traced to individual participants will not be provided to any other party.
SNP Genotyping
Four CYP genes were chosen based on the review of related literature, namely CYP3A7 (critical for sex steroid synthesis during embryogenesis and expressed mainly in fetal liver), and CYP11A1, CYP17A1, and CYP19A1 (the genes of 3 rate-limiting enzymes catalyzing sequential steps in steroidogenesis). SNPs for the CYP genes were selected using the 1000 Genomes Browser database and Haploview 4.2 software, per the following criteria: (1) Functional SNPs from regions including 1000bp upstream and downstream of target genes were systematically screened using the online tools ENSEMBL (https://asia.ensembl.org) and SNPinfo Web Server (https://snpinfo.niehs.nih.gov/); (2) an r2 cut-off (0.8) and minor allele frequency cut-off (0.05) for the Han Chinese in Beijing population of the SNPs were defined; (3) the SNPs have been studied or reported in other diseases. Finally, we selected 8 SNPs, CPY3A7 (rs2687133 C>T), CYP11A1 (rs7173655 C>T), CYP17A1 (rs1004467 A>G, rs17115149 C>T, rs2486758 T>C), and CYP19A1 (rs4646 C>A, rs2255192 C>T, rs4275794 T>C).
Genomic DNA was extracted from peripheral blood samples using DNA extraction kits, according to the manufacturer’s instructions (DP304 TIANamp Genomic DNA Kit, TIANGEN Biotech Co. Ltd., Beijing, China). Genotyping of each variant was performed using the multiplex polymerase chain reaction (PCR) technique developed by Novogene Biotech Co., Ltd. (Beijing, China). The PCR program was performed in 2 rounds: round 1 consisted of 1 cycle at 95°C for 15 min; 4 cycles at 94°C for 30 s, 60°C for 10 min, and 72°C for 30 s; and 24 cycles at 94°C for 30s, 60°C for 1 min, and 72°C for 30 s; and round 2 consisted of 1 cycle at 95°C for 15 min; 5 cycles at 94°C for 30 s, 60°C for 4 min, and 72°C for 30 s; and 10 cycles at 94°C for 30 s, 65°C for 1 min, and 72°C for 30 s. After PCR, the amplified products were mixed in a centrifuge tube and shaken overnight. The PCR products were then purified using a CA2 absorption column. The purified PCR product was loaded and run on the Illumina X-10 sequencer. Illumina RTA software was used to perform quality control and raw data analysis.
Data Collection
The participants put their 2 hands flat against the same white background with the palms facing up and fingers straight. A digital camera was used to collect the images of both hands. The photos were not used when the fine structures of each digit were not clearly visible. The Image-Pro Plus 6.0 (Media Cybernetics, Inc., USA) was used to measure the digit lengths of both hands (from the fingertip to the middle point of the most proximal crease to the palm). The measurement of each digit was performed 3 times by 3 different individuals who were well trained in the procedure, and the mean value was used for further analysis. The length ratios of both hands between the second and fourth digits were calculated.
Statistical Analyses
The Statistical Package for the Social Sciences (SPSS) 22.0 software (IBM, Armonk, NY, USA) was used to conduct the statistical analyses. The intra-class correlation coefficients (r1) were used for evaluating the digit length measurement credibility. The difference comparisons were performed using the t test for 2 groups and ANOVA for more than 2 groups. The Benjamini-Hochberg method was applied for multiple comparisons to control the false discovery rate. The χ2 test and Fisher’s exact test were used to evaluate the genotype and allele frequency distributions between groups. The statistical tests were 2-tailed and P<0.05 indicated statistical significance. The mean±standard deviation was used to represent all 2D: 4D data.
The Hardy-Weinberg equilibrium, minor allele frequency, and linkage disequilibrium were analyzed using Haploview software (Broad Institute of Harvard and MIT, MA, USA). The haplotype block was defined using the method of the Four Gamete Rule in Haploview.
Snpstats (https://www.snpstats.net/start.htm) was used to analyze the differences in haplotype, genotype, and allele frequencies between sexes. The linear regression analysis in the Plink package (v.1.9) was used to verify the associations between 2D: 4D and CYP SNPs. We assessed the interactions among SNPs of CYP genes using the generalized multifactor dimensionality reduction (GMDR) method [33]. The box-shaped scatter composite figure plotted by R was used to visualize the 2D: 4D genotype association characteristics.
Results
Description and distribution of CYP SNPs in our population
A total of 8 SNPs separately located in CPY3A7, CYP11A1, CYP17A1, and CYP19A1 were selected. The information and locations of the associated SNPs are listed in Table 1. The genotype frequencies of all the SNPs met the requirement of Hardy-Weinberg equilibrium in our population (P>0.05). The results indicated that the genotype and allele frequencies of all SNPs were not significantly different between men and women. A good linkage disequilibrium was found in the 2 groups of the following SNPs: 3 SNPs (rs1004467, rs17115149, rs2486758) of CYP17A1 and 3 SNPs (rs4646, rs2255192, rs4275794) of CYP19A1 (Figure 1). The haplotypes in the blocks were established and analyzed for differences between sexes, but no statistically significant differences were observed.
Table 1.
Specific information on candidate SNPs of CYP genes.
| Gene | SNP | Chromosome | Position | Location | Allele | MAF | HWpval |
|---|---|---|---|---|---|---|---|
| CPY3A7 | rs2687133 | 7 | 99170019 | Intron | C>T | 0.254 | 0.9280 |
| CYP11A1 | rs7173655 | 15 | 72419415 | Intron | C>T | 0.350 | 0.4637 |
| CYP17A1 | rs17115149 | 10 | 104587708 | 5′-UTR | C>T | 0.189 | 0.1475 |
| rs1004467 | 10 | 104584497 | Intron | A>G | 0.329 | 0.9660 | |
| rs2486758 | 10 | 104587470 | 5′-UTR | T>C | 0.190 | 0.1097 | |
| CYP19A1 | rs4646 | 15 | 49290136 | 3′-UTR | C>A | 0.297 | 0.6116 |
| rs2255192 | 15 | 49288127 | 3′-UTR | C>T | 0.188 | 0.1793 | |
| rs4275794 | 15 | 49288409 | 3′-UTR | T>C | 0.189 | 0.1940 |
SNPs – single-nucleotide polymorphisms.
Figure 1.
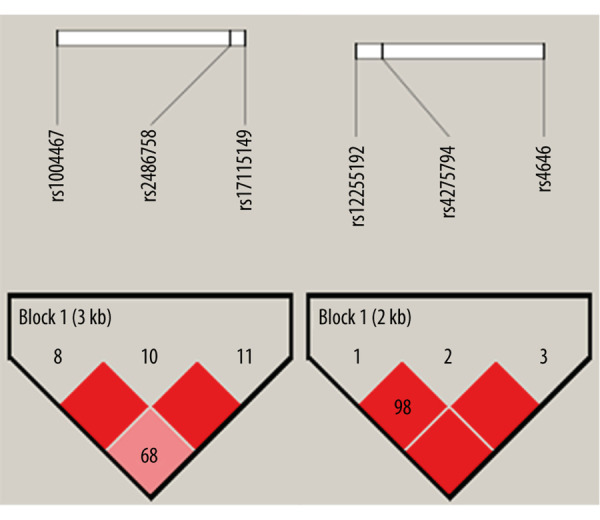
Linkage disequilibrium (LD) map of single-nucleotide polymorphisms (SNPs) in genes CYP17A1 and CYP19A1. A good LD was found among rs1004467, rs2486758, and rs17115149 of CYP17A1(rs1004467 – rs2486758: D’=0.9993, r2=0.1153; rs1004467 – rs17115149: D’=0.6885, r2=0.0202; rs2486758 – rs17115149: D’=0.9964, r2=0.0203); and rs2255192, rs4275794, and rs4646 of CYP19A1(rs2255192 – rs4275794: D’=0.9883, r2=0.9728; rs2255192 – rs4646: D’=0.9992, r2=0.0974; rs4275794 – rs4646: D’=0.9992, r2=0.0978). The haplotypes in the blocks were established based on the LD values.
2D: 4D Characterizations in Our Population
Remeasurement reliability of all digit lengths were acceptable for both hands in the intra-class correlation coefficient test (digit length of left hand: F=44.620, P<0.001; digit length of right hand: F=44.484, P<0.001). The significant difference was observed between different sexes in the left hand (L)2D: 4D (t=−2.951, P=0.003) and the right hand (R)2D: 4D) (t=−2.353, P=0.019). The L2D: 4D and R2D: 4D were both higher in women than in men, which was consistent with previous reports. However, the R2D: 4D minus L2D: 4D (DR-L) was not different between men and women (Table 2).
Table 2.
The summary of 2D: 4D results for men and women.
| L2D: 4D | R2D: 4D | DR-L | ||||
|---|---|---|---|---|---|---|
| Range | Mean±SD | Range | Mean±SD | Range | Mean±SD | |
| Total | 0.8271–1.0788 | 0.964±0.033 | 0.8551–1.0598 | 0.961±0.033 | −0.1185-0.0833 | −0.002±0.028 |
| Male | 0.8271–1.0595 | 0.960±0.036 | 0.8551–1.0524 | 0.959±0.033 | −0.1061-0.0833 | −0.002±0.027 |
| Female | 0.8726–1.0788 | 0.967±0.032 | 0.8811–1.0598 | 0.964±0.034 | −0.1185-0.0675 | −0.003±0.028 |
| t | −2.951 | −2.353 | 0.687 | |||
| p | 0.003 | 0.019 | 0.492 | |||
P<0.05;
P<0.01.
2D: 4D – second-to-fourth digit ratio.
Correlation of Single CYP SNPs to 2D: 4D in the Study Population
The L2D: 4D, R2D: 4D, and DR-L of CYP SNP genotypes were analyzed to explore the relationship between the CYP gene polymorphisms and the prenatal sex hormones. From the analytical results, the R2D: 4D was found to be significantly different in the 3 genotypes of SNP rs1004467 (F=4.113, P=0.017) (Table 3). The existence of minor allele G decreased the R2D: 4D, suggesting an increased fetal androgen or decreased fetal estrogen level. However, no genotype difference was observed in L2D: 4D and DR-L.
Table 3.
The difference of the digit ratios among SNP genotypes of CYP genes.
| Genotype | L2D: 4D | R2D: 4D | DR-L | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean±SD | F | P | Mean±SD | F | P | Mean±SD | F | P | |
| rs2687133 | |||||||||
| CC | 0.9676±0.0344 | 0.925 | 0.397 | 0.9634±0.0360 | 0.282 | 0.754 | −0.0043±0.0275 | 2.138 | 0.119 |
| TT | 0.9626±0.0331 | 0.9620±0.0337 | −0.0005±0.0271 | ||||||
| TC | 0.9652±0.0327 | 0.9605±0.0323 | −0.0047±0.0282 | ||||||
| rs7173655 | |||||||||
| CC | 0.9645±0.0336 | 0.187 | 0.830 | 0.9629±0.0351 | 0.739 | 0.478 | −0.0016±0.0273 | 0.301 | 0.740 |
| TT | 0.9645±0.0346 | 0.9619±0.0314 | −0.0026±0.0309 | ||||||
| TC | 0.9631±0.0322 | 0.9600±0.0320 | −0.0032±0.0269 | ||||||
| rs17115149 | |||||||||
| GG | 0.9636±0.0334 | 0.76 | 0.468 | 0.9615±0.0329 | 0.874 | 0.418 | −0.0021±0.0272 | 0.295 | 0.744 |
| TT | 0.9771±0.0367 | 0.9755±0.0480 | −0.0016±0.0508 | ||||||
| TG | 0.9645±0.0306 | 0.9603±0.0344 | −0.0042±0.0275 | ||||||
| rs1004467 | |||||||||
| AA | 0.9661±0.0336 | 2.097 | 0.123 | 0.9649±0.0331 | 4.113 | 0.017* | −0.0012±0.0279 | 0.625 | 0.535 |
| GG | 0.9587±0.0356 | 0.9553±0.0356 | −0.0034±0.0287 | ||||||
| AG | 0.9630±0.0318 | 0.9596±0.0326 | −0.0034±0.0270 | ||||||
| rs2486758 | |||||||||
| CC | 0.9705±0.0384 | 1.647 | 0.193 | 0.9629±0.0333 | 1.677 | 0.187 | −0.0076±0.0318 | 0.879 | 0.415 |
| TT | 0.9626±0.0319 | 0.9600±0.0339 | −0.0026±0.0273 | ||||||
| TC | 0.9659±0.0346 | 0.9647±0.0318 | −0.0012±0.0276 | ||||||
| rs4646 | |||||||||
| AA | 0.9656±0.0348 | 2.485 | 0.084 | 0.9654±0.0343 | 0.889 | 0.411 | −0.0001±0.0256 | 1.527 | 0.218 |
| CC | 0.9613±0.0327 | 0.9602±0.0328 | −0.0012±0.0269 | ||||||
| AC | 0.9666±0.033 | 0.9622±0.0337 | −0.0043±0.0287 | ||||||
| rs2255192 | |||||||||
| CC | 0.9637±0.0332 | 0.259 | 0.772 | 0.9605±0.0343 | 0.91 | 0.403 | −0.0032±0.0277 | 1.786 | 0.168 |
| TT | 0.9609±0.0315 | 0.9665±0.0294 | 0.0056±0.0245 | ||||||
| TC | 0.9649±0.0330 | 0.9630±0.0314 | −0.0018±0.0277 | ||||||
| rs4275794 | |||||||||
| CC | 0.9609±0.0315 | 0.154 | 0.857 | 0.9665±0.0294 | 0.542 | 0.582 | 0.0056±0.0245 | 1.723 | 0.179 |
| TT | 0.9640±0.0331 | 0.9609±0.0341 | −0.0031±0.0278 | ||||||
| TC | 0.9641±0.0332 | 0.9621±0.0320 | −0.0020±0.0274 | ||||||
P<0.05.
SNP – single-nucleotide polymorphism.
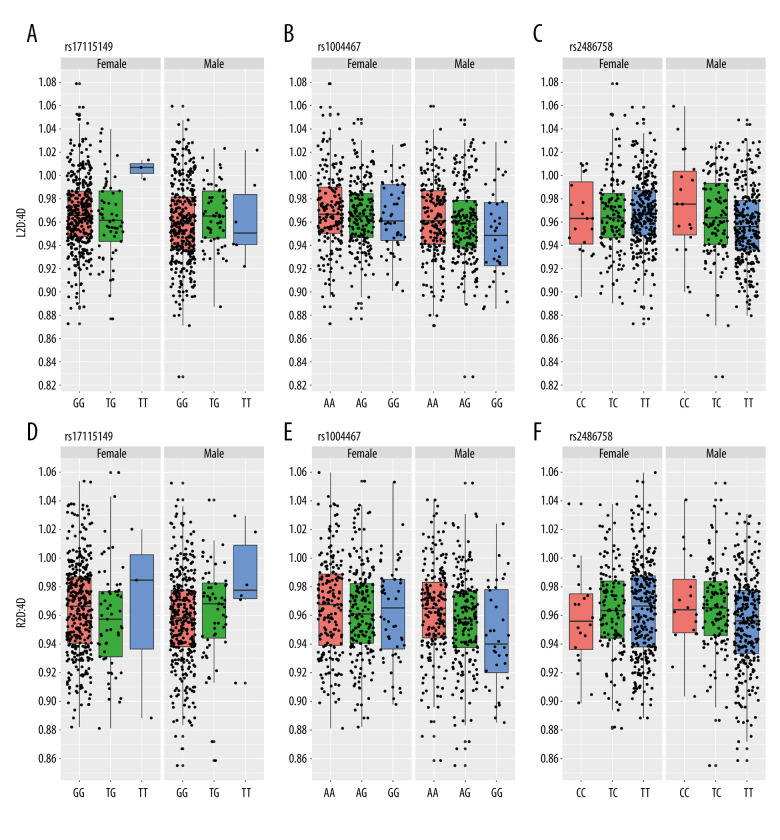
The associations between CYP SNPs and 2D: 4D by sex were then assessed, and only SNPs of CYP17A1 showed a relationship (Figure 2). For men, the L2D: 4D showed a statistically significant difference among different genotypes of SNP rs2486758 (F=4.771, P=0.009) and the genotypes with minor allele C had a higher L2D: 4D. The R2D: 4D was significantly different among different genotypes of SNPs rs1004467 (F=5.997, P=0.003) and rs2486758 (F=4.973, P=0.007). The minor allele G of rs1004467 was associated with a decrease in the R2D: 4D, while the minor allele C of rs2486758 was associated with an increase in the R2D: 4D. For women, a statistically significant difference in the L2D: 4D was observed in SNP rs4646 (F=3.142, P=0.044), and the L2D: 4D was higher in genotypes with minor allele A. Also, the DR-L in women was higher in genotypes containing the minor allele T of rs17115149 (F=3.260, P=0.039).
Figure 2.
Second-to-fourth digit ratios (2D: 4D) of different sexes distributed in different genotypes of the CYP17A1 single-nucleotide polymorphisms (SNPs). (A–C) Left hand 2D: 4D of rs17115149, rs1004467, and rs2486758 genotypes; (D–F) Right hand 2D: 4D of rs17115149, rs1004467, and rs2486758 genotypes. The black scatter points are individual 2D: 4D observations (including 400 men and 404 women). The box along with the upper and lower vertical lines represent the quartile range of 2D: 4D. Different colors of boxes represent different phenotypes. The horizontal line inside the box represents the mean value of the 2D: 4D. Remeasurement reliability of all digit lengths among 3 operators were acceptable for both hands in the intra-class correlation coefficient test.
Further, the linear regression model was used to verify the causal relationship between CYP SNPs and 2D: 4D. As shown in Table 4, rs2486758 was correlated with the L2D: 4D (P=0.026) in men, and rs1004467 was correlated with the R2D: 4D in men (P=0.008) and the whole population (P=0.032). However, no relationship was found between CYP SNPs and the 2D: 4D in women. Additionally, there was no correlation between CYP SNPs and DR-L for either men or women.
Table 4.
The relationship between CYP SNPs and L2D: 4D, R2D: 4D, and DR-L in men and women.
| Genotype | L2D: 4D | R2D: 4D | DR-L | ||||||
|---|---|---|---|---|---|---|---|---|---|
| BETA | STAT | P-adjusted | BETA | STAT | P-adjusted | BETA | STAT | P-adjusted | |
| rs2687133 | |||||||||
| Total | 0.0025 | 1.361 | 0.298 | −0.0004 | −0.195 | 0.845 | −0.0029 | −1.869 | 0.372 |
| Male | 0.0049 | 1.773 | 0.288 | 0.0008 | 0.319 | 0.818 | −0.0040 | −1.856 | 0.291 |
| Female | 0.0005 | 0.215 | 0.954 | −0.0013 | −0.503 | 0.820 | −0.0018 | −0.844 | 0.946 |
| rs7173655 | |||||||||
| Total | −0.0004 | −0.237 | 0.887 | −0.0012 | −0.723 | 0.627 | −0.0008 | −0.589 | 0.613 |
| Male | −0.0020 | −0.808 | 0.566 | −0.0025 | −1.073 | 0.426 | −0.0005 | −0.279 | 0.851 |
| Female | 0.0011 | 0.472 | 0.954 | −1.6×10−5 | −0.006 | 0.995 | −0.0011 | −0.537 | 0.946 |
| rs17115149 | |||||||||
| Total | 0.0024 | 0.822 | 0.617 | 0.0009 | 0.302 | 0.832 | −0.0015 | −0.620 | 0.613 |
| Male | 0.0046 | 1.119 | 0.566 | 0.0073 | 1.871 | 0.200 | 0.0027 | 0.849 | 0.595 |
| Female | 0.0004 | 0.094 | 0.954 | −0.0062 | −1.439 | 0.603 | −0.0066 | −1.829 | 0.618 |
| rs1004467 | |||||||||
| Total | −0.0035 | −2.038 | 0.176 | −0.0050 | −2.863 | 0.032* | −0.0014 | −1.005 | 0.613 |
| Male | −0.0055 | −2.082 | 0.228 | −0.0086 | −3.426 | 0.008** | −0.0031 | −1.482 | 0.291 |
| Female | −0.0023 | −1.045 | 0.889 | −0.0023 | −0.9664 | 0.790 | 3.69×10−5 | 0.019 | 0.985 |
| rs2486758 | |||||||||
| Total | 0.0036 | 1.808 | 0.176 | 0.0032 | 1.59 | 0.337 | −0.0004 | −0.246 | 0.805 |
| Male | 0.0091 | 3.087 | 0.026 | 0.0087 | 3.075 | 0.013 | −0.0004 | −0.174 | 0.862 |
| Female | −0.0017 | −0.645 | 0.954 | −0.0021 | −0.7386 | 0.790 | −0.0004 | −0.159 | 0.985 |
| rs4646 | |||||||||
| Total | 0.0034 | 1.921 | 0.176 | 0.0024 | 1.319 | 0.375 | −0.0011 | −0.708 | 0.613 |
| Male | 0.0025 | 0.934 | 0.566 | 0.0019 | 0.7177 | 0.568 | −0.0007 | −0.313 | 0.851 |
| Female | 0.0045 | 1.916 | 0.336 | 0.0030 | 1.215 | 0.675 | −0.0015 | −0.694 | 0.946 |
| rs2255192 | |||||||||
| Total | 6.1x10-5 | 0.030 | 0.976 | 0.0027 | 1.344 | 0.375 | 0.0027 | 1.588 | 0.400 |
| Male | −0.0003 | −0.109 | 0.913 | 0.0034 | 1.166 | 0.419 | 0.0038 | 1.554 | 0.291 |
| Female | −0.0001 | −0.057 | 0.954 | 0.0017 | 0.599 | 0.820 | 0.0019 | 0.782 | 0.946 |
| rs4275794 | |||||||||
| Total | −0.0006 | −0.300 | 0.887 | 0.0020 | 0.944 | 0.518 | 0.0025 | 1.502 | 0.400 |
| Male | −0.0007 | −0.223 | 0.899 | 0.0028 | 0.968 | 0.445 | 0.0035 | 1.459 | 0.291 |
| Female | −0.0011 | −0.407 | 0.954 | 0.0007 | 0.243 | 0.881 | 0.0018 | 0.750 | 0.946 |
P value was adjusted by Benjamini-Hochberg method;
P<0.05,
P<0.01.
SNPs – single-nucleotide polymorphisms; L2D: 4D – left hand second-to-fourth digit ratio; R2D: 4D – right hand second-to-fourth digit ratio; DR-L – R2D: 4D minus L2D: 4D.
Correlation of CYP SNPs with 2D: 4D Under Different Genetic Models and Haplotypes
The digit ratios of dominant and recessive models of SNP genotypes were compared. For the whole population, L2D: 4D was significantly lower in C/C than in A/C and A/A genotypes under the dominant model of rs4646 (t=−2.218, P=0.027), R2D: 4D was significantly higher in A/A than in A/G and G/G genotypes under the dominant model of rs1004467 (t=2.618, P=0.009), and DR-L was significantly different between the genotypes of SNP rs2687133 under the dominant model (t=2.067, P=0.039). In men, there were significant differences in L2D: 4D in the dominant model of rs2486758 (t=2.831, P=0.005), recessive models of rs1004467 (t=2.109, P=0.036) and rs2486758(t=−2.028, P=0.043), in R2D: 4D in dominant models of rs1004467 (t=2.878, P=0.004) and rs2486758 (t=−3.132, P=0.002), and the recessive model of rs1004467 (t=2.644, P=0.009). In women, the optimal models were the recessive model of rs17115149 (t=2.128, P=0.034) and the dominant model of rs4646 (t=2.419, P=0.016) for L2D: 4D, but there were no statistically significant genetic models for R2D: 4D and DR-L.
Next, 2D: 4D in different haplotypes of CYP17A1 and CYP19A1 were analyzed. As shown by the data summarized in Table 5, only rs17115149-rs1004467-rs2486758 in CYP17A1 showed significant differences in male R2D: 4D values (F=4.382, P=0.005). Multiple comparisons were performed, and the results indicated that R2D: 4D was significantly lower in the haplotype G-G-T.
Table 5.
The association between CYP17A1 SNPs and 2D: 4D under different haplotypes for men and women.
| rs17115149-rs1004467-rs2486758 | L2D: 4D | R2D: 4D | DR-L | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean±SD | F | p | Mean±SD | F | p | Mean±SD | F | p | ||
| Total | G-A-T | 0.9639±0.0331 | 1.116 | 0.341 | 0.962±0.0328 | 1.790 | 0.147 | −0.0019±0.0272 | 0.239 | 0.869 |
| G-G-T | 0.9618±0.0338 | 0.9589±0.0331 | −0.0030±0.0275 | |||||||
| G-A-C | 0.9662±0.0370 | 0.9644±0.0320 | −0.0018±0.0292 | |||||||
| T-A-T | 0.9657±0.0305 | 0.9622±0.0355 | −0.0035±0.0290 | |||||||
| Male | G-A-T | 0.9605±0.0343 | 2.135 | 0.094 | 0.9593±0.0321 | 4.382 | 0.005** | −0.0013±0.0277 | 0.389 | 0.761 |
| G-G-T | 0.9577±0.0374 | 0.9544±0.0332 | −0.0034±0.0275 | |||||||
| G-A-C | 0.9666±0.0404 | 0.9659±0.0313 | −0.0006±0.0307 | |||||||
| T-A-T | 0.9662±0.0257 | 0.9657±0.0339 | −0.0004±0.0279 | |||||||
| Female | G-A-T | 0.9673±0.0316 | 0.197 | 0.898 | 0.9648±0.0333 | 0.645 | 0.586 | −0.0025±0.0268 | 0.396 | 0.756 |
| G-G-T | 0.9656±0.0296 | 0.9630±0.0326 | −0.0026±0.0275 | |||||||
| G-A-C | 0.9659±0.0337 | 0.9630±0.0327 | −0.0028±0.0278 | |||||||
| T-A-T | 0.9652±0.0351 | 0.9586±0.0371 | −0.0066±0.0300 | |||||||
P<0.01.
SNPs – single-nucleotide polymorphisms; 2D: 4D – second-to-fourth digit ratio.
Implications of SNP-SNP Interactions Within CYP Genes in 2D: 4D
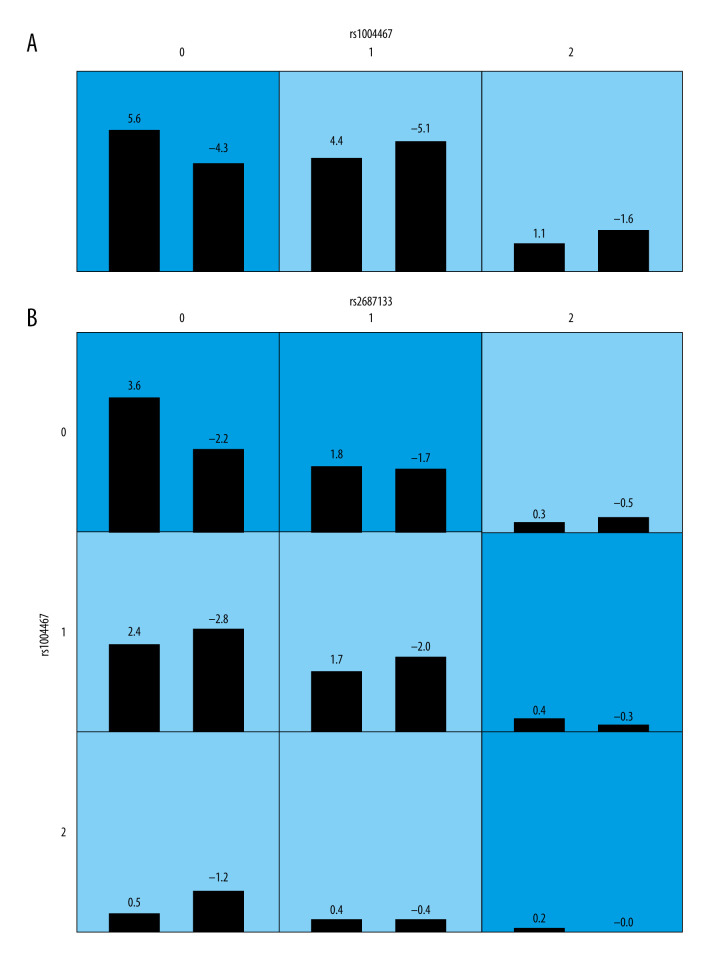
Next, we used GMDR analysis to detect the potential high-dimension interactions among the CYP SNPs, with the interaction confirmed by cross-validation consistency. Key variables were defined as L2D: 4D, R2D: 4D, and DR-L for each participant, with SNP variables ranging from 0 to 2, indicating the number of minor alleles in an individual participant (0=0 minor allele, 1=1 minor allele, 2=2 minor alleles). Considering the effect on the R2D: 4D, CYP17A1 SNP rs1004667 had the highest testing balanced accuracy and CV consistency among the 8 SNPs, but without statistical significance. On testing, SNPs rs2687133 (CYP3A7) and rs1004467 (CYP17A1) had the highest accuracy in a 2-way interaction (P=0.0107) (Table 6, Figure 3).
Table 6.
Generalized multifactor dimensionality reduction analysis for prediction of R2D: 4D.
| Model | Training Bal.Acc | Testing Bal.Acc | Sign Test(p) | CV consistency |
|---|---|---|---|---|
| rs1004467 | 0.5568 | 0.5558 | 8 (0.0547) | 10/10 |
| rs2687133-rs1004467 | 0.5786 | 0.5379 | 9 (0.0107*) | 9/10 |
P<0.05.
R2D: 4D – right hand second-to-fourth digit ratio.
Figure 3.
Single-nucleotide polymorphism (SNP) interaction graphs analyzed by generalized multifactor dimensionality reduction for second-to-fourth digit ratio (2D: 4D). (A, B) Optimal models as determined by generalized multifactor dimensionality reduction (GMDR) for CYP SNPs (0 = 0 minor allele; 1=1 minor allele; 2=2 minor alleles). The numbers within each small square represent positive scores (left) and negative scores (right). For each square, dark blu cells indicate the high effect on the 2D: 4D, and light blue cells represent the low effect on the 2D: 4D. The test was adjusted for sex.
Discussion
The current study was designed to help us understand the genetic polymorphisms of CYP genes and the association of these polymorphisms with the prenatal sex hormone balance in a northwestern Chinese population. Our results support the hypothesis that polymorphisms in CYP genes are correlated with the 2D: 4D; that is, variants of CYP genes might affect the synthesizing and metabolizing of sex hormones in utero and further result in the difference of digit ratio formation during early embryo development.
Genetic polymorphisms in CYP genes might lead to changes in CYP gene expression and thereby affect steroid synthetization and endocrine regulation [18]. Studies have found that the allele frequencies of many CYP SNPs vary between different ethnic groups [34] and diseases [35], but differences due to sex have been rarely investigated. For example, the CYP17 34 T>C allele distribution was found to be different between male (n=756) and female (n=915) healthy control participants in a study in an Austrian population, in which men had a statistically significant higher mutant C allele frequency than did women [36]. Our present study analyzed the genotype and allele frequencies of 8 SNPs of CYP genes in a population of northwestern China. However, the results indicated that there were no statistically significant differences in the genotype and allele frequencies of any SNPs between the men and women included in the study.
Research in the last decades has shown that unbalanced exposure to prenatal testosterone (PT) or prenatal estrogen (PE) may cause differences in 2D: 4D. High PT or low PE might lead to a low 2D: 4D, while low PT or low PE might result in a high 2D: 4D [37]. Many studies have demonstrated that women have a higher 2D: 4D than men, which is consistent with our findings. In addition, the DR-L or DL-R of 2D: 4D can reveal 2D: 4D asymmetry between the left and right hands [38]. Several reports have suggested that PT influences R2D: 4D and DR-L more than it influences L2D: 4D, and the DR-L of 2D: 4D is negatively correlated with prenatal exposure and sensitivity to testosterone [39]. However, in our present study, we did not observe marked differences in the range of DL-R between the sexes. Also, a study found that the number of TA(n) repeats in the promoter region of the ESR1 gene was significantly correlated with 2D: 4D in male left hands [40], but rs9340799 of the ESR1 gene might be related to the 2D: 4D of the right hands in school-aged boys [41]. Our research showed that some CYP gene polymorphisms were associated with 2D: 4D in the left or right hand, while others were associated with both left and right hands, indicating that the association between CYP gene polymorphisms and 2D: 4D had no left or right hand preference. Although the underlying mechanisms for this phenomenon are unclear, we assume that some SNP variants probably affect the CYP gene functions weakly and result in the weak effect on the associations between CYP SNPs and 2D: 4D.
CYP3A7, the most abundant CYP3A isoform in the fetus, is not typically expressed in adults. The SNP rs2687133 is an intron variant of CYP3A7. In the present study, the DR-L of T/C and C/C was higher than that of T/T genotypes in rs2687133, indicating that the minor allele C was possibly related to the prenatal sex hormone level. The SNP rs2687133 has been studied in research on drug metabolizing enzyme function and congenital adrenal hyperplasia, but no associations were observed [42]. Additional research may be required to confirm the relationship between rs2687133 and other sex steroid-related phenotypes and diseases. CYP11A1, the initial rate-limiting enzyme for steroidogenesis, is expressed mainly in the adrenal glands and gonads and is involved in the hypothalamus-pituitary-adrenal regulatory axis [43]. Because of its physiological importance, many studies have focused on the single-nucleotide polymorphism of CYP11A1. Women carrying the variant allele for SNPs rs7173655 have a higher risk of endometrial cancer than do women with 2 wild-type alleles in CYP11A1 [44]. Further, rs7173655 in CYP11A1 showed a statistically significant association with acrylamide intake for breast cancer risk [45]. Although 2D: 4D has been reported to be linked with cancers, especially breast cancer [46], no significant relationship between rs7173655 and 2D: 4D was observed in our study. We believe that the correlations between other SNPs in CYP11A1 and 2D: 4D are worthy of further study.
Three SNPs, namely rs1004467, rs17115149, and rs2486758 of CYP17A1, a gene encoding a key cytochrome P450 enzyme in the steroidogenic pathway, were analyzed to explore their roles in digit ratio formations. Our present data indicated that the R2D: 4D differed markedly in different genotypes and genetic models (dominant and recessive) of SNP rs1004467. In addition, we verified an association between rs1004467 and R2D: 4D by generalized linear analysis in men and the entire study population. The existence of the G allele significantly decreased the R2D: 4D, suggesting an increased PT or decreased PE. As previously reported, men carrying the GG genotype for SNP rs1004467 have significantly elevated concentrations of dihydrotestosterone and testosterone, which probably leads to a higher susceptibility for the development of prostate cancer [47]. Interestingly, it was found that low 2D: 4D is strongly associated with prostate cancer [6]. Additionally, the rs1004467 minor G allele of rs1004467 is statistically significantly correlated with a decreased risk of hypertension [48], and 2D: 4D is related to blood pressure. In terms of SNP rs2486758, which is mapped to the intergenic section near the 5′ UTR of the CYP17A1 gene, L2D: 4D and R2D: 4D showed significant correlations with SNP rs2486758 in men under different statistical or genetic models, and the minor allele C of rs2486758 increased the 2D: 4D values. The C allele of rs2486758 has been reported to be linked with a high serum estrogen level, prostate cancer, and coronary artery disease [49], which are also related to 2D: 4D [6,50]. In addition, rs17115149 showed a significant association with the development of prostate cancer in Korean men [51] and with testosterone levels of men with infertility [29]. In our present study, the DR-L in women was higher in minor allele T genotypes of rs17115149, and a significant difference of L2D: 4D was observed between TT and TC/CC in women. However, we did not find any association in men and failed to verify an association by linear analysis, indicating that a larger sample size and a more diverse ethnic population might be needed. It has been implied that rs1004467 and rs2486578 of CYP17A1 are important SNP loci for explaining digit ratio development and elucidating the fetal origin and organizational effect of disorders including prostate cancer, hypertension, and coronary artery disease, especially for men.
The product of the CYP19A1 gene, aromatase, is the key enzyme participating in converting androgen to estrogen. In human placental tissues, expression levels of CYP19A1 are elevated dramatically during pregnancy. A previous study on the relationship between 6 SNPs of CYP19A1 and 2D: 4D indicated that an association between rs4775936 and DR-L was found only in women [52]. In our study, we analyzed 3 SNPs which were also of clinical significance but we found no relationship between 2D: 4D and rs2255192 or rs4275794 of CYP19A1. However, we found rs4646 was correlated with L2D: 4D in women, under the dominant model. The L2D: 4D of genotype A/C and A/A was higher than that of genotype C/C, suggesting the A allele was associated with low PT or high PE. Researchers discovered that the rs4646 C/C genotype is correlated with higher estrogen levels in patients with breast cancer [53]. The homozygous minor allele (AA) of rs4646 is significantly related to the improved therapy outcomes of patients with early breast cancer [54]. It was found that a high 2D: 4D is linked with elevated breast cancer risk and bad prognosis [6]. Also, rs4775936 has been linked to both 2D: 4D and breast cancer in the study by Zhanbing et al [52]. For this reason, SNPs of CYP19A1 are valuable for identifying the 2D: 4D variances in women.
Heredity and GWAS studies have demonstrated that digit ratio is a polygenic genetic trait; that is, a single gene seldomly contributes to its development. The formation of 2D: 4D is influenced by multiple genes and environmental factors. Hence, the combined function and interaction among CYP genes involved in the synthesizing and metabolizing of sex steroids were studied to elucidate the relationship between CYP SNPs and 2D: 4D. The haplotype of rs17115149-rs1004467-rs2486758 in CYP17A1 showed remarkable differences compared with other haplotypes in R2D: 4D values in men, suggesting that multiple SNPs of CYP17A1 and the effect of their interaction might alter the digit ratios more obviously. As we know, progesterone is a common substrate for both CYP3A7 and CYP17A1 in producing downstream sex steroids [55]. It was found that the SNPs rs2687133 (CYP3A7) and rs1004467 (CYP17A1) have a strong interaction with R2D: 4D, which better explains the formation of 2D: 4D under the SNP-SNP interaction model between CYP genes.
Our study has the following limitations: 1) the SNPs of CYP genes selected in our research were not numerous enough to cover all potential function locations, and therefore more SNPs, as well as their interactions, might be required in future studies; 2) the source of all participants was limited to the Ningxia Medical University, and the small sample size of this study probably resulted in weak statistical significance; 3) the factor of a limited ethnic group might affect the results of SNP distributions and digit ratios; and 4) the study lacks functional validation. Therefore, an association study with more CYP SNP loci, a larger sample size, more ethic groups with different genetic backgrounds, and molecular function validation might be conducted in future.
Conclusions
Our research demonstrated that the SNPs rs1004467 and rs2486758 of the CYP17A1 gene are of great significance in the evaluation of the relationship between 2D: 4D and CYP gene polymorphisms under different conditions. SNP interactions between CYP genes also probably impact the association with 2D: 4D. Further, our study results suggest that the correlation between 2D: 4D and some sex hormone-related diseases is possibly due to the common effect of CYP variants on the 2 different phenotypes.
Acknowledgments
We appreciate the cooperation of all individuals who participated in this study.
Footnotes
Conflict of Interest
None.
Source of support: This study was supported by grants from the National Natural Science Foundation of China (Grant No. 31960195, 31560293)
References
- 1.Manning J, Cook C, Crewther B. Digit ratio (2D:4D) and testosterone supplementation. Early Hum Dev. 2019;139:104843. doi: 10.1016/j.earlhumdev.2019.104843. [DOI] [PubMed] [Google Scholar]
- 2.Zheng Z, Cohn MJ. Developmental basis of sexually dimorphic digit ratios. Proc Natl Acad Sci USA. 2011;108(39):16289–94. doi: 10.1073/pnas.1108312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu L, Yao R, Zhang Y, et al. The association between digit ratio (2D:4D) and overweight or obesity among Chinese children and adolescents:A cross-sectional study. Early Hum Dev. 2019;136:14–20. doi: 10.1016/j.earlhumdev.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Nitschke JP, Bartz JA. Lower digit ratio and higher endogenous testosterone are associated with lower empathic accuracy. Horm Behav. 2020;119:104648. doi: 10.1016/j.yhbeh.2019.104648. [DOI] [PubMed] [Google Scholar]
- 5.Eklund E, Ekström L, Thörngren JO, Ericsson M, et al. Digit ratio (2D:4D) and physical performance in female olympic athletes. Front Endocrinol (Lausanne) 2020;11:292. doi: 10.3389/fendo.2020.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bunevicius A. The association of digit ratio (2D:4D) with cancer:A systematic review and meta-analysis. Dis Markers. 2018;2018:7698193. doi: 10.1155/2018/7698193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richards G, Browne WV, Aydin E, et al. Digit ratio (2D:4D) and congenital adrenal hyperplasia (CAH):Systematic literature review and meta-analysis. Horm Behav. 2020;126:104867. doi: 10.1016/j.yhbeh.2020.104867. [DOI] [PubMed] [Google Scholar]
- 8.Vélez MP, Arbuckle TE, Monnier P, Fraser WD. Female digit length ratio (2D:4D) and time-to-pregnancy. Hum Reprod. 2016;31(9):2128–34. doi: 10.1093/humrep/dew164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tegin C, Canan F, El-Mallakh RS. The 2nd to 4th digit ratios (2D:4D) in patients with bipolar disorder. J Affect Disord. 2019;259:27–30. doi: 10.1016/j.jad.2019.08.011. [DOI] [PubMed] [Google Scholar]
- 10.Myers L, Van’t Westeinde A, Kuja-Halkola R, et al. 2D:4D Ratio in neurodevelopmental disorders:A twin study. J Autism Dev Disord. 2018;48(9):3244–52. doi: 10.1007/s10803-018-3588-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slama N, Warner M, Mocarelli P, et al. The 2nd to 4th digit length ratio (2D:4D) among children of Seveso women exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Early Hum Dev. 2019;131:45–50. doi: 10.1016/j.earlhumdev.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Medland SE, Zayats T, Glaser B, et al. A variant in LIN28B is associated with 2D:4D finger-length ratio, a putative retrospective biomarker of prenatal testosterone exposure. Am J Hum Genet. 2010;86(4):519–25. doi: 10.1016/j.ajhg.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrance-Owen AJ, Bargary G, Bosten JM, et al. Genetic association suggests that SMOC1 mediates between prenatal sex hormones and digit ratio. Hum Genet. 2013;132(4):415–21. doi: 10.1007/s00439-012-1259-y. [DOI] [PubMed] [Google Scholar]
- 14.Warrington NM, Shevroja E, Hemani G, et al. Genome-wide association study identifies nine novel loci for 2D:4D finger ratio, a putative retrospective biomarker of testosterone exposure in utero. Hum Mol Genet. 2018;27(11):2025–38. doi: 10.1093/hmg/ddy121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang K, Yang X, Zhang M, et al. Revisiting the relationships of 2D:4D with androgen receptor (AR) gene and current testosterone levels:Replication study and meta-analyses. J Neurosci Res. 2020;98(2):353–70. doi: 10.1002/jnr.24502. [DOI] [PubMed] [Google Scholar]
- 16.Vaillancourt KL, Dinsdale NL, Hurd PL. Estrogen receptor 1 promoter polymorphism and digit ratio in men. Am J Hum Biol. 2012;24(5):682–89. doi: 10.1002/ajhb.22297. [DOI] [PubMed] [Google Scholar]
- 17.Munro AW, McLean KJ, Grant JL, Makris TM. Structure and function of the cytochrome P450 peroxygenase enzymes. Biochem Soc Trans. 2018;46(1):183–96. doi: 10.1042/BST20170218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manikandan P, Nagini S. Cytochrome P450 structure, function and clinical significance:A review. Current Drug Targets. 2018;19(1):38–54. doi: 10.2174/1389450118666170125144557. [DOI] [PubMed] [Google Scholar]
- 19.Luo B, Chen C, Wu X, et al. Cytochrome P450 2U1 is a novel independent prognostic biomarker in breast cancer patients. Front Oncol. 2020;10:1379. doi: 10.3389/fonc.2020.01379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maksymchuk OV, Kashuba VI. Altered expression of cytochrome P450 enzymes involved in metabolism of androgens and vitamin D in the prostate as a risk factor for prostate cancer. Pharmacol Rep. 2020;72(5):1161–72. doi: 10.1007/s43440-020-00133-y. [DOI] [PubMed] [Google Scholar]
- 21.Salehi Z, Gholizadeh L, Vaziri H, Madani AH. Analysis of GSTM1, GSTT1, and CYP1A1 in idiopathic male infertility. Reprod Sci (Thousand Oaks, Calif) 2012;19(1):81–85. doi: 10.1177/1933719111413302. [DOI] [PubMed] [Google Scholar]
- 22.Munawar Lone N, Babar S, Sultan S, et al. Association of the and gene polymorphisms in women with polycystic ovary syndrome from Punjab, Pakistan. Gynecol Endocrinol. 2020 doi: 10.1080/09513590.2020.1822803. [Online ahead of print] [DOI] [PubMed] [Google Scholar]
- 23.Wang C, Pallan PS, Zhang W, et al. Functional analysis of human cytochrome P450 21A2 variants involved in congenital adrenal hyperplasia. J Biol Chem. 2017;292(26):10767–78. doi: 10.1074/jbc.M117.792465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill M, Paskova A, Kanceva R, et al. Steroid profiling in pregnancy:A focus on the human fetus. J Steroid Biochem Mol Biol. 2014;139:201–22. doi: 10.1016/j.jsbmb.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Lampe JN. Neonatal cytochrome P450 CYP3A7:A comprehensive review of its role in development, disease, and xenobiotic metabolism. Arch Biochem Biophys. 2019;673:108078. doi: 10.1016/j.abb.2019.108078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reinen J, Vermeulen NPE. Biotransformation of endocrine disrupting compounds by selected phase I and phase II enzymes – formation of estrogenic and chemically reactive metabolites by cytochromes P450 and sulfotransferases. Curr Med Chem. 2015;22(4):500–27. doi: 10.2174/0929867321666140916123022. [DOI] [PubMed] [Google Scholar]
- 27.Burton GJ, Fowden AL, Thornburg KL. Placental origins of chronic disease. Physiol Rev. 2016;96(4):1509–65. doi: 10.1152/physrev.00029.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson N, De Ieso P, Migliorini G, et al. Cytochrome P450 allele CYP3A7*1C associates with adverse outcomes in chronic lymphocytic leukemia, breast, and lung cancer. Cancer Res. 2016;76(6):1485–93. doi: 10.1158/0008-5472.CAN-15-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park JH, Lee J, Kim CH, Lee S. The polymorphism (−600 C>A) of CpG methylation site at the promoter region of CYP17A1 and its association of male infertility and testosterone levels. Gene. 2014;534(1):107–12. doi: 10.1016/j.gene.2013.09.088. [DOI] [PubMed] [Google Scholar]
- 30.Hosono S, Ito H, Oze I, et al. Polymorphisms in CYP19A1, HSD17B1 and HSD17B2 genes and serum sex hormone level among postmenopausal Japanese women. Maturitas. 2015;82(4):394–401. doi: 10.1016/j.maturitas.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Bethke L, Webb E, Sellick G, et al. Polymorphisms in the cytochrome P450 genes CYP1A2, CYP1B1, CYP3A4, CYP3A5, CYP11A1, CYP17A1, CYP19A1 and colorectal cancer risk. BMC Cancer. 2007;7:123. doi: 10.1186/1471-2407-7-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Shaughnessy PJ, Monteiro A, Bhattacharya S, et al. Steroidogenic enzyme expression in the human fetal liver and potential role in the endocrinology of pregnancy. Mol Hum Reprod. 2013;19(3):177–87. doi: 10.1093/molehr/gas059. [DOI] [PubMed] [Google Scholar]
- 33.Xu HM, Xu LF, Hou TT, et al. GMDR:Versatile software for detecting gene-gene and gene-environment interactions underlying complex traits. Curr Genomics. 2016;17(5):396–402. doi: 10.2174/1389202917666160513102612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hizbullah, Ahmed S, Noor Mumtaz M, et al. Genetic variations in drug-metabolizing enzyme CYP2C9 among major ethnic groups of Pakistani population. Gene. 2020;746:144659. doi: 10.1016/j.gene.2020.144659. [DOI] [PubMed] [Google Scholar]
- 35.Al-Eitan LN, Rababa’h DM, Alghamdi MA, Khasawneh RH. Association of CYP gene polymorphisms with breast cancer risk and prognostic factors in the Jordanian population. BMC Med Genet. 2019;20(1):148. doi: 10.1186/s12881-019-0884-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bentz EK, Hefler LA, Kaufmann U, et al. A polymorphism of the CYP17 gene related to sex steroid metabolism is associated with female-to-male but not male-to-female transsexualism. Fertil Steril. 2008;90(1):56–59. doi: 10.1016/j.fertnstert.2007.05.056. [DOI] [PubMed] [Google Scholar]
- 37.Manning JT, Fink B. Sexual dimorphism in the ontogeny of second (2D) and fourth (4D) digit lengths, and digit ratio (2D:4D) Am J Hum Biol. 2018;30(4):e23138. doi: 10.1002/ajhb.23138. [DOI] [PubMed] [Google Scholar]
- 38.Swaddle JP. Digit ratio:A pointer to fertility, behavior, and health. Heredity. 2002;89(5):403. [Google Scholar]
- 39.Coyne SM, Manning JT, Ringer L, Bailey L. Directional asymmetry (right–left differences) in digit ratio (2D:4D) predict indirect aggression in women. Pers Individ Differ. 2007;43(4):865–72. [Google Scholar]
- 40.Vaillancourt KL, Dinsdale NL, Hurd PL. Estrogen receptor 1 promoter polymorphism and digit ratio in men. Am J Hum Biol. 2012;24(5):682–89. doi: 10.1002/ajhb.22297. [DOI] [PubMed] [Google Scholar]
- 41.Nishimura Y, Moriya K, Kobayashi S, et al. Association between ESR1 polymorphisms and second to fourth digit ratio in school-aged children in the Hokkaido Study. Steroids. 2019;141:55–62. doi: 10.1016/j.steroids.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 42.Nebesio TD, Renbarger JL, Nabhan ZM, et al. Differential effects of hydrocortisone, prednisone, and dexamethasone on hormonal and pharmacokinetic profiles:A pilot study in children with congenital adrenal hyperplasia. Int J Pediatr Endocrinol. 2016;2016:17. doi: 10.1186/s13633-016-0035-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu MC, Hsu HJ, Guo IC, Chung BC. Function of Cyp11a1 in animal models. Mol Cell Endocrinol. 2004;215(1–2):95–100. doi: 10.1016/j.mce.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 44.Terry K, McGrath M, Lee IM, et al. Genetic variation in CYP11A1 and StAR in relation to endometrial cancer risk. Gynecol Oncol. 2010;117(2):255–59. doi: 10.1016/j.ygyno.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hogervorst JGF, van den Brandt PA, Godschalk RWL, et al. Interaction between dietary acrylamide intake and genetic variants for estrogen receptor-positive breast cancer risk. Eur J Nutr. 2019;58(3):1033–45. doi: 10.1007/s00394-018-1619-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hong L, Zhan-Bing M, Zhi-Yun S, et al. Digit ratio (2D:4D) in Chinese women with breast cancer. Am J Hum Biol. 2014;26(4):562–46. doi: 10.1002/ajhb.22546. [DOI] [PubMed] [Google Scholar]
- 47.Poniah P, Mohamed Z, Apalasamy YD, et al. Genetic polymorphisms in the androgen metabolism pathway and risk of prostate cancer in low incidence Malaysian ethnic groups. Int J Clin Exp Med. 2015;10(8):8. [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang N, Jia J, Ding Q, et al. Common variant rs11191548 near the CYP17A1 gene is associated with hypertension and the serum 25(OH) D levels in Han Chinese. J Hum Genet. 2018;63(6):731–37. doi: 10.1038/s10038-018-0435-x. [DOI] [PubMed] [Google Scholar]
- 49.Crucitta S, Del Re M, Paolieri F, et al. CYP17A1 polymorphism c.-362T>C predicts clinical outcome in metastatic castration-resistance prostate cancer patients treated with abiraterone. Cancer Chemother Pharmacol. 2020;86(4):527–33. doi: 10.1007/s00280-020-04133-w. [DOI] [PubMed] [Google Scholar]
- 50.Wang L, Huo Z, Lu H, et al. Digit ratio (2D:4D) and coronary artery disease in north Chinese women. Early Hum Dev. 2018;116:64–67. doi: 10.1016/j.earlhumdev.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 51.Myung S, Han J, Lee Y, et al. Association between cytochrome CYP17A1, CYP3A4, and CYP3A43 polymorphisms and prostate cancer risk and aggressiveness in a Korean study population. Asian J Androl. 2015;17(2):285–91. doi: 10.4103/1008-682X.133320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhanbing M, Jie D, Chunyue B, et al. Association of CYP19A1 single-nucleotide polymorphism with digit ratio (2D:4D) in a sample of men and women from Ningxia (China) Early Hum Dev. 2019;132:58–65. doi: 10.1016/j.earlhumdev.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 53.Armamento-Villareal R, Shah VO, et al. The rs4646 and rs12592697 polymorphisms in CYP19A1 are associated with disease progression among patients with breast cancer from different racial/ethnic backgrounds. Front Genet. 2016;7:211. doi: 10.3389/fgene.2016.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johansson H, Aristarco V, Gandini S, et al. Prognostic impact of genetic variants of CYP19A1 and UGT2B17 in a randomized trial for endocrine-responsive postmenopausal breast cancer. Pharmacogenomics J. 2020;20(1):19–26. doi: 10.1038/s41397-019-0087-z. [DOI] [PubMed] [Google Scholar]
- 55.Morel Y, Roucher F, Plotton I, et al. Evolution of steroids during pregnancy:Maternal, placental and fetal synthesis. Ann Endocrinol (Paris) 2016;77(2):82–89. doi: 10.1016/j.ando.2016.04.023. [DOI] [PubMed] [Google Scholar]